Abstract
To monitor the incidence of Plasmodium knowlesi infections and determine whether other simian malaria parasites are being transmitted to humans, we examined 1,047 blood samples from patients with malaria at Kapit Hospital in Kapit, Malaysia, during June 24, 2013–December 31, 2017. Using nested PCR assays, we found 845 (80.6%) patients had either P. knowlesi monoinfection (n = 815) or co-infection with other Plasmodium species (n = 30). We noted the annual number of these zoonotic infections increased greatly in 2017 (n = 284). We identified 6 patients, 17–65 years of age, with P. cynomolgi and P. knowlesi co-infections, confirmed by phylogenetic analyses of the Plasmodium cytochrome c oxidase subunit 1 gene sequences. P. knowlesi continues to be a public health concern in the Kapit Division of Sarawak, Malaysian Borneo. In addition, another simian malaria parasite, P. cynomolgi, also is an emerging cause of malaria in humans.
Keywords: Malaria, zoonoses, Plasmodium cynomolgi, Plasmodium knowlesi, parasitic diseases, Malaysian Borneo, parasites
Plasmodium spp. were identified in the late 1800s, and >30 species have been described in primates, including humans, apes, and monkeys (1,2). Of these, humans are natural hosts to 4 species: P. falciparum, P. malariae, P. vivax, and P. ovale. Human infections with simian malaria parasites were thought to be extremely rare until P. knowlesi was identified as a major cause of malaria in humans in Kapit, Malaysian Borneo (3). Subsequent human cases of infection with P. knowlesi have been reported across Southeast Asia (4–9). Most cases are reported in the Malaysian Borneo states of Sarawak and Sabah (4,10,11). The zoonotic capability of this parasite was confirmed with the aid of molecular techniques because P. knowlesi is morphologically similar to P. malariae (12). Molecular detection methods also were used to identify other zoonotic malaria parasites infecting humans, such as P. simium (13,14) and P. brasilianum (15) in South America and P. cynomolgi (16,17) in Southeast Asia.
After the large focus on human P. knowlesi infections in the Kapit Division of Sarawak state in 2004 (3), extended studies on wild long-tailed macaques (Macaca fascicularis) and pig-tailed macaques (M. nemestrina) in the area found these species harbor 6 simian malaria parasites: P. inui, P. knowlesi, P. cynomolgi, P. coatneyi, P. fieldi, and P. simiovale (12,18). Among 108 macaques examined, P. inui was the most prevalent parasite (82%), along with P. knowlesi (78%) and P. cynomolgi (56%). Besides P. knowlesi, 2 other simian parasites, P. inui and P. cynomolgi, have zoonotic capabilities that have been proven through accidental and experimental infections (1,19–21). P. inui was experimentally reported to infect humans in 1938, with a subsequent report in 1966 (20,21). P. cynomolgi was reported to infect humans during an accidental transmission in a laboratory in the United States in 1956 and later by experimental trials (1,19). A single infection of naturally acquired P. cynomolgi in a human was described in Peninsular Malaysia in 2014 (16) and was confirmed through molecular characterization.
Successful transmission of zoonotic malaria is highly dependent on the bionomics and distribution of competent vectors (22). After the 2004 report on human P. knowlesi cases in Kapit, Anopheles latens mosquitoes were incriminated as the only vector for P. knowlesi in the area and were found to harbor sporozoites of other species of simian malaria parasites (23). Because wild macaques in Kapit harbored potentially zoonotic malaria parasites and vectors were transmitting P. knowlesi to humans, we aimed to determine whether human infections with P. cynomolgi and P. inui also occurred in the Kapit Division of Sarawak, Malaysian Borneo.
Materials and Methods
Study Area, Participants, and Detection of Plasmodium spp.
Kapit is the largest administrative division in Sarawak and has an area of 38,934 km2. Most residents in Kapit belong to indigenous ethnic groups living in longhouses close to the forests. Inhabitants who are ill travel to Kapit Hospital for diagnosis and treatment. We enrolled 1,047 patients who had malaria from the Kapit and Song districts who were admitted to Kapit Hospital during June 24, 2013–December 31, 2017 (Figure 1).
Figure 1.
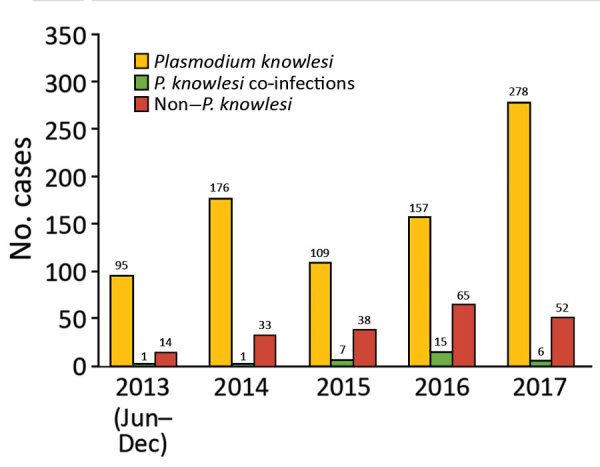
Number of patients admitted to Kapit Hospital with malaria during June 24, 2013–December 31, 2017, Malaysian Borneo. Non–P. knowlesi includes P. falciparum, P. malariae, P. ovale, and P. vivax. Infections with Plasmodium spp. other than P. knowlesi each year included the following. In 2013, P. knowlesi coinfections included 1 P. cynomolgi co-infection; non–P. knowlesi included 9 P. falciparum, 4 P. vivax, and 1 P. ovale. In 2014, P. knowlesi mixed included 1 P. falciparum coinfection; non–P. knowlesi included 14 P. falciparum, 4 P. malariae, 12 P. vivax, 3 P. ovale. In 2015, P. knowlesi co-infections included 3 P. falciparum and 4 P. vivax co-infections; non–P. knowlesi included 16 P. falciparum, 16 P. vivax, 1 P. malariae, 3 P. falciparum/P. vivax co-infections, 1 P. falciparum/P. ovale co-infection, and 1 P. vivax/P. ovale co-infection. In 2016, P. knowlesi co-infections included 8 P. falciparum, 6 P. vivax, and 1 P. cynomolgi co-infections; non–P. knowlesi included 41 P. falciparum, 14 P. vivax, 1 P. ovale, 3 P. falciparum/P. vivax co-infections, 1 P. falciparum/P. malariae co-infection, 4 P. falciparum/P. ovale co-infections, and 1 P. vivax/P. ovale co-infection. In 2017, P. knowlesi co-infections included 4 P. cynomolgi, and 2 P. vivax co-infections; non–P. knowlesi included 26 P. falciparum, 19 P. vivax, 3 P. malariae, 2 P. ovale, 1 P. falciparum/P. vivax co-infection, and 1 P. falciparum/P. malariae co-infection.
