Abstract
PURPOSE
Currently available human papillomavirus (HPV) detection devices are expensive, requiring a continuous power supply, high-priced reagents, skilled laboratory personnel, and infrastructure. These make it difficult to implement primary HPV screening in high-risk (HR) populations, particularly in low-income settings such as in India. The objective of our study was to evaluate the diagnostic performance of a point-of-care, portable, battery-operated device called Truenat, which detects 4 HR HPV genotypes (16, 18, 31, and 45), as a potentially cost-effective alternative to conventional HPV diagnostic tests.
PATIENTS AND METHODS
This was a single-site, blinded, cross-sectional study that evaluated the performance of the Trunat HPV-HR using cervical samples collected from nonpregnant women > 30 years old via consecutive sampling. The comparison was conducted against the Hybrid Capture 2 (HC2) method. All the positive samples were validated by 14 Real-TM Quant Kit.
RESULTS
Of 615 cervical samples, the HR-HPV DNA test was positive in 78 women (12.7%) by HC2 and in 49 (8%) by Truenat. With the consideration of limited genotype inclusivity, the sensitivity and specificity of Truenat HPV-HR were 97.7% and 98.9%, respectively.
CONCLUSION
The performance of Truenat HPV-HR test was comparable to that of HC2 in the 4 HPV genotypes and would be appropriate to consider for use in primary HR cervical cancer screening and particularly in low-income settings.
INTRODUCTION
The worldwide incidence and mortality (age-standardized ratio per 100,000 persons) of cervical cancer are 13.1 and 6.9, respectively.1 India accounts for 19.3% of total cervical cancer deaths in the world, with an incidence of 16.5% (n = 96,922 new diagnoses) and mortality rate of 16.2% (n = 60,078 deaths).1 The strong association of high-risk human papillomavirus (HR-HPV) infection with cervical cancer is well established.2,3 Approximately 60 HPV types are known to infect the human genital tract, including the uterine cervix. These are further categorized into HR-HPV and low-risk HPV types. HR-HPV types 16 and 18 are the most common strains, accounting for 70% to 80% of the total subtypes.4-6 HPV prevalence and genotype distribution are not well documented in the Indian subcontinent, and available scattered studies show a wide variation in the prevalence of HPV positivity ranging from 6% to 38% in the general population from different geographic regions.7-9 This has led to the implementation of primary cervical cancer screening by HPV DNA testing, which is more sensitive than cytology for the detection of high-grade cervical intraepithelial neoplasia.10
CONTEXT
Key Objective
To determine if the diagnostic performance of Truenat, a point-of-care, portable, battery-operated HPV DNA testing device that detects 4 high-risk HPV genotypes (16, 18, 31, and 45) is comparable to the reference standard test, Hybrid Capture 2 (HC2) for cervical HPV DNA testing.
Knowledge Generated
The results of Truenat HPV DNA test are comparable to HC2 for 4 high-risk genotypes, with sensitivity and specificity of 97.7% and 98.9%, respectively.
Relevance
This point-of-care device with testing facility for 4 high-risk HPV genotypes would be a suitable option for primary cervical cancer screening in low-resource settings and population-based cancer screening.
Primary HPV testing has been evaluated extensively as a cervical cancer screening tool and results from randomized controlled trials have shown testing increases protection significantly against the development of invasive cervical cancer, compared with cytology-based screening.11 On the basis of these results, WHO has recommended incorporating HPV testing wherever resources are available.12,13 In 2009, India adopted the visual inspection using acetic acid test as a method for initial cervical cancer risk assessment in population-based cancer screening programs8,14-16 because of the high cost involved in primary HPV-based diagnostic tests, which require skilled manpower and elaborate infrastructure. Also, the turnaround time for results could take a few days to weeks, likely leading to losses to follow-up.
Currently available HPV diagnostic devices are expensive, requiring a continuous power supply, highly-priced reagents, skilled laboratory personnel, and infrastructure. These factors make it difficult to implement cost-effective primary diagnosis in population health screening programs.17,18 They also have led to the exploration of affordable point-of-care (POC) or near-care devices to detect HPV infection to increase access to diagnosis, thereby reducing the number of visits and decreasing potential loss to follow-up.19,20
The Truelab (Molbio Diagnostics, Goa, India) device detects 4 HR-HPV types: 16, 18, 31, and 45, and is commercially available for diagnosis of > 15 diseases, including tuberculosis, hepatitis B, dengue, H1N1 influenza, chikungunya, and malaria.
The aim of this study was to validate the diagnostic performance of the Truenat HPV-HR assay performed on the Truelab Uno Dx Real Time Micro PCR Analyzer to detect HPV DNA against 2 reference standard tests, Hybrid Capture 2 (HC2; Digene, Germany) and 14 Real-TM Quant Kit (Sacace Biotechnologies, Como, Italy) in cervical samples collected from women attending a cancer-screening clinic.
