Abstract
Diarrheal diseases caused by intestinal protozoan parasites are a major food-borne public health problem across the world. Vegetables and fruits provide important nutrients and minerals, but are also common sources of some food-borne human pathogenic microorganisms. The contamination of raw vegetables and fruits with human pathogenic parasites are now a global public health threat, despite the health benefits of these foods in non-pharmacological prophylaxes against diseases. A large number of reports have documented the contamination of vegetables or fruits with human pathogenic microorganisms. In this paper, we reviewed the contamination and detection methods of human pathogenic intestinal protozoans that are frequently recovered from raw vegetables and fruits. The protozoan parasites include Cryptosporidium spp., Giardia duodenalis, Cyclospora cayetanensis, Entamoeba spp., Toxoplasma gondii, Balantioides coli, Blastocystis sp., Cystoisospora belli and Enterocytozoon bieneusi. The risk factors involved in the contamination of vegetables and fruits with parasites are also assessed. 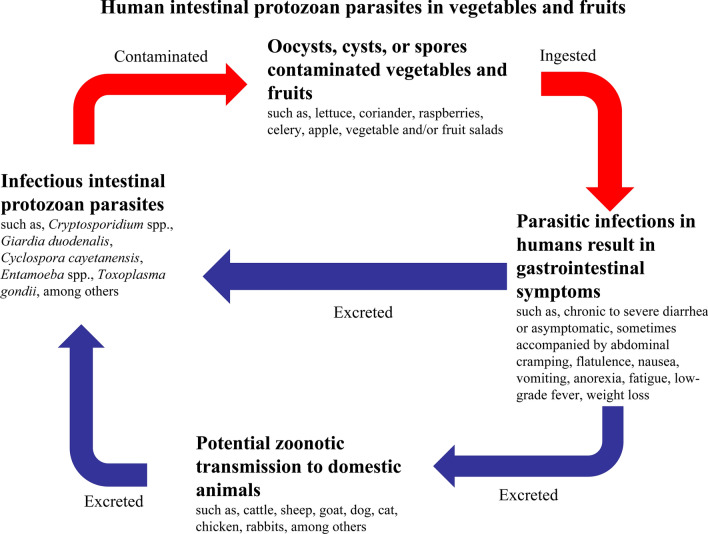
Keywords: Intestinal protozoans, Detection methods, Vegetables, Fruits, Contamination
Background
Nearly 1.7 billion cases of diarrheal disease are reported globally every year, imposing an annual socioeconomic burden on health services of 72.8 million disability-adjusted life years [1, 2]. A number of pathogens are responsible for causing diarrheal diseases, among which intestinal protozoan parasites are important contributors that can be transmitted by ingestion of the contaminated food [3, 4]. The intestinal protozoan infections are characterized by chronic to severe diarrhea, sometimes accompanied by abdominal cramping, flatulence, nausea, vomiting, anorexia, fatigue, low-grade fever and weight loss [5–7].
Vegetables and fruits provide important nutrients to humans, including various essential vitamins and minerals [8]. The ingestion of raw vegetables and fruits appear to be a quick, easy, and healthy source of nutrition. However, these fresh vegetables and fruits can be an important source of some food-borne pathogenic microorganisms, if they are contaminated [9, 10]. The contamination of raw vegetables and fruits with human parasites has recently been recognized as a global threat, despite the health benefits of these foods in non-pharmacological prophylaxes against diseases.
A number of studies documented the contamination of vegetables and fruits with human pathogenic microorganisms [11–15]. In this paper, we reviewed the detection methods and contamination of some human pathogenic intestinal protozoans that are frequently recovered from raw vegetables and fruits. The protozoan parasites include Cryptosporidium spp., Giardia duodenalis, Cyclospora cayetanensis, Entamoeba spp., Toxoplasma gondii, Balantioides coli, Blastocystis sp., Cystoisospora belli and Enterocytozoon bieneusi.
We searched PubMed and Web of Science databases, with no language restrictions, using the following search terms: ‘Cryptosporidium’ or ‘Giardia’ or ‘Cyclospora’ or ‘Entamoeba’ or ‘Toxoplasma gondii’ or ‘Balantioides coli’ or ‘Blastocystis sp.’ or ‘Cystoisospora belli’ or ‘Isospora belli’ or ‘microsporidian’ and ‘vegetable’ or ‘fruit’. Articles were screened using Endnote X9. For articles whose full text was unavailable or that were published in other languages, the titles and abstracts in English were screened. Articles published up to December 31st 2019 were included in this review.
Detection methods of intestinal protozoan parasites contaminating vegetables and fruits
The recovery of parasitic eggs/oocysts/cysts from contaminated vegetables and fruits with proper methods is the first and an important way for the detection of contaminating intestinal protozoa. The methods or techniques for the detection of Cryptosporidium in food samples were well reviewed by Ahmed and Karanis in 2018 [16].
Generally, a washing procedure is the first step in any recovery process. Several elution strategies have been used to isolate the parasites from vegetables and fruits. A portion (usually 50–250 g) of each vegetable or fruit sample is washed separately in a container containing some chemical solutions. The most widely used solutions are normal saline [14, 17–20] and phosphate-buffered saline [12, 21–24]. The commonly used solutions are glycine [11, 25], sodium dodecyl sulfate [26], Alconox® [27], and Tween 80 [28]. Other unusual solutions, such as 10% formal saline [29] and 0.1% peptone water [30] are also reported to isolate the contaminating parasites. Different elution methods can lead to variable recovery rates for parasites from contaminated vegetables or fruits, however, the Alconox® solution was reported to be more effective than the other commonly used solutions [27, 31].
The isolation of the detergent solution sediments is the second key step in parasite detection. Two methods are commonly used to obtain these concentrated sediments. One is the overnight sedimentation of the washing solution [19, 30]. The supernatant is discarded and the sediment is then transferred to a new tube to remove any unwanted material [32]. The other is membrane filtration (more commonly and effectively used), in which the deposit is collected by centrifugation. Membrane filtration devices include stomacher bags [23, 30], zipper bags [22, 24], sieves [18], gauze [21], or cellulose acetate membranes [28].
Finally, the sediment or deposit is screened with light microscopy, staining, immunofluorescence microscopy, or PCR to detect any parasite. More than one smear slide is usually prepared for each specimen to allow its precise detection [12, 26]. Oocysts or cysts can be detected microscopically based on their morphological features [14, 17, 20, 29], using Lugol’s iodine [12, 14, 29] or modified Ziehl-Neelsen staining (or any other staining technique) [14, 19, 26]. The extraction of the parasitic DNA from the sediment, followed by the PCR amplification of specific genes, is also efficiently used for the protozoan detection in vegetable and fruit samples [22, 24].
Contamination of vegetables and fruits with intestinal protozoan parasites
Cryptosporidium contamination
Cryptosporidium spp. are widespread protozoan parasites that infect humans and animals, and the second commonest cause of diarrhea in children after rotavirus [9]. Cryptosporidium is characterized by its extensive genetic variation that results in the existence of 38 species and more than 60 genotypes of this parasite [33]. At least 20 distinct species cause moderate or severe infections in humans, of which C. hominis and C. parvum are the major causative agents [34].
The detection of Cryptosporidium oocysts in vegetable and fruit samples with light microscopy is simple, convenient, and direct [13, 16], but it requires a high level of expertise to interpret the slides, while an immunofluorescence assay is standard practice and more sensitive [16]. Immunomagnetic separation (IMS) is used to concentrate Cryptosporidium oocysts for the efficient detection by microscopy or PCR [12, 25, 35]. The PCR amplification and sequencing of specific genes of Cryptosporidium recovered from contaminated vegetables and fruits is the most precise method of identification of human pathogenic and zoonotic species (e.g., [13, 23–25]. However, PCR is commonly used in developed countries, but most surveillance studies in developing countries involve microscopy.
The contamination of vegetables and fruits with Cryptosporidium spp. has been documented in many countries (Table 1), and the average prevalence is calculated as 6.0% (375/6210; 95% confidence interval, CI: 5.4–6.6%). Among the Cryptosporidium species, C. parvum, C. hominis, and C. ubiquitum were detected in the contaminated vegetable and fruit samples [12, 23, 25, 36]. The Cryptosporidium species are important human pathogens and major causes of human cryptosporidiosis, representing a threat to public health through food as a vehicle.
Table 1.
Contamination of vegetables and fruits by Cryptosporidium spp.