Malaria was diagnosed at Kapit Hospital by using blood film examination. Approximately 2 mL of venous blood was collected from each patient. Blood spots on filter paper and thick and thin films were prepared, and 500 µL aliquots of blood were stored frozen. We received samples at the Malaria Research Centre, Universiti Malaysia Sarawak, for further analyses. We extracted Plasmodium DNA from dried blood spots by using Instagene Matrix (BioRad Laboratories, https://www.bio-rad.com). We used nested PCR assays to test for the presence of different Plasmodium species by using primers specific for P. falciparum, P. vivax, P. malariae, P. ovale, P. knowlesi, P. inui, P. cynomolgi, P. fieldi, and P. coatneyi as described previously (3,24). We extracted genomic DNA from frozen blood samples by using the QIAamp DNA Mini Kit (QIAGEN, https://www.qiagen.com) and used it for molecular characterization of malaria parasites.
We obtained written consent from all enrolled patients or their parents or guardians for patients <17 years of age. Ethics clearance for this study was obtained from the medical research ethics committee of Universiti Malaysia Sarawak (approval nos. UNIMAS/TNC(AA)-03.02/06-Jld.2[24] and UNIMAS/NC-21.02/03-02 Jld.2[19]) and from the medical research and ethics committee of the Ministry of Health Malaysia (approval nos. NMRR-12-1086-13607[IIR] and NMRR-16-943-31224[IIR]).
PCR Amplification and Sequencing of Partial Cytochrome c Oxidase Subunit 1 Gene
Sequencing of the partial cytochrome c oxidase subunit 1 (COXI) gene of P. cynomolgi involved a 3-step PCR rather than the 2-step PCR used for P. knowlesi. First, we amplified the complete mitochondrial genome by PCR using Plasmodium-specific primers Pkmt F1 and Pkmt R1 (24). We performed PCR amplification for each sample in a 25-µL reaction mixture containing 1× PCR buffer for KOD FX Neo, 0.4 mmol dNTP mix (Toyobo, https://www.toyobo-global.com), 0.3 µmol of each primer (Pkmt F1 and Pkmt R1), 1 U/25 µL KOD FX Neo polymerase (Toyobo), and 3 µL of purified genomic DNA under the following conditions: 98°C for 30 s for first denaturation; 35 cycles at 98°C for 30 s, 55°C for 30 s, and 72°C for 5 min; then a final extension for 10 min at 72°C. We used S.N.A.P UV Gel Purification Kit (Invitrogen, https://www.thermofisher.com) to perform gel purification of the PCR amplified mitochondrial DNA (mtDNA) whole genome.
We then used the purified mtDNA amplicons as a template for the heminested touchdown PCR (TD-PCR) with P. cynomolgi-specific primers: cox1_F1 (5′-CCAAGCCTCACTTATTGTTAAT-3′), cox1_R1 (5′-ACCAAATAAAGTCATTGTTGATCC-3′), and cox1_R3 (5′-ATGGAAATGAGCAATTACATAG-3′). The heminested 1 (N1) TD-PCR amplification for each sample was performed in an 11-µL reaction mixture containing 1× colorless PCR buffer, 2 mmol MgCl2, 0.2 mmol dNTP mix, 0.25 µmol of each primer (cox1_F1 and cox1_R1), 0.275 U GoTaq G2 Flexi DNA Polymerase (Promega, https://www.promega.com), and 0.5 µL of purified mtDNA template under the following conditions: 94°C for 3 min for first denaturation; 10 cycles at 94°C for 30 s, 62°C for 45 s, and 72°C for 85 s; 25 cycles at 94°C for 30 s, 53°C for 45 s, and 72°C for 85 s; then a final extension for 5 min at 72°C. After N1 TD-PCR, we performed a heminested 2 (N2) TD-PCR amplification for each of the PCR products in a 22 µL reaction mixture containing similar concentration of PCR master-mix recipe with cox1_F1 and cox1_R1 primers and 1 µL of purified mtDNA template under the following conditions: 94°C for 3 min for first denaturation, 10 cycles at 94°C for 30 s, 59°C for 45 s, and 72°C for 85 s; 25 cycles at 94°C for 30 s, 53°C for 45 s, and 72°C for 85 s; then a final extension for 5 min at 72°C. We used the S.N.A.P UV Gel Purification Kit (Invitrogen) to gel purify the amplicon.
