Abstract
Research characterizing arthropod-associated microbiota has revealed that microbial dynamics can have an important impact on host phenotypic traits. The influence of fungi on these interactions are emerging as targets for research, especially in organisms associated with global human health. A recent study demonstrated colonization of a widespread gut fungus (Zancudomyces culisetae) in a larval mosquito (Aedes aegypti) digestive tract affected microbiomes in larvae and newly emerged adult females (Frankel-Bricker et al. Appl Environ Microbiol, 2020. 10.1128/AEM.02334-19) but did not investigate these processes in males. The objective of the study presented here was to assess fungal influences on adult male mosquito microbiomes to enable a more complete assessment of fungal–bacterial–host interactions in the A. aegypti–Z. culisetae system. Sequencing of 16S rRNA gene amplicons from microbiomes harbored in adult males directly after emerging from pupae revealed larval fungal exposure significantly decreased overall microbial community diversity, altered microbiome composition and structure, and decreased within-group microbiome variation across individuals. Further, bacteria in the family Burkholderiaceae were present in high abundance in fungal-exposed males, likely contributing to the disparate microbiota between treatment groups. Comparisons between male and the female microbiomes analyzed in Frankel-Bricker et al. (2020), showed distinct shifts in bacterial communities incurred by larval exposure to fungi, potentially revealing sex-specific fungal–bacterial–host dynamics in A. aegypti. These findings highlight the complex role a gut fungus can play in influencing the microbial communities harbored in an important insect and emphasize the significance of accounting for an organism’s sex when studying fungal–bacterial–host dynamics.
Subject terms: Applied microbiology, Metagenomics, Microbial ecology, Microbiome, Fungal biology
Introduction
Mosquitoes harbor communities of microbes that impact host phenotypic traits1,2. Studies investigating factors that contribute to microbial community dynamics provide valuable biological insights into these insects of global significance. Research analyzing mosquito-associated microbiota often investigate bacteria harbored in females1–11, in part, due to the distinct capacity of adult females to contract and transmit (vector competence) a variety of human pathogens. Importantly, studies show that certain microbes influence adult female-pathogen interactions12–16 and alter vector competence17–19, demonstrating how host-microbe interactions affect the proliferation of mosquito-borne diseases. While further analyses of female microbiomes are essential, studies of male-associated microbiota are also necessary. Males have the capacity to transmit certain viruses to females via venereal transmission20,21, demonstrating complex, sex-specific host–pathogen interactions occur in this system. Research investigating factors influencing male-associated microbiota are necessary to provide complete assessments of these intricate dynamics.
Several studies comparing male- and female-associated microbiota have revealed distinct characteristics of bacterial communities between the sexes22,23. These differences are potentially influenced by disparate ecological behaviors24,25, environmental microbiota9,26–28, and sources of nutrients11,29. In addition, unique microbiomes in sex-specific anatomical regions may also contribute to these divergent microbial community profiles30. Male testes and female ovaries harbor similar microbial taxa; however, these microbiomes have different distribution structures31,32. Further, the same bacteria exhibit distinct interactions dependent on their presence in either the testes or ovaries33. These sex-specific dynamics are also affected by temporal factors during larval development34, suggesting unique features of these organs, such as morphological or physiological characteristics, influence certain microbial interactions. Importantly, large amounts of bacteria are expelled from the larval digestive tract during and after pupation35,36, indicating the initial microbiota inherited by newly emerged adults originate from other anatomical regions37,38 and are transmitted transstadially (from larvae to adults). Differences in the microbiota harbored in adult males and females may be driven by disparate microbiomes harbored in the developing larval sex organs prior to pupation. If this is the case, factors that impact larval microbiomes may have downstream, sex-specific effects on the adult microbiota.
Biological factors such as fungal–bacterial–host interactions are known to affect host-associated microbiomes39. Fungi are found in natural mosquito populations4,7,40 and impact host fitness41–43. Recently, it was demonstrated that a mosquito-associated gut fungus, Zancudomyces culisetae, significantly affected microbiome dynamics in larvae and female adults of the yellow fever mosquito (Aedes aegypti)44. The fungus is an obligate gut fungal symbiont of larval A. aegypti and several other dipteran hosts in nature45,46, and its life cycle is bound to that of its larval host. Larvae ingest fungal spores from the aquatic environment, fungal growth in the larval hindgut is mediated by specific physiological cues47,48, and the fungus acquires nutrients from the host digestive tract45. Larval fungal colonization was found to reduce the variation of bacterial community diversity and distribution across individuals, differentially affect the transference efficacy of certain bacterial taxa from larvae to adults (transstadial transmission), and increase the overall diversity of initial microbiomes acquired by newly emerged adult females. Although complex fungal–bacterial–host interactions were revealed, the study did not investigate whether similar dynamics took place in male mosquitoes. A key interpretation of these patterns was that morphological (expansion of fungal tissue) and physiological (altered nutrient dynamics in the digestive tract) disturbances resulting from fungal colonization of the larval hindgut47,48 could have altered microbial interactions and spatial distributions of bacteria49, leading to differential transstadial transmission outcomes of certain bacteria and disparate initial adult female microbiomes in the presence or absence of the fungus. Male-associated microbial communities may respond differently to fungal colonization of the larval digestive tract due to unique anatomical characteristics, however, this concept has never been studied.
To further investigate fungal–bacterial–host dynamics in the A. aegypti–Z. culisetae system, comparative analyses were performed on a previously collected but unpublished dataset of sequenced 16S rRNA gene amplicons from microbiomes extracted from newly emerged adult male mosquitoes that developed from larvae with and without exposure to the fungus. I hypothesized that larval exposure to the fungus would alter adult male microbiomes and predicted that these microbial communities would increase in overall diversity and decrease in variation across individuals as previously shown in females. Additionally, I also predicted that different bacterial taxa would be impacted by the fungus due to the known differences between male- and female-associated microbiota.
In this study, the effect of larval fungal exposure on newly emerged adult male microbiomes was determined. These results were compared to those previously found for females, and potential processes contributing to the observed patterns are discussed.
