Abstract
In this study we performed a genotype-phenotype association analysis of meiotic stability in 10 autotetraploid Arabidopsis lyrata and A. lyrata/A. arenosa hybrid populations collected from the Wachau region and East Austrian Forealps. The aim was to determine the effect of eight meiosis genes under extreme selection upon adaptation to whole genome duplication. Individual plants were genotyped by high-throughput sequencing of the eight meiosis genes (ASY1, ASY3, PDS5b, PRD3, REC8, SMC3, ZYP1a/b) implicated in synaptonemal complex formation and phenotyped by assessing meiotic metaphase I chromosome configurations. Our results reveal that meiotic stability varied greatly (20–100%) between individual tetraploid plants and associated with segregation of a novel ASYNAPSIS3 (ASY3) allele derived from A. lyrata. The ASY3 allele that associates with meiotic stability possesses a putative in-frame tandem duplication (TD) of a serine-rich region upstream of the coiled-coil domain that appears to have arisen at sites of DNA microhomology. The frequency of multivalents observed in plants homozygous for the ASY3 TD haplotype was significantly lower than in plants heterozygous for ASY3 TD/ND (non-duplicated) haplotypes. The chiasma distribution was significantly altered in the stable plants compared to the unstable plants with a shift from proximal and interstitial to predominantly distal locations. The number of HEI10 foci at pachytene that mark class I crossovers was significantly reduced in a plant homozygous for ASY3 TD compared to a plant heterozygous for ASY3 ND/TD. Fifty-eight alleles of the 8 meiosis genes were identified from the 10 populations analysed, demonstrating dynamic population variability at these loci. Widespread chimerism between alleles originating from A. lyrata/A. arenosa and diploid/tetraploids indicates that this group of rapidly evolving genes may provide precise adaptive control over meiotic recombination in the tetraploids, the very process that gave rise to them.
Author summary
Whole genome duplication can promote adaptability, but is a dramatic mutation usually resulting in meiotic catastrophe and genome instability. Here we focus on a case of coordinated stabilization of meiotic recombination in 10 autotetraploid Arabidopsis lyrata and A. lyrata/A. arenosa hybrid populations from the Wachau region and East Austrian Forealps. We fuse population genomic data with a genotype-phenotype association study, concentrating on the effects of eight meiosis genes (ASY1, ASY3, PDS5b, PRD3, REC8, SMC3, ZYP1a and ZYP1b) implicated in synaptonemal complex formation in the tetraploids under extreme selection. Our analysis demonstrates that segregation of a novel allele of the meiotic chromosome axis protein ASYNAPSIS3 is associated with male meiotic stability. Adaptive restabilisation of meiotic recombination appears to be achieved by a reduction in the number of crossovers as well as a shift in their positioning towards the chromosome ends. Of the eight genes, 58 alleles were identified, indicating dynamic population variability at these loci under extreme selection. In addition, widespread allelic chimerism between alleles originating from A. lyrata/A. arenosa and diploid/tetraploids indicates that this group of rapidly evolving genes may provide precise adaptive control over meiotic recombination in the tetraploids, the very process that gave rise to them.
Introduction
Whole genome duplication (WGD) occurs in all eukaryotic kingdoms, and is associated with adaptability, speciation and evolvability [1, 2]. At the same time, it is also one of the most dramatic mutations observed, usually resulting in catastrophic problems during meiosis, when ensuring stable chromosome segregation and genome integrity is paramount [3]. Because efficient meiosis is required for the formation of euploid gametes during sexual reproduction, selection acts strongly on standing variation from the progenitor diploids in newly arisen polyploids.
In allopolyploids (formed by both genome duplication and interspecies hybridization), loci required for correct chromosome pairing and recombination have been identified in wheat [4], oil seed rape [5, 6] and Arabidopsis suecica [7]. However, in autopolyploids (which form within-species, without hybridization), there has been little functional confirmation of any gene controlling correct chromosome pairing, synapsis and crossing over (CO), although clear signatures of extreme selection have been detected in eight meiosis genes associated with the synaptonemal complex (SC) (ASY1, ASY3, PDS5b, PRD3, REC8, SMC3, ZYP1a, ZYP1b) in the young autotetraploid Arabidopsis arenosa [8].
The SC is a tripartite protein structure consisting of two lateral elements and a central element, specific to meiotic prophase I that is required for normal levels of COs in the majority of sexually reproducing eukaryotes [9]. In Arabidopsis, the chromosome axes (which come to form the SC lateral elements) consist of a scaffold of cohesin proteins (SMC1, SMC3, PDS5, REC8 and SCC3)[10–14] that organise sister chromatids into a loop/base conformation [15]. PRD3, the budding yeast MER2 homolog, is required for double-strand break (DSB) formation and is not an SC protein per se but may juxtapose the potential DSB site with the chromosome axis to promote inter-homolog recombination [16, 17]. In Sordaria, MER2 also transfers and releases recombination complexes to and from the SC central region [16]. The meiosis specific proteins ASY1, ASY3 and ASY4 load onto the chromosome axis defined by the cohesin scaffold, to promote inter-homolog recombination [18–20]. ASY1 and ASY3 are the functional homologs of budding yeast HOP1 and RED1 and, HORMAD1/2 and SCP2 in mammals, respectively, that facilitate correct chromosome pairing and synapsis, required for wild-type COs [19, 21, 22]. In Arabidopsis, synapsis is initiated by installation of the transverse filament proteins ZYP1a and ZYP1b between homologous chromosomes, thus ensuring appropriate levels of COs [23].
Arabidopsis lyrata and A. arenosa represent powerful models for investigating adaptation to autopolyploidy, particularly their populations from the eastern Austrian Forealps where interspecific hybridization and introgression is frequent [8] [24–26]. Extensive demographic analysis and coalescent modelling demonstrated that each of these species first underwent WGD and then later hybridised intraspecifically, only between their autotetraploid cytotypes [27]. Independent studies of the ploidy specific hybridisation barrier [28] functionally confirm that these species only hybridise following WGD, after each is established in its autopolyploid form. As a consequence, these populations represent ‘natural mapping experiments’ that can be studied to understand the relative contributions of the suite of alleles known to exhibit strong signatures of selection. These eight meiosis loci displaying highly differentiated alleles in A. arenosa were also reported in A. lyrata autotetraploids, along with signatures of extensive bidirectional gene flow [25]. At meiotic metaphase I in A. arenosa, chiasma frequency was reduced in autotetraploids carrying the derived alleles compared to the diploids, indicating an ongoing adaptive consequence of their evolution [8]. Recently ASY1 and ASY3 have been shown to have an effect in stabilising autotetraploid A. arenosa [29], but thus far, no formal confirmation of an association between meiotic stabilisation has been attributed to these evolved alleles in tetraploid A. lyrata.
In this study we fuse genomic, genetic and cytological approaches to investigate the effects of rapidly evolving adaptive haplotypes in these meiosis genes under strong selection. We measure the consequences of alternative haplotypes at these loci in autotetraploid A. lyrata, A. arenosa, and natural introgressants of these species across a hybrid zone. Our analysis reveals an association between a novel ASY3 haplotype and meiotic chromosome stability.
