Abstract
After a sharp decrease of influenza A(H7N9) virus in China in 2018, highly pathogenic H7N9 viruses re-emerged in 2019. These H7N9 variants exhibited a new predominant subclade and had been cocirculating at a low level in eastern and northeastern China. Several immune escape mutations and antigenic drift were observed in H7N9 variants.
Keywords: evolution, H7N9 viruses, antigenic drift, influenza virus, high pathogenicity, low pathogenicity, HPAI, viruses, China, influenza, highly pathogenic avian influenza
Since emerging in China in 2013, influenza A(H7N9) viruses have continued to circulate in mainland China, sporadically causing human infection (1–3). As of February 2020, a total of 1,568 laboratory-confirmed human cases and 616 related deaths had been reported, for a fatality rate of ≈40% (http://www.fao.org/ag/againfo/programmes/en/empres/H7N9/situation_update.html). In mid-2016, a highly pathogenic avian influenza (HPAI) virus of subtype H7N9 emerged, and the number of cases in humans began to rise sharply during a fifth wave (4,5). Animal studies indicated that these HPAI H7N9 viruses are highly virulent in chickens and have gained transmissibility among ferrets (5–7). Also, the cocirculation of HPAI (H7N9) viruses caused high genetic diversity and host adaption (8), posing public health concerns.
Although HPAI H7N9 viruses spread widely across China in 2017 (8,9), after an influenza H5/H7 bivalent vaccine for poultry was introduced in September 2017, the prevalence of the H7N9 viruses in birds and humans decreased dramatically (6,10). In early 2019, when the novel HPAI H7N9 viruses re-emerged, the isolation of HPAI H7N9 viruses from birds revealed them to be responsible for continuous epidemics in northeastern China (11). In March 2019, a human death in Gansu, China, was confirmed to have been caused by an H7N9 virus (12). To explore the prevalence and evolution of influenza A(H7N9) viruses, we sequenced 28 hemagglutinin (HA) and neuraminidase (NA) genes of poultry-origin H7N9 viruses circulating in China during 2019.
The Study
During January–December 2019, we conducted poultry surveillance for influenza virus at live poultry markets in 15 provinces of China (Appendix Figure 9). We isolated 28 H7N9 viruses from tracheal and cloacal swab samples of chickens in Shandong, Hebei, and Liaoning Provinces (Figure 1, panel C; Appendix Table 1). Vaccination of all chickens in China was compulsory according to the Ministry of Agriculture and Rural Affairs of the People’s Republic of China. We sequenced the HA and NA genes of 28 H7N9 viruses and submitted the sequences to GISAID (https://www.gisaid.org) (Appendix Table 2). All H7N9 viruses had 4 continuous basic amino acids at cleavage sites (i.e., KRKRTAR/G and KRKRIAR/G), suggestive of high pathogenicity. Phylogenic analysis demonstrated that the HA and NA genes of all of these HPAI H7N9 viruses belonged to the Yangtze River Delta lineage and formed a new subclade (Figure 1, panel A), which exhibited a long genetic distance to the HPAI H7N9 viruses that persisted during 2017–2018. In particular, the HA and NA genes of A/chicken/northeast China/19376-E5/2019(H7N9), A/chicken/northeast China/19254/2019(H7N9), and A/chicken/northeast China/LN190408A/2019(H7N9) were genetically closely related to the human-infecting influenza A(H7N9) viruses from Gansu (Figure 1, panel B; Appendix Figures 1–3), implying the potential risk for the reemerged HPAI H7N9 viruses to infect humans.
Figure 1.
