Highlights
-
•
There were significant differences in lncRNAs, 18 miRNAs, and 20 mRNAs between breast cancer with and without bone metastasis.
-
•
JGB3, CAMGV, PTPRZ1, and FBN3 mRNA were related to prognosis of patients with metastatic breast cancer of bone.
-
•
The proportions of plasma cells and follicular helper T cells were significantly higher in breast cancer with bone metastasis.
-
•
The model composed of activated mast cells, gamma delta T cells, activated dendritic cells, follicular helper T cells, eosinophils and neutrophils had the smallest Akaike information criterion.
-
•
DLX6-AS1, Wnt6, and GABBR2 expression may be related to bone metastasis in patients with breast cancer.
Abbreviations: ceRNA, competing endogenous RNA; TCGA, The Cancer Genome Atlas; lncRNA, long non-coding RNA; miRNA, microRNA; mRNA, messenger RNA; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; FPKM, fragments per kilobase per million mapped reads; DE, Differentially expressed; GO, Gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PCC, Pearson correlation coefficient; ROC curve, receiver operating characteristic curve; AUC, Area under curve; AIC, Akaike information criterion; EMT, epithelial-mesenchymal transition; Runx2, runt related transcription factor 2; DEmRNA, differentially expressed messenger RNA; TNM, Tumor, Node, Metastases
Keywords: Bone metastasis, Breast cancer, ceRNA network, Immune infiltration, Nomogram, Prognosis
Abstract
Purpose
Advanced breast cancer commonly metastasises to bone; however, the molecular mechanisms underlying the affinity for breast cancer cells to bone remains unclear. Thus, we developed nomograms based on a competing endogenous RNA (ceRNA) network and analysed tumour-infiltrating immune cells to elucidate the molecular pathways that may predict prognosis in patients with breast cancer.
Methods
We obtained the RNA expression profile of 1091 primary breast cancer samples included in The Cancer Genome Atlas database, 58 of which were from patients with bone metastasis. We analysed the differential RNA expression patterns between breast cancer with and without bone metastasis and developed a ceRNA network. Cibersort was employed to differentiate between immune cell types based on tumour transcripts. Nomograms were then established based on the ceRNA network and immune cell analysis. The value of prognostic factors was evaluated by Kaplan-Meier survival analysis and a Cox proportional risk model.
Results
We found significant differences in long non-coding RNAs (lncRNAs), 18 microRNAs (miRNAs), and 20 messenger RNAs (mRNAs) between breast cancer with and without bone metastasis, which were used to construct a ceRNA network. We found that the protein-coding genes GJB3, CAMMV, PTPRZ1, and FBN3 were significantly differentially expressed by Kaplan-Meier analysis. We also observed significant differences in the abundance of plasma cell and follicular helper T cell populations between the two groups. In addition, the proportion of mast cells, gamma delta T cells, and plasma cells differed depending on disease location and stage. Our analysis showed that a high proportion of follicular helper T cells and a low proportion of eosinophils promoted survival and that DLX6-AS1, Wnt6, and GABBR2 expression may be associated with bone metastasis in breast cancer.
Conclusions
We developed a bioinformatic tool for exploring the molecular mechanisms of bone metastasis in patients with breast cancer and identified factors that may predict the occurrence of bone metastasis.
1. Background
Breast cancer is the most prevalent of all cancer types and is leading the cause of cancer death in women [1]. In the advanced stages, breast cancer often metastasises to bone and studies have shown that 47–85% of patients with breast cancer will experience bone metastasis [2]. The lowest rate of bone metastasis has been found in tumours negative for the estrogen receptor (ER) and human epidermal growth factor receptor 2 (HER2) [3], [4]. Conversely, increased bone metastasis rates are observed in patients with HER2+, ER+/HER2−/Ki67Hi, and ER+/HER2−/Ki67Low tumours [3], [4]. Bone metastases are frequently found in the spine, ribs, pelvis, proximal femur, and skull. Bone deterioration in these areas lead to serious complications such as pain, fractures, hypercalcaemia, spinal cord compression, and other nerve compression events, which significantly reduce quality of life and may be fatal [1], [2], [3], [4], [5]. The process of bone metastasis is complex and includes multiple stages. Firstly, separation of breast cancer cells from the primary tumour site is required, followed by transport to the bone via blood or lymph, resulting in survival and proliferation in target bone tissue [6], [7], [8]. Genomics research has shown that every step of metastasis is composed of a series of sub-events. However, the molecular mechanisms underlying bone metastasis in breast cancer are not fully understood [9], [10], [11].
In this study, we developed a complete protein interaction map by constructing a competing endogenous RNA (ceRNA) network and gene expression profile of patients with breast cancer, with or without bone metastasis, to examine the underlying molecular mechanisms. Prediction of bone metastasis and subsequent prognosis of breast cancer may be related to both the ceRNA network and the type of tumour-infiltrating immune cells, thus we obtained the gene expression profiles of patients with breast cancer from The Cancer Genome Atlas (TCGA) and applied the Cibersort algorithm. This was used to establish a predictive nomogram. In addition, we evaluated the relationship between tumour-infiltrating immune cells and ceRNA networks, which provided insight into the molecular mechanisms and clinical predictors of breast cancer metastasis. A flow chart explaining this process is provided in Fig. 1.
Fig. 1.
A flow chart depicting the analytical process.
2. Materials and methods
2.1. Data collection
Breast cancer RNA-seq raw count and fragments per kilobase per million mapped reads (FPKM) and miRNA-seq data were downloaded from the TCGA database. Annotation information was downloaded from genecode. The microRNA (miRNA) expression profile included 503 genes from 1078 samples (bone metastasis: 58; non-metastasis: 1020). The messenger RNA (mRNA) expression profile included 1925 genes from 1091 samples (bone metastasis: 58; non-metastasis: 1033). The long non-coding RNA (lncRNA) expression profile included 1925 genes from 1091 samples (bone metastasis: 58; non-metastasis: 1033).
2.2. Differential gene acquisition, expression analysis and functional annotation
We collected the RNA-seq raw count and FPKM fragments of 1091 primary breast cancer samples, of which 58 had bone metastasis (detailed clinical information of all patients is summarised in Table 1). We also retrieved demographic information and survival data from all patients. The genes that were not specific to breast cancer were filtered out, and differential RNA expression between bone metastatic breast cancer and non-bone metastasis breast cancer was analysed using differentially expressed seq2 (DEseq2). Up or downregulated genes were defined as false discovery rate (FDR)-adjusted P < 0.05 and log fold change >1.0 or <-1.0. DEseq2 was specially designed to analyse raw RNA-seq expression data to calculate differential gene expression between bone metastasis samples and non-bone metastasis samples (threshold: logFC = 1, adj.P = 0.05). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DEmRNAs were carried out using the database for annotation, visualisation and integrated discovery (DAVID) [12]. Terms and pathways with adj.P < 0.05 were selected.
Table 1.
