Abstract
Autophagy is an evolutionary conserved physiological process with a fundamental role during development, differentiation, and survival of eukaryotic cells. On the other hand, autophagy dysregulation is observed in many pathological conditions, including cancer. In particular, tumor growth and progression are accompanied and promoted by increased autophagy that allows cancer cells to escape apoptosis and to proliferate also in harsh microenvironments. It is, therefore, clear that the impairment of the autophagic process may represent a valid strategy to inhibit or reduce cancer growth and progression. Among the plethora of molecular players controlling cancer growth, a group of small endogenous noncoding RNAs called microRNAs (miRNAs) has recently emerged. In fact, miRNAs can act as either oncogenes or oncosuppressors depending on their target genes. Moreover, among miRNAs, miRNA-34a has been connected with both tumor repression and autophagy regulation, and its expression is frequently lost in many cancers. Therefore, enforced expression of miRNA-34a in cancer cells may represent a valid strategy to reduce cancer growth. However, such strategy is limited by the fast biodegradation and short half-life of miRNA-34a and by the lack of an efficient intracellular delivery system. The following review describes the autophagic process and its role in cancer as well as the role of miRNAs in general and miRNA-34a in particular in regulating tumor growth by modulating autophagy. Finally, we describe the use of nanoparticles as a promising strategy to selectively deliver miRNA-34a to tumor cells for therapeutic and diagnostic purposes.
Keywords: miRNA-34a, miRNAs, autophagy, cancer treatment, nanoparticles
Introduction
Autophagy is an intracellular process with a role in several pathological conditions including cancer, neurodegenerative disorders, and infectious diseases (Mizushima et al., 2011; Deretic et al., 2013; Dikic and Elazar, 2018), all processes in which cytosolic wastes are delivered to the lysosomes for degradation and recycling (Mizushima et al., 2011). Recently, Yoshinori Ohsumi was awarded the Nobel prize for his studies on the basic mechanisms of autophagy in yeast (Tooze and Dikic, 2016). However, the understanding of the molecular mechanisms controlling autophagy induction and modulation in the physiopathology of mammalian cells is still far from being elucidated.
MiRNAs are endogenous small noncoding RNAs that participate in the regulation of gene expression, whose dysregulation is mechanistically implicated in many pathological processes, including autophagy and tumorigenesis (Wang et al., 2013; Peng and Croce, 2016). Among the plethora of miRNAs analyzed, miRNA-34a expression is abnormally regulated in many cellular processes, including cell division, senescence, apoptosis, autophagy, and tumorigenesis (Huang et al., 2014). Hence, since the two latter processes are tightly connected, the re-establishment of endogenous intracellular levels of miRNAs and, especially, of miRNA-34a may block pathologic autophagy and counteract cancer progression. However, the low stability of miRNAs and their difficult delivery into cells restrict their application in clinical practice (Chen et al., 2015).
In this sense, advances in nanotechnology have led to the development of many different nanostructures, which can be employed as drug delivery systems to improve the properties of a variety of bioactive compounds, including nucleic acids (Mei et al., 2019). For these reasons, nanocarriers are being explored to convey therapeutic miRNAs in order to interfere with key biological processes involved in tumor progression, including autophagy.
In this review, we summarize recent studies describing the use of nanoparticles for the delivery of miRNA-34a in cancer cells, alone or in combination with traditional chemotherapy drugs for the inhibition of different oncogenic pathways, with emphasis on autophagy regulation. Hence, the delivery of miRNA-34a combined to nanostructures may improve its biodistribution and accumulation at the target site, with some papers showing encouraging results, both in in vitro and in vivo studies.
Autophagy: When the Cells Eat Themselves
Autophagy is an evolutionarily conserved cellular homeostatic process that consists in the formation of double-membrane structures that engulf cytosolic wastes, including damaged organelles, protein aggregates, or invading pathogens, and fuse with lysosomes to degrade their content (Mizushima et al., 2011). Autophagy is further subdivided into several categories depending on its site of action, for instance, when it takes place in pathogens is called xenophagy (Levine, 2005), mitophagy in the mitochondria (Narendra et al., 2008), pexophagy in the peroxisomes (Nazarko et al., 2007), ERphagy in the endoplasmic reticulum (Chino et al., 2019. Khaminets et al., 2015), or lysophagy in the lysosomes (Maejima et al., 2013). Other relatively less studied forms of autophagy, called chaperon mediated autophagy involve molecular chaperones, such as HSP70 (Kaushik et al., 2006). Chaperone mediated autophagy does not require the whole autophagy machinery but involves the recruitment of the lysosomal membrane protein LAMP2 to substrate such as protein aggregates, damaged organelles and invading pathogens through chaperons and ultimately the degradation of the cargo (Kaushik et al., 2006). Chaperon mediated autophagy has several medical implications, including a role in cancer (Kaushik et al., 2011). Microautophagy is another form of autophagy that is mediated by the direct engulfment of cytoplasm and its components by membrane invaginations into lysosomes. A role of microautophagy has been demonstrated in various pathologies (Li et al., 2012). Another form of autophagy called LC3 dependent phagocytosis (LAP) has also been reported (Sanjuan et al., 2007). Unlike conventional autophagy, LAP involves the formation of a single membrane structure positive for autophagy marker LC3 around the phagocytic substrate and ultimately its fusion with the lysosome. Like conventional autophagy, LAP requires autophagy conjugation machinery (discussed in the next section) but does not require components of autophagy initiation machinery (Martinez et al., 2015). Another major difference between LAP and conventional autophagy is that LAP requires RUBICON (Run domain Beclin-1 interacting and cysteine-rich containing protein), which is an inhibitor of the conventional autophagy (Martinez et al., 2015). Like all other forms of autophagy, the role of LAP has been studied in various pathologies, such as inflammation related disorders (Martinez et al., 2016), neurodegeneration (Heckmann et al., 2019), and cancer (Cunha et al., 2018).
Role of Autophagy in Cancer
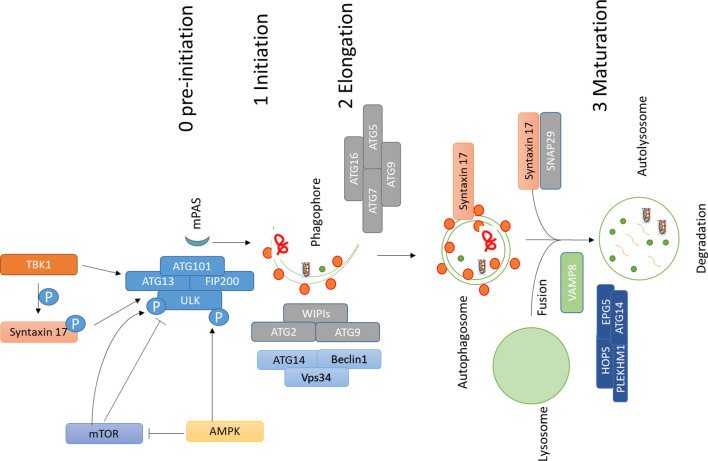
The role of autophagy in cancer is somewhat controversial. Earliest reports suggested a role of autophagy against tumorigenesis (Liang et al., 1999) and afterwards, several reports found a role of autophagy favoring cell death of tumor cells in an apoptotic-dependent or independent way (Levine and Yuan, 2005; Pattingre et al., 2005; Kumar et al., 2013; Sierra et al., 2015). On the other hand, it was demonstrated that inhibition of autophagy could also hamper the antitumor T cell response, which is required for immunogenic cell death (Townsend et al., 2012; Ma et al., 2013). However, the observation that in a different setting the inhibition of autophagy does not suppress the immune response suggests the existence of autophagy-independent mechanisms of the immune response (Starobinets et al., 2016). Therefore, the role of autophagy in immunogenic cell death in cancer may vary from one condition to other, and no conclusion can be drawn based on the present literature. All these studies suggest that autophagy plays a dual role in cancer: its induction may protect against cancer at earlier stages of tumorigenesis, whereas at later stages cancer cells may utilize this process to escape from apoptosis and to promote uncontrolled growth, as well as resistance to stress”. However, to better clarify the proproliferative role of autophagy in cancer, many efforts have been made by using pharmacological modulators for cancer therapy (Amaravadi et al., 2007; Levy et al., 2017; Amaravadi et al., 2019). For instance, autophagy inhibitors chloroquine (CHQ) and hydroxychloroquine (HCQ) are being tested against different types of cancer (Levy et al., 2017; Amaravadi et al., 2019). Both CHQ and HCQ inhibit the fusion between autophagosomes and lysosomes occurring at the final stage of the autophagic process (Klionsky et al., 2016). Moreover, proteins active in different stages of autophagy are being analyzed as potential therapeutic targets in cancer. Autophagy is initiated by the ULK1 complex, and inhibition of ULK1 has shown encouraging results in cancer therapy (Egan et al., 2015; Martin et al., 2018). Likewise, suppression of FIP200, another component of autophagy initiation complex, also induces apoptosis, and inhibition of FIP200 limits early tumorigenesis and also the progression of breast cancer in a mouse model (Wei et al., 2011; Xia et al., 2017). Inhibition of VPS34 has also shown promising results (Dowdle et al., 2014; Ronan et al., 2014). ATG4B, which is required for the processing of mAtg8s and consequently, for the progression of autophagy, is also being targeted to inhibit cancer progression (Levy et al., 2017). Metabolism is one of the targets in cancer therapy. Autophagy also plays an important role in maintaining cancer cell metabolism (Rabinowitz and White, 2010; Guo et al., 2011). More recently, it was shown that autophagy is required for maintaining high circulating arginine levels necessary to promote cancer progression (Poillet-Perez et al., 2018). Also, proteins with multiple roles in autophagy could be targeted. An example is Stx17, which acts in different steps of autophagy: it regulates the formation of earliest autophagosomal structures (Kumar et al., 2019) and consequently the initiation of autophagy (Hamasaki et al., 2013; Sugo et al., 2018; Kumar et al., 2019). Moreover, Stx17 is a key regulator of fusion between autophagosomes and lysosomes (Diao et al., 2015), and this step is being targeted in various cancer using chloroquine and hydroxychloroquine (Levy et al., 2017). All these reports put forward the role of autophagy regulators like Stx17 as a potential new target in cancer therapy. Figure 1 .
Figure 1.
