Abstract
Streptococcus thermophilus is widely used for producing fermented dairy products such as yogurt and cheese. Some S. thermophilus strains possessing the cell-wall protease PrtS show high proteolytic activity and fast acidification properties, which are very useful in industrial starters. However, few S. thermophilus strains possessing the prtS gene have been isolated from the environment. To clarify whether or not S. thermophilus strains possessing the prtS gene are present in Japan, we isolated S. thermophilus from raw milk collected in Japan from 2011 to 2017 and investigated the strains for the presence of prtS by PCR. A total of 172 S. thermophilus strains were isolated, and 59 strains were confirmed to possess prtS. We measured fermentation times of 59 prtS-positive strains in skim milk broth and found that 53 strains showed fast acidification properties, finishing fermentation within 10 hr. However, the remaining 6 prtS-positive strains showed slow acidification properties, and they had several amino acid mutations in PrtS compared with fast acidifying S. thermophilus LMD-9 and 4F44. These results demonstrate that S. thermophilus strains possessing prtS are prevalent in Japan and that some prtS-positive strains could lose their fast acidifying properties through mutations in PrtS.
Keywords: Streptococcus thermophilus, prtS, yogurt fermentation, mutation
INTRODUCTION
Yogurt is traditionally fermented by a symbiotic starter culture of Lactobacillus delbrueckii subsp. bulgaricus (L. bulgaricus) and Streptococcus thermophilus [1]. Both bacteria provide metabolites which promote mutual growth, as the nutrients in milk are not sufficient for their optimal growth [2, 3]. For example, L. bulgaricus produce peptides by degrading milk protein using the cell-wall protease PrtB, supporting the growth of weakly proteolytic S. thermophilus [4, 5]. However, a minority of S. thermophilus strains have been reported to show high proteolytic activity and fast acidification properties [6, 7]. Such S. thermophilus strains possess the prtS gene coding the cell-wall protease PrtS, which hydrolyzes casein into peptides [8]. Since S. thermophilus possessing prtS (prtS+) can acidify milk fast without the support of L. bulgaricus, prtS+ strains have a wide range of applications as industrial starters.
Despite its growth advantage in milk, only a small number of prtS+ strains have been reported so far. For example, only 21 among 135 S. thermophilus strains of the INRA historical collection were found to be prtS+ [9]. The first prtS+ strain was isolated from yogurt in Japan in 1971, and the second was isolated from fermented food in Mongolia in 1974 [7], suggesting the possibility that prtS+ strains have been prevalent in East Asia. However, to the best of our knowledge, no studies on the isolation of prtS+ strains from the environment have been carried out in these areas.
The aim of the present study was to clarify the prevalence of prtS+ strains in Japan, an island country in East Asia, by isolating S. thermophilus from raw milk collected from its 8 regions. Furthermore, we also measured the fermentation time and investigated the amino acid sequence of prtS in the isolated prtS+ strains to evaluate the effect of prtS on acidification properties.