Results
A metaphase I analysis to determine meiotic stability in A. lyrata/A. arenosa populations
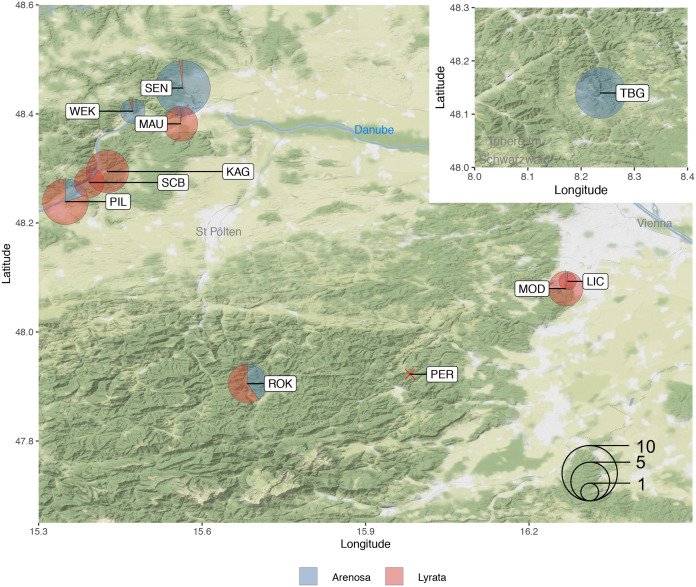
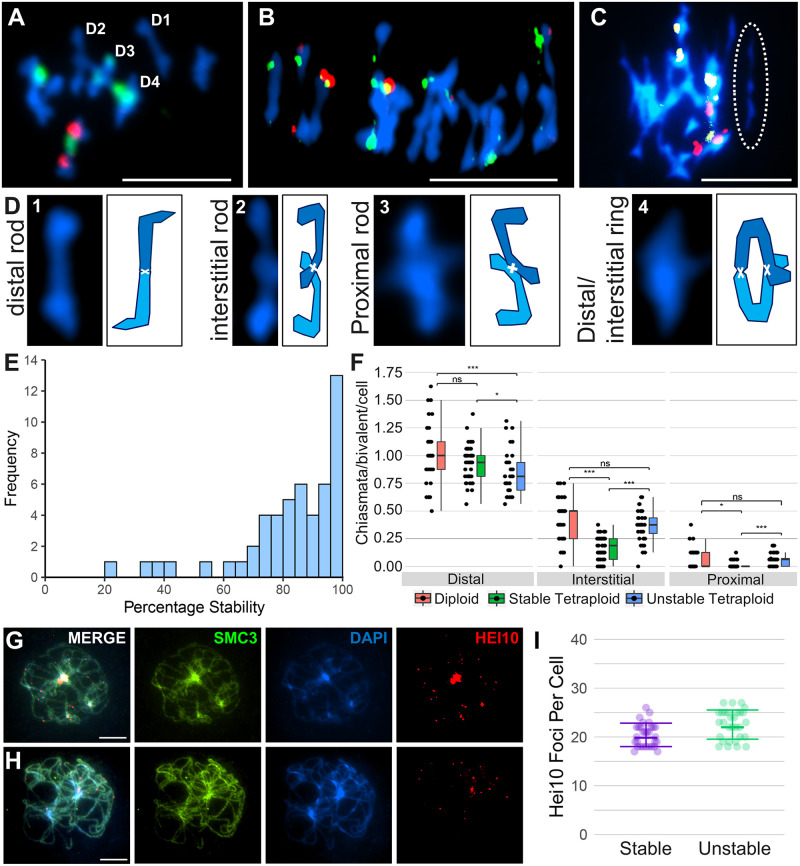
A thorough examination of meiotic stability was performed on 52 plants obtained from individual maternal seed lines sampled from tetraploid populations covering a range of known genomic backgrounds and demographic histories from the Wachau region and east Austrian Forealps. Relative genomic contributions (proportions of admixture) from A. lyrata and A. arenosa from sampled populations had previously been determined [25]. Sampled populations were: LIC, MAU, MOD, PIL, SCB, KAG, ROK (A. lyrata and A. lyrata-like hybrids that contain >50% genomic contribution from A. lyrata) and, TBG, SEN and WEK (A. arenosa/A. lyrata introgressants that contain >50% genomic contribution from A. arenosa) (Fig 1). Meiotic stability was assessed in individual plants by performing cytological analyses on metaphase I (MI) chromosome spreads of pollen mother cells (PMCs). A rod bivalent forms when only one chiasma (the cytological manifestation of a CO) connects a homologous chromosome pair (Fig 2A and 2D1–2D3). A ring bivalent forms when chiasmata occur in both chromosome arms of homologous pairs (Fig 2A and 2D4). Quadrivalents are structures formed of four chromosomes, usually two pairs of homologous chromosomes and, multivalents form between multiple chromosomes either connected by chiasmata or interlocks (Fig 2C). As a control, chiasma frequency and distribution were scored at MI in diploid A. lyrata PMCs (Fig 2A). For the tetraploids, MI nuclei were scored as stable when 16 individual bivalents could be observed aligned on the MI plate and unstable if quadrivalents or multivalents were observed (Fig 2B and 2C and S1 Fig). For each maternal line we scored blind the percentage of stable versus unstable nuclei per plant, revealing a range from 20–100% (Fig 2E). Furthermore, we scored chiasmata as distal, interstitial or proximal to the centromere based on chromosome bivalent shapes in sampled populations (8 x diploid plants, 5 x predominantly stable LIC plants and, 2 x KAG and 2 x MAU predominantly unstable plants) (Fig 2D1–2D4). In unstable nuclei, only a proportion of individual bivalents could be scored per nucleus (ranging from 1–11 per cell), that were not associated with other chromosomes (Fig 2C, dashed ellipse). A FISH analysis utilizing the 5S and 45S rDNA probes revealed that all chromosomes that could be scored in the samples of unstable nuclei were observed either as individual bivalents or associated with multivalents. From the total 16 pairs of chromosomes per nucleus, seven did not hybridize with the 5S and 45S rDNA probes, five with the 5S only, two with 45S only, and two with both 5S and 45S. However, the FISH analysis revealed a chromosome bias for those that could be scored in the unstable nuclei with an underrepresentation of chromosomes containing the 45S rDNA nucleolar organizing regions (NOR) and an overrepresentation of chromosomes without the 45S rDNA (Table 1). A Chi-squared test revealed that these values were significantly different (χ[3]2 = 9.54, P < 0.05), indicating that chromosomes with the 45S rDNA NOR were more likely to form multivalents (Fig 2C).
Fig 1. Map of maternal seed lines collection sites from the eastern Austrian fore Alps and Wachau valley.
Collection sites are indicated as circles for tetraploid plants and an X for diploids. Pie chart circle size represents the numbers of plants sampled and analysed from each site and the proportion indicates the relative amounts of admixture between A. lyrata (red) and A. arenosa (blue) populations determined in [25].
Fig 2. Cytological analysis of meiotic metaphase I stability, chiasma frequency and distribution.
(A) Diploid A. lyrata bivalents stained with DAPI (blue) 5S rDNA (red) and 45S rDNA (green). (B) Stable autotetraploid meiotic metaphase I, stained with DAPI (blue) 5S rDNA (red) and 45S rDNA (green). (C) Unstable autotetraploid meiotic metaphase I, stained with DAPI (blue) 5S rDNA (red) and 45S rDNA (green) exhibiting one multivalent and one bivalent highlighted by the ellipse with white dashes that can be scored. (D) Four bivalents from A, with diagrammatic representations of homologous chromosomes in different shades of blue physically linked by chiasmata, shown by white crosses. (E) Histogram of the frequency of meiotic stability in individual plants from all populations. (F) Comparison of chiasmata position and frequency of A. lyrata bivalents from diploid (red), stable tetraploid (green) and, unstable tetraploid (blue). (G) Representative image of HEI10 foci at late pachytene in a stable nucleus, immunolocalised with SMC3 and counterstained with DAPI. (H) Representative image of HEI10 foci at late pachytene in an unstable nucleus, immunolocalised with SMC3 and counterstained with DAPI. (I) HEI10 counts from stable and unstable pachytene nuclei. Bars for A-C, G and H = 10μm.
Table 1. Frequencies of expected and observed bivalents that were scored for chiasmata in unstable nuclei labelled with ribosomal DNA FISH probes.
| Bivalents with no rDNA FISH probes | Bivalents with 5S probe only | Bivalents with 45S probe only | Bivalents with 5S and 45S probe | |
|---|---|---|---|---|
| Expected | 34 | 24 | 9.5 | 9.5 |
| Observed | 46 | 21 | 6 | 4 |
All bivalents from stable cells were grouped into a set of 16, however only a proportion of bivalents from unstable cells could be scored. Therefore, these were randomly grouped into a set of 16 pseudo-nuclei with the caveat that these were less likely to form multivalents. Overall, significantly more chiasmata were observed in diploid A. lyrata MI bivalents than those from tetraploid stable or unstable nuclei (1.52±0.3, n = 312; versus 1.12±0.2, n = 960 and 1.26±0.3, n = 590, respectively, Mann Whitney Test, P < 0.001). The frequency of distal chiasmata was not significantly different between bivalents in diploid and stable tetraploid nuclei but was reduced in bivalents from unstable tetraploid nuclei (Fig 2F). Bivalents from stable tetraploid nuclei had significantly fewer interstitial and proximal chiasmata compared to bivalents from diploids and those from unstable nuclei, whereas interstitial and proximal chiasmata were not significantly different between bivalents from diploids and unstable tetraploid nuclei (Fig 2F).
A HEI10 immunocytological analysis was performed at late pachytene to confirm whether there was a difference in CO frequency between stable and unstable nuclei (Fig 2G–2I). MAU8.11 (phenotyped at MI with 98% stability) and SEN2.2 (phenotyped at MI with 21% stability) were selected as extreme examples of meiotic stability (S1 Table). During pachytene HEI10 marks class I CO sites that ensure each chromosome pair receives at least one ‘obligate’ CO/chiasma, and accounts for ~85% of all COs [30]. In our analysis HEI10 also marked heterochromatic DNA, which was not scored as designated class I CO sites. In MAU8.11 nuclei, an average of 20.4 HEI10 foci per pachytene (n = 30) were scored and SEN2.2 nuclei, an average of 22.5 HEI10 (n = 30) were scored, revealing that the predominantly unstable nuclei had significantly greater numbers of HEI10 foci (Wilcoxon rank sum test, P<0.05) (Fig 2I). Stable nuclei contained an average of 1.28 HEI10 foci/bivalent, whereas the predominantly unstable nuclei contained an average of 1.4 HEI10 foci/bivalent.
