Abstract
Aversive learning is fundamental for animals to increase chances of survival. In addition to classical neurotransmitters, neuropeptides have emerged to modulate such complex behaviors. Among them, neuropeptide Y (NPY) is well known to promote aversive memory acquisition in mammals. Here we identify an NPY/neuropeptide F (NPF)-related neuropeptide system in Caenorhabditis elegans and show that this FLP-34/NPR-11 system is required for learning negative associations, a process that is reminiscent of NPY signaling in mammals. The Caenorhabditis elegans NPY/NPF ortholog FLP-34 displays conserved structural hallmarks of bilaterian-wide NPY/NPF neuropeptides. We show that it is required for aversive olfactory learning after pairing diacetyl with the absence of food, but not for appetitive olfactory learning in response to butanone. To mediate diacetyl learning and thus integrate the aversive food context with the diacetyl odor, FLP-34 is released from serotonergic neurons and signals through its evolutionarily conserved NPY/NPF GPCR, NPR-11, in downstream AIA interneurons. NPR-11 activation in the AIA integration center results in avoidance of a previously attractive stimulus. This study opens perspectives for a deeper understanding of stress conditions in which aversive learning results in excessive avoidance.
SIGNIFICANCE STATEMENT Aversive learning evolved early in evolution to promote avoidance of dangerous and stressful situations. In addition to classical neurotransmitters, neuropeptides are emerging as modulators of complex behaviors, including learning and memory. Here, we identified the evolutionary ortholog of neuropeptide Y/neuropeptide F in the nematode Caenorhabditis elegans, and we discovered that it is required for olfactory aversive learning. In addition, we elucidated the neural circuit underlying this avoidance behavior, and we discovered a novel coordinated action of Caenorhabditis elegans neuropeptide Y/neuropeptide F and serotonin that could aid in our understanding of the molecular mechanisms underlying stress disorders in which excessive avoidance results in maladaptive behaviors.
Keywords: aversive learning, Caenorhabditis elegans, GPCR, neuropeptide Y, serotonin
Introduction
Learning and memory processes evolved early in evolution to benefit survival of animals in a dynamic environment. Fundamental insights into the molecular substrates of learning and memory have first been described in the mollusc Aplysia californica (Pinsker et al., 1970; Frost et al., 1985). Subsequent studies in vertebrate and invertebrate models revealed the conservation of these molecular principles across animal phyla (Ardiel and Rankin, 2010; Kandel, 2012).
Caenorhabditis elegans has proven to be a powerful model for dissecting molecular mechanisms involved in experience-dependent plasticity (Hobert, 2003). The C. elegans nervous system is compact and displays a broad range of experience-dependent behaviors, such as nonassociative and associative learning (Ardiel and Rankin, 2010; Sasakura and Mori, 2013). Most classical conditioning paradigms rely on pairing aversive or favorable feeding states with gustatory, thermal, and odorant cues to shape the animals' innate attraction or repulsion toward these stimuli, leading to experience-dependent remodeling of neural circuits (Stetak et al., 2009; Ardiel and Rankin, 2010). In addition to classic neurotransmitter systems, the C. elegans genome encodes a large number of neuropeptide precursor proteins that are predicted to generate over 300 bioactive peptides (Van Bael et al., 2018a). These evolutionarily ancient molecules mainly act through GPCRs, most of which are also conserved across animal phyla (Jékely, 2013; Mirabeau and Joly, 2013; Elphick et al., 2018). The structural diversity of neuropeptidergic systems underlies a broad range of physiological and behavioral functions, including learning and memory (Beets et al., 2012; Taghert and Nitabach, 2012; Peymen et al., 2019).
One highly conserved neuropeptidergic system involved in learning and memory in both vertebrates and invertebrates is the neuropeptide Y/neuropeptide F (NPY/NPF) system (Krashes et al., 2009; Gøtzsche and Woldbye, 2016). Vertebrate NPY and invertebrate NPF, as well as their receptors (NPYRs/NPFRs), were found in nearly all bilaterian phyla investigated to date (Fadda et al., 2019). In mammals, NPY is abundantly expressed in the CNS where it has varying effects on learning and memory formation depending on the brain region, the type of memory, and the NPYR subtypes activated (Gøtzsche and Woldbye, 2016). NPYRs are Gi/0-coupled receptors that are mainly postsynaptically located on glutamatergic neurons of the limbic system, where they exert antiexcitatory actions by decreasing glutamate release (Vollmer et al., 2016). In addition to a role in learning and memory formation, disruption of NPY signaling has been shown to decrease serotonin levels, leading to behavioral inflexibility and territorial aggressive behavior (Karl et al., 2004). In Drosophila, the NPY ortholog NPF regulates appetitive learning in accordance with feeding state (Krashes et al., 2009). Food deprivation induces the release of NPF, which promotes appetitive olfactory learning by inhibiting dopaminergic neurons that project to the mushroom body (Krashes et al., 2009), an important integration center in flies.
In C. elegans, several neuropeptide receptors (NPRs) have been predicted as putative NPY/NPF-like receptor orthologs based on sequence similarities (Keating et al., 2003). A large-scale phylogenetic analysis of neuropeptide systems across bilaterian animals revealed NPR-11 to be the closest C. elegans ortholog of the NPY/NPF receptor family (Mirabeau and Joly, 2013). This receptor has been shown to modulate local search behavior and chemotaxis (CTX) in response to the attractive odor isoamyl alcohol, sensed by AWC neurons. NPR-11 acts in AIA interneurons postsynaptic to AWC and its activation reduces AWC responses to isoamyl alcohol, a feedback mechanism that promotes odor adaptation (Chalasani et al., 2010). Given that NPF signaling is known to regulate learning in mammals (Gøtzsche and Woldbye, 2016) and appetitive memory in Drosophila (Krashes et al., 2009), we hypothesized that the NPYR/NPFR ortholog NPR-11 is involved in food-dependent learning and memory in C. elegans. Here, we demonstrate that npr-11 is not required for appetitive olfactory learning but regulates aversive learning in response to the odor diacetyl. NPY/NPF-related neuropeptides, encoded by flp-34, signal from serotonergic neurons through NPR-11 in AIA interneurons of the olfactory circuit to mediate aversive learning.
Materials and Methods
Strains and maintenance
All C. elegans strains were cultivated at 20°C on nematode growth medium (NGM) plates seeded with Escherichia coli OP50 bacteria. All experiments were performed using young adult hermaphrodites. Detailed information regarding genotypes, source, and figures in which each strain was used is provided in Table 1.
Table 1.
Strains used in this paper and corresponding figuresa
| Strain | Genotype | Source | Figures |
|---|---|---|---|
| N2 | WT Bristol strain | CGC | Figure 1B; Figure 2B-G; Figure 3B,C; Figure 6B-D; Figure 7B; Figure 8A |
| LSC882 | npr-11(ok594) X (outcrossed 6 times) | This paper | Figure 1B; Figure 2E-G; Figure 3B; Figure 6B; Figure 8A |
| LSC1667 | flp-34(lst1666) V (CRISPR KO) | This paper | Figure 6B-D; Figure 7B |
| VM487 | nmr-1(ak4) II | CGC | Figure 2C |
| MT15434 | tph-1(mg280) II | CGC | Figure 8A |
| RB1341 | nlp-1(ok1470) X | CGC | Figure 3C |
| CX7894 | kyIs408 [srsx-3::gfp;str-2::dsRed2;elt-2::gfp] II | Kindly provided by C. Bargmann, Rockefeller University, New York | as kyIs408 in Figure 1C |
| LSC1935 | kyIs408 [srsx-3::gfp;str-2::dsRed2;elt-2::gfp] II outcrossed 4 times to the WT strain used in positive butanone association assays | This paper | as kyIs408 x WT in Figure 1C |
| CX8912 | npr-11(ok594) X; kyEx1686 [gcy-28.dp::npr-11::SL2::gfp, elt-2p::gfp] | Kindly provided by C. Bargmann, Rockefeller University, New York | as npr-11[AIAp::npr-11] in Figure 3B |
| LSC1807 | flp-34(lst1666) V; lstEx1010 [tph-1p::flp-34; unc-122p::gfp] | This paper | as flp-34[tph-1p::flp-34] in Figure 7B |
| LSC1906 | flp-34(lst1666) V; lstEx1049 [ops-1p::flp-34; lin-44p::gfp] | This paper | as flp-34[ops-1p::flp-34] in Figure 7B |
| LSC1920 | flp-34(lst1666) V; lstEx1053 [gcy-17p::flp-34; myo-2p::mCherry] | This paper | as flp-34[gcy-17p::flp-34] in Figure 7B |
| LSC1925 | flp-34(lst1666) V; lstEx1055 [gcy-27p::flp-34; lin-44p::gfp] | This paper | as flp-34[gcy-27p::flp-34] in Figure 7B |
| LSC1812 | flp-34(lst1666) V; npr-11(ok594) X | This paper | as flp-34;npr-11 in Figure 6B |
| LSC1809 | tph-1(mg280) II; npr-11(ok594) X | This paper | as tph-1;npr-11 in Figure 8A |
| AQ3955 | ljEx1067[WRM0616cB05 npr-11(fos)::SL2::gfp] | This paper | as fosmid npr-11::sl2::gfp in Figure 3A |
| AX6999 | dbEx1028 [WRM0615cD01 flp-34(fos)::SL2::gfp] | This paper | as fosmid flp-34::sl2::gfp in Figure 7A |
| LSC1885 | dbEx1028 [WRM0615cD01 flp-34(fos)::SL2::gfp]; lin-15B&lin-15A(n765)X; vsIs97[tph-1p::DsRed2 + lin-15(+)] | This paper | as fosmid flp-34::sl2::gfp; tph-1p::DsRed2 in Figure 7A |
| LSC1905 | dbEx1028 [WRM0615cD01 flp-34(fos)::SL2::gfp]; [nlp-44p::TeTx::mCherry; unc-122p::mCherry] | This paper | as fosmid flp-34::sl2::gfp; ASGp::DsRed2 in Figure 7A |
aCGC, Caenorhabditis Genetics Centre at the University of Minnesota.
Appetitive olfactory learning in response to butanone
The butanone learning assay was performed as previously reported (Kauffman et al., 2011) in a climate-controlled room at 20 ± 0.5°C and 35%–55% relative humidity. Briefly, synchronized 1-d-old hermaphrodites were washed with M9 buffer (Brenner, 1974) from high growth media cultivation plates and collected into a 15 ml conical tube. After three washes, half of the population was immediately tested for naive CTX toward butanone, while the rest was kept for 1 h in the tube. Next, these starved worms were transferred to 90 mm NGM conditioning plates seeded with 1 ml of OP50 E. coli bacteria, and 2 µl of 10% butanone (diluted in 100% ethanol) was pipetted on the lid. After 1 h, a proportion of the conditioned worms was tested for CTX toward butanone (t0), whereas the rest of the population was placed on 90 mm NGM hold plates seeded with 1 ml OP50 for 30, 60, or 120 min. At the end of each interval, the CTX toward butanone for the specific resting group was quantified (t30, t60, and t120). CTX was tested on 90 mm unseeded NGM plates where 100–200 worms were pipetted at the origin spot. Worms were given a choice between two equidistant spots containing 1 µl of NaN3 + 10% butanone and 1 µl of NaN3 + ethanol, respectively. After 1 h, worms were counted, and the CTX index (CI) was calculated as follows:
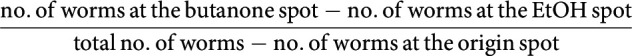 |
Diacetyl learning assay
The diacetyl learning assay is based on the assay described by Vukojevic et al. (2012) with some modifications. Animals were exposed to 0.1% diluted diacetyl both during conditioning and CTX assays, and the conditioning time was extended to 3 h. The assay was performed in a climate-controlled room at 20°C ± 0.5 and 35-55% relative humidity. A population of well-fed, synchronized young adult worms was washed 3 times with CTX buffer (Vukojevic et al., 2012), and the naive attraction toward diacetyl was immediately tested on CTX plates (Vukojevic et al., 2012). Approximately 100 worms were pipetted at the origin spot of a 90 mm CTX plate and given a choice between a spot of 1 µl of 0.1% diacetyl (Sigma Millipore, diluted in 100% ethanol) and an equidistant control spot of 1 µl of 100% ethanol (EtOH). Both spots also contained 1 µl of 1 m NaN3. After 1 h, the distribution of the worms on the CTX plate was checked as reported (Bargmann et al., 1993), and a CI was calculated as follows:
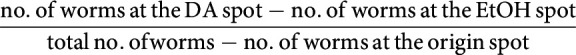 |
For aversive olfactory conditioning, worms were exposed for 3 h to 2 µl of 0.1% diacetyl pipetted directly on the center of the lid of an unseeded CTX plate. To prevent a change in diacetyl concentration due to evaporation during conditioning, the lids of the CTX plates were replaced every 30 min with new lids and fresh 0.1% diacetyl. The CI of the conditioned worms was calculated as described above.
Uncoupled conditioning was performed by exposing the worms for 3 h to only 1 cue as previously reported (Vukojevic et al., 2012). The diacetyl-only control was conditioned on standard NGM plates seeded with 250 µl of E. coli OP50 with 2 µl of 0.1% diacetyl pipetted on the lid. The no food-only control was conditioned on unseeded CTX plates without any odorant presentation.
To test for desensitization, a shock treatment to disrupt short-term learning was applied, consisting of a combination of thermal and mechanical perturbations (Bernhard and van der Kooy, 2000). Conditioning plates were first kept at 4°C for 15 min. Worms were then collected, vortexed intermittently for 30 s, and centrifuged twice at 500 × g (1 min in the first trial and 2 min in the second trial). Control worms for this experiment, showing desensitization, were conditioned for 90 min on CTX plates with 2 µl of undiluted diacetyl pipetted on the lid.
Molecular biology and generation of transgenic worms
The cell-specific rescue construct for flp-34 was cloned using NEBuilder HiFi DNA Assembly (NEB) in a pSM vector (kindly provided by C. Bargmann, Rockefeller University). The cDNA of flp-34 was amplified by PCR and cloned at the KpnI site. The tph-1p, ops-1p, gcy-17p, and gcy-27p promoters were amplified by PCR from N2 WT genomic DNA and cloned upstream of the flp-34 cDNA fragment. A long tph-1 promoter sequence (3124 nucleotides upstream of the translation start site) was chosen to drive expression in all three serotonergic neurons (NSM, HSN, and ADF) (Sze et al., 2002). Primer sequences used to clone the flp-34 cDNA, and the promoters are reported in Table 2. The constructs were injected into the syncytial gonad of young adult worms at the following concentrations: tph-1p::flp-34 at 5 ng/µl with 50 ng/µl of unc-122p::gfp coinjection marker and 48 ng/µl of a 1 kb DNA ladder (Thermo Fisher Scientific) as carrier DNA, ops-1p::flp-34 at 25 ng/µl with 50 ng/µl of lin-44p::gfp coinjection marker and 25 ng/µl of a 1 kb DNA ladder, gcy-17p::flp-34 at 25 ng/µl with 20 ng/µl of myo-2p::mCherry coinjection marker and 26 ng/µl of a 1 kb DNA ladder and gcy-27p::flp-34 at 40 ng/µl with 50 ng/µl of lin-44p::gfp coinjection marker and 10 ng/µl of a 1 kb DNA ladder.
