Abstract
To provide insights into the biology of opioid dependence (OD) and opioid use (i.e., exposure, OE), we completed a genome-wide analysis comparing 4,503 OD cases, 4,173 opioid-exposed controls, and 32,500 opioid-unexposed controls, including participants of European and African descent (EUR and AFR, respectively). Among the variants identified, rs9291211 was associated with OE (exposed vs. unexposed controls; EUR z=−5.39, p=7.2×10−8). This variant regulates the transcriptomic profiles of SLC30A9 and BEND4 in multiple brain tissues and was previously associated with depression, alcohol consumption, and neuroticism. A phenome-wide scan of rs9291211 in the UK Biobank (N>360,000) found association of this variant with propensity to use dietary supplements (p=1.68×10−8). With respect to the same OE phenotype in the gene-based analysis, we identified SDCCAG8 (EUR+AFR z=4.69, p=10−6), which was previously associated with educational attainment, risk-taking behaviors, and schizophrenia. In addition, rs201123820 showed a genome-wide significant difference between OD cases and unexposed controls (AFR z=5.55, p=2.9×10−8) and a significant association with musculoskeletal disorders in the UK Biobank (p=4.88×10−7). A polygenic risk score (PRS) based on a GWAS of risk-tolerance (N=466,571) was positively associated with OD (OD vs. unexposed controls, p=8.1×10−5; OD cases vs. exposed controls, p=0.054) and OE (exposed vs. unexposed controls, p=3.6×10−5). A PRS based on a GWAS of neuroticism (N=390,278) was positively associated with OD (OD vs. unexposed controls, p=3.2×10−5; OD vs. exposed controls, p=0.002) but not with OE (p=0.67). Our analyses highlight the difference between dependence and exposure and the importance of considering the definition of controls in studies of addiction.
Keywords: Opioids, Substance, Abuse, Genetics, Genome-wide association study
INTRODUCTION
The prevalence of opioid dependence (OD) is at epidemic levels and significantly affects public health and social and economic well-being. The use of opioid medications for analgesia is common, and opioids are considered a gold standard for pain control. However, they are also highly addictive, and are, along with heroin1, the leading contributors to the ongoing epidemic of opioid misuse and the high rate of fatal overdoses2–4.
Understanding the biology of human responses to opioids may lead to effective preventive strategies and treatments to reduce OD and its harmful consequences. Human genetic research has the potential to dissect the basis of inter-individual variability in the response to opioid exposure (i.e., whether an individual develops dependence on opioids). Genome-wide association studies (GWAS) of large cohorts have identified a number of risk loci and molecular pathways involved in the predisposition to numerous psychiatric disorders and behavioral traits5, 6. Previous OD GWAS included up to 10,000 participants and identified genome-wide significant (GWS) associations in KCNG2, KCNC1, APBB2, CNIH3, and RGMA7–10. However, there was no consistency across the previous OD GWAS with respect to the individual GWS loci, probably due to the limited statistical power and differences in case and control definitions in the context of polygenic architecture (thousands of causal loci with small effect).
Another key potential contributor to the lack of consistency in findings from prior GWAS is that different study designs were used. The most relevant design variation is related to the assessment of opioid exposure in controls. Two different control definitions have been considered: i) individuals exposed to opioids (OE) medically or illegally who did not develop OD; or ii) individuals without an OD diagnosis who were not assessed for opioid exposure. Although including individuals not exposed to opioids in the control group increases the overall sample size, it also potentially adds noise by including individuals who would have been likely to become OD if exposed, given the highly addictive nature of opioid drugs. Furthermore, exposure to opioids is a behavioral trait per se, and likely to be associated with its own specific genetic architecture, which may be different between licit and illicit exposure. Opioid use is rarer than the use of many other substances and it is often observed in individuals affected by severe mental and physical illnesses11, 12. Comparisons of OD cases with predominantly unexposed controls is likely to confound genetic risk for exposure to opioids with genetic factors specific to the transition to OD. Indeed, at least one prior smaller GWAS7 found that comparisons of OD cases to controls with significant exposure and from similar neighborhoods resulted in a GWS finding while comparisons with general population controls did not identify any GWS variants.
We leveraged genotypic and phenotypic information from 41,176 participants from 11 studies that are part of the Psychiatric Genomics Consortium Substance Use Disorder working group (PGC-SUD) to investigate genetic differences between OD cases (n=4,503), OE controls (n=4,173), and opioid-unexposed (OU) controls (n=32,500) using GWAS and polygenic risk score (PRS) analyses. In addition to identifying loci related to OD and OE, we also examined whether OD and OE could be differentiated with respect to their relationship with genetic liability to risk-taking behaviors and negative personality features (i.e., neuroticism), to provide further insights into the genetic architecture underlying opioid use and misuse.
MATERIALS AND METHODS
Study Design
This study leveraged the individual genotypic and phenotypic data available from the cohorts participating in the PGC-SUD workgroup. There is growing support for the idea that the genetic architecture of substance exposure is different from that of substance dependence13, 14. Based on these previous findings, we hypothesized that OD and OE are biologically different and therefore focused the present opioid study on three association tests: i) difference between opioid dependent (DSM-IV) and opioid exposed; ii) difference between opioid dependent and opioid unexposed, and iii) difference between opioid exposed and opioid unexposed. The results of these analyses provided information regarding the genetic differences between OD and OE.
Cohorts and Phenotype Definitions
Of the 11 studies from the PGC-SUD workgroup, seven were case-control studies and four were family-based studies (Supplementary Table 1; Supplementary Methods). Lifetime OD diagnoses was based on DSM-IV OD criteria15 and were derived either from clinician ratings or semi-structured interviews. The two control groups included OE controls (individuals without a lifetime OD diagnosis who were exposed to opioids at least once) and OU controls (individuals with no lifetime OD diagnosis who were not exposed to opioids). Lifetime opioid exposure included both licit, prescribed opioids and those used outside appropriate medical care. Some, but not all, studies distinguished between these forms of exposure. This study, which involved the analysis of de-identified data, was approved by the institutional review board (IRB) at Yale University School of Medicine and was conducted in accordance with all relevant ethical regulations. Each contributing study obtained informed consent from participants and ethics approvals for their study protocols from their respective review boards in accordance with applicable regulations.
Quality control and imputation
Individual genotype information was available for each subject. The Ricopili pipeline16 (https://github.com/Nealelab/ricopili) was used for the QC and imputation of the case-control cohorts. Most family-based cohorts were analyzed with the Picopili pipeline17 (https://github.com/Nealelab/picopili), which is designed to conduct genome-wide meta-analyses accounting for family structure. The genetic data from the Collaborative Studies on the Genetics of Alcoholism were imputed independently as previously described18 because of the need in that study to merge data on members of large multiplex families who were genotyped across multiple genotyping arrays.