| Location | Detection method | Vegetable or fruit item | No. of samples tested | No. of positive samples (%) | Cryptosporidium species (n) | References |
|---|---|---|---|---|---|---|
| Brazil | PCR | Vegetables | 21 | 2 (9.5) | Cryptosporidium spp. (1); C. parvum (1) | [45] |
| China | PCR | Lettuce | 200 | 0 | [36] | |
| Coriander | 152 | 0 | ||||
| Celery | 70 | 0 | ||||
| Baby bok choy | 59 | 0 | ||||
| Chinese cabbage | 47 | 0 | ||||
| Leaf lettuce | 44 | 0 | ||||
| Water spinach | 28 | 0 | ||||
| Crown daisy | 27 | 0 | ||||
| Fennel plant | 26 | 0 | ||||
| Endive | 25 | 0 | ||||
| Spinach | 20 | 0 | ||||
| Schizonepeta | 20 | 0 | ||||
| Cabbage | 18 | 0 | ||||
| Leaf mustard | 11 | 0 | ||||
| Chinese chive | 132 | 1 (0.8) | C. parvum (1) | |||
| Chive | 128 | 0 | ||||
| Cucumber | 41 | 0 | ||||
| Watermelon | 15 | 0 | ||||
| Potato | 3 | 0 | ||||
| Bean (kidney/French bean) | 28 | 0 | ||||
| Green chili | 5 | 0 | ||||
| Costa Rica | Direct smear, followed by light microscopy | Cilantro leaves | 80 | 4 (5.0) | Cryptosporidium spp. (4) | [79] |
| Cilantro roots | 80 | 7 (8.7) | Cryptosporidium spp. (7) | |||
| Lettuce | 80 | 2 (2.5) | Cryptosporidium spp. (2) | |||
| Radish, tomato, cucumbers, carrots | 80 | 1 (1.2) | Cryptosporidium spp. (1) | |||
| Costa Rica | Zielh-Nielsen stain, Weber stain | Lettuce | 50 | 7 (14.0) | Cryptosporidium spp. (7) | [71] |
| Parsley | 50 | 1 (2.0) | Cryptosporidium spp. (1) | |||
| Cilantro | 50 | 1 (2.0) | Cryptosporidium spp. (1) | |||
| Strawberries | 50 | 0 | ||||
| Blackberries | 50 | 3 (6.0) | Cryptosporidium spp. (3) | |||
| Egypt | Wet mount, Weber modified trichrome, modified Ziehl-Neelsen stains | Fresh fruit juices | 61.3 | Cryptosporidium spp. | [80] | |
| Ethiopia | Modifed Zeihl-Neelsen stain | Fruits and vegetables | 360 | 46 (12.8) | Cryptosporidium spp. (46) | [19] |
| Ethiopia | Modified Ziehl-Neelsen stain | Fruits and vegetables | 360 | 17 (4.7) | Cryptosporidium spp. (17) | [32] |
| Ethiopia | Modified Zeihl-Neelsen stain | Tomato | 100 | 9 (9.0) | Cryptosporidium spp. (9) | [14] |
| Cabbage | 96 | 0 | ||||
| Green pepper | 66 | 2 (3.0) | Cryptosporidium spp. (2) | |||
| Carrot | 62 | 7 (11.3) | Cryptosporidium spp. (7) | |||
| Salad | 23 | 2 (8.7) | Cryptosporidium spp. (2) | |||
| Ghana | Ziehl-Neelsen stain | Cabbage | 90 | 18 (20.0) | Cryptosporidium parvum (18) | [12] |
| Green pepper | 55 | 12 (21.8) | Cryptosporidium parvum (12) | |||
| Carrot | 47 | 6 (12.8) | Cryptosporidium parvum (6) | |||
| Onion | 70 | 9 (12.9) | Cryptosporidium parvum (9) | |||
| Tomato | 31 | 4 (12.9) | Cryptosporidium parvum (4) | |||
| Lettuce | 102 | 18 (17.6) | Cryptosporidium parvum (18) | |||
| Ghana | Sediment smears and fluorescence stain | Cabbage | 72 | 12 (16.7) | Cryptosporidium spp. (12) | [67] |
| Lettuce | 72 | 15 (20.8) | Cryptosporidium spp. (15) | |||
| Carrot | 72 | 4 (5.6) | Cryptosporidium spp. (4) | |||
| Spring onion | 72 | 8 (11.1) | Cryptosporidium spp. (8) | |||
| Tomatoes | 72 | 1 (1.4) | Cryptosporidium spp. (1) | |||
| Ghana | Direct wet mount, Trichrome, modified Zielh-Nielsen stain | Tiger nuts | 40 | 12 (30.0) | Cryptosporidium parvum (12) | [81] |
| India | DAPI-stain followed by fluorescence microscopy, and PCR | Cabbage | 47 | 3 (6.4) | Cryptosporidium parvum (3) | [13] |
| Chili | 42 | 2 (4.8) | Cryptosporidium spp. (2) | |||
| Coriander | 28 | 2 (7.1) | Cryptosporidium spp. (2) | |||
| Cucumber | 52 | 3 (5.8) | Cryptosporidium parvum (3) | |||
| Radish | 14 | 1 (7.1) | Cryptosporidium spp. (1) | |||
| Tomatoes | 56 | 6 (10.7) | Cryptosporidium spp. (6) | |||
| Iran | Modified Ziehl-Neelsen acid-fast stain | Mint | 82 | 7 (8.5) | Cryptosporidium spp. (7) | [26] |
| Leek | 90 | 3 (3.3) | Cryptosporidium spp. (3) | |||
| Cress | 90 | 8 (8.9) | Cryptosporidium spp. (8) | |||
| Green onion | 54 | 8 (14.8) | Cryptosporidium spp. (8) | |||
| Coriander | 90 | 6 (6.7) | Cryptosporidium spp. (6) | |||
| Basil | 90 | 1 (1.1) | Cryptosporidium spp. (1) | |||
| Iran | Modified Ziehl-Neelsen satin | Vegetables | 34 | 3 (8.8) | Cryptosporidium spp. (3) | [72] |
| Italy | modified Ziehl-Neelsen stain and PCR | Ready-to-eat packaged salads | 648 | 6 (0.9) | Cryptosporidium parvum/C. ubiquitum (6) | [23] |
| Korea | qPCR | Carrots | 3 | 1 (33.3) | Cryptosporidium parvum (1) | [22] |
| Cabbages | 3 | 1 (33.3) | Cryptosporidium parvum (1) | |||
| Blue berries | 3 | 1 (33.3) | Cryptosporidium parvum (1) | |||
| Korea | Multiplex qPCR | Perilla leaves | 72 | 5 (6.9) | Cryptosporidium spp. (5) | [24] |
| Winter-grown cabbage | 70 | 4 (5.7) | Cryptosporidium spp. (4) | |||
| Chives | 73 | 13 (17.8) | Cryptosporidium spp. (13) | |||
| Sprouts | 72 | 1 (1.4) | Cryptosporidium spp. (1) | |||
| Blueberries | 44 | 3 (6.8) | Cryptosporidium spp. (3) | |||
| Cherry tomatoes | 73 | 5 (6.8) | Cryptosporidium spp. (5) | |||
| Norway | Concentrated by IMS, and screening by light microscopy | Alfalfa sprouts | 16 | 0 | [35] | |
| Dill | 7 | 0 | ||||
| Lettuce | 125 | 5 (4.0) | Cryptosporidium spp. (5) | |||
| Mung bean sprouts | 149 | 14 (9.4) | Cryptosporidium spp. (14) | |||
| Mushrooms | 55 | 0 | ||||
| Parsley | 7 | 0 | ||||
| Precut salad mix | 38 | 0 | ||||
| Radish sprouts | 6 | 0 | ||||
| Raspberries | 10 | 0 | ||||
| Strawberries | 62 | 0 | ||||
| Norway | Concentrated by IMS, and screening by light microscopy | Alfalfa | 16 | 0 | [82] | |
| Mung bean | 149 | 14 (9.4) | Cryptosporidium spp. (14) | |||
| Radish | 6 | 0 | ||||
| Peru | Direct microscopic observation, acid-fast staining, and immunofluorescent assays | Vegetables | 14.5 | Cryptosporidium parvum | [83] | |
| Poland | Separated by IMS and identified by immunofluorescence and DIC microscopy, and PCR identified | Fresh vegetables | 128 | 6 (4.7) | Cryptosporidium parvum or C. hominis (6) | [25] |
| Fruits | 35 | 0 | ||||
| Spain | Concentrated by IMS and stain oocysts for immunofluorescence assay | Chinese cabbage | 6 | 2 (33.3) | Cryptosporidium spp. (2) | [11] |
| Lollo rosso lettuce | 4 | 3 (75.0) | Cryptosporidium spp. (3) | |||
| Romaine lettuce | 9 | 7 (77.8) | Cryptosporidium spp. (7) | |||
| Total | 6210 | 375 (6.0) |
Giardia duodenalis contamination
Giardia duodenalis (synonyms: G. intestinalis, G. lamblia) is a non-invasive protozoan parasite that adhere to and colonize the upper small intestine, causing acute watery diarrhea in humans and animals [37]. It is an important zoonotic protozoan and the main cause of human giardiasis, which therefore represents a threat to public health [38]. Eight genetically distinct assemblages (A to H) of G. duodenalis have been defined, with the occurrence of zoonotic assemblages A and B in both humans and animals. However, the other assemblages are mostly specific to animal hosts [38]. This parasite is estimated to cause ~28.2 million cases of diarrhea annually through the ingestion of contaminated foods [7]. The outbreaks of giardiasis have also been associated with a variety of processed foods. Human infections of G. duodenalis are often associated with the consumption of contaminated raw vegetables and fruits [39–41].
Giardia duodenalis cysts can be detected with light microscopy based on their morphological features [19, 42, 43], and staining with typical Lugol’s iodine is universally used for the detection of G. duodenalis cysts [12, 14, 17, 18, 29]. However, an immunofluorescence assay is usually applied for the detection of Giardia cysts in food items with more sensitivity [7]. The IMS method is also applied to concentrate G. duodenalis cysts for further detection [11, 35]. The PCR amplification and sequencing of specific G. duodenalis genes recovered from contaminated food are also commonly used for the confirmatory detection of this parasite (e.g. [28, 39, 44]).
The contamination of vegetables and fruits with G. duodenalis cysts has been reported in many countries (Table 2), and the average prevalence is estimated as 4.8% (276/5739; 95% CI: 4.2–5.4%). In contaminated vegetable and fruit samples, G. duodenalis zoonotic assemblages A and B were commonly detected [23, 28, 39, 44, 45].
Table 2.