Table 2.
Primers used to amplify and clone tph-1p, ops-1p, gcy-17p, gcy-27p and flp-34 cDNA constructs in the pSM vectora
| Plasmid | PCR fragment | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|---|
| pSM-flp-34 cDNA | flp-34 cDNA | ttggctagcgtcgacggtaccATGCAGTTCCAATTTTTGATG | gtaggatgagacagcggtaccTTATTTACGGCCAAATCG |
| pSM-tph-1p::flp-34 cDNA | tph-1p | tcactcattaggcaccccagATATGGCTGCAATACTATTTTTC | gcttggcgtaatcatggtcaCTAAAAACCAAAGAAAATCATGAAATG |
| pSM-ops-1p::flp-34 cDNA | ops-1p | tcactcattaggcaccccagGGAGATGAGCCAATATTTTTAAAAG | gcttggcgtaatcatggtcaATTTATTTACAGGTAGCTTTTAGAG |
| pSM-gcy-17p::flp-34 cDNA | gcy-17p | tcactcattaggcaccccagCAGTTGAACATCTCCCTG | gcttggcgtaatcatggtcaTAAAAGAAGCTTGCCCAATG |
| pSM-gcy-27p::flp-34 cDNA | gcy-27p | tcactcattaggcaccccagGCATTGTGTCAATTGCTC | gcttggcgtaatcatggtcaTTGGTAGAAAATAAATAAAATGAAAGTTG |
aUppercase nucleotides indicate the gene-specific sequence of the primer; lowercase nucleotides indicate the pSM vector overlapping sequence of the primer.
The flp-34(lst1666) V KO strain was generated by C. elegans optimized CRISPR/Cas9-mediated deletion of exons 2 and 3 (612 bp) (see Extended Data Fig. 6-1). The enhanced Cas9 (eCas9) was in-house purified from BL21 bacteria transfected with pGH496-Cas9-expressing vector (kindly provided by G. Hollopeter, Cornell University). Two guide RNA (gRNA) sequences were designed (https://eu.idtdna.com/site/order/designtool/index/CRISPR_SEQUENCE) as well as a repair template that induced a frameshift and included a premature stop codon at the beginning of exon 2 (signal peptide region), to prevent further translation of both mature peptides encoded by flp-34. To simplify selection of the successfully edited worms, we used a Co-CRISPR technique in which the dpy-10 gene was also knocked out by CRISPR editing (Arribere et al., 2014; Paix et al., 2015). A mix containing all the required components was injected into C. elegans young adult hermaphrodites (Table 3).
Table 3.
gRNA sequences used to target flp-34 and dyp-10, and the respective repair template sequencesa
| Gene | gRNA/template | Sequence |
|---|---|---|
| flp-34 | gRNA 1 | CAGCGTACTATCCCTGCCACTGG |
| flp-34 | gRNA 2 | GATAACGTCCTTCCGAGTAGAGG |
| flp-34 | Repair template | TAGCTGTCGCTCCCTCCGAGGGTCGATACC |
| dpy-10 | gRNA | GCTACCATAGGCACCACGAG |
| dpy-10 | Repair template | CACTTGAACTTCAATACGGCAAGATGAGAATGACTGGAAACCGTACCGCATGCGGTGCCTATGGTAGCGGAGCTTCACATGGCTTCAGACCAACAGCCTAT |
aThe dpy-10 gRNA and repair template have been previously reported (Arribere et al., 2014). Nucleotides in bold indicate a stop codon.
The plasmid for heterologous expression of npr-11 in CHO cells was obtained by directionally cloning the PCR-amplified npr-11 cDNA into the pcDNA3.1/V5-His-TOPO TA vector (Invitrogen).
The npr-11::sl2::gfp and flp-34::sl2::gfp fosmid-based transgenes were generated by cloning the sl2::GFP sequence C-terminally of the npr-11 and flp-34 open reading frame in the fosmid npr-11-WRM0616cB05 and flp-34-WRM0615cD01 as previously reported (Tursun et al., 2009). The npr-11 and flp-34 fosmids were microinjected into the syncytial gonad of young adult worms at 30 ng/µl.
Locomotion assay
The locomotion activity of 15-20 young adult worms was recorded in well-fed condition (on 250 µl OP50-seeded plates) or after 1 h of food deprivation (on NGM-unseeded plates). Locomotion was recorded for 10 min at 2 fps using an in-house tracking system with a 10-megapixel camera (GigEPRO GP11004M NET), a 12 inch × 12 inch diffuse LED light source to illuminate animals in trans (Rosco LitePad) and two consumer privacy filters (3 M PF17.0) placed perpendicularly on the glass stage where the assay plates were positioned to enhance the contrast. The StreamPix Multicamera 6 software was used to record and acquire video streams that were successively converted into frames. A custom MATLAB R2016a (MathWorks) script was used to track individual worms over consecutive frames. Each track was then visually evaluated to discard immobile background artifacts. The average speed of the worms along the recorded 10 min was obtained as output of the analysis and used to compare the locomotion activity of different worm strains. The custom MATLAB script is available on request.
Expression pattern analysis
Hermaphrodite transgenic worms were mounted on a 2% agarose pad and immobilized with 1 m NaN3 (Sigma Millipore) in M9 buffer. The expression pattern of the transgenic animals was visualized using an Olympus FluoView FV1000 (IX81) confocal microscope, and the z-stack projections were analyzed with Imaris 7.2 (Bitplane) software. For fosmid-based npr-11::sl2::gfp and flp-34::sl2::gfp transgenes, cell identifications were based on the following: position and morphology, colocalization with DiI (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate, Invitrogen) staining (Tong and Bürglin, 2010) and crossing with marker strains (tph-1p::DsRed2, nlp-44p::mCherry, and srsx-3::gfp;str-2::dsRed2, kindly provided by C. Bargmann, Rockefeller University).
Receptor activation assay
The in vitro GPCR activation assay for NPR-11 was performed using an aequorin-based luminescence assay as reported previously (Beets et al., 2012; Van Sinay et al., 2017; Peymen et al., 2019). Briefly, CHO cells stably expressing the aequorin calcium indicator and the promiscuous Gα16 protein (ES-000-A24, PerkinElmer) were transfected with pcDNA3.1/npr-11 or pcDNA3.1 empty vector at 40%–50% confluency using Lipofectamine LTX and Plus Reagent (Invitrogen). After 24 h at 37°C, the cells were moved to 28°C O/N. On the day of the screening, cells were collected at a density of 5 × 106 cells/ml in DMEM/F12 medium without phenol red (Invitrogen) supplemented with 0.1% BSA and loaded with coelenterazine H (Invitrogen) to reconstitute the Ca2+-sensitive photoprotein aequorin. Compound plates containing a library of over 350 synthetic peptides in a final concentration of 10 μm (for the identification of NPR-11 ligands) or serial dilutions of FLP-34 or NLP-1 peptides (for the dose–response analysis) were placed in a MicroBeta2 LumiJET (PerkinElmer) for screening. The transfected cells were then loaded at a density of 25,000 cells/well, and the luminescence was measured for 30 s at 469 nm. Upon receptor activation, Gα16 directs GPCR signaling to the PLCβ pathway, promoting intracellular Ca2+ release and a luminescence signal. To normalize the luminescence signal based on the total Ca2+ amount in each well, 0.2% Triton X-100 (Merck) was added to lyse the cells, and the maximal luminescence response was measured for another 30 s. The potency of FLP-34 peptides to activate NPR-11 was determined by a dose–response analysis (from 10 μm to 100 fm). The plotted Ca2+ responses were calculated as percentage of the highest peptide-evoked response (100% activation) after subtraction of the negative control values (BSA cell medium without peptide) and normalization to the total Ca2+ response. A nonlinear regression analysis was used to create sigmoidal dose–response curves, and the EC50 values were calculated as median of the 95% confidence interval. The comparison between FLP-34 and NLP-1 activation of NPR-11 was assessed using 10-fold serial dilutions (from 10 μm to 1 nm) of the peptides and the ratio of the normalized Ca2+ responses was represented for each peptide concentration.
Peptides
The GPCR activation screen was performed using an in-house C. elegans library composed of over 350 peptides of the RFamide (FLP) and neuropeptide-like protein (NLP) family. The library was compiled based on in silico predictions and peptidomics data (Husson et al., 2005; Van Bael et al., 2018b), and was custom-synthesized by Thermo Fisher Scientific and GL Biochem. FLP-34 and NLP-1 peptides used for dose–response experiments were purified by reversed-phase high-performance liquid chromatography on a Symmetry-C18 column (4.6 × 250 mm HPLC cartridge with pore size of 5 μm) and quantified with the bicinchoninic acid protein assay. The mass of peptides was verified by MALDI-TOF mass spectrometry (matrix assisted laser desorption/ionization time-of-flight analyzer) on a Reflex IV instrument (Bruker Daltonic).
NPF/NPY alignment and comparison of the exon-intron structure of genes encoding NPF/NPY peptides
The NPF/NPY alignment was performed using ClustalX 2.1 Multiple Sequence Alignment with default parameters (Jeanmougin et al., 1998; Larkin et al., 2007). The source of the bilaterian representative NPF/NPY mature peptides is indicated in the Table 4, together with species name abbreviations. The software BOXSHADE (www.ch.embnet.org/software/BOX_form.html) was used to highlight conserved amino acids: identical residues with a minimum of 70% conservation are highlighted in black, amino acid groups with strongly similar properties and a minimum of 70% conservation are highlighted in dark gray, and amino acid groups with strongly similar properties and a minimum of 55% conservation are highlighted in light gray.
Table 4.
Species and references of the sequences used for the mature peptide alignment
| Species | Abbreviation | Reference |
|---|---|---|
| Schmidtea mediterranea | Schme | Nuss et al., 2010 |
| Moniezia expansa | Monex | Maule et al., 1991 |
| Drosophila melanogaster | Drome | Brown et al., 1999 |
| Aedes aegypti | Aedae | Stanek et al., 2002 |
| Bombyx mori | Bommo | Deng et al., 2014 |
| Daphnia pulex | Dappu | Elphick and Mirabeau, 2014 |
| Aplysia californica | Aplca | Rajpara et al., 1992 |
| Lottia gigantea | Lotgi | Elphick and Mirabeau, 2014 |
| Caenorhabditis elegans | Caeel | Husson et al., 2014 |
| Pristionchus pacificus | Pripa | a |
| Capitella telata | Capte | Nuss et al., 2010 |
| Capitella capitata | Capca | a |
| Saccoglossus kowalevskii | Sacko | Mirabeau and Joly, 2013 |
| Branchiostoma floridae | Brafl | Mirabeau and Joly, 2013 |
| Danio rerio | Danre | Söderberg et al., 2000 |
| Gallus gallus | Galga | Blomqvist et al., 1992 |
| Mus musculus | Musmu | a |
| Homo sapiens | Homsa | Minth et al., 1986 |
aMature peptides predicted according to preprotein convertases cleavage sites.
Analysis of the exon-intron structure of genes encoding NPF/NPY was performed by comparing nucleotides, transcripts, and precursor proteins encoding for representative NPF/NPY peptides across bilaterians. Nucleotide and amino acid sequences were obtained from the NCBI, WormBase, and the Joint Genome Institute databases. Accession numbers of the sequences used for the analysis, together with species name abbreviations, are listed in the Table 5.
Table 5.
Species, accession numbers of the genomes, mRNAs and precursor proteins used for the exon-intron structure analysis
| Species name | Abbreviation | Genome | mRNA | Protein |
|---|---|---|---|---|
| Caenorhabditis elegans | Caeel | NC_003283.11a | R09A1.5a.1b | R09A1.5a.1b |
| Pristionchus pacificus | Pripa | ABKE00000000.3c | PPA29058.1b | PPA29058.1b |
| Drosophila melanogaster | Drome | AC011616.5c | NM_080493.3a | NP_536741a |
| Aedes aegypti | Aedae | NC_035107.1a | XM_021854183a | XP_021709875.1a |
| Daphnia pulex | Dappu | GCA_900092285.2c | FE283179.1c | EFX80468.1c |
| Bombyx mori | Bommo | HQ651090.1c | NM_001130883.2a | NP_001124355.1a |
| Lottia gigantea | Lotgi | NZ_AMQO00000000.1a | cJQ646097.1c | AFW19651.1c |
| Capitella capitata | Capca | GCA_001875675.1c | 204022d | 204022d |
| Danio rerio | Danre | NC_007130.7a | NM_131074.2a | NP_571149.1a |
| Gallus gallus | Galga | NC_006089.5a | NM_205473.1a | NP_990804.1a |
| Mus musculus | Musmu | NC_000072.6a | NM_023456.3a | NP_075945.1a |
| Homo sapiens | Homsa | NC_000007.14a | NM_000905.4a | NP_000896.1a |
NCBI reference sequence.
WormBase ID.
GenBank.
Joint Genome Institute Genome Portal.
Experimental design and statistical analysis
Data analysis and statistical comparisons were performed using Prism 8 software (GraphPad), setting the statistical significance to p < 0.05. Detailed information about the statistical significance between experimental groups and the total number of replicates (indicated by n for learning assays and receptor activation assays) or worms (indicated by w for locomotion assays) for each experiment are reported in the figure legends.
Learning assays were performed on at least two independent days, including each time two to four replicates per condition for the diacetyl leaning assays and 5 replicates for the butanone-positive association assay. For diacetyl learning assays, comparing different conditions for one genotype, data were analyzed using a Kruskal–Wallis test and Dunn's post hoc test for multiple comparison (nonparametric data) or a one-way ANOVA and Tukey's multiple comparisons test (for parametric data). For diacetyl learning and butanone learning assays comparing different conditions for multiple genotypes, data were analyzed using two-way ANOVA and Sidak's or Tukey's multiple comparison tests. Locomotion assays were performed in triplicate on 2 independent days, and the difference in average speed was analyzed with an unpaired t test. Receptor activation assays for dose–response measurements were done in triplicate on at least 2 independent days. The comparison between calcium responses of FLP-34 and NLP-1 peptides in NPR-11-expressing cells was performed in duplicate in three independent experiments. Data were analyzed using Kruskal–Wallis test and Dunn's post hoc test.