Association of haplotypes with meiotic stability in A. lyrata/A. arenosa tetraploids
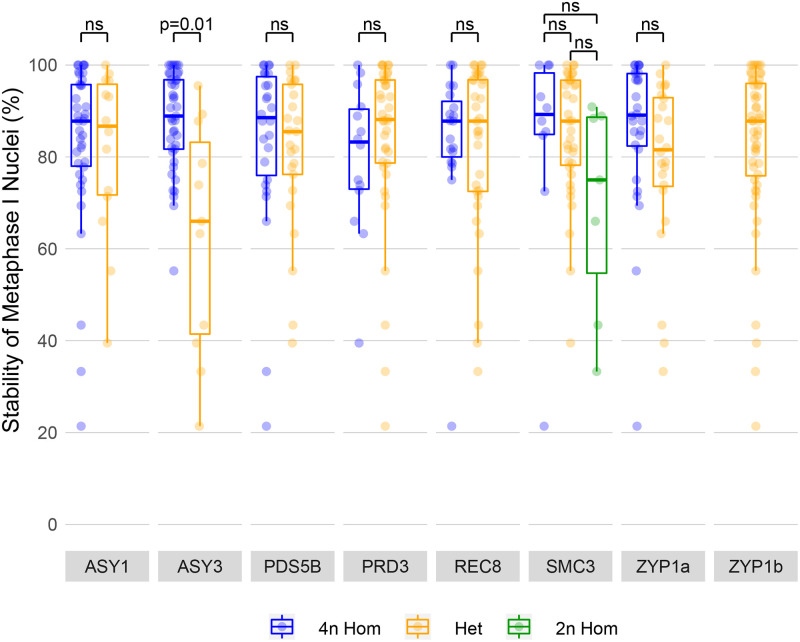
The 52 tetraploid plants phenotyped for male meiotic stability were then genotyped for the proportion of each meiosis gene haplotype by high-throughput sequencing. Accurate genotyping required obtaining precise population reference sequences from published genomic data from these populations [25]. Degenerate primers were used to amplify full length gene amplicons of the eight meiosis genes from the 52 tetraploid plants (including exons and introns) for construction of Nextera LITE libraries. Libraries were barcoded per plant and sequenced by MiSeq, generating an average sequence depth across all loci of >2000x, from which we determined the proportion of each haplotype per plant by SNP frequency. Because the coding regions of all eight meiosis genes from representative populations were cloned and Sanger sequenced (described below in ‘Adaptive polymorphisms in meiosis genes’), it was possible to resolve individual haplotypes. As these plants were drawn from a diversity of wild populations it was not surprising that an average of 7.25 alleles were identified for each of the eight meiosis genes. Given the limited sample size of 52 individuals it was not possible to statistically interrogate associations between all individual haplotypes for the eight genes. Consequently, similar haplotypes were collapsed together and classified into two groups: haplotypes with derived tetraploid alleles, and those with ancestral diploid alleles, as determined through phylogenetic analysis (S2–S5 Figs). Derived tetraploid haplotypes were those possessing conserved polymorphisms relative to the diploid reference sequences. For example, with Asynapsis3 ASY3, phylogenetic analysis identified three derived alleles (TD1, TD2 and TD3) that were very highly related and clearly different from the diploid alleles (S2 and S4 Figs). For each gene (except ZYP1b, which was heterozygous in all populations tested) a large proportion of the individuals carried four derived tetraploid haplotypes, whilst the others carried a mixture of derived tetraploid and diploid haplotypes. We therefore tested whether the presence of diploid haplotypes influenced meiotic stability. To do this, we classified the allele state at each of the eight meiosis genes in each individual tetraploid as either homozygous (i.e. exclusively either derived tetraploid haplotypes, or alternatively ancestral diploid haplotypes) or heterozygous (individuals harbouring both ancestral and derived alleles together at a given locus), and tested for any associations between these genotypes and meiotic stability by cytological analysis (Bonferroni corrected pairwise Mann-Whitney-Wilcoxon; S1 Table). This revealed that only the meiotic chromosome axis gene ASY3 had a significant effect on meiotic stability (Fig 3). Plants that were heterozygous for the ASY3 ancestral diploid haplotype and the derived tetraploid haplotype had significantly more unstable male metaphase I nuclei than those in plants homozygous for the derived ASY3 haplotype (4n Hom , IQR = 15.1, n = 41, , IQR = 41.7, n = 11, p = 0.008). There was a large range of meiotic stability within the ASY3 heterozygotes, so smaller effects from the other seven meiosis genes cannot be excluded. For example, there was a trend for the ancestral diploid SMC3 allele to associate with lower meiotic stability than the tetraploid homozygotes or diploid/tetraploid allele heterozygotes, although sampling sizes were not great enough to statistically confirm this trend. In addition, MiSeq sequencing revealed that the diploid ASY3 allele in the PIL population had an amino acid substitution near the N-terminus that may have had an effect on meiotic stability.
Fig 3. Association analysis of meiotic chromosome stability at metaphase I with meiosis gene haplotypes.
Individual plants are collapsed into groups containing either homozygous 4n derived (blue), 2n/4n heterozygous (orange) or 2n homozygous ancestral (green) haplotypes from all populations tested. Each data point represents the male meiotic stability of an individual plant for each specific haplotype.
Adaptive polymorphisms in meiosis genes
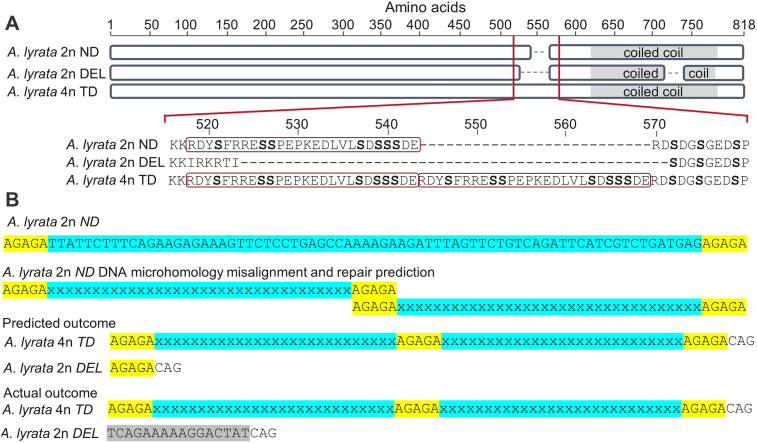
Previous studies have detected polymorphic amino acids in meiosis genes between diploids and tetraploids in A. arenosa and A. lyrata by aligning short read sequences to the A. lyrata reference, but could not infer contiguous autotetraploid alleles [8, 25, 31]. To overcome this and resolve individual haplotypes, we amplified, cloned and Sanger sequenced the coding regions of the eight meiosis genes from diploid A. arenosa SNO and A. lyrata PER populations, tetraploid A. arenosa SEN, TBG and WEK populations, as well as A. lyrata KAG and MAU populations. This approach provided high-resolution sequence polymorphism data for a total of 320 cDNA transcripts and by incorporating MiSeq data from all populations we were able to resolve 58 alleles with an average of seven alleles for each of the eight meiosis genes, thus allowing us to detect both structural variation including indels and divergent SNP variation (S6–S13 Figs, S2–S4 Tables, Dryad Digital Depository (https://doi.org/10.5061/dryad.x69p8czfb)). The eight encoded proteins associated with the synaptonemal complex are conserved at the secondary and tertiary amino acid level, rather than at the primary sequence [32]. Phosphorylation of synaptonemal complex proteins is known to be important for their activity [33][34], so software was used to determine if the identified polymorphisms may affect amino acids with important phosphorylation sites. Overall, 45% of polymorphisms led to either gains or losses of putative serine/threonine phosphorylation sites and 55% were non-phosphorylation sites. The ASY3 derived adaptive allele and ZYP1b exhibited the greatest quantity of residue changes compared to the diploids that were conserved in tetraploid populations (45 and 44, respectively) (S14 Fig), whilst SMC3 had none (although there were population specific SMC3 SNPs). Of the 45 polymorphic residues between the ancestral A. lyrata diploid ASY3 allele and the derived tetraploid ASY3 allele, 27 were due to a tandem duplication (TD allele) in a serine-rich region of the protein. The three ASY3 alleles (TD1, TD2 and TD3) all share this same tandem duplication, although population specific polymorphisms also occur outside this region (S6 Fig). The serine-rich region is upstream of the coiled-coil domain, possessing putative ATM/CKII phosphorylation motifs and a predicted SUMO site (K556, GPS-SUMO), and the TD allele contains 14 serines in this region, compared to 7 in the ancestral A. lyrata non-duplicated allele (ND) (Fig 4A). As the ASY3 TD allele had highest sequence similarity to the diploid A. lyrata ASY3 ND allele, to investigate the provenance of the adaptive tandem duplication, we exhaustively screened the local diploid A. lyrata population (PER) geographically adjacent to the A. lyrata autotetraploid LIC and MOD populations (Fig 1). 128 plants from the diploid A. lyrata PER population were screened, but the ASY3 TD allele was not identified, indicating the current absence (or vanishingly low frequency) of the TD allele, although it did reveal the presence of a deletion (DEL) allele at 7% frequency, where the entire serine-rich region is absent. We therefore cloned and sequenced genomic DNA from ASY3 ND, TD and DEL and aligned them. This revealed a 78 bp region of exon 2 is duplicated in-frame in the ASY3 TD allele that is missing in the ASY3 DEL allele between two AGAGA sites that possess DNA microhomology. We speculate that this DNA microhomology may have been instrumental in the formation of the ASY3 TD allele by homologous DNA repair through a replication error or during meiotic recombination (Fig 4B).
Fig 4. Structural variants of the three major A. lyrata ASY3 alleles.
(A) The serine-rich region (red box) with serines highlighted in bold in the non-duplicated (ND) ancestral diploid allele, absence in the deletion (DEL) diploid allele and tandemly duplicated (TD) in the derived tetraploid allele possessing two serine-rich regions (red boxes) as well as putative SUMO sites at K517, K531 and K556. (B) Major structural variation at the ASY3 locus suggesting that misalignment by DNA microhomology (highlighted yellow) and recombinational repair between the ancestral ND alleles may have led to the formation of the TD allele (blue = region of duplication). The outcome does not quite match the prediction as there are bases of unknown origin in the DEL allele (shaded in grey), but formation of these alleles may be independent events. The x’s highlighted in blue represent bases between the AGAGA sites as shown for A. lyrata 2n ND.
