Abstract
SARS-CoV-2 infection has emerged as a serious global pandemic. Because of the high transmissibility of the virus and the high rate of morbidity and mortality associated with COVID-19, developing effective and safe vaccines is a top research priority. Here, we provide a detailed evaluation of the immunogenicity of lipid nanoparticle-encapsulated, nucleoside-modified mRNA (mRNA-LNP) vaccines encoding the full-length SARS-CoV-2 spike protein or the spike receptor binding domain in mice. We demonstrate that a single dose of these vaccines induces strong type 1 CD4+ and CD8+ T cell responses, as well as long-lived plasma and memory B cell responses. Additionally, we detect robust and sustained neutralizing antibody responses and the antibodies elicited by nucleoside-modified mRNA vaccines do not show antibody-dependent enhancement of infection in vitro. Our findings suggest that the nucleoside-modified mRNA-LNP vaccine platform can induce robust immune responses and is a promising candidate to combat COVID-19.
Keywords: SARS-CoV-2, COVID-19, mRNA vaccine, nucleoside-modified mRNA, mRNA-LNP
Graphical Abstract

SARS-CoV-2 mRNA-based vaccines are among the most promising vaccine candidates against COVID-19. Laczkó et al. evaluate two nucleoside-modified mRNA vaccine candidates in mice and demonstrate that they induce potent T and B cell immune responses and high levels of neutralizing antibodies after administration of a single vaccine dose.
Introduction
SARS-CoV-2, the causative agent of COVID-19, causes significant mortality and morbidity worldwide and was declared a pandemic by the World Health Organization in March, 2020 (Cucinotta and Vanelli, 2020). The rapid spread of the virus has caused not only a significant health care burden but also an economic crisis. Governments around the world have introduced strict social distancing measures to keep transmission under control. However, a vaccine will ultimately be required to fully suppress the SARS-CoV-2 pandemic. A wide variety of vaccine designs are being developed against SARS-CoV-2, and there has been promising preclinical and clinical efficacy data published for at least four different vaccine candidates (Gao et al., 2020; Jackson et al., 2020; Smith et al., 2020; Yu et al., 2020).
Messenger RNA (mRNA)-based vaccines have recently demonstrated great promise in the fight against infectious diseases (Alameh et al., 2020). One of the most widely studied of these vaccine platforms uses antigen-encoding nucleoside-modified mRNA encapsulated into lipid nanoparticles (mRNA-LNP) (Pardi et al., 2015). Nucleoside-modified mRNA-LNP vaccines have induced protective immune responses against various pathogens in preclinical studies (Awasthi et al., 2019; Espeseth et al., 2020; Freyn et al., 2020; Meyer et al., 2018; Pardi et al., 2017, 2018a, 2018c; Richner et al., 2017; Roth et al., 2019), in many cases, after administration of a single dose (Freyn et al., 2020; Pardi et al., 2017, 2018a, 2018c). This vaccine type has been shown to elicit particularly strong CD4+ T cell, germinal center B cell and long-lived plasma cell responses associated with durable, protective neutralizing antibody responses (Lindgren et al., 2017; Pardi et al., 2018a). Of note, a very limited amount of published human clinical data are available on the safety and efficacy of mRNA-LNPs (Bahl et al., 2017; Feldman et al., 2019), and no mRNA vaccines for humans have been licensed to date.
In this study, we assessed the immunogenicity of two nucleoside-modified mRNA-LNP vaccines targeting the spike (S) glycoprotein of SARS-CoV-2: one is encoding the full-length S protein with deleted furin cleavage site and the other is encoding the S protein receptor binding domain (RBD). We evaluated immune responses after a single intramuscular (i.m.) injection with the SARS-CoV-2 mRNA-LNP or control vaccines in BALB/c mice. We found that both mRNA vaccines induced potent CD4+ and CD8+ T cell responses in the spleen and lung. Moreover, we measured strong long-lived plasma cell and memory B cell responses associated with high titers of neutralizing antibodies. Importantly, our nucleoside-modified (1-methylpseudouridine-containing) SARS-CoV-2 mRNA-LNP vaccines are very similar to clinical vaccine candidates utilized by Moderna Therapeutics (Jackson et al., 2020) and BioNTech RNA Pharmaceuticals in partnership with Pfizer (Mulligan et al., 2020); thus, we believe that the studies described within this manuscript may inform ongoing clinical trials and the design of future human trials.
Results
In Vitro Characterization of SARS-CoV-2 Nucleoside-Modified mRNA Constructs
We designed and produced mRNAs encoding three potential SARS-CoV-2 vaccine antigens: full-length S protein (wild-type [WT]), full-length S protein with a deleted furin cleavage site (Δfurin), and a short construct encoding the soluble RBD of S protein. The Δfurin mutant was included as a potential way to stabilize the full-length S and to maintain the covalent association of the S1 and S2 subunits (Kirchdoerfer et al., 2016), while the RBD was investigated as it is a critical target of neutralizing antibodies against SARS-CoV-2. Protein expression from mRNAs was confirmed by in vitro cell transfection studies. RBD protein secretion was demonstrated by ELISA using supernatant from RBD mRNA-transfected 293F cells (Figure 1 A). Because the full-length WT and Δfurin S proteins contain the transmembrane domain, they were expressed on the surface of transfected 293F cells. Thus, we used flow cytometry to assess binding of full-length WT and Δfurin S proteins by an anti-RBD monoclonal antibody, D001, and a human ACE2-Fc (hACE2-Fc) fusion protein. Interestingly, we found that the full-length Δfurin S protein showed higher binding capacity to D001 and hACE2-Fc compared to its WT counterpart, indicating that it may be a better vaccine antigen, due either to higher expression or favorable antigenicity (Figure 1B). Therefore, we selected the full-length Δfurin construct to evaluate in immunization studies along with RBD.
Figure 1.
In Vitro Characterization of SARS-CoV-2 Nucleoside-Modified mRNA Constructs
(A) Supernatant from 293F cells transfected with RBD-encoding mRNA or mock was tested for binding reactivity to D001 and hACE2-Fc by ELISA. Data shown are area under curve of the log-transformed concentrations (log AUC). Symbols represent independent experiments.
(B) 293F cells were transfected with mRNA encoding SARS-CoV-2 full-length WT and Δfurin S protein. Binding reactivity of full-length WT and Δfurin S proteins to D001, hACE2-Fc, and negative control CH65 (an anti-influenza neutralizing antibody) was measured by flow cytometry. Binding capacity was expressed in mean fluorescence intensity (MFI). Each dot represents an independent experiment. p value indicates a paired t test; ∗p < 0.05. Data represent mean plus SEM.
SARS-CoV-2 mRNA Vaccines Induce Strong T Cell Responses in the Spleen and Lungs
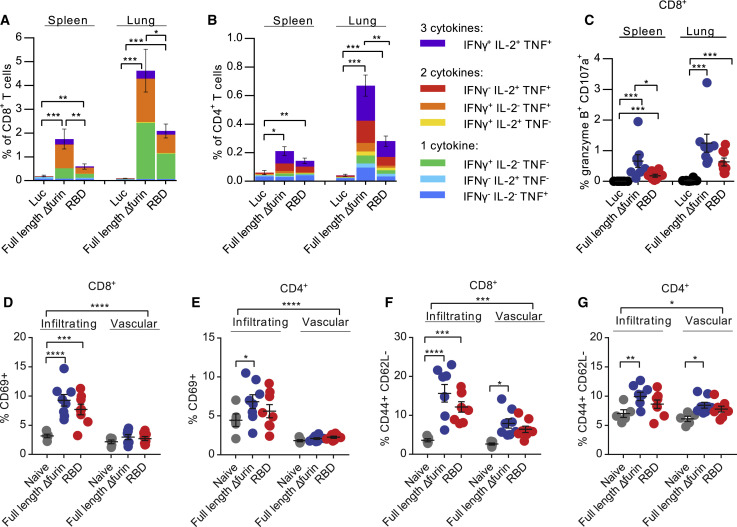
BALB/c mice were injected with a single i.m. dose of 30 μg of mRNA-LNPs encoding full-length Δfurin, RBD, or firefly luciferase (Luc, negative control) mRNA-LNPs, and S protein-specific CD4+ and CD8+ T cell responses were evaluated after 10 days by intracellular cytokine staining (Figures 2 , S1, and S2). Both spike mRNA constructs elicited antigen-specific, polyfunctional CD8+ (Figure 2A) and CD4+ (Figure 2B) T cells expressing type 1 (Th1) immune response cytokines (interferon [IFN]-γ, tumor necrosis factor [TNF], and interleukin [IL]-2) as well as CD8+ T cells with cytotoxic markers (granzyme B+ CD107a+) (Figure 2C) in both the spleen and lungs. These responses were particularly robust in the lungs, especially for CD8+ T cells. We also noted that the vast majority of the CD8+ T cell response in BALB/c mice was directed at epitopes in the N-terminal half of the S protein, while CD4+ T cells recognized epitopes in both halves of the protein (Figures S2A and S2B). Because S protein-specific lung-infiltrating T cell responses may contribute to SARS-CoV-2 vaccine protection as seen with SARS-CoV-1 (Zhao et al., 2016), we next examined whether vaccine-induced lung T cells were truly infiltrating into the lung parenchyma. We performed intravenous (i.v.) labeling with a CD45-specific antibody in order to differentiate between vascular (i.v. label-positive) and tissue-infiltrating (i.v. label-negative) lung CD4+ and CD8+ T cells (Figures 2D–2G, S1C, S2C, and S2D). SARS-CoV-2 mRNA-LNP vaccines elicited significant increases in activated (CD69+ or PD-1+) and antigen-experienced (CD44+CD62L−) CD8+ and CD4+ T cells that were tissue-infiltrating, with comparatively modest increases in the vasculature, suggesting that activated vaccine-induced T cells readily exit the vasculature and enter the lung parenchyma (Figures 2D–2G, S2C, and S2D). Of note, in each of the above assays, we found that the full-length Δfurin vaccine induced greater T cell responses compared to the RBD vaccine; this might be explained by the presence of additional T cell epitopes in the longer protein product produced by the full-length Δfurin construct.
