Abstract
Background
Understanding local Anopheles species compositions and bionomic traits are vital for an effective malaria vector intervention strategy. Though eight malaria vectors, including species complexes, have been documented across the island of Sulawesi, Indonesia, a comprehensive survey linking morphological and molecular species identification has not been conducted in this global hotspot of biodiversity.
Results
Eighteen distinct species of Anopheles were molecularly identified in a 1 km2 area in Karama village, West Mamuju Province, Sulawesi. Known species included An. aconitus, An. karwari, An. peditaeniatus, An. vagus, An. barbirostris, An. tessellatus, An. nigerrimus, An. crawfordi, An. maculatus, An. flavirostris and An. kochi. Of the 18 distinct sequence groups identified through both ribosomal DNA internal transcribed spacer region 2, and mitochondrial DNA cytochrome c oxidase subunit 1 loci, 8 could not be identified to species through comparison to published sequences. The comparison of morphological and molecular identities determined that interpretations of local species compositions for primary and expected species in Karama (An. barbirostris and An. vagus) had the highest rate of accuracy (92.1% and 87.6%, respectively) when compared to molecular analysis. However, the remaining distinct sequences molecularly identified to species were identified correctly by morphological methods less frequently, from 0 to 83%.
Conclusions
Karama, Indonesia has a high diversity of Anopheles spp. The unexpected high number of Anopheles species in a small area points to possible complex transmission dynamics and limitations with vector control based on possible varying behaviors and interactions with both humans and interventions. Morphological identification of Anopheles spp. in this study was more accurate for primary and expected species than secondary or unexpected species. Finally, the inability to identify seven sequence groups to species with consensus sequences implies that future studies employing sequencing are required to clarify species compositions in the Nigerrimus Subgroup, among others, as well as their distribution and vector status. Use of molecular methods in conjunction with morphological investigations for analysis of species composition, population dynamics and bionomic characteristics is directly implicated in understanding drivers of malaria transmission, intervention effectiveness, and the pursuit of malaria elimination. 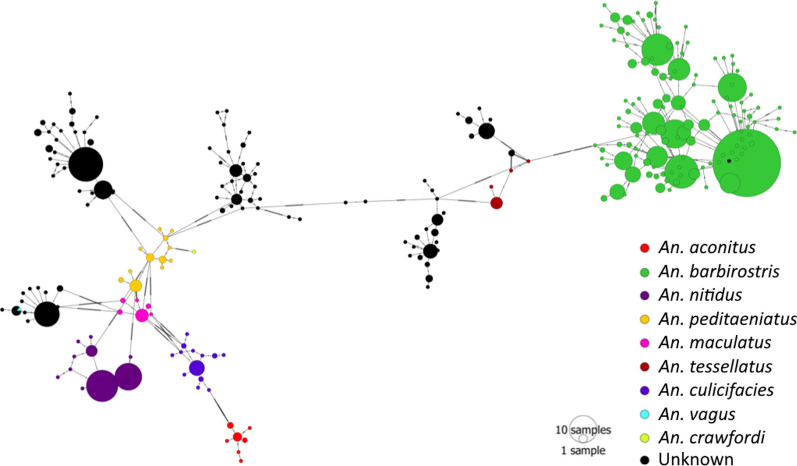
Keywords: Anopheles, Molecular identification, Sulawesi, Malaria vectors, Indonesia
Background
The World Health Organization (WHO) has set the ambitious goals of reducing malaria incidence and mortality by 90% by 2030 [1]. Although vector control, primarily through the use of long-lasting insecticide treated nets (LLINs) and indoor residual spraying (IRS), has led to a reduction of malaria cases worldwide [1], WHO reported a stall in progress in 2017 [2]. Despite global elimination efforts, almost half of the Republic of Indonesia’s population lives in malaria-endemic areas [3]. The highest rates of malaria cases are found in the eastern provinces of Papua, West Papua, East Nusa Tenggara, Maluku, North Maluku and Bengkulu [4, 5]. West Sulawesi has low to moderate endemicity (annual parasite index (API) of 0.99 per 1000 cases) and falls under the intensification and elimination phases of malaria control [4, 5].
Interventions target mosquito behaviors that overlap with how the intervention functions in both time and space. For example, LLINs target indoor and anthropophagic behaviors, while IRS targets indoor resting mosquitoes [6–8]. Regional or local vector populations usually consist of a variety of primary and secondary vectors, each with differing bionomic traits and seasonal population shifts, affecting the temporal and spatial protective efficacy of interventions accordingly. Data on local vector temporal composition and their behavior are crucial to the comprehension of transmission dynamics, the designing of intervention strategies, interpreting intervention efficacy, determining residual transmission, and understanding current gaps in protection from infectious bites.
Malaria is transmitted by mosquitoes of the genus Anopheles, which includes 475 recognized species and more un-named members of species complexes [9, 10]. Approximately 70 of these 475 species are competent as vectors of malaria parasites, while 41 species are considered dominant vectors [11]. The Indonesian archipelago has high Anopheles diversity, with 21 species being confirmed as vectors of malaria [5, 10–13]. Indonesia is a geographically diverse country, with differing island ecosystems, Sulawesi being a biodiversity hotspot [14]. This island, close to the Wallace line, has both Asian and Australian documented vector species and species complexes including An. barbirostris, An. barbumbrosus, An. flavirostris, An. kochi, An. nigerrimus, An. parangensis, An. sinensis and An. subpictus (s.l.) [12, 15–17]. There is documentation for multiple species in some complexes, such as the Barbirostris Complex [16, 18–21]. Additionally, there is molecular evidence for four cytological forms of An. nigerrimus in Southeast Asia [22, 23].
Published literature records indicate that multiple vectors and varied transmission dynamics are present across Indonesia [24–26] pointing to the need for a current, basic, and better understanding of species distributions, bionomic traits, and vector status, particularly relevant when considering that Indonesia aims to achieve malaria elimination by 2030 [3, 5, 10–12, 24–26].
Correct mosquito species identification is essential to understanding local mosquito species composition and associated bionomic traits that impact transmission. Identification of field-collected specimens is primarily based on morphological characteristics of adult males and females, as well as immatures [27, 28]. Although presently the most available and usually effective tool, morphological identification may be complicated by outdated, contradictory, and difficult to interpret keys [24, 26], particularly in non-African areas. Damage to crucial identifying characteristics such as the loss of scales, can occur to field-caught specimens, resulting in misidentifications. Additional issues with morphological identification include human error, presence of new or cryptic species, species with overlapping or non-documented characteristics, and intraspecific morphological variation [26]. Furthermore, accurate morphological identification requires comprehensive and rigorous training. Molecular identification allows for greater supporting granularity and may be more precise in regions of high diversity such as Southeast Asia, with an abundance of vectors, and novel, cryptic and sibling species [24–26].
To the best of our knowledge, this is the first comprehensive survey towards understanding Anopheles species diversity and species compositions, in and around human habitation, with molecular identification, in any area in West Sulawesi.
