Abstract
Salient experiences are often relived in the mind. Human neuroimaging studies suggest that such experiences drive activity patterns in visual association cortex that are subsequently reactivated during quiet waking. Yet, the circuit-level consequences of such reactivations remain unclear. Here, we imaged hundreds of neurons in visual association cortex across days as mice learned a visual discrimination task. Distinct patterns of neurons were activated by different visual cues. These same patterns were subsequently reactivated during quiet waking in darkness, with higher reactivation rates during early learning and for food-predicting vs. neutral cues. Reactivations involving ensembles of neurons encoding both the food cue and the reward predicted strengthening of next-day functional connectivity of participating neurons, while the converse was observed for reactivations involving ensembles encoding only the food cue. We propose that task-relevant neurons strengthen, while task-irrelevant neurons weaken their dialogue with the network via participation in distinct flavors of reactivation.
Introduction
Sensory experiences activate brain-wide patterns of neurons. During subsequent quiet periods, memories of salient and unexpected recent experiences may become consolidated via synchronous reactivation of these patterns throughout sensory cortex, amygdala, and hippocampus (e.g. 1–7). Reactivation of recent experiences has been observed in prefrontal, motor, and primary sensory cortices3,6,8–10 as well as basolateral amygdala11, and is often synchronized with moments of increased sharp-wave ripple activity and associated replay of the experience in hippocampus. Furthermore, the content of replay in the hippocampus and amygdala is often biased to experiences that culminate in rewarding or aversive outcomes (e.g. 4,11). Such distributed reactivations of recent salient experiences have been hypothesized to promote memory consolidation (for a review, see 5), in part by selectively strengthening connections between neurons representing task-relevant information12 while globally weakening other connections13. Disrupting hippocampal sharp-wave ripples or their coupling with cortical activity impairs memory consolidation14–17, while increasing their coupling with cortical activity enhances learning18.
A key hub that links the hippocampus, sensory cortex, and amygdala is the lateral visual association cortex, a region that integrates cue and outcome information (e.g. 19) and that is necessary for offline memory consolidation and remote recall of salient cue-outcome associations20–22. Recently, human neuroimaging studies reported preferential reactivation of salient experiences in lateral visual association cortex23,24. Lateral visual association cortex becomes activated in synchrony with hippocampal ripples during quiet waking25, including during voluntary recall26. Successful recent encoding of cue-outcome associations correlates with higher correlations in ongoing activity between this region and hippocampus27. However, the circuit-level effects of reactivation are not well understood, as previous studies of reactivation have not tracked large-scale activity patterns across days.
Here, we used two-photon calcium imaging to track the same neurons in visual association cortex across days during learning of a behaviorally constrained visual task. In this way, we could characterize offline reactivations of sensory cues following each training session throughout learning, and the changes in the response properties and functional connectivity of cells that participate in these reactivations. During quiet waking, we observed brief reactivations of patterns of cortical activity that matched those previously evoked by specific sensory cues. These cortical reactivation events were synchronized with hippocampal ripple activity. The rate of reactivations was higher for salient cues and following sessions with poor task performance early in learning, and predicted behavioral improvement in the following session. Critically, our long-term imaging approach revealed that cells that participated in cue reactivations exhibited bidirectional changes in their next-day functional connectivity with the local network. Our findings support the hypothesis that different flavors of reactivation of previous cue presentations may selectively strengthen relevant ensembles of neurons encoding both the cue and the associated reward, while weakening intermingled ensembles of putatively task-irrelevant neurons.
Results
Food-restricted mice gradually learned to associate visual gratings drifting in one of three directions with rewarding liquid food delivery, aversive quinine delivery, or no outcome. Mice then learned a new set of associations following a subsequent switch in cue-outcome contingencies22 (Fig. 1a–c; Extended Data Fig. 1a). During and following each training session on this operant discrimination task, we performed chronic two-photon calcium imaging of hundreds of layer 2/3 excitatory neurons in lateral visual association cortex (Fig. 1c–d; in GCaMP6f transgenic mice22,28; see Methods), a region required for performance of this task22. In each session, we first tracked population activity during 90 minutes of task engagement. We then tracked spontaneous activity during a subsequent 90 minutes in darkness, which included periods of quiet waking (particularly following a satiation protocol involving ad libitum access to liquid food19). During quiet waking, we observed discrete, transient (~100–200 ms) reactivations of population activity patterns previously observed upon presentation of a specific visual cue (‘cue reactivations’; see Fig. 1e, Fig. 2a–c, Extended Data Fig. 1b–d, Extended Data Fig. 2c, and Supplementary Fig. 1).
Fig. 1: Two-photon calcium imaging of visual association cortex during and following task engagement.

a-b. We used two-photon calcium imaging to record activity of hundreds of cells in retinotopically identified lateral visual association cortex (a) in hungry mice during and following engagement in a task (b). c. Mice gradually learned across sessions that licking in the 2-s period following presentation of one of three visual drifting gratings resulted in delivery of milkshake, quinine, or no outcome (see also Extended Data Fig. 1a). Upon task performance at greater than 90% accuracy for three days, the cue-outcome associations were changed. We recorded visual responses during the two-hour training period, and subsequently assessed the presence of reactivations of cue representations in darkness, before and after satiation. d. Example field of view (depth: 190 μm, from one of 109 imaging sessions). e. Top: example activity pattern during a single trial (from among the 180 trials during the session) in response to presentation of an aversive cue (peak activity during 2-s stimulus presentation). Bottom: example activity pattern observed two hours later in darkness during a single aversive-cue reactivation event (peak activity during 260 ms surrounding reactivation, selected from 30 reactivation events identified during this recording session; see also Fig. 2). Activity is represented on a scale from no activity (white) to high activity (red).
Fig. 2: Visual association cortex exhibits reactivation of visual cue representations in awake mice.
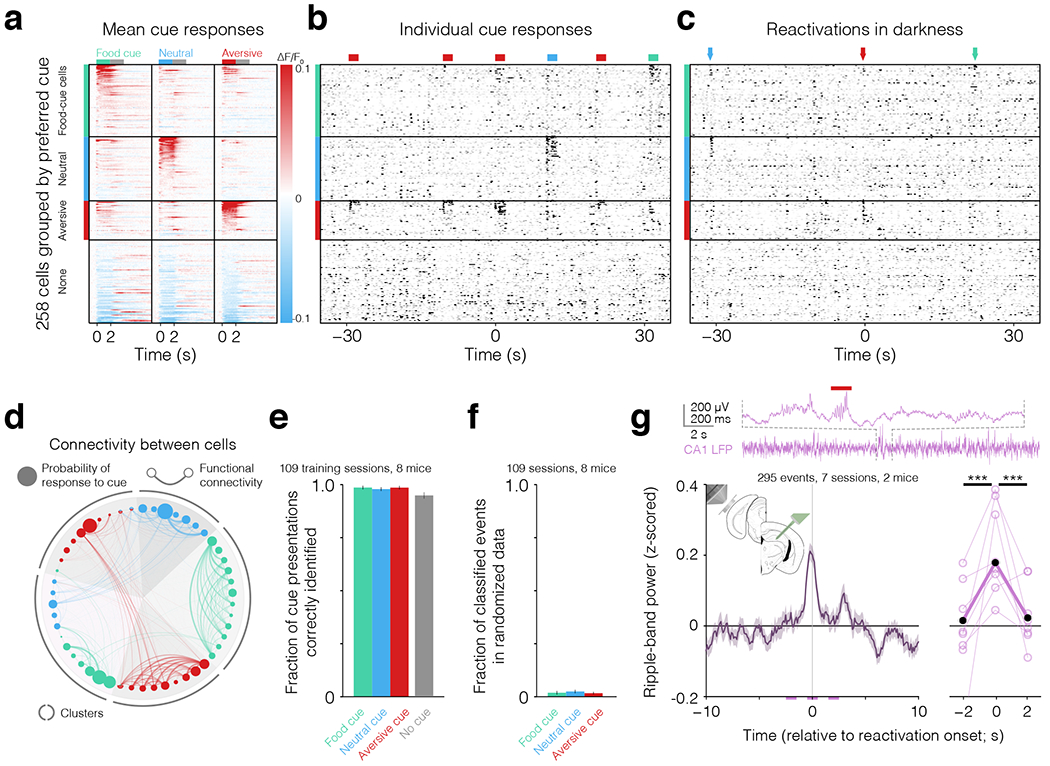
a. Mean activity time courses (fractional change in fluorescence, ΔF/F0) in response to food cues, neutral cues, and aversive cues (columns) for all simultaneously recorded neurons (rows) from one session, organized by preferred response. b. Example deconvolved activity (scaled by mean activity per cell) across individual cue presentations during task performance. The cue-evoked response at t=0 s is the same as in Fig. 1e, top. c. Example deconvolved activity during darkness. Arrows indicate reactivation events. The reactivation event at t=0 s is the same as in Fig. 1e, bottom. d. Example single-session probability estimates used by the averaged one-dependence estimator (AODE) classifier. Estimates of response probabilities of cue-driven neurons (proportional to thresholded, deconvolved calcium activity, see Methods) are represented by the diameter of circles along outer ring (gray circle diameter indicates a probability of 1), and estimates of joint response probabilities (related to co-variation in deconvolved activity of pairs of cells) are proportional to the thickness of the connecting lines. Cue-responsive cells in panel a cluster into five groups (shaded triangle wedges) based on trial-by-trial response co-fluctuations, defined using noise correlations (see Methods). e. Fraction of cue presentations and inter-trial intervals correctly identified by our AODE classifier when trained on two-thirds of the data and tested on the remaining third (mean ± SEM across 109 sessions; see also Extended Data Fig. 3a). f. Fraction of cue reactivation events in real data that were identified in data randomized by cell identity (see also Extended Data Fig. 3d). False positive rates were <5%. g. In 7 sessions from 2 mice, we imaged visual association cortex while recording from hippocampal area CA1 using a silicon multi-electrode probe (see inset). Top: purple trace and zoom-in show an example hippocampal CA1 local field potential (LFP) trace containing a sharp-wave and ripple immediately prior to an imaged aversive-cue reactivation event (red bar; see also Extended Data Fig. 4a–b). Bottom: reactivation events correlate with a transient increase in hippocampal sharp-wave ripple band power. Left: mean ± SEM across 295 reactivation events. Right: thin lines: mean of each session, thick line: mean across sessions. A generalized linear mixed-effects model (GLMM) comparing power between 1-s bins (marked by purple horizontal lines below left panel) confirmed an increase in ripple-band power around reactivation events (*** p < 0.001; accounting for shared variance within mouse and within day).
Identification of cue reactivation events
To identify cue reactivation events, we first designed a probabilistic classifier that could decode the presence and identity of visual cue presentations using single-trial population responses during task engagement, and then applied this classifier to subsequent recordings of spontaneous activity. We found that cells preferentially driven by a given cue were often correlated in their trial-to-trial cue-evoked responses, and that subsets of cells could be clustered based on high within-cluster pairwise correlations (Fig 2d). Thus, to capture information not only from the activity of individual cells but also from transient co-activation of pairs of cells during individual presentations of a given cue, we developed a classifier that employed an averaged one-dependence estimator (AODE, an extension to the Naïve Bayes classifier that does not assume that the activity of each cell is independent; Extended Data Fig. 2a–c; see Methods and Supplementary Information). We confirmed that our classifier could accurately identify cue presentations (97% accuracy; Fig. 2e and examples in Fig. 2b). By applying this classifier to recordings during quiet waking in darkness, we identified cue reactivations (~0.6 events/min; Fig. 2c, Extended Data Fig. 1b–d, Extended Data Fig. 2c–d, and Supplementary Fig. 1) consisting of patterns of population activity that were compressed in time but otherwise similar to those observed during cue presentations. Only half as many reactivation events were detected using a simpler, modified Naïve Bayes classifier. This finding demonstrates that accounting for co-fluctuations in neurons’ trial-by-trial food-cue-evoked activity can reveal additional food-cue reactivation events involving smaller yet particularly correlated subsets of food-cue-driven neurons (Extended Data Fig. 2e–f; cf. 29). Critically, our classifier had a low false positive rate, identifying far fewer cue reactivation events when applied to data for which the activity time course of each cell had been randomly shifted in time, or had been shuffled across cell identities (both with or without normalization of overall activity levels across cells; Fig. 2f, Extended Data Fig. 3).
Previous electrophysiological studies used hippocampal local field potential activity in the “ripple” band (150-300 Hz) as a temporal anchor for examining cortical reactivations3,6,8–11,14,16 and hippocampal reactivations30. To assess whether the cue reactivation events we identified solely from calcium imaging data were similar to these previously reported reactivations, we imaged visual association cortex while simultaneously recording contralateral hippocampal field potentials using a laminar silicon multi-electrode array. We found a significant increase in the mean ripple band power during cortical cue reactivations (Fig. 2g, Extended Data Fig. 4a–b). We confirmed that these cortical cue reactivations were not related to cortical or hippocampal epileptiform events (Extended Data Fig. 4c–e and Supplementary Information). We also analyzed pupil area, as hippocampal ripples are more common during moments of pupil constriction during quiet waking31. We found that cue reactivation events occurred during quiet waking periods involving pupil constriction and minimal eye or body movement (Extended Data Fig. 4f–h). Thus, cortical cue reactivations are associated with hippocampal ripple activity during normal states of quiet waking, raising the possibility that they may contribute to learning of recent experiences.
Cue reactivation rates are inversely correlated with recent task performance
We reasoned that if cortical cue reactivations were associated with learning, they should be (i) relatively rare prior to any pairing of cues with outcomes, (ii) more common during sessions with poor task performance, when learning rates are high, and (iii) rare in sessions in which mice have successfully learned the cue-outcome associations. Indeed, behavioral discrimination between stimuli (d’) was inversely correlated with rates of reactivation of the food, neutral, and aversive cues (Fig. 3a), using a generalized linear mixed-effects regression model accounting for different offsets per mouse. The inverse correlations between reactivation rates and behavioral performance were not due to greater novelty of the cues during early sessions with poor performance, as reactivation rates were lower in naïve mice during initial passive viewing of the same cues, prior to any pairing of cues with salient outcomes (Fig. 3a and Extended Data Fig. 5a). In addition, these effects were not due to a performance-dependent change in either overall population activity (Extended Data Fig. 5b–c) or in overall rate of transient, synchronous population events (Fig. 3b), of which cue reactivations are a small subset. In contrast to the correlation between the average performance across a training session and the rate of subsequent cue reactivations, we did not find a correlation between change in performance from the start to the end of the training session and the rate of subsequent cue reactivations (Extended Data Fig. 5d–f). These data suggest that reactivation rates scale with overall error rates during the preceding behavioral session.
Fig. 3: Cue reactivation rates in lateral association cortex decline with learning and exhibit a food-cue bias.