Main machinery and steps involved in autophagy. Autophagy involves different steps; assembly of ULK1 complex, which is negatively regulated by mTOR, which phosphorylates ULK1 and inhibits autophagy; AMPK phosphorylates ULK1 and activates autophagy initiation, AMPK might also affect this process by inhibiting mTOR activity by phosphorylating RAPTOR and TSC2. Autophagy initiation and assembly of ULK1 complex are controlled by TBK1 and its substrate Stx17. TBK1 and Stx17 also affect the formation of mPAS, which is then elongated to a cup-like phagophore by the action of WIPIs, ATG2, ATG9, and PI3K complex which comprises of VPS34, ATG14, and Beclin1. ATG conjugation machinery also helps in elongation and lipidation of the membrane. Once autophagosome is completed SNARE protein Stx17 gets recruited to autophagosomes and works together with SNAP29 and VAMP8 to regulate fusion between autophagosomes and lysosomes. Additional factors like HOPS, PLEKHM1, EPG5, and ATG14 help Stx17 in this process. Abbreviations: ATG, Autophagy related; AMPK, 5′ AMP-activated protein kinase; TBK1, TANK Binding Kinase 1; mTOR, mammalian Target of Rapamycin; SNARE, soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein receptors).
miRNAs in Cancer
MicroRNAs (miRNAs) are a class of small endogenous noncoding RNAs of about 21–25 nucleotides that modulate gene expression by binding to the 3′-UTR region of a specific mRNA target. The main effect of the miRNA binding is the decreased expression of specific RNA sequences with consequences on several biological processes, including cell proliferation, differentiation, apoptosis, development, and tumorigenesis (Esquela-Kerscher and Slack, 2006). Indeed, the alteration of miRNA expression pattern has been shown to support cancer initiation, progression and dissemination (Berindan-Neagoe and Calin, 2014). Increasing evidences have shown that, depending on their target genes, miRNAs can sustain tumor, by acting as either oncogene or tumor suppressor, thus favoring evasion from growth suppressors, support of proliferation, resistance to cell death, angiogenesis, and activation of invasion and metastasis (Peng and Croce, 2016). In many cancer types, miRNA profile is abnormal due to gene amplifications or deletions, alteration of their transcriptional control, epigenetic dysregulations and defects in the miRNA biogenesis machinery. In particular, miRNA-34a, a tumor-suppressor miRNA that belongs to an evolutionarily conserved family of miRNAs, is deregulated in many human tumor types, including breast, lung, prostate cancer, cholangiocarcinoma, acute myeloid leukemia (AML), and multiple myeloma (MM). MiRNA-34a plays a key role in many cellular processes, including the regulation of cell division, senescence, apoptosis, and proliferation (Huang et al., 2014). Indeed, in prostate cancer, one of the most frequently diagnosed tumors, it has been shown that, among the 50 altered miRNAs that act in carcinogenesis, miR-34a expression is strongly downregulated (Liao et al., 2016). MiRNA-34a decreased expression may lead to a reduction in cell cycle arrest and apoptosis and to the enhancement of chemoresistance and autophagy. Also, in cholangiocarcinoma miR-34a downregulation is implicated in drug resistance and in the control of epithelial–mesenchymal transition (EMT) through an effect on Smad4 (Sun et al., 2017). Interestingly, the enforced expression of miR-34a in MM cells or its delivery in the tumor xenografts by direct intratumor or intravenous injection mimics the effect of other anticancer agents without producing toxicity, thus representing a new therapeutic strategy (Di Martino et al., 2014). Thus, all this evidence supports the analysis in tumors of miRNAs, and in particular of miRNA-34a, as well as the artificial restoration of their physiological levels for both diagnostic and therapeutic purposes.
miRNA-34a in Autophagy Regulation
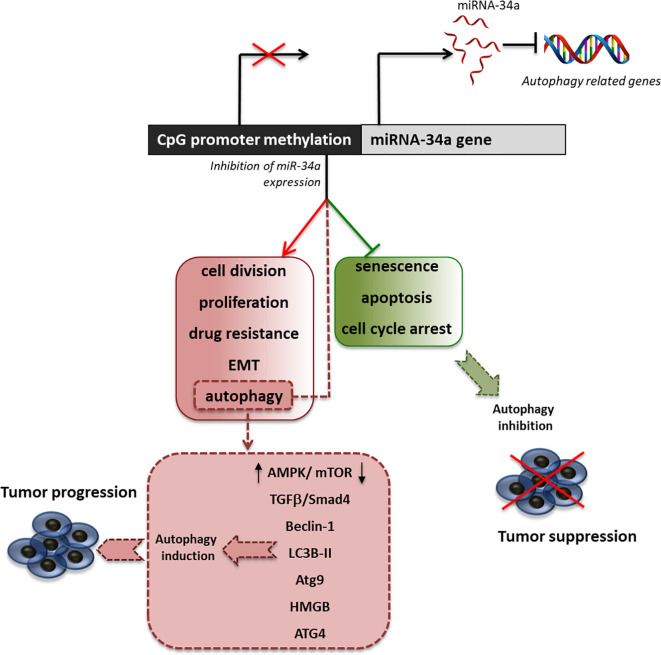
Many studies have investigated the multiple effects associated with the decreased expression of miRNA-34a in support of tumor growth, showing that this miRNA is also an important regulator of autophagy ( Figure 2 ). Indeed, Liao et al. demonstrated that in prostate cancer, miRNA-34a expression is decreased due to gene hypermethylation, correlating with higher cell proliferation, apoptosis abrogation, enhanced chemoresistance, and autophagy induction (Liao et al., 2016). By overexpressing miR-34a, they showed that the autophagy-related proteins, ATG4B, Beclin-1, and LC3B II/I, were downregulated. This led to enhanced chemosensitivity to the drugs doxorubicin and topotecan. Furthermore, they showed that the downregulation of miRNA-34a expression parallels the upregulation of ATG4B-induced autophagy through the AMPK/mTOR pathway regulation (Liao et al., 2016). Another study on colorectal cancer showed that the treatment with oxaliplatin decreased the level of miRNA-34a, with the consequent increase of drug resistance due to the activation of autophagy. This activation has been shown to be mediated by the regulation of the TGF-β/Smad4 pathway. Indeed, 34a in colorectal cancer patients the expression of Smad4 and miRNA-34a show a significant inverse correlation and the overexpression of miRNA-34a inhibits autophagy activation by directly targeting Smad4 through the TGF-β/Smad4 pathway (Sun et al., 2017).
Figure 2.
Effect of miRNA34a inhibition on autophagy. Schematic representation of promoter methylation, which represents the most common cause of miRNA34a decreased expression in tumors and of the principal effects of miRNA34a low expression in the sustainment of cancer proliferation. Particularly, we highlighted the main pathway involved in the regulation of autophagy by miRNA-34a.
The high mobility group box 1 (HMGB1) is an ubiquitous nuclear protein that regulates several DNA-related activities such as transcription, replication, recombination, and repair (Liu et al., 2014). HMGB1 is overexpressed in different tumor types and has been proposed as a target for cancer therapy (Lugrin et al., 2014). Among its multiple functions supporting tumor proliferation, HMGB1 improves chemotherapy resistance through the induction of autophagy in human myeloid leukemia cells (Liu et al., 2011). Interestingly, in acute myeloid leukemia (Liu et al., 2017) and in retinoblastoma cells (Liu et al., 2014), the induction of HMGB1 parallels a decreased expression of miRNA-34a, which targets HMGB1 mRNA, leading to reduced apoptosis and induction of autophagy.
As described previously, the ATG protein family is composed of 35 autophagy-related (Atg) genes: Atg1-10, 12-14, 16-18, 29, and 31 are essential for the formation of autophagosomes (Mizushima et al., 2011). Among these, Atg9 has attracted much attention because it is the only transmembrane protein among the core Atg proteins required for autophagosome formation. Fan et al. have shown that the inhibition of miR-34a restored the expression of Atg9a and significantly decreased ethanol-induced inhibition of autophagy and neural differentiation of neural crest cells (Fan et al., 2019). Huang et al. showed that miR-34a also modulates the expression of Atg9a during myocardial hypertrophy (Huang et al., 2014). These studies offer the cue to further investigations directed to the exploration of Atg9a mRNA regulation by miRNA-34a also in cancer cells.
Delivery of Autophagy-Related miRNA-34a with Nanoparticles
As described before, the ability of miRNA-34a to target autophagy-related genes, such as ATG4 (Wu et al., 2017), ATG5 (Cheng et al., 2019), ATG9 (Yang et al., 2013), and HMGB1 (Liu et al., 2014; Liu et al., 2017) has attracted considerable interest as a novel tool for anticancer therapy.
Therefore, the restoration of miRNA-34a physiological levels is now a promising tool to counteract tumor progression (Farooqi et al., 2017).
However, miRNAs share some properties, such as rapid biodegradation and short half-life in systemic circulation, poor biocompatibility, low membrane penetrability, and excessive off-target accumulation, that have greatly limited their application in in vivo systems so far (Chen et al., 2015). Thus, due to the growing awareness regarding the potential role of miRNA-34a in cancer therapy, many efforts have been addressed to optimize strategies for the targeted delivery of this miRNA.
In this sense, nanomedicine has emerged as a promising technology allowing the accumulation of systemically administered chemotherapeutics in the tumor tissues via the enhanced permeability and retention effect (EPR) due to leaky tumor vasculature and poor lymphatic drainage (Wilhelm et al., 2016). Extensive research focusing on developing cancer nanomedicine has generated nanostructures capable of overcoming biological barriers and transport chemotherapeutic drugs to the targeted sites while minimizing harmful effects on healthy tissues (Jain and Stylianopoulos, 2010). Moreover, the surface of nanoparticles (NPs) can be chemically modified by conjugating functional moieties, such as nucleic acids and targeting ligands, for increasing targeted delivery to the tumor sites, maximizing chemotherapy efficacy (Jabir et al., 2012).
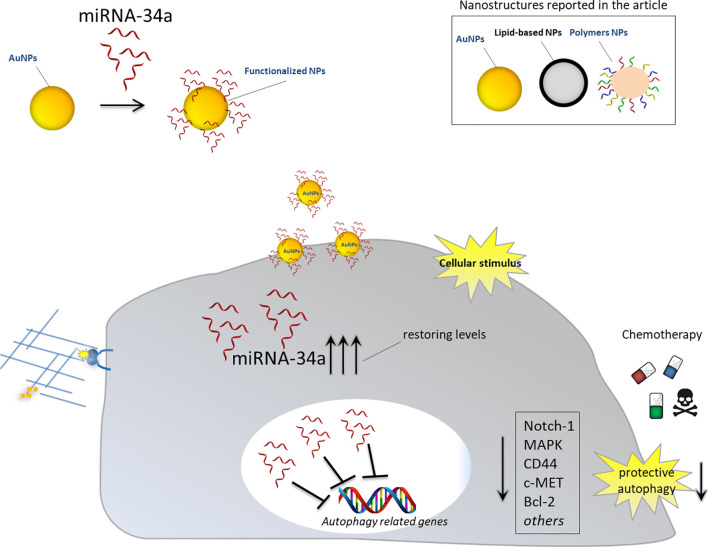
Thus, the delivery of miRNA-34a combined to nanostructures may overcome the weakness described above and function as antisense strategy to inhibit oncogenic mRNAs, involved in autophagy induction, or to restore the physiological levels of tumor suppressor miRNA-34a. Different examples regarding the employment of different nanomaterials to deliver this specific miRNA, alone or in combination with other anticancer drugs, are commented below ( Figure 3 ).
Figure 3.
Nanoparticles for delivery of miRNA-34a for autophagy inhibition. Schematic representation showing the effects of nanoparticles functionalized with anti-autophagy miRNA-34a in cancer cells. Upon intracellular stimulus, NPs can release therapeutic miRNA-34a and restore its cellular physiological levels. miRNA-34a can exert its tumor suppressor activity by downregulating autophagy-related genes and inhibiting protective autophagy. Blocking cancer autophagy leads to cancer cell death either directly or by sensitizing them to chemotherapy treatment.