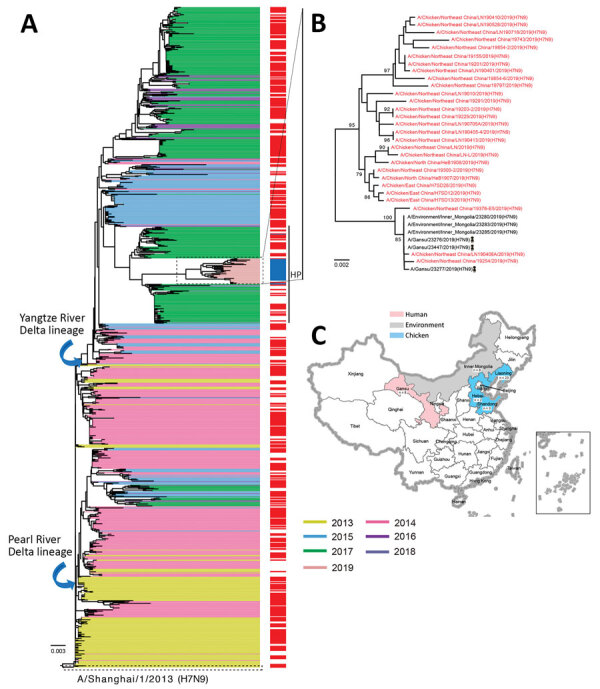
Evolutionary history of influenza A(H7N9) viruses, China, 2017–2019. A) Phylogenic tree of the hemagglutinin gene of H7N9 viruses. Colors indicate reference H7N9 viruses (n = 1,038) from each wave together with the H7N9 isolates from this study (panel B). Red on the right of the tree indicates isolates from humans. All branch lengths are scaled according to the numbers of substitutions per site. The tree was rooted by using A/Shanghai/1/2013(H7N9), which was collected in February 2013. B) Hemagglutinin gene tree revealing a single cluster of highly pathogenic H7N9 viruses circulating during 2019. Red indicates the H7N9 isolates from this study. Scale bar represents number of nucleotide substitutions per site. C) Distribution of highly pathogenic influenza A(H7N9) viruses during 2019. The backgrounds indicate the sampling spaces of highly pathogenic influenza A(H7N9) viruses during 2019 in humans (red), environment (gray), and chickens (blue). The map was designed by using ArcGIS Desktop 10.4 software (ESRI, http://www.esri.com).
A root-to-tip regression analysis of temporal structure revealed aspects of the clock-like structure of 189 H7N9 viruses (correlation coefficient 0.89; R2 0.95) during 2013–2019 (Figure 2, panel A). The epidemic HPAI H7N9 viruses had circulated in China since 2017 and can be classified into 2 sublineages, A and B. The HA and NA genes of the HPAI H7N9 viruses in 2019 belonged to a new sublineage B, whereas the HPAI H7N9 viruses circulating in 2017–2018 grouped into sublineage A (Figure 2, panel B; Appendix Figures 4, 5). Using the evolutionary rates of HA and NA, we estimated the times of origin (95% highest population density) of HPAI H7N9 viruses in sublineage B, which were September 2017–June 2018 for HA and April 2017–May 2018 for NA. Our HPAI H7N9 isolates exhibited traits of sublineages B-1 and B-2. We observed that the HPAI H7N9 viruses in eastern and northeastern China belonged to sublineage B-2 (Figure 2, panel B). However, in mid-2019, the HPAI H7N9 viruses continued to evolve and formed sublineage B-1, which suggested that the estimated times to the most recent common ancestors were May 2019 for HA genes and February 2019 for NA genes. Also, the human- and chicken-origin HPAI H7N9 viruses from Liaoning, Gansu, and Inner Mongolia clustered together in sublineage B-1. These results indicate that the poultry-origin H7N9 virus in sublineage B-1 emerged before the human spillover event in March 2019.
Figure 2.