Meiosis gene flow between A. lyrata/A. arenosa tetraploid populations
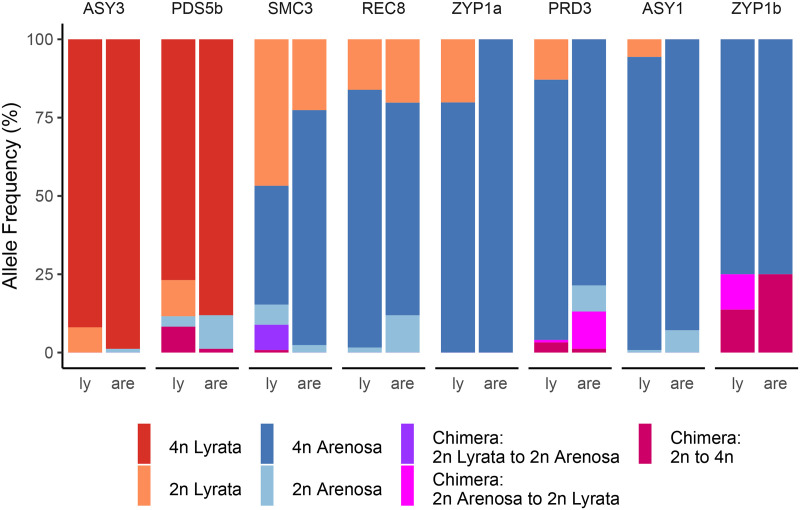
We categorized the admixture proportions of the tetraploid populations into predominantly A. lyrata (LIC, MOD, SCB, KAG, PIL and ROK) or predominantly A. arenosa (WEK, SEN and TBG), based on a demographic analysis [25]. The Sanger sequenced coding regions of the eight meiosis genes provided high quality reference data to determine allelic origin and to infer the direction of gene flow. Maximum likelihood and Bayesian phylogenetic analyses showed that each of these gene sequences clustered cleanly according to ploidy (S2–S5 Figs). The ancestral diploid sequences were further divided into A. lyrata or A. arenosa. Tetraploid sequences clustered into separate alleles irrespective of species, although population differences persisted (S2–S5 Figs). The ASY3 TD allele that associates with meiotic stability in tetraploids clusters with the A. lyrata diploid sequence (S2 and S4 Figs), as does PDS5b (S2 and S4 Figs). Conversely, derived tetraploid alleles for ASY1, PRD3, REC8, SMC3, ZYP1a and ZYP1b had highest homology with diploid A. arenosa (S2 and S4 Figs). The A. lyrata ASY3 TD allele is present at 99% frequency in tetraploid A. arenosa populations tested and 95% in tetraploid A. lyrata populations (Fig 5). The analysis revealed a small number of ancestral diploid A. arenosa and A. lyrata alleles in these populations. In contrast, the ASY1 allele derived from diploid A. arenosa had a frequency of 94% in A. arenosa tetraploid and 93% in A. lyrata tetraploid, indicating bidirectional gene flow (Fig 5). More surprisingly, bidirectional gene flow between the ancestral SMC3 and REC8 diploid alleles was observed in the tetraploids (Fig 5).
Fig 5. Contrasting origins and polymorphisms of adaptive meiosis gene haplotypes in tetraploid introgressants of A. lyrata/A. arenosa.
Haplotype frequencies of adaptive meiosis alleles from A. lyrata (Ly) and A. arenosa (Ar) populations.
Widespread chimerism in meiosis gene alleles
Our analysis of Sanger and MiSeq data identified a novel chimeric allele of ZYP1b in all tetraploid populations; chimeric PRD3 in SCB, SEN and WEK; and chimeric PDS5b in ROK. While rare chimeras could potentially result from PCR artefacts [35], these alleles have the same breakpoints in multiple individuals which suggests alterations in the genomic DNA through homologous DNA repair in planta [36]. At the 3’ end of all ZYP1b tetraploid alleles we detect evidence for a 1474 bp gene conversion (GC) to ZYP1a (S7 Fig). There is also evidence of GC in PRD3 between A. arenosa and A. lyrata ancestral diploid alleles (S5 Fig). In 11 plants from A. arenosa and A. lyrata populations, the first 740 bp of the diploid PRD3 allele is more similar to the diploid A. arenosa than to A. lyrata (7 vs 21 SNPs), while the remaining 625 bp of coding sequence has a higher similarity to diploid A. lyrata than to A. arenosa (5 vs 24 SNPs). In PDS5b, at the 5’ end of tetraploid alleles from five A. arenosa plants we observe evidence for a GC of the first two exons to the diploid allele, providing evidence of GC (or CO) between ploidy levels (S5C Fig). In addition, analysis of genome resequencing data [25] revealed a 3’ GC from a diploid ASY1 A. lyrata allele to a tetraploid A. arenosa allele in the KAG population (S5D Fig). The apparent widespread presence of gene conversion products in these loci suggests a mechanism by which genes may be modified in a narrow genetic window to confer adaptative advantages.
Discussion
Here we aimed to determine the impact of strongly selected meiosis gene alleles that underwent recent selective sweeps on the rapid evolution of autotetraploid meiotic stability in A. lyrata/A. arenosa hybrids and introgressants and to trace their evolutionary origin. By associating genotypic and cytological phenotypic data our analysis suggests that ASY3 is the major locus currently stabilising autotetraploid male meiosis in these populations. We identified structural variation of meiosis alleles including a novel derived ASY3 allele with a tandem duplication (TD) in a serine-rich region of the protein that is associated with stable meiotic chromosome phenotypes in the tetraploids, as well as novel ASY1, PDS5b, PRD3 and ZYP1b chimeric alleles originating between diploid and tetraploid A. arenosa and A. lyrata.
A cytological metaphase I (MI) analysis revealed that chiasmata in the autotetraploids were significantly reduced in both stable and unstable nuclei compared to diploid A. lyrata. Moreover, chiasma frequencies in meiotically stable nuclei were significantly reduced in regions proximal and interstitial to the centromere. A shift in chiasma distribution may reflect a fundamental mechanism for meiotic adaptation to autopolyploidy [37]. The unstable nuclei occur due to unregulated meiotic recombination between multiple chromosomes, either homologous or non-homologous. All chromosomes that could be distinguished were present in multivalents, although there was a bias for the 45S rDNA containing chromosomes to occur more frequently, which may be due to the NORs clustering during prophase I, thus bringing non-homologous chromosomes into close proximity. A reduction in numbers of chiasmata in stable nuclei was supported by an immunocytological approach counting HEI10 foci that mark designated class I COs. HEI10 foci numbers were significantly lower in pachytene nuclei from a stable plant (20.4 HEI10 foci per cell) compared to nuclei from the most unstable plant (22.5 HEI10 foci per cell)(P<0.01). This contrasts to autotetraploid A. arenosa where HEI10 counts in plants homozygous for ASY1 or ASY3 diploid ancestral alleles were not significantly different to plants homozygous for the derived alleles [29]. However, our analysis utilised genotypes with extreme meiotic stabilities. In [29], MI meiotic instability was less extreme, possibly reflecting more homogeneity of derived allelic combinations of PDS5b, PRD3, REC8, SMC3, ZYP1 and ZYP1b in plants tested.
Our genotype-phenotype association study revealed that among these eight meiosis genes, the allele state of the structurally variable meiotic chromosome axis protein ASY3 was the major factor associating with stable nuclei. Our data also indicated that REC8 and SMC3 may have had additive effects, but sample size was not great enough for statistical significance. We hypothesise that the ancestral diploid ASY3 ND allele promotes high levels of proximal and interstitial chiasmata, but in a tetraploid background it acts dominantly over the evolved ASY3 TD allele, promoting interstitial and proximal chiasmata as well as complex chromosome structures including multivalents. Such multivalents have previously been observed in diploid A. thaliana ZYP1RNAi lines where the authors postulated that chiasmata may have formed between extensive duplications with high sequence similarity on non-homologous chromosomes [23]. The tandemly duplicated serine-rich region in the ASY3 TD allele may function in a manner similar to the budding yeast N-terminus MSH4 degron that destabilizes the protein until it is phosphorylated [38]. Further studies are required to determine if the serine-rich region destabilizes the ASY3 protein thus creating a hypomorphic variant or whether these sites are phosphorylated. The analysed brassica SC phosphoproteome [34] did not recover peptides for ASY3 in the serine-rich region, although similar (serine-aspartic acid) residues recovered from ASY1 were phosphorylated. However, in a recent analysis of the A. thaliana proteome, Serine 526 of the ASY3 serine-rich region was phosphorylated [39]. The ASY3 TD allele also contains 19 derived residues outside the serine-rich region, of which 10 are predicted serine/threonine phosphorylation site gains or losses, that cannot be ruled out as functionally important along with unknown phosphorylation-related trans effects. Cis-regulatory variation outside the coding region in ASY3 could also contribute to effects on meiotic stability. Chiasmata are distalized in A. thaliana chromosome axis mutants asy1, asy3 and asy4, presumably due to telomere proximity enabling sufficient inter-homolog pairing, whereas a complete meiotic axis is required to promote high levels of recombination between spatially separated regions in nuclei along the chromosome arms [19, 20, 40]. We hypothesise that the ASY3 TD protein may be hypomorphic and act similarly to asy1, asy3 and asy4 mutants in distalizing chiasmata [19, 20, 37]. However, when heterozygous with the dominant ancestral ASY3 ND allele, rates of inter-homolog and non-homologous recombination increase. Axis components that favour interstitial and proximal recombination in diploids may promote associations with non-homologous chromosomes in the tetraploids, especially in regions with high sequence homology. However, for stable bivalents once a distal CO site is designated, CO interference may prevent further COs forming.