a. Left: mean food-cue reactivation rate per session across behavioral performance (discriminability, d’). Error bars: 95% confidence intervals of the fit. Rates were highest following low performance and decreased as the mice improved. Data were from imaging runs across 8 mice (115, 109, 112 classifiable runs for the food cue, neutral cue, and aversive cue, respectively) following satiation, and were pooled across sessions prior to and following a change in cue-outcome associations. *** p < 0.001, effect of behavioral performance using a generalized linear mixed-effects regression model (GLMM) accounting for shared variance within mice. r: Pearson correlations. Similar findings were observed for neutral (blue, middle) and aversive (red, right) cue reactivation rates. Compared to the above sessions during training, mean reactivation rates were lower in sessions involving naïve mice that viewed the same visual stimuli but in the absence of salient outcomes (gray dots at right, pooled across cue types). ††† p < 0.001 using a GLMM categorizing data into naïve vs. engaged sessions and accounting for shared variance within mice (see Extended Data Fig. 5a, Methods, and Supplementary Information). Horizontal dashed gray line: mean event rate across sessions from naïve mice. b. The overall rate of synchronous population events (see Methods), of which cue reactivations are a small subset, did not vary across learning (same 109 sessions as in a; Pearson correlation). c. On average, cue reactivation rates (normalized per session to neutral reactivation rate) were highest for the food cue, intermediate for the avoidable aversive cue, and lowest for the neutral cue. This ordering was evident in both hungry and sated states (*** p < 0.001, ** p < 0.01; using a GLMM comparing the effect of cue on the rates of reactivation, accounting for shared variance within mice and days; error bars: ±SEM). d. The bias in food-cue reactivation rates was evident when separately considering sessions prior to or following a change in cue-outcome associations in which the stimulus orientation initially associated with the food cue becomes associated with the neutral cue (*** p < 0.001; using a GLMM comparing the effect of cue on the rates of reactivation, accounting for shared variance within mice and days; error bars: ±SEM). e. Fraction of responsive cells driven by a particular cue. For each session, the fraction of responsive cells driven by a particular cue was normalized to the overall number of cue-responsive cells. These did not vary by cue type (p = 0.10; two-tailed Kruskal Wallis test). Data reflect mean ± SEM across sessions (see also Extended Data Fig. 5i). Thus, the observed enhancement in food cue reactivation rates was not due to a difference in the number of neurons responsive to any given cue. f. Food-cue reactivation rates predicted changes in behavioral performance between the end of the preceding training session and the beginning of the following session (across 102 pairs of days within 8 mice with fixed cue-outcome associations; green dots), which could occur one or several days later. We used a GLMM that accounts for shared variance within mice and for number of days elapsed between the two training sessions (** p < 0.01, effect of food-cue reactivation rate on change in performance). Green line: GLMM fit across mice when considering 1 day of elapsed time between sessions. Gray lines: similar fit as green line but using the estimate for each individual mouse. r: Spearman correlation. Inset: the association between reactivation rate and change in performance (same axes as in main panel) is stronger for shorter elapsed time between sessions. Lines show GLMM fits at 1 day, 3 days, and 5 days of elapsed time.
Cue reactivation rates reflect cue salience
Given that overall reactivation rates increased when certain stimuli became paired with outcomes, we wondered whether the behavioral salience of a given cue might bias its rate of reactivation. On average, we found that food-cue reactivations occurred ~30% more frequently than neutral-cue reactivations, both prior to and following satiation (Fig. 3c; Extended Data Fig. 5g). Reactivation of the mildly aversive cue predicting avoidable quinine delivery occurred at an intermediate rate. This order of relative reactivation rates was evident both in sessions prior to and following a reassignment of the outcomes associated with each of the three visual stimulus orientations (Fig. 3d and Extended Data Fig. 5h; see also Fig. 1c). Together with the learning-dependent changes in reactivation rates, these findings suggest that cue reactivation rates are related to salient recent experiences and not merely to the random co-activation of neurons with similar visual feature tuning29,32. These findings could not be explained by differential classifier sensitivity to patterns of neurons responsive to salient vs. neutral cues (Fig. 2e, Fig. 3e, Extended Data Fig. 3 and Extended Data Fig. 5i). In addition, these reactivation events did not reflect cue-specific activity patterns extending across longer timescales. Specifically, we did not observe a bias in cortical activity patterns towards any given cue representation in the seconds prior to or following the onset of a cue reactivation event (Supplementary Fig. 2). Together, these data suggest that the cue-specific bias in reactivation rates in sensory association cortex reflects the relatively higher motivational salience of the food cue during prior behavioral performance, similar to biases in hippocampal replay of previously visited locations with high motivational salience4,8,9,11.
Food-cue reactivation rates predict next-day changes in behavioral performance
The above findings suggest that food-cue reactivations are more common than other cue reactivations, particularly when animals are initially learning the task. This led us to ask: does the food-cue reactivation rate during quiet waking predict subsequent changes in behavioral performance? To answer this, we estimated the behavioral performance (d’) on each trial using a dynamic psychophysical model (Extended Data Fig. 5d–e) which predicts the trial choice (lick/no lick) based on estimates of internal ‘cognitive weights’ given to the identity of the various visual stimuli as well as the confounding influences of previous choice and previous outcome. This model allowed us to determine the change in behavioral performance both within a day and between days. Remarkably, we found that the rate of food-cue reactivations predicted improvements in behavioral performance from the end of the training session preceding the reactivations to the start of the following session (Fig. 3f, using a linear mixed-effects regression model accounting for non-independence of samples within mice and for the number of days between training sessions; see also Extended Data Fig. 5j). This effect decreased as the number of elapsed days to the following training session increased (Fig. 3f). We confirmed that these effects were not simply due to the dependence of reactivation rate on performance during the prior training session (Fig. 3a) by additionally including d’ in the mixed-effects model (p = 0.016). We compared a model assessing the association of reactivation rates with changes in behavioral performance within the prior training session vs. a model assessing association of reactivation rates with changes between the prior session and the following session, and found that the latter model fit the data substantially better (evidence ratio of 494; see Methods and Supplementary Information). Together, these findings show that salient cue reactivation rates are correlated with future performance, possibly due to reactivation-related plastic changes in the participating neurons. We therefore examined cross-day functional changes in reactivated neurons in greater detail.
Participation in cue reactivations is associated with bidirectional changes in functional connectivity
The above findings suggested that cue reactivations may be associated with network-wide changes as the mouse gradually learns associations between salient cues and associated outcomes. Thus, we tracked the same population of neurons across days (Fig. 4a and Supplementary Fig. 3), and considered how cue reactivations following a training session might modify network connectivity during the next day’s training session. For example, reactivations might drive changes in the synaptic strength of a common input encoding reward-related information to a subset of the food-cue-driven cells (e.g. long-range axonal input from lateral amygdala19), and/or changes in local synaptic connectivity between food-cue-driven cells and reward-related cells. Such changes should be reflected in reactivation-dependent increases in a cell’s net functional connectivity with other cells in the following session, as measured by the correlated variability in food-cue-evoked responses across pairs of neurons (i.e. noise correlations; 22,28). To this end, we estimated the ‘total functional connectivity’ of each food-cue-driven cell, defined as the clustering coefficient (i.e. the geometric mean across food-cue-evoked noise correlations between the cell and each of the other simultaneously recorded food-cue-driven cells; Fig. 4b, Extended Data Fig. 6, and Methods).
Fig. 4: Cue reactivations are associated with bidirectional changes in functional connectivity.
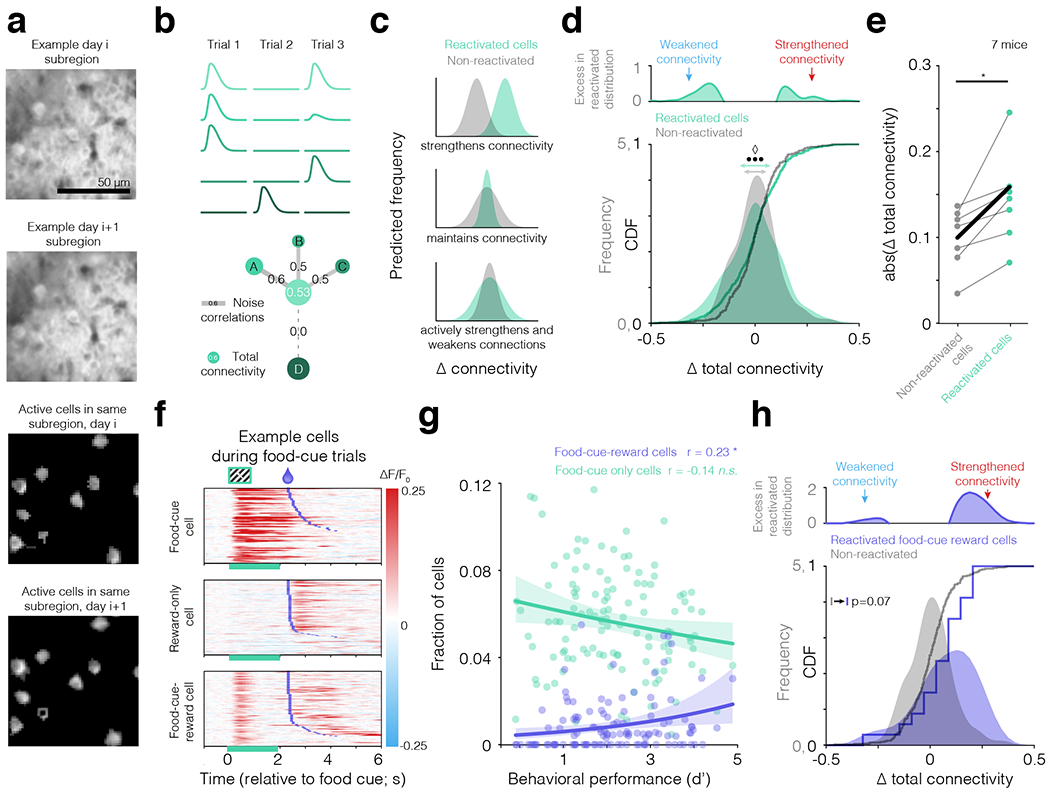
a. Top panel: example sub-region of a field of view (top: “day i”; mean contrast-adjusted GCaMP6f fluorescence image). Second-from-top: same field of view, imaged one day later (i.e. “day i+1”). Bottom two panels: analog masks of individual neurons across days, obtained using an ICA/PCA algorithm (see Methods and Supplementary Fig. S3). b. We estimated functional connectivity between two neurons as the shared variability in fluctuations of food-cue-evoked responses across trials (noise correlations). Schematic response time courses across three presentations of the food cue are shown (top) for each of five food-cue-driven cells (green circles at bottom). The lightest green cell (middle cell, bottom panel) had positive noise correlations with cells A-C but not with cell D. The total functional connectivity of the middle cell was defined as the graph-theoretical clustering coefficient across positive noise correlations between that cell and all other food-cue-driven cells (see also Extended Data Fig. 6). c. We estimated each cell’s change in total functional connectivity across two successive daily training sessions. We then considered whether this cross-day change in connectivity differed between cells that participated in offline reactivation following the first session (green model distributions) and cells that did not participate (gray model distributions). We predicted a net increase in total functional connectivity (upper panel) if local connections and/or long-range common inputs between a cell and its neighbors were strengthened following participation in reactivation. Alternatively, reactivated cells could be protected from increases or decreases in total functional connectivity (middle panel). Finally, if participation in reactivations had strengthening or weakening effects on different subsets of cells, we would expect a broader distribution of changes for cells that participated in reactivations vs. those that did not (lower panel). d. Bottom: cumulative distribution functions (CDFs) and kernel density estimates (KDEs) of the distributions of cross-day changes in total functional connectivity. Food-cue-driven cells that participated in food-cue reactivation events had a broader range of next-day increases or decreases in total functional connectivity (green distribution; horizontal arrows indicate 2σ) than those that did not participate (gray; ••• evidence ratio of 950 for a GLMM model that separately fit variances of reactivated categories of 0, 1, or 2+ reactivations and accounted for shared variance within days and mice relative to a GLMM that fit each category with the same variance, and evidence ratio of 8.03 for a GLMM model that separately fit variances of reactivated categories of 0 or 1+ reactivations and accounted for shared variance within days and mice relative to a GLMM that fit each category with the same variance; ◊ p < 0.05 using a model fitting the absolute value of the change in connectivity with a GLMM accounting for shared variance within days and mice using a permutation test, see Methods and Supplementary Information; N = 456 reactivated cells and 238 non-reactivated cells from 102 pairs of consecutive sessions in across 8 mice). Top: excess frequency of reactivated cells exhibiting a given change in next-day connectivity above that observed for non-reactivated cells (i.e. rectified difference between green and gray distributions). e. Similar results were observed when considering the mean absolute change in total connectivity in reactivated or non-reactivated cells per mouse (* p = 0.011, two-tailed Wilcoxon signed-rank test of 7 mice with sufficient data; one mouse was excluded from this analysis because it did not have at least two pairs of sessions for which at least one food-cue-driven cell from the first session was also driven by the food cue on the following session but did not participate in any reactivation events). f. Heatmaps of single-trial food cue response time courses (rows; ΔF/F: fractional change in fluorescence) for three example cells that encode only the food cue (top, ‘Food-cue cell’), only the reward (middle, ‘Reward-only cell’), or both (bottom, ‘Food-cue-reward cell’). Green bars mark food cue presentation and purple ticks indicate reward delivery. g. The fraction of Food-cue-reward cells increased with behavioral performance (* p < 0.05 across 115 imaging days in 8 mice, using a GLMM accounting for shared variance within mice), while the fraction of cells that only respond to the food cue did not change. r: Spearman correlation. Error bars: 95% confidence intervals. Thus, the increase in the proportion of Food-cue-reward cells is not simply due to a proportional increase in the overall number of food-cue-driven cells. h. For Food-cue-reward cells (a small subset of the food-cue-driven cells in d: 17/456 reactivated cells, 8/238 non-reactivated cells), participation in food-cue reactivations trended towards an asymmetric strengthening in next-day total functional connectivity (same axes as in d). p < 0.07 (GLMM accounting for shared variance within days and mice).
We considered three possible scenarios for changes in total connectivity following participation in cue reactivation events. First, if a cell’s participation in reactivation events between sessions strengthens its connections with other local cells or strengthens connections from common, reactivated inputs, then the cell’s total functional connectivity might increase (Fig. 4c, top). Second, if participation in reactivation stabilizes connection strengths, we might observe a narrower range of changes in total functional connectivity (Fig. 4c, middle). Third, if some cells that participate in reactivation events actively strengthen their connections while others actively weaken their connections, we might observe a broader range of changes in total functional connectivity (Fig. 4c, bottom)33.
Consistent with this third hypothesis, we found that food-cue-driven cells that participated in food-cue reactivations were more likely to show larger increases or decreases in next-day total functional connectivity than those that did not participate (Fig. 4d). This finding was not dependent on the specific metric used to quantify functional connectivity, as we observed a similar broadening in the distribution of cross-day changes in noise correlations between individual pairs of cells that both participated in reactivations vs. those that did not (p < 0.001, using a generalized linear mixed-effects regression model to account for non-independence within days and within mice). The consistency of these results across individual mice was also evident when comparing the mean absolute change in connectivity in the following session for food-cue driven cells that participated in food-cue reactivations between sessions vs. those that did not (Fig. 4e).
To better understand the diversity of changes in functional connectivity, we further categorized each cell based on its response to cues and outcomes using a generalized linear regression model. As expected, we found many cells driven only by the food cue19 (‘Food-cue cells’; Fig. 4f, top), as well as cells with activity that was consistently locked to reward delivery (i.e. encoding rewards or reward-related stimuli), but not to times of motor behavior (see Methods). This reward-related activity could occur following reward delivery (Fig. 4f, middle) or preceding (and thus anticipating) reward delivery. These cells either encoded only the reward (‘Reward-only cells’, Fig. 4f, middle) or jointly encoded both the reward and the food cue (‘Food-cue-reward cells’, Fig. 4f, bottom; Extended Data Fig. 7; far fewer cells responded to both the reward and to non-food-predicting visual cues, Extended Data Fig. 7c). We found that the fraction of cells classified as Food-cue-reward cells increased with improved performance on the task, while the fraction of cells classified as Food-cue cells did not significantly change (Fig. 4g). Food-cue-reward cells that participated in food-cue reactivations trended towards a significant next-day increase in total functional connectivity with other food-cue-driven cells, as compared to those that did not participate (Fig. 4h).
These data indicate that the total population of food-cue-driven cells, when reactivated, exhibits bidirectional changes in total functional connectivity (Fig. 4d–e). In contrast, the task-relevant but small subset of Food-cue-reward cells (4.1% of food-cue-driven cells) appear to show reactivation-dependent increases in total functional connectivity. This finding is consistent with a learning-related increase in excitatory feedback input from sources carrying reward-related information (e.g. 19,22,34,35) to a subset of neurons including both Food-cue-reward neurons and some of the other food-cue-driven cells. We next examined this diversity of cross-day changes in network connectivity in greater detail.