Preparation Techniques to Load miRNA in Nanoparticles
Several techniques such as emulsion based techniques (Cohen-Sela et al., 2009), nanoprecipitation (Alshamsan, 2014), interfacial polymerization (Singha et al., 2011) are used for preparation of NPs to load miRNAs. Some methods are more efficient than others based on the surface characteristics. Emulsion based nanoparticles are most commonly used methods for delivery of NPs. Emulsion based delivery method utilizes ultrasonication (Shi et al., 2014) and homogenization followed by purification at high speed centrifugation (Cosco et al., 2015). Single or double emulsion techniques are used for delivery of NPs (Jenjob et al., 2019). Oil in water emulsion is an example of single emulsion technique which can be used to encapsulate hydrophobic and hydrophilic drugs in micro- or nanoscale form. In single emulsion technique Poly(lactic-co-glycolic acid) (PLGA), one of the ideal reagent for nanoscale delivery (McCall and Sirianni, 2013), is dissolved into an organic phase (oil) that is emulsified with a surfactant or stabilizer (water). Hydrophobic drugs are added directly to the oil phase, whereas hydrophilic drugs (water) may be first emulsified with the polymer solution prior to the formation of particles (McCall and Sirianni, 2013; Jenjob et al., 2019). Double emulsion-solvent evaporation method has potential for encapsulation of both hydrophilic and hydrophobic payloads with high encapsulation efficiency and utilizes two emulsification steps to obtain water-in-oil-in-water (w/o/w) or oil-in water-in-oil (o/w/o) emulsions (McCall and Sirianni, 2013; Jenjob et al., 2019).
Another technique used for delivery of NPs is nanoprecipitation. It is a less complex and widely applicable method for hydrophobic drug molecules (Salatin et al., 2017). It is a solid displacement method which requires solvent (organic) and nonsolvent (inorganic) phase separation followed by addition of one phase to another (Salatin et al., 2017). Both the drug and the polymer for the delivery must be dissolved in solvent; once the solvent is mixed to a nonsolvent rapid desolvation and precipitation of polymer starts leading to drug entrapment (Fessi et al., 1989; Martínez Rivas et al., 2017). Solvents miscible with water and nonhalogenated solvents are commonly used; however, immiscible solvents such as dichloromethane can also be used (Salatin et al., 2017). One example is mixing of cucurbitacin I and PLGA in acetone. The organic phase is added dropwise to deionized water containing 1% Pluronic F-68 followed by acetone evaporation (Alshamsan, 2014). Another example in which miRNA-loaded PLGA/chitosan (PLGA/CS) NPs of 150–180 nm size are prepared via the nanoprecipitation method is by dropwise addition of PLGA solution into a water solution of CS and miR-34s in the presence of a Poloxamer surfactant (Salatin et al., 2017).
Interfacial polymerization is another technique used to deliver NPs. Using this approach hydrophilic substances such as miRNAs are incorporated into biodegradable lignin nanocontainers (Wurm and Weiss, 2014). This technique involves copolymerization of hydrophobic and hydrophilic monomers to constrain the polymerization at interfaces, but its free-radical mechanism allows precise control of initiation, which makes it possible to finely disperse the immiscible phases prior to polymerization (Scott et al., 2005). The interfacial polymerization falls into different types of interfaces: liquid–solid interfaces, liquid–liquid interfaces, and liquid-in-liquid emulsion interfaces (Song et al., 2017). There are also other interface categories, rarely used, including liquid–gas, solid–gas, and solid–solid (Salatin et al., 2017; Song et al., 2017).
Inorganic Nanoparticles
Recent advancement in nanotechnology has led to the introduction of various inorganic nanomaterials, such as silica dioxide nanoparticles (SiO2-NPs), calcium phosphate nanoparticles (CaP-NPs), gold nanoparticles (AuNPs), gold nanoshells (AuNShs), and magnetic nanoparticles (MNPs) that have been exploited as excellent nanocarriers for the delivery of nucleic acids both in vitro and in vivo models (Naz et al., 2019). These nanomaterials share some properties such as high bioactivity, biocompatibility, and chemical stability that make them efficient delivery systems (Cheng and Kuhn, 2007; Ghosh et al., 2008; Park et al., 2009; Tang et al., 2012; Xu et al., 2014; Xiong et al., 2018). Recently, it has been reported that SiO2-NPs can efficiently deliver active miR-34a into breast cancer cells leading to the reduction in mammosphere formation (Panebianco et al., 2019) and the downregulation of Notch-1, a well-established miR-34a target with an important role in regulating stem cell functions and autophagy process (Bouras et al., 2008; Marcel and Sarin, 2016). Notably, in vivo miR-34a delivery by SiO2-NPs was able to exert a tumor-suppressive function leading to the reduction of breast cancer in engrafted mice (Panebianco et al., 2019) ( Table 1 ).
Table 1.
Nanoparticles for delivery of autophagy-related miRNA-34a as cancer treatment.
| Entry | Nanoparticles | Compound carried/co-treatment | Tested cell lines (tissue of origin) | In vivo models | Biological effects | Molecular mechanisms | Refs |
|---|---|---|---|---|---|---|---|
| 1 | SiO2-NPs | None | SUM159pt; mammospheres (breast cancer) |
mutant miR34-TKO mice; NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ mice | Reduction of mammosphere formation; Reduction in tumor growth in vivo. |
Notch-1↓ | (Panebianco et al., 2019) |
| 2 | CaPs | Linear polyethyleneimine (LPEI) | PC3 (prostate cancer) | None | Inhibition of cell proliferation; Inhibition of cell migration |
Not reported | (Jung et al., 2015) |
| 3 | AuNPs | 7-ethyl-10-hidroxycamptothecin (SN-38) microRNA-182, microRNA-137 and microRNA-144 |
Mel-202 (uveal melanoma) | none | Synergistic increase in cytotoxicity; Sensitization to SN-38 |
c-MET↓ | (Milán Rois et al., 2018) |
| 4 | AuNSsh | None | MDA-MB-231 (breast cancer) | None | Inhibition of cell proliferation; Inhibition of metabolic activity |
SIRT1↓; Bcl2↓ | (Li et al., 2013) |
| 5 | MNPs | Nitrosonium tetrafluoroborate (NOBF4); Polyethylene glycol (PEG) | Not reported | None | Inhibition of cell migration and invasion | CD44↓ | (Lee et al., 2016) |
| 6 | CPP-9 (CRGDKGPDC) | b-cyclodextrin-polyethylenimine (PC) |
PANC-1 (pancreatic cancer) |
Athymic female mice (BALB/c strain) | Cell cycle arrest; apoptosis; suppression tumor cell migration; reduction tumor growth in vivo. | E2F3↓; Bcl-2↓; c-myc↓; cyclin D1↓ |
(Hu et al., 2013) |
| 7 | CPP Nona arginine (R9) | c(RGDfK) and Folic acid (targeting ligands) |
U87MG and HeLa (Breast and ovarian cancer) | None | Not reported | Not reported | (Xiao et al., 2018) |
| 8 | SLNs | Dimethyldioctadecylammonium bromide (DDAB) |
B16F10-CD44+ (Lung metastasis of melanoma) | Female C57BL/6 mice | Induction of apoptosis; Reduction of cell migration; Reduction of spheroids formation in vitro; Inhibition of tumor growth and metastasis in vivo |
CD44↓ | (Shi et al., 2013) |
| 9 | SLNs | Paclitaxel (PTX) | B16F10-CD44+ (Lung metastasis of melanoma) | Female C57BL/6 mice | Synergistic inhibition of cancer development and metastasis | CD44↓ | (Shi et al., 2014) |
| 10 | LPH | Polyethylene glycol (PEG), Single-chain antibody fragment (scFv) |
B16F10-CD44+ (Lung metastasis of melanoma) | None | Induction of apoptosis | Survivin↓ MAPK↓ |
(Chen et al., 2010) |
| 11 | PAMAMs | Phenylboronic acid (PBA) | BGC-823 (Gastric cancer) |
BALB/c-nude mice | Induction of apoptosis; Reduction of cancer cells invasion and migration; Reduction of tumor growth in vivo. |
Notch-1↓ | (Song et al., 2019) |
| 12 | PAMAMs | S6 aptamer | A549 (Lung cancer) |
None | Induction of apoptosis; Inhibition of cell growth, cell migration and invasion |
P53↓ Bcl-2↓ |
(Wang et al., 2015a) |
| 13 | DSS-BEN | 1,N11-bisethylnorspermine (BENSpm). |
HCT116 (colorectal carcinoma) |
Xenograft athymic nude mice |
Inhibition of polyamines metabolism; Cancer cell death; Inhibition of tumor growth in vivo. |
SMOX↑ SSAT↑ Bcl-2↓ |
(Xie et al., 2017) |
| 14 | CBSA | Docetaxel (DTX) | 4T1, A549 (breast and lung cancer) |
4T1 tumor-bearing mice | Induction of apoptosis; Cytotoxicity; Inhibition of cell migration; Inhibition of tumor growth in vivo. |
Bcl-2↓ | (Zhang et al., 2017) |
| 15 | HP-IPECs | None | MDAMB-231 and MCF-7 (breast cancer) |
Female athymic nude Balb/c mice | Induction of apoptosis; Inhibition of cell migration; inhibition of cell proliferation; reduction of tumor growth in vivo. |
Notch-1↓ CD44↓ |
(Wang et al., 2015b) |
| 16 | Chitosan-NPs | Poly(D,L-lactide-co-glycolide) (PLGA) | SKMM1 and RPMI8226 (multiple myeloma) |
Immunodeficient NOD–SCID mice | Reduction cell proliferation and toxicity in cell lines; Reduction of tumor growth in vivo. |
Bcl-2↓ CDK6↓ |
(Cosco et al., 2015) |
| 17 | Chitosan-NPs | Hyaluronic Acid (HA) Doxorubicin (DOX) |
MDA-MB-231 (triple negative breast cancer) |
Female athymic nude Balb/c | Induction of apoptosis; Reduction of cell proliferation; Inhibition of cancer cell migration; Inhibition of tumor growth in vivo. |
Bcl-2↓ Notch-1↓ |
(Deng et al., 2014) |
| 18 | Hyaluronic acid-poly(ethylene imine) (HA-PEI)-NPs HA-poly(ethylene glycol) (HA-PEG) |
None | A549 (cisplatin sensitive) and A549 DDP (cisplatin resistant) (lung cancer) |
None | Increased ATP levels; Reduction glycolysis; Elevation of mitochondrial complexes activity; Gene-Specific Changes in mtDNA encoded genes; Alteration of mitochondrial epigenetic enzymes; Induction of apoptosis. |
Bax↑ Apaf-1↑ PUMA↑ caspase3↑ Bcl-2↓ survivin↓ mt-TET1-3↑ Complex I ↑ Complex IV↑ |
(Trivedi et al., 2017) |
| 19 | β- cyclodextrin-PEI600-NPs |
polyethylene glycol (PEG) | B16F10 (melanoma) |
B16F10-CD44+ xenograft model of female C57BL/6 mice | Inhibition of tumor progression; Inhibition of cell invasion; Inhibition of sphere formation; Reduction of tumor growth in vivo. |
CD44↓ | (Fan et al., 2017) |
| 20 | poly(L-lysine-graft-imidazole) (PLI) | polyethylene glycol (PEG) | MKN-74 (gastric cancer) |
Orthotopic and subcutaneous xenograft model of male BALB/c-nude mice | Reduction metastasis and cell invasion; Induction of apoptosis; Inhibition of tumor relapse and growth in vivo. |
CD44↓ Bcl-2↓ Oct-3/4↓ Nanog↓ |
(Jang et al., 2016) |
In a study performed by Mok et al., a long chain miRNA-34a (lc-miRNA-34a) was prepared by chemical crosslinking in order to improve encapsulation efficiency into linear polyethyleneimine (LPEI)-coated CaP-NPs (LPEI-CaP) and intracellular delivery (Jung et al., 2015). These nanoformulations were successfully delivered into PC-3 cancer cells, where miRNA-34a was efficiently released, suppressing cancer cell proliferation as well as cell migration (Jung et al., 2015).