Contamination of vegetables and fruits with Giardia duodenalis
| Location | Detection method | Vegetable or fruit item | No. of samples tested | No. of positive samples (%) | Giardia duodenalis assemblages identified (n) | References |
|---|---|---|---|---|---|---|
| Bangladesh | Iodine and normal saline wet mount | Vegetables | 200 | 2 (1.0) | [52] | |
| Brazil | PCR | Lettuce and chicory | 11 | 2 (18.2) | Assemblage BIV (2) | [39] |
| Brazil | Immunofluorescence, PCR | Arugula | 4 | 2 (50.0) | Assemblage AII (2) | [28] |
| Chives | 12 | 1 (8.3) | Assemblage AII (1) | |||
| Crisp lettuce | 32 | 4 (12.5) | Assemblage AII (4) | |||
| Greens collard | 24 | 1 (4.2) | Assemblage AI (1) | |||
| Parsley | 12 | 2 (16.7) | Assemblage AII (2) | |||
| Watercress | 12 | 4 (33.3) | Assemblage AII (4) | |||
| Wild chicory | 12 | 2 (16.7) | Assemblage AII (2) | |||
| Brazil | Semi-nested PCR | Regular lettuce | 60 | 8 (13.3) | Assemblage AI (4); Assemblage B (1); Assemblage E (1); N/D (2) | [44] |
| Crisp lettuce | 100 | 5 (5.0) | Assemblage AI (2); N/D (3) | |||
| Chicory | 60 | 5 (8.3) | Assemblage AI (3); N/D (2) | |||
| Rocket | 20 | 1 (5.0) | N/D (1) | |||
| Kale | 20 | 0 | ||||
| Brazil | PCR | Vegetables | 21 | 10 (47.6) | Assemblage E (2); N/D (8) | [45] |
| Brazil | Sediment being stained in Lugolʼs solution | Lettuce | 100 | 0 | [15] | |
| Coriander | 100 | 1 (1.0) | ||||
| Costa Rica | Direct smear, followed by light microscopy | Cilantro leaves | 80 | 4 (5.0) | [79] | |
| Cilantro roots | 80 | 2 (2.5) | ||||
| Egypt | Lugol’s iodine stain | Lettuce | 101 | 16 (15.8) | [18] | |
| Watercress | 116 | 13 (11.2) | ||||
| Parsley | 102 | 12 (11.8) | ||||
| Green onion | 103 | 4 (3.9) | ||||
| Leek | 108 | 2 (1.9) | ||||
| Ethiopia | Lugol’s iodine stain | Fruits and vegetables | 360 | 27 (7.5) | [19] | |
| Ethiopia | Sediment smear under light microscope | Fruits and vegetables | 360 | 36 (10.0) | [32] | |
| Ethiopia | Sediment smear under light microscope | Tomatoes | 45 | 1 (2.2) | [43] | |
| Lettuce | 45 | 4 (8.8) | ||||
| Carrot | 45 | 7 (15.6) | ||||
| Cabbage | 45 | 8 (17.8) | ||||
| Green pepper | 45 | 6 (13.3) | ||||
| Avocado | 45 | 0 | ||||
| Ethiopia | Sediment smear and Lugol’s iodine stain | Tomato | 100 | 0 | [14] | |
| Cabbage | 96 | 16 (16.7) | ||||
| Green pepper | 66 | 4 (6.1) | ||||
| Carrot | 62 | 4 (6.5) | ||||
| Salad | 23 | 0 | ||||
| Ghana | Lugol’s iodine stain | Cabbage | 90 | 5 (5.6) | [12] | |
| Green pepper | 55 | 3 (5.5) | ||||
| Carrot | 47 | 4 (8.5) | ||||
| Onion | 70 | 3 (4.3) | ||||
| Tomato | 31 | 2 (6.5) | ||||
| Lettuce | 102 | 5 (4.9) | ||||
| India | DAPI-stain followed by fluorescence microscopy, and PCR | Cabbage | 47 | 1 (2.1) | [13] | |
| Carrot | 25 | 1 (4.0) | ||||
| Chili | 42 | 4 (9.5) | ||||
| Coriander | 28 | 3 (10.7) | ||||
| Cucumber | 52 | 1 (1.9) | Assemblage D (1) | |||
| Tomatoes | 56 | 2 (3.6) | Assemblage A (2) | |||
| Turnip | 3 | 1 (33.3) | ||||
| Iran | Lugol’s iodine stain | Vegetables | 141 | 11 (7.8) | [84] | |
| Iran | Sediment smear under light microscopy | Leek | 30 | 3 (10.0) | [42] | |
| Spring onion | 22 | 0 | ||||
| Basil | 15 | 1 (6.7) | ||||
| Parsley | 21 | 0 | ||||
| Lettuce | 23 | 0 | ||||
| Cress | 17 | 0 | ||||
| Spearmint | 18 | 0 | ||||
| Tarragon | 19 | 0 | ||||
| Coriander | 24 | 0 | ||||
| Radish | 29 | 0 | ||||
| Italy | Lugolʼs iodine satin and PCR | Ready-to-eat packaged salad | 648 | 4 (0.6) | Assemblage A (4) | [23] |
| Jordan | Lugol’s iodine stain | Lettuce | 30 | 7 (23.3) | [20] | |
| Tomato | 33 | 2 (6.1) | ||||
| Parsley | 42 | 0 | ||||
| Cucumber | 28 | 0 | ||||
| Norway | Concentrated by IMS, and screening by light microscopy | Alfalfa sprouts | 16 | 0 | [35] | |
| Dill | 7 | 2 (28.6) | ||||
| Lettuce | 125 | 2 (1.6) | ||||
| Mung bean sprouts | 149 | 3 (2.0) | ||||
| Mushrooms | 55 | 0 | ||||
| Parsley | 7 | 0 | ||||
| Precut salad mix | 38 | 0 | ||||
| Radish sprouts | 6 | 1 (16.7) | ||||
| Raspberries | 10 | 0 | ||||
| Strawberries | 62 | 2 (3.2) | ||||
| Norway | Concentrated by IMS, and screening by light microscopy | Alfalfa | 16 | 0 | [82] | |
| Mung bean | 149 | 3 (2.0) | ||||
| Radish | 6 | 1 (16.7) | ||||
| Saudi Arabia | Lugol’s iodine stain | Green onion | 50 | 0 | [17] | |
| Watercress | 50 | 0 | ||||
| Lettuce | 50 | 0 | ||||
| Cucumber | 50 | 0 | ||||
| Cabbage | 50 | 0 | ||||
| Pea | 50 | 0 | ||||
| Tomato | 50 | 0 | ||||
| Carrot | 50 | 4 (8.0) | ||||
| Spain | Concentrated by IMS and stain cysts for immunofluorescence assay | Chinese cabbage | 6 | 2 (33.3) | [11] | |
| Lollo rosso lettuce | 4 | 3 (75.0) | ||||
| Romaine lettuce | 9 | 5 (55.6) | ||||
| Sudan | Lugol’s iodine stain | Tomatoes | 36 | 1 (2.8) | [29] | |
| Cucumber | 12 | 0 | ||||
| Armenian cucumber | 16 | 0 | ||||
| Green pepper | 25 | 1 (4.0) | ||||
| Cayenne pepper | 7 | 0 | ||||
| Radish | 24 | 1 (4.2) | ||||
| Beet | 19 | 0 | ||||
| Watercress | 23 | 2 (8.7) | ||||
| Lettuce | 11 | 1 (9.1) | ||||
| Green onion | 36 | 1 (2.8) | ||||
| Carrot | 50 | 1 (2.0) | ||||
| Total | 5739 | 276 (4.8) |
Giardia duodenalis, G. intestinalis, G. lamblia
Cyclospora cayetanensis contamination
Cyclospora cayetanensis is another important protist parasite, usually transmitted via food that causes human gastrointestinal cyclosporiasis [5, 46]. Globally, C. cayentanesis is an important food-borne human protozoan [5, 46]. Many reports have documented the food-borne cyclosporiasis outbreaks that were associated with the consumption of contaminated raw vegetables or fruits.
Cyclospora cayetanensis oocysts can be detected simply and directly with light microscopy provided that there are a large number of oocysts present in the vegetables and fruits [23, 37]. Modified Ziehl-Neelsen staining, and autofluorescence or immunofluorescence assays are also commonly used for their detection [12, 14, 19, 47]; however, there are no immunofluorescence assays commercially available for Cyclospora. Furthermore, PCR amplification and sequencing of C. cayetanensis genes have currently been used for the specific detection of this organism in contaminated food samples [23, 24, 48].
The contamination of vegetables and fruits with C. cayetanensis oocysts have been documented in many countries (Table 3). The average prevalence of C. cayetanensis contamination is counted as 3.9% (180/4628; 95% CI: 3.3–4.5%).
Table 3.
Contamination of vegetables and fruits with Cyclospora cayetanensis
| Location | Detection method | Vegetable or fruit item | No. of samples tested | No. of positive samples (%) | References |
|---|---|---|---|---|---|
| Cameroon | Sediment smear, followed by light microscopy | Green cabbage | 30 | 0 | [66] |
| Red cabbage | 30 | 0 | |||
| Lettuce | 30 | 10 (33.3) | |||
| Cucumber | 30 | 0 | |||
| Carrots | 30 | 0 | |||
| Green pepper | 30 | 20 (66.7) | |||
| China | PCR | Lettuce | 200 | 1 | [36] |
| Coriander | 152 | 0 | |||
| Celery | 70 | 0 | |||
| Baby bok choy | 59 | 0 | |||
| Chinese cabbage | 47 | 0 | |||
| Leaf lettuce | 44 | 1 (2.3) | |||
| Water spinach | 28 | 0 | |||
| Crown daisy | 27 | 0 | |||
| Fennel plant | 26 | 0 | |||
| Endive | 25 | 0 | |||
| Spinach | 20 | 0 | |||
| Schizonepeta | 20 | 0 | |||
| Cabbage | 18 | 0 | |||
| Leaf mustard | 11 | 0 | |||
| Chinese chive | 132 | 0 | |||
| Chive | 128 | 0 | |||
| Cucumber | 41 | 0 | |||
| Watermelon | 15 | 0 | |||
| Potato | 3 | 0 | |||
| Bean (kidney/French bean) | 28 | 0 | |||
| Green chili | 5 | 0 | |||
| Costa Rica | Zielh-Nielsen and Weber stain | Lettuce | 50 | 2 (4.0) | [71] |
| Parsley | 50 | 0 | |||
| Cilantro | 50 | 0 | |||
| Strawberries | 50 | 0 | |||
| Blackberries | 50 | 0 | |||
| Egypt | Weber modified trichrome and modified Ziehl-Neelsen stains | Fresh fruit juices | 14.5 | [80] | |
| Ethiopia | Modifed Zeihl-Neelsen stain | Fruits and vegetables | 360 | 18 (5.0) | [19] |
| Ethiopia | Modified Ziehl-Neelsen stain | Fruits and vegetables | 360 | 25 (6.9) | [32] |
| Ethiopia | Modified Zeihl-Neelsen stain | Tomato | 100 | 4 (4.0) | [14] |
| Cabbage | 96 | 0 | |||
| Green pepper | 66 | 2 (3.0) | |||
| Carrot | 62 | 0 | |||
| Salad | 23 | 1 (4.5) | |||
| Ghana | Direct wet mount, trichrome modified Ziehl-Neelsen stain | Tiger nuts | 40 | 9 (22.5) | [81] |
| Ghana | Ziehl-Neelsen stain | Cabbage | 90 | 5 (5.6) | [12] |
| Green pepper | 55 | 3 (5.5) | |||
| Carro | 47 | 3 (6.4) | |||
| Onion | 70 | 3 (4.3) | |||
| Tomato | 31 | 3 (9.7) | |||
| Lettuce | 102 | 3 (2.9) | |||
| Italy | qPCR | Vegetables | 49 | 6 (12.2) | [48] |
| Italy | modified Ziehl-Neelsen stain and PCR | Ready-to-eat packaged salad | 648 | 8 (1.2) | [23] |
| Korea | Multiplex qPCR | Perilla leaves | 72 | 0 | [48] |
| Winter-grown cabbage | 70 | 4 (5.7) | |||
| Chives | 73 | 0 | |||
| Sprouts | 72 | 1 (1.4) | |||
| Blueberries | 44 | 1 (2.3) | |||
| Cherry tomatoes | 73 | 1 (1.4) | |||
| Peru | Direct microscopic observation, acid-fast staining, and immunofluorescent assay | Vegetables | 1.8 | [83] | |
| Vietnam | Modified acid-fast smear by light and UV epifluorescence microscopy | Basil | 96 | 10 (10.4) | [47] |
| Coriander sativum | 80 | 3 (3.8) | |||
| Coriander | 86 | 10 (11.6) | |||
| Lettuce | 79 | 8 (10.1) | |||
| Vietnamese mint | 61 | 6 (9.8) | |||
| Marjoram | 26 | 2 (7.7) | |||
| Persicaria | 68 | 7 (10.3) | |||
| Total | 4628 | 180 (3.9) |
Entamoeba contamination
Among the Entamoeba spp., E. histolytica is responsible for most cases of human amebiasis and remains one of the top three causes of parasitic mortality worldwide [49]. Although some of the E. histolytica infections are asymptomatic, many infections may lead to severe amoebic colitis and disseminated disease [50]. Entamoeba spp. infections are significantly associated with the consumption of contaminated vegetables and fruits [17, 41, 51, 52].
