Abstract
The aim of this study was to verify the effect of treatment with isoxanthohumol (IX) on the metabolomics profile of mouse feces to explore the host-intestinal bacterial interactions at the molecular level. The fecal contents of several amino acids in the high-fat diet (HFD) + 0.1% IX group (treated with IX mixed in diets for 12 weeks) were significantly lower than in the HFD group. The fecal contents of the secondary bile acid deoxycholic acid (DCA) in the HFD + 180 mg/kg IX group (orally treated with IX for 8 weeks) were significantly lower than in the HFD group; the values in the HFD group were higher than those in the normal diet (ND) group. Administration of IX changed the fecal metabolomics profile. For some metabolites, IX normalized HFD-induced fluctuations.
Keywords: metabolomics, amino acids, bile acids, feces, isoxanthohumol
INTRODUCTION
Intestinal flora, which comprise 100 trillion microorganisms, are closely associated with health and disease [1]. Progression of inflammatory bowel disease, colon cancer, and metabolic syndrome, including obesity and diabetes, has been reported to be related to a change in the intestinal microbiome in humans [2,3,4,5,6]. A genomics approach, such as 16S rRNA gene sequencing technology, can reveal the composition of colonic bacteria from feces, implying an association between the abundance of certain bacteria and healthy status. For example, obese subjects exhibit a higher ratio of Firmicutes to Bacteroidetes compared with nonobese subjects, which inversely decreases with weight loss via a low-calorie diet [5]. The abundance of Fusobacterium nucleatum is related to the development of colorectal cancer [7]. However, the molecular mechanism of the interaction between intestinal bacteria and the host is unclear.
More recently, thanks to the progress in analytic technology, a metabolomics approach has allowed for the simultaneous determination and quantification of various low-molecular-weight compounds that intestinal bacteria produce or degrade [8]. Short-chain fatty acids (SCFAs), including butyric acid, propionic acid, and acetic acid, are typical metabolites produced in colonic fermentation [9]. SCFAs enhance energy harvest via activation of G protein-coupled receptors expressed in intestinal epithelial cells [10]. They are also involved in inter- and intra-cellular pH regulation and act as energy sources for colonic epithelium cells [11]. Although it is too complicated to associate each metabolite with a specific intestinal bacterium, Daniel et al. reported that the bacterial abundances of the Lachnospiraceae and Ruminococcaceae families showed a positive correlation with the production of SCFAs [12]. Because of their beneficial effect on the regulation of metabolic health, including glucose homeostasis and obesity, it is important to consider strategies to generate SCFAs [13].
Bile acids are other typical low-molecular-weight compounds affected by intestinal bacteria. Some intestinal bacteria like Clostridium cluster XI and XIVa transform primary bile acids into secondary bile acids via 7-α-dehydroxylase activity [14,15,16,17]. In humans, cholic acid (CA) and chenodeoxycholic acid (CDCA) are the primary bile acids that exist in free or conjugated forms with glycine or taurine. In rodents, α/β-muricholic acid (MCA) is also a primary bile acid [18]. The primary bile acids CA, CDCA, and α/β-MCA are transformed into the secondary bile acids deoxycholic acid (DCA), lithocholic acid (LCA) and ursodeoxycholic acid (UDCA), and ω-MCA, respectively [19, 20]. DCA is cytotoxic and causes DNA damage [15]. It has been reported that intake of a high-fat diet (HFD) enhances the production of DCA and that enterohepatic circulation of DCA promotes obesity-associated hepatocellular carcinoma via secretion of various inflammatory and tumor-promoting factors [21]. Secondary bile acids also have adverse effects on the metabolism of glucose and lipids [22, 23]. Kuno et al. revealed that serum glucose and triglyceride levels were improved by short-term treatment with nonabsorbable antibiotics, which reduced DCA and LCA levels in the liver. Moreover, supplementation of DCA and LCA worsened the metabolism of glucose and lipids [23]. Therefore, it is physiologically important to prevent the generation of such secondary bile acids in the gastrointestinal tract.
Intestinal bacteria also contribute to the metabolism of amino acids, most of which are derived from dietary proteins [24]. For example, Clostridia and Gammaproteobacteria bacteria have been reported to deaminate multiple amino acids [25]. One of the essential amino acids, tryptophan, is degraded to indole by Bacteroides and Enterobacteriaceae, which subsequently increases the expression of intestinal tight junction proteins and suppresses the expression of pro-inflammatory cytokines [26]. Acidic amino acids like aspartate and glutamate are ultimately converted to SCFAs by intestinal bacteria via complicated reactions [26, 27]. Previously, dynamic alterations in fecal amino acids as well as the composition of bacteria were observed as a result of HFD-induced pre-obesity in rats [28]. Taken together, fluctuations in fecal amino acids could be an indicator of health status.
We have demonstrated that isoxanthohumol (IX), a hop-derived stable prenylflavonoid, had an anti-obesity effect in an HFD-induced obese mouse model, with microbial changes at both the phylum and genus levels (submitted for publication). Similarly, repeated administration of IX suppressed the progression of insulin resistance and maintained glucose metabolism [29], suggesting that it may be a functional ingredient for metabolic health. Although we have demonstrated that 8 weeks of administration of IX increased the relative abundance of Akkermansia muciniphila and decreased that of Clostridium cluster XI in mice, it is still unclear whether the contents of fecal metabolites dynamically change. Some food ingredients can influence the contents of fecal metabolites. For example, long-term intake of green tea extract by healthy postmenopausal females changed the fecal metabolites of aromatic amino acids (phenylacetylglutamine, hippuric acid, and indoxyl sulfate) [30]. Short-term intake of a grape seed extract increased the contents of fecal bile acids and sterols in pigs [31]. From these reports, we assume that IX may change the metabolomics profile in feces.
