Highlights
-
•
Saliva has been recommended as an alternative sample for detection of SARS CoV-2 infection, but data are limited.
-
•
In this study, SARS CoV-2 was detected by RT-PCR in 35 of 124 patients, 30 (85.7 %) by saliva and 33 (94.3 %) by NP swab.
-
•
The median cycle threshold value was significantly lower for NPS than for saliva.
-
•
A third of pure saliva samples were difficult to pipet, which slowed processing.
Keywords: SARS CoV-2, COVID-19, Saliva, Nasopharyngeal swab, Real-time RT-PCR
Abstract
Background
A major expansion in SARS CoV-2 testing is urgently needed. Saliva is an attractive option as an alternative for nasopharyngeal swabs (NPS), since saliva can be self-collected, is non-invasive, and sample quality is not dependent on the expertise of the collector.
Objective
To compare SARS CoV-2 positivity on paired NPS and saliva samples.
Study design
NPS and paired saliva samples were prospectively collected from symptomatic outpatients suspected of having COVID-19 and were tested by real-time RT-PCR.
Results
In total, 35/124 (26.6 %) samples were RT-PCR positive, with 33/35 positive by NPS (sensitivity = 94.3 % (95 % CI 81.4%–99.0%)) and 30/35 by pure saliva (sensitivity = 85.7 % (95 % CI 70.6%–93.7%)), for an overall agreement of 117/124 (94.4 %). The median cycle threshold value was significantly lower for NPS than for saliva (p = 0.0331). A third or more of pure saliva samples from symptomatic patients were thick, stringy, and difficult to pipet.
Conclusions
Real-time RT-PCR of pure saliva had an overall sensitivity for SARS CoV-2 RNA detection of 85.7 % when compared to simultaneously collected NPS. Our study highlighted the need to optimize collection and processing before saliva can be used for high volume testing.
1. Introduction
A major expansion in SARS CoV-2 testing is urgently needed. In the U.S., in addition to symptomatic patients, testing may be undertaken for all hospital admissions, prior to immunosuppression or invasive medical procedures, and may be considered for asymptomatic nursing home residents, healthcare workers, first responders, residential college students, correctional facilities, and employees in various work settings.
Although nasopharyngeal swabs (NPS) are widely considered to be the preferred specimen for SARS-CoV-2 testing [1], obstacles to collection include lack of swabs and transport media, the need for skilled staff and personal protective equipment (PPE) for NPS collection, risk of exposure for the collector if coughing or sneezing is induced, and discomfort for the patients that may preclude recurrent testing. In addition, the quality of NPS specimen obtained can be highly variable and in some cases, falsely negative. Thus, alternative sample types are being explored to meet the diagnostic and public health goals for COVID-19 testing.
Saliva is an attractive option as an alternative specimen for NPS. It can be self-collected non-invasively, and sample collection is not dependent on the expertise the collector. However, data on the utility of saliva for SARS CoV-2 are minimal. In reviewing the literature, many different collection methods are reported, including swabbing the mouth, coughing to generate saliva-sputum mixtures, and dilution in viral transport media (VTM). The recently issued Infectious Diseases Society of America (IDSA) guidelines state that saliva as the sole sample source for COVID-19 diagnosis cannot be recommended due to a paucity of studies of pure saliva [1].
2. Methods
To investigate the utility of saliva as an alternate sample type, during a two week period April 16 to April 28, 2020, 124 NPS in 3 mL universal transport medium (Becton Dickinson) and paired saliva samples were prospectively collected at Yale New Haven Hospital drive-through testing sites from symptomatic outpatients suspected of having COVID-19. Patients were asked to not eat or drink for 30 min, let saliva pool in their mouths and then spit into sterile containers. Samples were kept in a cooler and delivered within 2 h to the laboratory. NPS were tested on the day of collection. Residual NPS samples and their paired saliva samples were frozen on day of receipt at -70 C. Within 2 weeks, positive NPS and all 124 saliva samples were thawed and tested. Nucleic acid was extracted from 200 μL of sample and eluted in 55 μL using EasyMag (bioMerieux, Durham, NC). Reverse-transcriptase PCR was performed using a Emergency Use Authorized laboratory developed assay based on the Centers for Disease Control and Prevention protocol. Cycle threshold (Ct) values were recorded for N1, N2 and RNAse P for each sample. Saliva specimens that were viscous and difficult to pipette were treated with sputasol (Thermo Scientific) to liquify the sample.