In another study by Milán-Rois et al., a therapeutic mixture containing SN-38(7-ethyl-10-hidroxycamptothecin), a topoisomerase inhibitor (Palakurthi, 2015), and miRNA-34a has been delivered in uveal melanoma cells using AuNPs (Milán Rois et al., 2018). The nanoformulations led a synergistic cytotoxicity effect in cancer cells and were able to reprogram their oncogenic phenotype by downregulating c-MET tyrosine kinase, making Mel-202 cancer cells more susceptible to SN-38 (Milán Rois et al., 2018).
Other inorganic nanostructures widely used in biomedical applications are gold nanoshells (AuNShs), which are composed of an inorganic core coated with a thin layer of gold (Bardhan et al., 2011).
In a recent study, Goyal et al. reported that layer-by-layer assembled gold nanoshells (LbL-AuNShs) were able to efficiently deliver miRNA-34a to MDA-MB-231 breast cancer cells and release it upon intracellular stimulus (Goyal et al., 2018). Notably, the cancer cells treated with these inorganic nanostructures downregulated the expression of the autophagy-modulators SIRT1 and Bcl2, which are known downstream targets of miRNA-34a (Li et al., 2013). Moreover, treatment with LbL-AuNSsh affected the proliferation and metabolic activity of triple-negative breast cancer cells without negatively impacting noncancerous MCF10A breast epithelial cells (Li et al., 2013).
MNPs are ideal candidates for surface modifications generating functional nanostructures that can be employed in a variety of biomedical applications, including drug delivery for cancer therapy (Xiong et al., 2018). In this regard, MNPs modified with nitrosonium tetrafluoroborate (NOBF4) and polyethylene glycol (PEG) have been successfully employed for the delivery of miR-34a in cancer cells. The miRNA-34a conjugated nanostructures led to the downregulation of CD44 (Lee et al., 2016), a protein upregulated by autophagy and involved in chemoresistance in oral squamous cell carcinoma (Naik et al., 2018). The inhibition of CD44 expression showed therapeutic effects, such as the reduction of cell migration and invasion, suggesting that magnetic nanostructures have the potential for miRNA-based cancer therapy.
Lipid-Based Nanoparticles
Among the many lipid-based nanocarriers that have been developed during the last years, solid lipid nanoparticles (SLNs) have attracted considerable interest for drug delivery and targeting (Mishra et al., 2018). SLNs can include a cationic component that facilitates association with anionic miRNAs and are able to protect them from degradation during systemic circulation (Müller et al., 2000). Interestingly, Shy et al. developed a lipid nanoparticle system containing SLNs and cationic dimethyl dioctadecyl ammonium bromide (DDAB) to carry miRNA-34a in cancerous lung for CSC therapy (Shi et al., 2013). These miRNA-34a-SLNP nanocomplexes induced B16F10-CD44+ cell apoptosis and inhibited cell migration by negatively regulating the cell surface protein CD44.
Moreover, the nanoformulations were also effective in inhibiting B16F10-CD44+ tumor development and tumorigenicity in vivo (Shi et al., 2013).
However, when miRNA-34a and paclitaxel (PTX), an inhibitor of microtubules dynamic (Jordan and Wilson, 2004), were coincorporated into SLNs, the combination of these two drugs could cooperatively and more efficiently induce cell apoptosis and inhibit B16F10-CD44+ development in in vitro and in vivo models through different mechanisms (Shi et al., 2014).
Apart from SLNs, it has been shown that liposome-polycation-hyaluronic acid (LPH) nanoparticles could systemically deliver therapeutic siRNA into the tumor site with relatively low toxicity (Li et al., 2008). In this regard, Huang et al. employed LPH nanoparticles modified with PEG and tumor-targeting single-chain antibody fragment (scFv) for systemic delivery of the anti-autophagic miRNA-34a in experimental lung metastasis of murine B16F10 melanoma (Chen et al., 2010). Interestingly, miRNA-34a was found to inactivate the MAPK pathway, leading to therapeutic activity in B16F10 melanoma cells (Chen et al., 2010). MAPK pathway has been shown to induce autophagy through phosphorylation of c-Jun/c-Fos transcription factors, which in turn lead transcription of autophagy-related genes (Zhou et al., 2015; Lee et al., 2019).
Polymeric Nanoparticles
Amine terminated polyamidoamines (PAMAMs) are a class of synthetic polymers that have been used as nanocarriers because of their unique properties, such as highly branched structure, water-solubility, high charge density, and numerous amine groups for further modification (Han et al., 2018). Besides, conjugation with specific tumor-targeted ligands is an effective way for facilitating the intracellular delivery of PAMAMs in a receptor-mediated endocytosis manner (Chen et al., 2017). Phenylboronic acid (PBA) exhibits a high affinity with sialic acid (SA), which is overexpressed in various types of tumor cells (Narayanan, 1994), making it a promising tumor-targeted ligand (Jia et al., 2014).
In a recent work, PBA was successfully attached to a PAMAM surface to obtain a functional PAMAM (PPP), which has been employed for miRNA-34a delivery in gastric carcinoma cell line BGC-823 (Song et al., 2019). Interestingly, PPP/miRNA-34a nanocomplexes were found to counteract the autophagic Notch-1 signaling pathway resulting in the induction of apoptosis and the inhibition of cell migration and invasion (Song et al., 2019).
Alternatively, PAMAM may be conjugated with aptamers to generate nanocomplexes, which display several properties, such as high affinity and specificity to the target molecules, less toxicity, and rapid tissue penetration (Tan et al., 2011). In this regard, miRNA-34a has been encapsulated into S6 aptamer conjugated dendrimer to form lung cancer-targeted delivery nanoparticles (PAM-Ap/pMiRNA-34a NPs) (Wang et al., 2015a). The aptamer conjugation improved cellular uptake of miRNA-34, which targeted important genes involved in autophagy regulation, such as Bcl-2 and p53 (Lindqvist et al., 2014; Mrakovcic and Fröhlich, 2018). Thus, PAM-Ap/pMiRNA-34a-NPs showed therapeutic activity by inhibiting cell growth, migration, invasion and by inducing apoptosis of lung cancer cells (Wang et al., 2015a).
These studies provide novel therapeutic strategies, based on dendrimer nanoparticles, to deliver tumor suppressor miRNA-34 in cancer cells to target autophagy-related mechanisms and counteract tumor progression and cancer growth.
Natural polyamines, such as spermidine and spermine, are key regulators of cell growth, differentiation, and survival (Pegg and Casero, 2011). Recently, spermidine was found to induce autophagy by inhibiting the lysine acetyl transferase E1A-binding protein p300 (EP300) (Pietrocola et al., 2015). In cancer, dysregulation of polyamine metabolism promotes tumor development and progression (Lu et al., 2011) and represents a promising target in cancer chemotherapy (Basuroy and Gerner, 2006).
In this sense, in a study performed by Oupický et al., biodegradables nanoparticles synthesized from a polyamine analog N1, N11-bisethylnorspermine (BENSpm) were employed to efficiently deliver miR-34a in colorectal cancer cells (Xie et al., 2017). After treatment with an intracellular stimulus, these nanostructures could disassemble in the cytosol releasing both the miRNA-34a and BENSpm, which exerted a therapeutic function by downregulating Bcl-2 and inducing the expression of enzymes involved in polyamine catabolism, such as SMOX and SSAT (Stewart et al., 2018).
The ability of these biodegradable nanocarriers to deliver therapeutic miRNA-34a and inhibit polyamine metabolism may provide an efficient approach to combination nanomedicines for autophagy inhibition in cancer therapy.
Recent studies have evidenced that self-assembly strategies are useful tools to generate various nanomaterials for drug delivery (Declerck et al., 1990; Naito et al., 2012). In this regard, interpolyelectrolyte complexes (IPECs) can be formed by self-assembly mixing two oppositely charged polyelectrolytes in aqueous solution (Pergushov et al., 2012). By using the IPEC-based approach, nucleic acids have been efficiently absorbed on the cationic surface of the nanocomplexes and successfully delivered in cells (Cheng et al., 2009; Kim et al., 2009).
In a work by Wang et al, miRNA-34a has been delivered into breast cancer cells using nanocapsules prepared through IPECs-based approach and composed of cationic protamine sulfate (PS) and anionic Hyaluronic Acid (HA) (Wang et al., 2015b). Notably, nanocomplex-assisted delivery of miRNA-34a induced apoptosis and suppressed migration and proliferation of breast cancer cells as well as reduced tumor growth in a xenograft mouse model via targeting signaling pathways autophagy-related, as CD44 and Notch-1 (Wang et al., 2015b). Therefore, this biodegradable nanoplatform provides a great potential for miRNA-34a based therapy against triple-negative breast cancer.
Amiji et al. developed a novel drug delivery system incorporating self-assembling hyaluronic acid-poly(ethylene imine) (HA-PEI) and HA-poly(ethylene glycol) (HA-PEG) nanoparticles to efficiently deliver tumor suppressor miR-34a inA549 lung cancer cells for redox-epigenetic modifications (Ganesh et al., 2013; Trivedi et al., 2017). The authors reported that miRNA-34a HA-NPs treatment resulted in decreased glycolytic flux and antioxidant response element Nrf-2 resulting in depleted glutathione levels and ultimately, increased in several pro-apoptotic factors in both cisplatin-sensitive and cisplatin-resistant lung adenocarcinoma cells (Trivedi et al., 2017). Remarkably, molecular changes in epigenetic status on both mitochondrial (mt) and nuclear (nc)DNA and transcription of mtDNA-encoded genes were also observed after treatment with miRNA-34aHA-NPs. Interestingly, some studies report the existence of a functional interplay between glycolytic metabolism and autophagy in a positive loop that sustains tumor progression (Watson et al., 2015; Fan et al., 2018; Jiao et al., 2018). Therefore, the delivery of miRNA-34a through HA-PEI/HA-PEG nanoparticles may represent a novel challenge to counteract the Warburg effect and autophagy of cancer cells by promoting a glycolytic to oxidative metabolism switch and inducing deep changes in their metabolic settings, correlating with alterations of cell proliferation.
Other studies reported that polymeric nanostructures composed of biocompatible cationic β-cyclodextrin-PEI600 (CDP) or poly(L-lysine-graft-imidazole) (PLI) were loaded with anionic miRNA-34a and subsequently PEGylated to protect them from degradation, thus allowing a safe and efficient intracellular delivery (Jang et al., 2016; Fan et al., 2017).