A metabolomics approach using capillary electrophoresis mass spectrometry with time-of-flight (CE-TOFMS) makes it possible to quantify charged low-molecular-weight compounds produced by intestinal microbiota with high resolution and high throughput, providing a detailed explanation for the host-intestinal bacterial interactions at the molecular level [32]. Combined with liquid chromatography mass spectrometry with time-of-flight (LC-TOFMS), which is adequate for relatively lipophilic compounds such as fatty acids and bile acids, various biological compounds can be determined and quantified [33]. Such a metabolomics approach can be used to elucidate the mechanism of functional foods. Azuma et al. used LC-TOFMS to show that oral administration of chitin nanofiber affects the metabolism of acyl-carnitines and fatty acids, confirming the effectiveness of the technology [33].
In the present study, we attempted to verify the possibility that IX changes the metabolomics profile in mouse feces. First, we assessed the fecal contents of metabolites in HFD-induced obese mice by taking advantage of CE-TOFMS. We quantified 3 major categories of compounds: amino acids, SCFAs, and nucleic acids. Second, we used ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC-QTOFMS) to quantify the absolute amount of fecal bile acids, which could be relevant to maintaining health and slowing the progression of diseases.
MATERIALS AND METHODS
Preparation of IX
The IX-enriched product named Isoxanthoflav (Hopsteiner, Mainburg, Germany) was used for the animal experiment in HFD mice treated with IX, mixed into diets, for 12 weeks. We confirmed that the purity of the IX was over 90% using an analytical standard (Tokyo Chemical Industry Co., Ltd., Tokyo, Japan).
Purified IX was used for the animal experiment in HFD mice treated with oral administration of IX for 8 weeks. The purified IX was prepared as follows: IX was isolated and purified from the hop extract using normal and reverse phase column chromatography. The final purity of the IX was confirmed by its ultraviolet absorbance at 280 nm via high-performance liquid chromatography, specifically a peak area of IX over 95% of the overall absorbance.
Animal experiments
Collection of fresh feces from HFD mice after 12 weeks of treatment with IX mixed into diets
Male C57BL/6 J mice (7 weeks of age) were purchased from Japan SLC, Inc. (Shizuoka, Japan). Throughout the experiment, the mice had free access to food and water and were housed at 25 ± 1°C and 60 ± 5% humidity under a 12 hr light-dark cycle. The mice were fed a normal diet (ND) (D12450JN, Research Diets, New Brunswick, NJ, USA) for 1 week. After that, they were randomly divided into 5 groups (n=10 per group) based on body weight and allocated to the following conditions: ND, HFD that consisted of 60% kcal from fat (D12492NM, Research Diets), HFD + 0.01% IX (HFD containing a final concentration of 0.01% IX), HFD + 0.03% IX (HFD containing a final concentration of 0.03% IX), and HFD + 0.1% IX (HFD containing a final concentration of 0.1% IX). Mice were allowed free access to food and water for 12 weeks, and then fresh feces were collected. The number of mice whose fresh feces were successfully collected within a short period was 10 per group. Mice were then euthanized under anesthesia with isoflurane. Fecal samples were collected separately, freeze-dried, stored at around −80°C until analysis, and then subjected to metabolite extraction and metabolomics analysis using CE-TOFMS (sections ‘Metabolite extraction from feces’ and ‘Metabolomics analysis using CE-TOFMS’ of MATERIALS AND METHODS). All experiments were conducted in 2017–2018. All protocols for the animal procedures were approved by the Ethics Committee for Animal Experiments in accordance with the Internal Regulations on Animal Experiments at Suntory and were based on the Law for the Humane Treatment and Management of Animals (Law No. 105, 1 October 1973, as amended on 2 June 2017).
Collection of fresh feces from HFD mice after 8 weeks of oral treatment with IX
Male C57BL/6 J mice (7 weeks of age) were purchased from CLEA Japan, Inc. (Tokyo, Japan). Throughout the experiment, mice were allowed access to food and water ad libitum and were maintained at 25 ± 1°C and 60 ± 5% humidity under a 12 hr light-dark cycle. Animals were fed an ND (D12450B, Research Diets) for 1 week. After that, the mice were randomly divided into 5 groups (n=8 per group) based on body weight and allocated to the following conditions: ND, HFD that consisted of 60% kcal from fat (D12492, Research Diets), HFD + 20 mg/kg IX, HFD + 60 mg/kg IX, or HFD + 180 mg/kg IX.
IX was diluted into 0.5 w/w% carboxymethyl cellulose sodium salt aqueous solution (Kanto Chemical Co., Inc., Tokyo, Japan) and orally given to C57BL/6 J mice once a day using a feeding needle. After 8 weeks of administration, fresh feces were collected. The number of mice whose fresh feces were successfully collected within a short period was 4–8 per group. Mice were then euthanized under anesthesia with isoflurane. Fecal samples were collected separately, freeze-dried, stored at around −80°C until analysis, and then subjected to bile acid extraction and quantification using UPLC-QTOFMS (sections ‘Bile acid extraction from feces’ and ‘Quantification of bile acids using UPLC-QTOFMS’ of MATERIALS AND METHODS). The experiments were conducted in 2017.
Metabolite extraction from feces
For CE-TOFMS measurements, ionic metabolites were extracted as follows. Approximately 50 mg of feces were dissolved in Milli-Q water with a ratio of 1:9 (w/v). After centrifugation, 80 µL of the supernatant was mixed with 20 µL of internal standard solution (H3304-1002, Human Metabolome Technologies, Inc. [HMT], Yamagata, Japan). The solution was then centrifugally filtered through a Millipore 5-kDa cutoff filter (UltrafreeMC-PLHCC, HMT) to remove macromolecules (9,100 × g, 4°C, 60 min) for subsequent analysis with CE-TOFMS. Metabolome measurements were carried out through a facility service at HMT.