Figure 2.
SARS-CoV-2 mRNA Vaccines Induce S Protein-Specific Type 1 Cellular Responses
BALB/c mice were vaccinated i.m. with a single dose of 30 μg of mRNA-LNP vaccines.
(A–C) Spleen and lungs were harvested and stimulated with SARS-CoV-2 S protein peptide pools 10 days after immunization. (A) CD8+ and (B) CD4+ T cells were stained for type 1 intracellular cytokine expression and (C) CD8+ T cells for cytolytic markers granzyme B and CD107a as well.
(D–G) Cells were stained directly ex vivo for activation markers, showing the proportion of i.v.-label negative (tissue-“infiltrating”) and i.v.-label positive (“vascular”) T cells that are (D and E) CD69+ and (F and G) CD44+ CD62L− in lung. n = 8 mice per vaccine group and n = 5 naive mice, pooled from two independent experiments. Naive mice were age matched, non-immunized BALB/c mice. (C–G) Symbols represent individual animals. Data shown are mean plus SEM. Statistical analysis: (A–C) Kruskal-Wallis and post hoc Mann-Whitney U tests with Bonferroni correction and (D–G) two-way repeated-measures ANOVA test with multiple post hoc comparisons with Dunnett’s correction. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
See also Figures S1 and S2.
SARS-CoV-2 mRNA Vaccines Elicit Potent and Sustained Humoral Immune Responses without In Vitro Antibody-Dependent Enhancement Activity
Mice were immunized i.m. with a single dose of 30 μg of full-length Δfurin, RBD, and Luc mRNA-LNPs, and antibody responses were evaluated. Both SARS-CoV-2 vaccines induced high levels of S protein-specific IgG by 4 weeks post immunization, and IgG titers further increased by week 9 (Figure 3 A). Using a vesicular stomatitis virus (VSV)-based pseudovirus neutralization assay, we demonstrated that nucleoside-modified SARS-CoV-2 mRNA-LNP vaccines induced high and sustained levels of neutralizing antibodies after administration of a single vaccine dose, with week 9 sera showing higher neutralization activity than week 4 (Figure 3B). Importantly, induction of antibodies with high neutralization titers was also demonstrated by microneutralization assay using live SARS-CoV-2 with week 9 post immunization samples (Figure 3C). Both assays indicated that the full-length Δfurin mRNA-LNPs generated slightly higher levels of neutralizing antibodies than the RBD vaccine at 9 weeks post immunization (Figures 3B and 3C). Importantly, the levels of neutralization we observed are similar to those mediated by SARS-CoV-2 immune human convalescent plasma samples analyzed by the same laboratory in the same assay format (Amanat et al., 2020).
Figure 3.
Humoral Immune Responses after SARS-CoV-2 mRNA Vaccination
BALB/c mice received a single i.m. immunization with 30 μg of SARS-CoV-2 or Luc mRNA-LNP vaccines.
(A and B) S protein-specific IgG levels were determined by endpoint dilution ELISA (A) and neutralizing antibody (Nab) levels were measured by a VSV-based pseudovirus neutralization assay (B) before immunization and 4 and 9 weeks post immunization.
(C) Nab levels were further confirmed by microneutralization assay using serum obtained 9 weeks post vaccination. n = 10 mice/group. Naive mice were age matched, non-immunized BALB/c mice. (A–C) Symbols represent individual animals. Horizontal lines represent the limit of detection. End-point dilution ELISA, FRNT50, and IC50 titers below the limit of detection are reported as half of the limit of detection. Data shown are mean plus SEM.
(D) HEK293T cells transfected to express mFcγR1 were infected with SARS-CoV-2 pseudovirus or ZIKV virus-like particles preincubated with serially diluted anti-SARS-CoV-2 sera obtained 9 weeks post immunization or anti-ZIKV sera, respectively. Serum samples were pooled from 5 mice belonging to the same experimental group. Infection level was measured by luciferase assays. Mean ± SEM of three independent experiments is presented. Statistical analysis: (A and B) two-way ANOVA and (C) one-way ANOVA with Tukey’s multiple comparison on log-transformed data. (D) SARS-CoV-2: there are no significant differences when analyzed by two-way ANOVA with Tukey’s multiple comparisons test; ZIKV: two-way ANOVA with Sidak’s multiple comparisons test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
See also Figure S3.
Antibody-dependent enhancement (ADE) of infection by virus-specific antibodies is a potential serious concern for several vaccines including those for Zika and dengue viruses and coronaviruses (Smatti et al., 2018). Thus, we investigated whether or not the SARS-CoV-2 mRNA vaccine-elicited antibodies induced ADE of infection in HEK293T cells expressing mouse FcγR1 (mFcγR1-293T cells). We demonstrated that none of the mRNA-vaccinated mouse immune sera mediated SARS-CoV-2 ADE under in vitro conditions. As a positive control to validate the mFcγR1-293T cells as generally susceptible to ADE, we show robust ADE of Zika virus (ZIKV) infection by sera derived from ZIKV-infected mice (Figure 3D) using the same system. Although ADE assays are typically conducted in the absence of the bona fide viral entry receptor, to examine whether the viral receptor affects ADE efficiency, cells expressing hACE2 as well as mFcγR1 were also used in ADE assays. SARS-CoV-2 pseudovirus infection of hACE2/mFcγR1-293T cells was efficiently neutralized as expected by sera derived from mice vaccinated with the full-length Δfurin or RBD mRNAs at low dilutions (Figure S3), and there was no enhanced infection observed at any serum dilution. As in the mFcγR1-293T cells, ZIKV-immune mouse sera mediated robust ADE in the hACE2/mFcγR1-293T cells. These results demonstrate that neither the full-length Δfurin nor RBD mRNA-LNP vaccines generate antibodies with mFcγR1-dependent ADE activity in vitro.
SARS-CoV-2 mRNA Vaccines Induce Strong Long-Lived Plasma Cell and Memory B Cell Responses
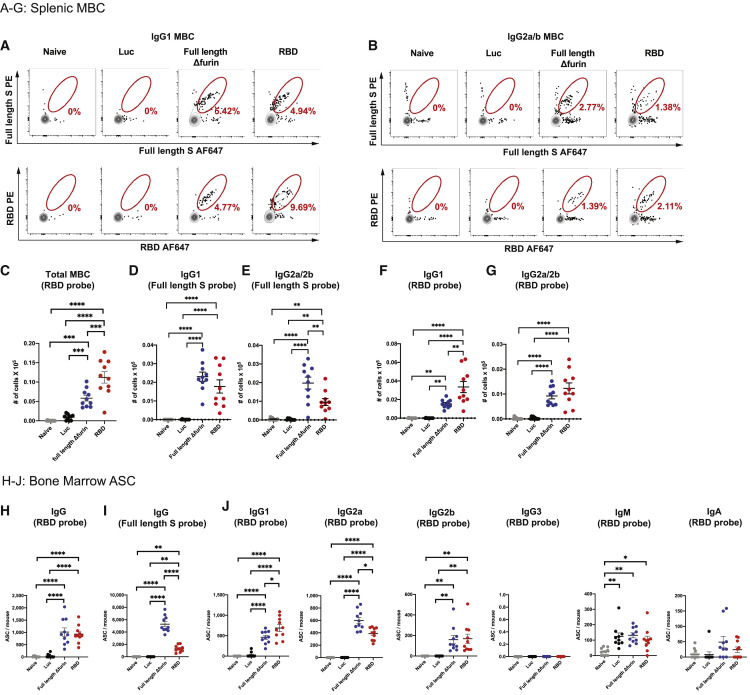
Most successful vaccine approaches rely on the generation of memory B cells (MBC) and long-lived plasma cells (LLPC) (Sallusto et al., 2010). While MBCs can mount rapid recall responses upon a secondary exposure, LLPCs residing in the bone marrow contribute to protection from infection by a persistent production of antigen-specific antibodies. To examine the magnitude and quality of antigen-specific LLPC and MBC responses, mice were immunized i.m. with a single dose of 30 μg of full-length Δfurin, RBD, or Luc mRNA-LNPs and sacrificed 9 weeks post vaccination (Figures 4 and S4). Splenic full-length S protein and RBD-specific IgG1 (Figures 4A, 4D, 4F, S4A, and S4C) and IgG2a/b (Figures 4B, 4E, 4G, and S4C) expressing MBCs were identified by flow cytometry. Of note, we found that a single immunization with the SARS-CoV-2 mRNA vaccines resulted in the generation of antigen-specific class-switched MBCs. We also observed a significant increase of full-length S protein and RBD-specific IgM B cells (Figures S4B–S4D). To assess antigen-specific LLPC responses, bone marrow was collected from vaccinated mice, and the number of full-length S protein and RBD-specific antibody secreting cells (ASC) was determined (Figures 4H and 4I). Both SARS-CoV-2 mRNA vaccines induced high levels of antigen-specific ASCs after administration of a single vaccine dose. Various subsets of RBD-specific Ig-producing cells were further characterized by ELISPOT (Figure 4J). We found that ASCs primarily produce antigen-specific IgG1, IgG2a, and IgG2b.