MATERIALS AND METHODS
Isolation of Streptococcus thermophilus
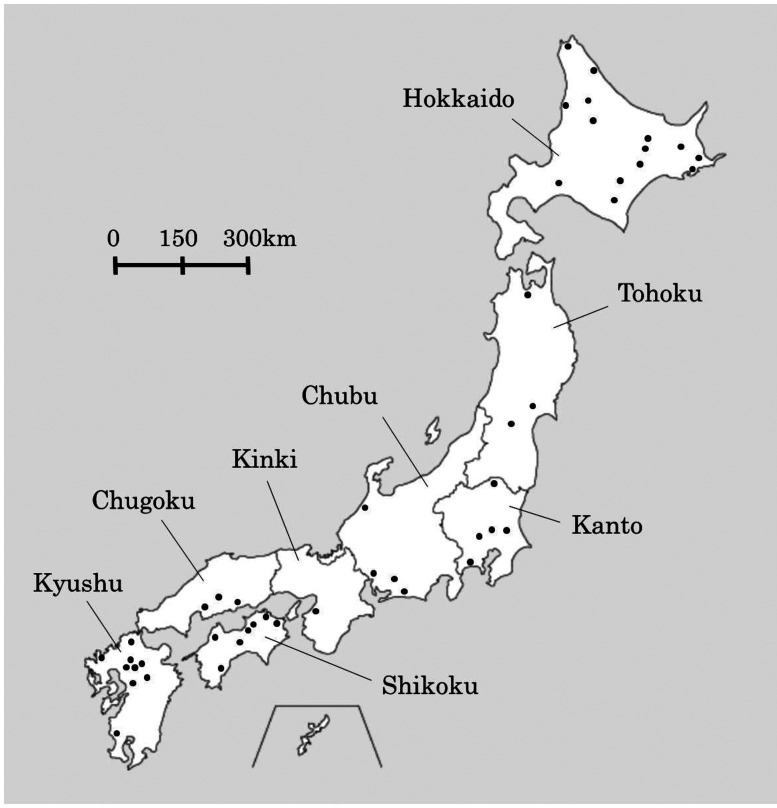
Raw milk samples were collected from 8 regions of Japan (Fig. 1) from 2011 to 2017, mainly from factories of Meiji Co., Ltd. (Tokyo, Japan). Collected samples were incubated at 48°C for 24 hr under anaerobic conditions. Each sample was diluted with sterile 0.85% (w/w) NaCl solution, and 0.1 mL aliquots were spread on milk agar plates containing 7% (w/w) skimmed milk powder (Meiji Co., Ltd., Tokyo, Japan), 1.9% (w/w) β-glycerophosphate disodium salt (Cayman Chemical, Ann Arbor, MI, USA), and 1.5% Bacto Agar (Becton Dickinson and Company, Franklin Lakes, NJ, USA). β-glycerophosphate disodium salt was added to prevent the growth of Lactobacillus species [10]. The plates were incubated anaerobically at 48°C for 24 hr. Obtained colonies were isolated by streaking on the abovementioned milk agar plates and incubated anaerobically at 48°C for 24 hr. Colonies were gram stained, and cocci-shaped gram-positive colonies were further incubated on BL agar plates (Nissui Pharmaceutical, Tokyo, Japan) anaerobically at 48°C for 24 hr. DNA was extracted from each colony using InstaGene Matrix (Bio-Rad, Hercules, CA, USA) and identified as S. thermophilus by a BLAST search of 16S rDNA and phosphoserine phosphatase gene (serB) sequences [11]. Then, RAPD (Random Amplification of Polymorphic DNA) PCR was carried out with strains isolated from the same raw milk sample using the primers RAPD1 (5′-CGTAAATTGC-3′) and RAPD2 (5′-CGTACATTGC-3′) to remove genetically identical strains. The primers used in this study were purchased from Eurofins Japan (Tokyo, Japan). The PCR reaction mixture was prepared as follows: 1 µL genomic DNA from each isolated S. thermophilus strain was mixed with 0.4 µL of TaKaRa Ex Taq (Takara Bio, Kusatsu, Shiga, Japan), 2.5 µL of 10×Ex Taq buffer, 2 µL of 2.5 mM dNTP, 0.4 µL of 100 µM primer, and 18.7 µL distilled water. The PCR reaction was performed using a Veriti Thermal Cycler (Thermo Fisher Scientific, Waltham, MA, USA) as follows: 4 cycles of 5 min at 94°C, 5 min at 36°C, and 5 min at 72°C followed by 30 cycles of 1 min at 94°C, 1 min at 36°C, and 2 min at 72°C. Amplified products were then resolved by electrophoresis using 1% agarose L03 gel (Takara Bio, Kusatsu, Shiga, Japan) containing 0.5 µg/mL ethidium bromide (Nippon Gene, Tokyo, Japan) and visualized under UV light by ChemiDoc (Bio-Rad, Hercules, CA, USA). Isolates with the same band pattern were regarded as the same strain, and only isolates showing different band patterns were selected for further investigation.