Results
Fungal effect on overall microbiome diversity
Measures of alpha diversity were calculated for initial microbiomes to assess whether larval fungal exposure affected the overall diversity of microbial communities acquired by newly emerged adult males (Table S1). Significantly lower mean values were calculated for fungal mosquitoes for both Simpson (P < 0.01, Fig. 1a) and Shannon (P < 0.01, Fig. 1b) diversity indices, indicating larval exposure to the fungus decreased the overall diversity of microbial communities acquired by newly emerged adults. No significant difference was detected in the variation of alpha diversity measures for either metric (Table 1), however, the CV value for Shannon diversity was higher in non-fungal (29.09%) than fungal (15.47%) groups with P < 0.1 calculated with an asymptotic and MSLRT, suggesting a marginal reduction of variation in overall microbial community diversity.
Figure 1.
Box plots of alpha diversity measures calculated for microbiomes harbored in newly emerged males developed from larvae in the absence (non-fungal, blue) or presence (fungal, red) of fungal exposure during larval development. (a) Comparative analysis of Simpson diversity index shows significantly higher values for non-fungal relative to fungal mosquito microbiomes. (b) Comparative analysis of Shannon diversity index shows significantly higher values for non-fungal relative to fungal mosquito microbiomes. Upper and lower limits of boxes represent quartiles around the mean and horizontal lines within boxes represent median values within each treatment group. Significant differences of mean alpha diversity measures were calculated with a linear mixed effects model (N = 22, **P < 0.01).
Table 1.
Results from comparative statistical analyses for measures of alpha and beta diversity across treatment groups.
| Metric | DF Num | DF Den | F value | P (treatment) | Non-fungal CV | Fungal CV | Test statistic (asymptotic) | CV P (asymptotic) | Test statistic (MSLRT) | CV P (MSLRT) | DF Num | DF Den | F value | P (homogeneity) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Simpson | 1 | 5.967 | 14.489 | 0.009 | 12.28% | 7.05% | 3.009 | 0.083 | 2.459 | 0.117 | ||||
| Shannon | 1 | 5.975 | 14.457 | 0.009 | 29.09% | 15.47% | 3.598 | 0.058 | 3.187 | 0.074 | ||||
| Bray–Curtis | 1 | 14.000 | 6.591 | 0.027 | 1 | 20 | 135.560 | < 0.001 | ||||||
| weighted UniFrac | 1 | 14.000 | 12.635 | 0.027 | 1 | 20 | 60.766 | < 0.001 | ||||||
| Jaccard | 1 | 14.000 | 5.449 | 0.027 | 1 | 20 | 74.719 | < 0.001 | ||||||
| Unweighted UniFrac | 1 | 14.000 | 2.811 | 0.043 | 1 | 20 | 0.040 | 0.863 |
Significant differences of mean alpha diversity measures were calculated with linear mixed effects models, beta diversity measures with nested permutational analysis of variance, and within-group variations of beta diversity measures with permutational statistical tests for the homogeneity of group dispersions. Values in boldface are statistically significant (P < 0.05). These data are presented and described following the format provided in Frankel-Bricker et al.44.
Distinct microbiome composition and structure across treatments
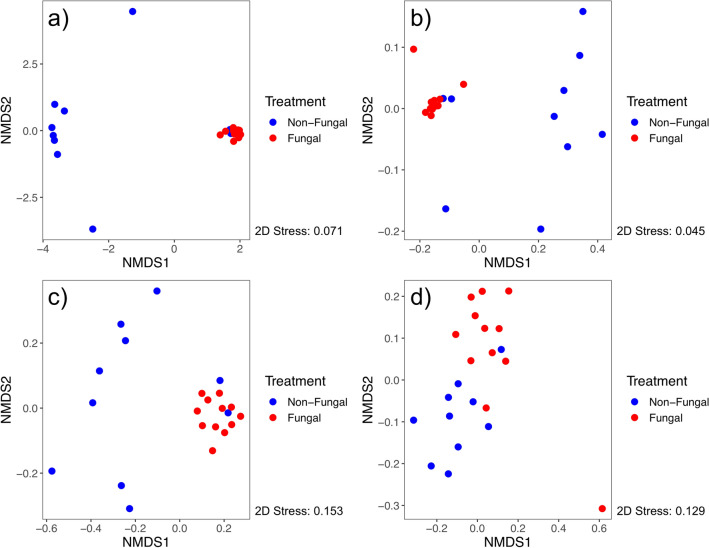
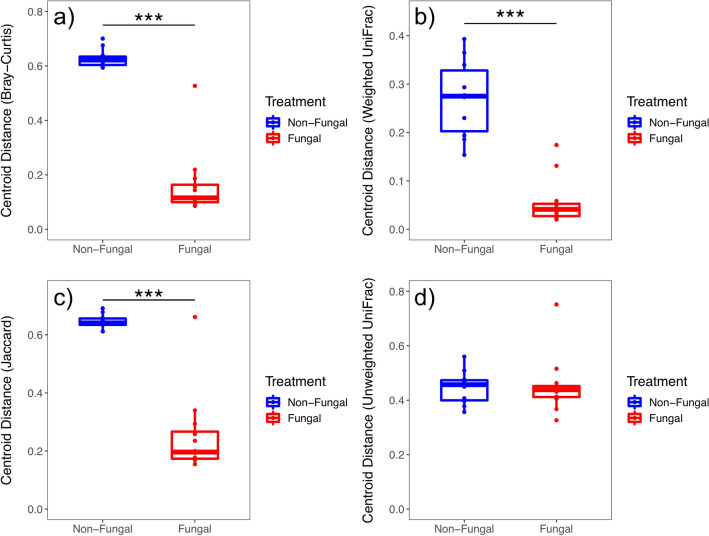
After read processing, 444 ASVs were identified from 22 samples. Differences in beta diversity measures across treatment groups were detected with PERMANOVA for all metrics tested (Table 1), showing larval exposure to the fungus contributed to disparate microbial communities acquired by newly emerged adults. Permutational analysis of variance tests are sensitive to differences in variation across group comparisons and may produce false detections of significance, however, NMDS plots show separation between treatment groups for all metrics tested (Fig. 2). Non-fungal mosquitoes had higher within-group variation for Bray–Curtis dissimilarity (P < 0.001, Fig. 3a), weighted UniFrac distance (P < 0.001, Fig. 3b), and Jaccard index (P < 0.001, Fig. 3c), indicating the fungus reduced structural variation of the microbiota across individuals in the group. However, no differential within-group variation was detected for unweighted UniFrac distance (P = 0.863, Fig. 3d), demonstrating taxonomic composition variation of the microbial communities was not affected by the fungus. These significant patterns were reproduced by analyses conducted on a non-rarefied data set (Table S2), demonstrating that the implementation of rarefaction did not affect these results.
