Abstract
Endometriosis is a common and benign gynecological disorder but exhibits malignant features. However, the underlying pathogenesis and pathophysiology of endometriosis remain unclear. Circular RNAs have been demonstrated to participate in the occurrence and progression of multiple diseases. This study was aimed to explore the roles of circATRNL1 in endometriosis in vitro. Based on the results of reverse transcription-quantitative polymerase chain reaction analysis, we found significant upregulation of circATRNL1 and Yes-associated protein 1 (YAP1), while downregulation of miR-141-3p and miR-200a-3p in ectopic tissues compared to eutopic tissues. The immunohistochemistry and western blot analysis showed differentially expressed epithelial–mesenchymal transition (EMT) markers between EuEM and EcEM tissues. The in vitro assays indicated that overexpression of circATRNL1 could promote the proliferation, migration, and invasion of Ishikawa cells, and induce EMT process, while circATRNL1 silencing showed the opposite effect. The mechanical investigation indicated that circATRNL1 upregulated YAP1 by sponging miR-141-3p and miR-200a-3p. Gain-of-function assays validated the inhibitory function of miR-141-3p and miR-200a-3p in endometriosis. The results of rescue assays confirmed the function of circATRNL1–miR-141-3p/miR-200a-3p–YAP1 axis on Ishikawa cells. Our findings demonstrate that abnormal upregulation of circATRNL1 regulates cell proliferation and motility and promotes EMT process via the miR-141-3p/miR-200a-3p–YAP1 axis in vitro, which could contribute to the progression of endometriosis.
Subject terms: Epithelial-mesenchymal transition, Mechanisms of disease, miRNAs
Introduction
Endometriosis is a common and refractory estrogen-dependent gynecological disorder associated with dysmenorrhea, pelvic pain, and infertility1–4. Although benign, this chronic and multifactorial disease exhibits tumor-like biological behaviors and affects the physical and mental health of affected women5. However, the underlying pathogenesis and pathophysiology of endometriosis remain unclear.
The epithelial–mesenchymal transition (EMT) is a special biological process in which immotile epithelial cells convert into highly motile mesenchymal cells with migratory and invasive properties during embryonic development, chronic inflammation, tissue construction, fibrosis formation, and cancer metastasis6–9. The EMT is characterized by decreased expression of epithelial markers, such as E-cadherin, and increased expression of mesenchymal markers, such as N-cadherin and vimentin10. Many studies have documented the involvement of the EMT in the pathogenesis and development of endometriosis7–9,11. Chen et al.12 first found decreased E-cadherin and increased vimentin expression in ectopic glandular epithelial cells in adenomyosis. Another study showed that E-cadherin-negative cells were frequently observed in endometriotic tissues, whereas N-cadherin, Twist, Slug, and Snail were all upregulated in endometriosis compared with that in the healthy endometrium7. The mesenchymal marker zinc finger E-box-binding homeobox 1 (ZEB1), a transcriptional repressor of E-cadherin, is also upregulated in endometriotic lesions13. Several signal pathways, including the Wnt/β-catenin pathway11, the Notch pathway14, and the transforming growth factor (TGF)-β/Smad pathway15, have been reported to regulate the EMT in endometriosis. Moreover, other factors, including estrogen11,12 and hypoxia-inducible factor-1α16, may act as powerful inducers of the EMT in endometriosis. Changes in EMT marker proteins have been detected in ectopic epithelial cells, resulting in enhanced migration and invasion as a prerequisite for the establishment of endometriotic lesions.
Recent studies have highlighted the regulatory mechanisms through which noncoding RNAs (ncRNAs) participate in the occurrence and progression of multiple diseases17–19. Circular RNAs (circRNAs), as a distinct type of ncRNAs, have a covalently closed continuous loop and display resistance to exonuclease digestion20,21. CircRNAs show highly stable expression in eukaryotic cells, are predominantly localized in the cytoplasm, and are evolutionarily conserved across species, suggestive of their important regulatory roles17. As competing endogenous RNAs (ceRNAs), circRNAs may sequester microRNAs (miRNAs) with their own miRNA response elements and function as miRNA “sponges” to strongly suppress miRNA activity, thereby increasing the levels of miRNA target genes and mediating cellular biological behaviors involved in human diseases17,22. For example, ciRS-7 contains more than 70 selectively conserved miRNA target sites and regulates the initiation and progression of various malignancies in an miR-7-dependent manner23,24. In addition, circ-PVT1 facilitates paclitaxel resistance by upregulating ZEB1 via miR-124-3p in gastric cancer25. Overexpression of circ-0001649 inhibits the proliferation and migration of hepatocellular carcinoma (HCC) in vitro and in vivo by serving as a ceRNA to sponge miR-127-5p, miR-612, and miR-468826. These findings suggest that circRNAs may be potential biomarkers and therapeutic targets in the diagnosis and treatment of multiple diseases. However, our understanding of the relationships between circRNAs and endometriosis remains limited, and translating current circRNA-related research into clinical practice is challenging.
Our previous study showed that circRNAs are aberrantly expressed in the ectopic endometrium (EcEM) compared with that in paired eutopic endometrium (EuEM) samples and demonstrated that hsa-circ-0020093, derived from the ATRNL1 gene locus (thus, designated circATRNL1), was upregulated in ovarian endometriosis27. Accordingly, in this study, we aimed to investigate the roles of circATRNL1 in endometriosis in vitro. Our findings provided important insights into the roles of circATRNL1 in the EMT in endometriosis and may aid in developing suitable therapeutic strategies.
Materials and methods
Ethics
This study was approved by the Ethics Committees of Shengjing Hospital of China Medical University (ethics no. 2018PS504K), and written informed consent was obtained from all patients before surgical procedures and sample collection.
Clinical specimens and cell culture
Snap-frozen cyst walls of ovarian endometriomas and matched EuEM samples from the same patient were collected from 60 women (20–44 years old) with a laparoscopic and histological diagnosis of stage III/IV endometriosis according to the Revised American Society for Reproductive Medicine classification system (rASRM; 1997). All patients had regular menstrual cycles (21–35 days), and none had received any hormone therapy for at least 6 months prior to the operation. All EuEM samples were collected during the proliferative phase of the menstrual cycle, as confirmed by both the date of the last menstrual period and histological diagnosis.
Ishikawa cells (a well-differentiated endometrial adenocarcinoma cell line) and HEK-293T cells were purchased from Huiying Biological Technology Co. Ltd (Shanghai, China) and cultured in modified Eagle’s medium (Gibco, USA) with 10% fetal bovine serum (FBS; Bioind, Israel), 100 μg/mL streptomycin, and 100 IU/mL penicillin (Beyotime, Shanghai, China) in a humidified atmosphere with 5% CO2 at 37 °C.
Immunohistochemistry (IHC)
Immunohistochemical staining was performed on paraffin-embedded tissues. Three-micrometer-thick sections were cut and placed on glass slides. Tissues were deparaffinized in xylene and rehydrated in an ethanol gradient. Antigen retrieve was performed in 10 mM Sodium Citrate Buffer (pH 6.0). Overall, 3% H2O2 was used to block endogenous peroxidase for 15 min. Nonspecific background staining was then blocked by incubation with goat serum (Solarbio, China) for 15 min. The slices were incubated with primary antibodies overnight at 4 °C and horseradish peroxidase (HRP)-conjugated secondary antibodies at 37 °C for 1 h. All primary and secondary antibodies were as follows: rabbit polyclonal anti-E-cadherin (P12830) (Cat. No.WL01482, 1:200; Wanleibio, China), mouse monoclonal anti-N-cadherin (P19022) (1:300; Proteintech, Wuhan, China), rabbit polyclonal anti-vimentin (P08670) (1:200; Proteintech), mouse monoclonal anti-YAP1 (P46937) (1:200; Proteintech), rabbit polyclonal anti-ZEB1 (P37275) (1:200; Proteintech), goat anti-mouse IgG H&L antibody (Product # 31321, 1:500; Thermo Fisher, USA), and goat anti-rabbit IgG H&L antibody (Product # 31460, 1:500; Thermo Fisher, USA). All slides were incubated with DAB (Solarbio, China) and then counterstained with hematoxylin (Solarbio, China) before they were dehydrated and mounted. Finally, the staining was visualized under a light microscopy (Olympus, Japan). Three randomly selected microscopic fields (×400 magnification) were photographed and the mean optic density in each field was counted and analyzed using Image-Pro Plus software (Media Cybernetics, USA).
