Abstract
The MosaiQ® COVID-19 Antibody test fulfills the minimal requirements for serological testing according to the French regulation.
Keywords: Serology, COVID-19, SARS-CoV-2
1. Background
Antibody testing may be useful in the management of patients. SARS-CoV-2-specific antibodies (Abs) can be detected in serum of approximately 40 % of COVID-19 patients as early as seven days after the onset of symptoms, with seroconversion rates rapidly increasing to >90 % by day 14 [1]. In recent studies, antibody testing has been shown to be more sensitive than viral nucleic acid detection after approximately eight days of COVID-19 illness duration [2]. While the combination of PCR and antibody tests is optimal for accurate diagnosis [3], antibody detection will be particularly relevant for the later stages of infection where the virus has been eliminated [4]. In addition to the diagnostic value, antibody testing enables identification of individuals who developed immunity after infection that may protect against subsequent re-infection [5], as well as define and monitor the extent of virus spread and a population’s herd immunity on a societal level. To date, the French Authorities have identified two situations in which serological testing may be prescribed: (i) when the diagnosis is suspected but PCR failed to confirm or was not available and (ii) for epidemiological purposes in support of developing Public Health Policy[6]. This authority also gathers and publishes steps to validate current serological testing platforms[7]. Here, we describe the evaluation of a microarray-based serological assay for the detection of Abs against SARS-CoV-2. In accordance with the French guidelines of assay validation, we report the validation of a previously unreleased CE-marked commercial platform
2. Materials and methods
2.1. Study design and serum samples
We conducted a retrospective study evaluating the sensitivity and specificity of a proprietary microarray-based assay for the detection of Abs specific to the SARS-CoV-2 virus. Serum samples used in this study were obtained from the Blood Donation Screening Laboratory, from the Medical Laboratory of the Military Medical Centers Percy (Clamart, France), Bégin (Saint-Mandé, France) and Laveran (Marseille, France) and from the Military Biomedical Research Institute (Marseille, France).
Repeatability and reproducibility were assessed on 3 quality controls levels (high, low and negative). Those quality controls were obtained from plasma collected for convalescent plasma therapy purposes which were tested by a licensed and validated assay (SARS-CoV-2 IgG/A assay, Euroimmun AG, Lübeck, Germany, positive threshold > 1.1). Repeatability was assessed by testing the same samples repeatedly during the same day (n = 30, each level). Reproducibility was assessed by testing quality control levels, twice a day, for 4–5 days (n = 8–9, each). As the Quotient assay is a qualitative assay, a surrogate marker was calculated to address coefficient of variation. This index was blindly calculated by Quotient engineer and provided to the investigator.
Accuracy was determined using 68 serum samples tested by the Euroimmun assay and neutralization (see below). Positive threshold was compared with manufacturer’s threshold using 100 samples from archived anonymous sera obtained from healthy donors in March 2019. Cross-reactivity assessment was performed using control sera obtained from patients positive for IgG and IgM against Dengue virus (n = 5) and Chikungunya virus (n = 5), for HBsAg or anti−HCV (n = 5), rheumatoid factor (n = 5), monoclonal proteins (n = 10), Abs against malaria (n = 10), Abs against syphilis (n = 10), IgG and IgM against EBV (n = 4) and IgG against CMV (n = 5).
Clinical specificity was assessed using archived serum samples from healthy donors, obtained in March 2019 (n = 500). Finally, clinical sensitivity was assessed using case serum obtained from COVID-19 patients (n = 101) admitted to the Military Medical Center Percy, France. SARS-CoV-2 infection was confirmed by PCR in samples from the respiratory tract according to French guidelines [8]. Date of symptoms onset was assessed by the physician in charge of the patients and recorded in the medical file.
