Abstract
Riboswitch, a part of regulatory mRNA (50–250nt in length), has two main classes: aptamer and expression platform. One of the main challenges raised during the classification of riboswitch is imbalanced data. That is a circumstance in which the records of a sequences of one group are very small compared to the others. Such circumstances lead classifier to ignore minority group and emphasize on majority ones, which results in a skewed classification. We considered sixteen riboswitch families, to be in accord with recent riboswitch classification work, that contain imbalanced sequences. The sequences were split into training and test set using a newly developed pipeline. From 5460 k-mers (k value 1 to 6) produced, 156 features were calculated based on CfsSubsetEval and BestFirst function found in WEKA 3.8. Statistically tested result was significantly difference between balanced and imbalanced sequences (p < 0.05). Besides, each algorithm also showed a significant difference in sensitivity, specificity, accuracy, and macro F-score when used in both groups (p < 0.05). Several k-mers clustered from heat map were discovered to have biological functions and motifs at the different positions like interior loops, terminal loops and helices. They were validated to have a biological function and some are riboswitch motifs. The analysis has discovered the importance of solving the challenges of majority bias analysis and overfitting. Presented results were generalized evaluation of both balanced and imbalanced models, which implies their ability of classifying, to classify novel riboswitches. The Python source code is available at https://github.com/Seasonsling/riboswitch.
Author summary
Machine learning application has been used in many ways in bioinformatics and computational biology. Its use in riboswitch classification is still limited. Existing attempts showed challenges due to imbalanced sequences. Algorithms can classify sequences with majority and minority groups, but they tend to ignore minority group and emphasize on majority class, consequential return a skewed classification. We used a new pipeline including SMOTE for balancing sequences that showed better-classified riboswitch as well as improved performance of algorithms selected. Statistically significant difference observed between balanced and imbalanced in sensitivity, specificity, accuracy and F-score, this proved balanced sequences better for classification of riboswitch. Biological functions and motif search of k-mers in riboswitch families revealed their presence in interior loops, terminal loops and helices. Some of the k-mers were reported to be riboswitch motifs of aptamer domains and critical for metabolite binding. The pipeline can be used in machine learning and deep learning study in other domains of bioinformatics and computational biology suffering from imbalanced sequences. Finally, scientific community can use python source code, the work done and flow to develop packages.
This is a PLOS Computational Biology Methods paper.
Introduction
Riboswitches, primarily discovered in bacteria [1], are parts of regulatory noncoding mRNA [2]. Riboswitches are predominantly present in the 5’ untranslated region [3,4]. They have complex folded structure [5,6]. They act as a switch to transform the transcription or translation of the genes. In transcription, they turn a downstream gene ‘off’ or ‘on’ [7] in changing concentration of specific metabolites or ligands [8] and allow microbes to quickly react to change degrees of metabolites [7]. A high-throughput platform showed how RNA makes structural transitions [9] kinetically compete during transcription in a new mechanism for riboswitch.
A riboswitch (50–250 nt in length) has two main classes aptamer and an expression platform [10]. The aptamer region is a highly conserved domain, which is a site for binding of ligands (metabolites) and the latter one alters conformation on the binding of metabolite and hence regulates the expression of related genes [5,6]. Recently, almost over twenty diverse classes of riboswitches have been found in bacteria, archaea [11,12] and eukaryote. The majority of the riboswitch classes are in bacteria [12,13]. Thiamine pyrophosphate (TPP) is the only eukaryotic riboswitch. It is detected in Arabidopsis thaliana. TPP was also found in some fungi [13] for instance Neurospora crassa, in algae [14,15].
The last two decades have revealed incredible advancement in big and complex omics data due to emerged novel high-throughput experimental technologies such as next-generation sequencing [16,17]. Numerous bioinformatics databases are available to gather data for riboswitches analyses and assemble the information regarding diverse functionality of RNA molecules [18], including GenBank, National Center for Biotechnology Information (NCBI), Rfam [19], Protein Data Bank (PDB), RiboD [20] and European Bioinformatics Institute (EMBL-EBI).
Many efforts have been made to develop suitable bioinformatics tools to predict the presence of riboswitches in ribonucleic acid sequences [18]. The most commonly used computation tools for the analysis of riboswitches are: RiboD [20], Riboswitch finder [21], RibEx [22], RiboSW [23], mFold [24] and RegRNA [18]. These available bioinformatics tools use Covariance Model (CM), Support Vector Machine (SVM) and Hidden Markov Model (HMM) algorithm. Most research exists mainly depending on the principal of multiple sequence alignment to investigate conserved sequences in already reported riboswitch. The attempt was to find out the conserved sequence of previously reported riboswitches in a targeted manner. Most reported studies depend on multiple sequence and thus limited for the classes of riboswitches in a family [21–24]. However, research conducted on frequency-dependent revealed its importance in the classification of riboswitch [25,26]. Frequency-dependent classification uses k-mers counts. K-mers counts have many application like, building de Bruijn graphs [27] in case of de novo assembly from very big number of short reads, generated from next-generation sequencing (NGS), used in case of multiple sequence alignment [28], and repeat detection [29].
A tremendous amount of data are generated every day that create the demand for learning algorithms that can classify, predict and analyze data more accurately [30]. There are two classification categories: classification of binary format [31] and multi-class classification [32,33].
The concept of imbalanced sequences is defined as follows. Each family in classes of riboswitch with majority groups has more than two thousand class and minority group below thousands, which is considered as an imbalanced sequence. Whereas, the imbalanced group used and treated with Synthetic Minority Over-Sampling Technique (SMOTE) and thereafter it is called a balanced sequences. The classification with imbalanced data gives favors for a sample with the majority class [30]. Classifiers trained by balanced sequences are defined as balanced classifiers. Imbalanced data occur as a circumstance where the records of a sequences of one class are very little in relation to the other classes’ sequences. This leads classifier algorithms to ignore minority groups and emphasize on majority class, which can result in skewed accuracy of the classifier. The value of the accuracy of the classifier might be high, but minority class misclassified. Several findings have been done for riboswitch classification [25,26] based on imbalanced data. However, data resampling can be a solution to handle the class imbalance problems [30]. Synthetic Minority Over-Sampling Technique (SMOTE) has been discovered in 2002, which is a sampling-based algorithm. Synthetic Minority Over-Sampling Technique [34] balances the class distribution of imbalanced sequences through an incrementing approach on some virtual samples.
To address the needs for riboswitch prediction, nucleotide frequency counts are considered. SMOTE was used for resampling. Different machine learning algorithms are used for evaluation such as: Random forest (RF) randomizes the variables (columns) and data (rows), generating thousands of classification trees, and then summarizing the results of the classification tree [35]. Gradient boosting (GB) is a boosting algorithm, which belongs to ensemble learning as well as random forest and proved to have great performance in imbalance problem. It builds the model in a stage-wise fashion, and generalizes them by allowing optimization of an arbitrary differentiable loss function. Support vector machine (SVM) is a simple and efficient method for solving the quadratic programming problem through computing the maximum marginal hyper-plane. In SVM, the kernel function implicitly defines the feature space for linear partitioning, which means the choice of kernel function is the largest variable of SVM [35]. K-Nearest Neighbors (KNN) is classifier offers numerous choices to speed up the undertaking to locate nearest neighbors, Naïve Bayes (NB) classifier based on Bayes' theorem [25]. This is a probability-based model in Bayesian networks. Multilayer perceptron (MLP) is a commonly used machine learning algorithm. It is a deep, artificial neural network. A neural network is comprised of layers of nodes which activate at various levels depending on the previous layer’s nodes [25]. The performances of each algorithm on classification were derived from the confusion matrix, which reveals the number of matches correctly and mismatched instances of riboswitches. Specificity, sensitivity, accuracy, and macro F-score were calculated. That parameters are the main performance evaluation criteria for machine learning algorithms [35–38].
Results
Sequences preprocessing and feature selection
Riboswitch families considered for this analysis and their corresponding details were presented and analyzed in Fig 1 and features where clustered in Fig 2 (see detail in S1 Fig). Looking into instances in riboswitch, there were differences in representation between families range in distribution from Cobalamin riboswitch (4,826 sequence classes) to PreQ1-II (39 sequence classes). Out of 16 riboswitch class, Cobalamin riboswitch, TPP riboswitch (THI element), and Glycine riboswitch contributed for 68% and the remaining 13 riboswitch family has 32% instances. The performances of algorithms and methods were computed and evaluated based on training and test set (details in the methodological approach part). We produced 5460 k-mers (1≤k≤6) by R script and exported a matrix containing all riboswitch sequences and their corresponding k-mers value. A Sequences preprocessing and feature selection afterward, 156 features were calculated based on the Correlation-based Feature Subset Selection algorithm (CfsSubsetEval) and Best First Search (BestFirst) in WEKA 3.8 [39] (Fig 3 and detail in S2 Fig), which was consistent with previous research [26].
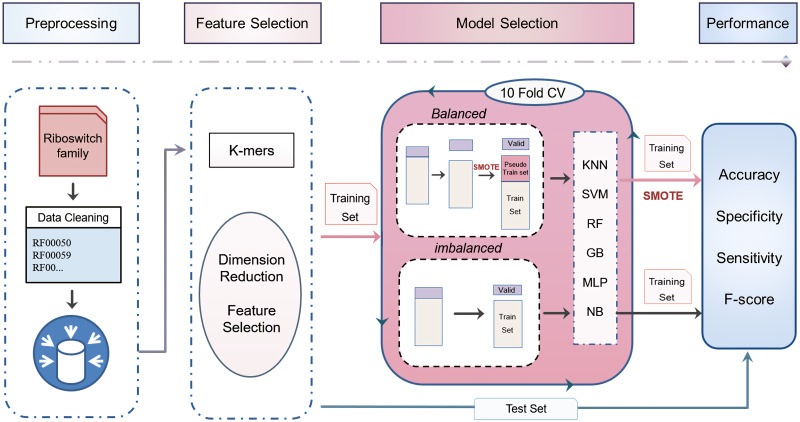
Fig 1. The workflow used to analyze imbalanced and balanced sequences.