Metabolomics analysis using CE-TOFMS
To determine and quantify the fecal contents of 20 amino acids, 2 SCFAs (butyric acid and propionic acid), and nucleic acids (nucleobases, nucleosides, nucleotides), metabolomics analysis was conducted by Dual Scan package (HMT) using CE-TOFMS based on previously described methods [34, 35]. Briefly, CE-TOFMS analysis was carried out using an Agilent CE capillary electrophoresis system equipped with an Agilent 6210 TOFMS, Agilent 1100 isocratic HPLC pump, Agilent G1603A CE-MS adapter kit, and Agilent G1607A CE-ESI-MS sprayer kit (Agilent Technologies, Waldbronn, Germany). The systems were controlled by the Agilent G2201AA ChemStation software, version B.03.01, for CE (Agilent Technologies) and connected by a fused silica capillary (50 μm i.d. × 80 cm total length) with a commercial electrophoresis buffer (H3301-1001 and H3302-1021 for cation and anion analyses, respectively, HMT) as the electrolyte. The spectrometer was scanned from m/z 50 to 1,000 [34]. Peaks were extracted using the automatic integration software MasterHands (Keio University, Yamagata, Japan) to obtain peak information, including m/z, peak area, and migration time (MT) [36]. Signal peaks corresponding to isotopomers, adduct ions, and other product ions of known metabolites were excluded, and the remaining peaks were annotated according to the HMT metabolite database based on their m/z values with the MTs determined by TOFMS. Areas of the annotated peaks were then normalized based on internal standard levels and sample amounts to obtain relative levels of each metabolite.
Bile acid extraction from feces
For UPLC-QTOFMS measurement, bile acids were extracted from feces using a previously described method [37] with small modifications. Approximately 100 mg of feces were placed in a 2 mL tube with zirconia beads, suspended in 900 µL of 50 mM cold sodium acetate buffer (pH 5.6)/ethanol mixture (1:3, v/v), vortexed (5 m/sec, 45 sec) using FastPrep 24G (MP Biomedicals, Santa Ana, CA, USA), and heated at 80°C for 30 min. After centrifugation (18,400 × g, 10 min), the supernatant was diluted 4 times with Milli-Q and applied to a Bond Elute C 18 cartridge (500 mg/6 mL, Agilent Technologies). The cartridge was washed with 10% ethanol (5 mL) and then bile acids were eluted with ethanol (5 mL). The solvent was evaporated, and the residue was dissolved in 1 mL of 50% ethanol. The extracted solution was diluted with 50% ethanol including an internal standard and transferred to a vial after filtration using a 0.2 μm filter (Ultrafree-MC, MilliporeSigma, Burlington, MA, USA).
Quantification of bile acids using UPLC-QTOFMS
Quantification of bile acids was performed on a Waters Acquity UPLC system with an Acquity UPLC BEH C18 column (2.1 × 150 mm, pore size 1.7 μm; Waters, Milford, MA, USA) coupled with a Waters Xevo G2-S QTOF mass spectrometer with an electrospray ionization probe. The injection volume was 4 µL. Mobile phase A was water and mobile phase B was acetonitrile, both containing 0.1% formic acid. The flow rate was 0.5 mL/min. The column and autosampler temperatures were kept at 65°C and 10°C, respectively. The Waters Xevo G2-S QTOF was run in negative mode (scan 50–850 amu at a rate of 0.3 scans per second). The following instrument conditions were used: capillary, 0.5 kV; source temperature, 150°C; sampling cone, 20 V; cone gas, 100 L/hr; desolvation gas flow, 1,000 L/hr at 450°C. To ensure mass accuracy and reproducibility, leucine enkephalin was used as the reference lock mass (m/z 554.2615) with a lock-mass spray. Data analyses were performed using the TargetLynx software (Waters).
Statistical analysis
All data are presented as means ± SE. IBM SPSS Statistics version 23 was used for statistical analysis (IBM, Armonk, NY, USA). Dunnett’s test was used for comparisons of more than 2 groups. Student’s t-test was used for comparisons between 2 independent groups (e.g., ND and HFD). Differences were considered significant at p<0.05.
