Abstract
A key mechanism of Pseudomonas spp. adaptation to environmental stressors is their ability to convert the cis-unsaturated fatty acids of the membrane lipids to their trans-isomers to rigidify the membrane and thereby resist stresses. Although this Cti-catalyzed enzymatic isomerization has been well investigated in the P. putida paradigm, several bacterial species have been found to produce trans-unsaturated fatty acids. Although cti orthologs have only been reported in Gram-negative bacteria, we report that E. faecalis FA2–2 cultures synthesize trans-unsaturated fatty acids during growth by a mechanism similar of P. putida. Although the role of trans-unsaturated fatty acids (trans-UFAs) in E. faecalis remains obscure, our results indicate that organic solvents, as well as the membrane altering antibiotic, daptomycin, had no effect on trans-UFA formation in E. faecalis FA2–2. Moreover trans-UFA production in E. faecalis FA2–2 membranes was constant in oxidative stress conditions or when metal chelator EDTA was added, raising the question about the role of heme domain in cis-trans isomerization in E. faecalis FA2–2. Although growth temperature and growth phase had significant effects on cis-trans isomerization, the bulk physical properties of the membranes seems unlikely to be altered by the low levels of trans-UFA. Hence, any effects seems likely to be on membrane proteins and membrane enzyme activities.
We also report investigations of cti gene distribution in bacteria was and suggest the distribution to be triggered by habitat population associations. Three major Cti clusters were defined, corresponding to Pseudomonas, Pseudoalteromonas and Vibrio Cti proteins.
Keywords: cis-trans isomerase, E. faecalis, fatty acid, membrane, adaptation, heme
1. Introduction
In response to stress many bacteria have evolved different mechanisms in order to adapt and survive the changing environment. The cell membrane of bacteria is typically the first barrier between the environment and the bacterial cell. Several stressors, such as temperature increase or addition of organic solvents directly increase membrane fluidity, leading to the disruption of essential membrane functions (Hermann J. Heipieper et al., 2003; Heipieper and de Bont, 1994). As a result, bacteria have developed mechanisms to alter cell membrane fluidity in order to maintain constant fluidity in the presence of environmental stress (Sinensky, 1974). One such mechanism is the post-synthetic transformation of membrane cis-unsaturated fatty acids (cis-UFA) into their trans isomers (trans-UFA) (von Wallbrunn et al., 2003). The benefit of this conversion is based on steric differences between cis- and trans-UFA. The double bonds of a cis-UFA form a kinked steric structure resulted in highly fluid membranes. In contrast, the more extended trans-UFA orders the membrane decreasing fluidity relative to their cis-isomers (McDonough et al., 1983; Seelig and Waespe-Sarcevic, 1978). Enzymatic isomerization of cis-UFA to trans-UFA is catalyzed by Cti and does not depend on de novo FA and protein synthesis, ATP or any other cofactor (Heipieper et al., 2003; Pedrotta and Witholt, 1999), presenting an efficient means to rigidify the membrane in the response to changing environments. Cti is a periplasmic cytochrome c-type protein (Pedrotta and Witholt, 1999) and containing a covalently bound heme essential for the cis to trans isomerization reaction (Holtwick et al., 1999). The iron provided by heme domain was proposed to remove the electrons from the cis double bond and then reconstitute the double bond in lower energy trans configuration without its transient saturation (Heipieper et al., 2003; von Wallbrunn et al., 2003).
The production of trans-UFA has been well described in various Pseudomonas putida strains where trans-UFA are reported to play an important role in adaptation of diverse P. putida strains to temperature increase, presence of organic solvents and heavy metals, as well as osmotic stress and addition of membrane-active antibiotics (Heipieper et al., 1996; Isken et al., 1997; Neumann et al., 2003). However, as summarized in a recent review, cis-trans-isomerization was shown in strains of Pseudomonas sp., Vibrio sp., Methylococcus capsulatus, Alcanivorax borkumensis, and Colwellia psychrerythraea. However, the presence of Cti orthologs have been found in other microorganisms (Eberlein et al., 2018; Heipieper et al., 2010).
Enterococcus faecalis is a Firmicute (Gram-positive) bacterium, found in the gastrointestinal tract of mammals and is one of the leading causes of surgical wound infections (Huycke et al., 1998). Unlike Gram-negative bacteria, Gram-positive bacteria lack an outer layer (wall) and periplasm (Silhavy et al., 2010) and have not been reported to produce trans-UFA. Here, we have investigated the cti gene distribution among bacterial species and found that only 5.5% of tested bacterial strains code for cti orthologs and all were Gram-negative bacteria found in environments membrane altering stressors are present. However, our results showed that E. faecalis FA2–2 forms trans-UFA during growth using the pattern similar to seen in P. putida. Nonetheless, the oxidative stress and the metal chelation had no effect on E. faecalis FA2–2 trans-UFA production. Whereas the role of these FA in E. faecalis membrane is unclear, organic solvents, as well as daptomycin, had no effect on trans-UFA formation in E. faecalis membranes.
2. Material and Methods
2.1. Strains and growth conditions
Bacterial strains used in this study are listed in Table S1. P. putida F1 and E. faecalis FA2–2 were grown at 30°C and 37°C, respectively, in M17 (BD Difco) fatty acid free broth, obtained by three chloroform (v/v) extractions of FA from the medium. When indicated, M17-FA free was supplemented with 9,10-D2-oleic acid or 0.09% sodium [1-13C]acetate both from Cambridge Isotope Laboratories. For P. putida F1, 9,10-D2-oleic acid was neutralized with KOH, solubilized with Tergitol NP-40 and added at final concentrations of 0.01%. For E. faecalis FA2–2, 9,10-D2-oleic acid was added at a final concentration of 100 μM. When treated with daptomycin, bacterial cultures were supplemented with 50 mg of Ca2+ /liter. Minimum inhibitory concentrations (MICs) were determined by the broth microdilution method achieved in M17 FA free. Briefly, overnight cultures were diluted to OD600 of 0.08 and added to a 96-well test plate (Nunc) containing different concentrations of stressors in triplicate. The test plates were incubated at 30°C or 37°C for overnight. MIC was defined as the lowest antibiotic concentration that inhibited bacteria growth as determined by turbidimetry at OD600.
2.2. Fatty acid analyses
Phospholipids were extracted (Bligh and Dyer, 1959) and fatty acid methyl esters (FAME) were prepared according to a standard protocol (Zhu et al., 2010). Briefly, the phospholipids were dissolved in 1.2 mL of dry methanol. Esterification reaction was conducted by incubation with 0.2 mL of 25% (v/v) sodium methoxide at room temperature for 15 min and stopped by addition of 1.2 mL of 2 M HCl. FAME were then obtained by three extractions each with 1.2 mL of hexanes. The solvent was removed under a nitrogen stream. Fatty acid methyl esters were analyzed using a GC-MS system (Agilent Inc, CA, USA) consisting of an Agilent 7890B gas chromatograph, an Agilent 5977A MSD.
The cis- and trans-isomers were separated on a CP-Sil88 (50 m×0.25 mm I.D. and 0.2 μm film thickness) capillary column (Agilent J&W, CA, USA). The inlet temperature was 220°C, MSD interface temperature – 230°C, and the ion source temperature adjusted to 230°C. An aliquot of 1 μL was injected in a split mode (20:1). The helium carrier gas was kept at a constant flow rate of 1.9 mL min-1. The temperature program was: 2 min isothermal heating at 80°C followed by temperature increase of 10°C min−1 to 165°C, then 20°C min−1 to 180°C, 10°C min−1 to 210°C. The mass spectrometer was operated in positive electron impact mode (EI) at 69.9 eV ionization energy at m/z 33–500 scan range. Mass spectra were recorded in combined scan/SIM mode. For a SIM mode, following m/z fragments were tracked: 242 (C14:0), 236 (C16:1cis,trans), 250 (C17:0 cyclo), 264 (C18:1cis,trans), 270 (C16:0), 278 (C19:0 cyclo), 298 (C18:0). Obtained retention time was confirmed by authentic standards (Sigma, USA). Target peaks were evaluated by the Mass Hunter Quantitative Analysis B.08.00 (Agilent Inc., CA, USA) software. The results are presented as a % of total FA. All experiments were performed in three biological replicates.
2.3. Double bond localization
To locate double bonds dimethyldisulfide adducts the protocol previously reported (Feng and Cronan, 2009) was used. Fatty acid methyl esters in hexane (100 μl) were converted to their dimethyldisulfide adducts by treatment with 75 μl of dimethyldisulfide and one drop of 6%, iodine solution in diethyl ether for 14 h at 50°C. Samples were cooled, and 50 μl of 10% aqueous Na2S2O3 were added to remove iodine. The hexane layer was pooled and concentrated to 50 μl under N2. Gas chromatography-mass spectroscopy analyses were done on an Agilent system consisting of a 5975C mass selective detector, a 7683B autosampler, and a 7890A gas chromatograph equipped with ZB-5MS (60 m×0.32 mm I.D. and 0.25μm film thickness) capillary column (Phenomenex, CA, USA). Injection temperature and the mass selective detector transfer line were set to 250 °C, the ion source and MS quadrupole were adjusted to 230 and 150°C, respectively. The helium carrier gas was set at a constant flow rate of 2 ml/min. The temperature program was: 2 min at 100°C, followed by an oven temperature increase of 8°C/min until 300°C. A 1 μl sample was injected with a split ratio of 10:1. The spectra acquired were recorded in the m/z 50–500 scanning range and processed using the Mass Hunter Quantitative Analysis B.08.00 (Agilent Inc., CA, USA) software.
2.4. Detection of trans-UFA in different E. faecalis FA2–2 lipid species
Lipids were extracted from E. faecalis FA2–2 pellets as described above and separated on HPTLC plates (silica gel 60 F254, Sigma) using CHCl3/CH3OH/CH3COOH (65/25/10, v/v/v) as the solvent system. Individual lipid spots were visualized by UV fluorescence at 365 nm after spraying a primulin dye solution (0.05% in acetone/H2O, 8/2, v/v). The identification of individual HPTLC spots was made in comparison to control HPTLC plates of known lipid standards. High purity lipid standards were obtained from Avanti Polar Lipids (Alabaster, AL).
