Visual Abstract
Key Words: aortic valve interstitial cell, apolipoprotein B, calcific aortic valve stenosis, LDL cholesterol, lipoprotein(a), proprotein convertase subtilisin/kexin type 9
Abbreviations and Acronyms: Ad DMEM, advanced Dulbecco’s modified Eagle’s medium; apoB, apolipoprotein B; CAD, coronary artery disease; CAVS, calcific aortic valve stenosis; HDL-C, high-density lipoprotein cholesterol; IQR, interquartile range; LDL-C, low-density lipoprotein cholesterol; Lp(a), lipoprotein(a); PBS, phosphate-buffered saline; PBST, 1× phosphate-buffered saline with 0.1% Triton; PCSK9, proprotein convertase subtilisin/kexin type 9; SNP, single nucleotide polymorphism; TC, total cholesterol; VIC, valve interstitial cell; VLDL-C, very-low-density lipoprotein cholesterol; wGRS, weighted genetic risk score
Highlights
-
•
Aortic stenosis was less prevalent in carriers of the PCSK9 R46L variant.
-
•
Variation at the PCSK9 locus influences LDL-C levels, but not Lp(a).
-
•
PCSK9 is produced and secreted by aortic valves.
-
•
In vitro, PCSK9 inhibition might lower calcification in aortic valve cells.
-
•
PCSK9 inhibition could represent a therapeutic strategy for aortic stenosis.
Summary
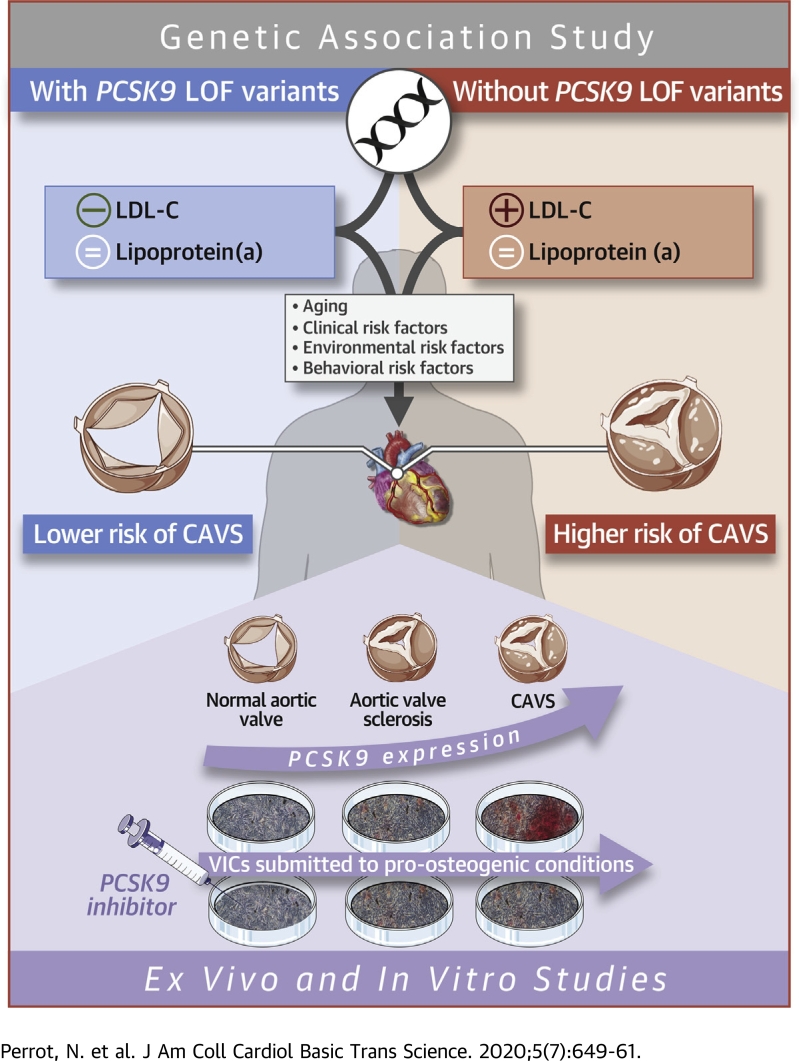
The authors investigated whether PCSK9 inhibition could represent a therapeutic strategy in calcific aortic valve stenosis (CAVS). A meta-analysis of 10 studies was performed to determine the impact of the PCSK9 R46L variant on CAVS, and the authors found that CAVS was less prevalent in carriers of this variant (odds ratio: 0.80 [95% confidence interval: 0.70 to 0.91]; p = 0.0011) compared with noncarriers. PCSK9 expression was higher in the aortic valves of patients CAVS compared with control patients. In human valve interstitials cells submitted to a pro-osteogenic medium, PCSK9 levels increased and a PCSK9 neutralizing antibody significantly reduced calcium accumulation.