PATIENTS AND METHODS
Study Site and Population
The study was conducted at the cancer-screening clinic of the National Institute of Cancer Prevention and Research, Noida, India. Consecutive sampling was performed. The inclusion criteria included sexually active or married women aged ≥ 30 years; the exclusion criteria included pregnant women, women reporting no previous sexual activity, and women who had had a hysterectomy.
Two endocervical samples were obtained from each woman; the same health care provider used 2 sterile brushes. The cervical sample collected by the first brush was stored in the viral lysis medium provided in the pretreatment tube and was used for DNA isolation on the Trueprep AUTO sample preparation device (Molbio Diagnostics). The second cervical brush was stored in the specimen transport medium provided by the manufacturer and was used for HR-HPV DNA assay by HC2. The sequence of the collection of samples was alternately taken for HC2 and Truenat HPV testing and documented in the pro forma document. HC2 and Truenat HPV testing were performed in different laboratories and the results were blinded to each laboratory.
Study Design
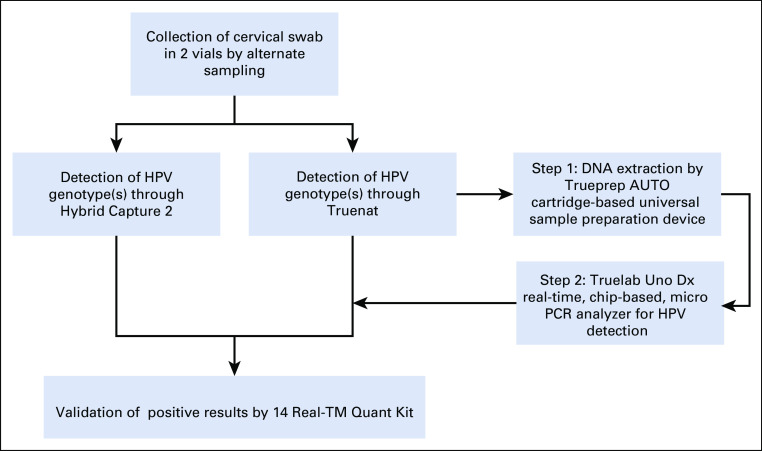
This was a single-site, blinded, cross-sectional study for HPV detection by Truenat HPV-HR in comparison with the reference standard testing methods, HC2 and 14 Real-TM Quant Kit (Fig 1).
FIG 1.
Study design for evaluation of Truenat performance. HPV, human papillomavirus; PCR, polymerase chain reaction.
Truelab Workstation
The Truelab workstation comprises a sample processing device (Trueprep AUTO) and a real-time quantitative micro polymerase chain reaction (PCR) analyzer (Truelab Uno Dx), along with accessories such as a cartridge and microtip holding stand. Both the devices are portable, powered by a rechargeable battery, developed for POC application, and can run continuously for ≥ 8 hours on a single charge. Trueprep AUTO is fully automated and uses a disposable fluidic cartridge to extract and enrich total DNA from the specimen, typically within 20 minutes. It can perform 16 sample extractions with 1 recharge and requires minimal hands-on time. Real-time PCR is performed on Truenat HPV-HR, which is a ready-to-use microchip that carries test- and batch-related information. The microchip is run on the Truelab Uno Dx analyzer, which has 3-wavelength fluorescence detection. The PCR takes approximately 40 minutes. One channel is used to detect amplification of HPV 16 and 31, a second channel for HPV 18 and 45, and a third channel for an internal positive control (IPC). The IPC is coextracted with the sample from the Trueprep Autocartridge and serves to validate the run conditions. The Truelab device can transfer data to a centralized server for remote monitoring and surveillance.
Ethical Approval
The authors followed the standards of the World Medical Association’s Declaration of Helsinki for ethical approval, which was obtained from the Institutional Ethics Committee of the Indian Council of Medical Research, National Institute of Cancer Prevention and Research, Noida, India, before the initiation of the study. Written informed consent was obtained from all participants prior to enrollment.
Sample Size
To estimate an assumed sensitivity of 90% sensitivity with a relative error margin of 10% and an absolute error margin of 9 (ie, 10% of 90% = 9), we needed 43 positive cases of HPV. Assuming the prevalence of HR-HPV infection in the study population as 8%, to get 43 positive cases, we required ≥ 538 women in the study. Assuming 10% wastage of samples during processing, storing, and transporting, the sample size was adjusted to a final count of 598.