Details regarding the QC criteria were reported previously17. Briefly, after initial sample and variant QC, population outlier samples were excluded, and each retained individual was assigned to a specific ancestry on the basis of the principal components derived from genome-wide data. The 1000 Genomes Project Phase 3 reference panel19 was used as a reference for the ancestry assignment. Based on genetic information, we identified 9,591 and 31,585 individuals of African and European descent, respectively. Other ancestry groups were not investigated due to the limited number of informative subjects. The final QC criteria included variant filters for call rate, heterozygosity, and departure from Hardy–Weinberg equilibrium expectations (HWE), performed within each ancestry group in each cohort stratified by genotyping array. We also used sample QC filters for cryptic relatedness and for departures from reported pedigree structures. Imputation was performed using SHAPEIT220 and IMPUTE221, and the 1000 Genomes Project Phase 3 reference panel, which includes five continental groups19. High-quality imputed SNPs were retained for the association analysis, filtering for imputation INFO score > 0.8 and minor allele frequency (MAF) > 0.01 before analysis. After imputation, we tested for duplicated samples and cryptic relatedness among the cohorts analyzed. The association analysis was conducted considering variants present in at least 80% of the cohorts investigated (Supplementary Table 2).
Data analysis
The association analysis was conducted stratifying each cohort by ancestry (i.e., African and European ancestries) and genotyping array. For case–control studies, imputed dosages were entered in a logistic regression. For family-based studies, logistic mixed models were used to analyze hard-called best-guess genotypes. The association analyses were adjusted for sex and the within-ancestry top 10 principal components to account for possible confounding by population stratification. To investigate differences between OE and OD, three phenotype definitions were considered: i) OD cases vs. OE controls (ODexposed; n=4,503 and 4,173, respectively); ii) OD cases vs. OU controls (ODunexposed; n=4,238 and 17,700, respectively; iii) OE controls vs. OU controls (OEcontrols; n=4,173 and 32,500). As explained in the Supplementary Methods, we removed some of the cohorts from the ODunexposed meta-analysis due to the deflation (λGC<0.9) caused by the low number of cases and the small case-control ratio. For each phenotype, meta-analyses of the results across the different cohorts were conducted in METAL with weights proportional to the square-root of the sample size for each study22. The effective sample size of each cohort was calculated based on the case-control ratio and the relatedness matrix. Ancestry-specific (African-ancestry and European-ancestry) and trans-ancestry meta-analyses were conducted. Heterogeneity was evaluated across all cohorts and between study designs.
To investigate the loci identified in the individual GWAS further, we performed a phenome-wide scan considering 4,082 traits assessed in up to 361,194 participants from the UK Biobank using previously generated GWAS association summary data23. Details regarding QC criteria and GWAS methods of this previous analysis are available at https://github.com/Nealelab/UK_Biobank_GWAS/tree/master/imputed-v2-gwas. Briefly, the association analyses for all phenotypes were conducted using regression models available in Hail (available at https://github.com/hail-is/hail) including the first 20 ancestry principal components, sex, age, age2, sex×age, and sex×age2 as covariates. We applied a false discovery rate (FDR) multiple testing correction (q<0.05) to account for the number of variants and phenotypes tested. Additionally, we investigated the associations of the loci we identified here with respect to 27 non-duplicated traits related to mental and behavioral disorders attributable to use of alcohol, cannabis, and tobacco (Supplementary Table 3). This information was derived from large-scale summary association data collected by the GWAS Atlas (available at https://atlas.ctglab.nl/)24. We also conducted a gene-based phenome-wide scan across 4,756 available datasets in the GWAS Atlas. A Bonferroni correction accounting for the number of traits tested was applied to this gene-based analysis (p<1.05×10−5).
Linkage disequilibrium (LD) score regression25 was performed to estimate the heritability explained by common SNPs (h2g) in the European-ancestry meta-analysis of case-control and family-based cohorts. The inclusion of related subjects may affect the LD score regression results due to the residual effect of family structure on the summary association data. To limit this potential confounder, the analyses was limited to variants assessed in more than 80% of the total sample and considering the effective sample size adjusted for both case-control ratio and family structure. The heritability analysis was not conducted on African-specific and trans-ancestry meta-analyses, because LD score regression is not suitable when analyzing GWAS summary data derived from admixed populations25. LD score regression analysis was performed considering HapMap3 SNPs26 and LD scores computed from the 1000 Genomes Project reference for European populations. Conversion of h2g estimates from observed scale to liability scale was performed accounting for the difference between population prevalence (ODexposed=1%, ODunexposed=1%, and OEcontrols=5%) and sample prevalence (ODexposed=55%, ODunexposed=22%, and OEcontrols=12%).
Gene-based association, enrichment analysis for molecular pathways, Gene Ontologies (GO) annotations and tissue-specific transcriptomic profiles were conducted using the MAGMA tool27 implemented in the FUMA platform28. Information regarding molecular pathways and GO annotations was derived from MsigDB v6.229. Tissue-specific transcriptomic profiles were derived from GTEx V730 and BrainSpan31. A Bonferroni multiple testing correction was used to control for the number of tests conducted in each enrichment analysis. GTEx data were also used to verify whether the GWS loci identified affect the transcriptomic regulation of the surrounding genes. To evaluate the effect across multiple tissues, we considered multi-tissue expression quantitative trait locus (eQTL) data. These were calculated using Meta-Tissue 32. This meta-analytic approach calculates a posterior probability (m value) that an effect exists in each of the tissues tested assuming that the eQTL effect is consistent across the affected tissues. M values>0.9 indicate that the tissue was predicted to show the eQTL association.
A PRS analysis was conducted to test the genetic overlap with behavioral traits that could differentiate between OD and OE status using the PRSice software33. Risk-taking and neuroticism were selected as we expected they would capture genetic susceptibility to early versus later stages of opioid use and misuse. For polygenic profile scoring, we used summary statistics generated from large-scale GWAS of risk tolerance (N= 466,571)34 and neuroticism (N=390,278)35. We considered multiple association P value thresholds (PT < 5×10−8, 10−7, 10−6, 10−5, 10−4, 0.001, 0.05, 0.1, 0.3, 0.5, 1) for SNP inclusion to identify the best-fit for each target phenotype tested. The PRS were calculated after using P-value-informed clumping with a LD cut-off of R2 = 0.3 within a 500 kb window and excluding the major histocompatibility complex region of the genome because of its complex LD structure. The PRS were calculated considering unrelated subjects of European descent available in both case-control and family-based cohorts (ODexposed Neffective=3,038; ODunexposed Neffective=4,728; and OEcontrols Neffective=5,376). The PRS were fitted in regression models with adjustments for sex and the top 10 within-ancestry principal components. We applied FDR multiple testing correction (q<0.05) to correct for the number of thresholds tested.
RESULTS
SNP-Heritability Estimates Comparing OD and OE Traits
The GWAS meta-analyses of ODexposed, ODunexposed, and OEcontrols phenotypes included up to 4,503 OD cases (African-ancestry=1,231; European-ancestry=3,272), 4,173 OE controls (African-ancestry=1,297; European-ancestry=2,876), and 32,500 OU controls (African-ancestry=7,063; European-ancestry=25,437). Significant SNP-heritability was observed for ODunexposed (liability-scale h2g=0.28, SE=0.1; population prevalence=0.01, sample prevalence=0.22), but not for ODexposed (liability-scale h2g=−0.08, SE=0.08; population prevalence=0.01, sample prevalence=0.55) and OEcontrols (liability-scale h2g=0.05, SE=0.1; population prevalence=0.05, sample prevalence=0.12). Moderate genome-wide inflation was observed in the European-specific meta-analyses, but genomic control using lambda or the LD score regression intercept did not affect the significance of any variants in the GWAS meta-analyses (Supplementary Table 4; Supplementary Methods).
