Abstract
Fast excitatory synaptic transmission in the mammalian brain is largely mediated by AMPA-type ionotropic glutamate receptors (AMPARs), which are activated by the neurotransmitter glutamate. In synapses, AMPAR function is tuned by their auxiliary subunits, a diverse set of membrane proteins associated with the core pore forming subunits of AMPARs. Each auxiliary subunit provides distinct functional modulation of AMPARs, ranging from trafficking regulation to shaping ion channel gating kinetics. Understanding the molecular mechanism for the function of these complexes is key to decoding synaptic modulation and their global roles in cognitive activities, such as learning and memory. Here we review the structural and molecular complexity of AMPAR-auxiliary subunit complexes, as well as their functional diversity in different brain regions. We suggest that the recent structural information provides new insights into the molecular mechanisms underlying synaptic functions of AMPAR-auxiliary subunit complexes.
Keywords: synaptic transmission, synaptic plasticity, ionotropic glutamate receptors, AMPA receptors, AMPA type glutamate receptors, auxiliary subunits, TARP, stargazin, GSG1L, cornichon, CKAMP44, Shisa, SynDIG, cryo-electron microscopy, structural biology, electrophysiology, ion channel, ion channel gating modulation
Graphical Abstract
Native AMPAR complexes are composed of core GluA subunits and various structurally unrelated auxiliary subunits. In recent years, the cryo-EM structures of the receptor complexes are producing new insights into their function as well as their roles at the synapses. PDB models 5WEO, 5WEL, 6PEQ, 6U6I were used.
Introduction
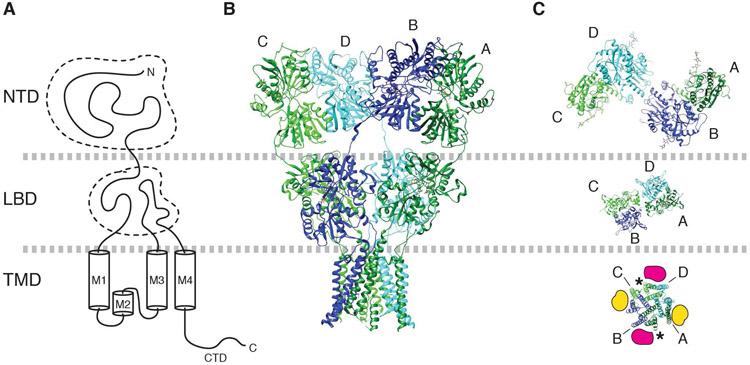
The ionotropic glutamate receptors (iGluRs) are ligand gated ion channels activated by the neurotransmitter glutamate. Among these, a workhorse of fast excitatory synaptic transmission is the AMPA-type iGluR (AMPAR) that transduces signals at millisecond timescales (Traynelis et al., 2010; Huganir & Nicoll, 2013; Greger et al., 2017). The pore forming subunits of AMPARs, known as GluA1-4 (Collingridge et al., 2009), consist of four domains (Fig. 1A). The N-terminal domain (NTD) in the extracellular space is most distantly positioned relative to the membrane. The function of the NTD is least understood, but it is critical for subunit assembly (Ayalon & Stern-Bach, 2001; Rossmann et al., 2011) as well as receptor clustering and synaptic localization (Sia et al., 2007; Garcia-Nafria et al., 2016; Diaz-Alonso et al., 2017; Watson et al., 2017). Region C-terminal to the NTD forms a short linker that connects the NTD to the ligand-binding domain (LBD). Upon glutamate binding, the LBD undergoes conformational changes that result in channel gating (Armstrong et al., 1998; Armstrong & Gouaux, 2000; Chen et al., 2017; Twomey et al., 2017a). The LBD connects to the transmembrane domain (TMD), which consists of three membrane spanning segments (M1, M3, and M4) and a re-entrant helix-loop (M2) (Hollmann et al., 1994). Namely, in the primary structure the LBD is divided by the M1-3 of the TMD into two fragments, S1 and S2. The TMD forms an ion channel in the membrane that when open, conducts cations. Finally, there is a cytoplasmic C-terminal domain (CTD) that regulates anchoring, signaling, and trafficking (Kim & Sheng, 2004; Huganir & Nicoll, 2013).
Figure 1. Architecture of AMPAR.
A. Domain organization of a GluA subunit of AMPAR.
B. Architecture of tetrameric assembly of GluA subunits in the canonical ‘Y’ shape. Subunits are labeled as A (dark green), B (blue), C (green), and D (cyan). PDB model 3KG2 (Sobolevsky et al., 2009) is displayed.
C. The NTD, LBD, and TMD layers are viewed from top. The dimer pairs formed at NTD and LBD layer are different; A/B and C/D at NTD, whereas A/D and B/C at LBD. The TMD is pseudo four-fold symmetric. Labels A-D point to the M4 helix of each subunit. In the TMD, the A/B surface is equivalent to the C/D surface (magenta). Similarly, the B/D surface and A/D surface are equivalent (yellow). The locations indicated by magenta and yellow are the binding site for TARPs and CNIH3. The asterisk indicates the locations of lipids found in AMPAR/TARP γ-8 complex (Herguedas et al., 2019).
The core architecture made of pore forming subunits of AMPAR
The pore forming subunits of AMPAR assemble into homo- and hetero-tetramers (Traynelis et al., 2010). A distinguishing structural feature of AMPARs, and iGluRs in general, from other tetrameric cation permeating ligand gated ion channels is the symmetry mismatch between the extracellular domains and TMD (Sobolevsky et al., 2009); the NTD and LBD each form dimers (Armstrong & Gouaux, 2000; Jin et al., 2009), whereas the TMD is tetrameric. The linkers connecting the LBD to TMD, which form part of the gating machinery, accommodate this symmetry mismatch. In addition, a domain swap occurs at the transition between NTD and LBD; in each subunit the NTD dimer partners differ from LBD dimer partners. The loose NTD-LBD linkers accommodate this domain swap (Fig. 1B). The architecture of the GluD1 is distinct among the iGluRs and lacks the domain swap, while retaining the flexibility of the NTD-LBD linker (Burada et al., 2020).
To accurately describe the architecture, AMPAR subunits within a tetramer are often referred to as A, B, C, and D (Fig. 1C) (Sobolevsky et al., 2009). The NTD dimers are formed between A/B and C/D subunits, whereas the LBD dimers are formed between A/D and B/C subunits. The TMD is pseudo four-fold symmetric, where the diagonally located A/B and C/D interfaces (Fig. 1C yellow) are geometrically equivalent relative to the overall two-fold symmetric extracellular architecture. Similarly, the B/D and A/C interfaces (Fig.1C magenta) of the TMD are equivalent.
In the majority of structural studies, mature AMPARs in the resting state are described as two-fold symmetric tetramers that have dimer-of-dimers organization of subunits, resembling a ‘Y’ shape (Sobolevsky et al., 2009; Chen et al., 2014; Meyerson et al., 2014; Yelshanskaya et al., 2014). Variations in the ‘Y’ shapes were reported in the GluA2-GSG1L complexes, which are introduced by a small twist between the NTD layer relative to the LBD layer (Twomey et al., 2017a). However, the canonical ‘Y’ shape is not the only configuration. For example, asymmetric global architectures (Nakagawa et al., 2005; Nakagawa et al., 2006; Durr et al., 2014) and more compact ‘O’ shape configuration (Herguedas et al., 2016) have been reported. The NTDs adopt asymmetric and symmetric configurations in the GluA2-CNIH3 complex (Nakagawa, 2019). These results suggest that the NTD layer of AMPAR has the capacity to reorganize into different global conformations, in part caused by the relatively flexible NTD-LBD linker (Fig. 2). The gap between the NTD and LBD layer is large enough to accommodate 20kDa cone snail toxin binding (Chen et al., 2014). To understand the dynamics of the NTDs, it is necessary to study the AMPAR without engineering the NTD-LBD linker. Alternative global conformation of the NTD layer is likely to have functional consequences. In fact, the extent of positional fluctuation of the NTD measured by atomic force microscopy is correlated with different gating behavior of the subunit isoforms (Dawe et al., 2019). The way the NTD layer is arranged relative to the LBD is potentially a significant issue, since its direct contact with the LBD (Herguedas et al., 2019; Nakagawa, 2019) and/or intrinsic dynamics (Dawe et al., 2019) may modulate gating. NTD mediated allosteric gating modulation is observed in NMDA receptors (NMDARs) (Hansen et al., 2010; Tajima et al., 2016) but remains elusive in AMPARs.
Figure 2. The variety of global conformations sampled by the NTD layer.
The NTD and LBD are connected by a relatively flexible linker. As a result the NTD layer is positioned differently depending on the length of the linker and subunit composition. The GluA subunits are color coded in red, blue, yellow, and green.
A. Y-shape global architecture of GluA2cryst homotetramer (PDB: 3KG2).
B. GluA2/GluA3 heterotetramer (PDB: 5IDE).
C. GluA2 homotetramer in complex with CNIH3 adopting asymmetric NTD layer (PDB: 6U6I and 6UD4).
D. Same as C, but adopting pseudo-symmetric NTD layer (PDB: 6U5S and 6UD8). The gap between the NTD and LBD layer is greater in D than in A. In A, the linker is engineered and shorter than wild type. Similar gap was also observed in GluA2 homotetramer without auxiliary subunits (Meyerson et al., 2014).