It was used to compare the computational performance of machine learning algorithms for classification.
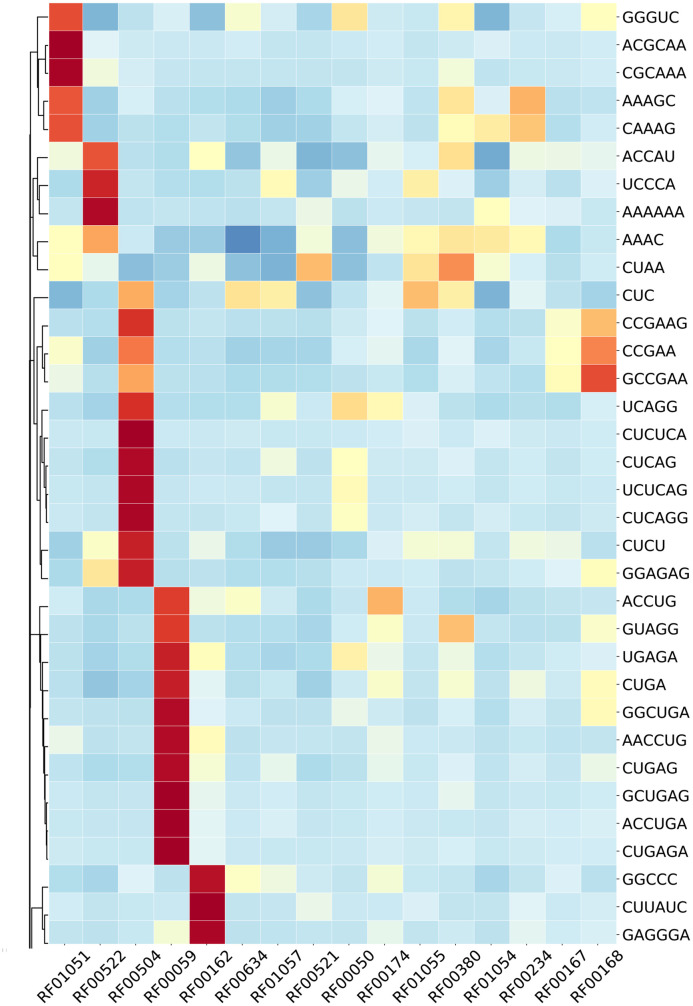
Fig 2. Heat-map in this figure represented as row-normalized k-mer counting distribution.
Rows correspond to the k-mers, and columns revealed 16 families of riboswitch. The clustering heatmap depicts feature clustering, clustered features were essential for classification in that family. Red means a high relatively counting number while blue means lower (see details in S1 Fig).
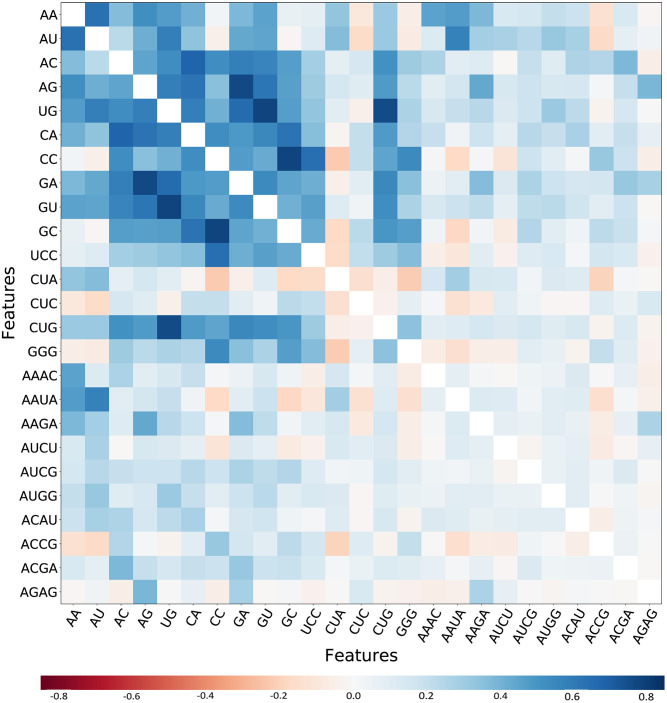
Fig 3. Heat-map showed features correlation.
It depicts the diagonal white line represented their correlation factor equals to one. Blue means a positive correlation, while red means a negative correlation (see details in S2 Fig).
Imbalanced class on classification performance
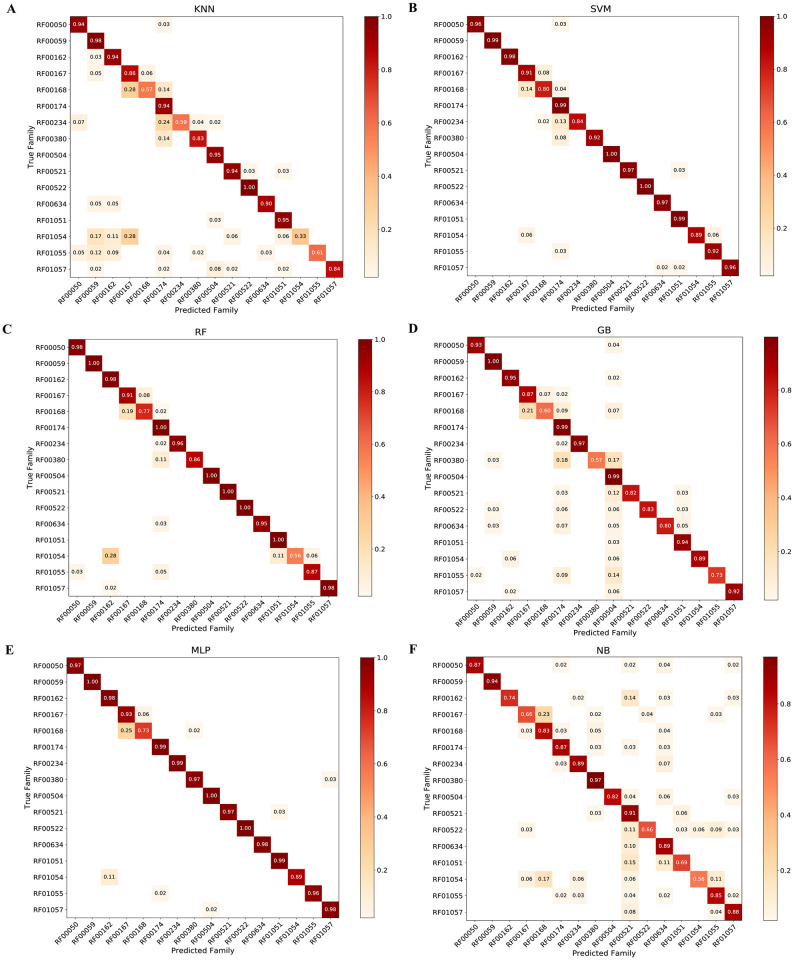
After feature selection, sequences containing 156 k-mers values were split into 70% training dataset and 30% test dataset. Improved cross-validation method in training dataset was used to validate both imbalanced models and balanced models, while the remaining test set was applied to test generalizations of those models. All the following results are results based on the test set, which can demonstrate their ability to classify novel sequences. Classifiers on minority class resulted in F-score value from 0.50 (NB) to 0.94 (MLP), while on majority class, the range is from 0.91 to 1.00, as indicated in Table 1 and Fig 4. Riboswitch families considered for classification were present in S1 Table. The average performance of each classifier is computed using mean and standard deviation for parameters: accuracy, specificity, sensitivity and macro F-score.
Table 1. Accuracy, sensitivity, specificity and F-score.
This parameters were used for Naïve Bayes(NB), Multilayer Perceptron(MLP), Random Forest(RF), Gradient Boosting(GB), Support Vector Machine(SVM) and K-Nearest Neighbors(KNN) algorithms evaluation when applied on the imbalanced sequences. The color trend of F-score from blue to red indicates performance from the best to the poorest. Accuracy, sensitivity, specificity, and F-score are represented in the table as Acc, Sen, Spec, and F-sco, respectively.