3. Results
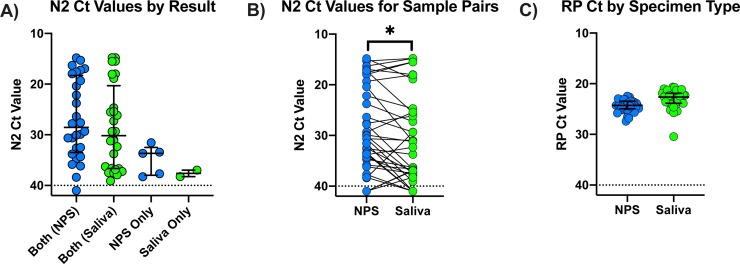
Overall, 33/124 NPS (26.6 %) were PCR positive, and saliva was also positive for 28 of these 33 (84.8 %) NPS-saliva pairs. For the 91 negative NPS, 2 (2.2 %) of the paired saliva samples were PCR-positive. These two NPS negative/ saliva positive pairs were reextracted and retested and results were confirmed (Table 1 ). In total, 35 samples were RT-PCR positive, with 33/35 positive by NPS (sensitivity = 94.3 % (95 % CI 81.4%–99.0%)) and 30/35 by pure saliva (sensitivity = 85.7 % (95 % CI 70.6%–93.7%)), for an overall agreement of 117/124 (94.4 %) between the two sample types. These data give a Cohen’s kappa of 0.851 (95 % CI 0.745 to 0.958). The Ct values for N2 for each result type are shown in Fig. 1 A. For pairs in which both NPS and saliva were positive, Ct values ranged from 14.8 to 40, but samples testing positive only in one specimen had Ct values above 30. For 23 (69.7 %) of 33 pairs, Ct values were lower for NPS samples than for saliva (Fig. 1B), and the difference in median Ct value was significant (p = 0.0331). Despite the high variability of N2 Ct values seen in these samples, there was little variation in Ct values for RNAse P, a marker of human cellular content and a surrogate for sample quality (Fig. 1C).
Table 1.
SARS CoV-2 real-time RT PCR results for paired NPS and saliva in symptomatic outpatients.
| NP Swab |
||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| Saliva | Positive | 28 | 2 | 30 |
| Negative | 5 | 89 | 94 | |
| Total | 33 | 91 | 124 | |
Fig. 1.
Cycle threshold (Ct) values for N2 and RNAse P (RP) for NPS and saliva specimens. A) N2 Ct values by testing concordance. The N2 Ct value was set to 41 for samples in which only the N1 target was detected. The horizontal dashed line is at Ct = 40, the assay cut-off. Horizontal lines indicate the median and interquartile range (IQR). The median and IQR for each group are: 28.56 (18.3 to 33.5), 30.18 (20.32 to 36.66), 33.68 (32.5 to 37.99), and 37.62 (36.98 to 38.26) for Both (NPS), Both (Saliva), NPS Only, and Saliva Only, respectively. B) N2 Ct values for paired NPS and saliva samples. Pairs are connected by a line. The N2 Ct was set to 41 for samples in which N2 was not detected including those negative for SARS-CoV-2 RNA. The horizontal dashed line is at Ct = 40, the assay cut-off. Ct values were significantly lower for NPS when compared by Wilcoxon matched-pairs signed rank test (p = 0.0331). C) RP Ct values for NPS and saliva specimens. Median and IQR are 24.27 (23.46 to 24.96) and 22.63 (21.88 to 23.85), respectively.
4. Discussion
There is tremendous interest in using saliva for detection of SARS CoV-2 RNA, especially for large scale and repeated testing of high risk populations. Our results were virtually identical to those obtained from a similar prospective study in symptomatic outpatients by Williams et al. in Australia [2]. In that study, 33/39 (84.6 %) of patients with positive NPS had SARS CoV-2 RNA detected in saliva, and 1/50 (2%) of saliva samples from patients with negative NPS were also positive. Similar to our findings, Ct values were lower in NPS than in saliva.
Reports by Azzi et al. in a subset of known NPS-positive hospitalized patients in Italy have found saliva collected by the drooling technique can be positive when a subsequent NP swab is negative [3,4]. Another study of 12 hospitalized patients in Hong Kong using coughed saliva found 11/12 NPS-positive patients were also saliva PCR-positive [5].