AMPAR-auxiliary subunit complexes
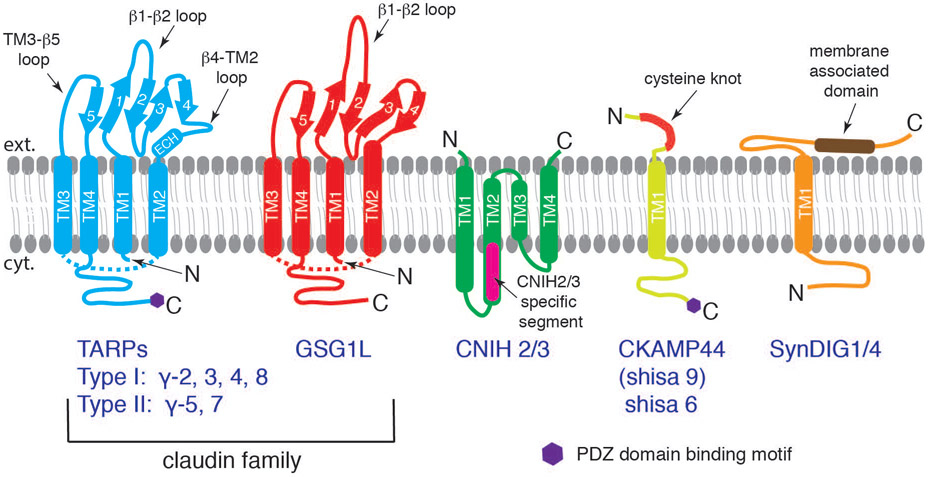
The native AMPAR complexes are made of combinations of ion channel forming core GluA subunits and various structurally unrelated transmembrane auxiliary subunits, such as TARPs (Chen et al., 2000; Tomita et al., 2003; Tomita et al., 2005; Kato et al., 2008), cornichons (CNIHs) (Schwenk et al., 2009), CKAMP44 (Shisa9) (von Engelhardt et al., 2010), Shisa6 (Klaassen et al., 2016), SynDIG1 and SynDIG4 (Kalashnikova et al., 2010; Matt et al., 2018), and GSG1L (Schwenk et al., 2012; Shanks et al., 2012) (Fig. 3). The two most abundant AMPAR auxiliary subunits in the hippocampus, cortex, and striatum are TARPs and CNIHs (Schwenk et al., 2014), although other auxiliary subunits may be enriched in smaller sub-populations of neurons within these regions. The association of auxiliary subunits with the receptor core adds functional diversity to channel gating and receptor localization (Jackson & Nicoll, 2011). For example, in the cerebellar granule neurons, association of different subfamily of TARPs alters synaptic AMPAR kinetics to varying degrees (Cho et al., 2007; Milstein et al., 2007). CNIH2 is responsible for the slow EPSC decay kinetics in hilar mossy cells in the dentate gyrus (Boudkkazi et al., 2014). TARP γ-2 is essential for surface expression of AMPARs in cerebellar granule cells (Chen et al., 2000), whereas TARP γ-8 is important for basal AMPAR activity and LTP in hippocampal CA1 neurons (Rouach et al., 2005). In recent years, cryo-EM structures of AMPARs in complex with auxiliary subunits are providing molecular foundations to understand their biology at high precision.
Figure 3. Schematic illustrations of structurally unrelated AMPAR auxiliary subunits.
TARPs (Type I: γ-2, 3, 4, 8 with a canonical PDZ binding motif; Type II: γ-5, 7 with an atypical PDZ binding motif) and GSG1L belong to the claudin superfamily. The α-helices (thick tubes) and β-strands (thick arrows) are depicted in the topology diagram. Both subclasses contain four transmembrane domains, two extracellular loops, and a cytoplasmic C-terminal tail. The β1-β2 loop is significantly longer in GSG1L. TM1~TM4 fold as a helical bundle, and therefore, the cytoplasmic TM2-TM3 loops of TARP and GSG1L (dashed lines) are shorter than how they are illustrated. CNIH is also a four-pass transmembrane protein, which is mostly embedded within the membrane. The N- and C-termini of CNIH are both extracellular. The CNIH2/3 specific segment, absent in CNIH1/4, is indicated (magenta). CKAMP44 (Shisa9) and Shisa6 have one transmembrane domain with an extracellular cysteine-knot motif and a long intracellular C-terminal tail with type II PDZ binding motif. SynDIG1 and 4 have one transmembrane domain and a long extracellular domain containing a membrane-associated domain.
Architectures of recombinant AMPAR-TARP complexes
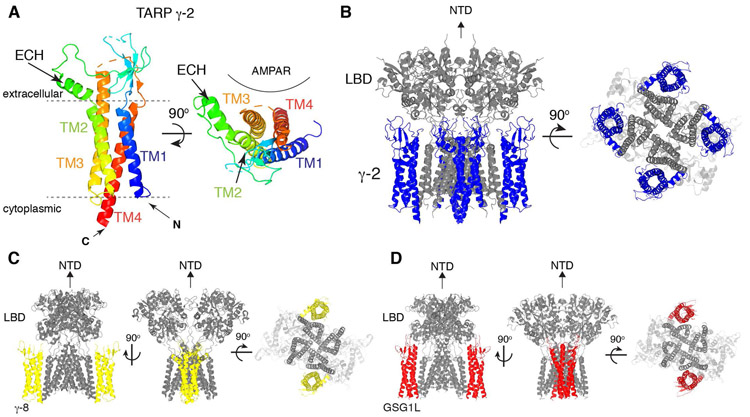
The recombinant AMPAR-TARP complexes were studied extensively using cryo-EM, generating detailed information about their architectures (Twomey et al., 2016; Zhao et al., 2016; Chen et al., 2017; Twomey et al., 2017a; Herguedas et al., 2019; Twomey et al., 2019). The overall architecture of TARPs is similar to those of claudins (Suzuki et al., 2014; Saitoh et al., 2015; Nakamura et al., 2019). The topology of TARPs comprises four transmembrane helices (TM1-4) with the N-terminus in the cytoplasm, immediately preceding TM1 (Fig. 3 and 4). TM1-4 forms a twisted bundle in the membrane. The extracellular extensions connecting the transmembrane helices fold into an extracellular domain (ECD), while a short cytoplasmic loop is formed between TM2 and TM3. The ECD overall resembles a shape of a hand palm, which is made of five β-strands (β1-5), an extracellular helix (ECH), and four flexible loops (β1-β2, β3-β4, β4-TM2, and TM3-β5). A cytoplasmic C-terminal tail (C-tail) extends from the TM4 and terminates with a PDZ domain ligand peptide. Multiple sites in the C-tail mediate binding to the synaptic scaffold protein PSD-95 and the formation of the complex induces liquid phase separation (Zeng et al., 2019). Consistently, the C-tail is unresolved in the available cryo-EM structures, possibly due to its flexibility or truncation from the constructs.
Figure 4. Architecture of TARPs and AMPAR/TARP complexes.
A. A ribbon diagram of TARP γ-2 shows the 3D arrangement of the 4-helix bundle made of TM1-4. An extracellular helix (ECH) is attached to TM2. TARP γ-2 from PDB: 5WEO is displayed at two different views, one parallel to the membrane (left) and another from the extracellular side (right). Location of AMPAR is indicated.
B. A ribbon diagram of the AMPAR(GluA2)/TARP γ-2 complex. GluA2(gray):TARP γ-2(blue) =4:4 stoichiometry. 5WEO is displayed at two different views (side and bottom), but the NTD was removed for clarity.
C. A ribbon diagram of the heteromeric AMPAR(GluA1 and 2)/TARP γ-8 complex. GluA1(gray):GluA2(gray):TARP γ-8(yellow) =2:2:2 stoichiometry. 6QKC is displayed at three different views (side and bottom), but the NTD was removed for clarity.
D. A ribbon diagram of the AMPAR(GluA2)/GSG1L complex. GluA2(gray):GSG1L(red)=4:2 stoichiometry. 5WEL is displayed at three different views (side and bottom), but the NTD was removed for clarity.
TARPs bind preferentially to the tetrameric AMPARs (Vandenberghe et al., 2005; Shanks et al., 2010), and consistently, the TARP binding site spans across adjacent subunits of AMPARs. Specifically, the M1 and M4 of adjacent GluA subunits interface with the TM3 and TM4 of TARP. The structurally confirmed stoichiometries of the complexes are four, two, and one TARPs per tetrameric AMPAR (Twomey et al., 2016; Zhao et al., 2016; Chen & Gouaux, 2019; Herguedas et al., 2019) (see Fig. 4 for GluA2:TARP γ-2 = 4:4 stoichiometry). Studies have compared functional signatures of neuronal AMPARs with heterologously expressed AMPAR-TARP complexes made of defined stoichiometry and suggested the presence of AMPARs with two and four TARPs (Shi et al., 2009; Dawe et al., 2019).
Due to the two-fold symmetry of the extracellular domain of the AMPAR, the A/B and C/D interfaces (Fig. 1C, yellow) are equivalent. Similarly, A/D and B/C interfaces (Fig. 1C, magenta) are also equivalent. Accordingly, the TARP ECD interacts with different elements of the AMPAR depending on its binding site. The β4-TM2 loop interfaces with the LBD when the TARP is bound at A/B and C/D interface, whereas the TM3-β5 loop interfaces with the LBD-M4 (i.e. S2-M4) linker when the TARP is bound at the A/D and B/C interface (Chen et al., 2017; Twomey et al., 2017a). In the heterotetrameric AMPAR made of GluA1 and GluA2, two TARP γ-8 preferentially bind to the A/B and C/D interfaces (Herguedas et al., 2019) (Fig. 4C). In this complex, the β1-β2 loop of TARP γ-8 interacts with the LBD of AMPARs (Herguedas et al., 2019). Interfaces proposed from these structures are suggested to be functionally critical for gating (or modulation) (Chen et al., 2017; Twomey et al., 2017a). Receptors with mutations located at these interfaces exhibit abnormal gating behaviors, indicating the physiological relevance of these structurally defined interaction sites (Priel et al., 2005; Tomita et al., 2005; Dawe et al., 2016; Hawken et al., 2017; Riva et al., 2017).
The residue contacts between the transmembrane helices of AMPAR and TARP are also critical for gating modulation, because the mutations in this region abolish slowing of desensitization imposed by TARPs (Ben-Yaacov et al., 2017; Hawken et al., 2017). How this interaction regulates gating behavior remains elusive. The initial functional mappings of TARP γ-2 also suggested that the CTD is also critical for gating modulation (Priel et al., 2005; Tomita et al., 2005; Milstein & Nicoll, 2009), but the underlying mechanism is unclear. Further investigations that combine structural biology, channel physiology, and molecular dynamic simulation, may provide information to improve our understanding of the mechanism of TARP action.
Architecture of recombinant AMPAR-GSG1L complex
GSG1L is a distant homolog of TARPs within the claudin family (Schwenk et al., 2012; Shanks et al., 2012) (Fig. 3). Accordingly, GSG1L folds like TARPs and binds to the identical surface on AMPAR as TARPs bind (Twomey et al., 2017b) (Fig. 4D). Unlike the TARPs, which can simultaneously bind up to four molecules per AMPAR tetramer, current cryo-EM structures contain only two GSG1L per AMPAR tetramer (Twomey et al., 2017a). GSG1L slows AMPAR desensitization, as well as recovery from desensitization, which is functionally different compared to TARPs. GSG1L lacks the ECH and instead has a longer TM2 extending to the extracellular space. In addition, the β1-β2 loop is longer than that of the TARPs and densities of this loop reach towards the LBD. These architectural differences contribute to the drastic functional differences between GSG1L and TARPs. In fact, transplanting the β1-β2 loop of TARP γ-2 into the corresponding location in GSG1L is sufficient for GSG1L to acquire the TARP γ-2-like gating modulation characteristics (Riva et al., 2017; Twomey et al., 2017b). While critical structural elements responsible for gating modulation have been identified, how these elements function during gating remain elusive. Interestingly, when GSG1L and TARP γ-2 are co-expressed with AMPAR in HEK cells, the gating modulation characteristic of TARP γ-2 dominates (Schwenk et al., 2012). This may imply that two types auxiliary subunits compete for the same binding site on AMPAR but one is preferred over the other.