Figure 4.
SARS-CoV-2 mRNA Vaccines Elicit Antigen-Specific MBC and LLPC Responses
BALB/c mice received a single i.m. immunization with 30 μg of SARS-CoV-2 or Luc mRNA-LNP vaccines and sacrificed 9 weeks post immunization.
(A and B) Representative flow cytometry staining of full-length Δfurin and RBD-specific splenic (A) IgG1 and (B) IgG2a/2b memory B cells (MBC).
(C) Quantification of total splenic RBD-specific MBC.
(D and E) Quantification of splenic full-length Δfurin-specific (D) IgG1 and (E) IgG2a/2b MBC.
(F and G) Quantification of RBD-specific splenic (F) IgG1 and (G) IgG2a/2b MBC.
(H and I) Quantification of bone marrow (H) RBD and (I) full-length Δfurin-specific IgG antibody secreting cells (ASC).
(J) Quantification of bone marrow RBD-specific IgG1, IgG2a, IgG2b, IgG3, IgM and IgA ASCs. n = 10 mice per group, pooled from two independent experiments. Naive mice were age-matched, non-immunized BALB/c mice. Symbols represent individual animals. Data shown are mean plus SEM. Statistical analysis: one-way ANOVA with Bonferroni correction, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
See also Figure S4.
Discussion
The rapid spread of COVID-19 has caused a global health tragedy and significant economic loss. Safe, effective, and widely available vaccines could end the pandemic; thus, generating such vaccines is an urgent unmet clinical need. Currently, multiple SARS-CoV-2 vaccine candidates are under development using conventional immunogens (live attenuated virus, inactivated virus, and adjuvanted protein subunit vaccines), as well as viral and genetic (DNA and mRNA) approaches (Amanat and Krammer, 2020). mRNA-based vaccines are among the lead candidates because they have been shown to be generally highly effective in small and large animal models (Pardi et al., 2018b) and have additional beneficial features over other vaccine platforms. First, mRNA is non-infectious, cannot integrate into the genome, and incorporation of modified nucleosides into the mRNA sequence along with proper purification (Baiersdörfer et al., 2019; Karikó et al., 2005, 2008; Weissman et al., 2013) can decrease its inflammatory capacity; thus mRNA vaccines may have a stronger safety profile than virus-based (potentially integrating, replicating, or contagious) vaccine platforms. Second, mRNA vaccine antigens can be very rapidly designed and produced, as illustrated by Moderna Therapeutics who produced a SARS-CoV-2 nucleoside-modified mRNA-LNP vaccine for human use in just 42 days after obtaining the sequence of the target antigen. Third, nucleoside-modified mRNA-LNP vaccines have been demonstrated to induce extremely potent and protective immune responses against other viruses after administration of a single dose in animal models (Pardi et al., 2017, 2018a, 2018c). If reproducible in humans, this one-time immunization feature of the vaccine platform would be particularly important in a pandemic or epidemic setting, by inducing immune protection after a single point-of-care visit.
Despite active research, no peer-reviewed preclinical studies have been published on nucleoside-modified SARS-CoV-2 mRNA-LNP vaccines to date. Here, we provide a detailed evaluation of two nucleoside-modified SARS-CoV-2 mRNA-LNP vaccine formulations (full-length Δfurin and RBD) in mice. Animals were immunized only once in all experiments presented in this study. Both the full-length Δfurin and RBD mRNA vaccines induced potent CD4+ and CD8+ T cell responses, particularly in the lungs. The production of Th1 cytokines was strong, with approximately one half of detectable S protein-specific CD4+ T cells producing the hallmark Th1 cytokine IFN-γ, along with a high frequency of IFN-γ-producing CD8+ T cells. This result is noteworthy, as a major safety consideration of SARS-CoV-2 vaccine design is the elicitation of a strong Th1-biased immune response, instead of a Th2-biased response that might induce vaccine-associated enhanced respiratory disease (VAERD) (Graham, 2020). A potential protective role of lung-homing T cells has been suggested by the SARS-CoV-1 literature (Zhao et al., 2016) and prompted us to examine T cell responses in the lung. Interestingly, we found that S protein-specific CD4+ and CD8+ T cell responses were substantially higher in the lung compared to the spleen, leading us to hypothesize that T cells raised by this format of nucleoside-modified mRNA-LNP vaccine may preferentially home to the lungs, or at least to mucosal surfaces in general. Probing further, we found that a large fraction of these cells could be detected in the lung parenchyma, indicating that these cells might be well positioned to contribute to immune protection. Future studies should examine the role of T cells in immune protection from SARS-CoV-2 infection and diseases, particularly at later time points.
Induction of long-lived plasma cells (LLPCs) and memory B cells (MBCs) is very important to achieve durable protective humoral immune responses (Sallusto et al., 2010). We found that both SARS-CoV-2 mRNA-LNP vaccines elicited potent LLPC and MBC responses, as well as the rapid generation of neutralizing antibodies that persisted at a high level until at least week 9 after immunization. ADE of disease results from antibody-mediated enhancement of infection, which has been described for some flaviviruses (Dowd and Pierson, 2011) and has been raised as a potential concern for coronaviruses. ADE has only been demonstrated in in vitro studies for SARS-CoV-1 (Jaume et al., 2011); thus, it is uncertain if it poses a substantial risk in the setting of SARS-CoV-2. We demonstrated that neither mRNA-LNP vaccine elicited antibodies with ADE activity in in vitro experiments. Thus, we reason that ADE may not be a critical safety issue for SARS-CoV-2 mRNA-based vaccines, although this assumption needs to be directly investigated in future in vivo (animal) experiments and may differ between animals and humans.
Nucleoside-modified mRNA-LNP vaccines for SARS-CoV-2 are currently under clinical development by Moderna (Jackson et al., 2020) and Pfizer/BioNTech (Mulligan et al., 2020). A recent publication provided preliminary data from a clinical phase I dose-escalation study led by Moderna (Jackson et al., 2020). The company used a nucleoside-modified mRNA-LNP vaccine platform (similar or identical to ours) encoding SARS-CoV-2 spike stabilized in its prefusion conformation and immunized participants two times each with 25, 100, or 250 μg of mRNA. In agreement with our mouse data, they found that nucleoside-modified mRNA-LNPs induced Th1-biased T cell responses and robust humoral immune responses in people. Adverse events included typical local and systemic reactions, were clearly dose-dependent, and were typically worse following the second immunization. A second nucleoside-modified mRNA-LNP vaccine platform (also similar or identical to ours), encoding the SARS-CoV-2 spike receptor binding domain and developed by Pfizer/BioNTech, was recently shared as a pre-print and showed similar dose-dependent immunogenicity and adverse events (Mulligan et al., 2020). The data presented here could potentially inform the evaluation of these two vaccines in humans. For example, our data prompt the question of whether these vaccines would induce T cells that home to the airways in humans and whether these might contribute to protection from COVID-19 disease or otherwise alter the inflammatory environment during SARS-CoV-2 infection. Finally, our observation of strong CD8+ T cell responses with cytotoxic markers suggests a potentially important way that mRNA-LNP vaccines may differ from protein subunit and inactivated virus vaccines, which are not expected to generate strong CD8+ T cell responses. However, it remains to be seen whether CD8+ T cell responses, lung-resident or otherwise, contribute meaningfully to immune protection in SARS-CoV-2 animal models or in humans.
Limitations of Study
Our study has some limitations, the most critical one being the absence of protective efficacy studies that could assess whether our nucleoside-modified mRNA-LNP vaccines induce protection from SARS-CoV-2 viral replication or disease in an animal model. We were unable to perform these studies in the BALB/c mouse model used here, because BALB/c mice are not susceptible to robust SARS-CoV-2 replication or disease. Instead, transgenic mice carrying the human angiotensin-converting enzyme 2 (hACE2) gene are a more appropriate animal model for future protective efficacy studies for SARS-CoV-2 (Bao et al., 2020; McCray et al., 2007). Another suitable mouse model was recently reported (Hassan et al., 2020), involving the intranasal delivery of an adenoviral vector encoding hACE2 to WT mice prior to viral challenge. These useful tools were not available as of the time of this publication but are the subject of ongoing and future protection studies.
One key consideration for human SARS-CoV-2 vaccines is to determine the ideal vaccine dose and vaccination schedule. While important, our study was not designed to address this dose optimization question. Additionally, it is unclear how mouse vaccine doses can be extrapolated to human immunization, i.e., whether it is at all proportional to body weight or affected by immunogenetic differences between species. As a result, the dose and schedule will likely need to be optimized through human clinical trials. The major aim of our studies was to provide detailed analyses of immune responses to two nucleoside-modified SARS-CoV-2 mRNA vaccines in mice as a proof-of-principle that may guide the exploration of similar mRNA-LNP vaccines in human clinical trials. We used a single immunization of 30 μg mRNA-LNPs in these studies because we found that this vaccine dose induced durable protective immune responses against other viruses in mice (Pardi et al., 2018a; Pardi et al., 2017; Pardi et al., 2018c). In agreement with previous studies, we demonstrated that a single immunization with 30 μg of nucleoside-modified SARS-CoV-2 vaccines elicited potent T and B cell responses and generated high and sustained levels of neutralizing antibodies in 100% of mice. In future experiments, it may be useful to systematically scale down dosing in order to determine the minimal protective dose after one or two immunizations. However, as cautioned above, this would require confirmation in non-human primate models or human clinical trials.