Results
Npr-11 is not required for appetitive olfactory learning in response to butanone
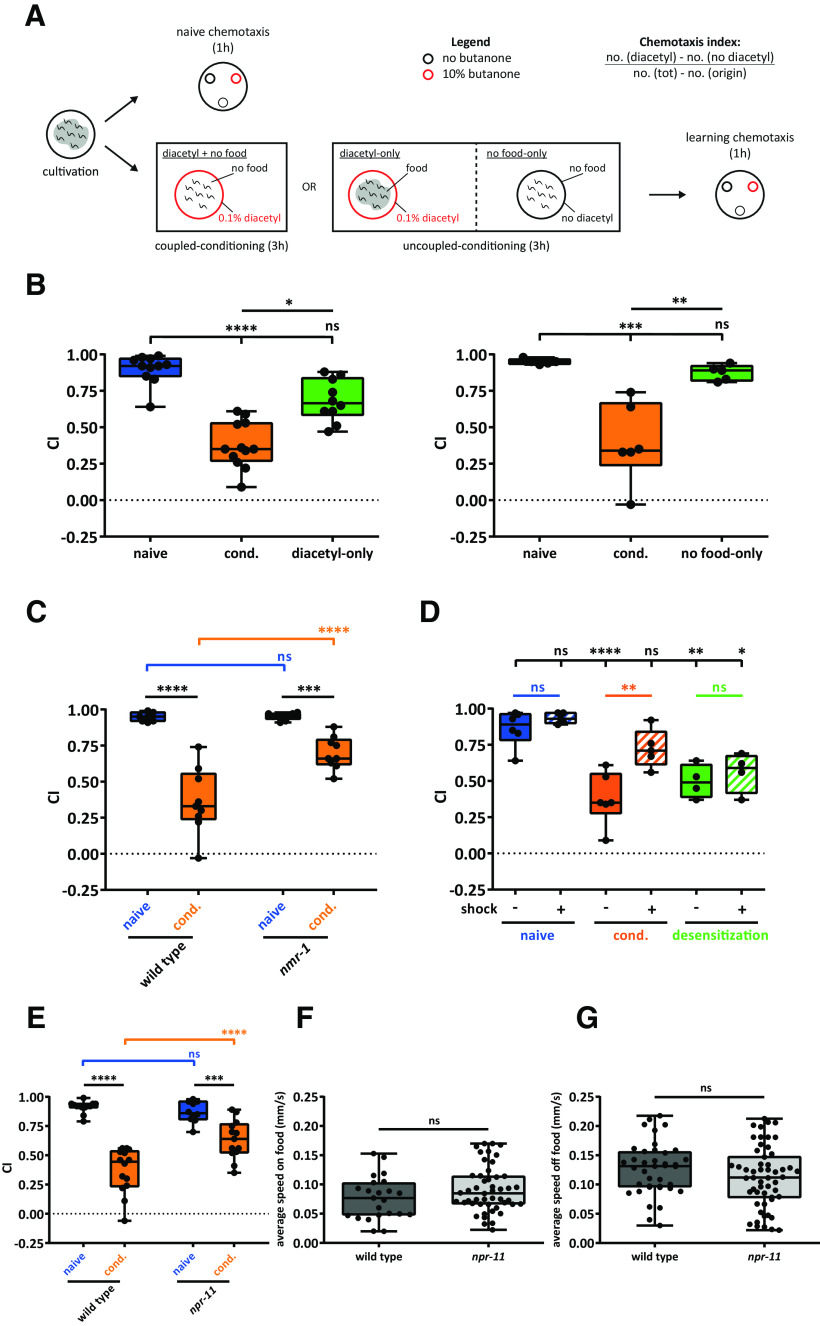
In Drosophila melanogaster, RNAi knockdown of the npfr gene impairs appetitive olfactory learning (Krashes et al., 2009). In C. elegans, the ortholog of this receptor, encoded by the npr-11 gene (Mirabeau and Joly, 2013), mediates adaptation to the attractive odor isoamyl alcohol, sensed by olfactory AWC neurons (Chalasani et al., 2010). Based on these findings, we hypothesized that NPR-11 is involved in AWC-mediated appetitive learning. To test this, we quantified the performance of npr-11 mutants in an established appetitive learning assay referred to as positive butanone association in which worms learn to associate food with butanone, an odorant sensed by AWC (Kauffman et al., 2011). We exposed the worms to food deprivation for 1 h, conditioned them for the same amount of time with butanone in the presence of food, and quantified CTX behavior to butanone before and after conditioning by calculating a CI (Fig. 1A). Untrained WT C. elegans were moderately attracted to butanone (Fig. 1B). As expected, conditioned worms showed a strongly increased CI and thus enhanced attraction to the odorant immediately after conditioning (time 0, t0). This increased attraction to butanone gradually declined over 2 h when animals were kept on food in the absence of butanone (Fig. 1B). Since the naive attraction of WT animals to butanone was slightly lower compared with CI values reported previously (Kauffman et al., 2011), we validated the identities of AWC neurons in this WT strain by investigating the asymmetric expression of str-2, encoding a receptor that senses the butanone odour (Troemel et al., 1999). Previous studies demonstrated the asymmetric identities of AWCON and AWCOFF neurons derived by the mutually exclusive expression of STR-2 and SRSX-3 receptors, respectively (Troemel et al., 1999; Lesch et al., 2009). This mutually exclusive expression pattern was conserved in the WT strain showing positive butanone association (Fig. 1C), suggesting that AWC identities are not impaired in this strain and minor deviations in CI most likely result from differences in experimental conditions.
Figure 1.
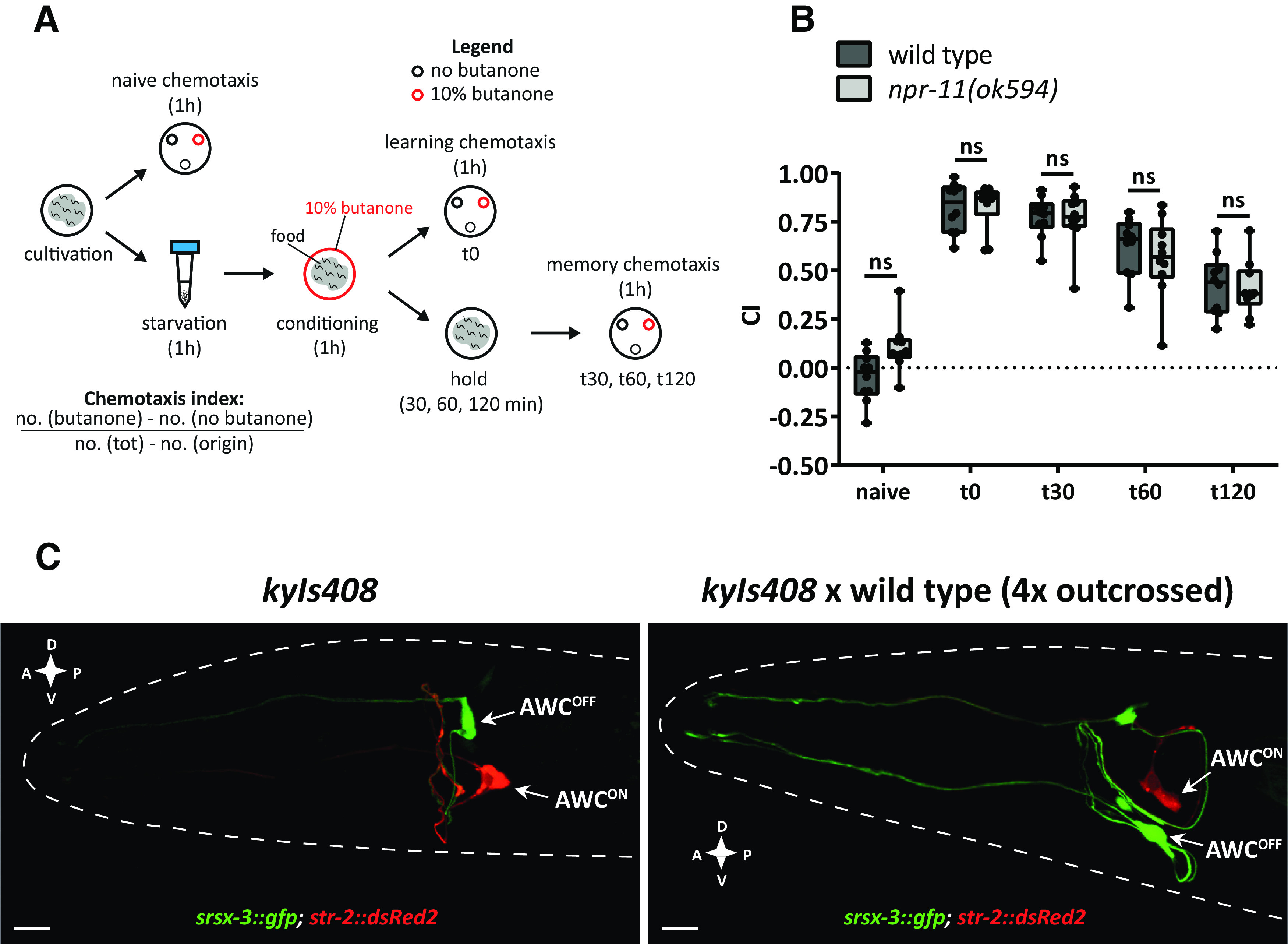
npr-11 mutants show normal appetitive olfactory learning in response to butanone. A, Schematic of the positive butanone association paradigm (Kauffman et al., 2011). Age-synchronized worms are washed from the cultivation plates and immediately tested for naive CTX to butanone. The remaining animals undergo 1 h of food deprivation followed by 1 h of conditioning with food and butanone. A part of the population is immediately tested for CTX to butanone (learning CTX, t0), while the rest of the animals are kept on food-seeded plates for 30, 60, or 120 min before testing CTX to butanone (memory CTX). B, npr-11 mutants are not defective in positive butanone association. For each time point (t0 = 0, t30 = 30, t60 = 60, and t120 = 120 min after conditioning), the CI values of npr-11 mutants are similar to those of WT. Data were analyzed by two-way ANOVA (F(4,90) = 102.5, p < 0.0001) and Sidak's multiple comparison test (n = 10). Boxplots represent 25th (lower boundary) and 75th (upper boundary) percentiles. The 50th percentile (line) shows the median. Whiskers plot the minimum and maximum values. Black dots represent individual CIs. C, WT animals show asymmetric expression of str-2 and srsx-3 receptor-encoding genes in AWCON and AWCOFF neurons. Representative confocal z-stack projections of AWC neurons in transgenic adult hermaphrodites expressing a srsx-3::gfp; str-2::dsRed2 transgene (kyIs408) in a reporter strain (CX7894, kindly provided by C. Bargmann, Rockefeller University) before (left) and after (right) crossing with the WT strain used in positive butanone association assays. A, Anterior; P, posterior; D, dorsal; V, ventral. Scale bars, 10 µm. ns, not significant.
Like WT animals, mutants of npr-11 showed enhanced attraction to butanone after conditioning (t0), suggesting that NPR-11 does not mediate this type of appetitive olfactory learning (Fig. 1B). In addition, the CI progressively decreased in both WT and npr-11 mutant animals over time (t30, t60, to t120 after conditioning), which indicates normal extinction of the appetitive memory after training (Fig. 1B). Together, our findings show that learning and retention of the positive association between butanone and food do not require npr-11.
Npr-11 mutants are defective in aversive olfactory learning
Since NPR-11 signaling is not required for appetitive olfactory learning in response to butanone, we asked whether it is involved in learning negative associations, a process that is reminiscent of NPY signaling in mammals (Gøtzsche and Woldbye, 2016). To investigate whether npr-11 mutants are defective in aversive olfactory learning, we modified an established conditioning protocol (Stetak et al., 2009; Vukojevic et al., 2012) pairing short-term food deprivation with exposure to diluted diacetyl, which we refer to as diacetyl learning (Fig. 2A). After pairing diacetyl with the absence of food, the behavioral preference of worms toward the odorant diacetyl is switched from attractive to repulsive.
Figure 2.
npr-11 mutants are defective in diacetyl learning. A, Schematic of the diacetyl learning assay, modified from Vukojevic et al., (2012). Age-synchronized worms are washed from cultivation plates and immediately tested for naive CTX to diacetyl, while the remaining animals undergo 3 h of coupled (diacetyl + no food) or uncoupled (diacetyl-only or no food-only) conditioning. CTX to diacetyl is then quantified on CTX plates; and after 1 h, a CI is calculated. B, WT worms after coupled conditioning (cond.) show a significant drop in the CI compared with animals conditioned with diacetyl-only (left) or no food-only (right). Data were analyzed by Kruskal–Wallis (KW(2) = 24.15, p < 0.0001) and Dunn's multiple comparison tests (left, n ≥ 10) or one-way ANOVA (F(2,13) = 17.37, p = 0.0002) and Tukey's post hoc test (right, n ≥ 5). C, nmr-1 mutants show impaired diacetyl learning compared with WT. Two-way ANOVA revealed a significant effect of genotypes (F(1,30) = 13.83, p = 0.0008), of behavioral treatments (F(1,30) = 83.83, p < 0.0001), and of interaction between genotypes and behavioral treatments (F(1,30) = 13.14, p = 0.0011). Data were analyzed by Sidak's multiple comparison test (n ≥ 7). D, Shock treatment reverses the reduced attraction to diacetyl in animals conditioned for diacetyl learning (cond.), but not in a control group for desensitization exposed to undiluted diacetyl. Data were analyzed by one-way ANOVA (F(7,33) = 11.50, p < 0.0001) and Tukey's post hoc test (n ≥ 4). E, npr-11 mutants show normal attraction to diacetyl before conditioning but are defective in diacetyl learning. Two-way ANOVA revealed a significant effect of genotypes (F(1,46) = 7.666, p = 0.0081), of behavioral treatments (F(1,46) = 94.92, p < 0.0001), and of interaction between genotypes and behavioral treatments (F(1,46) = 14.61, p = 0.0004). Data were analyzed by Sidak's post hoc test (n ≥ 10). F, G, npr-11 mutants display normal locomotion speed on and off food. Activity is recorded for 10 min on E. coli OP50-seeded (F) or unseeded (G) NGM plates. The average speed of mutants is not significantly different from that of WT in both conditions. Data were analyzed by unpaired t test (F, w ≥ 23; G, w ≥ 39). B–G, Boxplots represent 25th (lower boundary) and 75th (upper boundary) percentiles. The 50th percentile (line) indicates the median. Whiskers represent the minimum and maximum values. Black dots represent individual CIs or worms. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, not significant.
