Abstract
In the midst of the severe acute respiratory syndrome coronavirus 2 pandemic and its attendant morbidity and mortality, safe and efficacious vaccines are needed that induce protective and long-lived immune responses. More than 120 vaccine candidates worldwide are in various preclinical and phase 1 to 3 clinical trials that include inactivated, live-attenuated, viral-vectored replicating and nonreplicating, protein- and peptide-based, and nucleic acid approaches. Vaccines will be necessary both for individual protection and for the safe development of population-level herd immunity. Public-private partnership collaborative efforts, such as the Accelerating COVID-19 Therapeutic Interventions and Vaccines mechanism, are key to rapidly identifying safe and effective vaccine candidates as quickly and efficiently as possible. In this article, we review the major vaccine approaches being taken and issues that must be resolved in the quest for vaccines to prevent coronavirus disease 2019. For this study, we scanned the PubMed database from 1963 to 2020 for all publications using the following search terms in various combinations: SARS, MERS, COVID-19, SARS-CoV-2, vaccine, clinical trial, coronavirus, pandemic, and vaccine development. We also did a Web search for these same terms. In addition, we examined the World Health Organization, Centers for Disease Control and Prevention, and other public health authority websites. We excluded abstracts and all articles that were not written in English.
Abbreviations and Acronyms: ACE2, angiotensin-converting enzyme 2; ADE, antibody-dependent enhancement; COVID-19, coronavirus disease 2019; IL, interleukin; MERS, Middle East respiratory syndrome; MVA, modified vaccinia virus Ankara; NIH, National Institutes of Health; RBD, receptor-binding domain; S, spike; SARS, severe acute respiratory syndrome; SARS-CoV, SARS coronavirus; TLR, Toll-like receptor; VLP, virus-like particle; WHO, World Health Organization
Article Highlights.
-
•
This review briefly summarizes what is currently known about severe acute respiratory syndrome coronavirus 2 and outlines the implications that knowledge may have on vaccine development.
-
•
This review summarizes current coronavirus disease 2019 (COVID-19) vaccine approaches and issues that must be resolved as we work toward developing safe and effective vaccines to prevent COVID-19.
-
•
In particular, the vaccines in advanced phase 3 clinical trials are reviewed, and we outline the rationale for their use.
-
•
We describe potential challenges and a research agenda critical to COVID-19 vaccine development.
Severe acute respiratory syndrome coronavirus (SARS-CoV) 2 is the second virus to cause a human pandemic in the 21st century and the third novel betacoronavirus to emerge as a human pathogen in the past 18 years.1 As of this writing, over 13 million cases have been identified worldwide, and almost 600,000 deaths have been reported.2 These numbers are certainly underestimates, in part related to the now-apparent wide spectrum of disease ranging from asymptomatic individuals to severe disease to death, as well as the dearth of diagnostic testing and standardized reporting. In a mere 28 weeks, we have learned much, but far more remains to be learned. The genetic sequence of SARS-CoV-2 was solved very quickly, and within weeks the sequence was widely available, identifying it as a betacoronavirus with close genetic similarity to SARS-CoV-1. We have learned the cellular receptor used by this virus and the concomitant inflammatory cytokine storm that can result from infection. We have identified the approximate reproductive number and the infection fatality rate of SARS-CoV-2 and are witnessing the wide spectrum of clinical and human immune responses to infection with the virus. We have also learned that severe disease and death vary by patient age, comorbidities, smoking status, body mass index, when in the context of a localized epidemic one presents for medical care, and other factors. At this time, there are no validated point-of-care assays for rapid diagnostics that have been widely deployed, no licensed antivirals, and no licensed vaccines for use in civilian populations. Absent a safe and effective vaccine, safe achievement of herd immunity will prove elusive. Although multiple vaccine candidates were developed against the SARS-CoV-1 and Middle East respiratory syndrome (MERS) viruses, no vaccine candidates progressed past phase 1 studies. This review will focus on the current development of SARS-CoV-2 vaccines and address issues relevant to devising a safe and effective vaccine. To gather material for this review, we scanned the PubMed database from 1963 to 2020 for all publications using the following search terms in various combinations: SARS, MERS, COVID-19, SARS-CoV-2, vaccine, clinical trial, coronavirus, pandemic, and vaccine development. We also did a Web search for these same terms. In addition, we examined the World Health Organization (WHO), Centers for Disease Control and Prevention (CDC), and other public health authority websites.
SARS-CoV-2 Disease/Outbreaks
In December 2019, a cluster of atypical viral pneumonia cases in Wuhan, China, was reported to the WHO. It was reported that most patients exhibited respiratory symptoms consistent with severe acute respiratory disease (SARS) and had previously visited the Huanan seafood wholesale market, suggesting an animal origin of SARS-CoV-2. Subsequently, person-to-person transmission through droplets or direct contact occurred, resulting in the early January 2020 declaration by Chinese health authorities that a novel coronavirus (2019-nCoV) had been identified and isolated from patients in Wuhan.3, 4, 5 Within a month, the full-length genomic sequence of the new coronavirus was made available to the WHO and became publicly available.6
SARS-CoV-2 has rapidly spread across the globe.7 On January 30, 2020, the WHO declared the SARS-CoV-2 epidemic a global health emergency. By March 11, 2020, the WHO declared a pandemic. The rapid transmission of SARS-CoV-2 has caused fear, panic, economic disruption, morbidity and mortality, and significant public health concerns.8 Recent data have revealed that the clinical characteristics of coronavirus disease 2019 (COVID-19) can be very heterogeneous with a broad spectrum of severity, including illness resulting in death.9 Data document that asymptomatic individuals can transmit SARS-CoV-2 infection.10 Individuals over 65 years of age, individuals of all ages who have serious underlying medical conditions, and those who are immunocompromised are at higher risk of serious COVID-19 illness and complications. Although COVID-19 has been detected in equal numbers of confirmed cases in males and females, there appear to be sex-based differences in severity and mortality of the disease (ie, higher mortality in older male patients).11 , 12 Considering the alarming outcomes of the current COVID-19 pandemic, it is critical to develop safe and effective vaccines and antiviral agents to prevent, control, and treat COVID-19.13
The SARS-CoV-2 virus shares up to 82% nucleotide identity with human SARS-CoV-1 and utilizes the same host cellular receptor as SARS-CoV-1, angiotensin-converting enzyme 2 (ACE2),14 as an entry receptor into host cells.15 Data from initial COVID-19 studies revealed that ACE2 is differentially expressed in many human tissues, such as lung (type 2 alveolar cells), liver (cholangiocytes), stomach (epithelial cells), ileum (enterocytes), kidney (proximal tubules), and colon (colonocytes).16 Notably, the cellular serine protease TMPRSS2 (also a host cell factor for influenza A and other coronaviruses) has recently been documented as critical for activation of the SARS-CoV-2 transmembrane spike (S) glycoprotein (ie, the main target of neutralizing antibodies), priming, and viral cell entry.17 , 18 Wang et al19 found that SARS-CoV-2 can invade host cells (Vero E6 cells) via a new CD147-S protein route. This observation indicates that CD147 receptor-targeted antivirals might also be a useful therapeutic strategy against COVID-19.
Little is known about SARS-CoV-2–specific immune responses during COVID-19 infection. Histopathology, immunohistochemistry, in situ hybridization, and electron microscopy data from SARS-CoV-1–infected human lung tissue revealed that SARS-CoV-1 can infect and replicate in alveolar macrophages, type I and type II pneumocytes, and bronchiolar epithelial cells.20 , 21 Both Th1-type (interferon-γ, interleukin [IL] 1β, inducible protein 10, monocyte chemotactic protein 1, and IL-6) and Th2-type (IL-4 and IL-10) cytokines are produced in high concentrations in plasma in response to SARS-CoV-2 infection, indicating that the host immune response itself is involved in disease progression and pathogenesis.22
In silico study results reveal that SARS-CoV-2 S protein induces an innate inflammatory immune response via nuclear factor κB activation and possibly through Toll-like receptor (TLR) 4 ligand.23 High concentrations of proinflammatory and anti-inflammatory cytokines (eg, IL-2R, IL-6, IL-10, and tumor necrosis factor α) have been detected in serum samples from severe cases of COVID-19 compared with levels in serum from moderate cases. This finding suggests that a massive cytokine storm likely contributes to disease severity.24 Other factors that have been reported to be associated with disease severity outcomes (eg, lymphopenia, decrease in CD4+ and CD8+ T lymphocyte counts, suppressed interferon-γ secretion by CD4+ T lymphocytes, and lower counts of CD16+CD14+ monocytes) may also be potential significant immunologic markers of severe and moderate COVID-19.24 , 25 As per a recent case report, the increased frequency of antibody-secreting cells, follicular helper T cells, activated CD38+ HLA-DR+ CD8+ and CD4+ T lymphocytes, together with SARS-CoV-2–specific IgG and IgM antibodies, detected in the blood of a patient with nonsevere COVID-19 prior to symptomatic recovery, suggests that early adaptive immune-related biomarkers may be predictors of better clinical outcomes.25 Given SARS-CoV-2 pathogenesis and tissue tropism, and the significant morbidity and mortality at the public health level, it is essential to develop an effective vaccine to protect against SARS-CoV-2.
