Abstract
C-Type Lectin Receptors (CTLR) are involved in the activation of innate and adaptative immune responses. Among these receptors, the Dendritic Cell-Specific ICAM-3-Grabbing nonintegrin (DC-SIGN/CD209) has become a hot topic due to its ability to bind and facilitate the infections processes of several pathogens. Although well characterized in mammals, little documentation exists about the receptor in salmonid fishes. Here, we report the sequence and expression analysis of eight DC-SIGN-like genes in Salmo salar. Each receptor displays structural similarities to DC-SIGN molecules described in mammals, including internalization motifs, a neck region with heptad repeats, and a Ca+2-dependent carbohydrate recognition domain. The receptors are expressed in multiple tissues of fish, and fish cell lines, with differential expression upon infection with viral and bacterial pathogens. The identification of DC-SIGN-like receptors in Salmo salar provides new information regarding the structure of the immune system of salmon, potential markers for cell subsets, as well as insights into DC-SIGN conservation across species.
Keywords: Salmo salar, Immune receptor, C-Type lectic receptor, DC-SIGN
Abbreviations: CTL, C-Type Lectin; CTLD, C-Type Lectin Domain; ASGR, Asialoglycoprotein Receptors; DC, Dendritic Cell; APC, Antigen Presenting Cell; ITAM, Immunoreceptor Tyrosine-based Activation Motif; HIV, Human Immunodeficiency Virus; SsSIGN, Salmo salar SIGN; TM, Transmembrane Domain; SERNAPESCA, Chilean National Fisheries and Aquaculture Service; FBS, Fetal Bovine Serum; CBD, Carbohydrate-Binding Domain; dpi, days post infection; MOI, multiplicity of infection
1. Introduction
The C-Type Lectin (CTL) superfamily includes a large number of members throughout the animal kingdom. Characterized by Ca+2-dependent carbohydrate-binding, they are functionally involved in cell adhesion, cell communication, pathogen recognition and activation of immune responses, among others (Dambuza and Brown, 2015; Weis et al., 1998; Zelensky and Gready, 2005). This superfamily has been classified in 14 groups of proteins, based on their C-type Lectin Domain (CTLD) architecture and phylogeny (Drickamer and Fadden, 2002). Group II contains Asialoglycoprotein Receptors (ASGR) and Dendritic Cell (DC), Macrophage, Langerin, and Kupffer cells receptors. They are type II transmembrane proteins, containing a short cytoplasmatic tail and an extracellular neck region, which varies significantly among different members, which connects to the C-terminal CTLD.
CD209, also known as DC-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN), and its homolog DC-SIGNL/CD209R, are members of the Group II CTL superfamily. DC-SIGN has been identified as an adhesion molecule, involved in the attachment of antigen-presenting cells (APC) to resting T cells, and the aggregation and migration of APCs, as well as inflammatory responses, concomitantly participating in innate and adaptative immunity in mammals (Geijtenbeek et al., 2000; Khoo et al., 2008; Rappocciolo et al., 2008, 2006). Similar to Toll-like receptors (TLRs), DC-SIGN also acts as a pattern recognition receptor (PRR), promoting phagocytosis in macrophages and DCs (Montoya et al., 2009; Serrano-Gómez et al., 2004). Intracellular signaling pathways can be activated indirectly via association with other receptors, or directly, through their own Immunoreceptor Tyrosine-based Activation Motif (ITAM, YxxL/I) (Hoving et al., 2014).
Even though they play an essential role in the defense against a broad range of pathogens, viral recognition by CTLRs can favor infection. Most notably, recognition of the Human Immunodeficiency Virus (HIV) by DC-SIGN in dendritic cells facilitates the viral infection of CD4+ cells. CTLRs also aid in the infective process of other viruses, such as Influenza A, Cytomegalovirus, Dengue, Ebola, Hepatitis C, Coronavirus, West Nile, and Measles (Avota et al., 2013; Gillespie et al., 2016; Hillaire et al., 2013; Mesman et al., 2012). Moreover, there is also evidence that these receptors interact with bacterial pathogens and parasites (Appelmelk et al., 2003; Cambi et al., 2003). This condition makes CD209 all the more relevant not only in the context of the immune response but in the identification of susceptibilities and the design of prevention strategies (de Witte et al., 2008).
DC-SIGN genes have been described in at least three species of fish, including, Fugu rubripes, Danio rerio, and Cynoglossus semilaevis, based on sequence homology to mammal genes and different functional assays (Jiang and Sun, 2017; Lin et al., 2009; Zelensky and Gready, 2004). In fugu, eight different copies of DC-SIGN were found, a similar condition to the eight mouse genes that encode SIGNs (Powlesland et al., 2006). A significant number of putative gene sequences in the available salmon genome are annotated as DC-SIGN-like genes, with no dedicated or functional analyses involved. Furthermore, even though DC has not been fully isolated in Salmo salar, a putative CD209 sequence was detected in and used to characterize a DC-like subtype of cells in this species (Iliev et al., 2019). A dedicated curation of available data may help to identify sequences corresponding to proprietary Salmo salar DC-SIGN genes.
In the present study, we describe the characterization of eight novel DC-SIGN/CD209 orthologs from Salmo salar, based on available sequences from the genome and ESTs data, as well as expression analysis assays. We termed the genes SsSIGN1 - 8 (Salmo salar SIGN1 to 8), retaining the original SIGN acronym, but removing the DC-limited component. The eight genes code for proteins that display remarkable structural similarities to mammalian DC-SIGN proteins, including internalization motifs, a neck region with conserved heptad repeats, and a CTLD. The identified genes are distributed in two groups, containing four genes each, and are located in discrete regions of chromosomes 4 and 8. Differential gene expression in fish tissues and cell lines was detected, as well as specific responses to viral and bacterial pathogens. The presence of similar genes in other salmonid fishes was also identified and further discussed. Our work provides fundamental knowledge about the Salmo salar immune system, its response to specific pathogens, as well as novel insights into the DC-SIGN function and conservation across species.
2. Materials and methods
2.1. Database screening and sequence analyses
The ICSASG_v2 RefSeq genome records for Salmo salar, annotated by the NCBI Eukaryotic Genome Annotation Pipeline, was screened using “CD209”, “DC-SIGN” and “C-Type Lectin” as queries (Davidson et al., 2010). Non-redundant gene results were analyzed for transcription variant, using the Salmo salar ESTs database to identify effectively transcribed sequences. Identified protein sequences were further analyzed for the presence of a Transmembrane Domain (TM) (TMHMM Server 2.0, CBS), Heptad Repeats (RADAR, EMBL-EBI), coiled-coil domains (Parcoil2, MIT) and a CTLD at the carboxy end (NCBI, CD-search) (Madeira et al., 2019; Marchler-Bauer et al., 2017; McDonnell et al., 2006; Sonnhammer et al., 1998). Eight genes were identified, coding for DC-SIGN-like proteins in Salmo salar. The Oncorhynchus mykiss (Rainbow trout) Omyk_1.0 RefSeq, Salmo trutta (River trout) fSalTru1.1 RefSeq, Danio rerio (Zebrafish) GRCz11 RefSeq genome records, were used for similar analyses, to identify DC-SIGN genes on those species (Pasquier et al., 2016; Zardoya et al., 1995). Putative promoter sequences for each gene were analyzed for the presence of transcription factor binding sites using the TRANSFAC 8.3 database in PROMO (Farré et al., 2003; Messeguer et al., 2002). Sequence alignment was performed using Clustal Omega and visualized in Jalview (Madeira et al., 2019; Waterhouse et al., 2009). Protein structure homology modeling was performed for SsSIGN5 using the Swiss-Model server, with 1fih as a template (Waterhouse et al., 2018).
2.2. Expression analysis of Salmo salar SIGN genes
The expression of Salmo salar SIGN genes was assessed in vivo using healthy fishes, as well as in infected cell lines.
2.2.1. Animal ethics
Experiments involving live animals were conducted following the regulations of Chile, according to the Chilean National Fisheries and Aquaculture Service (SERNAPESCA). Salmon were cultivated in Centro de Investigación en Acuicultura Curauma at Pontificia Universidad Católica de Valparaíso. Fish were purchased from authorized centers of Sernapesca, and the same organization approved the transfer and the experiment itself.
2.2.2. Tissue collection
Selected organs were recovered from clinically healthy juvenile Salmo salar specimens (average weight of 30 g), that previously tested free of bacterial, viral and fungal pathogens, and were cultivated in saltwater conditions. Fish were euthanized with an overdose of benzocaine. Samples from 5 fish were recovered, including blood, kidney, spleen, gill, liver, and brain. Peripheral blood leukocytes (PBL) were recovered from blood samples using a discontinuous Percoll gradient, as previously described (Pettersen et al., 1995).
2.2.3. Cell culture
Atlantic salmon kidney (ASK) cells (ATCC CRL2747), were cultured at 20 °C in Leibovitz's L-15 media with 4 mM glutamine (Gibco) and supplemented with 200 U/ml penicillin, 200 μg/ml streptomycin, 0.5 μg/ml amphotericin, and 20% Fetal Bovine Serum (FBS) (Gibco). Atlantic Salmon Head Kidney (SHK-1) cells (ECACC 97111106) were cultured at 20 °C in Leibovitz's L-15 media with 4 mM glutamine (Gibco), 10% FBS (Gibco) and 40 μM 2-Me (Gibco). Cells were grown to 80% confluence and sub-cultured accordingly.
2.2.4. In vitro infection assays
To evaluate the expression profile of SsSIGN genes upon infection with relevant pathogens, we selected experimental models of Infectious Salmon Anemia Virus (ISAV) infection of ASK cells, and Piscirickettsia salmonis infection of SHK-1 cells (Castillo-Cerda et al., 2014; Gómez et al., 2013).