Methods
Site description
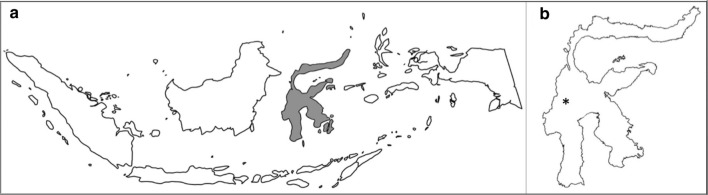
Karama, Indonesia is a single km2 village in the northwestern regency of Mamuju, West Sulawesi (Fig. 1). This isolated village bordered by the Sungai Karana River, is located on its flood-plain, and reaches into the foothills. The main economic activity in the area is agriculture, with the primary crop being rice. Other activities include fishing and hunting in the surrounding forest. Houses in this area are made of wood or concrete with thatched roofs. Low-lying houses are elevated on stilts due to flooding episodes. The open construction of these primarily wood houses allow for mosquito entry from all directions. This remote area has stable, year-round, malaria transmission with high incidences during the rainy season (November-March) [5].
Fig. 1.
Map of Indonesia field collection sites. a Map of Indonesia outlining the island of Sulawesi. b Location of Karama village, West Sulawesi. The map was created using Google: imagery 2019 DigitalGlobe, Map data 2019
Mosquito collections and trap description
Mosquitoes were intermittently collected in Karama during 2013 to 2015 (Fig. 1, Table 1). Multiple sampling methods were utilized to enable the comprehensive sampling of anophelines over several years. Sampling methods included human landing catches, barrier screens, barrier screens with eaves, and kelambu traps (IW, unpublished data) [29, 30].
Table 1.
Overview of Anopheles mosquito collections. Four collection methods were utilized in Karama, Indonesia during 2013–2015
| Trapping method | 2013 | 2014 | 2015 | Total | |||
|---|---|---|---|---|---|---|---|
| April–May | September | December | May | January | March | ||
| Barrier screens | 1074 | 566 | 1523 | 680 | – | 1062 | 4905 |
| Barrier screens with eaves | 710 | – | – | – | – | 1283 | 1993 |
| Indoor human landing catches | 1435 | 985 | 1094 | 1087 | 153 | 762 | 5516 |
| Outdoor human landing catches | 1712 | 1000 | 995 | 322 | 190 | 902 | 5121 |
| Kelambu trap | 1851 | 853 | 1486 | 789 | 126 | 1869 | 6974 |
| Totals | 6782 | 3404 | 5098 | 2878 | 469 | 5878 | 24,509 |
The barrier screens with eaves were not performed in the 2014 (indicated by –)
Human landing catches (HLCS)
HLCs [29] were performed between 18:00 h and 06:00 h. Collections were performed both inside and outside houses. Collections were done in 2-h shifts, with a single collector indoors and a single collector outdoors in each sampled house (n = 8). After each 2-h period, the two collectors swapped positions to reduce collector bias. Location and time of collection were recorded for all mosquitoes.
Barrier screens
Barrier screens were constructed with 2 m high, untreated bednet material secured to wooden poles at 2 m intervals for a length of 10 m. Barrier screens were set up and located as described [30]. Barrier screens were examined for mosquitoes hourly between 18:00 h and 06:00 h. Two collectors walked along each side of the trap for 15–20 min every hour, using a flashlight to spot, and mouth aspirator to collect, resting intercepted mosquitoes. Location, time of collection, and flight direction (determined by the side of the barrier screen) were recorded for all mosquitoes.
Barrier screens with eaves
Barrier screens with eaves were constructed in the same manner as barrier screens, but with untreated bed net material eaves at the top to prevent mosquitoes from escaping over the vertical netting (NL, unpublished data). Mosquitoes were collected off barrier screens with eaves and labeled in the same manner as barrier screens.
Kelambu traps
Kelambu traps (IW, unpublished data) are attractant-free, modified bednet traps that target free-flying mosquitoes. The trap is divided diagonally to give 4 quadrants, which allows for the determination of mosquito flight direction. Mosquitoes were collected by aspiration every hour from 18:00 h to 06:00 h from each quadrant.
Mosquitoes sampled from all traps (n = 19,467) were morphologically identified in the field to species, group, or complex level [28]. Anophelines were individually stored on desiccant and shipped to the University of Notre Dame, USA, for molecular identification and data analysis.
Molecular processing and sequence analysis
A subset of morphologically identified mosquitoes (n = 4102 out of 24,509) were sequenced at the ribosomal DNA internal transcribed spacer region 2 (ITS2) and/or cytochrome c oxidase subunit 1 (cox1) loci towards species identification [24, 26]. Samples were first sequenced at the ITS2 locus, and then a subset of samples with successful ITS2 sequences were also sequenced at the cox1 locus. Samples sequenced were randomly chosen across trapping method and year collected. Approximately 10% of samples from each trap type were randomly selected and sequenced. Only 4.5% of the samples from barrier screens with eaves were randomly selected and sequenced due to collections only being performed during two collection periods.
Species identification
Molecular identification was conducted blind to morphological identity to prevent any bias in the analysis. Final species confirmation required high sequence identity (≥ 98%) to voucher sequences in multiple databases [24–26, 31, 32]. cox1 and ITS2 database comparisons for each sample were paired to determine species when either cox1 or ITS2 alone did not produce significant results to voucher sequences [24–26, 31, 32]. Consensus sequences were manually inspected for insertions, deletions, and repeat regions to ensure these sequence differences did not inflate divergence and decrease identity scores. Consensus sequences of each sequence group were compared (BLASTn) to the NCBI nr and BOLD databases to identify species. Sequences mapping to the Funestus Group, Hyrcanus Group, Barbirostris Complex, Sundaicus Complex and Maculatus Group were compared to voucher sequences and primers used in diagnostic PCR [17, 33–37].
Phylogenetic analysis
For cox1, 2034 sequences were queried against the nucleotide non-redundant database (date of download: 31 January 2018) using the BLASTn algorithm in the BLAST + v.2.2.18 command line application [38]. Sequences with a percent identity greater than 90% over 75% of the query length matching any Anopheles cox1 were retained for further analysis. This resulted in 2013 cox1 sequences.
cox1 sequences were aligned using MAFFT v7.394 [39] with parameters: –auto –quiet –preserve-case. Aligned sequences were formatted into a nexus alignment using seqmagick (https://fhcrc.github.io/seqmagick/). Nexus-formatted sequences were used to create a haplotype network in PopArt [40] with the inclusion of a trait file indicating the identity of each sequence from the previous BLAST query. PopArt parameters were selected to construct a minimum spanning network with an epsilon value of 0. The final network nodes were colored to reflect species identity.