Plasmid construction and cell transfection
Three interference sequences targeting circATRNL1 were constructed and inserted into the EcoRI–BamHI site of the pHBLV-U6-MCS-CMV-ZsGreen-PGK-Puro vector. The sequences were as follows: #1, 5′-ATAGTTGGCAAGGTTAACAGAACCT-3′; #2, 5′-TTGGCAAGGTTAACAGAACCTTCTG-3'; #3, 5'-CAATGATAGTTGGCAAGG TTAACAG-3′; For lentivirus-mediated overexpression of circATRNL1 in Ishikawa cells, full-length circATRNL1 was inserted into the pHBLV-CMV-circ-MCS-EF1-zsgreen-T2A-puro vector. Lentivirus packaging, cell infection, and selection of puromycin-resistant cells were performed according to the instruction manual provided by Hanbio Biotechnology (Shanghai, China). The sequences of miRNA mimics, miRNA inhibitors, and siRNAs were as follows: miR-141-3p mimic, forward 5′-UAACACUGUCUGGUAAAGAUGG-3′ and reverse 5′-AUCUUUACCAGACAGUGUUAUU-3′; miR-141-3p inhibitor, 5′-CCAUCUUUACCAGACAGUGUUA-3′; miR-200a-3p mimic, forward 5′-UAACACUGUCUGGUAAACG AUGU-3′ and reverse 5′-AUCGUUACCAGACAGUGUUAUU-3′; miR-200a-3p inhibitor, 5′-AC AUCGUUACCAGACAGUGUUA-3′; Yes-associated protein 1 (YAP1) siRNA (#1), forward 5′-CUGCCACCAA GCUAGAUAATT-3′ and reverse 5′-UUAUCUAGCUUGGUGGCAGTT-3′; (#2), forward 5′-GGUGAUACUAUCAACCAAATT-3′ and reverse 5′-UUUGGUUGAUAGUAUCACCTT-3′; (#3), forward 5′-GACGACCAAUAGCUCAGAUTT-3′ and reverse 5′-AUCUGAGCUAUUGGUCGUCTT-3′; negative control (NC), forward 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse 5′-ACGUGACACGUUCGGAG AATT-3′; inhibitor negative control (si-NC), 5′-CAGUACUUUUGUGUAGUACAA-3′. All of these RNA oligos were purchased from GenePharma (Suzhou, China) and were transfected into Ishikawa cells using Lipofectamine 3000 Reagent (Invitrogen, USA).
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from human tissues and cultured cells with TRIzol reagent (TaKaRa, Japan). RNA quantity and quality were measured using a Nanophotometer N50 (Implen, Germany). Only when the ratio of the absorbance at 260 and 280 nm was between 1.8 and 2.2 was the total RNA sample considered acceptable. All RNA samples were stored at −80 °C until further use. The expression levels of circRNAs were evaluated by RT-qPCR using a One-step SYBR PrimeScript RT-PCR kit (cat. no. RR660A; TaKaRa) on an Applied Biosystems 7500 FAST instrument (Applied Biosystems, Foster City, CA, USA). The cycling program was initiated at 42 °C for 5 min and 95 °C for 10 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s. An Mir-X miRNA First-strand Synthesis Kit (cat. no. 638315; TaKaRa) and PrimeScript RT Reagent Kit with gDNA Eraser (cat. no. RR047A; TaKaRa) were used for first-strand synthesis of miRNAs and mRNAs, respectively. Then, the amplification conditions were carried out using SYBR Premix Ex TaqII (cat. no. RR820A; TaKaRa) according to the manufacturer’s instructions, as follows: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s. All PCR primers were designed and synthesized by Sangon Biotech (Shanghai, China) and were as follows: circATRNL1, forward 5′-GCAATGATAGTTGCAAGGTTAAC-3′, reverse 5′-GCCTTCAATGAGC CAAGTACA-3′; miR-141-3p, forward 5′-GCGCTAACACTGTCTGGTAAAGATGG-3′; miR-200a-3p, forward 5′-GCGTAACACTGTCTGGTAACGATGT-3′; YAP1, forward 5′-CCGTTTCCCAGACTACCTT-3′, reverse 5′-TTGGCATCAGCTCCTCTC-3′. Each sample had three individual technical replicates. The threshold cycle method (2−ΔΔCT) was used to calculate relative quantification of expression levels compared with the internal standard.
Cell proliferation assay
After transfection, Ishikawa cells were seeded on 96-well flat-bottomed microplates at a density of 5000 cells/well. The culture medium was regularly replaced. For analysis of cell proliferation, 100 μL of Cell Counting Kit-8 (CCK-8) reagent (Dojindo, Japan) was added into each well at different time points (0, 24, 48, 72, and 96 h), followed by incubation for 1 h at 37 °C. The absorbance of each well at 490 nm was observed and measured with a Universal Microplate Spectrophotometer (Bio-Tek Instruments, Inc., Winooski, VT, USA).
Cell invasion and migration assays
Cell invasion assays were performed using a Transwell system (Corning, NY, USA). After dilution with serum-free medium, 100 μL Matrigel (cat. no. 356234; Corning) was added to each transwell chamber and coagulated for 4 h at 37 °C. After treatment for 48 h, Ishikawa cells were washed and cultured with serum-free medium for 12 h. Next, a 200-μL aliquot of the above single-cell suspension (2 × 104 cells) was placed into each upper chamber of a transwell plate. The lower chamber was filled with 700 mL of medium containing 10% FBS. After 24 h of incubation, a cotton swab was used to remove cells remaining in the upper chamber. The cells that invaded into the membrane were fixed in 4% formaldehyde (Solarbio, Beijing) for 30 min, and then stained with 0.1% crystal violet solution (Solarbio). Eight randomly selected microscopic fields were photographed with an Eclipse Ti-s microscope (Nikon, Japan), and the number of cells in each field was counted using Image-Pro Plus software (Media Cybernetics, USA). Cell migration assays were performed according to the same procedure, except that the Matrigel was omitted.
Fluorescence in situ hybridization (FISH)
Hybridization was performed overnight at 37 °C with specific circATRNL1 probes, sections were washed, and DAPI staining was performed for 20 min in the dark. The sections were observed under a Leica TCS SP2 AOBS Confocal (upright) fluorescence microscope (×40 objective, ×10 ocular), and images were acquired. Nuclei stained with DAPI appeared blue under UV excitation and contained circATRNL1, which was labeled with Cy3 (red emission), and miRNAs (miR-141-3p and miR-200a-3p) which were labeled with FITC (green emission). The sequence of probes used for FISH were as follows: circATRNL1, 5′-TTCTGTTAACCTTGCCAACTA-3′; miR-141-3p, 5′-CCATCTTTACCAGACAGTGTTA-3′; miR-200a-3p, 5′-CATCGTTACCAGACAGTGTTA-3′. All the probes synthesis and FISH detection were performed by Geneseed (Guangzhou, China).