A field isolate of ISAV corresponding to the HPR7b type was obtained from the Laboratorio de Genética e Inmunología Molecular strain collection (Pontificia Universidad Católica de Valparaíso, PUCV). For viral infection and propagation, an 80% confluent ASK cell monolayer was washed twice with L-15 media and covered with a viral dilution prepared in L-15 media. After 4 h, the virus was removed, and the cells were washed twice with L-15 media and cultured in 2% FBS L-15 media with antibiotics at 17 °C. After 7 days, the cell supernatant was filtered (0.45 μm), and the virus was collected. A plaque assay was used for virus tittering 12 days post infection (dpi), as previously described (Castillo-Cerda et al., 2014).
A Piscirickettsia salmonis field isolate, termed Psal-104, was obtained from the Chilean National Piscirickettsia salmonis Strain Collection (PUCV), and cultured in BM3 media, with 100 rpm. agitation, at 19 °C, as previously described (Henríquez et al., 2013). Exponentially growing bacteria (O.D.600 = 0.6) was recovered by centrifugation at 5000 x g, washed twice, and resuspended in L-15 media. Bacteria were counted using a Petroff-Hausser chamber.
ASK cells were seeded in 6 well plates, at 250.000 cells/well, and incubated for 24 h before removing the culture media and infecting with an ISAV inoculum at a multiplicity of infection (MOI) of 0.5. After a 4-h incubation, the monolayer was washed twice with L-15 media and incubated in antibiotics and 2% FBS supplemented L-15 at 17 °C. Three days post infection culture media was removed, and cells were recovered applying TRIzol™ solution (Invitrogen™) for RNA extraction. An aliquot of the virus, corresponding to the same viral load, was inactivated by incubation at 56 °C for 30 min and used to infect ASK cells; uninfected cells were used as controls (Falk et al., 1997). All infections were performed in triplicates.
SHK-1 cells were seeded in 6 well plates, at 200.000 cells/well, and incubated for 24 h before removing the culture media and infecting with a Piscirickettsia salmonis inoculum at a MOI of 100. After a 4-h incubation, cells were washed five times with L-15 media, and fresh SFB supplemented L-15 media was added. Infected cells were incubated at 20 °C. Five dpi, media was removed, and cells were recovered, applying TRIzol™ solution (Invitrogen™) for RNA extraction. An aliquot of the bacteria, corresponding to the same bacterial load, was inactivated by incubation at 56 °C for 15 min, and used to infect SHK-1 cells; uninfected cells were used as controls (Álvarez et al., 2016). All infections were performed in triplicates.
2.2.5. RNA extraction and cDNA synthesis
RNA was obtained from tissue samples using a combination of TRIzol™ buffer, and E.Z.N.A.® Total RNA Kit I (OMEGA Bio-Tek). 50 mg of tissue, or cells from 1 well from a 6-well plate, were resuspended in 1 ml of TRIzol buffer and disaggregated using a syringe. After following the manufacturer's indications, the aqueous phase from the TRIzol-Chloroform extraction was mixed with 1 volume of 70% ethanol, and RNA extraction proceeded according to the column's recommended protocol. RNA was resuspended in nuclease-free water and quantified using a Nanodrop spectrophotometer (Thermo Scientific). All samples had A260/A280 ratios over 1.8. Samples corresponding to 1 μg of total RNA were treated with 1 U of DNase I, RNase-free (Thermo Scientific) for 45 min at 37 °C. After DNase inactivation, cDNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit, using Random Hexamers as primers (Thermo Scientific).
2.2.6. Quantitative PCR (qPCR)
Specific primers were designed to assess the level of expression of each selected Salmo salar SIGN mRNA. Expression of Elongation factor 1 alpha was used as a house-keeping gene for normalization (Table 1 ) (Salazar et al., 2016).
Table 1.
Primers sequences for RT-qPCR of Salmo salar DC-SIGN-like receptors. Elongation factor 1 alpha was used as a house-keeping gene for normalization. Chromosomal location is indicated by Chr04 (chromosome 04) and Chr08 (chromosome 08).
| Chro. | RefSeq mRNA | Forward | Reverse | Obs. |
|---|---|---|---|---|
| Chr04 | XM_014194757.1 | TCAACGATAGGACCAGAACCTG | AGGGTGTCTGCACTGACGTA | SsSIGN1 |
| XM_014194638.1 | GGAGGAGGACTGTGTTGAGC | TGCGTTTGATACCGGTCCAT | SsSIGN2 | |
| XM_014194726.1 | AGGCTGGTGGAAGTCATGTG | CTTCTCCCTGCACTGTCCTG | SsSIGN3 | |
| XM_014194753.1 | TATATGGCAATGTGGGAGCCT | AGACACACTGCAACAACTAGGA | SsSIGN4 | |
| Chr08 | XM_014209668.1 | TACTGACCACCCCAAGGTACTG | TTGAGTTTGCTGTCGCATGAC | SsSIGN5 |
| XM_014209793.1 | ACCACTGACCACAAAGTATTGGA | GTTGAGAGAGTACCGAGGAGG | SsSIGN6 | |
| XM_014209821.1 | GGCTTACCAATCTCGGCTGG | ATCAGGCTGCTTGTCATTCCA | SsSIGN7 | |
| XM_014209791.1 | GTCATCAGACACCGGAGCAT | CAGCGAGTTGTCCCCTCTTT | SsSIGN8 | |
| NM_001173967.1 | GCTTACAAAATCGGCGGTAT | CTTGACGGACACGTTCTTGA | EF-1a |
qPCR reactions were performed using the Brilliant III Ultra-Fast SYBR® Green QPCR Master Mix (Agilent Technologies), according to the manufacturer's instructions, with 200 nM of each primer, and 2 μl of cDNA as the template for a 20 μl reaction. Reactions were performed in a Bio-Rad CFX96 thermal cycler, with cycling conditions as follows: 1 cycle of 3 min at 95 °C and 40 cycles of 5 s at 95 °C and 10 s at 60 °C. Finally, a dissociation curve was performed according to the instrument settings.
2.2.7. Expression analysis
The fold change of gene expression levels, relative to controls, was assessed using the 2−ΔΔCT method (Livak and Schmittgen, 2001). The results are presented as the means ± standard deviations of replicas. The statistical significance of the data was determined by using a Student's t-test. P-values < 0.05, <0.01 and < 0.001, are indicated by *, ** and *** respectively, and were considered significant as properly indicated.
2.3. Phylogenetic analysis
Amino acid sequences corresponding to the Carbohydrate-Binding Domains (CBD) of selected DC-SIGN/CD209 molecules were recovered from NCBI (Table 2 ). Analysis of Oncorhynchus mykiss, Salmo trutta and Danio rerio genomes, as previously described, allowed for the identification of DC-SIGN/CD209 orthologous in these species; CBDs were recovered from these sequences and included in further analyses. A phylogenetic tree was constructed by Neighbor-Joining method, using MEGA X (Stecher et al., 2020), based on the amino acid alignment of all these sequences (Clustal Omega) (Madeira et al., 2019). Bootstrap values were calculated from 2000 repetitions. The phylogenetic tree was rendered using iTOL (Letunic and Bork, 2007).
Table 2.
Sequences used for phylogenetic analyses. Species, GenBank accession number, and the region corresponding to CTLD are displayed.
| Species | Access nº | CTLD | Species | Access nº | CTLD | Species | Access nº | CTLD |
|---|---|---|---|---|---|---|---|---|
| H.sapiens | >Q9NNX6-2 | 251..398 | G.gallus | >NP_990815.1 | 85..203 | O.mykiss | >XP_021475407.1 | 156..280 |
| H.sapiens | >AAK20998.1 | 207..343 | X.tropicalis | >XP_031755207.1 | 91..211 | O.mykiss | >XP_021475400.1 | 167..291 |
| M.musculus | >AAL13235.1 | 162..297 | O.niloticus | >XP_025758281.1 | 84..224 | O.mykiss | >XP_021472964.1 | 136..264 |
| M.musculus | >AAL13236.1 | 105..240 | S.scrofa | >NP_001123444.1 | 112..240 | O.mykiss | >XP_021475402.1 | 167..291 |
| M.musculus | >AAL13237.1 | 101..237 | D.rotundus | >XP_024416423.1 | 131..259 | O.mykiss | >XP_021428956.1 | 175..297 |
| M.musculus | >AAL13238.1 | 72..208 | P.walrus | >XP_004412214.1 | 121..249 | O.mykiss | >XP_021428954.1 | 193..315 |
| M.musculus | >AAL13234.1 | 103..238 | O.orca | >XP_004277712.1 | 150..278 | O.mykiss | >XP_021428962.1 | 24..146 |
| M.musculus | >XP_889104.1 | 72..208 | L.africana | >XP_023396298.1 | 130..261 | O.mykiss | >XP_021428969.1 | 193..315 |
| M.musculus | >NP_081619.3 | 61..194 | C.Hircus | >XP_005682468.1 | 131..259 | O.kisutch | >XP_020327037.1 | 133..251 |
| M.musculus | >NP_081232.2 | 122..255 | E.asinus | >XP_014717459.1 | 107..238 | O.kisutch | >XP_020312192.1 | 169..295 |
| P.troglodytes | >AAL89545.1 | 236..358 | O.cuniculus | >XP_017193467.1 | 109..239 | S.trutta | >XP_029622272.1 | 166..294 |
| P.troglodytes | >AAL89535.1 | 314..437 | C.griseus | >XP_027271607.1 | 110..238 | S.trutta | >XP_029621727.1 | 161..286 |
| P.pygmaeus | >AAL89542.1 | 256..379 | U.maritimus | >XP_008709536.1 | 121..249 | S.trutta | >XP_029622357.1 | 179..302 |
| P.paniscus | >ABW34403.1 | 256..379 | V.pacos | >XP_015096456.1 | 121..249 | S.trutta | >XP_029622352.1 | 195..321 |
| P.paniscus | >ABW34400.1 | 268..391 | L.obliquidens | >XP_026941610.1 | 177..305 | S.trutta | >XP_029622368.1 | 146..272 |
| H.lar | >AAL89538.1 | 302..425 | P.vampyrus | >XP_023380689.1 | 62..193 | S.trutta | >XP_029622359.1 | 177..301 |
| H.lar | >AAL89528.1 | 268..391 | S.salar | >XP_014050232.1 | 176..310 | S.trutta | >XP_029622481.1 | 191..315 |
| N.leucogenys | >ABW34404.1 | 256..379 | S.salar | >XP_014050113.1 | 231..367 | S.trutta | >XP_029622476.1 | 317..441 |
| N.leucogenys | >ABW34402.1 | 245..368 | S.salar | >XP_014050200.1 | 163..287 | D.rerio | >XP_003197805.3 | 196..308 |
| S.syndactylus | >AAL89539.1 | 233..356 | S.salar | >XP_014050228.1 | 134..262 | D.rerio | >XP_017211404.1 | 195..306 |
| S.syndactylus | >AAL89529.1 | 291..414 | S.salar | >XP_014065143.1 | 137..272 | D.rerio | >XP_009293464.2 | 196..307 |
| C.l. familiaris | >NP_001124304.1 | 118..238 | S.salar | >XP_014065268.1 | 146..275 | D.rerio | >NP_001186302.2 | 220..343 |
| O.latipes | >ADB55614.1 | 75..203 | S.salar | >XP_014065296.1 | 187..311 | C.semilaevis | >XP_024920596.1 | 122..265 |
| T.nigroviridis | >ADB55615.1 | 47..168 | S.salar | >XP_014065266.1 | 373..502 |
3. Results
3.1. Sequence analysis of DC-SIGN-like genes in Salmo salar
Potential Salmo salar orthologs of mammalian DC-SIGN genes were identified on the fish genome. We screened the ICSASG_v2 RefSeq genome for non-redundant gene sequences coding for transmembrane proteins containing canonical features of DC-SIGN receptors, that is: a CTLD, a neck region with heptad repeats, and internalization motifs in the cytoplasmatic region. The genes encoding each of the identified DC-SIGN homologs, termed SsSIGN 1–8, are located in discrete regions (~2 megabases) of chromosomes 4 and 8 (Table 3 ). Transcriptional variants were selected upon analysis of the Salmo salar EST database.