In a recent study, known NPS-positive hospitalized patients had saliva and repeat NPS collected every 3 days, and more saliva samples were positive than repeat NPS [6]. In the same report, serial paired samples in asymptomatic healthcare workers found two individuals with SARS CoV-2 RNA at low levels in saliva, but not in self-collected NPS. In contrast to other reports, viral loads were lower and more variable in NPS than in saliva samples. However, the Ct values for the RNase P (RP) control were significantly higher and more variable for NPS samples than for saliva, suggesting that variable NPS sample quality in both patient groups could have contributed to poor NPS results. We found very little variation in RP Ct values in NPS samples in our study (Fig. 1C).
In a recent prospective study of NPS-saliva pairs using Cepheid Xpert Xpress SARS- CoV-2 RT-PCR (Sunnyvale, CA), only the liquid, non-viscous components of each specimen were drawn into the disposable pipets for test cartridge inoculation [7]. Overall 50/156 samples were positive, 47 by both NPS and saliva, 2 by NPS only and 1 by saliva only. However, the GeneXpert platform has limited product availability and is also not suitable to high volume testing.
In our study, a significant finding was that a third or more of saliva samples from sick outpatients were thick, stringy, and difficult to pipet, requiring sputasol treatment. These obstacles greatly slowed processing, and risked contamination of the workspace during pipetting. For high volume testing, liquid samples that are readily aspirated by robotic instruments are essential to an efficient workflow. While thick saliva may be more frequently encountered in potentially dehydrated symptomatic outpatients, from our experience it can also occur in about 10 % of asymptomatic volunteers. Three laboratories in the U.S. have now received EUA for saliva testing by SARS CoV-2 PCR, and use saliva collection devices commonly used for genetic testing. These devices facilitate collection of a standard sample volume by filling up to a line on the device, add a preservative to prevent viral RNA degradation during transport, and promote liquefaction of the sample to facilitate processing.
In summary, our prospective study in symptomatic outpatients found an overall sensitivity for SARS CoV-2 detection of 85.7 % for pure saliva when compared to simultaneously collected NPS. However, our study highlights the difficulty in using pure saliva for high volume testing and the need to optimize saliva collection and processing. Thus, studies are currently in progress to evaluate saliva collection devices on ease of pipetting using liquid handlers and sensitivity of detection.
Research funding
No research support was received for this work. The work was performed as part of routine clinical laboratory operations.
CRediT authorship contribution statement
Marie L. Landry: Conceptualization, Methodology, Validation, Resources, Data curation, Writing - original draft, Writing - review & editing, Supervision, Project administration. Jody Criscuolo: Investigation, Validation, Data curation. David R. Peaper: Writing - review & editing, Formal analysis.
Declaration of Competing Interest
The authors have no conflicts to declare.
Acknowledgements
The authors would like to thank Ourania Ouzounidis for organizing sample collection and Alicia Cimino, Sandra Cohen and the Clinical Virology Laboratory staff for processing and testing the samples for this study.
References
- 1.Hanson K.E., Caliendo A.M., Arias C.A. Infectious diseases society of America guidelines on the diagnosis of COVID-19 [published online ahead of print, 2020 Jun 16] Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa760. ciaa760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams E., Bond K., Zhang B., Putland M., Williamson D.A. Saliva as a non-invasive specimen for detection of SARS-CoV-2 [published online ahead of print, 2020 Apr 21] J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00776-20. JCM.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azzi L., Carcano G., Gianfagna F. Saliva is a reliable tool to detect SARS-CoV-2. J. Infect. 2020;81(1):e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azzi L., Carcano G., Dalla Gasperina D., Sessa F., Maurino V., Baj A. Two cases of COVID-19 with positive salivary and negative pharyngeal or respiratory swabs at hospital discharge: a rising concern [published online ahead of print, 2020 Apr 25] Oral Dis. 2020 doi: 10.1111/odi.13368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyllie A.L., Fournier J., Casanovas-Massana A., Campbell M., Tokuyama M., Vijayakumar P. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. medRxiv. 2020;(April (22)) doi: 10.1101/2020.04.16.20067835. [DOI] [Google Scholar]
- 7.McCormick-Baw C., Morgan K., Gaffney D. Saliva as an alternate specimen source for detection of SARS-CoV-2 in symptomatic patients using Cepheid Xpert Xpress SARS-CoV-2 [published online ahead of print, 2020 May 15] J. Clin. Microbiol. 2020 doi: 10.1128/JCM.01109-20. JCM.01109-01120. [DOI] [PMC free article] [PubMed] [Google Scholar]