Architecture of recombinant AMPAR-CNIH complex
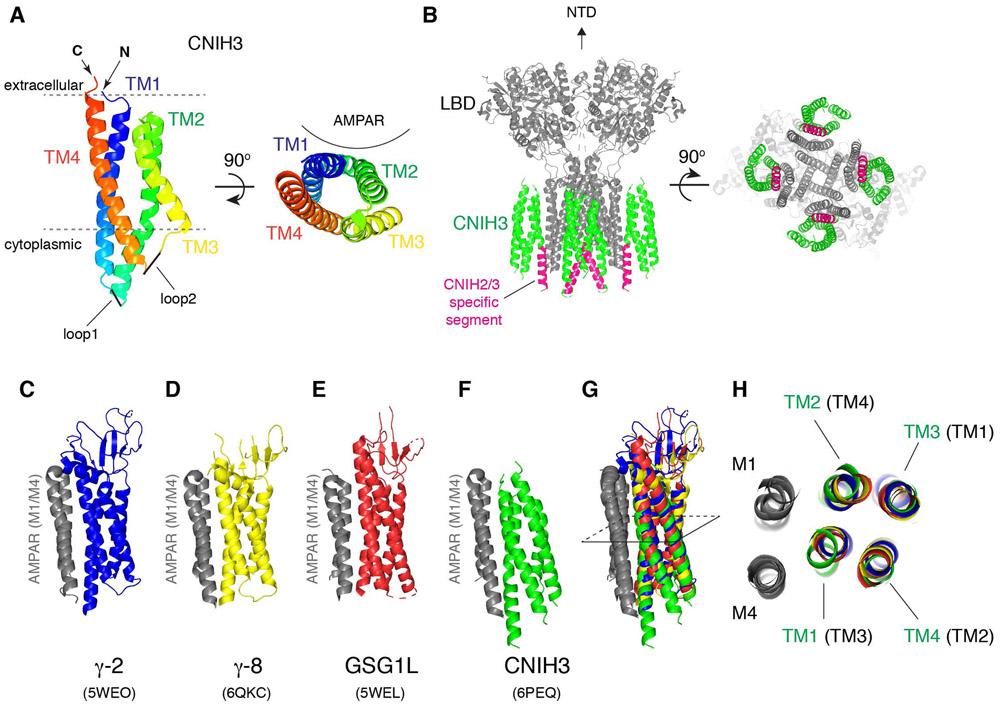
The membrane topology of CNIHs has long been thought to be three transmembrane segments with a cytoplasmic amino terminus; CNIHs lack a canonical signal peptide and topology prediction programs would always specify three hydrophobic helices (Hoshino et al., 2007; Schwenk et al., 2009; Greger et al., 2017; Wudick et al., 2018; Chen & Gouaux, 2019). Unexpectedly, the cryo-EM structure of GluA2/CNIH3 complex at GluA2: CNIH3=4:4 stoichiometry bound to antagonist ZK200775 revealed that CNIH3 has, in fact, four hydrophobic helices (TM1 to TM4) and an extracellular amino terminal (Nakagawa, 2019) (Fig. 5A-C). The majority of the side chains were clearly resolved in the calculated map, which enabled building a molecular model of CNIH3 de novo. In this architecture, the 2nd hydrophobic helix (TM2) starts in the cytoplasm but does not penetrate into the extracellular space. Instead, a short hydrophobic loop at the end of TM2 bends 180 degrees inside the membrane and connects to the 3rd hydrophobic helix (TM3). The continuous hydrophobicity throughout TM2 and TM3 must have caused the topology prediction algorithm to misinterpret them as a single transmembrane helix. Despite the absence of a canonical signal peptide, the hydrophobic N-terminal segment appears to possess the ability to insert into the membrane.
Figure 5. Architecture of CNIH3 and AMPAR/CNIH3 complex.
A. A ribbon diagram of CNIH3 shows the 3D arrangement of the 4-helix bundle made of TM1-4. CNIH3 from PDB: 6PEQ is displayed at two different views, one parallel to the membrane (left) and another from the extracellular side (right). Location of AMPAR is indicated.
B. A ribbon diagram of an AMPAR/CNIH3 complex.
GluA2(gray):CNIH3(green)=4:4 stoichiometry. 6PEQ is displayed at two different views (side and bottom), but the NTD was removed for clarity. The CNIH2/3 specific segment is highlighted in magenta.
C and F. The ribbon diagram of the M1 and M4 of AMPAR (gray) bound to various auxiliary subunits. The binding interface is viewed from the side in parallel to the membrane. C=TARP γ-2, D= TARP γ-8, E= GSG1L, F=CNIH3.
G. Superposition of the structures in C-F. Specifically, when the M4 of AMPAR in C-F is aligned, the M1 and the transmembrane helices of the auxiliary subunits superimpose. The M1/M4 of AMPAR form the auxiliary subunit binding module for TARPs, GSG1L, and CNIH3, which are geometrically conserved.
H. Cross section at the slice shown in G is viewed from the top. The TM3 and TM4 (of TARP and GSG1L) interface with M1 and M4 of adjacent subunit of AMPAR. In contrast, the TM1 and TM2 of CNIH3 interface with M1 and M4. The TM1 to TM4 of CNIH3 are labeled in green, whereas the corresponding TM in TARPs and GSG1L are labeled in black and in brackets.
The 3D architectures of transmembrane domains of the TARP, GSG1L, and CNIH are very similar (Fig. 5). The transmembrane helices of CNIH3 superimpose well with those of TARPs, but the correspondence between the helices is different. The TM1 of CNIH3 overlay with TM3 of TARPs. TM2 of CNIH3 overlay with TM4 of TARPs, and so forth. Moreover, both CNIHs and TARPs bind to the same location on AMPARs. The binding interface is made by the M1 and M4 of adjacent subunits of AMPARs. The TM1 and TM2 of CNIH3 interface with AMPAR, whereas the TM3 and TM4 of TARPs interface with AMPAR. There may be functional or structural constraints that require the molecular envelope of the AMPAR-CNIH and AMPAR-TARP complexes to remain conserved. Another distinguishing structural feature of the GluA2/CNIH3 complex is the more compact TMD, characterized by translation of cytoplasmic half of M1 and M4 towards the center of the channel induced by CNIH3 binding. The smaller space under the M3 helix bundle and the lipid appear to rearrange the architecture of the reentrant loop that contains the M2 and filter. The M2 and filter are important for polyamine sensitivity of AMPAR/CNIH3 complex (Coombs et al., 2012; Brown et al., 2018; Twomey et al., 2018), but the current structure falls short in resolving these elements. The structural reorganization of the TMD was not observed in the AMPARs in complex with TARPs.
There are key structural differences between TARPs and CNIHs. Each TARP has a small extracellular domain that reaches towards the LBD. On the other hand, CNIH3 hardly has an extracellular domain; the majority of the protein is embedded in the membrane. A cytoplasmic α-helix unique to CNIH2 and 3 (Fig. 3 and Fig. 5B, magenta), but absent in CNIH1 and 4, interfaces the M4 of GluA2 and plays a role in gating modulation but not in stabilizing the complex (Shanks et al., 2014). The residues critical for gating modulation by CNIH3 were at the interaction interface between GluA2 and CNIH3; the phenylalanine 3, 5, and 8 of CNIH3, which are also present in CNIH2, interact with several residues in the M1 and M4 of GluA2. These residues that are critical for the interaction are fully conserved in GluA1 subunit. However, in hippocampal neurons there appears to be a mechanism that enables CNIH2 to distinguish between GluA1 and GluA2, which ultimately regulates the number of GluA1 containing AMPARs in synapses (Herring et al., 2013). In mammalian neurons CNIH2/3 regulates both AMPAR gating and trafficking (Kato et al., 2010; Shi et al., 2010; Gill et al., 2011; Boudkkazi et al., 2014), while in C. elegans cornichon homolog CNI regulates ER export of AMPARs (Brockie et al., 2013). Future experiments, based on the molecular information on AMPAR/CNIH interaction interface and topology, may further clarify the roles of CNIH2/3 in synaptic transmission and plasticity, as well as whether binding of CNIH2/3 and TARPs to AMPAR is regulated. In addition, how the key residues at the interaction interface transduce their effects during gating modulation remains elusive. The detailed residue arrangements that occur at the interaction interface with AMPAR appear different between CNIH3, GSG1L, and TARPs (Chen et al., 2017; Twomey et al., 2017a; Herguedas et al., 2019; Nakagawa, 2019), but how these differences translate into functional differences is unclear. The structure of the AMPAR/CNIH complex bound to an agonist together with blockers of desensitization, which may mimic the channel open state, could provide clues to these questions.
Lipids may be critical molecular components of native AMPAR complexes
A biochemical property that distinguishes AMPARs and kainate receptors (KARs) from NMDARs is the detergent solubility (Blackstone et al., 1992). In order to solubilize them, NMDARs require much harsher detergents than AMPARs and KARs (Sheng et al., 1994; Shanks et al., 2012). Properties of the membrane surrounding the receptors and receptor anchoring mechanism are likely very different among iGluR subtypes. Lipids are frequently found in structures of ion channels (Long et al., 2007; Tang et al., 2016; McGoldrick et al., 2018; Zhang et al., 2018). Indeed, cholesterol and fatty acids modulate ion channel function of NMDARs and KARs, respectively (Wilding et al., 1998; Korinek et al., 2015). Cholesterol depletion in cultured hippocampal neurons results in redistribution of synaptic AMPARs (Hering et al., 2003). However, it was not until recently that lipids were found to be associated with AMPARs.
Lipid-like densities were observed in the architecture of heterotetrameric AMPAR made of GluA1 and GluA2 in complex with TARP γ-8 (Herguedas et al., 2019) (Fig. 1C asterisk). These lipids must be carried over from HEK cells in which the receptor complex was expressed. Interestingly, lipids surrounding the GluA2-CNIH3 complexes (Nakagawa, 2019) were arranged differently from what was observed in the GluA1/GluA2/TARP γ-8 complex. These observations lead to a hypothesis that lipids may play functional roles in auxiliary subunit assembly and action, and that they may have distinct roles in different classes of AMPAR-auxiliary subunit complexes.
TARPs were essential for trapping the channel gate open in detergent (Chen et al., 2017; Twomey et al., 2017a), as no structures without TARPs maintained an open gate architecture despite being bound to an agonist plus a blocker of desensitization, or a potentiating toxin (Chen et al., 2014; Yelshanskaya et al., 2014). AMPAR/TARP complexes have higher open probability and longer dwell-time at higher conductance level compared to without TARPs (Tomita et al., 2005; Shelley et al., 2012; Zhang et al., 2014; Carrillo et al., 2020), suggesting that TARPs stabilize the open and activated channel conformation. The allosteric coupling between agonist binding and gating may be disrupted in detergent, which is known to happen in nicotinic acetylcholine receptors (Sun et al., 2017). Therefore, it is possible that TARPs recruit lipids to the complex and create an environment mimicking the membrane.