Sample Processing on Trueprep AUTO
A 1-mL cervical sample pretreated with the lysis buffer was added to the sample chamber of the cartridge, which was then placed in the cartridge holder of the device. Sample processing was initiated by pressing the start button on the device, through an automatic preprogrammed process wherein nucleic acids released by chemical and thermal lysis of cells bind to the proprietary matrix. In subsequent steps, the matrix was again washed with buffers to remove the PCR inhibitors, and bound nucleic acids were finally eluted from the matrix using the elution buffer. On completion of the process, which typically took < 20 minutes, the cartridge was automatically ejected and the elute containing purified DNA was collected from the elute chamber for analysis.
HPV Detection on Truelab HPV-HR
Truenat HPV-HR detects 4 HR subtypes of HPV. It can differentiate the sample as HPV 16 or 31 and HPV 18 or 45. The test was initiated by selecting the profile name, entering sample details, and loading the master mix. Elute (6 µL) collected from the cartridge in the previous step was added to a microtube containing lyophilized PCR master mix; subsequently, the reconstituted solution was transferred to the chip well. During thermal cycling, fluorescent signals from 3 wavelengths were captured by the optoelectronic system, and data were visualized as a graph on the graph-user interface of the device. Results were auto-interpreted by the system and visualized as a simple readout form (Fig 2). Results were displayed as “not detected” if only the IPC showed amplification and both the 16/31 and 18/45 channels did not show amplification. A positive result was indicated by amplification in either fluorescent channel for 16/31 or 18/45 or both (indicating mixed infection). When there was no amplification in target channels and an absence of or shift of IPC cycle threshold beyond a preset, the run was considered as invalid. The results can be printed using the Truelab Micro PCR printer or transferred to the laboratory computer or any remote computer via Wi-Fi or Bluetooth. A minimum of 20,000 test results can be stored on the analyzer for future recall and reference.
FIG 2.
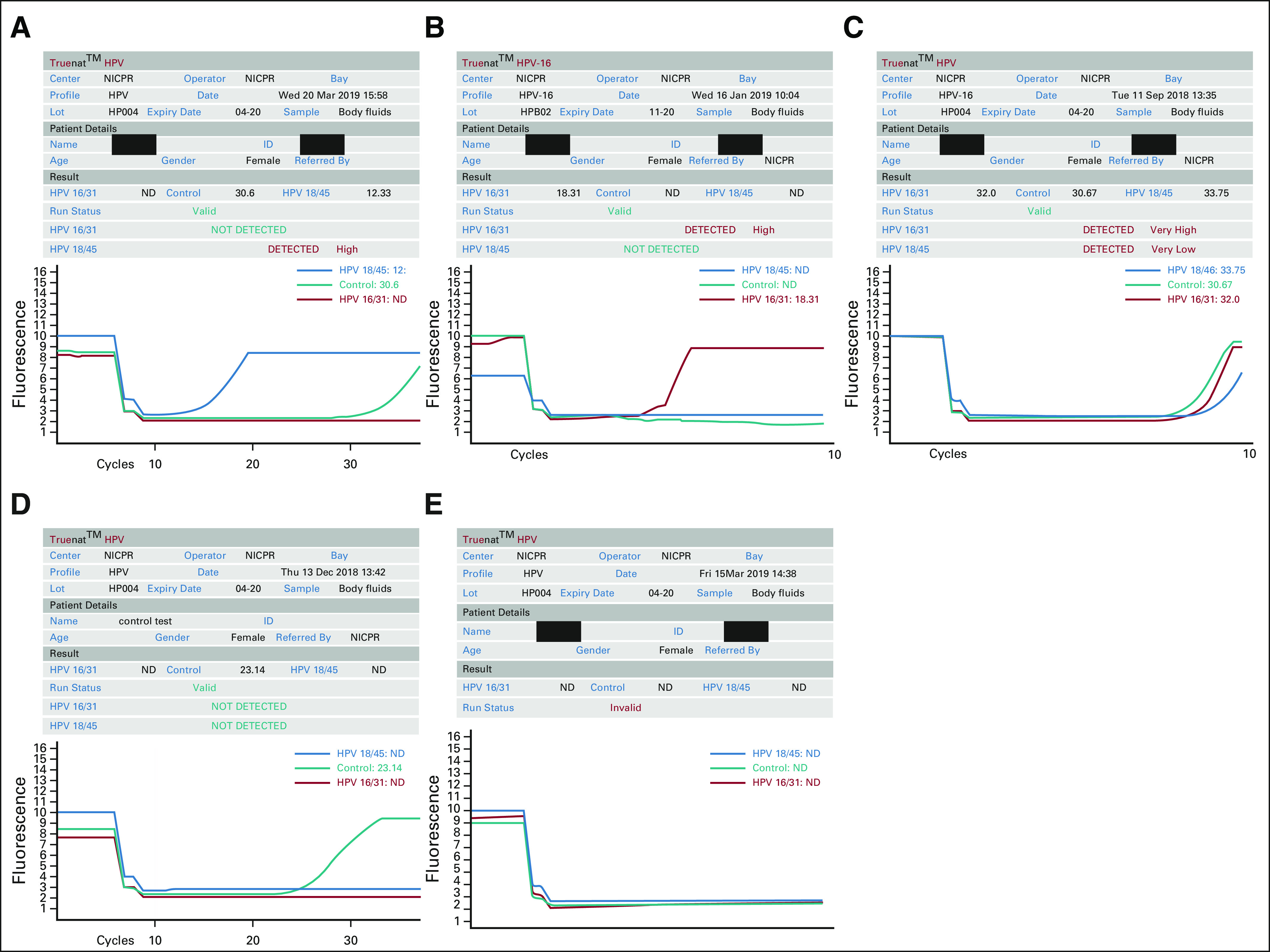
Illustration of human papillomavirus (HPV) genotype results by Truelab Uno Dx Real-Time Micro PCR Analyzer. (A) Graph showing positive amplification for HPV 18 and 45 genotypes. (B) Graph showing positive amplification for HPV 16 and 31genotypes. (C) Graph showing positive amplification for both HPV 16/31 and 18/45 genotypes. (D) Graph showing internal positive control amplification only. Results are displayed as “Not Detected.” (E) Graph showing no amplification. Results are displayed as “Invalid.”