Calcific aortic valve stenosis (CAVS) is the most common form of heart valve disease, and its prevalence is steadily rising in Western societies, affecting almost 3% of the population older than 65 years of age (1). Identification of the risk factors for CAVS could facilitate the development of novel and innovative treatment strategies. To date, the only effective treatments for CAVS are surgical or transcatheter aortic valve replacement; no pharmacological agents have been proven effective for the treatment of CAVS. We and others have shown that drugs that lower low-density lipoprotein cholesterol (LDL-C), such as statins and ezetimibe, do not impede CAVS progression (2, 3, 4) or decrease CAVS incidence (5). Coronary artery disease (CAD) and CAVS share many risk factors and pathophysiological mechanisms (6). Whether other cardiovascular drugs could be effective for the treatment of CAVS is unknown.
Proprotein convertase subtilisin/kexin type 9 (PCSK9) is an enzyme secreted by the liver that binds to the LDL receptor (LDLR) and targets it for lysosomal degradation (7). Genetic association studies have shown that natural variations at the PCSK9 locus (present in 2% to 3% of the population) are associated with lifelong exposure to low LDL-C levels, and cardiovascular protection (8,9). PCSK9 inhibitors have been shown to substantially lower LDL-C levels in various populations and reduce the risk of adverse cardiovascular outcomes in patients at high cardiovascular risk (10,11).
Lifelong low LDL-C levels has also been linked to lower aortic valve calcium accumulation and protection against CAVS (12). Recently, investigators of the Copenhagen General Population Study, the Copenhagen City Heart Study, and the Copenhagen Ischemic Heart Disease Study observed that individuals carrying the R46L variant in PCSK9 are characterized by lower levels of LDL-C, lipoprotein(a) (Lp(a)), and a lower risk of CAVS (13). However, these results have not been replicated, and it is still unclear whether the cardiovascular benefits are due to changes in LDL-C, Lp(a), or both. It also remains unclear whether PCSK9 is expressed in human aortic valves and whether it contributes to valve interstitial cell (VIC) calcification.
The objectives of our study were to determine whether variation at the PCSK9 locus are associated with plasma lipoprotein-lipid levels and CAVS. We also sought to determine which parameter(s) of the lipoprotein-lipid profile (LDL-C and or Lp[a]) best explained the potential benefits of genetically mediated PCSK9 inhibition for CAVS prevention. We also evaluated whether PCSK9 was present in the aortic valves, whether isolated human VICs could secrete PCSK9 under pro-osteogenic conditions, and whether blocking PCSK9 could mitigate the impact of pro-osteogenic conditions on human VIC calcification.
Methods
Genetic association study of PCSK9 R46L variant and CAVS
We investigated the association between the R46L PCSK9 variant and CAVS in a meta-analysis of 1 published (Copenhagen General Population Study, Copenhagen City Heart Study, and Copenhagen Ischemic Heart Disease Study, totaling 1,437 cases and 99,040 control patients [13]) and 9 unpublished studies (UK Biobank 1,350 cases and 349,043 control patients; EPIC-Norfolk [European Prospective Investigation into Cancer and Nutrition–Norfolk] 508 cases and 20,421 control patients; MDCS [Malmo Diet and Cancer Study] 682 cases and 5,963 control patients; GERA [Genetic Epidemiology Research on Aging], 3,469 cases and 41,234 control patients, the Estonian Biobank 481 cases and 7,223 control patients, the QUEBEC-CAVS study 1,009 cases and 11,625 control patients, and 3 French cohorts 3,123 cases and 6,532 control patients). This meta-analysis was performed after each study had tested the impact of this variant on CAVS using logistic regression adjusted for age and sex, and the first 10 ancestry-based principal components when available. We performed a fixed-effect meta-analysis using the inverse-variance weighted method as implemented in the rmeta package (version 3.0) in R version 3.5.1 software (R Foundation for Statistical Computing, Vienna, Austria). The design of each study and the definition of CAVS is presented in the Supplemental Appendix. All study protocols were approved by local ethics committees, and all patients provided informed consent. A summary of the definition of CAVS in each cohort is described in Supplemental Table 1.
Genetic association studies in the EPIC-norfolk study
We selected independent (in low linkage disequilibrium) single nucleotide polymorphisms (SNPs) (r2 < 0.10) at the PCSK9 locus (within 100 Kb of the gene) associated with LDL-C levels at a genome-wide level of significance in the Global Lipids Genetics Consortium (GLGC) (14). This approach yielded 11 SNPs independently associated with LDL-C levels. Of these, 10 were successfully genotyped in the EPIC-Norfolk study (described in Supplemental Table 2). The design of the EPIC-Norfolk prospective population study has been published previously (15,16). We built a weighted genetic risk score (wGRS) using these 10 SNPs weighted by the effect of each SNP on LDL-C levels in the GLGC. We then assessed the relationship between each of the 10 SNPs individually and the wGRS with plasma lipoprotein-lipid levels (total cholesterol [TC], LDL-C, high-density lipoprotein cholesterol [HDL-C], very-low-density lipoprotein cholesterol [VLDL-C], triglycerides, apolipoprotein B [apoB], apolipoprotein A-1 [apoA-I], and Lp[a]) in 9,692 participants of the EPIC-Norfolk study using linear regression, adjusted for age and sex. We considered a p value of 0.00625 (0.05 of 8 lipid traits) as statistically significant. Because lipoprotein-lipid levels are not normally distributed, the associations between the wGRS and natural log-transformed lipoprotein-lipid levels were assessed. We also constructed a wGRS scaled to a 1 mmol/l reduction in LDL-C levels. In the EPIC-Norfolk study, serum levels of total cholesterol, HDL-C, and triglycerides were measured in fresh samples with the RA 1000 (Bayer Diagnostics, Basingstoke, United Kingdom). LDL-C levels were calculated using the Friedewald formula, and VLDL-C levels were calculated as TC minus LDL-C minus HDL-C. Lp(a) and apoB levels were measured in a subset of the cohort with available stored frozen blood samples. Lp(a) levels were measured with an immunoturbidimetric assay using polyclonal antibodies directed against epitopes in apolipoprotein(a) (Denka Seiken, Coventry, United Kingdom), as previously described (17). This assay has been shown to be apolipoprotein(a) isoform-independent. Serum levels of apoB were measured by rate immunonephelometry (Behring Nephelometer BNII, Marburg, Germany) with calibration traceable to the International Federation of Clinical Chemistry primary standards.