Opioid Dependence vs. Exposed and Unexposed Controls
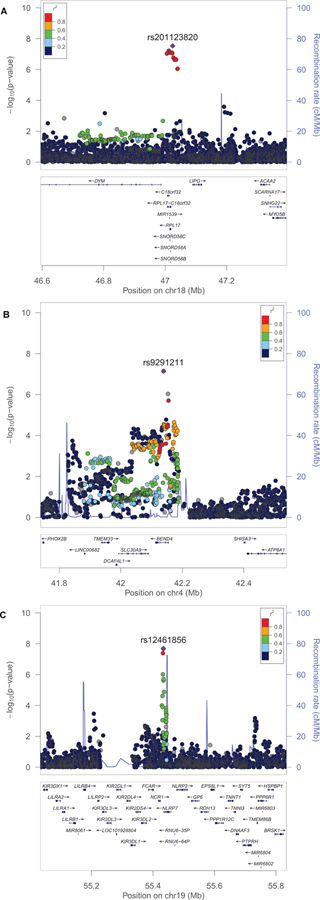
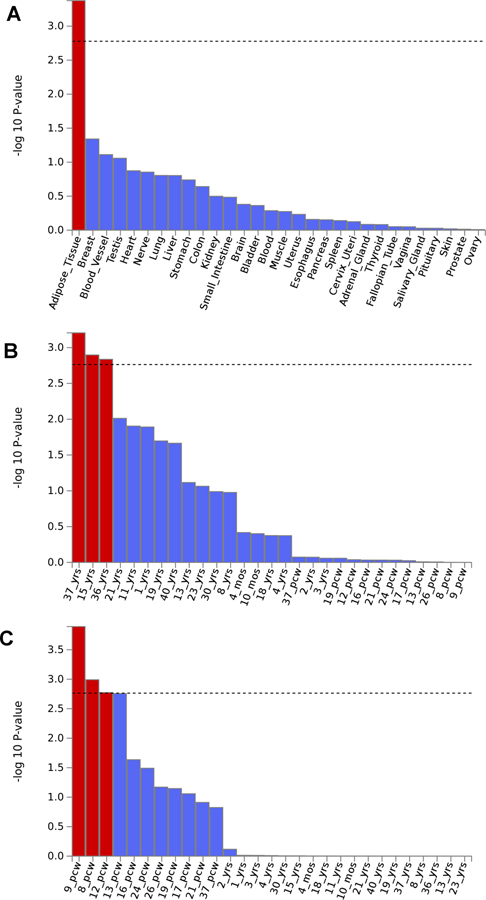
In the ODexposed analysis, which is the comparison most relevant to dependence liability given exposure but also the one that most constricted the sample size, no association survived the genome-wide significance threshold (p=5×10–8). Additionally, there were no significant enrichments for GO annotations, molecular pathways, nor tissue-specific regulation. The ODunexposed comparison identified a GWS association in the African-ancestry meta-analysis, rs201123820 on chromosome 18 (z=5.55, p=2.9×10–8; Figure 1A; Table 1; see Supplementary Table 5 for ancestry-specific results for each genome-wide significant variant). With respect to this locus, no heterogeneity was observed among the cohorts included in the meta-analysis (heterogeneity: I2=0, p=0.473; Supplementary Table 6). This variant did not show significant genetic associations with traits related to other addictive substances (Supplementary Table 3). The gene-based association analysis identified a GWS gene in the same genomic region, C18orf32 (p=1.8×10–6; Table 1; Supplementary Figure 1A). Additionally, in the African-ancestry meta-analysis, we also observed an enrichment for adipose tissue (beta=0.04, p=4.21×10–4; Figure 2A) and GO:0034498 – early endosome to Golgi transport (beta=1.01, p=5.1×10–8). In the trans-ancestry meta-analysis, we observed significant enrichment for specific adult stages of brain development (37 y; beta=0.06, p=6.22×10–4; 15 y; beta=0.06, p=0.001; 36yrs: beta=0.06, p=0.002; Figure 2B) and GO:0007143~female meiotic division (beta=0.73, p=1.08×10–7). In the European-ancestry ODunexposed GWAS meta-analysis, no result survived multiple testing correction.
Figure 1:
Regional Manhattan plots of the genetic association identified: rs201123820 in the African-ancestry ODunexposed GWAS meta-analysis (A); rs92911211 in the European-ancestry OEcontrols GWAS meta-analysis (B); rs12461856 in the trans-ancestry OEcontrols GWAS meta-analysis (C).
Table 1:
Top loci identified considering different OD and OE phenotypic definitions.
| Single-variant Associations | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Phenotype | Meta-analysis | rsid | Chromosome | Location (bp) | Effect Allele | Other Allele | Effect Allele Frequency | Z score | P value |
| ODunexposed | AFR | rs201123820 | 18 | 47025347 | T | TAAACAAAAACA | 0.019 | 5.547 | 2.90E-08 |
| OEcontrols | EUR | rs9291211 | 4 | 42139132 | A | G | 0.782 | −5.387 | 7.16E-08 |
| Trans-ancestry | rs12461856 | 19 | 55433852 | A | G | 0.8402 | −5.606 | 2.07E-08 | |
| Gene-based Associations | |||||||||
| Phenotype | Meta-analysis | Gene | Chromosome | Location-Start (bp) | Location-End (bp) | SNP (n) | Z score | P value | |
| ODunexposed | AFR | C18orf32 | 18 | 47008028 | 47013622 | 18 | 4.633 | 2E-06 | |
| OEcontrols | EUR | BEND4 | 4 | 42112955 | 42154895 | 81 | 4.756 | 1E-06 | |
| Trans-ancestry | SDCCAG8 | 1 | 243419320 | 243663394 | 407 | 4.686 | 1E-06 | ||
Figure 2:
Significant tissue enrichments identified in the African-ancestry ODunexposed meta-analysis (A); the trans-ancestry ODunexposed GWAS meta-analysis (B), and the European-ancestry OEcontrols GWAS meta-analysis (C).