SARS-CoV-2 Virus
SARS-CoV-2 is an emerging, enveloped, nonsegmented, approximately 30-kilobase, positive-sense RNA virus of global significance. It belongs to the subfamily Orthocoronavirinae, in the family Coronaviridae (group betacoronavirus). 26 , 27 Among coronaviruses that can infect humans, 6 types have been previously described: alphacoronaviruses HCoV-229E and HCoV-NL63 and betacoronaviruses HCoV-OC43, HCoV-HKU1, SARS-CoV-1, and MERS coronavirus.28 Current evidence demonstrates that SARS-CoV-2 and MERS coronavirus are highly transmissible, pathogenic, and associated with significant morbidity and mortality in humans.7
Phylogenetic analysis has indicated that SARS-CoV-1, MERS coronavirus, and SARS-CoV-2 most likely originated from bats, with transmission to human populations happening via intermediary animal hosts.29 Genome composition studies, which have yielded significant insights into the divergence of SARS-CoV-2, resulted in the identification of 380 amino acid substitutions between amino acid sequences of SARS-CoV-2 (Wuhan/HB01 strain) and the equivalent sequences of SARS-CoV-1.26 A recent phylogenetic network study of 160 SARS-CoV-2 genome nucleotide sequences from COVID-19 cases around the world identified multiple mutations in SARS-CoV-2 viral genomes (ie, nonsynonymous C28144T, synonymous T29095C, and synonymous T8782C), which may help track COVID-19 infection sources.30 Examination of 247 sequences of SARS-CoV-2 genomes found 4 viral clusters demonstrating a high mutation rate and each becoming prevalent in various countries.31 It is imperative to understand if and how nonsynonymous and synonymous variations, as well as recombination events in the SARS-CoV-2 genome, alter viral binding to the ACE2 receptor, affect virulence, alter transmissibility, or potentially alter the efficacy of antivirals, monoclonal therapeutic antibodies, or vaccines. The estimated mutation rate in the SARS-CoV-1 genome appears to be moderate (0.80 to 2.38 × 10−3 nucleotide substitution per base per year).32 , 33 The SARS-CoV-2 genome exhibits a mutation rate of less than 25 mutations per year, which is much slower than seasonal influenza virus, but it is virtually certain that further mutations and recombination events will be identified.
The receptor-binding domain (RBD) of the S protein is a critical factor for binding to ACE2 and to determine tropism and infectivity of SARS-CoV-2.14 Previous mutagenesis studies have suggested that the cross-neutralization resistance between SARS-CoV and palm civet-CoV may result in mutations within the RBD of the S protein.34 Recent structural analysis of S protein-ACE2 receptor complexes identified several amino acid substitutions and deletions in the SARS-CoV-2 RBD of S protein (ie, S1 subunit) compared with those of SARS-CoV and bat coronavirus.35 These mutations resulted in a higher affinity of the SARS-CoV-2 S protein for the human ACE2 in comparison with SARS-CoV and bat coronavirus.35 This difference is likely associated with the dynamic of viral spreading. Another genetic study examining amino acid mutations in circulating SARS-CoV-2 RBDs has found 8 mutation types (from a total of 18 mutant strains) that were divided into 2 different groups of amino acid mutations in SARS-CoV-2 RBDs based on human ACE2 affinity for the S protein (ie, the “similar affinity” group—V341I, F342L, R408I, A435S, and V483A—and the significantly “higher affinity” group—N354D, D364Y, V367F, and W436R).36 The study investigators proposed that the “higher affinity” group of mutated amino acids (specifically, amino acid mutation V367F) demonstrated an enhancement of SARS-CoV-2 binding affinity to human ACE2 and may have allowed for increasingly significant infectivity and more severe virus transmission.37 A pipeline data analysis of real-time mutations in SARS-CoV-2 identified 14 mutations in S protein (including mutation D614G) and a viral recombination event. Recent reports have revealed that the D614G mutation increases infection of human cell lines,38, 39, 40 with mounting evidence that it also influences disease severity.41
A study by Fehr and Perlman42 found that the human SARS-CoV-2 genome is similar to that of other RNA viruses and encodes for 4 major structural proteins, including the surface S, small envelope (E), membrane (M), and nucleocapsid (N) proteins. The coronavirus genome also encodes for 5′ nonstructural (n=16) and lineage-specific accessory genes (n=6, functionally not well characterized). The betacoronaviruses HCoV-OC43 and HCoV-HKU1 have also been found to encode for an additional structural hemagglutinin esterase protein that may enhance the S protein–mediated viral infection and generation of infectious virions.43 The S protein is implicated in host cell invasion and is cleaved by furin-like enzymes into 2 functional subunits or regions, S1 and S2, which are responsible for host cell receptor binding and host receptor membrane fusion, respectively.44 , 45 The S protein of SARS-CoV-2 is primed/activated by the cellular serine protease TMPRSS2, which is essential for viral entry and spread in the infected host.17 The main factor determining SARS-CoV-2 tropism is the RBD of S protein, which binds to the host receptor ACE2 and can only interact with the RBS when it is in the hinge-like “up” conformation in SARS-CoV-2.46 Notably, SARS-CoV-2 and SARS-CoV share a conserved epitope in the RBD that may be an important consideration in SARS-CoV-2 vaccine antigenicity and cross-protective antibody responses. This theory remains to be further elucidated.
Issues in Vaccine Development
The WHO is coordinating an international group of experts (eg, scientists, physicians, and industry leaders) who are working to create vaccine candidates and has released a target product profile47 that includes both critical and preferred characteristics—the ideal vaccine will have the features outlined in Table 1 . The US Food and Drug Administration has recently issued a guidance document on COVID-19 vaccines.48 This document is primarily focused on regulatory requirements and key considerations for licensure data such as the following: preclinical data, characterization of immune responses in animal models, toxicity, minimum efficacy requirements, and the potential for vaccine-associated enhanced respiratory disease. Despite the relatively mild disease seen in most infants and children and given the relative ease of transmission of SARS-CoV-2, vaccines targeting this audience must also be developed.
Table 1.
Ideal SARS-CoV-2 Vaccine Characteristicsa
The ideal vaccine should
|
SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2; WHO = World Health Organization.
Indicates a characteristic included in the WHO target product profile.47
It is likely that multiple vaccine candidates, each geared toward specific population groups at increased risk, will be necessary. Because we have never had a coronavirus vaccine, it is also likely that we will need long-term data on multiple vaccines in order to identify products that meet most of those desired characteristics. This process requires thoughtful consideration and collaborations such as that proposed by the Accelerating COVID-19 Therapeutic Interventions and Vaccines public-private partnership.49 Ethical considerations regarding vaccine testing in the elderly, in children, in pregnant women, and in other vulnerable populations must also be carefully considered. To accelerate vaccine development, calls for human challenge models have also emerged. Thus far, a clear and compelling ethical framework for making such decisions has not reached consensus.
Data from studies on SARS and MERS vaccine candidates have shaped much of the early vaccine development efforts to SARS-CoV-2.50 Preclinical studies, animal models, and other data have been used to accelerate vaccine development for SARS-CoV-2. These data indicate the following: (1) the S protein is the major target of neutralizing antibodies,51 (2) many of these antibodies target the RBD of S protein,52, 53, 54 (3) neutralizing antibodies generated by vaccination or adoptively transferred are protective in animal models (eg, mice, rabbits, and nonhuman primates),55 , 56 (4) clinical trials of 2 MERS vaccines, a DNA-based vaccine consisting of the S protein and a replication-deficient chimpanzee adenovirus expressing the S protein, both elicit robust antibody responses, (5) a modified vaccinia virus Ankara (MVA)–based vaccine expressing the S protein has been used to vaccinate camels and significantly reduces viral loads and virus secretion,57 and (6) most of the vaccine candidates also induced cellular immunity, which is thought to be critical to viral clearance.58, 59, 60 Work in these areas continues with an expanded scope that now includes this latest novel coronavirus.