Genetic association studies in the UK biobank
The design of the UK Biobank has also been previously published (18). The present analyses were conducted under UK Biobank data application number 25205. The associations between the wGRS and prevalent CAVS in the UK Biobank were tested using logistic regression adjusted for age, sex, and the first 10 ancestry-based principal components using R version 3.5.1 software.
Human aortic valve collection
Patients requiring aortic valve replacement due to CAVS were prospectively enrolled in this study at Centro Cardiologico Monzino IRCCS. Control valves were harvested from people deceased for noncardiovascular causes, and the valves were provided by the Cardiovascular Tissue Bank at Centro Cardiologico Monzino. Inclusion criteria for CAVS patients were elective surgery, age more than 18 years, ejection fraction >30%, and normal sinus rhythm. Exclusion criteria were prior cardiac surgery, rheumatic heart disease, endocarditis, active malignancy, chronic liver or kidney diseases, calcium regulation disorders (hyperparathyroidism, hyperthyroidism, and hypothyroidism), and chronic or acute inflammatory states (sepsis, autoimmune disease, and inflammatory bowel disease). Aortic valve specimens were collected from the operating room and processed within 30 min. The demographic and clinical characteristics of enrolled patients are reported in Supplemental Table 3. This study was approved by the IRCCS Centro Cardiologico Monzino Institutional Review Board and by the IRCCS Istituto Europeo di Oncologia and Centro Cardiologico Monzino Ethics Committee, and informed consent was obtained from all the participants.
Histological evaluation of PCSK9 expression and aortic leaflet calcification
Tissues were washed 3 times in phosphate-buffered saline (PBS) 1× and were fixed in 4% formalin, dehydrated, included in paraffin, and cut into 5- to 7-μm slides. Before staining, slides were rehydrated. Von Kossa staining was carried out with silver nitrate added on top of each section and exposed to ultraviolet illumination at room temperature for 20 min. Slides were washed in deionized H2O and then incubated in sodium thiosulfate solution 2 g/dl (Sigma-Aldrich, St. Louis, Missouri) at room temperature for 5 min. Slides were washed 3 times in deionized H2O and then incubated for 10 s with hematoxylin. Slides were dehydrated, mounted with EUKITT (O. Kindler/Orsatec, Bobingen, Germany), and images were taken with AxioScope (Carl Zeiss, Oberkochen, Germany). For PCSK9 staining, slides were incubated in antigen retrieval solution (Target Retrieval Solution Citrate [pH 6], Dako Cytomation, Glostrup, Denmark) at 98°C for 20 min and then cooled at room temperature for 20 min. Slides were treated with 3% H2O2 to block endogenous peroxidases and then washed 3 times with 1× PBS with 0.1% Triton (PBST). Blocking solution (PBST and 5% bovine serum albumin) was added and kept for 45 min at room temperature. Slides were incubated with primary mouse monoclonal anti-PCSK9 (Abcam, Cambridge, United Kingdom) at 4°C overnight. Slides were washed 3 times with PBST and incubated with biotinylated anti-mouse secondary antibody (Vector Laboratories, Burlingame, California) for 60 min at room temperature. Slides were washed 3 times and ABC complex (ABC kit, Vector Laboratories) was added and incubated for 30 min at room temperature. Slides were washed 3 times, and ImmPACT DAB Peroxidase (HRP) Substrate (Vector Laboratories) was added and let react for 25 s. Slides were immediately rinsed in distilled H2O and then incubated for 10 s with hematoxylin. Slides were dehydrated, mounted with EUKITT (O. Kindler/Orsatec), and images were taken with AxioScope (Carl Zeiss). The quantification of PCSK9 immunohistochemistry has been performed implementing the ImageJ v1.50i software (NIH, Bethesda, Maryland) with the plugin IHC Tool box. The automated color detection, which allows the generation of deconvoluted images, has been performed with the default model H-DAB for brown color detection. The integrate pixel density has been calculated on binary panoramic images and normalized with the total area of the section (pixel2).