In most cases, the samples were processed on the day of the collection. On days when the sample numbers were high, however, samples were stored in a refrigerator at 2°C to 8°C, per the package instructions. In such cases, processing was completed within a week.
Analysis Using HC2 and 14 Real-TM Quant Kit
HC2 was performed according to manufacturer’s instructions on the Digene Hybrid Capture system (model No. DML-2000), which detects 13 HR-HPV types: 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68. The collected cervical samples were processed and the relative light units > 1 were reported as HR-HPV DNA positive.
All the samples positive by either HC2 or Truenat were analyzed using 14 Real-TM Quant Kit according to the protocol mentioned by the manufacturer. This in vitro real-time amplification test is used for quantitative detection and genotyping of 14 HPV strains: 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68. The kit contains 4 PCR mixes, Hot Start DNA Polymerase, PCR-buffer-FRT, 4 positive controls specific to 4 different PCR mixes, and a negative control. Each PCR mix detects different HPV strains. PCR mix 1 detects HPV genotypes 16, 18, and 31 (also labeled with an internal control); PCR mix 2 detects genotypes 39, 45, and 59 (also labeled with an internal control); PCR mix 3 detects genotypes 33, 35, 56, and 68; and PCR mix 4 detects genotypes 51, 52, 58, and 66.
For each sample, 4 tubes each for PCR mix 1, 2, 3, and 4 were prepared along with 4 tubes for a positive control and 4 tubes for a negative control. The reaction mix was prepared with 10 µL of every PCR mix and 5 µL of PCR-buffer-FRT with Hot Start DNA Polymerase. An aliquot of 15 µL reaction mix was added to each well after adding 10 µL of the extracted DNA sample to an appropriate tube.
Statistical Analysis
The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and 95% CIs were calculated using STATA, version 15.1 (StataCorp, College Station, TX).
RESULTS
A total of 647 cervical samples were collected in duplicate by alternate sampling for both HC2 and Truenat HPV-HR. Figure 3 shows the details of the steps involved in the testing along with the number of samples processed at each step to arrive at the final analyzable data and also provides the details of samples excluded because of errors. On the Truelab workstation, 3.7% of samples showed errors (n = 18 errors during sample preparation; n = 6 tests invalidated during PCR). The cartridge-based device gave the following errors: clogged cartridge (n = 2), where no elute was collected; valve issue (n = 3), in which no elute was collected; and no elute collected at the end of the procedure (n = 13), despite no apparent error. However, this was attributed to user error, where the user did not retrieve the cartridge (with elute) after the alert beep. In such cases, the vacuum created in the chamber sucked the elute back into its microchannels, resulting in loss of the elute. In the Uno Dx device, the cycle gave invalid results (n = 6) due to 2 types of errors: error 1, which was categorized as “incorrect thermal cycle”; and error 2, which was classified as “invalid maneuver” by the device manufacturers. HC2 had 8 lapses during the sample collection. All the aforementioned samples were categorized as errors (n = 32) and excluded from the study, resulting in total analyzable data from 615 samples.
FIG 3.