The program ABGD [41] was used to identify molecular operational taxonomic units (MOTU) for delineating species. Values of intraspecific genetic diversity of 0.03 and interspecific genetic diversity of 0.10 were used to separate clusters. After 10 permutations, 14 clusters were identified following the methods in Puillandre et al. [41]. From each cluster, 2 sequences were randomly chosen for phylogenetic analysis. A phylogenetic tree was constructed in Geneious v 11.1.2 using PhyML with 100 bootstrap samples [42].
A phylogenetic analysis was performed on ITS2 consensus sequences [43]. A multiple alignment of consensus ITS2 sequences was performed with MUSCLE v3.8.31 [44]. After alignment, Gblocks v0.91b was used to eliminate poorly aligned positions and divergent regions [45]. The phylogenetic tree was reconstructed using the maximum likelihood method implemented in the PhyML v3.1/3.0 aLRT program [42, 46] was used to calculate the unrooted tree. Reliability for internal branch was assessed using the bootstrapping method (100 bootstrap replicates). Graphical representation and edition of the phylogenetic tree were performed with TreeDyn v198.3 [47].
Results
Approximately 24,509 morphologically identified Anopheles mosquitoes were sampled in a comprehensive trapping effort. Sampling over almost 3 years ensured the capture of seasonal population variations. Trapping habitats included inside and outside domestic spaces, open fields, rice fields, farms, as well as areas that faced the jungle, marshes, river and mountains. Collections using several different methods ensured that multiple behaviors were solicited in mosquito capture. These included HLCs, both inside and outside houses, capturing human seeking mosquitoes. Barrier screens and Kelambu traps were placed to capture free-flying mosquitoes flying to oviposition sites (forest, river, marsh and ponds) as well as towards multiple hosts (humans, goats, cows and chickens). A set of 4102 specimens, randomly sampled over all trapping types, spaces and periods were processed molecularly.
Molecular species identification
ITS2 and cox1 sequences representing 2616 and 2163 Anopheles mosquitoes, respectively, were processed. Approximately 677 specimens were sequenced for both ITS2 and cox1 loci while the remaining had only one sequence (either ITS2 or cox1).
ITS2 sequences representing 2616 Anopheles mosquitoes were aligned into 18 sequences with a stringency of greater than 98% identity within each sequence (Table 2). The cox1 sequences representing 2163 Anopheles mosquitoes and 19 morphologically identified species were aligned into 20 distinct sequence groups (Table 2). The parameters used to produce the final sequence groups resulted in no hyper-variable regions present in any specific sequence group. Similarly, there was little (under 2%) variation in sequences within a sequence group, i.e. none with insertions or deletions more than 2 base pairs (bp). Distinct sequence groups were arbitrarily called sequence groups (AN) 1 through 18 prior to a more in-depth database comparison and species level identification. High similarity (≥ 98% identity), the presence of voucher specimens in multiple databases, concordant ITS2-cox1 pairs, and/or sequence alignment to those used in PCR diagnostic assays allowed the identification of eleven species: An. aconitus (ANI); An. karwari (AN2); An. peditaeniatus (AN3); An. vagus (AN4 and AN5); An. barbirostris (AN6); An. tessellatus (AN7); An. nigerrimus (AN8); An. flavirostris (AN9), An. crawfordi (AN10); An. maculatus (AN11); and An. kochi (ANI2) (Table 2).
Table 2.
Overview of molecular identifications
| Sequence group | No. of samples (ITS2; cox1) | Sequence length in bp (ITS2; cox1) | ITS2 homology (%ID) | Cox1 homology (%ID) | Final ID |
|---|---|---|---|---|---|
| AN1 | 23; 20 | 515; 651 | An. aconitus (98.6) | An. aconitus (95.9) | An. aconitus |
| AN2 | 1; 1 | 495; 327 | An. karwari (98.7) | An. karwari (97.7) | An. karwari |
| AN3 | 113; 29 | 524; 698 | An. peditaeniatus (100) | An. peditaeniatus (99.5) | An. peditaeniatus |
| AN4 | 29; 29 | 658; 697 | An. vagus (98.6) | An. vagus (96.6) | An. vagus |
| AN5 | 275; 35 | 640; 697 | An. vagus (99.8) | An. vagus (96.6) | An. vagus |
| AN6 | 1305; 222 | 1470; 704 | An. barbirostris (97.2) | An. barbirostris (98.6) | An. barbirostris |
| AN7 | 35; 28 | 592; 643 | An. tessellatus (96.8) | An. tessellatus (95.6) | An. tessellatus |
| AN8 | 233; 43 | 592; 652 | An. nigerrimus (96.7) | An. nitidus (96.1) | An. nigerrimus |
| AN9 | 47; 39 | 503; 628 | An. flavirostris (99.8) | An. flavirostris (98.9) | An. flavirostris |
| 503; 643 | An. flavirostris (98.95) | ||||
| AN10 | 5; 4 | 534; 630 | An. sinensis (97.9) | An. crawfordi (97.1) | An. crawfordi |
| AN11 | 28; 25 | 450; 643 | An. maculatus (95.4) | An. maculatus (95.9) | An. maculatus |
| AN12 | 7; 4 | 491; 631 | An. kochi (99.4) | An. kochi (100) | An. kochi |
| AN13 | 32; 29 | 617; 679 | An. saeungae (9.0) | An. sp. 14 (91.7) | Myzorhynchus Series |
| AN14 | 140; 40 | 420; 689 | An. bancroftii genotype B (93.3) | An. coustani (92.1) | Myzorhynchus Series |
| 420; 636 | An. farauti (92.3) | Neomyzomyia Series | |||
| AN15 | 5; 4 | 381; 689 | An. bancroftii genotype B (91.5) | An. coustani (92.0) | Myzorhynchus Series |
| AN16 | 1; 1 | 615; 616 | An. subpictus (78.6) | An. albitarsis E (91.5) | Genus Anopheles |
| AN17 | 277; 72 | 569; 717 | An. sundaicus (80.7) | An. albitarsis (91.4) | Genus Anopheles |
| AN18 | 60; 52 | 589; 689 | An. tessellatus (88.1) | An. lutzii (92.0) | Genus Anopheles |
Table represents the 2616 total ITS2 sequences and the corresponding 677 cox1 sequences. Final species identifications are based on both ITS2 and cox1 comparisons. % ID is the percentage identity based on BLAST database comparison. Final species identification, when not to specific species, was based on the lowest common taxonomic identity for the paired ITS2 and cox1 sequences
Sequence comparisons of AN1 ITS2 to those used to develop PCR diagnostic assays for the Funestus Group [33] confirmed the identification of An. aconitus. Sequence comparisons of AN3, AN8 and AN10 to the species diagnostic sequences and primers [34] further clarified species identities. AN3, AN8 and AN9 were confirmed to be An. peditaeniatus, An. nigerrimus and An. flavirostris, respectively. Even though the cox1 sequence had the highest match to An. nitidus for AN8, we have used the ITS2 result for the species identification as the species diagnostic PCR would be positive for this species. The AN10 cox1 sequence aligned with primers and sequence confirming An. crawfordi. However, the AN10 ITS2 top database hit was to a sequence identified as An. sinensis, also identical to the An. crawfordi sequence in the Hempolchom PCR diagnostic assay [34]. We have designated AN10 as An. crawfordi using the results from this PCR, along with support from the cox1 hit indicating An. crawfordi. The AN11 ITS2 demonstrated the highest similarity to specimens used in the Maculatus Group diagnostic assay [37]. In addition, comparison of these sequences to those in Ali et al. [36] and Garjito et al. [48] demonstrated a 100% match thus confirming An. maculatus with the added support from the cox1 sequence (Table 2).