Because no other peer-reviewed preclinical experiments are available on SARS-CoV-2 nucleoside-modified mRNA-LNP vaccines, we believe that our work will provide valuable information about the magnitude and quality of immune responses induced by this new and promising vaccine type and will inform ongoing and future clinical trials.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Fixable Viability Dye eFluor780 | eBioScience | 65-0865-14 |
| CD3 clone 17A2, APC-Fire750 | Biolegend | 100248 |
| Ter-119 clone Ter-119, APC-Fire750 | Biolegend | 116250 |
| CD19 clone 6D5, BV605 | Biolegend | 115540 |
| B220 clone RA3-6B2, Alexa Fluor 700 | eBioScience | 56-0452-82 |
| CD38 clone 90, PE-Cy7 | Biolegend | 102718 |
| FAS clone JO2, BV510 | BD Biosciences | 563646 |
| IgG1 clone A85-1, V450 | BD Biosciences | 562107 |
| IgG2a/2b clone R2-40, Brilliant Blue 700 | BD Biosciences | 745969 |
| IgM polyclonal, FITC | Jackson ImmunoResearch | 115-095-020 |
| IgG polyclonal, HRP | Jackson ImmunoResearch | 115-035-003 |
| IgM polyclonal, biotin | Southern Biotech | 1020-08 |
| Igκ polyclonal, biotin | Southern Biotech | 1050-08 |
| Igλ polyclonal, biotin | Southern Biotech | 1060-08 |
| IgG1 polyclonal, biotin | Southern Biotech | 1070-08 |
| IgG2a polyclonal, biotin | Southern Biotech | 1080-08 |
| IgG2b polyclonal, biotin | Southern Biotech | 1090-08 |
| IgG3 polyclonal, biotin | Southern Biotech | 1100-08 |
| IgA clone RMA-1, biotin | Biolegend | 400703 |
| B220 clone RA36B2, BUV496 | BD Biosciences | 612950 |
| CD19 clone ID3, BUV661 | BD Biosciences | 612971 |
| CD138 clone 281-2, BUV737 | BD Biosciences | 564430 |
| PD-L2 clone TY25, BV711 | BD Biosciences | 740818 |
| CD4 clone H129.19, PE-Cy5 | BD Biosciences | 553654 |
| CD8α clone 53-6.7, PE-Cy5 | BD Biosciences | 553034 |
| CD86 clone GL-1, BV421 | Biolegend | 105031 |
| CXCR4 clone I.276F12, PE-Dz594 | Biolegend | 146514 |
| IgD clone 11-26c.2a, APC-Cy7 | Biolegend | 405716 |
| GL7 clone GL-7, AF488 | Biolegend | 144612 |
| streptavidin, BV650 | Biolegend | 405231 |
| Ter-119 clone Ter-119, PE-Cy5 | eBioScience | 15-5921-82 |
| F4/80 clone BM8, PE-Cy5 | eBioScience | 15-4801-82 |
| CD73 clone TY/11.8, PE-Cy7 | eBioScience | 25-0731-82 |
| CD38 clone 90 AF700 | Invitrogen | 56-0381-82 |
| anti-RBD chimeric monoclonal antibody, D001 | Sino Biologicals | 40150-D001 |
| anti-human IgG Fc secondary antibody, PE | Invitrogen | 12-4998-82 |
| LIVE/DEAD Fixable Aqua Dead Cell Stain Kit | Invitrogen | L34966 |
| CD45 clone 30-F11 Alexa Fluor 700 | Biolegend | 103128 |
| Live/Dead Aqua | Thermo Fisher | L34957 |
| CD16/CD32 clone 2.4G2 | bioXcell | BE0307 |
| CD3, clone 145-2C11, APC-Cy7 | BD Biosciences | 557596 |
| CD4 clone RM4-5, Pacific Blue | Biolegend | 100531 |
| CD8, clone 53-6.7 PerCP-Cy5.5 | BD Biosciences | 551162 |
| PD-1 clone RMP1-30, APC | Biolegend | 109112 |
| CD69 clone H1.2F3, BV605 | Biolegend | 104530 |
| CD44, clone IM7, BV785 | Biolegend | 103059 |
| CD62L clone MEL-14, PE-Cy7 | Biolegend | 104418 |
| CD28 clone 37.51 | Tonbo | 40-0281-M001 |
| CD107a clone 1D4B, Alexa Fluor 647 | Biolegend | 121610 |
| CD4 clone RM4-5, BV786 | BD Biosciences | 563727 |
| CD8 clone 53-6.7, BUV395 | BD Biosciences | 563786 |
| IFN gamma clone XMG1.2, Alexa Fluor 488 | Biolegend | 505813 |
| TNF alpha clone MP6-XT22, BV605 | Biolegend | 506329 |
| IL-2 clone JES6-5H4, PE | Biolegend | 503808 |
| granzyme B clone GB11, Pacific Blue | Biolegend | 515408 |
| anti-guinea pig IgG Fc | Jackson ImmunoResearch | 106-035-008 |
| anti-human IgG Fc | Sigma-Aldrich | I2136 |
| rabbit anti-RBD antibody R007 | Sino Biologicals | 40150-R007 |
| anti-rabbit IgG H&L, HRP | Abcam | 97080 |
| anti-mouse IgG, HRP | Jackson ImmunoResearch | 715-035-150 |
| anti-VSV Indiana G | Absolute Antibody | Ab01401-2.0 |
| SARS-CoV-1 nucleoprotein antibody | Thomas Moran | N/A |
| Chemicals and Recombinant Protein | ||
| m1-pseudouridine-5′-triphosphate | TriLink | N-1081 |
| CleanCap | TriLink | N-7413 |
| Cellulose | Sigma-Aldrich | 11363-250G |
| Recombinant RBD and Full Spike | Florian Krammer and Raffael Nachbagauer | N/A |
| SARS-CoV-2 spike peptide pools | JPT | PM-WCPV-S |
| brefeldin A | Biolegend | 420601 |
| monensin | Biolegend | 420701 |
| Cytofix/Cytoperm kit | BD Biosciences | 554714 |
| ELISPOT AEC Substrate Set | BD Biosciences | 551951 |
| BCIP/NBT | Sigma | B1911-100mL |
| KPL TMB 2-Component Microwell Peroxidase Substrate | Seracare | 5120-0050 |
| SureBlue Reserve TMB 1-Component Microwell Peroxidase Substrate | Seracare | 5120-0083 |
| Luc-Pair Firefly Luciferase HS Assay Kit | Genecopoeia | LF007 |
| Luc-Pair Renilla Luciferase HS Assay Kit | Genecopoeia | LF010 |
| TransIT-mRNA Transfection Kit | Mirus | MIR 2250 |
| Collagenase D | Sigma | 11088866001 |
| ACK Lysing buffer | Lonza | 10-548E |
| Lightning-Link® R-Phycoerythrin | Expedeon | 336-0005 |
| Lightning-Link (R) Rapid Alexa Fluor 647 | Expedeon | 703-0010 |
| BD Brilliant Buffer | BD Biosciences | 563794 |
| ExpiFectamine 293 transfection kit | GIBCO | A14525 |
| Ni-NTA resin | QIAGEN | 30230 |
| Bradford reagent | Bio-Rad | 5000201 |
| Experimental Models: Organisms/Strains | ||
| BALB/c mouse (T cell studies) | Jackson Laboratory | N/A |
| BALB/c mouse (all other studies) | Charles River Laboratories | N/A |
| Experimental Models: Cell Lines | ||
| HEK293T/17 cells | ATCC | CRL11268 |
| FreeStyle 293 (293F) cells | GIBCO | R79007 |
| Vero E6 cells stably expressing TMPRSS2 | Dr. Stefan Pohlmann | N/A |
| Bacterial and Virus Strains | ||
| SARS-CoV-2 | GenBank | MT020880 |
| ZIKV | Strain PB-81 | N/A |
| Recombinant DNA | ||
| pCMV-SPORT6-mFcγR1 | Dharmacon | MMM1013-202708624 |
| SARS-CoV-2 S protein | GenBank | MN908947.3 |
| SARS-CoV-2 S delta18 | Dr. Stefan Pohlmann | N/A |
| ZIKV replicon (FSS13025) | GenBank | KU955593.1 |
| Softwares and Algorithms | ||
| S6 FluoroSpot Analyzer | CTL | N/A |
| FlowJo software | FlowJo LLC | N/A |
| FACSDIVA software | BD Biosciences | N/A |
| GraphPad Prism | GraphPad | N/A |
Resource Availability
Lead Contact
Further information and requests for supporting data, resources, and reagents should be directed to and will be fulfilled upon request by the Lead Contact, Norbert Pardi (pnorbert@pennmedicine.upenn.edu).
Materials Availability
Reagents from this study are available upon request.