This same body of work with SARS and MERS has revealed that there are also obstacles that we must carefully navigate. One serious issue is antibody-dependent enhancement (ADE) of infection and disease that has been noted in SARS.61 Interestingly, antibodies targeting the S protein were found to mediate ADE,62 which results in enhanced infection of macrophages and B cells63 , 64; therefore, S only protein-based vaccines must be carefully evaluated in terms of safety. Another obstacle facing vaccine development is that animal studies of SARS and MERS vaccines (including formulations that moved into phase 1 clinical trials) found evidence of lung and/or liver pathology after live-virus challenge.65, 66, 67, 68 Eosinophilic infiltration, enhanced Th2 responses, and increased infectivity have been noted with both whole-virus vaccines and with full-length S protein–based vaccines.69 The ability of a vaccine to elicit a robust, Th1-type helper T cell response is considered ideal, given the antiviral properties of this type of response and its suppressive effect on Th2 responses.70 Yet another issue is the observed lack of durable protective immunity to seasonal coronaviruses. It remains to be seen if the appropriate use of adjuvants and highly immunogenic vaccination platforms are able to overcome this problem. A number of studies have begun to examine immune responses to SARS-CoV-2 and have found that antibody responses (IgM, IgG, IgA) appear 1 to 2 weeks after infection, peak several weeks later, and then decline. Humoral immunity targets the S and nucleocapsid proteins, with neutralizing antibody primarily directed against the RBD of the S protein.71, 72, 73, 74, 75, 76 Similarly, infection induces T-cell responses (primarily Th1) against a broad range of viral proteins.77, 78, 79, 80, 81, 82 It has also been shown that follicular helper T cell responses occur and are correlated with the magnitude of the humoral response.83 T-cell responses have also been detected in individuals lacking humoral immunity.84 Despite these initial findings, our understanding of SARS-CoV-2 immunity is far from complete; therefore, further studies examining innate, humoral, and cellular immune responses to this new virus are necessary in order to fully understand mechanisms of protection that need to be activated by COVID-19 vaccines.
Computational Approaches to Accelerate Vaccine Development
Because the immune response to SARS-CoV-2 has not yet been fully characterized, we have a very limited understanding of the viral proteins that may be important targets of the immune system, which could be useful for developing effective vaccine candidates. In this respect, predictive computational algorithms may prove to be beneficial tools for the identification of immunogenic T-cell and B-cell epitopes that can accelerate the rational design of SARS-CoV-2 vaccine formulations. Computational algorithms offer the distinct advantage of rapidly screening the entire amino acid sequence of viral proteins to predict peptides with high antigenicity or binding affinity for HLA molecules—a task that would take countless hours to accomplish in the laboratory while consuming valuable biological specimens. Advancements in machine learning, artificial neural networks, and other computational fields have led to the continued development and refinement of epitope prediction algorithms with improved accuracy,85, 86, 87, 88, 89, 90, 91, 92, 93, 94 but performance gaps still exist. Studies of vaccinia virus infection have found that computer-based algorithms fail to identify up to 20% of peptides presented by HLA molecules,95 and the conformational nature of B-cell epitopes makes it difficult for computer-based methods to accurately predict them. Nevertheless, these approaches are state-of-the-art for epitope identification in the absence of biological data and, given the urgent need for a vaccine to combat the spread of COVID-19, should be used to guide experimental vaccine design where appropriate.
Computational algorithms have been previously applied for the identification of peptide epitopes and the design of experimental vaccines against MERS coronavirus. A recent study by Tahir Ul Qamar et al96 identified both T-cell and B-cell epitopes from the MERS coronavirus S protein that were conserved across clinical isolates, suggesting that these epitopes may be used to develop broadly protective vaccines. A similar study focused on the MERS coronavirus N protein as a potential vaccine target, identifying candidate B-cell (15 linear, 10 conformational) and T-cell (10 helper T cell, 10 cytotoxic) epitopes for further study.97 Once individual peptide epitopes have been identified by immunoinformatic approaches, they can be computationally modeled as larger polypeptide assemblies for immunologic evaluation. Srivastava et al98 employed such an approach to identify cytotoxic and helper T-cell epitopes from the MERS coronavirus proteome and design 2 vaccine constructs, both of which were predicted to provide broad global HLA population coverage (94%) and dock with TLRs. Applying similar immunoinformatic approaches, we comprehensively analyzed 10 SARS-CoV-2 proteins to identify potential targets for inclusion in COVID-19 vaccines.99
Similar studies detailing the in silico prediction of T-cell and B-cell epitopes from SARS-CoV-2 began to rapidly emerge following publication of the viral genome sequence. Grifoni et al100 reported the bioinformatic identification of T-cell and B-cell epitopes from SARS-CoV-2 structural proteins that possessed high homology with immunogenic epitopes from SARS-CoV-1. A number of other studies have identified SARS-CoV-2 T-cell and B-cell epitopes a priori based on B-cell antigenicity scoring or HLA binding affinity,101, 102, 103, 104, 105 with several designing polypeptide vaccine candidates and modeling their binding with HLA and TLR molecules.106, 107, 108 We have pursued a similar approach, stringently applying combinations of in silico approaches to identify subsets of T-cell (CD4+ and CD8+) and B-cell (linear and conformational) epitopes from the SARS-CoV-2 proteome to serve as candidates for peptide-based vaccine development.99 These studies illustrate the utility of bioinformatics and computer-based predictive modeling for designing vaccines against rare and emerging diseases when immunologic data and biological samples are limited.
Current Status of Vaccine Development
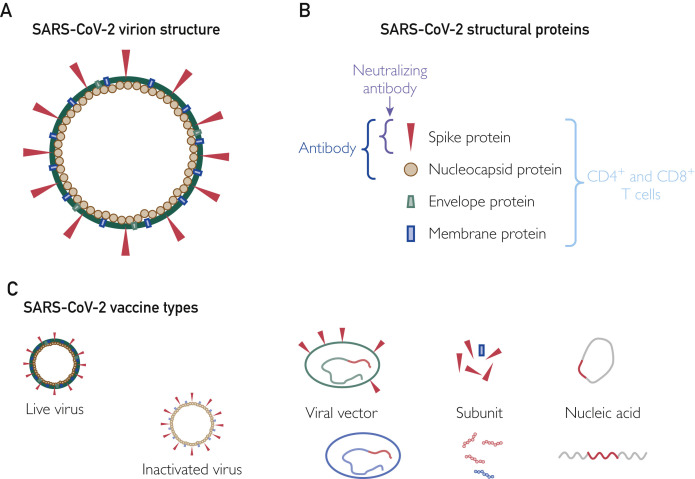
Some of the first vaccines are already in clinical trials 4 to 5 months after the start of the outbreak. As of the time of this writing, 1 vaccine has been licensed in China (only for use in the Chinese military), 3 vaccines are in phase 3 trials, 8 are in phase 2 trials, 11 are in phase 1 trials, and the remainder are in preclinical studies. This amazingly rapid development cycle is due to several factors: existing vaccine candidates, data, and animal models from SARS and MERS; the early publication of the full-length genome sequence of SARS-CoV-2; the striking sequence similarity in the S protein between SARS-CoV-1 and SARS-CoV-2; the use of DNA and RNA “plug and play” vaccine platforms; and reduced regulatory burdens due to the urgent nature of the outbreak (Figure ).
Figure.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccines. A, Diagram of the SARS-CoV-2 virion, including the lipid membrane and structural proteins. B, The 4 major structural proteins are targeted by immune response. Humoral responses are directed at both the spike protein and the nucleocapsid proteins. Neutralizing antibodies have been identified that target the receptor-binding domain of the spike protein. All of the structural (and many of the nonstructural) proteins have predicted T-cell epitopes within them, suggesting that the T-cell response is likely able to recognize most viral proteins. C, Representation of the major types of SARS-CoV-2 vaccines under development. Live-virus vaccines typically consist of a weakened version of the virus, while whole inactivated vaccines use chemicals or radiation to eliminate viral replication. Vector-based vaccines incorporate one or more viral genes (in red) into the genome of a viral vector. Some vectors are replicating (eg, measles), while others may be replication-defective but are capable of limited transcription and expression of the desired coronavirus antigen. Subunit vaccines typically consist of specific viral proteins or immunogenic peptides derived from those proteins. Nucleic acid vaccines contain DNA (top figure) or RNA (bottom figure) that are delivered using electroporation or liposomal delivery systems that enable the nucleic acid to enter target cells. Viral protein is then produced by the host cells.