Dual-luciferase activity assay
pmiR-GLO-YAP1 wild-type or mutated vectors, miRNA mimics or negative control (Genepharm), and wild-type circATRNL1 or mutated circATRNL1 vectors (Ribobio, Guangzhou, China) were transfected into 293T cells. Firefly and Renilla luciferase activities were measured consecutively 24 h after transfection using Dual-Luciferase Reporter Assays (cat. no. E1910; Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Western blot analysis
Tissues and cell lysates were prepared with RIPA buffer (Beyotime) containing 10% phenylmethylsulfonyl fluoride. Next, a BCA Kit (Beyotime) was used to quantify protein concentrations. Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 10% gels (Beyotime) and transferred onto polyvinylidene difluoride membranes (Millipore, USA). The membranes were blocked with 5% skim milk for 2 h and incubated with primary antibodies at room temperature for 1.5 h or overnight at 4 °C. All primary and secondary antibodies were purchased from Proteintech (Wuhan, China). The antibodies were as follows: mouse monoclonal anti-E-cadherin (Cat.No. 60335-1-Ig, 1:2500), mouse monoclonal anti-N-cadherin (Cat.No. 66219-1-Ig,1:2000), mouse monoclonal anti-vimentin (Cat.No. 60330-1-Ig, 1:6000), mouse monoclonal anti-YAP1 (Cat.No. 66900-1-Ig,1:1000), rabbit polyclonal anti-ZEB1 (Cat.No.21544-1-AP, 1:1000), mouse monoclonal anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (P04406) (Cat.No.60004-1-Ig, 1:20000), mouse monoclonal anti-β-tubulin (P04350) (Cat.No. 66240-1-Ig, 1:20000), goat anti-mouse IgG H&L (Cat.No. SA00001-1 HRP conjugated; 1:5000), and goat anti-rabbit IgG H&L antibodies (Cat.No. SA00001-2, HRP conjugated; 1:5000). The immunoblot signal was quantified with ImageJ software.
Statistical analysis
All experiments were performed in triplicate. The data are expressed as means ± standard errors of the means and analyzed by GraphPad Prism 7 (La Jolla, CA, USA). Differences between two groups were evaluated with Student’s t tests. Comparisons among three or more groups were performed by one-way analysis of variance and Tukey’s post hoc test. Each experiment was conducted triplicate. Correlations among circATRNL1, miR-141-3p/miR-200a-3p, and YAP1 were analyzed with Spearman rank correlation. P values of less than 0.05 were considered statistically significant.
Results
CircATRNL1 was upregulated in ovary endometriosis tissues
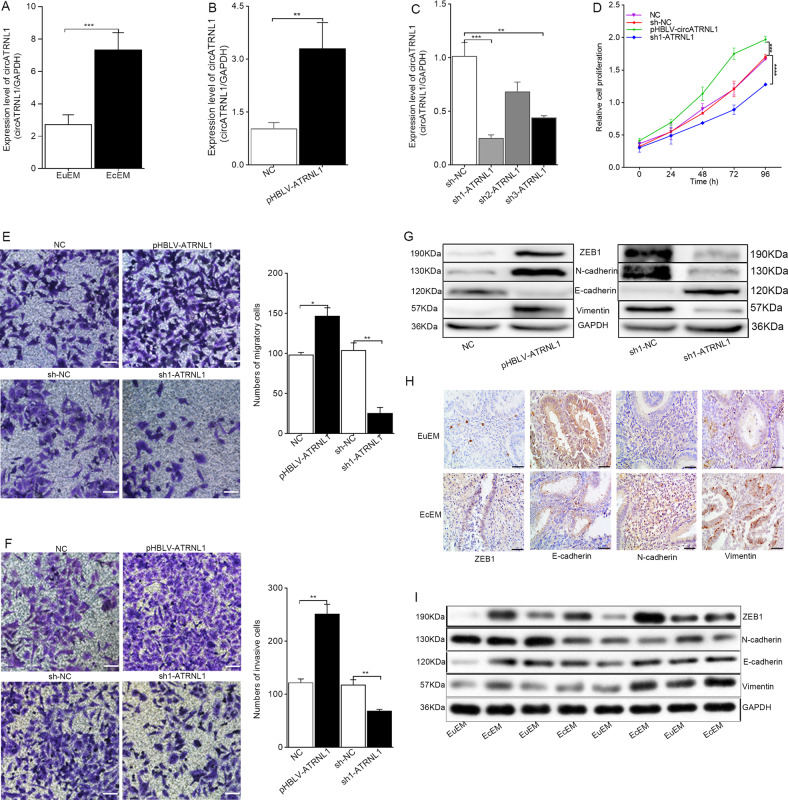
To validate the expression levels of circATRNL1, RT-qPCR was conducted in 60 EcEM and EuEM samples. The data showed that circATRNL1 expression was significantly increased in ovary EcEM tissues compared with that in EuEM tissues (Fig. 1a, P = 0.0002). This suggested that circATRNL1 may play a role in the development of endometriosis.
Fig. 1. circATRNL1 regulates proliferation, migration, invasion, and EMT process in endometriosis.
a The expression level of circATRNL1 in 60 ovary endometriosis tissue samples was examined by using qRT-PCR. bcircATRNL1 was overexpressed in Ishikawa cells by transfecting with lentivirus-mediated vector pHBLV-ATRNL1. NC was used as a control. ccircATRNL1 was silenced in Ishikawa cells by transfecting with specific sh-ATRNL1. shRNA-NC was used as a control. d CCK-8 assays were conducted to determine the influence of circATRNL1 on cell proliferation. e Transwell migration assays showed that circATRNL1 regulated the migrative potential of Ishikawa cells. Photographs were taken at ×200 magnification. Scale bar represents 100 μm. f Transwell invasion assays showed that circATRNL1 regulated the invasive potential of Ishikawa cells. g Western blot analysis was utilized to analyze the impacts of circATRNL1 overexpression or knockdown on EMT progress in Ishikawa cells. h Immunohistochemistry of EMT markers in 60 EuEM and EcEM tissues. Original magnification: ×400. Scale bars represent 50 μm. i Western blot analysis of EMT markers in 60 EuEM and EcEM tissues. Assays were performed in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 represent statistical difference.
Knockdown of circATRNL1 inhibited cell growth, migration, and invasion in vitro
To further explore the underlying functions of circATRNL1 in endometriosis, we performed a series of validation assays by overexpressing and silencing circATRNL1 in Ishikawa cells, respectively. The relative expression level of circATRNL1 was validated by RT-qPCR. Importantly, the pHBLV-ATRNL1 vector upregulated circATRNL1 in Ishikawa cells (Fig. 1b, P = 0.0071). Three specific lentivirus-mediated shRNAs (sh1-ATRNL1, sh2-ATRNL1, and sh3-ATRNL1) were transfected into Ishikawa cells to silence circATRNL1 expression; sh1 showed the highest efficiency and was selected for further cellular experiments (Fig. 1c, P = 0.0009 for sh-NC vs. sh1, P = 0.0815 for sh-NC vs. sh2, P = 0.003 for sh-NC vs. sh3).
Subsequently, CCK-8 assays were conducted to determine the influence of circATRNL1 on cell proliferation. Our results demonstrated that overexpression of circATRNL1 significantly promoted the proliferation of Ishikawa cells (P = 0.0010), whereas circATRNL1 knockdown decreased the growth of Ishikawa cells (Fig. 1d, P = 0.00005). We also observed that the migratory and invasive capabilities of Ishikawa cells were significantly enhanced by overexpression of circATRNL2 (Fig. 1e, P = 0.0108 for NC vs. pHBLV-ATRNL1, P = 0.0025 for sh-NC vs. sh1-ATRNL1), but significantly inhibited by silencing of circATRNL1 (Fig. 1f, P = 0.0025 for NC vs. pHBLV-ATRNL1, P = 0.0078 for sh-NC vs. sh1-ATRNL1). These results revealed that circATRNL1 knockdown may efficiently reduce endometriosis by inhibiting cell growth, migration, and invasiveness.