Distinct flavors of cue reactivations are associated with opposite changes in functional connectivity
To better understand why some food-cue-driven cells showed increases while others showed decreases in functional connectivity upon participation in food-cue reactivations, we returned to our finding that individual presentations of the same stimulus can activate distinct, correlated clusters of cells (Fig. 2d). We found that cells within the same cluster were preferentially reactivated together (Fig. 5a, confirmed using a Naïve Bayes classifier and further examined across cluster types in Extended Data Fig. 8a–e). This finding led to the following hypothesis: food-cue-driven cells that engage in a privileged dialogue with Food-cue-reward cells will show a similar reactivation-dependent increase in total functional connectivity as observed for Food-cue-reward cells in Fig. 4h. Relatedly, if the reactivation-dependent strengthening of functional connectivity in Food-cue-reward cells is due, for example, to increases in neuromodulatory drive during associated reactivation events, then other food-cue-driven cells that are co-active with Food-cue-reward cells during the same reactivation events (Fig. 5b) should also strengthen their functional connectivity with the network.
Fig. 5: Distinct types of food-cue reactivation events drive opposite changes in functional connectivity.

a. Left: while the same cue could drive multiple clusters of cells with strong intra-cluster correlations, reactivations were sometimes composed of cells mostly belonging to a single cluster, as for two example reactivation events (reactivated cells are shown, together with their pairwise joint response probabilities) from the session whose correlation structure and clusters are shown in Fig. 2d. Right: violin plots of shared reactivations per session for pairs of cells belonging to the same vs. different clusters (horizontal black lines: means). Cells preferring a given cue were more likely to participate in the same cue reactivation events when they belonged to the same cluster (*** p < 0.0001, Bonferroni-corrected two-tailed Wilcoxon rank sum test; similar results were observed using a Naïve Bayes classifier that did not consider pairwise correlations, see Extended Data Fig. 8d–e). b. Average deconvolved activity for food-cue-driven cells defined in Fig. 4f, triggered on food-cue reactivation events containing at least one participating Food-cue-reward cell, shows that some other food-cue-driven cells are also co-active during these events (lines and shaded regions: mean ± SEM; 143 Food-cue-reward cells, 991 other food-cue-driven cells. 567 events; *** p < 0.0001, Bonferroni-corrected two-tailed Wilcoxon signed-rank test comparing peri-event activity with activity 200 ms before and after event). c. Clustering of food-cue-driven neurons into groups based on their functional connectivity (food-cue-evoked noise correlations; c.f. Fig. 2d) for an example session. See also Extended Data Fig. 8g. The two ‘reward-related clusters’ (purple wedges) contained at least one Food-cue-reward cell, while the ‘non-reward-related cluster’ (orange wedge) did not. d. Individual food-cue reactivation events were separated into those with >80% of active cells belonging to non-reward-related clusters (‘reward-poor reactivations,’ see example in left column), with >80% belonging to reward-related clusters (‘reward-rich,’ right column), or ‘mixed’ (middle column). See also Extended Data Figs. 9. e. We reran the analyses in Fig. 4d separately on subsets of food-cue-driven cells that participated in these different flavors of food-cue reactivation events. Left: changes in total connectivity for cells that only participated in ‘mixed’ reactivations (dark gray; 240 cells) were not different from those of cells that were not reactivated (light gray, 191 cells), as assessed using a GLMM accounting for shared variance within mouse and day (see Supplementary Information). Right: comparison of three groups of cells that participated in (i) reward-poor reactivations (orange, 90 cells), (ii) reward-rich reactivations (purple, 240 cells), or (iii) cells that either did not participate in reactivations or that only participated in mixed reactivations (light gray). We modeled these three categories using a categorical GLMM that accounted for shared variability within days and mice. This showed that the type of reactivation a cell experienced significantly affected the direction and amount of change in total connectivity across pairs of days (* p < 0.05; see also Results and Methods). Bottom panels: CDFs and KDEs of the distributions of cross-day changes in total functional connectivity. Top: excess frequency of reactivated cells above that observed for non-reactivated cells (i.e. rectified difference between the pairs of distributions). f. We confirmed that these results were consistent across mice by averaging the changes in total connectivity per mouse. Cells that participated in reward-poor reactivations (1 orange dot per mouse) showed significant decreases in total connectivity in all 8 mice († p = 0.012, Bonferroni-corrected two-tailed Wilcoxon signed-rank test relative to 0). In addition, cells that participated in reward-rich reactivations showed the opposite trend (i.e. increases in 5/6 mice, p = 0.25, Wilcoxon sign-rank test vs. 0; datasets from two mice were excluded from this analysis because they did not contain at least two pairs of sessions for which at least one Food-cue-reward cell from the first session was also driven by the food cue on the following session). These two populations of cells were also significantly different from each other (* p = 0.028, two-tailed Wilcoxon rank-sum test). g. Summary. As shown in Fig. 4d, reactivated cells showed a broader distribution of cross-day changes in connectivity than non-reactivated cells. Surprisingly, this broadening could be ascribed to separate sets of cells that participated in reward-rich (strengthening) or reward-poor (weakening) food-cue reactivations, but not to those that only participated in mixed reactivations.
To test this hypothesis, we first separated food-cue-driven cells into those that belonged to the same cluster as at least one Food-cue-reward cell (e.g. a purple cluster in Fig. 5c), and those that did not (e.g. an orange cluster in Fig. 5c; see also Extended Data Fig. 8f–k). This allowed us to sort food-cue reactivation events into three flavors (Fig. 5d and Extended Data Fig. 9): (1) ‘reward-rich’ reactivations that contained predominantly reward-related cells (i.e. cells from purple clusters), (2) ‘reward-poor’ reactivations that contained predominantly reward-unrelated cells (i.e. cells from orange clusters), and (3) ‘mixed’ reactivations that contained a mixture of the two.
Cells that participated only in ‘mixed’ food-cue reactivations showed changes in connectivity that were indistinguishable from those of non-reactivated cells (Fig. 5e, left, using a regression model comparing changes in total connectivity by group, and by using a model comparison, accounting for differences in variances across groups, see Supplementary Information).
Cells that participated in ‘reward-rich’ or ‘reward-poor’ food-cue reactivation events both showed robust changes in next-day connectivity relative to the pooled group of cells that either did not participate in reactivations or that participated in mixed-reactivations only (Fig. 5e, right, p < 0.05 for both reward-rich and reward-poor reactivations using a linear mixed-effects regression model accounting for non-independence within days and within mice, see legend for Fig. 5e; see also Extended Data Fig. 9e; similar effects were also observed using pairwise rather than groupwise analyses of noise correlations, data not shown). Consistent with our hypothesis, food-cue-driven cells that participated in reward-rich reactivations showed increased next-day connectivity (Fig. 5e, right, p < 0.05; using a single model with a categorical variable reflecting cell membership in one of three categories: ‘reward-rich’ reactivated, ‘reward-poor’ reactivated, or ‘mixed and non-reactivated’; see Supplementary Information for details). Cells that participated in ‘reward-poor’ food-cue reactivations appeared to show decreases in next-day connectivity (Fig. 5e, right). We confirmed that these results were consistent across mice by averaging the changes in total connectivity per mouse. Specifically, cells that participated in reward-poor reactivations showed significant decreases in total connectivity in all 8 mice (Fig. 5f; p = 0.012, Wilcoxon sign-rank test vs. 0). In addition, cells that participated in reward-rich reactivations showed the opposite trend (Fig. 5f; i.e. increases in 5/6 mice, p = 0.25, Wilcoxon sign-rank test vs. 0; datasets from two mice were excluded from this analysis because they did not contain at least two pairs of sessions for which at least one Food-cue-reward cell from the first session was also driven by the food cue on the following session). These two populations of cells were also significantly different from each other across mice (p = 0.028, Wilcoxon rank-sum test).
These findings suggest that the cellular composition of a given food-cue reactivation event predicts the sign and magnitude of cross-day changes in the functional connectivity of the participating cells. In particular, reactivation-dependent increases and decreases in total connectivity (Fig. 4d–e) were largely carried by cells that participated in reward-rich and reward-poor food-cue reactivation events, respectively (summarized in Fig. 5g).
As discussed further below, our findings are consistent with a model of memory consolidation involving gradual formation and cross-day strengthening of ‘mental schemas’ (e.g. 5,36) that reflect a growing knowledge of the relationship between a specific cue and a reward. We propose that reward-related clusters in association cortex reflect task-relevant schemas whose functional network connectivity strengthens when neurons in these clusters reactivate together (reward-rich food-cue reactivations). This could be due to strengthening of local connectivity and/or of common feedback input from reward-related areas such as lateral amygdala (Fig. 6; 19). The commensurate weakening of functional connectivity in non-reward-related clusters when neurons in these clusters reactivate together (reward-poor reactivations) could ensure that task-irrelevant representations of the same visual stimulus are weakened and that a constant level of overall connectivity is maintained13,33,37. Indeed, these opposite reactivation-related changes in network connectivity did not result in an overall change in network connectivity when averaged across all food-cue-driven neurons (Fig. 4d).
Fig. 6: Model for how cue reactivations modify network functional connectivity.

a. Top left: in a typical session midway through learning, food-cue-driven cells (green and purple circles) have begun to separate into correlated clusters. Cells in reward-related clusters may receive stronger common top-down input from reward-coding sources such as lateral amygdala, while both clusters also receive input from earlier visual areas and local connections. Bottom: following that day’s training, reward-rich reactivations strengthen the functional connectivity of cells in reward clusters, while reward-poor reactivations weaken the functional connectivity of cells in non-reward clusters (which may be less task-relevant). Top right: these changes in functional connectivity may reflect further strengthening of reward-related top-down inputs or of specific local connections within reward clusters.
The finding that participation in reward-rich or reward-poor food-cue reactivations was selectively associated with increases or decreases in next-day functional connectivity, respectively (Fig. 5e–g), suggests that reward-rich reactivations strengthen network connectivity of participating cells, while reward-poor reactivations weaken network connectivity of participating cells. We speculate that these effects could be driven by reinforcement signals acting within association cortex, hippocampus, and elsewhere in the brain-wide network of reactivated cells. This reactivated network likely includes reinforcement-related ventral tegmental area (VTA) dopaminergic neurons38,39 and their targets in the nucleus accumbens (NAc).
As an initial examination of this hypothesis in a small number of mice (N=3), we combined cortical imaging with bulk recordings of calcium activity from mainly VTA axons in NAc (VTANAc axons, a large fraction of which are dopaminergic; Extended Data Fig. 10a). VTANAc axons responded selectively to the reward-related food cue but not to other cues, consistent with previous studies of VTA dopaminergic projections to NAc (see Extended Data Fig. 10b and associated legend). Aversive-cue and neutral-cue reactivations were associated with a drop in VTANAc activity (Extended Data Fig. 10c,d), potentially reflecting the lack of association between these cues and reward. VTANAc axons also showed increases and decreases in activity during reward-rich and reward-poor food-cue reactivations, respectively (Extended Data Fig. 10c,e), consistent with the strengthening and weakening effects of these two subtypes of food-cue reactivations. These preliminary observations suggest that VTANAc axons may be modulated during offline cue reactivations, and highlight a plausible experimental strategy for more detailed testing of the sign and magnitude of neuromodulatory responses associated with distinct reactivation content.
Discussion
We have demonstrated that ensembles in lateral visual association cortex that are driven by specific visual cues are briefly reactivated hours later during quiet waking. Cortical cue reactivations were synchronized with hippocampal ripple activity, and were more common for salient sensory experiences and following sessions with poor task performance. Critically, our long-term imaging approach revealed that cells that participated in cue reactivations exhibited larger next-day changes in network functional connectivity. Our findings support the hypothesis that different flavors of offline reactivations may drive further strengthening of ensembles of neurons encoding both the cue and the associated reward, while actively weakening intermingled ensembles of neurons encoding the same cue but not the reward. Specifically, our findings suggest that the joint reactivation of cortical ensembles encoding both a cue and an associated reward constitutes a possible substrate for plasticity mechanisms driving the consolidation of specific cue-outcome associations. Such consolidation may enhance existing functional connectivity of cells within these ensembles. At the same time, other reactivations of ensembles that do not additionally encode salient outcomes may actively weaken the functional connectivity and influence of the participating cells. The combination of these processes should enhance cue-outcome associations while maintaining overall functional connectivity within the network.
Bidirectional, reactivation-dependent changes in across-day functional connectivity
How are cortical memory representations formed and maintained? Models of cortical memory consolidation following learning have posited seemingly opposing roles for ongoing activity during quiet waking or sleep in strengthening40 or weakening33 synapses, respectively. One recent model41 suggests that coordinated offline reactivation of cortical and hippocampal neurons promotes strengthening of synapses between neurons within an ensemble encoding a given sensory experience, as well as weakening of synapses with other neurons. In this way, both the strengthening and weakening processes may ultimately fortify the selectivity of within-ensemble synaptic and structural connectivity.
The strengthening of functional connectivity (trial-to-trial correlations in food cue-evoked responses) of neurons in reward-related ensembles following participation in reward-rich reactivations may reflect strengthening of synaptic connectivity locally28,42 and/or in common long-range inputs (e.g. 19,22,34,35; Fig. 6). Although Fauth et al.41 propose that strengthening and weakening of synapses occur during a common flavor of reactivation, our findings suggest that participation in reward-poor reactivations can also weaken functional connectivity within task-irrelevant ensembles, possibly reflecting weakening of synaptic connectivity. Such bidirectional changes in network connectivity may facilitate accurate pattern completion41, thereby binding together representations of cues and expected outcomes. While reductions in pairwise correlations of trial-to-trial activity may be advantageous in early sensory brain areas in order to reduce redundancy of correlated sensory inputs (e.g. 43), increased within-ensemble trial-to-trial correlations in association cortex may reflect the linking together of previously unrelated features of an experience (e.g. representations of a particular drifting grating and of an expected food reward). Indeed, a recent study in primate lateral prefrontal cortex demonstrated that while removing correlated activity from all neurons increased coding accuracy, removing correlations between neurons within ensembles decreased accuracy44.
Prioritized reactivation of salient and unexpected recent experiences
Similar to previous findings in hippocampus and entorhinal cortex (e.g. 4,45–48) and amygdala11, cue reactivations in association cortex were more common following sessions with poor task performance and for salient vs. neutral cues. Theoretical studies suggest that prioritized reactivation of recent experiences based on prediction error magnitudes can promote efficient reinforcement learning via simulation of cues and unexpected outcomes49,50. The notion that cortical cue reactivations may be important for learning is consistent with the larger across-day changes in functional connectivity that we observed for reactivated neurons. Indeed, the preliminary findings from our recordings of putative VTA dopaminergic inputs to ventral striatum during cue reactivations further support a highly-distributed framework for learning from simulation of recent salient experiences during subsequent offline periods38.
Two-photon imaging of visual cue reactivation in lateral association cortex
Cue reactivations are likely widely distributed throughout the cortex and hippocampus. We chose to focus on lateral visual association cortex, a region that contains a substantial proportion of neurons with sharp visual orientation preference (compared, for example, to downstream entorhinal areas) (e.g.19). These properties, coupled with the use of simple visual gratings with large differences in orientation, allowed us to more easily decode which grating was being presented (or reactivated) than if we had used more complex stimuli. Equally important, a substantially larger proportion of neurons in lateral visual association cortex contain reward-related information than in early visual cortex19,22, likely due to strong feedback input from reward-coding regions such as lateral amygdala19.