We performed TD-PCR amplification of the P. knowlesi COXI fragment by using P. knowlesi–specific primers: cox1_Pk_F1 (5′-CATATCCAAGCCTCATTTATGA-3′) and cox1_Pk_R1 (5′-GTGAAAATGAGCAATTACATAA-3′). Amplification was performed in a 22 µL reaction mixture containing similar concentration of PCR master-mix recipe with cox1_F1 and cox1_R1 primers and 1 µL of purified mtDNA template. We used the following thermal cycling parameters: 94°C for 3 min for first denaturation; 10 cycles at 94°C for 30 s, 62°C for 45 s, and 72°C for 85 s; 25 cycles at 94°C for 30 s, 56°C for 45 s, and 72°C for 85 s; then a final extension for 5 min at 72°C. We purified the amplicon as we mentioned in the previous step.
We cloned the purified COXI fragments into TOPO TA Cloning Kit with pCR2.1-TOPO vector (Invitrogen) according to the manufacturer’s protocol. We purified the recombinant plasmids containing the COXI fragment by using the S.N.A.P. Plasmid DNA MiniPrep Kit (Invitrogen) and sent these to a commercial facility for DNA sequencing. For all samples, >3 clones each for P. cynomolgi and P. knowlesi were sequenced and both forward and reverse strands were sequenced for each clone.
Computational Analyses of COXI of Plasmodium spp.
We used ClustalX version 2 to align the COXI sequences (25). We inferred phylogenetic relationships by using the neighbor-joining method in MEGA7 (26) then by Bayesian Markov chain Monte Carlo (MCMC) method in the BEAST package version 1.7.5 (27). We confirmed the convergence of the chain by inspecting the MCMC samples using Tracer version 1.5 (https://tracer-1-5.software.informer.com), discarded the first 10% sampling of the MCMC chains as burn-in, and conformed the sample size to >200 for all continuous parameters. We used Tree Annotator to annotate the tree generated by BEAST and visualized the maximum clade credibility tree by using FigTree version 1.3.1 (https://figtree-1-3-1.software.informer.com). We deposited Plasmodium COXI sequences generated in this study in GenBank under accession nos. MN372324–61 (Appendix).
Microscopy
We stained thick and thin blood films with Giemsa by using pH 7.2 buffer and examined films under an Olympus BX53 microscope (Olympus, https://www.olympus-lifescience.com). We estimated parasitemia by examining thick blood films and counting parasites associated with <500 leukocytes by 2 independent microscopists. We converted the parasite count to a count per microliter of blood by using the actual leukocyte count of each patient measured with a Sysmex KX-21N Kobe hematology analyzer (Sysmex Corporation, https://www.sysmex.com). We used a light microscope to observe the morphologic characteristics of the parasites in thin blood films.
Results
Patient Demographics and Identification of Malaria Parasites
Of the 1,047 patients, 845 (80.7%) had P. knowlesi monoinfections or co-infections with other species of Plasmodium, based on nested PCR assays (Figure 1; Table 1). Most (730; 86.3%) patients were >20 years of age, 91 (11%) were 12–19 years of age, and 24 (3%) were <12 years of age; 540 (51.5%) were female and 507 (48.5%) were male. Six patients (codes K07, KT46, K199, K221, K222, and K229) had co-infections with P. knowlesi and P. cynomolgi (Table 2) without prior history of malaria, and Kapit Hospital initially diagnosed P. knowlesi by microscopy. Throughout the 5-year study, we observed a remarkable increase in Plasmodium knowlesi malaria cases. In 2017 alone, 284 cases were identified, >2 times that of the highest annual number of cases, 122, admitted during a previous clinical study at Kapit Hospital during 2006–2008 (28).
Table 1. Comparison of microscopy and PCR for identifying Plasmodia spp. among patients with malaria during June 24, 2013–December 31, 2017, Kapit Hospital, Kapit, Malaysian Borneo*.
| Microscopy | PCR |
Total | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pf | Pk | Pm | Pv | Po | Pf + Pv | Pf + Pm | Pf + Po | Pk + Pf | Pk + Pv | Pk + Pcy | Pv + Po | ||
| P. falciparum | 93 | 4 | 0 | 2 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 103 |
| P. knowlesi | 7 | 810 | 1 | 12 | 2 | 2 | 1 | 1 | 12 | 11 | 6 | 0 | 865 |
| P. malariae | 6 | 1 | 7 | 2 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 19 |
| P. ovale | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 5 |
|
P. vivax
|
0 |
0 |
0 |
48 |
3 |
1 |
0 |
1 |
0 |
1 |
0 |
1 |
55 |
| Total | 106 | 815 | 8 | 65 | 7 | 7 | 2 | 5 | 12 | 12 | 6 | 2 | 1,047 |
* Pcy, Plasmodium cynomolgi; Pf, P. falciparum; Pk, P. knowlesi; Pm, P. malariae; Po, P. ovale; Pv, P. vivax.
Table 2. Parasitemia and characteristics of patients with diagnosed Plasmodium cynomolgi and P. knowlesi coinfections, Malaysian Borneo.
| Case no. | Year detected | Age, y/sex | Parasites/µL blood |
|---|---|---|---|
| K222 | 2017 | 36/F | 213 |
| KT46 | 2013 | 18/M | 334 |
| K229 | 2017 | 63/F | 1,386 |
| K221 | 2017 | 50/F | 13,661 |
| K199 | 2017 | 17/M | 17,072 |
| K07 | 2016 | 65/M | 84,299 |
Despite the high number of P. knowlesi malaria cases, 19% (202) of malaria cases in Kapit Hospital were infected by human Plasmodium species (non–simian Plasmodium parasites). During 2013–2016, 2–3 indigenous infections were reported annually, but no indigenous infections were recorded in 2017. Based on travel history, 95% (192/202) of patients infected with nonsimian Plasmodium species were timber camp workers who had returned to Sarawak after working in Africa, Papua New Guinea, Brazil, and the Solomon Islands.