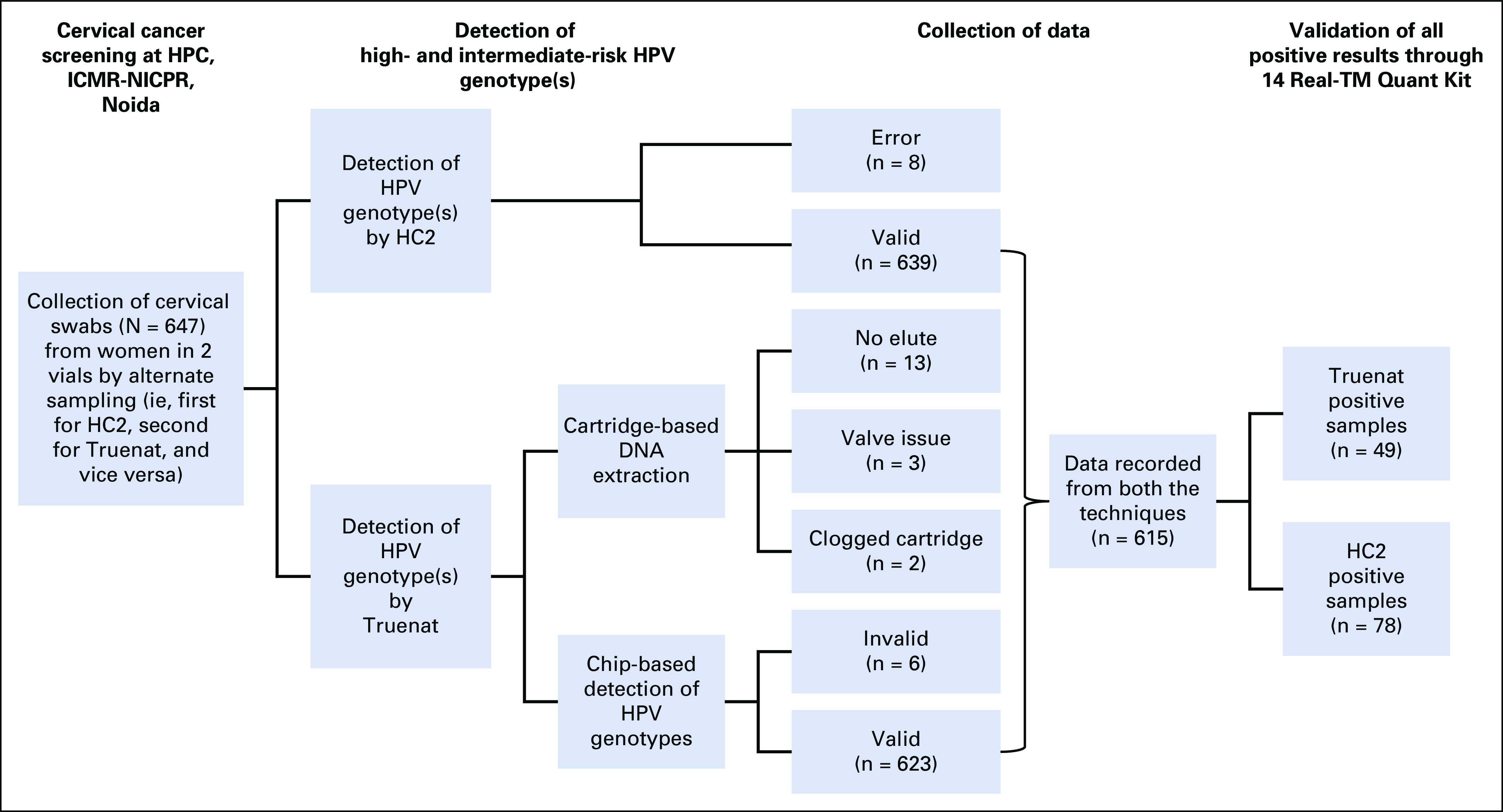
Flow diagram depicting the details of steps involved in the study. HC2, Hybrid Capture 2; HPC, health promotion clinic; HPV, human papillomavirus; ICMR, Indian Council of Medical Research; NICPR, National Institute of Cancer Prevention and Research.
Of the 615 cervical samples, the HR-HPV DNA test was positive in 78 (12.7%) by HC2 and 49 (8%) by Truenat (Table 1). The specificity of Truenat HPV-HR was 98.32% (n = 528 of 615; 95% CI, 96.8% to 99.2%; Table 2). Overall positivity observed in this study was 12.68% (n = 78 of 615) by HC2. Of 78 positive samples, 40 were detected positive by Truenat HPV-HR. This could be due to the difference in genotype inclusion range in both tests. To identify this, all positive samples were analyzed by the 14 Real-TM Quant Kit, which could identify the genotype present in the sample. Considering only samples with the genotypes included in Truenat HPV-HR, sensitivity was 97.5% (95% CI, 86.8% to 99.9%) with a PPV of 90.7% (95% CI, 77.9% to 97.4%). The sensitivity, specificity, PPV, NPV, and concordance data between HC2 and Truenat are listed in Table 2.