PCSK9 measurements in explanted aortic valves and VIC medium
Whole-protein extracts were obtained by pulverizing frozen aortic valve leaflets. The powder was suspend in 1× RIPA buffer with the addition of protease inhibitors (Sigma-Aldrich) and assayed as follows. VIC supernatant culture medium was collected at the end of the treatments and assayed as follows. The PCSK9 quantification was carried out with an enzyme-linked immunosorbent assay (R&D Systems, Minneapolis, Minnesota) following the manufacturer’s instructions. The intraplate and the interplate percent coefficients of variation for PCSK9 protein measurements were 3.2 ± 0.3% and 8.4 ± 0.9%, respectively.
Aortic valve interstitial cell isolation
Isolation of aortic VICs was performed using a modified method described by Poggio et al. (19). Briefly, aortic leaflets were placed in 2 mg/ml type II collagenase (Worthington Biochemical Corporation, Lakewood, New Jersey) in advanced Dulbecco’s modified Eagle’s medium (Ad DMEM) containing 10% fetal bovine serum, l-glutamine 2 mmol/l, and 1% penicillin/streptomycin solution and incubated at 37°C for 20 min. The loosened endothelial layer was removed, and tissues were finely minced and dissociated in type II collagenase (1 mg/ml) for 4 h at 37°C. The resulting VICs were seeded in tissue culture plates in Ad DMEM (Thermo Fisher Scientific, Waltham, Massachusetts) medium and maintained at 37°C with 5% CO2. All the experiments were performed on cultured cells between their second and fifth passages in Ad DMEM containing 10% fetal bovine serum, 1% penicillin, 1% streptomycin solution, and 1% l-glutamine (normal medium).
Impact of osteogenic stimuli on PCSK9 expression in VICs
To evaluate PCSK9 expression under osteogenic stimuli, VICs were treated with normal medium and osteogenic medium (+10 mmol/l β-glycerophosphate and +50 μg/ml ascorbic acid) with or without a neutralizing antibody anti-PCSK9 (NAb anti-PCSK9, 0.8 ng/μl, BPS Bioscience, San Diego, California) or immunoglobulin G1 as control (IgG1, 0.8 ng/μl, Novus Biologicals, Centennial, Colorado) for 7 days. Total cell RNA was isolated using a Total RNA Purification kit (Norgen Biotek Corp., Thorold, Ontario, Canada), following the manufacturer's protocol. RNA quantification was determined with a spectrophotometer (ND-1000, NanoDrop, EuroClone). Reverse transcription was performed with the TaqMan Reverse Transcription Reagent (Applied Biosystems, Foster City, California) following the manufacturer’s instructions. Quantitative real-time polymerase chain reaction was executed by the use of the SYBR Green PCR MasterMix (Applied Biosystems). All reactions were performed in a MicroAmp Fast 96-Well Reaction Plate (Applied Biosystems). The relative quantities of specific mRNA were obtained with the use of the comparative Ct method and were normalized to glyceraldehyde 3-phosphate dehydrogenase gene (GAPDH). Calcium quantification was performed by calcium colorimetric assay (BioVision, Milpitas, California) following the manufacturer’s instructions.
Statistical analysis
For continuous data, normally distributed values were compared between groups with Student’s t-test, and the results were expressed as mean ± SD; not normally distributed values were compared between groups with Mann-Whitney test, and the results were expressed as median (interquartile range [IQR]); whereas categorical values were compared with Pearson's chi-square test, and the results were expressed as mean and percentage. The measure of the linear correlation between in vitro PCSK9 secretion and calcium deposition was performed with the Pearson correlation coefficient (rp). Each value has been previously “mean-centered.” Data were analyzed with GraphPad Prism v7.00 software (GraphPad Prism, San Diego, California), and a value of p < 0.05 was considered statistically significant.
Results
Meta-analysis of the association between the PCSK9 R46L variant and CAVS
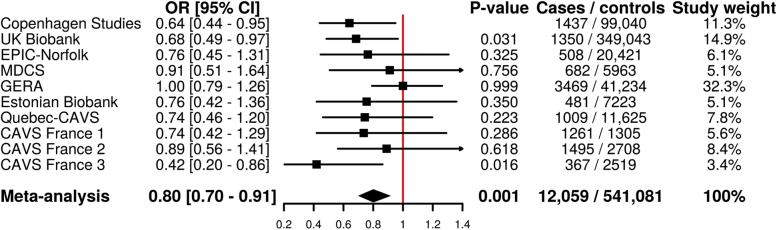
Figure 1 presents the association between the PCSK9 R46L variant and CAVS in 1 published and 9 previously unpublished studies totaling 12,059 CAVS cases and 541,081 control patients. Carriers of the R46L variant had lower odds of CAVS compared with noncarriers (odds ratio: 0.80 [95% confidence interval: 0.70 to 0.91]; p < 0.001). A funnel plot of the resulting meta-analysis is presented in Supplemental Figure 1. The p value for heterogeneity was 0.418.
Figure 1.
Association of the PCSK9 R46L Variant With CAVS
Association of the PCSK9 R46L variant with calcific aortic valve stenosis in a meta-analysis of 10 cohorts totaling 12,059 cases and 541,081 control subjects. CAVS = calcific aortic valve stenosis; CI = confidence interval; EPIC = European Prospective Investigation into Cancer and Nutrition; GERA = Genetic Epidemiology Research on Aging study; MDCS = Malmo Diet and Cancer Study; OR = odds ratio.