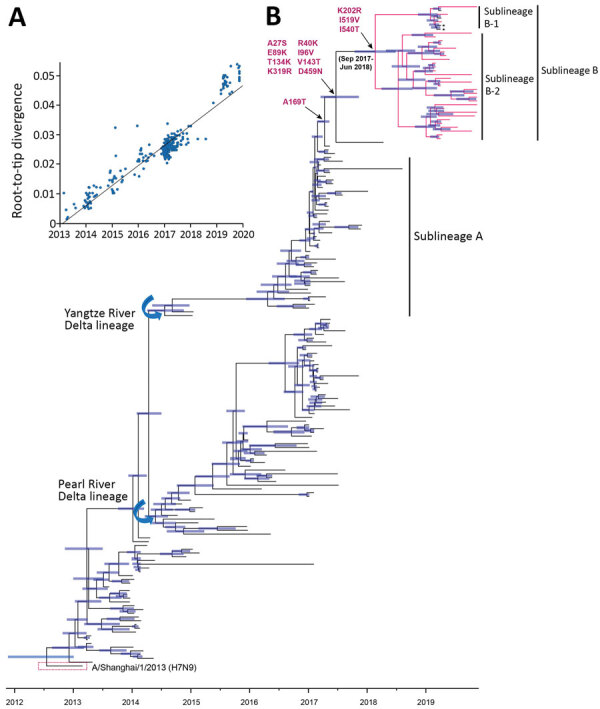
Time-scaled evolution of influenza A(H7N9) viruses, China. A) Analysis of root-to-tip divergence against sampling date for the hemagglutinin gene segment (n = 189). B) Maximum clade credibility tree of the hemagglutinin sequence of H7N9 viruses sampled in China (n = 189); the H7N9 viruses collected in this study are highlighted in red. Asterisk indicates viruses from a human with H7N9 infection within sublineage B during March 2019. Shaded bars represent the 95% highest probability distribution for the age of each node. Parallel amino acid changes along the trunk are indicated.
Although no substantial difference surfaced in the substitution rate of HA genes between H7N9 viruses during 2017–2018 and the viruses during 2019, the increased substitution rate occurred in the first and second codons of reemerged HPAI H7N9 viruses (Appendix Table 4). In a maximum clade credibility tree of the HA gene, 9 independently occurring mutations gave rise to the new sublineage-B circulating in 2019, including A9S, R22K, E71K, I78V, T116K, V125T, A151T, K301R, D439N (H7 numbering, https://www.fludb.org/brc/haNumbering.spg) (Figure 2, panel B), and only the V125T and A151T substitutions of the HA protein were reported as immune escape mutations (13). In addition, sublineage B-1 appeared to have acquired 3 parallel K184R, I499V, I520T (H7 numbering) mutations. The prevailing K184R substitutions of HPAI H7N9 viruses occurred during 2019. The K184R mutation was located in the antigenic site B and receptor binding region (Appendix Figure 6), suggesting that K184R was a potential mediator of viral antigenicity.
We used a hemagglutinin inhibition assay with an antigen of 15 H7N9 viruses circulating during 2017–2019, along with specific antiserum of 6 H7N9 viruses and 2 commonly used reassortant inactivated vaccines, H7N9-Re-2 and H7N9-rGD76, as controls. Antiserum from chickens vaccinated with H7N9-Re-2 strains showed high titers (9–10 log2) and with H7N9-rGD76 strains showed low titers (4–8 log2) to the HPAI H7N9 viruses circulating during 2018–2019 (Table 1). Moreover, the cross–hemagglutinin inhibition assay suggested statistically significant antigenic differences between the HPAI H7N9 viruses circulating during 2017 and during 2018–2019 (Table 2; Appendix Figure 7), indicative of antigenic drift of the reemerged HPAI H7N9 viruses. H7N9-Re-2 and H7N9-rGD76 inactivated vaccines have been widely used in chicken populations in mainland China since 2019 (14). Of note, we found that the virus shedding of chickens vaccinated with H7N9-Re-2 and H7N9-rGD76 against HPAI H7N9 viruses during 2019 ranged from 30% to 80% (Appendix Table 3); therefore, a timely update of H7N9 vaccine is needed.