Knockdown of circATRNL1 inhibited EMT progression in endometriosis
To further detect whether circATRNL1 promoted migration and invasion by facilitating the EMT in Ishikawa cells, we tested the expression of EMT hallmarks by western blot analysis. As shown in Fig. 1g, the levels of epithelial markers (E-cadherin) were enhanced, whereas the levels of mesenchymal markers (N-cadherin, vimentin, and ZEB1) were obviously decreased in circATRNL1 knockdown Ishikawa cells. In contrast, the levels of EMT hallmarks showed the opposite changes in response to circATRNL1 overexpression. Taken together, our findings demonstrated that circATRNL1 expression was correlated with the EMT process in endometriosis. In addition, we tested the expression of EMT hallmarks by IHC and western blot analysis in 60 ovary EcEM tissues and EuEM tissues. The data showed that, compared with that in EuEM tissues, the levels of epithelial markers (E-cadherin) were decreased in EcEM tissues, whereas the levels of mesenchymal markers (N-cadherin, vimentin, and ZEB1) were obviously enhanced (Fig. 1h, i).
miR-141-3p and miR-200a-3p were targets of circATRNL1
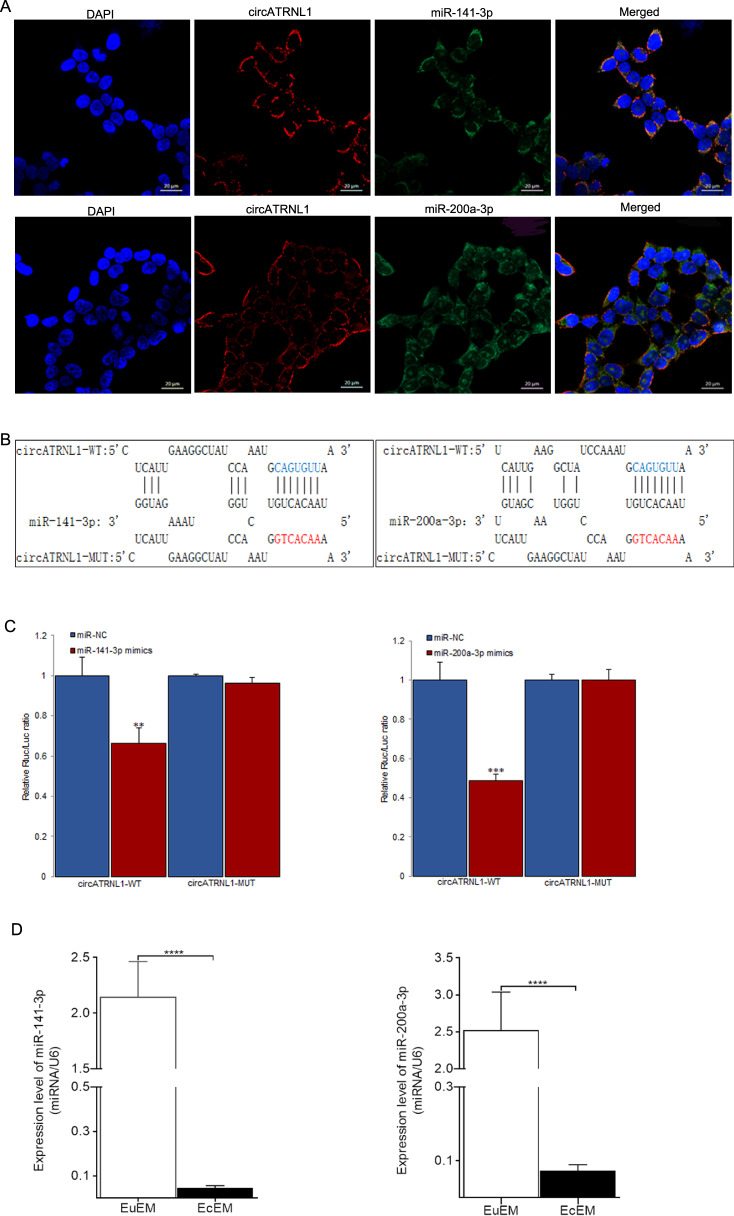
Accumulating evidence has demonstrated that circRNAs act as ceRNAs to sponge miRNAs and modulate the expression of target genes at the post-transcriptional level. To confirm that circATRNL1 exerted biological functions through acting as a ceRNA, we first performed FISH assays to identify the localization of circATRNL1 in Ishikawa cells. CircATRNL1 was mainly localized in the cytoplasm of Ishikawa cells (Fig. 2a). According to our previous bioinformatics prediction, miR-141-3p and miR-200a-3p may bind circATRNL1 to affect the pathogenesis of endometriosis27. Furthermore, a luciferase reporter vector was designed and transfected into HEK-293T cells to facilitate the combination between circATRNL1 and the two miRNAs (Fig. 2b). The results showed that both miR-141-3p mimics (P = 0.0078) and miR-200a-3p mimics (P = 0.0008) decreased the luciferase activity of wild-type circATRNL1 (circATRNL1-WT), whereas the luciferase activity of mutated circATRNL1 (circATRNL1-MUT) was not significantly altered (Fig. 2c). Subsequently, miR-141-3p (P < 0.00001) and miR-200a-3p (P < 0.00001) were shown to be downregulated in EcEM tissues compared with that in EuEM tissues (Fig. 2d). Accordingly, the expression correlation between circATRNL1 and miR-141-3p (r = −0.434, P = 0.001) and miR-200a-3p (r = −0.418, P = 0.001) was separately proved to be negative.
Fig. 2. miR-141-3p and miR-200a-3p are targets of circATRNL1.
a FISH assay was used to determine the location of circATRNL1, miR-141-3p, and miR-200a-3p in Ishikawa cells (blue, DAPI nuclear staining; red, circATRNL1; green, miR-141-3p and miR-200a-3p). Scale bars represent 20 μm. b The binding sites between wild-type circATRNL1 or mutated circATRNL1 and miR-141-3p/miR-200a-3p were predicted by using bioinformatics analysis. c Dual-luciferase reporter assays were carried out in HEK-293T cell to confirm the combination between circATRNL1 and miR-141-3p mimics or miR-200a-3p mimics. d qRT-PCR analysis was carried to determine the expression pattern of miR-141-3p and miR-200a-3p in ovary endometriosis tissues. ****P < 0.0001 represents statistical difference.