These pH-sensitive nanoparticles released therapeutic miRNAs in the acid tumor microenvironment, favoring its cytoplasmic uptake by gastric and melanoma cancer cells.
The miRNA-34 loaded NPs repressed the expression of oncogenic CD44 protein with decreased levels of Bcl-2, Oct-3/4 and Nanog genes, thus leading the suppression of CSCs-like characteristics, induction of apoptosis, reduction of cell invasion and metastasis and tumor growth inhibition in xenograft gastric cancer models (Jang et al., 2016; Fan et al., 2017).
These polymer-based strategies of miRNA-34a delivery might represent novel therapeutic challenges with highly selective tumor cell death and tumor growth inhibition in CD44-positive tumors, thus opening a therapeutic window for autophagy inhibition for cancer treatment.
Biopolymer-Based Nanoparticles
Nanomaterials based on bovine serum albumin (BSA) are nontoxic, nonantigenic, and biodegradable polymers and have been widely exploited in drug delivery (Rhaese et al., 2003). The anionic side-chain carboxylic groups can be modified with a cationic amino group to obtain cationic bovine serum albumin (CBSA) (Fischer et al., 2001). This cationic structure can be conjugated to nanoparticles and act as a safe drug delivery system for nucleic acids (Han et al., 2014).
In this regard, core–shell nanocarriers coated by CBSA were developed for delivery of miRNA-34a and the chemotherapeutic drug docetaxel (DTX) for a cosynergistic treatment of metastatic breast cancer (Zhang et al., 2017). The coloaded nanocarriers (CNCs) could internalize through the caveolae-mediated pathway and exerted therapeutic effect by inducing cytotoxicity in vitro and by inhibiting tumor growth and metastasis in 4T1-tumor-bearing mice models (Zhang et al., 2017). Thus, these nanocarriers represent new nanoplatforms for the delivery of therapeutic miRNA-34 and provide a promising strategy for the treatment of metastatic breast cancer.
Chitosan, a partially deacetylated derivative of chitin composed of N-acetylglucosamine, has emerged as significant biopolymer for drug delivery because of its unique chemical proprieties such as biocompatibility, biodegradability, low toxicity, and easy modification (Mohammed et al., 2017). In a study performed by Cosco et al., safe chitosan/PLGA nanocomplexes have been developed that were able to efficiently encapsulate and deliver miRNA-34a in order to provide a new tool for the treatment of multiple myeloma (Cosco et al., 2015). These nanostructures led to a significant in vitro antitumor effect by reducing the proliferative capabilities of multiple myeloma cells. Moreover, the systemic injection of miRNA-34a-loaded nanoparticles significantly inhibited tumor growth through the downregulation of Bcl-2 and CDK6 expression and improved the survival of multiple myeloma xenografts in NOD-SCID mice (Cosco et al., 2015).
Among the various polymeric nanocomplex systems, hyaluronic acid-chitosan nanoparticles (HA-CS NPs) have been extensively studied. In addition to its biocompatibility and biodegradability, HA backbone possesses tumor-targeting properties through specific binding to CD44, an integral membrane glycoprotein overexpressed on the surface of various tumor cells (Aruffo et al., 1990) that makes it as an ideal polymer carrier for systemic drug delivery applications (De La Fuente et al., 2008).
It has been reported that HA-CS nano-complexes were able to simultaneously encapsulate and deliver positively charged Doxorubicin (DOX) and negatively charged miRNA-34a mimics into triple-negative breast cancer cells for improved chemotherapeutic effects (Deng et al., 2014). Interestingly, through the restoration of endogenous miRNA-34a levels, these nanoformulations synergistically enhanced antitumor effects of DOX in both in vitro and in vivo models by suppressing the expression of nonpump resistance and anti-apoptotic Bcl-2 protein. In addition, the delivery of miRNA-34a inhibited breast cancer cell migration via targeting Notch-1 signaling (Deng et al., 2014). Hence, nanosystem-based codelivery of chemotherapeutic agents and tumor suppressor miRNA-34a may be a promising combined therapeutic strategy for enhanced antitumor therapy.
Conclusions
The discovery and characterization of noncoding RNAs in the last 20 years have opened a new layer in the understanding of gene expression physiology as well as novel therapeutic strategies. In particular, various miRNAs, and miRNA-34a in particular, have been characterized as powerful tools regulating the expression of key genes relevant in the development of a disease, such as the control of autophagy during carcinogenesis.
However, despite many years of intense study, the translational impact on this knowledge is not satisfactory. Specific biologic characteristic of miRNAs, such as poor chemical stability and low membrane permeability, may preclude their use in clinics. For this purpose, nanotechnology has developed a bunch of new approaches aimed at enhancing the in vivo stability and at facilitating the delivery of specific miRNAs to the site of disease.
Thus, the use of several nanomaterials to deliver therapeutic nucleic acids, such as miRNA-34a, in cancer cells may result in the inhibition of protective autophagy leading to a tumor suppressor phenotype such as apoptosis induction, increased chemosensitivity and therapeutic efficacy.
The high potentiality of the use of encapsulated miRNAs is supported by the generation of MRX34, a lipid nanoparticle filled with miR-34 mimics, that has been the first microRNA-associated therapeutic drug tested in a clinical trial (Zhang et al., 2019). Indeed, a study on adults affected by solid tumors refractory to standard treatment that have been treated twice weekly for three weeks in 4-week cycles with MRX34 showed that this formulation exerts an evident antitumoral activity (Beg et al., 2017). Interestingly, the administered MRX34 was also found to be present in various tissues, including liver, bone marrow, spleen, mammary gland, and lung (Kelnar and Bader, 2015), thus supporting its application in the treatment of numerous cancer types.
Another evidence of the therapeutic application of delivered miRNA has been proposed by the use of the coencapsulated miR-34a and let-7b to NSCLC mice resistant to conventional anticancer therapy. The obtained data showed that the dual treated animals have a reduced tumor burden and a prolonged survival (Kasinski et al., 2015). Taking into account these data and the here reported formulations and applications of miRNA-34a delivery, the therapeutic usefulness of the development of novel miR-34a formulations may help to successfully achieve the clinical trial.
However, despite the potential advantages of the described nanoparticles, it is necessary to remember that the use of these nanomedicines may display some toxicity. Indeed, in some cases, nanomaterials may overstimulate autophagy in healthy tissues leading to dangerous effects, including inflammation, oxidative stress, and neoplastic transformation (Cordani and Somoza, 2019). Hence, it should be kept in mind that such nanostructures can produce undesired side effects whereby even if miRNA-34a is delivered in cancer cells, autophagy might increase in the same way by eliminating the beneficial effect of that therapeutic molecule.
In conclusion, deeper integration of knowledge on the role of autophagy in tumors with the recent advances in nanomedicine may allow an effective use of microRNA in the therapy of tumors.
Author Contributions
All authors contributed to the article and approved the submitted version. MC conceived the article, supervised the study, and coordinated the writing of the manuscript.
Funding
This work was supported by the Spanish Ministry of Economy and Competitiveness (SAF2017-87305-R) and by Fondi di Ateneo 2019 (Sapienza University). IMDEA Nanociencia acknowledges support from the ‘Severo Ochoa’ Programme for Centres of Excellence in R&D (MINECO, Grant SEV-2016-0686).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Alshamsan A. (2014). Nanoprecipitation is more efficient than emulsion solvent evaporation method to encapsulate cucurbitacin I in PLGA nanoparticles. Saudi Pharm. J. 22, 219–222. 10.1016/j.jsps.2013.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaravadi R. K., Yu D., Lum J. J., Bui T., Christophorou M. A., Evan G., II, et al. (2007). Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J. Clin. Invest. 117, 326–336. 10.1172/JCI28833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaravadi R. K., Kimmelman A. C., Debnath J. (2019). Targeting autophagy in cancer: Recent advances and future directions. Cancer Discovery 9, 1167–1181. 10.1158/2159-8290.CD-19-0292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruffo A., Stamenkovic I., Melnick M., Underhill C. B., Seed B. (1990). CD44 is the principal cell surface receptor for hyaluronate. Cell 61, 1303–1313. 10.1016/0092-8674(90)90694-A [DOI] [PubMed] [Google Scholar]
- Bardhan R., Lal S., Joshi A., Halas N. J. (2011). Theranostic nanoshells: From probe design to imaging and treatment of cancer. Acc. Chem. Res. 44, 936–946. 10.1021/ar200023x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basuroy U. K., Gerner E. W. (2006). Emerging concepts in targeting the polyamine metabolic pathway in epithelial cancer chemoprevention and chemotherapy. J. Biochem. 139, 27–33. 10.1093/jb/mvj022 [DOI] [PubMed] [Google Scholar]
- Beg M. S., Brenner A. J., Sachdev J., Borad M., Kang Y. K., Stoudemire J., et al. (2017). Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest. New Drugs 35, 180–188. 10.1007/s10637-016-0407-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berindan-Neagoe I., Calin G. A. (2014). Molecular pathways: MicroRNAs, cancer cells, and microenvironment. Clin. Cancer Res. 20, 6247–6253. 10.1158/1078-0432.CCR-13-2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouras T., Pal B., Vaillant F., Harburg G., Asselin-Labat M. L., Oakes S. R., et al. (2008). Notch Signaling Regulates Mammary Stem Cell Function and Luminal Cell-Fate Commitment. Cell Stem Cell 3, 429–441. 10.1016/j.stem.2008.08.001 [DOI] [PubMed] [Google Scholar]
- Chen Y., Zhu X., Zhang X., Liu B., Huang L. (2010). Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol. Ther. 18, 1650–1656. 10.1038/mt.2010.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Gao D. Y., Huang L. (2015). In vivo delivery of miRNAs for cancer therapy: Challenges and strategies. Adv. Drug Deliv. Rev. 81, 128–141. 10.1016/j.addr.2014.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Liu Y., Liang X., Huang Y., Li Q. (2017). Chondroitin sulfate-functionalized polyamidoamine as a tumor-targeted carrier for miR-34a delivery. Acta Biomater. 57, 238–250. 10.1016/j.actbio.2017.05.030 [DOI] [PubMed] [Google Scholar]