TABLE 1.
Comparison of the positivity rates of Truenat Human Papillomavirus–High Risk Test and HC2
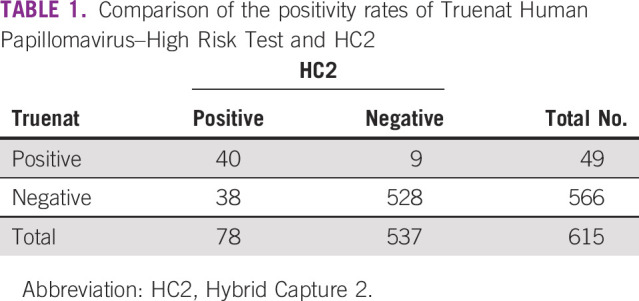
TABLE 2.
Comparison of HC2 and Truenat HPV-HR Test Results
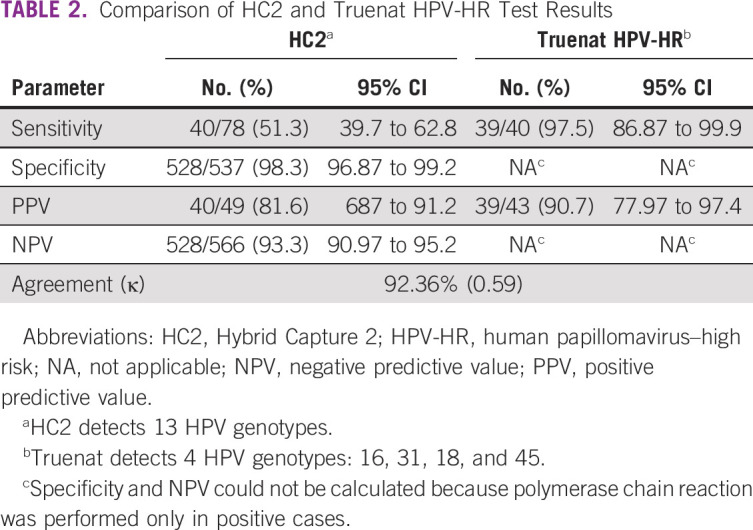
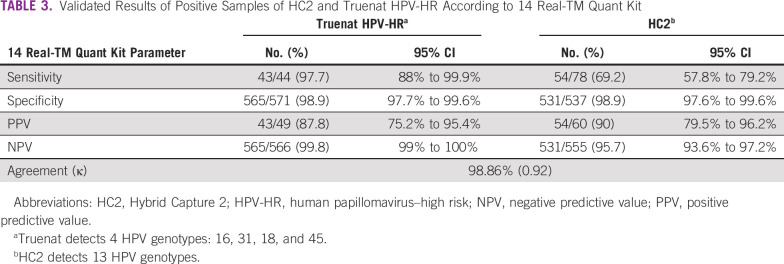
Table 3 lists observed performance parameters after validating all HPV positive samples and including only Truenat HPV-HR assay–claimed genotypes. The sensitivity, specificity, PPV, and NPV of Truenat HPV-HR were 97.7% (95% CI, 88% to 99.9%), 98.9% (95% CI, 97.7% to 99.6%), 87.8% (95% CI, 75.2% to 95.4%), and 99.8% (95% CI, 99% to 100%), respectively, thereby maintaining the consistency of the Truenat HPV-HR results (sensitivity and PPV) when they were compared with HC2 (Table 2).
TABLE 3.
Validated Results of Positive Samples of HC2 and Truenat HPV-HR According to 14 Real-TM Quant Kit
DISCUSSION
In this study, we evaluated the new POC Truenat HPV-HR test against the existing US Food and Drug Administration (FDA)-approved method, HC2. There were differences in genotype inclusivity for both the tests: Truenat HPV-HR was specific for 4 HR-HPV genotypes (ie, 16, 31, 18, and 45) and HC2 detects 13 genotypes. However, due to the availability of HC2 and because this is an FDA-approved reference standard, we selected this method.
With the consideration of genotype inclusivity, Truenat HPV-HR performed well in this study, with observed sensitivity of 97.6% and specificity of 99.4%. Although these 4 strains constitute the majority of the HPV infections found in the population, adding 4 additional prevalent types (ie, 33, 35, 58, and 59) would increase the sensitivity of this method.21
Our analysis showed good sensitivity and specificity, and when combined with results being reported within an hour, indicates this test is suitable for community-level cervical cancer screening in low-resource settings, particularly where patients who leave the clinic without knowing their test results may be lost to any potential follow-up. This device has high primer sensitivity and specificity and requires only 6 μL of the elute. The smart chip has preset data for quantitation of results. This device is resistant to contamination and evaporation at the reaction port. The PCR takes approximately 40 minutes.