To test the trans-UFA content in individual lipid types, the major lipid spots separated on the HPTLC plate were scrapped off and extracted using the Bligh and Dyer method. Next, FAME were prepared and analyzed using GC MS as described above.
2.5. Phylogenic analyses
The closest Cti orthologs were extracted from the MaGe Platform (http://www.genoscope.cns.fr/agc/mage, last accessed June 10, 2018), which carried out Blast analyses using the P. putida F1 Cti amino acid sequence (Pput_3319) as a query in BLAST-P searches. We thoroughly checked all available genomes of species lacking cti to ensure absence of the gene itself or lack of any significant remnant. Whenever possible, we also favored fully assembled genomes over draft or incomplete ones. The resulting sequences were analyzed using the neighbor-joining method using CLC Sequence Viewer 7 (CLC bio). Bootstrap values at nodes greater than 80% (1000 replicates) were chosen to construct the tree.
3. Results
3.1. E. faecalis FA2–2 forms trans-UFA during growth
Despite the absence of Cti orthologous genes in Gram-positive bacteria, we tested if a Gram-positive bacterium might produce trans-UFA. We analyzed the FA composition of early stationary growth cultures of E. faecalis FA2–2 grown in M17 FA free broth and found 1.6% of the total bacterial FA was trans-UFA (Table 1). In cultures of P. putida F1 grown under the same conditions, 20.2% trans-UFA from total FA was found. However, in cultures of E. coli MG1655, a bacterium known to be unable to produce trans-UFA, as well as a cti mutant of P. putida F1 (Kondakova and Cronan, 2019) trans-UFA were not detected.
Table 1.
Detection of trans-UFA in E. faecalis FA2–2 cultures
| Strain | % of total FA | |||
|---|---|---|---|---|
| trans-UFA | cyclo-FA | cis-UFA | SFA | |
| E. faecalis FA2–2 | 1.6±0.0 | 14.8±0.9 | 39.7±1.1 | 43.9±0.2 |
|
E. coli K-12 MG1655 |
0.0±0.0 | 9.2±0.1 | 32.6±0.5 | 58.3±0.4 |
| P. putida F1 WT | 20.2±0.4 | 1.4±0.1 | 44.9±0.2 | 33.5±0.1 |
| P. putida F1 Δcti | 0.0±0.0 | 4.8±0.1 | 53.1±0.7 | 42.1±0.6 |
Bacterial strains were grown in M17-FA free medium for 6.5 h. FAME samples were prepared according the standard protocol (Material and Methods). Mean ± SEM is shown. N=3. N indicates biological replicates corresponding to independent experiments. MG1655 is an E. coli wild type (WT) strain.
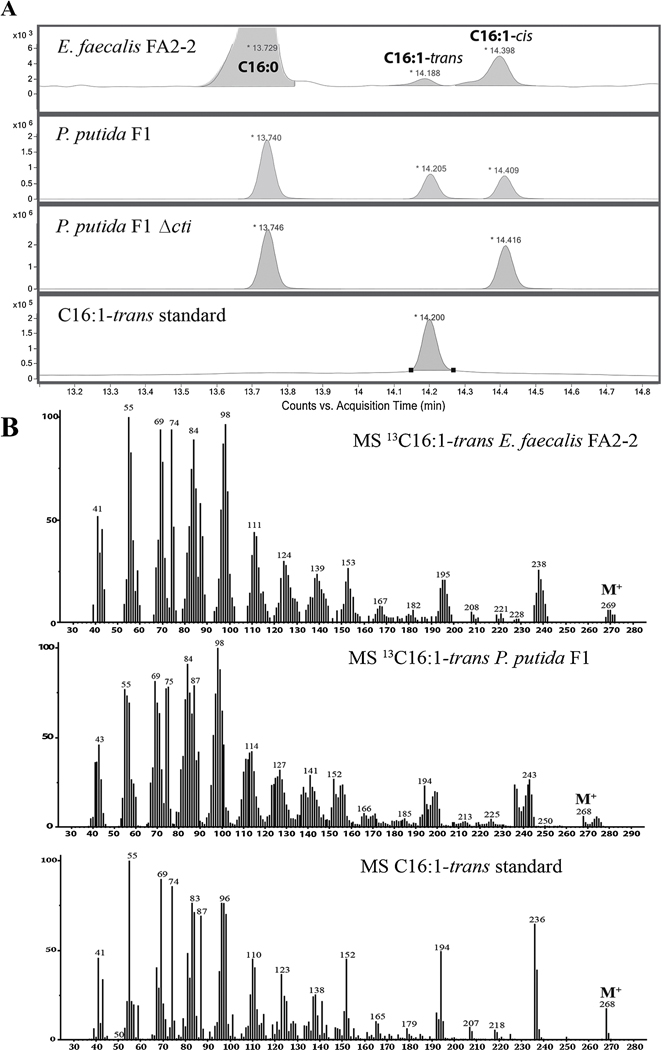
To ask if the trans-UFA found in E. faecalis FA2–2 cultures were synthesized by the bacterium rather than taken up from the medium, sodium [1-13C]acetate was added to bacterial cultures. Incorporation of 13C into C16:1-trans UFA would indicate that the fatty acid was synthesized from (ultimately) acetyl-CoA rather than derived from the medium. Indeed, 13C labeled C16:1-trans UFA were detected in E. faecalis FA2–2 and P. putida F1 cultures, but not in P. putida F1 Δcti cultures (Fig. 1A). The MS spectra of C16:1-trans showed the enrichment in 13C compared to the spectrum of C16:1-trans standard (Fig. 1B), demonstrating the production of trans-UFA by E. faecalis FA2–2.
Figure 1. E. faecalis FA2–2 contains trans-UFA.
(A) Representative total ion chromatograms of bacterial FAME showing that E. faecalis forms C16:1-trans fatty acids. Note the peak corresponding to C16:1-trans in E. faecalis FA2–2 and in P putida wild type, whereas this peak is absent in the P. putida F1 Δcti strain. Bacteria were grown in M17-FA free medium supplemented with 0.09% sodium [1-13C ]acetate for 6.5 h and FAME were obtained as described in Material and Methods. N=3.
(B) Representative MS spectra showing the incorporation of sodium [1-13C ]acetate into C16:1-trans in E. faecalis FA2–2 (top) and P. putida F1 (middle). Note the large carbon isotopic distribution of C16:1-trans extracted from cultures of E. faecalis FA2–2 and P. putida F1 comparing to the C16:1-trans standard (bottom), which does not contain 13C (N =3). For all panels, N indicates biological replicates corresponding to independent experiments.
We next tested if the production of trans-UFA occurred during E. faecalis FA2–2 growth or during the sample preparation, as was previously reported for several P. putida strains (Härtig et al., 2005). Bacterial cultures were treated with the powerful chaotrophic agent 10% trichloroacetic acid (TCA) as previously described (Kondakova and Cronan, 2019) (Fig. S1A). Fatty acid methyl esters (FAME) were extracted from the TCA precipitates and separated using a CP-Sil 88 column which gave a well-separated peak corresponding to C16:1-trans in both the P. putida F1 and E. faecalis FA2–2 samples (Fig. S1B). No trans-UFA was detected in E. coli K-12 MG1655 samples given the same treatment, indicating that the formation of C16:1-trans is not an artefact of TCA treatment. Altogether, these data indicated that E. faecalis FA2–2 synthesized trans-UFA during growth rather than during the stress of handling.
3.2. E. faecalis FA2–2 produces C16:1-trans Δ9 using cis-UFA as a substrate utilizing a mechanism similar to that of P. putida spp.
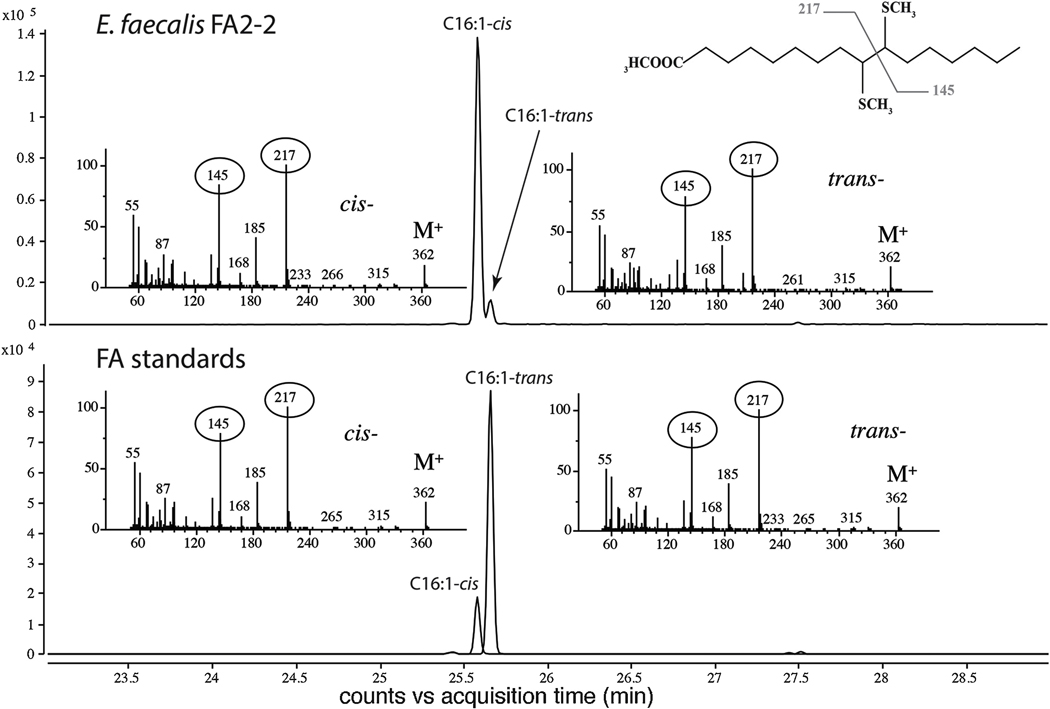
To determine the location of the E. faecalis C16:1-trans fatty acid double bond we performed derivatization of fatty acid methyl esters (FAMEs) with dimethyldisulfide as previously reported (Feng and Cronan, 2009). Both the C16:1-cis and C16:1-trans UFAs gave adduct peaks corresponding to the Δ9 position of the double bond (Fig. 2). These data confirmed the C16:1-trans identification in E. faecalis FA2–2 samples and showed that trans- and cis-double bonds are located in the same position of the acyl chain.