Several specimens with matched ITS2 and cox1 sequences had homology to different species in the database resulting in uncertain species identification. A conservative approach was used resulting in sequence groups AN14 and AN15 being identified to the lowest common taxonomic level based on the closest identified sequences, instead of a specific species.
Sequence groups AN6 and AN13 both had similarity to the Barbirostris Complex. The ITS2 and cox1 sequences of AN6 demonstrated high similarity to An. barbirostris. The ITS2 sequence was compared to voucher sequences and primer sites used in Brosseau et al. [17], as the highest homology was to these sequences. Although most similar to An. barbirostris, the presence of multiple SNPs, insertions and deletions would result in only a single primer being able to bind–rendering this multiplex assay producing no bands for these specimens.
The AN13 ITS2 sequence was closest to An. saeungae with the caveat that the similarity was only in the coding region (9% ID overall and 91% ID to the coding region) with no real homology seen in the intergenic spacer region rendering the Brosseau et al. [17], multiplex PCR incompatible. The cox1 sequence was closest to an unknown Anopheles species (92% ID) found in both Zambia and the Kenyan highlands. This may represent a new member of the Barbirostris Group but due to the conservative approach to identification and high percentage identity requirement for both ITS2 and cox1 loci, these sequences were identified to the Myzorhynchus Series.
Sequence groups AN16-AN18 did not share greater than 93% identity (when combined with sequence coverage) with any nr database sequences and were identified to the genus level only. The comparison of ITS2 sequences from AN16 and AN17, to those used to develop the Dusfour et al. [35], diagnostic PCRs, revealed the absence of any primer sites with low homology across the sequences. The combination of disparate ITS2 and cox1 sequence pairs also complicates this analysis. AN16, AN17 and AN18 remain unknown species.
Phylogeny
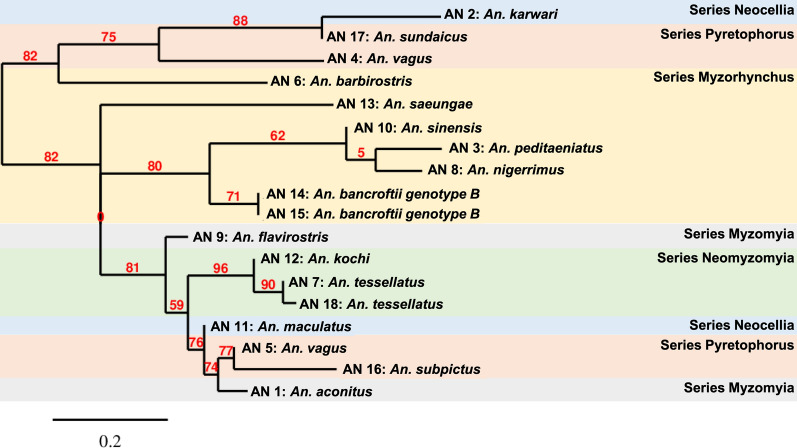
Consensus ITS2 sequences were aligned to construct a phylogenetic tree. The ITS2 tree (Fig. 2) groups putative species as expected based on their taxonomy. The An. barbirostris sequence groups (AN6 and AN13), An. nigerrimus (AN8), An. sinensis (AN10), An. peditaeniatus (AN3), and An. bancroftii sequence groups (AN14 and AN15) clustered separately as part of the Myzorhynchus Series. Members of the Barbirostris Complex, An. nigerrimus, An. sinensis, An. peditaeniatus, and An. bancroftii group all belong to the subgenus Anopheles, whereas the rest of the species identified here are members of the subgenus Cellia. Anopheles vagus, An. subpictus and An. sundaicus (i.e. AN4, AN5, AN16 and AN17, respectively) clustered together as part of the Pyretophorus Series. Anopheles vagus AN4 was more closely related to An. sundaicus AN17 while An. vagus AN5 is more closely related to An. subpictus. The Myzomyia Series cluster was composed of An. aconitus and An. flavirostris (AN1 and AN9, respectively) while the Neocellia Series cluster was composed of An. karwari and An. dispar (AN2 and AN11, respectively). Anopheles tessellatus and An. kochi clustered as part of the Neomyzomyia Series (AN7, AN12 and AN18, respectively). Species AN12-AN18 had low similarity (< 93% ID) to known sequences in the database and may represent new species.
Fig. 2.
Phylogenetic tree based on ITS2 sequences. A phylogenetic tree was constructed using 1985 ITS2 sequences in Geneious v 11.1.2 using PhyML with 100 bootstrap replicates. Bootstrap values are shown as percentages above the branches
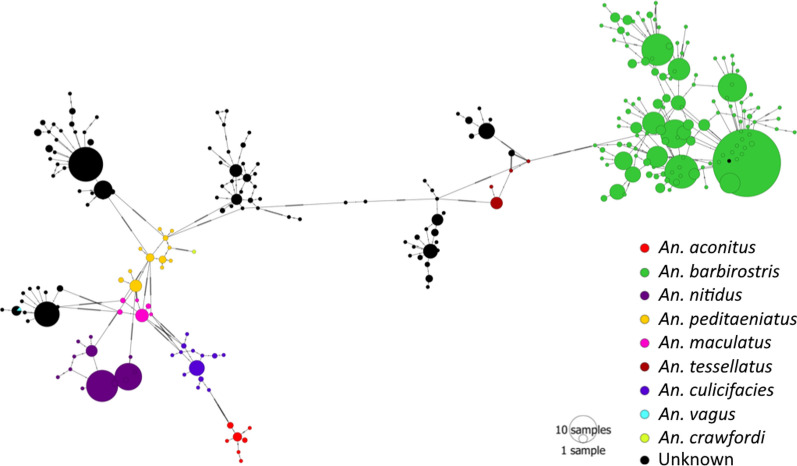
Consensus cox1 sequences were aligned to construct a haplotype network (Fig. 3). Among these cox1 samples, 9 haplotypes allowed species identification: An. aconitus; An. barbirostris; An. nitidus; An. peditaeniatus; An. maculatus; An. tessellatus; An. culicifacies; An. vagus; and An. crawfordi. Furthermore, black circles represent possible cryptic species in the dataset, numbering 5 to 8 species. The network indicates there is intermediate divergence between An. barbirostris and An. culicifacies nodes.