Table 2 lists clinical trials currently under way. The first vaccine in clinical trials in the United States was the mRNA-1273 vaccine. This is a nonreplicating RNA vaccine that induces S protein production in host cells, leading to an antibody response. This vaccine was developed as a collaboration between the National Institutes of Health (NIH) Vaccine Research Center and Moderna, Inc. The clinical trial initially enrolled 45 adults aged 18 to 50 years who received an initial priming vaccine and a booster 4 weeks later. Jackson et al109 reported a vaccine dose-dependent increase in serum antibodies to SARS-CoV-2 S2 and RBD regions of the S protein after the first dose and a significant boost on receipt of the second vaccination. Vaccinated recipients also developed antibodies capable of neutralizing both a pseudotyped lentivirus reporter and wild-type SARS-CoV-2. Examination of the T-cell responses in the 2 lower vaccine dose (25 μg and 100 μg) groups identified the presence of SARS-CoV-2–specific CD4+ T cells with a Th1 phenotype. Virus-specific CD8+ T cells were detected in the 100-μg vaccine group. With regard to safety, no serious adverse events were noted; however, fatigue, chills, headache, myalgia, and pain at the injection site were common (reported in >50% of recipients). Local adverse events were typically mild, although severity was more pronounced at higher doses. Of the 5 grade-2 adverse events noted, only 2 were deemed to be related to the vaccine. Both (elevated lipase and decreased hemoglobin) occurred 7 days after the second vaccination. The NIH is now recruiting 2 additional age groups (51 to 70 years and ≥71 years) to evaluate the vaccine in older populations in a phase 2 clinical trial and is seeking regulatory approval for a much larger-scale phase 3 trial that began during the summer of 2020. INOVIO Pharmaceuticals has developed a DNA-based vaccine that is injected and then electroporated into muscle cells in order to induce host cell production of the S protein.
Table 2.
Clinical Trials Involving SARS-CoV-2 Vaccines
| NCT number | Vaccine type | Sponsor/collaborators | Trial phase | Location |
|---|---|---|---|---|
| NCT04299724 | Artificial APCs expressing SARS-CoV-2 proteins | Shenzhen Geno-Immune Medical Institute | 1 | Guangdong, China |
| NCT04383574 | Alum-adjuvanted, formalin-inactivated vaccine | Sinovac Research and Development Co, Ltd | 1/2 | Hebei, China |
| NCT04352608 | Alum-adjuvanted, formalin-inactivated vaccine | Sinovac Research and Development Co, Ltd | 1/2 | Jiangsu, China |
| NCT04450004 | Virus-like particle vaccine | Medicago Inc | 1 | Not provided |
| NCT04412538 | Inactivated SARS-CoV-2 vaccine | Chinese Academy of Medical Sciences, West China Second University Hospital, Yunnan Center for Disease Control and Prevention | 1/2 | Sichuan, China |
| NCT04283461 | RNA vaccine: mRNA-1273 | National Institute of Allergy and Infectious Diseases | 1 | United States |
| NCT04405908 | Subunit vaccine: spike protein trimer | Clover Biopharmaceuticals AUS Pty Ltd | 1 | Australia |
| NCT04313127 | Vectored vaccine: adenovirus type 5 vector | CanSino Biologics Inc, Institute of Biotechnology, Academy of Military Medical Sciences, PLA of China, Jiangsu Provincial Center for Disease Control and Prevention, Hubei Provincial Center for Disease Control and Prevention, Tongji Hospital | 1 | Hubei, China |
| NCT04437875 | Vectored vaccine: adenovirus type 26 with spike protein | Gamaleya Research Institute of Epidemiology and Microbiology, Ministry of Health of the Russian Federation, Acellena Contract Drug Research and Development | 1/2 | Russia |
| NCT04368728 | RNA vaccines: BNT162a1, BNT162b1, BNT162b2, BNT162c2 | BioNTech SE, Pfizer Inc | 1/2 | United States |
| NCT04341389 | Vectored vaccine: adenovirus type 5 vector | Insitute of Biotechnology, Academy of Military Medical Sciences, PLA of China, CanSino Biologics Inc, Jiangsu Provincial Center for Disease Control and Prevention, Hubei Provincial Center for Disease Control and Prevention, Zhongnan Hospital | 2 | Hubei, China |
| NCT04386252 | Artificial APCs expressing SARS-CoV-2 proteins | AIVITA Biomedical, Inc | 1/2 | United States |
| NCT04368988 | Nanoparticle vaccine with Matrix-M adjuvant | Novavax, Inc | 1 | Australia |
| NCT04324606 | Vectored vaccine: chimpanzee adenovirus, ChAdOx1 | University of Oxford | 1/2 | United Kingdom |
| NCT04334980 | Oral vaccine: bacTRL-Spike | Symvivo Corporation | 1 | United States, Canada |
| NCT04405076 | RNA vaccine: mRNA-1273 | Moderna, Inc, Biomedical Advanced Research and Development Authority | 2 | United States |
| NCT04400838 | Vectored vaccine: chimpanzee adenovirus, ChAdOx1 | University of Oxford | 2/3 | United Kingdom |
| NCT04428073 | Vectored vaccine: adeno-associated virus | GeneCure Biotechnologies | 1 | Not provided |
| NCT04453852 | Recombinant protein vaccine with Advax-SM adjuvant | Vaxine Pty Ltd, Central Adelaide Local Health Network Incorporated | 1 | Australia |
| NCT04398147 | Vectored vaccine: adenovirus type 5 vector | CanSino Biologics Inc, Beijing Institute of Biotechnology, Canadian Center for Vaccinology | 1/2 | Canada |
| NCT04444674 | Vectored vaccine: chimpanzee adenovirus, ChAdOx1 | University of Witwatersrand, South Africa, South African Medical Research Council, Bill and Melinda Gates Foundation, University of Oxford | 1/2 | South Africa |
| NCT04449276 | Biological: CVnCoV vaccine; Drug: placebo | CureVac AG, Coalition for Epidemic Preparedness Innovations | 1 | Germany |
| NCT04447781 | DNA vaccine: INO-4800 | International Vaccine Institute, Coalition for Epidemic Preparedness Innovations, INOVIO Pharmaceuticals | 1/2 | Not provided |
| NCT04336410 | DNA vaccine: INO-4800 | INOVIO Pharmaceuticals, Coalition for Epidemic Preparedness Innovations | 1 | United States |
| NCT04380701 | RNA vaccines: BNT162a1, BNT162b1, BNT162b2, BNT162c2 | BioNTech RNA Pharmaceuticals GmbH, BioNTech SE | 1/2 | Germany |
APCs = antigen-presenting cells; NCT = National Clinical Trial; PLA = People’s Liberation Army; SARS-CoV-2 = severe acute respiratory syndrome coronavirus.
There are over 120 additional vaccines in various stages of preclinical development, and the number increases weekly.110, 111, 112 A wide variety of vaccine approaches are being used, including DNA and RNA vaccines, live coronavirus vaccines, inactivated virus vaccines, subunit vaccines (predominantly S protein), vectored vaccines (eg, vesicular stomatitis virus, adenovirus, MVA, measles virus), and peptide-based vaccines.
Live Virus Vaccines and Whole Inactivated Vaccines
Live virus and inactivated, whole-virus vaccine have an extensive history of success. They are the most immunogenic of the vaccine formulations; however, this comes at a price in terms of potential safety issues. Given the existing data that these vaccines can cause immunopathology and ADE, careful scrutiny of safety signals will be paramount during animal studies and clinical trials. Codagenix Inc and the Serum Institute of India are developing a live attenuated vaccine based on their CodaVax technology that uses codon-deoptimization to attenuate viruses. Influenza, respiratory syncytial virus, and DENV-2 vaccines based on this technology have documented both safety and immunogenicity in animal models.113, 114, 115 The University of Hong Kong is developing an intranasal vaccine using an attenuated influenza virus (similar to what is in FluMist [AstraZeneca]) expressing the SARS-CoV2 S protein.