Figure 2. E. faecalis FA2–2 forms C16:1-trans Δ9.
Representative total ion chromatograms of dimethyldisulfide adduct methyl esters showing that E. faecalis FA2–2 forms C16:1-cis,transΔ9. The localization of double bonds was assayed as reported in Material and Methods. The representative MS spectra of dimethyldisulfide adducts are shown by black arrows. The encircled fragments corresponding to methylsulfides were used to define the double bond position. Bacterial cultures in early stationary growth phase in M17-FA free medium were analyzed. N=3. N indicates biological replicates corresponding to independent experiments.
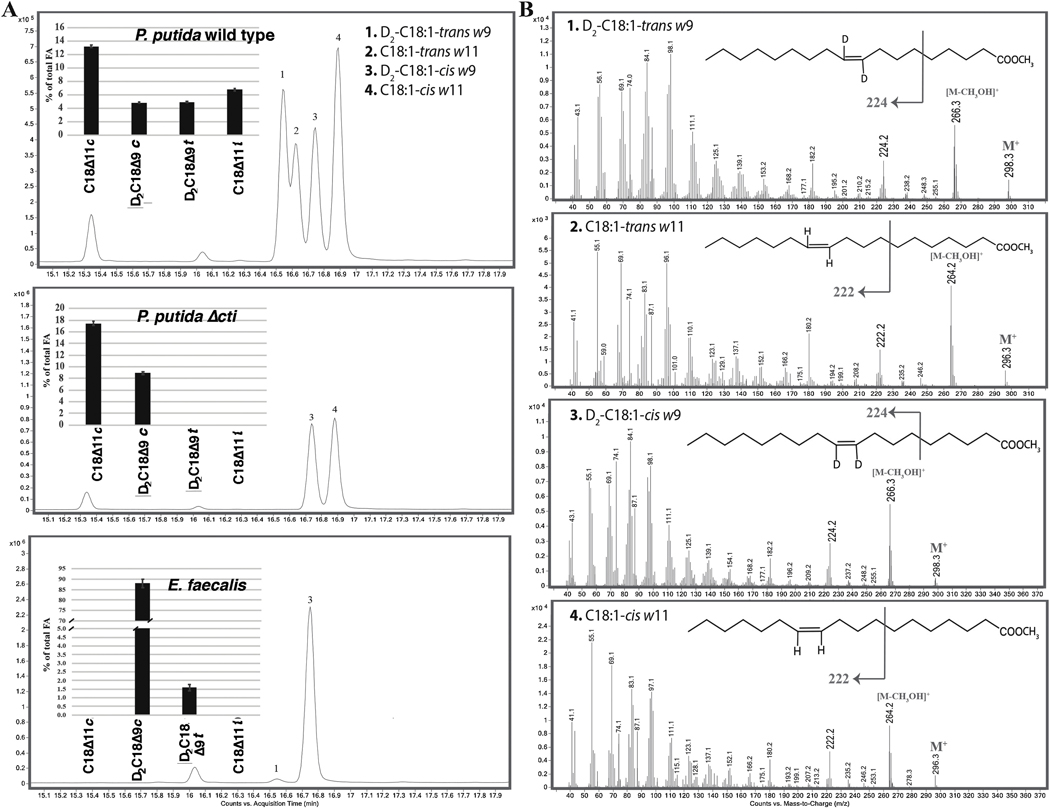
Previous reports investigated the pattern of the cis- to trans-UFA isomerization in P. putida S12 showing that Cti-mediated isomerization did not include the transient saturation of the double bond (von Wallbrunn et al., 2003). To test if in E. faecalis FA2–2 cis-UFA are the substrate for trans-UFA formation and if this isomerization reaction occurs without double bond saturation, bacterial cultures were supplemented with oleic acid (C18:1-cis Δ9) deuterated at both C atoms of the double bond (D2-C18:1-cis Δ9). In agreement with previous study (von Wallbrunn et al., 2003), P. putida F1 samples showed four C18:1 FA (Fig. 3A top) species; two native species, C18:1-cis Δ11 and C18:1-trans Δ11 (Fig. 3B peaks 4 and 2, respectively, m/z 296.3 indicating no deuteration), which are known to be formed using the anaerobic pathway for UFA synthesis (Cronan, 2006; Cronan and Thomas, 2009) and two nonnative doubly deuterated UFAs, D2-C18:1-cis Δ9 and D2-C18:1-trans Δ9 FA (Fig. 3B peaks 3 and 1, respectively, m/z 298.3, indicating double deuteration). No trans-UFA were detected in P. putida F1 Δcti cultures, showing the absence of sample contamination during the experiments. E. faecalis FA2–2 ΔfabI cultures fed with deuterated oleic acid did not produce any detectable amount of native C18:1-cis Δ11 and C18:1-trans Δ11 FA, due to the blockage of the FA biosynthetic pathway by the ΔfabI mutation (Zhu et al., 2013), but readily incorporated the supplemented D2-C18:1-cis Δ9 and converted it to the trans-isomer, D2-C18:1-trans Δ9 FA, (Fig. 3A bottom panel), indicating that E. faecalis FA2–2 directly converted cis-UFA to trans-UFA. The mass fragmentation pattern of D2-C18:1-trans Δ9 in these samples showed a C18:1-trans Δ9 product completely labeled with two deuterium atoms demonstrating that no deuterium was lost during the isomerization. These data demonstrate that E. faecalis FA2–2 produced trans-UFA using cis-UFA as a substrate without shift of double bond position or transient saturation of the double bond.
Figure 3. E. faecalis FA2–2 forms trans-UFA using a mechanism similar to that seen in P. putida.
(A) Representative total ion chromatograms and peak assignment of bacterial FAME showing that P. putida F1 and E. faecalis FA2–2 form trans-UFA using a similar mechanism. Bacteria were grown in M17-FA free medium supplemented with 9,10-D2-oleic acid for 6.5 h and FAME were obtained as described in Material and Methods. Note that the fatty acids extracted from P. putida F1 cultures (top panel) show the peaks corresponding to D2-C18:1-trans Δ9, C18:1-trans Δ11, D2-C18:1-cis Δ9 and C18:1-cis Δ11. In contrast (middle panel) the P. putida F1 Δcti strain was unable to synthetize trans-UFA and has only two peaks, corresponding to D2-C18:1-cis Δ9 and C18:1-cis Δ11 fatty acids, whereas the E. faecalis FA2–2 ΔfabI strain (an oleate auxotroph) has two peaks corresponding to D2-C18:1-trans Δ9 and D2-C18:1-cis Δ9 fatty acids (bottom panel). The percentages of C18:1 in total bacterial FA are superimposed onto the chromatograms, N=3.
(B) Representative MS spectra showing the fragmentation patterns of C18:1 fatty acid species. The shift of 2 atomic mass units was observed for D2-C18:1 and indicated no deuterium loss was observed during the isomerization reaction in E. faecalis FA2–2. Peak numbers and identifications are shown, N=3. For all panels, N indicates biological replicates corresponding to independent experiments.
3.3. trans-UFA are detected in all E. faecalis FA2–2 lipids, except lyso-phosphatidylglycerol.
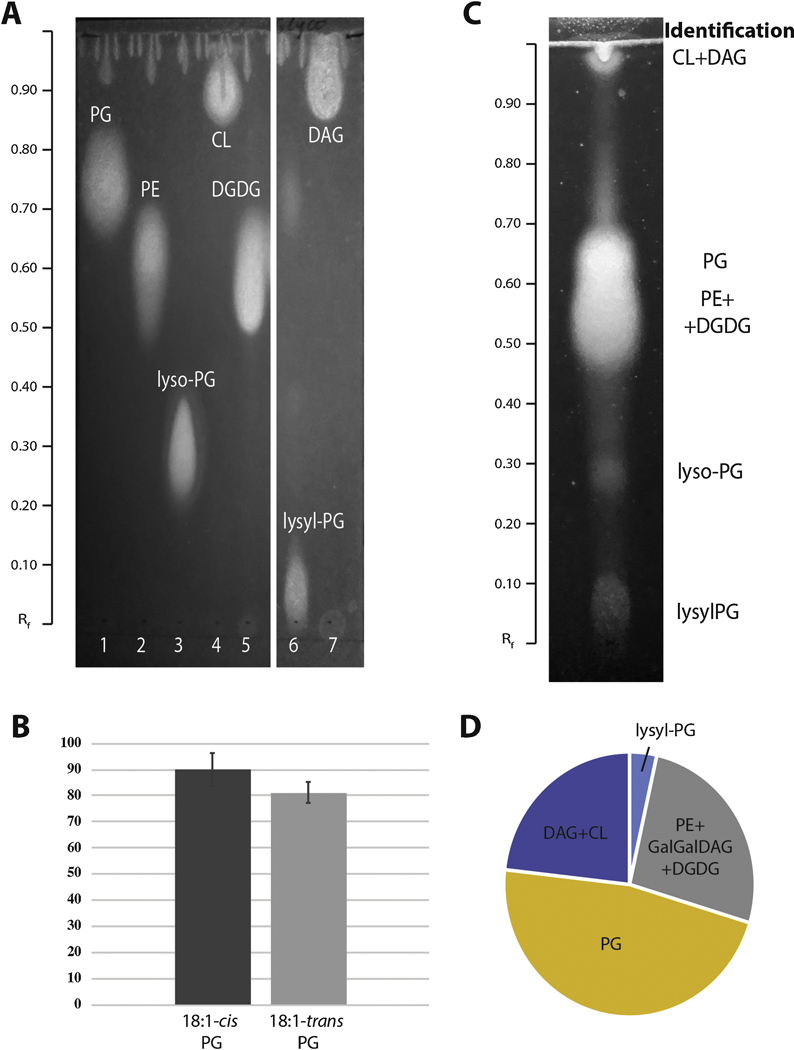
To ask in which lipid species trans-UFA are located, we first studied the E. faecalis FA2–2 lipid composition. Enterococcal lipid compositions have been previously described in detail (Bao et al., 2012; Mishra et al., 2012; Rashid et al., 2017) and the predominant E. faecalis lipid species are phosphatidylglycerol (PG) and cardiolipin (CL). However, small amounts of phosphatidylethanolamine (PE) and phosphatidic acid were also reported. This bacterium is also able to modify PG with lysine to produce lysyl-PG and thereby modify the membrane charge (Bao et al., 2012). In addition, E. faecalis has been shown to contain several glycolipids including phosphatidyldiglycosyldiglyceride and glycerophosphoryl-diglycosyl-diglyceride (Mishra et al., 2012), as well as diglycosyl-diacylglycerol (DGDAG), dyacylglycerols (DAG) and triacylglycerols (TAG) (Rashid et al., 2017). To identify the major E. faecalis FA2–2 lipid species, the total FA2–2 lipids were separated on silica gel HPTLC plates and identified by comparing with HPTLC plates of lipid standards (Fig 4A). Two major lipid spots were identified as PG (Rf of 0.76) and DGDG coeluted with PE (Rf of 0.60), (Fig. 4C). However, three smaller spots were also detected and identified as lysyl-PG (Rf of 0.07), lyso-PG (Rf of 0.28) and CL probably co-eluted with DAG (Rf of 0.91).