In our modified protocol, we conditioned animals with 0.1% diluted diacetyl for 3 h in the absence of food. We first tested whether this training elicits aversive associative learning. Before conditioning, WT worms were strongly attracted toward diacetyl and showed a highly positive CI (Fig. 2B). As expected for aversive learning, after pairing food deprivation with exposure to diacetyl, attraction toward the odorant was reduced, resulting in a significantly decreased CI (Fig. 2B). To test whether the effect on diacetyl attraction in conditioned animals resulted from the paired association of diacetyl and the absence of food, and not from exposure to one of the stimuli, we tested two unpaired controls in which animals were exposed only to the diacetyl in the presence of food or only to food deprivation without diacetyl. WT worms conditioned with both diacetyl and the absence of food were less attracted toward diacetyl. However, their CI for diacetyl was not significantly reduced after training with diacetyl-only or no food-only (Fig. 2B). Thus, paired exposure to diacetyl and food deprivation is required to reduce diacetyl attraction and indeed elicits aversive associative learning. To establish further whether diacetyl learning represents associative learning, we tested the performance of nmr-1 mutants that have a mutation in subunit 1 of the NMDA receptor. Loss of nmr-1 function causes defects in associative learning but not in nonassociative habituation (Lau et al., 2013). As expected for associative learning, NMDA receptor mutants were defective in diacetyl learning, as they were still strongly attracted to the odorant after conditioning (Fig. 2C). We also tested whether the reduced attraction to diacetyl after conditioning was elicited by learning and not by desensitization, a form of adaptation (Colbert and Bargmann, 1997; Nuttley et al., 2001). Adaptation is distinct from learning, as it is characterized by the temporary inability to sense and respond to diacetyl due to receptor saturation and internalization. To test this, we exposed animals after conditioning to a mechanical and thermal shock treatment that erases short-term memory, but not desensitization (Nuttley et al., 2001). As a control for desensitization, we included an experimental group exposed to undiluted diacetyl for 90 min (Colbert and Bargmann, 1997). Animals conditioned with 0.1% diacetyl showed a significant increase of CI after shock treatment, as expected for learning (Fig. 2D). The same treatment did not affect the CI of the control group exposed to undiluted diacetyl, characteristic of desensitization (Fig. 2D). Together, these results indicate that 3 h exposure to 0.1% diacetyl in the absence of food elicits aversive associative learning.
We next evaluated diacetyl learning of npr-11 loss-of-function mutants. In contrast to WT animals, npr-11 mutants were still strongly attracted toward diacetyl after conditioning (Fig. 2E). Since this diacetyl learning defect could be caused by a general defect in neural circuits involved in locomotion, we quantified the speed of npr-11 mutants in well-fed conditions and after 1 h of food deprivation, to test potential locomotory defects. Mutant animals moved at an average speed similar to that of WT worms in both fed and starved conditions (Fig. 2F,G). Untrained npr-11 mutants also showed normal attraction to diacetyl (Fig. 2E). Together, these results suggest that npr-11 signaling does not affect general locomotion or diacetyl sensing but is required for diacetyl learning.
NPR-11 is required in AIA interneurons for diacetyl learning
To shed light on the cells in which NPR-11 is required for diacetyl learning, we examined the expression pattern of the receptor. Worms expressing a fosmid-recombineered GFP transgene for npr-11 showed fluorescence in several head and tail neurons (Fig. 3A). This transgene confirmed the previously reported expression of npr-11 in AIA interneurons (Chalasani et al., 2010) (Fig. 3A). In addition, we observed npr-11 expression in ASK and ADL sensory neurons, validated by DiI staining of amphid neurons, as well as in several unidentified cells in the head (Fig. 3A). In the tail, the npr-11 reporter transgene was expressed in PVQ and most likely in DVA or DVC neurons (Fig. 3A).
Figure 3.
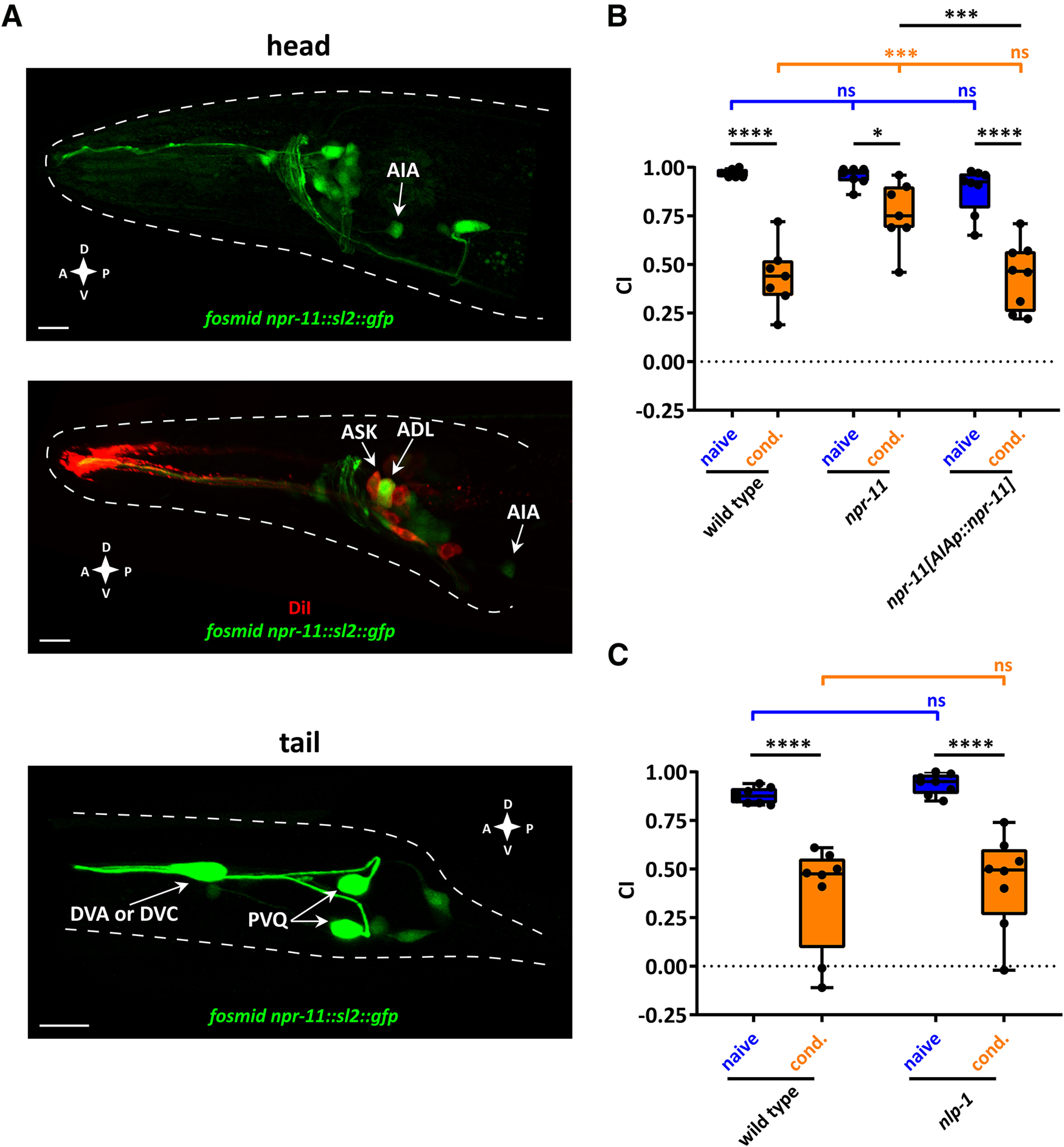
Diacetyl learning requires NPR-11 signaling in AIA interneurons. A, Representative confocal z-stack projections of head and tail neurons expressing a fosmid-based npr-11::sl2::gfp reporter transgene in adult hermaphrodites. Top, npr-11 expression in AIA interneurons. Middle, Expression of npr-11 in ASK and ADL sensory neurons is shown, colocalized with DiI staining of these amphid sensory neurons. Bottom, Expression of npr-11 in tail neurons, including PVQ and most likely DVA or DVC. A, Anterior; P, posterior; D, dorsal; V, ventral. Scale bars, 10 µm. B, npr-11 is required in AIA for diacetyl learning. Expressing WT copies of npr-11 from the AIA-specific gcy-28d promoter rescues the diacetyl learning defect of npr-11 mutants. Two-way ANOVA revealed a significant effect of genotypes (F(2,38) = 8.668, p = 0.0008), of behavioral treatments (F(1,38) = 95.49, p < 0.0001), and of interaction between genotypes and behavioral treatments (F(2,38) = 6.154, p = 0.0048). Data were analyzed by Sidak's and Tukey's post hoc test (n = 7). C, nlp-1 mutants are not defective in diacetyl learning. CTX of nlp-1 mutants toward diacetyl before and after conditioning resembles that of WT worms. Two-way ANOVA revealed only a significant effect of behavioral treatments (F(1,28) = 60.95, p < 0.0001). Data were analyzed by Sidak's post hoc test (n = 8). B, C, Boxplots represent 25th (lower boundary) and 75th (upper boundary) percentiles. The 50th percentile (line) indicates the median. Whiskers represent the minimum and maximum values. Black dots represent individual CIs. *p < 0.05, ***p < 0.001, ****p < 0.0001. ns, not significant.
AIA interneurons play a well-described role as sensory integration centers involved in aversive learning (Chalasani et al., 2010; Dobosiewicz et al., 2019). We therefore asked whether npr-11 is required in these neurons for diacetyl learning. Restoring npr-11 expression in AIA, under control of the gcy-28.d promoter, was sufficient to rescue diacetyl learning of npr-11 mutants to the level of WT worms, indicating that NPR-11 acts in AIA to mediate this type of learning (Fig. 3B).
Nlp-1 mutants display normal diacetyl learning
One of the buccalin-related NLP-1 neuropeptides (MDANAFRMSFa) has been reported to activate NPR-11 and thereby inhibit local search behavior (Chalasani et al., 2010). Since npr-11 is also required for diacetyl learning, we tested whether this effect is mediated by NLP-1. If NLP-1 neuropeptides modulate diacetyl learning by activating NPR-11, we expect nlp-1 mutants to recapitulate the learning defect observed for npr-11 in the diacetyl assay. However, after 3 h of conditioning with diacetyl in the absence of food, nlp-1 mutants showed a reduced attraction to diacetyl similar to the behavior of WT animals (Fig. 3C). These results suggest that NLP-1 is not required for diacetyl learning and NPR-11 may be activated by other ligands in the aversive olfactory learning circuit.
The C. elegans NPY/NPF ortholog FLP-34 dose-dependently activates NPR-11 in vitro
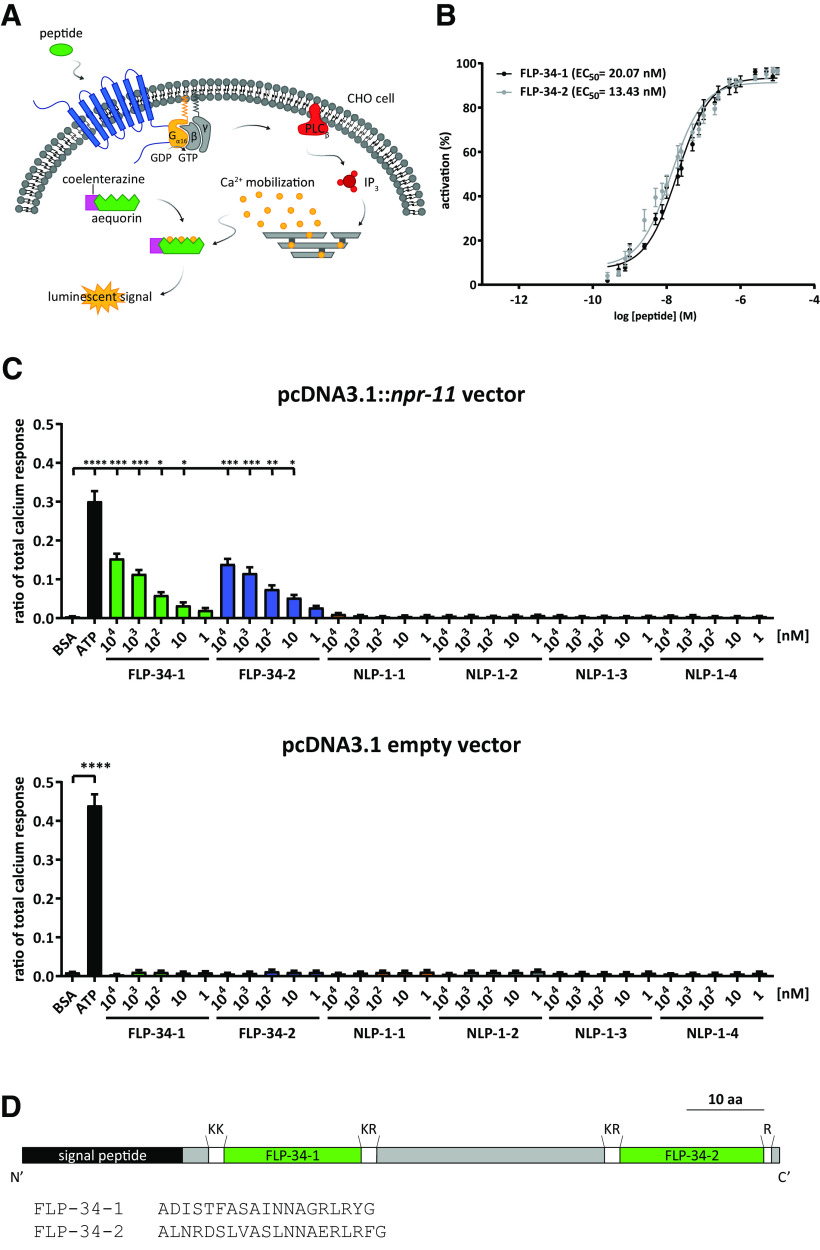
To search for neuropeptide ligands of NPR-11 involved in diacetyl learning, we used a reverse pharmacology approach and tested the functional response of this GPCR to a synthetic library of C. elegans peptides in a calcium-based reporter assay (Fig. 4A). In this cellular assay, Chinese Hamster Ovary (CHO) cells expressing NPR-11 were challenged with a library of over 350 FLP and NLP peptides. Two peptides derived from the flp-34 neuropeptide precursor gene dose-dependently activated NPR-11 with EC50 values in the nanomolar range (Fig. 4B), typical for neuropeptide-GPCR interactions, whereas cells transfected with a control vector showed no calcium responses (Fig. 4C). NLP-1 peptides did not activate NPR-11 when expressed in CHO cells, using our calcium-based receptor activation readout (Fig. 4C).
Figure 4.