Fig. 3.
Haplotype network based on 2034 cox1 sequences from this study. Each observed haplotype is indicated by a filled circle, sized according to its frequency and colored according to the Anopheles species represented. Haplotype relationships are indicated by lines; mutational steps between haplotypes are represented by the number of lines
Comparison of molecular and morphological identifications
Morphologically derived species identities revealed that no species were correctly identified with 100% accuracy when compared with molecular identifications (Table 3). Morphological identifications had the highest rate of accuracy for the most abundant species in the area, An. barbirostris and An. vagus, 92.1% and 87.6%, respectively, when compared to molecular analysis (Table 3). For the remaining four distinct sequences molecularly identified to species, the percentage of correctly identified morphological specimens ranged from 0% to 83% (Table 3). Sequencing demonstrated the presence of 18 distinct sequence groups while morphology indicated the presence of 19 species. Specimens morphologically identified as Aedes albopictus, An. barbumbrosus, An. hyrcanus, An. indefinitus, An. parangensis, An. pseudobarbirostris, An. schueffneri, An. sulawesi, An. umbrosus and Culex spp., did not have sequences that reflected these identifications (Table 3). Of these 10 species, 6 have ITS2 and/or cox1 sequences in the databases indicating that these morphological results were probably misidentifications. Morphological identification did not identify any An. karwari, An. peditaeniatus or An. sinensis specimens, all identified molecularly. Members of sibling species cannot be differentiated morphologically; so, the specimens morphologically identified as An. barbirostris were considered correct since molecular identification indicating these as being in the Barbirostris Complex.
Table 3.
Molecular identification of species employing both ITS2 and cox1 comparisons
| Molecular species ID | Morphology species ID | No. of sequences identified molecularly | No. of correctly identified specimens based on morphology | % of correctly identified specimens based on morphology |
|---|---|---|---|---|
| An. aconitusa | a, b, c, e, r | 28 | 8 | 28.6 |
| An. barbirostrisa | c, d, f, g, j, k, p, q, r, x | 2396 | 2207 | 92.1 |
| An. karwaria | m | 1 | 0 | 0 |
| An. peditaeniatusa | c, d, f, j, k, q, x | 133 | 0 | 0 |
| An. tessellatusa | b, c, d, p | 41 | 34 | 83.0 |
| An. vagusa | c, d, g, j, k, p, r | 331 | 290 | 87.6 |
| Total | – | 2931 | 2539 | 86.6 |
aSpecies previously confirmed as malaria vectors in Indonesia
Morphologically-based species identifications included: aAedes albopictus; bAnopheles aconitus; cAn. barbirostris; dAn. barbumbrosus; eAn. flavirostris; fAn. hyrcanus; gAn. indefinitus; hAn. kochi; iAn. maculatus; jAn. nigerrimus; kAn. parangensis; lAn. pseudobarbirostris; mAn. schueffneri; nAn. subpictus; oAn. sulawesi; pAn. tessellatus; qAn. umbrosus; rAn. vagus; xCulex. Number and percentage of correctly identified morphological specimens are calculated from the number of sequences that were molecularly identified per species
Discussion
For Indonesia to achieve its malaria elimination goal, intervention strategies need to cater to varied and complex transmission dynamics with multiple local mosquito vectors. Understanding temporal vector compositions along with their bionomic traits may allow for better and more targeted intervention strategies as well as understanding important gaps in protection.
This study represents an initial foray into characterization of the Anopheles species in a single village within an area known for its biodiversity [5, 10–13]. Multiple trapping methods were utilized over multiple seasons and years to ensure capture of as many species as possible. Eighteen separate species sequences were identified in this single 1 km2 area over the span of almost 3 years. Molecular identification using both ITS2 and cox1 sequences was used for species identification with the conservative algorithm outlined. The unexpected high number of novel sequences, combined with ITS2 and cox1 sequences from the same specimen matching separate (though closely related species) in the database, resulted in multiple specimens from each sequence group being re-sequenced to eliminate the possibility of contamination or mislabeling.
Of the molecularly identified sequences, only a subset could be identified to species based on present available data and the conservative algorithm for species identification. Of these molecularly identified species, ten are previously confirmed malaria vectors from Indonesia: An. aconitus; An. barbirostris; An. karwari; An. peditaeniatus; An. tessellatus; An. vagus; An. kochi; An. flavirostris; An. nigerrimus; and An. maculatus [12, 13, 49–55]. Although these Barbirostris Complex species have unclear vector status, they are likely vectors since specimens identified morphologically as An. barbirostris have been considered medically important vectors in Sulawesi [12, 18]. Although it was impossible to identify to the species level the other molecularly identified sequences, their likelihood of containing confirmed vectors is plausible. The diversity of malaria vectors in Indonesia with suboptimal morphological identifications highlights the importance of integrating molecular identification into vector studies.
Anopheles vagus has been previously suspected of being a species complex [12]. In Timor-Leste on Timor Island, an island south of Sulawesi, a putative species An. vagus genotype B has been found positive with Plasmodium CS protein [56]. Furthermore, two forms of mitotic karyotypes have been found from An. vagus in Thailand [57], indicating intraspecies. However, further studies were unable to determine if the two forms were sibling species [58]. In this study, ITS2 sequence groups indicated two distinct groups identified as An. vagus (AN4 and AN5). Additionally, the ITS2 phylogenetic tree indicates that there are two genetically distinct An. vagus-like species (Fig. 2). These results corroborate the previous literature elucidating that An. vagus is likely a part of a species complex.
Sequence groups AN6 and AN13, both members of the Barbirostris Complex, demonstrate the complexity present in this group of species. Comparisons to multiple databases, voucher specimens and species identification assays [17] resulted AN6 being identified as a sequence variants of An. barbirostris. These differences in the non-coding ITS2 spacer region in AN6 may be reflective of the Sulawesi island population of An. barbirostris, supported by the high An. barbirostris homology with the cox1 sequence.
Sequence group AN13 had low ITS2 homology (9%) to known members of the Barbirostris Complex. This is not surprising, since previous studies have indicated that the Barbirostris Subgroup ITS2 region is large in size and has internal repeats, characteristics that make species-diagnostic PCR based on ITS2 difficult [17, 23]–also relevant to AN6. Furthermore, the 16 corresponding cox1 sequences identified AN13 as an unknown Anopheles species (92% ID) demonstrating closest similarity to an unknown African species previously reported from the Western Kenyan Highlands and Zambia [24, 26]. The combination of the novel ITS2 spacer region combined with a cox1 sequence closest to an unknown African species points to a novel species in this complex.
Similar results were seen for sequence groups AN10 and AN11 in which ITS2 and cox1 results had differing sequence homology with high percentage identity while similar associated ITS2 and cox1 sequences were present in the database. Comparison to voucher specimens and primers in diagnostic PCRs [34, 36, 37] enabled their species identification (Table 2). The sequence differences observed with those in PCR diagnostics as well as those in the NCBI database highlight how morphological misidentification may perpetuate species level misidentifications even in published databases.