RESULTS
Fecal amino acids from HFD mice after 12 weeks of treatment with IX mixed into diets
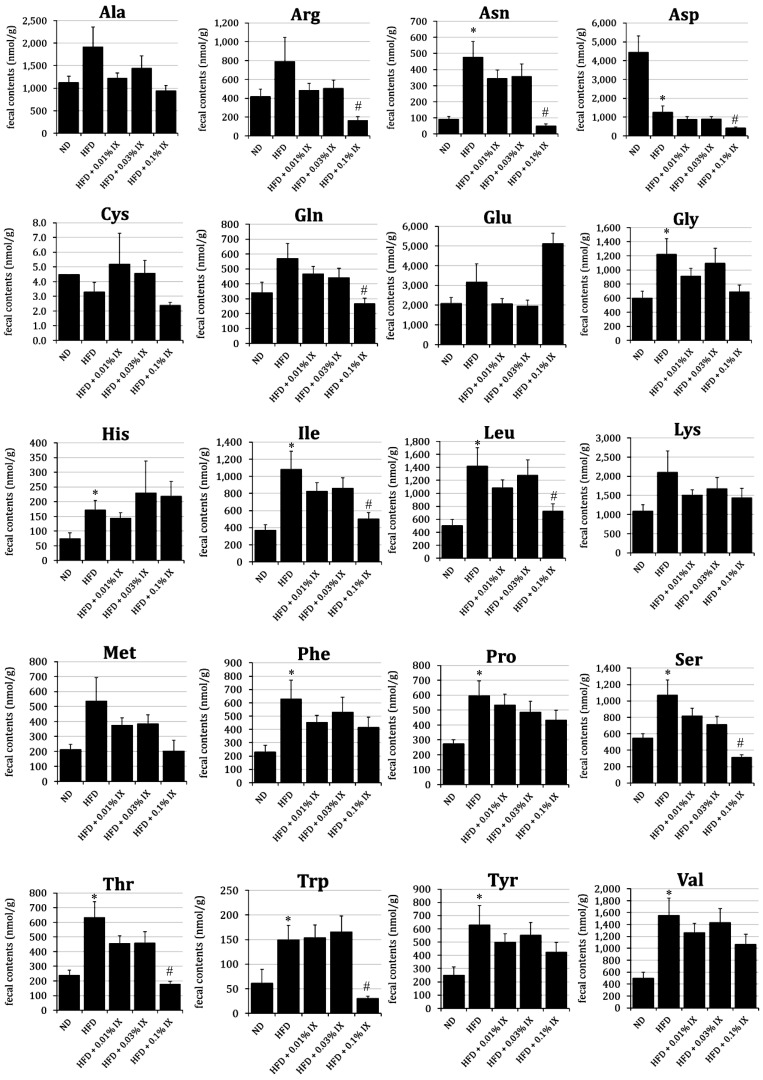
Fecal contents of Asparagine (Asn), Glycine (Gly), Histidine (His), Isoleucine (Ile), Leucine (Leu), Phenylalanine (Phe), Proline (Pro), Serine (Ser), Threonine (Thr), Tryptophan (Trp), Tyrosine (Tyr), and Valine (Val) were significantly higher in the HFD group than those in the ND group. Only the content of Aspartic acid (Asp) in the HFD was significantly lower than that in the ND. At the same time, the fecal contents of Arginine (Arg), Asn, Asp, Glutamine (Gln), Ile, Leu, Ser, Thr, and Trp in the HFD + 0.1% IX group were significantly lower than those in the HFD group (Fig. 1). There were no significant differences in any amino acids between the HFD and HFD + 0.01% IX groups or the HFD and HFD + 0.03% IX groups.
Fig. 1.
Profile of fecal amino acids from HFD mice after 12 weeks of treatment with IX mixed into diets.
Effect of the treatment with IX for 12 weeks on the profile of fecal amino acids in HFD-fed mice. Fecal contents of each amino acid are presented as means ± SE of 10 mice. *p<0.05 between the ND and HFD groups (Student’s t-test). #p<0.05 between the HFD and other groups (Dunnett’s test). IX: isoxanthohumol; ND: normal diet; HFD: high-fat diet; HFD + 0.01% IX: HFD containing 0.01% IX as final concentration; HFD + 0.03% IX: HFD containing 0.03% IX as final concentration; HFD + 0.1% IX: HFD containing 0.1% IX as final concentration.
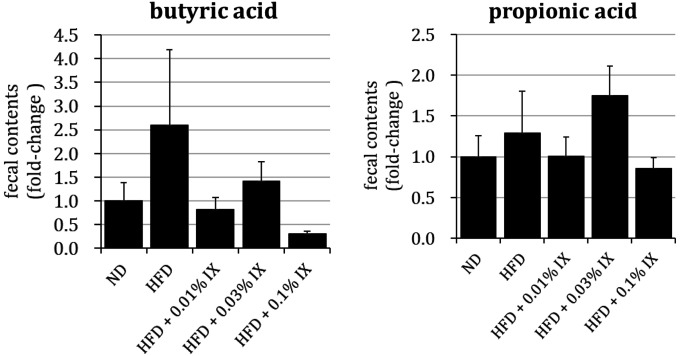
Fecal SCFAs (butyric acid and propionic acid) from HFD mice after 12 weeks of treatment with IX mixed in diets
There were no significant differences in the fecal contents of SCFAs (butyric acid and propionic acid) between the ND and HFD groups, HFD and HFD + 0.01% IX groups, HFD and HFD + 0.03% IX groups, or HFD and HFD + 0.1% IX groups (Fig. 2). The fecal contents of SCFAs were quantified and described as relative values divided by those of the ND group.
Fig. 2.
Profile of fecal SCFA (butyric acid and propionic acid) from HFD mice after 12 weeks of treatment with IX mixed into diets.
Effect of the treatment with IX for 12 weeks on the profile of SCFA in HFD-fed mice. Fecal contents of each SCFA are quantified and described as relative values divided by those of the ND group and presented as means ± SE of 10 mice. *p<0.05 between the ND and HFD groups (Student’ t-test). #p<0.05 between the HFD and other groups (Dunnett’s test). SCFA: short-chain fatty acid; IX: isoxanthohumol; ND: normal diet; HFD: high-fat diet; HFD + 0.01% IX: HFD containing a final concentration of 0.01% IX; HFD + 0.03% IX: HFD containing a final concentration of 0.03% IX; HFD + 0.1% IX: HFD containing a final concentration of 0.1% IX.