Table 1. Results of hemagglutinin inhibition assay in study of evolution and antigenic drift of influenza A(H7N9) viruses, China, 2019*.
| Antigen | Antiserum, titer |
|||||||
|---|---|---|---|---|---|---|---|---|
| H7N9-Re-2† | H7N9-rGD76† | 181115 | H7SD12 | H71903‡ | LN19010 | 19225 | 19294 | |
| H7N9-Re-2 | 1,024 | 2,048 | 256 | 512 | 512 | 1,024 | 1,024 | 2,048 |
| H7N9-rGD76 | 128 | 1,024 | 128 | 512 | 256 | 256 | 256 | 128 |
| 181115 | 64 | 256 | 1,024 | 256 | 256 | 256 | 128 | 128 |
| H7SD12 | 64 | 256 | 256 | 1,024 | 1,024 | 2048 | 1,024 | 1,024 |
| H71903 | 64 | 512 | 256 | 1,024 | 1,024 | 1,024 | 2048 | 1,024 |
| LN19010 | 32 | 256 | 32 | 512 | 512 | 512 | 512 | 512 |
| 19225 | 32 | 256 | 64 | 1,024 | 512 | 1,024 | 512 | 1,024 |
| 19294 | 32 | 256 | 64 | 512 | 512 | 1,024 | 512 | 512 |
| 19300–1 | 32 | 256 | 64 | 1,024 | 512 | 1,024 | 512 | 512 |
| 19743 | 16 | 64 | 16 | 512 | 256 | 256 | 256 | 128 |
| 19797 | 16 | 32 | 16 | 256 | 128 | 128 | 64 | 128 |
| 19854–2 | 16 | 64 | 16 | 512 | 256 | 256 | 256 | 256 |
| 19854–6 | 16 | 64 | 16 | 256 | 256 | 512 | 256 | 256 |
| LN191012 | 16 | 64 | 16 | 512 | 256 | 256 | 128 | 128 |
| AH191005 | 32 | 128 | 32 | 1,024 | 512 | 256 | 256 | 256 |
*181115, A/chicken/northeast China/181115/2018(H7N9); H7SD12, A/chicken/east China/H7SD12/2019(H7N9); HeB1908, A/chicken/north China/HeB1908/2019(H7N9); LN19010, A/chicken/northeast China/LN19010/2019(H7N9); 19225, A/chicken/northeast China/19225/2019(H7N9); 19294, A/chicken/northeast China/19294/2019(H7N9); 19300–1, A/chicken/northeast China/19300–1/2019(H7N9); 19743, A/chicken/northeast China/19743/2019(H7N9); 19797, A/chicken/northeast China/197971/2019(H7N9); 19854–2, A/chicken/northeast China/19854–2/2019(H7N9); 19854–6, A/chicken/northeast China/19854–6/2019(H7N9); LN19010, A/chicken/northeast China/LN19010/2019(H7N9); LN191012, A/chicken/northeast China/LN191012/2019(H7N9); AH191005, A/chicken/east China/AH191005/2019(H7N9). †H7N9-Re-2 and H7N9-rGD76 are vaccine strains widely used in China; both antigen and antiserum of H7N9-Re-2 were purchased from the Harbin Weike Biotechnology Development Company (www.hvriwk.com), the antigen of H7N9-Re-2 was available from reassortant avian influenza virus trivalent vaccine. The antigen and antiserum of H7N9-rGD76 were available from Guangzhou South China Biologic Medicine (http://www.gzscbm.com). ‡H71903 is candidate vaccine strain containing the hemagglutinin and neuraminidase genes from H7SD12 and 6 internal genes from A/duck/Guangdong/D7/2007(H5N2). H7SD12, 181115, HeB1908, LN19010, 19225, 19294, 19300–1, 19743, 19797, 19854–2, 19854–6, LN19010, LN191012, and AH191005 are highly pathogenic H7N9 strains in this study.
Table 2. r values of cross-hemagglutinin inhibition assay in study of evolution and antigenic drift of influenza A(H7N9) viruses, China, 2019*.