Some lipids that stabilize the complex may be absent in non-neuronal cells but present in brain lipids. This is implicated from the observation that different detergent conditions were optimal for dissolving AMPARs from brain versus recombinant expression systems, such as Sf9 and HEK cells (Nakagawa, 2010). Identifying the lipid composition of native AMPARs would be a challenge but maybe critical for understanding the function of AMPAR-auxiliary subunit complexes.
Molecular and structural diversity of native AMPAR complexes
The architectures of native AMPAR complex purified from rat brains were investigated using single particle EM before the resolution revolution of cryo-EM (Nakagawa et al., 2005; Nakagawa et al., 2006). At a resolution of ~30Å, the studies provided the first glimpse of global domain organization of brain-derived heterotetrameric AMPARs (Note; evidence for heterotetramer is based on anti-GluA1-NTD antibody labeling native AMPARs that were purified using anti-GluA2 antibody), their conformational flexibility induced between the NTD and LBD, and conformational changes of the NTDs that occur in desensitized receptors. The majority of native AMPAR complexes adopted asymmetric global architecture. Another key finding from this work was that auxiliary subunits co-purify with AMPARs and contribute to enlargement of transmembrane density of the complex; fab fragments located TARPs in the transmembrane density of the particles (Nakagawa et al., 2005; Nakagawa et al., 2006).
A recent study investigated the architectures of native AMPARs at higher resolution, using modern cryo-EM technology (Zhao et al., 2019). Interestingly, this study revealed mostly ‘Y’ shaped global architecture, where two NTD dimers appear pseudo symmetric, even without imposing symmetry during image processing. The global conformation adopted by the NTD layer was less diverse than previous works on native and recombinant AMPARs. However, the Fabs that were used to label the NTDs appear to make contacts between each other (see EMD-9388 and -9389) and may have restricted the freedom of conformational flexibility of the NTDs, and therefore the repertoire of global conformations remains to be an open question. Fab and scFv that specifically label the NTD of different GluA subunits were used to determine the molecular composition of the complex, detecting previously uncharacterized tri-heteromers formed of GluA1/2/3 subunits and the heterotetramers made of GluA1/2 subunits, which were the two most abundant classes among other subunit compositions. Furthermore, the study showed more than one type of auxiliary subunit binding to a single AMPAR. The first type of auxiliary subunit was interpreted as TARPs but the identity of the other one was unclear. Considering that the TARPs and CNIHs are the two most abundant auxiliary subunits, and that the CNIHs have four membrane spanning helices, it is speculated that the second auxiliary subunit density derives from CNIHs (Schwenk & Fakler, 2019). Although single particle cryo-EM studies on native AMPAR architectures provide important information, less frequently occurring complexes must be escaped from being detected.
Despite the molecular diversity of binding partners (Schwenk et al., 2009; Kang et al., 2012; Schwenk et al., 2012; Shanks et al., 2012; Schwenk et al., 2014), the structural repertoire of the TMDs of native AMPAR complexes is surprisingly small (Nakagawa et al., 2005; Zhao et al., 2019). This could be explained in two ways. First, the most abundant auxiliary subunits, TARPs and CNIHs, adopt a similar fold in the transmembrane helix bundles and bind to a similar location on the AMPAR, and thus the molecular envelope is not very different between complexes containing TARPs and CNIHs, especially at an intermediate resolution (~4-5Å) (Nakagawa, 2019). Secondly, other auxiliary subunits are less abundant than TARPs and CNIHs, and thus they are difficult to detect from gross purification of AMPAR subunits (Schwenk et al., 2014). However, these less abundant auxiliary subunits are sometimes enriched in specific neuronal cell types and play specialized roles (Chen et al., 2018). Elucidating the function and mechanism of different AMPAR auxiliary subunits seems critical for understanding the modulation of excitatory circuits. Therapeutically, focally enriched AMPAR auxiliary subunits should serve as ideal targets for brain region specific drugs that control AMPAR activity.
Functional co-expression of auxiliary subunits
In many cases, multiple auxiliary subunits function within a given neuronal population (Tomita et al., 2003; Fukaya et al., 2005; Lein et al., 2007; Jackson & Nicoll, 2011; Schwenk et al., 2014). In various brain regions, there is a considerable functional redundancy among the TARPs (Jackson & Nicoll, 2011). Here we focus our discussion on functional interactions that are present across different classes of AMPAR auxiliary subunits. CKAMP44 and TARP γ-8 are co-expressed in the dentate gyrus (DG) granule cells (von Engelhardt et al., 2010). The two have opposing effects on the recovery from AMPAR desensitization, with CKAMP44 significantly slowing this parameter in DG neurons. AMPAR desensitization itself is known to contribute to short-term plasticity (Chen et al., 2000). Consistently, the paired pulse ratio (PPR) of AMPAR mediated EPSCs was increased in CKAMP44 KO mice and decreased in γ-8 KO mice (Khodosevich et al., 2014). Interestingly, γ-8 slows the recovery from desensitization in HEK cells (Cais et al., 2014). This discrepancy between heterologous cells and mouse slice data could be due to other co-existing auxiliary subunits or differing composition of GluA subunits in the same neuron.
While multiple studies have identified functional interactions between auxiliary subunits at the synapse, the signaling pathways that control auxiliary subunit composition remain largely unknown. Gating properties of AMPARs could change significantly if switching of an auxiliary subunit takes place within the synapse, although currently there is no evidence in support of this hypothesis. Functionally, co-expression of multiple auxiliary subunits with AMPAR leads to composite gating phenotypes in HEK cells. It is becoming clear that more than one type of auxiliary subunit could simultaneously bind to an AMPAR (Kato et al., 2010; Gill et al., 2011; Khodosevich et al., 2014; Zhao et al., 2019). Revealing the regulation of AMPAR complexes in the context of biogenesis may provide critical information. A specialized set of proteins interacts with AMPARs in the early biogenic pathway (Schwenk et al., 2014; Erlenhardt et al., 2016; Schwenk et al., 2019). Whether and how the assembly is regulated is an important question to address in the future.
Role of AMPAR auxiliary subunits in synaptic plasticity
Changes in synaptic AMPAR trafficking is one of the primary mechanisms for synaptic plasticity (Huganir & Nicoll, 2013). γ-8 and CNIH2/3 are co-expressed in hippocampal CA1 cells and each contributes to regulating AMPAR mediated response (Rouach et al., 2005; Herring et al., 2013). In the hippocampus, removal of γ-8 or CNIH2/3 results in reduced AMPAR-mediated synaptic transmission and attenuated LTP (Rouach et al., 2005; Herring et al., 2013).
One of the key roles of γ-8 in hippocampal CA1 is to mediate molecular interaction via its cytoplasmic C-tail with synaptic scaffold protein PSD-95, whereby facilitating recruitment of γ-8 containing AMPAR complexes into synapses (Sumioka et al., 2011). This interaction is dispensable for LTP, as LTP was normal in knock in (KI) mice that lack the PDZ domain ligand of the γ-8 (Sumioka et al., 2011). Instead of the PDZ domain interaction phosphorylation of the C-tail of γ-8 by CaMKII plays a critical role, as LTP was impaired in KI mice whose phospho-substrate residues in the C-tail of γ-8 were mutated to alanine (Park et al., 2016). The events downstream of the phosphorylation remain unclear. In one view, phosphorylation releases the C-terminal from the membrane and facilitates its interaction with PSD-95 (Park et al., 2016), but such mechanism appears unimportant for LTP (recall that interaction of γ-8 with PSD-95 is dispensable for LTP). On the other hand, based on quantitative binding assay between the C-tail of γ-8 and the PDZ domain of PSD-95, introducing phosphomimetic mutation to the phospho-substrate residues in the C-terminal tail of γ-8 was shown to decrease the affinity (Zeng et al., 2019). In two recent studies, which used molecular replacement at the cellular resolution, the interaction between the C-tail of γ-8 and PSD-95 was suggested to be critical for LTP in CA1 cells (Sheng et al., 2018; Zeng et al., 2019), which is a completely opposite model from what was described previously. The different conclusions may be due to different experimental system used in these studies.
In hippocampal CA1 cells, CNIH2/3 regulate the number of synaptic heteromeric AMPARs that contain GluA1 (Herring et al., 2013). However, as described earlier, the residues in GluA1 and GluA2 critical for binding CNIH2/3 are fully conserved, and thus it is unclear how CNIH2 can distinguish between GluA1 and GluA2. Considering that γ-8 and CNIH2/3 bind to the same site on AMPAR (Herguedas et al., 2019; Nakagawa, 2019) and co-assemble (Kato et al., 2010; Gill et al., 2011; Schwenk & Fakler, 2019; Zhao et al., 2019), it will be important to evaluate in more detail the functional contribution of each auxiliary subunit belonging to the same AMPAR complex (containing both TARP and CNIH).
Gating modulation imposed by auxiliary subunits contribute to short-term plasticity mechanisms in some neurons. While most commonly seen mechanisms for short-term plasticity are presynaptic (Zucker, 1999), postsynaptic mechanisms, such as changes in the rate of desensitization, also occur in the brain (Chen et al., 2002; Kielland & Heggelund, 2002). AMPAR auxiliary subunits affect short-term plasticity in a complex manner. For example, members of the CKAMP family of proteins, CKAMP44 and Shisa6, slow recovery from desensitization in HEK cells, and thus one would expect that both affect short term plasticity in a similar way. However, genetic deletion studies show opposing effects on short-term plasticity in the hippocampus. CKAMP44 KO mice show decreased short-term depression at the lateral perforant path to DG granule cell synapses (Khodosevich et al., 2014). Shisa6 KO mice, on the other hand, have increased synaptic depression at Schaffer collateral-CA1 synapses (Klaassen et al., 2016). It is predicted that this effect is due to Shisa6 slowing down the entry into the desensitized state. Another possible explanation for the discrepancy between results in HEK cells and neurons could be the presence of other auxiliary subunits or other unknown factors specific to the neuronal cell types. Shisa7, another member of CKAMP family, also slows the recovery from desensitization. However, it has been shown not to regulate short-term plasticity in CA1 neurons (Schmitz et al., 2017; Han et al., 2019). A more recent study established how changes in short term dynamics imposed by an auxiliary subunit, CKAMP44, directly affects in vivo firing during behavior (Chen et al., 2018). Retinogeniculate synapses exhibit high release probability and pronounced short-term depression, which is partly due to AMPAR desensitization (Chen & Regehr, 2000; Chen et al., 2002). CKAMP44 regulates short-term depression in these synapses by slowing the recovery from desensitization. CKAMP44 KO mice have increased spike probability and in vivo firing of dorsal lateral geniculate neurons (Chen et al., 2018). Future structural analyses of AMPAR/CKAMP44 complex may produce new functional insights into their role in synaptic AMPAR regulation.