Table 3.
Identified genes coding for SsSIGN proteins. Chromosomal locations for each of them, reveals two clusters of genes located on chromosome 4 and 8, respectively. Internalization Motifs (Int.Motif) are annotated as single (+), double (++) or triple (+++). Predicted transmembrane domains (TM), and Heptad repeats (Hept. Rep.) are indicated. CTLD denotes the position of the C-Type Lectin Domain.
| Chro. | Location | RefSeq mRNA | RefSeq Protein | Size (aa) | Int. Motif. | TM | Hept. Rep. | CTLD | Name |
|---|---|---|---|---|---|---|---|---|---|
| Chr04 | 2.240.891..2.244.441 | XM_014194757.1 | XP_014050232.1 | 310 | ++ | 83..105 | 117..170 | 176..310 | SsSIGN1 |
| 3.468.837..3.485.920 | XM_014194638.1 | XP_014050113.1 | 367 | + | 40..62 | No | 231..367 | SsSIGN2 | |
| 3.596.403..3.600.814 | XM_014194726.1 | XP_014050201.1 | 287 | +++ | 64..86 | 117..156 | 163..287 | SsSIGN3 | |
| 3.706.690..3.710.679 | XM_014194753.1 | XP_014050228.1 | 280 | ++ | 57..79 | 109..148 | 152..280 | SsSIGN4 | |
| Chr08 | 19.257.147..19.259.043 | XM_014209668.1 | XP_014065143.1 | 272 | +++ | 63..85 | 92..131 | 137..272 | SsSIGN5 |
| 20.942.043..20.948.139 | XM_014209793.1 | XP_014065268.1 | 309 | ++ | 59..81 | 82..177 | 180..309 | SsSIGN6 | |
| 22.609.016..22.616.088 | XM_014209821.1 | XP_014065296.1 | 311 | +++ | 60..82 | 99..175 | 187..311 | SsSIGN7 | |
| 22.870.914..22.885.313 | XM_014209791.1 | XP_014065266.1 | 502 | ++ | 50..72 | 103..198/260..357 | 373..502 | SsSIGN8 |
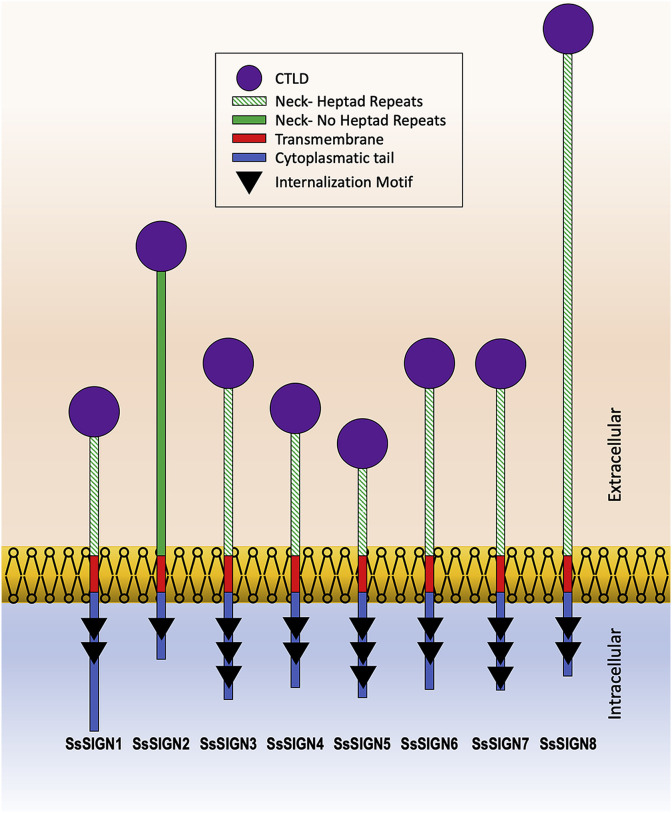
Fig. 1 shows the projected domain organization of the SsSIGN proteins. The sizes of the proteins range from 272 to 502 amino acids, with notable differences among the neck and cytoplasmatic regions of different receptors. All of them display typical internalization signals corresponding to either double leucine, triple acid, or ITAM motifs (Fig. 2 ). Heptad repeats are absent only on the SsSIGN2 gene, but the neck does conform a theoretical coiled-coil domain (data not shown), which is probably involved in oligomerization. The cytoplasmatic domains contain internalization signals, typical of this type of receptor. Notably, the repeats on the neck region correspond to a semi-conserved sequence ERDQLQYNNLTK, forming a heptad pattern of hydrophobic residues. SsSIGNs feature conserved residues implicated in carbohydrate and Ca+2 binding, compared to Homo sapiens and Mus musculus sequences; the fish CTLD contains only six cysteine residues, possibly involved in three disulfide bridges, unlike mammals DC-SIGNs which are characterized by a fourth bridge.
Fig. 1.
Domain organization of SsSIGN proteins. CTLD is located at the carboxyl end and connected to the transmembrane domain trough a neck region containing, in most cases, heptad repeats possibly involved in oligomerization. Neck and cytoplasmatic regions primary structure lengths are displayed proportionally for each homolog.
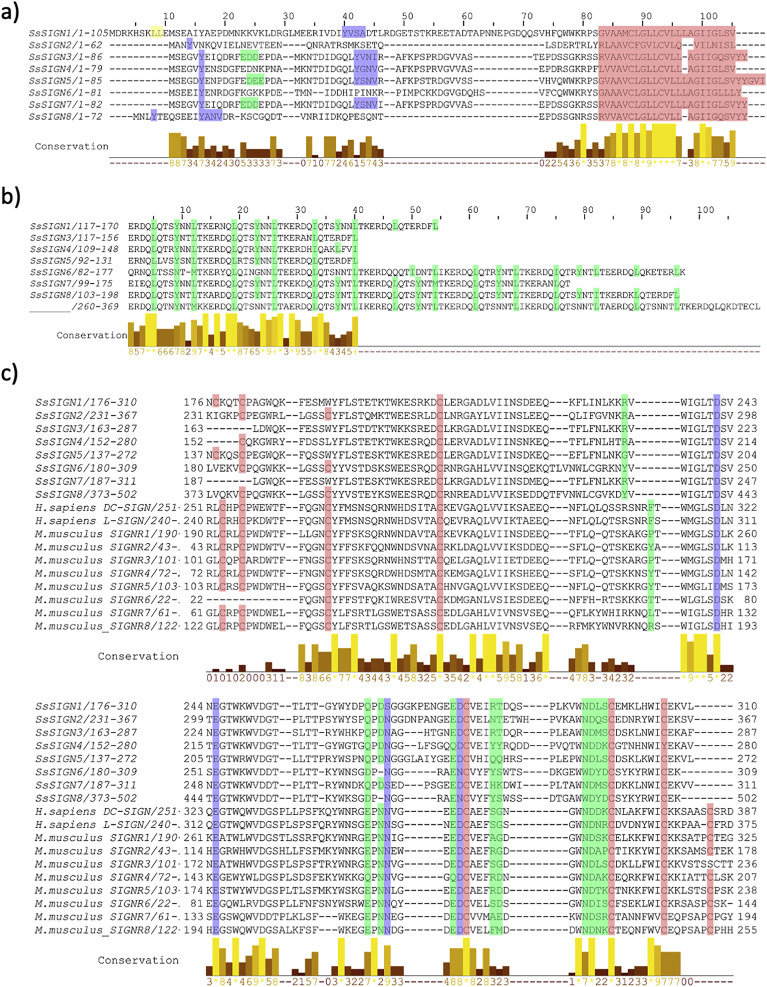
Fig. 2.