2.2. Immunoassays
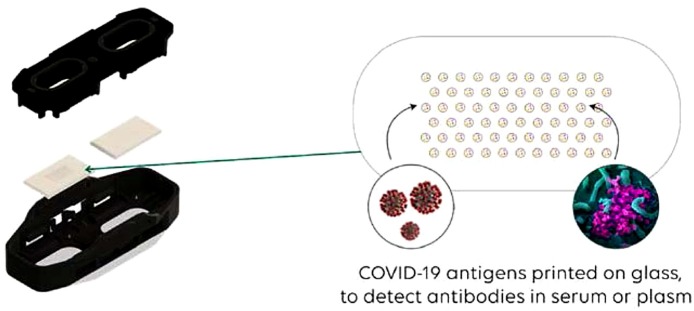
The MosaiQ® COVID-19 Antibody Microarray is used in combination with the MosaiQ® System for the automated determination of the presence of Abs against SARS-CoV-2 antigens in whole blood, plasma or serum specimens. It is a serological disease screening assay that enables the detection of Abs directed to the spike protein specific to SARS-CoV-2, on preprinted single use microarrays containing the antigen probes. The MosaiQ® test combines antigen-antibody interactions with automated image capture and analysis of the reaction. Each microarray is composed of the following panels: COVID-19 panel for the qualitative determination of human Abs against SARS-CoV-2 and empty panel without any printed probes (Fig. 1 ). The use of a magazine assembly containing the microarrays in association with the MosaiQ® instrument supports the high throughput multiplex screening of samples within a clinical laboratory setting.
Fig. 1.
MosaiQ COVID-19 micro-array.
Blood specimen are dispensed into the COVID-19 panel containing the printed antigens and controls. An incubation step allows Abs to bind the printed antigens and is followed by removal of unbound Abs using a wash buffer, which is then followed by addition of a detection reagent containing gold-conjugated secondary antibody formulation that binds to human IgG and IgM. Finally, the addition of enhancement reagent enables silver to nucleate on the gold nanoparticles, and subsequent washing with purified water reveals an optically detectable probe ready for imaging and analysis by the instrument.
2.3. Microneutralization assay
Virus neutralization assay was performed upon sera after filtration. Sera were 2-fold serially diluted from 1:10 to 1:80 on 96 well plate. One hundred μL of diluted sera were added to 100 μL of virus suspension containing 100 TCID50 (human SARS-CoV-2 strain BavPat1/2020, European Virus Archive global), incubated during 1 h at 37 °C and then added on Vero cells (ATCC CCL-81, 1.3 × 105 cells/well). After 4 days of incubation, then cytopathogenic effects were observed under a light microscope. The serum neutralizing titer was calculated as the inverse of the highest dilution resulting in an infectious reduction of 50 %. All experiences were performed in a BSL-3 facility.
2.4. Statistical analyses
Results are presented as mean +/- standard deviation. Confidence interval 95 % were calculated to estimate percentage when required. Sensitivity (positive percent agreement) was defined as the detection of serological response in the proportion of patients correctly diagnosed with SARS-CoV-2 infections using nucleic acid detection of SARS-CoV-2 in respiratory samples. Specificity (negative percent agreement) was defined as the proportion of SARS-CoV-2 immune naïve study participants accurately identified as negative for COVID-19. The analytical accuracy of the ELISA assays was examined as compared to two assays considered as gold standard (seroneutralization assays and commercial assay). GraphPad Prism version 8.0.2 (GraphPad Software, San Diego, CA, USA) was used to analyze the data.
2.5. Protocol approval and patient consent
All blood donors (March 2019 and convalescent plasma donors) provided consent for the use of their samples for research purposes at the time of pre-donation medical interview. Samples were obtained from unused laboratory samples and were fully anonymized before experimentation. Patients were informed of results according to French regulation through medical book and hospital policy, and by printed laboratory results sheets.
3. Results
3.1. Repeatability and reproducibility
Quality control (QC) “level high” was a serum with a 160 titer of neutralizing Abs. Quality control “level low” was a serum with a 40 titer of neutralizing Abs. Negative quality control was a plasma without neutralizing Abs (< 20). All positive QC resulted repeatably positive and negative QC repeatably negative. Reproducibility assessed at 2 days, 4 days or 5 consecutive days showed concordant results. Data are presented in Table 1 .
Table 1.