Figure 4. Distribution of trans-UFA in E. faecalis FA2–2 lipids.
(A) Lipid standard profiles by HPTLC silica gel chromatography. Individual lipids were separated on HPTLC plates using CHCl3/CH3OH/CH3COOH (65/25/10, v/v/v) as solvent system and detected by UV fluorescence at 365 nm after spraying with a primulin dye solution.
(B) About 81% and 90% recovery of C18:1-cis and C18:1-trans fatty acids, respectively, from phosphatidylglycerol (PG) after separation on HPTLC plates. The PG-C18:1-cis and PG-C18:1-trans samples were first run on HPTLC plates. The lipid spots were scrapped off and extracted by the Bligh and Dyer procedure. Esterification reaction was conducted as shown in material and methods. The recovery from HPTLC plate was calculated using the peak areas. The recovery in positive control sample (no HPTLC) was taken as 100%, and all other recoveries were expressed as percent of this value. Values shown are means ± SEM of three separate analyses.
(C) E. faecalis FA2–2 lipids separated and visualized on HPTLC plate as described for lipid standards (see Fig. 4A). The identification of individual HPTLC spots was made in comparison to control HPTLC plates of known standards.
(D) The content of trans-UFA in E. faecalis FA2–2 lipids showing that the bulk of trans-UFA was found in phosphatidylglycerol (PG) whereas no trans-UFA was found in lyso-PG. The trans-UFA content was assayed using GC MS as described in Fig. 4B and Materials and Methods. The negative control (silica gel from the used HPTLC plate outside of the lipid spots) was used to measure the noise rate. The peak areas obtained from the negative control samples were subtracted from the peak areas of lipid spots. The fractions of trans-UFA in each lipid spot were calculated using the peak areas. The total amount of trans-UFA was taken as 100%, and trans-UFA contents in lipid spots were expressed as percent of this value.
Designations for all panels, PG, phosphatidylglycerol; PE, phosphatidylethanolamine; lyso-PG, lyso-phosphatidylglycerol; CL, cardiolipin; DGDG, digalactosyldiacylglycerol; lysyl-PG, lysyl-phosphatidylglycerol; DAG, diacylglycerol; Rf, retention factor.
To ask which lipid types contain trans-UFA, the appropriate areas of silica gel were scrapped from the HPTLC plates, extracted and hydrolyzed to produce FA. The FA were then methylated and analyzed by GC MS. This method was first tested for extraction of cis- and trans-UFA from PG standard separated or not on HPTLC plate. About 90% and 81% of trans-UFA and cis-UFA (non-HPTLC treated control was 100%) were extracted from HPTC plate and detected by GC MS (Fig. 4B). Next, assayed for trans-UFA in all E. faecalis FA2–2 lipids. Interestingly, trans-UFA were detected in all tested spots, except that of lyso-PG, indicating that E. faecalis FA2–2 lyso-PG does not contain trans-UFA. PG is the major PL and as expected this PL contained most of the trans-UFA (Fig. 4D). Together these data indicated that trans-UFA is present in all the tested major E. faealis FA2–2 lipids, excepting lyso-PG.
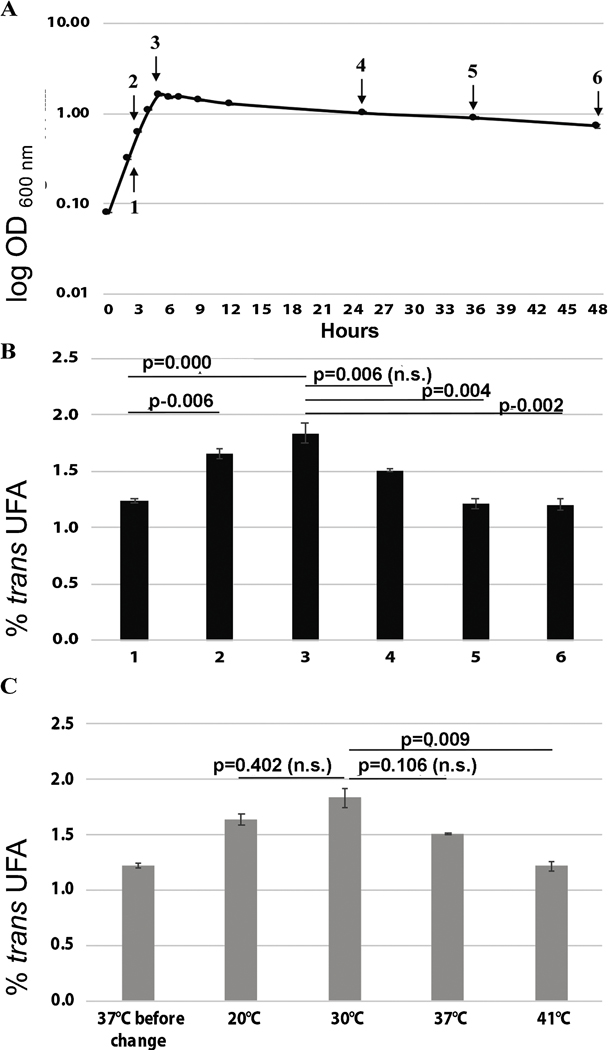
3.4. Effect of growth phase and temperature on trans-UFA production in E. faecalis FA2–2
We evaluated the effects of growth phase and temperature on E. faecalis FA2–2 trans-UFA production. The fraction of trans-UFA in E. faecalis FA2–2 membrane significantly increased from exponential to early stationary growth phase, where the maximum trans-UFA levels were seen (Fig. 4A&B). The fraction of trans-UFA in E. faecalis FA2–2 membranes then modestly decreased to become stable after 36 h of growth. The same trend has been observed in P. putida F1 cultures (Kondakova and Cronan, 2019), as well as in Vibrio sp. strain no. 5710 cultures, which showed maximal of trans-UFA levels in early stationary growth phase (Hamamoto et al., 1994). This indicated a possible correlation between the production of trans-UFA and growth phase in these bacterial species.
Previous studies showed that the production of trans-UFA plays an important role in P. putida and Vibio spp. adaptation to temperature increase (Diefenbach et al., 1992; Holtwick et al., 1997; Okuyama et al., 1991, 1990) and is considered as a fast-adaptive response of bacterial membranes to temperature change. Thus, we tested the temperature shift effect on production of trans-UFA in E. faecalis FA2–2. As reported by (Diefenbach et al., 1992) and our previous study (Kondakova and Cronan, 2019), bacterial cultures were grown at optimal growth temperature 37°C until the early stationary growth phase (point #3 Fig. 4A) and then the growth temperature was shifted to 20°C, 30°C or 42°C for 2 hr. Whereas the fraction of trans-UFA did not change when temperature was shifted at 20°C and 30°C, the amount of trans-UFA in E. faecalis FA2–2 cultures grown at 30°C was significantly higher that seen in cultures grown at 42°C (Fig. 4C).
3.5. The levels of trans-UFA in E. faecalis FA2–2 is independent of de novo protein synthesis
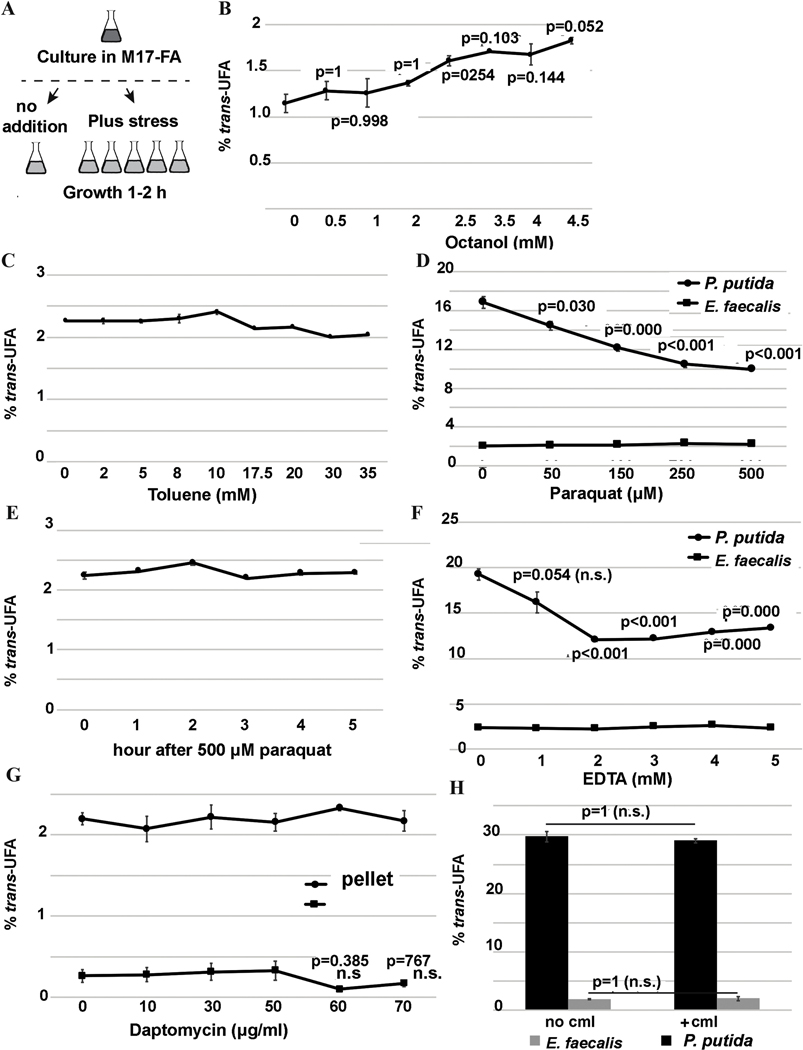
It was previously proposed that trans-UFA synthesis in P. putida does not depend on the de novo protein synthesis (Heipieper et al., 1992). To ask if the production of trans-UFAs in E. faecalis FA2–2 depends on de novo protein biosynthesis, we exposed E. faecalis FA2–2 and P. putida F1 cultures to chloramphenicol (Aakra et al., 2010) for 2 h in concentrations above the minimal inhibitory concentration (10 μg/mL for E. faecalis FA2–2 and 550 μg/mL for P. putida F1). In agreement with previous study (Heipieper et al., 1992), there was no significant change in trans-UFA production in either strain (Fig. 5H), indicating that production of trans-UFAs in E. faecalis FA2–2 does not depend on de novo protein biosynthesis. It should be noted that after 2 h exposure both bacterial strains remained viable (Fig. S2G).