FLP-34 neuropeptides of the NPY/NPF neuropeptide family activate NPR-11 in vitro. A, Schematic of the calcium-based receptor activation assay to identify ligands of NPR-11. The receptor is expressed in CHO cells that stably coexpress the promiscuous Gα16 subunit and the luminescent calcium indicator aequorin. Upon receptor activation, NPR-11 signaling via Gα16 activates phospholipase Cβ (PLCβ), resulting in intracellular Ca2+ mobilization monitored by the luminescent biosensor aequorin. B, Two peptides of the FLP-34 precursor dose-dependently activate NPR-11 with nanomolar EC50. The Ca2+ responses of CHO cells expressing NPR-11 are shown as percentage of the highest peptide-evoked response (100% activation). Error bars indicate SEM (n ≥ 7). C, FLP-34, but not NLP-1, peptides dose-dependently activate NPR-11. Bar graphs represent the ratio of total calcium response of CHO cells expressing NPR-11 (top) or pcDNA3.1 empty vector (bottom) challenged with five 10-fold serial dilutions of peptides (from 104 to 1 nm), BSA (negative control), or ATP (positive control). Peptide sequences are as follows: FLP-34-1 (ADISTFASAINNAGRLRYa), FLP-34-2 (ALNRDSLVASLNNAERLRFa), NLP-1-1 (MDANAFRMSFa), NLP-1-2 (MDPNAFRMSFa), NLP-1-3 (VNLDPNSFRMSFa), NLP-1-4 (STVGYNLDARNYFVGL). Ratio of total calcium response is calculated as peptide-evoked response normalized to the total calcium response. Data were analyzed by Kruskal–Wallis test, pcDNA3.1::npr-11 (KW(31) = 176.4, p < 0.0001), and pcDNA3.1 empty vector (KW(31) = 78.69, p < 0.0001), and Dunn's post hoc test. Error bars indicate SEM (n = 8). D, Structure of the FLP-34 neuropeptide precursor. Green represents regions of the precursor encoding the mature peptides. The peptide sequences (before amidation) are shown. Top, Cleavage sites for proprotein convertases. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
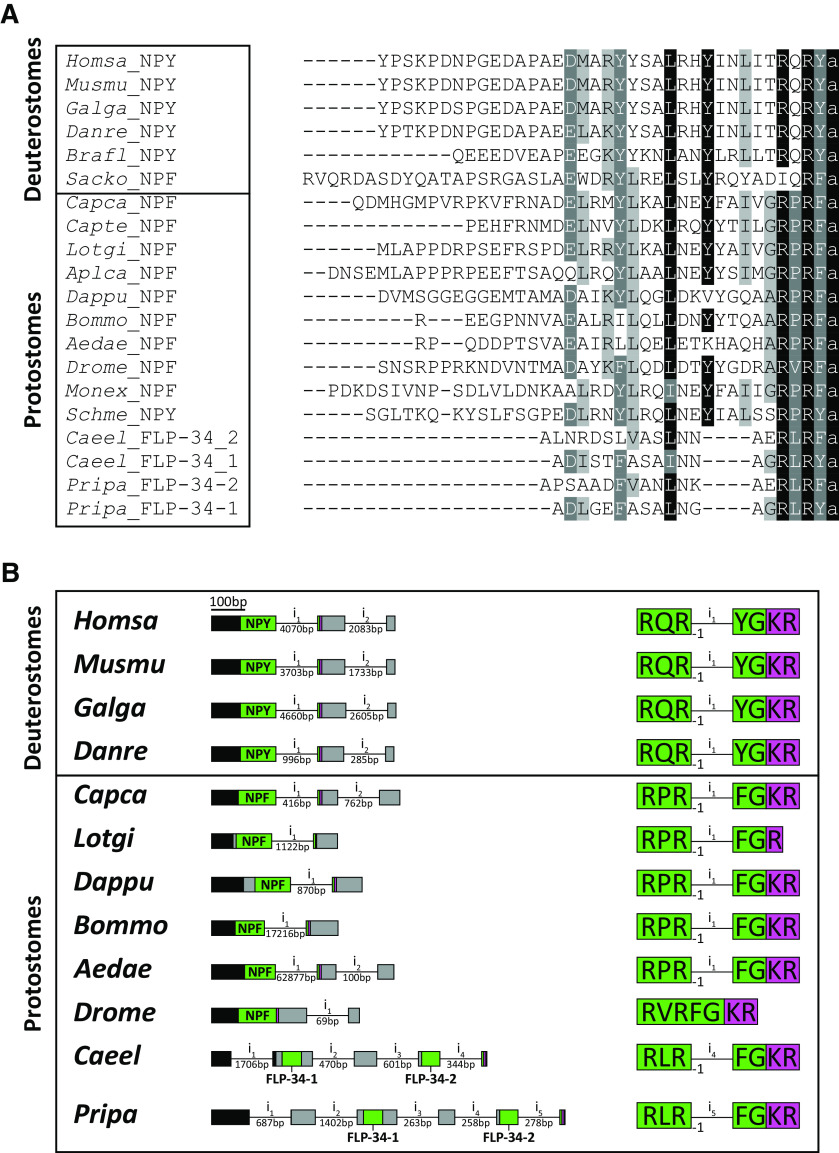
Post-translational processing of the FLP-34 neuropeptide precursor yields two mature peptides, FLP-34-1 and FLP-34-2 (Fig. 4D), both of which have been confirmed to be present in vivo by mass spectrometry (Husson et al., 2014). Interestingly, FLP-34-1 and FLP-34-2 neuropeptides display the RXRF/Yamide carboxyterminal motif typical of all bilaterian NPY/NPF neuropeptides (Fadda et al., 2019) (Fig. 5A). In addition, the flp-34 precursor gene contains a conserved exon-intron junction after the second nucleotide of the second Arginine (R) codon within the C-terminal RXRF/Ya motif of the FLP-34-2 peptide, which is conserved in the npy/npf genes of several protostomes and deuterostomes in the exact same position (Fig. 5B). The conservation of this exon-intron junction together with the conserved NPY/NPF sequence motif and ligand interaction of FLP-34 with the NPYR/NPFR homolog NPR-11 suggests that flp-34 is orthologous to the npy and npf precursor genes found in other animals.
Figure 5.
FLP-34 is orthologous to the bilaterian NPF/NPY peptides. A, Amino acid sequence alignment of representative NPF/NPYs from different phyla. Black represents identical residues with a minimum of 70% conservation. Dark gray represents amino acid groups with strongly similar properties and a minimum of 70% conservation. Light gray represents amino acid groups with strongly similar properties and a minimum of 55% conservation. FLP-34-1 and FLP-34-2 display the typical RXRF/Yamide carboxyterminal motif of all bilaterian NPY/NPF neuropeptides. FLP-34-2 has a conserved leucine residue in position 13 (counted from the last C-terminal amino acid in this alignment), whereas FLP-34-1 has an isoleucine residue at this position. Species abbreviations and references are listed in the Table 4. B, The flp-34 gene shows a conserved exon-intron junction typical of npy/npf precursors. Left, Schematic of the npf/npy gene structures. Exons are represented as boxes (scale indicated in the figure), whereas introns (i) are shown as lines (not in scale, intron length indicated underneath). The precursor protein-coding exons are color-coded to indicate the signal peptide (black), the neuropeptide (green, with name indicated), dibasic or monobasic cleavage site for proprotein convertases (magenta), and the rest of the precursor regions (gray). Right, Detailed position of the exon-intron junction in the npf/npy coding sequences. Bilaterian npf/npy genes show a conserved exon-intron junction positioned between the second and the third nucleotide of the second arginine codon (frame represented as −1) of the C-terminal RXRF/YG motif. In nematodes, only the sequence encoding for FLP-34-2 is interrupted by the intron, whereas in Drosophila the exon-intron junction is not conserved in the npf coding region. Representation of exon-intron junctions (as -1) was based on Yanez-Guerra et al., 2019. Species abbreviations and accession numbers of sequences are listed in the Table 5.
Loss-of-function mutants of flp-34 phenocopy the diacetyl learning defect of npr-11 mutants
Because FLP-34 neuropeptides activate NPR-11 in vitro, we asked whether, like NPR-11, they are involved in diacetyl learning. Thus, flp-34 mutants should display the same learning defect as npr-11 mutants. As no null mutants for flp-34 were available, we used CRISPR/Cas9 genome editing to generate a flp-34 loss-of-function mutant. For this, we designed two gRNAs to delete exon 2 and a major part of exon 3. In addition, the repair template recoded a premature stop codon in the signal peptide region to prevent further translation of the remaining sequence, including both FLP-34 peptides (Fig. 6A, Extended Data Fig. 6-1). Like npr-11 mutants, mutants defective in flp-34 still showed strong attraction to diacetyl after training (Fig. 6B), whereas locomotion behavior was unaffected (Fig. 6C,D). Moreover, a double mutant of npr-11 and flp-34 displayed a learning defect similar to that of single npr-11 and flp-34 mutants (Fig. 6B). The absence of an additive effect in the double mutant indicates that flp-34 and npr-11 act in the same genetic pathway. Together, these results suggest that FLP-34 neuropeptides signal through NPR-11 in vivo to mediate diacetyl learning.
Figure 6.
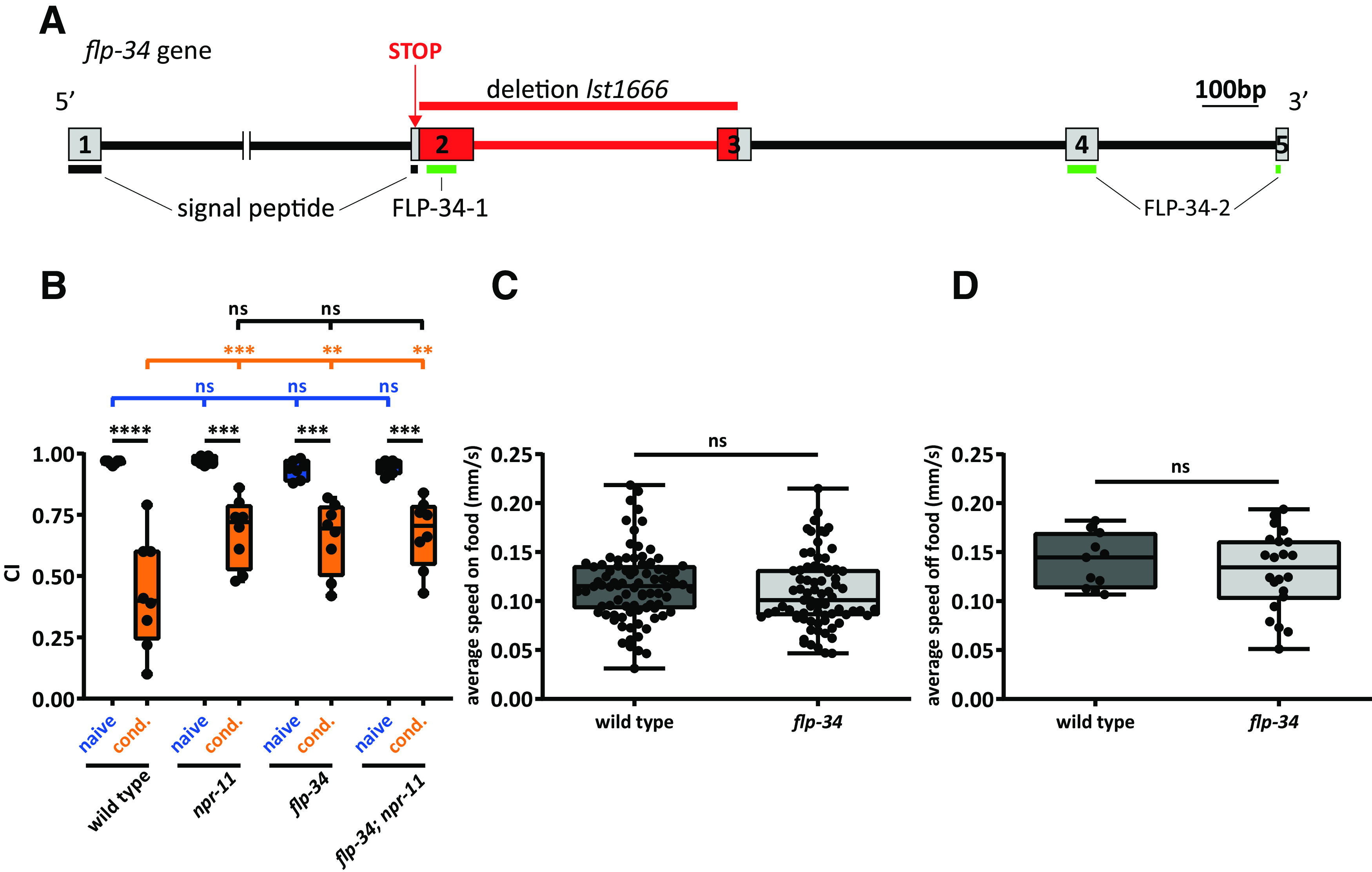
flp-34 mutants phenocopy the defect of npr-11 mutants in diacetyl learning. A, CRISPR/Cas9-induced deletion in a flp-34 loss-of-function mutant. Boxes represent exons encoding FLP-34, numbered 1-5. Lines between boxes indicate introns. Part of the intron between exons 1 and 2 is cropped. Red represents the CRISPR/Cas9 deletion (lst1666). A stop codon introduced in exon 2 interrupts translation of the signal peptide region, preventing the synthesis of downstream sequences containing both mature peptides. Horizontal lines indicate signal peptide (black) and mature peptide sequences (green). Additional information about the CRISPR/Cas9-edited deletion can be found in Extended Data Figure 6-1. B, flp-34 mutants are defective in diacetyl learning. The CI of flp-34 mutants after conditioning is similar to that of conditioned npr-11 mutant animals and significantly different from WT. An npr-11;flp-34 double mutant does not display additive learning defects. Two-way ANOVA revealed a significant effect of genotypes (F(3,52) = 3.262, p = 0.0286), of behavioral treatments (F(1,52) = 116.1, p < 0.0001), and of interaction between genotypes and behavioral treatments (F(3,52) = 4.048, p = 0.0116). Data were analyzed by Sidak's post hoc test (n ≥ 7). C, D, flp-34 mutants display normal locomotion speed on and off food. Activity is recorded for 10 min on OP50-seeded (C) or unseeded (D) NGM plates. The average speed of mutants is not significantly different from that of WT in both conditions. Data were analyzed by unpaired t test (C, w ≥ 75; D, w ≥ 11). B–D, Boxplots represent 25th (lower boundary) and 75th (upper boundary) percentiles. The 50th percentile (line) indicates the median. Whiskers represent the minimum and maximum values. Black dots represent individual CIs or worms. **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, not significant.