miR-141-3p and miR-200a-3p inhibited EMT progression in endometriosis
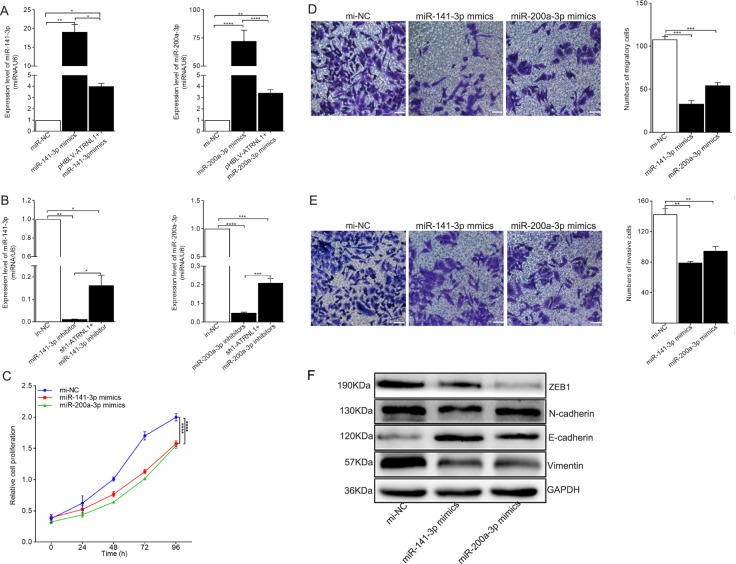
miR-141-3p and miR-200a-3p belong to the miR-200 family, which modulates EMT progression in several diseases. Nevertheless, the specific roles of miR-141-3p and miR-200a-3p in endometriosis are still unknown. Accordingly, miR-141-3p mimics/inhibitors and miR-200a-3p mimics/inhibitors were transfected into Ishikawa cells for overexpression or knockdown of the corresponding target gene, with mi-NC and in-NC used as the control group. RT-qPCR was used to measure transfection efficiency after 48 h. Co-transfection with pHBLV-ATRNL1 or sh1-ATRNL1 rescued the efficiency of miRNA mimics (Fig. 3a, P = 0.0018 for mi-NC vs. miR-141-3p mimics, P = 0.0129 for miR-141-3p mimics vs. pHBLV-ATRNL1+ miR-141-3p mimics, P = 0.0111 for mi-NC vs. pHBLV-ATRNL1+ miR-141-3p mimics; P < 0.00001 for mi-NC vs. miR-200a-3p mimics, P < 0.00001 for miR-200a-3p mimics vs. pHBLV-ATRNL1+ miR-200a-3p mimics, P = 0.0025 for mi-NC vs. pHBLV-ATRNL1+ miR-200a-3p mimics) or inhibitors (Fig. 3b, P = 0.002 for in-NC vs. miR-141-3p inhibitors, P = 0.0105 for miR-141-3p inhibitors vs. sh1-ATRNL1+ miR-141-3p inhibitors, P = 0.0328 for in-NC vs. sh1-ATRNL1 + miR-141-3p inhibitors; P < 0.00001 for in-NC vs. miR-200a-3p inhibitors, P = 0.0002 for miR-200a-3p inhibitors vs. sh1-ATRNL1+ miR-200a-3p inhibitors, P = 0.0001 for in-NC vs. sh1-ATRNL1+ miR-200a-3p inhibitors). Similarly, CCK-8 assays and transwell migration and invasion assays were carried out to evaluate the influence of miR-14-3p/miR-200a-3p mimics on Ishikawa cells. As showed in Fig. 3c, cell proliferation was obviously inhibited by miR-141-3p mimics (P < 0.00001) and miR-200a-3p mimics (P < 0.00001). The results of transwell experiments demonstrated that the migratory (Fig. 3d, P = 0.0001 for mi-NC vs. miR-141-3p, P = 0.0001 for mi-NC vs. miR-200a-3p) and invasive (Fig. 3e, P = 0.0013 for mi-NC vs. miR-141-3p, P = 0.0072 for mi-NC vs. miR-200a-3p) capabilities of Ishikawa cells were dramatically suppressed in the context of miR-141-3p or miR-200a-3p overexpression. Finally, the results of western blot analysis demonstrated that EMT progression was inhibited in miR-141-3p- or miR-200a-3p-overexpressing Ishikawa cells (Fig. 3f). Overall, these results suggested that both miR-141-3p and miR-200a-3p may exert inhibitory effects during the development of endometriosis.
Fig. 3. miR-141-3p and miR-200a-3p inhibited EMT process in endometriosis.
amiR-141-3p and miR-200a-3p were overexpressed with miR-141-3p or miR-200a-3p mimics in Ishikawa cells, respectively, and be partly rescued by pHBLV-ATRNL1 co-transfection. mi-NC was taken as the control group. bmiR-141-3p and miR-200a-3p were downregulated with miR-141-3p or miR-200a-3p inhibitors in Ishikawa cells, respectively, and be partly rescued by sh1-circATRNL1 co-transfection. in-NC was taken as the control group. c–e CCK-8 and transwell assays were separately carried out to identify the influence of miR-141-3p and miR-200a-3p mimics on cell proliferation, migration, and invasion. Photographs were taken at ×200 magnification. Scale bar represents 100 μm. f The effect of miR-141-3p and miR-200a-3p mimics on EMT progress in Ishikawa cells was evaluated through detecting the EMT marker proteins with western blot analysis. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 represent statistical difference.
YAP1 was a common target of miR-141-3p and miR-200a-3p
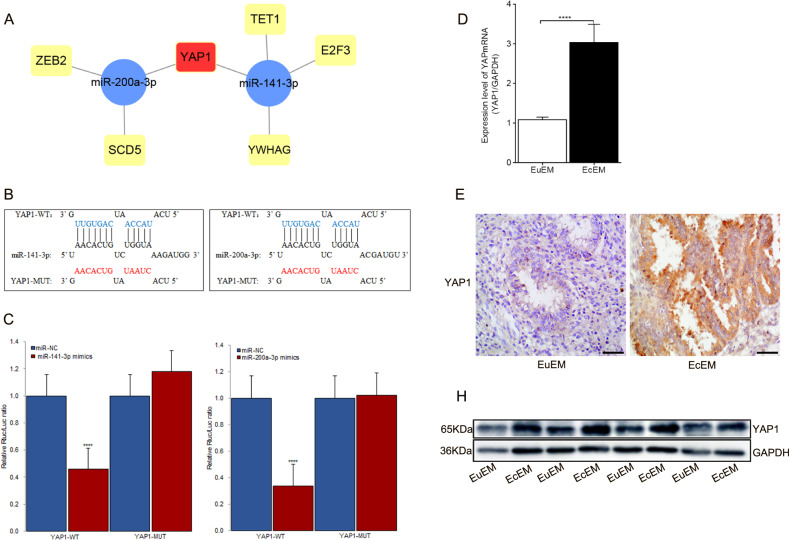
Based on the ceRNA hypothesis, we used MiRWalk v3.0 (http://mirwalk.umm.uni-heidelberg.de/), with strict screening conditions and three prediction algorithms (Targetscan, MiRDB, and mirTarbase), to predict downstream target mRNAs of miR-141-3p and miR-200a-3p. Four mRNAs were found to be potential targets of miR-141-3p, whereas three mRNAs were found to be potential targets of miR-200a-3p (Fig. 4a). Among these seven mRNAs, YAP1 was the only common target and was chosen for subsequent analyses. The binding sites for miR-141-3p and miR-200a-3p in the 3′-untranslated region (UTR) of YAP1 were predicted (Fig. 4b). Similarly, luciferase reporter assays were conducted to demonstrate the interactions between the two miRNAs and YAP1. The luciferase activity of wild-type YAP1 (YAP1-WT) was significantly reduced in HEK-293T cells transfected with miR-141-3p mimics (P = 0.00001) or miR-200a-3p mimics (P = 0.00001), whereas that of mutated YAP1 (YAP1-MUT) was not significantly altered (Fig. 4c). Subsequently, high expression levels of YAP1 mRNA (P = 0.00004) and protein (P = 0.013) were detected in EcEM tissues compared with that in EuEM tissues (Fig. 4d–f). Spearman correlation analysis proved negative relevance between YAP1 and miR-141-3p (r = −0.326, P = 0.012) as well as negative relevance between YAP1 and miR-200a-3p (r = −0.312, P = 0.015). On the contrary, YAP1 expression correlation with circATRNL1 was proved to be positive (r = 0.950, P < 0.0001).
Fig. 4. YAP1 is a common target mRNA of miR-141-3p and miR-200a-3p.
a The downstream target mRNAs of miR-141-3p/miR-200a-3p were predicted by three bioinformatics tools (Targetscan, MiRDB, and mirTarbase). b The binding sites between 3′UTR of YAP1 and miR-141-3p/miR-200a-3p were predicted by using bioinformatics analysis. c Dual-luciferase reporter assays were carried in HEK-293T cells which were co-transfected with miR-141-3p mimics or miR-200a-3p mimics and pmiR–pmiR-GLO-YAP1. d The expression levels of YAP1 mRNA were tested in ovary endometriosis tissues through using qRT-PCR. e, f Immunohistochemistry and western blot analysis were used to test YAP1 protein in ovary endometriosis tissues. Immunohistochemistry photographs were taken at ×400 magnification. Scale bars represent 50 μm. ****P < 0.0001 represents statistical difference.