| Strain |
Antiserum, r value |
|||||||
| H7N9-Re-2 |
H7N9-rGD76 |
181115 |
H7SD12 |
H71903 |
LN19010 |
19225 |
19294 |
|
| H7N9-Re-2 | 1 | 0.5 | 0.25 | 0.18 | 0.18 | 0.25 | 0.25 | 0.35 |
| H7N9-rGD76 | 0.5 | 1 | 0.18 | 0.35 | 0.35 | 0.35 | 0.35 | 0.25 |
| 181115 | 0.25 | 0.18 | 1 | 0.25 | 0.25 | 0.13 | 0.13 | 0.13 |
| H7SD12 | 0.18 | 0.35 | 0.25 | 1 | 1 | 1.41 | 1.41 | 1 |
| H71903 | 0.18 | 0.35 | 0.25 | 1 | 1 | 1 | 1.41 | 1 |
| LN19010 | 0.25 | 0.35 | 0.13 | 1.41 | 1 | 1 | 1.41 | 1.41 |
| 19225 | 0.25 | 0.35 | 0.13 | 1.41 | 1.41 | 1.41 | 1 | 1.41 |
| 19294 | 0.35 | 0.25 | 0.13 | 1 | 1 | 1.41 | 1.41 | 1 |
*H7N9-Re-2 and H7N9-rGD76 are vaccine strains widely used in China; both antigen and antiserum of H7N9-Re-2 were purchased from the Harbin Weike Biotechnology Development Company (http://www.hvriwk.com). The antigen of H7N9-Re-2 was available from reassortant avian influenza virus trivalent vaccine. The antigen and antiserum of H7N9-rGD76 were available from Guangzhou South China Biologic Medicine (http://www.gzscbm.com). H71903 is candidate vaccine strain containing the hemagglutinin and neuraminidase genes from H7SD12 and 6 internal genes from A/duck/Guangdong/D7/2007(H5N2). 181115, H7SD12, LN19010, 19225, and 19294 are highly pathogenic H7N9 strains in this study. r values indicate antigenic relatedness. r>1 indicates no significant antigenic difference between the strains; r = 1 indicates the same antigenicity; r<0.5 indicates a statistically significant antigenic difference between the strains. 181115, A/chicken/northeast China/181115/2018(H7N9); H7SD12, A/chicken/east China/H7SD12/2019(H7N9); LN19010, A/chicken/northeast China/LN19010/2019(H7N9); 19225, A/chicken/northeast China/19225/2019(H7N9); 19294, A/chicken/northeast China/19294/2019(H7N9).
Next, we evaluated the protective efficacy of the new candidate H7N9 inactivated vaccine (H71903)—that is, reverse genetic recombinant carrying HA and NA of A/chicken/east China/H7SD12/2019(H7N9) with internal genes of A/duck/Guangdong/D7/2007(H5N2)—in chickens against the challenge of 4 HPAI H7N9 viruses prevailing in sublineage B in 2019. All of the control chickens challenged with the H7N9 viruses died within 6 days of challenge (Appendix Figure 8). However, virus shedding was not detected from any of the vaccinated chickens challenged with H7N9 viruses (Appendix Table 3), indicating that the new candidate H7N9 vaccine could provide sound protection for chickens against challenge with these reemerged H7N9 variants.
Conclusions
Our findings highlight that the HPAI H7N9 viruses that reemerged during 2019 had been cocirculating at a low level in eastern and northeastern China after the vaccination strategy was implemented. These HPAI H7N9 viruses continued to evolve and showed antigenic drift, posing a public health concern. Although vaccination can largely control the occurrence of H7N9 virus outbreaks, it can also accelerate the generation of novel variants. Therefore, comprehensive surveillance and enhancement of biosecurity precautions should be undertaken immediately to prevent the influenza virus epidemic from becoming a pandemic.
Supplemental results from study of evolution and antigenic drift of influenza A(H7N9) viruses, China, 2019.
Acknowledgments
We acknowledge all contributors who submitted the sequence data on which this research is based to the GISAID EpiFlu Database. All submitters of data may be contacted directly through the GISAID website (http://www.gisaid.org).
This work was supported by the Key Research and Development Program of Guangdong Province (2019B020218004), the National Natural Science Foundation of China (31672586, 31830097, and 319410014), the Earmarked Fund for China Agriculture Research System (CARS-41-G16), the Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2018, to W.Q.), and the Young Scholars of Yangtze River Scholar Professor Program (2019, to W.Q.).
Biography
Dr. Zhang is a PhD student at South China Agricultural University, Guangzhou, China. His research interests are the epidemiology and pathogenesis of emerging and re-emerging infectious diseases.