| NB | MLP | RF | GB | SVM | KNN | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family | Acc | Sen | Spec | F-sco | Acc | Sen | Spec | F-sco | Acc | Sen | Spec | F-sco | Acc | Sen | Spec | F-sco | Acc | Sen | Spec | F-sco | Acc | Sen | Spec | F-sco |
| RF00521 | 0.95 | 0.91 | 0.95 | 0.24 | 1.00 | 0.97 | 1.00 | 0.97 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.82 | 1.00 | 0.89 | 1.00 | 0.97 | 1.00 | 0.99 | 1.00 | 0.94 | 1.00 | 0.94 |
| RF00522 | 1.00 | 0.66 | 1.00 | 0.69 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.83 | 1.00 | 0.89 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 0.97 |
| RF00059 | 0.98 | 0.94 | 1.00 | 0.97 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 | 0.99 | 0.98 | 0.98 | 0.98 | 0.96 |
| RF00174 | 0.95 | 0.87 | 0.99 | 0.91 | 1.00 | 0.99 | 1.00 | 0.99 | 0.99 | 1.00 | 0.99 | 0.99 | 0.99 | 0.99 | 0.98 | 0.98 | 0.99 | 0.99 | 0.99 | 0.98 | 0.97 | 0.94 | 0.98 | 0.94 |
| RF00504 | 0.97 | 0.83 | 1.00 | 0.90 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.98 | 0.99 | 0.98 | 0.93 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.95 | 0.99 | 0.96 |
| RF01051 | 0.98 | 0.69 | 1.00 | 0.81 | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 | 0.94 | 1.00 | 0.96 | 1.00 | 0.99 | 1.00 | 0.98 | 0.99 | 0.95 | 1.00 | 0.95 |
| RF01057 | 0.98 | 0.88 | 0.98 | 0.53 | 1.00 | 0.98 | 1.00 | 0.96 | 1.00 | 0.98 | 1.00 | 0.99 | 1.00 | 0.92 | 1.00 | 0.96 | 1.00 | 0.96 | 1.00 | 0.97 | 1.00 | 0.84 | 1.00 | 0.87 |
| RF00050 | 0.99 | 0.87 | 1.00 | 0.92 | 1.00 | 0.97 | 1.00 | 0.98 | 1.00 | 0.98 | 1.00 | 0.98 | 0.99 | 0.93 | 1.00 | 0.96 | 1.00 | 0.96 | 1.00 | 0.98 | 0.99 | 0.94 | 0.99 | 0.92 |
| RF00162 | 0.98 | 0.74 | 1.00 | 0.84 | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 | 0.98 | 1.00 | 0.98 | 0.99 | 0.95 | 1.00 | 0.97 | 1.00 | 0.98 | 1.00 | 0.99 | 0.99 | 0.94 | 0.99 | 0.91 |
| RF00234 | 0.99 | 0.89 | 0.99 | 0.79 | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 | 0.96 | 1.00 | 0.98 | 1.00 | 0.97 | 1.00 | 0.98 | 1.00 | 0.84 | 1.00 | 0.91 | 0.99 | 0.59 | 1.00 | 0.70 |
| RF00634 | 0.99 | 0.89 | 1.00 | 0.38 | 1.00 | 0.98 | 1.00 | 0.98 | 0.99 | 0.95 | 0.99 | 0.98 | 0.98 | 0.80 | 0.99 | 0.87 | 0.99 | 0.97 | 0.99 | 0.97 | 0.98 | 0.90 | 0.99 | 0.84 |
| RF01055 | 0.99 | 0.85 | 1.00 | 0.81 | 1.00 | 0.96 | 1.00 | 0.96 | 1.00 | 0.87 | 1.00 | 0.93 | 0.99 | 0.73 | 1.00 | 0.83 | 1.00 | 0.93 | 1.00 | 0.95 | 0.99 | 0.61 | 1.00 | 0.74 |
| RF00380 | 0.97 | 0.97 | 0.97 | 0.62 | 0.99 | 0.97 | 0.99 | 0.96 | 0.99 | 0.86 | 0.99 | 0.92 | 0.99 | 0.57 | 0.99 | 0.84 | 0.99 | 0.92 | 0.99 | 0.95 | 0.98 | 0.83 | 0.99 | 0.82 |
| RF00167 | 0.98 | 0.66 | 0.98 | 0.78 | 0.99 | 0.93 | 1.00 | 0.91 | 0.99 | 0.91 | 1.00 | 0.91 | 0.99 | 0.87 | 1.00 | 0.88 | 0.99 | 0.91 | 1.00 | 0.92 | 0.98 | 0.86 | 1.00 | 0.84 |
| RF00168 | 0.97 | 0.83 | 0.98 | 0.59 | 0.99 | 0.73 | 1.00 | 0.77 | 0.99 | 0.77 | 1.00 | 0.78 | 0.99 | 0.60 | 1.00 | 0.68 | 0.99 | 0.80 | 1.00 | 0.80 | 0.99 | 0.57 | 1.00 | 0.66 |
| RF01054 | 1.00 | 0.56 | 1.00 | 0.50 | 1.00 | 0.89 | 1.00 | 0.94 | 1.00 | 0.56 | 1.00 | 0.71 | 1.00 | 0.89 | 1.00 | 0.80 | 1.00 | 0.89 | 1.00 | 0.94 | 1.00 | 0.33 | 1.00 | 0.50 |
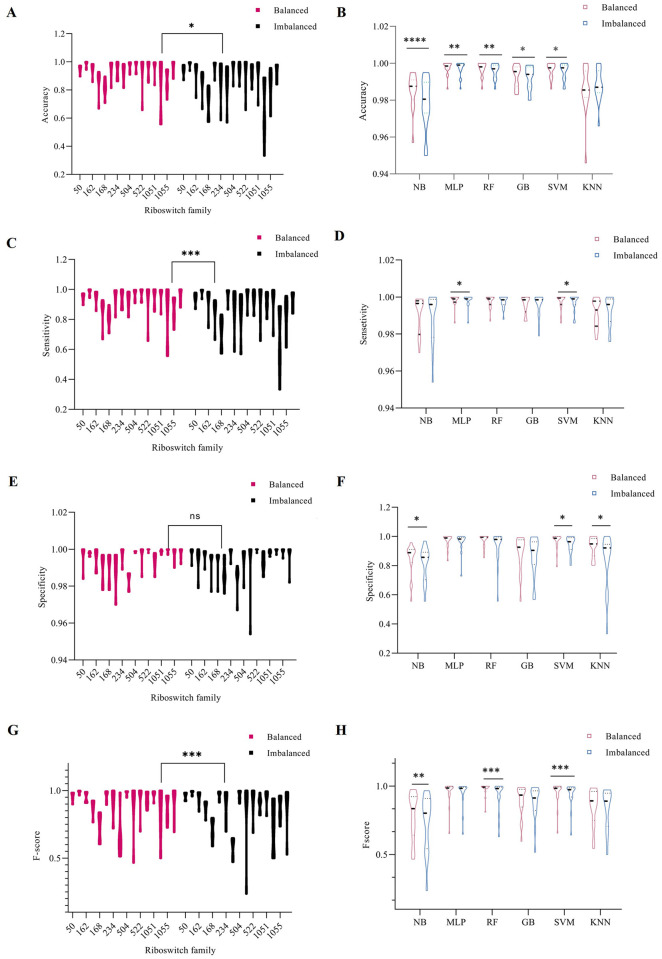
Fig 4. The figures showed a comparison of the balanced and imbalanced sequences and performance of classifiers.
It has been done using the Wilcoxon rank test, A) Accuracy showed significant difference between balanced and imbalanced sequences (p < 0.05) C) Sensitivity showed very significant difference between balanced and imbalanced sequences (p < 0.001) E) Specificity revealed no significant differences at all levels G) F-score showed very significant difference between balanced and imbalanced sequences (p < 0.001). Classifiers performance evaluation on imbalanced and imbalanced sequences shown as B) Accuracy resulted to have significant difference in all classifiers except KNN (p < 0.05, p < 0.01, p < 0.001) D) Sensitivity observed to have significant difference in only MLP and SVM (p < 0.05) whereas the remaining algorithms showed no differences F) Specificity depicted significant differences in NB, SVM and KNN (p < 0.05) on the other hand MLP, RF and GB showed no differences in both sequences group H) F-score depicted very significance differences in NB (p < 0.01), RF (p < 0.001) and SVM (p < 0.001) whereas KNN and MLP showed no differences. Violin box was used to depict the statistical differences between two group were provided as the plots. (* indicated significant difference of p < 0.05, ** denoted very significant difference of p < 0.01, and *** showed very significant difference p < 0.001).
The comparative analysis of six algorithms has revealed that MLP performs best, while NB performed the poorest results (S2 Table). RF00174, RF00059, RF00504, RF00522 classified better than others with minority classes like RF01054, RF00634, RF00380 (Table 1). F-scores of MLP and RF for the majority group (RF00174) were 0.997 and 0.996, respectively. In the minority group, classifiers with high accuracy had F-score up to 0.50 in the case of NB. The computed minimum value in overall NB analysis in RF01054, RF00634, and RF00521 were 0.50, 0.38, and 0.24, respectively. Accuracy of all algorithms across all riboswitch families showed values greater than 0.97. In confusion matrix, predicted family and true family exhibited performance of classifiers and riboswitch classification (Fig 5).
Fig 5. Confusion matrix for imbalanced sequences from independent test experiments depicted true family and predicted family.
For the classifiers such as: A) K-Nearest Neighbors, B) Support Vector Machine, C) Random Forest, D) Gradient Boosting, E) Multilayer Perceptron and F) Naïve Bayes.