Figure 5. Effect of growth temperature and phase on production of trans-UFA in E. faecalis FA2–2.
(A) Growth curve of E. faecalis FA2–2 at 37°C in M17 FA free medium. The arrows show the growth stages at which samples of cultures were taken for assays. N=3.
(B) Effect of growth phase on trans-UFA production in E. faecalis FA2–2. The numbers of the X axis show the growth stages at which samples of cultures were taken for GC MS analyses (see Fig. 4A). N=3.
(C) Effect of temperature shift on production of trans-UFA in E. faecalis FA2–2. Bacterial cultures grown in M17 FA free medium at 37°C with vigorous shaking (250 rpm) and constant aeration for OD600=1.4 (corresponding to the point #3 on Fig. 4A) were incubated for 2 h at 20°C, 30°C, 37°C and 37°C. N=3. For all panels, mean ± SEM is shown. N indicates biological replicates corresponding to independent experiments. Statistical significance was determined by using one-way ANOVA (Turkey multiple comparison). n.s., not significant.
3.6. Oxidative stress and addition of the EDTA chelating agent does not modify trans-UFA production in the E. faecalis FA2–2 membrane
In P. putida Cti is a cytochrome c-type protein that has the characteristic covalently bound heme-binding motif (Holtwick et al., 1997), the iron of which (probably Fe3+) was proposed to be essential for the cis- to trans-isomerization (Okuyama et al., 1998). When added to bacterial cultures, paraquat is known to produce oxidative stress by generating reactive oxygen species (Lascano et al., 2012; Mancini and Imlay, 2015), which reduce cytochrome c (Koppenol et al., 1976; Lascano et al., 2012) and could inhibit Cti-dependent production of trans-UFA. Thus, we tested 2 h treatments of both P. putida F1 and E. faecalis FA2–2 cultures with increasing concentrations of paraquat. E. faecalis FA2–2 showed no significant decrease in trans-UFA production as compared to P. putida F1 which showed a significant decrease in trans-UFA production (Fig. 5D). Paraquat is a cation which cannot passively diffuse across the membrane and needs to be taken into the cell via transporters (Lascano et al., 2012). To insure the paraquat penetration into E. faecalis FA2–2 cells, we analyzed the time course of 500 μM paraquat effect on trans-UFA production in this bacterium. Even after 5 h of incubation time there was no significant change in trans-UFA production (Fig. 5E). This indicates that in contrast to P. putida F1, in E. faecalis FA2–2 reactive oxygen species did not affect trans-UFA production.
Ethylenediaminetetraacetic acid (EDTA) is one of the most effective chelating agents and forms an open complex with Fe3+ ion (Flora and Pachauri, 2010; Maketon et al., 2008) and thus EDTA could compete with a heme binding site for the metal. To ask if the EDTA-mediated metal chelation alters the production of trans-UFA in E. faecalis FA2–2, we exposed E. faecalis FA2–2 and P. putida F1 cultures to increasing concentrations of EDTA for 2 h. There was a significant decrease in trans-UFA production in P. putida F1, however there was no decrease in trans-UFA production in E. faecalis FA2–2 (Fig. 5F), indicating that chelating agent such as EDTA does not affect trans-UFA production in E. faecalis FA2–2.
Together these data showed that in contrast to P. putida F1, trans-UFA production in E. faecalis FA2–2 did not decrease in oxidative stress conditions or in presence of the chelating agent, EDTA, suggesting that the E. faecalis isomerization enzyme(s) did not have P. putida Cti characteristics.
3.7. The levels of trans-UFA in E. faecalis FA2–2 membrane unaffected by exposure to organic solvents or daptomycin
Next, we investigated the role of trans-UFA production in E. faecalis FA2–2. The importance of membrane trans-UFA in maintaining constant membrane fluidity was well documented for several P. putida strains, which overproduce trans-UFAs to decrease membrane fluidity in the response of organic solvents, such as toluene or octanol (Heipieper et al., 1996, 1995; Heipieper and de Bont, 1994; Junker and Ramos, 1999; Pedrotta and Witholt, 1999). To test whether E. faecalis FA2–2 trans-UFA production would increase when exposed to organic solvents, we subjected it to octanol and toluene treatment. Hence, cultures in early stationary growth phase were incubated for 1 h with increasing concentrations of octanol and toluene (Fig. 5A). No significant increase in trans-UFA production was observed when bacterial cultures were incubated with toluene or octanol (Fig. 5B&C) indicating that exposure to these organic solvents did not affect E. faecalis FA2–2 trans-UFA production. Surprisingly, E. faecalis FA2–2 was found to tolerate high concentrations of octanol and toluene. In fact, it grew even after 1 h exposure to 4.5 mM and 35 mM octanol and toluene, respectively (Fig. S2A&B).
Daptomycin (DAP) is a lipopeptide antibiotic effective against Gram-positive bacteria such as E. faecalis, which interacts with the cell membrane lipids (Jung et al., 2004; Straus and Hancock, 2006). In complex with Ca2+, DAP has an increasing affinity for the negatively charged phospholipids, including phosphatidylglycerol, which is a major component of the E. faecalis membrane lipids (Mishra et al., 2012; Rashid et al., 2017). In addition, DAP binds and clusters fluid lipids, such as cis-UFA (Müller et al., 2016). To test if DAP treatment of E. faecalis FA2–2 would change the level of trans-UFA in the membrane, we treated E. faecalis FA2–2 for 1 h with increasing DAP concentrations and measured the trans-UFA content in both the bacterial pellet and supernatant. The supernatant was analyzed because E. faecalis is known to release phospholipids to inactivate DAP (Mishra et al., 2012). DAP concentrations below and above the MIC (60 μg/mL) were tested. Neither the supernatant nor the pellet treated with DAP showed any change in trans-UFA production compared to the untreated cells (Fig. 5G), indicating that DAP has no effect on trans-UFA production in E. faecalis FA2–2.
Together these results indicated that the production of trans-UFA in E. faecalis FA2–2 was independent of the addition of organic solvents or membrane altering agents, such as daptomycin, suggesting that trans-UFA are not involved in the protection of bacterial membranes against the tested stress factors and likely play another role in E. faecalis.
3.8. Cti distribution among bacterial kingdoms
As summarized in a recent review (Eberlein et al., 2018), the presence of cti orthologous genes have been found in various bacterial strains. To investigate Cti distribution among bacterial species, we performed BLASTP analysis on translated reading frames of 3912 bacterial genomes available in MaGe database using the P. putida F1 Cti sequence (Pput_3319) as the query. Only 217 candidates were found to code for P. putida F1 cti orthologs (Table S2). All Cti encoding bacteria are Gram-negative bacteria belonging to Alpha-, Beta-, Delta- or Gamma subdivisions and use either aerobic or anaerobic respiration (Table S3). The majority of cti coding bacteria were found in high osmolarity marine environments, as well as environments contaminated by hydrocarbons, heavy metals or other toxic membrane-altering compounds, indicating that the environment could be an important driving force for cti distribution among bacterial kingdom.