Multiple sequence alignment, displaying typical features of SsSIGN. A) Alignment for cytoplasmatic regions of salmon SIGN, with color-coded internalization motifs: yellow – double leucine, green-triple acid, and blue ITAM (YXX-I/L or YXXΦ). Residues in red correspond to the transmembrane domain. B) Alignment of neck regions of salmon SIGNs. The sequence ERDQLQYNNLTK appears highly conserved among the different SsSIGNs. Hydrophobic residues, forming the heptad repeats, are highlighted in green. C) Alignment of SsSIGN with sequences from Homo sapiens and Mus musculus homologous, with highlighted conserved residues. In red, cysteines involved in disulfide bridges in mammals. In green, residues involved in carbohydrate binding. In blue, residues involved in Ca+2 binding. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
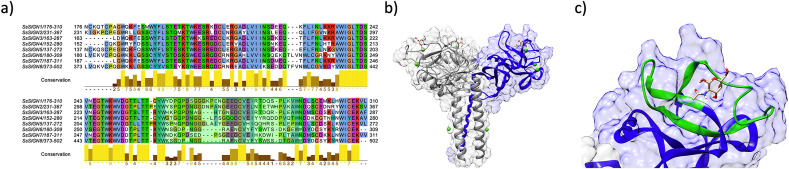
The CTLD sequence on the SsSIGNs appears highly conserved except for a 35–40 amino acid stretch, which displays high variability among different receptors (Fig. 3 ). Structural modeling locates these residues in the putative CBD, suggesting different specificities/avidities for each one of the receptors.
Fig. 3.
Features of the CTLD of SsSIGNs. A) Multiple sequence alignment for the different salmon DC-SIGN homologous CTLD. Sequences are conserved among different genes, with a discrete region of particular variability highlighted in green. B) A structural model for a monomer of SsSIGN5 (blue), in the context of a tetramer formed by the template protein, with mannose molecules located at the CBD. C) Structural detail of the putative CBD of SsSIGN5. The variable region from A is highlighted in green and corresponds to residues interacting with the carbohydrate ligand. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
A 1000 bp gene sequence upstream of the start codon was analyzed to identify possible Transcription Factor (TF) binding sites described in mammalian homologs (Fig. 4 ). The promoter regions contain binding sites for Activator Protein 1 (AP-1), Nuclear Factor kappa B (NF- κB), and Transcription Factor SP1 (SP1). Each gene contains different number and distribution of sites, with discrete conservancy between SsSIGN3 and 7.
Fig. 4.
Promoter regions for SsSIGN genes. Predicted binding sites for transcription factors are highlighted in red for AP-1 sites, green for NF-κB, and blue for SP1. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Expression analyses of SsSIGNs
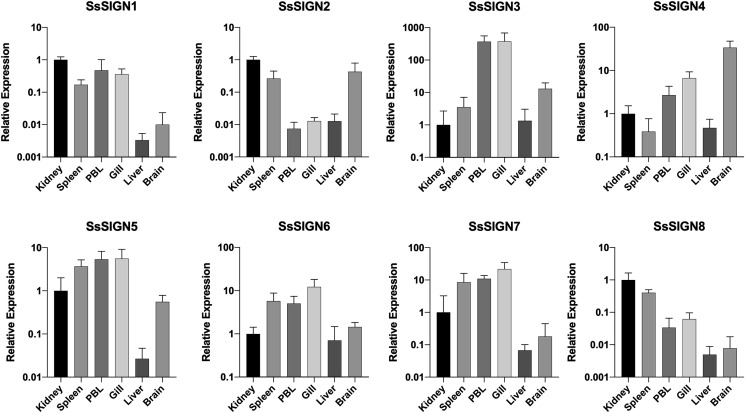
The expression of each receptor was evaluated in immuno-relevant tissues of healthy fish, using RT-qPCR. Expression was effectively detected in all samples, with unique distribution patterns for each (Fig. 5 ). SsSIGN1, 2, 7, and 8, had in general higher expression levels in organs directly involved in immune response (kidney and spleen). Interestingly, SsSIGN 3 was highly expressed in PBL and gill, compared to the kidney, and SsSIGN 4 had the highest levels in the brain. Differential expression of the genes in each tissue suggests specific roles for the receptors in different organs.
Fig. 5.
Expression analysis of SsSIGN in fish organs. Expression levels for each gene in different organs relative to the expression in the kidney. Each gene displays a different pattern of distribution. Results correspond to an n = 5.
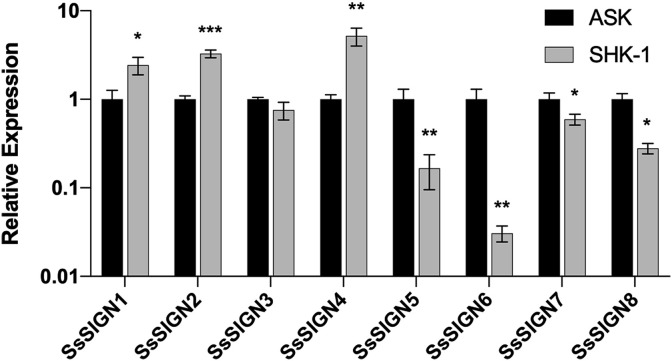
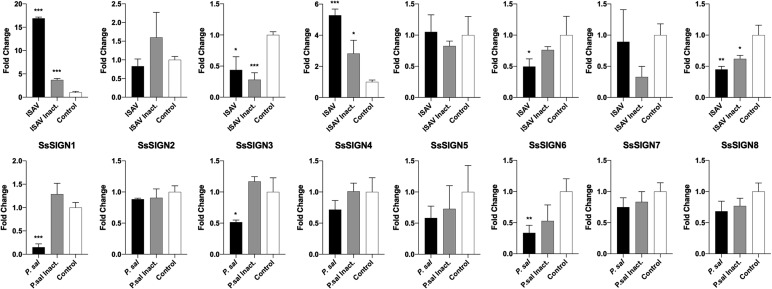
Two Salmo salar cell lines were tested for expression of the SsSIGN genes. ASK cells correspond to epithelial cells, highly susceptible to ISAV infection; SHK-1 cells display properties of fish macrophages and are susceptible to infection with Piscirickettsia salmonis. The expression of the receptors was detected in both cell lines, previous to infection assays (Fig. 6 ). Both pathogens regulated the expression of the SsSIGNs differentially. ISAV infection induced the expression of SsSIGN 1 and 4 and reduced the expression of SsSIGN 3, 6, and 8. The effects were dependent on the active infection (i.e., inactivated virus did not render the effect in the same magnitude), which suggests that the regulation is not only related to the receptor activation but may be enhanced by an integrated immune response to the viral infection. On the other hand, Piscirickettsia salmonis infection of the SHK-1 cell line downregulated SsSIGN 1, 3, and 6, with the effect being absent in cells with the inactivated bacteria (Fig. 7 ).
Fig. 6.
Expression of SsSIGN genes on two salmon cell lines. Levels are relative to the expression in the ASK line. Results correspond to an n = 3.
Fig. 7.
Expression of SsSIGN genes during viral and bacterial infection in vitro. The upper panel displays the expression levels of SsSIGNs in ASK cells after 72 h infection with ISAV. The lower panel shows gene expression on SHK-1 cells after a 5-day infection with Piscirickettsia salmonis. Levels are relative to the uninfected controls. Results correspond to an n = 3.
3.3. Phylogenetic analyses of SsSIGNs
To explore the conservation of DC-SIGN-Like genes in other fishes, we screened the genomes of two other salmonid species, Oncorhynchus mykiss and Salmo trutta, and the cyprinid Danio rerio, for DC-SIGN-like sequences, using the same approach described for Salmo salar. Both salmonid species display a similar array of DC-SIGN-like genes, located in discrete regions of their genomes. Particularly, O. mykiss has eight homologous, evenly distributed between chromosome 10 a chromosome 19. On the other hand, the 8 DC-SIGN-Like genes of S. trutta are located in chromosome 11, with one, 4-gene cluster at ~18 Mb, and two pairs of genes at ~21 and ~16 Mb. The homologous, display the conserved features of this type of gene, with a single copy in both salmonid genomes lacking the heptad repeat domain, similar to SsSIGN2. Moreover, one gene in O. mykiss and two in S. trutta, lack a TM domain, suggesting a soluble nature, similar to the structure of M. musculus SIGNR2 and 6. Finally, we identified only three homologous DC-SIGN genes in the Danio rerio genome, located in a discrete region of chromosome 1 (Table 4 ).
Table 4.