Characteristics and distribution of breast cancer patients in the TCGA-BRCA cohort.
| Variables | mRNA (n = 1091) | miRNA (n = 1078) | Bone metastasis (n = 58) |
|---|---|---|---|
| Age (y) | |||
| >60 | 490 | 485 | 25 |
| ≤60 | 600 | 592 | 33 |
| NA | 1 | 1 | 0 |
| Pathologic stage, n | |||
| Stage I | 181 | 181 | 4 |
| Stage II | 620 | 609 | 18 |
| Stage III | 248 | 245 | 23 |
| Stage IV | 20 | 20 | 11 |
| NA | 22 | 23 | 2 |
| T | |||
| T1 | 279 | 279 | 7 |
| T2 | 631 | 620 | 29 |
| T3 | 137 | 135 | 16 |
| T4 | 40 | 40 | 6 |
| NA | 4 | 4 | 0 |
| M | |||
| M0 | 907 | 893 | 39 |
| M1 | 22 | 21 | 12 |
| NA | 162 | 164 | 7 |
| N | |||
| N0 | 514 | 508 | 10 |
| N1 | 360 | 356 | 28 |
| N2 | 120 | 118 | 7 |
| N3 | 76 | 75 | 11 |
| NA | 21 | 21 | 2 |
| ER status, n | |||
| positive | 803 | 795 | 41 |
| negative | 237 | 232 | 11 |
| NA | 51 | 51 | 6 |
| PR status, n | |||
| positive | 694 | 689 | 33 |
| negative | 343 | 335 | 20 |
| NA | 54 | 54 | 5 |
| HER2 status, n | |||
| Positive | 164 | 159 | 4 |
| Negative | 559 | 554 | 18 |
| Equivocal | 178 | 177 | 9 |
| NA | 190 | 188 | 27 |
| Histological type | |||
| Infiltrating ductal carcinoma | 779 | 768 | 32 |
| Infiltrating lobular carcinoma | 203 | 200 | 14 |
| Infiltrating carcinoma of no special type (NOS) | 1 | 1 | 0 |
| Medullary carcinoma | 6 | 6 | 0 |
| Metaplastic carcinoma | 9 | 9 | 1 |
| Mucinous carcinoma | 17 | 17 | 3 |
| Mixed histology | 29 | 29 | 4 |
| Other | 45 | 46 | 4 |
| NA | 2 | 2 | 0 |
TCGA: The Cancer Genome Atlas; BRCA: Breast cancer; ER: estrogen receptor; PR: progesterone receptor; HER2: human epidermal growth factor receptor 2; NA: not available.
2.3. Construction of a ceRNA network in breast cancer bone metastasis
miRNA-mRNA and lncRNA-miRNA interaction were predicted using miRTarBase and LncBase v.2 experimental modules, respectively [13], [14]. We searched for miRNA with DElncRNA as the target in LncBase database, and for target genes of these miRNAs in miRTarBase, which were intersected with DEmRNA. Then, the Pearson correlation coefficient (PCC) was calculated and the correlation between lncRNA and mRNA was screened. We used Cytoscape v.3.5.1 [15] to select the miRNAs that regulate lncRNAs and mRNAs, which were significant in the hypergeometric test and correlation analysis. These were used for visualising the ceRNA network (Sankey plot by Ggalluvial package).
2.4. Clinical significance of ceRNA network for bone metastasis in breast cancer
Survival analysis and Cox risk regression analyses were performed for the genes in the ceRNA network to identify key genes related to prognosis. The differences in gene expression from samples of different TNM stage were analysed.
2.5. Model building
We randomised 1091 breast cancer samples into two groups. Overall, 75% of the samples were used as the training set and 25% as the test set. A Lasso regression model was constructed from the training set, and the best lambda value and gene set were obtained. Then, the lifetime of the test set was predicted by the constructed model, and receiver operating characteristic (ROC) curve and calibration curves were drawn. A Lasso regression model was then constructed using the glmnet package. A ROC curve was drawn using the timeROC package.
The Cox risk regression model was constructed with these genes, and the nomogram was drawn with the rms package. In order to evaluate the prediction effect of the nomogram, a calibration curve was developed.
2.6. Cibersort estimation
Cibersort software (http://cibersort.stanford.edu/) [16] estimated the abundance of 22 different types of immune cells based on RNA-seq count data. In order to explore the clinical significance of differing proportions of immune cells, all samples were sorted according to cell proportion, with the median used as the dividing line. The samples were categorised as high- or low-proportion and included in the Kaplan-Meier survival analysis.
2.7. Survival analysis and nomogram of central cells in tumour immunity
The impact of the immune cell types on the prognosis of breast cancer patients was examined by Kaplan-Meier survival analysis and Cox regression. The Wilcoxon rank-sum test evaluated the relationship between immune cell subtypes, the occurrence of metastasis, and TNM stage. In the original Cox model, immune cells showed a significant correlation with prognosis and were selected to establish a nomogram. We quantified the ROC and area under the curve (AUC) to evaluate the sensitivity and specificity of our diagnostic and prognostic models. The accuracy of the predictive capabilities of the nomogram was evaluated using a calibration curve and consistency index. The relationship between ceRNAs and the 22 types of immune cells were investigated using Pearson’s correlation coefficient. Statistical significance was determined by a two-sided P < 0.05. R version 3.5.1 (Institute of Statistics and Mathematics, Austria) with the following packages was used for statistical analyses: ggplot2, rms, glmnet, survminer, and timeROC.
3. Results
3.1. Identification and expression analysis of genes with significant differences
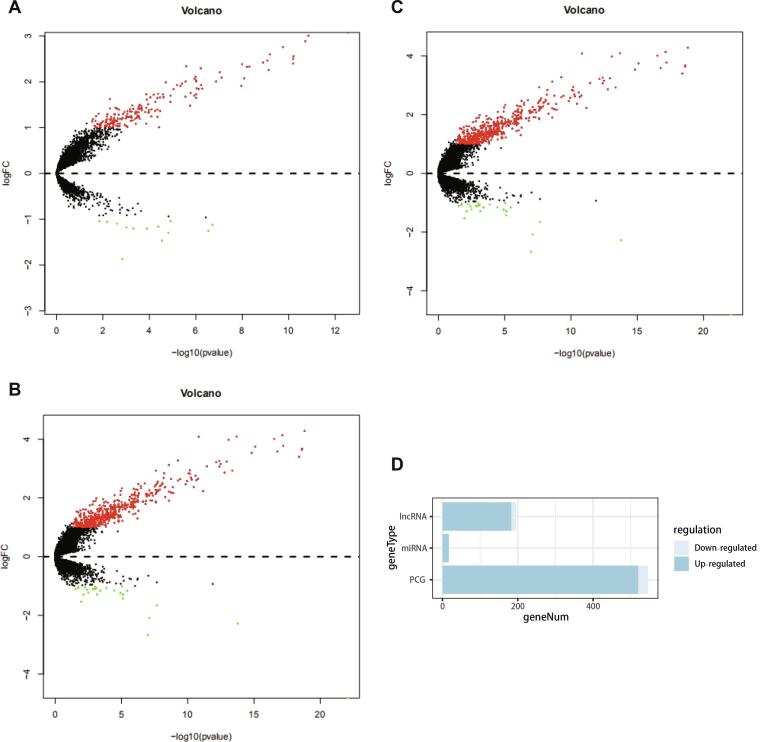
We identified differentially expressed genes with a log fold change of >1.0 or <-1.0 and a FDR of <0.05. The differential expression of lncRNAs, miRNAs and mRNAs (Fig. 2A–C) in breast cancer samples with and without bone metastasis was calculated using DESeq2 package. Overall, there were 183 upregulated lncRNAs, 13 downregulated lncRNAs; 17 upregulated miRNAs, 0 downregulated miRNAs; 520 up-regulated mRNAs and 26 down regulated mRNAs (Fig. 2D).
Fig. 2.
(A–C) Volcano map of differential expression of lncRNA, miRNA and mRNA. Red dots indicate genes significantly upregulated in bone metastasis samples, green indicates genes significantly downregulated, and black dots indicate genes without a significant difference. (D) The composition of differentially expressed genes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Functional enrichment analysis
Fig. 3 shows the enrichment of the top 10 GO terms and pathways of differentially expressed mRNA (DEmRNA). We found that the main functional areas of gene enrichment were: go: 0007267 ~ cell signalling, go: 0070098 ~ chemokine mediated signalling, go: 0010628 ~ position regulation of gene expression, go: 0010942 ~ position regulation of cell death, go: 0010737 ~ protein kinase A signalling, and go: 0008284 ~ position regulation of cell promotion. The main pathways of gene enrichment were: hsa04080: neuroactive living receptor interaction, hsa04062: chemokine signalling, hsa05204: chemical carcinogenesis, hsa00982: drug metabolism – cytochrome P450, and hsa00350: tyrosine metabolism and other tumour related signalling pathways.
Fig. 3.
(A) Gene ontology function enrichment circle; (B) Pathway enrichment cycle. On the left side of each circle is the gene, which is sorted according to the multiple of expression difference between bone metastasis and non-bone metastasis.