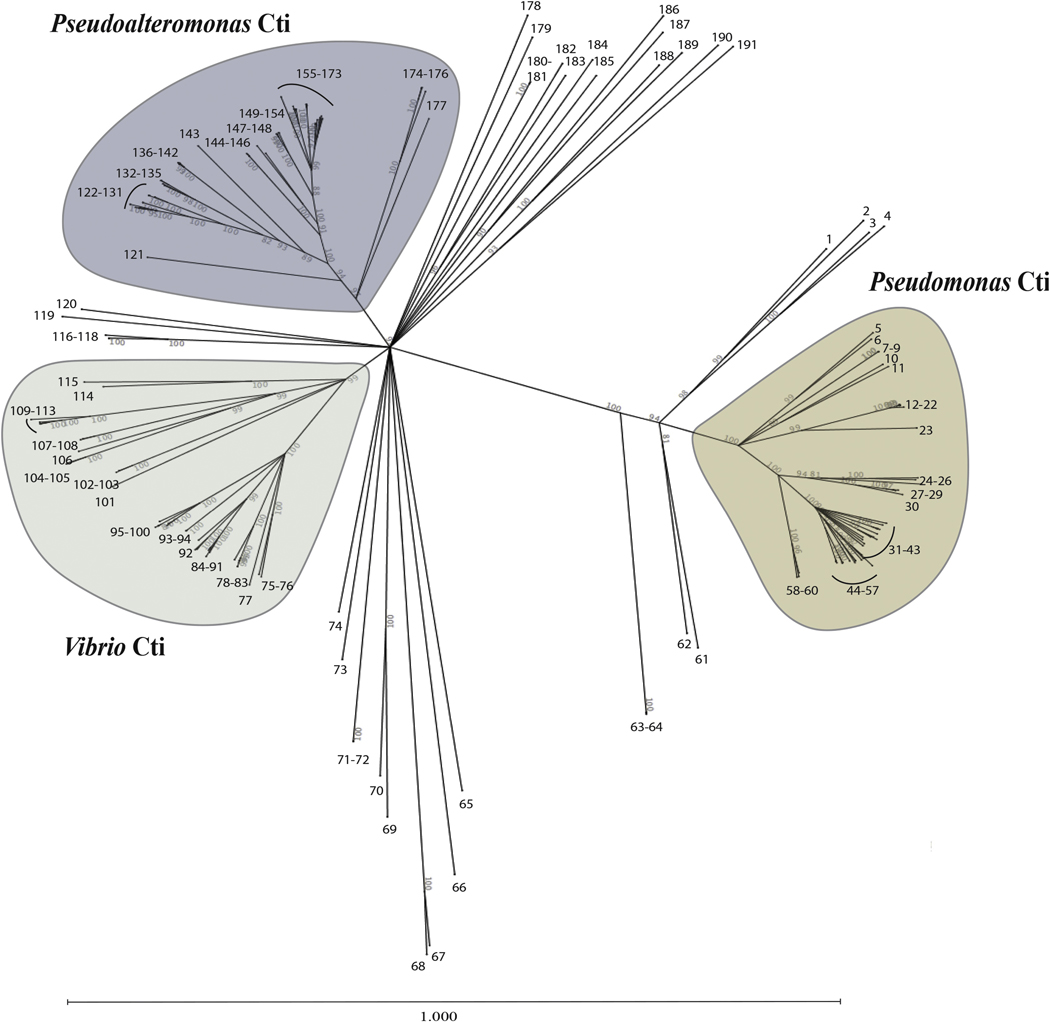
To map the phyletic patterns of Cti proteins, we reconstructed a phylogenic tree from a concatenated alignment of all found sequences (Fig. 6 and Table S2). Three major Cti clusters were found representing Pseudomonas spp. Cti; Pseudoalteromonas spp. Cti and Vibrio spp. Cti. Whereas Pseudoalteromonas and Vibrio clusters had about 50% Cti amino acid sequence identity between each other, Pseudomonas Cti has only 35–39% identity with Pseudoalteromonas and Vibrio Cti clusters. The Pseudomonas Cti cluster was homogenous, including all available in MaGe database Pseudomonas spp. Cti species, as well as Cti from Azotobacter vinelandii, which is phylogenetically closely related to Pseudomonas genus (Özen and Ussery, 2012). All available into the database Pseudoalteromonas species coded for cti, which were clustered in Pseudoalteromonas Cti cluster. Only Cti from Pseudalteromonas atlantica T6c and Pseudoalteromonas sp. PLSV appeared in the Vibrio Cti cluster. Whereas the Vibrio Cti cluster was manly composed of Vibrio spp. Cti, not all Vibrio species available in the database were found to code for cti. For instance, Cti was not found in several V. cholerae, V. fischeri, V. vulnificus and V. harveyi strains, indicating the heterogenic distribution of Cti among Vibrio genus. Instead, the Vibrio Cti cluster contained Cti encoding genes of two Photobacterium spp., five Alteromonas spp., two Shewanella spp. and Rheinheimera sp. EpRS3. Although Photobacterium belongs to the order Vibrionales, the presence of Alteromonas and Shewanella Cti coding genes in the Vibrio cluster rather than the phylogenetically closer Pseudoalteromonas cluster (Pseudoalteromonas and Shewanella belong to the order Alteromonadales) indicates the possible occurrence of horizontal gene transfer. This is supported by the fact that only a few Alteromonas spp. (6 from 23 strains available into the database), Photobacterium spp. (2 from 10 strains) and Shewanella spp. (2 from 10 strains) were found to possess Cti coding sequences.
Figure 6. Addition of several stress inducing molecules does not modify trans-UFA production in E. faecalis FA2–2.
(A) Workflow. Prior to treatment bacterial cultures were grown in M17 FA free at 37°C (for E. faecalis FA2–2) or at 30°C (for P. putida F1) with vigorous shaking (250 rpm) and constant aeration to the early stationary phase (OD600 of 1.4 and 5.5 for E. faecalis FA2–2 and P. putida F1, respectively). After treatment with different concentrations of stressors, bacterial samples were taken for GC MS assays. The viability of bacterial cultures after treatment was also monitored (see Fig. S2).
(B) Fractions of trans-UFA from total bacterial FA treated with 1-octanol. N=3.
(C) Fractions of trans-UFA from total bacterial FA treated with toluene. N=3.
(D) Fractions of trans-UFA from total bacterial FA treated with paraquat showing that in contrast to P. putida F1, production of trans-UFA in E. faecalis FA2–2 is independent on the oxidative stress caused by paraquat. N=3.
(E) Time-dependent fractions of trans-UFA from total bacterial FA treated with 500 μM paraquat showing that production of trans-UFA in E. faecalis FA2–2 is independent on the oxidative stress caused by paraquat. N=3.
(F) Fractions of trans-UFA from total bacterial FA treated with a metal chelator EDTA showing that in contrast to P. putida F1, production of trans-UFA in E. faecalis FA2–2 is independent on the EDTA addition. N=3.
(G) Fractions of trans-UFA from total bacterial FA treated with daptomycin. N=6.
(H) Fractions of trans-UFA from total bacterial FA treated with MIC concentrations of chloramphenicol (cml) N=3.
Several species that encode putative Cti proteins were not included in the three major clusters and formed individual branches or small groups of Cti orthologous proteins. Several of these Cti encoding genes were found in poorly studied bacterial genera, suggesting that with the steadily increasing number of sequenced bacterial genomes several additional Cti clusters could form. However, from the ten Nitromonas spp. available in the database, only two were found to code for cti, indicating that Nitromonas spp. probably obtained Cti through horizontal gene transfer and Nitromonas Cti proteins should probably be clustered with Cti species belonging to another genera. Interestingly, two strains, Methylococcus capsulatus Texas and Cycloclasticus zancles 7-ME, each had two cti gene copies. The Methylococcus capsulatus Texas two Cti sequences (#180 and #189) are similar to each other, whereas Cycloclasticus zancles 7-ME encoded Cti proteins having only 36% identity (#63 and #190, Fig. 6 and Table S1). Although Cti #63 was close to Pseudomonas Cti and had 45% identity with P. putida F1 Cti, Cti #190 neighbored the Pseudoalteromonas Cti cluster with 42% identity with Cti of Pseudoalteromonas tunicate D2 (#177).
Together, these data indicate that only 5.5% of sequenced bacterial genomes encode a Cti candidate and these form three major clusters.
4. Discussion
Our analyses of FA composition of Gram-positive commensal bacterium E. faecalis FA2–2 have shown the presence of trans-UFA in its membrane lipids, a first since trans-UFA had previously been reported only in Gram-negative bacteria. This bacterium synthetized C16:1-trans-UFA during growth using cis-UFA as a substrate. The trans- and cis-double bonds were located at Δ9 position, indicating that no shift in double bond position nor transient saturation of the double bond occurred during isomerization. These data agree with previous findings obtained with P. putida cultures (von Wallbrunn et al., 2003), indicating that E. faecalis probably uses an enzymatic mechanism similar to that of P. putida Cti to form trans-UFA, although no apparent Cti homologue is encoded in the E. faecalis genome. Trans-UFA were detected in all major E. faecalis FA2–2lipids. Only lyso-PG lacked trans-UFA. Together with the prior report that trans-UFA were located in the sn-2 position of the PL glycerol moiety in Vibrio ABE-1 (Okuyama et al., 1991) suggests that E. faecalis FA2–2 cis/trans isomerization occurs at the sn-2 position of PLs.
Since Cti in P. putida was reported have be a covalently attached cytochrome c type heme essential for the isomerization reaction (Holtwick et al., 1997; Okuyama et al., 1998), we tested the effect of oxidative stress caused by paraquat, as well as that of a chelating agent, EDTA, on the trans-UFA formation in E. faecalis FA2–2. Although, P. putida F1 cultures showed a decreased rate of trans-UFA into the membrane lipids when treated with either compound, E. faecalis trans-UFA rate was constant despite the treatment. One possible explanation could be heme inaccessibility in E. faecalis cis-trans isomerase.
Enterococci use an electron transport chain for aerobic respiration when heme is provided or, alternatively, can perform fermentation in the absence of heme. E. faecalis does not synthesize heme de novo and in contrast to pseudomonads has little or no requirement for nutritional iron (Cornelis and Dingemans, 2013; Keogh et al., 2017). However, in presence of heme E. faecalis cells were found to assemble two heme proteins, a membrane-bound cytochrome bd (Winstedt et al., 2000), and a cytoplasmic typical catalase (Baureder et al., 2014; Baureder and Hederstedt, 2012; Frankenberg et al., 2002). Thus, these proteins could be involved in cis-trans-isomerization in E. faecalis. We tested the FA composition of E. faecalis FA2–2 cultures, which grew in TSBG medium containing less than 0.05 μM heme and reported to unable E. faecalis V583 catalase activity (Frankenberg et al., 2002). These preliminary tests did not show any decrease of trans-UFA production in TSBG cultures comparing to heme supplemented cultures (data not shown). Although future investigations are needed to find the enzyme catalyzing cis-trans isomerization in E. faecalis FA2–2, we suggest these proteins are unlikely to be involved. This indicates that trans-UFA in E. faecalis FA2–2 could be formed by using a heme or other metal-dependent mechanism.
Although there are statistically significant differences in trans-UFA production between measurements at different temperatures and in different growth phases (Fig. 5), given the very modest trans-UFA content (ca. 2% of the total acyl chains) these differences are too small to affect bulk membrane physical properties. However, this does not preclude specific interactions with proteins or in signaling processes where the trans-UFA could play a physiological role(s).