Entamoeba spp. cysts can be detected with light microscopy based on their morphological features [29, 42, 43]. Staining with Lugol’s iodine is widely used to detect the Entamoeba spp. cysts (e.g. [12, 14, 17, 19, 52]). The PCR technique is also commonly used to detect Entamoeba spp. in food items based on amplification and sequencing of specific genes [23, 53].
Many reports have documented the contamination of raw vegetables and fruits with Entamoeba spp. cysts worldwide (Table 4). The average prevalence of Entamoeba contamination is calculated as 3.5% (199/5647; 95% CI: 3.0–4.0%). Entamoeba histolytica, E. dispar and E. coli were the most commonly detected species among the isolates from contaminated vegetables and fruits [12, 17, 29, 42].
Table 4.
Contamination of vegetables and fruits with Entamoeba spp.
| Location | Detection method | Vegetable or fruit item | Number of samples tested | Number of positive samples (%) | Entamoeba species identified (n) | References |
|---|---|---|---|---|---|---|
| Bangladesh | Wet mount | Vegetables | 200 | 17 (8.5) | Entamoeba histolytica | [52] |
| Brazil | Direct smear, followed by light microscopy | Lettuce | 30 | 3 (10.0) | Entamoeba coli (3) | [85] |
| Brazil | Lugol’s iodine stain | Loose leaf lettucea | 1 | 1 | Entamoeba sp. | [30] |
| Red lettucea | 1 | 1 | Entamoeba sp. | |||
| Curly lettucea | 1 | 1 | Entamoeba sp. | |||
| Iceberg lettucea | 1 | 1 | Entamoeba sp. | |||
| Parsleya | 1 | 1 | Entamoeba sp. | |||
| Chivea | 1 | 1 | Entamoeba sp. | |||
| Coriandera | 1 | 1 | Entamoeba sp. | |||
| Basila | 1 | 1 | Entamoeba sp. | |||
| Arugulaa | 1 | 1 | Entamoeba sp. | |||
| Chicorya | 1 | 1 | Entamoeba sp. | |||
| Kalea | 1 | 1 | Entamoeba sp. | |||
| Bean sproutsa | 1 | 1 | Entamoeba sp. | |||
| Brazil | Sediment smear, followed by light microscopy | Vegetables | 100 | 32 (32.0) | Entamoeba spp. (32) | [86] |
| Brazil | Sediment being stained in Lugolʼs solution | Lettuce | 100 | 9 (9.0) | Entamoeba histolytica (9) | [15] |
| Lettuce | 100 | 4 (4.0) | Entamoeba coli (4) | |||
| Coriander | 100 | 11 (11.0) | Entamoeba histolytica (11) | |||
| Coriander | 100 | 4 (4.0) | Entamoeba coli (4) | |||
| Cameroon | Lugol’s iodine stain | Green cabbage | 30 | 5 (16.7) | Entamoeba spp. (5) | [66] |
| Red cabbage | 30 | 3 (10.0) | Entamoeba spp. (3) | |||
| Lettuce | 30 | 9 (30.0) | Entamoeba spp. (9) | |||
| Cucumber | 30 | 5 (16.7) | Entamoeba spp. (5) | |||
| Carrots | 30 | 3 (10.0) | Entamoeba spp. (3) | |||
| Green pepper | 30 | 5 (16.7) | Entamoeba spp. (5) | |||
| Costa Rica | Direct smear, followed by light microscopy | Cilantro leaves | 80 | 5 (6.2) | Entamoeba histolytica (5) | [79] |
| Cilantro roots | 80 | 2 (2.5) | Entamoeba histolytica (2) | |||
| Lettuce | 80 | 3 (3.8) | Entamoeba histolytica (3) | |||
| Radish | 80 | 2 (2.5) | Entamoeba histolytica (2) | |||
| Egypt | Lugol’s iodine stain | Lettuce | 101 | 14 (13.9) | Entamoeba spp. (14) | [18] |
| Watercress | 116 | 9 (7.8) | Entamoeba spp. (9) | |||
| Parsley | 102 | 8 (7.8) | Entamoeba spp. (8) | |||
| Green onion | 103 | 2 (1.9) | Entamoeba spp. (2) | |||
| Leek | 108 | 3 (2.8) | Entamoeba spp. (3) | |||
| Ethiopia | Lugol’s iodine stain | Fruits and vegetables | 360 | 19 (5.3) | Entamoeba histolytica/E.dispar (19) | [19] |
| Ethiopia | Sediment smear | Fruits and vegetables | 360 | 52 (14.4) | E. histolytica/dispar (52) | [32] |
| Ethiopia | Lugol’s iodine stain | Tomato | 100 | 22 (22.0) | E. histolytica (22) | [14] |
| Cabbage | 96 | 0 | ||||
| Green pepper | 66 | 0 | ||||
| Carrot | 62 | 7 (11.3) | E. histolytica (7) | |||
| Salad | 23 | 0 | ||||
| Ethiopia | Sediment smear under light microscope | Tomatoes | 45 | 1 (2.2) | E. histolytica/E. dispar (1) | [43] |
| Lettuce | 45 | 4 (8.8) | E. histolytica/E. dispar (4) | |||
| Carrot | 45 | 6 (13.3) | E. histolytica/E. dispar (6) | |||
| Cabbage | 45 | 7 (15.6) | E. histolytica/E. dispar (7) | |||
| Green pepper | 45 | 5 (11.1) | E. histolytica/E. dispar (5) | |||
| Avocado | 45 | 5 (11.1) | E. histolytica/E. dispar (5) | |||
| Ghana | Lugol’s iodine stain | Cabbage | 90 | 5 (5.6) | Entamoeba coli (5) | [12] |
| Green pepper | 55 | 4 (7.3) | Entamoeba coli (4) | |||
| Onion | 70 | 2 (2.9) | Entamoeba coli (2) | |||
| Tomato | 31 | 2 (6.5) | Entamoeba coli (2) | |||
| Lettuce | 102 | 4 (3.9) | Entamoeba coli (4) | |||
| Ghana | Lugol’s iodine stain | Cabbage | 90 | 11 (12.2) | Entamoeba histolytica (11) | |
| Carrot | 47 | 4 (8.5) | Entamoeba histolytica (4) | |||
| Onion | 70 | 2 (2.9) | Entamoeba histolytica (2) | |||
| Tomato | 31 | 4 (12.9) | Entamoeba histolytica (4) | |||
| Lettuce | 102 | 6 (5.9) | Entamoeba histolytica (6) | |||
| Iran | Lugol’s iodine stain | Vegetables | 141 | 18 (12.8) | Entamoeba coli (18) | [84] |
| Iran | Sediment smear under light microscopy | Leek | 30 | 0 | [42] | |
| Spring onion | 22 | 2 (9.1) | Entamoeba coli (2) | |||
| Basil | 15 | 0 | ||||
| Parsley | 21 | 0 | ||||
| Lettuce | 23 | 0 | ||||
| Cress | 17 | 1 (5.9) | Entamoeba coli (1) | |||
| Spearmint | 18 | 0 | ||||
| Tarragon | 19 | 1 (5.3) | Entamoeba coli (1) | |||
| Coriander | 24 | 2 (8.3) | Entamoeba coli (2) | |||
| Radish | 29 | 0 | ||||
| Iran | Sediment smear under light microscopy | Leek | 30 | 2 (6.7) | Entamoeba histolytica (2) | [42] |
| Spring onion | 22 | 0 | ||||
| Basil | 15 | 0 | ||||
| Parsley | 21 | 0 | ||||
| Lettuce | 23 | 0 | ||||
| Cress | 17 | 0 | ||||
| Spearmint | 18 | 1 (5.6) | Entamoeba histolytica (1) | |||
| Tarragon | 19 | 0 | ||||
| Coriander | 24 | 0 | ||||
| Radish | 29 | 0 | ||||
| Iran | Lugol’s iodine stain | Vegetables | 34 | 1 (2.9) | Entamoeba coli (1) | [72] |
| Jordan | Lugol’s iodine stain | Lettuce | 30 | 3 (10.0) | Entamoeba histolytica (3) | [20] |
| Tomato | 33 | 2 (6.1) | Entamoeba histolytica (2) | |||
| Parsley | 42 | 0 | ||||
| Cucumber | 28 | 0 | ||||
| Saudi Arabia | Lugol’s iodine stain | Green onion | 50 | 6 (12.0) | Entamoeba spp. (6) | [17] |
| Watercress | 50 | 8 (16.0) | Entamoeba spp. (8) | |||
| Lettuce | 50 | 6 (12.0) | Entamoeba spp. (6) | |||
| Cucumber | 50 | 7 (14.0) | Entamoeba spp. (7) | |||
| Cabbage | 50 | 6 (12.0) | Entamoeba spp. (6) | |||
| Pea | 50 | 5 (10.0) | Entamoeba spp. (5) | |||
| Tomato | 50 | 0 | ||||
| Carrot | 50 | 6 (12.0) | Entamoeba spp. (6) | |||
| Saudi Arabia | Lugol’s iodine stain | Green onion | 50 | 3 (6.0) | Entamoeba coli (3) | |
| Watercress | 50 | 4 (8.0) | Entamoeba coli (4) | |||
| Lettuce | 50 | 2 (4.0) | Entamoeba coli (2) | |||
| Cucumber | 50 | 2 (4.0) | Entamoeba coli (2) | |||
| Cabbage | 50 | 4 (8.0) | Entamoeba coli (4) | |||
| Pea | 50 | 3 (6.0) | Entamoeba coli (3) | |||
| Tomato | 50 | 2 (4.0) | Entamoeba coli (2) | |||
| Carrot | 50 | 3 (6.0) | Entamoeba coli (3) | |||
| Sudan | Lugol’s iodine stain | Tomatoes | 36 | 1 (2.8) | Entamoeba coli (1) | [29] |
| Cucumber | 12 | 0 | ||||
| Armenian cucumber | 16 | 0 | ||||
| Green pepper | 25 | 0 | ||||
| Cayenne pepper | 7 | 0 | ||||
| Radish | 24 | 1 (4.2) | Entamoeba coli (1) | |||
| Beet | 19 | 1 (5.3) | Entamoeba coli (1) | |||
| Watercress | 23 | 1 (4.3) | Entamoeba coli (1) | |||
| Lettuce | 11 | 1 (9.1) | Entamoeba coli (1) | |||
| Green onion | 36 | 0 | ||||
| Carrot | 50 | 0 | ||||
| Sudan | Lugol’s iodine stain | Tomatoes | 36 | 1 (2.8) | Entamoeba spp. (1) | [29] |
| Cucumber | 12 | 0 | ||||
| Armenian cucumber | 16 | 2 (12.5) | Entamoeba spp. (2) | |||
| Green pepper | 25 | 1 (4.0) | Entamoeba spp. (1) | |||
| Cayenne pepper | 7 | 0 | ||||
| Radish | 24 | 0 | ||||
| Beet | 19 | 1 (5.3) | Entamoeba spp. (1) | |||
| Watercress | 23 | 1 (4.3) | Entamoeba spp. (1) | |||
| Lettuce | 11 | 2 (18.2) | Entamoeba spp. (2) | |||
| Green onion | 36 | 4 (11.1) | Entamoeba spp. (4) | |||
| Carrot | 50 | 3 (6.0) | Entamoeba spp. (3) | |||
| Total | 5647 | 199 (3.5) |
aSingle sample in a case report
Toxoplasma gondii contamination
Toxoplasma gondii is a ubiquitous protozoan parasite capable of infecting virtually all warm-blooded animals [54]. According to a new nomenclature system, T. gondii genotypes are classified as Type I, Type II or Type III. Other atypical or exotic genotypes include Chinese 1, Type Br I, Type Br II, Type Br III, Type IV and Type 12 [55, 56]. Among the three principal routes of toxoplasmosis transmission, consumption of unwashed vegetables and fruits contaminated with cat feces is an important one that sometimes may lead to food-borne outbreaks [57]. The significant association of T. gondii infections with the consumption of contaminated raw vegetables is also observed in previous studies [58–60].