Our task involving controlled presentation of cues across hundreds of trials per day in head-fixed mice was essential for training a Bayesian classifier that could identify reactivation events in real data but not in shuffled data. Our classifier has three benefits: first, its assumptions are minimal and explicit. Second, unlike classifiers based on a Naïve Bayes estimator, our classifier based on an Average One-Dependence Estimator does not make the assumption that the activity of each cell is independent. Rather, it gains sensitivity by considering the co-activation of pairs of cells during any given cue presentation. Third, in contrast to classifiers built to identify spatiotemporal sequences of activity across neurons during hippocampal navigation tasks (e.g. 7), our classifier was designed to decode momentary patterns of neurons that simultaneously respond to one of several discrete visual stimuli.
Future studies can combine chronic imaging of reactivation of cell bodies with longitudinal imaging of the activity, molecular signaling, and structure of dendritic spines or long-range synaptic boutons. Additionally, the approach described here of combining imaging of reactivation with contralateral laminar multi-electrode recordings may be extended to high-density electrophysiology probes for combined analysis of these widely distributed cue reactivation events. These approaches should help dissect the long-term effects of reactivation events on cortical microcircuitry at unprecedented resolution.
Methods
All animal care and experimental procedures were approved by the Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee. Animals were housed with standard mouse chow and water provided ad libitum, unless specified otherwise. Mice used for two-photon imaging (n = 11, 3 females and 8 males, [8 were recorded across naïve conditions and prior to and following reversal of cue-outcome contingencies, 2 were recorded during naïve conditions only, and 1 was excluded due to possible seizure-like activity], Emx1-Cre;Ai93 (TITL-GCaMP6f)-D;CaMK2a-tTA51) were instrumented with a headpost and a 3-mm cranial window centered over lateral visual association cortex, and were allowed to recover for at least one week. See 52 for detailed methods.
Behavioral training:
See 19 for extended methods. Briefly, mice were food restricted to 85% of their ad libitum weight and were head-fixed on a plastic running wheel for habituation prior to training. Mice were introduced to the taste of the calorically dense liquid Ensure and were then shown Pavlovian stimuli predicting a droplet of Ensure using MonkeyLogic53. Mice were habituated to long task durations prior to being introduced to quinine by slowly increasing the session length across days while rewarding the animal throughout. After habituation to the non-operant Pavlovian task, we slowly introduced operant cues by increasing the fraction of trials with reward delivery contingent on licking during the response period (see below) until the animal’s cue-evoked behavioral response rate exceeded 80%. At this point, low numbers of trials containing neutral and aversive cues were introduced for two days, after which the mouse was imaged across initial training and across a subsequent change in cue-outcome associations.
Mice were trained to discriminate full-field square-wave drifting gratings at a spatial frequency of 0.04 cycles/degree and a temporal frequency of 2 Hz. Each outcome (delivery of 5 μl of Ensure milkshake, 5 μl of 0.1 mM quinine, or no outcome) was randomly assigned per animal to the 0°, 135°, and 270° drifting grating stimuli. These cues were displayed on a calibrated 20” 3:4 LCD screen (Dell) positioned 20 cm from the mouse. Cues were presented for 2 s, followed by a 2-s response window during which licks were detected using a capacitance-sensing lickspout (3D printed with a metal-containing filament connected to the MPR121 capacitance sensor; Adafruit). Licking outside of the response window did not affect the task outcome, and a lick response in the response window yielded a single 5 μl delivery of Ensure or quinine.
After the mice performed the task at >90% success rate across all three trial types for at least three days, the cue-outcome association was changed. In three mice, the food cue became the neutral cue, the neutral cue became the aversive cue, and the aversive cue became the food cue. In two mice, the food cue and neutral cues were switched while the aversive cue was held constant. In each case, performance across all three trial types was equally high following initial learning, and in each case dropped to chance (indiscriminately licking to stimuli) immediately following the switch in cue-outcome contingencies. For these reasons, the datasets were combined.
Randomization and blinding:
As the experiments were performed, the experimenter was blind to the number of reactivations (the measured predictive factor). Reactivation rates were determined either by a different individual from the one interacting with the mice or after completion of the experiments for an individual mouse. Because reactivations occurred in each mouse, there was no single variable to which the authors performing data analysis could be blind to. Most comparisons were made within-mouse, rather than across experimental groups.
Mapping of retinotopic areas:
To initially map the locations of lateromedial area (LM) and retinotopically identified visual postrhinal cortex (visPOR54), we used epifluorescence imaging of the entire 3-mm window while presenting 20-degree patches of noise in nine retinotopic locations. A 470-nm LED was passed through a long-pass emission filter (500 nm cutoff) and images were recorded with an EMCCD camera (Rolera EM-C2 QImaging, 251 x 250 pixels at 4 Hz). Retinotopy was compared to 54, and the imaging field was centered over an area encompassing the lateral portion of LM, laterointermediate area (LI), and visPOR. Note that single-trial population responses to visual gratings drifting in different directions are easily distinguishable from population imaging data in area visPOR19.
Two-photon imaging:
Imaging was performed using a Nikon 16x 0.8 NA objective on a resonant-scanning two-photon microscope (Neurolabware; 15 frames/second; 796 x 512 pixels covering an area of 1150 x 800 microns). Laser power of a Mai Tai DeepSee laser was set at 25-60 mW at 960 nm. Imaging fields in layer 2/3 of lateral visual association cortex (centered on visPOR) were adjusted to be parallel with the surface of the cranial window and were approximately 180 μm below the surface. The microscope was placed in a light- and sound-attenuating chamber in a dark, quiet room to prevent unexpected visual or auditory stimuli. The chamber was kept at a steady temperature of 20° C and the objective was heated to 33° C so that the mouse would not lose heat.
Data were collected for a 30-minute session in darkness (while the mouse typically ran on the running wheel), followed by three 30-minute training sessions of 180 trials each. This was followed by a single 30-min “hungry” spontaneous session in the dark. In all but two animals, friction was applied to the running wheel to limit motion during this and subsequent imaging periods in darkness. In previous studies (e.g. 31) and in our data, the frequency of hippocampal sharp-wave ripples decreased with locomotion. Because hippocampal replay and cortical reactivation have been linked to sharp-wave ripples, decreasing mouse motility might increase the probability of reactivation events. Mice were given access to milkshake (Ensure) until sated (operationally defined by a cessation of licking for at least 5 min), or for roughly 45 min. Finally, mice were imaged for three 40-min sessions in which the first 10 min involved stimulus presentations (with little to no task engagement by the sated animals) and the remaining 30 min involved recordings of spontaneous data in the dark with friction applied to the running wheel to enhance stability and further promote quiet waking. From these latter runs, only data from periods of darkness were analyzed further.
Electrophysiology preparation:
In a subset of animals (n = 2), a small craniotomy (~1 mm2) was marked on the surface of the skull for targeting hippocampal area CA1 (coordinate centered on AP −2.54 mm ML −3.1 mm from Bregma) contralateral to the imaging window craniotomy. Following training, this craniotomy was prepared for acute recording. The craniotomy was opened, flushed with sterile saline, and covered in a thin layer of petrolatum ointment (Puralube, Dechra). The craniotomy was closed with a silicone plug (Kwik-Cast, World Precision Instruments) anchored to the skull with dental cement (Flow-It ALC, Pentron). Animals were allowed one week to recover before recordings began. Before each day of recording, the silicone plug was removed, and the craniotomy flushed with sterile saline. A camera (Flea3, FLIR) was used to visually guide the multielectrode silicon probe into the craniotomy using a motorized micromanipulator (PatchStar, Scientifica; see below for silicon probe details). First, the micromanipulator was zeroed with the electrode resting on the surface of the brain. The electrode was then retracted so that the surface of the brain could be hydrated with a thin layer of petrolatum ointment, and then the electrode was lowered ~1.0–2.4 mm into the brain depending on the precise angle (−8 to −17 degrees from horizontal along the coronal plane, depending on the level of tilt of the mouse headpost). In addition to stereotaxic targeting, the probe was determined to be within hippocampal area CA1 when clear spiking from single units were apparent on multiple adjacent electrode contacts. The probe was left in place for at least 45 minutes before recording started, to allow the tissue to stabilize. Following each recording session, the craniotomy was once again flushed with sterile saline, coated thinly in petrolatum ointment, and closed with a silicone plug and dental cement. Following termination of the experiments, electrode placement was validated histologically to be located in hippocampal area CA1, and in one animal fluorescent latex beads (Red Retrobeads IX, Lumafluor) were used to coat the silicon probe to further verify probe placement.
Electrophysiology recordings and analysis:
Acute hippocampal recordings were performed using a 16-channel linear silicon probe (A1x16-5mm-50-177-A16, NeuroNexus) or a 16-channel silicon tetrode rake (A2x2-tet-3mm-150-150-121-A16, NeuroNexus). Neural signals were recorded with a Digital Lynx SX recording system (Neuralynx). Broadband signals from each contact were filtered between 0.1 and 9000 Hz and recorded continuously at 32kHz. Following acquisition, signals were down-sampled to 800 Hz, bandpass filtered between 1 and 399 Hz, and common-mode referenced to remove motion artifacts. Broken channels were discarded from further analysis. Multitaper analysis (Chronux) was used to generate power spectra from all contacts. The channel with the highest ripple power (150-300 Hz frequency band) for each recording session was selected and power across the ripple band was z-scored and averaged. Signed-rank statistical analysis was performed using one-second bins centered either at the time of peak population activity during each reactivation event (AODE classifier output > 0.1, see below), at 1.5 seconds before each reactivation event, or at 1.5 seconds after each reactivation event. These datasets were also used to assess the presence of epileptiform activity (see “Analysis of possible epileptiform events” section of Supplementary Information).
Image registration and source extraction:
Alignment and registration was performed in Matlab 2016a (Mathworks). For each run, a reference stack of 500 images was created and aligned using subpixel translation alignment55. This resulted in one reference image per run. Each reference image was aligned to the reference image from the first run of the session, using affine alignment from the TurboReg plugin for ImageJ56. We used affine alignment because of slow brain volume changes and associated stretching following satiation. Next, we averaged data down to ~1 frame/s to limit noise (15 frames averaged), and these averaged images were aligned using affine alignment. The alignment parameters were interpolated (back to the 15.5 Hz frame rate), combined with the cross-session alignment and applied to each frame (at 15.5 Hz). Finally, we ran a subpixel translational alignment on each frame to remove remaining high-frequency translations.
We used PCA/ICA to extract masks of pixels with correlated activity57. By default, we used only the top 75% of pixels58, but users screened each prospective region of interest (ROI) and could edit the size of the mask, selectively removing the lowest probability pixels. Pixels found in more than one mask were excluded from further analyses. Time courses were extracted by averaging each of the pixels within each binarized mask. We calculated neuropil activity surrounding each cell as the median value of an annulus surrounding each ROI (inner radius: 20 μm; outer radius: 25 μm; all other ROIs excluded from these annulus masks). This time course of neuropil activity was then subtracted from the activity time course of the associated ROI to create a fluorescence time course, F(t), where t is time of each imaging frame. From this, we calculated a running baseline-subtracted time course, (F(t) – F0(t))/F0(t), where F0(t) is a running estimate of baseline fluorescence calculated as the 10th percentile of F(t) in the previous 32-second sliding window59. Finally, for input to the classifier, we deconvolved this baseline-subtracted time course using standard methods60 (using a 4th order regressive model with the cvx method, scaling the output with an estimate of a unitary change in fluorescence from 61).
Alignment of ROI masks across days:
We aligned recording sessions on sequential pairs of days. For each pair of overlapping ROIs from two sequential recording sessions, we calculated the two-dimensional correlation of the PCA/ICA masks. Any ROIs correlated at less than 0.05 were ignored. The remaining PCA/ICA masks were shown to a user for confirmation, along with an average image of the 200 frames containing the brightest ΔF/F0 values for that ROI, keeping only cells for which we could identify matching processes. The vast majority of ROI masks had only a single possible match across a pair of days. We kept only those ROIs with high correlations in which we could identify similar shapes of soma and processes in the maximum-intensity image on both days. To confirm these results, we used the CellReg software62 to match the locations of ROIs across pairs of days.
Display of activity time courses:
ROIs were sorted by their mean stimulus-evoked ΔF/F0 response from 0-2 seconds after cue onset after subtracting the mean activity of the ROI in the 1 second prior to cue onset (e.g. Fig. 2a). Any ROI with an increase in activity in response to any of the three cues was assigned to the cue that evoked the largest response, with non-responsive cells assigned to the group of non-cue preferring cells. Deconvolved traces (e.g. Fig. 2b) are shown with a sliding maximum filter of 4 frames, matching the sliding window used by the classifier (see above), and each cell was scaled by the maximum activity level across the trace. In the case of Extended Data Fig. 2c, the resulting value was further scaled by the band-pass filtered scale factor used to generate the temporally varying prior (defined above).
Classifier:
To identify reactivation events, we wanted to create a classifier with a minimal number of hidden assumptions. To do so, we initially considered the Naïve Bayes classifier. However, the Naïve Bayes classifier makes the assumption that the activity of each cell is independent, which may introduce errors in representing the network of visual association cortex neurons given that the many local connections and sources of common input to this region can drive correlated activity. Therefore, we used an extension to the Naïve Bayes classifier called an Averaged one-dependence estimator (AODE), which accounts for pairwise probabilities63,64 and does not make the assumption that the activity of each neuron in a field of view is independent (classifier training code available at https://github.com/asugden/flow/blob/master/flow/classifier/aode.py). We describe the classifier in detail in Supplemental Information.
Classifier event rate:
A single reactivation event involved a set of one or more contiguous frames that (i) all had estimated classifier output exceeding the classifier match threshold and (ii) that were separated by a least two frames with probabilities below this threshold (enforcing a minimum time between reactivations of 90 ms). From this set of contiguous frames, we chose the precise time of the reactivation event to be the frame with the highest mean population activity across all cells. We only assigned reactivation events during epochs that did not include brain or body motion (excluding all data ±3 s surrounding any period of brain motion greater than four standard deviations above the mean, ±10 s surrounding each lick, and ±10 s surrounding each epoch involving locomotion at > 4 cm/s). The priors and matching threshold are complementary variables; given the priors defined above, we varied the matching threshold and selected a value of 0.1, above which the rate of identified events in randomized data (see below) was approximately 5% of the rate of identified events in real data. The event rate of sharp transient events was determined as the event rate of the band-pass filtered, time-varying scale factor of the prior probabilities described above (i.e. the rate of threshold crossings of this timeseries of probabilities above a threshold of 0.1).
Validation of classifier:
We validated the classifier in two ways: using held-out cue presentations and using spontaneous data from quiet waking animals following randomization in time or in cell identity.
First, we tested whether the classifier accurately identified single cue presentations. For this test, because the classifier was being applied to data during task engagement, we did not scale down the output of each deconvolution (as was otherwise done when applying the classifier to spontaneous population activity in darkness). In addition, we set the priors for all five classes to be constant across all frames and of equal value (0.2). This first validation confirmed that the classifier could specifically identify instantaneous patterns of population activity during individual presentations of each of the three cues, and could distinguish these patterns from non-cue activity patterns (Fig. 2e).
To confirm that, when applied to spontaneous population activity, the classifier was identifying reactivations of cues (i.e. patterns highly similar to those during cue presentations) but not random fluctuations in population activity, we tested the classifier on data randomized in three ways. First, we randomized data from spontaneous recording in the dark by circularly shifting each cell’s time course by a random offset (drawn from a uniform distribution of all possible shifts). We performed classification on 10 such randomizations and detected very few events in randomized data (Extended Data Fig. 3f–g). Second, we randomized the identity of individual cells, focusing on those moments in the recording of spontaneous activity in which a reactivation of any one of the three cues was identified in non-randomized data. To do so, we permuted the identities of each cell 100 times and applied the original classifier to the permuted vector of cell activity at each of these frames (Extended Data Fig. 3b–c). Third, we repeated the randomization in identity, but performed the measurements on data in which the activity of each cell across the entire day was scaled to the median activity across all cells. This ensured that classification was not dominated by the activity of a small subset of cells (Extended Data Fig. 3d–e). Finally, we asked how frequently a cue reactivation event of any type would be identified in this permuted data. Again, we detected very few events in randomized data (Fig. 2f). These control analyses strongly suggest that the identified cue reactivation events would not be found by chance. This is further supported by the alignment of cue reactivations with hippocampal ripple and constricted pupil size (Fig. 2g, Extended Data Fig. 4f), together with the learning-dependent changes in cue reactivation rates (Fig. 3a) and the enhanced level of food-cue reactivations both prior to and following a change in cue-outcome associations (Fig. 3d, Extended Data Fig. 5h).