SMOTE balancing on classifiers performance
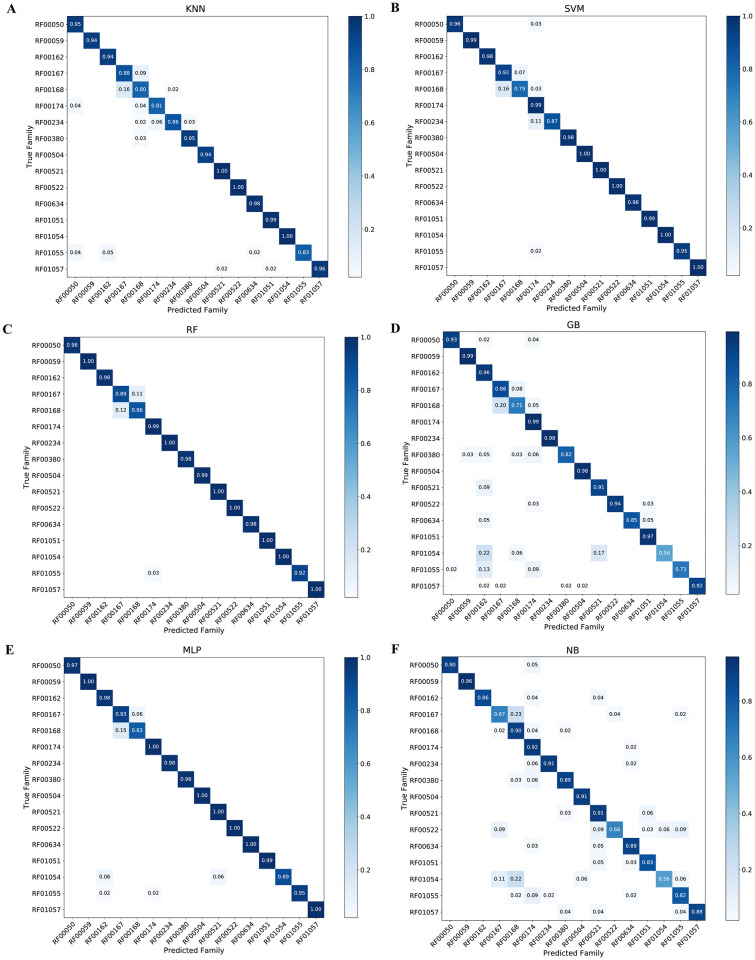
The overall analysis computed for frequency counts of all families had discovered improved performances of classifiers (S2 Table, Table 2 and Fig 4). RF00059 and RF00174 results showed F-score between 0.93 and 1.00. In the case of NB and KNN, results of the F-score indicated their poorer performance with a value less than 0.84. Performance evaluations have revealed that KNN, NB, SVM, MLP, RF and GB can be used for classification of riboswitch (Fig 6).
Table 2. Performances of Naïve Bayes (NB), Multilayer Perceptron (MLP), Random Forest (RF), Gradient Boosting (GB), Support Vector Machine (SVM) and K- Nearest Neighbors (KNN).
These algorithms were evaluated using the balanced sequences from 16 riboswitch families measured by using accuracy, sensitivity, specificity and F-score. The color trend of F-score from blue to red indicates performance from the best to the poorest. Accuracy, sensitivity, specificity, and F-score are represented in the table as Acc, Sen, Spec, and F-sco, respectively.
| NB | MLP | RF | GB | SVM | KNN | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family | Acc | Sen | Spec | F-sco | Acc | Sen | Spec | F-sco | Acc | Sen | Spec | F-sco | Acc | Sen | Spec | F-sco | Acc | Sen | Spec | F-sco | Acc | Sen | Spec | F-sco |
| RF00059 | 0.99 | 0.96 | 1.00 | 0.98 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 | 0.99 | 0.98 | 0.94 | 1.00 | 0.96 |
| RF00234 | 1.00 | 0.92 | 1.00 | 0.88 | 1.00 | 0.98 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.98 | 1.00 | 0.98 | 1.00 | 0.87 | 1.00 | 0.92 | 0.99 | 0.86 | 0.99 | 0.72 |
| RF00521 | 0.98 | 0.91 | 0.99 | 0.47 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.91 | 1.00 | 0.89 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.96 |
| RF00522 | 1.00 | 0.66 | 1.00 | 0.70 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.94 | 1.00 | 0.97 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 0.99 |
| RF01054 | 1.00 | 0.56 | 1.00 | 0.50 | 1.00 | 0.89 | 1.00 | 0.94 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.56 | 1.00 | 0.71 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.84 |
| RF01057 | 0.99 | 0.88 | 0.99 | 0.69 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.92 | 1.00 | 0.96 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 0.96 | 1.00 | 0.82 |
| RF00162 | 0.99 | 0.86 | 1.00 | 0.91 | 1.00 | 0.98 | 1.00 | 0.98 | 1.00 | 0.99 | 1.00 | 0.99 | 0.98 | 0.96 | 0.99 | 0.91 | 1.00 | 0.99 | 1.00 | 0.99 | 0.98 | 0.94 | 0.99 | 0.91 |
| RF00174 | 0.96 | 0.92 | 0.97 | 0.93 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 | 0.99 | 0.99 | 0.99 | 0.99 | 0.98 | 0.99 | 0.99 | 0.99 | 0.98 | 0.95 | 0.81 | 1.00 | 0.89 |
| RF00504 | 0.99 | 0.91 | 1.00 | 0.95 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 | 0.98 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 | 0.94 | 1.00 | 0.96 |
| RF01051 | 0.99 | 0.83 | 1.00 | 0.91 | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 0.97 | 1.00 | 0.97 | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 | 0.97 |
| RF00050 | 0.99 | 0.90 | 1.00 | 0.94 | 1.00 | 0.97 | 1.00 | 0.98 | 1.00 | 0.97 | 1.00 | 0.98 | 0.99 | 0.93 | 1.00 | 0.96 | 1.00 | 0.96 | 1.00 | 0.98 | 0.98 | 0.95 | 0.98 | 0.90 |
| RF00380 | 0.98 | 0.89 | 0.98 | 0.69 | 0.99 | 0.99 | 0.99 | 0.96 | 0.99 | 0.94 | 0.99 | 0.98 | 0.99 | 0.82 | 0.99 | 0.84 | 0.99 | 0.99 | 0.99 | 0.98 | 0.98 | 0.95 | 0.98 | 0.73 |
| RF00634 | 0.99 | 0.89 | 1.00 | 0.64 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1.00 | 1.00 | 0.98 | 0.98 | 0.85 | 0.99 | 0.87 | 0.99 | 0.98 | 0.99 | 0.99 | 0.99 | 0.98 | 0.99 | 0.86 |
| RF01055 | 0.99 | 0.82 | 0.99 | 0.79 | 1.00 | 0.95 | 1.00 | 0.95 | 1.00 | 0.93 | 1.00 | 0.96 | 1.00 | 0.73 | 1.00 | 0.85 | 1.00 | 0.95 | 1.00 | 0.96 | 0.99 | 0.83 | 0.99 | 0.73 |
| RF00167 | 0.98 | 0.67 | 0.98 | 0.77 | 0.99 | 0.93 | 1.00 | 0.93 | 0.99 | 0.89 | 0.99 | 0.91 | 0.99 | 0.89 | 1.00 | 0.88 | 0.99 | 0.93 | 1.00 | 0.93 | 0.99 | 0.88 | 0.98 | 0.85 |
| RF00168 | 0.98 | 0.90 | 0.98 | 0.62 | 0.99 | 0.83 | 1.00 | 0.84 | 0.99 | 0.85 | 0.99 | 0.81 | 0.99 | 0.71 | 1.00 | 0.72 | 0.99 | 0.79 | 1.00 | 0.80 | 0.98 | 0.80 | 0.98 | 0.60 |
Fig 6. Confusion matrix for the balanced sequences from independent test experiments.
It showed True family and Predicted value with classifiers as: A) K-Nearest Neighbors, B) Support Vector Machine, C) Random Forest, D) Gradient Boosting, E) Multilayer Perceptron and F) Naïve Bayes.
As presented, Random Forest and MLP exhibited the consistently higher accuracy and F-score values compared to NB, GB, SVM and KNN. Fig 4 and Table 2 have shown that SMOTE improves riboswitch classification and algorithm performances.
The overall accuracy of classifiers trained with SMOTE analyzed sequences (balanced sequences) showed consistent and better results than with imbalanced sequences (S2 Table and Tables 1 and 2). The specificity of NB, MLP, RF, GB, SVM and KNN was better in the balanced classifiers than imbalanced sequences ones. Calculated sensitivity results were slightly better in balanced instances. Surprisingly, evidence discovered in that F-score value in all the models showed that balanced training sequences could improve the classification of riboswitches. When tested by independent test sequences, balanced sequences trained classifiers increased not only classification accuracy, but also algorithms performances than control groups. Balanced sequences increased not only classification accuracy but also algorithms performances. Table 2 has depicted F-score values increasing from 0.50 while in the case of the imbalanced sequences to 0.84.
Application of statistical significances
Statistical computation using the Wilcoxon rank test [39] between balanced and imbalanced sequences depicts significant differences between these two groups. In addition, the performance of NB, MLP, RF, GB, SVM and KNN statistically showed variation in accuracy, specificity, sensitivity and F-score values. Statistically very significant differences were noticed between balanced and imbalanced in F-score and sensitivity (p < 0.001) and accuracies were significantly different (p <0.05), whereas specificity showed no significant difference between the two groups (Fig 4, S3 Table).
SVM was a very significant difference in all parameters used for performance evaluation, F-score (p < 0.001) whereas accuracy, specificity and sensitivity were significantly different (p < 0.05). RF performance in both groups has shown very significant differences in F-score (p < 0.001) and accuracy (p < 0.01) (Fig 4 and S2 Table). In KNN we did not notice statistical significant differences in all used parameters, except significant differences in specificity (p < 0.05).
MLP of the balanced and imbalanced group depicted very significant differences in accuracy and sensitivity (p < 0.01). GB showed significant differences only in accuracy (p < 0.05). Finally, both imbalanced and balanced sequences in the case of NB have shown very significantly differences in F-score (p < 0.01), accuracy (p < 0.001), whereas specificity was a significant difference (p < 0.05). Accuracy of all classifiers is significantly different at different levels in both groups except in KNN (Fig 4 and S3 Table).