3.3. Construction of a protein interaction network based on differentially expressed genes
We input the DEmRNA into the STRING protein database with a score threshold of 0.9 (the highest confidence). The final protein interaction network had 183 nodes, 491 edges, and protein-protein interaction (PPI) enrichment of P < 1.0e−16. See Fig. 4 for details. The average degree of each node was 5.37. This suggests that there is a complex regulatory relationship between the different genes involved in breast cancer with bone metastasis.
Fig. 4.
Protein interaction network of different genes. In the network, point represents proteins, edges represent interactions, and a wider edge indicated a stronger interaction.
3.4. Construction and survival analysis of ceRNA network
The shared miRNAs of differentially expressed mRNA and lncRNA were obtained from the LncBase and miRTarBase databases, the Pearson’s correlation coefficient of lncRNA-mRNA pairs with shared miRNA was calculated, and the positive expressed lncRNA-mRNA pairs were screened (Table 2). Using Cytoscape software (Fig. 5A), we constructed a ceRNA network related to breast cancer with bone metastasis, including 20 mRNAs, 18 miRNAs, 2 lncRNAs, and 48 edges. We analysed gene batch survival in the ceRNA network and the results showed that JGB3, CAMGV, PTPRZ1, FBN3 mRNA were all related to prognosis (Fig. 5B–E).
Table 2.
lncRNA-miRNA-mRNA pairs.
| miRNA | lncRNA | mRNA | P value | PCC |
|---|---|---|---|---|
| hsa-let-7b-5p | DLX6-AS1 | IGF2BP2 | 3.62E-32 | 0.347 |
| hsa-let-7b-5p | DLX6-AS1 | IGF2BP3 | 2.00E-61 | 0.471 |
| hsa-let-7f-5p | DLX6-AS1 | KLK6 | 2.52E-27 | 0.32 |
| hsa-mir-1-3p | DLX6-AS1 | GJB3 | 7.91E-41 | 0.389 |
| hsa-mir-1-3p | DLX6-AS1 | SOX6 | 1.75E-85 | 0.545 |
| hsa-mir-124-3p | DLX6-AS1 | OCA2 | 1.92E-46 | 0.414 |
| hsa-mir-124-3p | DLX6-AS1 | GABBR2 | 3.18E-52 | 0.437 |
| hsa-mir-124-3p | DLX6-AS1 | BARX1 | 1.49E-26 | 0.315 |
| hsa-mir-124-3p | DLX6-AS1 | PRDM13 | 4.99E-29 | 0.329 |
| hsa-mir-124-3p | DLX6-AS1 | PTPRZ1 | 1.09E-31 | 0.344 |
| hsa-mir-132-3p | DLX6-AS1 | FBN3 | 7.32E-56 | 0.451 |
| hsa-mir-148b-3p | DLX6-AS1 | CCKBR | 1.28E-31 | 0.344 |
| hsa-mir-148b-3p | DLX6-AS1 | DLX6 | 0 | 0.884 |
| hsa-mir-155-5p | AFAP1-AS1 | SOX6 | 2.52E-25 | 0.308 |
| hsa-mir-16-5p | DLX6-AS1 | ALKAL2 | 7.95E-44 | 0.403 |
| hsa-mir-16-5p | DLX6-AS1 | SOX6 | 1.75E-85 | 0.545 |
| hsa-mir-16-5p | DLX6-AS1 | KLHL34 | 2.04E-28 | 0.326 |
| hsa-mir-16-5p | DLX6-AS1 | CAMKV | 4.92E-25 | 0.306 |
| hsa-mir-181a-5p | DLX6-AS1 | PTPRZ1 | 1.09E-31 | 0.344 |
| hsa-mir-181a-5p | DLX6-AS1 | OCA2 | 1.92E-46 | 0.414 |
| hsa-mir-195-5p | DLX6-AS1 | CAMKV | 4.92E-25 | 0.306 |
| hsa-mir-221-3p | DLX6-AS1 | FBN3 | 7.32E-56 | 0.451 |
| hsa-mir-26a-5p | DLX6-AS1 | CAMKV | 4.92E-25 | 0.306 |
| hsa-mir-27a-3p | DLX6-AS1 | RNF182 | 2.35E-44 | 0.405 |
| hsa-mir-30b-5p | DLX6-AS1 | CAMKV | 4.92E-25 | 0.306 |
| hsa-mir-30c-5p | DLX6-AS1 | CAMKV | 4.92E-25 | 0.306 |
| hsa-mir-320a | DLX6-AS1 | POLR2F | 2.15E-47 | 0.418 |
| hsa-mir-320a | DLX6-AS1 | IGF2BP3 | 2.00E-61 | 0.471 |
| hsa-mir-7-5p | DLX6-AS1 | IL12RB2 | 6.13E-50 | 0.428 |
| hsa-mir-9-5p | DLX6-AS1 | WNT6 | 2.16E-32 | 0.348 |
miRNA: microRNA; lncRNA: long non-coding RNA; mRNA: messenger RNA; PCC: Pearson correlation coefficient.
Fig. 5.
Construction of ceRNA network and prognosis analysis. (A) Sankey plot of ceRNA network. Each square represents a gene. The larger the square, the larger the degree of the gene node. (B–E) Significant survival curve in the ceRNA network of (B) JGB3, (C) CAMGV, (D) PTPRZ1, (E) FBN3.
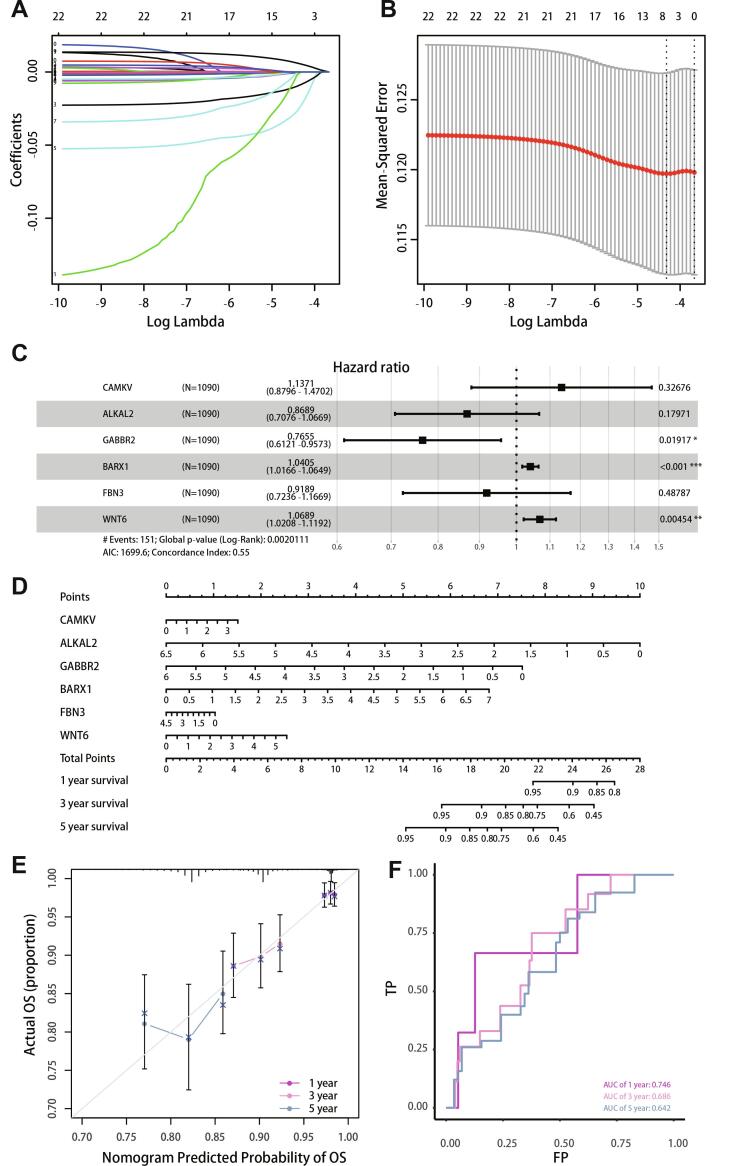
3.5. Lasso regression analysis and nomogram construction
LncRNA and mRNA in the ceRNA network of bone metastasis were used to calculate the optimal lambda value (Fig. 6A, B), which was 0.0133. Six genes (CAMGV, ALKAL2, GABBR2, BARX1, FBN3, and Wnt6) were used to construct a multiple Cox risk regression model (Fig. 6C). The six genes were used to construct a graph based on Cox regression (Fig. 6D), which predicted the 1-, 3-, and 5-year survival status. To evaluate the prediction effect of the nomogram model, the 1-, 3-, and 5-year calibration curves (Fig. 6E) were drawn, and the results showed that the nomogram performed well. The ROC curve (Fig. 6F) showed that the average ROC of the nomogram was 0.686, which represents a high prediction accuracy. Lasso regression results also showed that the six genes were necessary for modelling. In addition, ROC and calibration curves showed acceptable accuracy as demonstrated by AUC values of 0.746, 0.686, and 0.642 for 1-, 3-, and 5-year survival, respectively.