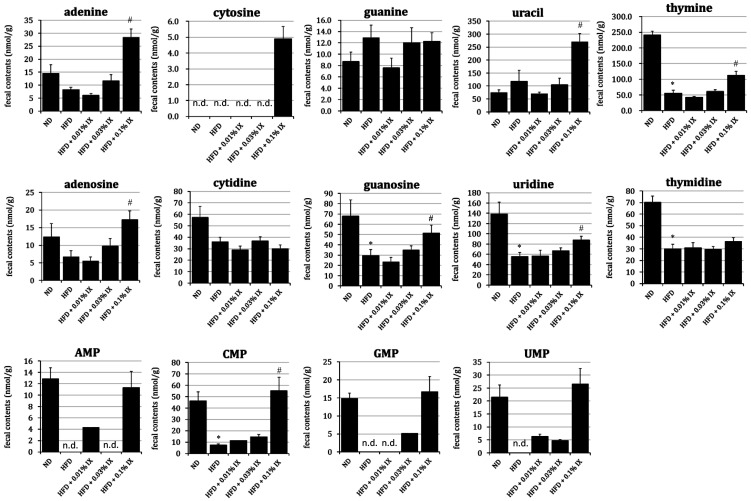
Fecal nucleic acids from HFD mice after 12 weeks of treatment with IX mixed into diets
The fecal contents thymine, guanosine, uridine, thymidine, and cytidine monophosphate (CMP) in the HFD group were significantly lower than those in the ND group. At the same time, the fecal contents of adenine, uracil, thymine, adenosine, guanosine, uridine, and CMP in the HFD + 0.1% IX group were significantly higher than those in the HFD group (Fig. 3). There were no significant differences in any nucleic acids between the HFD and HFD + 0.01% IX groups or the HFD and HFD + 0.03% IX groups.
Fig. 3.
Profile of fecal nucleic acids from HFD mice after 12 weeks of treatment with IX mixed into diets.
Effect of the treatment with IX for 12 weeks on the profile of nucleic acids in HFD-fed mice. Fecal contents of each nucleic acid are presented as means ± SE of 10 mice. *p<0.05 between the ND and HFD groups (Student’s t-test). #p<0.05 between the HFD and other groups (Dunnett’s test). n.d.: not detected in any mice; IX: isoxanthohumol; ND: normal diet; HFD: high-fat diet; HFD + 0.01% IX: HFD containing a final concentration of 0.01% IX; HFD + 0.03% IX: HFD containing a final concentration of 0.03% IX; HFD + 0.1% IX: HFD containing a final concentration of 0.1% IX; AMP: adenosine monophosphate; CMP: cytidine monophosphate; GMP: guanosine monophosphate; UMP: uridine monophosphate.
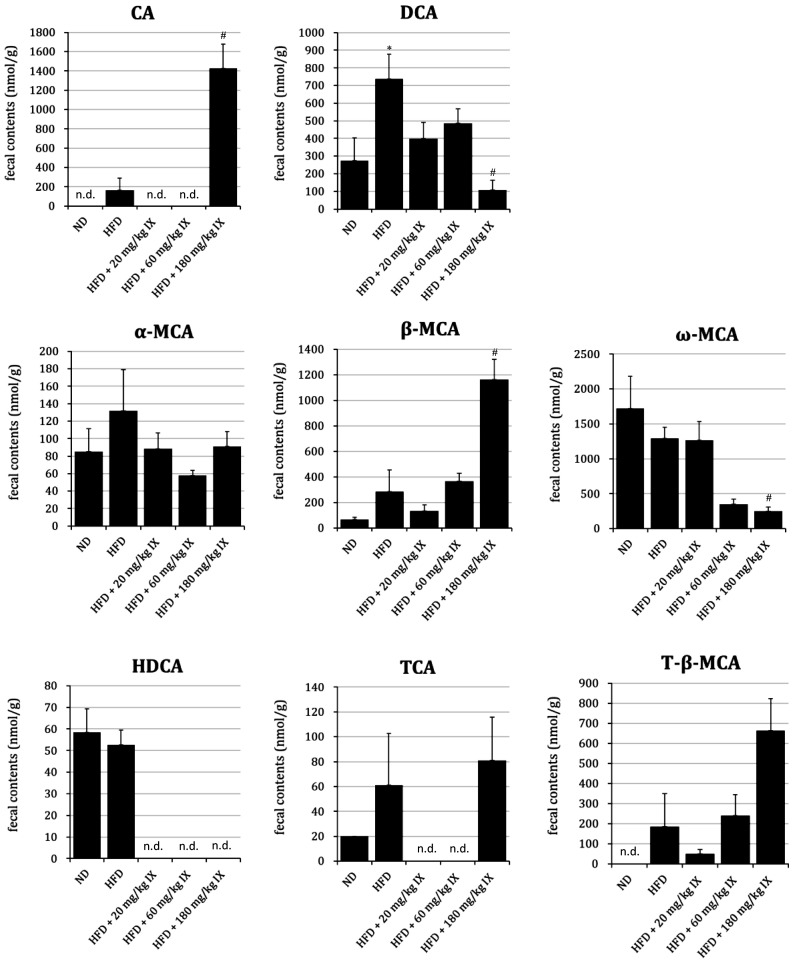
Fecal bile acids from HFD mice after 8 weeks of oral treatment with IX
In almost all mice in the ND, HFD, HFD + 20 mg/kg IX, and HFD + 60 mg/kg IX groups, the fecal contents of CA were under the limit of quantification (LOQ) of 40 nmol/g. In the HFD + 180 mg/kg IX group, the fecal contents of CA were significantly higher than the LOQ. The fecal contents of DCA in the HFD group were significantly higher than those in the ND group. At the same time, the fecal contents of DCA and ω-MCA in the HFD + 180 mg/kg IX group were significantly lower than those in the HFD group, whereas the fecal contents of β-MCA in the HFD + 180 mg/kg IX group were significantly higher than those in the HFD group (Fig. 4). Hyodeoxycholic acid (HDCA), a secondary bile acid transformed from β-MCA, was quantifiable in 4 of 6 mice in the HFD group; its contents were under the LOQ (40 nmol/g) for all mice in the HFD + 20 mg/kg IX, HFD + 60 mg/kg IX, and HFD + 180 mg/kg IX groups. The fecal contents of LCA and CDCA were under the LOQ for almost all mice. The mean ratios of DCA to DCA+CA (an index of 7-α-dehydroxylase activity) were 0.82 and 0.07 in the HFD and HFD + 180 mg/kg IX groups, respectively. When data were under the LOQ, they were replaced by the concentration at the LOQ.