Biological functions of clustered k-mers
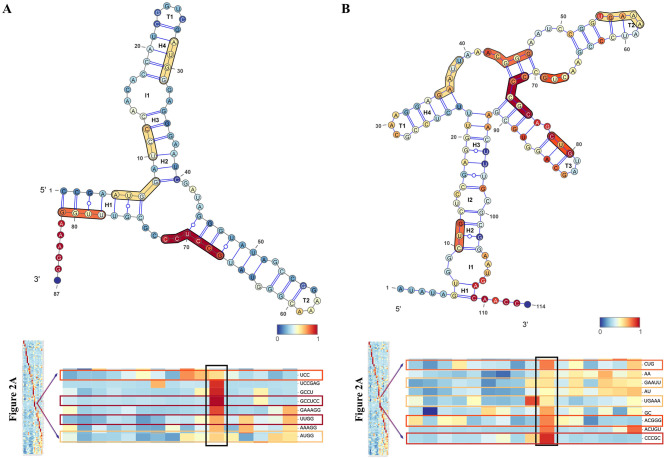
K-mers counting was extracted from distribution heat-map (Fig 2, S1 Fig), which depicted feature clustering and high relative count number. These clustered k-mers were used for biological function and motif searching. Accordingly, in Table 3 riboswitch families and their k-mers were used to verify their biological functions. Structural analysis from k-mers coverage results is depicted in the case of RF00174 (A) and RF01055 (B). In every individual base, the color gradient scale indicates a normalized count. Results depict different color scale in each region and their interior loops, helices, and terminal loops (Fig 7).
Table 3. Clustered k-mers from S1 Fig used for validation of their biological function and reported riboswitch motifs.
Nucleotide location designated refers to match with their position reported in reference.
| Rfam ID | K-mers | Position | Riboswitch function and motifs | Ref |
|---|---|---|---|---|
| RF00168 | UCAU | U57-U60 | Motifs predicted to interact with the Nova-1 protein | [40] |
| RF01051 | CAAAG | C22-G26 | Secondary structure representation of the crystallized c-di-GMP aptamer, necessary for c-di-GMP binding pocket formation | [41] |
| GGUC | G8-C11 | Found in Helices P1 area beginning of 5′UTR | [41] | |
| RF00522 | AAAAAA AAAC |
A27-A31 A30-C33 |
Overlaying K-mers in the 3′ aptamer domain, rich in A, which has unique folding pseudoknot that compresses PreQ1 | [42] |
| UCCCA | U24-A18 | Found in P2 of preQ1 riboswitch aptamer structure | [42] | |
| RF00504 | CCGAAG | C168-G173 | The glycine-mediated changes in spontaneous cleavage (GAA) | [43] |
| CUCU | C204-U207 | In glycine riboswitch, secondary structure and in-line decreasing cleavage pattern | [43] | |
| RF00059 | UGAGA | U39-A43 | The pyrimidine part of TPP is bound by bulge J3-2 located in the pyrimidine-sensor helix P2-P3 | [44] |
| RF00162 | GAGGGA | G19-A24 | It is a kink-turn motif that allows pseudoknot interaction. It interacts with SAM which helps to make stable formation, can cause the downstream expression platform to form a rho-independent TT (transcriptional terminator), turning off gene expression | [45] |
| RF00634 | CAACC CCCUC |
C54-C58 C57-C61 |
Overlapping k-mers in SAM-IV RNA binds SAM, last C in cleavage increased by SAM | [46] |
| RF01057 | AGGCUC | A61-C66 | In P1 SAH riboswitch control reporter gene Expression, ahcY 5'UTR | [47] |
| CGCU | C28-U31 | In SAH riboswitch hairpin loops of P4 | [47] | |
| RF00521 | GCUAAA | G42-A47 | Secondary structure of the Env12 metX SAM-II riboswitch its base-pairing reflecting the tertiary structure of the SAM-bound RNA | [48] |
| RF00050 | AGUC | A126-C129 | In the secondary structure of FMN, the first three AGU make hairpin loops and identified from B. cleavage. | [49] |
| ACAGU GGCGGU | A137-U141 G56-U61 |
Form Secondary-structure model of the 165 ribD RNA and side hairpin between P2 and 165 ribD RNA and side hairpin | [49] | |
| RF00174 | CCCGC AGUCAG |
C70-C74 | Predicted secondary structure of the cobalamin riboswitch in the btuB leader region of Synechococcus sp. Strain. The boxed bases represent the B12 box-P1 helix interface, where a CC-to-TT (UU in the RNA structure | [50] |
| RF01055 | GAAAGG | G120-G125 | Containing AGG at the site of Ribosomal Binding Site (RBS) located at multiple junction site. Region of the central multi-stem junction in Sequence of the 138 moaA Moco RNA. | [51] |
| GCCU | G18-U21 | Found in a Moco RNA at left multiple junction site | [51] | |
| GCCUCC | G106-C111 | In Moco RNA, the last UCC makes parts of multiple junctions in P4. | [51] | |
| RF00380 | UGAGG | U28-G32 | k-mers that found in part of a conserved bulge-stem region of Secondary structure of the 5`M-box portion | [52] |
| RF01054 | AAAGG | A83-G87 | Structural modulation and nucleotides comprise a conserved Shine–Dalgarno (SD) | [53] |
| AGCAU | A58-U62 | Unpaired Structural modulation containing constant cleavage | [53] | |
| AGAAAA | A88-A93 | Structural modulation, AGAA in decreasing cleavage and AA in constant cleavage | [53] | |
| RF00234 | AGCGC | A12-C16 | Downstream of the ribozyme cleavage site Ribozyme core site P2a | [54] |
| ACGAGG | A53-G56 | Ribozyme core region (Unpaired) | ||
| RF00167 | CUAC | C50-C53 | structural features of the guanine aptamer domain and critical for metabolite binding | [55] |
Fig 7. Secondary structure of RF00174 Cobalamin riboswitch (Acido bacterium) (A) and RF01055 MOCO riboswitch class (B).
In every individual base, the color gradient scale represents a normalized hit number from 156 features aligned to the sequence. The different color scale in each region represents its coverage of the k-mers in the family that it represents. Whereas, I, H, and T are abbreviations for Interior loops, Helices, and Terminal loops, respectively.
Discussion
Machine learning has an enormous capacity to boost our knowledge in the classification of riboswitch, an area that is still in the early stage of a comprehensive investigation. Numerous machine learning applications have been developed based on different methods to detect riboswitch. However, most riboswitch classification studies applied machine learning algorithms on the imbalanced sequences [25,26]. Several findings revealed the impact of imbalance sequences on correct classification and performance of algorithms [25,26,30]. Chawla and colleagues proposed SMOTE method of treating imbalanced sequences for better classification of majority and minority instances [30,34]. SMOTE based balancing of sequences improves the oversampling minority classes accurately and also produces sequences that do not influence majority class.
In this analysis, there are imbalances of instances in the riboswitch family. Comparative results revealed the reality of the impact of such an imbalance on classification which has widely been reported (S1 Table and Table 1). Imbalanced distribution exhibited variation from 4826 majority class (Cobalamin riboswitch) to 39 minority class (PreQ1-II riboswitch). General classifiers, when encountering such imbalanced data, favor class with majority instances [30,34]. The analysis also revealed in imbalanced and balanced confusion matrix the same problem (Figs 5 and 6). Out of 16 riboswitch class, cobalamin riboswitch, TPP riboswitch (THI element), and glycine riboswitch sum up contribution was 68% while the remaining 13 riboswitch family has 32% instances. In Table 2, full sequences grouped into two sets training (70%) and test set (30%) was selected and performances of classifiers were evaluated regarding sensitivity, accuracy, specificity and F-score. The correlation heat-map in Fig 3 (see detail in S2 Fig) indicates the relationships between k-mers.
Imbalanced sequences in riboswitch showed different performances of classifiers ranked as: MLP—the best and NB—the poorest regarding their mean scores that range from 0.771 to 0.961. In Table 2, individual score results of this method have shown best result in RF00234, RF00522, RF01057 (1.00 in RF): greater values than reported in other study using BLAST+ [26,56], which is most popular tools in analysis of sequence similarity [56] and others [25,26]. Conversion of sequences into vector revealed good results in both groups used for analysis (S2 Table and Tables 1 and 2). In protein study, protein sequence converted into feature vectors showed good performance in cases of SVM and KNN [57–60]. RF00174, RF00059, RF00504, RF00522 predicted better than others with minority classes like RF01054, RF00634, RF00380 (Tables 1 and 2). The class with maximum instances (RF00174) resulted in an F-score value greater than 0.94 in all classifiers except NB, which had a value less than 0.93 in both cases.
NB classifier depicted poor performance in imbalanced sequences compared to other classifiers. Its accuracy, sensitivity, specificity, and F-score had the following values 0.979, 0.989, 0.814 and 0.705, respectively (S2 Table). These results were improved to 0.985, 0.991, 0.841 and 0.771 when sequences were balanced. Compared with the F-score value reported by Hugo and colleagues (NB-HEXCFS- 0.525), the changes indicated the influence of imbalanced sequences on the performance of classifiers. Similarly, improved performance of NB on large sequences has been reported [61].
Table 2, S2 Table, and Fig 4 indicate that the proposed method of balancing instances increases classifier performances. The used approach was also reported as a solution for machine learning [62]. RF shows the best result followed by MLP, which revealed optimal performance. On the other hand, Naïve Bayes has poor performances in imbalanced sequences classification, which is in accordance with Mwagha and colleagues [63,64]. The overall comparison revealed that balanced classifiers are better for classification of riboswitch, their performances were compared to BLAST+ [26] and other finding (S3 Table and Tables 1 and 2).