All ITS2 and cox1 sequences with conflicting, high percentage identification are closely related taxonomically and the discrepancies may be due to variation in sequences based on these collections being from both a center of biodiversity as well as being an isolated island population and therefore, having diverged from other populations represented more frequently in the databases. Note that multiple randomly chosen samples from each of these groups were re-sequenced to confirm the results. For example, the majority of the research that focuses on the Funestus Group is concentrated in sub-Saharan Africa [59–61]. Furthermore, the sequences within each of these species may vary with the island of Sulawesi being a biodiversity hotspot, further complicating analyses. This study supports the likelihood that distributions and phylogenetic relationships between species in the Barbirostris Subgroup, Nigerrimus Subgroup, Hyrcanus Group and Maculatus Group, need further clarification and research. Therefore, additional research that implements nuclear and mtDNA sequencing within in Indonesia is necessary to accurately identify species that are malaria vectors.
Six ITS2 and paired cox1 sequences (AN13-AN18) could not be identified to species or species group because homology did not meet the conservative criteria (< 93% identity) when comparing to the databases [32, 62]. This may be due to these species not having the related sequences in the database, further stressing the need for more molecular analysis to be completed in areas with high diversity and unknown species composition. It is possible that the species have diverged sufficiently because Karama is an isolated habitat on a biodiverse island–Sulawesi. Furthermore, to the best of our knowledge, this study represents a first-time sample and molecular identification for any Sulawesi population. These six unidentified sequences may represent divergence from specimens in database, novel and/or unidentified sibling species, subspecies, and/or cryptic species.
This study highlights the importance of cross-referencing morphological identifications with molecular identifications, especially in areas of high vector diversity. Morphological identifications were most accurate, when compared to molecular identifications, for the most abundant species groups in the area, An. barbirostris and An. vagus (92.1% and 87.3%, respectively). However, when examining less common species, a comparison of molecularly and morphologically derived species identities demonstrated the inconsistency of relying solely on morphological identification. Finally, all molecularly identified species were mistaken for multiple species when utilizing morphological identification alone. Misidentifications resulting from morphological identification may have negative downstream effects when determining species’ bionomic traits, associations of vector status, entomological inoculation rates, and impacts on control [31]. The discrepancy between morphological and molecular identification underlies the importance of incorporating molecular tools to help distinguish vector species.
Conclusions
This study highlights the importance of cross-referencing morphological identifications with molecular identifications to determine mosquito species composition. Eleven distinct sequences were identified to species, with an additional seven sequences identified to either subgroup, group, or series. Three sequences could only be identified to the genus level, as the percentage identification was too low to identify them to a series. This is the first study to characterize species composition in Karama, West Sulawesi with molecular identification techniques. Future studies employing sequencing are required to clarify the species in several taxonomic groups, as well as their distributions and vector status. Identifying the primary and secondary malaria vectors in this area is vital for appropriate, targeted malaria control interventions and accurate monitoring of their effectiveness. Finally, this study design and analysis represents a dataset and methodologies that can be applied anywhere to enable Indonesia to move forward with their goal of malaria elimination.
Acknowledgements
This project has been funded in whole with funds from the Eck Center for Global Health, Notre Dame, IN, USA. The authors would like to thank the community of Karama. Thanks to Julie Niedbalski and Katie Cybulski for help and support at the University of Notre Dame.
Abbreviations
- WHO
World Health Organization
- PCR
Polymerase chain reaction
- HLC
Human landing catch
Authors’ contributions
IW, RS, PBA and NFL designed the study. IW, RS, HH, AA, MY, NDN, PBA and NFL implemented the study implementation and collected the data. JRD, SS, ALH, TAB, VM, HX, XY, ET, SL and NFL performed sample and study analysis and interpretations. JRD, RNB and NFL drafted and revised the manuscript. All authors read and approved the final manuscript.
Funding
This project has been funded in whole or in part with funds from the Eck Center for Global Health, Notre Dame, IN, USA. The funding body had no role in the design of the study, collection, analysis, and interpretation of data or in writing the manuscript.
Availability of data and materials
Data supporting the conclusions of this article are included within the article. Representative sequences were deposited in the GenBank database under the accession numbers MT740899-MT740916 (ITS2) and MT753033-MT753050 (cox1). The raw datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
The need for ethics approval for non-human subjects (which was determined to include performing human landing collections) was waived by the University of Notre Dame. All other Ethical approvals were received by Pusat Teknologi Intervensi Kesehatan Masyarakat, Badan Litbangkes Kemenkes (Center for Public Health Intervention Technology, Ministry of Health), Indonesia and Universitas Hasanuddin to perform HLCs under the supervision of IW.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jenna R. Davidson, Email: jdavids2@nd.edu
Isra Wahid, Email: israwahid@gmail.com.
Rusdiyah Sudirman, Email: dyah.chayank@gmail.com.
Scott T. Small, Email: ssmall2@nd.edu
Allison L. Hendershot, Email: Allison.L.Hendershot.1@nd.edu
Robert N. Baskin, Email: rnbaskin@outlook.com
Timothy A. Burton, Email: Timothy.A.Burton.39@nd.edu
Victoria Makuru, Email: Victoria.Makuru.1@nd.edu.
Honglin Xiao, Email: Honglin.Xiao.15@nd.edu.
Xiaoyu Yu, Email: Xiaoyu.Yu.68@nd.edu.
Emma V. Troth, Email: emma.troth@uga.edu
Daniel Olivieri, Email: daniel_olivieri@brown.edu.
Stephanny Lizarraga, Email: Stephanny.lizarraga95@gmail.com.
Hajar Hasan, Email: hajarfkm@gmail.com.
Andi Arfah, Email: andiarfah@gmail.com.
Muhammad Yusuf, Email: yusufkesling@gmail.com.
Nirwana Nur, Email: nana.antik@gmail.com.
Din Syafruddin, Email: din@eijkman.go.id.
Puji Asih, Email: puji@eijkman.go.id.
Neil F. Lobo, Email: nlobo@nd.edu
References
- 1.WHO. Eliminating malaria. Geneva: World Health Organization; 2016.
- 2.WHO. World malaria report 2017. Geneva: World Health Organization; 2017.
- 3.WHO. Indonesia. Malaria. Geneva: World Health Organization; 2018. http://www.searo.who.int/indonesia/topics/malaria/en/. Accessed 31 Jul 2018.