Data and Code Availability
The source of protein and nucleic acid sequences are indicated in the manuscript and are available from the corresponding author on request.
Experimental Model and Subject Details
Mice
BALB/c mice aged 8 weeks were purchased from Jackson Laboratory (T cell studies), or Charles River Laboratories (all other studies).
Cell lines
FreeStyle 293 (293F) cells (GIBCO #R79007) were cultured in Freestyle 293 Expression Medium (GIBCO). The 293F cell line was tested for mycoplasma contamination after receipt from Life Technologies and before expansion and cryopreservation.
HEK293T/17 cells (ATCC #CRL11268) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Mediatech #MT10-013-CM) containing 10% fetal calf serum (FCS). Vero E6 cells stably expressing TMPRSS2 were a gift from Stefan Pohlman and were cultured in DMEM +10% FCS.
Method Details
Ethics statement
The investigators faithfully adhered to the “Guide for the Care and Use of Laboratory Animals” by the Committee on Care of Laboratory Animal Resources Commission on Life Sciences, National Research Council. Mouse studies were conducted under protocols approved by the Institutional Animal Care and Use Committees (IACUC) of the University of Pennsylvania (UPenn), BIOQUAL Inc. (Rockville, MD), and the Children’s Hospital of Philadelphia (CHOP). All animals were housed and cared for according to local, state and federal policies in an Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC)-accredited facility.
mRNA-LNP vaccine production
mRNA vaccines were designed based on the SARS-CoV-2 spike (S) protein sequence (Wuhan-Hu-1, GenBank: MN908947.3). Coding sequences of full length WT S protein, full length Δfurin S protein (RRAR furin cleavage site abolished between amino acids 682-685), RBD (amino acids 1-14 fused with amino acids 319-541) and firefly luciferase (Luc) were codon-optimized, synthesized and cloned into the mRNA production plasmid as described (Freyn et al., 2020). mRNA production and LNP encapsulation was performed as described (Freyn et al., 2020). Briefly, mRNAs were transcribed to contain 101 nucleotide-long poly(A) tails. m1Ψ-5′-triphosphate (TriLink #N-1081) instead of UTP was used to generate modified nucleoside-containing mRNA. Capping of the in vitro transcribed mRNAs was performed co-transcriptionally using the trinucleotide cap1 analog, CleanCap (TriLink #N-7413). mRNA was purified by cellulose (Sigma-Aldrich # 11363-250G) purification, as described (Baiersdörfer et al., 2019). All mRNAs were analyzed by agarose gel electrophoresis and were stored frozen at −20°C. Cellulose-purified m1Ψ-containing RNAs were encapsulated in LNPs using a self-assembly process as previously described wherein an ethanolic lipid mixture of ionizable cationic lipid, phosphatidylcholine, cholesterol and polyethylene glycol-lipid was rapidly mixed with an aqueous solution containing mRNA at acidic pH (Maier et al., 2013). The RNA-loaded particles were characterized and subsequently stored at −80°C at a concentration of 1 μg μl-1. The mean hydrodynamic diameter of these mRNA-LNP was ~80 nm with a polydispersity index of 0.02-0.06 and an encapsulation efficiency of ~95%. Two or three batches from each mRNA-LNP formulations were used in these studies and we did not observe variability in vaccine efficacy.
mRNA transfection
293F cells were diluted to 1 × 106 cells/ml before transfection. 3 μg mRNA encoding full length WT and Δfurin S protein was transfected into 6 mL of cells. For soluble RBD, 30 mL of cells were transfected with 15 μg mRNA. TransIT-mRNA Transfection Kit (Mirus #MIR 2250) was used for mRNA transfection following the manufacturer’s instructions. Transfected cells were cultured at 37°C with 8% CO2 and shaking at 130 rpm for 48 hours (for full length WT and Δfurin S protein) or 72 hours (for soluble RBD).
Sample processing and flow cytometry
In vitro studies
Binding reactivity of anti-RBD chimeric mAb, D001 (Sino Biologicals #40150-D001) and hACE2-Fc fusion protein to full length S protein constructs (WT and Δfurin) was measured by flow cytometry. Briefly, mRNA-transfected 293F cells were harvested 48 hours after transfection and were washed once with 1% bovine serum albumin (BSA) in phosphate buffered saline (PBS). Next, cells were incubated with 10 μg/ml D001 or hACE2-mFc in V-bottom 96-well plates for 30 minutes at 4°C. Cells were then incubated with goat anti-human IgG Fc secondary antibody, PE (Invitrogen #12-4998-82) at final concentration of 2.5 μg/ml for 30 minutes at 4°C in dark. Following this, dead cells were stained with LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Invitrogen #L34966, used at 1:1000 in PBS) for 15 minutes at 4°C in dark, then washed twice and re-suspended with 1% BSA in PBS. Flow cytometric data were acquired on a LSRII with high-throughput system using FACSDIVA software (BD Biosciences).
T cell studies
In vivo antibody labeling: to distinguish lung-infiltrating and vascular T cells, mice were injected intravenously under isoflurane anesthesia with 2 μg of anti-CD45 Alexa Fluor 700 antibody (Biolegend #103128). After 5 minutes, mice were euthanized via cervical dislocation and organs were collected for analysis.
Lung and spleen isolation for flow cytometry: to isolate lung lymphocytes, the lung vasculature was first perfused with 5 mL 1% fetal bovine serum (FBS) in PBS by injecting into the cardiac right ventricle. Lungs were collected in gentleMACS C tubes containing 1% FBS in PBS on ice. Digest media was added to achieve a final concentration of 2.25 mg/ml sterile-filtered Collagenase D (Sigma #11088866001) and 0.15 mg/ml DNase I in 4 mL of 1% FBS in PBS. Lungs were disrupted using gentleMACS Dissociator program m_spleen_01.01, then incubated for 45 minutes at 37°C with shaking. Ten ml of complete RPMI media (10% FBS, 2 mM L-glutamine, 50 μM 2-mercaptoethanol, and penicillin/streptomycin) was then added to each tube, followed by further homogenization using gentleMACS Dissociator program m_lung_02.01. Digested lungs were then passed through a 70 μm strainer, incubated in ACK lysis buffer to remove RBCs, then passed through a 40 μm strainer to obtain a single cell suspension. Spleens were collected in PBS and homogenized through a 70 μm cell strainer using the hard end of a syringe plunger. Splenocytes were incubated in ACK lysis buffer to remove red blood cells (RBCs), then passed through a 40 μm strainer to obtain a single cell suspension.
T cell activation analysis: after preparing lung and spleen single cell suspensions, cells were immediately analyzed for activation markers. Cells were stained with Live/Dead Aqua (Thermo Fisher #L34957) in PBS, followed by Fc-receptor blockade with anti-CD16/CD32 (bioXcell #BE0307), and then stained for 30 minutes at 4°C with the following antibody panel each at 1:100 in 0.1% BSA in PBS: anti-CD3 APC-Cy7 (BD Biosciences #557596), anti-CD4 Pacific Blue (Biolegend #100531), anti-CD8 PerCP-Cy5.5 (BD Biosciences #551162), anti-PD-1 APC (Biolegend #109112), anti-CD69 BV605 (Biolegend #104530), anti-CD44 BV785 (Biolegend #103059), anti-CD62L PE-Cy7 (Biolegend #104418). Samples were analyzed on the CytoFLEX LX flow cytometer (Beckman Coulter).
Intracellular cytokine staining: to measure antigen-specific T cells, 1 million cells per well of lung or spleen cells were stimulated with SARS-CoV-2 spike peptide pools (JPT PM-WCPV-S) in a U-bottom plate for at 37°C, 6% CO2 with 2 μg/ml anti-CD28 (Tonbo #40-0281-M001) providing co-stimulation. Vial 1 (N-terminal) and vial 2 (C-terminal) of spike peptides were dissolved in DMSO at 666 μg/ml per peptide and used separately at a final concentration of 1.5 μg/ml. Stimulations proceeded for 1 hour before adding 5 μg/ml brefeldin A (Biolegend #420601), 2 μM monensin (Biolegend #420701), and 5 μg/ml anti-CD107a Alexa Fluor 647 (Biolegend #121610) for 5 hours more. DMSO served as a negative control and the combination of 50 μg/ml phorbol 12-myristate 13-acetate and 1 μg/ml ionomycin served as a positive control. After a total of 6 hours, samples were kept < 4°C and stained with Live/Dead Aqua and anti-CD16/CD32 blockade as above, fixed and permeabilized using the Cytofix/Cytoperm kit (BD Biosciences #554714), and stained intracellularly for 1 hour in 0.1% BSA in PBS with antibodies including (each at 2 μg/ml): anti-CD3 APC-Cy7 (BD Biosciences #557596), anti-CD4 BV786 (BD Biosciences #563727), anti-CD8 BUV395 (BD Biosciences #563786), anti-IFN gamma Alexa Fluor 488 (Biolegend #505813), anti-TNF BV605 (Biolegend #506329), anti-IL-2 (Biolegend #503808), and anti-granzyme B Pacific Blue (Biolegend #515408). Samples were analyzed on the Aurora flow cytometer (Cytek).