Among all of the auxiliary subunits, GSG1L stands out for having the strongest effect on slowing the recovery from AMPAR-desensitization in HEK cells (Schwenk et al., 2012; Shanks et al., 2012). However, overexpression of GSG1L in CA1 hippocampal neurons speeds the recovery from desensitization (Gu et al., 2016). It is predicted that this discrepancy is due to innate properties specific to neuronal systems, such as AMPAR subunit composition and the presence of other auxiliary subunits including CNIH2 (Gu et al., 2016). Association of GSG1L with AMPARs prevents CNIH2 regulation in heterologous systems (Gu et al., 2016). The functional competition between GSG1L and CNIH2 is in agreement with structural data indicating that the two bind to the same location on AMPARs (Twomey et al., 2017b; Nakagawa, 2019).
LTP at hippocampal CA1 synapses was diminished in GSG1L KO rats at P13-P19 (Gu et al., 2016). However, GSG1L overall has a very low expression in the hippocampus (Schwenk et al., 2014). In situ hybridization data also shows that the hippocampal expression is minimal and is only present in adult mice starting at P60 (Lein et al., 2007). Therefore, the observed phenotypes in the GSG1L KO rats could be due to the loss of GSG1L elsewhere in the brain. More studies looking at the direct postsynaptic role of GSG1L are needed to determine its physiological function.
Mutations in native AMPAR complexes and diseases
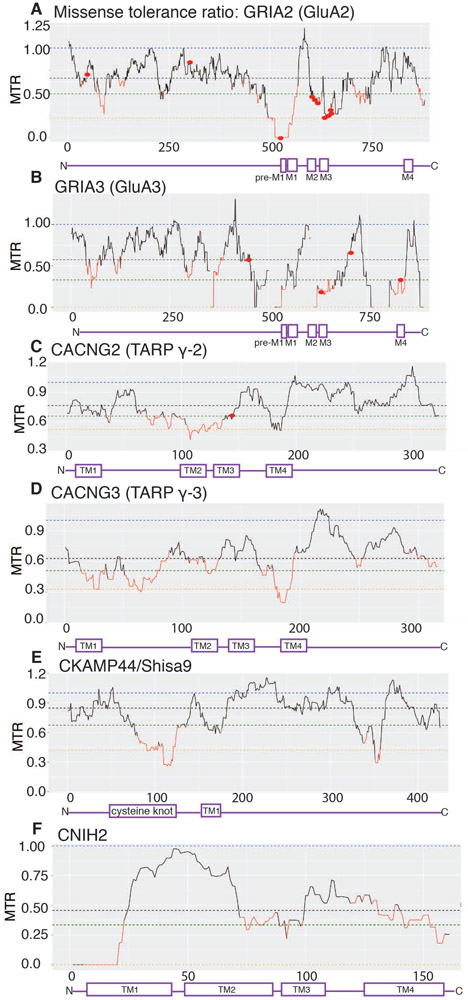
Mutations in the core AMPAR subunits have been linked to various neurological diseases. Namely, mutations in GluA2 (encoded by GRIA2) are linked to intellectual disability (ID) (Tzschach et al., 2010; Hackmann et al., 2013; Salpietro et al., 2019). In addition, multiple GluA3 (encoded by GRIA3) mutations are also associated with ID (Chiyonobu et al., 2007; Wu et al., 2007; Bonnet et al., 2009; Philippe et al., 2013). For instance, a missense A653T mutation within the ion conduction pore of GluA3 is associated with sleep deregulations as well as ID (Davies et al., 2017). Additionally, missense tolerance ratio (MTR) analysis indicates GluA2 and GluA3 are highly intolerant to genetic variation. The A653T mutation in GluA3 has an MTR=0.33 thereby indicating the relevance of MTR scores to real diseases (Fig. 6B). Missense mutation G833R in the M4 of GluA3 is associated with cognitive impairments (Wu et al., 2007). Mutations in the neighboring residues within the M4 of GluA2 are known to disrupt receptor tetramerization (Amin et al., 2017). Interestingly, the region immediately preceding M1 (pre-M1: GVFSFLDPLAYE) in GluA2, where lipids are predicted to bind (Herguedas et al., 2019), and the M1 (WMCIVFAYIGVSVVL), where TARPs and CNIHs are predicted to bind, are highly intolerant to variation (MTR=0). Mutations in the pre-M1 of NMDARs are also linked to epilepsy and developmental delay (Ogden et al., 2017). Overexpression of these mutants results in prolonged EPSCs and excitotoxicity, which can be attenuated by NMDAR inhibitor memantine (Ogden et al., 2017). In AMPARs the pre-M1 region may also tune gating (Shi et al., 2019).
Figure 6. Intolerance of AMPAR complexes to missense mutations.
Missense tolerance ratio (MTR) is an estimate of the extent of purifying selection or the removal of deleterious alleles (Traynelis et al., 2017). An MTR=1 indicates neutrality and the ratios below the 10th percentile (in red) are under purifying selection. AMPAR auxiliary subunits have varying degrees of tolerance for missense mutations (i.e. low MTR scores).
A. GRIA2, which encodes GluA2 subunit, has regions with MTR=0 (pre-M1 and part of M1), no tolerance to variation. Missense mutations associated with neurodevelopmental disorders (G47E, D302G, P528T, Q/R607G/E, G609R, D611N, A639S, F644L, T646L, V647L) are indicated with red dots (Salpietro et al., 2019).
B. GluA3 subunits also has multiple regions with MTR=0, highly intolerant to variation. The locations of mutations associated with cognitive impairment (G833R, M706T, R631S, R450Q) are indicated as red dots (Wu et al., 2007).
C. MTR distribution for γ-2 shows multiple regions of high intolerance (in red) including TM2. Missense mutation V143L associated with ID is indicated as a red dot.
D. Among TARPs, γ-3 has the highest number regions of intolerance including TM1, TM2, and TM4.
E. Cysteine knot domain within CKAM44 is highly intolerable to genetic variation.
F. CNIH2 has a stretch of amino acids that are highly intolerable to genetic variation. This region is mapped as the critical AMPAR binding site.
The disease associated mutations are not limited to core AMPAR subunits but can also be found in auxiliary subunits. A missense de novo mutation in the TM3 of TARP γ-2 (V143L) is associated with ID (Hamdan et al., 2011). The V143L mutation reduces AMPAR- γ-2 interaction, as well as mEPSC amplitude and frequency in cultured hippocampal neurons. De novo deletion of CNIH2 has also been linked to ID (Floor et al., 2012). In general, TARPs γ-3 and γ-2 have been identified as susceptibility loci for a subpopulation of patients with childhood absence epilepsies and schizophrenia respectively (Everett et al., 2007; Liu et al., 2008). MTR analysis indicates that AMPAR auxiliary subunits contain multiple regions that are mildly intolerant to genetic variation (as shown in Fig. 6B-E). Within the TARP sub-family, γ-3 has the highest degree of intolerance (Fig. 6D). Interestingly, the cysteine knot domain in the CKAMP44 is also intolerant to variation (Fig. 6E). Among all of the auxiliary subunits, CNIH2 stands out for having a stretch of amino acids that are highly intolerable to missense variation (Fig. 6F)
Dysregulation of AMPAR function has been associated with a myriad of neurological diseases (Chang et al., 2012). However, due to their ubiquitous expression and essential function, the pore forming subunits of AMPAR are not optimal targets for therapeutic compounds; such drugs may cause substantial off-target effects such as dizziness and sedation (Rogawski, 2011). On the other hand, because of their rich spatiotemporal diversity of expression, auxiliary subunits are potential targets for therapeutic compounds that could control AMPAR function in a region specific manner (Azumaya et al., 2017; Maher et al., 2017). Compounds specific for AMPARs associated with TARP γ-8 are being developed and proven to work in animal models of epilepsy (Gardinier et al., 2016; Kato et al., 2016; Lee et al., 2017). While the residues critical for drug action have been mapped, current structural data suggests that the proposed binding pocket is too small for drug binding (Herguedas et al., 2019). Establishing structural basis for its action is expected to reveal new information about strategies to control functions of AMPAR-auxiliary subunit complexes in various disease states.
Conclusion
Solving the mechanism of functional modulation of AMPARs by their auxiliary subunits would benefit from continuing efforts in order to reach a critical point where it would become useful for developing improved therapeutics. The lipids require attention, as they may play important roles in AMPAR-auxiliary subunit function. Structural studies are expected to produce only the snapshots of complexes in action, and therefore, functional investigation and molecular dynamic simulation approaches are expected to play equally important roles. Native AMPAR complexes contain more than one type of auxiliary subunits. Structural and functional investigation of AMPAR-auxiliary subunit with complex molecular composition, including lipids, would be necessary in the future. Given the robust functional modulation imposed on AMPARs by an individual auxiliary subunit, their regulation is predicted to have significant impact on circuit activity, cognition, learning and memory. The function of TARP γ-8 in hippocampal LTP is extensively studied, however roles of non-TARP auxiliary subunits in synaptic plasticity are only starting to become clear. Specifying the underlying molecular mechanisms that regulate circuit dynamics will be important problems to solve in the future.
Acknowledgments
Funding: The work performed in author’s laboratory is funded by Vanderbilt University (to T.N.) and NIH grant R01HD061543 and R21MH102546 (to T.N.)
Footnotes
Competing interests: The authors have no competing interests and conflict of interests to declare.