Data analysis and statistics:
Data was analyzed in Matlab 2016a, Python 2.7, and R 3.5, with the classifier written as a Numpy extension in C. Statistics were calculated using the scipy.stats and statsmodels packages for Python in addition to the lme4 and afex packages in R. Correlation values were determined using Spearman correlations except where otherwise noted.
Matlab code for converting raw calcium imaging movies into cellular activity time courses is available at https://github.com/asugden/pipe. Python and C code used for computing the classifier and all resulting analyses is available at https://github.com/asugden/flow and https://github.com/asugden/pool.
To account for possible non-independence within mice and within a recording session, we used generalized linear mixed-effects models (GLMMs, see 65) in addition to simpler comparisons such as non-parametric tests. For all GLMMs modeling normal distributions, p-values were calculated using the Kenward-Roger approximation for degrees of freedom. For Poisson-distributed GLMMs (i.e. those modeling Poisson distributions), p-values were calculated using parametric bootstrapping, as implemented by the afex R package 66. In short, we performed the following separately for each parameter in the model: we generated a restricted model by removing that parameter from the model and generating 1000 simulated datasets using this restricted model matching the distributions fit by the original full GLMM. Next, we fit the simulated datasets using both the restricted and full models, and calculated the likelihood ratio between the full model and the restricted model, resulting in a distribution of 1000 likelihood-ratio test values. Finally, we estimated a likelihood ratio for the real data, and compared it to the distribution of likelihood ratios estimated from the 1000 simulated datasets. This yielded a p-value based on the estimated probability that the likelihood ratio for the real data is larger than a fraction of the likelihood ratios estimated from simulated data. In other words, we generated simulated datasets in which the parameter in question is irrelevant by design, to obtain a ‘null distribution’ that allowed us to determine how often a model that includes the parameter in question is better than a model that does not include it.
No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in 22,35,67 and were chosen to measure experimental parameters while remaining in compliance with ethical guidelines for minimizing animal use.
We have described the statistics for each figure in Supplementary Information. Non-parametric tests that did not assume specific distributions were used in most cases. In the cases where specific distributions were assumed (only for GLMMs), these assumptions were addressed.
Example images of stimulus and reactivation:
We defined the activity of cell ROI masks as above, and used a variable shading within the outlines of these ROIs to represent the activity of the cells (Fig. 1e). For each ROI, we started with deconvolved, neuropil-corrected fluorescence traces (see above), which we used to determine whether a cell was active or not during the stimulus or reactivation (conservatively, defined as containing at least one timepoint with deconvolved activity > 0.2 during the 0–1 second period after stimulus onset, or from −130 to 130 ms around the reactivation event). For those cells that were active, we calculated the mean ΔF/F0 with the F0 defined as the average over the 1 second prior to the cue onset, or the 1 second prior to the time of initial crossing of the classifier output threshold of a reactivation event (as defined above) for Fig. 1e, top and bottom, respectively. We then displayed the mean ΔF/F0 value for each active cell from 0–1 seconds after cue onset or after the beginning of the reactivation (i.e. time of initial crossing of the classifier matching threshold, see above). Red and white colors indicate fractional changes in fluorescence of 0.5 and 0, respectively.
Connectivity and clustering:
The specific questions we asked in analysis of cross-day changes in functional connectivity were: how do a given cell’s functional properties change across two days during visual stimulus presentation, how do these changes depend on the cell’s participation in reactivation events in the previous session, and how do these changes depend on the cell’s functional identity. Thus, the analyses required cell-based metrics of functional properties, as opposed to pairwise metrics. We chose to use “total functional connectivity” because, faced with the challenge of assessing the net connectivity between a cell and a set of other cells, we chose a metric commonly used in graph theory for describing such connectivity – the clustering coefficient – which is the geometric mean of all connections between a node (here, a food-cue-driven cell) and all other nodes (i.e. all other food-cue-driven cells)68. This metric is described in more detail below, and the step-by-step procedure is also described in Extended Data Fig. 6a. This metric is robust relative to changes in ΔF/F0 levels across days, as it (i) only depends on cells with a sufficient signal-to-noise ratio to be determined as driven by the food cue, and (ii) depends on correlations rather than the magnitude of response.
To assess functional connectivity, we first estimated the noise correlations between pairs of cells 22,28,69. To determine noise correlations for pairs of cells (shared variability in trial-to-trial responses of two cells across presentations of a single type of cue; Fig. 4, 5 and Extended Data Fig. 8), we measured the mean correlation between each pair of neurons of responses averaged from 0-2 seconds after the cue onset, and subtracted the mean correlation obtained from 100 per-cell randomizations of trial order (e.g. 22, similar to removing the signal correlation). For all analyses of connectivity, we removed any cell with a motor component, as assessed by the cell-response GLM. We further controlled for motor artifacts by recalculating the noise correlation estimates used in Fig. 5, following removal of time points in each trial following onset of licking, and confirmed similar results (data not shown). In particular, data from a given trial was limited to times earlier than 100 ms before the first lick, and 100 ms before the median lick latency across all food-cue trials. To determine noise correlations across multiple cues in Figs. 2d, 5a, and Extended Data Fig. 2f (which were for display purposes only and serve only to provide additional graphical intuition into how the AODE classifier works), we determined the correlations between neurons’ responses from 0-2 seconds after the stimulus onset, after subtracting the mean response across all trials to remove the signal component (as in 70).
Graphs were made using Python 2.7 and the package networkx71 (https://networkx.github.io). Nodes were defined as neurons and edges were defined as the noise correlations between pairs of neurons (restricted to cases with positive noise correlations); clusters were defined using Louvain community detection72. This clustering technique does not require the user to estimate the number of clusters, but rather uses a greedy approach to optimize the density of links within clusters relative to those between clusters. To determine the total connectivity of a cell, we used a clustering coefficient that accounted for weighted edges (defined as the geometric mean of the edges with positive noise correlation values, using the function ‘clustering’ in the networkx package).
Pupil tracking and brain motion:
During imaging sessions, an infrared-sensitive camera was directed at the mouse’s pupil, contralateral to the cranial window and below the stimulus display. Infrared light from the two-photon laser illuminated the pupil. To identify the pupil, a user created a mask around the eye (to exclude stray reflections off of hairs near the eye) and selected the center of the pupil on the first frame. The center and area of the pupil was fit using starburst pupil detection from the openEyes toolkit and a ransac algorithm, and the center position from the previous frame was used to initialize the following frame. Frames in which the algorithm returned a low quality score were replaced with interpolated data, and only sessions with high-quality fits were used in the analysis. Brain motion was calculated as the root-mean-square translation of the image, as estimated during image registration.
Behavioral Performance:
The overall behavioral performance per session, d’, was estimated using z(hit rate) – z(false alarm rate)73. To obtain more accurate estimates of d’ that did not include epochs with sustained disengagement from the task, we excluded long periods of inactivity by using a hidden Markov model to estimate engaged/disengaged transitions. For this model, we estimated that the animal would have an 80% hit rate and 40% false alarm rate when engaged and a 2% hit rate and a 2% false alarm rate when disengaged. This eliminated at most a third of trials in any single session, and eliminated no trials at all in two thirds of all sessions.
For Fig. 3f and Extended Data Fig. 5d–f,j, we created a dynamic trial-by-trial estimate of behavioral performance using the PsyTrack Python package74. We used the code associated with the Roy et al. paper, and fit seven variables to predict behavioral outcome (Go-NoGo). In particular, we fit the likelihood that the animal would lick given each of the three drifting grating orientations, the lick/no-lick choice from the previous trial (to capture perseverative behavior), whether the previous trial was rewarded or punished (to collectively capture a win-stay/lose-switch strategy), and an offset (i.e. baseline licking rate). This model separately estimates the possible changes in performance between trials and between sessions using all available data. After fitting the model, we can use the fits to predict the probability of the mouse licking on each depending on which of the three drifting gratings were presented (and excluding the incorrect perseverance, win-stay/lose-switch strategies, and a baseline rate of licking). The predicted lick rate for the food-cue stimulus is the predicted hit rate and the combined predicted lick rates across the neutral and aversive stimuli is the predicted false alarm rate, which we then combined to estimate a per-trial estimate of behavioral performance, d’, as described above. Thus, we could estimate within-day and across-day changes in behavioral performance by using this estimated d’ for the first and last trials of each day (note that, due to the nature of this model, these estimates incorporated information spanning many trials).
Identification of cell responses using a generalized linear model (GLM):
To classify cell responses, we fit a Poisson GLM to the deconvolved activity of each cell, accounting for task and behavioral variables67,75 using the glmnet package. We first downsampled the data by a factor of 2 and convolved the results with a Gaussian kernel of width 260 ms. We created a series of basis functions to describe task events and behavioral variables. These basis functions were a series of Gaussian curves separated by 260 ms or 4 frames, and with a full-width at half maximum of 260 ms. The basis functions spanned different time ranges surrounding each variable. We included the basis functions representing each cue type (food cue, neutral cue, and aversive cue), separated by trial type (hit, miss, correct reject, false alarm), and tiled across the entire cue duration (0-2 s). We also included basis functions representing times relative to (i) any stimulus offset (−1 to 2 s surrounding offset), (ii) the presentation of reward or quinine (−1 to 2 s), (iii) the onset of a lick bout (−1 to 2 s from bout onset, with lick bouts separated by ≥ 2 s), (iv) all other individual licks (one kernel at the time of each lick), (v) brain motion (the kernel convolved with the analog vector) and (vi) locomotion (the kernel convolved with the running speed). We reproduced all results from Fig. 4d and 5e with a second GLM in which basis functions ranged from 0-3 s following cue onset, 0-2 s following reward or quinine onset, and adding a reward-anticipation category. Reward anticipation was defined as the 1 second prior to the reward or quinine onset on both hit and false alarm trials, the time at which the animal might anticipate a reward (data not shown).
The GLM was fit on two thirds of the data for each cell with elastic net regularization (α= 0.01). We then used the GLM coefficients to measure the deviance explained on the remaining one third of the data. Cells were determined to have activity that was locked to a given variable if (i) the deviance explained by the model was greater than 0.01 (note that the deviance explained is limited by the small number of behavioral variables) and (ii) the category of basis sets (i.e. food-cue, neutral-cue, aversive-cue, reward, quinine, or licking basis sets) made up at least 5% of that explanatory value. Cells with any activity locked to licking were excluded from analysis in Fig. 4, 5, and Extended Data Fig. 7c–f, 8–9.
We determined if a cell was “food-cue-driven” (visually responsive to food cue presentation as described previously19), i.e. for Food-cue cells, and Food-cue-reward cells. Specifically, we assessed whether a neuron had a significant increase in fluorescence (ΔF/F0) during the cue presentation (0-2 s from cue onset) relative to baseline (−1 to 0 s prior to cue onset) within at least one 0.33-s bin. The significance was computed using a Wilcoxon signed-rank p-value < 0.05, Bonferroni-corrected for the number of cell comparisons, cue comparisons, and time comparisons. At this value, roughly one third of the recorded cells respond to any cue. For the data in Figs. 4–5 and associated Extended Data figures, we used a more conservative p-value threshold that limited analyses to roughly the third of those cells with the smallest p-values (specifically, p-values < 1.3 × 10−18).
Distributions and kernel density estimations:
To display changes in connectivity across days, we plotted the distribution as the kernel density estimation rather than a histogram because histograms are sensitive to the bin sizes. The software package used was KDEUnivariate from the statsmodels plugin in Python. The grid size of the distribution was set to the min of (number of entries, 1000). Using continuous distributions whose areas were normalized to 1 (i.e. the kernel density estimates) allowed us to subtract distributions to determine the areas in which there was an excess of total connectivity for reactivated neurons (as in Fig. 4d,h, and 5e). These plots show the amount of reactivation beyond what is expected by the controls defined in the figure legend.
Types of reactivation events:
Food-cue reactivation events were determined to be “reward-rich”, “reward-poor”, or “mixed”, solely based on the cluster membership (see Connectivity and clustering, above) of active food-cue-driven cells in lateral visual association cortex. If >80% of active food-cue-driven cells were from reward-related clusters, the event was determined to be “reward-rich”. If >80% of active cells were in from non-reward-related clusters, the event was determined to be “reward-poor”. All other food-cue reactivation events were considered “mixed”. All events contained multiple active cells (as otherwise, they would not have been classified as a food-cue reactivation event). We reran the analyses in Fig. 5 using only days in which there were both reward-rich and reward-poor reactivations and found similar effects (not shown).
Fiber photometry recordings:
On the same day as the cranial window surgery, AAV1-hSyn-FLEX-GCaMP6s (University of Pennsylvania Vector Core) was injected into the contralateral ventral tegmental area (VTA) of 7/11 mice (300 nl, Bregma: AP: −3.2 mm, DV: −4.5 mm, ML 0.3 mm). Optic fibers with a metal ferrule (400 μm diameter core; multimode; NA 0.48; 5.0 mm length; Doric Fibers) were implanted over the medial core and shell of the nucleus accumbens (NAc; Bregma: AP: 1.45 mm; DV: −4.3 mm; ML: 0.75 mm; injection locations were confirmed histologically following the end of each experiment.). During imaging, a fiber optical cable (1-m long; 400-μm core; 0.48 NA; Doric Lenses) was coupled to the implanted optic fiber with zirconia sleeves (Precision Fiber Products). Black heat shrink material was slid around the fiber coupling to prevent outside light such as the visual stimulus monitor from interfering. Excitation and emission light was passed through a separated two-fluorophore fluorescence minicube (FMC5_E1(465-480)_F1(500-540)_E2(555-570)_F2(580-680)_S); Doric Lenses). This allowed stray infrared light from imaging to pass through the red channel. Excitation light (~60 μW, kept low to prevent bleedthrough into the imaging PMTs) was provided by a 465 nm LED (Plexon LED and driver). Emission light was collected by a femtowatt photoreceiver (Newport 2151) and digitized at a 2 kHz sample rate by Neurolabware’s Scanbox software.
We calculated the ΔF/F0 in the same manner as described in “Image registration and source extraction,” above. We set the mean activity of the baseline from −1 to 0 seconds prior to a cue or reactivation event to be zero and calculated the mean across events.
For additional information, please see “Life Sciences Reporting Summary.”
Data Availability
The data that support the findings of this study are available from the corresponding author upon request.
Code Availability
The code that support the findings of this study is available on Github at https://github.com/asugden/pipe, https://github.com/asugden/flow , and https://github.com/asugden/pool . For specific links, see above.
Extended Data
Extended Data Fig. 1: Correlation of behavioral performance and session number and examples of offline cue reactivation events in darkness.