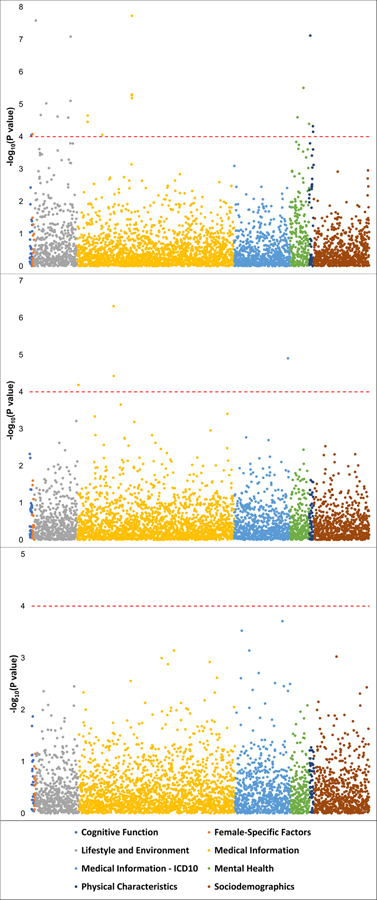
To extend the phenotypic breadth of our findings, we conducted a phenome-wide scan (4,082 traits tested; Supplementary Table 7) identified in 361,194 participants of European descent from the UK Biobank. Rs201123820 was identified in the African-ancestry ODunexposed GWAS meta-analysis. Although this variant was not significant in the European-ancestry meta-analysis (Supplementary Table 5), Rs201123820 has reasonably similar MAF in African and European populations (1000 Genomes Project: AFR MAF=0.024; EUR MAF=0.042). In UK Biobank cohort, we observed several associations for rs201123820 (FDR q<0.05; Figure 3, center panel; Supplementary Table 7) including postprocedural musculoskeletal disorders (UK Biobank Field ID: 41202 “Diagnoses – main ICD10 [M96]”, p=4.88×10−7), other disorders of the musculoskeletal system and connective tissue (UK Biobank Field ID: 41270 “Diagnoses - ICD10 [M13_MUSCULOSKELEOTH]”, p=1.25×10−5), postpartum care and examination (UK Biobank Field ID: 41202 “Diagnoses - main ICD10 [Z39]”, p=3.74×10−5), and auto-refraction measurements for eye prescription (UK Biobank Field ID: 5159 “3mm asymmetry index (right)”, p=6.54×10−5).
Figure 3:
Manhattan plot of the phenome-wide scan conducted in the UK Biobank with respect to rs12461856, rs201123820, and rs9291211 (bottom, center, and upper panels, respectively). As indicated in the legend, the phenotypic categories are color coded. Red dashed line indicates the significance threshold accounting for the number of variants and phenotypes tested (FDR q < 0.05).
Exposed vs. Unexposed Controls
In GWAS meta-analysis of OEcontrols in the European-ancestry cohort, we observed a gene-based association for the BEND4 locus that was GWS in the gene-based test (p=9.9×10−6; Table 1; Supplementary Figure 1B). In the BEND4 gene region, we identified a genetic association that nearly reached GWS: rs9291211 on chromosome 4 (z=−5.38, p=7.2×10–8; Figure 1B; Table 1). With respect to this locus, no heterogeneity was observed among the cohorts (heterogeneity: I2=0, p=0.879; Supplementary Table 6). This variant (or LD proxies in the same ancestry group) was identified in previous GWAS of behavioral traits: alcohol consumption (rs4501255, LD proxy r2=0.94, p=5×10–10)36; neuroticism (rs9291211, p=2×10–8)35; and helping behavior (rs2880666, LD-proxy r2=0.77, p=5×10–7)37. Additionally, rs9291211 is an eQTL for SLC30A9 and BEND4 in multiple tissues (GTEx multi-tissue eQTL p = 1.2×10−26 and 2.88×10−9, respectively). The rs9291211×SLC30A9 eQTL (i.e., rs9291211 regulating the SLC30A9 expression) showed a posterior probability>90% in seven brain tissues (amygdala m=0.99; anterior cingulate cortex m=1; caudate m=1; cortex m=0.99; hypothalamus m=1; nucleus accumbens m=1, putamen m=0.99; Supplementary Figure 2A). The rs9291211×BEND4 eQTL showed posterior probabilities>90% in two brain tissues (caudate m=0.9; cortex m=1; Supplementary Figure 2B). We found significant association of rs9291211 with additional traits from the GWAS Atlas24 that were related to addictive substances including “alcohol usually taken with meals” (p=3.02×10−8), “frequency of consuming six or more units of alcohol” (p=2.78×10−4), “alcohol intake frequency” (p=5.91×10−4), and “cigarettes per day” (p=0.004) (Supplementary Table 3). The European-ancestry OEcontrols meta-analysis also showed enrichment for several brain development stages: post-conception weeks 9 (beta=0.04, p=1.28×10−4), 8 (beta=0.032, p=0.001), and 12 (beta=0.04, p=0.002) (Figure 2C). No result in the association and the enrichment analyses based on the African-ancestry OEcontrols GWAS meta-analysis survived multiple testing correction.
Although rs9291211 showed only a “suggestive” GWS association with the OEcontrols phenotype, it was the strongest signal responsible for the significant BEND4 gene-based association. Accordingly, we tested its phenotypic spectrum in UK Biobank. This variant was associated with 22 phenotypes (FDR q<0.05; Figure 3, upper panel; Supplementary Table 7), which included dietary habits (e.g., UK Biobank Field ID: 6179 “Mineral and other dietary supplements [None of the above]”, p=1.68×10−8), anthropometric traits (e.g., UK Biobank Field ID: 1687 “Comparative body size at age 10”, p=7.58×10−8), behavioral traits (e.g., UK Biobank Field ID: 20127 “Neuroticism score”, p=3.12×10−6), physical outcomes (e.g., UK Biobank Field ID: 6152 “Hay fever, allergic rhinitis or eczema”, p=3.49×10−5), reproductive function (UK Biobank Field ID: 3581 Age at menopause [last menstrual period], p=8.40×10−5), and cognitive tests (e.g., UK Biobank Field ID: 404 “Reaction time [Duration to first press of snap-button in each round]”, p= 8.82×10−5).
Based on the gene-based and eQTL analyses, we also investigated the phenotypic spectrum of SLC30A9 and BEND4. In line with the shared effect of rs9291211, we observed associations surviving multiple-testing correction (p<1.05×10−5) in both gene-based phenome-wide scans (Supplementary Table 8). Among the 14 common associations, we observed: “alcohol usually taken with meals” (BEND4 p=6.5×10−11; SLC30A9 p=3.59×10−6), “neuroticism sum score” (BEND4 p=1.92×10−8; SLC30A9 p=3.63×10−7), and “depressive symptoms” (BEND4 p=6.24×10−7; SLC30A9 p=3.58×10−6).
In the trans-ancestry GWAS meta-analysis of OEcontrols, we observed an additional single-variant GWS association, rs12461856 on chromosome 19 (z=−5.61, p=2.1×10−8; Figure 1C; Table 1). With respect to this locus, no heterogeneity was observed among the cohorts included in the meta-analysis (heterogeneity: I2=0, p=0.554; Supplementary Table 6). This variant did not show significant genetic associations with traits related to other addictive substances (Supplementary Table 3). In the UK Biobank, no novel phenotypic associations with rs12461856 survived multiple testing correction (FDR q<0.05; Figure 3, bottom panel; Supplementary Table 7). A gene-based GWS association was identified for SDCCAG8 on chromosome 1 (p=1.4×10−6, Table 1; Supplementary Figure 1C). In the gene-based phenome-wide scan, we observed 77 traits associated with SDCCAG8 that survived multiple testing correction (p<1.05×10−5; Supplementary Table 8). Among them, we observed strong associations for schizophrenia (p=2.5×10−12) and risk taking (p=1.1×10−9). With respect to the molecular pathways, significant enrichments were also observed for GO:0017069~small RNA binding (beta=0.65, p=5.4×10−7) and a curated gene set related to genes downregulated 6 h after induction of HoxA5 expression in a breast cancer cell line (Standard name: CHEN_HOXA5_TARGETS_6HR_DN; beta=1.37, p=8.6×10−6).