References
- Amin JB, Salussolia CL, Chan K, Regan MC, Dai J, Zhou HX, Furukawa H, Bowen ME & Wollmuth LP. (2017). Divergent roles of a peripheral transmembrane segment in AMPA and NMDA receptors. J Gen Physiol 149, 661–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N & Gouaux E. (2000). Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron 28, 165–181. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Sun Y, Chen GQ & Gouaux E. (1998). Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature 395, 913–917. [DOI] [PubMed] [Google Scholar]
- Ayalon G & Stern-Bach Y. (2001). Functional assembly of AMPA and kainate receptors is mediated by several discrete protein-protein interactions. Neuron 31, 103–113. [DOI] [PubMed] [Google Scholar]
- Azumaya CM, Days EL, Vinson PN, Stauffer S, Sulikowski G, Weaver CD & Nakagawa T. (2017). Screening for AMPA receptor auxiliary subunit specific modulators. PLoS One 12, e0174742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yaacov A, Gillor M, Haham T, Parsai A, Qneibi M & Stern-Bach Y. (2017). Molecular Mechanism of AMPA Receptor Modulation by TARP/Stargazin. Neuron. [DOI] [PubMed] [Google Scholar]
- Blackstone CD, Moss SJ, Martin LJ, Levey AI, Price DL & Huganir RL. (1992). Biochemical characterization and localization of a non-N-methyl-D-aspartate glutamate receptor in rat brain. J Neurochem 58, 1118–1126. [DOI] [PubMed] [Google Scholar]
- Bonnet C, Leheup B, Beri M, Philippe C, Gregoire MJ & Jonveaux P. (2009). Aberrant GRIA3 transcripts with multi-exon duplications in a family with X-linked mental retardation. Am J Med Genet A 149A, 1280–1289. [DOI] [PubMed] [Google Scholar]
- Boudkkazi S, Brechet A, Schwenk J & Fakler B. (2014). Cornichon2 dictates the time course of excitatory transmission at individual hippocampal synapses. Neuron 82, 848–858. [DOI] [PubMed] [Google Scholar]
- Brockie PJ, Jensen M, Mellem JE, Jensen E, Yamasaki T, Wang R, Maxfield D, Thacker C, Hoerndli F, Dunn PJ, Tomita S, Madsen DM & Maricq AV. (2013). Cornichons Control ER Export of AMPA Receptors to Regulate Synaptic Excitability. Neuron 80, 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P, McGuire H & Bowie D. (2018). Stargazin and cornichon-3 relieve polyamine block of AMPA receptors by enhancing blocker permeation. J Gen Physiol 150, 67–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burada AP, Vinnakota R & Kumar J. (2020). Cryo-EM structures of the ionotropic glutamate receptor GluD1 reveal a non-swapped architecture. Nat Struct Mol Biol 27, 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cais O, Herguedas B, Krol K, Cull-Candy SG, Farrant M & Greger IH. (2014). Mapping the interaction sites between AMPA receptors and TARPs reveals a role for the receptor N-terminal domain in channel gating. Cell Rep 9, 728–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo E, Shaikh SA, Berka V, Durham RJ, Litwin DB, Lee G, MacLean DM, Nowak LM & Jayaraman V. (2020). Mechanism of modulation of AMPA receptors by TARP-gamma8. J Gen Physiol 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PK, Verbich D & McKinney RA. (2012). AMPA receptors as drug targets in neurological disease--advantages, caveats, and future outlook. Eur J Neurosci 35, 1908–1916. [DOI] [PubMed] [Google Scholar]
- Chen C, Blitz DM & Regehr WG. (2002). Contributions of receptor desensitization and saturation to plasticity at the retinogeniculate synapse. Neuron 33, 779–788. [DOI] [PubMed] [Google Scholar]
- Chen C & Regehr WG. (2000). Developmental remodeling of the retinogeniculate synapse. Neuron 28, 955–966. [DOI] [PubMed] [Google Scholar]
- Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS & Nicoll RA. (2000). Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature 408, 936–943. [DOI] [PubMed] [Google Scholar]
- Chen L, Durr KL & Gouaux E. (2014). X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism. Science 345, 1021–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S & Gouaux E. (2019). Structure and mechanism of AMPA receptor - auxiliary protein complexes. Curr Opin Struct Biol 54, 104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhao Y, Wang Y, Shekhar M, Tajkhorshid E & Gouaux E. (2017). Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM. Cell 170, 1234–1246 e1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Aslam M, Gollisch T, Allen K & von Engelhardt J. (2018). CKAMP44 modulates integration of visual inputs in the lateral geniculate nucleus. Nat Commun 9, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiyonobu T, Hayashi S, Kobayashi K, Morimoto M, Miyanomae Y, Nishimura A, Nishimoto A, Ito C, Imoto I, Sugimoto T, Jia Z, Inazawa J & Toda T. (2007). Partial tandem duplication of GRIA3 in a male with mental retardation. Am J Med Genet A 143A, 1448–1455. [DOI] [PubMed] [Google Scholar]
- Cho CH, St-Gelais F, Zhang W, Tomita S & Howe JR. (2007). Two families of TARP isoforms that have distinct effects on the kinetic properties of AMPA receptors and synaptic currents. Neuron 55, 890–904. [DOI] [PubMed] [Google Scholar]
- Collingridge GL, Olsen RW, Peters J & Spedding M. (2009). A nomenclature for ligand-gated ion channels. Neuropharmacology 56, 2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs ID, Soto D, Zonouzi M, Renzi M, Shelley C, Farrant M & Cull-Candy SG. (2012). Cornichons modify channel properties of recombinant and glial AMPA receptors. J Neurosci 32, 9796–9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B, Brown LA, Cais O, Watson J, Clayton AJ, Chang VT, Biggs D, Preece C, Hernandez-Pliego P, Krohn J, Bhomra A, Twigg SRF, Rimmer A, Kanapin A, Consortium WGS, Sen A, Zaiwalla Z, McVean G, Foster R, Donnelly P, Taylor JC, Blair E, Nutt D, Aricescu AR, Greger IH, Peirson SN, Flint J & Martin HC. (2017). A point mutation in the ion conduction pore of AMPA receptor GRIA3 causes dramatically perturbed sleep patterns as well as intellectual disability. Hum Mol Genet 26, 3869–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe GB, Kadir MF, Venskutonyte R, Perozzo AM, Yan Y, Alexander RPD, Navarrete C, Santander EA, Arsenault M, Fuentes C, Aurousseau MRP, Frydenvang K, Barrera NP, Kastrup JS, Edwardson JM & Bowie D. (2019). Nanoscale Mobility of the Apo State and TARP Stoichiometry Dictate the Gating Behavior of Alternatively Spliced AMPA Receptors. Neuron 102, 976–992 e975. [DOI] [PubMed] [Google Scholar]
- Dawe GB, Musgaard M, Aurousseau MR, Nayeem N, Green T, Biggin PC & Bowie D. (2016). Distinct Structural Pathways Coordinate the Activation of AMPA Receptor-Auxiliary Subunit Complexes. Neuron 89, 1264–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Alonso J, Sun YJ, Granger AJ, Levy JM, Blankenship SM & Nicoll RA. (2017). Subunit-specific role for the amino-terminal domain of AMPA receptors in synaptic targeting. Proc Natl Acad Sci U S A 114, 7136–7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr KL, Chen L, Stein RA, De Zorzi R, Folea IM, Walz T, McHaourab HS & Gouaux E. (2014). Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States. Cell 158, 778–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlenhardt N, Yu H, Abiraman K, Yamasaki T, Wadiche JI, Tomita S & Bredt DS. (2016). Porcupine Controls Hippocampal AMPAR Levels, Composition, and Synaptic Transmission. Cell Rep 14, 782–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett KV, Chioza B, Aicardi J, Aschauer H, Brouwer O, Callenbach P, Covanis A, Dulac O, Eeg-Olofsson O, Feucht M, Friis M, Goutieres F, Guerrini R, Heils A, Kjeldsen M, Lehesjoki AE, Makoff A, Nabbout R, Olsson I, Sander T, Siren A, McKeigue P, Robinson R, Taske N, Rees M & Gardiner M. (2007). Linkage and association analysis of CACNG3 in childhood absence epilepsy. Eur J Hum Genet 15, 463–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floor K, Baroy T, Misceo D, Kanavin OJ, Fannemel M & Frengen E. (2012). A 1 Mb de novo deletion within 11q13.1q13.2 in a boy with mild intellectual disability and minor dysmorphic features. Eur J Med Genet 55, 695–699. [DOI] [PubMed] [Google Scholar]
- Fukaya M, Yamazaki M, Sakimura K & Watanabe M. (2005). Spatial diversity in gene expression for VDCCgamma subunit family in developing and adult mouse brains. Neurosci Res 53, 376–383. [DOI] [PubMed] [Google Scholar]
- Garcia-Nafria J, Herguedas B, Watson JF & Greger IH. (2016). The dynamic AMPA receptor extracellular region: a platform for synaptic protein interactions. J Physiol 594, 5449–5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardinier KM, Gernert DL, Porter WJ, Reel JK, Ornstein PL, Spinazze P, Stevens FC, Hahn P, Hollinshead SP, Mayhugh D, Schkeryantz J, Khilevich A, De Frutos O, Gleason SD, Kato AS, Luffer-Atlas D, Desai PV, Swanson S, Burris KD, Ding C, Heinz BA, Need AB, Barth VN, Stephenson GA, Diseroad BA, Woods TA, Yu H, Bredt D & Witkin JM. (2016). Discovery of the First alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptor Antagonist Dependent upon Transmembrane AMPA Receptor Regulatory Protein (TARP) gamma-8. J Med Chem 59, 4753–4768. [DOI] [PubMed] [Google Scholar]
- Gill MB, Kato AS, Roberts MF, Yu H, Wang H, Tomita S & Bredt DS. (2011). Cornichon-2 modulates AMPA receptor-transmembrane AMPA receptor regulatory protein assembly to dictate gating and pharmacology. J Neurosci 31, 6928–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger IH, Watson JF & Cull-Candy SG. (2017). Structural and Functional Architecture of AMPA-Type Glutamate Receptors and Their Auxiliary Proteins. Neuron 94, 713–730. [DOI] [PubMed] [Google Scholar]