Repeatability and reproducibility of high, low and negative quality control levels (COVID Index).
| Repeatability | Negative QC | Low QC | High QC |
|---|---|---|---|
| Number of values | 29 | 29 | 28 |
| Minimum | 025 | 100 | 176 |
| Maximum | 025 | 664 | 859 |
| Mean | 025 | 419 | 509 |
| Std. Deviation | 000 | 163 | 183 |
| Coefficient of variation | 0,00 % | 38,92 % | 35,97 % |
| Reproducibility | Negative QC | Low QC | High QC |
| Number of values | 9 | 9 | 8 |
| Minimum | 025 | 160 | 386 |
| Maximum | 025 | 557 | 623 |
| Mean | 025 | 313 | 469 |
| Std. Deviation | 000 | 116 | 090 |
| Coefficient of variation | 0,00 % | 37,14 % | 19,24 % |
3.2. Accuracy
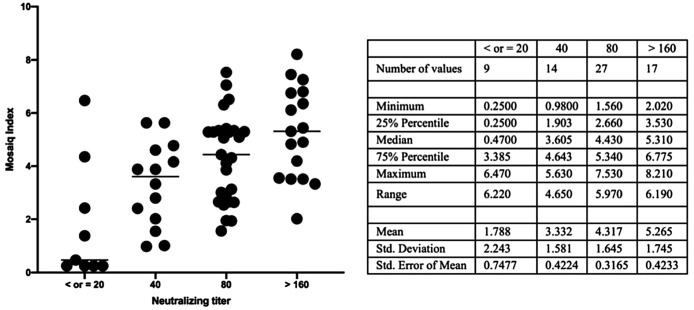
Serum (n = 68) tested with the 3 assays showed a good correlation. In comparison with the microneutralization assay, the MosaiQ® test revealed 3 discrepancies among the 7 negative expected samples (<20) and 2 among the 61 positive expected samples (one with a titer of 80 and one with a titer of 40). In comparison with the Euroimmun assay, the MosaiQ® test yielded 4 positive results among 8 expected negative samples: discrepancies showed that 3 samples had detectable neutralizing Abs as one exhibited titer under the threshold (<20). Among expected positive samples (n = 60), two samples were not correlated and founded negative with the MosaiQ® test: one exhibited neutralizing Abs at the threshold (20) and the other one was negative (<20). Finally, we addressed the correlation between MosaiQ® test index and seroneutralization titer and showed a significant correlation between titer and index with an increase of the index where the titer is higher (p < 0.001). Results are summarized in Table 2 and Fig. 2 .
Table 2.
Accuracy in comparison with EuroImmun and Seroneutralizing test.
| Seroneutralizing antibodies |
|||
|---|---|---|---|
| Positive | Negative | ||
| MosaiQ COVID-19 Antibody test | Positive | 59 | 3 |
| Negative | 2 | 4 | |
| Euroimmun | |||
| Positive | Negative | ||
| MosaiQ COVID-19 Antibody test | Positive | 58 | 4 |
| Negative | 2 | 4 | |
Fig. 2.
Correlation betweenCOVID-19 MosaiQ Index and seroneutralization titer.
3.3. Positive threshold
All samples were negative (n = 95), including one which was close to the index threshold (0.27) with a negative qualitative result, and 5 samples were flagged with a data reduction error (DRE) making the result unavailable. No medical information was available regarding the sample above the manufacturer detection threshold.
3.4. Cross-reactivity
Assay specificity was determined using a panel of 59 samples representing typical cross-reactivity situations. One sample, seropositive for Abs to Treponema pallidum (syphilis), showed a positive reaction with the COVID-19 antibody test. According to transfusion guidelines, this sample was twice repeated and found negative on both occasions and finally concluded as negative. All other samples were negative. Results are presented in Table 3 .
Table 3.