Polygenic Risk Score Analysis
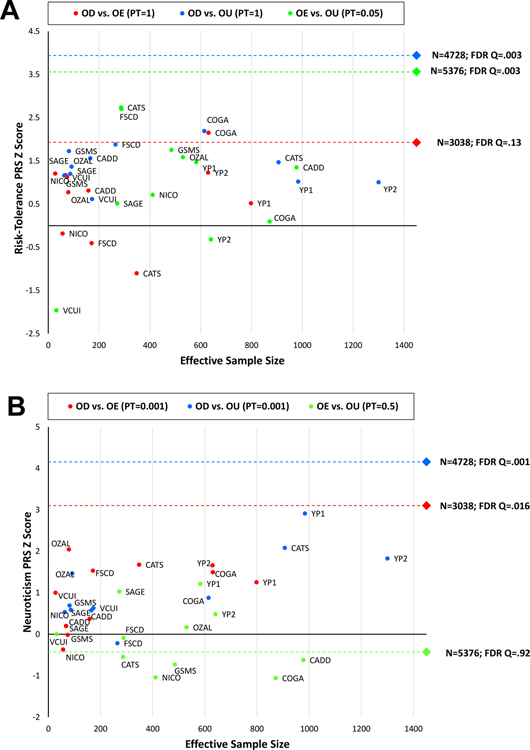
We also used PRS to compare the three opioid-related phenotypes – dependence (with exposed controls – ODexposed – and with unexposed controls – ODunexposed) and exposure in non-dependent individuals (OEcontrols). Similarly to other addictive substances, illicit opioid users would be expected to have greater propensity to risk-taking behaviors, impulsivity, and stress responsivity than unexposed subjects38. Accordingly, we derived a PRS from the large-scale GWAS (N=466,571) conducted by the Social Science Genetic Association Consortium (SSGAC) on risk tolerance, which was defined as the tendency, preparedness, or willingness to take risks in general34. The PRS analysis was conducted on European-ancestry subjects only due to the well-known lack of large-scale GWAS in other ancestry groups39. The risk-tolerance PRS was positively associated with OD when contrasted with unexposed controls (ODunexposed: Neffective=4,728, PT=1, z=3.94, p=8.1×10−5, FDR q=0.003), whereas OD contrasted with exposed controls displayed only a trend (p<0.1; ODexposed: Neffective=3,038, PT=1, z=1.93, p=0.054, FDR q=0.13). OE (OEcontrol: Neffective=5,376, PT=0.05, z=3.57, p=3.6×10−5, FDR q=0.003) was also significant for the risk-tolerance PRS.
We also tested PRS derived from a large-scale GWAS of neuroticism (N=390,278)35. This behavioral trait represents a tendency to negative affect and was previously observed to be genetically correlated with several psychiatric disorders, including SUDs17, 40, major depression41, and posttraumatic stress disorder42. Consistent with our expectation of genetic liability to negative affect being related to dependence but not exposure alone, the neuroticism PRS was associated with dependence compared with unexposed (ODunexposed: Neffective=4,728, PT=0.001, z=4.16, p=3.2×10−5, FDR q=5.76×10−4) and dependence compared with exposed controls (ODexposed: Neffective=3,038, PT=0.001, z=3.1, p=0.002, FDR q=0.016), but not with exposed vs unexposed controls (OEcontrols: Neffective=5,376, PT=0.5, z=−0.42, p=0.671, FDR q=0.919).
Figure 4 shows the association of risk-taking and neuroticism PRS (Panels A and B, respectively) across the cohorts included in the meta-analyses. We did not observe significant heterogeneity (Supplementary Table 9), which indicates that the meta-analytic results were driven by the sizes of the samples investigated and not by the different recruitment strategies.
Figure 4:
Relationship between PRS z scores (A: risk tolerance; B: neuroticism) and effective sample size across the opioid-related phenotypes tested. Each circle represents an individual cohort; the diamond represents the results from the meta-analysis with respect to the phenotypes tested.
DISCUSSION
We investigated the genetic architecture of opioid-related traits in informative cohorts. Our comparison of opioid dependence (OD) compared with unexposed (ODunexposed) and exposed (ODexposed) controls, as well as of opioid exposure among controls (OEcontrols) provides new insights into opioid addiction. We identified GWS loci and genes for ODunexposed and OEcontrols, and found these variants to be associated with health relevant traits in the UK Biobank. Most critically, our PRS analyses highlighted distinctions between exposed and unexposed controls, as well as the progression from exposure to dependence. Use and dependence are behaviors with different relationships to other genetically influenced traits, as has been shown for alcohol use vs. alcohol dependence13, 14. To our knowledge, no previous study investigated the specific genetic differences between OD and OE; our current findings provide the first insights based on genome-wide data into the molecular mechanisms by which OE and OD differ. The lack of sufficiently large numbers of OD cases and OE controls is a fundamental barrier to facilitating our understanding of the biological underpinnings of this serious public health epidemic, as it limited the power of what we would regard as the most informative comparison.
With respect to the single-variant associations observed, the strongest bioinformatics support from other studies was observed for rs9291211, identified in the European-ancestry GWAS meta-analysis of the OEcontrols phenotype. Although it reached only a suggestive GWS threshold (p=7.2×10–8), this variant was the leading signal in the significant BEND4 gene-based association and also showed strong regulatory effects on the brain-specific transcriptomic profile of BEND4 and SLC30A9. The function of BEND4 gene is unclear, but previous GWAS identified several variants at this locus (including rs9291211) that were associated with depression43, alcohol consumption36, autism spectrum disorder43, neuroticism35, height44, and helping behavior37. SLC30A9 encodes a zinc transporter involved in intracellular zinc homeostasis, which also plays a role in transcriptional activation of Wnt-responsive genes45. In previous GWAS, variants located in SLC30A9 gene (rs9291211 is located in BEND4 but also affects SLC30A9 gene expression) were associated with neuroticism44 and depression46. The phenome-wide scan of rs9291211 in the UK Biobank showed an effect of this SNP on a wide range of complex traits, some with an easy-to-conceptualize relationship to OD such as alcohol consumption, neuroticism, depression, and anxious feelings. Considering traits related to mental and behavioral disorders attributable to use of alcohol, cannabis, and tobacco, we observed that rs9291211 is associated with alcohol consumption and tobacco smoking and not with dependence-relevant phenotypes. This support that this variant may have pleiotropic effects across consumption of multiple substancs. In our phenome-wide analysis, the strongest results were observed with respect to dietary habits: rs9291211*A was positively associated with reduced OE risk in the PGC-SUD cohorts and with increased propensity to use dietary supplements, such as vitamin and mineral supplements in the UK Biobank. A recent GWAS identified several loci associated with dietary habits and indicated a causal relationship between educational attainment and healthy eating47. With respect to rs9291211, we also observed a nominally significant association with traits related to educational attainment (e.g., UK Biobank Field ID: 6138 Qualification [College or University degree], p=0.033). Accordingly, we hypothesize that rs9291211 could be involved in the individual variability to consume chemicals ranging from dietary supplements to opioids, independent from educational attainment.