DC-SIGN-Like sequences identified in selected fish species.
| Species | Chro. | Location | RefSeq Protein | Int. Motif | TM | Hept. Motif | CTLD |
|---|---|---|---|---|---|---|---|
| Oncorhynchus mykiss | Chr10 | 68,316,695..68,351,882 | XP_021475407.1 | +++ | 58..80 | 94..145 | 156..278 |
| 68,437,277..68,440,058 | XP_021475400.1 | ++ | 60..82 | 106..157 | 167..289 | ||
| 68,449,595..68,452,375 | XP_021472964.1 | ++ | 7..29 | 44..124 | 136..262 | ||
| 68,502,845..68,505,833 | XP_021475402.1 | ++ | 60..82 | 106..157 | 167..289 | ||
| Chr19 | 3,359,438..3,415,338 | XP_021428956.1 | + | 63..85 | 107..160 | 175..295 | |
| 3,418,729..3,424,484 | XP_021428954.1 | + | 62..84 | 107..179 | 193..313 | ||
| 3,426,261..3,427,566 | XP_021428962.1 | – | – | – | 24..144 | ||
| 3,615,353..3,621,025 | XP_021428969.1 | ++ | 62..84 | 107..179 | 193..313 | ||
| Salmo trutta | Chr11 | 16,730,161..16,733,194 | XP_029622272.1 | + | 59..81 | 77..158 | 166..292 |
| 16,911,497..16,937,038 | XP_029621727.1 | – | – | – | 161..284 | ||
| 18,643,399..18,648,784 | XP_029622357.1 | ++ | 65..87 | 106..156 | 179..300 | ||
| 18,740,308..18,756,197 | XP_029622352.1 | ++ | 86..108 | 134..185 | 195..319 | ||
| 18,775,740..18,784,853 | XP_029622368.1 | + | 39..61 | 83..133 | 146..270 | ||
| 18,918,066..18,936,039 | XP_029622359.1 | + | 58..80 | 92..168 | 177..297 | ||
| 21,422,649..21,430,498 | XP_029622481.1 | – | – | 3..167 | 191..315 | ||
| 21,527,965..21,532,327 | XP_029622476.1 | + | 32..54 | 97..309 | 317..437 | ||
| Danio rerio | Chr01 | 55,903,808..55,906,907 | XP_003197805.3 | ++ | 43..65 | 103..184 | 196..308 |
| 55,908,572..55,919,463 | XP_017211404.1 | ++ | 42..64 | 102..183 | 195..306 | ||
| 55,920,864..55,922,977 | XP_009293464.2 | ++ | 43..65 | 103..184 | 196..307 |
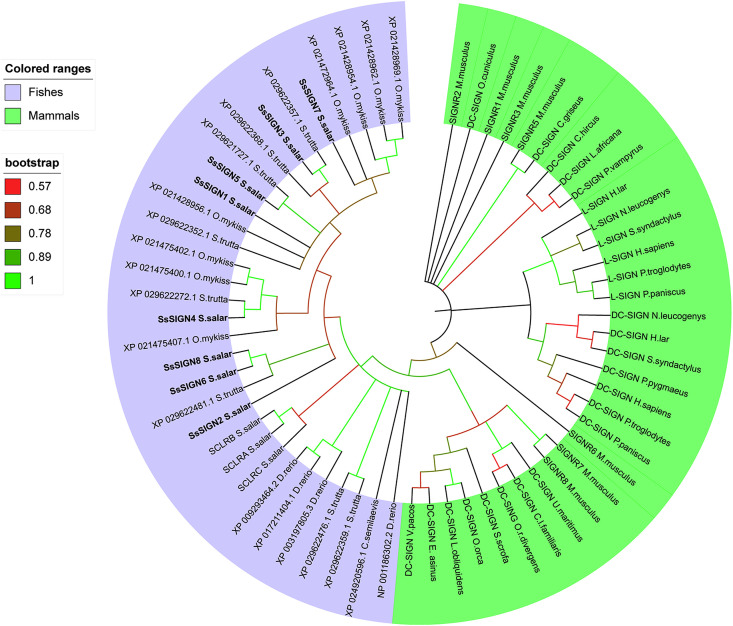
For phylogenetic analysis, a total of 33 DC-SIGN sequences described for mammal species, the 19 DC-SIGN-Like homologous from O. mykiss, S. trutta and D. rerio, 3 CTL Receptors previously described by Soanes (Soanes et al., 2004), DC-SIGN sequences described for Danio rerio and Cynoglosus semilaevis (Jiang and Sun, 2017; Lin et al., 2009), and the eight SsSIGN sequences, were used to construct an unrooted phylogenetic tree using the neighbor-joining method. The analysis showed that SsSIGN sequences are classified in a cluster formed by salmonid species, except for two S. trutta sequences, and separated from the other CTLR from salmon. The three sequences we identified in D. rerio form an independent cluster, separated from the previously reported sequence. Among mammals, primates are clustered in a defined group, and M. musculus sequences are distributed in all clades. The phylogenetic analysis reflects the variety and diversity of DC-SIGN genes, and its distribution across species (Fig. 8 ).
Fig. 8.
Phylogenetic tree showing the relationship between SsSIGN amino acid sequences and other species of the DC-SIGN family. The unrooted phylogenetic tree was constructed by the neighbor-joining method, based on the amino acid alignment (Clustal Omega) of CBD of protein sequences. Bootstrap values were calculated from 2000 repetitions.
4. Discussion
4.1. DC-SIGN receptors
Initially recognized as a receptor for HIV, present in human placenta, DC-SIGN was further characterized as a broad range pathogen-binding receptor, as well as an adhesion molecule that facilitates attachment of Dendritic Cells (DC) to T cells, supporting primary immune responses (Curtis et al., 1992; Garcia-Vallejo and van Kooyk, 2013). DC-SIGN homologs have been described in various species of mammals, where there are at least three family members with conserved functional domains; interestingly, mouse has eight DC-SIGN homologs, clustered in a discrete genomic region (Liu et al., 2004; Powlesland et al., 2006).
Even though DC-SIGN homologs have been described for other fish species, including zebrafish, fugu, and tongue sole, no ortholog has been described in salmonid species. Salmo salar is a commercially important farmed fish species, with a continually growing, worldwide industry (Little et al., 2015). Robust expansion on fish farming is based on the development of sanitary measures, which in turn relies on a proper understanding of the fish immune system (Andresen et al., 2020). In this work, we sought to identify and describe putative salmon orthologs of mammalian DC-SIGN receptors, focused on conserved structural features and expression patterns in response to viral and bacterial infections.
4.2. Sequences and features of SsSIGN
Our analysis of the Salmo salar genome and EST sequences led us to identify eight putative proteins sequences with characteristics of DC-SIGN receptors, including the canonical CTLD at the carboxy end connected to the transmembrane domain by a neck region, containing heptad repeats, and a cytoplasmatic tail with internalization signals. Even though our screening revealed the presence of several other proteins containing CTLD, only these eight sequences carry the features described for DC-SIGN genes (Table 3).
DC-SIGN molecules are type II transmembrane proteins, characterized by the presence of a C-Type Lectin Domain (CTLD), which interacts with glycans in a Ca+2 dependent fashion. The CTLD in SsSIGN displays conserved amino acid residues involved in the interaction with carbohydrates and Ca+2, compared to the Homo sapiens and Mus musculus sequences (Fig. 2) (Feinberg, 2001). The non-conserved residues in the Receptor Binding Domain (RBD) display high diversity among the salmon DC-SIGN sequences; this is similar to the mouse SIGN receptors, were the eight homologs feature different ligand-binding specificities, with this divergence being, arguably, a product of evolutionary pressure related to the exposure to species-specific pathogens (Garcia-Vallejo and van Kooyk, 2013) (Fig. 3). Furthermore, DC-SIGN typically recognizes fucosylated and high-mannose structures, modulating different cellular responses depending on the bound ligand: high-mannose glycans, expected to occur in higher mammals only in proteins during maturation, trigger a proinflammatory response (i.e., cells damage caused by pathogens). In contrast, fucosylated glycans suppress proinflammatory cytokines (inter-cellular signaling) (Gringhuis et al., 2009). Similarly, SsSIGNs could display different specificities, with relevant functional implications: on the one hand, allowing for the interaction with a broad range of microorganisms (from virus to fungus), and on the other, discriminating between different types of signals coming from other cells.
The heptad repeats in the neck domain in DC-SIGN consist of a heptad pattern of hydrophobic residues in a helical region, which mediates the packing of helices from different DC-SIGN monomers to form a 4-stranded coiled-coil in the neck domain of the DC-SIGN tetramer (dos Santos et al., 2017). According to our analyses, SsSIGN 2 lacks a heptad repeat motif in its neck region, where all the other homologs carry a repeated, semi-conserved sequence (Fig. 2). The lack of heptad repeats is also present in the CD209L receptor, found in some non-human primates, as well as in the zebrafish and tongue sole DC-SIGNs; it has been suggested that this type of SIGN receptor corresponds to the ancestor of the DC-SIGN family (Lin et al., 2009; Ortiz et al., 2008). Furthermore, a monomeric conformation, due to the lack of heptad repeats, may correlate with a specific function for SsSIGN2.
Dendritic Cells (DC) are specialized in presenting antigens for the activation of T cells to initiate an immune response. In these cells, DC-SIGN is involved in the internalization of antigens and pathogens upon ligand binding, and the complex is targeted to late endosomes/lysosomes (Engering et al., 2002). The cytoplasmatic tails of all SsSIGNs possess different internalization signals, including di-leucine motifs and tri-acid clusters which can be involved in the internalization process (Engering et al., 2002; Lin et al., 2009). On the other hand, even though all of the receptors carry tyrosine residues in this region, only SsSIGN3 displays a canonical Hemi ITAM motif (i.e., YxxI/L), with SsSIGN1,4 and 8 displaying a YxxΦ motif (where Φ is a hydrophobic residue), all of which may be involved in intracellular signaling (Guo et al., 2004) (Fig. 2). Concomitantly, DC-SIGN molecules are characterized by the lack of typical ITAM or ITIM motifs but do interact with a sophisticated signalosome inside of DCs (Gringhuis et al., 2009). Moreover, homo or heterotetramerization may play a role in signal transduction, where multiple phosphorylated tyrosine residues are necessary for signaling from a homotetramer, or the activation signal is conducted using an ITAM/ITIM carrying protein, in the context of a heterotetramer (Garcia-Vallejo and van Kooyk, 2013; Haining et al., 2017). As their homologs in mammals, SsSIGNs could be involved in the modulation of the responses initiated by other receptors (Hovius et al., 2008; Rodríguez et al., 2017).