Fig. 1.
Raw milk sampling areas in Japan. The 8 regions of Japan are differentiated by black lines, and each black dot (•) represents a sampling location.
Analysis of prtS
Primers used to check for possession of prtS and to sequence the entire coding region of prtS are listed in Table 1. The primer set of prtS-1F and prtS-1R was used to amplify 684 bp inside prtS. The primer set of prtS-2F and prtS-2R was used to amplify the entire coding region of prtS, and the primers prtS-3, 4, 5, 6, 7, 8, and 9 were used for sequencing. The PCR reaction mixture was prepared as follows: 0.5 µL genomic DNA from each isolated S. thermophilus strain was mixed with 0.1 µL of Phusion High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA, USA), 2 µL of 5×HF buffer, 0.8 µL of 2.5 mM dNTP, 1 µL of 5 µM each primer, and 4.6 µL distilled water. The PCR reaction was performed using a Veriti Thermal Cycler (Thermo Fisher Scientific, Waltham, MA, USA) as follows: 30 sec at 98°C, followed by 30 cycles of 5 sec at 98°C, 20 sec at 63°C, and 3 min at 72°C, and then finally 5 min at 72°C. PCR products were resolved by electrophoresis, and strains showing an amplification product of 684 bp by using the prtS-1F and prtS-1R primers were judged to be S. thermophilus possessing prtS. For sequencing the entire coding region of prtS, PCR products were purified using ExoSAP-IT Express PCR Product Cleanup Reagent (Thermo Fisher Scientific, Waltham, MA, USA) and cycle sequenced by BigDye Terminator v1.1 Cycle Sequencing Kit (Thermo Fisher Scientific, Waltham, MA, USA) using the prtS-3 to 9 primers. Cycle-sequenced products were sequenced using an ABI 3130xl Genetic Analyzer (Applied Biosystems, Foster, CA, USA) according to the manufacturer’s instructions. Amino acid sequences were predicted and aligned using the GENETYX ver.14 software (Genetyx, Tokyo, Japan) to identify mutations.
Table 1. Oligonucleotide primers used in this study.
| Primer name | Sequene (5´-3´) |
|---|---|
| prtS-1F | GACTTGAAGAAACAGCGCGT |
| prtS-1R | TAGGTGGAGGCGTTACAGTG |
| prtS-2F | AAGGACGGAGCCATCATGAA |
| prtS-2R | CGTCCAGTGATGACTTTCCTC |
| prtS-3 | AGAAGAGGAGAAGCTTTCCGT |
| prtS-4 | ATGTGACTGCAGCAATCGAG |
| prtS-5 | TGGCGAGCTAAAACCAGACT |
| prtS-6 | TAGCCGTTCTAGAAGAGCCG |
| prtS-7 | GCAAGCGGTGTGACAACTAT |
| prtS-8 | GACTTGAAGAAACAGCGCGT |
| prtS-9 | ATGACCAGGCAGTTGAAGCA |
Measurement of acidification properties of the isolated strains
Each strain tested was anaerobically precultured twice at 37°C for 18 hr in skimmed milk powder broth containing 10% skimmed milk powder (Meiji Co., Ltd., Tokyo, Japan) and 0.1% yeast extract (Asahi Food and Healthcare, Tokyo, Japan) which was autoclaved at 121°C for 7 min. The preculture was inoculated at 2% (w/w) in 10% skimmed milk powder broth and incubated at 43°C. When fermented under peptide-rich conditions, 0.2% Hyvital Casein CMA 500 (FrieslandCampina, Amersfoort, The Netherlands) was added to 10% skimmed milk powder broth. During fermentation, pH was measured every 5 min using a multiple electrode measuring device (Horiba, Kyoto, Japan) with pH sensor SE 555 (Knick, Berlin, Germany). The time needed for the pH to decline to 4.7 was defined as the fermentation time.