Cross-reactivity results for sera from controls diagnosed with conditions other than SARS-CoV-2 (n = 59).
| MosaiQ COVID-19 Antibody test |
|||
|---|---|---|---|
| Condition detected | # sera tested | Negative | Borderline |
| Dengue (IgG and IgM) | 5 | 5 | |
| Chikungunya (IgG and IgM) | 5 | 5 | |
| Hepatitis C virus | 3 | 3 | |
| HBs Antigen | 2 | 2 | |
| Malaria | 10 | 10 | |
| Syphilis | 10 | 9 | 1 |
| Cytomegalovirus (IgG) | 5 | 5 | |
| Epstein-Barr Virus (IgG and IgM) | 4 | 4 | |
| Rhumatoid factor | 5 | 5 | |
| Monoclonal Ig and free light chain | 10 | 10 | |
3.5. Clinical specificity
The clinical specificity refers to the probability to report a negative result among non−COVID-19 patients. All 500 sera reported negative, thereby demonstrating assay specificity of 100 % (95 % CI: 99–100).
3.6. Clinical sensitivity
The ability of the assay to detect IgG and IgM against SARS-CoV-2 in samples from patients with positive nasopharyngeal SARS-CoV-2 RT-PCR, performed in accordance with guidelines [8], was assessed on 101 patients samples. The overall sensitivity was 88 % (CI95: 82–94). However, taking into account the kinetics of seroconversion in infected patients, sensitivity was calculated according to time, in days, between the onset of symptoms and the sampling. Three groups of samples were therefore defined according to guidelines: samples taken fewer than 14 days post symptoms onset (PSO), between day 14 days and 20 days, and greater than 20 days. Among the 38 samples available from patients less than 14 days PSO, 27 were positive and 11 were negative. The average age and the average PSO were not significantly different between the two groups (75+/- 13 vs. 68+/-13 years and 9+/-2 vs. 10+/-3, p = 0.096 and 0.25, respectively). Sensitivity in this group was 71 % (95 % CI: 57–96). Among the 33 samples available from patients 14–20 days PSO, 32 were positive (average age: 64+/- 10 years, average PSO: 16+/-2 days). Only one patient showed no seroconversion: this patient suffers from severe hematological disorders and after a 30 day-long follow-up, did not exhibit seroconversion. Sensitivity in this group was 97 % (95 % CI: 91−100). However, discordance management took into account the persistently seronegative patient, the assay reported positive 100 % of samples with an immune response. Finally, for the 30 samples available from patients after 20 days PSO, all samples were positive (average age 60+/-15, average PSO 26+/-6 days), and therefore sensitivity in this group was 100 % (95 % CI: 90−100). Adding to the calculation, samples which were used to assess accuracy, where 60 samples were obtained from patients positive by RT-PCR and more than 3 weeks after the symptoms onset, then among the 90 available samples, 88 were positive and 2 remain negative: sensitivity increases to 98 % (95 % CI: 95–100). We then focused on hospitalized patients to assess sensitivity in the same time groups. Before 14 days PSO, sensitivity was 80 % (95 % CI: 60−100, n = 15), between day 14 and 20, 100 % (95 % CI: 86−100, n = 22) and 100 % (95 % CI:86−100, n = 21) after day 20. To compare our results to previously published results with high-throughput assays, we calculated sensitivity after day 14 PSO: sensitivity was 98 % (95 % CI: 96−100) and 100 % (95 % CI: 93−100) in non-severe/severe patients and severe patients, respectively (Table 4 ).
Table 4.
Clinical sensitivity in testing patient samples according to post symptoms onset and disease severity.
| Severe and non-severe patients (n = 101) | |||
|---|---|---|---|
| Day < 14 | D14−20 | D > 20 | |
| Positive | 27 | 32 | 30 |
| Negative | 11 | 1 | 0 |
| Sensitivity | |||
| % | 71 | 97 | 100 |
| 98 | |||
| 95 % CI | 57−86 | 91−100 | 90−100 |
| 96−100 | |||
| Severe patients (n = 58) | |||
| Day < 14 | D14−20 | D > 20 | |
| Positive | 12 | 22 | 21 |
| Negative | 3 | 0 | 0 |
| Sensitivity | |||
| % | 80 | 100 | 100 |
| 100 | |||
| 95 % CI | 60−100 | 86−100 | 86−100 |
| 93−100 | |||
4. Discussion
Serological assays and indications for use are well defined in the French guidelines relative to SARS−COV-2 antibody assays. Regardless of the indication, the sensitivity and specificity are key points to choose the most appropriate assay. Antibody response against SARS−COV-2 is incompletely known but the majority of Abs seem to be typically produced against the N-protein (which therefore might be the most sensitive target protein), whereas Abs produced against the S-protein are expected to be more specific and potentially neutralizing. S1-protein is considered as the most specific, in comparison with S2, which cross-react with antisera directed to SARS−COV-1.