Knockdown of YAP1 inhibited EMT progression in endometriosis
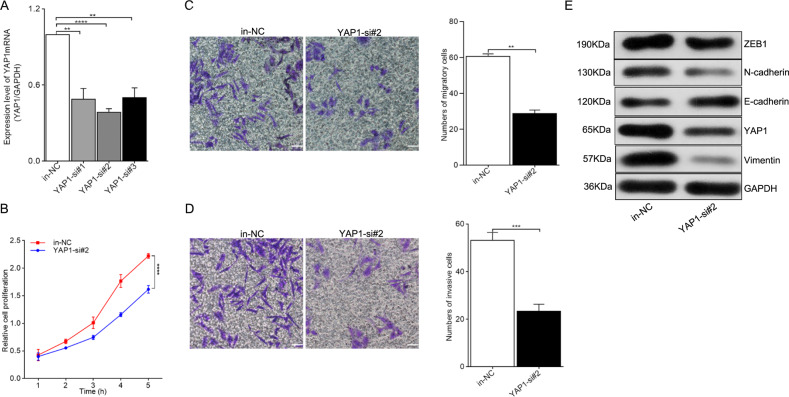
Three siRNA sequences were constructed and transfected into Ishikawa cells to knockdown YAP1 mRNA expression; YAP1-si#2 showed the highest inhibitory efficiency as demonstrated through RT-qPCR results (Fig. 5a, P = 0.0034 for in-NC vs. YAP1-si#1, P = 0.00002 for in-NC vs. YAP1-si#2, and P = 0.0026 for in-NC vs. YAP1-si#3) and was selected for further cellular experiments. CCK-8 assays and transwell migration and invasion assays were carried out to evaluate the influence of YAP1 siRNA on Ishikawa cells. As depicted in Fig. 5b, cell proliferation was obviously inhibited by YAP1-si#2 (P < 0.0001). The results of transwell experiments demonstrated that the migratory (Fig. 5c, P = 0.0001) and invasive (Fig. 5d, P = 0.0013) capabilities of Ishikawa cells were significantly inhibited in the context of YAP1 knockdown. Finally, the results of western blot analysis demonstrated that EMT progression was inhibited in YAP1-silencing Ishikawa cells (Fig. 5e). Overall, these results suggested that YAP1 may promote EMT progression in endometriosis.
Fig. 5. YAP1 silencing inhibited EMT process in endometriosis.
a Knockdown of YAP1 expression with YAP1 siRNA in Ishikawa cells. in-NC was taken as the control group. b–d CCK-8 and transwell assays were separately carried out to identify the influence of YAP1 siRNA on cell proliferation, migration, and invasion. Photographs were taken at ×200 magnification. Scale bar represents 100 μm. e The effect of YAP1 siRNA on EMT progress in Ishikawa cells was evaluated through detecting the EMT marker proteins with western blot analysis. **P < 0.01, ****P < 0.0001 represent statistical difference.
Regulation of the circATRNL1/miR-141-3p/miR-200a-3p/YAP1 axis in EMT progression in endometriosis
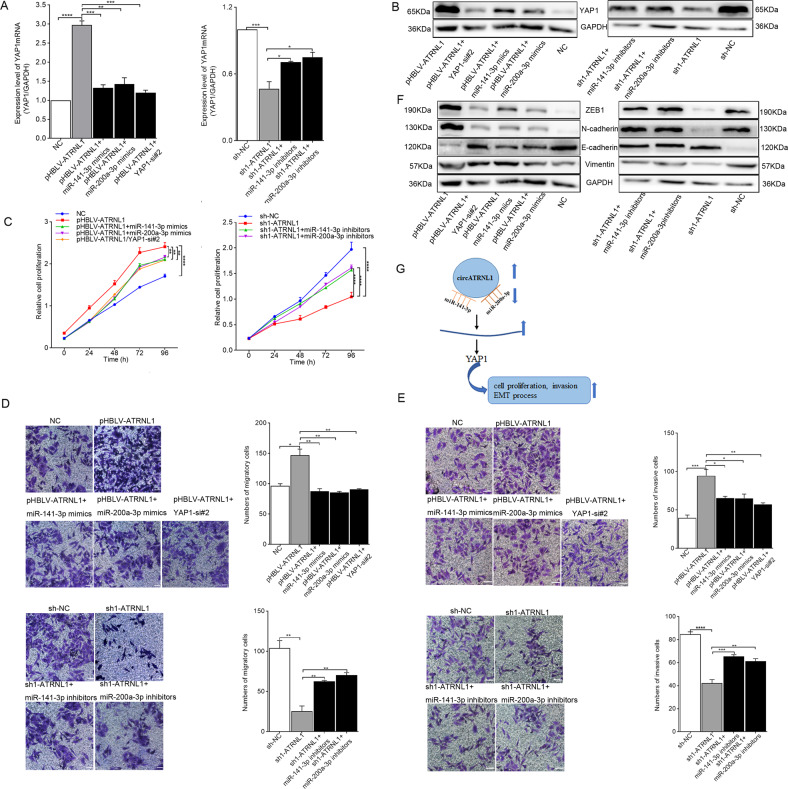
To analyze the regulatory relationships among circATRNL1, miR-141-3p/miR-200a-3p, and YAP1, mRNA and protein levels of YAP1 were measured, and rescue assays were performed in Ishikawa cells. As depicted in Fig. 6a, b, the increased mRNA and protein levels of YAP1 caused by pHBLV-ATRNL1 was rescued by miR-141-3p or miR-200a-3p mimics or YAP1-si#2, and meanwhile, miR-141-3p or miR-200a-3p inhibitors could rescue effects of sh1-ATRNL1 on YAP1. According to the results of CCK-8 and transwell assays, the effects of pHBLV-ATRNL1 on promoting proliferation, migration, and invasion in Ishikawa cells could be partly rescued by miR-141-3p or miR-200a-3p mimics or YAP1-si#2. In addition, miR-141-3p or miR-200a-3p inhibitors could partly rescue the suppressive effects of silencing circATRNL1 (Fig. 6c–e). Similarly, western blot results showed that the enhancing effects of pHBLV-ATRNL1 on EMT progression were partly reversed by miR-141-3p or miR-200a-3p mimics or YAP1-si#2, whereas the inhibitory effects of sh1-ATRNL1 on EMT progression were partly reversed by miR-141-3p or miR-200a-3p inhibitors (Fig. 6f). Based on these data, we confirmed that circATRNL1 acted as a ceRNA to modulate endometriosis progression via the miR-141-3p/miR-200a-3p/YAP1 axis (Fig. 6g).
Fig. 6. Function of circATRNL1–miR-141-3p/miR-200a-3p–YAP1 axis in endometriosis progression.
a, b mRNA level and protein level of YAP1 were measured by using qRT-PCR and western blot in Ishikawa cells which were co-transfected with miR-141-3p/miR-200a-3p mimics and pHBLV-ATRNL1 or miR-141-3p/miR-200a-3p inhibitors and sh-ATRNL1 or YAP1-si and pHBLV-ATRNL1. c–e CCK-8 and transwell assays were separately performed to detect proliferation and motility ability of indicated Ishikawa cells. Photographs were taken at ×200 magnification. Scale bar represents 100 μm. f The effect of circATRNL1–miR-141-3p/miR-200a-3p-YAP1 axis on EMT protein markers in Ishikawa cells was evaluated through western blot analysis. g The mechanism diagram was generated to illustrate the mechanism of circATRNL1–miR-141-3p/miR-200a-3p–YAP1 axis in endometriosis. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 represent statistical difference.
Discussion
In recent years, a growing body of evidence has indicated that circRNAs play crucial regulatory roles in the pathogenesis and development of various diseases, including both malignant and benign diseases18,19,22–27. The circRNA circAF4 functions as an oncogene to regulate the expression of the MLL-AF4 fusion protein and inhibit MLL leukemia progression by sponging miR-128-3p28. CircRNAs and miRNAs function together to serve as miRNA sponges through a ceRNA network. A recent study revealed that upregulation of circRNA-100338 was associated with poor prognosis in patients with hepatitis B-related HCC by activating the mammalian target of rapamycin signaling pathway in HCC via the circRNA-100338/miR-141-3p/RHEB axis29. In a diabetic mouse myocardial fibrosis model, functional experimental results demonstrated that circRNA-010567 silencing upregulated miR-141 and downregulated TGF-β1 expression, thus suppressing myocardial fibrosis30. Circ-ZEB1.33 is an oncogene that promotes HCC cell proliferation by enhancing the expression of cyclin-dependent kinases 6 through sponging miR-200a-3p31. This inspired us to explore the underlying mechanisms and functions of circRNAs in endometriosis.