In this study we also investigated possible physiological roles for trans-UFA production in E. faecalis FA2–2. In contrast to P. putida, E. faecalis FA2–2 did not increase trans-UFA production when organic solvents, such as octanol and toluene were added to bacterial cultures. This indicated that in this bacterium trans-UFA are not involved in membrane response to organic solvents. However, E. faecalis FA2–2 was found to exhibit surprising for Gram-positive bacteria solvent tolerance. Due to the inherent disadvantage of lacking an outer membrane, only a few Firmicute bacteria have been previously reported to exhibit solvent tolerance, including species of Bacillus, Rhodococcus, Clostridium, Arthrobacter, Lactobacillus, Staphylococcus and Enterococcus (Isken and de Bont, 1998; Na et al., 2005; Nielsen et al., 2005; Paje et al., 1997; Sardessai and Bhosle, 2002; Torres and Castro, 2003; Zahir et al., 2006). Some organic solvent tolerance mechanisms in Gram-positive bacteria have been proposed such as induction of general stress regulon; production of organic solvent emulsifying or deactivating enzymes; active solvent efflux pumps, as well as cell morphology alterations and filamentous growth (Torres et al., 2011). However E. faecalis is a ubiquitous commensal of mammalian gastrointestinal flora (Lebreton et al., 2014) and thus unlikely to be naturally exposed to organic solvents. Thus, we tested the effect of daptomycin on the production of trans-UFA in E. faecalis FA2–2. Although daptomycin was reported to decrease membrane fluidity in B. subtilis and E. faecalis cells (Mishra et al., 2012; Müller et al., 2016), it had no effect on production of trans-UFA in E. faecalis FA2–2 cultures. It remains possible that trans-UFA production in E. faecalis could be in response to a specific stressor(s). that we have not tested, and the possibility that E. faecalis trans-UFA synthesis is not linked to membrane stress cannot be excluded.
Our analysis of Cti distribution among bacterial species showed that only about 5.5% of tested bacterial genomes contained cti coding genes. The detailed analysis of all bacterial species showed that the majority of them were found in environments containing membrane fluidity stressors suggesting that environment is an important factor in cti distribution. Among three Cti clusters reported in our study, the Pseudomonas and Pseudoalteromonas Cti clusters were homogenous whereas the Vibrio Cti cluster contained several Cti species from other bacteria species consistent with the known ability of these bacteria to take up DNA molecules This indicated that Cti could be evolutionary acquired via horizontal gene transfer, triggered by association in differential population habitats (Oliveira et al., 2017; Polz et al., 2013) which leads to a model of ecological Cti speciation. Nonetheless, in agreement with previous studies (Okuyama et al., 1998; Pedrotta and Witholt, 1999) we found cti orthologous genes only in Gram-negative bacteria.
5. Conclusions
In this study we investigated the distribution of cti coding sequence among bacterial species, showing that cti orthologs presented in only 5.5% tested bacterial strains. There are Gram-negative bacteria, the majority of which were found in contaminated or highly osmotic environments. However, we found that a Gram-positive bacterium E. faecalis FA2–2 formed trans-UFA using an undetermined pathway. The C16:1-trans Δ9 were found to be formed during growth of the bacterium from C16:1-cis Δ9. The role of trans-UFA in E. faecalis membrane remains a puzzle, organic solvents, as well as daptomycin did not have significant effect on trans-UFA formation.
Supplementary Material
Figure 7. Neighbor-joining tree based on Cti protein sequences.
Cti protein sequences were extracted from the MaGe Platform (http://www.genoscope.cns.fr/agc/mage). Node bootstrap values greater than 80% were used to construct the tree. The scale bar represents the average number of substitutions per site. Each number corresponds to one Cti sequence, whose reference and bacteria strains are reported in Table S3. Three major Cti clusters Pseudomonas Cti, Vibrio Cti and Pseudoalteromonas Cti are shown in orange, green and blue, respectively.
Highlights.
For the first time we demonstrate the production of trans-unsaturated fatty acids by a Gram-positive bacterium
The mechanism of trans-UFA production was investigated.
Although the role of trans-unsaturated fatty acids in E. faecalis FA2–2 remains unresolved, organic solvents, as well as daptomycin were unable to activate trans-UFA production in E. faecalis FA2-
This work defines cis-trans isomerase (Cti) distribution among bacterial species. Three major Cti clusters were defined and a habitat associated cti distribution is proposed.
Acknowledgments
We thank Dr. Alex Ulanov of the Metabolomics Center, University of Illinois for help with GC-MS analyses. This work was supported by National Institutes of Health Grant AI15650 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Conflict of Interest
We have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aakra Å, Vebø H, Indahl U, Snipen L, Gjerstad O, Lunde M, Nes IF, 2010. The response of Enterococcus faecalis V583 to chloramphenicol treatment. Int. J. Microbiol 2010 10.1155/2010/483048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baureder M, Barane E, Hederstedt L, 2014. In vitro assembly of catalase. J. Biol. Chem 289, 28411–28420. 10.1074/jbc.M114.596148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baureder M, Hederstedt L, 2012. Genes important for catalase activity in Enterococcus faecalis. PLoS One 7, e36725. 10.1371/journal.pone.0036725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ, 1959. A rapid method of total lipid extraction and purification. Biochem. Cell Biol 37, 911–917. 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- Cornelis P, Dingemans J, 2013. Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front. Cell. Infect. Microbiol 3, 75 10.3389/fcimb.2013.00075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE, 2006. A bacterium that has three pathways to regulate membrane lipid fluidity. Mol. Microbiol 60, 256–259. 10.1111/j.1365-2958.2006.05107.x [DOI] [PubMed] [Google Scholar]
- Cronan JE, Thomas J, 2009. Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways. Methods Enzymol. 459, 395–433. 10.1016/S0076-6879(09)04617-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbach R, Heipieper H-J, Keweloh H, 1992. The conversion of cis into trans unsaturated fatty acids in Pseudomonas putida P8: evidence for a role in the regulation of membrane fluidity. Appl. Microbiol. Biotechnol 38, 382–387. 10.1007/BF00170090 [DOI] [Google Scholar]
- Eberlein C, Baumgarten T, Starke S, Heipieper HJ, 2018. Immediate response mechanisms of Gram-negative solvent-tolerant bacteria to cope with environmental stress: cis-trans isomerization of unsaturated fatty acids and outer membrane vesicle secretion. Appl. Microbiol. Biotechnol 102, 2583–2593. 10.1007/s00253-018-8832-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Cronan JE, 2009. Escherichia coli unsaturated fatty acid synthesis. J. Biol. Chem 284, 29526–29535. 10.1074/jbc.M109.023440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flora SJS, Pachauri V, 2010. Chelation in metal intoxication. Int. J. Environ. Res. Public. Health 7, 2745–2788. 10.3390/ijerph7072745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankenberg L, Brugna M, Hederstedt L, 2002. Enterococcus faecalis heme-dependent catalase. J. Bacteriol 184, 6351–6356. 10.1128/JB.184.22.6351-6356.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto T, Takata N, Kudo T, Horikoshi K, 1994. Effect of temperature and growth phase on fatty acid composition of the psychrophilic Vibrio sp. strain no. 5710. FEMS Microbiol. Lett 119, 77–81. [DOI] [PubMed] [Google Scholar]
- Härtig C, Loffhagen N, Harms H, 2005. Formation of trans fatty acids is not involved in growth-linked membrane adaptation of Pseudomonas putida. Appl. Environ. Microbiol 71, 1915–1922. 10.1128/AEM.71.4.1915-1922.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heipieper HJ, de Bont JA, 1994. Adaptation of Pseudomonas putida S12 to ethanol and toluene at the level of fatty acid composition of membranes. Appl. Environ. Microbiol 60, 4440–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heipieper HJ, Diefenbach R, Keweloh H, 1992. Conversion of cis unsaturated fatty acids to trans, a possible mechanism for the protection of phenol-degrading Pseudomonas putida P8 from substrate toxicity. Appl. Environ. Microbiol 58, 1847–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heipieper HJ, Fischer J, Meinhardt F, 2010. cis–trans Isomerase of unsaturated fatty acids: an immediate bacterial adaptive mechanism to cope with emerging membrane perturbation caused by toxic hydrocarbons, in: Handbook of Hydrocarbon and Lipid Microbiology. Springer, Berlin, Heidelberg, pp. 1605–1614. 10.1007/978-3-540-77587-4_112 [DOI] [Google Scholar]
- Heipieper HJ, Loffeld B, Keweloh H, de Bont JAM, 1995. The cis/trans isomerization of unsaturated fatty acids in Pseudomonas putida S12: An indicator for environmental stress due to organic compounds. Chemosphere 30, 1041–1051. 10.1016/0045-6535(95)00015-Z [DOI] [Google Scholar]
- Heipieper HJ, Meinhardt F, Segura A, 2003. The cis–trans isomerase of unsaturated fatty acids in Pseudomonas and Vibrio: biochemistry, molecular biology and physiological function of a unique stress adaptive mechanism. FEMS Microbiol. Lett 229, 1–7. 10.1016/S0378-1097(03)00792-4 [DOI] [PubMed] [Google Scholar]