RESULTS
Isolation of S. thermophilus from raw milk in Japan
From 2011 to 2017, we collected 313 raw milk samples from all regions of Japan, and a total of 172 colonies were identified as S. thermophilus. The number of isolated strains and their isolated regions in each sampling year are summarized in Table 2. We obtained at least 1 strain of S. thermophilus from each region in Japan and, of note, over 100 strains of S. thermophilus from the Hokkaido region.
Table 2. Number of S. thermophilus strains isolated from raw milk.
| Sampling year | Numbef of raw milk samples | Number of isolated strains |
Total number of isolated strains | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hokkaido | Tohoku | Kanto | Chubu | Kinki | Chugoku | Shikoku | Kyushu | |||
| 2011 | 92 | 16 | 16 | |||||||
| 2012 | 88 | 4 | 6 | 12 | 3 | 1 | 1 | 3 | 7 | 37 |
| 2014 | 89 | 71 | 1 | 3 | 5 | 80 | ||||
| 2016 | 36 | 7 | 5 | 7 | 6 | 3 | 28 | |||
| 2017 | 8 | 11 | 11 | |||||||
| Total | 313 | 109 | 11 | 19 | 9 | 1 | 2 | 6 | 15 | 172 |
Detection of the S. thermophilus strains possessing prtS
To examine whether the 172 isolated S. thermophilus strains possessed prtS, extracted DNA from each strain was amplified by PCR using the prtS-1F and prtS-1R primers. Among the 172 strains, 59 strains showed one amplification product of 684 bp, and the remaining 113 strains showed no products, indicating that 59 strains were prtS+ and that 113 strains did not possess prtS (prtS-). The numbers and percentages of prtS+ strains isolated from each region of Japan are summarized in Table 3. The total percentage of prtS+ strains among the 172 isolated S. thermophilus strains was 34.3%.
Table 3. Number and percentage of prtS+ strains.
| Region | Number of isolated strains | Number of prtS+ strains | Percentage of prtS+ |
|---|---|---|---|
| Hokkaido | 109 | 31 | 28.4 |
| Tohoku | 11 | 6 | 54.5 |
| Kanto | 19 | 7 | 36.8 |
| Chubu | 9 | 5 | 55.6 |
| Kinki | 1 | 0 | 0.0 |
| Chugoku | 2 | 1 | 50.0 |
| Shikoku | 6 | 0 | 0.0 |
| Kyushu | 15 | 9 | 60.0 |
| Total | 172 | 59 | 34.3 |
Acidification properties of prtS+ strains
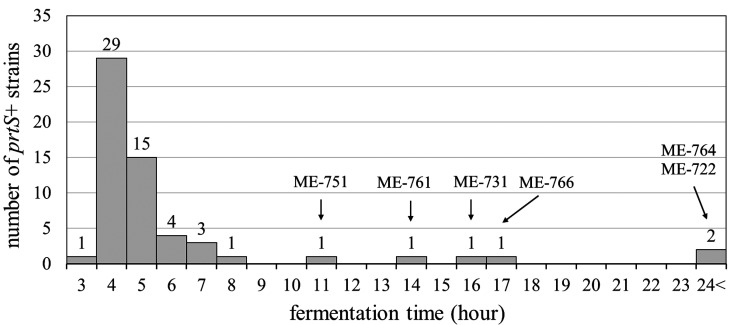
To examine the acidification properties of the 59 prtS+ strains, the fermentation time at 43°C in skimmed milk broth was measured. The number of prtS+ strains showing each fermentation time is shown in Fig. 2. Fifty-three prtS+ strains showed fast acidification properties, reaching pH4.7 within 10 hr. However, 6 prtS+ strains (ME-722, ME-731, ME-751, ME-761, ME-764, and ME-766) needed over 10 hr to reach pH4.7. We also measured the fermentation times of 11 prtS- strains isolated in this study and confirmed that all strains needed over 10 hr to reach pH4.7 (data not shown). These results demonstrated that the majority of prtS+ strains in Japan were able to acidify milk fast but that a few prtS+ strains showed slow acidification properties like prtS- strains.