In this study, we found that compared with the EuEM in patients with endometriosis, circATRNL1 was significantly elevated in the EcEM. According to the results of IHC and western blot assays, we found decreased E-cadherin and increased ZEB1, N-cadherin as well as vimentin expression in EcEM tissues compared with that in EuEM tissues, which was consistent with previous research results7,11–13. To further investigate the influence of circATRNL1 dysregulation on endometrial epithelial cell activities, a series of functional assays were conducted in Ishikawa cells. The results demonstrated that knockdown of circATRNL1 inhibited cell proliferation, migration, and invasion, whereas overexpression of circATRNL1 promoted these cellular bioactivities. Concomitantly, knockdown of circATRNL1 reduced the protein expression of N-cadherin, vimentin, and ZEB1, but increased the expression of E-cadherin. Therefore, circATRNL1 may exhibit oncogene-like properties in endometriosis by regulating the EMT process.
Through FISH assays, we first showed that circATRNL1 was mainly localized in the cytoplasm of Ishikawa cells, which further confirmed our hypothesis that circATRNL1 may act as a ceRNA by binding with certain miRNAs. Next, bioinformatics analysis and luciferase reporter assays were utilized to identify the target miRNAs of circATRNL1. As a result, miR-141-3p and miR-200a-3p were identified. Previous studies have revealed a large number of miRNAs showing expression level alterations between EuEM and EcEM tissues, indicating their involvement in the establishment and progression of endometriosis, such as angiogenesis, inflammation, and immune regulation32–36. For example, a recent cell type-specific analysis identified 149 abnormally expressed miRNAs in endometriotic lesions, including extensive upregulation of miR-139-5p and downregulation of miR-375 compared with eutopic stromal cells, which are potentially involved in endometriosis-associated infertility or in the regulation of invasive growth and cell proliferation in endometriosis development36. In our study, we detected downregulation of both miR-141-3p and miR-200a-3p in ectopic endometriotic tissues, consistent with previous research34–36. Furtherly, a negative expression correlation between circATRNL1 and miR-141-3p as well as circATRNL1 and miR-200a-3p in endometriosis tissues was separately proved.
Importantly, miR-141-3p and miR-200a-3p belong to the miR-200 family, which has been reported to regulate the EMT and metastasis by targeting a cohort of target genes, such as ZEB1 and E-cadherin8,16,37. Moreover, miR-200a-3p and miR-141-3p are downregulated in the plasma samples of patients with endometriosis compared with that in healthy controls, suggesting that these miRNAs could be used as putative noninvasive diagnostic biomarkers of endometriosis38. Li et al.39 found that inhibition of miR-200a could promote migration and high mobility group box 1 expression, while decreasing E-cadherin expression in HCC cell lines. Another study showed that EMT progression was inhibited in miR-200a-3p-overexpressing papillary thyroid cancer (PTC) cells40. Similarly, in this study, we also observed the inhibitory effects of miR-141-3p and miR-200a-3p on the EMT in endometriosis, and the effects of circATRNL1 knockdown on cell proliferation, invasion, and EMT suppression could be partially attenuated by miR-141-3p/miR-200a-3p inhibition.
MiRNAs regulate gene expression post-transcriptionally by interacting with the 3′-UTRs of mRNAs and triggering either translational repression or mRNA degradation36. Through bioinformatics prediction, YAP1 was found to be the common target of miR-141-3p and miR-200a-3p. YAP1 is a downstream oncogene of the Hippo pathway41. When activated, YAP1 localizes to the nucleus and binds to transcription factors, such as TEA domain DNA-binding family of transcription factors (TEAD), and then drives tumor growth, metastasis, and senescence in cancer cell lines and induces the EMT in many types of tumors. YAP1 induces the EMT in non-small cell lung cancer by regulating the transcription of Slug via interacting with TEAD42. In addition, YAP1 is highly associated with HCC and PTC and frequently upregulated during tumor formation40,43. Song et al.44 demonstrated that elevated expression of YAP1 promotes endometrial stromal cell proliferation and blocks apoptosis in vitro and in vivo. In the current study, we found that YAP1 mRNA and protein levels were positively regulated by circATRNL1 in endometrial cell lines. The functional assay results furtherly demonstrated that knockdown of YAP1 expression in Ishikawa cells could significantly inhibited cell proliferation, migration and invasion, and reversed the EMT process. Finally, rescue assays demonstrated that circATRNL1 improved the EMT by modulating miR-141-3p/miR-200a-3p/YAP1 in endometriosis.
In conclusion, our findings suggested that circATRNL1 promoted the EMT in endometriosis by upregulating YAP1 through sponging miR-141-3p and miR-200a-3p. However, there were some limitations to this study. First, the sample size was relatively small for RT-qPCR, and only rASRM stage III/IV cases were included. Second, the tissue samples were only derived from ectopic cyst walls and EuEM and not from other types of endometriosis or healthy controls. Accordingly, further validation involving in larger cohorts of patients with different types of endometriosis and healthy controls is warranted to confirm the clinical value of circATRNL1. Moreover, we used Ishikawa cells rather than primary endometrial epithelial cells for functional experiments, which may lead to unreliable conclusions. Further detailed studies of the mechanisms and functions of circATRNL1 are needed. With the deepening exploration of circRNAs, we expect to provide new ideas on pathogenesis and find new promising therapeutic targets for endometriosis.
Supplementary information
Acknowledgements
This study was supported by grants from the Research Foundation for the Science and Welfare Career of Liaoning Province (No. 20170017), the Local Technology and Development Key Program of Liaoning Province (No. 2019416020), and the Support Program for Youth Backbone of China Medical University (No. QGZD2018062). We thank all of the members of the Biological Specimen Bank of Shengjing Hospital for providing tissue samples.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by N. Barlev
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-020-02784-4).