Phylogenetic analysis revealed that the ASY3 TD allele most likely originated from diploid A. lyrata, consistent with [25]. A screen of the diploid A. lyrata population geographically adjacent to the tetraploids for the ASY3 TD did not detect the allele, but instead identified one with the same region deleted (DEL). The ASY3 DEL allele coding region is in-frame and contains a second deletion in the coiled-coil. It may be a coincidence that the same region is lost and gained in the ASY3 alleles, or that this sequence is more susceptible to DNA replicative errors. The region of interest is flanked by DNA microhomology (AGAGA) that is positioned at the putative exchange points and may have led to DNA polymerase slippage or aberrant replication fork repair. It is unlikely to have occurred during meiotic recombination due to the low level of sequence homology. However, we observe numerous examples of apparent gene conversion that appear to have arisen during meiotic recombination in ASY1, PRD3, PDS5b and ZYP1a/b between both species and ploidies. As chimeric alleles are present at high frequencies and have single origins, we hypothesise that these polymorphisms are under positive selection. We were unable to test the effect of ZYP1b on meiotic stability as there were no homozygotes in our sampled populations. This is surprising and may reflect that another ZYP1b locus may have arisen when the apparent gene conversion occurred. When we cross ASY3 ND/DEL diploid A. lyrata plants, in the progeny we only recover the same genotype as the parents, suggesting that another locus may have arisen that has not been mapped. The bidirectional gene flow between A. arenosa and A. lyrata tetraploids has enlarged the gene pool for beneficial alleles to be borrowed and selected upon and the novel gene converted chimeric alleles may precisely coalesce advantageous sequences from differing origins for adaptation, although this would require further testing with a larger sample size.
Evidence suggests that the origin of the adaptive ASY3 TD allele in the tetraploid populations is relatively recent but has spread extensively and introgressed within tight boundaries in the A. lyrata/A. arenosa hybrid genomes tested [25]. Our current analysis provides haplotype-level sequence data that supports this hypothesis. The orthologous diploid ASY3 alleles from A. arenosa and A. lyrata are highly divergent, and yet the tetraploid ASY3 TD allele is very similar among tetraploid populations. We speculate that preceding the origin of the ASY3 TD allele, gene flow of adaptive alleles from A. arenosa (ASY1, PRD3, REC8, SMC3, ZYP1a and ZYP1b) may have been necessary to establish meiotic stability in the nascent A. lyrata tetraploids, but has since been relaxed due to the presence of the predominant ASY3 TD allele.
The A. lyrata autotetraploid populations contain plants with variable levels of meiotic stability as well as relatively high frequencies of diploid-like alleles. The diploid alleles may persist due to: a) continuous gene flow via unreduced gametes [41]; b) being beneficial in certain environmental conditions e.g. high altitudes [42]; c) being advantageous during female meiosis (COs are reduced in A. thaliana female compared to male) [43]; d) inability to purge genetic load in autotetraploids [44] and; e) a limited effect on overall male pollen fecundity, due to an excess of grains transmitted, despite variable quality.
Taken together, our data indicate multiple mechanisms for rapid meiotic evolution in autotetraploid A. lyrata. They reveal an association between MI stability and a duplication in the serine-rich region in the ASY3 TD allele. Furthermore, tetraploid A. lyrata has introgressed ASY1, PRD3, REC8, SMC3, ZYP1a and ZYP1b alleles from A. arenosa by gene flow. Finally, novel chimeric genes of ASY1, PDS5b, PRD3, ZYP1a and ZYP1b have arisen evidently through gene conversion, suggesting highly dynamic mechanisms to generate variation that may be selected upon during evolution to ensure meiotic success in these populations.
Material and methods
Cloning and sequencing of meiosis gene transcripts
Plants were grown from seed to obtain fresh flower buds from diploid A. arenosa SNO and A. lyrata PER populations, and tetraploid A. arenosa, SEN, TBG and WEK and A. lyrata KAG and MAU populations [25]. These buds were collected, flash frozen in liquid nitrogen and stored at -80C until RNA extraction. Total RNA was extracted using a Bioline ISOLATE II RNA Plant Kit (Bioline Ltd, London, UK), following manufacturer’s instructions, eluting into a final volume of 60 μl nuclease free water. Concentration and purity were determined using a NanoDrop spectrophotometer (LabTech International, Lewes, UK) and one microgram of total RNA was electrophoresed on a non-denaturing 1% (w/v) agarose gel to check for degradation. First strand cDNA was reverse transcribed from 0.5 μg of total RNA using a Quantitect Reverse Transcription Kit (Qiagen, Hilden, Germany) that incorporates genomic DNA removal prior to reverse transcription. The coding regions of 8 meiosis genes (ASY1, ASY3, PRD3, PDS5b, REC8, SMC3, ZYP1a and ZYP1b) were amplified by PCR using 0.2 μM primers (S5 Table) designed using HiSeq data [45] and Platinum Taq DNA Polymerase High Fidelity (ThermoFisher Scientific, MA, USA). PCR conditions were as follows: 94 °C for 2 min, followed by 35 cycles of 94 °C for 15 s, 60–65 °C for 30 s and 68 °C for 2–5 min (see S5 Table), with a final extension of 68 °C for 5–10 min. PCR products were electrophoresed on a 2% (w/v) agarose gel, and single bands of the expected size were excised and purified with a Monarch DNA Gel Extraction kit (New England Biolabs, MA, USA). Purified PCR products were cloned into pCR-XL-TOPO vector using a TOPO XL PCR Cloning Kit (ThermoFisher Scientific) following the manufacturer’s instructions. For each gene a total of 8 clones from each plant were isolated from overnight LB cultures using an ISOLATE II Plasmid Mini Kit (Bioline) prior to sequencing with universal M13F and M13R primers by GATC Biotech (Konstanz, Germany). Nucleotide sequences of the cDNAs were processed in Geneious 11.1.2 (https://www.geneious.com) to remove vector and low-quality sequence before using BLASTN to search the June 2010 (v.1.0/INSDC) assembly of the North American A. lyrata reference genome [46] and NCBI nonredundant (nr) database for confirmation that the obtained cDNAs were the expected gene transcripts. Primer walking was then used to sequence the entire length of the transcript. For each meiosis gene, cDNAs from each population were aligned with the respective Ensembl gene predictions from the A. lyrata reference genome (S6 Table), and to act as outgroups, A. thaliana transcripts obtained from The Arabidopsis Information Resource (TAIR) using the MUSCLE 3.8.425 plugin in Geneious 11.1.2 with default settings [47]. Phylogenetic trees were constructed using a maximum likelihood (ML) method with PhyML v3.3 [48] and bootstrap testing (1000 replicates). The ML phylogenetic trees were confirmed by a Bayesian approach with MrBayes v3.2.6 [49] with 1.1 x 106 generations of Markov Chain Monte Carlo searches containing 4 heated chains, a heated chain temperature of 0.2 and a burn-in of 100,000 generations. MrBayes trees were subsampled every 200 generations and the confidence values presented as posterior probabilities. The best nucleotide substitution method for both ML and MrBayes was determined with the Find Best-Fit Substitution Model in Mega v10.0 [50].
Meiotic haplotype genotyping
Genomic DNA was extracted from leaf material of each of the 52 plants in the study using a DNeasy Plant Mini Kit (Qiagen) and eluting into 100 μl nuclease free water. Full length coding regions of each of the 8 meiosis genes (including introns) were amplified from this genomic DNA by PCR using Platinum SuperFi Green PCR Master Mix (ThermoFisher Scientific) and 0.5 μM primers (S5 Table) designed against the Sanger sequenced cDNA of the 8 meiosis genes (see above). PCR conditions were as follows: 98 °C for 30 s, followed by 35 cycles of 98 °C for 30 s, 60–63 °C for 10 s and 72 °C for 2.5–10 min (see S5 Table), with a final extension of 72 °C for 5–10 min. PCR products were electrophoresed on a 1% (w/v) agarose gel, and single bands of the expected size were excised and purified with a Monarch DNA Gel Extraction kit (New England Biolabs). Libraries were constructed using 1ng of input DNA in a Low Input, Transposase Enabled (LITE) pipeline developed at the Earlham Institute (Norwich, UK) and based on the Illumina Nextera kits (Illumina, San Diego, CA, USA) [51]. Each library was constructed using unique 9 bp dual index combinations allowing samples to be multiplexed. Pooled libraries were size selected between 600 and 750 bp on a BluePippin (Sage Science, Beverly, MA, USA) 1.5% Cassette and then sequenced with a 2x250 bp read metric on an Illumina MiSeq sequencer.
MiSeq fastq files were imported into Geneious 11.1.2 (https://www.geneious.com) and R1 and R2 reads paired. Quality trimming of reads was performed with the BBDuk Adaptor/Quality Trimming v.37.64 plugin with default settings (Min quality:20; Min overlap:20; Min length:20). For each of the 52 individuals, trimmed reads were mapped to each of the 8 meiosis genes (S6 Table) from the North American A. lyrata reference genome [46] using Geneious 11.1.2 Read Mapper (Medium sensitivity; 5 iterations and default settings). SNPs relative to the reference genome genes were then called (Minimum Variant Frequency 0.25; Maximum Variant P-value 6x10-6) and used to identify and determine the proportion of 2n and 4n alleles for each gene per plant using a set of allele specific SNPs (as revealed from the Sanger sequencing of meiosis gene transcripts described above). Allele specific indels (eg ASY3 TD) were identified by associated SNPs.