B cell studies
Sample Processing: spleens were mashed in complete DMEM [(Corning #T10014CV) containing 10% heat inactivated Fetal Bovine Serum (FBS Corning #35-015), 1% Glutamax (GIBCO #35050-061) and 1% Penicillin/Streptomycin (GIBCO #5070063)] on ice and filtered through a 40 μm cell strainer. RBCs were lysed with ACK Lysing buffer (Lonza #10-548E) for 5 minutes on ice and the reaction was stopped with ten times the volume PBS. Bone marrow (BM) was harvested from femurs and tibia from each mouse using a 23.5g X ¾” needle and syringe into FACS buffer and filtered through a 63 μm Nitex mesh. RBCs were lysed in ACT buffer for 5 minutes on ice. After RBC lysis, splenocytes and BM cells were resuspended in cold media and immediately used for cell counting, culture or staining.
Staining and flowcytometry: single cell suspensions of murine splenocytes were incubated with anti-CD16/CD32 (BioXCell #BE0307) in FACS buffer (PBS with 2% heat inactivated FBS) prior to staining with all surface anti-mouse antibodies, labeled recombinant proteins (probes) and viability dye for 30 minutes at 4°C. Recombinant SARS-CoV-2 Receptor Binding Domain (RBD) or Full length S proteins were independently conjugated to both R-PE and Alexa Fluor 647 using Lightning-Link® R-Phycoerythrin (R-PE) (Expedeon #336-0005) and Lightning-Link (R) Rapid Alexa Fluor 647 (Expedeon #703-0010) kits according to the manufacturer’s instructions. For immunophenotyping of antigen-specific memory B cells (MBC), splenocytes were stained with: anti-mouse CD19 BV605 (Biolegend #115540), B220 AF700 (eBioScience #56-0452-82), CD3 APC-Fire750 (Biolegend #100248), Ter119 APC-Fire750 (Biolegend #116250), CD38 PE-Cy7 (Biolegend #102718), FAS BV510 (BD Biosciences #563646), IgG1 eFluor450 (BD Biosciences #562107), and IgG2a/2b BB700 (BD Biosciences #745969) and IgM FITC (Jackson ImmunoResearch #115-095-020) antibodies, together with RBD- or full length S-PE, RBD- or full length S AlexaFluor 647, and Fixable Viability Dye eFluor780 (eBioscience #115-095-020). The excess of antibodies were washed away with FACS buffer and cells were fixed with 1% paraformaldehyde (PFA) for 30 minutes at 4°C prior to acquisition on a 5 laser Cytoflex LX (Beckman Coulter).
Alternatively, MBC were assessed without respect to isotype as follows: 5 million splenocytes or BM cells were prepared as above stained with fixable live dead aqua (Biolegend Zombie Aqua #423101) for 15 minutes at room temperature. Cells were then washed with FACS buffer and stained with the following dilutions of antibodies: B220-BUV496 (BD Biosciences #612950), CD19-BUV661 (BD Biosciences #612971), CD138-BUV737 (BD Biosciences #564430), PD-L2-BV711 (BD Biosciences #740818), CD4-PE-Cy5 (BD Biosciences #553654), CD8α-PE-Cy5 (BD Biosciences #553034), CD86-BV421 (Biolegend #105031), IgA-biotin (Biolegend #400703), CXCR4-PE-Dz594 (Biolegend #146514), IgD-APC-Cy7 (Biolegend #405716), GL7-AF488 (Biolegend #144612), SA-BV650 (Biolegend #405231), Ter-199-PE-Cy5 (eBioScience #15-5921-82), F4/80-PE-Cy5 (eBioScience #15-4801-82), CD73-PE-Cy7 (eBioScience #25-0731-82), and CD38-AF700 (Invitrogen #56-0381-82) in BD Brilliant Buffer (BD Biosciences #563794) for 15 minutes at 4°C. Cells were then washed and stained streptavidin BV650 for 10 minutes at 4°C prior to wash and resuspension in FACS buffer. ~2 million events per sample were acquired on a BD Symphony A3 Lite.
All flow cytometry data were analyzed with FlowJo software (FlowJo LLC).
Full-length S and RBD protein production
The RBD and full-length S proteins were produced in 293F cells, as described previously (Amanat et al., 2020; Stadlbauer et al., 2020). Briefly, 600 million cells were transfected with 200 μg of purified DNA encoding codon-optimized RBD of SARS-CoV-2 using ExpiFectamine 293 transfection kit (GIBCO #A14525). The manufacturer’s protocol was followed and cells were harvested on day 3. Cells were spun at 4000 g for 10 minutes and sterile-filtered with a 0.22 μm filter. Supernatant was incubated with Ni-NTA resin (QIAGEN #30230) for 2 hours. After 2 hours, this mixture was loaded onto columns and the protein was eluted using elution buffer with high amounts of imidazole. Protein was concentrated using 10 kDa Amicon centrifugal units (Millipore Sigma #UFC901024) and re-constituted in PBS. Concentration was measured using Bradford reagent (Bio-Rad #5000201) and a reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was run to check the integrity of the protein.
ELISPOT assay
MultiScreenHTS IP Filter Plate, 0.45 μm (Millipore Sigma #MSIPS4W10) were coated overnight at 4°C with 2.5 μg/ml recombinant SARS-CoV-2 RBD or Full Spike proteins in bicarbonate buffer (35 mM NaHCO3 and 15 mM Na2CO3). Plates were washed three times with PBS and blocked with complete DMEM for at least 1 hour at 37°C. Single cell suspensions of murine BM cells were diluted serially in complete DMEM with halving dilutions starting at 1 × 106 cells. Following overnight incubation at 37°C and 5% CO2, plates were washed three times with 0.05% Tween-20 in PBS. Membranes were incubated with IgG-HRP (Jackson ImmunoResearch #115-035-003) diluted in complete DMEM for 2 hours at room temperature. Following incubation with the detection antibody, plates were washed three times with 0.05% Tween-20 in PBS. Spots corresponding to antigen-specific antibody-secreting cells were developed using BD ELISPOT AEC Substrate Set (#551951) and counted using a CTL Immunospot analyzer. Isotype specific ELISPOT plates were coated and incubated with cells as above and then were washed 4x with 0.1% Tween-20 in PBS. Membranes were then incubated with 1:3000 isotype-specific biotinylated antibodies (Southern Biotech #1020-08, #1050-08, #1060-08, #1080.08, #1090-08, #1100-08, Biolegend #400703) for 1 hour. Membranes were then washed 4x and incubated in 1:20,000 streptavidin-alkaline phosphatase (Sigma # E2636) for 30 minutes. Membranes were then washed 4x and incubated with 50 μL BCIP/NBT (Sigma #B1911-100mL) for ~10 minutes or until spots developed at which time reaction was quenched with 100 μL 1M sodium phosphate monobasic solution. Membranes were then dried and counted above.
Mouse immunizations
mRNA-LNPs were diluted in PBS and injected into the gastrocnemius muscle (40 μL injection volume) with a 3/10cc 29½G insulin syringe (Covidien #8881600145).
Blood collection
Blood was collected from the orbital sinus under isoflurane anesthesia. Blood was centrifuged for 5 minutes at 13,000 rpm and the serum was stored at −20°C and used for ELISA, virus neutralization assays, and ADE assays.
Enzyme linked immunosorbent assay (ELISA)
Samples from cell transfections
Supernatant from 293F cells transfected with RBD-encoding mRNA was harvested 72 hours after transfection and concentrated 60x with Vivaspin 20 kDa molecular weight cut-off concentrator (GE Healthcare #20-9323-60). The expression and binding of soluble RBD were measured by indirect ELISA. RBD samples were added to capture antibody D001 (2 μg/ml)-coated plates for one hour, followed by detection with serum from a SARS-CoV S protein-immunized guinea pig for 1 hour. Serum binding was detected via horseradish peroxidase-conjugated goat anti-guinea pig IgG (Fc) (Jackson ImmunoResearch #106-035-008, used at 1:10,000). Plates were developed with SureBlue Reserve TMB 1-Component Microwell Peroxidase Substrate (Seracare #5120-0083). Absorbance at 450 nm were measured by a SpectraMax Plus 384 microplate reader (Molecular Devices) and log area under curve (logAUC) were calculated.
To test RBD sample binding to ACE2, plates were first coated with goat anti-human IgG (Fc) antibody (Sigma-Aldrich #I2136) (2 μg/ml), in order to capture the hACE2-mFc construct (5 μg/ml, one hour). Next, the RBD samples were incubated for one hour. The RBD was detected by a rabbit anti-RBD antibody R007 (Sino Biologicals #40150-R007, used at 1:4000) followed by Goat Anti-Rabbit IgG H&L (HRP) (Abcam #97080). The detection of RBD and development procedure were the same as described above.
Samples from mouse immunizations
Corning 96 Well Clear Polystyrene High Bind Stripwell Microplates (Corning #2592) were coated with 1 μg/ml purified RBD in PBS overnight at 4°C. The plates were blocked with 2% BSA in PBS for 2 hours and washed four times with wash buffer (0.05% Tween-20 in PBS). Mouse sera were diluted in blocking buffer and incubated for 2 hours at room temperature, followed by four washes. HRP-conjugated anti-mouse secondary antibody (Jackson Immunoresearch #715-035-150) was diluted 1:10,000 in blocking buffer and incubated for 1.5 hours, followed by 4 washes. KPL 2-component TMB Microwell Peroxidase Substrate (Seracare #5120-0050) was applied to the plate and the reaction was stopped with 2 N sulfuric acid. The absorbance was measured at 450 nm using a SpectraMax 190 microplate reader. RBD-specific IgG end-point dilution titer was defined as the highest dilution of serum to give an OD greater than the sum of the background OD plus 0.01 units. All samples were run in technical duplicates.