The detection of Toxoplasma gondii in contaminated vegetables and fruits is usually performed by PCR amplification [23, 61–63]. The contamination of vegetables and fruits with T. gondii was observed in Brazil, China, Italy and Poland (Table 5), and the average prevalence of the contamination was estimated as 3.8% (63/1676; 95% CI: 2.9–4.7%). The T. gondii isolates obtained from vegetables and fruits belonged to genotypes Type I and II [23, 61, 64].
Table 5.
Contamination of vegetables and fruits with Toxoplasma gondii
| Location | Detection method | Vegetable or fruit item | No. of samples tested | No. of positive samples (%) | Toxoplasma gondii genotypes identified (n) | References |
|---|---|---|---|---|---|---|
| Brazil | PCR | Smooth lettuce | 62 | 1 (0.6) | Toxo4-5 D (1) | [62] |
| Crisp head lettuce | 106 | 4 (3.7) | B22-23 D (4) | |||
| Chicory | 40 | 2 (5.0) | B22-23 D (1); Toxo4-5 D (1) | |||
| Rocket | 7 | 1 (14.3) | B22-23 D (1) | |||
| Parsley | 5 | 1 (20.0) | B22-23 D (1) | |||
| Brazil | PCR | Vegetables | 21 | 3 (14.3) | N/A (3) | [45] |
| China | Quantitative real-time PCR (qPCR) | Lettuce | 71 | 5 (7.0) | Type I (4); Type II (1) | [63] |
| Spinach | 50 | 2 (4.0) | Type I (2) | |||
| Pak choi | 34 | 1 (2.9) | Type I (1) | |||
| Chinese cabbage | 26 | 0 | ||||
| Rape | 22 | 1 (4.5) | Type II (1) | |||
| Asparagus | 18 | 0 | ||||
| Chrysanthemum coronarium | 16 | 0 | ||||
| Endive | 14 | 0 | ||||
| Chinese chives | 11 | 0 | ||||
| Cabbage | 9 | 0 | ||||
| Red cabbage | 8 | 1 (12.5) | Type II (1) | |||
| Czech Republic | Triplex real time PCR | Carrots | 93 | 7 (7.5) | [64] | |
| Cucumbers | 109 | 13 (11.9) | Type II (5) | |||
| Salads | 90 | 8 (8.9) | Type II (2) | |||
| Italy | qPCR | Ready-to-eat packaged salad | 648 | 5 (0.8) | Type I (5) | [23] |
| Poland | qPCR | Strawberries | 60 | 0 | [61] | |
| Radish | 60 | 3 (5.0) | Type I (2); Type II (1) | |||
| Carrot | 46 | 9 (19.6) | Type I (3); Type II (1) | |||
| Lettuce | 50 | 9 (18.0) | Type I (1) | |||
| Total | 1676 | 63 (3.8) |
Other intestinal protozoan contaminations
Fresh vegetables and fruits are occasionally contaminated with some other intestinal protozoans, such as Balantioides coli, Cystoisospora belli, Blastocystis sp. and Enterocytozoon bieneusi.
Several reports have documented B. coli contamination of vegetables, leading to global public health concerns [65]. Balantioides coli is usually detected on vegetables and fruits with light microscopy [14, 30, 52, 66, 67]. The contamination of vegetables with B. coli has been reported in Bangladesh, Brazil, Cameroon, Ethiopia, and Ghana (Table 6) and the average prevalence of the contamination is calculated as 9.3% (72/907; 95% CI: 7.6–11.0%).
Table 6.
Contamination of vegetables and fruits with Balantidium coli, Cystoisospora belli, Blastocystis sp. and Enterocytozoon bieneusi
| Location | Detection method | Vegetable or fruit item | No. of samples tested | No. of positive samples (%) | Identified species or genotypes (n) | References |
|---|---|---|---|---|---|---|
| Balantidium coli | ||||||
| Bangladesh | Sediment smears, followed by light microscopy | Vegetables | 200 | 8 (4.0) | B. coli | [52] |
| Brazil | Sediment smears, followed by light microscopy | Loose leaf lettucea | 1 | 1 | B. coli | [30] |
| Red lettucea | 1 | 1 | B. coli | |||
| Curly lettucea | 1 | 1 | B. coli | |||
| Iceberg lettucea | 1 | 1 | B. coli | |||
| Parsleya | 1 | 1 | B. coli | |||
| Chivea | 1 | 1 | B. coli | |||
| Coriandera | 1 | |||||
| Cameroon | Sediment smears, followed by light microscopy | Green cabbage | 30 | 3 (10.0) | B. coli (3) | [66] |
| Red cabbage | 30 | 7 (23.3) | B. coli (7) | |||
| Lettuce | 30 | 8 (26.7) | B. coli (8) | |||
| Cucumber | 30 | 5 (16.7) | B. coli (5) | |||
| Carrots | 30 | 4 (13.3) | B. coli (4) | |||
| Green pepper | 30 | 2 (6.7) | B. coli (2) | |||
| Ethiopia | Sediment smears, followed by light microscopy | Tomato | 100 | 0 | [14] | |
| Cabbage | 96 | 4 (4.2) | B. coli-like (4) | |||
| Green pepper | 66 | 6 (9.1) | B. coli-like (6) | |||
| Carrot | 62 | 4 (6.5) | B. coli-like (4) | |||
| Salad | 23 | 1 (4.3) | B. coli-like (1) | |||
| Ghana | Sediment smears, followed by light microscopy | Cabbage | 72 | 21 (29.2) | B. coli (21) | [67] |
| Lettuce | 72 | 3 (4.2) | B. coli (3) | |||
| Carrot | 72 | 2 (2.8) | B. coli (2) | |||
| Spring onion | 72 | 1 (1.4) | B. coli (1) | |||
| Tomatoes | 72 | 22 (30.6) | B. coli (22) | |||
| Subtotal | 1087 | 101 (9.3) | ||||
| Cystoisospora belli | ||||||
| Ethiopia | Modified Ziehl-Neelsen stain | Fruits and vegetables | 360 | 11 (3.1) | I. belli (11) | [32] |
| Ethiopia | Modified Ziehl-Neelsen stain | Tomatoes | 45 | 0 | [43] | |
| Lettuce | 45 | 1 (2.2) | C. belli (1) | |||
| Carrot | 45 | 2 (4.4) | C. belli (2) | |||
| Cabbage | 45 | 4 (8.8) | C. belli (4) | |||
| Green pepper | 45 | 0 | ||||
| Avocado | 45 | 0 | ||||
| Ghana | Ziehl-Neelsen stain | Cabbage | 90 | 0 | [12] | |
| Green pepper | 55 | 0 | ||||
| Carro | 47 | 0 | ||||
| Onion | 70 | 0 | ||||
| Tomato | 31 | 1 (3.2) | I. beli (1) | |||
| Lettuce | 102 | 0 | ||||
| Subtotal | 1025 | 19 (1.9) | ||||
| Blastocystis sp. | ||||||
| Brazil | Sediment being stained in Lugolʼs solution | Lettuce | 100 | 15 (15.0) | B. hominis (15) | [15] |
| Coriander | 100 | 19 (19.0) | B. hominis (19) | |||
| Italy | Lugolʼs stain, Giemsa Stain, and PCR | Ready-to-eat packaged salad | 648 | 3 (0.5) | B. hominis (3) | [23] |
| Subtotal | 848 | 37 (4.4) | ||||
| Enterocytozoon bieneusi | ||||||
| China | PCR | Lettuce | 200 | 14 (7.0) | E. bieneusi genotype CM8 (2); CD6 (7); EbpA (3); Henan-IV (1) | [36] |
| Coriander | 152 | 1 (0.7) | E. bieneusi genotype CM8 (1) | |||
| Celery | 70 | 1 (1.4) | E. bieneusi genotype EbpA (1) | |||
| Baby bok choy | 59 | 1 (1.7) | E. bieneusi genotype CHV3 (1) | |||
| Chinese cabbage | 47 | 0 | ||||
| Leaf lettuce | 44 | 2 (4.5) | E. bieneusi genotype CHG19 (1) | |||
| Water spinach | 28 | 3 (10.7) | E. bieneusi genotype CD6 (1); BEB8 (1); CTS3 (1) | |||
| Crown daisy | 27 | 0 | ||||
| Fennel plant | 26 | 1 (3.9) | E. bieneusi genotype EbpC (1) | |||
| Endive | 25 | 1 (4.0) | E. bieneusi genotype Henan-IV (1) | |||
| Spinach | 20 | 0 | ||||
| Schizonepeta | 20 | 0 | ||||
| Cabbage | 18 | 0 | ||||
| Leaf mustard | 11 | 0 | ||||
| Chinese chive | 132 | 6 (4.5) | E. bieneusi genotype CD6 (1); EbpA (2); EbpC (1); CHV1 (1) | |||
| Chive | 128 | 4 (1.4) | E. bieneusi genotype CD6 (2); CHV2 (1); CTS3 (1) | |||
| Cucumber | 41 | 1 (2.4) | E. bieneusi genotype CD6 (1) | |||
| Watermelon | 15 | 1 (6.7) | E. bieneusi genotype CD6 (1) | |||
| Potato | 3 | 1 (33.3) | E. bieneusi genotype CHV4 (1) | |||
| Bean (kidney/French bean) | 28 | 4 (14.3) | E. bieneusi genotype CD6 (4) | |||
| Green chili | 5 | 0 | ||||
| Costa Rica | Zielh-Nielsen stain | Lettuce | 50 | 16 (32.0) | E. bieneusi (16) | [71] |
| Parsley | 50 | 0 | ||||
| Cilantro | 50 | 2 (4.0) | E. bieneusi (2) | |||
| Strawberries | 50 | 1 (2.0) | E. bieneusi (1) | |||
| Blackberries | 50 | 0 | ||||
| Poland | Conventional stain and FISH | Berries | 25 | 6 (24.0) | E. intestinalis (4); E. bieneusi (2) | [21] |
| Sprouts | 20 | 1 (5.0) | E. bieneusi (1) | |||
| Vegetables | 35 | 2 (5.7) | E. cuniculi (1); E. bieneusi (1) | |||
| Sub-total | 1429 | 52 (3.6) | ||||
aSingle sample in a case report
Cystoisospora belli infection is commonly reported in tropical and subtropical areas of the world [68]. Cystoisosporiasis can be acquired through the ingestion of contaminated food. Cystoisospora belli is commonly detected with modified Ziehl-Neelsen staining, followed by microcopy [32, 43]. There are three reports on Cystoisospora belli contamination in vegetables and fruits in Ethiopia and Ghana (Table 6). The average prevalence of the contamination is estimated as 1.9% (19/1025; 95% CI: 1.1–2.7%).