Figure 2.
Nonmetric multidimensional scaling plots of beta diversity measures across treatment groups (non-fungal, blue; fungal, red) for (a) Bray–Curtis dissimilarity, (b) weighted UniFrac distance, (c) Jaccard index, and (d) unweighted UniFrac distance.
Figure 3.
Box plots of within treatment group variation of beta diversity measures for (a) Bray–Curtis dissimilarity, (b) weighted UniFrac distance, (c) Jaccard index, and (d) unweighted UniFrac distance. Significant differences were calculated with permutational statistical tests for the homogeneity of group dispersions (***P < 0.001).
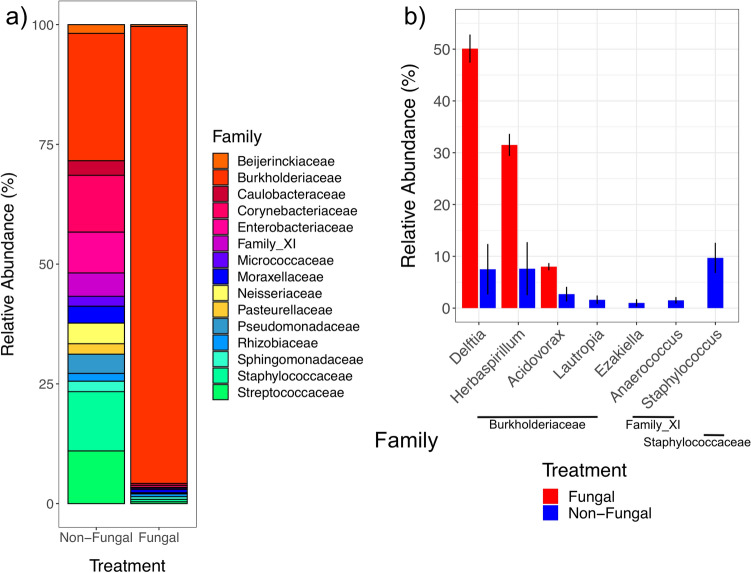
Relative abundances of bacterial families were calculated to investigate whether certain taxa were differentially impacted by the fungus (Fig. 4a, Table S3). The bacterial family Burkholderiaceae was present at moderate levels in non-fungal adults (21.06%) but was significantly more abundant in fungal adults (89.75%, P < 0.01). Conversely, Staphylococcaceae, an intermediate community member in non-fungal adults (9.81%), was found at significantly lower levels in fungal adults (0.48%, P = 0.013). Family_XI followed a similar trend and was present at low amounts in non-fungal (3.91%) but was nearly undetectable in fungal adults (0.01%, P = 0.036). These significant families were further analyzed at the genus level to assess whether genera abundances followed similar patterns within families (Fig. 4b; Table S4). Within Burkholderiaceae, three of the four predominant genera detected had higher abundance in fungal relative to non-fungal adults (Delftia, Herbaspirillum, Acidovorax), whereas the genus Staphylococcus (Staphylococcaceae) was higher in non-fungal adults, demonstrating genera followed similar trends as their associated families.
Figure 4.
Mean relative abundances of prominent bacterial taxa across treatment groups. (a) Bar plots of the 15 most abundant families shared between treatment groups. (b) Bar plots of genera within families identified to significantly differ across treatment groups. Only genera present at greater than 1% relative abundance within each treatment group are shown. Line segments represent standard errors of the mean.
Supplementary analyses of female-associated microbiomes
Measures of alpha diversity were calculated for initial microbiomes harbored in newly emerged females and compared with previously published findings44 (Table S1; Fig. S1) to assess general similarities or differences between the frozen samples analyzed here and those not frozen in the earlier study. Measures calculated in the current experiment fell within the previously calculated ranges for non-fungal mosquitoes and in the lower ranges of fungal mosquitoes for Simpson and Shannon diversity indices. General trends were conserved, with non-fungal mosquitoes having larger variation than fungal mosquitoes for both metrics (Fig. S1) and similar mean values across groups. Female sample sizes collected for the current study were small (n = 9) and did not represent all replicate containers in the experiment, preventing a more in-depth statistical analysis of these data. However, the detection of similar trends for females between the two experiments for alpha diversity measures could indicate a reproducible result and suggests comparisons between these experiments may be appropriate.
Discussion
The results presented herein provide additional evidence that a widespread gut fungus played a significant role in its mosquito host’s microbiome. Substantial differences in the composition and structure of bacterial communities harbored across treatment groups were found, supporting the hypothesis that larval fungal exposure would impact initial adult male microbiomes. Fungal microbiomes were dominated by bacteria in the family Burkholderiaceae, with nearly 90% of sequencing reads assigned to genera within this family (Delftia [50.1%], Herbaspirillum [31.5%], Acidovorax [8.0%]). The differential abundance of these microbes likely defined the overall characteristics and differences in microbiome composition and structure. Fungal mosquitoes had significantly lower overall diversity than non-fungal mosquitoes (Fig. 1) and different taxonomic composition and community structure (Figs. 2, 4; Table 1). Low within-group variation of beta diversity measures for fungal mosquitoes (Fig. 3) indicated that the high relative abundance of Burkholderiaceae was a conserved characteristic of these microbial communities. While this bacterial family was present at intermediate levels in non-fungal adults (21.1%), it was more evenly distributed with the other families and corresponded with higher community diversity, reflected by significantly higher values for Simpson (Fig. 1a) and Shannon (Fig. 1b) diversity indices (Table S1). Morphological and physiological characteristics of fungal colonization in the larval hindgut47,48,50 may have driven these disparities in the microbiota. The spatial distribution of fungal tissue, physiological shifts in the digestive tract microecosystem47,48, and depletion of nutrients from the host digestive tract incurred by the fungus45 could have promoted the proliferation of Burkholderiaceae if taxa in this family were better adapted to these fungal-induced changes49,51. Future research could investigate these concepts through inoculation experiments of larvae with experimentally controlled microbiota to assess whether similar taxa persist after larval fungal colonization. In addition, studies of these dynamics could be performed on wild-caught mosquitoes to assess whether the patterns described here are conserved in nature.