- Gu X, Mao X, Lussier MP, Hutchison MA, Zhou L, Hamra FK, Roche KW & Lu W. (2016). GSG1L suppresses AMPA receptor-mediated synaptic transmission and uniquely modulates AMPA receptor kinetics in hippocampal neurons. Nat Commun 7, 10873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackmann K, Matko S, Gerlach EM, von der Hagen M, Klink B, Schrock E, Rump A & Di Donato N. (2013). Partial deletion of GLRB and GRIA2 in a patient with intellectual disability. Eur J Hum Genet 21, 112–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Gauthier J, Araki Y, Lin DT, Yoshizawa Y, Higashi K, Park AR, Spiegelman D, Dobrzeniecka S, Piton A, Tomitori H, Daoud H, Massicotte C, Henrion E, Diallo O, Group SD, Shekarabi M, Marineau C, Shevell M, Maranda B, Mitchell G, Nadeau A, D’Anjou G, Vanasse M, Srour M, Lafreniere RG, Drapeau P, Lacaille JC, Kim E, Lee JR, Igarashi K, Huganir RL, Rouleau GA & Michaud JL. (2011). Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet 88, 306–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Li J, Pelkey KA, Pandey S, Chen X, Wang YX, Wu K, Ge L, Li T, Castellano D, Liu C, Wu LG, Petralia RS, Lynch JW, McBain CJ & Lu W. (2019). Shisa7 is a GABAA receptor auxiliary subunit controlling benzodiazepine actions. Science 366, 246–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Furukawa H & Traynelis SF. (2010). Control of assembly and function of glutamate receptors by the amino-terminal domain. Mol Pharmacol 78, 535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawken NM, Zaika EI & Nakagawa T. (2017). Engineering defined membrane-embedded elements of AMPA receptor induces opposing gating modulation by cornichon 3 and stargazin. J Physiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herguedas B, Garcia-Nafria J, Cais O, Fernandez-Leiro R, Krieger J, Ho H & Greger IH. (2016). Structure and organization of heteromeric AMPA-type glutamate receptors. Science 352, aad3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herguedas B, Watson JF, Ho H, Cais O, Garcia-Nafria J & Greger IH. (2019). Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP gamma8. Science 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering H, Lin CC & Sheng M. (2003). Lipid rafts in the maintenance of synapses, dendritic spines, and surface AMPA receptor stability. J Neurosci 23, 3262–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring BE, Shi Y, Suh YH, Zheng CY, Blankenship SM, Roche KW & Nicoll RA. (2013). Cornichon proteins determine the subunit composition of synaptic AMPA receptors. Neuron 77, 1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmann M, Maron C & Heinemann S. (1994). N-glycosylation site tagging suggests a three transmembrane domain topology for the glutamate receptor GluR1. Neuron 13, 1331–1343. [DOI] [PubMed] [Google Scholar]
- Hoshino H, Uchida T, Otsuki T, Kawamoto S, Okubo K, Takeichi M & Chisaka O. (2007). Cornichon-like protein facilitates secretion of HB-EGF and regulates proper development of cranial nerves. Mol Biol Cell 18, 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huganir RL & Nicoll RA. (2013). AMPARs and synaptic plasticity: the last 25 years. Neuron 80, 704–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AC & Nicoll RA. (2011). The expanding social network of ionotropic glutamate receptors: TARPs and other transmembrane auxiliary subunits. Neuron 70, 178–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y & Gouaux E. (2009). Crystal structure and association behaviour of the GluR2 amino-terminal domain. Embo J 28, 1812–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalashnikova E, Lorca RA, Kaur I, Barisone GA, Li B, Ishimaru T, Trimmer JS, Mohapatra DP & Diaz E. (2010). SynDIG1: an activity-regulated, AMPA-receptor-interacting transmembrane protein that regulates excitatory synapse development. Neuron 65, 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MG, Nuriya M, Guo Y, Martindale KD, Lee DZ & Huganir RL. (2012). Proteomic analysis of AMPA receptor complexes. J Biol Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato AS, Burris KD, Gardinier KM, Gernert DL, Porter WJ, Reel J, Ding C, Tu Y, Schober DA, Lee MR, Heinz BA, Fitch TE, Gleason SD, Catlow JT, Yu H, Fitzjohn SM, Pasqui F, Wang H, Qian Y, Sher E, Zwart R, Wafford KA, Rasmussen K, Ornstein PL, Isaac JT, Nisenbaum ES, Bredt DS & Witkin JM. (2016). Forebrain-selective AMPA-receptor antagonism guided by TARP gamma-8 as an antiepileptic mechanism. Nat Med 22, 1496–1501. [DOI] [PubMed] [Google Scholar]
- Kato AS, Gill MB, Ho MT, Yu H, Tu Y, Siuda ER, Wang H, Qian YW, Nisenbaum ES, Tomita S & Bredt DS. (2010). Hippocampal AMPA Receptor Gating Controlled by Both TARP and Cornichon Proteins. Neuron 68, 1082–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato AS, Siuda ER, Nisenbaum ES & Bredt DS. (2008). AMPA receptor subunit-specific regulation by a distinct family of type II TARPs. Neuron 59, 986–996. [DOI] [PubMed] [Google Scholar]
- Khodosevich K, Jacobi E, Farrow P, Schulmann A, Rusu A, Zhang L, Sprengel R, Monyer H & von Engelhardt J. (2014). Coexpressed auxiliary subunits exhibit distinct modulatory profiles on AMPA receptor function. Neuron 83, 601–615. [DOI] [PubMed] [Google Scholar]
- Kielland A & Heggelund P. (2002). AMPA and NMDA currents show different short-term depression in the dorsal lateral geniculate nucleus of the rat. J Physiol 542, 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E & Sheng M. (2004). PDZ domain proteins of synapses. Nat Rev Neurosci 5, 771–781. [DOI] [PubMed] [Google Scholar]
- Klaassen RV, Stroeder J, Coussen F, Hafner AS, Petersen JD, Renancio C, Schmitz LJ, Normand E, Lodder JC, Rotaru DC, Rao-Ruiz P, Spijker S, Mansvelder HD, Choquet D & Smit AB. (2016). Shisa6 traps AMPA receptors at postsynaptic sites and prevents their desensitization during synaptic activity. Nat Commun 7, 10682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek M, Vyklicky V, Borovska J, Lichnerova K, Kaniakova M, Krausova B, Krusek J, Balik A, Smejkalova T, Horak M & Vyklicky L. (2015). Cholesterol modulates open probability and desensitization of NMDA receptors. J Physiol 593, 2279–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MR, Gardinier KM, Gernert DL, Schober DA, Wright RA, Wang H, Qian Y, Witkin JM, Nisenbaum ES & Kato AS. (2017). Structural Determinants of the gamma-8 TARP Dependent AMPA Receptor Antagonist. ACS Chem Neurosci 8, 2631–2647. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA & Jones AR. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. [DOI] [PubMed] [Google Scholar]
- Liu YL, Fann CS, Liu CM, Chen WJ, Wu JY, Hung SI, Chen CH, Jou YS, Liu SK, Hwang TJ, Hsieh MH, Chang CC, Yang WC, Lin JJ, Chou FH, Faraone SV, Tsuang MT & Hwu HG. (2008). RASD2, MYH9, and CACNG2 genes at chromosome 22q12 associated with the subgroup of schizophrenia with non-deficit in sustained attention and executive function. Biol Psychiatry 64, 789–796. [DOI] [PubMed] [Google Scholar]
- Long SB, Tao X, Campbell EB & MacKinnon R. (2007). Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382. [DOI] [PubMed] [Google Scholar]
- Maher MP, Matta JA, Gu S, Seierstad M & Bredt DS. (2017). Getting a Handle on Neuropharmacology by Targeting Receptor-Associated Proteins. Neuron 96, 989–1001. [DOI] [PubMed] [Google Scholar]
- Matt L, Kirk LM, Chenaux G, Speca DJ, Puhger KR, Pride MC, Qneibi M, Haham T, Plambeck KE, Stern-Bach Y, Silverman JL, Crawley JN, Hell JW & Diaz E. (2018). SynDIG4/Prrt1 Is Required for Excitatory Synapse Development and Plasticity Underlying Cognitive Function. Cell Rep 22, 2246–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGoldrick LL, Singh AK, Saotome K, Yelshanskaya MV, Twomey EC, Grassucci RA & Sobolevsky AI. (2018). Opening of the human epithelial calcium channel TRPV6. Nature 553, 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML & Subramaniam S. (2014). Structural mechanism of glutamate receptor activation and desensitization. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein AD & Nicoll RA. (2009). TARP modulation of synaptic AMPA receptor trafficking and gating depends on multiple intracellular domains. Proc Natl Acad Sci U S A 106, 11348–11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milstein AD, Zhou W, Karimzadegan S, Bredt DS & Nicoll RA. (2007). TARP subtypes differentially and dose-dependently control synaptic AMPA receptor gating. Neuron 55, 905–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T (2010). The biochemistry, ultrastructure, and subunit assembly mechanism of AMPA receptors. Mol Neurobiol 42, 161–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T (2019). Structures of the AMPA receptor in complex with its auxiliary subunit cornichon. Science 366, 1259–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Cheng Y, Ramm E, Sheng M & Walz T. (2005). Structure and different conformational states of native AMPA receptor complexes. Nature 433, 545–549. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Cheng Y, Sheng M & Walz T. (2006). Three-dimensional structure of an AMPA receptor without associated stargazin/TARP proteins. Biol Chem 387, 179–187. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Irie K, Tanaka H, Nishikawa K, Suzuki H, Saitoh Y, Tamura A, Tsukita S & Fujiyoshi Y. (2019). Morphologic determinant of tight junctions revealed by claudin-3 structures. Nat Commun 10, 816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden KK, Chen W, Swanger SA, McDaniel MJ, Fan LZ, Hu C, Tankovic A, Kusumoto H, Kosobucki GJ, Schulien AJ, Su Z, Pecha J, Bhattacharya S, Petrovski S, Cohen AE, Aizenman E, Traynelis SF & Yuan H. (2017). Molecular Mechanism of Disease-Associated Mutations in the Pre-M1 Helix of NMDA Receptors and Potential Rescue Pharmacology. PLoS Genet 13, e1006536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Chavez AE, Mineur YS, Morimoto-Tomita M, Lutzu S, Kim KS, Picciotto MR, Castillo PE & Tomita S. (2016). CaMKII Phosphorylation of TARPgamma-8 Is a Mediator of LTP and Learning and Memory. Neuron 92, 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe A, Malan V, Jacquemont ML, Boddaert N, Bonnefont JP, Odent S, Munnich A, Colleaux L & Cormier-Daire V. (2013). Xq25 duplications encompassing GRIA3 and STAG2 genes in two families convey recognizable X-linked intellectual disability with distinctive facial appearance. Am J Med Genet A 161A, 1370–1375. [DOI] [PubMed] [Google Scholar]
- Priel A, Kolleker A, Ayalon G, Gillor M, Osten P & Stern-Bach Y. (2005). Stargazin reduces desensitization and slows deactivation of the AMPA-type glutamate receptors. J Neurosci 25, 2682–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva I, Eibl C, Volkmer R, Carbone AL & Plested AJ. (2017). Control of AMPA receptor activity by the extracellular loops of auxiliary proteins. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogawski MA. (2011). Revisiting AMPA receptors as an antiepileptic drug target. Epilepsy Curr 11, 56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M, Sukumaran M, Penn AC, Veprintsev DB, Babu MM & Greger IH. (2011). Subunit-selective N-terminal domain associations organize the formation of AMPA receptor heteromers. EMBO J 30, 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouach N, Byrd K, Petralia RS, Elias GM, Adesnik H, Tomita S, Karimzadegan S, Kealey C, Bredt DS & Nicoll RA. (2005). TARP gamma-8 controls hippocampal AMPA receptor number, distribution and synaptic plasticity. Nat Neurosci 8, 1525–1533. [DOI] [PubMed] [Google Scholar]
- Saitoh Y, Suzuki H, Tani K, Nishikawa K, Irie K, Ogura Y, Tamura A, Tsukita S & Fujiyoshi Y. (2015). Tight junctions. Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin. Science 347, 775–778. [DOI] [PubMed] [Google Scholar]