4.3. Expression of SsSIGN and response to infection
Analysis of the upstream sequences of the 8 SsSIGN, revealed the presence of potential binding sites for transcription factors described in the mammalian homologs, including NF-κB, Sp1 and AP-1 sites (Fig. 4) (Liu et al., 2003). DC cells activate distinct sets of transcription factors upon maturation, which will lead to the transcription of different sets of genes as well; furthermore, expression of SsSIGNs in a wider variety of cell types would lead to a much more sophisticated expression profile, effectively mediated by its promoters (Mizumoto et al., 2005). Interestingly, polymorphism in mammalian DC-SIGN promoters is associated with susceptibility to viral infections (Wang et al., 2011).
SsSIGNs were expressed in all the analyzed salmon tissues, with differential expression patterns for each of them (Fig. 5). Immune system-related tissues (kidney and spleen) were, in general, enriched for the expression of the receptors. The liver displays discrete levels of most genes compared to the kidney. SsSIGN3 was highly expressed in PBL and Gill (over 300 times compared to the expression in the kidney), which suggest homing of a specific subset of SsSIGN3-expressing cells; macrophages and microfold-like cells have been described in rainbow trout gills, featuring antigen-sampling capabilities (Kato et al., 2018). Interestingly, the brain displays relatively high levels of most SsSIGN genes, particularly of SsSIGN4; both macrophages and mast cells, which act as APCs, have been detected in fish brains and could account for SsSIGN4 expression in that organ (Herbomel et al., 2001; Kordon et al., 2018).
Two salmon cell lines were analyzed for the expression of SsSIGN genes. ASK cells correspond to epithelial cells and SHK-1 to macrophage-like cells. Both cell lines displayed expression of all genes, with differential levels for each of them (Fig. 6). Although DC-SIGN has been canonically associated with APC like macrophages and DCs, expression of SsSIGN in salmon epithelial cells may be related to a role in antigen processing and presentation on this species. Furthermore, the expression of a specific profile of these receptors could increase differential susceptibility to infection and differential responses to specific pathogens to each cell line.
Infectious Salmon Anemia Virus (ISAV) produces an aggressive disease, primarily affecting Salmo salar (Vike et al., 2014). The virus is part of the Orthomyxoviridae family, with a segmented single-stranded negative-sense RNA genome and a viral envelope (Krossøy et al., 1999). Attachment to the cell surface is mediated by a viral glycoprotein termed hemagglutinin-esterase (HE), which binds to specific sialic acids (sia) on glycan chains present in the cellular surface proteins; endocytosis of the virion leads to membrane fusion and infection (Aamelfot et al., 2012). Furthermore, the virus codes for at least two proteins with interferon (IFN) antagonistic activities (McBeath et al., 2006; Olsen et al., 2016). We sought to determine the effect of ISAV infection on the expression of SsSIGN genes in vitro, considering the role that these receptors could play in viral binding and the regulation that viral proteins could have over them. ISAV infection of the permissive ASK cell line leads to significant upregulation of SsSIGN1 and 4, with a more pronounced effect on the former. The promoter for SsSIGN1 contains an NF-κB binding site, which is canonically activated by Influenza Virus infection in mammals, suggesting a similar effect for ISAV infection in salmon (Alexopoulou et al., 2001; Schmitz et al., 2014) (Fig. 7).
On the other hand, SsSIGN genes containing AP-1 sites display a tendency to be downregulated, a process that could be mediated by ISAV NS1, in parallel to what is observed in Influenza A Virus (IAV) infection (Ludwig et al., 2002). Moreover, differential effects are observed in ISAV infected and mock (inactivated virus) infected cells, which suggest a direct connection between active infection and cellular responses. Regulation of expression of SsSIGN genes during ISAV infection could play a direct role in cellular susceptibility: IAV is capable of infecting DC-SIGN/L-SIGN expressing cells, in a sia-independent fashion. In that context, SIGN molecules interact with IAV hemagglutinin (HA) glycosylations, acting as actual receptors for the virus (Hillaire et al., 2013). ISAV HE possesses at least two glycosylation sites, and infection has proven to be Ca+2 dependent, suggesting a role for CTLRs in the infective process (Fourrier et al., 2015).
Piscirickettsia salmonis (P. sal) is a facultative intracellular gram-negative bacteria, the etiological agent of the disease known as piscirickettsiosis, which causes significant economic losses in the aquaculture industry (Rozas and Enríquez, 2014). The bacteria produces an imbalance in the interleukin (IL) 10–12 equilibrium in infected macrophages, leading to an anti-inflammatory response and successful, productive infection in intracellular vesicles (Álvarez et al., 2016). We assessed the effect of Piscirickettsia salmonis in the expression of SsSIGN genes in the macrophage-like SHK-1 cell line. Most notably, the SsSIGN1 receptor was significantly downregulated in the productive infection, an opposite effect to what was observed with ISAV (Fig. 7). It has been shown that P. sal. induces IκBα expression, inhibiting NF-κB translocation to the nucleus, which would lead to IL-12 and SsSIGN1 downregulation (Soto et al., 2016). On the other hand, Ap-1 and Sp1 binding sites may also be involved in the downregulation of SsSIGN genes, as it has been described for the phylogenetically-related Francisella tularensis (Walters et al., 2015).
4.4. Phylogenetic analysis
DC-SIGN genes have been described in a variety of mammalian species, with recent reports for fish homologs, revealing the wide distribution of this type of gene. Even though they share structural features, namely the presence of a CTLD, distinct differences identify specific homologs. Our phylogenetic analysis was based on the sequence corresponding to the CBD of SIGN genes, which represents the pathogen-interaction domain for the receptor. The phylogenetic tree revealed a clade distribution, with mammals and fishes separated, and primates grouped in a defined cluster (Fig. 8). The structure of the CBD is related to the glycan with which it interacts; as previously discussed, species-specific pathogens may influence the evolution of SIGN genes (Garcia-Vallejo and van Kooyk, 2013; Powlesland et al., 2006). Salmon, like what it has been described in mouse, may display multiple versions of SIGN genes in response to exposure to a wide variety of pathogens. Concomitantly, several immune parameters in teleost fish display more diversity than their mammalian homologs (Rebl et al., 2010). Furthermore, the presence of multiple SIGN homologs in salmon may contribute to fine-tuning of the immune response, regulating mechanisms triggered by other PRRs (i.e., TLRs); fish live in intimate contact with a potentially high amount of microorganisms, so a tightly regulated immune response is a must to avoid deleterious inflammatory responses (Novoa et al., 2009).
To complement our observations, we extended our sequence analyses to other fish species with published assembled genomes. We identified SIGN-like genes in the three analyzed genomes (Oncorhynchus mykiss, Salmo trutta, and Danio rerio) coding for proteins with structural features present in mammalian and Salmo salar SIGN genes (Table 4). Interestingly, the identified sequences are located in discrete regions on the fish genomes, with both rainbow and brown trout having eight SIGN homologs, identical to what we describe for Salmo salar. These findings reinforce our proposal of a “SIGN cluster” in fish species.
5. Conclusions
We described eight homologs for DC-SIGN receptors in Salmo salar. The proteins possess conserved structural features compared to their mammalian counterparts and are differentially expressed and induced during infection. Our work is not only relevant for a better description and knowledge of the salmon immune system, but it can also offer new perspectives regarding prophylaxis development for this species: DC-SIGN-targeted vaccines has become a promising strategy to improve antigen immunogenicity (Hossain and Wall, 2019; van Kooyk et al., 2013).
Functional analyses are still necessary to assess the effective interaction of SsSIGNs with specific pathogens, their sub-cellular localization, and induction/regulation of immune responses; these are immediate objectives to our research group.
Funding
This work was supported by the National Commission for Scientific and Technological Research (CONICYT) [FONDECYT 3180609].