The MosaiQ® test, enabling the qualitative detection of IgG and IgM against the Spike S1 protein, demonstrated a high specificity and clinical sensitivity. These data were assessed on well characterized samples: positive samples were obtained from PCR confirmed cases with a medical record of the date of symptomatic onset and negative samples obtained from blood donors. In terms of sensitivity, our data are at least as good as previously reported: before day 14 after the symptom’s onset, the assay detect SARS-CoV-2 IgG Abs in 71 % and even 80 % in hospitalized patients. The sensitivity increased to almost 100 % in samples from ≥15 days [9,10]. An important finding of our study is, that (with exception of 2 samples, which were also negative for anti-SARS-CoV-2 IgG with the EuroImmun assay) all detected SARS-CoV-2 IgG Abs in the analyzed cohort, using the commercially available assays examined, demonstrated neutralizing (potentially protective) properties in the microneutralization assay.
As compared with other high-throughput SARS-CoV-2 antibody assays, sensitivity of the MosaiQ® test was higher than the SARS-CoV-2 assays available from Abbott (overall sensitivity: 78 %, sensitivity after day 14 PSO: 94 %, 95 % CI: 83–99), Roche (overall sensitivity: 76 %, sensitivity after day 14 PSO: 85 %, 95 % CI: 77–98) or EuroImmun (overall sensitivity: 71 %, sensitivity after day 14 PSO: 97 %, 95 % CI: 90–98), as recently reported [[11], [12], [13]]. The specificity of the assay reports as 100 %, as none of the 500 samples archived in March 2019 were positive. This was superior as compared with the Abbott (99 %, 95 % CI: 96−100), Roche (99 % (95 %, CI: 96−100) or EuroImmun (95 %, 95 % CI: 90–98) SARS-CoV-2 assays [12,13]. However, our study does not include samples positive for other coronaviruses that could cross-react with the MosaiQ® test. It must be underlined that as no human case of SARS-CoV-1 infection was reported since 2003, and that anti-SARS-CoV-1 Abs are unlikely to sustain more than 6 years, cross-reaction with this virus appears of theoretical relevance [14]. Cross-reaction with others coronaviruses is limited since the MosaiQ® test is a Spike S1-based assay, which is sufficiently divergent in its amino acid sequence to enable effective use as a specific antigen and avoid cross-reactivity with seasonal coronaviruses, where homology between SARS-CoV-2 and SARS-CoV-1 virus is clearly evident in the N protein (91 %) [15].
One of the characteristics of the MosaiQ® test is its qualitative output. Where this makes little difference where used in the diagnosis of the previous or current infection, this may limit utility in surveying the longitudinal immunity in convalescent patients. We showed that the calculated index, though its weak repeatability and reproducibility and a coefficient of variation above 19 %, is significantly correlated with the neutralizing titer. Updated software may be required for further examination of this correlation and enable a formal examination of accuracy in a cohort of convalescent patients.
5. Conclusion
Altogether, our findings showed that the MosaiQ® COVID-19 Antibody test fulfills the minimal requirements for serological testing according to the French regulation: clinical specificity of the test is 100 %, thereby exceeding the requirement for >90 % in the French regulations. Moreover, this system enabled rapid throughput of samples, and the single-use microarray is of specific interest to manage high numbers of samples in a limited time with low levels of laboratory technician support.