PCSK9 variants, lipoprotein-lipid levels, and CAVS
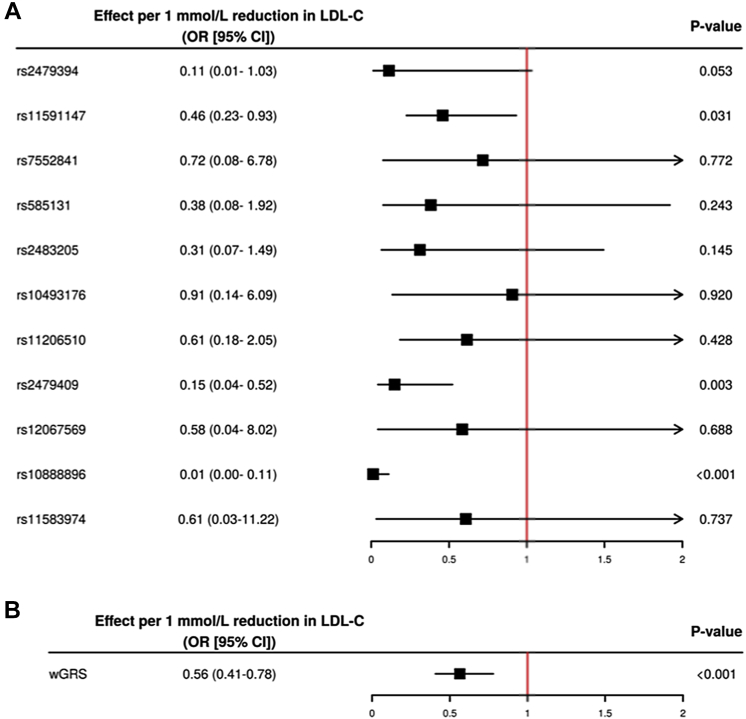
The association between the 10 PCSK9 SNPs as well as the wGRS and natural log-transformed lipoprotein-lipid levels in 9,692 participants of the EPIC-Norfolk study are presented in Table 1. The wGRS was significantly associated with TC, LDL-C, and apoB (all p < 0.001), but not with VLDL-C (although the R46L variant [rs11591147] was associated with lower VLDL-C levels), HDL-C, triglycerides, apoA-I, or Lp(a). In the UK Biobank, the wGRS associated with lower LDL-C was associated with lower odds of CAVS (odds ratio: 0.55 [95% confidence interval: 0.39 to 0.77]; p < 0.001) per 1 mmol/l reduction in LDL-C (Figure 2).
Table 1.
Association of 10 Independent SNPs at the PCSK9 Locus and of a wGRS With Natural Log-Transformed Lipoprotein-Lipid Levels in 9,692 Participants of the EPIC-Norfolk Study
| TC | p Value∗ | LDL-C | p Value∗ | HDL-C | p Value∗ | VLDL-C | p Value∗ | TG | p Value∗ | apoB | p Value∗ | apoA-I | p Value∗ | Lp(a) | p Value∗ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs585131 | −1.2 (0.75) | 0.092 | −1.3 (0.69) | 0.046 | −0.017 (0.0068) | 0.013 | 0.11 (0.28) | 0.738 | 3.8 (1.3) | 0.003 | −0.0047 (0.0043) | 0.248 | −0.0051 (0.0055) | 0.269 | −0.27 (0.43) | 0.877 |
| rs2483205 | −0.15 (0.60) | 0.764 | −0.52 (0.55) | 0.352 | 0.00013 (0.0054) | 0.971 | 0.37 (0.22) | 0.089 | 1.9 (1.0) | 0.064 | −0.0010 (0.0034) | 0.949 | 0.0046 (0.0043) | 0.333 | −0.086 (0.34) | 0.901 |
| rs2479394 | −0.41 (0.69) | 0.541 | −0.79 (0.63) | 0.201 | 0.012 (0.0062) | 0.088 | 0.37 (0.25) | 0.173 | −0.32 (1.2) | 0.906 | −0.0088 (0.0039) | 0.033 | 0.0077 (0.0050) | 0.135 | −0.42 (0.39) | 0.909 |
| rs10493176 | −2.2 (1.1) | 0.037 | −2.0 (0.97) | 0.043 | 0.00082 (0.0096) | 0.958 | −0.18 (0.39) | 0.520 | −1.2 (1.8) | 0.579 | −0.0060 (0.0060) | 0.540 | 0.014 (0.0077) | 0.042 | −0.72 (0.60) | 0.410 |
| rs11206510 | −1.7 (0.74) | 0.011 | −1.2 (0.68) | 0.029 | −0.0099 (0.0067) | 0.101 | −0.46 (0.27) | 0.053 | −0.65 (1.3) | 0.660 | −0.0051 (0.0042) | 0.213 | −0.0016 (0.0054) | 0.805 | −0.19 (0.42) | 0.952 |
| rs2479409 | −1.6 (0.64) | 0.010 | −1.7 (0.58) | 0.003 | 0.0030 (0.0057) | 0.590 | 0.12 (0.23) | 0.557 | −0.024 (1.1) | 0.613 | −0.0024 (0.0036) | 0.698 | 0.011 (0.0046) | 0.014 | −0.17 (0.36) | 0.669 |
| rs12067569 | −0.33 (1.8) | 0.866 | −1.9 (1.7) | 0.189 | 0.016 (0.017) | 0.363 | 1.6 (0.68) | 0.021 | 4.4 (3.1) | 0.052 | 0.0057 (0.010) | 0.767 | 0.013 (0.013) | 0.372 | −0.85 (1.0) | 0.381 |