Fig. 2.
Number of prtS+ strains showing each fermentation time. The names of the 6 strains which needed over 10 hr to reach pH4.7 are described above the bars.
Sequencing of the prtS of slow acidifying prtS+ strains
To investigate the factors affecting the slow acidification properties of the 6 prtS+ strains, amino acid sequences of PrtS were predicted using sequence data of prtS and compared with those of two fast acidifying prtS+ strains, S. thermophilus LMD-9 and S. thermophilus 4F44 [12]. Amino acid sequences of the 6 prtS+ strains that differed from those of both LMD-9 and 4F44 are listed in Table 4. The presence of a stop codon was confirmed for 4 strains, 3 of which (ME-751, ME-761, and ME-764) had a frameshift mutation and 1 of which (ME-766) had a nonsense mutation. The remaining 2 strains (ME-722 and ME-731) had several missense mutations. These results indicated that slow acidifying prtS+ strains had several mutations in PrtS compared with fast acidifying prtS+ strains, which might result in the loss of protease activity of PrtS.
Table 4. Mutation of PrtS amino acid sequences in slow acidifying prtS+ strains.
| Slow acidifying prtS+ strains | Accession No. of prtS | Domain | Mutation type | Predicted amino acid change |
|---|---|---|---|---|
| ME-722 | LC514594 | H | misssense | Thr1140Ile, Leu1189Ser, Asp1191Glu, Ile1211Val, Ala1232Thr, |
| Glu1294Lys, Ala1342Thr, Tyr1380phe, Asn1424Gly, Val1475Ala | ||||
| W | misssense | Ile1533Lys, Glu1548Gly | ||
| AN | misssense | Lys1593Arg, Pro1598Leu, Ile1611Lys | ||
| ME-731 | LC514595 | PP | misssense | Leu38Val, Ala53Val, Gly59Ala |
| ME-751 | LC514596 | PP | misssense | Leu38Val, Ala53Val, Gly59Ala |
| PR | frameshift | Ala316Serfs*13 | ||
| ME-761 | LC514597 | PP | frameshift | Leu26Phefs*12 |
| ME-764 | LC514598 | SS | misssense | Val19Ile |
| A | frameshift | Thr650Asnfs*41 | ||
| ME-766 | LC514599 | A | misssense | Asn937Asp, Asp944Asn |
| A | nonsense | Trp958* |
Predicted PrtS amino acid sequences of 6 slow acidifying prtS+ strains (accession Nos. are listed in the table) were compared with those of the PrtS of S. thermophilus LMD-9 (accession No. ABJ66087) and 4F44 (accession No. ADB77872). Domain names are abbreviated as follows: PP: signal sequence; PR: catalytic domain; A: globular domain; H: helical domain; W: wall domain; AN: anchored domain [8].
Fermentation properties of slow acidifying prtS+ strains under milk peptide-rich conditions
To verify the hypothesis that the amino acid mutations in PrtS of slow acidifying prtS+ strains resulted in the loss of protease activity and the lower availability of peptides, the fermentation times of the 6 slow acidifying prtS+ strains were measured with the addition of casein peptide. As shown in Table 5, the fermentation times of all 6 strains were shortened by adding casein peptide to the medium. Of note, 4 strains (ME-751, ME-761, ME-764, and ME-766) finished fermentation within 10 hr, showing acidification properties like fast acidifying prtS+ strains. However, two strains (ME-722 and ME-731) needed over 10 hr even when casein peptide was added to the medium.