Fig. 6.
Lasso regression analysis and nomogram construction. (A, B) Selection of important coefficient lambda in lasso regression. (C) Forest map of multiple Cox regression results. (D) Nomogram based on multiple Cox regression. (E) Calibration curve for 1-, 3-, and 5-year survival. The closer to the diagonal, the better the prediction effect. (F) ROC curve analysis for 1-3-, and 5-year survival.
3.6. Relationship between gene expression and TNM (Tumor, Node, Metastases) stage
The relationship between mRNA and different TNM stages were analysed by t-test. We found that the expression level of genes in the ceRNA network decreased in later TNM stages. We observed that the average expression levels of FBN3 in T1 and N1 stages were 0.270 and 0.228, respectively, while those in T4 and N3 stages were significantly decreased (average expression values 0.116 and 0.186, respectively; T stage, P = 0.006; N stage, P = 0.005, Fig. 7A, B). The average expression levels of GABBR2 in T1 and N1 stages were 0.226 and 0.211 respectively, while those in T4 and N3 stages were significantly decreased (average expression values were 0.072 and 0.140, respectively; T stage, P = 0.0004; N stage, P = 0.0006, Fig. 7D, E). Fig. 7C shows that the average expression level of ALKAL2 in T1 stage was 0.228, while in T4 stage was 0.186. Fig. 7F shows that the average CAMKV expression level in N1 stage was 0.057, while that in N3 stage was reduced to 0.048).
Fig. 7.
Relationship between gene expression level and TNM stage. (A) Expression of FBN3 in different T stages. (B) Expression of FBN3 in different N stages. (C) Expression of ALKALL2 in different T stages. (D) Expression of GABBR2 in different T stages. (E) Expression of GABBR2 in different N stages. (F) Expression of CNMKV in different N stages.
3.7. Distribution of tumour-infiltrating immune cells in breast cancer with bone metastasis
Fig. 8A shows the immune cell components of 58 bone metastasis samples. The split violin diagram reveals the different immune cell proportions between breast cancer with and without bone metastasis (Fig. 8B). Among them, the abundance of plasma cell and follicular helper T cells were significantly different between the two samples.
Fig. 8.
Component analysis of immune cells. (A) Proportion of lymphocytes in 58 bone metastasis samples. (B) Difference in the proportions of 22 types of immune cell in bone metastasis and non-bone metastasis samples.
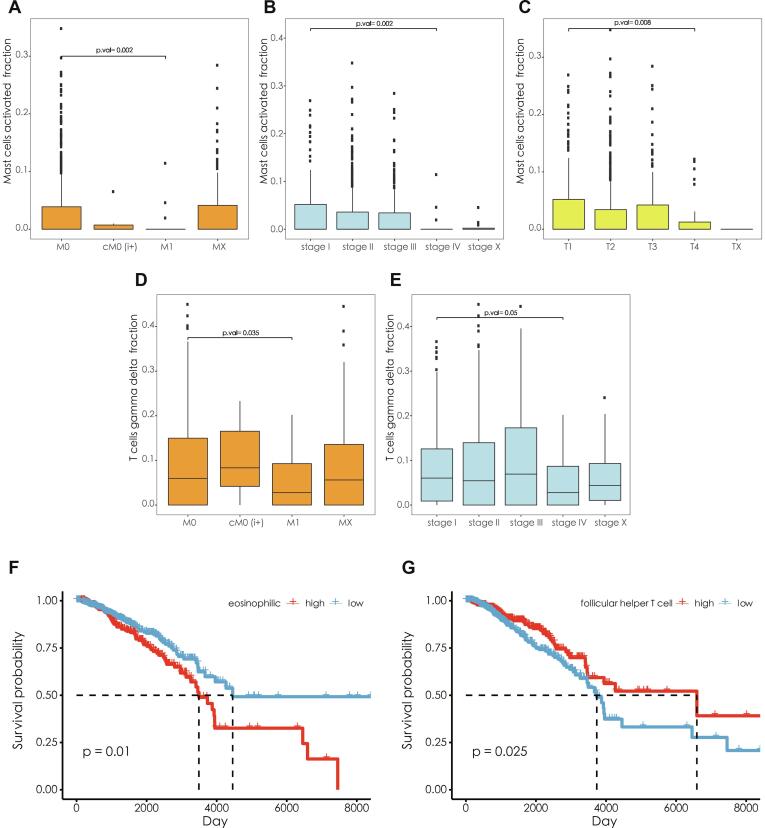
3.8. Clinical significance of immune cell components
We analysed the clinical correlation and prognosis of 22 types of immune cell. The results showed that there were significantly more mast cells present between M1 and M0 (Fig. 9A, P = 0.002), stage IV and stage I (Fig. 9B, P = 0.002), T4 and T1 samples (Fig. 9C, P = 0.008). Similarly, gamma delta T cells were significantly overrepresented in M1 samples compared with M0 samples (Fig. 9D, P = 0.035), and stage IV samples compared with stage I samples (Fig. 9E, P = 0.05). Survival analysis revealed that the samples with a low proportion of eosinophils had better survival status (Fig. 9F, log-rank test, P = 0.01); the samples with a high proportion of follicular helper T cells had better survival status (Fig. 9G, log-rank test, P = 0.025).
Fig. 9.
(A–E) Different cell contents at different stages. (F) Differences between the survival states of samples with high eosinophilic content and those with low eosinophilic content. (G) Differences in survival status between the high-content follicular helper T cell samples and the low-content samples.
3.9. Clinical correlation of immune cells and nomogram multiple Cox risk regression analysis
We performed multivariate Cox regression analysis for the 22 different types of immune cells, and showed that the model composed of activated mast cells, gamma delta T cells, activated dendritic cells, follicular helper T cells, eosinophils and neutrophils had the smallest Akaike information criterion (AIC) (Fig. 10A). According to the above clinical correlation analysis, six types of immune cells were selected to construct the multiple Cox risk regression model and calculate the risk ratio. Then, the nomogram (Fig. 10B) for 1-, 3- and 5-year survival was developed. The calibration curve (Fig. 10C) shows that our nomograms had an adequate predictive capacity and the product under the average ROC curve was 0.616 (Fig. 10D). Selection of the best threshold for 5-year survival status and division of the samples into a high-risk group and low-risk group showed a significant difference in survival status (P < 0.001, Fig. 10E, F) ROC curve and calibration curve analysis showed that the nomogram was consistent and had good accuracy with 1-, 3-, and 5-year survival AUC values of 0.549, 0.616, and 0.644, respectively. Multiple Cox regression analysis showed that the high-risk group differed significantly from the low-risk group.
Fig. 10.
(A) Forest map of multiple Cox regression results. (B) Nomogram based on multiple Cox regression for predicting 1-, 3-, and 5-year survival. (C) Calibration curves for predicting 1-, 3- and 5-year survival. (D) ROC curve analysis for predicting the 1-, 3-, and 5-year survival. (E) Survival curve of the high-risk group and low-risk groups. (F) Thermogram of six lymphocyte contents in the high-risk group and low-risk group.