Pseudovirus neutralization assay
Production of VSV pseudotype with SARS-CoV-2 S: 293T cells plated 24 hours previously at 5 X 106 cells per 10 cm dish were transfected using calcium phosphate with 35 μg of pCG1 SARS-CoV-2 S delta18 expression plasmid encoding a codon optimized SARS-CoV S gene with an 18 residue truncation in the cytoplasmic tail (kindly provided by Stefan Pohlmann). Twelve hours post transfection the cells were fed with fresh media containing 5mM sodium butyrate to increase expression of the transfected DNA. Thirty hours after transfection, the SARS-CoV-2 spike expressing cells were infected for 2-4 hours with VSV-G pseudotyped VSVΔG-RFP at an MOI of ~1-3. After infection, the cells were washed twice with media to remove unbound virus. Media containing the VSVΔG-RFP SARS-CoV-2 pseudotypes was harvested 28-30 hours after infection and clarified by centrifugation twice at 6,000 g then aliquoted and stored at −80°C until used for antibody neutralization analysis.
Antibody neutralization assay using VSVΔG-RFP SARS-CoV-2: Vero E6 cells stably expressing TMPRSS2 were seeded in 100 μL at 2.5x104 cells/well in a 96 well collagen coated plate. The next day, 2-fold serially diluted serum samples were mixed with VSVΔG-RFP SARS-CoV-2 pseudotype virus (50-200 focus forming units/well) and incubated for 1 hour at 37°C. Also included in this mixture to neutralize any potential VSV-G carryover virus was 8G5F11, a mouse anti-VSV Indiana G, at a concentration of 100 ng/ml (Absolute Antibody #Ab01401-2.0). The antibody-virus mixture was then used to replace the media on VeroE6 TMPRSS2 cells. 20 hours post infection, the cells were washed and fixed with 4% PFA before visualization on an S6 FluoroSpot Analyzer (CTL, Shaker Heights OH). Individual infected foci were enumerated and the values compared to control wells without antibody. The focus reduction neutralization titer 50% (FRNT50) was measured as the greatest serum dilution at which focus count was reduced by at least 50% relative to control cells that were infected with pseudotype virus in the absence of mouse serum. FRNT50 titers for each sample were measured in two technical replicates performed on separate days.
Microneutralization assay
Neutralization assays with live SARS-CoV-2 (USA-WA1/2020; GenBank: MT020880) were performed in a biosafety level 3 (BSL3) facility with strict adherence to institutional regulations. Twenty thousand Vero.E6 cells per well were seeded in a 96-well cell culture plate one day before the neutralization assay and this protocol has been published earlier (Amanat et al., 2020). Mouse serum samples were heat-inactivated at 56°C for 1 hour. Various dilutions of the serum samples in duplicates were prepared in 1X minimal essential medium (MEM) supplemented with FBS and each dilution was mixed with 600 TCID50 of SARS-CoV-2 for 1 hour at room temperature. Cell culture medium from Vero.E6 cells was removed and each dilution was added to the 96-well plate. Cells were incubated with this serum-virus mixture for 1 hour at 37°C. After 1 hour, the serum-virus mixture was removed, and the same respective dilutions were added with an equal amount of 1X MEM supplemented with 2% FBS. Cells were incubated for 48 hours at 37°C after which cells were fixed with 10% formaldehyde. Following fixation for 24 hours at 4°C, cells were stained with a SARS-CoV-1 nucleoprotein antibody (mouse 1C7, obtained from Dr. Thomas Moran, Icahn School of Medicine at Mount Sinai). Further details have been described earlier (Amanat et al., 2020). Each plate had six controls wells that were not infected and six wells that were infected but had no serum. The background from uninfected control wells was averaged and subtracted from all the wells. Percent inhibition at each well was calculated by the following formula: 100-(((X-[average of “no virus” wells])/[average of “virus only” wells])∗100) whereby ‘X’ is the read for each well. Non-linear regression curve fit analysis over the dilution curve was performed to calculate IC50.
SARS-CoV-2 pseudovirus (PV) and Zika virus (ZIKV) virus-like particle (VLP) production
SARS-CoV-2 PV was produced as previously described (Moore et al., 2004) with a minor modification. HEK293T cells were transfected by calcium-phosphate transfection method at a ratio of 5:5:1 with a plasmid encoding murine leukemia virus (MLV) gag/pol proteins, a retroviral vector pQCXIX expressing firefly luciferase, and a plasmid expressing the spike protein of SARS-CoV-2 (GenBank YP_009724390). Cells were washed 6 hours later, and the culture supernatant containing PV was harvested at 43 hours post transfection. ZIKV VLP was produced by transfecting HEK293T cells by the calcium transfection method with a ZIKV replicon (strain FSS13025, GenBank KU955593.1), whose expression is controlled by tetracycline, a plasmid encoding ZIKV capsid, prM, and E proteins (strain FSS13025, GenBank KU955593.1), and the pTet-On plasmid expressing a reverse Tet-responsive transcriptional activator (rtTA) at a ratio of 2:1:1. Cells were washed 6 hours later and replenished with fresh media containing 1 μg/ml Doxycycline. The VLP-containing culture supernatant was harvested 48 hours post transfection. ZIKV replicon was generated by replacing the region spanning 39th through 763rd amino acids of the polyprotein of a ZIKV molecular clone (Zhang et al., 2018) with Renilla luciferase with the 2A self-cleaving peptide fused at its C terminus. This construct contains the tetracycline-responsive Ptight promoter that derives ZIKV RNA transcription. The PV- and VLP-containing culture supernatants were cleared by 0.45 μm filtration and immediately frozen in aliquots at −80°C.
Antibody-dependent enhancement (ADE) assay
The ability of SARS-CoV-2 immune sera to mediate ADE was measured using HEK293T cells or those stably expressing hACE2 transfected with pCMV-SPORT6-mFcγR1 (Dharmacon #MMM1013-202708624). Mouse immune sera used for ADE assays were obtained 9 weeks post vaccination or from naive mice. Sera of ten mice per vaccine group were pooled in two groups (five per pool) and assessed separately in ADE assays but combined afterward for data analysis. We have previously shown efficient ZIKV ADE (Shim et al., 2019), and thus used ZIKV VLP as a positive control in ADE assays. ZIKV immune sera were prepared by intraperitoneally injecting mice with ZIKV (strain PB-81) and bled at 5 weeks post infection. The immune and naive sera samples, heat inactivated for 30 minutes at 56°C, were serially diluted in DMEM containing 10% heat-inactivated FBS. SARS-CoV-2 PV expressing firefly luciferase or ZIKV VLP expressing Renilla luciferase in 50 μL was preincubated for 1 hour at 37°C with 50 μL of diluted plasma and added to cells plated on the 96 well plates. Twenty four hours later, infection levels were assessed using Luc-Pair Firefly Luciferase HS Assay Kit (Genocopia #LF007) for SARS2-CoV-2 PV and Luc-Pair Renilla Luciferase HS Assay Kit (Genocopia #LF010) for ZIKV VLP.
Quantification and Statistical Analysis
GraphPad Prism was used to perform Kruskal-Wallis and Mann-Whitney tests for non-parametric data and one-way or two-way ANOVA corrected for multiple comparisons for parametric data to compare immune responses in vaccinated and control mice.
Acknowledgments
We thank Lisa Mistretta and Dalia Benetiene for helping with animal procedures (Bioqual). We thank Florin Tuluc and Jennifer Murray of the CHOP Flow Cytometry core facility for technical assistance. We thank Alec W. Freyn, Allen Zheng, Katarzyna Janowska, and Priyamvada Acharya for providing SARS-CoV-2 reagents used in the study. The graphical abstract was created with BioRender. N.P. was supported by the National Institute of Allergy and Infectious Diseases (1R01AI146101). M.L. was supported by the National Institute of Allergy and Infectious Diseases (R01AI123738). Work in the Krammer laboratory was supported by the National Institute of Allergy and Infectious Diseases Collaborative Influenza Vaccine Innovation Centers (CIVIC; 75N93019C00051), the NIAID Centers of Excellence for Influenza Research and Surveillance (CEIRS; HHSN272201400008C), institutional funding, and philanthropic donations. M.F. and H.C. were supported by an administrative supplement to NIH R01 AI129868 for coronavirus research. P.H. and T.B.M were supported by NIH (T32AI055400 and R21AI129531, respectively). P.B. was supported by a Peer Reviewed Medical Research Program (PR182551) and grants from the NIH (R21AI129531 and R21AI142638). B.F.H. was supported by NIAID (AI142596) and by a contract from the State of North Carolina from the CARES Act Funds. D.W. was supported by grants from the NIAID (AI142596 and AI124429) and BioNTech. M.J.H. is a Cancer Research Institute Irvington Fellow supported by the Cancer Research Institute. S.A.T was supported by the National Heart, Lung, And Blood Institute (F30HL145907).