- 4.Elyazar IRF, Gething PW, Patil AP, Rogayah H, Kusriastuti R, Wismarini DM, et al. Plasmodium falciparum malaria endemicity in Indonesia in 2010. PLoS One. 2011;6:e21315. doi: 10.1371/journal.pone.0021315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elyazar IRF, Hay SI, Baird JK. Malaria distribution, prevalence, drug resistance and control in Indonesia. Adv Parasitol. 2011;74:41–175. doi: 10.1016/B978-0-12-385897-9.00002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Killeen GF. Characterizing, controlling and eliminating residual malaria transmission. Malar J. 2014;13:330. doi: 10.1186/1475-2875-13-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enayati A, Hemingway J. Malaria management: past, present, and future. Annu Rev Entomol. 2010;55:569–591. doi: 10.1146/annurev-ento-112408-085423. [DOI] [PubMed] [Google Scholar]
- 8.Bugoro H, Cooper RD, Butafa C, Iroofa C, Mackenzie DO, Chen CC, et al. Bionomics of the malaria vector Anopheles farauti in Temotu Province, Solomon Islands: issues for malaria elimination. Malar J. 2011;10:133. doi: 10.1186/1475-2875-10-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harbach R. Genus Anopheles Meigen, 1818. Mosquito Taxonomic Inventory. 2011. http://mosquito-taxonomic-inventory.info/genus-anopheles-meigen-1818. Accessed 19 May 2020.
- 10.Service MW, Townson H . The Anopheles vector. In: Gilles HM, Warrell DA, editors. Essential malariology. 4. London: Arnold; 2002. pp. 59–84. [Google Scholar]
- 11.Hay SI, Sinka ME, Okara RM, Kabaria CW, Mbithi PM, Tago CC, et al. Developing global maps of the dominant Anopheles vectors of human malaria. PLoS Med. 2010;7:e1000209. doi: 10.1371/journal.pmed.1000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elyazar IRF, Sinka ME, Gething PW, Tarmidzi SN, Surya A, Kusriastuti R, et al. The distribution and bionomics of Anopheles malaria vector mosquitoes in Indonesia. Adv Parasitol. 2013;83:173–266. doi: 10.1016/B978-0-12-407705-8.00003-3. [DOI] [PubMed] [Google Scholar]
- 13.Sugiarto, Kesumawati Hadi U, Soviana S, Hakim L. Confirmation of Anopheles peditaeniatus and Anopheles sundaicus as malaria vectors (Diptera: Culicidae) in Sungai Nyamuk Village, Sebatik Island North Kalimantan, Indonesia using an enzyme-linked immunosorbent assay. J Med Entomol. 2016;53:1422–1424. doi: 10.1093/jme/tjw100. [DOI] [PubMed] [Google Scholar]
- 14.Sodhi NS, Koh LP, Brook BW, Ng PKL. Southeast Asian biodiversity: an impending disaster. Trends Ecol Evol. 2004;19:654–660. doi: 10.1016/j.tree.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Sinka ME, Bangs MJ, Manguin S, Chareonviriyaphap T, Patil AP, Temperley WH, et al. The dominant Anopheles vectors of human malaria in the Asia–Pacific region: occurrence data, distribution maps and bionomic précis. Parasit Vectors. 2011;4:89. doi: 10.1186/1756-3305-4-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Townson H, Dyer N, Mcalister E, Satoto TBT, Bangs MJ, Harbach RE. Systematics of Anopheles barbirostris van der Wulp and a sibling species of the barbirostris complex (Diptera: Culicidae) in eastern Java Indonesia. Syst Entomol. 2013;38:180–191. [Google Scholar]
- 17.Brosseau L, Udom C, Sukkanon C, Chareonviriyaphap T, Bangs MJ, Saeung A, Manguin S. A multiplex PCR assay for the identification of five species of the Anopheles barbirostris complex in Thailand. Parasit Vectors. 2019;12:223. doi: 10.1186/s13071-019-3494-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paredes-Esquivel C, Donnelly MJ, Harbach RE, Townson H. A molecular phylogeny of mosquitoes in the Anopheles barbirostris subgroup reveals cryptic species: implications for identification of disease vectors. Mol Phylogenet Evol. 2009;50:141–151. doi: 10.1016/j.ympev.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 19.Satoto TTB. Cryptic species within Anopheles barbirostris Van der Wulp, 1884: inferred from nuclear and mitochondrial gene sequence variation. PhD thesis, University of Liverpool, UK; 2001. http://europepmc.org/abstract/eth/250419. Accessed 31 Jul 2018.
- 20.Saeung A, Otsuka Y, Baimai V, Somboon P, Pitasawat B, Tuetun B, et al. Cytogenetic and molecular evidence for two species in the Anopheles barbirostris complex (Diptera: Culicidae) in Thailand. Parasitol Res. 2007;101:1337–1344. doi: 10.1007/s00436-007-0645-1. [DOI] [PubMed] [Google Scholar]
- 21.Saeung A, Baimai V, Otsuka Y, Rattanarithikul R, Somboon P, Junkum A, et al. Molecular and cytogenetic evidence of three sibling species of the Anopheles barbirostris Form A (Diptera: Culicidae) in Thailand. Parasitol Res. 2008;102:499–507. doi: 10.1007/s00436-007-0788-0. [DOI] [PubMed] [Google Scholar]
- 22.Songsawatkiat S, Baimai V, Saeung A, Thongsahuan S, Otsuka Y, Srisuka W, et al. Cytogenetic, hybridization and molecular evidence of four cytological forms of Anopheles nigerrimus (hyrcanus group) in Thailand and Cambodia. J Vector Ecol. 2013;38:266–276. doi: 10.1111/j.1948-7134.2013.12040.x. [DOI] [PubMed] [Google Scholar]
- 23.Paredes-Esquivel C, Harbach RE, Townson H. Molecular taxonomy of members of the Anopheles hyrcanus group from Thailand and Indonesia. Med Vet Entomol. 2011;25:348–352. doi: 10.1111/j.1365-2915.2010.00937.x. [DOI] [PubMed] [Google Scholar]
- 24.St Laurent B, Cooke M, Krishnankutty SM, Asih P, Mueller JD, Kahindi S, et al. Molecular characterization reveals diverse and unknown malaria vectors in the Western Kenyan Highlands. Am J Trop Med Hyg. 2016;94:327–335. doi: 10.4269/ajtmh.15-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.St Laurent B, Supratman S, Asih PBS, Bretz D, Mueller J, Miller HC, et al. Behaviour and molecular identification of Anopheles malaria vectors in Jayapura district, Papua Province Indonesia. Malar J. 2016;15:192. doi: 10.1186/s12936-016-1234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lobo NF, Laurent BS, Sikaala CH, Hamainza B, Chanda J, Chinula D, et al. Unexpected diversity of Anopheles species in eastern Zambia: implications for evaluating vector behavior and interventions using molecular tools. Sci Rep. 2015;5:17952. doi: 10.1038/srep17952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erlank E, Koekemoer LL, Coetzee M. The importance of morphological identification of African anopheline mosquitoes (Diptera: Culicidae) for malaria control programmes. Malar J. 2018;17:43. doi: 10.1186/s12936-018-2189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Connor C, Soepanta A. Illustrated key to female anophelines of Indonesia. Indonesia: Directorate of Communicable Disease, MoH and US Naval Medical Research, Jakarta; 1989. [Google Scholar]