- Cheng X., Kuhn L. (2007). Chemotherapy drug delivery from calcium phosphate nanoparticles. Int. J. Nanomedicine 2, 667–674. [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Li Y. Y., Zeng X., Sun Y. X., Zhang X. Z., Zhuo R. X. (2009). Protamine sulfate/poly(l-aspartic acid) polyionic complexes self-assembled via electrostatic attractions for combined delivery of drug and gene. Biomaterials 30, 1246–1253. 10.1016/j.biomaterials.2008.11.002 [DOI] [PubMed] [Google Scholar]
- Cheng X., Xu Q., Zhang Y., Shen M., Zhang S., Mao F., et al. (2019). miR-34a inhibits progression of neuroblastoma by targeting autophagy-related gene 5. Eur. J. Pharmacol. 850, 53–63. 10.1016/j.ejphar.2019.01.071 [DOI] [PubMed] [Google Scholar]
- Chino H., Hatta T., Natsume T., Mizushima N. (2019). Intrinsically Disordered Protein TEX264 Mediates ER-phagy. Mol. Cell. 74, 909–921. 10.1016/j.molcel.2019.03.033 [DOI] [PubMed] [Google Scholar]
- Cohen-Sela E., Teitlboim S., Chorny M., Koroukhov N., Danenberc H. D., Gao J., et al. (2009). Single and double emulsion manufacturing techniques of an amphiphilic drug in PLGA nanoparticles: Formulations of mithramycin and bioactivity. J. Pharm. Sci. 98, 1452–1462. 10.1002/jps.21527 [DOI] [PubMed] [Google Scholar]
- Cordani M., Somoza Á. (2019). Targeting autophagy using metallic nanoparticles: a promising strategy for cancer treatment. Cell. Mol. Life Sci. 76, 1215–1242. 10.1007/s00018-018-2973-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosco D., Cilurzo F., Maiuolo J., Federico C., Di Martino M. T., Cristiano M. C., et al. (2015). Delivery of miR-34a by chitosan/PLGA nanoplexes for the anticancer treatment of multiple myeloma. Sci. Rep. 5:17579. 10.1038/srep17579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha L. D., Yang M., Carter R., Guy C., Harris L., Crawford J. C., et al. (2018). LC3-Associated Phagocytosis in Myeloid Cells Promotes Tumor Immune Tolerance. Cell 175, 429–441. 10.1016/j.cell.2018.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Fuente M., Seijo B., Alonso M. J. (2008). Novel hyaluronic acid-chitosan nanoparticles for ocular gene therapy. Investig. Ophthalmol. Vis. Sci. 49, 2016–2024. 10.1167/iovs.07-1077 [DOI] [PubMed] [Google Scholar]
- Declerck P. J., Lijnen H. R., Verstreken M., Moreau H., Collen D. (1990). A monoclonal antibody specific for two-chain urokinase-type plasminogen activator. Application to the study of the mechanism of clot lysis with single-chain urokinase-type plasminogen activator in plasma. Blood 75, 1794–1800. 10.1182/blood.v75.9.1794.bloodjournal7591794 [DOI] [PubMed] [Google Scholar]
- Deng X., Cao M., Zhang J., Hu K., Yin Z., Zhou Z., et al. (2014). Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer. Biomaterials 35, 4333–4344. 10.1016/j.biomaterials.2014.02.006 [DOI] [PubMed] [Google Scholar]
- Deretic V., Saitoh T., Akira S. (2013). Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 13, 722–737. 10.1038/nri3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino M. T., Campani V., Misso G., Gallo Cantafio M. E., Gullà A., Foresta U., et al. (2014). In vivo activity of MiR-34a mimics delivered by stable nucleic acid lipid particles (SNALPs) against multiple myeloma. PloS One 9, e90005. 10.1371/journal.pone.0090005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao J., Liu R., Rong Y., Zhao M., Zhang J., Lai Y., et al. (2015). ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 520, 563–566. 10.1038/nature14147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I., Elazar Z. (2018). Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349–364. 10.1038/s41580-018-0003-4 [DOI] [PubMed] [Google Scholar]
- Dowdle W. E., Nyfeler B., Nagel J., Elling R. A., Liu S., Triantafellow E., et al. (2014). Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat. Cell Biol. 16, 1069–1079. 10.1038/ncb3053 [DOI] [PubMed] [Google Scholar]
- Egan D. F., Chun M. G. H., Vamos M., Zou H., Rong J., Miller C. J., et al. (2015). Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Mol. Cell 59, 285–297. 10.1016/j.molcel.2015.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A., Slack F. J. (2006). Oncomirs - MicroRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269. 10.1038/nrc1840 [DOI] [PubMed] [Google Scholar]
- Fan M., Zeng Y., Ruan H., Zhang Z., Gong T., Sun X. (2017). Ternary Nanoparticles with a Sheddable Shell Efficiently Deliver MicroRNA-34a against CD44-Positive Melanoma. Mol. Pharm. 14, 3152–3163. 10.1021/acs.molpharmaceut.7b00377 [DOI] [PubMed] [Google Scholar]
- Fan Q., Yang L., Zhang X., Ma Y., Li Y., Dong L., et al. (2018). Autophagy promotes metastasis and glycolysis by upregulating MCT1 expression and Wnt/β-catenin signaling pathway activation in hepatocellular carcinoma cells. J. Exp. Clin. Cancer Res. 37, 9. 10.1186/s13046-018-0673-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Yuan F., Yun Y., Wu T., Lu L., Liu J., et al. (2019). MicroRNA-34a mediates ethanol-induced impairment of neural differentiation of neural crest cells by targeting autophagy-related gene 9a. Exp. Neurol. 320:112981. 10.1016/j.expneurol.2019.112981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqi A. A., Tabassum S., Ahmad A. (2017). MicroRNA-34a: A versatile regulator of myriads of targets in different cancers. Int. J. Mol. Sci. 18, 2089. 10.3390/ijms18102089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessi H., Puisieux F., Devissaguet J. P., Ammoury N., Benita S. (1989). Nanocapsule formation by interfacial polymer deposition following solvent displacement. Int. J. Pharm. 55, R1–R4. 10.1016/0378-5173(89)90281-0 [DOI] [Google Scholar]
- Fischer D., Bieber T., Brüsselbach S., Elsässer H. P., Kissel T. (2001). Cationized human serum albumin as a non-viral vector system for gene delivery? Characterization of complex formation with plasmid DNA and transfection efficiency. Int. J. Pharm. 225, 97–111. 10.1016/S0378-5173(01)00765-7 [DOI] [PubMed] [Google Scholar]
- Ganesh S., Iyer A. K., Morrissey D. V., Amiji M. M. (2013). Hyaluronic acid based self-assembling nanosystems for CD44 target mediated siRNA delivery to solid tumors. Biomaterials 34, 3489–3502. 10.1016/j.biomaterials.2013.01.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P., Han G., De M., Kim C. K., Rotello V. M. (2008). Gold nanoparticles in delivery applications. Adv. Drug Deliv. Rev. 60, 1307–1315. 10.1016/j.addr.2008.03.016 [DOI] [PubMed] [Google Scholar]
- Goyal R., Kapadia C. H., Melamed J. R., Riley R. S., Day E. S. (2018). Layer-by-Layer Assembled Gold Nanoshells for the Intracellular Delivery of miR-34a. Cell. Mol. Bioeng. 11, 383–396. 10.1007/s12195-018-0535-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. Y., Chen H. Y., Mathew R., Fan J., Strohecker A. M., Karsli-Uzunbas G., et al. (2011). Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 25, 460–470. 10.1101/gad.2016311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., et al. (2013). Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393. 10.1038/nature11910 [DOI] [PubMed] [Google Scholar]
- Han J., Wang Q., Zhang Z., Gong T., Sun X. (2014). Cationic bovine serum albumin based self-assembled nanoparticles as siRNA delivery vector for treating lung metastatic cancer. Small 10, 524–535. 10.1002/smll.201301992 [DOI] [PubMed] [Google Scholar]
- Han H., Chen W., Yang J., Zhang J., Li Q., Yang Y. (2018). 2-Amino-6-chloropurine-modified polyamidoamine-mediated p53 gene transfection to achieve anti-tumor efficacy. New J. Chem. 42, 13375–13381. 10.1039/c8nj01870g [DOI] [Google Scholar]
- Heckmann B. L., Teubner B. J. W., Tummers B., Boada-Romero E., Harris L., Yang M., et al. (2019). LC3-Associated Endocytosis Facilitates β-Amyloid Clearance and Mitigates Neurodegeneration in Murine Alzheimer’s Disease. Cell 178, 536–551. 10.1016/j.cell.2019.05.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q. L., Jiang Q. Y., Jin X., Shen J., Wang K., Li Y. B., et al. (2013). Cationic microRNA-delivering nanovectors with bifunctional peptides for efficient treatment of PANC-1 xenograft model. Biomaterials 34, 2265–2276. 10.1016/j.biomaterials.2012.12.016 [DOI] [PubMed] [Google Scholar]
- Huang J., Sun W., Huang H., Ye J., Pan W., Zhong Y., et al. (2014). MiR-34a modulates angiotensin II-induced myocardial hypertrophy by direct inhibition of ATG9A Expression and Autophagic Activity. PloS One 9, e94382. 10.1371/journal.pone.0094382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabir N. R., Tabrez S., Ashraf G. M., Shakil S., Damanhouri G. A., Kamal M. A. (2012). Nanotechnology-based approaches in anticancer research. Int. J. Nanomedicine 7, 4391–4408. 10.2147/IJN.S33838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R. K., Stylianopoulos T. (2010). Delivering nanomedicine to solid tumors. Nat. Rev. Clin. Oncol. 7, 653–664. 10.1038/nrclinonc.2010.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang E., Kim E., Son H. Y., Lim E. K., Lee H., Choi Y., et al. (2016). Nanovesicle-mediated systemic delivery of microRNA-34a for CD44 overexpressing gastric cancer stem cell therapy. Biomaterials 105, 12–24. 10.1016/j.biomaterials.2016.07.036 [DOI] [PubMed] [Google Scholar]
- Jenjob R., Phakkeeree T., Seidi F., Theerasilp M., Crespy D. (2019). Emulsion Techniques for the Production of Pharmacological Nanoparticles. Macromol. Biosci. 19, e1900063. 10.1002/mabi.201900063 [DOI] [PubMed] [Google Scholar]
- Jia H. z., Zhu J., Wang X., Cheng H., Chen G., Zhao Y. f., et al. (2014). A boronate-linked linear-hyperbranched polymeric nanovehicle for pH-dependent tumor-targeted drug delivery. Biomaterials 35, 5240–5249. 10.1016/j.biomaterials.2014.03.029 [DOI] [PubMed] [Google Scholar]
- Jiao L., Zhang H. L., Li D. D., Yang K. L., Tang J., Li X., et al. (2018). Regulation of glycolytic metabolism by autophagy in liver cancer involves selective autophagic degradation of HK2 (hexokinase 2). Autophagy 14, 671–684. 10.1080/15548627.2017.1381804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan M. A., Wilson L. (2004). Microtubules as a target for anticancer drugs. Nat. Rev. Cancer. 4, 253–265. 10.1038/nrc1317 [DOI] [PubMed] [Google Scholar]
- Jung H., Kim S. A., Yang Y. G., Yoo H., Lim S. J., Mok H. (2015). Long chain microRNA conjugates in calcium phosphate nanoparticles for efficient formulation and delivery. Arch. Pharm. Res. 38, 705–715. 10.1007/s12272-014-0451-0 [DOI] [PubMed] [Google Scholar]