The k-mers position in the secondary structure illustrated riboswitch biological function and motif (Table 3 and Fig 7). In RF00174, CCCGC k-mers had predicted the secondary structure of the cobalamin riboswitch in the btuB leader region of Synechococcus. In cases like RF00168, UCAU k-mer had motifs predicted to interact with the Nova-1 protein, overlaying K-mers in the 3′ aptamer domain, rich in A, which has unique folding pseudoknot that compresses PreQ1 [40]. Turning off gene expressing observed in RF00162 with GAGGGA k-mer, is a kink-turn motif which allows pseudoknot interaction. It interacts with SAM which helps to make stable formation, and can cause the downstream expression platform to form a rho-independent TT (transcriptional terminator), turning off gene expression [45]. Overall, k-mers and their biological function for this study are summarized and described in Table 3.
The pipeline can be used in machine learning and deep learning study in other domains of bioinformatics and computational biology that suffer from imbalanced sequences. Finally, the scientific community can use the python source code for analysis of interest as well as to develop suitable software packages.
Methodology
We showed a complete evaluation of different machine learning approaches for classification and predicting regulatory riboswitches. First of all, we present the benchmark sequences and data mining approach followed by feature engineering that was done through testing. Besides, model selection methods were used to model and compare balanced and imbalanced sequences problem, as well as determine the best combination of hyperparameters for each classifier [65]. These methods are implemented in an open-source machine learning platform called WEKA 3.8 [66,67], SMOTE [31] and Python 3 [68], which allow evaluating different parameters and algorithms for classification and prediction of the riboswitch. Lastly, we described the results of classifications from the learned models. The workflow for the analysis of imbalanced and balanced sequences used for performance evaluation of different machine learning algorithms found in Fig 1. This workflow can be used for other research areas that suffer from challenges of imbalanced sequences. The python source codes are available at https://github.com/Seasonsling/riboswitch.
Data preprocessing
Sequences for investigation were collected from Rfam 13.0 [19] and other sequences that were already produced [26], intended for comparison of our new methods. Rfam is a source that collects RNA families including riboswitch [19]. There is a need to use a machine learning approach to train algorithms to classify riboswitch as it has been happening in other areas of bioinformatics. Only 16 families have been used to compare with previous research work and they clearly show the impact of imbalanced training sequences on the performance of classifiers. Preprocessing, cleaning and filtering were done, as well as handling missing values, noisy data, redundant features and irrelevant features to affect the accuracy of the model [67]. The sequences that contain sequences per family are shown in S1 Table.
Feature selection
FASTA format sequences were used for k-mer (1 ≤ k ≤ n) frequency counts through executing in the R package called kcount [69]. In order to obtain a sufficiently informative k-mer counting matrix for the task [70], we set k value to 6 and finally got 5,460 features. This k-mers composition was used to make frequencies of each riboswitch. This avoids unnecessary computing power consumption and dimensional disaster caused by extremely sparse matrices due to high k values as well.
Attribute evaluators CfsSubsetEval and BestFirst were used for dimensionality reduction and searching of the space of attribute subsets by greedy hill-climbing augmented with a backtracking facility [71], which was consistent with some other researchers [26]. WEKA 3.8 was used to implement the task [66,67]. Feature selection was done for the dimensionality reduction and thus for decrease processing load [72,73].
Imbalanced data
The sequences for this finding contains the imbalanced sequences ranging from 4,826 instances (RF00174) to 39 (RF01051) instances (S1 Table). Learning from the imbalanced sequences that become critical concerns nowadays, particularly when minority class contains small instances in its sequences [25,26,74]. Mainstream methods of dealing with imbalance data can be roughly divided into two categories. The first category considers the difference in the cost of different misclassifications [75], while the second one mainly focuses on training data sampling strategies. Here over-sampling and under-sampling were conventional techniques used to adjust class distribution. However, traditional random oversampling adopts the strategy of merely copying samples to increase the minority samples, which is prone to the problem of overfitting that makes the information learned by the model over-fitted and not generalized [76].
SMOTE improved scheme based on random oversampling was applied [59]. The basic idea of the SMOTE algorithm is to analyze a small number of samples and to add new samples to the data set based on a small number of samples.
The used algorithm flow is as follows:
For each sample x in a few classes, calculate the distance from all samples in a few samples sets by Euclidean distance, and get its k-nearest neighbors.
Set a sampling ratio according to the sample imbalance ratio to determine the sampling magnification N. For each minority sample x, randomly select several samples from its k-nearest neighbors, assuming that the selected neighbor is .
For each randomly selected neighbor , construct a new sample with the original sample according to the following formula:
| (1) |
SMOTE was deployed through importing “imblearn.over_sampling” module in Python 3 and it was applied both in the corresponding training set of 10-fold cross-validation and building final model processes, as shown in Fig 1.
Machine learning models
A crucial step in machine learning is model selection, as the performance of algorithms is sensitive to the calibration parameters. Configuration and choice of the hyper-parameters are found to be crucial. For our data, we calibrated a model using 10-fold cross-validation. Firstly, the complete feature selection of k-mers sequences was divided into two parts randomly: 70% of data were training set, while 30% of data as the test set. The 70% training set was used to build multiclass classification models and determine the hyper-parameters through 10-fold cross-validation. Then, the test set was used to test the final generalization performance of the balanced and imbalanced models. In order to increase the credibility of comparison results and to ensure the repeatability of the results, all sequences were chosen randomly. Input data and model parameters except for the step of SMOTE processing were strictly consistent for both balanced and imbalanced models. This task was left to make pipeline module and Pipeline object in Python package imblearn (0.5.0), which ensures that in cross-validation or generalization testing, SMOTE only treats the training data used to build the cross-validation model or the final model. By this means, the validation set in each fold cross-validation was consistent in all models just as in the case of the 30% test set.
During the model selection process, for each algorithm, the grid search method was applied to traverse all hyper-parameter combinations, while 10-fold cross-validation method for evaluating each parameter combination. Specifically, the program randomly divided the 70% training set into ten straight sections. During each cycle of the model training step, nine of those sections were treated by SMOTE (the control group not), and then for model training. Subsequent that, the remaining section of the training set will test the model and obtain a series of test indicators, including macro F-score, macro recall and macro precision. The valid score was calculated through the below formula:
| (2) |
Running the above cycle ten times independently, we take the average of ten valid scores as the overall performance index of the model under this parameter combination. After evaluating all the parameter combinations with the grid-search method, we pick the model with the highest comprehensive performance index as the final model.
Experimentation classifiers
Random Forest is a commonly used machine learning algorithm [77] with different successful function in computing and bioinformatics [77–79]. It randomizes the variables (columns) and data (rows), generating thousands of classification trees, and then summarizing the results of the classification tree. In this research, the mean decrease impurity method was used.
SVM is a simple and efficient method for solving the quadratic programming problem [80] through computing the maximum marginal hyper-plane [66]. In SVM, the kernel function implicitly defines the feature space for linear partitioning, which means the choice of kernel function is the largest variable of SVM.
Gradient boosting is a boosting algorithm, which belongs to ensemble learning as well as random forest and proved to have great performance in imbalance problem. It builds the model in a stage-wise fashion, and generalizes them by allowing optimization of an arbitrary differentiable loss function [81].
Another classifier is k-Nearest Neighbors (KNN) which also named IBK (instant-based learning with parameter k). This classifier offers numerous choices to speed up the undertaking to locate nearest neighbors [67], NB (Naïve Bayes) classifier based on Bayes' theorem [49]. This is a probability-based model in Bayesian networks [82]. MLP is another commonly used machine learning algorithms [83]. ncRNA classification and prediction problems have been widely conducted based on the six selected algorithms for this analysis [84–86] and riboswitch classification and prediction [3,26].
The tuning of KNN, SVM, RF, GB and MLP was carried out on the training set by evaluating the macro F-score in Python 3. The configurations of their parameters are as follows:
KNN: number of k = {2, 4, 6, 8, 10, 12, 14, 16}
SVM: type of kernel function = {linear, poly, rbf, sigmoid}
RF: with the method of GridSearchCV and kfold = 10, the number of trees in the forest = {500, 1000, 2000}, the maximum depth of the tree = {10, 15, 20}
GB: with the method of GridSearchCV and kfold = 10, the number of trees in the forest = {500, 1000, 2000}, learning rate = {0.01, 0.1, 0.05}, the maximum depth of the tree = {7, 9, 11, 15}
MLP: with the method of GridSearchCV and kfold = 10, hidden layer size = {{80, 80, 80}, {100, 100, 100}, {150, 150, 150}}, L2 penalty (regularization term) parameter = {1e-3, 1e-4}, the solver for weight optimization = {‘adam’, ‘sgd’}, tolerance for the optimization = {1e-8, 1e-7, 1e-6}
Gaussian NB: portion of the largest variance of all features that is added to variances for calculation stability = {1e-16, 1e-14, 1e-12}
Evaluation
In order to evaluate the performance of the classifiers, the confusion matrices were used to compute sensitivity, specificity, accuracy and F-score [32,87]. Most researchers used a weighted F-score to evaluate the classifier’s overall performance. However, it leads to assessment bias between majority families and minority families. In this evaluation, we used macro F1 instead, which gives an arithmetic mean of the per-class F1-scores and avoids assessment bias to some extent. A statistical test was carried out in GraphPad Prism 8.3.0 using the Wilcoxon rank test and multiple Wilcoxon rank test at p < 0.05, 0.01, 0.001 level (“Wilcoxon rank test were performed using GraphPad Prism version 8.3.0 for Windows, GraphPad Software, La Jolla California USA, www.graphpad.com”).