Table 5. Fermentation times of prtS+ strains with or without casein peptide.
| Slow acidifying prtS+ strains | Fermentation time to pH4.7 |
|
|---|---|---|
| No addition | Addition of casein peptide | |
| ME-722 | >24 hr | 15 hr 50 min |
| ME-731 | 16 hr 45 min | 12 hr 20 min |
| ME-751 | 11 hr 55 min | 3 hr 45 min |
| ME-761 | 14 hr 05 min | 3 hr 45 min |
| ME-764 | >24 hr | 5 hr 15 min |
| ME-766 | 17 hr 05 min | 9 hr 45 min |
DISCUSSION
The main purpose of this study was to isolate and clarify the prevalence of prtS+ strains in Japan and to characterize the acidification properties of each prtS+ strain. We isolated 172 S. thermophilus strains from 313 raw milk samples in Japan and revealed that 59 strains were prtS+. Among the 59 prtS+ strains, 53 showed fast acidification properties, whereas the remaining 6 strains needed over 10 hr to finish fermentation. These results demonstrated that prtS+ strains could be isolated from raw milk in Japan and that most of them had the ability to ferment milk fast without the support of other proteolytic bacteria.
The percentage of prtS+ strains isolated from 2011 to 2017 in this study was 34.3% (Table 3). This value is higher than that in the INRA historical collection collected from 1956 to 2008 (15.6%) [9]. This is also higher than that in the report of Urshev et al., who demonstrated that only 4 strains were prtS+ among 20 samples of homemade yogurt, 8 industrial starters, and 80 culture strains collected from 1970 to 1997 [13]. Nowadays, prtS+ strains are widely used as commercial starters for fermented foods such as yogurt and cheese due to their fast acidification properties. To obtain fast acidifying prtS+ strains, Dandoy et al. proposed not only selection but also a natural transformation method in which a 15 kb prtS genomic island is transferred to a prtS− strain [14]. The higher percentage of prtS+ strains in this study than in the previous reports could be attributed to the sampling years (2011–2017), as selection and creation of prtS+ strains for industrial starters would have been carried out after the first characterization of prtS+ strains in 1991 [6]. It is possible that selected or created prtS+ strains somehow made their way into the natural environment and have been increasing due to their fast-growth properties. Moreover, it could be suggested that some prtS− strains acquired the prtS genomic island from prtS+ strains through naturally occurring horizontal gene transfer, since the prtS genomic island is flanked by two tandem sequences of IS elements [9]. Continuous isolation of S. thermophilus from the environment and examination for possession of the prtS gene would give us important information about the spread of prtS+ strains.
Although 53 prtS+ strains isolated in this study finished fermentation within 10 hr, the fermentation times varied among the strains (Fig. 2). Galia et al. reported that two strains with the same allele of prtS showed different acidification rates in milk [12] and confirmed that the expression of prtS was higher in the fast acidifying strain than in the slow acidifying strain [15]. They also reported that the expression levels of other genes known to be involved in carbon and nitrogen metabolism were also high in the fast acidifying strain. It can thus be suggested that the variety of acidification properties among the 53 prtS+ strains originated from not only the sequence variety of prtS but also differences at the transcriptional level of prtS and/or other genes involved in carbon and nitrogen metabolism. Though we acquired prtS sequence data of slow acidifying prtS+ strains, analysis of fast acidifying prtS+ strains will be needed to comprehend the differences in acidification properties among strains.
Among the 59 prtS+ strains, 6 strains showed slow acidification properties and needed over 10 hr to finish fermentation (Fig. 2), and they had several amino acid mutations in PrtS (Table 4). The acidification properties were recovered by the addition of casein peptides in the strains with a nonsense mutation (ME-751, ME-761, ME-764, and ME-766). Fernandez-Espla et al. demonstrated that PR (catalytic domain), followed by A (globular domain), was the best conserved domain between PrtS and other cell-wall proteases of lactic acid bacteria [8]. All 4 strains with a nonsense mutation had a stop codon in PR or A, indicating that defects in these domains led to the dysfunction of PrtS. However, peptide addition was not so effective for 2 strains with missense mutations (ME-722 and ME-731; Table 5). That is, strains with a nonsense mutation in PrtS resulted in lowered protease activity, but strains with missense mutations in PrtS probably had another factor affecting the fermentation properties. One possible factor affecting the fermentation properties of ME-722 and ME-731 might be functional defects in the peptide transporting process or peptide metabolizing process. Three oligopeptide-binding proteins involved in transporting oligopeptide [16] and 15 different peptidase activities [17] were reported in S. thermophilus, and these might affect the usage of an added peptide. Another possible explanation is that the metabolic pathways of other nutrients indispensable for growth are impaired in ME-722 and ME-731. Derzelle et al. demonstrated that enzymes involved in the synthesis of purines as well as enzymes involved in the supply of amino acids were upregulated in S. thermophilus grown in milk [18], which indicated that purines were also important nutrients for growth. It might be possible that lowered acidification properties of ME-722 and ME-731 were caused by factors other than peptide utility, such as glycolysis or purine metabolism.