- Heipieper HJ, Meulenbeld G, van Oirschot Q, de Bont J, 1996. Effect of environmental factors on the trans/cis ratio of unsaturated fatty acids in Pseudomonas putida S12. Appl. Environ. Microbiol 62, 2773–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtwick R, Keweloh H, Meinhardt F, 1999. cis/trans Isomerase of unsaturated fatty acids of Pseudomonas putida P8: Evidence for a heme protein of the cytochrome c type. Appl. Environ. Microbiol 65, 2644–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtwick R, Meinhardt F, Keweloh H, 1997. cis-trans Isomerization of unsaturated fatty acids: cloning and sequencing of the cti gene from Pseudomonas putida P8. Appl. Environ. Microbiol 63, 4292–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huycke MM, Sahm DF, Gilmore MS, 1998. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg. Infect. Dis 4, 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken S, de Bont JA, 1998. Bacteria tolerant to organic solvents. Extrem. Life Extreme Cond 2, 229–238. [DOI] [PubMed] [Google Scholar]
- Isken S, Santos PMAC, de Bont JAM, 1997. Effect of solvent adaptation on the antibiotic resistance in Pseudomonas putida S12. Appl. Microbiol. Biotechnol 48, 642–647. 10.1007/s002530051109 [DOI] [Google Scholar]
- Jung D, Rozek A, Okon M, Hancock REW, 2004. Structural transitions as determinants of the action of the calcium-dependent antibiotic daptomycin. Chem. Biol 11, 949–957. 10.1016/j.chembiol.2004.04.020 [DOI] [PubMed] [Google Scholar]
- Junker F, Ramos JL, 1999. Involvement of the cis/trans isomerase Cti in solvent resistance of Pseudomonas putida DOT-T1E. J. Bacteriol 181, 5693–5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh D, Lam LN, Doyle L, Matysik A, Pavagadhi S, Umashankar S, Dale JL, Boothroyd CB, Dunny GM, Swarup S, Williams RBH, Marsili E, Kline K, 2017. Extracellular electron transfer powers Enterococcus faecalis biofilm metabolism. bioRxiv 130146. 10.1101/130146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondakova T, Cronan JE, 2019. Transcriptional regulation of fatty acid cis-trans isomerization in the solvent-tolerant soil bacterium, Pseudomonas putida F1. Environ. Microbiol 10.1111/1462-2920.14546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol WH, Van Buuren KJH, Butler J, Braams R, 1976. The kinetics of the reduction of cytochrome c by the superoxide anion radical. Biochim. Biophys. Acta BBA - Bioenerg. 449, 157–168. 10.1016/0005-2728(76)90130-4 [DOI] [PubMed] [Google Scholar]
- Lascano R, Muñoz N, Robert G, Rodriguez M, Melchiorre M, Trippi V, Quero G, 2012. Paraquat: an oxidative stress inducer. Herbic. - Prop. Synth. Control Weeds. 10.5772/32590 [DOI] [Google Scholar]
- Lebreton F, Willems RJL, Gilmore MS, 2014. Enterococcus diversity, origins in nature, and gut colonization, in: Gilmore MS, Clewell DB, Ike Y, Shankar N. (Eds.), Enterococci: From commensals to leading causes of drug resistant infection. Massachusetts Eye and Ear Infirmary, Boston.MA, USA: [PubMed] [Google Scholar]
- Maketon W, Zenner CZ, Ogden KL, 2008. Removal efficiency and binding mechanisms of copper and copper-EDTA complexes using polyethyleneimine. Environ. Sci. Technol 42, 2124–2129. [DOI] [PubMed] [Google Scholar]
- Mancini S, Imlay JA, 2015. The induction of two biosynthetic enzymes helps Escherichia coli sustain heme synthesis and activate catalase during hydrogen peroxide stress. Mol. Microbiol 96, 744–763. 10.1111/mmi.12967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough B, Macdonald PM, Sykes BD, McElhaney RN, 1983. Fluorine-19 nuclear magnetic resonance studies of lipid fatty acyl chain order and dynamics in Acholeplasma laidlawii B membranes. A physical, biochemical, and biological evaluation of monofluoropalmitic acids as membrane probes. Biochemistry 22, 5097–5103. 10.1021/bi00291a008 [DOI] [PubMed] [Google Scholar]
- Mishra NN, Bayer AS, Tran TT, Shamoo Y, Mileykovskaya E, Dowhan W, Guan Z, Arias CA, 2012. Daptomycin resistance in enterococci is associated with distinct alterations of cell membrane phospholipid content. PLoS One 7, e43958. 10.1371/journal.pone.0043958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A, Wenzel M, Strahl H, Grein F, Saaki TNV, Kohl B, Siersma T, Bandow JE, Sahl H-G, Schneider T, Hamoen LW, 2016. Daptomycin inhibits cell envelope synthesis by interfering with fluid membrane microdomains. Proc. Natl. Acad. Sci 113, E7077–E7086. 10.1073/pnas.1611173113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na K, Kuroda A, Takiguchi N, Ikeda T, Ohtake H, Kato J, 2005. Isolation and characterization of benzene-tolerant Rhodococcus opacus strains. J. Biosci. Bioeng 99, 378–382. 10.1263/jbb.99.378 [DOI] [PubMed] [Google Scholar]
- Neumann G, Kabelitz N, Heipieper HJ, 2003. The regulation of the cis-trans isomerase of unsaturated fatty acids in Pseudomonas putida: correlation between cti activity and K+-uptake systems. Eur. J. Lipid Sci. Technol 105, 585–589. 10.1002/ejlt.200300803 [DOI] [Google Scholar]
- Nielsen LE, Kadavy DR, Rajagopal S, Drijber R, Nickerson KW, 2005. Survey of Extreme solvent tolerance in Gram-positive cocci: membrane fatty acid changes in Staphylococcus haemolyticus grown in toluene. Appl. Environ. Microbiol 71, 5171–5176. 10.1128/AEM.71.9.5171-5176.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama H, Ueno A, Enari D, Morita N, Kusano T, 1998. Purification and characterization of 9-hexadecenoic acid cis-trans isomerase from Pseudomonas sp. strain E-3. Arch. Microbiol 169, 29–35. [DOI] [PubMed] [Google Scholar]
- Okuyama H, Okajima N, Sasaki S, Higashi S, Murata N, 1991. The cis/trans isomerization of the double bond of a fatty acid as a strategy for adaptation to changes in ambient temperature in the psychrophilic bacterium, Vibrio sp. strain ABE-1. Biochim. Biophys. Acta BBA - Lipids Lipid Metab. 1084, 13–20. 10.1016/0005-2760(91)90049-N [DOI] [PubMed] [Google Scholar]
- Okuyama H, Sasaki S, Higashi S, Murata N, 1990. A trans-unsaturated fatty acid in a psychrophilic bacterium, Vibrio sp. strain ABE-1. J. Bacteriol 172, 3515–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira PH, Touchon M, Cury J, Rocha EPC, 2017. The chromosomal organization of horizontal gene transfer in bacteria. Nat. Commun 8 10.1038/s41467-017-00808-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özen AI, Ussery DW, 2012. Defining the Pseudomonas genus: where do we draw the line with Azotobacter? Microb. Ecol 63, 239–248. 10.1007/s00248-011-9914-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paje ML, Neilan BA, Couperwhite I, 1997. A Rhodococcus species that thrives on medium saturated with liquid benzene. Microbiol. Read. Engl 143 ( Pt 9), 2975–2981. 10.1099/00221287-143-9-2975 [DOI] [PubMed] [Google Scholar]
- Pedrotta V, Witholt B, 1999. Isolation and characterization of the cis-trans-unsaturated fatty acid isomerase of Pseudomonas oleovorans GPo12. J. Bacteriol 181, 3256–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polz MF, Alm EJ, Hanage WP, 2013. Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet. TIG 29, 170–175. 10.1016/j.tig.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid R, Cazenave-Gassiot A, Gao IH, Nair ZJ, Kumar JK, Gao L, Kline KA, Wenk MR, 2017. Comprehensive analysis of phospholipids and glycolipids in the opportunistic pathogen Enterococcus faecalis. PLoS ONE 12 10.1371/journal.pone.0175886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardessai Y, Bhosle S, 2002. Tolerance of bacteria to organic solvents. Res. Microbiol 153, 263–268. 10.1016/S0923-2508(02)01319-0 [DOI] [PubMed] [Google Scholar]
- Seelig J, Waespe-Sarcevic N, 1978. Molecular order in cis and trans unsaturated phospholipid bilayers. Biochemistry 17, 3310–3315. 10.1021/bi00609a021 [DOI] [PubMed] [Google Scholar]
- Silhavy TJ, Kahne D, Walker S, 2010. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol 2 10.1101/cshperspect.a000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinensky M, 1974. Homeoviscous adaptation-a homeostatic process that regulates the viscosity of membrane lipids in Escherichia coli. Proc. Natl. Acad. Sci 71, 522–525. 10.1073/pnas.71.2.522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus SK, Hancock REW, 2006. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: Comparison with cationic antimicrobial peptides and lipopeptides. Biochim. Biophys. Acta BBA - Biomembr., Membrane Biophysics of Antimicrobial Peptides 1758, 1215–1223. 10.1016/j.bbamem.2006.02.009 [DOI] [PubMed] [Google Scholar]
- Torres S, Castro GR, 2003. Organic solvent resistant lipase produced by thermoresistant bacteria, in: Roussos S, Soccol CR, Pandey A, Augur C. (Eds.), New Horizons in Biotechnology. Springer Netherlands, pp. 113–122. [Google Scholar]
- Torres S, Pandey A, Castro GR, 2011. Organic solvent adaptation of Gram-positive bacteria: Applications and biotechnological potentials. Biotechnol. Adv 29, 442–452. 10.1016/j.biotechadv.2011.04.002 [DOI] [PubMed] [Google Scholar]
- von Wallbrunn A, Richnow HH, Neumann G, Meinhardt F, Heipieper HJ, 2003. Mechanism of cis-trans isomerization of unsaturated fatty acids in Pseudomonas putida. J. Bacteriol 185, 1730–1733. 10.1128/JB.185.5.1730-1733.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winstedt L, Frankenberg L, Hederstedt L, von Wachenfeldt C, 2000. Enterococcus faecalis V583 contains a cytochrome bd-type respiratory oxidase. J. Bacteriol 182, 3863–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahir Z, Seed KD, Dennis JJ, 2006. Isolation and characterization of novel organic solvent-tolerant bacteria. Extremophiles 10, 129–138. 10.1007/s00792-005-0483-y [DOI] [PubMed] [Google Scholar]
- Zhu L, Bi H, Ma J, Hu Z, Zhang W, Cronan JE, Wang H, 2013. The two functional enoyl-acyl carrier protein reductases of Enterococcus faecalis do not mediate triclosan resistance. mBio 4 10.1128/mBio.00613-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Lin J, Ma J, Cronan JE, Wang H, 2010. Triclosan resistance of Pseudomonas aeruginosa PAO1 is due to FabV, a triclosan-resistant enoyl-acyl carrier protein reductase. Antimicrob. Agents Chemother 54, 689–698. 10.1128/AAC.01152-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.