Subunit Vaccines
Subunit vaccines consist of viral proteins or protein fragments. The absence of infectious virus increases the safety profile and eliminates issues with viral inactivation or virulence reversion. The vast majority of the SARS-CoV-2 subunit vaccines have focused on the S protein or specific domains within the S protein, such as the RBD.27 Other groups have focused on the N protein because studies with SARS-CoV-1 and MERS coronavirus have revealed that it is targeted by antibodies and contains HLA-restricted T-cell epitopes.116 , 117 The proteins selected for use are often combined with adjuvants to boost immunogenicity. Large-scale production of the antigen can be problematic, although a variety of improved expression platforms, including plant-based systems, may provide high-throughput and scalable solutions.118 Baylor College of Medicine is evaluating whether a SARS-CoV-1 recombinant protein vaccine provides protection against SARS-CoV-2. Novavax, Inc has received funding from the Coalition for Epidemic Preparedness Innovations to move its protein nanoparticle vaccine into clinical trials. This vaccine uses a saponin-based adjuvant, which is a formulation that has been found to enhance adaptive immune responses to recombinant Ebola virus glycoprotein vaccines and MVA-based influenza vaccines.119 , 120 The Coalition for Epidemic Preparedness Innovations has also partnered with the University of Queensland to develop a protein-based vaccine that uses a “molecular clamp” to lock the coronavirus proteins into the correct 3-dimensional shape, allowing humoral immune responses to develop against appropriate conformational epitopes.121 Vaxart, Inc is developing an oral tablet-based vaccine that uses a replication-deficient adenovirus type 5 vector to deliver recombinant S protein and a TLR-3 adjuvant to the mucosal epithelium.122 An Israeli company, MigVax Ltd, is also developing an oral subunit vaccine against COVID-19. This product is based on their existing vaccine against poultry coronaviruses123 causing infectious bronchitis. The Mayo Clinic Vaccine Research Group is working on a peptide-based vaccine using naturally processed and presented epitopes from multiple SARS-CoV-2 proteins identified through mass spectrometry.95 , 124, 125, 126, 127 Virus-like particle (VLP) vaccines are a type of subunit vaccine consisting of an empty virus shell that lacks nucleic acid and is therefore noninfectious. The VLPs retain the 3-dimensional structure and repetitive antigenic nature of viral particles and have been found to be extremely immunogenic.128 Nearly a dozen groups are working on VLP platforms expressing S protein or RBD. The University of Pittsburgh Medical Center has developed a microneedle skin patch vaccine for SARS-CoV-2 that induced neutralizing antibody production in mice.129
Nucleic Acid Vaccines
Nucleic acid vaccines can be rapidly and inexpensively produced and contain no live virus; however, DNA vaccines require complicated delivery systems and generally higher doses and are more difficult to produce. RNA vaccines may suffer from transfection efficiency issues in vivo. In addition to the INO-4800 and mRNA-1273 vaccines currently in clinical trials, Sanofi and the Biomedical Advanced Research and Development Authority are also working on a DNA vaccine. The NIH’s Rocky Mountain Laboratories is also working with CureVac AG and with the University of Washington on additional RNA vaccine candidates. Tongji University in China has partnered with Stermirna Therapeutics Co, Ltd to develop an RNA-based vaccine.130 The Imperial College London is developing a self-amplifying RNA vaccine. The Karolinska Institute and Cobra Biologics are also collaborating on a DNA vaccine.131 Pfizer Inc and BioNTech SE have developed 4 mRNA-based formulations including 2 nucleoside-modified mRNAs, a uridine-containing mRNA, and a self-amplifying RNA. Results from a clinical trial of the BNT162b1 vaccine (encoding the RBD domain of the S protein) involving 45 participants aged 19 to 54 years was recently reported on medRxiv.132 The authors indicate that the most common adverse effects were pain at the injection site, fatigue, and headache. The vaccine elicited RBD-binding antibody at similar titers to those seen in COVID-19–convalescent patients. The vaccine also elicited modest increases in SARS-CoV-2–neutralizing antibody titers. Studies evaluating the durability of the humoral response are ongoing.
Vectored Vaccines
Vector-based vaccines are a form of live attenuated vaccines that adapt existing successful and safe viral vectors (eg, vesicular stomatitis virus, adenovirus, MVA, measles) to express coronavirus proteins on immunization. Many of these vectors are not replication-competent in human cells, while others are only capable of limited replication and have defined safety profiles. Recombinant versions of their viral vectors can be rapidly produced, protein expression verified, and vaccines quickly developed. These platforms also have existing safety and immunogenicity data for other pathogens, which can further accelerate their development. CanSino Biologics Inc has multiple clinical trials investigating their adenovirus type 5 vectored vaccine. The initial phase 1 trial (NCT04313127) included 108 participants and tested 3 doses of the vaccine; the follow-up phase 1/2 trial (NCT04398147) includes 696 participants and is a randomized, observer-blind, dose-escalation trial in individuals 18 to 85 years of age. The third trial (phase 2: NCT04341389) includes 508 participants across 2 different doses of vaccine. The NIH’s Rocky Mountain Laboratories in Hamilton, Montana, is collaborating with the University of Oxford to develop and test a chimpanzee adenovirus (serotype Y25)–vectored SARS-CoV-2 vaccine.133 Clinical trials, including a phase 1/2 trial (NCT04324606) involving 1090 participants and a phase 2/3 trial (NCT04400838) involving 10,260 participants are under way, as are additional trials in Brazil and South Africa (NCT04444674) aimed at studying the immunogenicity and efficacy in HIV-infected participants. Oxford University has partnered with AstraZeneca to produce hundreds of millions of vaccine doses.134 The Biomedical Advanced Research and Development Authority and Janssen Research and Development, LLC have an adenovirus 26–vectored vaccine expressing the S protein. This vaccine is based on a platform that was used to rapidly create an investigational vaccine for Ebola virus. The Pasteur Institute, Themis Bioscience GmbH, and the University of Pittsburgh Center for Vaccine Research are developing a measles virus–vectored vaccine that expresses the SARS-CoV-2 S protein.135 Investigators at Mayo Clinic have an adenovirus-vectored vaccine and a recombinant measles vaccine in preclinical development.
Expert Commentary/Look Ahead
Looking into the future, we see several issues relevant to COVID-19 vaccine development:
-
•
Concerns over an “S-only” vaccine approach for an RNA virus and the possibility of viral mutation and recombination events13 that could diminish or negate the efficacy of first generation vaccines
-
•
Ongoing research into the optimal balance of vaccine-induced immunity (innate, humoral, cellular) is needed
-
•
Discussion regarding controlled human challenge models and emergency use authorization approaches to development and use of candidate vaccines
-
•
Concerns about antibody/vaccine–enhanced disease, as was observed in initial animal studies of SARS-CoV-1 vaccine candidates in both mice and ferrets
-
•
The likely need for more than one vaccine type: those for immunoimmature (intranasal?); immunosenescent (adjuvanted or high dose?); immunocompromised (immunostimulant?); and pregnant (inactivated?) individuals
-
•
The unknown efficacy and durability of vaccine-induced protection must be determined and inform vaccine administration regimens; will a “prime-boost” 2-dose strategy be needed? Periodic booster doses? Will vacinees need to be screened for preexisting antibody? Will annual boosters be needed?
-
•
SARS-CoV-2 will very likely not be the last coronavirus to cause widespread and important human infections. Governments and funders must develop mechanisms for virus surveillance, as well as ongoing antiviral and vaccine development—even in the absence of current infections and beyond normal organizational attention spans
-
•
A correlate of protection for immunity must be defined, whether for wild virus or vaccine-induced immunity
-
•
Vaccine manufacturing and distribution capacity must be developed to provide ongoing immediate capacity for vaccine manufacture of new vaccines against novel human pathogens
Conclusion
SARS-CoV-2 is now circulating in both the Northern and Southern Hemispheres. Given the likelihood of severe disease due to risk factors, and less medical and public health infrastructure in the Southern Hemisphere compared with the Northern Hemisphere, the virus is likely to recirculate back to the Northern Hemisphere in the fall/winter of 2020-2021. For this reason, and due to the severity of the disease at the population level, a safe and efficacious vaccine against COVID-19 is imperative. While accelerated vaccine development must occur, it must do so without compromising safety when used in a variety of subpopulations. Much remains to be learned in regard to SARS-CoV-2 and vaccine development.
Acknowledgments
We thank Caroline L. Vitse for her editorial assistance with the submitted manuscript.
Author contributions: Drs Poland, Ovsyannikova, Crooke, and Kennedy conducted literature searches for this review. Dr Kennedy created the figure and the figure legend. All authors contributed equally to drafting and editing the submitted manuscript.
Footnotes
Potential Competing Interests: Dr Poland is the chair of a Safety Evaluation Committee for novel investigational vaccine trials being conducted by Merck Research Laboratories and is a consultant on vaccine development for Merck & Co, Inc, Avianax LLC, Adjuvance Technologies Inc, Valneva SE, Medicago Inc, GlaxoSmithKline plc, Sanofi Pasteur, Emergent BioSolutions Inc, Dynavax Technologies, Genentech, Inc, Eli Lilly and Company, Janssen Global Services, LLC, Kentucky BioProcessing, Inc, and Genevant Sciences Corporation. Drs Poland, Ovsyannikova, and Kennedy hold patents related to vaccinia, influenza, and measles peptide vaccines and have received grant funding from ICW Healthcare Ventures for preclinical studies on a peptide-based COVID-19 vaccine. Dr Kennedy has received funding from Merck Research Laboratories to study waning immunity to mumps vaccine. These activities have been reviewed by the Mayo Clinic Conflict of Interest Review Board and are conducted in compliance with Mayo Clinic Conflict of Interest policies. Dr Crooke reports no competing interests.