Comparisons with previous findings for females44 uncovered possible evidence of sex-specific fungal–bacterial–host dynamics in the system. I originally predicted that shifts in the initial male microbiome in response to larval fungal exposure would follow similar patterns as those previously described in females, which were characterized by lower within-group variation but similar means for alpha diversity measures relative to non-fungal adults. Contrary to this prediction, within-group variation in alpha diversity measures for male samples did not statistically differ across treatments and non-fungal males harbored microbiomes with significantly higher overall diversity relative to fungal males (Fig. 1; Table 1). Further, strong differences in microbiome composition and structure were detected across treatments in males (Figs. 2, 3) that were not previously found in females. These differences could reflect sex-specific fungal–bacterial–host dynamics, possibly driven by the influence of the fungus on the transmission of bacterial taxa in the family Burkholderiaceae. Adult females were previously shown to have higher prevalence of Burkholderiaceae in the absence of larval fungal colonization. Conversely, adult males from the fungal group were dominated by this taxon, possibly suggesting differential sex-specific interactions between the fungus and these bacteria (Fig. 4). Spatial disturbances in the larval hindgut during fungal colonization and expansion50 may have redistributed the microbiota. Indeed, fungal displacement of microbial communities was observed in a pathogenic mosquito-fungus system and impacted host fitness43. Mosquitoes expel large quantities of bacteria from the digestive tract during and after pupation35,36, indicating that bacteria inherited in newly emerged adults may be transferred transstadially through anatomical regions other than the digestive tract37,38. It is possible that the differential abundance of Burkholderiaceae in newly emerged adult males is driven by these mechanisms. Male and female reproductive organs harbor microbiomes composed of similar bacterial taxa but with different community distributions31,32. In the absence of fungal colonization, microbiota in the testes may be characterized with relatively high overall diversity. During larval fungal infestation, bacteria may spatially redistribute to the testes, or to other anatomical regions, altering existing microbial interactions and resulting in favorable conditions for genera within Burkholderiaceae. Different processes related to unique microbiomes in female ovaries may have led to distinct microbial dynamics and resulted in the disparate patterns between fungal male and female microbiomes. Future studies could perform experiments to investigate the spatial distribution of bacteria in the family Burkholderiaceae in males and females prior to and after pupation in the presence and absence of larval fungal colonization by utilizing previously described fluorescence-based assays37,38. Dissections isolating the testes and ovaries could also be performed on larvae throughout development to provide temporal information of these potentially sex-specific microbiomes. Results from these experiments could help clarify the mechanisms leading to differential transstadial transmission outcomes of Burkholderiaceae. In addition, future research could investigate whether bacteria in this family impact adult male fitness or have downstream impacts on life history traits. Experiments in this field may reveal distinct, sex-specific microbiome dynamics in mosquito populations contingent on the presence or absence of the fungus in the local environment.
Alpha diversity measures were calculated for a small subset of females to compare with previous findings44. Values fell within or near previous calculations and within-treatment variation followed similar trends. Non-fungal mosquitoes exhibited higher within-group variation than fungal mosquitoes, increasing confidence that comparisons of the primary results between experiments are appropriate. However, I acknowledge that the small samples sizes used for comparative analyses of females limits the power of these comparisons. Therefore, a future experiment should be designed to fully analyze the microbiomes harbored in males and females under standardized laboratory protocols and with larger sampling sizes. Reproducibility remains a fundamental, yet elusive element in microbial ecology and microbiome research52. Further research should be conducted under independent laboratory conditions to corroborate the results presented herein, as proposed and conducted in human studies and arthropod systems53–55.
Conclusion
Here, a significant effect of larval fungal exposure on host-associated microbiota in newly emerged adult male mosquitoes is revealed, providing further support that fungi are important microbial community members in A. aegypti. Preliminary evidence for sex-specific fungal–bacterial–host dynamics in a mosquito-fungus symbiotic system merits further investigation. Comprehensively studying all components of microbiomes is essential to understand biotic interactions contributing to an organism’s biology. These findings suggest studies of host-associated microbiomes should account for the presence of fungi, the host organism’s sex, and potentially distinct sex-specific host-microbe relationships.
Materials and methods
The majority of the protocols conducted for this experiment were carried out in tandem with those presented in Frankel-Bricker et al.44. Consequently, there is extensive overlap in the description of experimental design and protocols.
Male samples evaluated in this experiment were reared and collected alongside the females, however, these were separately frozen prior to DNA extraction and the subsequent sequencing analyses were conducted independently. This difference in collection and storage method may have altered downstream detection of certain bacterial taxa56,57, warranting independent analyses for these experimental samples.
Zancudomyces culisetae culture maintenance
A culture of Z. culisetae (USDA-ARS Collection of Entomopathogenic Fungal Cultures, Ithaca, New York, USA, ARSEF 9012, Smittium culisetae, COL-18-3) was grown at room temperature on a 1/10 brain heart infusion agar plate with 3 milliliters (ml) autoclaved Nanopure water (Barnstead Thermolyne Corp., Dubuque, IA, USA). Antibiotics were added to the overlay to mitigate bacterial contamination of the fungal culture (2 mg/ml of penicillin and 7 mg/ml of streptomycin). The overlay was filtered through a sheet of Miracloth (EMD Millipore, Burlington, MA, USA) and into 1.5 ml microcentrifuge tubes (Eppendorf, Hamburg, Germany) to collect fungal spores. Spores were concentrated by centrifugation at 900 g for 10 minutes (min) and the supernatant was discarded. The resulting pellets were resuspended in 1 ml autoclaved Arrowhead bottled spring water (Nestle, Vevey, Switzerland) and spore concentrations were estimated with a Neubauer Improved C-Chip Hemocytometer (SKC Inc., Covington, GA, USA) visualized with phase optics light microscopy.