Fig. 4.
Profile of fecal bile acids from HFD mice after 8 weeks of oral treatment with IX.
Effect of the oral administration of IX for 8 weeks on the profile of bile acids in HFD-fed mice. Fecal contents of each bile acid are presented as means ± SE of 4–8 mice whose feces were successfully collected. When data were under the limit of quantification (LOQ), they were replaced by the concentration at the LOQ. *p<0.05 between the ND and HFD groups (Student’s t-test). #p<0.05 between the HFD and other groups (Dunnett’s test). n.d.: not detected (under LOQ) in any mice; IX: isoxanthohumol; ND: normal diet; HFD: high-fat diet; CA: cholic acid; DCA: deoxycholic acid; α-MCA: α-muricholic acid; β-MCA: β-muricholic acid; ω-MCA: ω-muricholic acid; HDCA: hyodeoxycholic acid; TCA: taurocholic acid; T-β-MCA: tauro-β-muricholic acid.
DISCUSSION
Previously, we confirmed that long-term administration of IX changed the composition of intestinal bacteria, as well as that it suppressed the progression of insulin resistance (submitted for publication). In HFD-induced obese mice, administration of IX for 8 weeks increased the relative abundance of A. muciniphila, Blautia, and Bacteroides at the genus level (submitted for publication). In contrast, IX decreases the Clostridium cluster XI after treatment with IX for 12 weeks [29], which generates secondary bile acids [38]. However, it is still not clear how intestinal bacteria interact with the host at the molecular level. Therefore, in this study, we used a metabolomics approach to verify the possibility that IX could change the profile of fecal metabolites. Using CE-TOFMS and UPLC-QTOFMS, we quantified the contents of fecal metabolites in HFD-induced obese mice and compared them with those in ND mice.
In our analysis, the fecal contents of amino acids, including Asn, Gly, His, Ile, Leu, Phe, Pro, Ser, Thr, Trp, Tyr, and Val, were significantly higher in the HFD group than those in the ND group. Of the 20 amino acids that make up proteins, only Asp showed a significantly lower content in the HFD group than in the ND group. This finding indicates an HFD-induced fluctuation of fecal amino acids. When we focused on the effect of IX, the contents of Arg, Asn, Asp, Gln, Ile, Leu, Ser, Thr, and Trp in the HFD + 0.1% IX group were significantly lower than those in the HFD group. At least for several amino acids, IX normalized the HFD-induced changes. The decrease in the contents of these amino acids caused by IX can be partly explained by enhanced metabolism via certain intestinal bacteria, as intestinal bacteria contribute to the metabolism of amino acids [24,25,26,27]. For example, Matsumoto et al. compared the contents of colonic luminal metabolites between germ-free (GF) and ex-GF mice using CE-TOFMS and showed that 4 of the amino acids (Pro, Thr, Arg, and Asn) were more abundant in GF mice, while the other amino acids were almost equal between groups [32]. Their results imply the potential of the microbiome to change the contents of amino acids in the colon. In addition, we have observed a significant increase in the relative abundance of Bacteroides after treatment with IX (submitted for publication). The bacterium Bacteroides can metabolize such amino acids as Asn, Asp, Gln, Glu, Ser, and Thr [25]. Still, the effect of an HFD on the fecal contents of amino acids is controversial. A previous report showed that an HFD increased the contents of the amino acids Phe and Tyr compared with an ND [28], which is consistent with our findings. Those authors pointed out the potential involvement of Lachnospiraceae with metabolic functions associated with the aromatic amino acids Phe, Tyr, and Trp [28]. In contrast, an HFD-induced decrease in amino acids, including Ile, Thr, Val, Trp, and His, was observed in their study [28], which is inconsistent with our results. Although a metabolomics approach revealed dynamic changes in the fecal contents of amino acids, further study is needed to identify the bacterium responsible for the metabolism of each amino acid. All amino acids quantified were L-type amino acids. Therefore, D-type amino acids produced by microbiota were not quantified. This is a limitation of the present study.
In terms of SCFAs, we quantified the fecal contents of butyric acid and propionic acid as relative values divided by those of the ND group. No significant differences were observed between the HFD group and any of the groups treated with IX. There is some possibility that IX did not change the abundance of intestinal bacteria that produce SCFAs. Although A. muciniphila is reported to produce SCFAs as metabolites [39], no significant differences were observed in the present study. Because SCFAs are metabolites of multiple food ingredients, such as carbohydrates and amino acids [40, 41], the interpretation of fecal SCFA contents is complex.
For nucleic acids, different colonic contents between GF and ex-GF mice have been reported [30]. The colonic luminal contents of nucleobase adenine, guanine, cytosine, and uracil in ex-GF mice were significantly higher than those in GF mice, perhaps reflecting the existence of intestinal bacteria. In our study, the fecal contents of thymine, guanosine, uridine, thymidine, and CMP in the HFD group were lower than those in the ND group, suggesting HFD-induced fluctuations. At the same time, the HFD + 0.1% IX group exhibited higher amounts of adenine, uracil, thymine, adenosine, guanosine, uridine, and CMP than the HFD group. For some nucleic acids, IX normalized the HFD-induced fluctuations. Because there is limited information on the fecal contents of nucleic acids in disease models, we need to accumulate metabolomics data to determine the physiological meanings of our findings.