References
- 1.Giudice LC. Clinical practice: endometriosis. N. Engl. J. Med. 2014;362:2389–2398. doi: 10.1056/NEJMcp1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borghese B, Zondervan KT, Abrao MS, Chapron C, Vaiman D. Recent insights on the genetics and epigenetics of endometriosis. Clin. Genet. 2017;91:254–264. doi: 10.1111/cge.12897. [DOI] [PubMed] [Google Scholar]
- 3.Shafrir AL, et al. Risk for and consequences of endometriosis, a critical epidemiologic review. Best. Pract. Res. Clin. Obstet. Gynaecol. 2018;51:1–15. doi: 10.1016/j.bpobgyn.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Tomassetti C, D’Hooghe T. Endometriosis and infertility, Insights into the causal link and management strategies. Best. Pract. Res. Clin. Obstet. Gynaecol. 2018;51:25–33. doi: 10.1016/j.bpobgyn.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Matias-Guiu X, Stewart CJR. Endometriosis-associated ovarian neoplasia. Pathology. 2018;50:190–204. doi: 10.1016/j.pathol.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Hay ED. An overview of epithelial-mesenchymal transformation. Acta Anat. 1995;154:8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- 7.Bartley J, Jülicher A, Hotz B, Mechsner S, Hotz H. Epithelial to mesenchymal (EMT) seems to be regulated differently in endometriosis and the endometrium. Arch. Gynecol. Obstet. 2014;289:871–881. doi: 10.1007/s00404-013-3040-4. [DOI] [PubMed] [Google Scholar]
- 8.Liang Z, et al. miR-200c suppresses endometriosis by targeting MALAT1 in vitro and in vivo. Stem. Cell. Res. Ther. 2017;8:251. doi: 10.1186/s13287-017-0706-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin D, et al. Long non-coding RNA AFAP1-AS1 promoting epithelial mesenchymal transition of endometriosis is correlated with transcription factor ZEB1. Am. J. Reprod. Immunol. 2019;81:e13074. doi: 10.1111/aji.13074. [DOI] [PubMed] [Google Scholar]
- 10.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Xiong W, et al. E2-mediated EMT by activation of β-catenin/Snail signalling during the development of ovarian endometriosis. J. Cell. Mol. Med. 2019;23:8035–8045. doi: 10.1111/jcmm.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen YJ, et al. Oestrogen-induced epithelial-mesenchymal transition of endometrial epithelial cells contributes to the development of adenomyosis. J. Pathol. 2010;222:261–270. doi: 10.1002/path.2761. [DOI] [PubMed] [Google Scholar]
- 13.Furuya M, et al. ZEB1 expression is a potential indicator of invasive endometriosis. Acta Obstet. Gynecol. Scand. 2017;96:1128–1135. doi: 10.1111/aogs.13179. [DOI] [PubMed] [Google Scholar]
- 14.Yu MM, Zhou QM. 3,6-dihydroxyflavone suppresses the epithelial-mesenchymal transition migration and invasion in endometrial stromal cells by inhibiting the Notch signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018;22:4009–4017. doi: 10.26355/eurrev_201806_15287. [DOI] [PubMed] [Google Scholar]
- 15.Shen M, Liu X, Zhang H, Guo SW. Transforming growth factor β1 signaling coincides with epithelial-mesenchymal transition and fibroblast-to-myofibroblast trans-differentiation in the development of adenomyosis in mice. Hum. Reprod. 2016;31:355–369. doi: 10.1093/humrep/dev314. [DOI] [PubMed] [Google Scholar]
- 16.Xiong Y, et al. Hypoxia-inducible factor 1a induced epithelial- mesenchymal transition of endometrial epithelial cells may contribute to the development of endometriosis. Hum. Reprod. 2016;31:1327–1338. doi: 10.1093/humrep/dew081. [DOI] [PubMed] [Google Scholar]
- 17.Greene J, et al. Circular RNAs, biogenesis, function and role in human diseases. Front. Mol. Biosci. 2017;4:38. doi: 10.3389/fmolb.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lekka E, Hall J. Noncoding RNAs in disease. Febs. Lett. 2018;5:2884–2900. doi: 10.1002/1873-3468.13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu KS, Pan F, Mao XD, Liu C, Chen YJ. Biological functions of circular RNAs and their roles in occurrence of reproduction and gynecological diseases. Am. J. Transl. Res. 2019;11:1–15. [PMC free article] [PubMed] [Google Scholar]
- 20.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeck WR, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdollahzadeh R, et al. Competing endogenous RNA (ceRNA) cross-talk and language in ceRNA regulatory networks: a new look at hallmarks of breast cancer. J. Cell. Physiol. 2019;234:10080–10100. doi: 10.1002/jcp.27941. [DOI] [PubMed] [Google Scholar]
- 23.Liu L, et al. Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary. Pancreat. Dis. Int. 2019;18:580–586. doi: 10.1016/j.hbpd.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Huang H, et al. Circular RNA ciRS-7 triggers the migration and invasion of esophageal squamous cell carcinoma via miR-7/KLF4 and NF-κB signals. Cancer Biol. Ther. 2019;20:73–80. doi: 10.1080/15384047.2018.1507254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu YY, Zhang LY, Du WZ. Circular RNA circ-PVT1 contributes to paclitaxel resistance of gastric cancer cells through regulates ZEB1 expression by sponging miR-124-3p. Biosci. Rep. 2019;39:BSR20193045. doi: 10.1042/BSR20193045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su Y, Xu C, Liu Y, Hu Y, Wu H. Circular RNA has-circ-0001649 inhibits hepatocellular carcinoma progression via multiple miRNAs sponge. Aging. 2019;11:3362–3375. doi: 10.18632/aging.101988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang DD, Luo YJ, Wang GW, Yang Q. Circular RNA expression profiles and bioinformatics analysis in ovarian endometriosis. Mol. Genet. Genom. Med. 2019;7:e756. doi: 10.1002/mgg3.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang W, et al. circRNA circAF4 functions as an oncogene to regulate MLL-AF4 fusion protein expression and inhibit MLL leukemia progression. J. Hematol. Oncol. 2019;12:103. doi: 10.1186/s13045-019-0800-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang XY, et al. CircRNA-100338 is associated with mTOR signaling pathway and poor prognosis in hepatocellular carcinoma. Front. Oncol. 2019;9:392. doi: 10.3389/fonc.2019.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou B, Xu HY. A novel identified circular RNA, circRNA-010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-β1. Biochem. Biophys. Res. Commun. 2017;487:769–775. doi: 10.1016/j.bbrc.2017.04.044. [DOI] [PubMed] [Google Scholar]
- 31.Gong YH, et al. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell. Int. 2018;18:116. doi: 10.1186/s12935-018-0602-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohlsson TEM, et al. MicroRNA-regulated pathways associated with endometriosis. Mol. Endocrinol. 2009;23:265–275. doi: 10.1210/me.2008-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nothnick WB. MicroRNAs and endometriosis, distinguishing drivers from passengers in disease pathogenesis. Semin. Reprod. Med. 2017;35:173–180. doi: 10.1055/s-0037-1599089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saare M, et al. Challenges in endometriosis miRNA studies: from tissue heterogeneity to disease specific miRNAs. Biochim. Biophys. Acta. 2017;1863:2282–2292. doi: 10.1016/j.bbadis.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 35.Zhao L, et al. Integration analysis of microRNA and mRNA paired expression profiling identifies deregulated microRNA-transcription factor-gene regulatory networks in ovarian endometriosis. Reprod. Biol. Endocrinol. 2018;16:4. doi: 10.1186/s12958-017-0319-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rekker K, et al. Differentially-expressed miRNAs in ectopic stromal cells contribute to endometriosis development: the plausible role of miR-139-5p and miR-375. Int. J. Mol. Sci. 2018;19:E3789. doi: 10.3390/ijms19123789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhardwaj M, et al. miRNA-200c and miRNA-141 as potential prognostic biomarkers and regulators of epithelial-mesenchymal transition in eyelid sebaceous gland carcinoma. Br. J. Ophthalmol. 2017;101:536–542. doi: 10.1136/bjophthalmol-2016-309460. [DOI] [PubMed] [Google Scholar]
- 38.Rekker K, et al. Circulating miR-200-family micro-RNAs have altered plasma levels in patients with endometriosis and vary with blood collection time. Fertil. Steril. 2015;104:938–946.e2. doi: 10.1016/j.fertnstert.2015.06.029. [DOI] [PubMed] [Google Scholar]
- 39.Li S, et al. Circular RNA 101368/miR-200a axis modulates the migration of hepatocellular carcinoma through HMGB1/RAGE signaling. Cell. Cycle. 2018;217:2349–2359. doi: 10.1080/15384101.2018.1526599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu DM, et al. LncRNA SNHG15 acts as a ceRNA to regulate YAP1-Hippo signaling pathway by sponging miR-200a-3p in papillary thyroid carcinoma. Cell. Death. Dis. 2018;9:947. doi: 10.1038/s41419-018-0975-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat. Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu MX, et al. YAP1 contributes to NSCLC invasion and migration by promoting Slug transcription via the transcription co-factor TEAD. Cell. Death. Dis. 2018;9:464. doi: 10.1038/s41419-018-0515-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu GL, et al. COX-2 forms regulatory Loop with YAP to promote proliferation and tumorigenesis of hepatocellular carcinoma cells. Neoplasia. 2018;20:324–334. doi: 10.1016/j.neo.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song Y, et al. Activated Hippo/Yes-associated protein pathway promotes cell proliferation and anti-apoptosis in endometrial stromal cells of endometriosis. J. Clin. Endocrinol. Metab. 2016;101:1552–1561. doi: 10.1210/jc.2016-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.