In the trans-ancestry GWAS meta-analysis of the OEcontrols phenotype comparison, we identified GWS loci in the single-variant and the gene-based analyses. No external validations were observed for rs12461856 and further studies will be needed to confirm this finding. Conversely, SDCCAG8 identified in the gene-based analysis (but not related to any individual GWS variants) was shown in studies available in the GWAS catalog48 to have 49 single-variant associations with educational attainment49, blood-related parameters50, risk-taking behaviors34, anthropometric traits44, kidney function51, and schizophrenia52. The previous associations with behavioral traits support SDCCAG8 as potentially associated with behaviors that, in turn, associated with increased risk of OE.
With respect to the ODunexposed phenotype comparison, we identified rs201123820 in the African-ancestry meta-analysis. This is a non-coding deletion located 2 kb upstream of LOC101928144, an uncharacterized long intergenic non-protein coding RNA. The gene-based association analysis identified a GWS locus in the same region, C18orf32, a gene involved in the activation of the NF-kappaB and MAPK signaling pathways, which play a key role in immune and inflammatory responses53. The phenome-wide analysis in the UK Biobank, despite its being in a predominantly European cohort, showed a significant association of rs201123820 with physical conditions, particularly musculoskeletal disorders. This is particularly interesting as opioids are commonly prescribed for pain management in musculoskeletal disorders and early use is associated with prolonged work disability54, which may be related to the consequences of opioid abuse and/or the severity of the underlying disorder that required treatment. While these associations merit replication, this result highlights how human genetic research can also be relevant to improve pain management protocols.
The GWS risk loci identified by previous OD GWAS (e.g., KCNG2, CNIH3, and RGMA)7–10 were not concordant with the present investigation (Supplementary Table 10). Such discrepancies are not unexpected, given that these analyses were underpowered, and the reported findings are likely to be affected by phenotypic heterogeneity and the random variation allowing for discovery of alternate subsets of risk loci in small datasets55, 56.
The available genome-wide data also permitted us to compare the opioid-related phenotypes with respect to shared genetic risk of relevant behavioral traits. Although there is variability in the effective sample size and therefore statistical power of the phenotypes tested (ODunexposed Neffective=4,728; OEcontrols: Neffective=5,376; ODexposed Neffective=3,038), the PRS results showed an interesting pattern. The risk-tolerance PRS was positively associated with all three phenotypes with the strength of association mostly related to the effective sample size of the target sample. The association between OEcontrols and genetic liability to risk-taking highlights the importance of accounting for the genetic factors related to the individual differences in exposure when examining those contribution to dependence. Such a finding would support the hypothesis that the inclusion of exposed controls can “fine-tune” our ability to separate loci related to generalized risk-taking from those specific to repeated use that lead to opioid dependence. The neuroticism PRS showed positive associations with ODunexposed and ODexposed phenotype comparisons. Although it was non-significant, we observed a negative association of neuroticism PRS with the OEcontrols phenotype. This suggestive negative relationship parallels the rs9291211 result where the allele A was associated with reduced OE in the PGC GWAS meta-analysis and increased neuroticism score in the UK Biobank. Genetic liability to neuroticism may thus overlap with genetic liability to OD but not to OE.
Although several putative single-variant, gene-based, and PRS associations were identified based on the different OD and OE phenotypes, the sample size of the current investigation is still small, given the polygenic architecture of psychiatric disorders57. Novel studies specifically targeting SUDs and assessing opioid-related behaviors will be necessary to recruit cohorts informative for OD and OE GWAS. Another important limitation is the phenotypic heterogeneity within the opioid exposure sample, which included individual exposed to opioids via licit use (i.e., medical prescriptions) and illicit use. There may be important differences between these two subgroups (e.g., risk-taking may be more strongly associated with illicit exposure). However, several of the cohorts investigated lacked this information, and, due to the limited sample size, we were not able to make this comparison. In addition, this may have resulted in heterogeneity in the OU controls (i.e., those who reported not using opioids illicitly were unassessed for medical exposure). Future large opioid-informative datasets will be needed to determine whether illicit and licit opioid exposure have distinct effects on the molecular basis of opioid dependence. Finally, while the phenome-wide investigation in the UK Biobank provides encouraging support for the plausibility of our findings, it may reflect complex pleiotropic effects of these variants on multiple traits, including an unmeasured third variable. Replication of these association signals with opioid dependence and exposure phenotypes will be required.
In conclusion, we provide a comprehensive genome-wide investigation of opioid-related traits, highlighting different molecular mechanisms that could underlie exposure and dependence. These findings draw attention to challenges associated with the use of unexposed controls in genetic association studies for OD and potentially for other SUDs (where exposure is not widespread, as is the case for alcohol, or more recently marijuana). This information should be used to guide the next generation of human genetic studies of opioid-related behaviors.
Supplementary Material
ACKNOWLEDGMENTS
The Psychiatric Genomics Consortium Substance Use Disorders Working Group receives support from the National Institute on Drug Abuse and the National Institute of Mental Health via U01 MH109532 and U01 MH109528. We gratefully acknowledge prior support from the National Institute on Alcohol Abuse and Alcoholism. Statistical analyses for the PGC were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org) hosted by SURFsara and financially supported by the Netherlands Scientific Organization (NWO 480-05-003) along with a supplement from the Dutch Brain Foundation and the VU University Amsterdam.
AA acknowledges DA032573; ACH acknowledges support from NIH grants AA07535, AA07729, AA13320, AA13321, and AA11998; AEA acknowledges support from AA011408 and AA017828; AMG acknowledges support from U10 AA08401; BPR was supported by AA011408, AA017828 and AA022537; BTW acknowledges support from AA011408, AA017828 and AA022537; CJH acknowledges DA032555, DA035804, DA011015, and DA042755; DBH acknowledges support from R01DA036583; EJC acknowledges support from DA023026, DA011301, and DA024413; EOJ acknowledges support from R01 DA044014; HM acknowledges support from DA025109, DA024413, DA016977; JG acknowledges support from DA12690, DA047527; JKH acknowledges support from DA011015; KSK acknowledges support from AA011408, AA017828 and AA022537; LD is supported by an Australian National Health and Medical Research Council (NHMRC) Principal Research Fellowship; LJB acknowledges support from R01DA036583; LMH acknowledges support from AA011408 and AA017828; LMH acknowledges support from AA011408 and AA017828; MCS acknowledges support from DA035804; PAFM acknowledges funding support from NIH grants: DA012854 and R25DA027995; RAG acknowledges support from AA017444; REP is supported by NIH K01 grant MH113848; RP acknowledges support from DA12690 and DA047527; SAB acknowledges support from AA011408, AA017828, AA022537 and AA022717; SMH acknowledges support from R21AA024888 and K08DA032680; TBB acknowledges support from MH100549; TLW acknowledges support from R01 DA021905 and R01 DA035804; WEC acknowledges support from R01HD093651, R01DA036523, and P30DA023026, R01MH117559.