We have also confirmed the changes in the fecal contents of bile acids. Bile acids are derivatives of steroids and are ubiquitously contained in the bile of vertebrates [42]. After being endogenously synthesized from cytochrome P450-mediated cholesterol metabolism in the liver and excreted into the duodenum, bile acids are mixed with phospholipids such as phosphatidylcholine and cholesterols to form micelles. Micelles help with absorption of dietary fats and lipid-soluble nutrients such as vitamins [17]. Approximately 95% of bile acids are reabsorbed in the ileum and transported back into the liver. The rest are transformed into secondary bile acids by intestinal bacteria and excreted in the feces [16, 43]. In our study, we quantified the fecal contents of bile acids after 12 weeks of treatment with IX. This is because the relative abundance of the Clostridium cluster XI with 7-α-dehydroxylase activity was decreased under the same conditions in a previous study [29]. This time, the fecal contents of the secondary bile acids DCA and ω-MCA in the HFD + 180 mg/kg IX group were significantly lower than those in the HFD group. Similarly, HDCA, another secondary bile acid generated from β-MCA [44], was quantifiable only in the ND and HFD groups, suggesting a suppressive effect of IX on intestinal bacteria with 7-α-dehydroxylase. This hypothesis is reasonably explained by the amounts of their precursors, CA and β-MCA. The fecal contents of CA were much higher than the LOQ in the HFD + 180 mg/kg IX group, while they were almost negligible in other groups. Similarly, the fecal contents of β-MCA in the HFD + 180 mg/kg IX group were significantly higher than those in the HFD group. We have also confirmed that long-term treatment with IX decreased the relative abundance of Clostridium cluster XI with 7-α-dehydroxylase activity [29]. All things considered, IX has the potential to prevent the generation of secondary bile acids. From a physiological viewpoint, secondary bile acids like DCA worsen the metabolism of glucose and lipids [22, 23]. Therefore, IX may suppress the progression of glucose tolerance partly by inhibiting the generation of secondary bile acids in the gastrointestinal tract. As for conjugated bile acids, taurocholic acid (TCA) and tauro-β-muricholic acid (T-β-MCA) were detected. However, no significant differences in the fecal contents of these conjugated bile acids were observed between the HFD and other groups.
In conclusion, we demonstrated that administration of IX could change the contents of fecal metabolites, including amino acids, nucleic acids, and bile acids, in HFD-induced obese mice. For several amino acids, nucleic acids, and the secondary bile acid DCA, HFD-induced fluctuations were observed compared with mice fed the ND. Overall, administration of IX normalized these HFD-induced fluctuations. IX suppressed the generation of the secondary bile acid DCA, probably due to the change in intestinal bacteria composition, which could improve the metabolism of glucose and lipids. Although IX tended to normalize the HFD-induced fluctuations, there is much to be learned about the physiological meaning of IX-induced changes in the contents of each fecal metabolite.
REFERENCES
- 1.Guinane CM, Cotter PD. 2013. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Therap Adv Gastroenterol 6: 295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tannock GW. 2008. Molecular analysis of the intestinal microflora in IBD. Mucosal Immunol 1 Suppl 1: S15–S18. [DOI] [PubMed] [Google Scholar]
- 3.Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, Corthier G, Tran Van Nhieu J, Furet JP. 2011. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One 6: e16393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gagnière J, Raisch J, Veziant J, Barnich N, Bonnet R, Buc E, Bringer MA, Pezet D, Bonnet M. 2016. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol 22: 501–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444: 1022–1023. [DOI] [PubMed] [Google Scholar]
- 6.Upadhyaya S, Banerjee G. 2015. Type 2 diabetes and gut microbiome: at the intersection of known and unknown. Gut Microbes 6: 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M, Jenab M, Prehn JH, Hughes DJ. 2014. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis 33: 1381–1390. [DOI] [PubMed] [Google Scholar]
- 8.Soga T, Ueno Y, Naraoka H, Ohashi Y, Tomita M, Nishioka T. 2002. Simultaneous determination of anionic intermediates for Bacillus subtilis metabolic pathways by capillary electrophoresis electrospray ionization mass spectrometry. Anal Chem 74: 2233–2239. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Shimizu Y, Kimura I. 2017. Gut microbial metabolite short-chain fatty acids and obesity. Biosci Microbiota Food Health 36: 135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cani PD, Delzenne NM. 2011. The gut microbiome as therapeutic target. Pharmacol Ther 130: 202–212. [DOI] [PubMed] [Google Scholar]
- 11.Cook SI, Sellin JH. 1998. Review article: short chain fatty acids in health and disease. Aliment Pharmacol Ther 12: 499–507. [DOI] [PubMed] [Google Scholar]
- 12.Daniel H, Gholami AM, Berry D, Desmarchelier C, Hahne H, Loh G, Mondot S, Lepage P, Rothballer M, Walker A, Böhm C, Wenning M, Wagner M, Blaut M, Schmitt-Kopplin P, Kuster B, Haller D, Clavel T. 2014. High-fat diet alters gut microbiota physiology in mice. ISME J 8: 295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chambers ES, Preston T, Frost G, Morrison DJ. 2018. Role of gut microbiota-generated short-chain fatty acids in metabolic and cardiovascular health. Curr Nutr Rep 7: 198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. 2014. Bile acids and the gut microbiome. Curr Opin Gastroenterol 30: 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ridlon JM, Kang DJ, Hylemon PB. 2006. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47: 241–259. [DOI] [PubMed] [Google Scholar]