Molecular Characterization of P. cynomolgi and P. knowlesi Coinfections
To confirm the P. cynomolgi and P. knowlesi coinfections, we obtained Plasmodium DNA sequences of 38 partial COXI genes (1,088–1,144 bp) from the 6 patients. The Bayesian phylogenetic inference of these DNA sequences showed 20 sequences formed a monophyletic clade with P. cynomolgi referral sequences and was distinct from the P. vivax clade. The remaining 18 sequences, however, formed 2 distinct subclades with the P. knowlesi referral sequences (Figure 2), confirming the presence of P. knowlesi and P. cynomolgi parasites in these patients.
Figure 2.
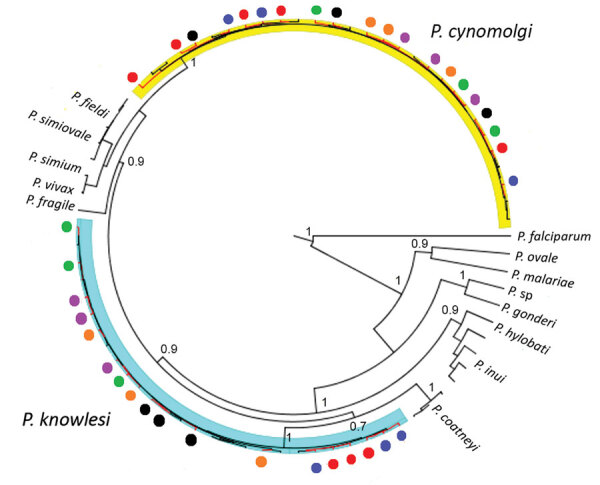
Maximum clade credibility tree for Plasmodium cytochrome c oxidase subunit 1 (COXI) sequences from samples from patients admitted to Kapit Hospital with malaria during June 24, 2013–December 31, 2017, Malaysian Borneo. Tree was generated by using strict clock model and Bayesian skyline coalescent tree prior. Circles indicate COXI sequences derived from patients: red indicates patient KT46; black indicates patient K07; orange indicates patient K199; purple indicates patient K221; blue indicates patient K222; and green indicates patient K229.
Morphological Characterization of P. cynomolgi and P. knowlesi Co-infections
Parasitemia in the 6 patients with P. cynomolgi and P. knowlesi co-infections ranged from 213 to 84,299 parasites/μL blood (Table 2). Only patients K07 and K199 had blood films of high enough quality for morphologic characterization of Plasmodium species. On examination, we noted P. cynomolgi–infected erythrocytes constituted a small proportion of the malaria-infected erythrocytes, consisting of ≈1.5% of infected erythrocytes for patients K07 and 4.7% for patient K199. We estimated the percentage of P. cynomolgi–infected erythrocytes by using the number of early trophozoite stages with Schüffner’s stippling because we only observed the trophozoite stage for P. cynomolgi in these 2 patients. We distinguished P. cynomolgi from P. knowlesi parasites based on morphologic characteristics of infected erythrocytes, as previously described (1). We observed Schüffner’s stippling in early trophozoite-infected erythrocytes, which were enlarged and distorted at times (Figures 3,4). The trophozoites of P. cynomolgi also had either single, double, or triple chromatin dots. The growing trophozoites of P. knowlesi did not cause erythrocyte enlargement and we did not observe Schüffner’s stippling (Figure 4, panels K, L). Furthermore, we observed band-form trophozoites of P. knowlesi in certain erythrocytes (Figure 3, panel I; Figure 4, panel M), and we noted 16 merozoites in 1 schizont (Figure 3, panel J). For patient K07, we examined 4,154 erythrocytes; 12 (0.3%) had prominent Schüffner’s stippling, but we did not observe malaria parasites within these erythrocytes (Figure 5).
Figure 3.
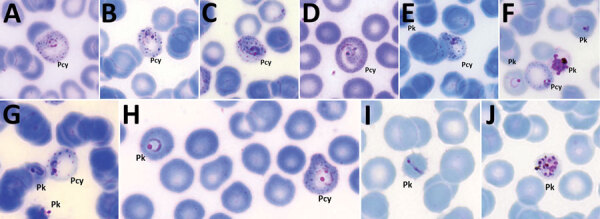
Plasmodium cynomolgi and P. knowlesi parasites in patient K07, admitted to Kapit Hospital, Kapit, Malaysia, with malaria in 2016. A–G) Early trophozoites of P. cynomolgi in enlarged and, at times, distorted erythrocytes, with Schüffner’s stippling and either single, double, or triple chromatin dots. E–H) P. knowlesi and P. cynomolgi early trophozoites. I) Band form trophozoite of P. knowlesi. J) Schizont of P. knowlesi. Pcy, P. cynomolgi; Pk, P. knowlesi. Original magnification ×100.
Figure 4.