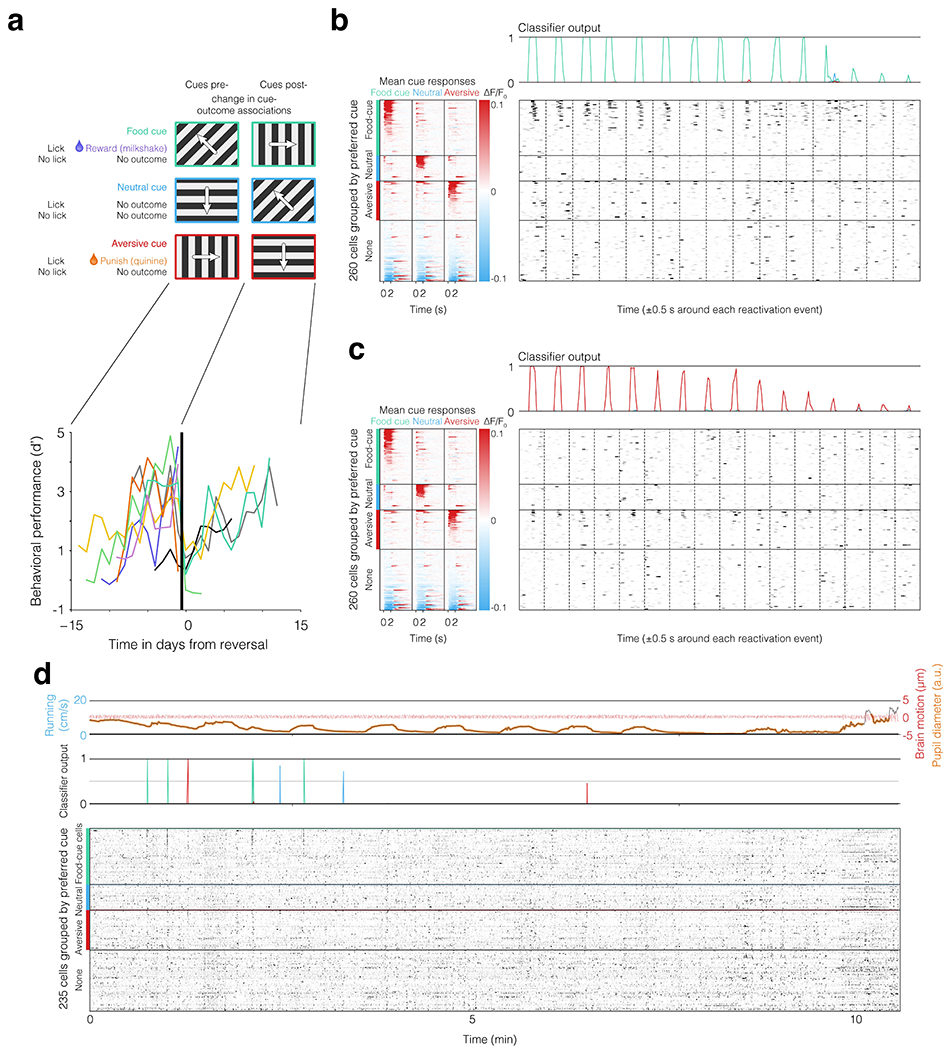
a. Top: Go-NoGo visual discrimination task, and description of the one-time change in cue-outcome contingencies (“reversal”). Bottom: behavioral performance as a function of number of training sessions relative to day of reversal of cue-outcome contingencies (‘day 0’). Data from each of the eight mice are plotted using a unique color. See also Ramesh et al., 2018. b. Left: average cue-evoked response time courses (rows) from each simultaneously recorded cell from a single session. Data are aligned to the onset of the visual cues, each of which was 2 s in duration. ΔF/F0: fractional change in fluorescence. Bottom right: peri-reactivation deconvolved activity (black) of all cells during the post-training and post-satiation ~1 hour period of recording in darkness. Data correspond to the 15 food-cue reactivation events with the highest classifier matches (classifier output shown on top), and are sorted in order of decreasing classifier output (top panel; roughly similar to a matching probability, see Methods and Supplementary Information). c. Same plot as in b, but for aversive-cue reactivations during the same example session. d. Portion of an example recording from a different mouse from that shown in Fig. 2a–c, from a sated mouse. Bottom: deconvolved activity of 235 simultaneously recorded cells. Behavioral and other variables are also shown: brain motion (root-mean-squared motion in the imaging plane; red, top), running speed (blue, top), pupil diameter (orange, top), licking (purple ticks, top), output of the classifier for each cue (colored lines, middle; red: aversive cue; blue: neutral cue; green: food cue).
Ramesh, R. N., Burgess, C. R., Sugden, A. U., Gyetvan, M. & Andermann, M. L. Intermingled Ensembles in Visual Association Cortex Encode Stimulus Identity or Predicted Outcome. Neuron 100, 900–915.e9 (2018).
Extended Data Fig. 2: Further characterization of our classifier, including temporal components and those related to pairwise co-activation of neurons.

We designed a classifier that uses an averaged one-dependence estimator (AODE) – an extension to Naïve Bayes that accounts for pairwise probabilities (Webb et al., 2005; Sugden et al., 2018). a. Mean deconvolved population activity, centered on cue reactivation events found with a version of this classifier that did not include a temporally varying prior. b. Construction of the temporally varying prior. We used three high-pass difference-of-Gaussians kernels to generate the population activity scale factor that modulates the prior probability employed to estimate the probability of a reactivation event. We took the minimum value across the convolutions of each of these three kernels with the mean deconvolved activity across cells, in order to define a temporally varying prior at each time point (see Methods and Supplementary Information for additional details). Each filter is constructed by subtracting from a narrow Gaussian (sigma: 41 imaging frames, 260 ms) one of three broad Gaussians (sigmas: 42, 43, and 44 imaging frames, corresponding to 1 s, purple; 4 s, blue; and 16 s, green respectively), to account for background fluctuations at multiple slower timescales than the timescales expected for cue reactivations (based on both previous studies and on the results of panel a). c. Visualization of the effects of the temporally varying prior – a high-pass temporal filter applied to each cell’s activity time course that biases classification towards brief, synchronous events. High-pass filtering the deconvolved data in Fig. 2c by applying this filter separately to each cell time course shows that the temporally varying prior selectively enhances transient events. Transient events that are not marked as reactivation events by the classifier may be associated with hippocampal sharp-wave ripples, but were not classified as reactivations of one of the three cues, as they were not sufficiently similar to cue responses (right panel; see also Fig. 2a–b). Note that the same temporal filter is applied to all cell time courses, and thus does not modify the relative activity across neurons at the time of cue reactivation. d. Cue reactivation events can occur in bursts with inter-event intervals of ~1 s. These short inter-event intervals in the true distribution of inter-event intervals (black line) are beyond what is expected by chance (95% confidence interval of shuffle is shaded in gray, estimated by assigning each event to a time drawn uniformly at random within the session in which the event was identified). e. Violin plot of rate of cue reactivation events in darkness (width is proportional to relative incidence), for a classifier using Naïve Bayes (left) vs. AODE (right). Black horizontal lines indicate means across sessions, and black vertical lines indicate SEM across sessions. Accounting for pairwise probabilities of co-active neurons using the AODE doubles the number of identified reactivation events for the same classifier output threshold (p < 0.0001, two-tailed Wilcoxon rank-sum test). f. Clusters were defined using the functional connectivity metric of noise correlations, measured across all three cues. Circles represent the activity of neurons (represented as in Fig. 2d; response probabilities of cue-driven neurons are represented by the diameter of circles along outer ring, and joint response probabilities are proportional to the thickness of connecting lines). Cue reactivation events detected by the classifier using AODE but not by the classifier using Naïve Bayes (right column) were often dominated by active cells belonging to a small number of clusters (gray shaded wedges). In contrast, events detected by both the classifier using AODE and the classifier using Naïve Bayes (left column) included active neurons spanning multiple clusters.
Sugden, L. A. et al. Localization of adaptive variants in human genomes using averaged one-dependence estimation. Nat Commun 9, 703 (2018).
Webb, G. I., Boughton, J. R. & Wang, Z. Not So Naive Bayes: Aggregating One-Dependence Estimators. Machine Learning 58, 5–24 (2005).
Extended Data Fig. 3: The classifier detects few reactivation events following randomization in cell identity or in time.

a. Fraction of cue presentations (colored lines) and inter-trial intervals (gray line) correctly identified by our AODE classifier when trained on two-thirds of the data and tested on the remaining third (mean ± SEM across 109 sessions), as a function of classifier output threshold. Green, blue and red lines correspond to food cue, neutral cue and aversive cue, respectively. b. To demonstrate that the classifier is not identifying random fluctuations in activity during quiet waking periods as cue reactivation events, we plotted the number of reactivation events for each cue type that would still be found following shuffling of cell identities in identified reactivation events compared to real data, as a function of the classifier output threshold (see also Methods and Supplementary Information). There are few events found in randomized data at any classifier output threshold greater than 0.05. c. The fraction of cue reactivation events found in data randomized by identity above our chosen classifier output threshold (same as Fig. 2f, replicated for comparison). Compared to the number of events that exceeded the classifier output threshold of 0.1 in the real data (which was the threshold we used in our analyses), we found a relatively small proportion in the randomized data. Results were similar for a modified classifier using Naïve Bayes (not shown). d. Similarly, we plotted the fraction of these events that would still be found at times of classifier-identified cue reactivation events, but in data randomized across cell identities in which we matched activity levels across cells. Specifically, at time points identified as cue reactivation events, we randomized the cell identities, and determined how many cue reactivation events (across all three cue types) were found above our chosen classifier output threshold of 0.1. e. As in c, the fraction of cue reactivation events found in data randomized in identity with matched activity levels. f. As in b, the fraction of events identified in data randomized in time relative to real data, as a function of classifier output threshold. This was estimated by randomizing data in time via circularly shifting each cell’s time course with random relative delays (i.e. offsetting each time course in time by a random amount for each cell, and wrapping the clipped portion of the time course beyond the end of the recording back to the beginning). g. As in c, the fraction of cue reactivation events identified in data randomized in time. h. Distribution of the number of identified events (across all sessions from all mice, in runs following satiation), for events of each classifier output threshold (from 0.05-1 in bins of 0.05). Dashed gray line represents our chosen classifier output threshold for defining an activity pattern as a cue reactivation event. Note that food-cue reactivation events were more common than neutral-cue reactivation events across a range of classifier output thresholds. All randomizations were performed using 109 sessions across 8 mice, and bars and lines represent the mean ± SEM.
Extended Data Fig. 4: Additional data on combined imaging and hippocampal electrophysiology; reactivation events are not a result of epileptiform events, transient brain motion, or eye motion.

a. Schematic for two-photon calcium imaging in visual association cortex while recording from contralateral hippocampal area CA1 using a silicon multi-electrode probe. b. Same example aversive-cue reactivation event as in Fig. 2g, but with additional data shown. Bottom left: mean activity time courses (fractional change in fluorescence, ΔF/F0) in response to food cues, neutral cues, and aversive cues (columns) for all simultaneously recorded neurons (rows) from the same session, organized by preferred response. Bottom right: deconvolved activity traces of all recorded cortical cells in the period surrounding the aversive-cue reactivation event (red arrow). Top: purple traces: CA1 local field potential trace and zoom-in. When band-pass filtered in the ripple band (150-300 Hz; gray trace), the LFP shows a transient increase just before the detected reactivation event. c-e. The Emx1-Cre;Ai93;CaMK2a-tTA mouse line (Madisen et al., 2015) used in our work has been reported to exhibit epileptiform events in some cases (Steinmetz et al., 2017). Early experiments in our lab using mice obtained directly from the Allen Brain Institute did appear to exhibit visible epileptiform events as detected in the neuropil signal in cortical imaging. While none of these events overlapped with any cue reactivation events, we excluded this mouse from the study. We then acquired mice from Jackson labs (line 024108 rather than 024103 used in (Steinmetz et al., 2017)) which may have been further back-crossed. To further address the possibilities that epileptiform events occurred in the 8 mice used in our study and that these might overlap with the cue reactivations described in our study, we characterized the amplitude and width of all transient events in the neuropil from spontaneous activity recordings in all of our mice (as in (Steinmetz et al., 2017)). As described below, we found that 7/8 mice did not show any evidence of epileptiform events, while 1/8 mice only demonstrated a small number of epileptiform events. Panel c: scatter plot of peak width vs. event peak prominence (height above local background), combined across 7/8 mice included in this study. Typical low-amplitude or high-width peaks (Steinmetz et al., 2017) are shown as blue dots, whereas brief, high-amplitude peaks that may reflect epileptiform events are shown as black dots (defined using a conservative manual threshold as in (Steinmetz et al., 2017)). In these 7 mice, we found almost no epileptiform events total across over a dozen sessions per mouse (0,0,0,0,1,2,3 events total per mouse; events rates ranged between 0 and 0.00003 epileptiform events/second). Panel d: same as c, but for the eighth mouse included in the current study. In this mouse, we found 87 epileptiform events total across all sessions, amounting to 0.0014 epileptiform events/second, or approximately one such ~150 ms event every 12 minutes. Across all sessions, a total of two epileptiform events were observed within 1 second of any reactivation event, and both of these were neutral-cue reactivation events. Panel e: same as c, but for the mouse excluded from this dataset. Detectable epileptiform events were very rare in this mouse (0.008 events/s). Critically, we found that across all 8 included mice, 0/1444 food cue reactivations, 0/1258 aversive cue reactivations, and 2/1040 neutral cue reactivations occurred within 1 second of an epileptiform event (and the two overlapping neutral cue reactivations were from a mouse with 2 epileptiform events detected in total across all sessions). Thus, epileptiform events are exceptionally rare in the mice used in this study, and those that occurred did not occur near cue reactivation events. To further confirm these observations, we re-analyzed the 7 imaging sessions from two mice in which we imaged from the identical mouse line while recording cortical and hippocampal local field potentials using a 16-channel silicon probe acutely implanted contralateral to the imaging window (see Fig. 2g). The epileptiform events previously described using cortical electrophysiology (Steinmetz et al., 2017) appear as large local field potential (LFP) spikes, characterized by high amplitude (>1000 μV; normal LFP amplitudes are 5–15x smaller) and long duration (>10 ms). Briefly, we used the exact methods as in Steinmetz et al., 2017, and analyzed 20-s LFP traces surrounding 250 cue reactivation events identified using contralateral imaging (5000 seconds of data). We found a total of 5 epileptiform events from our LFP data (average rate of 1 events / 1000 s). Critically, all 5 of these events occurred >2 seconds away from any cue reactivation event. f. Pupil area (normalized to mean area estimated during locomotion in darkness, a state involving dilated pupils) was constricted throughout reactivation events. g. There was no transient increase in brain motion (root-mean-squared motion in the imaging plane) in the moments surrounding a reactivation event (green: food-cue reactivations; blue: neutral-cue reactivations; red: aversive-cue reactivations). h. There was no transient increase in eye motion (root-mean-squared motion) in the moments surrounding a reactivation event. Error bars are ± SEM across sessions (f) and across events (g-h).
Madisen, L. et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958 (2015).
Steinmetz, N. A. et al. Aberrant Cortical Activity in Multiple GCaMP6-Expressing Transgenic Mouse Lines. eNeuro 4, ENEURO.0207–17.2017–33 (2017).
Extended Data Fig. 5: Additional evidence supporting a learning-related decrease and a food-cue bias in reactivation rates, and a reactivation-dependent change in future behavioral performance.