Footnotes
Suggested citation for this article: Zhang J, Ye H, Li H, Ma K, Qiu W, Chen Y, et al. Evolution and antigenic drift of influenza A(H7N9) viruses, China, 2017–2019. Emerg Infect Dis. 2020 Aug [date cited]. https://doi.org/10.3201/eid2608.200244
These authors contributed equally to this article.
References
- 1.Kang M, Lau EHY, Guan W, Yang Y, Song T, Cowling BJ, et al. Epidemiology of human infections with highly pathogenic avian influenza A(H7N9) virus in Guangdong, 2016 to 2017. Euro Surveill. 2017;22:22. 10.2807/1560-7917.ES.2017.22.27.30568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Jiang H, Wu P, Uyeki TM, Feng L, Lai S, et al. Epidemiology of avian influenza A H7N9 virus in human beings across five epidemics in mainland China, 2013-17: an epidemiological study of laboratory-confirmed case series. Lancet Infect Dis. 2017;17:822–32. 10.1016/S1473-3099(17)30323-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013;381:1926–32. 10.1016/S0140-6736(13)60938-1 [DOI] [PubMed] [Google Scholar]
- 4.Shi J, Deng G, Kong H, Gu C, Ma S, Yin X, et al. H7N9 virulent mutants detected in chickens in China pose an increased threat to humans. Cell Res. 2017;27:1409–21. 10.1038/cr.2017.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi W, Jia W, Liu D, Li J, Bi Y, Xie S, et al. Emergence and adaptation of a novel highly pathogenic H7N9 influenza virus in birds and humans from a 2013 human-infecting low-pathogenic ancestor. J Virol. 2018;92:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi J, Deng G, Ma S, Zeng X, Yin X, Li M, et al. Rapid evolution of H7N9 highly pathogenic viruses that emerged in China in 2017. Cell Host Microbe. 2018;24:558–568.e7. 10.1016/j.chom.2018.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao L, Bi Y, Wong G, Qi W, Li F, Lv Q, et al. Diverse biological characteristics and varied virulence of H7N9 from Wave 5. Emerg Microbes Infect. 2019;8:94–102. 10.1080/22221751.2018.1560234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quan C, Shi W, Yang Y, Yang Y, Liu X, Xu W, et al. New threats from H7N9 influenza virus: spread and evolution of high- and low-pathogenicity variants with high genomic diversity in wave five. J Virol. 2018;92:92. 10.1128/JVI.00301-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J, Raghwani J, Pryce R, Bowden TA, Thézé J, Huang S, et al. Molecular evolution, diversity, and adaptation of influenza A(H7N9) viruses in China. Emerg Infect Dis. 2018;24:1795–805. 10.3201/eid2410.171063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J, Ke C, Lau EHY, Song Y, Cheng KL, Zou L, et al. Influenza H5/H7 virus vaccination in poultry and reduction of zoonotic infections, Guangdong Province, China, 2017–18. Emerg Infect Dis. 2019;25:116–8. 10.3201/eid2501.181259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang W, Hou G, Li J, Peng C, Wang S, Liu S, et al. Antigenic variant of highly pathogenic avian influenza A(H7N9) virus, China, 2019. Emerg Infect Dis. 2020;26:26. 10.3201/eid2602.191105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu D, Xiang G, Zhu W, Lei X, Li B, Meng Y, et al. The re-emergence of highly pathogenic avian influenza H7N9 viruses in humans in mainland China, 2019. Euro Surveill. 2019;24:13–21. 10.2807/1560-7917.ES.2019.24.21.1900273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henry Dunand CJ, Leon PE, Huang M, Choi A, Chromikova V, Ho IY, et al. Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe. 2016;19:800–13. 10.1016/j.chom.2016.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ministry of Agricultural and Rural Affairs of the People’s Republic of China. No. 99 announcement [in Chinese] [cited 2018 Dec 13]. http://www.moa.gov.cn/gk/tzgg_1/gg/201812/t20181213_6164848.htm
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental results from study of evolution and antigenic drift of influenza A(H7N9) viruses, China, 2019.