- 16.Ridlon JM, Harris SC, Bhowmik S, Kang DJ, Hylemon PB. 2016. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7: 22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li T, Chiang JY. 2014. Bile acid signaling in metabolic disease and drug therapy. Pharmacol Rev 66: 948–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mertens KL, Kalsbeek A, Soeters MR, Eggink HM. 2017. Bile acid signaling pathways from the enterohepatic circulation to the central nervous system. Front Neurosci 11: 617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudling M. 2016. Understanding mouse bile acid formation: Is it time to unwind why mice and rats make unique bile acids? J Lipid Res 57: 2097–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minato K, Suzuki M, Nagao H, Suzuki R, Ochiai H. 2015. Development of analytical method for simultaneous determination of five rodent unique bile acids in rat plasma using ultra-performance liquid chromatography coupled with time-of-flight mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 1002: 399–410. [DOI] [PubMed] [Google Scholar]
- 21.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, Honda K, Ishikawa Y, Hara E, Ohtani N. 2013. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499: 97–101. [DOI] [PubMed] [Google Scholar]
- 22.Kuno T, Hirayama-Kurogi M, Ito S, Ohtsuki S. 2018. Reduction in hepatic secondary bile acids caused by short-term antibiotic-induced dysbiosis decreases mouse serum glucose and triglyceride levels. Sci Rep 8: 1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujisaka S, Ussar S, Clish C, Devkota S, Dreyfuss JM, Sakaguchi M, Soto M, Konishi M, Softic S, Altindis E, Li N, Gerber G, Bry L, Kahn CR. 2016. Antibiotic effects on gut microbiota and metabolism are host dependent. J Clin Invest 126: 4430–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergen WG, Wu G. 2009. Intestinal nitrogen recycling and utilization in health and disease. J Nutr 139: 821–825. [DOI] [PubMed] [Google Scholar]
- 25.Dai ZL, Wu G, Zhu WY. 2011. Amino acid metabolism in intestinal bacteria: links between gut ecology and host health. Front Biosci 16: 1768–1786. [DOI] [PubMed] [Google Scholar]
- 26.Oliphant K, Allen-Vercoe E. 2019. Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome 7: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neis EP, Dejong CH, Rensen SS. 2015. The role of microbial amino acid metabolism in host metabolism. Nutrients 7: 2930–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin H, An Y, Hao F, Wang Y, Tang H. 2016. Correlations of fecal metabonomic and microbiomic changes induced by high-fat diet in the pre-obesity state. Sci Rep 6: 21618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamashita M, Fukizawa S, Nonaka Y. 2020. Hop-derived prenylflavonoid isoxanthohumol suppresses insulin resistance by changing the intestinal microbiota and suppressing chronic inflammation in high fat diet-fed mice. Eur Rev Med Pharmacol Sci 24: 1537–1547. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y, Zhang N, Arikawa AY, Chen C. 2019. Inhibitory effects of green tea polyphenols on microbial metabolism of aromatic amino acids in humans revealed by metabolomic analysis. Metabolites 9: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quifer-Rada P, Choy YY, Calvert CC, Waterhouse AL, Lamuela-Raventos RM. 2016. Use of metabolomics and lipidomics to evaluate the hypocholestreolemic effect of Proanthocyanidins from grape seed in a pig model. Mol Nutr Food Res 60: 2219–2227. [DOI] [PubMed] [Google Scholar]
- 32.Matsumoto M, Kibe R, Ooga T, Aiba Y, Kurihara S, Sawaki E, Koga Y, Benno Y. 2012. Impact of intestinal microbiota on intestinal luminal metabolome. Sci Rep 2: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azuma K, Izumi R, Kawata M, Nagae T, Osaki T, Murahata Y, Tsuka T, Imagawa T, Ito N, Okamoto Y, Morimoto M, Izawa H, Saimoto H, Ifuku S. 2015. Effects of oral administration of chitin nanofiber on plasma metabolites and gut Microorganisms. Int J Mol Sci 16: 21931–21949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohashi Y, Hirayama A, Ishikawa T, Nakamura S, Shimizu K, Ueno Y, Tomita M, Soga T. 2008. Depiction of metabolome changes in histidine-starved Escherichia coli by CE-TOFMS. Mol Biosyst 4: 135–147. [DOI] [PubMed] [Google Scholar]
- 35.Ooga T, Sato H, Nagashima A, Sasaki K, Tomita M, Soga T, Ohashi Y. 2011. Metabolomic anatomy of an animal model revealing homeostatic imbalances in dyslipidaemia. Mol Biosyst 7: 1217–1223. [DOI] [PubMed] [Google Scholar]
- 36.Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. 2010. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 6: 78–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kakiyama G, Muto A, Takei H, Nittono H, Murai T, Kurosawa T, Hofmann AF, Pandak WM, Bajaj JS. 2014. A simple and accurate HPLC method for fecal bile acid profile in healthy and cirrhotic subjects: validation by GC-MS and LC-MS. J Lipid Res 55: 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitahara M, Takamine F, Imamura T, Benno Y. 2001. Clostridium hiranonis sp. nov., a human intestinal bacterium with bile acid 7alpha-dehydroxylating activity. Int J Syst Evol Microbiol 51: 39–44. [DOI] [PubMed] [Google Scholar]
- 39.Puertollano E, Kolida S, Yaqoob P. 2014. Biological significance of short-chain fatty acid metabolism by the intestinal microbiome. Curr Opin Clin Nutr Metab Care 17: 139–144. [DOI] [PubMed] [Google Scholar]
- 40.Bilotta AJ, Cong Y. 2019. Gut microbiota metabolite regulation of host defenses at mucosal surfaces: implication in precision medicine. Precis Clin Med 2: 110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasmussen HS, Holtug K, Mortensen PB. 1988. Degradation of amino acids to short-chain fatty acids in humans. An in vitro study. Scand J Gastroenterol 23: 178–182. [DOI] [PubMed] [Google Scholar]
- 42.Šarenac TM, Mikov M. 2018. Bile acid synthesis: from nature to the chemical modification and synthesis and their applications as drugs and nutrients. Front Pharmacol 9: 939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russell DW. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem 72: 137–174. [DOI] [PubMed] [Google Scholar]
- 44.Eyssen HJ, De Pauw G, Van Eldere J. 1999. Formation of hyodeoxycholic acid from muricholic acid and hyocholic acid by an unidentified gram-positive rod termed HDCA-1 isolated from rat intestinal microflora. Appl Environ Microbiol 65: 3158–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]