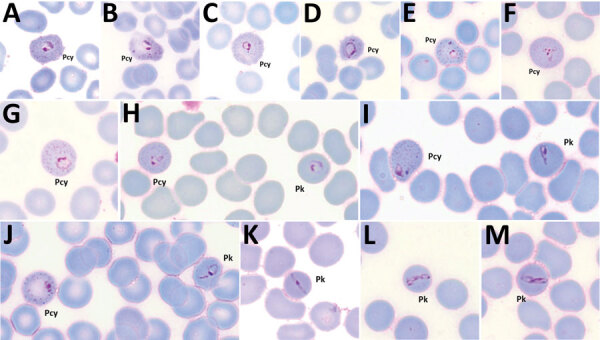
Plasmodium cynomolgi and P. knowlesi parasites in patient K199, admitted to Kapit , Kapit, Malaysia, with malaria in 2017. A–G) Early trophozoites of P. cynomolgi within enlarged and, at times, distorted erythrocytes, with Schüffner’s stippling and single, double, or triple chromatin dots. H–J) Early trophozoites of P. knowlesi and P. cynomolgi. K–M) Band form trophozoites of P. knowlesi. Pcy, P. cynomolgi; Pk, P. knowlesi. Magnification ×100.
Figure 5.
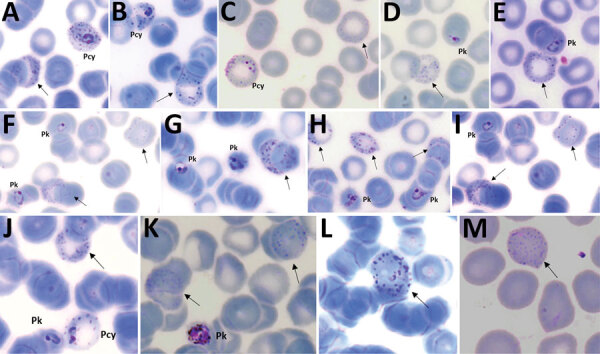
Plasmodium cynomolgi and P. knowlesi parasites in patient K07, admitted to Kapit , Kapit, Malaysia, with malaria in 2016. Arrows indicate Schüffner’s stippling in erythrocytes without parasites. A–C) Early trophozoites of P. cynomolgi; D–I) early trophozoites of P. knowlesi; J) early trophozoites of P. knowlesi and P. cynomolgi; K) gametocyte of P. knowlesi. Pcy, P. cynomolgi; Pk, P. knowlesi. Magnification ×100.
Discussion
The increasing number of patients admitted to Kapit Hospital with P. knowlesi infections represents a true increase in infections, not increased awareness, because we used nested PCR to examine Plasmodium DNA from blood spots on filter paper from all patients during the study period. The increasing incidence of P. knowlesi in the Kapit Division, particularly in 2017; the few patients infected with human malaria parasites, P. falciparum, P. vivax, P. ovale, and P. malariae, during June 2013–December 2016; and the lack of indigenous cases in Kapit in 2017 reflect the recent malaria situation in Malaysian Borneo and Peninsular Malaysia (11). The Ministry of Health, Malaysia, reported 7,745 cases of P. knowlesi malaria in Malaysia in 2017 and 2018, and 87% (6,743) occurred in the Malaysian Borneo states of Sarawak and Sabah (29). In 2017, only 85 cases of indigenous malaria caused by the human malaria parasites, P. falciparum, P. vivax, P. ovale, or P. malariae were reported, and none were reported in 2018 (30).
The increase in P. knowlesi malaria cases, particularly in Malaysian Borneo, is multifactorial and driven partly by anthropogenic land-use factors leading to changes in the transmission pattern of the parasite between humans, vectors, and macaque reservoirs (31). Both states in Malaysian Borneo have undergone substantial deforestation, primarily for palm oil plantations; the total area used for palm oil plantations in Sarawak and Sabah in 2018 was 3.12 million hectares (32) compared with just 2.3 million hectares in 2010 (33). Such extensive forest clearing activities result in loss of natural habitat for macaques and could have caused them to move closer to human settlements. In Sabah, an association between deforestation and increased incidence of human cases of P. knowlesi malaria has been demonstrated (34). Furthermore, predictive spatial scale analysis of data from a case-controlled study, a cross-sectional survey, and satellite imagery showed that landscape fragmentation influences P. knowlesi transmission to humans (35). However, similar studies need to be undertaken in Sarawak to determine whether a direct relationship between land change use and number of P. knowlesi malaria cases exists because the vectors in Sarawak are different from those in Sabah.
Entomologic studies also need to be undertaken because vector bionomics is influenced by landscape factors, and abrupt land use conversion or modifications can change vector distribution, invasion, and behavior (36,37). One such study conducted in the Kinabatangan area of Sabah in Malaysian Borneo demonstrated that, in a 2-year period Anopheles donaldi mosquitoes displaced An. balabacensis mosquitoes, the previously dominant vector in that area (38). Changes in host preference, biting behavior, and adaptation of the mosquitoes to habitat changes could affect the transmission of zoonotic malaria in Malaysian Borneo. However, determining whether there have been changes in mosquito bionomics in Sarawak is difficult because only 1 study on vector bionomics was conducted in Kapit District in 2004. New detailed entomologic studies at various locations clearly are necessary.
In the 2 patients whose blood films we examined, P. cynomolgi parasites comprised ≈1.5% and 4.7% of the total malaria parasites, which explains why microscopy only detected single P. knowlesi infections in these patients. The dominance of P. knowlesi over P. cynomolgi is probably due to the differences in the period of development in the liver and the duration of the erythrocytic cycle between the 2 species. Studies undertaken on the Mulligan (M) strain, and the B strain of P. cynomolgi indicated that the incubation period in the liver varied from 15 to 20 days for the B strain and 16 to 37 days for the M strain (1), which is much longer than the estimated incubation period for P. knowlesi of 9–12 days (1). Furthermore, the asexual erythrocytic cycle of P. knowlesi is only 24 hours but is 48 hours for P. cynomolgi (1,4). Hence, if P. knowlesi and P. cynomolgi co-infection occur simultaneously through a single mosquito bite, P. knowlesi parasites would emerge from the liver before P. cynomolgi and replicate rapidly to become the dominant species infecting the person.