- 29.Gimnig JE, Walker ED, Otieno P, Kosgei J, Olang G, Ombok M, et al. Incidence of malaria among mosquito collectors conducting human landing catches in western Kenya. Am J Trop Med Hyg. 2013;88:301–308. doi: 10.4269/ajtmh.2012.12-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burkot TR, Russell TL, Reimer LJ, Bugoro H, Beebe NW, Cooper RD, et al. Barrier screens: a method to sample blood-fed and host-seeking exophilic mosquitoes. Malar J. 2013;12:49. doi: 10.1186/1475-2875-12-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stevenson J, St Laurent B, Lobo NF, Cooke MK, Kahindi SC, Oriango RM, et al. Novel vectors of malaria parasites in the western highlands of Kenya. Emerging Infect Dis. 2012;18:1547–1549. doi: 10.3201/eid1809.120283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ratnashingham S, Hebert PDN. bold: the Barcode of Life Data System (http://www.barcodinglife.org). Mol Ecol Notes. 2007;7:355–64. [DOI] [PMC free article] [PubMed]
- 33.Garros C, Koekemoer LL, Coetzee M, Coosemans M, Manguin S. A single multiplex assay to identify major malaria vectors within the African Anopheles funestus and the Oriental An. minimus groups. Am J Trop Med Hyg. 2004;70:583–590. [PubMed] [Google Scholar]
- 34.Hempolchom C, Otsuka Y, Baimai V, Thongsahuan S, Saeung A, Taai K, et al. Development of a multiplex PCR assay for the identification of eight species members of the Thai Hyrcanus Group (Diptera: Culicidae) Appl Entomol Zool. 2013;48:469–476. [Google Scholar]
- 35.Dusfour I, Blondeau J, Harbach RE, et al. Polymerase chain reaction identification of three members of the Anopheles sundaicus (Diptera: Culicidae) complex, malaria vectors in Southeast Asia. J Med Entomol. 2007;44:723–731. doi: 10.1603/0022-2585(2007)44[723:pcriot]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 36.Ali RSM, Wahid I, Saingamsook J, et al. Molecular identification of mosquitoes of the Anopheles maculatus group of subgenus Cellia (Diptera: Culicidae) in the Indonesian Archipelago. Acta Trop. 2019;199:105124. doi: 10.1016/j.actatropica.2019.105124. [DOI] [PubMed] [Google Scholar]
- 37.Walton C, Somboon P, O’Loughlin SM, et al. Genetic diversity and molecular identification of mosquito species in the Anopheles maculatus group using the ITS2 region of rDNA. Infect Genet Evol. 2007;7:93–102. doi: 10.1016/j.meegid.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST + : architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leigh JW, Bryant D. popart: full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6:1110–1116. [Google Scholar]
- 41.Puillandre N, Lambert A, Brouillet S, Achaz G. ABGD, automatic barcode gap discovery for primary species delimitation. Mol Ecol. 2012;21:1864–1877. doi: 10.1111/j.1365-294X.2011.05239.x. [DOI] [PubMed] [Google Scholar]
- 42.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 43.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 46.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 47.Chevenet F, Brun C, Bañuls A-L, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinform. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garjito TA, Widiastuti U, Mujiyono M, Prihatin MT, Widiarti W, Setyaningsih R, et al. Genetic homogeneity of Anopheles maculatus in Indonesia and origin of a novel species present in Central Java. Parasit Vectors. 2019;12:351. doi: 10.1186/s13071-019-3598-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bangs M, Rusmiarto S. Malaria vector incrimination in Indonesia using CSP-ELISA from 1986 to 2007. Unpublished report. Jakarta, Indonesia: U.S. Naval Medical Research Unit No. 2; 2007.
- 50.Van Hell J. The anopheline fauna and malaria vectors in South Celebes. Med Geogr Trop. 1952;4:45–46. [Google Scholar]
- 51.Wigati R, Mardiana, Arianti Y, Mujiono. The incrimination of An. vagus Donitz, 1902 (Diptera: Culicidae) as a malaria vector in Kecamatan Kokap, Kulonprogo Regency. Science Kes. 2006;19:503–16.
- 52.Van Thiel PH, Metselaar D. A pilot project of residual-insecticide spraying to control malaria transmitted by the Anopheles punctulatus group in Netherlands New Guinea. Bull World Health Organ. 1954;11:521–524. [PMC free article] [PubMed] [Google Scholar]
- 53.Boewono D. Verification of malaria vectors in Teluk Dalam Subdistrict, Nias Island. Maj Parasitol Indon. 1997;10:23–32. [Google Scholar]
- 54.Marwoto H, Richie T, Atmosoedjono S, Sekartuti, Tumewu M. Local malaria transmission in Kodya Manado. Bull Penelitian Kes. 1996;24:60–8.
- 55.Overbeek J. Malaria research in the Balitang colonization (Palembang Residence) Geneesk Tijdschr Nederl Indie. 1940;80:2166–2177. [Google Scholar]
- 56.Cooper RD, Edstein MD, Frances SP, Beebe NW. Malaria vectors of Timor-Leste. Malar J. 2010;9:40. doi: 10.1186/1475-2875-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baimai V, Kijchalao U, Rattanarithikul R. Metaphase karyotypes of Anopheles of Thailand and Southeast. J Am Mosq Control Assoc. 1996;12:669–675. [PubMed] [Google Scholar]
- 58.Choochote W, Jitpakdi A, Sukontason K, Chaithong U, Wongkamchai S, Pitasawat B, et al. Intraspecific hybridization of two karyotypic forms of Anopheles vagus (Diptera: Culicidae) and the related egg surface topography. SE Asian J Trop Med. 2002;33:29–35. [PubMed] [Google Scholar]
- 59.Vezenegho SB, Chiphwanya J, Hunt RH, Coetzee M, Bass C, Koekemoer LL. Characterization of the Anopheles funestus group, including Anopheles funestus-like, from northern Malawi. Trans R Soc Trop Med Hyg. 2013;107:753–762. doi: 10.1093/trstmh/trt089. [DOI] [PubMed] [Google Scholar]
- 60.Coetzee M, Fontenille D. Advances in the study of Anopheles funestus, a major vector of malaria in Africa. Insect Biochem Molec. 2004;34:599–605. doi: 10.1016/j.ibmb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 61.Derua YA, Alifrangis M, Magesa SM, Kisinza WN, Simonsen PE. Sibling species of the Anopheles funestus group, and their infection with malaria and lymphatic filarial parasites, in archived and newly collected specimens from northeastern Tanzania. Malar J. 2015;14:104. doi: 10.1186/s12936-015-0616-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2014;42:D32–D37. doi: 10.1093/nar/gkt1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data supporting the conclusions of this article are included within the article. Representative sequences were deposited in the GenBank database under the accession numbers MT740899-MT740916 (ITS2) and MT753033-MT753050 (cox1). The raw datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.