| rs10888896 | −1.8 (0.69) | 0.0057 | −2.0 (0.63) | <0.001 | −0.0046 (0.0062) | 0.489 | 0.18 (0.25) | 0.459 | 1.9 (1.2) | 0.503 | −0.0039 (0.0039) | 0.413 | 1.9e−05 (0.0050) | 0.711 | −0.44 (0.39) | 0.600 |
| rs11591147 | −19.8 (2.5) | <0.001 | −18.2 (2.3) | <0.001 | −0.030 (0.023) | 0.183 | −1.6 (0.92) | 0.035 | −2.6 (4.3) | 0.602 | −0.068 (0.014) | <0.001 | −0.0042 (0.018) | 0.888 | −0.33 (1.4) | 0.585 |
| rs7552841 | −2.8 (0.62) | <0.001 | −2.5 (0.57) | <0.001 | −0.0037 (0.0056) | 0.514 | −0.32 (0.23) | 0.146 | −0.81 (1.1) | 0.476 | −0.0093 (0.0035) | 0.022 | −0.0015 (0.0045) | 0.837 | −0.23 (0.35) | 0.742 |
| wGRS | −16.9 (2.4) | <0.001 | −16.5 (2.2) | <0.001 | −0.027 (0.022) | 0.190 | −0.41 (0.90) | 0.463 | 2.9 (4.1) | 0.369 | −0.056 (0.014) | <0.001 | 0.018 (0.018) | 0.202 | −1.7 (1.4) | 0.435 |
Values are beta (SE). All associations were adjusted for age and sex.
apoA-I = apolipoprotein A-I; apoB = apolipoprotein B; Lp(a) = lipoprotein(a); HDL-C = high-density lipoprotein cholesterol; LDL-C = low-density lipoprotein cholesterol; SNP = single nucleotide polymorphisms; TC = total cholesterol; TG = triglycerides; VLDL-C = very-low-density lipoprotein cholesterol.
Level of statistical significance was set as 0.00625 (0.05/8 lipid traits) on log-transformed values.
Figure 2.
Impact of Individual SNPs and a wGRS on CAVS
Impact of individual single nucleotide polymorphisms (SNPs) (A) and of a weighted genetic risk score of single nucleotide polymorphisms at the PCSK9 locus associated with low-density cholesterol (LDL-C) levels only (B) on calcific aortic valve stenosis in the UK Biobank. wGRS = weighted genetic risk score; other abbreviations as in Figure 1.
PCSK9 expression and CAVS
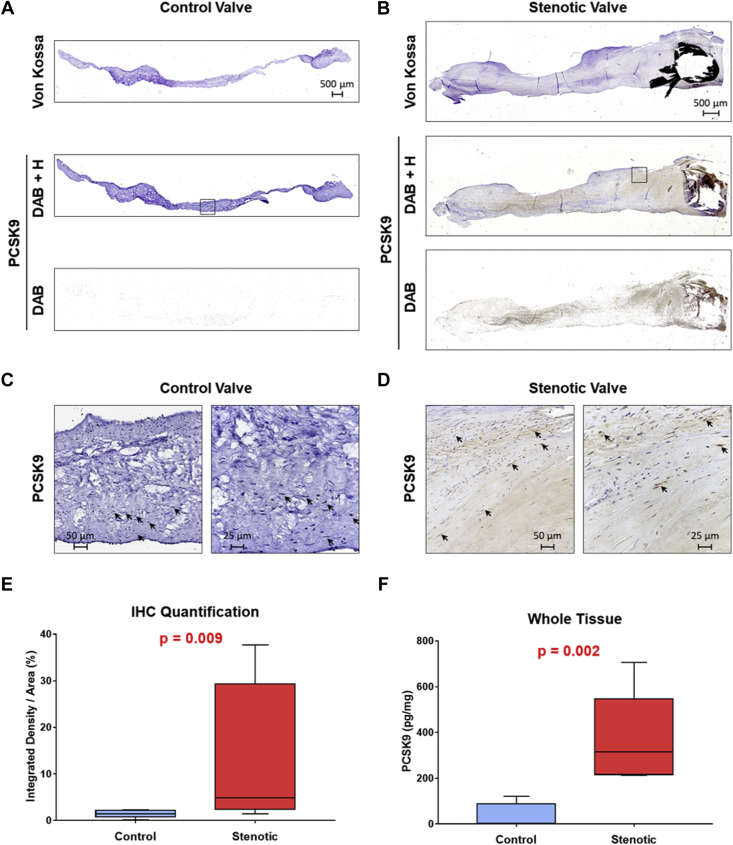
Immunohistochemical analysis identified PCSK9 in both calcified and noncalcified leaflets. However, PCSK9 was highly abundant in calcified ones (fold change +3.5 [IQR: +1.7 to +20.9]; p = 0.009) compared with non-calcified leaflets (Figure 3A, 3B, and 3E and Supplemental Figures 2 and 3). In healthy aortic valves, PCSK9 is detectable only in close proximity to cell nuclei, whereas in stenotic ones, PCSK9 is present also within the extracellular matrix (Figures 3C and 3D and Supplemental Figure 2). Enzyme-linked immunosorbent assay confirmed that PCSK9 is overexpressed in calcified leaflets compared with healthy ones (315.8 [IQR: 216.4 to 547.9] vs. 2.3 [IQR: 0.0 to 88.1] pg of PCSK9/mg of total proteins, respectively; p = 0.002) (Figure 3F).