Delorme et al. hypothesized that S. thermophilus acquired the prtS genomic island by horizontal gene transfer from other streptococcal strains [9]. It remains unclear how and when prtS+ strains in Japan acquired prtS and why some strains lost the proteolytic function of PrtS by mutation despite the fact that it would result in weak growth. We conducted a phylogenetic analysis of obtained 172 S. thermophilus strains by multilocus sequence analysis using 6 genes (ddlA, glcK, proA, ptsI, serB, tkt) by referring to a previous report [19], but there were no cluster correlations with the sampling area, possession of prtS, or sampling year (data not shown). We noticed that not only prtS− but also prtS+ strains were isolated together with proteolytic lactic acid bacteria such as Lactococcus lactis subsp. lactis and Lactobacillus helveticus, indicating that peptide availability might not be the key factor for acquiring prtS. Bassi et al. reported another function of PrtS, that is, a function which mediates the adhesion of S. thermophilus to surfaces conditioned by milk and the involvement in biofilm formation in the dairy environment [20]. To obtain deep insight into environmental conditions affecting gain or loss of prtS, further research will be necessary in the future.
In conclusion, we demonstrated that fast acidifying S. thermophilus possessing prtS were prevalent in Japan. To the best of our knowledge, this is the first report on the isolation of prtS+ strains from the environment in Japan. Since fast acidifying S. thermophilus strains can be used as important industrial dairy starters, future work will be needed to examine detailed fermentation properties such as urease activity, production of EPS, and organic acids involved in flavor.
Acknowledgments
The authors are grateful to Chinami Mizoguchi, Mariko Takeda, and Saori Takahashi for isolation, identification, and storage of lactic acid bacteria. The authors are also thank Hanae Tsuchihashi for a valuable discussion.
REFERENCES
- 1.Moon NJ, Reinbold GW. 1976. Commensalism and competition in mixed cultures of Lactobacillus bulgaricus and Streptococcus thermophilus. J Milk Food Technol 39: 337–341. [Google Scholar]
- 2.Zourari A, Accolasa J, Desmazeaud MJ. 1992. Metabolism and biochemical characterisitics of yogurt bacteria. Lait 72: 1–34. [Google Scholar]
- 3.Sieuwerts S, de Bok FA, Hugenholtz J, van Hylckama Vlieg JE. 2008. Unraveling microbial interactions in food fermentations: from classical to genomics approaches. Appl Environ Microbiol 74: 4997–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajagopal S, Sandine W. 1990. Associative growth and proteolysis of Streptococcus thermophilus and Lactobacillus bulgaricus in skim milk. J Dairy Sci 73: 894–899. [Google Scholar]
- 5.Gilbert C, Atlan D, Blanc B, Portailer R, Germond JE, Lapierre L, Mollet B. 1996. A new cell surface proteinase: sequencing and analysis of the prtB gene from Lactobacillus delbruekii subsp. bulgaricus. J Bacteriol 178: 3059–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shahbal S, Hemme D, Desmazeaud M. 1991. High cell wall-associated proteinase activitiy of some Streptococcus thermophilus strains (H-strains) correlated with a high acidification rate in milk. Lait 71: 351–357. [Google Scholar]