However, there are a few limitations of the device used in the current study. The Truelab Uno Dx device used in this study could process 1 sample at a time with a total turn-around time of 1 hour. This has been overcome by the newer-generation devices, Truelab Duo (which can perform independent, random-access PCR runs for 2 samples at a time) and Truelab Quattro (which can run 4 samples at a time). Another limitation from the public health perspective would be that, currently, only 4 HPV types can be identified. Hence, Truenat can be best used as a screening device for these 4 HR-HPV types and not for all HR-HPV types.
We also noticed some procedural limitations at the time of performing the HR- HPV assay. Trueprep AUTO Cartridge–based universal system, although functional by minimal user interference, indicates the end of the cycle by a beep. However, we noticed that the alert beep was not loud enough and the laboratory technician, provider, or other user often missed the alert or was not able to respond in a timely fashion, which resulted in the elute remaining in the collection chamber. When the elute was not retrieved immediately from the collection chamber, it was sucked back into the microchannels of the cartridge by the vacuum created in the chamber, thus leading to loss of the elute and “no elute” in the errors listed. A simple solution to this could be to increase the volume and frequency of the alert beep and to eject the cartridge all the way out of the collection chamber, thus nullifying the effect of the vacuum created in this chamber that sucks back the elute. This would enable the user to carry on with other activities, especially while in a health camp, without having to worry about the loss of elute. Some of the errors listed in the PCR cycle of the Uno DX device were labeled “invalid maneuver,” which could have arisen due to the introduction of an air bubble while loading the chip. This could be eliminated if the either the PCR mix or the microchip well could be colored for easier detection of air bubbles.
More errors occurred with the experimental device (n = 24) than with the reference standard method (n = 8), which could be because the experimental device is still a prototype in the process of being improved on the basis of users’ feedback and suggestions before it becomes commercially available.
With the aforementioned modifications, this POC device could provide an opportunity to incorporate self-sampling into cervical cancer screening algorithms where screening coverage is low because of women’s inability to participate in the facility-based screening programs. The availability of the result within 1 hour gives providers a chance to integrate facility-based HR-HPV testing into same-day “screen-and-treat” cervical screening programs in low-resource setups, particularly if self-collected specimens by this method are proven to be as accurate as clinician-collected specimens for the detection of HR-HPV infection.19
In conclusion, our results showed that with modification to include additional genotypes and other elements, the Trunat HPV-HR test may be appropriate for use in primary cervical cancer screening programs or services when used with the portable, rechargeable battery–powered, sample-processing device (Trueprep AUTO) and a real-time quantitative micro PCR analyzer (Truelab Uno Dx), along with accessories such as a cartridge and microtip holding stand.
ACKNOWLEDGMENT
The authors are thankful for the intramural funding received from the Institute Indian Council of Medical Research, National Institute of Cancer Prevention and Research to carry out this study. We also acknowledge the help and support of social workers Amita Kumar, Chandresh Pragya Verma, and Reena Dwivedi working in the Division of Clinical Oncology, National Institute of Cancer Prevention and Research, Noida, for collecting the samples.
SUPPORT
Supported by intramural funding received from the Indian Council of Medical Research, National Institute of Cancer Prevention and Research (R.H.).
AUTHOR CONTRIBUTIONS
Conception and design: Roopa Hariprasad, Ravi Mehrotra
Administrative support: Shalini Singh, Ravi Mehrotra
Provision of study material or patients: Roopa Hariprasad, Kavitha Dhanasekaran
Collection and assembly of data: Roopa Hariprasad, Sonam Tulsyan, Kavitha Dhanasekaran, Nisha Thakur, Richa Tripathi, Latha Sriram, Shalini Singh, Roshani Babu
Data analysis and interpretation: Roopa Hariprasad, Sonam Tulsyan, Nisha Thakur, Richa Tripathi, Vishnubhatla Sreenivas, Shashi Sharma, Shalini Singh, Ravi Mehrotra
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/go/site/misc/authors.html.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
No potential conflicts of interest were reported.