CRISPR/Cas9-edited deletion in the flp-34 gene. Local alignment of flp-34 WT gene with the flp-34(lst1666) mutant allele. Exons flanking the deletion are indicated in gray. The recoded regions in the lst1666 allele are highlighted in yellow and a stop codon is coloured in red. The two gRNAs used to generate the deletion are depicted in blue. Download Figure 6-1, EPS file (1.9MB, eps)
Flp-34 expression in serotonergic neurons is required for diacetyl learning
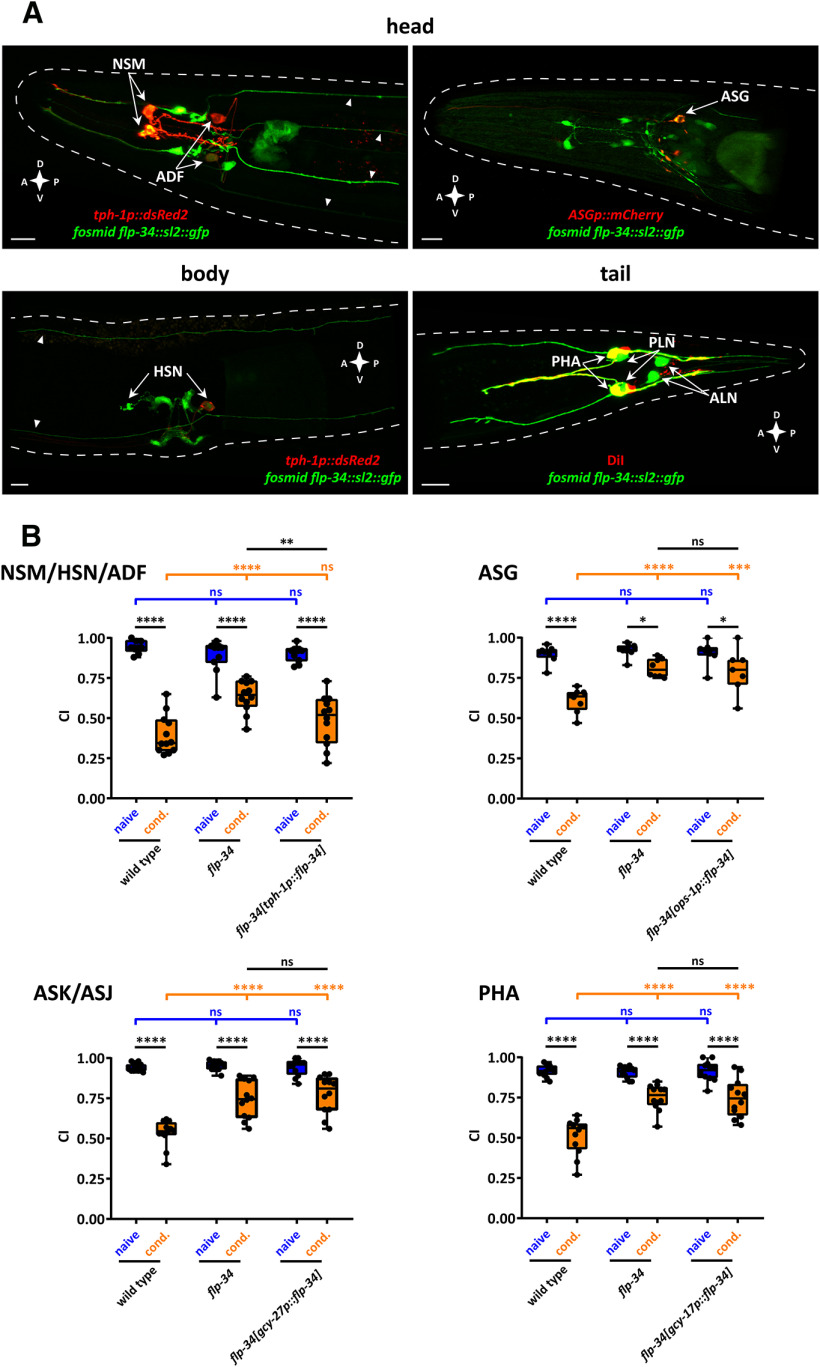
Next, we asked which cell(s) express flp-34 to understand further the NPY/NPF neural circuit modulating diacetyl learning. We generated a fosmid-based reporter transgene for flp-34 and found consistent fluorescence in several head and tail neurons, as well as in the vulval region (Fig. 7A). The flp-34 reporter transgene was expressed in all serotonergic neurons (NSM, HSN, and ADF) (Horvitz et al., 1982), validated by colocalization of the GFP signal with the expression of a red reporter transgene for tryptophan hydroxylase 1 (tph-1p::DsRed2) that encodes a serotonin biosynthesis enzyme (Sze et al., 2002) (Fig. 7A). The flp-34 transgene was also expressed in the ASG sensory neurons as confirmed by colocalization with an ASG specific reporter (nlp-44p::mCherry), in the tail PHA sensory neurons as confirmed by DiI staining, and in the PLN and ALN neurons as identified by their morphology and position (Fig. 7A).
Figure 7.
flp-34 is required in serotonergic neurons for diacetyl learning. A, Representative confocal z-stack projections of head, body, and tail neurons expressing a fosmid-based flp-34::sl2::gfp transgene in adult hermaphrodites. flp-34 expression in the serotonergic neurons NSM, ADF (head panel), and HSN (body panel) was identified by colocalization with a tph-1p::DsRed2 marker transgene. Arrowheads indicate head and body projections from tail neurons. flp-34 expression in ASG neurons (head panel) was identified by colocalization with nlp-44p::mCherry marker transgene. Expression of flp-34 in the tail neuron PHA was identified by colocalization with DiI staining, whereas ALN and PLN were determined by morphology and position (tail panel). A, Anterior; P, posterior; D, dorsal; V, ventral. Scale bars, 10 µm. B, flp-34 acts in serotonergic neurons to regulate diacetyl learning. Top left, Expressing WT copies of flp-34 under control of the tph-1 (NSM, HSN, ADF) promoter in serotonergic neurons partially rescues the learning defect of flp-34 mutant animals. The expression of flp-34 under control of ops-1 (ASG) (top right), gcy-27 (ASK, ASJ) (bottom left), and gcy-17 (PHA) (bottom right) promoters does not rescue the learning defect of flp-34 mutants. (NSM, HSN, ADF) Two-way ANOVA revealed a significant effect of genotypes (F(2,63) = 5.333, p = 0.0072), of behavioral treatments (F(1,63) = 267.4, p < 0.0001), and of interaction between genotypes and behavioral treatments (F(2,63) = 11.28, p < 0.0001). Data were analyzed by Sidak's and Tukey's post hoc test (n ≥ 11). (ASG) Two-way ANOVA revealed a significant effect of genotypes (F(2,40) = 9.418, p = 0.0004), of behavioral treatments (F(1,40) = 53.87, p < 0.0001), and of interaction between genotypes and behavioral treatments (F(2,40) = 6.521, p = 0.0035). Data were analyzed by Sidak's and Tukey's post hoc test (n = 8). (ASK, ASJ) Two-way ANOVA revealed a significant effect of genotypes (F(2,66) = 16.66, p < 0.0001), of behavioral treatments (F(1,66) = 194.3, p < 0.0001), and of interaction between genotypes and behavioral treatments (F(2,66) = 15.59, p < 0.0001). Data were analyzed by Sidak's and Tukey's post hoc test (n = 12). (PHA) Two-way ANOVA revealed a significant effect of genotypes (F(2,66) = 16.77, p < 0.0001), of behavioral treatments (F(1,66) = 161.6, p < 0.0001), and of interaction between genotypes and behavioral treatments (F(2,66) = 17.90, p < 0.0001). Data were analyzed by Sidak's and Tukey's post hoc test (n = 12). Boxplots represent 25th (lower boundary) and 75th (upper boundary) percentiles. The 50th percentile (line) indicates the median. Whiskers represent the minimum and maximum values. Black dots represent individual CIs. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, not significant.
Given the correlation between serotonin signaling and feeding state in C. elegans (Srinivasan et al., 2008), we hypothesized that NPY/NPF-like neuropeptides signal from these serotonergic cells to mediate diacetyl learning. We cell-specifically restored flp-34 expression in NSM, HSN, and ADF neurons using the tph-1 promoter (Sze et al., 2002), which rescued the diacetyl learning defect (Fig. 7B). By contrast, restoring flp-34 in other neurons that express the flp-34 gene, as evidenced by reporter transgene expression (Fig. 7A) or single-cell RNA-sequencing (Cao et al., 2017; Taylor et al., 2019), did not rescue this mutant phenotype. Restoring expression of flp-34 in ASK/ASJ, ASG, or PHA neurons did not restore the learning defect of flp-34 mutants (Fig. 7B). This suggests that flp-34 is specifically required in serotonergic cells to facilitate diacetyl learning rather than that learning is mediated by unregulated neuropeptide secretion. Together, these data show that NPY/NPF-related neuropeptides signal from serotonergic neurons, and possibly other unidentified cells, to NPR-11 in AIA interneurons regulating diacetyl learning.
NPY/NPF-like FLP-34 neuropeptides and serotonin act in the same pathway to mediate diacetyl learning
The requirement of flp-34 in serotonergic neurons for diacetyl learning suggests that NPY/NPF-like neuropeptides and serotonin act together to mediate aversive learning. To investigate this, we tested diacetyl learning for tph-1 mutants, defective in serotonin biosynthesis. After conditioning, tph-1 mutants displayed a learning defect, which was more severe than that of npr-11 mutants (Fig. 8A). This suggests that serotonin, in addition to NPY/NPF-like signaling, mediates diacetyl learning. We next generated a tph-1;npr-11 double mutant to simultaneously block serotonin and NPYR/NPFR signaling, and compared diacetyl learning of the single and double mutants. The learning defect displayed by tph-1;npr-11 mutants was not significantly different from that of tph-1 mutant animals (Fig. 8A), suggesting that serotonin and NPR-11 act in the same pathway to regulate diacetyl learning (Fig. 8B).
Figure 8.
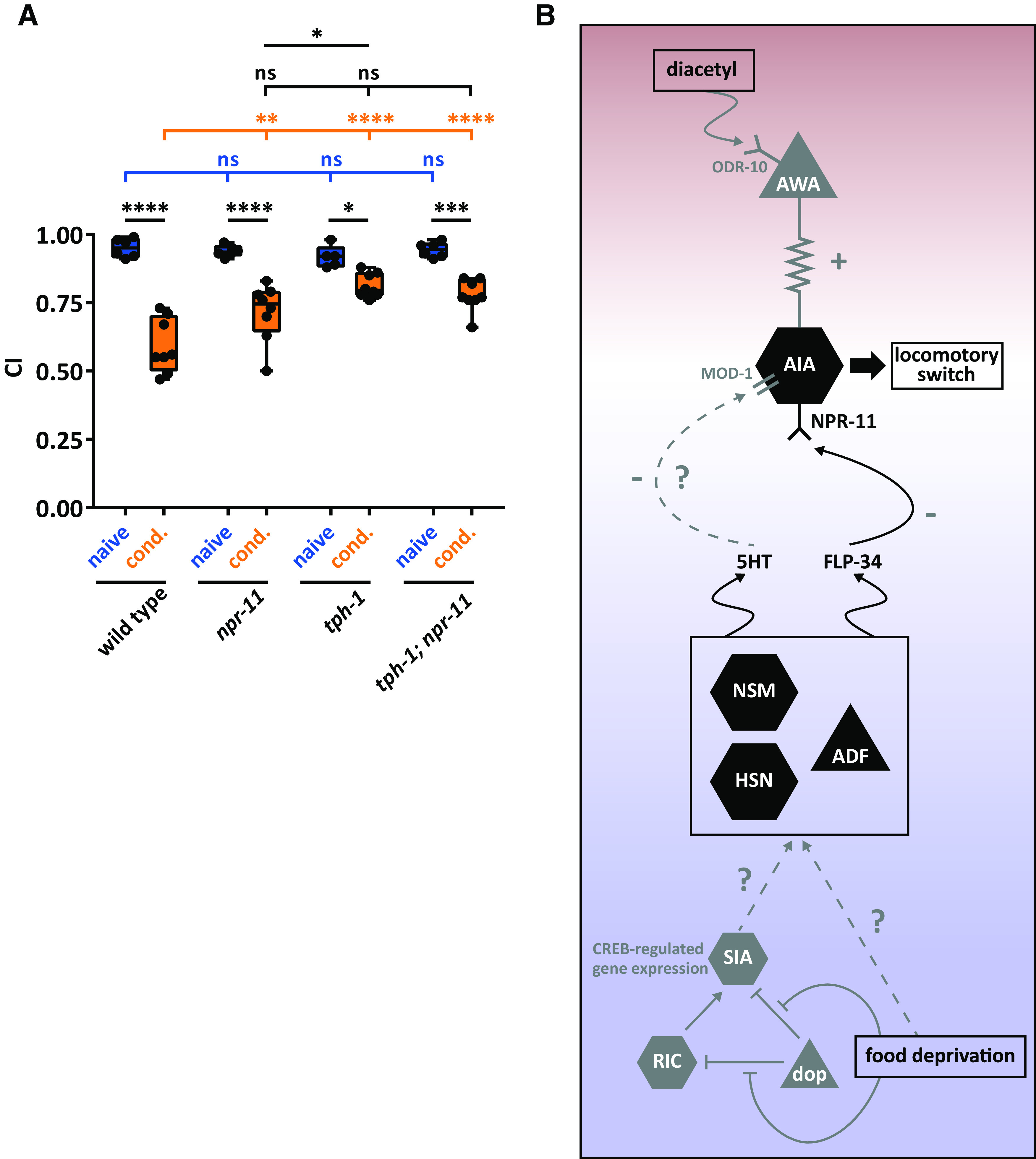
NPY/NPF-related FLP-34 neuropeptides and serotonin act in the same genetic pathway to regulate diacetyl learning. A, Both npr-11 and tph-1 mutants display learning defects compared with WT animals, the defect of tph-1 being more severe. An npr-11;tph-1 double mutant does not display an additive learning defect compared with the single mutants, suggesting that npr-11 and serotonin signaling act in the same pathway to mediate diacetyl learning. Two-way ANOVA revealed a significant effect of genotypes (F(3,47) = 5.888, p = 0.0017), of behavioral treatments (F(1,47) = 140.2, p < 0.0001), and of interaction between genotypes and behavioral treatments (F(3,47) = 8.950, p < 0.0001). Data were analyzed by Sidak's and Tukey's post hoc test (n ≥ 5). Boxplots represent 25th (lower boundary) and 75th (upper boundary) percentiles. The 50th percentile (line) indicates the median. Whiskers represent the minimum and maximum values. Black dots represent individual CIs. *p < 0.5, **p < 0.01, ***p < 0.001, ****p < 0.0001. ns, not significant. B, Model for NPR-11-mediated diacetyl learning based on data from this paper (black) and previously reported findings (gray; see Discussion). Diacetyl is sensed by the ODR-10 receptor in AWA sensory neurons (Sengupta et al., 1996). AWA activation is transmitted via electrical synapses to the AIA interneurons that express NPR-11 (Larsch et al., 2015). The absence of food leads to a food-deprived state that is perceived by a three-neuron-type circuit consisting of dopaminergic (dop), RIC, and SIA neurons (Suo et al., 2009). The absence of food leads to dopamine signaling inhibition and, in turn, to CREB-regulated gene expression in SIA neurons (Suo et al., 2009). By unknown mechanisms, food deprivation may trigger the release of serotonin and FLP-34 neuropeptides from one or more serotonergic neurons. The NPY/NPF-like FLP-34 neuropeptides signal to AIA by activating NPR-11 in these neurons, which are required for diacetyl learning. Serotonin (5HT) may activate the MOD-1 serotonin-gated chloride channel (Zhang et al., 2005). Red shading represents the part of the circuit activated by diacetyl perception. Blue shading represents the part activated by food deprivation. The two circuits converge on AIA, where diacetyl aversive memory is probably formed.
Discussion
Several molecular mechanisms for learning and memory have shown to be conserved during evolution (Glanzman, 2008; Ardiel and Rankin, 2010; Kandel, 2012). Mounting evidence implies neuropeptides to be key modulators of experience-dependent behaviors (Bargmann, 2012). Here, we show that NPY/NPF-related neuropeptides from serotonergic neurons signal through the NPYR/NPFR ortholog NPR-11 to mediate aversive olfactory learning in C. elegans.