- 7.Shahbal S, Hemme D, Renault P. 1993. Characterization of a cell envelope-associated proteinase activity from Streptococcus thermophilus H-strains. Appl Environ Microbiol 59: 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Espla MD, Garault P, Monnet V, Rul F. 2000. Streptococcus thermophilus cell wall-anchored proteinase: release, purification, and biochemical and genetic characterization. Appl Environ Microbiol 66: 4772–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delorme C, Bartholini C, Bolotine A, Ehrlich SD, Renault P. 2010. Emergence of a cell wall protease in the Streptococcus thermophilus population. Appl Environ Microbiol 76: 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shankar PA, Davies FL. 1977. Recent developments in yogurt starters: a note on the supression of Lactobacillus bulgaricus in media containing β-glycerophosphate and application of such media to selective isolation of Streptococcus thermophilus from yogurt. Int J Dairy Technol 30: 28–30. [Google Scholar]
- 11.El-Sharoud WM, Delorme C, Darwish MS, Renault P. 2012. Genotyping of Streptococcus thermophilus strains isolated from traditional Egyptian dairy products by sequence analysis of the phosphoserine phosphatase (serB) gene with phenotypic characterizations of the strains. J Appl Microbiol 112: 329–337. [DOI] [PubMed] [Google Scholar]
- 12.Galia W, Perrin C, Genay M, Dary A. 2009. Variability and molecular typing of Streptococcus thermophilus strains displaying different proteolytic and acidifying properties. Int Dairy J 19: 89–95. [Google Scholar]
- 13.Urshev Z, Ninova-Nikolova N, Ishlimova D, Pashova-Baltova K, Michaylova M, Savova T. 2014. Selection and characterization of naturally occurring high acidification rate Streptococcus thermophilus strains. Biotechnol Biotechnol Equip 28: 899–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dandoy D, Fremaux C, de Frahan MH, Horvath P, Boyaval P, Hols P, Fontaine L. 2011. The fast milk acidifying phenotype of Streptococcus thermophilus can be acquired by natural transformation of the genomic island encoding the cell-envelope proteinase PrtS. Microb Cell Fact 10 Suppl 1: S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galia W, Jameh N, Perrin C, Genay M, Dary-Mourot A. 2016. Aquisition of PrtS in Streptococcus thermophilus is not enough in certain strains to achieve rapid milk acidification. Dairy Sci Technol 96: 623–636. [Google Scholar]
- 16.Garault P, Le Bars D, Besset C, Monnet V. 2002. Three oligopeptide-binding proteins are involved in the oligopeptide transport of Streptococcus thermophilus. J Biol Chem 277: 32–39. [DOI] [PubMed] [Google Scholar]
- 17.Rul F, Monnet V. 1997. Presence of additional peptidases in Streptococcus thermophilus CNRZ 302 compared to Lactococcus lactis. J Appl Microbiol 82: 695–704. [DOI] [PubMed] [Google Scholar]
- 18.Derzelle S, Bolotin A, Mistou MY, Rul F. 2005. Proteome analysis of Streptococcus thermophilus grown in milk reveals pyruvate formate-lyase as the major upregulated protein. Appl Environ Microbiol 71: 8597–8605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delorme C, Legravet N, Jamet E, Hoarau C, Alexandre B, El-Sharoud WM, Darwish MS, Renault P. 2017. Study of Streptococcus thermophilus population on a world-wide and historical collection by a new MLST scheme. Int J Food Microbiol 242: 70–81. [DOI] [PubMed] [Google Scholar]
- 20.Bassi D, Cappa F, Gazzola S, Orrù L, Cocconcelli PS. 2017. Biofilm formation on stainless steel by Streptococcus thermophilus UC8547 in milk environments is mediated by the proteinase PrtS. Appl Environ Microbiol 83: e02840-16. [DOI] [PMC free article] [PubMed] [Google Scholar]