Figure 3.
PCSK9 Is Highly Abundant in Calcified Leaflets
(A) Representative explanted aortic valve leaflet with normal structure (control valve) and (B) representative calcified aortic valve leaflet (stenotic valve), both stained with Von Kossa to visualize calcium deposits (upper panels) and with anti-PCSK9 antibody (lower panels). PCSK9 immunohistochemical (IHC) staining is presented as 3,3′-diaminobenzidine (DAB) and hematoxylin (H) counter staining (DAB + H) and as deconvoluted image visualizing only the DAB staining. Panoramic images were taken with a 10× magnification. Black boxes indicate the higher magnification areas. (C and D) High magnification areas of aortic valve leaflets (20× left and 40× right). Arrows indicate representative PCSK9 expression in close proximity to cell nuclei. (E) Box and whisker plots represent DAB quantification by ImageJ with IHC Tool box plugin on normal (control; n = 6) and calcified (stenotic; n = 6) leaflets. (F) Box and whisker plots represent PCSK9 levels measured by enzyme-linked immunosorbent assay on control (n = 6) and stenotic (n = 6) whole tissue extracts.
PCSK9 secretion and VIC calcification
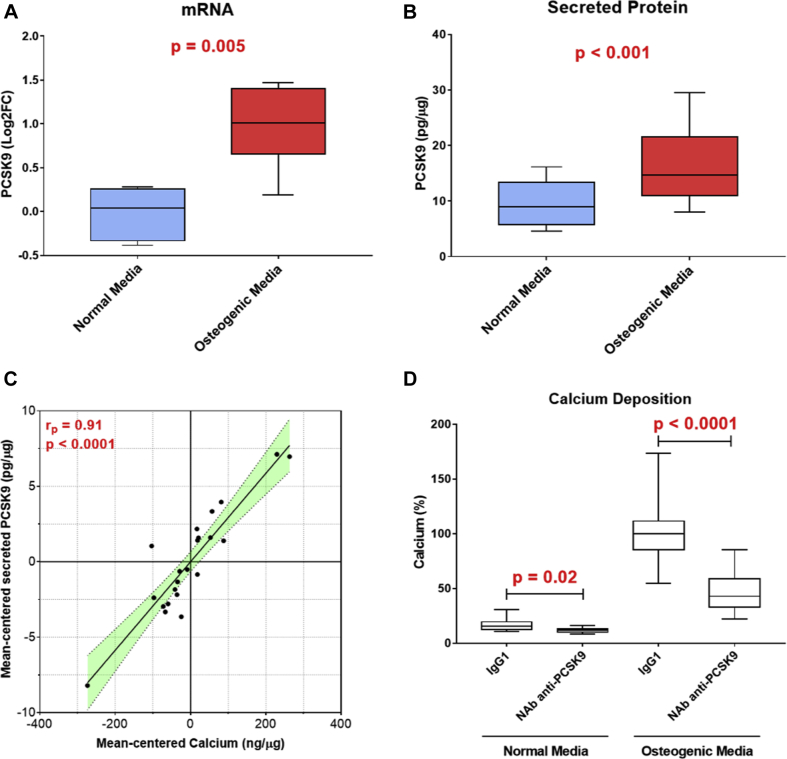
We sought to determine whether CAVS VICs could express PCSK9 under classic osteogenic stimulus. We treated human isolated VICs with osteogenic medium and then evaluated PCSK9 mRNA and secreted PCSK9 protein levels. PCSK9 transcript expression was induced by osteogenic medium treatment (log2 fold change = +1.0 [IQR: +0.66 to +1.4]; p = 0.005) compared with VICs in normal medium (Figure 4A). As expected, osteogenic medium induced VIC calcification compared with normal medium (472.2 [IQR: 316.4 to 701.1] ng vs. 20.7 [IQR: 15.8 to 28.6] ng of calcium/μg of total proteins, respectively; p < 0.0001) (Supplemental Figure 4). In culture, VICs were able to secrete a detectable amount of PCSK9 into the medium. In particular, VICs treated with osteogenic medium secreted significantly higher PCSK9 levels compared with VICs in normal one (14.6 [IQR: 11.0 to 21.6] vs. 8.9 [IQR: 5.8 to 13.4] pg of PCSK9/μg of total proteins, respectively; p < 0.001) (Figure 4B). We also found a significant and positive correlation (Pearson’s correlation coefficient rp = 0.91; p < 0.0001) between the extent of calcification and the amount of secreted PCSK9 (Figure 4C). Finally, inhibition of extracellular PCSK9 was able to significantly reduce the calcium deposition both in normal medium (−6.3% [IQR: −6.8% to −0.4%]; p = 0.02) and in osteogenic medium (−56.5% [IQR: −63.3% to −40.0%]; p < 0.0001) compared with their respective control medium (Figure 4D).