References
- 1.Decaro N., Lorusso A. Novel human coronavirus (SARS-CoV-2): a lesson from animal coronaviruses. Vet Microbiol. 2020;244:108693. doi: 10.1016/j.vetmic.2020.108693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization Coronavirus disease (COVID-19): Situation Report -162. https://www.who.int/docs/default-source/coronaviruse/20200630-covid-19-sitrep-162.pdf?sfvrsn=e00a5466_2 Published June 30, 2020. Accessed June 30, 2020.
- 3.Chan J.F.-W., Yuan S., Kok K.-H., et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q., Guan X., Wu P., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu P., Hao X., Lau E.H.Y., et al. Real-time tentative assessment of the epidemiological characteristics of novel coronavirus infections in Wuhan, China, as at 22 January 2020. Euro Surveill. 2020;25(3):2000044. doi: 10.2807/1560-7917.ES.2020.25.3.2000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan J.F.-W., Kok K.-H., Zhu Z., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan [published correction appears in Emerg Microbes Infect. 2020;9(1):540] Emerg Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poland G.A. SARS-CoV-2: a time for clear and immediate action. Lancet Infect Dis. 2020;20:531–532. doi: 10.1016/S1473-3099(20)30250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan W.-J., Ni Z.-Y., Hu Y., et al. China Medical Treatment Expert Group for Covid-19 Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang R., Xia J., Chen Y., Shan C., Wu C. A family cluster of SARS-CoV-2 infection involving 11 patients in Nanjing, China. Lancet Infect Dis. 2020;20(5):534–535. doi: 10.1016/S1473-3099(20)30147-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang L., Zhao L., Gong H., Wang L., Wang L. Severe acute respiratory syndrome coronavirus 2 RNA detected in blood donations. Emerg Infect Dis. 2020;26(7):1631–1633. doi: 10.3201/eid2607.200839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen N., Zhou M., Dong X., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poland G.A. Tortoises, hares, and vaccines: a cautionary note for SARS-CoV-2 vaccine development [editorial] Vaccine. 2020;38:4219–4220. doi: 10.1016/j.vaccine.2020.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7) doi: 10.1128/JVI.00127-20. e00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun. 2020;526(1):135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gierer S., Bertram S., Kaup F., et al. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J Virol. 2013;87(10):5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K., Chen W., Zhou Y.-S., et al. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. [preprint published online March 14, 2020]. bioRxiv. [DOI]
- 20.Shieh W.-J., Hsiao C.-H., Paddock C.D., et al. Immunohistochemical, in situ hybridization, and ultrastructural localization of SARS-associated coronavirus in lung of a fatal case of severe acute respiratory syndrome in Taiwan. Hum Pathol. 2005;36(3):303–309. doi: 10.1016/j.humpath.2004.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subbarao K., McAuliffe J., Vogel L., et al. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J Virol. 2004;78(7):3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China [published correction appears in Lancet. 2020;395(10223):496] Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choudhury A., Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. https://doi.org/10.1002/jmv.25987 [published online ahead of print May 8, 2020]. J Med Virol. [DOI] [PMC free article] [PubMed]
- 24.Chen G., Wu D., Guo W., et al. Clinical and immunological features in severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thevarajan I., Nguyen T.H.O., Koutsakos M., et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med. 2020;26(4):453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu A., Peng Y., Huang B., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang N., Shang J., Jiang S., Du L. Subunit vaccines against emerging pathogenic human coronaviruses. Front Microbiol. 2020;11:298. doi: 10.3389/fmicb.2020.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song Z., Xu Y., Bao L., et al. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11(1):59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L., Liu W., Zhang Q., et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg Microbes Infect. 2020;9(1):313–319. doi: 10.1080/22221751.2020.1725399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci U S A. 2020;117(17):9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X., Dong N., Chan E.W.-C., Chen S. Genetic cluster analysis of SARS-CoV-2 and the identification of those responsible for the major outbreaks in various countries. Emerg Microbes Infect. 2020;9(1):1287–1299. doi: 10.1080/22221751.2020.1773745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Z., Li H., Wu X., et al. Moderate mutation rate in the SARS coronavirus genome and its implications. BMC Evol Biol. 2004;4:21. doi: 10.1186/1471-2148-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu X., Cox N.J., Bender C.A., Regnery H.L., Shaw M.W. Genetic variation in neuraminidase genes of influenza A (H3N2) viruses. Virology. 1996;224(1):175–183. doi: 10.1006/viro.1996.0519. [DOI] [PubMed] [Google Scholar]
- 34.Liu L., Fang Q., Deng F., et al. Natural mutations in the receptor binding domain of spike glycoprotein determine the reactivity of cross-neutralization between palm civet coronavirus and severe acute respiratory syndrome coronavirus. J Virol. 2007;81(9):4694–4700. doi: 10.1128/JVI.02389-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortega J.T., Serrano M.L., Pujol F.H., Rangel H.R. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: an in silico analysis. EXCLI J. 2020;19:410–417. doi: 10.17179/excli2020-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ou J., Zhou Z., Zhang J., et al. RBD mutations from circulating SARS-CoV-2 strains enhance the structural stability and human ACE2 affinity of the spike protein. [preprint published online March 23, 2020]. bioRxiv. [DOI]
- 37.Ou J., Zhou Z., Dai R., et al. Emergence of RBD mutations in circulating SARS-CoV-2 strains enhancing the structural stability and human ACE2 receptor affinity of the spike protein. [preprint published online April 20, 2020]. bioRxiv. [DOI]
- 38.Korber B., Fischer W.M., Gnanakaran S., et al. Sheffield COVID-19 Genomics Group Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. [preprint published online April 30, 2020]. bioRxiv. [DOI]
- 39.Daniloski Z., Guo X., Sanjana N.E. The D614G mutation in SARS-CoV-2 spike increases transduction of multiple human cell types. [preprint published online June 15, 2020]. bioRxiv. [DOI]
- 40.Zhang L., Jackson C.B., Mou H., et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. [preprint published online June 12, 2020]. bioRxiv. [DOI]
- 41.Eaaswarkhanth M., Al Madhoun A., Al-Mulla F. Could the D614G substitution in the SARS-CoV-2 spike (S) protein be associated with higher COVID-19 mortality? Int J Infect Dis. 2020;96:459–460. doi: 10.1016/j.ijid.2020.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desforges M., Desjardins J., Zhang C., Talbot P.J. The acetyl-esterase activity of the hemagglutinin-esterase protein of human coronavirus OC43 strongly enhances the production of infectious virus. J Virol. 2013;87(6):3097–3107. doi: 10.1128/JVI.02699-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li F., Berardi M., Li W., Farzan M., Dormitzer P.R., Harrison S.C. Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. J Virol. 2006;80(14):6794–6800. doi: 10.1128/JVI.02744-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu L., Liu Q., Zhu Y., et al. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan M., Wu N.C., Zhu X., et al. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368(6491):630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.World Health Organization WHO target product profiles for COVID-19 vaccines: version 3. https://www.who.int/docs/default-source/blue-print/who-target-product-profiles-for-covid-19-vaccines.pdf?sfvrsn=1d5da7ca_5 Published April 29, 2020. Accessed July 1, 2020.
- 48.U.S. Department of Health and Human Services, Food and Drug Administration, Center for Biologics Evaluation and Research. Development and Licensure of Vaccines to Prevent COVID-19: Guidance for Industry. https://www.fda.gov/media/139638/download Published June 2020. Accessed July 2, 2020.