References
- Aamelfot M., Dale O.B., Weli S.C., Koppang E.O., Falk K. Expression of the infectious salmon anemia virus receptor on atlantic salmon endothelial cells correlates with the cell tropism of the virus. J. Virol. 2012;86:10571–10578. doi: 10.1128/JVI.00047-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Álvarez C.A., Gomez F.A., Mercado L., Ramírez R., Marshall S.H. Piscirickettsia salmonis imbalances the innate immune response to succeed in a productive infection in a salmonid cell line model. PloS One. 2016 doi: 10.1371/journal.pone.0163943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andresen A.M.S., Boudinot P., Gjøen T. Kinetics of transcriptional response against poly (I: C) and infectious salmon anemia virus (ISAV) in Atlantic salmon kidney (ASK) cell line. Dev. Comp. Immunol. 2020;110(103716) doi: 10.1016/j.dci.2020.103716. [DOI] [PubMed] [Google Scholar]
- Appelmelk B.J., van Die I., van Vliet S.J., Vandenbroucke-Grauls C.M.J.E., Geijtenbeek T.B.H., van Kooyk Y. Cutting edge: carbohydrate profiling identifies new pathogens that interact with dendritic cell-specific ICAM-3-grabbing nonintegrin on dendritic cells. J. Immunol. 2003 doi: 10.4049/jimmunol.170.4.1635. [DOI] [PubMed] [Google Scholar]
- Avota E., Koethe S., Schneider-Schaulies S. Membrane dynamics and interactions in measles virus dendritic cell infections. Cell Microbiol. 2013 doi: 10.1111/cmi.12025. [DOI] [PubMed] [Google Scholar]
- Cambi A., Gijzen K., de Vries I.J.M., Torensma R., Joosten B., Adema G.J., Netea M.G., Kullberg B.J., Romani L., Figdor C.G. The C-type lectin DC-SIGN (CD209) is an antigen-uptake receptor for Candida albicans on dendritic cells. Eur. J. Immunol. 2003 doi: 10.1002/immu.200310029. [DOI] [PubMed] [Google Scholar]
- Castillo-Cerda M.T., Cottet L., Toro-Ascuy D., Spencer E., Cortez-San Martin M. Development of plaque assay for Chilean Infectious Salmon Anaemia Virus, application for virus purification and titration in salmon ASK cells. J. Fish. Dis. 2014;37:989–995. doi: 10.1111/jfd.12198. [DOI] [PubMed] [Google Scholar]
- Curtis B.M., Scharnowske S., Watson A.J. Sequence and expression of a membrane-associated C-type lectin that exhibits CD4-independent binding of human immunodeficiency virus envelope glycoprotein gp120. Proc. Natl. Acad. Sci. 1992;89:8356–8360. doi: 10.1073/pnas.89.17.8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dambuza I.M., Brown G.D. C-type lectins in immunity: recent developments. Curr. Opin. Immunol. 2015 doi: 10.1016/j.coi.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson W.S., Koop B.F., Jones S.J.M., Iturra P., Vidal R., Maass A., Jonassen I., Lien S., Omholt S.W. Sequencing the genome of the atlantic salmon (Salmo salar) Genome Biol. 2010 doi: 10.1186/gb-2010-11-9-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Witte L., Nabatov A., Geijtenbeek T.B.H. Distinct roles for DC-SIGN+-dendritic cells and Langerhans cells in HIV-1 transmission. Trends Mol. Med. 2008 doi: 10.1016/j.molmed.2007.11.001. [DOI] [PubMed] [Google Scholar]
- dos Santos Á., Hadjivasiliou A., Ossa F., Lim N.K., Turgut A., Taylor M.E., Drickamer K. Oligomerization domains in the glycan-binding receptors DC-SIGN and DC-SIGNR: sequence variation and stability differences. Protein Sci. 2017;26:306–316. doi: 10.1002/pro.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drickamer K., Fadden A.J. Genomic analysis of C-type lectins. Biochem. Soc. Symp. 2002 doi: 10.1042/bss0690059. [DOI] [PubMed] [Google Scholar]
- Engering A., Geijtenbeek T.B.H., van Vliet S.J., Wijers M., van Liempt E., Demaurex N., Lanzavecchia A., Fransen J., Figdor C.G., Piguet V., van Kooyk Y. The dendritic cell-specific adhesion receptor DC-SIGN internalizes antigen for presentation to T cells. J. Immunol. 2002;168:2118–2126. doi: 10.4049/jimmunol.168.5.2118. [DOI] [PubMed] [Google Scholar]
- Falk K., Namork E., Rimstad E., Mjaaland S., Dannevig B.H. Characterization of infectious salmon anemia virus, an orthomyxo-like virus isolated from Atlantic salmon (Salmo salar L.) J. Virol. 1997;71:9016–9023. doi: 10.1128/jvi.71.12.9016-9023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré D., Roset R., Huerta M., Adsuara J.E., Roselló L., Albà M.M., Messeguer X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003 doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg H. Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science. 2001;294:2163–2166. doi: 10.1126/science.1066371. [DOI] [PubMed] [Google Scholar]
- Fourrier M., Lester K., Markussen T., Falk K., Secombes C.J., McBeath A., Collet B. Dual Mutation Events in the Haemagglutinin-Esterase and Fusion Protein from an Infectious Salmon Anaemia Virus HPR0 Genotype Promote Viral Fusion and Activation by an Ubiquitous Host Protease. PLoS One. 2015;10 doi: 10.1371/journal.pone.0142020. e0142020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Vallejo J.J., van Kooyk Y. The physiological role of DC-SIGN: a tale of mice and men. Trends Immunol. 2013;34:482–486. doi: 10.1016/j.it.2013.03.001. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek T.B.H., Torensma R., Van Vliet S.J., Van Duijnhoven G.C.F., Adema G.J., Van Kooyk Y., Figdor C.G. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000:80693–80695. doi: 10.1016/S0092-8674(00). [DOI] [PubMed] [Google Scholar]
- Gillespie L., Roosendahl P., Ng W.C., Brooks A.G., Reading P.C., Londrigan S.L. Endocytic function is critical for influenza A virus infection via DC-SIGN and L-SIGN. Sci. Rep. 2016;6(19428) doi: 10.1038/srep19428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez F.A., Tobar J.A., Henríquez V., Sola M., Altamirano C., Marshall S.H. Evidence of the presence of a functional dot/icm type IV-B secretion system in the fish bacterial pathogen Piscirickettsia salmonis. PLoS One. 2013 doi: 10.1371/journal.pone.0054934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gringhuis S.I., den Dunnen J., Litjens M., van der Vlist M., Geijtenbeek T.B.H. Carbohydrate-specific signaling through the DC-SIGN signalosome tailors immunity to Mycobacterium tuberculosis, HIV-1 and Helicobacter pylori. Nat. Immunol. 2009;10:1081–1088. doi: 10.1038/ni.1778. [DOI] [PubMed] [Google Scholar]
- Guo Y., Feinberg H., Conroy E., Mitchell D.A., Alvarez R., Blixt O., Taylor M.E., Weis W.I., Drickamer K. Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat. Struct. Mol. Biol. 2004;11:591–598. doi: 10.1038/nsmb784. [DOI] [PubMed] [Google Scholar]
- Haining E.J., Cherpokova D., Wolf K., Becker I.C., Beck S., Eble J.A., Stegner D., Watson S.P., Nieswandt B. CLEC-2 contributes to hemostasis independently of classical hemITAM signaling in mice. Blood. 2017 doi: 10.1182/blood-2017-03-771907. [DOI] [PubMed] [Google Scholar]
- Henríquez M., González E., Marshall S.H., Henríquez V., Gómez F.A., Martínez I., Altamirano C. A novel liquid medium for the efficient growth of the salmonid pathogen Piscirickettsia salmonis and optimization of culture conditions. PloS One. 2013 doi: 10.1371/journal.pone.0071830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbomel P., Thisse B., Thisse C. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev. Biol. 2001;238:274–288. doi: 10.1006/dbio.2001.0393. [DOI] [PubMed] [Google Scholar]
- Hillaire M.L.B., Nieuwkoop N.J., Boon A.C.M., de Mutsert G., Vogelzang-van Trierum S.E., Fouchier R.A.M., Osterhaus A.D.M.E., Rimmelzwaan G.F. Binding of DC-SIGN to the hemagglutinin of influenza A viruses supports virus replication in DC-SIGN expressing cells. PloS One. 2013;8:1–10. doi: 10.1371/journal.pone.0056164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M., Wall K. Use of dendritic cell receptors as targets for enhancing anti-cancer immune responses. Cancers. 2019;11(418) doi: 10.3390/cancers11030418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoving J.C., Wilson G.J., Brown G.D. Signalling C-type lectin receptors, microbial recognition and immunity. Cell Microbiol. 2014 doi: 10.1111/cmi.12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovius J.W.R., de Jong M.A.W.P., den Dunnen J., Litjens M., Fikrig E., van der Poll T., Gringhuis S.I., Geijtenbeek T.B.H. Salp15 binding to DC-SIGN inhibits cytokine expression by impairing both nucleosome remodeling and mRNA stabilization. PLoS Pathog. 2008;4:e31. doi: 10.1371/journal.ppat.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev D.B., Lagos L., Thim H.L., Jørgensen S.M., Krasnov A., Jørgensen J.B. CpGs induce differentiation of Atlantic salmon mononuclear phagocytes into cells with dendritic morphology and a proinflammatory transcriptional profile but an exhausted allostimulatory activity. Front. Immunol. 2019 doi: 10.3389/fimmu.2019.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Sun L. Tongue sole CD209: a pattern-recognition receptor that binds a broad range of microbes and promotes phagocytosis. Int. J. Mol. Sci. 2017 doi: 10.3390/ijms18091848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato G., Miyazawa H., Nakayama Y., Ikari Y., Kondo H., Yamaguchi T., Sano M., Fischer U. A novel antigen-sampling cell in the teleost gill epithelium with the potential for direct antigen presentation in mucosal tissue. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.02116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo U.S., Chan K.Y.K., Chan V.S.F., Lin C.L.S. DC-SIGN and L-SIGN: the SIGNs for infection. J. Mol. Med. 2008;86:861–874. doi: 10.1007/s00109-008-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordon A.O., Karsi A., Pinchuk L.M. Innate immune responses in fish: antigen presenting cells and professional phagocytes. Turk. J. Fish. Aquat. Sci. 2018;18:1123–1139. [Google Scholar]