Cloning and sequencing of ASY3 alleles
Genomic DNA was extracted from diploid A. lyrata PER plants as above and a short section of ASY3 was PCR amplified using 0.5 μM primers (S5 Table) and MyTaq Red Mix (Bioline). PCR conditions were as follows: 95 °C for 2 min, followed by 35 cycles of 95 °C for 30 s, 69 °C for 30 s and 72 °C for 30 s, with a final extension of 72 °C for 5 mins. PCR products were electrophoresed on a 2% (w/v) agarose gel and the lower band (~125 bp) corresponding to a partial ASY3 DEL allele was excised and purified with a Monarch DNA Gel Extraction kit (New England Biolabs). Purified PCR products were cloned into pCR 4-TOPO vector using a TOPO TA Cloning for Sequencing Kit (ThermoFisher Scientific) following manufacturer’s instructions. A total of 12 clones were isolated, purified and Sanger sequenced as above. Nucleotide sequences were processed in Geneious 11.1.2 (https://www.geneious.com) to remove vector and low-quality sequence before aligning with ASY3 ND/TD transcript alleles as described above. Primers designed against the partial DEL sequence were used to obtain the 3’ end of the transcript from 3 μg ASY3 ND/DEL heterozygous 2n A. lyrata (PER) floral bud total RNA using a GeneRacer Kit (ThermoFisher Scientific) following manufacturer’s instructions. Purified PCR products were cloned into pCR4-TOPO vector, sequenced and processed as above.
ASY3 ND, TD and DEL alleles were PCR amplified from genomic DNA extracted from diploid and tetraploid A. lyrata respectively using 0.2 μM primers designed against ASY3 cDNA sequences obtained above (S5 Table) and Q5 High-Fidelity DNA Polymerase (New England Biolabs). PCR conditions were as follows: 98 °C for 2 min, followed by 35 cycles of 98 °C for 10 s, 63°C for 30 s and 72 °C for 4 min, with a final extension of 72 °C for 10 min. PCR products were electrophoresed on a 1% (w/v) agarose gel, and single bands of the expected size were excised and purified as above. Purified PCR products were cloned into pDrive (Qiagen) and sequenced by Eurofins Genomics (Ebersberg, Germany).
Cytology
Chromosome spreads were performed as in [52] [25] on all populations used in this study. The HEI10 immunocytological analysis was performed using the protocol [53] with anti-AtSMC3 rat and anti-AtHEI10 rabbit antibodies described in [54]. Nikon Eclipse Ci and Ni-E microscopes installed with NIS Elements software were used to capture images of chromosomes.
Protein predictions
Protein post-translational predictions were provided by KinasePhos2.0 [55] and NetPhos3.1 [56] and SUMO sites were predicted by GPS-SUMO [57].
Statistical analyses and Map drawing
Statistical analysis was performed using the R Stats package. Mann-Whitney Wilcoxon tests were performed with function wilcox.test. Bonferoni adjusted p values were calculated using the function p.adjust. The map was drawn with ggmap [55] using map tiles by Stamen Design, under CC BY 3.0. Data by OpenStreetMap, under ODbL.
Supporting information
Stable (A) and unstable (B). Scale bar = 10μM.
(TIF)
(A) ASY1, (B) ASY3, (C) PDS5b, (D) PRD3. Bayesian posterior probabilities are indicated at the internodes of each branch. The dissimilarity scale showing substitutions per nucleotide is located at the bottom of each tree. Diploid and tetraploid alleles are indicated by ‘2’ and ‘4’ respectively (for ASY3 diploid = ND, tetraploid = TD), while the putative origins are indicated by ‘r’ (A. arenosa) and ‘y’ (A. lyrata).
(TIF)
(A) REC8, (B) SMC3, (C) ZYP1a/ZYP1b 5’ end, (D) ZYP1a/ZYP1b 3’ end. Bayesian posterior probabilities are indicated at the internodes of each branch. The dissimilarity scale showing substitutions per nucleotide is located at the bottom of each tree. Diploid and tetraploid alleles are indicated by ‘2’ and ‘4’ respectively, while the putative origins are indicated by ‘r’ (A. arenosa) and ‘y’ (A. lyrata).
(TIF)
(A) ASY1, (B) ASY3, (C) PDS5b, (D) PRD3. Maximum likelihood bootstrap values are indicated at the internodes of each branch (1000 replicates). The dissimilarity scale showing substitutions per nucleotide is located at the bottom of each tree. Diploid and tetraploid alleles are indicated by ‘2’ and ‘4’ respectively (for ASY3 diploid = ND, tetraploid = TD), while the putative origins are indicated by ‘r’ (A. arenosa) and ‘y’ (A. lyrata).
(TIF)
(A) REC8, (B) SMC3, (C) ZYP1a/ZYP1b 5’ end, (D) ZYP1a/ZYP1b 3’ end. Maximum likelihood bootstrap values are indicated at the internodes of each branch (1000 replicates). The dissimilarity scale showing substitutions per nucleotide is located at the bottom of each tree. Diploid and tetraploid alleles are indicated by ‘2’ and ‘4’ respectively, while the putative origins are indicated by ‘r’ (A. arenosa) and ‘y’ (A. lyrata).
(TIF)
The location of diploid and autotetraploid allele specific SNPs are indicated. Coloured bars in each sequence represent base specific SNPs relative to the consensus sequence (Green = A, Blue = C, Black = G, Red = T).
(JPG)
ZYP1 gene conversion (or CO) between ZYP1a (grey) and ZYP1b (red) in autotetraploid ZYP1b alleles. The location of diploid and autotetraploid allele specific SNPs are indicated. Coloured bars in each sequence represent base specific SNPs relative to the consensus sequence (Green = A, Blue = C, Black = G, Red = T).
(JPG)
Gene conversion between PRD3 diploid A. lyrata (green) and A. arenosa (green) in tetraploid A. arenosa. The location of diploid and autotetraploid allele specific SNPs are indicated. Coloured bars in each sequence represent base specific SNPs relative to the consensus sequence (Green = A, Blue = C, Black = G, Red = T).
(JPG)
Gene conversion (or CO) between PDS5b diploid (yellow) and autotetraploid (green) alleles. The location of diploid and autotetraploid allele specific SNPs are indicated. The location of diploid and autotetraploid allele specific SNPs are indicated. Coloured bars in each sequence represent base specific SNPs relative to the consensus sequence (Green = A, Blue = C, Black = G, Red = T).
(JPG)
Gene conversion (or CO) between ASY1 diploid A. lyrata (yellow) and autotetraploid A. arenosa (green). Coloured bars in each sequence represent base specific SNPs relative to the consensus sequence (Green = A, Blue = C, Black = G, Red = T).
(JPG)
ASY1 (A), ASY3 (B), PDS5b (C), showing conserved amino acid polymorphisms in autotetraploids compared to ancestral diploid alleles. Gains, losses and no change of predicted phosphorylation sites are indicated in blue, yellow and green respectively.
(TIF)
PRD3 (A), REC8 (B), showing conserved amino acid polymorphisms in autotetraploids compared to ancestral diploid alleles. Gains, losses and no change of predicted phosphorylation sites are indicated in blue, yellow and green respectively.
(TIF)
ZYP1a (A) and ZYP1b (B), showing conserved amino acid polymorphisms in autotetraploids compared to ancestral diploid alleles. Gains, losses and no change of predicted phosphorylation sites are indicated in blue, yellow and green respectively.
(TIF)
The analysis includes gains and losses of predicted serine/threonine phosphorylation sites by KinasePhos2.0 and NetPhos3.1.
(TIF)
Diploid lyrata alleles = ly; diploid arenosa alleles = ar and ar/ly = diploid arenosa to diploid lyrata putative gene conversions.
(DOCX)
Putative addition of serine/threonine phosphosites are highlighted in blue and loss of phosphorylation sites highlighted in yellow.
(DOCX)
Putative addition of serine/threonine phosphorylation sites are highlighted in blue and loss of phosphorylation sites highlighted in yellow.
(DOCX)
Putative addition of serine/threonine phosphorylation sites are highlighted in blue and loss of phosphorylation sites highlighted in yellow.
(DOCX)
(DOCX)
(DOCX)
Raw data used for generating figures.
(XLSX)
Acknowledgments
We would like to thank Neelam Dave and Maria Spezzati for providing technical support and Chris Franklin and Holger Puchta for helpful discussions during the project. DEEPSeq at the University of Nottingham provided MiSeq data. We would also like to thank the anonymous reviewers who invested their time and effort to make this a better manuscript.
Data Availability
All sequences in this study including cDNA transcripts and genomic DNA sequences have been deposited in the DDBJ/EMBL/GenBank databases under accession numbers MN512718 - MN513026 and MN520243 - MN520257. MiSeq amplicon reads have been deposited in the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) database under BioProject ID PRJNA575228.