Unlike P. knowlesi infections, which can be severe and sometimes fatal (4), P. cynomolgi infections in humans are mild or even asymptomatic (1). In a series of experiments conducted in the United States in the 1960s, volunteers infected with P. cynomolgi had mild infections, and for 1 participant the infection persisted for 58 days (1,39,40). A natural infection was reported in a woman with fever in Peninsular Malaysia (16), and a tourist from Denmark also had a mild infection with P. cynomolgi after returning home from travels in Southeast Asia (41). Recently, naturally acquired P. cynomolgi and P. knowlesi co-infections in Cambodia (17) and P. cynomolgi monoinfections in Sabah, Malaysian Borneo (42), were all asymptomatic. However, our study is hospital-based, so all patients who had P. cynomolgi with P. knowlesi co-infections were symptomatic.
A recent study described the morphologic characteristics of young to near-mature trophozoites of P. cynomolgi in a naturally acquired human host (41). We observed only young trophozoites of P. cynomolgi in enlarged and at times distorted erythrocytes with Schüffner’s stippling. We observed erythrocytes with stippling without malaria parasites in 1 of the patients. This phenomenon, termed pitting, was initially described for P. knowlesi in the 1960s during animal experiments (43,44). Pitting is a process by which the malaria parasite is expelled from an infected erythrocyte as it passes through the spleen. Once deparasitized through pitting, the erythrocytes return to circulation in the blood. The presence of pitting also was discovered in acute P. falciparum infections in which erythrocytes with ring-infected erythrocyte surface antigen and no intracellular parasites in circulation were observed (45–47). The presence of enlarged deparasitized erythrocytes with Schüffner’s stippling from patient K07 suggests P. cynomolgi parasites might have been pitted, leaving the once-infected erythrocytes circulating in the patient’s blood.
The morphologic similarities between P. cynomolgi and P. vivax have been described previously (1,16), highlighting the difficulty in correctly identifying P. cynomolgi by microscopy. In our study and recent reports of naturally acquired human P. cynomolgi infections in Cambodia, Malaysia, and Southeast Asia, molecular methods were necessary to identify P. cynomolgi (17,41,42). Microscopy, or rapid diagnostic tests, which are the main methods used in malaria surveys, would have identified the P. cynomolgi monoinfections in our study as P. vivax. Therefore, the incidence of P. cynomolgi in Asia, the natural habitat for several monkey hosts (1), is probably much higher than what has been reported so far. Continued surveillance by using molecular methods is needed to obtain accurate data for the incidence of not only P. cynomolgi, but also P. knowlesi.
In conclusion, we observed that P. knowlesi continues to be of public health concern in the Kapit Division of Sarawak, Malaysian Borneo, and that another simian malaria parasite, P. cynomolgi, also is an emerging cause of malaria in humans. Further epidemiologic and entomologic studies using molecular tools and a multidisciplinary approach need to be undertaken in Southeast Asia to determine the incidence of P. cynomolgi and P. knowlesi. Such studies also will assist in determining whether these parasites continue to cause zoonotic infections or whether the loss of natural habitat of macaques, coupled with changes in mosquito composition, abundance, and feeding behavior and an increase in the human population, result in their switch to humans as the preferred hosts.
Appendix. Accession numbers of the cytochrome c oxidase subunit 1 gene sequences of Plasmodium included in the phylogenetic analysis, including those generated in this study.
Acknowledgments
We thank colleagues at the Malaria Research Centre, UNIMAS, and the medical laboratory technologists, doctors, and nurses at Kapit Hospital for assistance, and the Director-General of Health in Malaysia for permission to publish this paper.
This study was supported by a postgraduate scholarship from the Ministry of Higher Education in Malaysia to T.H.H., and grants from UNIMAS (nos. 05-FA052000-0606-0002, F05/DPD/1793/2019, and F05/DPP/1505/2016).
Biography
Dr. Nada Raja is a post-doctoral researcher at the Malaria Research Centre in UNIMAS, Malaysia. Her research interests include molecular evolution, population genetics, and genomics of malaria parasites.