The detection of Blastocystis sp. is usually based on microscopy and PCR [23]. Cell culture is also used for the detection of this parasite. The contamination of vegetables and fruits with Blastocystis sp. has only been documented in Brazil and Italy, with a prevalence of 4.4% (37/848; 95% CI: 3.0–5.8%) (Table 6).
Enterocytozoon bieneusi is an important microsporidian species infecting humans [69]. The genetic diversity of the pathogen is inferred by the analysis of single nucleotide polymorphisms (SNPs) in the internal transcribed spacer (ITS) that resulted in nearly 500 valid genotypes of the pathogen [70]. The phylogenetic analysis of the valid genotypes recognized eleven genetic groups (Groups 1 to 11), figuring out their host specificity and zoonotic potential. Food-borne transmission of E. bieneusi has been documented and the contamination of vegetables and fruits with this pathogen was reported in China, Costa Rica and Poland (Table 6). The parasite was successfully detected in contaminated vegetables and fruits by staining or with fluorescence in situ hybridization [21, 71], and PCR amplification [36]. The average prevalence of the reported contamination was estimated as 3.6% (52/1429; 95% CI: 2.6–4.6%).
Risk factors involved in the contamination of vegetables and fruits with parasites
Previous studies in Ethiopia, Ghana, Brazil and Iran reported a relatively higher prevalence of intestinal parasitic infections associated with the consumption of vegetables sold at open-aired markets than those associated with supermarkets [12, 14, 15]. The parasitic load in the raw vegetables of open markets was high and posed a high risk of parasitic infections. The high contamination rates recorded in the open-market samples indicate poor hygiene in these locations, which is suitable for the propagation and transmission of the parasites [72].
High risk of diarrhea among raw vegetable consumers in the Kathmandu valley of Nepal, mostly due to the use of river water by farmers for washing vegetables, suggests a need to avoid the use of river water for washing vegetables [73]. There are also many reports that highlight the contamination of surface water with parasitic infective stages in Brazil [74], Iran [75], Poland [76] and Spain [77]. The use of such contaminated surface water for washing fresh vegetables and fruits might cause parasitic contamination.
Another study in the Czech Republic reported a significantly higher contamination of T. gondii in vegetables collected from farm storage rooms than those from fields [64], indicating a higher chance of contamination of vegetables and fruits during processing and selling [78]. Therefore, the adaptation of good practices in every step between farm and fork, such as production, processing, storage and selling minimize the microbial contamination of vegetables and fruits.
Conclusions
The accidental ingestion of parasitic infective stages such as eggs, oocysts, cysts or spores with the contaminated raw vegetables or fruits causes varying intestinal diseases in humans that sometimes may lead to serious health problems. On many occasions, the contamination of vegetables and fruits results in outbreaks of the parasitic diseases. Globally, the occurrence of protozoan parasitic contamination in vegetables and fruits ranges from 1.9% to 9.3%. However, contamination with protozoans may be grossly underestimated, especially in regions with poor sanitation. Contamination of vegetables and fruits with parasites can occur in many ways. The common stages between farm and fork at which vegetables and fruits are contaminated include production, processing, storage and selling. Therefore, the implementation of hygienic practices at every step between production and consumption may eliminate the contamination. The appropriate local public health authority is recommended to establish a system for continuous monitoring of contamination of vegetables and fruits sold at local markets.
Acknowledgements
Not applicable.
Abbreviations
- CI
confidence interval
- ITS
internal transcribed spacer
- PCR
polymerase chain reaction
- SNP
single-nucleotide polymorphism
Authors’ contributions
LZ and JL conceived and designed the review. JL, ZW and MRK analyzed the data and wrote the original draft of the manuscript. LZ and JL revised the final manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (2019YFC1605700), National Natural Science Foundation of China (30600603, 31672548), the Natural Science Foundation of Henan Province (162300410129), and the Doctoral Scientific Research Start-up Foundation from Henan University of Chinese Medicine (KYQD021).
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Junqiang Li, Zhenzhen Wang and Md Robiul Karim contributed equally to this work
Contributor Information
Junqiang Li, Email: lijunqiangcool@126.com.
Zhenzhen Wang, Email: wzlinxi@163.com.
Md Robiul Karim, Email: vet_robiul@bsmrau.edu.bd.
Longxian Zhang, Email: zhanglx8999@henau.edu.cn.
References
- 1.Ryan U, Paparini A, Oskam C. New technologies for detection of enteric parasites. Trends Parasitol. 2017;33:532–546. doi: 10.1016/j.pt.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Julian TR. Environmental transmission of diarrheal pathogens in low and middle income countries. Environ Sci Process Impacts. 2016;18:944–955. doi: 10.1039/c6em00222f. [DOI] [PubMed] [Google Scholar]
- 3.Fletcher SM, Stark D, Harkness J, Ellis J. Enteric protozoa in the developed world: a public health perspective. Clin Microbiol Rev. 2012;25:420–449. doi: 10.1128/CMR.05038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dawson D. Foodborne protozoan parasites. Int J Food Microbiol. 2005;103:207–227. doi: 10.1016/j.ijfoodmicro.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 5.Giangaspero A, Gasser RB. Human cyclosporiasis. Lancet Infect Dis. 2019;19:e226–e236. doi: 10.1016/S1473-3099(18)30789-8. [DOI] [PubMed] [Google Scholar]
- 6.Ryan U, Hijjawi N, Xiao L. Foodborne cryptosporidiosis. Int J Parasitol. 2018;48:1–12. doi: 10.1016/j.ijpara.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Ryan U, Hijjawi N, Feng Y, Xiao L. Giardia: an under-reported foodborne parasite. Int J Parasitol. 2018;49:1–11. doi: 10.1016/j.ijpara.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Olza J, Aranceta-Bartrina J, González-Gross M, Ortega RM, Serra-Majem L, Varela-Moreiras G, et al. Reported dietary intake and food sources of zinc, selenium, and vitamins A, E and C in the Spanish population: findings from the anibes study. Nutrients. 2017;9:697. doi: 10.3390/nu9070697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouzid M, Kintz E, Hunter PR. Risk factors for Cryptosporidium infection in low and middle income countries: a systematic review and meta-analysis. PLoS Negl Trop Dis. 2018;12:e0006553. doi: 10.1371/journal.pntd.0006553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herman KM, Hall AJ, Gould LH. Outbreaks attributed to fresh leafy vegetables, United States, 1973–2012. Epidemiol Infect. 2015;143:3011–3021. doi: 10.1017/S0950268815000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amorós I, Alonso JL, Cuesta G. Cryptosporidium oocysts and Giardia cysts on salad products irrigated with contaminated water. J Food Prot. 2010;73:1138–1140. doi: 10.4315/0362-028x-73.6.1138. [DOI] [PubMed] [Google Scholar]
- 12.Duedu KO, Yarnie EA, Tetteh-Quarcoo PB, Attah SK, Donkor ES, Ayeh-Kumi PF. A comparative survey of the prevalence of human parasites found in fresh vegetables sold in supermarkets and open-aired markets in Accra, Ghana. BMC Res Notes. 2014;7:836. doi: 10.1186/1756-0500-7-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Utaaker KS, Kumar A, Joshi H, Chaudhary S, Robertson LJ. Checking the detail in retail: occurrence of Cryptosporidium and Giardia on vegetables sold across different counters in Chandigarh, India. Int J Food Microbiol. 2017;263:1–8. doi: 10.1016/j.ijfoodmicro.2017.09.020. [DOI] [PubMed] [Google Scholar]
- 14.Alemu G, Mama M, Misker D, Haftu D. Parasitic contamination of vegetables marketed in Arba Minch town, southern Ethiopia. BMC Infect Dis. 2019;19:410. doi: 10.1186/s12879-019-4020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodrigues AC, da Silva MDC, Pereira RÂS, Pinto LC. Prevalence of contamination by intestinal parasites in vegetables (Lactuca sativa L. and Coriandrum sativum L.) sold in markets in Belém, northern Brazil. J Sci Food Agric. 2020;100:2859–2865. doi: 10.1002/jsfa.10265. [DOI] [PubMed] [Google Scholar]
- 16.Ahmed SA, Karanis P. An overview of methods/techniques for the detection of Cryptosporidium in food samples. Parasitol Res. 2018;117:629–653. doi: 10.1007/s00436-017-5735-0. [DOI] [PubMed] [Google Scholar]
- 17.Gabre RM, Shakir A. Prevalence of some human enteroparasites in commonly consumed raw vegetables in Tabuk, Saudi Arabia. J Food Prot. 2016;79:655–658. doi: 10.4315/0362-028X.JFP-15-485. [DOI] [PubMed] [Google Scholar]