- Salpietro V, Dixon CL, Guo H, Bello OD, Vandrovcova J, Efthymiou S, Maroofian R, Heimer G, Burglen L, Valence S, Torti E, Hacke M, Rankin J, Tariq H, Colin E, Procaccio V, Striano P, Mankad K, Lieb A, Chen S, Pisani L, Bettencourt C, Mannikko R, Manole A, Brusco A, Grosso E, Ferrero GB, Armstrong-Moron J, Gueden S, Bar-Yosef O, Tzadok M, Monaghan KG, Santiago-Sim T, Person RE, Cho MT, Willaert R, Yoo Y, Chae JH, Quan Y, Wu H, Wang T, Bernier RA, Xia K, Blesson A, Jain M, Motazacker MM, Jaeger B, Schneider AL, Boysen K, Muir AM, Myers CT, Gavrilova RH, Gunderson L, Schultz-Rogers L, Klee EW, Dyment D, Osmond M, Parellada M, Llorente C, Gonzalez-Penas J, Carracedo A, Van Haeringen A, Ruivenkamp C, Nava C, Heron D, Nardello R, Iacomino M, Minetti C, Skabar A, Fabretto A, Group SS, Raspall-Chaure M, Chez M, Tsai A, Fassi E, Shinawi M, Constantino JN, De Zorzi R, Fortuna S, Kok F, Keren B, Bonneau D, Choi M, Benzeev B, Zara F, Mefford HC, Scheffer IE, Clayton-Smith J, Macaya A, Rothman JE, Eichler EE, Kullmann DM & Houlden H. (2019). AMPA receptor GluA2 subunit defects are a cause of neurodevelopmental disorders. Nat Commun 10, 3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz LJM, Klaassen RV, Ruiperez-Alonso M, Zamri AE, Stroeder J, Rao-Ruiz P, Lodder JC, van der Loo RJ, Mansvelder HD, Smit AB & Spijker S. (2017). The AMPA receptor-associated protein Shisa7 regulates hippocampal synaptic function and contextual memory. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk J, Baehrens D, Haupt A, Bildl W, Boudkkazi S, Roeper J, Fakler B & Schulte U. (2014). Regional diversity and developmental dynamics of the AMPA-receptor proteome in the mammalian brain. Neuron 84, 41–54. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Boudkkazi S, Kocylowski MK, Brechet A, Zolles G, Bus T, Costa K, Kollewe A, Jordan J, Bank J, Bildl W, Sprengel R, Kulik A, Roeper J, Schulte U & Fakler B. (2019). An ER Assembly Line of AMPA-Receptors Controls Excitatory Neurotransmission and Its Plasticity. Neuron. [DOI] [PubMed] [Google Scholar]
- Schwenk J & Fakler B. (2019). Folding unpredicted. Science 366, 1194–1195. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Brechet A, Zolles G, Berkefeld H, Muller CS, Bildl W, Baehrens D, Huber B, Kulik A, Klocker N, Schulte U & Fakler B. (2012). High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron 74, 621–633. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Zolles G, Bildl W, Kulik A, Heimrich B, Chisaka O, Jonas P, Schulte U, Fakler B & Klocker N. (2009). Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science 323, 1313–1319. [DOI] [PubMed] [Google Scholar]
- Shanks NF, Cais O, Maruo T, Savas JN, Zaika EI, Azumaya CM, Yates JR 3rd, Greger I & Nakagawa T. (2014). Molecular Dissection of the Interaction between the AMPA Receptor and Cornichon Homolog-3. J Neurosci 34, 12104–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks NF, Maruo T, Farina AN, Ellisman MH & Nakagawa T. (2010). Contribution of the global subunit structure and stargazin on the maturation of AMPA receptors. J Neurosci 30, 2728–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks NF, Savas JN, Maruo T, Cais O, Hirao A, Oe S, Ghosh A, Noda Y, Greger IH, Yates JR 3rd & Nakagawa T. (2012). Differences in AMPA and kainate receptor interactomes facilitate identification of AMPA receptor auxiliary subunit GSG1L. Cell Rep 1, 590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelley C, Farrant M & Cull-Candy SG. (2012). TARP-associated AMPA receptors display an increased maximum channel conductance and multiple kinetically distinct open states. J Physiol 590, 5723–5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Cummings J, Roldan LA, Jan YN & Jan LY. (1994). Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature 368, 144–147. [DOI] [PubMed] [Google Scholar]
- Sheng N, Bemben MA, Diaz-Alonso J, Tao W, Shi YS & Nicoll RA. (2018). LTP requires postsynaptic PDZ-domain interactions with glutamate receptor/auxiliary protein complexes. Proc Natl Acad Sci U S A 115, 3948–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi EY, Yuan CL, Sipple MT, Srinivasan J, Ptak CP, Oswald RE & Nowak LM. (2019). Noncompetitive antagonists induce cooperative AMPA receptor channel gating. J Gen Physiol 151, 156–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lu W, Milstein AD & Nicoll RA. (2009). The stoichiometry of AMPA receptors and TARPs varies by neuronal cell type. Neuron 62, 633–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Suh YH, Milstein AD, Isozaki K, Schmid SM, Roche KW & Nicoll RA. (2010). Functional comparison of the effects of TARPs and cornichons on AMPA receptor trafficking and gating. Proc Natl Acad Sci U S A 107, 16315–16319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia GM, Beique JC, Rumbaugh G, Cho R, Worley PF & Huganir RL. (2007). Interaction of the N-terminal domain of the AMPA receptor GluR4 subunit with the neuronal pentraxin NP1 mediates GluR4 synaptic recruitment. Neuron 55, 87–102. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP & Gouaux E. (2009). X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 745–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumioka A, Brown TE, Kato AS, Bredt DS, Kauer JA & Tomita S. (2011). PDZ binding of TARPgamma-8 controls synaptic transmission but not synaptic plasticity. Nat Neurosci 14, 1410–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Comeau JF & Baenziger JE. (2017). Probing the structure of the uncoupled nicotinic acetylcholine receptor. Biochim Biophys Acta Biomembr 1859, 146–154. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Nishizawa T, Tani K, Yamazaki Y, Tamura A, Ishitani R, Dohmae N, Tsukita S, Nureki O & Fujiyoshi Y. (2014). Crystal structure of a claudin provides insight into the architecture of tight junctions. Science 344, 304–307. [DOI] [PubMed] [Google Scholar]
- Tajima N, Karakas E, Grant T, Simorowski N, Diaz-Avalos R, Grigorieff N & Furukawa H. (2016). Activation of NMDA receptors and the mechanism of inhibition by ifenprodil. Nature 534, 63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Gamal El-Din TM, Swanson TM, Pryde DC, Scheuer T, Zheng N & Catterall WA. (2016). Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs. Nature 537, 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S, Adesnik H, Sekiguchi M, Zhang W, Wada K, Howe JR, Nicoll RA & Bredt DS. (2005). Stargazin modulates AMPA receptor gating and trafficking by distinct domains. Nature 435, 1052–1058. [DOI] [PubMed] [Google Scholar]
- Tomita S, Chen L, Kawasaki Y, Petralia RS, Wenthold RJ, Nicoll RA & Bredt DS. (2003). Functional studies and distribution define a family of transmembrane AMPA receptor regulatory proteins. J Cell Biol 161, 805–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis J, Silk M, Wang Q, Berkovic SF, Liu L, Ascher DB, Balding DJ & Petrovski S. (2017). Optimizing genomic medicine in epilepsy through a gene-customized approach to missense variant interpretation. Genome Res 27, 1715–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ & Dingledine R. (2010). Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev 62, 405–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J & Sobolevsky AI. (2016). Elucidation of AMPA receptor–stargazin complexes by cryo–electron microscopy. Science 353, 83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J & Sobolevsky AI. (2017a). Channel opening and gating mechanism in AMPA-subtype glutamate receptors. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J & Sobolevsky AI. (2017b). Structural Bases of Desensitization in AMPA Receptor-Auxiliary Subunit Complexes. Neuron 94, 569–580 e565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, Yelshanskaya MV & Sobolevsky AI. (2019). Structural and functional insights into transmembrane AMPA receptor regulatory protein complexes. J Gen Physiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twomey EC, Yelshanskaya MV, Vassilevski AA & Sobolevsky AI. (2018). Mechanisms of Channel Block in Calcium-Permeable AMPA Receptors. Neuron 99, 956–968 e954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzschach A, Menzel C, Erdogan F, Istifli ES, Rieger M, Ovens-Raeder A, Macke A, Ropers HH, Ullmann R & Kalscheuer V. (2010). Characterization of an interstitial 4q32 deletion in a patient with mental retardation and a complex chromosome rearrangement. Am J Med Genet A 152A, 1008–1012. [DOI] [PubMed] [Google Scholar]
- Vandenberghe W, Nicoll RA & Bredt DS. (2005). Stargazin is an AMPA receptor auxiliary subunit. Proc Natl Acad Sci U S A 102, 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Engelhardt J, Mack V, Sprengel R, Kavenstock N, Li KW, Stern-Bach Y, Smit AB, Seeburg PH & Monyer H. (2010). CKAMP44: a brain-specific protein attenuating short-term synaptic plasticity in the dentate gyrus. Science 327, 1518–1522. [DOI] [PubMed] [Google Scholar]
- Watson JF, Ho H & Greger IH. (2017). Synaptic transmission and plasticity require AMPA receptor anchoring via its N-terminal domain. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding TJ, Chai YH & Huettner JE. (1998). Inhibition of rat neuronal kainate receptors by cis-unsaturated fatty acids. J Physiol 513 ( Pt 2), 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Arai AC, Rumbaugh G, Srivastava AK, Turner G, Hayashi T, Suzuki E, Jiang Y, Zhang L, Rodriguez J, Boyle J, Tarpey P, Raymond FL, Nevelsteen J, Froyen G, Stratton M, Futreal A, Gecz J, Stevenson R, Schwartz CE, Valle D, Huganir RL & Wang T. (2007). Mutations in ionotropic AMPA receptor 3 alter channel properties and are associated with moderate cognitive impairment in humans. Proc Natl Acad Sci U S A 104, 18163–18168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wudick MM, Portes MT, Michard E, Rosas-Santiago P, Lizzio MA, Nunes CO, Campos C, Santa Cruz Damineli D, Carvalho JC, Lima PT, Pantoja O & Feijo JA. (2018). CORNICHON sorting and regulation of GLR channels underlie pollen tube Ca(2+) homeostasis. Science 360, 533–536. [DOI] [PubMed] [Google Scholar]
- Yelshanskaya MV, Li M & Sobolevsky AI. (2014). Structure of an agonist-bound ionotropic glutamate receptor. Science 345, 1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M, Diaz-Alonso J, Ye F, Chen X, Xu J, Ji Z, Nicoll RA & Zhang M. (2019). Phase Separation-Mediated TARP/MAGUK Complex Condensation and AMPA Receptor Synaptic Transmission. Neuron 104, 529–543 e526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Devi SP, Tomita S & Howe JR. (2014). Auxiliary proteins promote modal gating of AMPA- and kainate-type glutamate receptors. Eur J Neurosci 39, 1138–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Bulkley DP, Xin Y, Roberts KJ, Asarnow DE, Sharma A, Myers BR, Cho W, Cheng Y & Beachy PA. (2018). Structural Basis for Cholesterol Transport-like Activity of the Hedgehog Receptor Patched. Cell 175, 1352–1364 e1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chen S, Swensen AC, Qian WJ & Gouaux E. (2019). Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM. Science 364, 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chen S, Yoshioka C, Baconguis I & Gouaux E. (2016). Architecture of fully occupied GluA2 AMPA receptor-TARP complex elucidated by cryo-EM. Nature 536, 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker RS. (1999). Calcium- and activity-dependent synaptic plasticity. Curr Opin Neurobiol 9, 305–313. [DOI] [PubMed] [Google Scholar]