Footnotes
Suggested citation for this article: Nada Raja T, Hu TH, Kadir KA, Mohamad DSA, Rosli N, Wong LL, et al. Naturally acquired human Plasmodium cynomolgi and Plasmodium knowlesi infections, Malaysian Borneo. Emerg Infect Dis. 2020 Aug [date cited]. https://doi.org/10.3201/eid2608.200343
References
- 1.Coatney GR, Collins WE, Warren M, Contacos PG. The primate malarias. Washington (DC): US National Institute of Allergy and Infectious Diseases; 1971. [Google Scholar]
- 2.Ramasamy R. Zoonotic malaria - global overview and research and policy needs. Front Public Health. 2014;2:123. 10.3389/fpubh.2014.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh B, Kim Sung L, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–24. 10.1016/S0140-6736(04)15836-4 [DOI] [PubMed] [Google Scholar]
- 4.Singh B, Daneshvar C. Human infections and detection of Plasmodium knowlesi. Clin Microbiol Rev. 2013;26:165–84. 10.1128/CMR.00079-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaw MT, Lin Z. Human Plasmodium knowlesi infections in South-East Asian countries. J Microbiol Immunol Infect. 2019;52:679–84. 10.1016/j.jmii.2019.05.012 [DOI] [PubMed] [Google Scholar]
- 6.Maeno Y, Culleton R, Quang NT, Kawai S, Marchand RP, Nakazawa S. Plasmodium knowlesi and human malaria parasites in Khan Phu, Vietnam: Gametocyte production in humans and frequent co-infection of mosquitoes. Parasitology. 2017;144:527–35. 10.1017/S0031182016002110 [DOI] [PubMed] [Google Scholar]
- 7.Iwagami M, Nakatsu M, Khattignavong P, Soundala P, Lorphachan L, Keomalaphet S, et al. First case of human infection with Plasmodium knowlesi in Laos. PLoS Negl Trop Dis. 2018;12:e0006244. 10.1371/journal.pntd.0006244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pongvongsa T, Culleton R, Ha H, Thanh L, Phongmany P, Marchand RP, et al. Human infection with Plasmodium knowlesi on the Laos-Vietnam border. Trop Med Health. 2018;46:33. 10.1186/s41182-018-0116-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lubis IND, Wijaya H, Lubis M, Lubis CP, Divis PCS, Beshir KB, et al. Contribution of Plasmodium knowlesi to multispecies human malaria infections in North Sumatera, Indonesia. J Infect Dis. 2017;215:1148–55. 10.1093/infdis/jix091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox-Singh J, Davis TME, Lee KS, Shamsul SSG, Matusop A, Ratnam S, et al. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin Infect Dis. 2008;46:165–71. 10.1086/524888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hussin N, Lim YAL, Goh PP, William T, Jelip J, Mudin RN. Updates on malaria incidence and profile in Malaysia from 2013 to 2017. Malar J. 2020;19:55. 10.1186/s12936-020-3135-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee KS, Cox-Singh J, Singh B. Morphological features and differential counts of Plasmodium knowlesi parasites in naturally acquired human infections. Malar J. 2009;8:73. 10.1186/1475-2875-8-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brasil P, Zalis MG, de Pina-Costa A, Siqueira AM, Júnior CB, Silva S, et al. Outbreak of human malaria caused by Plasmodium simium in the Atlantic Forest in Rio de Janeiro: a molecular epidemiological investigation. Lancet Glob Health. 2017;5:e1038–46. 10.1016/S2214-109X(17)30333-9 [DOI] [PubMed] [Google Scholar]
- 14.de Alvarenga DAM, Culleton R, de Pina-Costa A, Rodrigues DF, Bianco C Jr, Silva S, et al. An assay for the identification of Plasmodium simium infection for diagnosis of zoonotic malaria in the Brazilian Atlantic Forest. Sci Rep. 2018;8:86. 10.1038/s41598-017-18216-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lalremruata A, Magris M, Vivas-Martínez S, Koehler M, Esen M, Kempaiah P, et al. Natural infection of Plasmodium brasilianum in humans: Man and monkey share quartan malaria parasites in the Venezuelan Amazon. EBioMedicine. 2015;2:1186–92. 10.1016/j.ebiom.2015.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ta TH, Hisam S, Lanza M, Jiram AI, Ismail N, Rubio JM. First case of a naturally acquired human infection with Plasmodium cynomolgi. Malar J. 2014;13:68. 10.1186/1475-2875-13-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imwong M, Madmanee W, Suwannasin K, Kunasol C, Peto TJ, Tripura R, et al. Asymptomatic natural human infections with simian malaria parasites Plasmodium cynomolgi and Plasmodium knowlesi. J Infect Dis. 2019;219:695–702. 10.1093/infdis/jiy519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nada Raja T, Hu TH, Zainudin R, Lee KS, Perkins SL, Singh B. Malaria parasites of long-tailed macaques in Sarawak, Malaysian Borneo: a novel species and demographic and evolutionary histories. BMC Evol Biol. 2018;18:49. 10.1186/s12862-018-1170-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheong WH, Coombs GL. Transmission of Plasmodium cynomolgi (Perlis strain) to man. Southeast Asian J Trop Med Public Health. 1970;1:302. [Google Scholar]
- 20.Gupta JMD. Transmission of Plasmodium inui to man. Proc Natl Inst Sci India. 1938;4:241–4. [Google Scholar]
- 21.Coatney GR, Chin W, Contacos PG, King HK. Plasmodium inui, a quartan-type malaria parasite of Old World monkeys transmissible to man. J Parasitol. 1966;52:660–3. 10.2307/3276423 [DOI] [PubMed] [Google Scholar]
- 22.Garamszegi LZ. Patterns of co-speciation and host switching in primate malaria parasites. Malar J. 2009;8:110. 10.1186/1475-2875-8-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan CH, Vythilingam I, Matusop A, Chan ST, Singh B. Bionomics of Anopheles latens in Kapit, Sarawak, Malaysian Borneo in relation to the transmission of zoonotic simian malaria parasite Plasmodium knowlesi. Malar J. 2008;7:52. 10.1186/1475-2875-7-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee KS, Divis PCS, Zakaria SK, Matusop A, Julin RA, Conway DJ, et al. Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques. PLoS Pathog. 2011;7:e1002015. 10.1371/journal.ppat.1002015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 26.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daneshvar C, Davis TME, Cox-Singh J, Rafa’ee MZ, Zakaria SK, Divis PCS, et al. Clinical and laboratory features of human Plasmodium knowlesi infection. Clin Infect Dis. 2009;49:852–60. 10.1086/605439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ministry of Health Malaysia. Annual report of malaria elimination progress and activities 2018. Kuala Lumpur: the Ministry; 2018.