References
- 1.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Fu Y., Ge S., Liu L., Zhang J., Xia N., Zhang Z. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Dela Cruz C.S., Wang Y., Wu C., Xiao Y., Zhang L., Han L., Dang S., Xu Y., Yang Q., Xu S., Zhu H., Xu Y., Jin Q., Sharma L., Wang L., Wang J. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020 doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., Yip C.C.-Y., Cai J.-P., Chan J.M.-C., Chik T.S.-H., Lau D.P.-L., Choi C.Y.-C., Chen L.-L., Chan W.-M., Chan K.-H., Ip J.D., Ng A.C.-K., Poon R.W.-S., Luo C.-T., Cheng V.C.-C., Chan J.F.-W., Hung I.F.-N., Chen Z., Chen H., Yuen K.-Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;0 doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.-L., Peiris M., Wu J. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao L., Deng W., Gao H., Xiao C., Liu J., Xue J., Lv Q., Liu J., Yu P., Xu Y., Qi F., Qu Y., Li F., Xiang Z., Yu H., Gong S., Liu M., Wang G., Wang S., Song Z., Zhao W., Han Y., Zhao L., Liu X., Wei Q., Qin C. Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. Microbiology. 2020 doi: 10.1101/2020.03.13.990226. [DOI] [Google Scholar]
- 6.Suzie D. 2020. Place des tests sérologiques rapides (TDR, TROD, autotests) dans la stratégie de prise en charge de la maladie COVID-19; p. 34. [Google Scholar]
- 7.Laëtitia L.G. 2020. Cahier des charges définissant les modalités d’évaluation des performances des tests sérologiques détectant les anticorps dirigés contre le SARS-CoV-2; p. 11. [Google Scholar]
- 8.Avis n°2020.0020/AC/SEAP du 6 mars 2020 du collège de la HAS relatif à l’inscription sur la LAP mentionnée à l’article L. 162-1-7 du CSS, de la détection du génome du coronavirus SARS-CoV-2 par technique de transcription inverse suivie d’une amplification, Haute Aut. Santé. (2020). https://www.has-sante.fr/jcms/p_3161218/fr/avis-n2020-0020/ac/seap-du-6-mars-2020-du-college-de-la-has-relatif-a-l-inscription-sur-la-lap-mentionnee-a-l-article-l-162-1-7-du-css-de-la-detection-du-genome-du-coronavirus-sars-cov-2-par-technique-de-transcription-inverse-suivie-d-une-amplification (accessed June 10, 2020).
- 9.Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S., Xiong Z., Zheng S. Evaluation of Nucleocapsid and Spike Protein-based ELISAs for detecting antibodies against SARS-CoV-2. MedRxiv. 2020;2020 doi: 10.1101/2020.03.16.20035014. 03.16.20035014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cassaniti I., Novazzi F., Giardina F., Salinaro F., Sachs M., Perlini S., Bruno R., Mojoli F., Baldanti F. Members of the San Matteo Pavia COVID-19 task force, performance of VivaDiag COVID-19 IgM/IgG rapid test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J. Med. Virol. 2020 doi: 10.1002/jmv.25800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohmer N., Westhaus S., Rühl C., Ciesek S., Rabenau H.F. Brief clinical evaluation of six high-throughput SARS-CoV-2 IgG antibody assays. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang M.S., Hock K.G., Logsdon N.M., Hayes J.E., Gronowski A.M., Anderson N.W., Farnsworth C.W. Clinical performance of two SARS-CoV-2 serologic assays. Clin. Chem. 2020 doi: 10.1093/clinchem/hvaa120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang M.S., Hock K.G., Logsdon N.M., Hayes J.E., Gronowski A.M., Anderson N.W., Farnsworth C.W. Clinical performance of the Roche sars-cov-2 serologic assay. Clin. Chem. 2020 doi: 10.1093/clinchem/hvaa132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okba N.M.A., Müller M.A., Li W., Wang C., GeurtsvanKessel C.H., Corman V.M., Lamers M.M., Sikkema R.S., de Bruin E., Chandler F.D., Yazdanpanah Y., Le Hingrat Q., Descamps D., Houhou-Fidouh N., Reusken C.B.E.M., Bosch B.-J., Drosten C., Koopmans M.P.G., Haagmans B.L. Severe acute respiratory syndrome Coronavirus 2-specific antibody responses in Coronavirus disease 2019 patients. Emerg. Infect. Dis. 2020;26 doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]