- 49.Corey L., Mascola J.R., Fauci A.S., Collins F.S. A strategic approach to COVID-19 vaccine R&D. Science. 2020;368(6494):948–950. doi: 10.1126/science.abc5312. [DOI] [PubMed] [Google Scholar]
- 50.Xu J., Jia W., Wang P., et al. Antibodies and vaccines against Middle East respiratory syndrome coronavirus. Emerg Microbes Infect. 2019;8(1):841–856. doi: 10.1080/22221751.2019.1624482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coleman C.M., Liu Y.V., Mu H., et al. Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine. 2014;32(26):3169–3174. doi: 10.1016/j.vaccine.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Du L., Zhao G., Kou Z., et al. Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development [published correction appears in J Virol. 2013;87(21):11963] J Virol. 2013;87(17):9939–9942. doi: 10.1128/JVI.01048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du L., Kou Z., Ma C., et al. A truncated receptor-binding domain of MERS-CoV spike protein potently inhibits MERS-CoV infection and induces strong neutralizing antibody responses: implication for developing therapeutics and vaccines. PLoS One. 2013;8(12):e81587. doi: 10.1371/journal.pone.0081587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He Y., Lu H., Siddiqui P., Zhou Y., Jiang S. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J Immunol. 2005;174(8):4908–4915. doi: 10.4049/jimmunol.174.8.4908. [DOI] [PubMed] [Google Scholar]
- 55.Munster V.J., Wells D., Lambe T., et al. Protective efficacy of a novel simian adenovirus vaccine against lethal MERS-CoV challenge in a transgenic human DPP4 mouse model. NPJ Vaccines. 2017;2:28. doi: 10.1038/s41541-017-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muthumani K., Falzarano D., Reuschel E.L., et al. A synthetic consensus anti-spike protein DNA vaccine induces protective immunity against Middle East respiratory syndrome coronavirus in nonhuman primates. Sci Transl Med. 2015;7(301):301ra132. doi: 10.1126/scitranslmed.aac7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haagmans B.L., van den Brand J.M.A., Raj V.S., et al. An orthopoxvirus-based vaccine reduces virus excretion after MERS-CoV infection in dromedary camels. Science. 2016;351(6268):77–81. doi: 10.1126/science.aad1283. [DOI] [PubMed] [Google Scholar]
- 58.Zhao J., Li K., Wohlford-Lenane C., et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci U S A. 2014;111(13):4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao J., Zhao J., Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J Virol. 2010;84(18):9318–9325. doi: 10.1128/JVI.01049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao J., Zhao J., Van Rooijen N., Perlman S. Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. PLoS Pathog. 2009;5(10):e1000636. doi: 10.1371/journal.ppat.1000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Q., Zhang L., Kuwahara K., et al. Immunodominant SARS coronavirus epitopes in humans elicited both enhancing and neutralizing effects on infection in non-human primates [published correction appears in ACS Infect Dis. 2020;6(5):1284-1285] ACS Infect Dis. 2016;2(5):361–376. doi: 10.1021/acsinfecdis.6b00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang S.-F., Tseng S.-P., Yen C.-H., et al. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem Biophys Res Commun. 2014;451(2):208–214. doi: 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yip M.S., Leung N.H.L., Cheung C.Y., et al. Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virol J. 2014;11:82. doi: 10.1186/1743-422X-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jaume M., Yip M.S., Cheung C.Y., et al. Anti-severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH- and cysteine protease-independent FcγR pathway. J Virol. 2011;85(20):10582–10597. doi: 10.1128/JVI.00671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bolles M., Deming D., Long K., et al. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J Virol. 2011;85(23):12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weingartl H., Czub M., Czub S., et al. Immunization with modified vaccinia virus Ankara-based recombinant vaccine against severe acute respiratory syndrome is associated with enhanced hepatitis in ferrets. J Virol. 2004;78(22):12672–12676. doi: 10.1128/JVI.78.22.12672-12676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Agrawal A.S., Tao X., Algaissi A., et al. Immunization with inactivated Middle East Respiratory Syndrome coronavirus vaccine leads to lung immunopathology on challenge with live virus. Hum Vaccin Immunother. 2016;12(9):2351–2356. doi: 10.1080/21645515.2016.1177688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Czub M., Weingartl H., Czub S., He R., Cao J. Evaluation of modified vaccinia virus Ankara based recombinant SARS vaccine in ferrets. Vaccine. 2005;23(17-18):2273–2279. doi: 10.1016/j.vaccine.2005.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Perlman S., Dandekar A.A. Immunopathogenesis of coronavirus infections: implications for SARS. Nat Rev Immunol. 2005;5(12):917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Graham B.S. Rapid COVID-19 vaccine development. Science. 2020;368(6494):945–946. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- 71.Ou X., Liu Y., Lei X., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wong S.K., Li W., Moore M.J., Choe H., Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem. 2004;279(5):3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang H., Li Y., Zhang H., et al. Global profiling of SARS-CoV-2 specific IgG/IgM responses of convalescents using a proteome microarray. [preprint published online March 27, 2020]. medRxiv. [DOI]
- 74.Okba N.M.A., Müller M.A., Li W., et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020;26(7):1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ni L., Ye F., Cheng M.-L., et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52(6):971–977.e973. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Padoan A., Sciacovelli L., Basso D., et al. IgA-Ab response to spike glycoprotein of SARS-CoV-2 in patients with COVID-19: a longitudinal study. Clin Chim Acta. 2020;507:164–166. doi: 10.1016/j.cca.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kuri-Cervantes L., Pampena M.B., Meng W., et al. Immunologic perturbations in severe COVID-19/SARS-CoV-2 infection. [preprint published online May 18, 2020]. bioRxiv. [DOI]
- 78.Braun J., Loyal L., Frentsch M., et al. Presence of SARS-CoV-2 reactive T cells in COVID-19 patients and healthy donors. [preprint published online April 22, 2020]. medRxiv. [DOI]
- 79.Grifoni A., Weiskopf D., Ramirez S.I., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peng Y., Mentzer A.J., Liu G., et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent COVID-19 patients. [preprint published online June 8, 2020]. bioRxiv. [DOI] [PMC free article] [PubMed]
- 81.Zhu F.-C., Li Y.-H., Guan X.-H., et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395(10240):1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Neidleman J., Luo X., Frouard J., et al. SARS-CoV-2-specific T cells exhibit unique features characterized by robust helper function, lack of terminal differentiation, and high proliferative potential. [preprint published online June 8, 2020]. bioRxiv. [DOI]
- 83.Juno J.A., Tan H.-X., Lee W.S., et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. https://doi.org/10.1038/s41591-020-0995-0 [published online ahead of print July 13, 2020]. Nat Med. [DOI] [PubMed]
- 84.Sekine T., Perez-Potti A., Rivera-Ballesteros O., et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. [preprint published online June 29, 2020]. bioRxiv. [DOI] [PMC free article] [PubMed]
- 85.Lundegaard C., Lund O., Nielsen M. Prediction of epitopes using neural network based methods. J Immunol Methods. 2011;374(1-2):26–34. doi: 10.1016/j.jim.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang H.-W., Pai T.-W. Machine learning-based methods for prediction of linear B-cell epitopes. Methods Mol Biol. 2014;1184:217–236. doi: 10.1007/978-1-4939-1115-8_12. [DOI] [PubMed] [Google Scholar]
- 87.Nielsen M., Lundegaard C., Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinformatics. 2007;8:238. doi: 10.1186/1471-2105-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Larsen M.V., Lundegaard C., Lamberth K., Buus S., Lund O., Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinformatics. 2007;8:424. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Larsen M.V., Lundegaard C., Lamberth K., et al. An integrative approach to CTL epitope prediction: a combined algorithm integrating MHC class I binding, TAP transport efficiency, and proteasomal cleavage predictions. Eur J Immunol. 2005;35(8):2295–2303. doi: 10.1002/eji.200425811. [DOI] [PubMed] [Google Scholar]