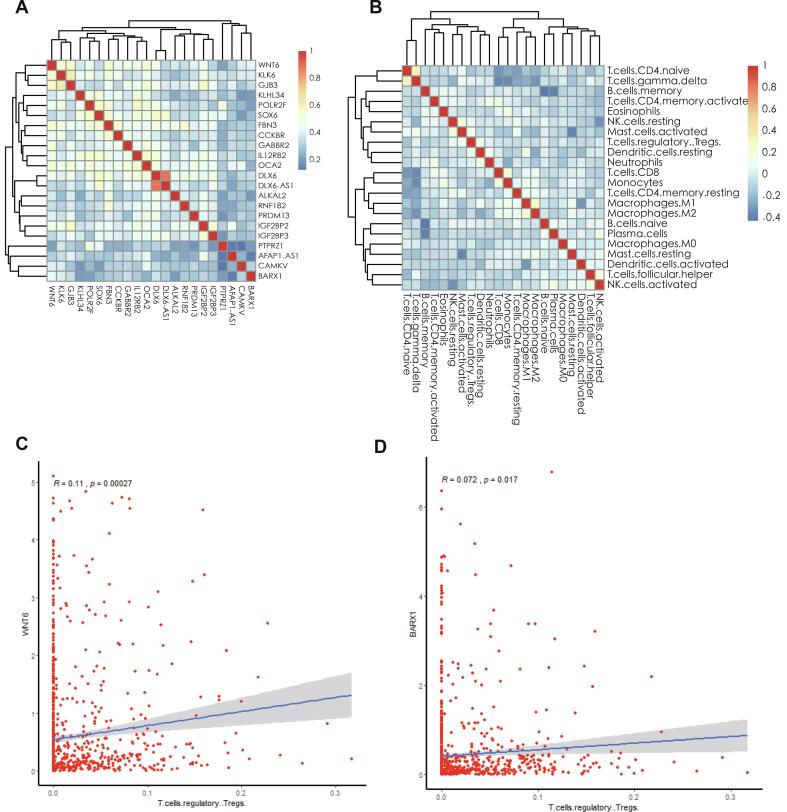
3.10. Co-expression analysis of immune cells and key genes
Pearson’s correlation coefficient indicated the correlation between the thermogram of RNAs (Fig. 11A) and 22 types of lymphocytes in breast cancer samples (Fig. 11B). Here, we specifically analyzed the correlation between T-regulatory cells and Wnt6/BARX1, and found that T-regulatory cells and Wnt6 (Fig. 11C, R = 0.11, P = 0.0027), and T-regulatory cells and BARX1 (Fig. 11D, R = 0.072, P = 0.0017) were positively correlated. We found that Wnt6 was positively correlated with KLK6 and GJB3. Wnt6 was positively correlated with FBN3 and GABBR2, and DLX6 and DLX6-AS1 were positively correlated. Naïve CD4+ T cells and gamma delta T cells were also positively correlated. Memory B cells negatively correlated with naïve B cells and plasma cells, and CD8+ T cells positively correlated with monocytes. We also showed that M1 macrophages positively correlated with M2 macrophages.
Fig. 11.
(A) Correlation thermogram of lncRNA and PCG in prognosis-related ceRNA network. (B) Correlation thermogram of 22 types of lymphocytes in breast cancer samples. (C) Correlation between regulatory T cells and Wnt6 expression. (D) Correlation between regulatory T cells and BARX1 expression.
4. Discussion
Breast cancer is one of the most common malignant tumors in women globally [1]. With continuing improvements in the prognosis of breast cancer, the incidence of bone metastasis has also increased. Epidemiological data showed that the incidence of bone metastasis in advanced breast cancer was 65–75%, and the initial symptoms of bone metastasis accounted for 27–50% in advanced breast cancer [1], [2], [3]. Patients with advanced breast cancer often develop distant metastasis in locations such as bone. However, the underlying molecular mechanisms are still unknown. There are several determining factors related to molecular and cellular features that are often used in the clinic to determine prognosis. Bone metastasis is detrimental to quality of life and survival in patients with breast cancer, and causes serious complications including pain, fracture, spinal cord compression, and malignant hypercalcemia [1], [2], [3], [4]. Due to the frequent occurrence of bone metastasis in patients with advanced breast cancer, the understanding of pathogenesis and clinical management of bone metastasis are important and challenging topics in basic research and clinical practice. The ceRNA network, including mRNA, miRNA, and lncRNA, and infiltrating immune cells may be critical to further understand this phenomenon [17], [18], [19], [20]. We observed that tumour samples with bone metastasis had significantly altered proportions of infiltrating cells compared with breast cancer without bone metastasis. We then developed two models to predict the prognosis of breast cancer patients with bone metastasis. The resulting AUC of the two nomograms demonstrated their value in a clinical setting.
LncRNA is a type of ncRNA with over 200 nucleotides, which is not related to protein-coding [17], [18], [19], [20], [21]. New evidence suggests that lncRNA imbalance occurs frequently in many malignant tumours, which is a key factor for carcinogenesis through post-transcriptional regulation and epigenetic modification [22], [23]. Here, we bioinformatically analysed the ceRNA network regulating bone metastasis in breast cancer with 20 protein encoded mRNAs, 2 lncRNAs and 18 miRNAs. We found a significant correlation between four protein-coding genes (GJB3, CAMGV, PTPRZ1, FBN3) and their associated miRNAs and lncRNAs and survival in patients with breast cancer patients and bone metastasis. The AUC values for 1-, 3-, and 5-year survival were 0.746, 0.686, and 0.642, respectively. Using a hypergeometric test and correlation analysis, we found a significant correlation between GJB3 (DLX6-AS1, hsa-mir-1-3p), FBN3 (DLX6-AS1, hsa-mir-132-3p), CAMKV (DLX6-AS1, hsa-mir-16-5p), PTPRZ1 (DLX6-AS1, hsa-mir-181a-5p), and survival rate in breast cancer patients with bone metastasis. These results indicated that DLX6-AS1 may be a key contributor to bone metastasis in patients with advanced breast cancer. We speculate that DLX6-AS1 may regulate the occurrence and progression of metastasis by interacting with Wnt/β-catenin signalling.
Recently, an increasing number of studies show that aberrant lncRNA expression leads to the development of many kinds of malignant tumours, including breast cancer [17], [18], [19], [20], [21], [22], [23]. DLX6-AS1 is a lncRNA believed to be carcinogenic by regulating the progression of renal cell carcinoma, liver cell carcinoma, glioma, pancreatic cancer and lung adenocarcinoma [24], [25], [26], [27], [28], [29], [30], [31]. Normal brain tissue expresses a high level of DLX6-AS1, which is involved in development regulation [29], [30], [31]. In recent years, it has been found that DLX6-AS1 is abnormally expressed in a variety of tumour tissues and is closely related to poor clinical outcomes [24], [25], [26], [27], [28], [29], [30], [31]. However, the molecular mechanisms of DLX6-AS1 and how it contributes to the pathogenesis of breast cancer are still unclear. Zhao et al. demonstrated significant upregulation of DLX6-AS1 in breast cancer tissues and cell lines [32]. Furthermore, high DLX6-AS1 expression is linked to poor outcomes with regards to tumour size, lymph node metastasis, TNM stage, and survival of breast cancer patients [24], [25], [26], [27], [28], [29], [30], [31], [32]. SiRNA knockout of DLX6-AS1 showed a reduction in proliferation, apoptosis, invasion, migration, and the epithelial-mesenchymal transition (EMT) of breast cancer cells [30], [31], [32]. These findings suggest that the progression of breast cancer could be partly due to overexpression of DLX6-AS1. Studies have indicated that mRNA expression may be regulated by lncRNAs through competitive communication between ceRNAs and miRNAs [33], [34]. For example, DLX6-AS1 silencing inhibits cell proliferation, migration, and invasion in non-small cell lung cancer by interacting with mir-144. In addition, DLX6-AS1 is important in the carcinogenesis of glioma by competing with mir-197-5p. In pancreatic cancer, knocking out the DLX6-AS1 gene led to inhibition of proliferation and metastasis of cancer cells through enhancement of mir-181b’s endogenous effects. The results of our study suggest that DLX6-AS1 may be important for the expression and regulation of miRNAs in breast cancer bone metastasis. Zhao et al. evaluated the expression of DLX6-AS1 in breast cancer and analysed the correlation between DLX6-AS1 expression and clinicopathological parameters. They identified increased DLX6-AS1 expression in tumour tissue compared with normal tissue, which was linked to poor prognosis [32]. Similar to pancreatic cancer, DLX6-AS1 gene knockout reduces the proliferation, invasion, and migration capacity of breast cancer cells and promotes apoptosis [32]. Furthermore, luciferase analysis confirmed that DLX6-AS1 is an endogenous mediator of mir-505-3p and negatively regulates its expression. In addition, mir-505-3p inhibits runt related transcription factor 2 (Runx2) expression by binding directly to the 3′ untranslated region. Partial reversal of the carcinogenic effects of mir-505-3p can be achieved by overexpressing Runx2. Wang et al. showed that DLX6-AS1 exerts its downstream effects on breast cancer cells by downregulating Fus [28]. These findings indicate that DLX6-AS1 promotes breast cancer progression, which is consistent with our results. Accordingly, targeting DLX6-AS1 may be beneficial for the treatment of breast cancer.