- Kasinski A. L., Kelnar K., Stahlhut C., Orellana E., Zhao J., Shimer E., et al. (2015). A combinatorial microRNA therapeutics approach to suppressing non-small cell lung cancer. Oncogene 34, 3547–3555. 10.1038/onc.2014.282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S., Massey A. C., Cuervo A. M. (2006). Lysosome membrane lipid microdomains: Novel regulators of chaperone-mediated autophagy. EMBO J. 25, 3921–3933. 10.1038/sj.emboj.7601283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S., Bandyopadhyay U., Sridhar S., Kiffin R., Martinez-Vicente M., Kon M., et al. (2011). Chaperone-mediated autophagy at a glance. J. Cell Sci. 124, 495–499. 10.1242/jcs.073874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelnar K., Bader A. G. (2015). A qRT-PCR method for determining the biodistribution profi le of a miR-34a mimic. Methods Mol. Biol. 1317, 125–133. 10.1007/978-1-4939-2727-2_8 [DOI] [PubMed] [Google Scholar]
- Khaminets A., Heinrich T., Mari M., Grumati P., Huebner A. K., Akutsu M., et al. (2015). Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 522, 354–358. 10.1038/nature14498 [DOI] [PubMed] [Google Scholar]
- Kim E. J., Shim G., Kim K., Kwon I. C., Oh Y. K., Shim C. K. (2009). Hyaluronic acid complexed to biodegradable poly L-arginine for targeted delivery of siRNAs. J. Gene Med. 11, 791–803. 10.1002/jgm.1352 [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., Abdelmohsen K., Abe A., Abedin M. J., Abeliovich H., Arozena A. A., et al. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222. 10.1080/15548627.2015.1100356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Guru S. K., Pathania A. S., Kumar A., Bhushan S., Malik F. (2013). Autophagy triggered by magnolol derivative negatively regulates angiogenesis. Cell Death Dis. 4, e889. 10.1038/cddis.2013.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Gu Y., Abudu Y. P., Bruun J. A., Jain A., Farzam F., et al. (2019). Phosphorylation of Syntaxin 17 by TBK1 Controls Autophagy Initiation. Dev. Cell 49, 130–144.e6. 10.1016/j.devcel.2019.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Kim H. O., Son H. Y., Lee S. B., Jang E., Kang B., et al. (2016). Magnetic nanovector enabling miRNA-34a delivery for CD44 suppression with concurrent MR imaging. J. Nanosci. Nanotechnol. 16, 12939–12946. 10.1166/jnn.2016.13797 [DOI] [Google Scholar]
- Lee C. S., Lee L. C., Yuan T. L., Chakka S., Fellmann C., Lowe S. W., et al. (2019). MAP kinase and autophagy pathways cooperate to maintain RAS mutant cancer cell survival. Proc. Natl. Acad. Sci. U. S. A. 116, 4508–4517. 10.1073/pnas.1817494116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Yuan J. (2005). Autophagy in cell death: An innocent convict? J. Clin. Invest. 115, 2679–2688. 10.1172/JCI26390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B. (2005). Eating oneself and uninvited guests: Autophagy-related pathways in cellular defense. Cell 120, 159–162. 10.1016/j.cell.2005.01.005 [DOI] [PubMed] [Google Scholar]
- Levy J. M. M., Towers C. G., Thorburn A. (2017). Targeting autophagy in cancer. Nat. Rev. Cancer 17, 528–542. 10.1038/nrc.2017.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. D., Chen Y. C., Hackett M. J., Huang L. (2008). Tumor-targeted delivery of siRNA by self-assembled nanoparticles. Mol. Ther. 16, 163–169. 10.1038/sj.mt.6300323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. W., Li J., Bao J. K. (2012). Microautophagy: Lesser-known self-eating. Cell. Mol. Life Sci. 69, 1125–1136. 10.1007/s00018-011-0865-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Yuan L., Luo J., Gao J., Guo J., Xie X. (2013). MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin. Exp. Med. 13, 109–117. 10.1007/s10238-012-0186-5 [DOI] [PubMed] [Google Scholar]
- Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., et al. (1999). Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676. 10.1038/45257 [DOI] [PubMed] [Google Scholar]
- Liao H., Xiao Y., Hu Y., Xiao Y., Yin Z., Liu L., et al. (2016). Methylation-induced silencing of miR-34a enhances chemoresistance by directly upregulating ATG4B-induced autophagy through AMPK/mTOR pathway in prostate cancer. Oncol. Rep. 35, 64–72. 10.3892/or.2015.4331 [DOI] [PubMed] [Google Scholar]
- Lindqvist L. M., Heinlein M., Huang D. C. S., Vaux D. L. (2014). Prosurvival Bcl-2 family members affect autophagy only indirectly, by inhibiting Bax and Bak. Proc. Natl. Acad. Sci. U. S. A. 111, 8512–8517. 10.1073/pnas.1406425111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Yang M., Kang R., Wang Z., Zhao Y., Yu Y., et al. (2011). HMGB1-induced autophagy promotes chemotherapy resistance in leukemia cells. Leukemia 25, 23–31. 10.1038/leu.2010.225 [DOI] [PubMed] [Google Scholar]
- Liu K., Huang J., Xie M., Yu Y., Zhu S., Kang R., et al. (2014). MIR34A regulates autophagy and apoptosis by targeting HMGB1 in the retinoblastoma cell. Autophagy 10, 442–452. 10.4161/auto.27418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Ren W., Chen K. (2017). MiR-34a promotes apoptosis and inhibits autophagy by targeting HMGB1 in acute myeloid leukemia cells. Cell. Physiol. Biochem. 41, 1981–1992. 10.1159/000475277 [DOI] [PubMed] [Google Scholar]
- Lu Y., Gu J., Jin D., Gao Y., Yuan M. (2011). Inhibition of telomerase activity by HDV ribozyme in cancers. J. Exp. Clin. Cancer Res. 30:1. 10.1186/1756-9966-30-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugrin J., Rosenblatt-Velin N., Parapanov R., Liaudet L. (2014). The role of oxidative stress during inflammatory processes. Biol. Chem. 395, 203–230. 10.1515/hsz-2013-0241 [DOI] [PubMed] [Google Scholar]
- Ma Y., Galluzzi L., Zitvogel L., Kroemer G. (2013). Autophagy and cellular immune responses. Immunity 39, 211–227. 10.1016/j.immuni.2013.07.017 [DOI] [PubMed] [Google Scholar]
- Maejima I., Takahashi A., Omori H., Kimura T., Takabatake Y., Saitoh T., et al. (2013). Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 32, 2336–2347. 10.1038/emboj.2013.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel N., Sarin A. (2016). Notch1 regulated autophagy controls survival and suppressor activity of activated murine T-regulatory cells. Elife 5, e14023. 10.7554/eLife.14023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K. R., Celano S. L., Solitro A. R., Gunaydin H., Scott M., O’Hagan R. C., et al. (2018). A Potent and Selective ULK1 Inhibitor Suppresses Autophagy and Sensitizes Cancer Cells to Nutrient Stress. iScience 8, 74–84. 10.1016/j.isci.2018.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Malireddi R. K. S., Lu Q., Cunha L. D., Pelletier S., Gingras S., et al. (2015). Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat. Cell Biol. 17, 893–906. 10.1038/ncb3192 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Martinez J., Cunha L. D., Park S., Yang M., Lu Q., Orchard R., et al. (2016). Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature 533, 115–119. 10.1038/nature17950 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Martínez Rivas C. J., Tarhini M., Badri W., Miladi K., Greige-Gerges H., Nazari Q. A., et al. (2017). Nanoprecipitation process: From encapsulation to drug delivery. Int. J. Pharm. 532, 66–81. 10.1016/j.ijpharm.2017.08.064 [DOI] [PubMed] [Google Scholar]
- McCall R. L., Sirianni R. W. (2013). PLGA nanoparticles formed by single- or double-emulsion with vitamin E-TPGS. J. Vis. Exp. 82, 51015. 10.3791/51015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y., Wang R., Jiang W., Bo Y., Zhang T., Yu J., et al. (2019). Recent progress in nanomaterials for nucleic acid delivery in cancer immunotherapy. Biomater. Sci. 7, 2640–2651. 10.1039/c9bm00214f [DOI] [PubMed] [Google Scholar]
- Milán Rois P., Latorre A., Rodriguez Diaz C., del Moral Á., Somoza Á. (2018). Reprogramming Cells for Synergistic Combination Therapy with Nanotherapeutics against Uveal Melanoma. Biomimetics 3:28. 10.3390/biomimetics3040028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra V., Bansal K., Verma A., Yadav N., Thakur S., Sudhakar K., et al. (2018). Solid Lipid Nanoparticles: Emerging Colloidal Nano Drug Delivery Systems. Pharmaceutics 10:191. 10.3390/pharmaceutics10040191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Ohsumi Y. (2011). The Role of Atg Proteins in Autophagosome Formation. Annu. Rev. Cell Dev. Biol. 27, 107–132. 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- Mohammed M., Syeda J., Wasan K., Wasan E. (2017). An Overview of Chitosan Nanoparticles and Its Application in Non-Parenteral Drug Delivery. Pharmaceutics 9:53. 10.3390/pharmaceutics9040053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrakovcic M., Fröhlich L. F. (2018). p53-Mediated Molecular Control of Autophagy in Tumor Cells. Biomolecules 8:14. 10.3390/biom8020014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R. H., Mäder K., Gohla S. (2000). Solid lipid nanoparticles (SLN) for controlled drug delivery - A review of the state of the art. Eur. J. Pharm. Biopharm. 50, 161–177. 10.1016/S0939-6411(00)00087-4 [DOI] [PubMed] [Google Scholar]
- Naik P. P., Mukhopadhyay S., Panda P. K., Sinha N., Das C. K., Mishra R., et al. (2018). Autophagy regulates cisplatin-induced stemness and chemoresistance via the upregulation of CD44, ABCB1 and ADAM17 in oral squamous cell carcinoma. Cell Prolif. 51, e12411. 10.1111/cpr.12411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito M., Ishii T., Matsumoto A., Miyata K., Miyahara Y., Kataoka K. (2012). A phenylboronate-functionalized polyion complex micelle for ATP-triggered release of siRNA. Angew. Chemie - Int. Ed. 51, 10751–10755. 10.1002/anie.201203360 [DOI] [PubMed] [Google Scholar]
- Narayanan S. (1994). Sialic acid as a tumor marker. Ann. Clin. Lab. Sci. 24, 376–384. [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008). Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803. 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz S., Shamoon M., Wang R., Zhang L., Zhou J., Chen J. (2019). Advances in Therapeutic Implications of Inorganic Drug Delivery Nano-Platforms for Cancer. Int. J. Mol. Sci. 20:965. 10.3390/ijms20040965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarko T. Y., Farré J. C., Polupanov A. S., Sibirny A. A., Subramani S. (2007). Autophagy-related pathways and specific role of sterol glucoside in yeasts. Autophagy 3, 263–265. 10.4161/auto.3907 [DOI] [PubMed] [Google Scholar]
- Palakurthi S. (2015). Challenges in SN38 drug delivery: Current success and future directions. Expert Opin. Drug Deliv. 12, 1911–1921. 10.1517/17425247.2015.1070142 [DOI] [PubMed] [Google Scholar]
- Panebianco F., Climent M., Malvindi M. A., Pompa P. P., Bonetti P., Nicassio F. (2019). Delivery of biologically active miR-34a in normal and cancer mammary epithelial cells by synthetic nanoparticles. Nanomedicine Nanotechnology Biol. Med. 19, 95–105. 10.1016/j.nano.2019.03.013 [DOI] [PubMed] [Google Scholar]