Experimental conditions and mosquito rearing
Aedes aegypti eggs, derived from the USDA-ARS Gainesville line (Benzon Research Inc., Carlisle, PA, USA) were stored at room temperature. Histology containers (Fisher Scientific, Pittsburgh, PA, USA) were filled with 350 ml of bottled spring water and subsequently autoclaved. Four containers were assigned to each of two experimental treatment groups: larvae not exposed to fungal spores (Non-Fungal), and larvae exposed to fungal spores (Fungal). Approximately 50 eggs were added to each rearing container, containers were covered with 4 layers of autoclaved Miracloth to mitigate airborne contamination and were separately placed in a vacuum chamber (SP Industries Inc., Warminster, PA, USA) for 30 min to synchronize egg hatch timing58. The larval mosquito food source was prepared by finely grinding Tetramin Fish Food (Tetra, Melle, Germany) with a mortar and pestle and suspending 0.2 g of fish food powder per 10 ml autoclaved bottled spring water. One ml of the slurry was added to each rearing container at the start of the experiment. Containers assigned to the fungal group were inoculated with approximately 400,000 fungal trichospores. Mosquitoes were reared at 24 °C ± 1 °C with a 16:8 h light/dark cycle in a low temperature refrigerated incubator (Fisher Scientific, model #3724), and 1–2 ml of fish slurry were added daily to each rearing container. All experimental protocols were performed on a sterilized laboratory workbench next to a Bunsen burner to minimize contamination.
Mosquito sample collection
Mosquitoes were reared to pupae, transferred to 1.5 ml microcentrifuge tubes, and surface-sterilized following an adult protocol described in Coon et al.5. Surface-sterilized pupae were transferred separately to 15 ml centrifuge tubes (Corning Inc., Corning, NY, USA) containing 7 ml autoclaved bottled spring water and reared axenically for 2–3 days until adult emergence. The sex of newly emerged adults was visually identified and individual adult males were transferred to 1.5 ml microcentrifuge tubes and stored at − 80 °C. In addition, 5 adult females were collected from the non-fungal group and 4 from the fungal group and frozen similarly.
Microbial DNA extraction
The Quick-DNA Fungal/Bacterial Kit (Zymo Research, Irvine, CA, USA) was used to extract microbial DNA from frozen male and female mosquitoes and other experimental sources following the protocol provided by the manufacturer with the following modifications previously described in Frankel-Bricker et al.44: Lysis buffer was added directly to the 1.5 ml microcentrifuge tubes containing harvested mosquitoes. Mosquitoes were manually ruptured with an autoclaved pellet pestle (DWK Life Sciences, Wertheim, Germany) for approximately 1–2 min. Homogenized tube mixtures were transferred to bead tubes supplied with the extraction kit and disrupted using a vortex mixer at maximum setting for 5 min. The elution buffer was heated to 45 °C prior to its application to the spin-filters supplied by the extraction kit and remained on the filter surface for 5 min prior to the final elution step. Extracted microbial DNA was stored at − 80 °C.
At least 2 DNA extractions were performed on mosquitoes collected from each replicate container for the non-fungal treatment group (10 total) and 3 from each replicate container for the fungal group (12 total). Extractions were also performed on a small subset of female mosquitoes (9 total) from 5 of the replicate containers (3 non-fungal, 2 fungal). Additional DNA extractions were performed on blank extraction kit reagents from the 2 kits used to account for possible contamination of kit reagents. Two blank polymerase chain reactions (PCR) were also carried out to check for contamination of PCR reagents. All PCR were performed using 5PRIME HotMasterMix (Quantabio, Beverly, MA, USA).
Bacterial 16S rRNA gene amplicon sequencing
The V3/V4 hypervariable regions of microbial 16S rDNA were amplified with primer pair 341f/785r59, with suggested linker sequences60, and adapter and spacer sequences provided by the University of Idaho Genomics Resources Core (GRC) (University of Idaho, Moscow, ID, USA). Targeted 16S PCR were performed on extracted experimental DNA samples using four primer pair variants containing spacer sequences of different lengths to mitigate amplification biases (Tables S5, S6: Rxn_1). PCR products were visualized on 1.5% agarose gels to confirm amplification of 16S rDNA.
Secondary PCR were performed (Table S6: Rxn_2) to attach barcode sequences provided by the University of Idaho GRC to PCR amplicons. Amplicon sequencing was performed with an Illumina MiSeq v3 (Illumina Inc., San Diego, CA, USA) at the University of Idaho GRC, producing 300 base pair paired end reads. Reads were demultiplexed by sample barcode sequences by the sequencing facility.
Raw read processing and ASV assignment
The bioinformatics pipeline and subsequent analyses were performed using the R programming language version 3.6.261. The DADA2 pipeline62 was used to process paired end reads. Forward and reverse reads were trimmed to 278 base pairs and 167 base pairs, respectively, and at the location of the first occurrence of a base call with a Phred score less than or equal to 15, filtered by discarding reads with any number of N base calls or containing greater than or equal to 6 estimated errors, and merged with a minimum overlap of 12 bases. Experimental samples with less than 100 reads after initial filtering were removed from the pipeline. Chimeric sequences were discarded and merged reads dereplicated. Taxonomy was assigned to amplicon sequence variants (ASVs) using the SILVA v132 database63,64. A neighbor-joining tree was inferred using the phangorn package in R65 and a generalized time-reversible with gamma rate variation maximum likelihood tree was fit using the neighbor-joining tree as the starting point. The phylogenetic tree, taxonomically assigned ASVs, read count data, and experimental sample metadata were combined into a single data object using the Phyloseq package in R66.
Contaminant ASV removal
Non-experimental sources, such as reagents from DNA extraction kits and PCR, can add contaminant sequences and mislead analyses of microbiomes if not properly accounted for67. Of the controls sequenced, both extraction kits used had over 100 reads after processing in DADA2. Reads from extraction kits were pooled and the decontam package in R68 was used to identify contaminant ASVs with the “prevalence” method and the threshold set to 0.5. Amplicon sequence variants classified as likely contaminants originating from the extraction kits were discarded from the sequencing dataset prior to downstream analyses.
Transformation and analyses of sequencing data
Non-transformed reads were used to calculate Simpson and Shannon alpha diversity indices (Table S1) in Phyloseq. Values for female samples were combined with those previously calculated in a larger experiment44. Box plots (Fig. S1) were generated using the ggplot2 package in R69. Coefficient of variation values (CV; the ratio of the standard deviation to the mean) were calculated with the sjstats package in R70 to assess variation of alpha diversity measures, and the R package cvequality71 was used to calculate significant differences in CV values with an asymptotic test72 and a modified signed-likelihood ratio test (MSLRT)73. Rarefaction curves were generated using the ranacapa package74 and ggplot2 in R (Fig. S2). Read coverage varied across samples (min: 1532 reads; max: 12,420 reads). Sequencing data were rarefied to 1532 reads (the lowest read count for a sequenced sample). Singletons were removed and ASVs not represented by at least 5 reads in one sample after rarefaction were discarded.