- 18.Eraky MA, Rashed SM, Nasr Mel S, El-Hamshary AM, Salah El-Ghannam A. Parasitic contamination of commonly consumed fresh leafy vegetables in Benha, Egypt. J Parasitol Res. 2014;2014:613960. doi: 10.1155/2014/613960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tefera T, Biruksew A, Mekonnen Z, Eshetu T. Parasitic contamination of fruits and vegetables collected from selected local markets of Jimma town, southwest Ethiopia. Int Sch Res Notices. 2014;2014:382715. doi: 10.1155/2014/382715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ismail Y. Prevalence of parasitic contamination in salad vegetables collected from supermarkets and street vendors in Amman and Baqa’a - Jordan. Pol J Microbiol. 2016;65:201–207. doi: 10.5604/17331331.1204480. [DOI] [PubMed] [Google Scholar]
- 21.Jedrzejewski S, Graczyk TK, Slodkowicz-Kowalska A, Tamang L, Majewska AC. Quantitative assessment of contamination of fresh food produce of various retail types by human-virulent microsporidian spores. Appl Environ Microbiol. 2007;73:4071–4073. doi: 10.1128/AEM.00477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong S, Kim K, Yoon S, Park WY, Sim S, Yu JR. Detection of Cryptosporidium parvum in environmental soil and vegetables. J Korean Med Sci. 2014;29:1367–1371. doi: 10.3346/jkms.2014.29.10.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caradonna T, Marangi M, Del Chierico F, Ferrari N, Reddel S, Bracaglia G. Detection and prevalence of protozoan parasites in ready-to-eat packaged salads on sale in Italy. Food Microbiol. 2017;67:67–75. doi: 10.1016/j.fm.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Sim S, Won J, Kim JW, Kim K, Park WY, Yu JR. Simultaneous molecular detection of Cryptosporidium and Cyclospora from raw vegetables in Korea. Korean J Parasitol. 2017;55:137–142. doi: 10.3347/kjp.2017.55.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rzezutka A, Nichols RA, Connelly L, Kaupke A, Kozyra I, Cook N, et al. Cryptosporidium oocysts on fresh produce from areas of high livestock production in Poland. Int J Food Microbiol. 2010;139:96–101. doi: 10.1016/j.ijfoodmicro.2010.01.027. [DOI] [PubMed] [Google Scholar]
- 26.Ranjbar-Bahadori Sh, Mostoophi A, Shemshadi B. Study on Cryptosporidium contamination in vegetable farms around Tehran. Trop Biomed. 2013;30:193–198. [PubMed] [Google Scholar]
- 27.Shields JM, Lee MM, Murphy HR. Use of a common laboratory glassware detergent improves recovery of Cryptosporidium parvum and Cyclospora cayetanensis from lettuce, herbs and raspberries. Int J Food Microbiol. 2012;153:123–128. doi: 10.1016/j.ijfoodmicro.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 28.Tiyo R, de Souza CZ, Arruda Piovesani AF, Tiyo BT, Colli CM, Marchioro AA, et al. Predominance of Giardia duodenalis assemblage AII in fresh leafy vegetables from a market in southern Brazil. J Food Prot. 2016;79:1036–1039. doi: 10.4315/0362-028X.JFP-15-306. [DOI] [PubMed] [Google Scholar]
- 29.Mohamed MA, Siddig EE, Elaagip AH, Edris AM, Nasr AA. Parasitic contamination of fresh vegetables sold at central markets in Khartoum state, Sudan. Ann Clin Microbiol Antimicrob. 2016;15:17. doi: 10.1186/s12941-016-0133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Machado ER, Maldonade IR, Riquette RFR, Mendes VS, Gurgel-Gonçalves R, Ginani VC. Frequency of enteroparasites and bacteria in the leafy vegetables sold in Brazilian public wholesale markets. J Food Prot. 2018;81:542–548. doi: 10.4315/0362-028X.JFP-17-358. [DOI] [PubMed] [Google Scholar]
- 31.Chandra V, Torres M, Ortega YR. Efficacy of wash solutions in recovering Cyclospora cayetanensis, Cryptosporidium parvum, and Toxoplasma gondii from basil. J Food Prot. 2014;77:1348–1354. doi: 10.4315/0362-028X.JFP-13-381. [DOI] [PubMed] [Google Scholar]
- 32.Bekele F, Tefera T, Biresaw G, Yohannes T. Parasitic contamination of raw vegetables and fruits collected from selected local markets in Arba Minch town, southern Ethiopia. Infect Dis Poverty. 2017;6:19. doi: 10.1186/s40249-016-0226-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng Y, Ryan UM, Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 2018;34:997–1011. doi: 10.1016/j.pt.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Khan A, Shaik JS, Grigg ME. Genomics and molecular epidemiology of Cryptosporidium species. Acta Trop. 2018;184:1–14. doi: 10.1016/j.actatropica.2017.10.023. [DOI] [PubMed] [Google Scholar]
- 35.Robertson LJ, Gjerde B. Occurrence of parasites on fruits and vegetables in Norway. J Food Prot. 2001;64:1793–1798. doi: 10.4315/0362-028x-64.11.1793. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Shi K, Sun F, Li T, Wang R, Zhang S, et al. Identification of human pathogenic Enterocytozoon bieneusi, Cyclospora cayetanensis, and Cryptosporidium parvum on the surfaces of vegetables and fruits in Henan, China. Int J Food Microbiol. 2019;307:108292. doi: 10.1016/j.ijfoodmicro.2019.108292. [DOI] [PubMed] [Google Scholar]
- 37.Einarsson E, Ma’ayeh S, Svärd SG. An up-date on Giardia and giardiasis. Curr Opin Microbiol. 2016;34:47–52. doi: 10.1016/j.mib.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 38.Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. 2011;24:110–140. doi: 10.1128/CMR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colli CM, Bezagio RC, Nishi L, Bignotto TS, Ferreira ÉC, Falavigna-Guilherme AL, et al. Identical assemblage of Giardia duodenalis in humans, animals and vegetables in an urban area in southern Brazil indicates a relationship among them. PLoS One. 2015;10:e0118065. doi: 10.1371/journal.pone.0118065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Figgatt M, Mergen K, Kimelstein D, Mahoney DM, Newman A, Nicholas D, et al. Giardiasis outbreak associated with asymptomatic food handlers in New York State, 2015. J Food Prot. 2017;12:837–841. doi: 10.4315/0362-028X.JFP-16-415. [DOI] [PubMed] [Google Scholar]
- 41.Sitotaw B, Mekuriaw H, Damtie D. Prevalence of intestinal parasitic infections and associated risk factors among Jawi primary school children, Jawi town, north-west Ethiopia. BMC Infect Dis. 2019;19:341. doi: 10.1186/s12879-019-3971-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shahnazi M, Jafari-Sabet M. Prevalence of parasitic contamination of raw vegetables in villages of Qazvin Province, Iran. Foodborne Pathog Dis. 2010;7:1025–1030. doi: 10.1089/fpd.2009.0477. [DOI] [PubMed] [Google Scholar]
- 43.Bekele F, Shumbej T. Fruit and vegetable contamination with medically important helminths and protozoans in Tarcha town, Dawuro zone, South West Ethiopia. Res Rep Trop Med. 2019;10:19–23. doi: 10.2147/RRTM.S205250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rafael K, Marchioro AA, Colli CM, Tiyo BT, Evangelista FF, Bezagio RC, et al. Genotyping of Giardia duodenalis in vegetables cultivated with organic and chemical fertilizer from street markets and community vegetable gardens in a region of southern Brazil. Trans R Soc Trop Med Hyg. 2017;111:540–545. doi: 10.1093/trstmh/try013. [DOI] [PubMed] [Google Scholar]
- 45.Ferreira FP, Caldart ET, Freire RL, Mitsuka-Breganó R, Freitas FM, Miura AC, et al. The effect of water source and soil supplementation on parasite contamination in organic vegetable gardens. Rev Bras Parasitol Vet. 2018;27:327–337. doi: 10.1590/S1984-296120180050. [DOI] [PubMed] [Google Scholar]
- 46.Ortega YR, Sanchez R. Update on Cyclospora cayetanensis, a food-borne and waterborne parasite. Clin Microbiol Rev. 2010;23:218–234. doi: 10.1128/CMR.00026-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tram NT, Hoang LM, Cam PD, Chung PT, Fyfe MW, Isaac-Renton JL, et al. Cyclospora spp. in herbs and water samples collected from markets and farms in Hanoi, Vietnam. Trop Med Int Health. 2008;13:1415–1420. doi: 10.1111/j.1365-3156.2008.02158.x. [DOI] [PubMed] [Google Scholar]
- 48.Giangaspero A, Marangi M, Koehler AV, Papini R, Normanno G, Lacasella V, et al. Molecular detection of Cyclospora in water, soil, vegetables and humans in southern Italy signals a need for improved monitoring by health authorities. Int J Food Microbiol. 2015;211:95–100. doi: 10.1016/j.ijfoodmicro.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 49.Cui Z, Li J, Chen Y, Zhang L. Molecular epidemiology, evolution, and phylogeny of Entamoeba spp. Infect Genet Evol. 2019;75:104018. doi: 10.1016/j.meegid.2019.104018. [DOI] [PubMed] [Google Scholar]
- 50.Kantor M, Abrantes A, Estevez A, Schiller A, Torrent J, Gascon J, et al. Entamoeba histolytica: updates in clinical manifestation, pathogenesis, and vaccine development. Can J Gastroenterol Hepatol. 2018;2018:4601420. doi: 10.1155/2018/4601420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anuar TS, Al-Mekhlafi HM, Abdul Ghani MK, Abu Bakar E, Azreen SN, Salleh FM, et al. Molecular epidemiology of amoebiasis in Malaysia: highlighting the different risk factors of Entamoeba histolytica and Entamoeba dispar infections among Orang Asli communities. Int J Parasitol. 2012;42:1165–1175. doi: 10.1016/j.ijpara.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 52.Azim A, Ahmed S, Paul SK, Nasreen SA, Sarkar SR, Ahmed MU, et al. Prevalence of intestinal parasites in raw vegetables consumed by inhabitants of Mymensingh City. Mymensingh Med J. 2018;27:440–444. [PubMed] [Google Scholar]