Several bioinformatic studies have classified C. elegans NPRs as putative NPYR/NPFR orthologs (Keating et al., 2003). NPR-1 was first annotated as an NPYR homolog based on its sequence similarity with human NPYRs (de Bono and Bargmann, 1998). Later on, 11 additional C. elegans NPRs (NPR-2 to −8 and NPR-10 to −13) were predicted as NPYR/NPFR homologs based on a phylogenetic analysis of C. elegans GPCRs (Keating et al., 2003). However, bilaterian-wide phylogenetic analyses of neuropeptide GPCRs revealed that the majority of these putative NPYR/NPFR in C. elegans cluster closely with insect short NPF receptors, suggesting that this family has largely expanded in nematodes (Jékely, 2013; Mirabeau and Joly, 2013). These studies also revealed NPR-11 as the genuine C. elegans ortholog of the bilaterian-wide NPYR/NPFR family (Jékely, 2013; Mirabeau and Joly, 2013). A recent study showed that pharmacological concentrations of human NPY activate C. elegans NPR-11 in vitro, whereas the same or higher concentrations of human NPY (10 μm) elicited weak or no responses by other tested C. elegans NPRs (Gershkovich et al., 2019).
Here we show that the NPY/NPF-like FLP-34 neuropeptides are cognate ligands of NPR-11. Several of our results support the orthology of these neuropeptides with the bilaterian NPY/NPF family: (1) the FLP-34 neuropeptide precursor contains two peptides, FLP-34-1 and FLP-34-2, which display structural hallmarks of bilaterian NPY/NPF neuropeptides. These include the evolutionarily conserved C-terminal RXRF/Yamide motif, which is distinct from the M/T/L/FRFamide motif of sNPF neuropeptides (Fadda et al., 2019). (2) Both FLP-34-1 and FLP-34-2 peptides dose-dependently activate the NPYR/NPFR ortholog NPR-11 in vitro with EC50 values in the nanomolar range. (3) The flp-34 gene also displays a conserved exon-intron junction that is retained in the same identical position in most of the analyzed bilaterian npf/npy genes, including insect (A. aegypti, B. mori, and D. pulex), nematode (P. pacificus), mollusc (L. gigantea), annelid (C. capitata), and vertebrate (D. rerio, G. gallus, M. musculus, and H. sapiens) homologs. Remarkably, the conservation of this exon-intron structure is not retained in Drosophila npf. Although the FLP-34 precursor contains two mature NPY/NPF-like peptides, only the C-terminal sequence of FLP-34-2 is interrupted by the conserved exon-intron junction, suggesting that FLP-34-1 may be a more recent duplication of FLP-34-2 in nematodes. This is supported by the fact that NPY/NPF precursors outside nematodes produce only a single mature peptide, whereas in nematodes they contain two NPFs (McCoy et al., 2014).
We found that FLP-34 is expressed in the three serotonergic neurons (NSM, HSN, and ADF), in the ASG sensory neurons and in the tail neurons ALN, PLN, and PHA, in agreement with published single cell RNA-seq data (Cao et al., 2017; Taylor et al., 2019). Both RNA-seq data and our expression profile evidenced FLP-34 expression in additional neurons, although we could not establish their identities in the fosmid reporter strain. We showed that FLP-34 neuropeptides signal from serotonergic neurons to NPR-11 in AIA interneurons to mediate diacetyl learning. AIA interneurons are important sensory integration centres (Chalasani et al., 2010; Shinkai et al., 2011; Dobosiewicz et al., 2019) required for aversive but not appetitive learning (Tomioka et al., 2006; Lin et al., 2010; Cho et al., 2016). In agreement with this, we showed that npr-11 in AIA is required for aversive olfactory learning. Aversive learning is an evolutionarily conserved trait that allows animals to avoid harmful situations and survive in a competitive environment. In rodents, avoidance learning also requires NPY signaling (Morley et al., 1990; Gøtzsche and Woldbye, 2016). In adult Drosophila, NPF signaling mediates appetitive learning under conditions of food deprivation and is thought to mimic the absence of food (Krashes et al., 2009). The absence of food might also trigger FLP-34 release from C. elegans serotonergic neurons in diacetyl learning. This is consistent with our findings that deficient NPYR/NPFR signaling in C. elegans does not affect appetitive learning in the positive butanone association paradigm, where worms are exposed to butanone in the presence of food.
How does FLP-34/NPR-11 signaling modulate aversive olfactory learning? Together with previous work, our results suggest the following model for diacetyl learning (Fig. 8B). The simultaneous presentation of diacetyl with food deprivation leads to the activation of neurons sensing odorants and feeding state. Diacetyl is sensed by the ODR-10 receptor in AWA neurons (Sengupta et al., 1996), which in turn depolarize AIA interneurons via gap junctions (Larsch et al., 2015). The absence of food, on the other hand, is signaled by a three-neuron-type circuit in which octopamine triggers CREB-regulated gene expression in SIA neurons to initiate a behavioral response to food deprivation (Suo et al., 2009). In addition, serotonergic neurons are well known for their role in signaling internal and external feeding cues (Horvitz et al., 1982; Srinivasan et al., 2008). We show that FLP-34 is required in serotonergic neurons for diacetyl learning, and food deprivation perceived by these neurons may trigger release of FLP-34. This role of FLP-34 neuropeptides would be consistent with the evolutionarily conserved role of NPY/NPF in hunger signaling and feeding (Sohn et al., 2013; Fadda et al., 2019). In mammals, modulation of feeding behavior by NPY is mainly restricted to hypothalamic nuclei, where Y1 and Y5 receptors stimulate orexigenic signaling under deprived conditions (Kohno and Yada, 2012). In Drosophila larvae, NPF signaling increases food-seeking behavior and inhibits locomotion to promote feeding under food-deprived circumstances (Wu et al., 2005). In C. elegans, NPR-11 in AIA has been shown to modulate local search behavior when worms are removed from food (Chalasani et al., 2010), which is reminiscent of NPF-regulated food-seeking behavior in starved Drosophila. Diacetyl and food deprivation cues thus seem to converge on AIA interneurons expressing the NPYR/NPFR ortholog NPR-11.
In addition to FLP-34, our results suggest that serotonin modulates diacetyl learning, which is in agreement with its role as a reinforcement signal in aversive associative learning both in Drosophila and C. elegans (Giurfa, 2006). Based on the reported role of serotonin in signaling feeding state (Srinivasan et al., 2008; Donovan and Tecott, 2013; Voigt and Fink, 2015) and its action as a reinforcement signal in negative associations (Giurfa, 2006; Sitaraman et al., 2017), serotonin signaling may also reinforce the food deprivation cue in diacetyl learning. Although its target in this pathway remains unknown, the serotonin-gated chloride channel MOD-1 (Ranganathan et al., 2000), expressed in AIA as well as in most of the first layer interneurons of C. elegans, may be a promising candidate (Fig. 8B) (Harris et al., 2009). In another aversive learning paradigm where pathogenic bacteria serve as the aversive cue, MOD-1 activation on serotonin release results in the inhibition of the first layer interneurons, including AIA, and in a change of olfactory preference (Zhang et al., 2005). A similar mechanism could take place in diacetyl learning. All bilaterian NPY/NPF neuropeptides investigated so far mainly have an antiexcitatory effect on their receptor-expressing neurons (Vollmer et al., 2016), in agreement with the coupling of NPYR/NPFR to a G0/i α inhibitory subunit (Gershkovich et al., 2019). This suggests that FLP-34 signaling through NPR-11 may have an antiexcitatory effect on AIA, thus inhibiting AIA on release in an environment deprived of qualitative food.
NPR-11 signaling in AIA also promotes local search behavior and adaptation to isoamyl alcohol, behaviors mediated by the AWC-released NLP-1 neuropeptides that act through NPR-11 in AIA (Chalasani et al., 2010). Our findings show that NLP-1 is not required for diacetyl learning. In addition, our receptor activation assay shows that flp-34-, but not nlp-1-derived, peptides activate NPR-11 in vitro. This discrepancy in NPR-11 activation could be related to the different cellular expression systems used in the two studies: we expressed npr-11 in CHO cells, whereas Chalasani et al. (2010) used human embryonic kidney (HEK)293 cells. Previous studies demonstrated that calcium responses elicited by receptor activation in heterologous expression systems can be affected by the endogenous cellular context (Hermansson et al., 2007; Zhang et al., 2018). Depending on the conditions, NPR-11 in AIA can thus be activated by FLP-34 neuropeptides released from serotonergic neurons, or by NLP-1 neuropeptides released from AWC. These findings suggest that different environmental stimuli trigger the release of distinct neuropeptides that in turn activate the same receptor in the same neuron, both resulting in a locomotory change, but overall distinct output behaviors.
In conclusion, we identify an NPY/NPF-related signaling system and neural mechanism for the control of aversive olfactory learning in C. elegans. This opens interesting perspectives for investigating the coordinated action of NPY/NPF neuropeptides and serotonin mediating aversive reinforcement in other learning paradigms, which could aid in our understanding of the molecular mechanisms underlying stress disorders in which excessive avoidance results in maladaptive behaviors (Krypotos et al., 2015).
Footnotes
The authors declare no competing financial interests.
This work was supported by European Research Council Grant 340318 and the Research Foundation–Flanders Grant G0C0618N. The Caenorhabditis Genetics Center, funded by National Institutes of Health Office of Research Infrastructure Programs P40 OD010440, for providing the Caenorhabditis elegans strains used in this study. We thank Cori Bargmann for providing the pSM vector, the CX8912 and CX7894 strains, and advice on the manuscript; and Gunther Hollopeter for providing the pGH496-Cas9 expressing vector.
References
- Ardiel EL, Rankin CH (2010) An elegant mind: learning and memory in Caenorhabditis elegans. Learn Mem 17:191–201. 10.1101/lm.960510 [DOI] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, Fire AZ (2014) Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198:837–846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI. (2012) Beyond the connectome: how neuromodulators shape neural circuits. BioEssays 34:458–465. 10.1002/bies.201100185 [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Hartwieg E, Horvitz HR (1993) Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74:515–527. 10.1016/0092-8674(93)80053-H [DOI] [PubMed] [Google Scholar]
- Beets I, Janssen T, Meelkop E, Temmerman L, Suetens N, Rademakers S, Jansen G, Schoofs L (2012) Vasopressin/oxytocin-related signaling regulates gustatory associative learning in C. elegans. Science 338:543–545. 10.1126/science.1226860 [DOI] [PubMed] [Google Scholar]
- Bernhard N, van der Kooy D (2000) A behavioral and genetic dissection of two forms of olfactory plasticity in Caenorhabditis elegans: adaptation and habituation. Learn Mem 7:199–212. 10.1101/lm.7.4.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomqvist AG, Soderberg C, Lundell I, Milner RJ, Larhammar D (1992) Strong evolutionary conservation of neuropeptide Y: sequences of chicken, goldfish, and Torpedo marmorata DNA clones. Proc Natl Acad Sci USA 89:2350–2354. 10.1073/pnas.89.6.2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MR, Crim JW, Arata RC, Cai HN, Chun C, Shen P (1999) Identification of a Drosophila brain-gut peptide related to the neuropeptide Y family. Peptides 20:1035–1042. 10.1016/s0196-9781(99)00097-2 [DOI] [PubMed] [Google Scholar]
- Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, Adey A, Waterston RH, Trapnell C, Shendure J (2017) Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357:661–667. 10.1126/science.aam8940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalasani SH, Kato S, Albrecht DR, Nakagawa T, Abbott LF, Bargmann CI (2010) Neuropeptide feedback modifies odor-evoked dynamics in Caenorhabditis elegans olfactory neurons. Nat Neurosci 13:615–621. 10.1038/nn.2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho CE, Brueggemann C, L'Etoile ND, Bargmann CI (2016) Parallel encoding of sensory history and behavioral preference during Caenorhabditis elegans olfactory learning. Elife 5:e14000 10.7554/eLife.14000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbert HA, Bargmann CI (1997) Environmental signals modulate olfactory acuity, discrimination, and memory in Caenorhabditis elegans. Learn Mem 4:179–191. 10.1101/lm.4.2.179 [DOI] [PubMed] [Google Scholar]
- de Bono M, Bargmann CI (1998) Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell 94:679–689. 10.1016/S0092-8674(00)81609-8 [DOI] [PubMed] [Google Scholar]
- Deng X, Yang H, He X, Liao Y, Zheng C, Zhou Q, Zhu C, Zhang G, Gao J, Zhou N (2014) Activation of Bombyx neuropeptide G protein-coupled receptor A4 via a Gαi-dependent signaling pathway by direct interaction with neuropeptide F from silkworm, Bombyx mori. Insect Biochem Mol Biol 45:77–88. 10.1016/j.ibmb.2013.12.007 [DOI] [PubMed] [Google Scholar]
- Dobosiewicz M, Liu Q, Bargmann CI (2019) Reliability of an interneuron response depends on an integrated sensory state. ELife 8:e50566 10.7554/eLife.50566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan MH, Tecott LH (2013) Serotonin and the regulation of mammalian energy balance. Front Neurosci 7:36. 10.3389/fnins.2013.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elphick MR, Mirabeau O (2014) The evolution and variety of RFamide-type neuropeptides: insights from deuterostomian invertebrates. Front Endocrinol (Lausanne) 5:93. 10.3389/fendo.2014.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elphick MR, Mirabeau O, Larhammar D (2018) Evolution of neuropeptide signalling systems. J Exp Biol 221:jeb151092 10.1242/jeb.151092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadda M, Hasakiogullari I, Temmerman L, Beets I, Zels S, Schoofs L (2019) Regulation of feeding and metabolism by neuropeptide F and short neuropeptide F in invertebrates. Front Endocrinol (Lausanne) 10:64. 10.3389/fendo.2019.00064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost WN, Castellucci VF, Hawkins RD, Kandel ER (1985) Monosynaptic connections made by the sensory neurons of the gill- and siphon-withdrawal reflex in Aplysia participate in the storage of long-term memory for sensitization. Proc Natl Acad Sci USA 82:8266–8269. 10.1073/pnas.82.23.8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershkovich MM, Groß VE, Kaiser A, Prömel S (2019) Pharmacological and functional similarities of the human neuropeptide Y system in C. elegans challenges phylogenetic views on the FLP/NPR system. Cell Commun Signal 17:123. 10.1186/s12964-019-0436-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giurfa M. (2006) Associative learning: the instructive function of biogenic amines. Curr Biol 16:R892–R895. 10.1016/j.cub.2006.09.021 [DOI] [PubMed] [Google Scholar]