Beta diversity measures [Bray–Curtis dissimilarity (compositional dissimilarity), unweighted UniFrac distance (qualitative compositional similarity weighted for phylogenetic distance of taxa), weighted UniFrac distance (quantitative compositional comparison weighted for phylogenetic distance), and Jaccard similarity index (compositional similarity)] were calculated in Phyloseq and non-metric multidimensional scaling (NMDS) plots were created in combination with ggplot2. Tests for significant differences between treatment groups were performed with permutational analysis of variance (PERMANOVA)75 with 999 permutations in the Vegan package in R76, along with the nested.npmanova function in the BiodiversityR package in R77. Nested PERMANOVA calculated the correct pseudo F and P values for the treatment effect while accounting for random effects across replicate rearing containers. Dispersions of beta diversity (the distance from an individual measure to the group’s centroid) were calculated for each beta diversity metric within each group in Vegan to estimate within-group variation78 across individuals. Significant differences in beta diversity variation were tested using permutational statistical tests for the homogeneity of group dispersions79 with 999 permutations in Vegan and box plots were constructed in ggplot2. These analyses were also performed on a non-rarefied data set to test whether rarefaction impacted results from these tests. Mean relative abundances of the top 15 bacterial families shared between groups (which accounted for greater than 79% of the total reads) were calculated (Table S3) and stacked bar plots were created in Phyloseq in combination with ggplot2. Relative abundances of genera within significant families were calculated (Table S4) and taxa found at greater than a mean of 1% in a group were plotted using ggplot2.
Linear mixed effects models
The statistical significance of the treatment effect on mean alpha diversity measures and relative abundances of each of the top 15 shared bacterial families were calculated by fitting a linear mixed effects model constructed with the lme4 package in R80, which accounted for random effects across replicate rearing containers. Models were tested with Type II Wald F tests with Kenward-Roger degrees of freedom using the car package in R81.
Approval for the use of animal samples
All experimental protocols for the rearing and harvesting of mosquito samples were in strict accordance with the guidelines and regulations set forth and approved by the Boise State University Institutional Biosafety Committee.
Supplementary information
Acknowledgements
Research reported in this publication was supported by Institutional Development Awards (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under Grants #P20GM103408 (Idaho INBRE) and #P20GM109095 (Idaho COBRE), the Robert W. Lichtwardt Student Research Award from the Mycological Society of America, and a College of Arts and Sciences Award from Boise State University. In addition, I thank Merlin White for providing funds from the NSF “ZyGoLife” Grant DEB1441677, which funded the previously published research used in comparison with the results presented in this work and for providing laboratory space. All sequencing data collection and preliminary analyses performed by the IBEST Genomics Resources Core at the University of Idaho were supported in part by NIH COBRE Grant #P30GM103324. I gratefully thank the collaboration and advice from colleagues at the University of Idaho GRC: Dan New, Sam Hunter, and Matt Fagnan. I would like to thank Laurie Frankel for extensive support with the scientific writing process and Terry Bricker for academic mentorship. Finally, I thank two anonymous reviewers and the handling editor for giving constructive critiques and suggested revisions that greatly improved the quality of the manuscript.
Author contributions
J.F.B designed the experiment, conducted research, analyzed data, and wrote the manuscript.
Data availability
Raw sequences are deposited in the NCBI Sequence Read Archive under accession number PRJNA606895.
Competing interests
The author declares no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-69828-9.
References
- 1.Coon KL, Brown MR, Strand MR. Gut bacteria differentially affect egg production in the anautogenous mosquito Aedes aegypti and facultatively autogenous mosquito Aedes atropalpus (Diptera: Culicidae) Parasite Vector. 2016;9:375. doi: 10.1186/s13071-016-1660-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaio ADO, et al. Contribution of midgut bacteria to blood digestion and egg production in Aedes aegypti (Diptera: Culicidae) (L.) Parasite Vector. 2011;4:105. doi: 10.1186/1756-3305-4-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bascuñán P, et al. Factors shaping the gut bacterial community assembly in two main Colombian malaria vectors. Microbiome. 2018;6:148. doi: 10.1186/s40168-018-0528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandler JA, Liu RM, Bennett SN. RNA shotgun metagenomic sequencing of Northern California (USA) mosquitoes uncovers viruses, bacteria, and fungi. Front. Microbiol. 2015;6:1–16. doi: 10.3389/fmicb.2015.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coon KL, Vogel KJ, Brown MR, Strand MR. Mosquitoes rely on their gut microbiota for development. Mol. Ecol. 2014;23:2727–2739. doi: 10.1111/mec.12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gimonneau G, et al. Composition of Anopheles coluzzii and Anopheles gambiae microbiota from larval to adult stages. Infect. Genet. Evol. 2014;28:715–724. doi: 10.1016/j.meegid.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 7.Muturi EJ, et al. Culex pipiens and Culex restuans mosquitoes harbor distinct microbiota dominated by few bacterial taxa. Parasite Vector. 2016;9:18. doi: 10.1186/s13071-016-1299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muturi EJ, Ramirez JL, Rooney AP, Kim CH. Comparative analysis of gut microbiota of mosquito communities in central Illinois. PLoS Negl. Trop. Dis. 2017;11:e0005377. doi: 10.1371/journal.pntd.0005377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muturi EJ, et al. Mosquito microbiota cluster by host sampling location. Parasite Vector. 2018;11:468. doi: 10.1186/s13071-018-3036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osei-Poku J, Mbogo CM, Palmer WJ, Jiggins FM. Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya. Mol. Ecol. 2012;21:5138–5150. doi: 10.1111/j.1365-294X.2012.05759.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, et al. Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS ONE. 2011;6:1–9. doi: 10.1371/journal.pone.0024767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carissimo G, et al. Antiviral immunity of Anopheles gambiae is highly compartmentalized, with distinct roles for RNA interference and gut microbiota. Proc. Natl. Acad. Sci. 2015;112:E176–E185. doi: 10.1073/pnas.1412984112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dennison NJ, Jupatanakul N, Dimopoulos G. The mosquito microbiota influences vector competence for human pathogens. Curr. Opin. Insect Sci. 2014;3:6–13. doi: 10.1016/j.cois.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegde S, Rasgon JL, Hughes GL. The microbiome modulates arbovirus transmission in mosquitoes. Curr. Opin. Virol. 2015;15:97–102. doi: 10.1016/j.coviro.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jupatanakul N, Sim S, Dimopoulos G. The insect microbiome modulates vector competence for arboviruses. Viruses. 2014;6:4294–4313. doi: 10.3390/v6114294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramirez JL, et al. Reciprocal tripartite interactions between the Aedes aegypti midgut microbiota, innate immune system and dengue virus influences vector competence. PLoS Negl. Trop. Dis. 2012;6:e1561. doi: 10.1371/journal.pntd.0001561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charan SS, et al. Comparative analysis of midgut bacterial communities of Aedes aegypti mosquito strains varying in vector competence to dengue virus. Parasitol. Res. 2013;112:2627–2637. doi: 10.1007/s00436-013-3428-x. [DOI] [PubMed] [Google Scholar]
- 18.Dickson LB, et al. Carryover effects of larval exposure to different environmental bacteria drive adult trait variation in a mosquito vector. Sci. Adv. 2017;3:e1700585. doi: 10.1126/sciadv.1700585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonçalves CM, et al. Distinct variation in vector competence among nine field populations of Aedes aegypti from a Brazilian dengue-endemic risk city. Parasite Vector. 2014;7:320. doi: 10.1186/1756-3305-7-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campos SS, et al. Zika virus can be venereally transmitted between Aedes aegypti mosquitoes. Parasite Vector. 2017;10:605. doi: 10.1186/s13071-017-2543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sánchez-Vargas I, et al. Demonstration of efficient vertical and venereal transmission of dengue virus type-2 in a genetically diverse laboratory strain of Aedes aegypti. PLOS Negl. Trop. D. 2018;12:e0006754. doi: 10.1371/journal.pntd.0006754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valiente Moro CV, et al. Diversity of culturable bacteria including Pantoea in wild mosquito Aedes albopictus. BMC Microbiol. 2013;13:70. doi: 10.1186/1471-2180-13-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zouache K, et al. Bacterial diversity of field-caught mosquitoes, Aedes albopictus and Aedes aegypti, from different geographic regions of Madagascar. FEMS Microbiol. Ecol. 2011;75:377–389. doi: 10.1111/j.1574-6941.2010.01012.x. [DOI] [PubMed] [Google Scholar]
- 24.Foster WA. Mosquito sugar feeding and reproductive energetics. Annu. Rev. Entomol. 1995;40:443–474. doi: 10.1146/annurev.en.40.010195.002303. [DOI] [PubMed] [Google Scholar]
- 25.Minard G, Mavingui P, Moro CV. Diversity and function of bacterial microbiota in the mosquito holobiont. Parasite Vector. 2013;6:146–158. doi: 10.1186/1756-3305-6-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coon KL, Brown MR, Strand MR. Mosquitoes host communities of bacteria that are essential for development but vary greatly between local habitats. Mol. Ecol. 2016;25:5806–5826. doi: 10.1111/mec.13877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponnusamy L, et al. Diversity of bacterial communities in container habitats of mosquitoes. Microb. Ecol. 2008;56:593–603. doi: 10.1007/s00248-008-9379-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yee DA, Allgood D, Kneitel JM, Kuehn KA. Constitutive differences between natural and artificial container mosquito habitats: vector communities, resources, microorganisms, and habitat parameters. J. Med. Entomol. 2012;49:482–491. doi: 10.1603/me11227. [DOI] [PubMed] [Google Scholar]
- 29.Rani A, et al. Bacterial diversity analysis of larvae and adult midgut microflora using culture-dependent and culture-independent methods in lab-reared and field-collected Anopheles stephensi-an Asian malarial vector. BMC Microbiol. 2009;9:96. doi: 10.1186/1471-2180-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guégan M, et al. The mosquito holobiont: fresh insight into mosquito-microbiota interactions. Microbiome. 2018;6:49. doi: 10.1186/s40168-018-0435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mancini MV, et al. Estimating bacteria diversity in different organs of nine species of mosquito by next generation sequencing. BMC Microbiol. 2018;18:126. doi: 10.1186/s12866-018-1266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Segata N, et al. The reproductive tracts of two malaria vectors are populated by a core microbiome and by gender-and swarm-enriched microbial biomarkers. Sci. Rep. 2016;6:24207. doi: 10.1038/srep24207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossi P, et al. Mutual exclusion of Asaia and Wolbachia in the reproductive organs of mosquito vectors. Parasite Vector. 2015;8:278. doi: 10.1186/s13071-015-0888-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tchioffo MT, et al. Dynamics of bacterial community composition in the malaria mosquito’s epithelia. Front. Microbiol. 2016;6:1500. doi: 10.3389/fmicb.2015.01500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moll RM, et al. Meconial peritrophic membranes and the fate of midgut bacteria during mosquito (Diptera: Culicidae) metamorphosis. J. Med. Entomol. 2001;38:29–32. doi: 10.1603/0022-2585-38.1.29. [DOI] [PubMed] [Google Scholar]
- 36.Moncayo AC, et al. Meconial peritrophic matrix structure, formation, and meconial degeneration in mosquito pupae/pharate adults: histological and ultrastructural aspects. J. Med. Entomol. 2005;42:939–944. doi: 10.1093/jmedent/42.6.939. [DOI] [PubMed] [Google Scholar]
- 37.Chavshin AR, et al. Escherichia coli expressing a green fluorescent protein (GFP) in Anopheles stephensi: a preliminary model for paratransgenesis. Symbiosis. 2013;60:17–24. [Google Scholar]
- 38.Chavshin AR, et al. Malpighian tubules are important determinants of Pseudomonas transstadial transmission and longtime persistence in Anopheles stephensi. Parasite Vector. 2015;8:36. doi: 10.1186/s13071-015-0635-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deveau A, et al. Bacterial-fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol. Rev. 2018;42:335–352. doi: 10.1093/femsre/fuy008. [DOI] [PubMed] [Google Scholar]
- 40.Alencar YB, Ríos-Velásquez CM, Lichtwardt RW, Hamada N. Trichomycetes (Zygomycota) in the digestive tract of arthropods in Amazonas Brazil. Mem. Inst. Oswaldo Cruz. 2003;98:799–810. doi: 10.1590/s0074-02762003000600016. [DOI] [PubMed] [Google Scholar]