a. Reactivation rates were significantly higher in sessions following task engagement (black dots: reactivation event counts per session for all 3 cues across 109 training sessions) than in sessions in which naïve animals viewed the same visual stimuli but in the absence of salient outcomes (i.e. sessions during initial habituation to the stimuli but prior to any conditioning; gray dots: reactivation event counts per session for all 3 cues across 11 sessions). Data were fit using a GLMM accounting for shared variance within mice (see Methods and Supplementary Information). b. The number of recorded cells did not change across learning (r and p: Pearson correlation; error bars: 95% confidence intervals; N = 109 sessions). c. The overall population activity across all neurons (measured as the mean deconvolved activity across quiet waking periods) did not decrease across learning (fit using the same 109 sessions as in b). d-e. Example plots from a single animal using PsyTrack (Roy et al., 2018) to dynamically determine behavioral variables. d. Individual behavioral variables fit across time in this animal. The vertical black bar represents the time of change in cue-outcome associations (i.e. reversal), and thin gray vertical bars separate different days. Colored lines for each angle (0°, 135°, 270°) reflect behavioral sensitivity to a grating drifting at that angle (colors of gratings in legend refer to the cue contingencies associated with that grating prior to and following reversal, as shown using the vertical black line; green: food cue; red: aversive cue; blue: neutral cue). Variables labeled ‘Prev choice’, ‘Prev punish’, and ‘Prev reward’ refer to effects on current performance related to choice (Go or NoGo) or outcome from the previous trial, and ‘Offset’ reflects the baseline licking rate. Error bars: 95% posterior credibility interval (the Bayesian equivalent of a confidence interval). e. Example of behavioral performance calculated per day (orange) and estimated dynamically (blue) across time in the same animal (Roy et al., 2018). Green dots indicate performance at the beginning of each day and red dots indicate performance at the end of each day. f. The change in behavioral performance from the beginning to the end of a training session is not associated with cue reactivation rate during the subsequent quiet waking period (for any of the three cues; same 109 sessions as a-c). r: Spearman correlation. p-values were determined using a GLMM accounting for shared variance within mice (see Methods and Supplementary Information). g. Cumulative distribution of single-session cue reactivation rates (same data as in Fig. 3a). h. Similar to Fig. 3c–d, the food-cue is preferentially reactivated when using the Naïve Bayes classifier instead of the AODE classifier (*** p < 0.001, ** p < 0.01; using a GLMM comparing the effect of cue on the rates of reactivation accounting for shared variance within mice and days), both for sessions prior to reversal (65 sessions) and following reversal (44 sessions). Inset at right shows relative reactivation rates for each session and cue type (normalized to the neutral rate for that session) using Naïve Bayes, colored by cue type. i. Fraction of responsive cells driven by a particular cue (same data as in Fig. 3e but with data points for each session shown). For each session, the fraction of responsive cells driven by a particular cue was normalized to the overall number of cue-responsive cells. Data reflect mean ± SEM across sessions. Thus, the observed enhancement in food cue reactivation rates was not due to a difference in the number of neurons responsive to any given cue (no significant difference, two-tailed Wilcoxon rank-sum test), as we reversed cue contingencies or stopped recording at the point in training where a food-cue bias typically emerges, after the mouse becomes very well trained; Burgess et al., 2016; data not shown). j. Unlike Fig. 3f, reactivation rates of neutral and aversive cues do not significantly predict changes in behavioral performance across sessions (though weak trends are evident). We used a GLMM that accounts for shared variance within mice and for number of days elapsed between the two training sessions. Line: GLMM fit across mice when considering 1 day of elapsed time between sessions. r: Spearman correlation p-values indicate the effect of reactivation rate from the GLMM. Error bars represent mean ± SEM across sessions in panels b-c (109 sessions) and h-i (65, 44, and 109 sessions).
Burgess, C. R. et al. Hunger-Dependent Enhancement of Food Cue Responses in Mouse Postrhinal Cortex and Lateral Amygdala. Neuron 91, 1154–1169 (2016).
Roy, N. A., Bak, J. H., Akrami, A., Brody, C. D. & Pillow, J. W. Efficient inference for time-varying behavior during learning. Adv Neural Inf Process Syst 31, 5695–5705 (2018).
Extended Data Fig. 6: Estimation and simulation of total functional connectivity.

a. A description of the steps taken to compute the change in total connectivity across sequential pairs of days. Step i. For a given day, we generate a matrix containing the mean response, from 0-2 seconds after the food cue onset, for each trial and each cell driven by the food cue on that day. Step ii. We then compute the matrix of pairwise noise correlations for these cells (described in Methods). Step iii. Correlation matrices can be represented as weighted graphs with edge weights defined by the correlation coefficients. As such, we create a weighted graph with food-cue cells as nodes, and using positive noise correlations as edge weights. Step iv. We compute the total connectivity per cell, defined as the clustering coefficient for each node (i.e. the geometric mean across connected edges; Hagberg et al., 2008). The measure of total connectivity thus reflects the notion that pairs of cells having stronger noise correlations are more strongly connected and/or receive stronger common input. Step v. For each cell identified on both of two consecutive days (i.e. cells lacking an ‘x’ symbol on both days), we calculate the difference in the cell’s total connectivity (clustering coefficient) between the two days. Further analysis (as in Fig. 4d and Fig. 5e) compares this difference in total connectivity between two different groups of cells -- cells that either did or did not participate in inter-session reactivation events. b. We considered whether the computation of the total connectivity metric might introduce correlations between estimates of connectivity of simultaneously recorded cells (as each cell’s estimate involved averaging across the pairwise correlations with the other cells), and thereby invalidate the assumption of conditional independence of data points from simultaneously recorded cells in the GLMM analyses used in Fig. 4d and 5e. Therefore, we simulated noise correlation matrices from two distributions (for the two groups of reactivated vs. non-reactivated cells, see above and further described in Methods) and computed changes in total connectivity (steps iii-v) between sequential pairs of these matrices. We found that this metric does introduce a random shift in the change in total connectivity that is common to all cells recorded within a given day-pair (top panel), but that these shifts are accounted for by using a generalized linear mixed regression model that includes fixed offsets for each day-pair. We demonstrated this by measuring, for each pair of simulated cells, the Pearson correlation of changes in total connectivity across multiple day pairs (after subtracting off the mean shift across cells for each day-pair; bottom panel). The resulting correlation was found to be significant in only approximately one out of every 20 simulations (i.e. p = 0.05), as expected if there was no underlying correlation introduced by using the total connectivity metric. Given this result, we concluded that there was no correlation introduced by the total connectivity metric above and beyond a common mean effect across cells recorded on any given day-pair, further justifying the use of the GLMM in these analyses. See Data analysis and statistics section of Methods for additional details.
Hagberg, A. A., Swart, P. J. & Schult, D. A. Exploring network structure, dynamics, and function using NetworkX. Proceedings of the th Python in Science Conference SciPy 11–15 (2008).
Extended Data Fig. 7: Generalized linear model for categorizing groups of lateral visual association cortex neurons that encode one or multiple task-related variables; further analyses of cells that also encode reward.

a. Mean deconvolved activity of cell groups identified by a generalized linear model (see Identification of cell responses using a generalized linear model section of Methods) surrounding times of task-related events (e.g. cue presentations, motor behavior, reward delivery). Rows, top to bottom: cell types that only respond to the food cue, to the neutral cue, and to the aversive cue, those that only respond in a time-locked manner at moments surrounding the reward presentation or surround9ing the punishment presentation, and those with activity time-locked to both the food cue and to the reward presentation. Time zero for each column in a corresponds, from left to right, to the time of food cue onset, neutral cue onset, aversive cue onset, Ensure reward delivery onset, quinine delivery onset, and onset of a lick bout (i.e. onset of a lick sequence after ≥ 2 s without any licks). N’s at left indicate numbers of neurons. b. Mean deconvolved activity of cell groups identified by a generalized linear model (see Methods) surrounding reactivation events. The peak cue amplitude for those cells that respond only to the food cue, neutral cue, or aversive cue (top 3 rows) has been scaled to match the amplitude during reactivation events (gray). The other rows (i.e. other cell categories) have had their cue-evoked responses scaled by an equal amount. As such, the level of activity during reactivations can be compared with the expected level given the response magnitude of cells of a given category during the cue period used by our classifier. Error bars are ± SEM across the same sets of cells as in a. c. Cells encoding both the reward and the food cue (i.e. Food-cue-reward cells) are more common than cells encoding both the reward and other cues (two-tailed Wilcoxon rank-sum test, Bonferroni corrected for 3 comparisons, *** p < 0.0001; mean ± SEM across sessions). d-f. Heatmaps of individual trials (row) from example reward-coding cells that increase their activity at different times relative to reward delivery. Display of data is identical to Fig. 4f.
Extended Data Fig. 8: Additional data demonstrating distinct clusters of cells with strong intra-cluster correlations in trial-by-trial cue responses; comparison of cells belonging to reward-related vs. non-reward-related clusters.

a. The clustering algorithm used in Fig. 2d and Fig. 5 (see Methods) clusters cells into groups that share high within-group, cue-driven, trial-by-trial noise correlations. This algorithm automatically chooses the optimal number of clusters for each session. The number of clusters per session did not differ between stimulus types (two-tailed Wilcoxon rank-sum test). b. Cluster sizes (number of cells per cluster) also did not differ between stimulus types (two-tailed Wilcoxon rank-sum test). c. Within-cluster noise correlations were substantially higher than between-cluster noise correlations (see Methods; two-tailed Wilcoxon rank-sum test; *** p < 0.0001). d. Same violin plots as Fig. 5a, but using the Naïve Bayes classifier. These data confirms that the finding in Fig. 5a is not due to the fact that the AODE classifier used in Fig. 5a includes pairwise activity as part of the information used for identification of reactivation events. e. Similar plots as in d, but separately examining subsets of pairs of cells considered in d that belong to reward-related clusters (pink) or to non-reward-related clusters (orange; two-tailed Wilcoxon rank-sum test; *** p < 0.0001). f. Bottom: CDFs and KDEs of the distributions of cross-day changes in total functional connectivity. Reactivated food-cue-driven cells from reward clusters (purple, 194 cells) increased their next-day total connectivity compared to all non-reactivated food-cue-driven cells (gray, 238 cells). Conversely, reactivated food-cue-driven cells from non-reward clusters (orange, 245 cells) decreased their next-day total functional connectivity. P-values were computed using a linear GLMM involving a categorical comparison involving 3 categories: non-reactivated cells, cells from reward clusters, and cells from non-reward clusters. Permutation tests within days demonstrated that cells from non-reward clusters had significantly decreased next-day connectivity when reactivated (* p < 0.05; see Methods). Top: excess frequency of reactivated cells with a given change in next-day connectivity above that observed for non-reactivated cells (i.e. rectified difference of colored vs. gray distributions). g. Data from Fig. 5c, presented as a correlation matrix between all pairs of cells and a hierarchical clustering dendrogram. The clusters identified on the left are labeled on the right. Note the distinct reward-related clusters and non-reward-related clusters. h. Cluster size (i.e. the number of cells per cluster), with 1 dot per cluster for all clusters from all recordings, separated by reward-related clusters (purple) or non-reward-related clusters (orange). i. Number of clusters of a given type per session. Approximately 44% of sessions contained at least one cluster of each type. j. Within both Day 1 and Day 2 of each pair of days (more specifically, on Day i relative to a pair of days [i, i+1]), the total functional connectivity was not different between cells belonging to reward-related clusters vs. those belonging to non-reward-related clusters. The overall responsivity of each cell group was also unchanged across days and did not differ between groups (two-tailed Wilcoxon rank-sum test). k. Total connectivity on Day 1 was indistinguishable between (i) all food-cue-driven cells in reward-related clusters, (ii) Food-cue cells in reward-related clusters, defined as cells that are responsive to food cues but not to rewards, and (iii) Food-cue cells in non-reward-related clusters (p = 0.81, Kruskal-Wallis test). This was true both when comparing the subsets of cells that did not participate in any food-cue reactivations on Day 1 (left bars), and when comparing the subsets of cells that participated in at least one food-cue reactivation on Day 1 (right bars, p = 0.96, Kruskal-Wallis test). All error bars are mean ± SEM across sessions, including in violin plots (some bars are too small to show).
Extended Data Fig. 9: Different flavors of food-cue reactivation events from an example session; additional analyses of cross-day changes in total connectivity.

a. Plot similar to that shown in Fig. 5c, but for a different example session. Response probabilities of cue-driven neurons are represented by the diameters of circles along the outer ring, and joint response probabilities are proportional to the thickness of connecting lines. Food-cue-driven neurons were clustered into groups based on their functional connectivity (food-cue-evoked noise correlations; c.f. Fig. 5c). The two ‘Reward-related clusters’ (purple wedges) contained at least one Food-cue-reward cell, while the ‘non-reward-related cluster’ (orange wedge) did not. b. Left: as also described in Fig. 5d, food-cue reactivation events could be composed primarily of active food-cue-driven cells from non-reward-related clusters (“#1: reward-poor reactivations”, top), from reward-related clusters (“#3: reward-rich reactivations”, bottom), or of similar numbers of active food-cue-driven cells of both types (“#2: mixed reactivations”, middle). Right: Across 1444 reactivations from 109 sessions in 8 mice, we found a substantial number of food-cue reactivation events of each flavor. Plotted is the kernel density estimate (KDE, or smoothed distribution of frequencies), across cue reactivation events, of the fraction of reactivated cells from reward-related clusters (a value of 1 indicates that all food-cue-driven cells active during the reactivation are from reward-related clusters; a value of 0 indicates that all food-cue-driven cells active during the reactivation are from non-reward-related clusters). Also plotted is the CDF. The large incidence of reactivation events with proportions equaling 0 or 1 is due to sessions in which food-cue-driven cells belonged either entirely to non-reward-related clusters, or to reward-related clusters, respectively. Vertical dashed lines indicate the thresholds that were chosen to categorize food-cue reactivations into one of the three groups defined above (compare circled numbers in left and right panels). c. Left: mean activity time courses (fractional change in fluorescence, ΔF/F0) in response to food cues, neutral cues, and aversive cues (columns) for all simultaneously recorded neurons (rows) from the same example session as in a and organized by preferred response. Right: deconvolved activity time courses (1 row/cell) in the 0.5 s surrounding each food-cue reactivation event (plotted in a similar manner as in Extended Fig. 1b–c). Reactivations are sorted, in descending order, by strength of classifier output (posterior estimates; green lines at top). Thick horizontal colored bars below the classifier output estimates are shaded from orange to purple and indicate the proportion of participating food-cue-driven cells that are from reward-related clusters (see color bar at bottom of panel b). Circular plots below each reactivation event indicate which cells participated in the event, and their membership in reward or in non-reward-related clusters. Note that the first 8 reactivations are mixed reactivations, while the ninth reactivation (at right) is a reward-rich reactivation, as most participating cells belong to reward-related clusters. d. Same as c but for the next nine food-cue reactivations from the same session (again in descending order of estimates of strength of classifier output). e. Left: changes in total connectivity for non-reactivated cells in Fig. 4d, plotted on the same axis. Right: changes in total connectivity for reactivated Food-cue cells in non-reward-related clusters, Food-cue cells in reward-related clusters, and Food-cue-reward cells. Note that while reactivated cells increased total connectivity more when they belonged to reward-related clusters, non-reactivated cells decreased connectivity more when they belonged to reward-related clusters.
Extended Data Fig. 10: Additional analyses and control experiments regarding changes in VTANAc axon activity during cue reactivations.