Author Contributions
N.P. conceptualized the study. M.L. and D.A. designed B cell studies. N.P., D.L., M.J.H., and S.A.T. wrote the paper with help from co-authors. N.P., D.W., and F.K. designed vaccine antigens. K.K. designed the untranslated region of the mRNA. N.P. and H.M. produced mRNA vaccine antigens. Y.K.T. and P.J.C.L. encapsulated mRNAs into LNPs. N.P., D.L., I.T., M.J.H., K.L., H.A., J.G., L.P., M.P., T.P-T., A.S., and T.-A.C. performed immunizations. D.L. performed ELISAs. M.L., K.L., D.C., D.A., and B.T.G. performed B cell analyses. L.C.E., M.J.H., and S.A.T. performed T cell studies. P.B., P.H., T.B.M., and B.R. performed pseudovirus neutralization assays. F.K. and R.N. produced recombinant RBD protein. F.K., F.A., and S.S. performed microneutralization studies. M.F., H.C., A.O., and L.Z. performed ADE studies. B.F.H., K.O.S., Z.M., R.P., T.H.O., L.A., and G.D.S. performed in vitro experiments and edited the paper.
Declaration of Interests
In accordance with the University of Pennsylvania policies and procedures and our ethical obligations as researchers, we report that D.W. and K.K. are named on patents that describe the use of nucleoside-modified mRNA as a platform to deliver therapeutic proteins. D.W. and N.P. are also named on a patent describing the use of nucleoside-modified mRNA in lipid nanoparticles as a vaccine platform. We have disclosed those interests fully to the University of Pennsylvania, and we have an approved plan for managing any potential conflicts arising from licensing of our patents in place. K.K. is an employee of BioNTech. P.J.C.L. and Y.K.T. are employees of Acuitas Therapeutics, a company involved in the development of mRNA-LNP therapeutics. Y.K.T. is named on patents that describe lipid nanoparticles for delivery of nucleic acid therapeutics including mRNA and the use of modified mRNA in lipid nanoparticles as a vaccine platform.
Published: July 30, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.immuni.2020.07.019.
Supplemental Information
References
- Alameh M.G., Weissman D., Pardi N. Messenger RNA-Based Vaccines Against Infectious Diseases. Curr. Top. Microbiol. Immunol. 2020 doi: 10.1007/82_2020_202. Published online April 17, 2020. [DOI] [PubMed] [Google Scholar]
- Amanat F., Krammer F. SARS-CoV-2 Vaccines: Status Report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Asthagiri Arunkumar G., Jurczyszak D., Polanco J. A serological assay to detect SARS-CoV-2 seroconversion in humans. medRxiv. 2020 doi: 10.1101/2020.03.17.20037713. Published online April 16, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi S., Hook L.M., Pardi N., Wang F., Myles A., Cancro M.P., Cohen G.H., Weissman D., Friedman H.M. Nucleoside-modified mRNA encoding HSV-2 glycoproteins C, D, and E prevents clinical and subclinical genital herpes. Sci. Immunol. 2019;4:eaaw7083. doi: 10.1126/sciimmunol.aaw7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., Laska M.E., Smith M., Almarsson Ö., Thompson J. Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines against H10N8 and H7N9 Influenza Viruses. Mol. Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baiersdörfer M., Boros G., Muramatsu H., Mahiny A., Vlatkovic I., Sahin U., Karikó K. A Facile Method for the Removal of dsRNA Contaminant from In Vitro-Transcribed mRNA. Mol. Ther. Nucleic Acids. 2019;15:26–35. doi: 10.1016/j.omtn.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020 doi: 10.1038/s41586-020-2312-y. Published online May 7, 2020. [DOI] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd K.A., Pierson T.C. Antibody-mediated neutralization of flaviviruses: a reductionist view. Virology. 2011;411:306–315. doi: 10.1016/j.virol.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeseth A.S., Cejas P.J., Citron M.P., Wang D., DiStefano D.J., Callahan C., Donnell G.O., Galli J.D., Swoyer R., Touch S. Modified mRNA/lipid nanoparticle-based vaccines expressing respiratory syncytial virus F protein variants are immunogenic and protective in rodent models of RSV infection. NPJ Vaccines. 2020;5:16. doi: 10.1038/s41541-020-0163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman R.A., Fuhr R., Smolenov I., Mick Ribeiro A., Panther L., Watson M., Senn J.J., Smith M., Almarsson Ӧ., Pujar H.S. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine. 2019;37:3326–3334. doi: 10.1016/j.vaccine.2019.04.074. [DOI] [PubMed] [Google Scholar]
- Freyn A.W., Ramos da Silva J., Rosado V.C., Bliss C.M., Pine M., Mui B.L., Tam Y.K., Madden T.D., de Souza Ferreira L.C., Weissman D. A Multi-Targeting, Nucleoside-Modified mRNA Influenza Virus Vaccine Provides Broad Protection in Mice. Mol. Ther. 2020;28:1569–1584. doi: 10.1016/j.ymthe.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369:77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham B.S. Rapid COVID-19 vaccine development. Science. 2020;368:945–946. doi: 10.1126/science.abb8923. [DOI] [PubMed] [Google Scholar]
- Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell. 2020 doi: 10.1016/j.cell.2020.06.011. Published online June 10, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J. An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2022483. Published online July 14, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaume M., Yip M.S., Cheung C.Y., Leung H.L., Li P.H., Kien F., Dutry I., Callendret B., Escriou N., Altmeyer R. Anti-severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH- and cysteine protease-independent FcγR pathway. J. Virol. 2011;85:10582–10597. doi: 10.1128/JVI.00671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren G., Ols S., Liang F., Thompson E.A., Lin A., Hellgren F., Bahl K., John S., Yuzhakov O., Hassett K.J. Induction of Robust B Cell Responses after Influenza mRNA Vaccination Is Accompanied by Circulating Hemagglutinin-Specific ICOS+ PD-1+ CXCR3+ T Follicular Helper Cells. Front. Immunol. 2017;8:1539. doi: 10.3389/fimmu.2017.01539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y.K., Ansell S.M., Kumar V., Qin J. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M., Huang E., Yuzhakov O., Ramanathan P., Ciaramella G., Bukreyev A. Modified mRNA-Based Vaccines Elicit Robust Immune Responses and Protect Guinea Pigs From Ebola Virus Disease. J. Infect. Dis. 2018;217:451–455. doi: 10.1093/infdis/jix592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J., Dorfman T., Li W., Wong S.K., Li Y., Kuhn J.H., Coderre J., Vasilieva N., Han Z., Greenough T.C. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 2004;78:10628–10635. doi: 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A. Phase 1/2 Study to Describe the Safety and Immunogenicity of a COVID-19 RNA Vaccine Candidate (BNT162b1) in Adults 18 to 55 Years of Age: Interim Report. medRxiv. 2020 doi: 10.1101/2020.06.30.20142570. Published online July 1, 2020. [DOI] [Google Scholar]
- Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Naradikian M.S., Parkhouse K., Cain D.W., Jones L., Moody M.A., Verkerke H.P., Myles A., Willis E. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018;215:1571–1588. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Parkhouse K., Kirkpatrick E., McMahon M., Zost S.J., Mui B.L., Tam Y.K., Karikó K., Barbosa C.J., Madden T.D. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018;9:3361. doi: 10.1038/s41467-018-05482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell. 2017;168:1114–1125. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth C., Cantaert T., Colas C., Prot M., Casadémont I., Levillayer L., Thalmensi J., Langlade-Demoyen P., Gerke C., Bahl K. A Modified mRNA Vaccine Targeting Immunodominant NS Epitopes Protects Against Dengue Virus Infection in HLA Class I Transgenic Mice. Front. Immunol. 2019;10:1424. doi: 10.3389/fimmu.2019.01424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F., Lanzavecchia A., Araki K., Ahmed R. From vaccines to memory and back. Immunity. 2010;33:451–463. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim B.S., Kwon Y.C., Ricciardi M.J., Stone M., Otsuka Y., Berri F., Kwal J.M., Magnani D.M., Jackson C.B., Richard A.S. Zika Virus-Immune Plasmas from Symptomatic and Asymptomatic Individuals Enhance Zika Pathogenesis in Adult and Pregnant Mice. MBio. 2019;10:e00758-19. doi: 10.1128/mBio.00758-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smatti M.K., Al Thani A.A., Yassine H.M. Viral-Induced Enhanced Disease Illness. Front. Microbiol. 2018;9:2991. doi: 10.3389/fmicb.2018.02991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.R.F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., Gary E.N., Walker S.N., Schultheis K., Purwar M. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat. Commun. 2020;11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlbauer D., Amanat F., Chromikova V., Jiang K., Strohmeier S., Arunkumar G.A., Tan J., Bhavsar D., Capuano C., Kirkpatrick E. SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr. Protoc. Microbiol. 2020;57:e100. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D., Pardi N., Muramatsu H., Karikó K. HPLC purification of in vitro transcribed long RNA. Methods Mol. Biol. 2013;969:43–54. doi: 10.1007/978-1-62703-260-5_3. [DOI] [PubMed] [Google Scholar]
- Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020:eabc6284. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ji W., Lyu S., Qiao L., Luo G. Tet-Inducible Production of Infectious Zika Virus from the Full-Length cDNA Clones of African- and Asian-Lineage Strains. Viruses. 2018;10:700. doi: 10.3390/v10120700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K., Agnihothram S., Baric R.S., David C.S., Perlman S. Airway Memory CD4(+) T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity. 2016;44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source of protein and nucleic acid sequences are indicated in the manuscript and are available from the corresponding author on request.