- 30.World Health Organization. World malaria report 2018. Geneva: The Organization; 2018. [cited 2020 Jan 28]. https://www.who.int/malaria/publications/world-malaria-report-2018/report
- 31.Davidson G, Chua TH, Cook A, Speldewinde P, Weinstein P. The role of ecological linkage mechanisms in Plasmodium knowlesi transmission and spread. EcoHealth. 2019;16:594–610. 10.1007/s10393-019-01395-6 [DOI] [PubMed] [Google Scholar]
- 32.Malaysian Palm Oil Board. Economic and Industry Development Division. Overview of Malaysian palm oil industry 2018. Bangi, Malaysia: The Board; 2018. [cited 2020 Jan 16]. http://bepi.mpob.gov.my/images/overview/Overview_of_Industry_2018.pdf
- 33.Malaysian Palm Oil Board. Economic and Industry Development Division. Overview of Malaysian palm oil industry 2010. Bangi, Malaysia: The Board; 2010. [cited 2020 Jan 16]. http://bepi.mpob.gov.my/images/overview/Overview_of_Industry_2010.pdf
- 34.Fornace KM, Herman LS, Abidin TR, Chua TH, Daim S, Lorenzo PJ, et al. Exposure and infection to Plasmodium knowlesi in case study communities in Northern Sabah, Malaysia and Palawan, The Philippines. PLoS Negl Trop Dis. 2018;12:e0006432. 10.1371/journal.pntd.0006432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brock PM, Fornace KM, Grigg MJ, Anstey NM, William T, Cox J, et al. Predictive analysis across spatial scales links zoonotic malaria to deforestation. Proc Biol Sci. 2019;286:20182351. 10.1098/rspb.2018.2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guerra CA, Snow RW, Hay SI. A global assessment of closed forests, deforestation and malaria risk. Ann Trop Med Parasitol. 2006;100:189–204. 10.1179/136485906X91512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lambin EF, Tran A, Vanwambeke SO, Linard C, Soti V. Pathogenic landscapes: interactions between land, people, disease vectors, and their animal hosts. Int J Health Geogr. 2010;9:54. 10.1186/1476-072X-9-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vythilingam I, Chan ST, Shanmugratnam C, Tanrang H, Chooi KH. The impact of development and malaria control activities on its vectors in the Kinabatangan area of Sabah, East Malaysia. Acta Trop. 2005;96:24–30. 10.1016/j.actatropica.2005.06.022 [DOI] [PubMed] [Google Scholar]
- 39.Kuvin SFK, Beye HK, Stohlman F Jr, Contacos PG, Coatney GR. Clinical and physiological responses in sporozoite-induced B strain Plasmodium cynomolgi and Plasmodium vivax infections in normal volunteers. Trans R Soc Trop Med Hyg. 1962;56:371–8. 10.1016/0035-9203(62)90007-X [DOI] [PubMed] [Google Scholar]
- 40.Coatney GR, Elder HA, Contacos PG, Getz ME, Greenland R, Rossan RN, et al. Transmission of the M strain of Plasmodium cynomolgi to man. Am J Trop Med Hyg. 1961;10:673–8. 10.4269/ajtmh.1961.10.673 [DOI] [PubMed] [Google Scholar]
- 41.Hartmeyer GN, Stensvold CR, Fabricius T, Marmolin ES, Hoegh SV, Nielsen HV, et al. Plasmodium cynomolgi as cause of malaria in tourist to Southeast Asia, 2018. Emerg Infect Dis. 2019;25:1936–9. 10.3201/eid2510.190448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grignard L, Shah S, Chua TH, William T, Drakeley CJ, Fornace KM. Natural human infections with Plasmodium cynomolgi and other malaria species in an elimination setting in Sabah, Malaysia. J Infect Dis. 2019;220:1946–9. 10.1093/infdis/jiz397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conrad ME, Dennis LH. Splenic function in experimental malaria. Am J Trop Med Hyg. 1968;17:170–2. 10.4269/ajtmh.1968.17.170 [DOI] [PubMed] [Google Scholar]
- 44.Schnitzer B, Sodeman T, Mead ML, Contacos PG. Pitting function of the spleen in malaria: ultrastructural observations. Science. 1972;177:175–7. 10.1126/science.177.4044.175 [DOI] [PubMed] [Google Scholar]
- 45.Angus BJ, Chotivanich K, Udomsangpetch R, White NJ. In vivo removal of malaria parasites from red blood cells without their destruction in acute falciparum malaria. Blood. 1997;90:2037–40. 10.1182/blood.V90.5.2037 [DOI] [PubMed] [Google Scholar]
- 46.Newton PN, Chotivanich K, Chierakul W, Ruangveerayuth R, Teerapong P, Silamut K, et al. A comparison of the in vivo kinetics of Plasmodium falciparum ring-infected erythrocyte surface antigen-positive and -negative erythrocytes. Blood. 2001;98:450–7. 10.1182/blood.V98.2.450 [DOI] [PubMed] [Google Scholar]
- 47.Buffet PA, Safeukui I, Deplaine G, Brousse V, Prendki V, Thellier M, et al. The pathogenesis of Plasmodium falciparum malaria in humans: insights from splenic physiology. Blood. 2011;117:381–92. 10.1182/blood-2010-04-202911 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix. Accession numbers of the cytochrome c oxidase subunit 1 gene sequences of Plasmodium included in the phylogenetic analysis, including those generated in this study.