We used the following abbreviations: True Positives (TP), False Positive (FP), True Negative (TN), and False Negative (FN). The used formulas are as follows:
| (3) |
| (4) |
| (5) |
| (6) |
Supporting information
The training (70%) and test sequences (30%) for classification and evaluation performance of machine learning algorithms. Feature distribution across different 16 riboswitch families using heat-map is shown in Fig 2.
(DOCX)
For a specific classifier, mean represents average sensitivity, specificity, accuracy and F-score value, while standard deviation (SD) depicted variation in different riboswitch families.
(DOCX)
Bolded p-values indicate the statistical difference (SD).
(DOCX)
A) row-normalized k-mer counting distribution, rows correspond to the k-mers, and columns revealed 16 families of riboswitch and B) the clustering heatmap depicts feature clustering, clustered features were essential for classification in that family. Red means a high relatively counting number while blue means lower.
(TIF)
The diagonal white line represented their correlation factor equals to one. Blue means a positive correlation, while red means a negative correlation.
(TIF)
Acknowledgments
We would like to thank Ming Chen's Bioinformatics Group members for assisting whenever needed.
Data Availability
All relevant data are within the manuscript and its Supporting Information files and accessable at https://github.com/Seasonsling/riboswitch.
Funding Statement
This work was supported by the National Key Research and Development Program of China [2018YFC0310602; 2016YFA0501704]; National Natural Science Foundation of China [31571366, 31771477]; the Chinese Government Scholarship for foreign students(MOFCOM), Jiangsu Collaborative Innovation Center for Modern Crop Production, the Fundamental Research Funds for the Central Universities; and the Ministry of Education and Science of the Republic of North Macedonia. Opinions, results, and conclusions articulated in this paper are those of the authors and do not necessarily reflect the views of the supporting organization. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jones CP, Ferré-D'Amaré AR. Long-range interactions in riboswitch control of gene expression. Annual review of biophysics. 2017;46:455–81. 10.1146/annurev-biophys-070816-034042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5(6):451–63. 10.1038/nrm1403 [DOI] [PubMed] [Google Scholar]
- 3.Chen Z, Zhao P, Li F, Marquez-Lago TT, Leier A, Revote J, et al. iLearn: an integrated platform and meta-learner for feature engineering, machine-learning analysis and modeling of DNA, RNA and protein sequence data. Brief Bioinform. 2019. [DOI] [PubMed] [Google Scholar]
- 4.Havill JT, Bhatiya C, Johnson SM, Sheets JD, Thompson JS. A new approach for detecting riboswitches in DNA sequences. Bioinformatics. 2014;30(21):3012–9. 10.1093/bioinformatics/btu479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43(6):867–79. 10.1016/j.molcel.2011.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152(1–2):17–24. 10.1016/j.cell.2012.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodgers ML, Hao Y, Woodson SA. A newborn RNA switches its fate. Nat Chem Biol. 2019;15(11):1031–2. 10.1038/s41589-019-0391-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–34. 10.1146/annurev.biochem.78.070507.135656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2015;43(Database issue):D30–5. 10.1093/nar/gku1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson CJ, Vincent HA, Wu MC, Lowe PT, Dunstan MS, Leys D, et al. Modular riboswitch toolsets for synthetic genetic control in diverse bacterial species. J Am Chem Soc. 2014;136(30):10615–24. 10.1021/ja502873j [DOI] [PubMed] [Google Scholar]
- 11.Abduljalil JM. Bacterial riboswitches and RNA thermometers: Nature and contributions to pathogenesis. Noncoding RNA Res. 2018;3(2):54–63. 10.1016/j.ncrna.2018.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Machtel P, Bakowska-Zywicka K, Zywicki M. Emerging applications of riboswitches—from antibacterial targets to molecular tools. J Appl Genet. 2016;57(4):531–41. 10.1007/s13353-016-0341-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9(6):644–7. 10.1261/rna.5090103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bocobza SE, Aharoni A. Small molecules that interact with RNA: riboswitch‐based gene control and its involvement in metabolic regulation in plants and algae. The Plant Journal. 2014;79(4):693–703. 10.1111/tpj.12540 [DOI] [PubMed] [Google Scholar]
- 15.Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR. Riboswitch control of gene expression in plants by splicing and alternative 3' end processing of mRNAs. Plant Cell. 2007;19(11):3437–50. 10.1105/tpc.107.053645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen M, Harrison A, Shanahan H, Orlov Y. Biological Big Bytes: Integrative Analysis of Large Biological Datasets. J Integr Bioinform. 2017;14(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Q, Meng X, Liao Q, Chen M. Versatile interactions and bioinformatics analysis of noncoding RNAs. Brief Bioinform. 2018. [DOI] [PubMed] [Google Scholar]
- 18.Chang TH, Huang HY, Hsu JB, Weng SL, Horng JT, Huang HD. An enhanced computational platform for investigating the roles of regulatory RNA and for identifying functional RNA motifs. BMC Bioinformatics. 2013;14 Suppl 2:S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalvari I, Argasinska J, Quinones-Olvera N, Nawrocki EP, Rivas E, Eddy SR, et al. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018;46(D1):D335–D42. 10.1093/nar/gkx1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mukherjee S, Das Mandal S, Gupta N, Drory-Retwitzer M, Barash D, Sengupta S. RiboD: a comprehensive database for prokaryotic riboswitches. Bioinformatics. 2019;35(18):3541–3. 10.1093/bioinformatics/btz093 [DOI] [PubMed] [Google Scholar]
- 21.Bengert P, Dandekar T. Riboswitch finder—a tool for identification of riboswitch RNAs. Nucleic Acids Res. 2004;32(Web Server issue):W154–9. 10.1093/nar/gkh352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abreu-Goodger C, Merino E. RibEx: a web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res. 2005;33(Web Server issue):W690–2. 10.1093/nar/gki445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang TH, Huang HD, Wu LC, Yeh CT, Liu BJ, Horng JT. Computational identification of riboswitches based on RNA conserved functional sequences and conformations. RNA. 2009;15(7):1426–30. 10.1261/rna.1623809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuker M. Calculating nucleic acid secondary structure. Curr Opin Struct Biol. 2000;10(3):303–10. 10.1016/s0959-440x(00)00088-9 [DOI] [PubMed] [Google Scholar]
- 25.Singh S, Singh R. Application of supervised machine learning algorithms for the classification of regulatory RNA riboswitches. Brief Funct Genomics. 2017;16(2):99–105. 10.1093/bfgp/elw005 [DOI] [PubMed] [Google Scholar]
- 26.Guillen-Ramirez HA, Martinez-Perez IM. Classification of riboswitch sequences using k-mer frequencies. Biosystems. 2018;174:63–76. 10.1016/j.biosystems.2018.09.001 [DOI] [PubMed] [Google Scholar]
- 27.Compeau PE, Pevzner PA, Tesler G. How to apply de Bruijn graphs to genome assembly. Nature biotechnology. 2011;29(11):987–91. 10.1038/nbt.2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32(5):1792–7. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurtz S, Narechania A, Stein JC, Ware D. A new method to compute K-mer frequencies and its application to annotate large repetitive plant genomes. BMC genomics. 2008;9(1):517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh ND, Dhall A. Clustering and learning from imbalanced data. arXiv preprint arXiv:181100972. 2018.