Funding Statement
This work was funded by UKRI through Biological and Biotechnological Sciences Research Council (BBSRC) New Investigator grant (BB/M01973X/1) to JDH. This work was supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (ERC-StG 679056 HOTSPOT), via a grant to LY; and the BBSRC (BB/P013511/1), via a grant to the John Innes Centre (LY). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Alix K, Gerard PR, Schwarzacher T, Heslop-Harrison JSP. Polyploidy and interspecific hybridization: partners for adaptation, speciation and evolution in plants. Ann Bot. 2017;120(2):183–94. 10.1093/aob/mcx079 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selmecki AM, Maruvka YE, Richmond PA, Guillet M, Shoresh N, Sorenson AL, et al. Polyploidy can drive rapid adaptation in yeast. Nature. 2015;519(7543):349–52. 10.1038/nature14187 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baduel P, Bray S, Vallejo-Marin M, Kolar F, Yant L. The "Polyploid Hop": Shifting Challenges and Opportunities Over the Evolutionary Lifespan of Genome Duplications. Front Ecol Evol. 2018;6 ARTN 117 10.3389/fevo.2018.00117 [DOI] [Google Scholar]
- 4.Rey MD, Martin AC, Higgins J, Swarbreck D, Uauy C, Shaw P, et al. Exploiting the ZIP4 homologue within the wheat Ph1 locus has identified two lines exhibiting homoeologous crossover in wheat-wild relative hybrids. Mol Breed. 2017;37(8):95 10.1007/s11032-017-0700-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalo A, Lucas MO, Charpentier C, Sandmann G, Lloyd A, Jenczewski E. Reducing MSH4 copy number prevents meiotic crossovers between non-homologous chromosomes in Brassica napus. Nat Commun. 2019;10(1):2354 10.1038/s41467-019-10010-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenczewski E, Eber F, Grimaud A, Huet S, Lucas MO, Monod H, et al. PrBn, a major gene controlling homeologous pairing in oilseed rape (Brassica napus) haploids. Genetics. 2003;164(2):645–53. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henry IM, Dilkes BP, Tyagi A, Gao J, Christensen B, Comai L. The BOY NAMED SUE quantitative trait locus confers increased meiotic stability to an adapted natural allopolyploid of Arabidopsis. Plant Cell. 2014;26(1):181–94. 10.1105/tpc.113.120626 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yant L, Hollister JD, Wright KM, Arnold BJ, Higgins JD, Franklin FCH, et al. Meiotic adaptation to genome duplication in Arabidopsis arenosa. Curr Biol. 2013;23(21):2151–6. Epub 2013/10/22. 10.1016/j.cub.2013.08.059 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol. 2004;20:525–58. 10.1146/annurev.cellbio.19.111301.155141 . [DOI] [PubMed] [Google Scholar]
- 10.Lam WS, Yang X, Makaroff CA. Characterization of Arabidopsis thaliana SMC1 and SMC3: evidence that AtSMC3 may function beyond chromosome cohesion. J Cell Sci. 2005;118(Pt 14):3037–48. 10.1242/jcs.02443 . [DOI] [PubMed] [Google Scholar]
- 11.Pradillo M, Knoll A, Oliver C, Varas J, Corredor E, Puchta H, et al. Involvement of the Cohesin Cofactor PDS5 (SPO76) During Meiosis and DNA Repair in Arabidopsis thaliana. Front Plant Sci. 2015;6:1034 10.3389/fpls.2015.01034 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhatt AM, Lister C, Page T, Fransz P, Findlay K, Jones GH, et al. The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J. 1999;19(4):463–72. 10.1046/j.1365-313x.1999.00548.x [DOI] [PubMed] [Google Scholar]
- 13.Cai X, Dong F, Edelmann RE, Makaroff CA. The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. J Cell Sci. 2003;116(Pt 14):2999–3007. 10.1242/jcs.00601 . [DOI] [PubMed] [Google Scholar]
- 14.Chelysheva L, Diallo S, Vezon D, Gendrot G, Vrielynck N, Belcram K, et al. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci. 2005;118(Pt 20):4621–32. 10.1242/jcs.02583 . [DOI] [PubMed] [Google Scholar]
- 15.Lambing C, Tock AJ, Topp SD, Choi K, Kuo PC, Zhao X, et al. Interacting genomic landscapes of REC8-cohesin, chromatin and meiotic recombination in Arabidopsis thaliana. Plant Cell. 2020. Epub 2020/02/07. 10.1105/tpc.19.00866 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tesse S, Bourbon HM, Debuchy R, Budin K, Dubois E, Liangran Z, et al. Asy2/Mer2: an evolutionarily conserved mediator of meiotic recombination, pairing, and global chromosome compaction. Genes Dev. 2017;31(18):1880–93. Epub 2017/10/13. 10.1101/gad.304543.117 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Muyt A, Pereira L, Vezon D, Chelysheva L, Gendrot G, Chambon A, et al. A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. Plos Genet. 2009;5(9):e1000654 10.1371/journal.pgen.1000654 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Armstrong SJ, Caryl AP, Jones GH, Franklin FC. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J Cell Sci. 2002;115(Pt 18):3645–55. 10.1242/jcs.00048 . [DOI] [PubMed] [Google Scholar]
- 19.Ferdous M, Higgins JD, Osman K, Lambing C, Roitinger E, Mechtler K, et al. Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. Plos Genet. 2012;8(2):e1002507 10.1371/journal.pgen.1002507 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chambon A, West A, Vezon D, Horlow C, De Muyt A, Chelysheva L, et al. Identification of ASYNAPTIC4, a Component of the Meiotic Chromosome Axis. Plant Physiol. 2018;178(1):233–46. 10.1104/pp.17.01725 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caryl AP, Armstrong SJ, Jones GH, Franklin FC. A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma. 2000;109(1–2):62–71. Epub 2000/06/16. 10.1007/s004120050413 . [DOI] [PubMed] [Google Scholar]
- 22.West AMV, Rosenbereg SC, Ur SN, Lehmern MK, Ye QZ, Hagemann G, et al. A conserved filamentous assembly underlies the structure of the meiotic chromosome axis. Elife. 2019;8 ARTN e40372 10.7554/eLife.40372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FC. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 2005;19(20):2488–500. 10.1101/gad.354705 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmickl R, Koch MA. Arabidopsis hybrid speciation processes. Proc Natl Acad Sci U S A. 2011;108(34):14192–7. Epub 2011/08/10. 10.1073/pnas.1104212108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marburger S, Monnahan P, Seear PJ, Martin SH, Koch J, Paajanen P, et al. Interspecific introgression mediates adaptation to whole genome duplication. Nat Commun. 2019;10(1):5218 Epub 2019/11/20. 10.1038/s41467-019-13159-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hohmann N, Koch MA. An Arabidopsis introgression zone studied at high spatio-temporal resolution: interglacial and multiple genetic contact exemplified using whole nuclear and plastid genomes. BMC Genomics. 2017;18(1):810 Epub 2017/10/24. 10.1186/s12864-017-4220-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnold B, Kim ST, Bomblies K. Single Geographic Origin of a Widespread Autotetraploid Arabidopsis arenosa Lineage Followed by Interploidy Admixture. Mol Biol Evol. 2015;32(6):1382–95. Epub 2015/04/12. 10.1093/molbev/msv089 . [DOI] [PubMed] [Google Scholar]
- 28.Lafon-Placette C, Köhler C. Endosperm-based postzygotic hybridization barriers: developmental mechanisms and evolutionary drivers. Molecular Ecology. 2016;25(11):2620–9. 10.1111/mec.13552 [DOI] [PubMed] [Google Scholar]
- 29.Morgan C, Zhang H, Henry CE, Franklin FCH, Bomblies K. Derived alleles of two axis proteins affect meiotic traits in autotetraploid Arabidopsis arenosa. Proceedings of the National Academy of Sciences. 2020;117(16):8980–8. 10.1073/pnas.1919459117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chelysheva L, Vezon D, Chambon A, Gendrot G, Pereira L, Lemhemdi A, et al. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. Plos Genet. 2012;8(7):e1002799 Epub 2012/07/31. 10.1371/journal.pgen.1002799 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright KM, Arnold B, Xue K, Surinova M, O'Connell J, Bomblies K. Selection on meiosis genes in diploid and tetraploid Arabidopsis arenosa. Mol Biol Evol. 2015;32(4):944–55. 10.1093/molbev/msu398 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grishaeva TM, Bogdanov YF. Conservation and variability of synaptonemal complex proteins in phylogenesis of eukaryotes. Int J Evol Biol. 2014;2014:856230 Epub 2014/08/26. 10.1155/2014/856230 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang C, Sofroni K, Wijnker E, Hamamura Y, Carstens L, Harashima H, et al. The Arabidopsis Cdk1/Cdk2 homolog CDKA;1 controls chromosome axis assembly during plant meiosis. The EMBO Journal. 2020;39(3):e101625 10.15252/embj.2019101625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osman K, Yang J, Roitinger E, Lambing C, Heckmann S, Howell E, et al. Affinity proteomics reveals extensive phosphorylation of the Brassica chromosome axis protein ASY1 and a network of associated proteins at prophase I of meiosis. Plant J. 2018;93(1):17–33. 10.1111/tpj.13752 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brakenhoff RH, Schoenmakers JG, Lubsen NH. Chimeric cDNA clones: a novel PCR artifact. Nucleic Acids Res. 1991;19(8):1949 Epub 1991/04/25. 10.1093/nar/19.8.1949 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen JM, Cooper DN, Chuzhanova N, Ferec C, Patrinos GP. Gene conversion: mechanisms, evolution and human disease. Nat Rev Genet. 2007;8(10):762–75. Epub 2007/09/12. 10.1038/nrg2193 . [DOI] [PubMed] [Google Scholar]