REFERENCES
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Tulay P, Serakinci N. The route to HPV-associated neoplastic transformation: A review of the literature. Crit Rev Eukaryot Gene Expr. 2016;26:27–39. doi: 10.1615/CritRevEukaryotGeneExpr.v26.i1.40. [DOI] [PubMed] [Google Scholar]
- 3.Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 4.Basu P, Roychowdhury S, Bafna UD, et al. Human papillomavirus genotype distribution in cervical cancer in India: Results from a multi-center study. Asian Pac J Cancer Prev. 2009;10:27–34. [PubMed] [Google Scholar]
- 5.Deodhar K, Gheit T, Vaccarella S, et al. Prevalence of human papillomavirus types in cervical lesions from women in rural Western India. J Med Virol. 2012;84:1054–1060. doi: 10.1002/jmv.23310. [DOI] [PubMed] [Google Scholar]
- 6.Akram Husain RS, Rajakeerthana R, Sreevalsan A, et al. Prevalence of human papilloma virus with risk of cervical cancer among south Indian women: A genotypic study with meta-analysis and molecular dynamics of HPV E6 oncoprotein. Infect Genet Evol. 2018;62:130–140. doi: 10.1016/j.meegid.2018.04.029. [DOI] [PubMed] [Google Scholar]
- 7.Gravitt PE, Paul P, Katki HA, et al. Effectiveness of VIA, Pap, and HPV DNA testing in a cervical cancer screening program in a peri-urban community in Andhra Pradesh, India. PLoS One. 2010;5:e13711. doi: 10.1371/journal.pone.0013711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sankaranarayanan R, Nene BM, Shastri SS, et al. HPV screening for cervical cancer in rural India. N Engl J Med. 2009;360:1385–1394. doi: 10.1056/NEJMoa0808516. [DOI] [PubMed] [Google Scholar]
- 9.Srivastava S, Gupta S, Roy JK. High prevalence of oncogenic HPV-16 in cervical smears of asymptomatic women of eastern Uttar Pradesh, India: A population-based study. J Biosci. 2012;37:63–72. doi: 10.1007/s12038-012-9181-y. [DOI] [PubMed] [Google Scholar]
- 10.Ruffin M. Combined HPV and cytology better than cytology for protection against cervical cancer. Evid Based Med. 2014;19:148. doi: 10.1136/eb-2014-101792. [DOI] [PubMed] [Google Scholar]
- 11.Ronco G, Dillner J, Elfström KM, et al. Efficacy of HPV-based screening for prevention of invasive cervical cancer: Follow-up of four European randomised controlled trials. Lancet. 2014;383:524–532. doi: 10.1016/S0140-6736(13)62218-7. [DOI] [PubMed] [Google Scholar]
- 12.Denny L, de Sanjose S, Mutebi M, et al. Interventions to close the divide for women with breast and cervical cancer between low-income and middle-income countries and high-income countries. Lancet. 2017;389:861–870. doi: 10.1016/S0140-6736(16)31795-0. [DOI] [PubMed] [Google Scholar]
- 13.Sherris J, Wittet S, Kleine A, et al. Evidence-based, alternative cervical cancer screening approaches in low-resource settings. Int Perspect Sex Reprod Health. 2009;35:147–154. doi: 10.1363/ifpp.35.147.09. [DOI] [PubMed] [Google Scholar]
- 14.Ankit J, Prakriti J, Lakshmaiah KC. HPV screening for cervical cancer in rural India. N Engl J Med. 2009;361:304–305, author reply 306. [PubMed] [Google Scholar]
- 15.Cremer ML, Conlisk E, Felix JC. HPV screening for cervical cancer in rural India. N Engl J Med. 2009;361:305. , author reply 306. [PubMed] [Google Scholar]
- 16.Suba EJ, Cibas ES, Raab SS. HPV screening for cervical cancer in rural India. N Engl J Med. 2009;361:304–306, author reply 306. doi: 10.1056/NEJMc090939. [DOI] [PubMed] [Google Scholar]
- 17.Einstein MH, Smith KM, Davis TE, et al. Clinical evaluation of the cartridge-based GeneXpert human papillomavirus assay in women referred for colposcopy. J Clin Microbiol. 2014;52:2089–2095. doi: 10.1128/JCM.00176-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levi AW, Bernstein JI, Hui P, et al. A comparison of the Roche Cobas HPV test with the Hybrid Capture 2 test for the detection of high-risk human papillomavirus genotypes. Arch Pathol Lab Med. 2016;140:153–157. doi: 10.5858/arpa.2015-0027-OA. [DOI] [PubMed] [Google Scholar]
- 19.Toliman P, Badman SG, Gabuzzi J, et al. Field evaluation of Xpert HPV point-of-care test for detection of human papillomavirus infection by use of self-collected vaginal and clinician-collected cervical specimens. J Clin Microbiol. 2016;54:1734–1737. doi: 10.1128/JCM.00529-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly H, Mayaud P, Segondy M, et al. A systematic review and meta-analysis of studies evaluating the performance of point-of-care tests for human papillomavirus screening. Sex Transm Infect. 2017;93(S4):S36–S45. doi: 10.1136/sextrans-2016-053070. [DOI] [PubMed] [Google Scholar]
- 21.Bhatla N, Lal N, Bao YP, et al. A meta-analysis of human papillomavirus type-distribution in women from South Asia: Implications for vaccination. Vaccine. 2008;26:2811–2817. doi: 10.1016/j.vaccine.2008.03.047. [DOI] [PubMed] [Google Scholar]