a. Setup for simultaneous two-photon calcium imaging in cortex and contralateral fiber photometry recording in NAc of putative VTANAc axons. b. Left: average time courses of bulk GCaMP6 fiber photometry responses in nucleus accumbens (mainly from dopaminergic VTANAc axons; Rodriguez-Lopez et al., 2017) to visual food-cue presentations during training. Responses are averages of ‘hit’ trials (green) in which the mice made an operant response in the response window (2-4 s after cue onset) and received a reward, or ‘miss’ trials in which the mice did not respond in the response window. Percent change in GCaMP6 fluorescence, ΔF/F0. Mean ± SEM across trials, pooled across 33 sessions from 3 mice (mouse #1: 16 sessions; mouse #2: 8 sessions; mouse #3: 9 sessions). Middle and right: same analysis but for neutral cue and aversive cue trials, separated by ‘correct-reject’ trials (no response during response window) and ‘false-alarm’ trials (lick response during response window). Gray bars represent the time of cue presentation and black bars represent the first second of the response window that follows offset of cue presentation. Licking in this response window results in reward delivery, no outcome, or quinine delivery during food-cue trials, neutral-cue trials, or aversive-cue trials, respectively. While some GCaMP6f-labeled axons from EMX1-Cre-expressing neurons do innervate the NAc in this mouse line (as we used this transgenic mouse to track cortical cue reactivations), and while the AAV-GCAMP6s infection of VTA likely led to expression in both dopaminergic and non-dopaminergic NAc-projecting neurons, the observed cue-evoked responses in these NAc recordings show a strong response bias to the rewarded food cue and a decrease in activity in the response window following aversive-cue trials (particularly for false-alarm trials), which closely match the activity profile expected for putative VTA dopaminergic neurons projecting to NAc (Roitman et al., 2008; Cohen et al., 2012; Parker et al., 2016; Lutas et al., 2019) . c. Left: reward-rich food-cue reactivations during quiet waking evoked increases in activity in putative VTANAc axons, while reward-poor food-cue reactivations during quiet waking evoked decreases. Mixed reactivations had little effect on the photometry signal. The fraction of reward-rich cells active during a food-cue reactivation predicted the direction and amplitude of the VTANAc fluorescence response between 0.5 and 1 seconds after reactivation event onset (* p < 0.05, categorical GLMM; see Methods and Supplementary Information). Middle and right: average change in putative VTANAc axon activity following offline neutral-cue reactivations and aversive-cue reactivations, respectively. Mean ± SEM across cue reactivations. Two-tailed Wilcoxon sign-rank tests mean evoked response in the 0.5–1 s after the reactivation event vs. 0 (p-values indicated in panels). d. Heatmaps showing all individual cue reactivation-triggered fractional changes in VTANAc fluorescence for the data plotted in the middle and right panels of c. Black horizontal lines separate groups of trials from each of the three mice. Note the consistent drop in activity (blue) following most reactivation events. e. Similar to d, but for activity of reward-rich, mixed, and reward-poor reactivations (cf. panel c, left).
Cohen, J. Y., Haesler, S., Vong, L., Lowell, B. B. & Uchida, N. Neuron-type-specific signals for reward and punishment in the ventral tegmental area. Nature 482, 85–88 (2012).
Lutas, A. et al. State-specific gating of salient cues by midbrain dopaminergic input to basal amygdala. Nat Neurosci 1–21 (2019). doi:10.1038/s41593-019-0506-0
Parker, N. F. et al. Reward and choice encoding in terminals of midbrain dopamine neurons depends on striatal target. Nat Neurosci (2016). doi:10.1038/nn.4287
Rodríguez-López, C., Clascá, F. & Prensa, L. The Mesoaccumbens Pathway: A Retrograde Labeling and Single-Cell Axon Tracing Analysis in the Mouse. Front Neuroanat 11, 406–15 (2017).
Roitman, M. F., Wheeler, R. A., Wightman, R. M. & Carelli, R. M. Real-time chemical responses in the nucleus accumbens differentiate rewarding and aversive stimuli. Nat Neurosci 11, 1376–1377 (2008).
Supplementary Material
Acknowledgments
We thank G. Goldey for performing initial surgeries, and N. Patel, M. Barbini, G. Niyazov, and E. Bamberg for help with mouse training. We thank B. Healy, R. Born, J. Drugowitsch, I. Witten, C. Harvey, Y. Livneh, and members of the Andermann lab for useful discussion. We thank Drs. V. Jayaraman, R. Kerr, D. Kim, L. Looger, and K. Svoboda and the GENIE Project at Janelia Farm Research Campus, Howard Hughes Medical Institute for use of GCaMP6f, and Dr. H. Zeng for sharing GCaMP6f transgenic mice prior to publication. Support was provided by NIH T32 5T32DK007516 (AUS), NSF GRFP 2016207224 (KLM), NIH F31 105678 (RNR), F32 DK112589-01 (AL), a Davis Family Foundation Postdoctoral Fellowship (CRB), an NIH New Innovator Award DP2 DK105570 and R01 DK109930, a McKnight Scholar Award, a Pew Scholar Award, a Smith Family Foundation Award, a Harvard Mind Brain Behavior Interfaculty Initiative Faculty Research Award, and grants from the Klarman Family Foundation, the American Federation for Aging Research, and the Harvard Brain Science Initiative Bipolar Disorder Seed Grant, supported by Kent and Liz Dauten (MLA).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Nádasdy Z, Hirase H, Czurkó A, Csicsvari J & Buzsáki G Replay and Time Compression of Recurring Spike Sequences in the Hippocampus. J. Neurosci 19, 9497–9507 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee AK & Wilson MA Memory of sequential experience in the hippocampus during slow wave sleep. Neuron 36, 1183–1194 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Ji D & Wilson MA Coordinated memory replay in the visual cortex and hippocampus during sleep. Nat Neurosci 10, 100–107 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Singer AC & Frank LM Rewarded outcomes enhance reactivation of experience in the hippocampus. Neuron 64, 910–921 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Squire LR, Genzel L, Wixted JT & Morris RG Memory Consolidation. Cold Spring Harb Perspect Biol 7, a021766–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothschild G, Eban E & Frank LM A cortical-hippocampal-cortical loop of information processing during memory consolidation. Nat Neurosci (2016). doi: 10.1038/nn.4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster DJ Replay Comes of Age. Annu. Rev. Neurosci. 40, 581–602 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Euston DR, Tatsuno M & McNaughton BL Fast-forward playback of recent memory sequences in prefrontal cortex during sleep. Science 318, 1147–1150 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Peyrache A, Khamassi M, Benchenane K, Wiener SI & Battaglia FP Replay of rule-learning related neural patterns in the prefrontal cortex during sleep. Nat Neurosci 12, 919–926 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Xu S, Jiang W, Poo M-M & Dan Y Activity recall in a visual cortical ensemble. Nat Neurosci 15, 449–55– S1–2 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girardeau G, Inema I & Buzsáki G Reactivations of emotional memory in the hippocampus–amygdala system during sleep. Nat Neurosci 20, 1634–1642 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Puentes-Mestril C & Aton SJ Linking Network Activity to Synaptic Plasticity during Sleep: Hypotheses and Recent Data. Front Neural Circuits 11, 61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norimoto H et al. Hippocampal ripples down-regulate synapses. Science 359, 1524–1527 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Girardeau G, Benchenane K, Wiener SI, Buzsáki G & Zugaro MB Selective suppression of hippocampal ripples impairs spatial memory. Nat Neurosci 12, 1222–1223 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Jadhav SP, Kemere C, German PW & Frank LM Awake hippocampal sharp-wave ripples support spatial memory. Science 336, 1454–1458 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jadhav SP, Rothschild G, Roumis DK & Frank LM Coordinated Excitation and Inhibition of Prefrontal Ensembles during Awake Hippocampal Sharp-Wave Ripple Events. Neuron 90, 113–127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia F et al. Parvalbumin-positive interneurons mediate neocortical-hippocampal interactions that are necessary for memory consolidation. Elife 6, 191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maingret N, Girardeau G, Todorova R, Goutierre M & Zugaro MB Hippocampo-cortical coupling mediates memory consolidation during sleep. Nat Neurosci 19, 959–964 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Burgess CR et al. Hunger-Dependent Enhancement of Food Cue Responses in Mouse Postrhinal Cortex and Lateral Amygdala. Neuron 91, 1154–1169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sacco T & Sacchetti B Role of secondary sensory cortices in emotional memory storage and retrieval in rats. Science 329, 649–656 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Grosso A et al. The higher order auditory cortex is involved in the assignment of affective value to sensory stimuli. Nat Commun 6, 8886 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramesh RN, Burgess CR, Sugden AU, Gyetvan M & Andermann ML Intermingled Ensembles in Visual Association Cortex Encode Stimulus Identity or Predicted Outcome. Neuron 100, 900–915.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deuker L et al. Memory Consolidation by Replay of Stimulus-Specific Neural Activity. J. Neurosci. 33, 19373–19383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murty VP, Tompary A, Adcock RA & Davachi L Selectivity in Postencoding Connectivity with High-Level Visual Cortex Is Associated with Reward-Motivated Memory. J Neurosci 37, 537–545 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Logothetis NK et al. Hippocampal-cortical interaction during periods of subcortical silence. Nature 491, 547–553 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Norman Y et al. Hippocampal sharp-wave ripples linked to visual episodic recollection in humans. Science 365, eaax1030–16 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Tambini A, Ketz N & Davachi L Enhanced brain correlations during rest are related to memory for recent experiences. Neuron 65, 280–290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ko H et al. Functional specificity of local synaptic connections in neocortical networks. Nature 473, 87–91 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller J-EK, Ayzenshtat I, Carrillo-Reid L & Yuste R Visual stimuli recruit intrinsically generated cortical ensembles. Proc Natl Acad Sci USA 111, E4053–E4061 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malvache A, Reichinnek S, Villette V, Haimerl C & Cossart R Awake hippocampal reactivations project onto orthogonal neuronal assemblies. Science 353, 1280–1283 (2016). [DOI] [PubMed] [Google Scholar]
- 31.McGinley MJ, David SV & McCormick DA Cortical Membrane Potential Signature of Optimal States for Sensory Signal Detection. Neuron 87, 179–192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kenet T, Bibitchkov D, Tsodyks M, Grinvald A & Arieli A Spontaneously emerging cortical representations of visual attributes. Nature 425, 954–956 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Tononi G & Cirelli C Sleep and the Price of Plasticity: From Synaptic and Cellular Homeostasis to Memory Consolidation and Integration. Neuron 81, 12–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paz R, Bauer EP & Paré D Learning-Related Facilitation of Rhinal Interactions by Medial Prefrontal Inputs. J. Neurosci. 27, 6542–6551 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makino H & Komiyama T Learning enhances the relative impact of top-down processing in the visual cortex. Nat Neurosci 18, 1116–1122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S-H & Morris RGM Hippocampal-neocortical interactions in memory formation, consolidation, and reconsolidation. Annu. Rev. Psychol 61, 49–79– C1–4 (2010). [DOI] [PubMed] [Google Scholar]
- 37.van de Ven GM, Trouche S, McNamara CG, Allen K & Dupret D Hippocampal Offline Reactivation Consolidates Recently Formed Cell Assembly Patterns during Sharp Wave-Ripples. Neuron 92, 968–974 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gomperts SN, Kloosterman F & Wilson MA VTA neurons coordinate with the hippocampal reactivation of spatial experience. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valdés JL, McNaughton BL & Fellous J-M Offline reactivation of experience-dependent neuronal firing patterns in the rat ventral tegmental area. J Neurophysiol 114, 1183–1195 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Timofeev I & Chauvette S Sleep slow oscillation and plasticity. Curr Opin Neurobiol 44, 116–126 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Fauth MJ & van Rossum MC Self-organized reactivation maintains and reinforces memories despite synaptic turnover. Elife 8, 568 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cossell L et al. Functional organization of excitatory synaptic strength in primary visual cortex. Nature 518, 399–403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graf ABA, Kohn A, Jazayeri M & Movshon JA Decoding the activity of neuronal populations in macaque primary visual cortex. Nat Neurosci 14, 239–245 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leavitt ML, Pieper F, Sachs AJ & Martinez-Trujillo JC Correlated variability modifies working memory fidelity in primate prefrontal neuronal ensembles. Proc Natl Acad Sci USA 114, E2494–E2503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atherton LA, Dupret D & Mellor JR Memory trace replay: the shaping of memory consolidation by neuromodulation. Trends Neurosci 38, 560–570 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ambrose RE, Pfeiffer BE & Foster DJ Reverse Replay of Hippocampal Place Cells Is Uniquely Modulated by Changing Reward. Neuron 91, 1124–1136 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ólafsdóttir HF, Carpenter F & Barry C Task Demands Predict a Dynamic Switch in the Content of Awake Hippocampal Replay. Neuron 96, 925–934.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jung MW, Lee H, Jeong Y, Lee JW & Lee I Remembering rewarding futures: A simulation-selection model of the hippocampus. Hippocampus 28, 913–930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ludvig EA, Mirian MS, Kehoe EJ & Sutton RS Associative Learning from Replayed Experience. bioRxiv 100800 (2017). doi: 10.1101/100800 [DOI] [Google Scholar]
- 50.Mattar MG & Daw ND Prioritized memory access explains planning and hippocampal replay. Nat Neurosci 1–14 (2018). doi: 10.1038/s41593-018-0232-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Madisen L et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goldey GJ et al. Removable cranial windows for long-term imaging in awake mice. Nature Protocols 9, 2515–2538 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Asaad WF & Eskandar EN A flexible software tool for temporally-precise behavioral control in Matlab. Journal of Neuroscience Methods 174, 245–258 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Q & Burkhalter A Area map of mouse visual cortex. J Comp Neurol 502, 339–357 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Bonin V, Histed MH, Yurgenson S & Reid RC Local Diversity and Fine-Scale Organization of Receptive Fields in Mouse Visual Cortex. J. Neurosci 31, 18506–18521 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thevenaz P, Ruttimann UE & Unser M A pyramid approach to subpixel registration based on intensity. IEEE Trans. on Image Process 7, 27–41 (1998). [DOI] [PubMed] [Google Scholar]
- 57.Mukamel EA, Nimmerjahn A & Schnitzer MJ Automated Analysis of Cellular Signals from Large-Scale Calcium Imaging Data. Neuron 63, 747–760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ziv Y et al. Long-term dynamics of CA1 hippocampal place codes. Nat Neurosci 16, 264 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petreanu L et al. Activity in motor–sensory projections reveals distributed coding in somatosensation. Nature 489, 299 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pnevmatikakis EA et al. Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron 89, 285–299 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen T-W et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sheintuch L et al. Tracking the Same Neurons across Multiple Days in Ca2+ Imaging Data. CellReports 21, 1102–1115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Webb GI, Boughton JR & Wang Z Not So Naive Bayes: Aggregating One-Dependence Estimators. Machine Learning 58, 5–24 (2005). [Google Scholar]
- 64.Sugden LA et al. Localization of adaptive variants in human genomes using averaged one-dependence estimation. Nat Commun 9, 703 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aarts E, Verhage M, Veenvliet JV, Dolan CV & van der Sluis S A solution to dependency: using multilevel analysis to accommodate nested data. Nat Neurosci 17, 491–496 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Singmann H, Bolker B, Westfall J, Højsgaard S & Fox J Package ‘afex’. (2015). [Google Scholar]
- 67.Driscoll LN, Pettit NL, Minderer M, Chettih SN & Harvey CD Dynamic Reorganization of Neuronal Activity Patterns in Parietal Cortex. Cell 170, 986–999 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saramäki J, Kivelä M, Onnela J-P, Kaski K & Kertész J Generalizations of the clustering coefficient to weighted complex networks. Phys. Rev. E 75, 027105–4 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Komiyama T et al. Learning-related fine-scale specificity imaged in motor cortex circuits of behaving mice. Nature 464, 1182–1186 (2010). [DOI] [PubMed] [Google Scholar]
- 70.Kim M-H, Znamenskiy P, Iacaruso MF & Mrsic-Flogel TD Exclusive functional subnetworks of intracortical projection neurons in primary visual cortex. 1–11 (2017). doi: 10.1101/153247 [DOI] [PubMed] [Google Scholar]
- 71.Hagberg AA, Swart PJ & Schult DA Exploring network structure, dynamics, and function using NetworkX. Proceedings of the th Python in Science Conference SciPy 11–15 (2008). [Google Scholar]
- 72.Blondel VD, Guillaume J-L, Lambiotte R & Lefebvre E Fast unfolding of communities in large networks. J. Stat. Mech 2008, P10008–13 (2008). [Google Scholar]
- 73.Green DM & Swets JA Signal detection theory and psychophysics. (Wiley). [Google Scholar]
- 74.Roy NA, Bak JH, Akrami A, Brody CD & Pillow JW Efficient inference for time-varying behavior during learning. Adv Neural Inf Process Syst 31, 5695–5705 (2018). [PMC free article] [PubMed] [Google Scholar]
- 75.Friedman J, Hastie T & Tibshirani R Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw 33, 1–22 (2010). [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.