- 31.McCormick TH, Raftery AE, Madigan D, Burd RS. Dynamic logistic regression and dynamic model averaging for binary classification. Biometrics. 2012;68(1):23–30. 10.1111/j.1541-0420.2011.01645.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Y, Kamel MS, Wong AK, Wang Y. Cost-sensitive boosting for classification of imbalanced data. Pattern Recognition. 2007;40(12):3358–78. [Google Scholar]
- 33.Wu T-F, Lin C-J, Weng RC. Probability estimates for multi-class classification by pairwise coupling. Journal of Machine Learning Research. 2004;5(Aug):975–1005. [Google Scholar]
- 34.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over-sampling technique. Journal of artificial intelligence research. 2002;16:321–57. [Google Scholar]
- 35.Rentzsch R, Deneke C, Nitsche A, Renard BY. Predicting bacterial virulence factors–evaluation of machine learning and negative data strategies. Briefings in Bioinformatics. 2019. [DOI] [PubMed] [Google Scholar]
- 36.Ribeca P, Valiente G. Computational challenges of sequence classification in microbiomic data. Briefings in Bioinformatics. 2011;12(6):614–25. 10.1093/bib/bbr019 [DOI] [PubMed] [Google Scholar]
- 37.Mei S, Li F, Leier A, Marquez-Lago TT, Giam K, Croft NP, et al. A comprehensive review and performance evaluation of bioinformatics tools for HLA class I peptide-binding prediction. Briefings in Bioinformatics. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Cai H, Wang X, Ao L, Guo Y, He J, et al. A rank-based algorithm of differential expression analysis for small cell line data with statistical control. Briefings in Bioinformatics. 2017;20(2):482–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woolson R. Wilcoxon signednedo Y, He J, et al. A rank-based algorithm of differential
- 40.Scott ML, John EE, Bellone RR, Ching JC, Loewen ME, Sandmeyer LS, et al. Redundant contribution of a Transient Receptor Potential cation channel Member 1 exon 11 single nucleotide polymorphism to equine congenital stationary night blindness. BMC veterinary research. 2016;12(1):121 10.1186/s12917-016-0745-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. Structural basis of ligand binding by a c-di-GMP riboswitch. Nature structural & molecular biology. 2009;16(12):1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang M, Peterson R, Feigon J. Structural insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Molecular cell. 2009;33(6):784–90. 10.1016/j.molcel.2009.02.019 [DOI] [PubMed] [Google Scholar]
- 43.Kwon M, Strobel SA. Chemical basis of glycine riboswitch cooperativity. Rna. 2008;14(1):25–34. 10.1261/rna.771608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miranda-Rios J. The THI-box riboswitch, or how RNA binds thiamin pyrophosphate. Structure. 2007;15(3):259–65. 10.1016/j.str.2007.02.001 [DOI] [PubMed] [Google Scholar]
- 45.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441(7097):1172 10.1038/nature04819 [DOI] [PubMed] [Google Scholar]
- 46.Weinberg Z, Regulski EE, Hammond MC, Barrick JE, Yao Z, Ruzzo WL, et al. The aptamer core of SAM-IV riboswitches mimics the ligand-binding site of SAM-I riboswitches. Rna. 2008;14(5):822–8. 10.1261/rna.988608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang JX, Lee ER, Morales DR, Lim J, Breaker RR. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Molecular cell. 2008;29(6):691–702. 10.1016/j.molcel.2008.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nature structural & molecular biology. 2008;15(2):177. [DOI] [PubMed] [Google Scholar]
- 49.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proceedings of the National Academy of Sciences. 2002;99(25):15908–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pérez AA, Rodionov DA, Bryant DA. Identification and regulation of genes for cobalamin transport in the cyanobacterium Synechococcus sp. strain PCC 7002. Journal of bacteriology. 2016;198(19):2753–61. 10.1128/JB.00476-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Regulski EE, Moy RH, Weinberg Z, Barrick JE, Yao Z, Ruzzo WL, et al. A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism. Molecular microbiology. 2008;68(4):918–32. 10.1111/j.1365-2958.2008.06208.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dann CE III, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130(5):878–92. 10.1016/j.cell.2007.06.051 [DOI] [PubMed] [Google Scholar]
- 53.Meyer MM, Roth A, Chervin SM, Garcia GA, Breaker RR. Confirmation of a second natural preQ1 aptamer class in Streptococcaceae bacteria. Rna. 2008;14(4):685–95. 10.1261/rna.937308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428(6980):281 10.1038/nature02362 [DOI] [PubMed] [Google Scholar]
- 55.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113(5):577–86. 10.1016/s0092-8674(03)00391-x [DOI] [PubMed] [Google Scholar]
- 56.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hong J, Luo Y, Zhang Y, Ying J, Xue W, Xie T, et al. Protein functional annotation of simultaneously improved stability, accuracy and false discovery rate achieved by a sequence-based deep learning. Briefings in bioinformatics. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li YH, Xu JY, Tao L, Li XF, Li S, Zeng X, et al. SVM-Prot 2016: a web-server for machine learning prediction of protein functional families from sequence irrespective of similarity. PloS one. 2016;11(8):e0155290 10.1371/journal.pone.0155290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu B, Qiu W, Chen C, Ma A, Jiang J, Zhou H, et al. SubMito-XGBoost: predicting protein submitochondrial localization by fusing multiple feature information and eXtreme gradient boosting. Bioinformatics. 2019. [DOI] [PubMed] [Google Scholar]
- 60.Yu C, Li X, Yang H, Li Y, Xue W, Chen Y, et al. Assessing the performances of protein function prediction algorithms from the perspectives of identification accuracy and false discovery rate. International journal of molecular sciences. 2018;19(1):183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Douglass S, Hsu SW, Cokus S, Goldberg RB, Harada JJ, Pellegrini M. A naive Bayesian classifier for identifying plant microRNAs. Plant J. 2016;86(6):481–92. 10.1111/tpj.13180 [DOI] [PubMed] [Google Scholar]
- 62.He H, Garcia EA. Learning from imbalanced data. IEEE Transactions on knowledge and data engineering. 2009;21(9):1263–84. [Google Scholar]
- 63.Mwagha SM, Muthoni M, Ochieg P. Comparison of nearest neighbor (ibk), regression by discretization and isotonic regression classification algorithms for precipitation classes prediction. International Journal of Computer Applications. 2014;96(21):44–8. [Google Scholar]
- 64.Gong H, Liu X, Wu J, He Z. Data construction for phosphorylation site prediction. Brief Bioinform. 2014;15(5):839–55. 10.1093/bib/bbt012 [DOI] [PubMed] [Google Scholar]
- 65.Japkowicz N, Stephen S. The class imbalance problem: A systematic study. Intelligent data analysis. 2002;6(5):429–49. [Google Scholar]
- 66.Witten IH, Frank E, Hall MA, Pal CJ. Data Mining: Practical machine learning tools and techniques: Morgan Kaufmann; 2016. [Google Scholar]
- 67.Han J, Kamber M, Pei J. Data mining concepts and techniques third edition. The Morgan Kaufmann Series in Data Management Systems. 2011:83–124. [Google Scholar]
- 68.Hamelryck T, Manderick B. PDB file parser and structure class implemented in Python. Bioinformatics. 2003;19(17):2308–10. 10.1093/bioinformatics/btg299 [DOI] [PubMed] [Google Scholar]
- 69.Edgar RC. Local homology recognition and distance measures in linear time using compressed amino acid alphabets. Nucleic Acids Research. 2004;32(1):380–5. 10.1093/nar/gkh180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fang J. A critical review of five machine learning-based algorithms for predicting protein stability changes upon mutation. Briefings in bioinformatics. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watkins AM, Rangan R, Das R. Using Rosetta for RNA homology modeling. Methods in enzymology. 2019;623:177–207. 10.1016/bs.mie.2019.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saghir H, Megherbi DB, editors. An efficient comparative machine learning-based metagenomics binning technique via using Random forest. 2013 IEEE International Conference on Computational Intelligence and Virtual Environments for Measurement Systems and Applications (CIVEMSA); 2013: IEEE.
- 73.Ditzler G, Morrison JC, Lan Y, Rosen GL. Fizzy: feature subset selection for metagenomics. BMC Bioinformatics. 2015;16:358 10.1186/s12859-015-0793-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin W, Xu D. Imbalanced multi-label learning for identifying antimicrobial peptides and their functional types. Bioinformatics. 2016;32(24):3745–52. 10.1093/bioinformatics/btw560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paper D, Paper D. Scikit-Learn Classifier Tuning from Complex Training Sets. Hands-on Scikit-Learn for Machine Learning Applications: Data Science Fundamentals with Python. 165-88.
- 76.He H, Garcia EA. Learning from Imbalanced Data IEEE Transactions on Knowledge and Data Engineering v. 21 n. 9. September; 2009.
- 77.Guyon I, Elisseeff A. An introduction to variable and feature selection. Journal of machine learning research. 2003;3(Mar):1157–82. [Google Scholar]
- 78.An Y, Wang J, Li C, Leier A, Marquez-Lago T, Wilksch J, et al. Comprehensive assessment and performance improvement of effector protein predictors for bacterial secretion systems III, IV and VI. Brief Bioinform. 2018;19(1):148–61. 10.1093/bib/bbw100 [DOI] [PubMed] [Google Scholar]
- 79.Song J, Li F, Takemoto K, Haffari G, Akutsu T, Chou K-C, et al. PREvaIL, an integrative approach for inferring catalytic residues using sequence, structural, and network features in a machine-learning framework. Journal of theoretical biology. 2018;443:125–37. 10.1016/j.jtbi.2018.01.023 [DOI] [PubMed] [Google Scholar]
- 80.Keerthi SS, Gilbert EG. Convergence of a generalized SMO algorithm for SVM classifier design. Machine Learning. 2002;46(1–3):351–60. [Google Scholar]
- 81.Friedman JH. Greedy function approximation: a gradient boosting machine. Annals of statistics. 2001:1189–232. [Google Scholar]
- 82.Cheng I. Hybrid Methods for Feature Selection. 2013. [Google Scholar]
- 83.Zhang GP. Neural networks for classification: a survey. IEEE Transactions on Systems, Man, and Cybernetics, Part C (Applications and Reviews). 2000;30(4):451–62. [Google Scholar]
- 84.Liu B, Zhang D, Xu R, Xu J, Wang X, Chen Q, et al. Combining evolutionary information extracted from frequency profiles with sequence-based kernels for protein remote homology detection. Bioinformatics. 2014;30(4):472–9. 10.1093/bioinformatics/btt709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Panwar B, Arora A, Raghava GP. Prediction and classification of ncRNAs using structural information. BMC genomics. 2014;15(1):127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kong L, Zhang Y, Ye Z-Q, Liu X-Q, Zhao S-Q, Wei L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic acids research. 2007;35(suppl_2):W345–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sokolova M, Lapalme G. A systematic analysis of performance measures for classification tasks. Information processing & management. 2009;45(4):427–37. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The training (70%) and test sequences (30%) for classification and evaluation performance of machine learning algorithms. Feature distribution across different 16 riboswitch families using heat-map is shown in Fig 2.
(DOCX)
For a specific classifier, mean represents average sensitivity, specificity, accuracy and F-score value, while standard deviation (SD) depicted variation in different riboswitch families.
(DOCX)
Bolded p-values indicate the statistical difference (SD).
(DOCX)
A) row-normalized k-mer counting distribution, rows correspond to the k-mers, and columns revealed 16 families of riboswitch and B) the clustering heatmap depicts feature clustering, clustered features were essential for classification in that family. Red means a high relatively counting number while blue means lower.
(TIF)
The diagonal white line represented their correlation factor equals to one. Blue means a positive correlation, while red means a negative correlation.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files and accessable at https://github.com/Seasonsling/riboswitch.