Alcohol Dependence in African Americans (ADAA) study was funded by NIH grant R01 AA017444. Funding support for the Comorbidity and Trauma Study (CATS) (dbGAP accession number: phs000277.v1.p1) was provided by the National Institute on Drug Abuse (R01 DA17305); GWAS genotyping services at the CIDR at The Johns Hopkins University were supported by the National Institutes of Health (contract N01-HG-65403). The data collection and analysis of the Center on Antisocial Drug Dependence (CADD) was supported by the following grants: DA011015, DA012845, DA021913, DA021905, DA032555, and DA035804. The Collaborative Study on the Genetics of Alcoholism (COGA) is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). Funding support for this GWAS genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the National Institute on Alcohol Abuse and Alcoholism, the NIH GEI (U01HG004438), and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). COGA Principal Investigators: B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut, includes eleven different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., T. Foroud); University of Iowa (S. Kuperman, J. Kramer); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, J. Rice, K. Bucholz, A. Agrawal); University of California at San Diego (M. Schuckit); Rutgers University (J. Tischfield, A. Brooks); Department of Biomedical and Health Informatics, The Children’s Hospital of Philadelphia; Department of Genetics, Perelman School of Medicine, University of Pennsylvania, Philadelphia PA (L. Almasy), Virginia Commonwealth University (D. Dick), Icahn School of Medicine at Mount Sinai (A. Goate), and Howard University (R. Taylor). Other COGA collaborators include: L. Bauer (University of Connecticut); J. McClintick, L. Wetherill, X. Xuei, Y. Liu, D. Lai, S. O’Connor, M. Plawecki, S. Lourens (Indiana University); G. Chan (University of Iowa; University of Connecticut); J. Meyers, D. Chorlian, C. Kamarajan, A. Pandey, J. Zhang (SUNY Downstate); J.-C. Wang, M. Kapoor, S. Bertelsen (Icahn School of Medicine at Mount Sinai); A. Anokhin, V. McCutcheon, S. Saccone (Washington University); J. Salvatore, F. Aliev, B. Cho (Virginia Commonwealth University); and Mark Kos (University of Texas Rio Grande Valley). A. Parsian are the NIAAA Staff Collaborators. M. Reilly was an NIAAA staff collaborator. We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, currently a consultant with COGA, P. Michael Conneally, Raymond Crowe, and Wendy Reich, for their critical contributions. The authors thank Kim Doheny and Elizabeth Pugh from CIDR and Justin Paschall from the NCBI dbGaP staff for valuable assistance with genotyping and quality control in developing the dataset available at dbGaP (phs000125.v1.p1; also: phs000763.v1.p1; phs000976.v1.p1). Support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI; U01 HG004422; dbGaP study accession phs000092.v1.p1]. SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center [U01 HG004446]. Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism [COGA; U10 AA008401], the Collaborative Genetic Study of Nicotine Dependence [COGEND; P01 CA089392; see also phs000404.v1.p1], and the Family Study of Cocaine Dependence [FSCD; R01 DA013423, R01 DA019963]. Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research (CIDR), was provided by the NIH GEI [U01HG004438], the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” [HHSN268200782096C]. The Gene–Environment Development Initiative: Great Smoky Mountains Study (phs000852.v1.p1) was supported by the National Institute on Drug Abuse (U01DA024413, R01DA11301), the National Institute of Mental Health (R01MH063970, R01MH063671, R01MH048085, K01MH093731 and K23MH080230), NARSAD, and the William T. Grant Foundation. We are grateful to all the GSMS and CCC study participants who contributed to this work. The following grants supported data collection and analysis of CADD: DA011015, DA012845, DA021913, DA021905, DA032555, and DA035804. Gene-Environment-Development Initiative -GEDI – Virginia Commonwealth University (VTSABD; dbGAP in progress) was supported by the National Institute on Drug Abuse (U01DA024413, R01DA025109), the National Institute of Mental Health (R01MH045268, R01MH055557, R01MH068521), and the Virginia Tobacco Settlement Foundation grant 8520012. We are grateful to all the VTSABD-YAFU-TSA study participants who contributed to this work. Yale-Penn (phs000425.v1.p1; phs000952.v1.p1) was supported by National Institutes of Health Grants RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, and R01 AA017535 and the Veterans Affairs Connecticut and Philadelphia Veterans Affairs Mental Illness Research, Educational, and Clinical Centers. Australian Alcohol and Nicotine studies (OZALC; phs000181.v1.p1) were supported by National Institutes of Health Grants AA07535,AA07728, AA13320, AA13321, AA14041, AA11998, AA17688,DA012854, and DA019951; by Grants from the Australian National Health and Medical Research Council (241944, 339462, 389927,389875, 389891, 389892, 389938, 442915, 442981, 496739, 552485,and 552498); by Grants from the Australian Research Council(A7960034, A79906588, A79801419, DP0770096, DP0212016, and DP0343921); and by the 5th Framework Programme (FP-5) GenomEUtwin Project (QLG2-CT-2002–01254). Genome-wide association study genotyping at Center for Inherited Disease Research was supported by a Grant to the late Richard Todd, M.D., Ph.D., former Principal Investigator of Grant AA13320.
Footnotes
Full list of Substance Use Disorder Working Group members appears in the Acknowledgments
CONFLICT OF INTEREST
Dr. Kranzler has been an advisory board member, consultant, or continuing medical education speaker for Indivior, Lundbeck, and Otsuka. He is a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which is sponsored by AbbVie, Alkermes, Ethypharm, Indivior, Lilly, Lundbeck, Pfizer, and Xenoport. Drs. Kranzler and Gelernter are named as inventors on PCT patent application #15/878,640 entitled: “Genotype-guided dosing of opioid agonists,” filed January 24, 2018. Drs. Beirut and Goate are listed as inventors on Issued U.S. Patent 8080,371, “Markers for Addiction” covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction. The spouse of Dr. Saccone is listed as an inventor on Issued U.S. Patent 8,080,371,“Markers for Addiction” covering the use of certain SNPs in determining the diagnosis, prognosis, and treatment of addiction. The other authors do not report any conflict of interest.