- 90.Hoof I., Peters B., Sidney J., et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61(1):1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jurtz V., Paul S., Andreatta M., Marcatili P., Peters B., Nielsen M. NetMHCpan-4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol. 2017;199(9):3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jensen K.K., Andreatta M., Marcatili P., et al. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology. 2018;154(3):394–406. doi: 10.1111/imm.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Larsen J.E.P., Lund O., Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2:2. doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haste Andersen P., Nielsen M., Lund O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006;15(11):2558–2567. doi: 10.1110/ps.062405906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Johnson K.L., Ovsyannikova I.G., Mason C.J., Bergen H.R., III, Poland G.A. Discovery of naturally processed and HLA-presented class I peptides from vaccinia virus infection using mass spectrometry for vaccine development. Vaccine. 2009;28(1):38–47. doi: 10.1016/j.vaccine.2009.09.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tahir Ul Qamar M., Saleem S., Ashfaq U.A., Bari A., Anwar F., Alqahtani S. Epitope-based peptide vaccine design and target site depiction against Middle East Respiratory Syndrome Coronavirus: an immune-informatics study. J Transl Med. 2019;17(1):362. doi: 10.1186/s12967-019-2116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shi J., Zhang J., Li S., et al. Epitope-based vaccine target screening against highly pathogenic MERS-CoV: an in silico approach applied to emerging infectious diseases. PLoS One. 2015;10(12):e0144475. doi: 10.1371/journal.pone.0144475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Srivastava S., Kamthania M., Singh S., Saxena A.K., Sharma N. Structural basis of development of multi-epitope vaccine against Middle East respiratory syndrome using in silico approach. Infect Drug Resist. 2018;11:2377–2391. doi: 10.2147/IDR.S175114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Crooke S.N., Ovsyannikova I.G., Kennedy R.B., Poland G.A. Immunoinformatic identification of B cell and T cell epitopes in the SARS-CoV-2 proteome. Sci Rep. 2020;10(1):14179. doi: 10.1038/s41598-020-70864-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27(4):671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Baruah V., Bose S. Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J Med Virol. 2020;92(5):495–500. doi: 10.1002/jmv.25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Santoni D., Vergni D. In the search of potential epitopes for Wuhan seafood market pneumonia virus using high order nullomers. J Immunol Methods. 2020;481-482:112787. doi: 10.1016/j.jim.2020.112787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee C.H., Koohy H. In silico identification of vaccine targets for 2019-nCoV. F1000Res. 2020;9:145. doi: 10.12688/f1000research.22507.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nguyen A., David J.K., Maden S.K., et al. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. J Virol. 2020;94(13):e00510–e00520. doi: 10.1128/JVI.00510-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Campbell K.M., Steiner G., Wells D.K., Ribas A., Kalbasi A. Prediction of SARS-CoV-2 epitopes across 9360 HLA class I alleles. [preprint published online April 1, 2020]. bioRxiv. [DOI]
- 106.Enayatkhani M., Hasaniazad M., Faezi S., et al. Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: an in silico study. https://doi.org/10.1080/07391102.2020.1756411 [published online ahead of print May 2, 2020]. J Biomol Struct Dyn. [DOI] [PMC free article] [PubMed]
- 107.Bhattacharya M., Sharma A.R., Patra P., et al. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS-COV-2): immunoinformatics approach. J Med Virol. 2020;92(6):618–631. doi: 10.1002/jmv.25736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jakhar R., Kaushik S., Gakhar S.K. 3CL hydrolase-based multiepitope peptide vaccine against SARS-CoV-2 using immunoinformatics. https://doi.org/10.1002/jmv.25993 [published online ahead of print May 7, 2020]. J Med Virol. [DOI] [PubMed]
- 109.Jackson L.A., Anderson E.J., Rouphael N.G., et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. https://doi.org/10.1056/NEJMoa2022483 [published online ahead of print July 14, 2020]. N Engl J Med. [DOI] [PMC free article] [PubMed]
- 110.Chen W.-H., Strych U., Hotez P.J., Bottazzi M.E. The SARS-CoV-2 vaccine pipeline: an overview. https://doi.org/10.1007/s40475-020-00201-6 [published online ahead of print March 3, 2020]. Curr Trop Med Rep. [DOI] [PMC free article] [PubMed]
- 111.Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52(4):583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pang J., Wang M.X., Ang I.Y.H., et al. Potential rapid diagnostics, vaccine and therapeutics for 2019 novel coronavirus (2019-nCoV): a systematic review. J Clin Med. 2020;9(3):623. doi: 10.3390/jcm9030623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mueller S., Stauft C.B., Kalkeri R., et al. A codon-pair deoptimized live-attenuated vaccine against respiratory syncytial virus is immunogenic and efficacious in non-human primates. Vaccine. 2020;38(14):2943–2948. doi: 10.1016/j.vaccine.2020.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stauft C.B., Yang C., Coleman J.R., et al. Live-attenuated H1N1 influenza vaccine candidate displays potent efficacy in mice and ferrets. PLoS One. 2019;14(10):e0223784. doi: 10.1371/journal.pone.0223784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stauft C.B., Shen S.H., Song Y., et al. Extensive recoding of dengue virus type 2 specifically reduces replication in primate cells without gain-of-function in Aedes aegypti mosquitoes. PLoS One. 2018;13(9):e0198303. doi: 10.1371/journal.pone.0198303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu S.-J., Leng C.-H., Lien S.-P., et al. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine. 2006;24(16):3100–3108. doi: 10.1016/j.vaccine.2006.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zheng N., Xia R., Yang C., et al. Boosted expression of the SARS-CoV nucleocapsid protein in tobacco and its immunogenicity in mice. Vaccine. 2009;27(36):5001–5007. doi: 10.1016/j.vaccine.2009.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shanmugaraj B., Malla A., Phoolcharoen W. Emergence of novel coronavirus 2019-nCoV: need for rapid vaccine and biologics development. Pathogens. 2020;9(2):148. doi: 10.3390/pathogens9020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bengtsson K.L., Song H., Stertman L., et al. Matrix-M adjuvant enhances antibody, cellular and protective immune responses of a Zaire Ebola/Makona virus glycoprotein (GP) nanoparticle vaccine in mice. Vaccine. 2016;34(16):1927–1935. doi: 10.1016/j.vaccine.2016.02.033. [DOI] [PubMed] [Google Scholar]
- 120.Magnusson S.E., Altenburg A.F., Bengtsson K.L., et al. Matrix-M™ adjuvant enhances immunogenicity of both protein- and modified vaccinia virus Ankara-based influenza vaccines in mice. Immunol Res. 2018;66(2):224–233. doi: 10.1007/s12026-018-8991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.University of Queensland UQ COVID-19 vaccine shown to induce potent protective response in pre-clinical trials. https://www.uq.edu.au/news/article/2020/04/uq-covid-19-vaccine-shown-induce-potent-protective-response-pre-clinical-trials Published April 29, 2020. Accessed Juiy 1, 2020.
- 122.Vaxart, Inc Five COVID-19 vaccine candidates in preclinical testing [press release] https://investors.vaxart.com/news-releases/news-release-details/vaxart-provides-update-its-oral-covid-19-vaccine-program Published March 31, 2020. Accessed Juiy 1, 2020.
- 123.Meir R., Krispel S., Simanov L., Eliahu D., Maharat O., Pitcovski J. Immune responses to mucosal vaccination by the recombinant A1 and N proteins of infectious bronchitis virus. Viral Immunol. 2012;25(1):55–62. doi: 10.1089/vim.2011.0050. [DOI] [PubMed] [Google Scholar]
- 124.Ovsyannikova I.G., Johnson K.L., Bergen H.R., III, Poland G.A. Mass spectrometry and peptide-based vaccine development. Clin Pharmacol.Ther. 2007;82(6):644–652. doi: 10.1038/sj.clpt.6100389. [DOI] [PubMed] [Google Scholar]
- 125.Ovsyannikova I.G., Johnson K.L., Muddiman D.C., Vierkant R.A., Poland G.A. Identification and characterization of novel, naturally processed measles virus class II HLA-DRB1 peptides. J Virol. 2004;78(1):42–51. doi: 10.1128/JVI.78.1.42-51.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Johnson K.L., Ovsyannikova I.G., Poland G.A., Muddiman D.C. Identification of class II HLA-DRB1∗03-bound measles virus peptides by 2D-liquid chromatography tandem mass spectrometry. J Proteome Res. 2005;4(6):2243–2249. doi: 10.1021/pr0501416. [DOI] [PubMed] [Google Scholar]
- 127.Ovsyannikova I.G., Johnson K.L., Naylor S., Muddiman D.C., Poland G.A. Naturally processed measles virus peptide eluted from class II HLA-DRB1∗03 recognized by T lymphocytes from human blood. Virology. 2003;312:495–506. doi: 10.1016/s0042-6822(03)00281-2. [DOI] [PubMed] [Google Scholar]
- 128.Mohsen M.O., Zha L., Cabral-Miranda G., Bachmann M.F. Major findings and recent advances in virus-like particle (VLP)-based vaccines. Semin Immunol. 2017;34:123–132. doi: 10.1016/j.smim.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 129.Kim E., Erdos G., Huang S., et al. Microneedle array delivered recombinant coronavirus vaccines: immunogenicity and rapid translational development. EBioMedicine. 2020;55:102743. doi: 10.1016/j.ebiom.2020.102743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.China fast-tracks novel coronavirus vaccine development. http://www.xinhuanet.com/english/2020-01/28/c_138739378.htm Published January 28, 2020. Accessed July 1, 2020.
- 131.World Health Organization DRAFT landscape of COVID-19 candidate vaccines – 23 April 2020. https://www.who.int/blueprint/priority-diseases/key-action/draft-landscape-COVID-19-candidate-vaccines-23-April-2020.pdf
- 132.Mulligan M.J., Lyke K.E., Kitchin N., et al. Phase 1/2 study to describe the safety and immunogenicity of a COVID-19 RNA vaccine candidate (BNT162b1) in adults 18 to 55 years of age: interim report. [preprint published online July 1, 2020]. medRxiv. [DOI]
- 133.Hillis W.D., Goodman R. Serologic classification of chimpanzee adenoviruses by hemagglutination and hemagglutination inhibition. J Immunol. 1969;103(5):1089–1095. [PubMed] [Google Scholar]
- 134.Kirka D. AstraZeneca agrees to make COVID-19 vaccine for Europe. Medical Xpress website. https://medicalxpress.com/news/2020-06-astrazeneca-covid-vaccine-europe.html Published June 13, 2020. Accessed July 1, 2020.
- 135.UPMC. Researchers in Pittsburgh, Paris and Vienna win grant for COVID-19 vaccine [press release] https://www.upmc.com/media/news/032020-cepi-grant Published March 20, 2020. Accessed July 1, 2020.