The regulatory mechanisms between lncRNAs and miRNAs are extraordinarily complex. Among them, lncRNA can be used as the combination of ceRNA and miRNA to compete and share mRNA, thus forming a complex lncRNA-miRNA-mRNA network [35]. Wnt/β-catenin signalling is critical in numerous cellular processes such as proliferation, invasion, and migration [36]. The involvement of Wnt/β-catenin signalling has been demonstrated in several malignant tumours including breast cancer, and Wnt/β-catenin activation promotes tumour cell growth and metastasis [36], [37]. Previous studies have shown that Wnt activation inhibits memory T cells by reducing key transcription factors produced by these cells [38]. In our study, we found that the activation of mast cells was correlated with Wnt6 expression. Zhang et al proved that Wnt signal is often activated under the action of DLX6-AS1 [39]. Therefore, we speculate that the Wnt pathway may be important for the impact of DLX6-AS1 on the composition of immune cells. However, the molecular mechanisms of DLX6-AS1-induced Wnt activation remain to be elucidated.
Guo et al. demonstrated DLX6-AS1 upregulation in cell lines and tissues from bladder cancer patients, which augmented the proliferation, invasion, and migration of these cells by regulating the EMT process and the activity of Wnt/β-catenin signalling [40]. The impact of DLX6-AS1 on the characteristics of cancer stem cells in osteosarcoma has also been investigated and was found to positively correlate with more advanced disease and poorer survival [39]. Similar findings have been demonstrated in prostate cancer patients. Recently, several studies have shown the participation of Wnt6, BARX1, PTPRZ1, and other genes in the regulation of Wnt/β-catenin signalling and their involvement in the development of malignant tumours [36], [37], [38], [39], [40], [41], [42], [43], [44], [45]. These findings support our hypothesis that DLX6-AS1 may affect the mRNA expression of Wnt6, BARX1, PTPRZ1 and other genes through the ceRNA network to regulate Wnt/β-catenin signalling and participate in the distant bone metastasis of breast cancer.
In this study, we incorporated six genes (CAMGV, ALKAL2, GABBR2, BARX1, FBN3, and Wnt6) into a multiple Cox risk regression model in the process of Lasso regression. We constructed a nomogram using these six genes to predict the 1-, 3- and 5-year survival status in patients with breast cancer and bone metastases, showing that the prediction effect of the model was satisfactory.
In the study of immune cell infiltration in breast cancer bone metastasis, we found that plasma cells and follicular helper T cells were significantly different in two different samples. We also observed that the proportion of mast cells, gamma delta T cells, plasma cells, follicular helper T cells, and eosinophils varied depending on disease stage and progression. These results suggest that plasma cells, follicular helper T cells, mast cells and gamma delta T cells have potential value for predicting bone metastasis, disease grading and staging of breast cancer, which is expected to be further verified and applied in clinical diagnosis and treatment. Subsequently, our correlation analysis showed that Wnt6 and GABBR2 positively correlated with mast cell activation.
Our research inevitably has several limitations. The data included in our study are from Western countries and may not be directly extrapolated to patients in Asian countries. We were unable to comprehensively analyse the clinical and pathological parameters due to limited public information; therefore, we minimised bias by investigating the gene and protein expression of key biomarkers at the cell and tissue level. The uneven distribution of clinical samples in different TNM stages in Cibersort and the application off Cibersort to the TCGA data may also have an impact on the conclusions drawn in this study, and this issue must be addressed in future research. However, these research models have been widely accepted and adopted by international scholars and still need to be further explored [46], [47]. In this study, we included as many clinicopathological types and samples as possible. One reason is that breast cancer bone metastasis samples represent numerous pathological types and clinical stages, thus the inclusion of these data can help to obtain a larger, and more representative sample size. Secondly, the inclusion of all breast cancer pathological types is more conducive to explore the common characteristics of bone metastasis in different breast cancer subtypes.
The tumour microenvironment often affects the process of tumour invasion and migration [30], [31], [48]. Invasion of tumour cells is largely dependent on the composition of the extracellular matrix as well as growth factors that are secreted by surrounding cells [30], [31]. Furthermore, metastasis of the tumour may have occurred in the early stages of development and is not directly related to proliferation. Therefore, it is necessary to determine the molecular mechanisms leading to aggressive breast cancer with bone metastasis. We developed a ceRNA network based on breast cancer samples, and our nomogram of tumour-infiltrating immune cells predicted prognosis in breast cancer patients with and without bone metastasis with high accuracy. The predictive nomogram can provide more comprehensive clinical data for the individualised treatment of breast cancer with bone metastasis. Further studies should be performed to show the interactions and communications between cancer cells and immune cells. In particular, the exosomes secreted by tumour cells contain ceRNAs and may also play a role mediating breast cancer metastasis.
Declarations
Ethics approval and consent to participate: Not applicable.
Consent for publication: Not applicable.
Data availability statement: The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Funding
This study was supported by National Natural Science Foundation of China (Project No. 81871746; Grant recipient: Y.W.), and Peking Union Medical College Graduate Student Innovation Fund (2018) (Project No. 2018-1002-02-08; Grant recipient: S.L.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
CRediT authorship contribution statement
Shuzhong Liu: Conceptualization, Formal analysis, Funding acquisition, Methodology, Writing - original draft, Writing - review & editing. An Song: Conceptualization, Formal analysis, Writing - original draft. Xi Zhou: Formal analysis, Software. Zhen Huo: Formal analysis. Siyuan Yao: Formal analysis, Methodology, Software. Bo Yang: Conceptualization, Formal analysis, Methodology, Supervision, Writing - review & editing. Yong Liu: Conceptualization, Formal analysis, Software, Supervision. Yipeng Wang: Conceptualization, Funding acquisition, Methodology, Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank our colleagues at the Department of Orthopaedic Surgery, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College.
Contributor Information
Bo Yang, Email: ybsurg@vip.sina.com.
Yong Liu, Email: liuyong_pumch@163.com.
Yipeng Wang, Email: ypwang_pumch@163.com.