- 53.M’rad S, Chaabane-Banaoues R, Lahmar I, Oumaima H, Mezhoud H, Babba H, et al. Parasitological contamination of vegetables sold in Tunisian retail markets with helminth eggs and protozoan cysts. J Food Prot. 2020;83:1104–1109. doi: 10.4315/JFP-19-559. [DOI] [PubMed] [Google Scholar]
- 54.Aguirre AA, Longcore T, Barbieri M, Dabritz H, Hill D, Klein PN, et al. The one health approach to toxoplasmosis: epidemiology, control, and prevention strategies. Ecohealth. 2019;16:378–390. doi: 10.1007/s10393-019-01405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharif M, Amouei A, Sarvi S, Mizani A, Aarabi M, Hosseini SA, et al. Genetic diversity of Toxoplasma gondii isolates from ruminants: a systematic review. Int J Food Microbiol. 2017;258:38–49. doi: 10.1016/j.ijfoodmicro.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 56.Hosseini SA, Amouei A, Sharif M, Sarvi Sh, Galal L, Javidnia J, et al. Human toxoplasmosis: a systematic review for genetic diversity of Toxoplasma gondii in clinical samples. Epidemiol Infect. 2018;147:e36. doi: 10.1017/S0950268818002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hussain MA, Stitt V, Szabo EA, Nelan B. Toxoplasma gondii in the food supply. Pathogens. 2017;6:21. doi: 10.3390/pathogens6020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teweldemedhin M, Gebremichael A, Geberkirstos G, Hadush H, Gebrewahid T, Asgedom SW, et al. Seroprevalence and risk factors of Toxoplasma gondii among pregnant women in Adwa district, northern Ethiopia. BMC Infect Dis. 2019;19:327. doi: 10.1186/s12879-019-3936-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sadaghian M, Amani S, Jafari R. Prevalence of toxoplasmosis and related risk factors among humans referred to main laboratories of Urmia city, north west of Iran, 2013. J Parasit Dis. 2016;40:520–523. doi: 10.1007/s12639-014-0537-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paul E, Kiwelu I, Mmbaga B, Nazareth R, Sabuni E, Maro A, et al. Toxoplasma gondii seroprevalence among pregnant women attending antenatal clinic in northern Tanzania. Trop Med Health. 2018;46:39. doi: 10.1186/s41182-018-0122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lass A, Pietkiewicz H, Szostakowska B, Myjak P. The first detection of Toxoplasma gondii DNA in environmental fruits and vegetables samples. Eur J Clin Microbiol Infect Dis. 2012;31:1101–1108. doi: 10.1007/s10096-011-1414-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marchioro AA, Tiyo BT, Colli CM, de Souza CZ, Garcia JL, Gomes ML, et al. First detection of Toxoplasma gondii DNA in the fresh leafs of vegetables in South America. Vector Borne Zoonotic Dis. 2016;16:624–626. doi: 10.1089/vbz.2015.1937. [DOI] [PubMed] [Google Scholar]
- 63.Lass A, Ma L, Kontogeorgos I, Zhang X, Li X, Karanis P. First molecular detection of Toxoplasma gondii in vegetable samples in China using qualitative, quantitative real-time PCR and multilocus genotyping. Sci Rep. 2019;9:17581. doi: 10.1038/s41598-019-54073-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Slany M, Dziedzinska R, Babak V, Kralik P, Moravkova M, Slana I. Toxoplasma gondii in vegetables from fields and farm storage facilities in the Czech Republic. FEMS Microbiol Lett. 2019;366:fnz170. doi: 10.1093/femsle/fnz170. [DOI] [PubMed] [Google Scholar]
- 65.Schuster FL, Ramirez-Avila L. Current world status of Balantidium coli. Clin Microbiol Rev. 2008;21:626–638. doi: 10.1128/CMR.00021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Akoachere JTK, Tatsinkou BF, Nkengfack JM. Bacterial and parasitic contaminants of salad vegetables sold in markets in Fako division, Cameroon and evaluation of hygiene and handling practices of vendors. BMC Res Notes. 2018;11:100. doi: 10.1186/s13104-018-3175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kudah C, Sovoe S, Baiden F. Parasitic contamination of commonly consumed vegetables in two markets in Ghana. Ghana Med J. 2018;52:88–93. doi: 10.4314/gmj.v52i2.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Legua P, Seas C. Cystoisospora and cyclospora. Curr Opin Infect Dis. 2013;26:479–483. doi: 10.1097/01.qco.0000433320.90241.60. [DOI] [PubMed] [Google Scholar]
- 69.Matos O, Lobo ML, Xiao L. Epidemiology of Enterocytozoon bieneusi infection in humans. J Parasitol Res. 2012;2012:981424. doi: 10.1155/2012/981424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Karim MR, Rume FI, Rahman ANMA, Zhang Z, Li J, Zhang L. Evidence for zoonotic potential of Enterocytozoon bieneusi in its first molecular characterization in captive mammals at Bangladesh National Zoo. J Eukaryot Microbiol. 2020;67:427–435. doi: 10.1111/jeu.12792. [DOI] [PubMed] [Google Scholar]
- 71.Calvo M, Carazo M, Arias ML, Chaves C, Monge R, Chinchilla M. Prevalence of Cyclospora sp., Cryptosporidium sp., microsporidia and fecal coliform determination in fresh fruit and vegetables consumed in Costa Rica. Arch Latinoam Nutr. 2004;54:428–432. [PubMed] [Google Scholar]
- 72.Taghipour A, Javanmard E, Haghighi A, Mirjalali H, Zali MR. The occurrence of Cryptosporidium sp., and eggs of soil-transmitted helminths in market vegetables in the north of Iran. Gastroenterol Hepatol Bed Bench. 2019;12:364–369. [PMC free article] [PubMed] [Google Scholar]
- 73.Shrestha S, Haramoto E, Shindo J. Assessing the infection risk of enteropathogens from consumption of raw vegetables washed with contaminated water in Kathmandu Valley, Nepal. J Appl Microbio. 2017;123:1321–1334. doi: 10.1111/jam.13573. [DOI] [PubMed] [Google Scholar]
- 74.de Araújo RS, Aguiar B, Dropa M, Razzolini MT, Zanoli Sato MI, de Souza Lauretto M, et al. Detection and molecular characterization of Cryptosporidium species and Giardia assemblages in two watersheds in the metropolitan region of São Paulo, Brazil. Environ Sci Pollut Res Int. 2018;25:15191–15203. doi: 10.1007/s11356-018-1620-3. [DOI] [PubMed] [Google Scholar]
- 75.Mahmoudi MR, Kazemi B, Haghighi A, Karanis P. Detection of Acanthamoeba and Toxoplasma in river water samples by molecular methods in Iran. Iran J Parasitol. 2015;10:250–257. [PMC free article] [PubMed] [Google Scholar]
- 76.Adamska M. Molecular characterization of Cryptosporidium and Giardia occurring in natural water bodies in Poland. Parasitol Res. 2015;114:687–692. doi: 10.1007/s00436-014-4234-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Castro-Hermida JA, González-Warleta M, Mezo M. Cryptosporidium spp. and Giardia duodenalis as pathogenic contaminants of water in Galicia, Spain: the need for safe drinking water. Int J Hyg Environ Health. 2015;218:132–138. doi: 10.1016/j.ijheh.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 78.Berrouch S, Escotte-Binet S, Harrak R, Huguenin A, Flori P, Favennec L, et al. Detection methods and prevalence of transmission stages of Toxoplasma gondii, Giardia duodenalis and Cryptosporidium spp. in fresh vegetables: a review. Parasitology. 2020;147:516–532. doi: 10.1017/S0031182020000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Monge R, Arias ML. Presence of various pathogenic microorganisms in fresh vegetables in Costa Rica. Arch Latinoam Nutr. 1996;46:292–294. [PubMed] [Google Scholar]
- 80.Mossallam SF. Detection of some intestinal protozoa in commercial fresh juices. J Egypt Soc Parasitol. 2010;40:135–149. [PubMed] [Google Scholar]
- 81.Ayeh-Kumi PF, Tetteh-Quarcoo PB, Duedu KO, Obeng AS, Addo-Osafo K, Mortu S, et al. A survey of pathogens associated with Cyperus esculentus L (tiger nuts) tubers sold in a Ghanaian city. BMC Res Notes. 2014;7:343. doi: 10.1186/1756-0500-7-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Robertson LJ, Johannessen GS, Gjerde BK, Loncarevic S. Microbiological analysis of seed sprouts in Norway. Int J Food Microbiol. 2002;75:119–126. doi: 10.1016/s0168-1605(01)00738-3. [DOI] [PubMed] [Google Scholar]
- 83.Ortega YR, Roxas CR, Gilman RH, Miller NJ, Cabrera L, Taquiri C, et al. Isolation of Cryptosporidium parvum and Cyclospora cayetanensis from vegetables collected in markets of an endemic region in Peru. Am J Trop Med Hyg. 1997;57:683–686. doi: 10.4269/ajtmh.1997.57.683. [DOI] [PubMed] [Google Scholar]
- 84.Daryani A, Ettehad GH, Sharif M, Ghorbani L, Ziaei H. Prevalence of intestinal parasites in vegetables consumed in Ardabil, Iran. Food Control. 2008;19:790–794. [Google Scholar]
- 85.Paula P, Rodrigues PS, Tórtora JC, Uchôa CM, Farage S. Microbiological and parasitological contamination of lettuce (Lactuca sativa) from self service restaurants of Niterói city, RJ. Rev Soc Bras Med Trop. 2003;36:535–537. doi: 10.1590/s0037-86822003000400019. [DOI] [PubMed] [Google Scholar]
- 86.Do Ramos NIC, RamosRamos RAN, Giannelli A, Lima VFS, Cringoli G, Rinaldi L, et al. An additional asset for the FLOTAC technique: detection of gastrointestinal parasites in vegetables. Acta Parasitol. 2019;64:423–425. doi: 10.2478/s11686-019-00059-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.