- 37.Hazarika MH, Rees H. Genotypic control of chromosome behaviour in rye X. Chromosome pairing and fertility in autotetraploids. Heredity. 1967;22(3):317–32. 10.1038/hdy.1967.44 [DOI] [Google Scholar]
- 38.He W, Rao H, Tang S, Bhagwat N, Kulkarni DS, Ma Y, et al. Regulated Proteolysis of MutSgamma Controls Meiotic Crossing Over. Mol Cell. 2020. Epub 2020/03/05. 10.1016/j.molcel.2020.02.001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mergner J, Frejno M, List M, Papacek M, Chen X, Chaudhary A, et al. Mass-spectrometry-based draft of the Arabidopsis proteome. Nature. 2020. 10.1038/s41586-020-2094-2 [DOI] [PubMed] [Google Scholar]
- 40.Sanchez-Moran E, Santos JL, Jones GH, Franklin FC. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 2007;21(17):2220–33. 10.1101/gad.439007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jorgensen MH, Ehrich D, Schmickl R, Koch MA, Brysting AK. Interspecific and interploidal gene flow in Central European Arabidopsis (Brassicaceae). BMC Evol Biol. 2011;11:346 10.1186/1471-2148-11-346 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bomblies K, Higgins JD, Yant L. Meiosis evolves: adaptation to external and internal environments. The New phytologist. 2015;208(2):306–23. 10.1111/nph.13499 . [DOI] [PubMed] [Google Scholar]
- 43.Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, Mezard C. Genome-Wide Crossover Distribution in Arabidopsis thaliana Meiosis Reveals Sex-Specific Patterns along Chromosomes. Plos Genet. 2011;7(11). ARTN e1002354 10.1371/journal.pgen.1002354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131(3):452–62. 10.1016/j.cell.2007.10.022 . [DOI] [PubMed] [Google Scholar]
- 45.Monnahan P, Kolar F, Baduel P, Sailer C, Koch J, Horvath R, et al. Pervasive population genomic consequences of genome duplication in Arabidopsis arenosa. Nat Ecol Evol. 2019;3(3):457–68. 10.1038/s41559-019-0807-4 . [DOI] [PubMed] [Google Scholar]
- 46.Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 2011;43(5):476–+. 10.1038/ng.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. 10.1093/nar/gkh340 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–21. Epub 2010/06/09. 10.1093/sysbio/syq010 . [DOI] [PubMed] [Google Scholar]
- 49.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–5. Epub 2001/08/29. 10.1093/bioinformatics/17.8.754 . [DOI] [PubMed] [Google Scholar]
- 50.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35(6):1547–9. Epub 2018/05/04. 10.1093/molbev/msy096 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, et al. A chromosome conformation capture ordered sequence of the barley genome. Nature. 2017;544(7651):427–33. 10.1038/nature22043 . [DOI] [PubMed] [Google Scholar]
- 52.Higgins JD, Wright KM, Bomblies K, Franklin FC. Cytological techniques to analyze meiosis in Arabidopsis arenosa for investigating adaptation to polyploidy. Front Plant Sci. 2014;4:546 10.3389/fpls.2013.00546 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chelysheva L, Grandont L, Vrielynck N, le Guin S, Mercier R, Grelon M. An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: immunodetection of cohesins, histones and MLH1. Cytogenet Genome Res. 2010;129(1–3):143–53. Epub 2010/07/16. 10.1159/000314096 . [DOI] [PubMed] [Google Scholar]
- 54.Lambing C, Osman K, Nuntasoontorn K, West A, Higgins JD, Copenhaver GP, et al. Arabidopsis PCH2 Mediates Meiotic Chromosome Remodeling and Maturation of Crossovers. Plos Genet. 2015;11(7):e1005372 10.1371/journal.pgen.1005372 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong YH, Lee TY, Liang HK, Huang CM, Wang TY, Yang YH, et al. KinasePhos 2.0: a web server for identifying protein kinase-specific phosphorylation sites based on sequences and coupling patterns. Nucleic Acids Research. 2007;35:W588–W94. 10.1093/nar/gkm322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4(6):1633–49. 10.1002/pmic.200300771 . [DOI] [PubMed] [Google Scholar]
- 57.Zhao Q, Xie YB, Zheng YY, Jiang S, Liu WZ, Mu WP, et al. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Research. 2014;42(W1):W325–W30. 10.1093/nar/gku383 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Stable (A) and unstable (B). Scale bar = 10μM.
(TIF)
(A) ASY1, (B) ASY3, (C) PDS5b, (D) PRD3. Bayesian posterior probabilities are indicated at the internodes of each branch. The dissimilarity scale showing substitutions per nucleotide is located at the bottom of each tree. Diploid and tetraploid alleles are indicated by ‘2’ and ‘4’ respectively (for ASY3 diploid = ND, tetraploid = TD), while the putative origins are indicated by ‘r’ (A. arenosa) and ‘y’ (A. lyrata).
(TIF)
(A) REC8, (B) SMC3, (C) ZYP1a/ZYP1b 5’ end, (D) ZYP1a/ZYP1b 3’ end. Bayesian posterior probabilities are indicated at the internodes of each branch. The dissimilarity scale showing substitutions per nucleotide is located at the bottom of each tree. Diploid and tetraploid alleles are indicated by ‘2’ and ‘4’ respectively, while the putative origins are indicated by ‘r’ (A. arenosa) and ‘y’ (A. lyrata).
(TIF)
(A) ASY1, (B) ASY3, (C) PDS5b, (D) PRD3. Maximum likelihood bootstrap values are indicated at the internodes of each branch (1000 replicates). The dissimilarity scale showing substitutions per nucleotide is located at the bottom of each tree. Diploid and tetraploid alleles are indicated by ‘2’ and ‘4’ respectively (for ASY3 diploid = ND, tetraploid = TD), while the putative origins are indicated by ‘r’ (A. arenosa) and ‘y’ (A. lyrata).
(TIF)
(A) REC8, (B) SMC3, (C) ZYP1a/ZYP1b 5’ end, (D) ZYP1a/ZYP1b 3’ end. Maximum likelihood bootstrap values are indicated at the internodes of each branch (1000 replicates). The dissimilarity scale showing substitutions per nucleotide is located at the bottom of each tree. Diploid and tetraploid alleles are indicated by ‘2’ and ‘4’ respectively, while the putative origins are indicated by ‘r’ (A. arenosa) and ‘y’ (A. lyrata).
(TIF)
The location of diploid and autotetraploid allele specific SNPs are indicated. Coloured bars in each sequence represent base specific SNPs relative to the consensus sequence (Green = A, Blue = C, Black = G, Red = T).
(JPG)
ZYP1 gene conversion (or CO) between ZYP1a (grey) and ZYP1b (red) in autotetraploid ZYP1b alleles. The location of diploid and autotetraploid allele specific SNPs are indicated. Coloured bars in each sequence represent base specific SNPs relative to the consensus sequence (Green = A, Blue = C, Black = G, Red = T).
(JPG)
Gene conversion between PRD3 diploid A. lyrata (green) and A. arenosa (green) in tetraploid A. arenosa. The location of diploid and autotetraploid allele specific SNPs are indicated. Coloured bars in each sequence represent base specific SNPs relative to the consensus sequence (Green = A, Blue = C, Black = G, Red = T).
(JPG)
Gene conversion (or CO) between PDS5b diploid (yellow) and autotetraploid (green) alleles. The location of diploid and autotetraploid allele specific SNPs are indicated. The location of diploid and autotetraploid allele specific SNPs are indicated. Coloured bars in each sequence represent base specific SNPs relative to the consensus sequence (Green = A, Blue = C, Black = G, Red = T).
(JPG)
Gene conversion (or CO) between ASY1 diploid A. lyrata (yellow) and autotetraploid A. arenosa (green). Coloured bars in each sequence represent base specific SNPs relative to the consensus sequence (Green = A, Blue = C, Black = G, Red = T).
(JPG)
ASY1 (A), ASY3 (B), PDS5b (C), showing conserved amino acid polymorphisms in autotetraploids compared to ancestral diploid alleles. Gains, losses and no change of predicted phosphorylation sites are indicated in blue, yellow and green respectively.
(TIF)
PRD3 (A), REC8 (B), showing conserved amino acid polymorphisms in autotetraploids compared to ancestral diploid alleles. Gains, losses and no change of predicted phosphorylation sites are indicated in blue, yellow and green respectively.
(TIF)
ZYP1a (A) and ZYP1b (B), showing conserved amino acid polymorphisms in autotetraploids compared to ancestral diploid alleles. Gains, losses and no change of predicted phosphorylation sites are indicated in blue, yellow and green respectively.
(TIF)
The analysis includes gains and losses of predicted serine/threonine phosphorylation sites by KinasePhos2.0 and NetPhos3.1.
(TIF)
Diploid lyrata alleles = ly; diploid arenosa alleles = ar and ar/ly = diploid arenosa to diploid lyrata putative gene conversions.
(DOCX)
Putative addition of serine/threonine phosphosites are highlighted in blue and loss of phosphorylation sites highlighted in yellow.
(DOCX)
Putative addition of serine/threonine phosphorylation sites are highlighted in blue and loss of phosphorylation sites highlighted in yellow.
(DOCX)
Putative addition of serine/threonine phosphorylation sites are highlighted in blue and loss of phosphorylation sites highlighted in yellow.
(DOCX)
(DOCX)
(DOCX)
Raw data used for generating figures.
(XLSX)
Data Availability Statement
All sequences in this study including cDNA transcripts and genomic DNA sequences have been deposited in the DDBJ/EMBL/GenBank databases under accession numbers MN512718 - MN513026 and MN520243 - MN520257. MiSeq amplicon reads have been deposited in the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) database under BioProject ID PRJNA575228.