References
- 1.DeSantis C.E., Ma J., Gaudet M.M., Newman L.A., Miller K.D., Goding Sauer A., Jemal A., Siegel R.L. Breast cancer statistics, 2019. CA A Cancer J. Clin. 2019;69(6):438–451. doi: 10.3322/caac.21583. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen O.S., Munro A.J., Tannock I.F. Bone metastases: Pathophysiology and management policy. J. Clin. Oncol. 1991;9:509–524. doi: 10.1200/JCO.1991.9.3.509. [DOI] [PubMed] [Google Scholar]
- 3.Hayashi N., Iwamoto T., Qi Y., Niikura N., Santarpia L., Yamauchi H., Nakamura S., Hortobagyi G.N., Pusztai L., Fraser Symmans W., Ueno N.T. Bone metastasis-related signaling pathways in breast cancers stratified by estrogen receptor status. J. Cancer. 2017;8(6):1045–1052. doi: 10.7150/jca.13690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arciero C.A., Guo Y., Jiang R., Behera M., O’Regan R., Peng L., Li X. ER+/HER2+ breast cancer has different metastatic patterns and better survival than ER−/HER2+ breast cancer. Clin. Breast Cancer. 2019;19(4):236–245. doi: 10.1016/j.clbc.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Kawaguchi T., Yan L., Qi Q., Peng X., Edge S.B., Young J., Yao S., Liu S., Otsuji E., Takabe K. Novel microRNA-based risk score identified by integrated analyses to predict metastasis and poor prognosis in breast cancer. Ann. Surg. Oncol. 2018;25(13):4037–4046. doi: 10.1245/s10434-018-6859-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.X. Chen, Z. Pei, H. Peng, Z. Zheng, Exploring the molecular mechanism associated with breast cancer bone metastasis using bioinformatic analysis and microarray genetic interaction network, Med. (United States). 97(37) (2018) e12032, https://doi.org/10.1097/MD.0000000000012032. [DOI] [PMC free article] [PubMed]
- 7.C.N. Johnstone, A.D. Pattison, K.L. Gorringe, P.F. Harrison, D.R. Powell, P. Lock, D. Baloyan, M. Ernst, A.G. Stewart, T.H. Beilharz, R.L. Anderson, Functional and genomic characterisation of a xenograft model system for the study of metastasis in triple-negative breast cancer, Dis. Model. Mech. 11(5) (2018) dmm032250, https://doi.org/10.1242/dmm.032250. [DOI] [PMC free article] [PubMed]
- 8.Erin N., Ogan N., Yerlikaya A. Secretomes reveal several novel proteins as well as TGF-β1 as the top upstream regulator of metastatic process in breast cancer. Breast Cancer Res. Treat. 2018;170(2):235–250. doi: 10.1007/s10549-018-4752-8. [DOI] [PubMed] [Google Scholar]
- 9.Gangoda L., Liem M., Ang C.S., Keerthikumar S., Adda C.G., Parker B.S., Mathivanan S. Proteomic profiling of exosomes secreted by breast cancer cells with varying metastatic potential. Proteomics. 2017;17(23–24) doi: 10.1002/pmic.201600370. [DOI] [PubMed] [Google Scholar]
- 10.Lawler K., Papouli E., Naceur-Lombardelli C., Mera A., Ougham K., Tutt A., Kimbung S., Hedenfalk I., Zhan J., Zhang H., Buus R., Dowsett M., Ng T., Pinder S.E., Parker P., Holmberg L., Gillett C.E., Grigoriadis A., Purushotham A. Gene expression modules in primary breast cancers as risk factors for organotropic patterns of first metastatic spread: a case control study. Breast Cancer Res. 2017;19(1):113. doi: 10.1186/s13058-017-0881-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.J.N. Li, R. Zhong, X.H. Zhou, Prediction of bone metastasis in breast cancer based on minimal driver gene set in gene dependency network, Genes (Basel). 10(6) (2019) 466, https://doi.org/10.3390/genes10060466. [DOI] [PMC free article] [PubMed]
- 12.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 13.Chou C.H., Shrestha S., Yang C.D., Chang N.W., Lin Y.L., Liao K.W., Huang W.C., Sun T.H., Tu S.J., Lee W.H., Chiew M.Y., Tai C.S., Wei T.Y., Tsai T.R., Huang H.T., Wang C.Y., Wu H.Y., Ho S.Y., Chen P.R., Chuang C.H., Hsieh P.J., Wu Y.S., Chen W.L., Li M.J., Wu Y.C., Huang X.Y., Ng F.L., Buddhakosai W., Huang P.C., Lan K.C., Huang C.Y., Weng S.L., Cheng Y.N., Liang C., Hsu W.L., Da Huang H. MiRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46(D1):D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paraskevopoulou M.D., Vlachos I.S., Karagkouni D., Georgakilas G., Kanellos I., Vergoulis T., Zagganas K., Tsanakas P., Floros E., Dalamagas T., Hatzigeorgiou A.G. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44(D1):D231–D238. doi: 10.1093/nar/gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q., Jin X., Shi W., Chen X., Pang W., Yu X., Yang L. A long non-coding RNA LINC00461-dependent mechanism underlying breast cancer invasion and migration via the miR-144-3p/KPNA2 axis. Cancer Cell Int. 2020;20:137. doi: 10.1186/s12935-020-01221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ni C., Fang Q.Q., Chen W.Z., Jiang J.X., Jiang Z., Ye J., Zhang T., Yang L., Meng F.B., Xia W.J., Zhong M., Huang J. Breast cancer-derived exosomes transmit lncRNA SNHG16 to induce CD73+γδ1 Treg cells. Signal Transduct. Target. Ther. 2020;5(1):41. doi: 10.1038/s41392-020-0129-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang B., Zhang Y., Zhang H., Lin F., Tan Q., Qin Q., Bao W., Liu Y., Xie J., Zeng Q. Long intergenic non-protein coding RNA 324 prevents breast cancer progression by modulating miR-10b-5p, Aging (Albany. NY) 2020;12(8):6680–6699. doi: 10.18632/aging.103021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen X., Gao L., Hu Y. LAceModule: identification of competing endogenous RNA modules by integrating dynamic correlation. Front. Genet. 2020;11:235. doi: 10.3389/fgene.2020.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hua K., Deng X., Hu J., Ji C., Yu Y., Li J., Wang X., Fang L. Long noncoding RNA HOST2, working as a competitive endogenous RNA, promotes STAT3-mediated cell proliferation and migration via decoying of let-7b in triple-negative breast cancer. J. Exp. Clin. Cancer Res. 2020;39(1):58. doi: 10.1186/s13046-020-01561-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan P., Tang L., Liu L., Tu G. Identification of candidate RNA signatures in triple–negative breast cancer by the construction of a competing endogenous RNA network with integrative analyses of gene expression omnibus and the Cancer Genome Atlas data. Oncol. Lett. 2020;19(3):1915–1927. doi: 10.3892/ol.2020.11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lou W., Ding B., Fu P. Pseudogene-derived lncRNAs and their miRNA sponging mechanism in human cancer. Front. Cell Dev. Biol. 2020;8:85. doi: 10.3389/fcell.2020.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X., Zhang H., Wu X. Long noncoding RNA DLX6-AS1 accelerates the glioma carcinogenesis by competing endogenous sponging miR-197-5p to relieve E2F1. Gene. 2019;686(686):1–7. doi: 10.1016/j.gene.2018.10.065. [DOI] [PubMed] [Google Scholar]
- 25.Tian W., Jiang C., Huang Z., Xu D., Zheng S. Comprehensive analysis of dysregulated lncRNAs, miRNAs and mRNAs with associated ceRNA network in esophageal squamous cell carcinoma. Gene. 2019;696:206–218. doi: 10.1016/j.gene.2019.02.051. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J.J., Xu W.R., Chen B., Wang Y.Y., Yang N., Wang L.J., Zhang Y.L. The up-regulated lncRNA DLX6-AS1 in colorectal cancer promotes cell proliferation, invasion and migration via modulating PI3K/AKT/mTOR pathway. Eur. Rev. Med. Pharmacol. Sci. 2019;23(19):8321–8331. doi: 10.26355/eurrev_201910_19143. [DOI] [PubMed] [Google Scholar]