- 41.Angleró-Rodríguez YI, et al. An Aedes aegypti-associated fungus increases susceptibility to dengue virus by modulating gut trypsin activity. Elife. 2017;6:e28844. doi: 10.7554/eLife.28844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramirez JL, et al. Entomopathogenic fungal infection leads to temporospatial modulation of the mosquito immune system. PLoS Negl. Trop. Dis. 2018;12:e0006433. doi: 10.1371/journal.pntd.0006433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei G, et al. Insect pathogenic fungus interacts with the gut microbiota to accelerate mosquito mortality. Proc. Natl. Acad. Sci. 2017;114:5994–5999. doi: 10.1073/pnas.1703546114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frankel-Bricker J, Buerki S, Feris KP, White MM. Influences of a prolific gut fungus (Zancudomyces culisetae) on larval and adult mosquito (Aedes aegypti)-associated microbiota. Appl. Environ. Microbiol. 2020;86:e02334-19. doi: 10.1128/AEM.02334-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lichtwardt RW. Species of Harpellales living within the guts of aquatic Diptera larvae. Mycotaxon. 1984;19:529–550. [Google Scholar]
- 46.Williams MC, Lichtwardt RW. Infection of Aedes aegypti larvae by axenic cultures of the fungal genus Smittium (trichomycetes) Am. J. Bot. 1972;59:189–193. [Google Scholar]
- 47.Horn BW. Ultrastructural changes in trichospores of Smittium culisetae and S. culicis during in vitro sporangiospore extrusion and holdfast formation. Mycologia. 1989;81:742–753. [Google Scholar]
- 48.Horn BW. Physiological changes associated with sporangiospore extrusion from trichospores of Smittium culisetae. Exp. Mycol. 1990;14:113–123. [Google Scholar]
- 49.Karl PJ, et al. Effects of psychological, environmental and physical stressors on the gut microbiota. Front. Microbiol. 2018 doi: 10.3389/fmicb.2018.02013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCreadie JW, Beard CE. The microdistribution of the trichomycete Smittium culisetae in the hindgut of the black fly host Simulium vittatum. Mycologia. 2003;95:998–1003. doi: 10.1080/15572536.2004.11833015. [DOI] [PubMed] [Google Scholar]
- 51.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 2010;8:15. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schloss PD. Identifying and overcoming threats to reproducibility, replicability, robustness, and generalizability in microbiome research. mBio. 2018;9:e00525-18. doi: 10.1128/mBio.00525-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frankel-Bricker J, Song MJ, Benner MJ, Schaack S. Variation in the microbiota associated with Daphnia magna across genotypes, populations, and temperature. Microb Ecol. 2019 doi: 10.1007/s00248-019-01412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sinha R, et al. The microbiome quality control project: Baseline study design and future directions. Genome Biol. 2015;16:276. doi: 10.1186/s13059-015-0841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sinha R, et al. Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nat. Biotechnol. 2017;35:1077–1086. doi: 10.1038/nbt.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bahl MI, Bergström A, Licht TR. Freezing fecal samples prior to DNA extraction affects the Firmicutes to Bacteroidetes ratio determined by downstream quantitative PCR analysis. FEMS Microbiol. Lett. 2012;329:193–197. doi: 10.1111/j.1574-6968.2012.02523.x. [DOI] [PubMed] [Google Scholar]
- 57.Cardona S, et al. Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol. 2012;12:158. doi: 10.1186/1471-2180-12-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foggie, T. & Achee, N. Standard operating procedures: rearing Aedes aegypti for the HITSS and box laboratory assays training manual (2009).
- 59.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takahashi S, et al. Development of a prokaryotic universal primer for simultaneous analysis of bacteria and archaea using next-generation sequencing. PLoS ONE. 2014;9:e105592. doi: 10.1371/journal.pone.0105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.R Core Team. R: A Language Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/ (2019).
- 62.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pruesse E, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics. 2011;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Salter SJ, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Davis NM, et al. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wickham H. Ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011;3:180–185. [Google Scholar]
- 70.Lüdecke, D. sjstats: statistical functions for regression models. https://cran.r-project.org/web/packages/sjstats/in (2017).
- 71.Marwick, B. & Krishnamoorthy, K. cvequality: tests for the equality of coefficients of variation from multiple groups. https://github.com/benmarwick/cvequality (2016).
- 72.Feltz CJ, Miller GE. An asymptotic test for the equality of coefficients of variation from k populations. Stat. Med. 1996;15:647–658. doi: 10.1002/(sici)1097-0258(19960330)15:6<647::aid-sim184>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 73.Krishnamoorthy K, Lee M. Improved tests for the equality of normal coefficients of variation. Comput. Stat. 2014;29:215–232. [Google Scholar]
- 74.Kandlikar GS, et al. ranacapa: an R package and Shiny web app to explore environmental DNA data with exploratory statistics and interactive visualizations. F1000 Res. 2018;7:1734. doi: 10.12688/f1000research.16680.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anderson MJ. Permutational multivariate analysis of variance (PERMANOVA) Wiley StatsRef Stat. Ref. Online. 2017 doi: 10.1002/9781118445112.stat07841. [DOI] [Google Scholar]
- 76.Dixon P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003;14:927–930. [Google Scholar]
- 77.Kindt, R. Package ‘ BiodiversityR .’ R Proj.https://CRAN.R-project.org/package=BiodiversityR (2016).
- 78.Anderson MJ, Ellingsen KE, McArdle BH. Multivariate dispersion as a measure of beta diversity. Ecol. Lett. 2006;9:683–693. doi: 10.1111/j.1461-0248.2006.00926.x. [DOI] [PubMed] [Google Scholar]
- 79.Anderson MJ. Distance-based tests for homogeneity of multivariate dispersions. Biometrics. 2006;62:245–253. doi: 10.1111/j.1541-0420.2005.00440.x. [DOI] [PubMed] [Google Scholar]
- 80.Bates, D. et al. Package “lme4”: linear mixed-effects models using “Eigen” and S4. J. Stat. Softw. https://CRAN.R-project.org/package=lme4 (2015).
- 81.Fox, J. et al. Package “car.” R Doc. https://CRAN.R-project.org/package=car (2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequences are deposited in the NCBI Sequence Read Archive under accession number PRJNA606895.