REFERENCES
- 1.Cicero TJ, Ellis MS. Nonmedical Prescription-Opioid Use and Heroin Use. N Engl J Med 2016; 374(13): 1295–1296. [DOI] [PubMed] [Google Scholar]
- 2.Volkow ND, Frieden TR, Hyde PS, Cha SS. Medication-assisted therapies--tackling the opioid-overdose epidemic. N Engl J Med 2014; 370(22): 2063–2066. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control Prevention. CDC grand rounds: prescription drug overdoses-a U.S. epidemic. MMWR Morb Mortal Wkly Rep 2012; 61(1): 10–13. [PubMed] [Google Scholar]
- 4.Rudd RA, Aleshire N, Zibbell JE, Gladden RM. Increases in Drug and Opioid Overdose Deaths--United States, 2000–2014. MMWR Morb Mortal Wkly Rep 2016; 64(50–51): 1378–1382. [DOI] [PubMed] [Google Scholar]
- 5.Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA et al. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am J Hum Genet 2017; 101(1): 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Borglum AD, Breen G et al. Psychiatric Genomics: An Update and an Agenda. Am J Psychiatry 2018; 175(1): 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson EC, Agrawal A, Heath AC, Bogdan R, Sherva R, Zhang B et al. Evidence of CNIH3 involvement in opioid dependence. Mol Psychiatry 2016; 21(5): 608–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelernter J, Kranzler HR, Sherva R, Koesterer R, Almasy L, Zhao H et al. Genome-wide association study of opioid dependence: multiple associations mapped to calcium and potassium pathways. Biol Psychiatry 2014; 76(1): 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Z, Zhou H, Sherva R, Farrer LA, Kranzler HR, Gelernter J. Genome-wide Association Study Identifies a Regulatory Variant of RGMA Associated With Opioid Dependence in European Americans. Biol Psychiatry 2018; 84(10): 762–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalsi G, Euesden J, Coleman JR, Ducci F, Aliev F, Newhouse SJ et al. Genome-Wide Association of Heroin Dependence in Han Chinese. PLoS One 2016; 11(12): e0167388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly TM, Daley DC, Douaihy AB. Treatment of substance abusing patients with comorbid psychiatric disorders. Addict Behav 2012; 37(1): 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark RE, Baxter JD, Aweh G, O’Connell E, Fisher WH, Barton BA. Risk Factors for Relapse and Higher Costs Among Medicaid Members with Opioid Dependence or Abuse: Opioid Agonists, Comorbidities, and Treatment History. J Subst Abuse Treat 2015; 57: 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL, andMe Research Team tSUDWGotPGC, Adams MJ et al. Genome-Wide Association Study Meta-Analysis of the Alcohol Use Disorders Identification Test (AUDIT) in Two Population-Based Cohorts. Am J Psychiatry 2019; 176(2): 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun 2019; 10(1): 1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition, Text Revision. APA Publishing: Washington DC, USA, 2000. [Google Scholar]
- 16.Lam M, Awasthi S, Watson HJ, Goldstein J, Panagiotaropoulou G, Trubetskoy V et al. RICOPILI: Rapid Imputation for COnsortias PIpeLIne. Bioinformatics 2019. [DOI] [PMC free article] [PubMed]
- 17.Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci 2018; 21(12): 1656–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai D, Wetherill L, Bertelsen S, Carey CE, Kamarajan C, Kapoor M et al. Genome-wide association studies of alcohol dependence, DSM-IV criterion count and individual criteria. Genes Brain Behav 2019; 18(6): e12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium. GP, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM et al. A global reference for human genetic variation. Nature 2015; 526(7571): 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Connell J, Gurdasani D, Delaneau O, Pirastu N, Ulivi S, Cocca M et al. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet 2014; 10(4): e1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009; 5(6): e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26(17): 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015; 12(3): e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe K, Stringer S, Frei O, Umicevic Mirkov M, de Leeuw C, Polderman TJC et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet 2019; 51(9): 1339–1348. [DOI] [PubMed] [Google Scholar]
- 25.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics C et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015; 47(3): 291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.International HapMap Consortium, Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA et al. Integrating common and rare genetic variation in diverse human populations. Nature 2010; 467(7311): 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015; 11(4): e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017; 8(1): 1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011; 27(12): 1739–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Consortium GTEx. Genetic effects on gene expression across human tissues. Nature 2017; 550(7675): 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci 2014; 17(10): 1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sul JH, Han B, Ye C, Choi T, Eskin E. Effectively identifying eQTLs from multiple tissues by combining mixed model and meta-analytic approaches. PLoS Genet 2013; 9(6): e1003491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Euesden J, Lewis CM, O’Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics 2015; 31(9): 1466–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karlsson Linner R, Biroli P, Kong E, Meddens SFW, Wedow R, Fontana MA et al. Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet 2019; 51(2): 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagel M, Jansen PR, Stringer S, Watanabe K, de Leeuw CA, Bryois J et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat Genet 2018; 50(7): 920–927. [DOI] [PubMed] [Google Scholar]
- 36.Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet 2019; 51(2): 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Primes G, Fieder M. Real-life helping behaviours in North America: A genome-wide association approach. PLoS One 2018; 13(1): e0190950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kreek MJ, Nielsen DA, Butelman ER, LaForge KS. Genetic influences on impulsivity, risk taking, stress responsivity and vulnerability to drug abuse and addiction. Nat Neurosci 2005; 8(11): 1450–1457. [DOI] [PubMed] [Google Scholar]
- 39.Sirugo G, Williams SM, Tishkoff SA. The Missing Diversity in Human Genetic Studies. Cell 2019; 177(1): 26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pasman JA, Verweij KJH, Gerring Z, Stringer S, Sanchez-Roige S, Treur JL et al. GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat Neurosci 2018; 21(9): 1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howard DM, Adams MJ, Clarke TK, Hafferty JD, Gibson J, Shirali M et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci 2019; 22(3): 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gelernter J, Sun N, Polimanti R, Pietrzak RH, Levey D, Bryois J et al. Genome-wide association study of post-traumatic stress disorder reexperiencing symptoms in >165,000 US veterans. Nature Neuro 2019. [DOI] [PMC free article] [PubMed]
- 43.Grove J, Ripke S, Als TD, Mattheisen M, Walters RK, Won H et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 2019; 51(3): 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kichaev G, Bhatia G, Loh PR, Gazal S, Burch K, Freund MK et al. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. Am J Hum Genet 2019; 104(1): 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez Y, Shorer Z, Liani-Leibson K, Chabosseau P, Kadir R, Volodarsky M et al. SLC30A9 mutation affecting intracellular zinc homeostasis causes a novel cerebro-renal syndrome. Brain 2017; 140(4): 928–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 2018; 50(5): 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cole JB, Florez JC, Hirschhorn JN. Comprehensive genomic analysis of dietary habits in UK Biobank identifies hundreds of genetic loci and establishes causal relationships between educational attainment and healthy eating. bioRxiv 2019: 662239.
- 48.MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 2017; 45(D1): D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet 2018; 50(8): 1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoffmann TJ, Ehret GB, Nandakumar P, Ranatunga D, Schaefer C, Kwok PY et al. Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat Genet 2017; 49(1): 54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yasukochi Y, Sakuma J, Takeuchi I, Kato K, Oguri M, Fujimaki T et al. Identification of CDC42BPG as a novel susceptibility locus for hyperuricemia in a Japanese population. Mol Genet Genomics 2018; 293(2): 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511(7510): 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsuda A, Suzuki Y, Honda G, Muramatsu S, Matsuzaki O, Nagano Y et al. Large-scale identification and characterization of human genes that activate NF-kappaB and MAPK signaling pathways. Oncogene 2003; 22(21): 3307–3318. [DOI] [PubMed] [Google Scholar]
- 54.Busse JW, Craigie S, Sadeghirad B, Couban R, Hong P, Oparin Y et al. Management of acute musculoskeletal pain (excluding low back pain): protocol for a systematic review and network meta-analysis of randomised trials. BMJ Open 2019; 9(4): e024441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang W, Yu W. Power estimation and sample size determination for replication studies of genome-wide association studies. BMC Genomics 2016; 17 Suppl 1: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zou J, Zhou J, Faller S, Brown R, Eskin E. Accurate modeling of replication rates in genome-wide association studies by accounting for winner’s curse and study-specific heterogeneity. bioRxiv 2019: 856898. [DOI] [PMC free article] [PubMed]
- 57.Sullivan PF, Geschwind DH. Defining the Genetic, Genomic, Cellular, and Diagnostic Architectures of Psychiatric Disorders. Cell 2019; 177(1): 162–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.