- 27.Wu D.M., Zheng Z.H., Zhang Y.B., Fan S.H., Zhang Z.F., Wang Y.J., Zheng Y.L., Lu J. Down-regulated lncRNA DLX6-AS1 inhibits tumorigenesis through STAT3 signaling pathway by suppressing CADM1 promoter methylation in liver cancer stem cells. J. Exp. Clin. Cancer Res. 2019;38(1):237. doi: 10.1186/s13046-019-1239-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Wang M., Li Y., Yang Y., Liu X., Zang M., Li Y., Yang K., Yang W., Zhang S. Long non-coding RNA DLX6-AS1 is associated with malignant progression and promotes proliferation and invasion in esophageal squamous cell carcinoma. Mol. Med. Rep. 2019;19(3):1942–1950. doi: 10.3892/mmr.2018.9786. [DOI] [PubMed] [Google Scholar]
- 29.Alcaraz-Sanabria A., Baliu-Piqué M., Saiz-Ladera C., Rojas K., Manzano A., Marquina G., Casado A., Cimas F.J., Pérez-Segura P., Pandiella A., Gyorffy B., Ocana A. Genomic signatures of immune activation predict outcome in advanced stages of ovarian cancer and basal-like breast tumors. Front. Oncol. 2020;9:1486. doi: 10.3389/fonc.2019.01486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Z., Mi M., Li X., Zheng X., Wu G., Zhang L. lncRNA OSTN-AS1 may represent a novel immune-related prognostic marker for triple-negative breast cancer based on integrated analysis of a ceRNA network. Front. Genet. 2019;10:850. doi: 10.3389/fgene.2019.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reddy S.M., Reuben A., Barua S., Jiang H., Zhang S., Wang L., Gopalakrishnan V., Hudgens C.W., Tetzlaff M.T., Reuben J.M., Tsujikawa T., Coussens L.M., Wani K., He Y., Villareal L., Wood A., Rao A., Woodward W.A., Ueno N.T., Krishnamurthy S., Wargo J.A., Mittendorf E.A. Poor response to neoadjuvant chemotherapy correlates with mast cell infiltration in inflammatory breast cancer. Cancer Immunol. Res. 2019;7(6):1025–1035. doi: 10.1158/2326-6066.CIR-18-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao P., Guan H., Dai Z., Ma Y., Zhao Y., Liu D. Long noncoding RNA DLX6-AS1 promotes breast cancer progression via miR-505-3p/RUNX2 axis. Eur. J. Pharmacol. 2019;865 doi: 10.1016/j.ejphar.2019.172778. [DOI] [PubMed] [Google Scholar]
- 33.Xiong H., Shen J., Chen Z., Yang J., Xie B., Jia Y., Jayasinghe U., Wang J., Zhao W., Xie S., Wang L., Zhou J. H19/let-7/Lin28 ceRNA network mediates autophagy inhibiting epithelial-mesenchymal transition in breast cancer. Int. J. Oncol. 2020;56(3):794–806. doi: 10.3892/ijo.2020.4967. [DOI] [PubMed] [Google Scholar]
- 34.Yang S.J., Wang D.D., Zhou S.Y., Zhang Q., Wang J.Y., Zhong S.L., Da Zhang H., Wang X.Y., Xia X., Chen W., Yang S.Y., Hu J.H., Zhao J.H., Tang J.H. Identification of circRNA-miRNA networks for exploring an underlying prognosis strategy for breast cancer. Epigenomics. 2020;12(2):101–125. doi: 10.2217/epi-2019-0058. [DOI] [PubMed] [Google Scholar]
- 35.Yao Y., Zhang T., Qi L., Zhou C., Wei J., Feng F., Liu R., Sun C. Integrated analysis of co-expression and ceRNA network identifies five lncRNAs as prognostic markers for breast cancer. J. Cell Mol. Med. 2019;23(12):8410–8419. doi: 10.1111/jcmm.14721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimomura T., Kawakami M., Tatsumi K., Tanaka T., Morita-Takemura S., Kirita T., Wanaka A. The role of the wnt signaling pathway in upper jaw development of chick embryo. Acta Histochem. Cytochem. 2019;52(1):19–26. doi: 10.1267/ahc.18038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matthews B.G., Roguljic H., Franceschetti T., Roeder E., Matic I., Vidovic I., Joshi P., Kum K.Y., Kalajzic I. Gene-expression analysis of cementoblasts and osteoblasts. J. Periodontal Res. 2016;51(3):304–312. doi: 10.1111/jre.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woo J., Miletich I., Kim B.M., Sharpe P.T., Shivdasani R.A. Barx1-Mediated inhibition of Wnt signaling in the mouse thoracic foregut controls Tracheo-Esophageal septation and epithelial differentiation. PLoS ONE. 2011;6(7) doi: 10.1371/journal.pone.0022493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.mou Zhang R., Tang T., Yu H.M., Yao X.D. LncRNA DLX6-AS1/miR-129-5p/DLK1 axis aggravates stemness of osteosarcoma through Wnt signaling. Biochem. Biophys. Res. Commun. 2018;507(1–4):260–266. doi: 10.1016/j.bbrc.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 40.Guo J., Chen Z., Jiang H., Yu Z., Peng J., Xie J., Li Z., Wu W., Cheng Z., Xiao K. The lncRNA DLX6-AS1 promoted cell proliferation, invasion, migration and epithelial-to-mesenchymal transition in bladder cancer via modulating Wnt/β-catenin signaling pathway. Cancer Cell Int. 2019;19:312. doi: 10.1186/s12935-019-1010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim B.M., Buchner G., Miletich I., Sharpe P.T., Shivdasani R.A. The stomach mesenchymal transcription factor barx1 specifies gastric epithelial identity through inhibition of transient Wnt signaling. Dev. Cell. 2005;8(4):611–622. doi: 10.1016/j.devcel.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 42.Yang J., Ye Z., Mei D., Gu H., Zhang J. Long noncoding RNA DLX6-AS1 promotes tumorigenesis by modulating miR-497-5p/FZD4/ FZD6/Wnt/β-catenin pathway in pancreatic cancer. Cancer Manag. Res. 2019;11:4209–4221. doi: 10.2147/CMAR.S194453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang P., Mokhtari R., Pedrosa E., Kirschenbaum M., Bayrak C., Zheng D., Lachman H.M. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells. Mol. Autism. 2017;8:11. doi: 10.1186/s13229-017-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue J., Zhu W., Song J., Jiao Y., Luo J., Yu C., Zhou J., Wu J., Chen M., Ding W.Q., Cao J., Zhang S. Activation of PPARα by clofibrate sensitizes pancreatic cancer cells to radiation through the Wnt/β-catenin pathway. Oncogene. 2018;37(7):953–962. doi: 10.1038/onc.2017.401. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y.T., Shang D., Akatsuka S., Ohara H., Dutta K.K., Mizushima K., Naito Y., Yoshikawa T., Izumiya M., Abe K., Nakagama H., Noguchi N., Toyokuni S. Chronic oxidative stress causes amplification and overexpression of ptprz1 protein tyrosine phosphatase to activate β-catenin pathway. Am. J. Pathol. 2007;171(6):1978–1988. doi: 10.2353/ajpath.2007.070741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu R., Hu R., Zeng Y., Zhang W., Zhou H.H. Tumour immune cell infiltration and survival after platinum-based chemotherapy in high-grade serous ovarian cancer subtypes: a gene expression-based computational study. EBioMedicine. 2020;51 doi: 10.1016/j.ebiom.2019.102602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ali H.R., Chlon L., Pharoah P.D.P., Markowetz F., Caldas C. Patterns of immune infiltration in breast cancer and their clinical implications: a gene-expression-based retrospective study. PLoS Med. 2016;13(12) doi: 10.1371/journal.pmed.1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao Y., Ma D., Zhao S., Suo C., Shi J., Xue M.Z., Ruan M., Wang H., Zhao J., Li Q., Wang P., Shi L., Yang W.T., Huang W., Hu X., Da Yu K., Huang S., Bertucci F., Jiang Y.Z., Shao Z.M. Multi-omics profiling reveals distinct microenvironment characterization and suggests immune escape mechanisms of triple-negative breast cancer. Clin. Cancer Res. 2019;25(16):5002–5014. doi: 10.1158/1078-0432.CCR-18-3524. [DOI] [PubMed] [Google Scholar]