- Krossøy B., Hordvik I., Nilsen F., Nylund A., Endresen C. The putative polymerase sequence of infectious salmon anemia virus suggests a new genus within the Orthomyxoviridae. J. Virol. 1999;73:2136–2142. doi: 10.1128/jvi.73.3.2136-2142.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive Tree of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007 doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- Lin A.-F., Xiang L.-X., Wang Q.-L., Dong W.-R., Gong Y.-F., Shao J.-Z. The DC-SIGN of zebrafish: insights into the existence of a CD209 homologue in a lower vertebrate and its involvement in adaptive immunity. J. Immunol. 2009 doi: 10.4049/jimmunol.0803955. [DOI] [PubMed] [Google Scholar]
- Little C., Felzensztein C., Gimmon E., Muñoz P. The business management of the Chilean salmon farming industry. Mar. Pol. 2015;54:108–117. doi: 10.1016/j.marpol.2014.12.020. [DOI] [Google Scholar]
- Liu H., Yu W., Liou L.-Y., Rice A.P. Isolation and characterization of the human DC-SIGN and DC-SIGNR promoters. Gene. 2003;313:149–159. doi: 10.1016/S0378-1119(03)00674-7. [DOI] [PubMed] [Google Scholar]
- Liu W., Tang L., Zhang G., Wei H., Cui Y., Guo L., Gou Z., Chen X., Jiang D., Zhu Y., Kang G., He F. Characterization of a novel C-type lectin-like gene, LSECtin: demonstration of carbohydrate binding and expression in sinusoidal endothelial cells of liver and lymph node. J. Biol. Chem. 2004 doi: 10.1074/jbc.M311227200. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ludwig S., Wang X., Ehrhardt C., Zheng H., Donelan N., Planz O., Pleschka S., García-Sastre A., Heins G., Wolff T. The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J. Virol. 2002;76:11166–11171. doi: 10.1128/JVI.76.21.11166-11171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C.J., Lu S., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Lu F., Marchler G.H., Song J.S., Thanki N., Wang Z., Yamashita R.A., Zhang D., Zheng C., Geer L.Y., Bryant S.H. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBeath A.J.A., Collet B., Paley R., Duraffour S., Aspehaug V., Biering E., Secombes C.J., Snow M. Identification of an interferon antagonist protein encoded by segment 7 of infectious salmon anaemia virus. Virus Res. 2006;115:176–184. doi: 10.1016/j.virusres.2005.08.005. [DOI] [PubMed] [Google Scholar]
- McDonnell A.V., Jiang T., Keating A.E., Berger B. Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics. 2006 doi: 10.1093/bioinformatics/bti797. [DOI] [PubMed] [Google Scholar]
- Mesman A.W., de Vries R.D., McQuaid S., Duprex W.P., de Swart R.L., Geijtenbeek T.B.H. A prominent role for DC-SIGN+ dendritic cells in initiation and dissemination of measles virus infection in non-human primates. PloS One. 2012;7 doi: 10.1371/journal.pone.0049573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messeguer X., Escudero R., Farré D., Núñez O., Martínez J., Albà M.M. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002 doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- Mizumoto N., Hui F., Edelbaum D., Ryan Weil M., Wren J.D., Shalhevet D., Matsue H., Liu L., Garner H.R., Takashima A. Differential activation profiles of multiple transcription factors during dendritic cell maturation. J. Invest. Dermatol. 2005;124:718–724. doi: 10.1111/j.0022-202X.2005.23616.x. [DOI] [PubMed] [Google Scholar]
- Montoya D., Cruz D., Teles R.M.B., Lee D.J., Ochoa M.T., Krutzik S.R., Chun R., Schenk M., Zhang X., Ferguson B.G., Burdick A.E., Sarno E.N., Rea T.H., Hewison M., Adams J.S., Cheng G., Modlin R.L. Divergence of macrophage phagocytic and antimicrobial programs in leprosy. Cell Host Microbe. 2009 doi: 10.1016/j.chom.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa B., Bowman T.V., Zon L., Figueras A. LPS response and tolerance in the zebrafish (Danio rerio) Fish Shellfish Immunol. 2009;26:326–331. doi: 10.1016/j.fsi.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen C.M., Markussen T., Thiede B., Rimstad E. Infectious salmon anaemia virus (ISAV) RNA binding protein encoded by segment 8 ORF2 and its interaction with ISAV and intracellular proteins. Viruses. 2016;8 doi: 10.3390/v8020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz M., Kaessmann H., Zhang K., Bashirova A., Carrington M., Quintana-Murci L., Telenti A. The evolutionary history of the CD209 (DC-SIGN) family in humans and non-human primates. Gene Immun. 2008;9:483–492. doi: 10.1038/gene.2008.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier J., Cabau C., Nguyen T., Jouanno E., Severac D., Braasch I., Journot L., Pontarotti P., Klopp C., Postlethwait J.H., Guiguen Y., Bobe J. Gene evolution and gene expression after whole genome duplication in fish: the PhyloFish database. BMC Genom. 2016 doi: 10.1186/s12864-016-2709-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen F.E., Fyllingen I., Kavlie A., Maaseide N.P., Glette J., Endresen C., Wergeland H.I. Monoclonal antibodies reactive with serum igm and leukocytes from atlantic salmon (salmo salarl.) Fish Shellfish Immunol. 1995 doi: 10.1006/fsim.1995.0027. [DOI] [Google Scholar]
- Powlesland A.S., Ward E.M., Sadhu S.K., Guo Y., Taylor M.E., Drickamer K. Widely divergent biochemical properties of the complete set of mouse DC-SIGN-related proteins. J. Biol. Chem. 2006 doi: 10.1074/jbc.M601925200. [DOI] [PubMed] [Google Scholar]
- Rappocciolo G., Piazza P., Fuller C.L., Reinhart T.A., Watkins S.C., Rowe D.T., Jais M., Gupta P., Rinaldo C.R. DC-SIGN on B lymphocytes is required for transmission of HIV-1 to T lymphocytes. PloS Pathog. 2006 doi: 10.1371/journal.ppat.0020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappocciolo G., Hensler H.R., Jais M., Reinhart T.A., Pegu A., Jenkins F.J., Rinaldo C.R. Human herpesvirus 8 infects and replicates in primary cultures of activated B lymphocytes through DC-SIGN. J. Virol. 2008 doi: 10.1128/jvi.01587-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebl A., Goldammer T., Seyfert H.M. Toll-like receptor signaling in bony fish. Vet. Immunol. Immunopathol. 2010 doi: 10.1016/j.vetimm.2009.09.021. [DOI] [PubMed] [Google Scholar]
- Rodríguez E., Kalay H., Noya V., Brossard N., Giacomini C., van Kooyk Y., García-Vallejo J.J., Freire T. Fasciola hepatica glycoconjugates immuneregulate dendritic cells through the Dendritic Cell-Specific Intercellular adhesion molecule-3-Grabbing Non-integrin inducing T cell anergy. Sci. Rep. 2017;7(46748) doi: 10.1038/srep46748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas M., Enríquez R. Piscirickettsiosis and Piscirickettsia salmonis in fish: a review. J. Fish Dis. 2014 doi: 10.1111/jfd.12211. [DOI] [PubMed] [Google Scholar]
- Salazar C., Haussmann D., Kausel G., Figueroa J. Molecular cloning of Salmo salar Toll-like receptors (TLR1, TLR22, TLR5M and TLR5S) and expression analysis in SHK-1 cells during Piscirickettsia salmonis infection. J. Fish Dis. 2016 doi: 10.1111/jfd.12354. [DOI] [PubMed] [Google Scholar]
- Schmitz M.L., Kracht M., Saul V.V. The intricate interplay between RNA viruses and NF-κB. Biochim. Biophys. Acta Mol. Cell Res. 2014;1843:2754–2764. doi: 10.1016/j.bbamcr.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Gómez D., Domínguez-Soto A., Ancochea J., Jimenez-Heffernan J.A., Leal J.A., Corbí A.L. Dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin mediates binding and internalization of Aspergillus fumigatus conidia by dendritic cells and macrophages. J. Immunol. 2004 doi: 10.4049/jimmunol.173.9.5635. [DOI] [PubMed] [Google Scholar]
- Soanes K.H., Figuereido K., Richards R.C., Mattatall N.R., Ewart K.V. Sequence and expression of C-type lectin receptors in Atlantic salmon (Salmo salar) Immunogenetics. 2004;56:572–584. doi: 10.1007/s00251-004-0719-5. [DOI] [PubMed] [Google Scholar]
- Sonnhammer E.L., von Heijne G., Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998 [PubMed] [Google Scholar]
- Soto L., Lagos A., Isla A., Haussmann D., Figueroa J. Immunostimulatory effects of prolactin on TLR1 and TLR5M in SHK-1 cells infected with Piscirickettsia salmonis. Dis. Aquat. Org. 2016;118:237–245. doi: 10.3354/dao02967. [DOI] [PubMed] [Google Scholar]
- Stecher G., Tamura K., Kumar S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 2020 doi: 10.1093/molbev/msz312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kooyk Y., Unger W.W.J., Fehres C.M., Kalay H., García-Vallejo J.J. Glycan-based DC-SIGN targeting vaccines to enhance antigen cross-presentation. Mol. Immunol. 2013;55:143–145. doi: 10.1016/j.molimm.2012.10.031. [DOI] [PubMed] [Google Scholar]
- Vike S., Duesund H., Andersen L., Nylund A. Release and survival of infectious salmon anaemia (ISA) virus during decomposition of Atlantic salmon (Salmo salar L.) Aquaculture. 2014;420–421:119–125. doi: 10.1016/j.aquaculture.2013.09.043. [DOI] [Google Scholar]
- Walters K.-A., Olsufka R., Kuestner R.E., Wu X., Wang K., Skerrett S.J., Ozinsky A. Prior infection with Type A Francisella tularensis antagonizes the pulmonary transcriptional response to an aerosolized Toll-like receptor 4 agonist. BMC Genom. 2015;16(874) doi: 10.1186/s12864-015-2022-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Chen R.-F., Liu J.-W., Lee I.-K., Lee C.-P., Kuo H.-C., Huang S.-K., Yang K.D. DC-SIGN (CD209) promoter −336 A/G polymorphism is associated with Dengue hemorrhagic fever and correlated to DC-SIGN expression and immune augmentation. PLoS Neglected Trop. Dis. 2011;5:e934. doi: 10.1371/journal.pntd.0000934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., De Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis W.I., Taylor M.E., Drickamer K. The C-type lectin superfamily in the immune system. Immunol. Rev. 1998 doi: 10.1111/j.1600-065X.1998.tb01185.x. [DOI] [PubMed] [Google Scholar]
- Zardoya R., Garrido-Pertierra A., Bautista J.M. The complete nucleotide sequence of the mitochondrial DNA genome of the rainbow trout, Oncorhynchus mykiss. J. Mol. Evol. 1995 doi: 10.1007/BF00173174. [DOI] [PubMed] [Google Scholar]
- Zelensky A.N., Gready J.E. C-type lectin-like domains in fugu rubripes. BMC Genom. 2004 doi: 10.1186/1471-2164-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelensky A.N., Gready J.E. The C-type lectin-like domain superfamily. FEBS J. 2005 doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]