- Glanzman DL. (2008) The cell biology of learning and memory in Aplysia. Adv Psychol 139:243–270. [Google Scholar]
- Gøtzsche CR, Woldbye DP (2016) The role of NPY in learning and memory. Neuropeptides 55:79–89. 10.1016/j.npep.2015.09.010 [DOI] [PubMed] [Google Scholar]
- Harris GP, Hapiak VM, Wragg RT, Miller SB, Hughes LJ, Hobson RJ, Steven R, Bamber B, Komuniecki RW (2009) Three distinct amine receptors operating at different levels within the locomotory circuit are each essential for the serotonergic modulation of chemosensation in Caenorhabditis elegans. J Neurosci 29:1446–1456. 10.1523/JNEUROSCI.4585-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermansson NO, Morgan DG, Drmota T, Larsson N (2007) Adenosine is not a direct GHSR agonist: artificial cross-talk between GHSR and adenosine receptor pathways. Acta Physiol (Oxf) 190:77–86. 10.1111/j.1365-201X.2007.01691.x [DOI] [PubMed] [Google Scholar]
- Hobert O. (2003) Behavioral plasticity in C. elegans: paradigms, circuits, genes. J Neurobiol 54:203–223. 10.1002/neu.10168 [DOI] [PubMed] [Google Scholar]
- Horvitz H, Chalfie M, Trent C, Sulston J, Evans P (1982) Serotonin and octopamine in the nematode Caenorhabditis elegans. Science 216:1012–1014. 10.1126/science.6805073 [DOI] [PubMed] [Google Scholar]
- Husson SJ, Clynen E, Baggerman G, De Loof A, Schoofs L (2005) Discovering neuropeptides in Caenorhabditis elegans by two dimensional liquid chromatography and mass spectrometry. Biochem Biophys Res Commun 335:76–86. 10.1016/j.bbrc.2005.07.044 [DOI] [PubMed] [Google Scholar]
- Husson SJ, Reumer A, Temmerman L, De Haes W, Schoofs L, Mertens I, Baggerman G (2014) Worm peptidomics. EuPA Open Proteom 3:280–290. 10.1016/j.euprot.2014.04.005 [DOI] [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23:403–405. 10.1016/s0968-0004(98)01285-7 [DOI] [PubMed] [Google Scholar]
- Jékely G. (2013) Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc Natl Acad Sci USA 110:8702–8707. 10.1073/pnas.1221833110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER. (2012) The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol Brain 5:14. 10.1186/1756-6606-5-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl T, Lin S, Schwarzer C, Sainsbury A, Couzens M, Wittmann W, Boey D, von Hörsten S, Herzog H (2004) Y1 receptors regulate aggressive behavior by modulating serotonin pathways. Proc Natl Acad Sci USA 101:12742–12747. 10.1073/pnas.0404085101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman A, Parsons L, Stein G, Wills A, Kaletsky R, Murphy C (2011) C. elegans positive butanone learning, short-term, and long-term associative memory assays. J Vis Exp 49:e2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating CD, Kriek N, Daniels M, Ashcroft NR, Hopper NA, Siney EJ, Holden-Dye L, Burke JF (2003) Whole-genome analysis of 60 G protein-coupled receptors in Caenorhabditis elegans by gene knockout with RNAi. Curr Biol 13:1715–1720. 10.1016/j.cub.2003.09.003 [DOI] [PubMed] [Google Scholar]
- Kohno D, Yada T (2012) Arcuate NPY neurons sense and integrate peripheral metabolic signals to control feeding. Neuropeptides 46:315–319. 10.1016/j.npep.2012.09.004 [DOI] [PubMed] [Google Scholar]
- Krashes MJ, DasGupta S, Vreede A, White B, Armstrong JD, Waddell S, Douglas AJ, Waddell S (2009) A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell 139:416–427. 10.1016/j.cell.2009.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krypotos AM, Effting M, Kindt M, Beckers T (2015) Avoidance learning: a review of theoretical models and recent developments. Front Behav Neurosci 9:189. 10.3389/fnbeh.2015.00189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Larsch J, Flavell SW, Liu Q, Gordus A, Albrecht DR, Bargmann CI (2015) A circuit for gradient climbing in C. elegans chemotaxis. Cell Rep 12:1748–1760. 10.1016/j.celrep.2015.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau HL, Timbers TA, Mahmoud R, Rankin CH (2013) Genetic dissection of memory for associative and non-associative learning in Caenorhabditis elegans. Genes Brain Behav 12:210–223. 10.1111/j.1601-183X.2012.00863.x [DOI] [PubMed] [Google Scholar]
- Lesch BJ, Gehrke AR, Bulyk ML, Bargmann CI (2009) Transcriptional regulation and stabilization of left-right neuronal identity in C. elegans. Genes Dev 23:345–358. 10.1101/gad.1763509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Tomioka M, Pereira S, Sellings L, Iino Y, van der Kooy D (2010) Insulin signaling plays a dual role in Caenorhabditis elegans memory acquisition and memory retrieval. J Neurosci 30:8001–8011. 10.1523/JNEUROSCI.4636-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule AG, Shaw C, Halton DW, Thim L, Johnston CF, Fairweather I, Buchanan KD (1991) Neuropeptide F: a novel parasitic flatworm regulatory peptide from Moniezia expansa (Cestoda: cyclophyllidea). Parasitology 102:309–316. 10.1017/S0031182000062648 [DOI] [Google Scholar]
- McCoy CJ, Atkinson LE, Zamanian M, McVeigh P, Day TA, Kimber MJ, Marks NJ, Maule AG, Mousley A (2014) New insights into the FLPergic complements of parasitic nematodes: informing deorphanisation approaches. EuPA Open Proteom 3:262–272. 10.1016/j.euprot.2014.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minth CD, Andrews PC, Dixon JE (1986) Characterization, sequence, and expression of the cloned human neuropeptide Y gene. J Biol Chem 261:11974–11979. [PubMed] [Google Scholar]
- Mirabeau O, Joly JS (2013) Molecular evolution of peptidergic signaling systems in bilaterians. Proc Natl Acad Sci USA 110:E2028–E2037. 10.1073/pnas.1219956110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley JE, Flood JF, Louis S (1990) Neuropeptide Y and memory processing. Ann NY Acad Sci 611:226–231. 10.1111/j.1749-6632.1990.tb48934.x [DOI] [PubMed] [Google Scholar]
- Nuss AB, Forschler BT, Crim JW, TeBrugge V, Pohl J, Brown MR (2010) Molecular characterization of neuropeptide F from the eastern subterranean termite Reticulitermes flavipes (Kollar) (Isoptera: rhinotermitidae). Peptides 31:419–428. 10.1016/j.peptides.2009.09.001 [DOI] [PubMed] [Google Scholar]
- Nuttley WM, Harbinder S, van der Kooy D (2001) Regulation of distinct attractive and aversive mechanisms mediating benzaldehyde chemotaxis in Caenorhabditis elegans. Learn Mem 8:170–181. 10.1101/lm.36501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Rasoloson D, Seydoux G (2015) High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics 201:47–54. 10.1534/genetics.115.179382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peymen K, Watteyne J, Borghgraef C, Van Sinay E, Beets I, Schoofs L (2019) Myoinhibitory peptide signaling modulates aversive gustatory learning in Caenorhabditis elegans. PLoS Genet 15:e1007945. 10.1371/journal.pgen.1007945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsker H, Kupfermann I, Castellucci V, Kandel E (1970) Habituation and dishabituation of the gill-withdrawal reflex in Aplysia. Science 167:1740–1742. 10.1126/science.167.3926.1740 [DOI] [PubMed] [Google Scholar]
- Rajpara SM, Garcia PD, Roberts R, Eliassen JC, Owens DF, Maltby D, Myers RM, Mayeri E (1992) Identification and molecular cloning of a neuropeptide Y homolog that produces prolonged inhibition in Aplysia neurons. Neuron 9:505–513. 10.1016/0896-6273(92)90188-j [DOI] [PubMed] [Google Scholar]
- Ranganathan R, Cannon SC, Horvitz HR (2000) MOD-1 is a serotonin-gated chloride channel that modulates locomotory behaviour in C. elegans. Nature 408:470–475. 10.1038/35044083 [DOI] [PubMed] [Google Scholar]
- Sasakura H, Mori I (2013) Behavioral plasticity, learning, and memory in C. elegans. Curr Opin Neurobiol 23:92–99. 10.1016/j.conb.2012.09.005 [DOI] [PubMed] [Google Scholar]
- Sengupta P, Chou JH, Bargmann CI (1996) odr-10 encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell 84:899–909. 10.1016/s0092-8674(00)81068-5 [DOI] [PubMed] [Google Scholar]
- Shinkai Y, Yamamoto Y, Fujiwara M, Tabata T, Murayama T, Hirotsu T, Ikeda DD, Tsunozaki M, Iino Y, Bargmann CI, Katsura I, Ishihara T (2011) Behavioral choice between conflicting alternatives is regulated by a receptor guanylyl cyclase, GCY-28, and a receptor tyrosine kinase, SCD-2, in AIA interneurons of Caenorhabditis elegans. J Neurosci 31:3007–3015. 10.1523/JNEUROSCI.4691-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaraman D, Kramer EF, Kahsai L, Ostrowski D, Zars T (2017) Discrete serotonin systems mediate memory enhancement and escape latencies after unpredicted aversive experience in Drosophila place memory. Front Syst Neurosci 11:92. 10.3389/fnsys.2017.00092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg C, Wraith A, Ringvall M, Yan YL, Postlethwait JH, Brodin L, Larhammar D (2000) Zebrafish genes for neuropeptide Y and peptide YY reveal origin by chromosome duplication from an ancestral gene linked to the homeobox cluster. J Neurochem 75:908–918. 10.1046/j.1471-4159.2000.0750908.x [DOI] [PubMed] [Google Scholar]
- Sohn JW, Elmquist JK, Williams KW (2013) Neuronal circuits that regulate feeding behavior and metabolism. Trends Neurosci 36:504–512. 10.1016/j.tins.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Sadegh L, Elle IC, Christensen AG, Faergeman NJ, Ashrafi K (2008) Serotonin regulates C. elegans fat and feeding through independent molecular mechanisms. Cell Metab 7:533–544. 10.1016/j.cmet.2008.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek DM, Pohl J, Crim JW, Brown MR (2002) Neuropeptide F and its expression in the yellow fever mosquito, Aedes aegypti. Peptides 23:1367–1378. 10.1016/s0196-9781(02)00074-8 [DOI] [PubMed] [Google Scholar]
- Stetak A, Hörndli F, Maricq AV, van den Heuvel S, Hajnal A (2009) Neuron-specific regulation of associative learning and memory by MAGI-1 in C. elegans. PLoS One 4:e6019. 10.1371/journal.pone.0006019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo S, Culotti JG, Van Tol HH (2009) Dopamine counteracts octopamine signalling in a neural circuit mediating food response in C. elegans. EMBO J 28:2437–2448. 10.1038/emboj.2009.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze JY, Zhang S, Li J, Ruvkun G (2002) The C. elegans POU-domain transcription factor UNC-86 regulates the tph-1 tryptophan hydroxylase gene and neurite outgrowth in specific serotonergic neurons. Development 129:3901–3911. [DOI] [PubMed] [Google Scholar]
- Taghert PH, Nitabach MN (2012) Peptide neuromodulation in invertebrate model systems. Neuron 76:82–97. 10.1016/j.neuron.2012.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SR, Santpere G, Reilly M, Glenwinkel L, Poff A, McWhirter R, Xu C, Weinreb A, Basavaraju M, Cook SJ, Barrett A, Abrams A, Vidal B, Cros C, Rafi I, Sestan N, Hammarlund M, Hobert O, Miller DM (2019) Expression profiling of the mature C. elegans nervous system by single-cell RNA-sequencing. bioRxiv 737577. doi: . [Google Scholar]
- Tomioka M, Adachi T, Suzuki H, Kunitomo H, Schafer WR, Iino Y (2006) The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron 51:613–625. 10.1016/j.neuron.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Tong YG, Bürglin TR (2010) Conditions for dye-filling of sensory neurons in Caenorhabditis elegans. J Neurosci Methods 188:58–61. 10.1016/j.jneumeth.2010.02.003 [DOI] [PubMed] [Google Scholar]
- Troemel ER, Sagasti A, Bargmann CI (1999) Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99:387–398. 10.1016/S0092-8674(00)81525-1 [DOI] [PubMed] [Google Scholar]
- Tursun B, Cochella L, Carrera I, Hobert O (2009) A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS One 4:e4625. 10.1371/journal.pone.0004625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bael S, Watteyne J, Boonen K, De Haes W, Menschaert G, Ringstad N, Horvitz HR, Schoofs L, Husson SJ, Temmerman L (2018a) Mass spectrometric evidence for neuropeptide-amidating enzymes in Caenorhabditis elegans. J Biol Chem 293:6052–6063. 10.1074/jbc.RA117.000731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bael S, Zels S, Boonen K, Beets I, Schoofs L, Temmerman L (2018b) A Caenorhabditis elegans mass spectrometric resource for neuropeptidomics. J Am Soc Mass Spectrom 29:879–889. 10.1007/s13361-017-1856-z [DOI] [PubMed] [Google Scholar]
- Van Sinay E, Mirabeau O, Depuydt G, Van Hiel MB, Peymen K, Watteyne J, Zels S, Schoofs L, Beets I (2017) Evolutionarily conserved TRH neuropeptide pathway regulates growth in Caenorhabditis elegans. Proc Natl Acad Sci USA 114:E4065–E4074. 10.1073/pnas.1617392114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt JP, Fink H (2015) Serotonin controlling feeding and satiety. Behav Brain Res 277:14–31. 10.1016/j.bbr.2014.08.065 [DOI] [PubMed] [Google Scholar]
- Vollmer LL, Schmeltzer S, Schurdak J, Ahlbrand R, Rush J, Dolgas CM, Baccei ML, Sah R (2016) Neuropeptide Y impairs retrieval of extinguished fear and modulates excitability of neurons in the infralimbic prefrontal cortex. J Neurosci 36:1306–1315. 10.1523/JNEUROSCI.4955-13.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukojevic V, Gschwind L, Vogler C, Demougin P, de Quervain DF, Papassotiropoulos A, Stetak A (2012) A role for α-adducin (ADD-1) in nematode and human memory. EMBO J 31:1453–1466. 10.1038/emboj.2012.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Zhang Y, Xu J, Shen P (2005) Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc Natl Acad Sci USA 102:13289–13294. 10.1073/pnas.0501914102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez-Guerra LA, Zhong X, Moghul P, Butts T, Zampronio CG, Jones AM, Mirabeau O, Elphick MR (2019) Urbilaterian origin and evolution of sNPF-type neuropeptide signalling. Biorxiv. doi: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu Y, Wu L, Fan C, Wang Z, Zhang X, Alachkar A, Liang X, Civelli O (2018) Receptor‐specific crosstalk between prostanoid E receptor 3 and bombesin receptor subtype 3. FASEB J 32:3184–3192. 10.1096/fj.201700337RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu H, Bargmann CI (2005) Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438:179–184. 10.1038/nature04216 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CRISPR/Cas9-edited deletion in the flp-34 gene. Local alignment of flp-34 WT gene with the flp-34(lst1666) mutant allele. Exons flanking the deletion are indicated in gray. The recoded regions in the lst1666 allele are highlighted in yellow and a stop codon is coloured in red. The two gRNAs used to generate the deletion are depicted in blue. Download Figure 6-1, EPS file (1.9MB, eps)