- Park J. H., Gu L., Von Maltzahn G., Ruoslahti E., Bhatia S. N., Sailor M. J. (2009). Biodegradable luminescent porous silicon nanoparticles for in vivo applications. Nat. Mater. 8, 331–336. 10.1038/nmat2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S., Tassa A., Qu X., Garuti R., Xiao H. L., Mizushima N., et al. (2005). Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122, 927–939. 10.1016/j.cell.2005.07.002 [DOI] [PubMed] [Google Scholar]
- Pegg A. E., Casero R. A. (2011). Current status of the polyamine research field. Methods Mol. Biol. 720, 3–35. 10.1007/978-1-61779-034-8_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Croce C. M. (2016). The role of MicroRNAs in human cancer. Signal Transduction Targeting Ther. 1, 15004. 10.1038/sigtrans.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergushov D. V., Müller A. H. E., Schacher F. H. (2012). Micellar interpolyelectrolyte complexes. Chem. Soc Rev. 41, 6888–6901. 10.1039/c2cs35135h [DOI] [PubMed] [Google Scholar]
- Pietrocola F., Lachkar S., Enot D. P., Niso-Santano M., Bravo-San Pedro J. M., Sica V., et al. (2015). Spermidine induces autophagy by inhibiting the acetyltransferase EP300. Cell Death Differ. 22, 509–516. 10.1038/cdd.2014.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poillet-Perez L., Xie X., Zhan L., Yang Y., Sharp D. W., Hu Z. S., et al. (2018). Autophagy maintains tumour growth through circulating arginine. Nature 563, 569–573. 10.1038/s41586-018-0697-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz J. D., White E. (2010). Autophagy and metabolism. Sci. (80-. ). 330, 1344–1348. 10.1126/science.1193497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhaese S., Von Briesen H., Rübsamen-Waigmann H., Kreuter J., Langer K. (2003). Human serum albumin-polyethylenimine nanoparticles for gene delivery. J. Control. Release 92, 199–208. 10.1016/S0168-3659(03)00302-X [DOI] [PubMed] [Google Scholar]
- Ronan B., Flamand O., Vescovi L., Dureuil C., Durand L., Fassy F., et al. (2014). A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 10, 1013–1019. 10.1038/nchembio.1681 [DOI] [PubMed] [Google Scholar]
- Salatin S., Barar J., Barzegar-Jalali M., Adibkia K., Kiafar F., Jelvehgari M. (2017). Development of a nanoprecipitation method for the entrapment of a very water soluble drug into Eudragit RL nanoparticles. Res. Pharm. Sci. 12, 1–14. 10.4103/1735-5362.199041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan M. A., Dillon C. P., Tait S. W. G., Moshiach S., Dorsey F., Connell S., et al. (2007). Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253–1257. 10.1038/nature06421 [DOI] [PubMed] [Google Scholar]
- Scott C., Wu D., Ho C. C., Co C. C. (2005). Liquid-core capsules via interfacial polymerization: A free-radical analogy of the nylon rope trick. J. Am. Chem. Soc 127, 4160–4161. 10.1021/ja044532h [DOI] [PubMed] [Google Scholar]
- Shi S., Han L., Gong T., Zhang Z., Sun X. (2013). Systemic delivery of microRNA-34a for cancer stem cell therapy. Angew. Chemie - Int. Ed. 52, 3901–3905. 10.1002/anie.201208077 [DOI] [PubMed] [Google Scholar]
- Shi S., Han L., Deng L., Zhang Y., Shen H., Gong T., et al. (2014). Dual drugs (microRNA-34a and paclitaxel)-loaded functional solid lipid nanoparticles for synergistic cancer cell suppression. J. Control. Release 194, 228–237. 10.1016/j.jconrel.2014.09.005 [DOI] [PubMed] [Google Scholar]
- Sierra H., Cordova M., Chen C. S. J., Rajadhyaksha M. (2015). Confocal imaging-guided laser ablation of basal cell carcinomas: An ex vivo study. J. Invest. Dermatol. 135, 612–615. 10.1038/jid.2014.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singha K., Namgung R., Kim W. J. (2011). Polymers in small-interfering RNA delivery. Nucleic Acid Ther. 21, 133–147. 10.1089/nat.2011.0293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Fan J. B., Wang S. (2017). Recent progress in interfacial polymerization. Mater. Chem. Front. 1, 1028–1040. 10.1039/c6qm00325g [DOI] [Google Scholar]
- Song Z., Liang X., Wang Y., Han H., Yang J., Fang X., et al. (2019). Phenylboronic acid-functionalized polyamidoamine-mediated miR-34a delivery for the treatment of gastric cancer. Biomater. Sci. 7, 1632–1642. 10.1039/c8bm01385c [DOI] [PubMed] [Google Scholar]
- Starobinets H., Ye J., Broz M., Barry K., Goldsmith J., Marsh T., et al. (2016). Antitumor adaptive immunity remains intact following inhibition of autophagy and antimalarial treatment. J. Clin. Invest. 126, 4417–4429. 10.1172/JCI85705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart T. M., Dunston T. T., Woster P. M., Casero R. A. (2018). Polyamine catabolism and oxidative damage. J. Biol. Chem. 293, 18736–18745. 10.1074/jbc.TM118.003337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugo M., Kimura H., Arasaki K., Amemiya T., Hirota N., Dohmae N., et al. (2018). Syntaxin 17 regulates the localization and function of PGAM5 in mitochondrial division and mitophagy. EMBO J. 37, e98899. 10.15252/embj.201798899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Wang F. J., Zhang H. G., Xu X. Z., Jia R. C., Yao L., et al. (2017). MIR-34a mediates oxaliplatin resistance of colorectal cancer cells by inhibiting macroautophagy via transforming growth factor-β/Smad4 pathway. World J. Gastroenterol. 23, 1816–1827. 10.3748/wjg.v23.i10.1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W., Wang H., Chen Y., Zhang X., Zhu H., Yang C., et al. (2011). Molecular aptamers for drug delivery. Trends Biotechnol. 29, 634–640. 10.1016/j.tibtech.2011.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Li L., Chen D. (2012). Mesoporous silica nanoparticles: Synthesis, biocompatibility and drug delivery. Adv. Mater. 24, 1504–1534. 10.1002/adma.201104763 [DOI] [PubMed] [Google Scholar]
- Tooze S. A., Dikic I. (2016). Autophagy Captures the Nobel Prize. Cell 167, 1433–1435. 10.1016/j.cell.2016.11.023 [DOI] [PubMed] [Google Scholar]
- Townsend K. N., Hughson L. R. K., Schlie K., Poon V., II, Westerback A., Lum J. J. (2012). Autophagy inhibition in cancer therapy: Metabolic considerations for antitumor immunity. Immunol. Rev. 249, 176–194. 10.1111/j.1600-065X.2012.01141.x [DOI] [PubMed] [Google Scholar]
- Trivedi M., Singh A., Talekar M., Pawar G., Shah P., Amiji M. (2017). MicroRNA-34a Encapsulated in Hyaluronic Acid Nanoparticles Induces Epigenetic Changes with Altered Mitochondrial Bioenergetics and Apoptosis in Non-Small-Cell Lung Cancer Cells. Sci. Rep. 7, 3636. 10.1038/s41598-017-02816-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Zhang L., Chen Z., Meng Z. (2013). MicroRNA targets autophagy in pancreatic cancer cells during cancer therapy. Autophagy 9, 2171–2172. 10.4161/auto.26463 [DOI] [PubMed] [Google Scholar]
- Wang H., Zhao X., Guo C., Ren D., Zhao Y., Xiao W., et al. (2015. a). Aptamer-dendrimer bioconjugates for targeted delivery of miR-34a expressing plasmid and antitumor effects in non-small cell lung cancer cells. PloS One 10, e0139136. 10.1371/journal.pone.0139136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Cao M., Deng X., Xiao X., Yin Z., Hu Q., et al. (2015. b). Degradable hyaluronic acid/protamine sulfate interpolyelectrolyte complexes as miRNA-delivery nanocapsules for triple-negative breast cancer therapy. Adv. Healthc. Mater. 4, 281–290. 10.1002/adhm.201400222 [DOI] [PubMed] [Google Scholar]
- Watson A., Riffelmacher T., Stranks A., Williams O., De Boer J., Cain K., et al. (2015). Autophagy limits proliferation and glycolytic metabolism in acute myeloid leukemia. Cell Death Discovery 1, 15008. 10.1038/cddiscovery.2015.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Wei S., Gan B., Peng X., Zou W., Guan J. L. (2011). Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 25, 1510–1527. 10.1101/gad.2051011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm S., Tavares A. J., Dai Q., Ohta S., Audet J., Dvorak H. F., et al. (2016). Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 1, 16014. 10.1038/natrevmats.2016.14 [DOI] [Google Scholar]
- Wu Y., Dai X., Ni Z., Yan X., He F., Lian J. (2017). The downregulation of ATG4B mediated by microRNA-34a/34c-5p suppresses rapamycin-induced autophagy. Iran. J. Basic Med. Sci. 20, 1125–1130. 10.22038/IJBMS.2017.9446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm F. R., Weiss C. K. (2014). Nanoparticles from renewable polymers. Front. Chem. 2:49:49. 10.3389/fchem.2014.00049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Wang W., Crespo J., Kryczek I., Li W., Wei S., et al. (2017). Suppression of FIP200 and autophagy by tumor-derived lactate promotes naïve T cell apoptosis and affects tumor immunity. Sci. Immunol. 2, eaan4631. 10.1126/sciimmunol.aan4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Wang X., Gao H., Chen X., Li J., Zhang Y. (2018). Cell-Selective Delivery of MicroRNA with a MicroRNA–Peptide Conjugate Nanocomplex. Chem. - Asian J. 13, 3845–3849. 10.1002/asia.201801396 [DOI] [PubMed] [Google Scholar]
- Xie Y., Murray-Stewart T., Wang Y., Yu F., Li J., Marton L. J., et al. (2017). Self-immolative nanoparticles for simultaneous delivery of microRNA and targeting of polyamine metabolism in combination cancer therapy. J. Control. Release 246, 110–119. 10.1016/j.jconrel.2016.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong F., Huang S., Gu N. (2018). Magnetic nanoparticles: recent developments in drug delivery system. Drug Dev. Ind. Pharm. 44, 697–706. 10.1080/03639045.2017.1421961 [DOI] [PubMed] [Google Scholar]
- Xu Z., Wang Y., Zhang L., Huang L. (2014). Nanoparticle-delivered transforming growth factor-β siRNA enhances vaccination against advanced melanoma by modifying tumor microenvironment. ACS Nano 8, 3636–3645. 10.1021/nn500216y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Chen D., He Y., Meléndez A., Feng Z., Hong Q., et al. (2013). MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age (Omaha). 35, 11–22. 10.1007/s11357-011-9324-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Yang X., Lv Y., Xin X., Qin C., Han X., et al. (2017). Cytosolic co-delivery of miRNA-34a and docetaxel with core-shell nanocarriers via caveolae-mediated pathway for the treatment of metastatic breast cancer. Sci. Rep. 7:46186. 10.1038/srep46186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Liao Y., Tang L. (2019). MicroRNA-34 family: A potential tumor suppressor and therapeutic candidate in cancer. J. Exp. Clin. Cancer Res. 38, 53. 10.1186/s13046-019-1059-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. Y., Li Y., Jiang W. Q., Zhou L. F. (2015). MAPK/JNK signalling: A potential autophagy regulation pathway. Biosci. Rep. 35, 1–10. 10.1042/BSR20140141 [DOI] [PMC free article] [PubMed] [Google Scholar]