Figure 4.
PCSK9 Expression and Secretion Is Induced by Osteogenic Milieu in Valve Interstitial Cells
(A) Box and whisker plots represent PCSK9 transcript levels in valve interstitial cells (VIC) cultured for 7 days in normal or osteogenic media (n = 6). RNA levels were normalized to GAPDH expression. (B) Box and whisker plots represent secreted PCSK9 levels from VICs cultured for 7 days in normal or osteogenic media (n = 4). PCSK9 levels were normalized to total protein content. (C) Mean-centered correlation between secreted PCSK9 and calcium levels in VICs. The linear correlation between the 2 variables was performed with the Pearson correlation coefficient. (D) Bar graph shows the calcification potential, after 7 days, in normal and osteogenic media (n = 3) of VICs treated with a neutralizing antibody anti-PCSK9 (NAb anti-PCSK9) or immunoglobulin G1 as control (IgG1).
Discussion
Previous genetic association studies observed that carriers of genetic variants in PCSK9 associated with low LDL-C levels are at lower risk of a broad range of atherosclerotic cardiovascular diseases (20). In this study, we confirmed that variants in PCSK9 associated with lower LDL-C levels are also associated with a lower risk of CAVS, and for the first time to our knowledge, we demonstrated that PCSK9 is present in human aortic valves. Investigating the potential parameters of the lipoprotein-lipid profile that might explain the benefits of carrying these variants, we found LDL-C, but not Lp(a), levels to be a key factor that may explain the benefits of carrying PCSK9 variants for CAVS prevention. The potential benefits of targeting PCSK9 for CAVS prevention were also supported by our in vitro experimental approach, showing that treating VICs with a neutralizing PCSK9 antibody reduced calcium accumulation in VICs under pro-osteogenic stimuli.
In a previous report of 2,373 individuals included in a nested case-control design of the EPIC-Norfolk study, we have shown that carriers of the PCSK9 R46L variant had lower levels of nuclear magnetic resonance spectroscopy-measured VLDL and LDL particle concentrations and lower Lp(a) (21). In another study, Sliz et al. (22) reported no impact of this variant on VLDL lipid measures. In our study, we found a significant association between PCSK9 variants and CAVS in the UK Biobank. However, given the relatively small number of CAVS cases in the UK Biobank (1,350), these results are not definitive, and additional studies are needed to confirm these findings and to determine whether the impact of reduced PCSK9 function is independent of the presence of concomitant CAD because approximately one-half of the patients with CAVS also have CAD. Our results confirm those of the Copenhagen cohorts (13) and extend our previous observations that Pcsk9−/− mice are less likely to develop aortic valve calcification than wild-type mice (23). Results of our previous study, confirmed by the present one, also suggest that there is a significant correlation between the amount of PCSK9 produced by VICs and the extent of VIC calcification (23). Our results are also in line with a report from the CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology) consortium and the MDCS study (Malmo Diet and Cancer Study) that has shown that a GRS associated with lifelong LDL-C reduction is associated with lower aortic valve calcium accumulation and lower CAVS incidence (12).
Study limitations
In the current study, we included additional participants and PCSK9 variants, and found no association between SNPs in PCSK9 and VLDL-C or Lp(a). Although our study population was not as large, our results for Lp(a) are in contrast with those of the Copenhagen investigators who have shown that carriers of the R46L variant (n = 1,293) have approximately 10% lower Lp(a) levels compared with noncarriers (n = 48,324). Consequently, the impact of PCSK9 variants on Lp(a) is at best modest. Also in support of this premise, results of the FOURIER (Further Cardiovascular Outcomes Research With PCSK9 Inhibition in Subjects With Elevated Risk) trial have shown that PCSK9 inhibition with evolocumab provides modest (16%) reduction in Lp(a) levels in patients with high Lp(a) levels (24), suggesting that PCSK9 inhibitors may not exert their cardiovascular benefits through Lp(a) reduction but rather via reductions in other apoB-containing lipoprotein particles such as LDL. These findings are also supported by the recently published ANITSCHKOW (Effects of Proprotein Convertase Subtilisin/Kexin Type 9 [PCSK9] Inhibition on Arterial Wall Inflammation in Patients With Elevated Lipoprotein(a) [Lp(a)]) trial that concluded that PCSK9 inhibition with evolocumab was associated with modest reductions in Lp(a) and did not influence arterial wall inflammation assessed by 18F-fluoro-deoxyglucose positron-emission tomography/computed tomography in patients with hyperlipoproteinemia(a) (25). Because circulating PCSK9 levels were not measured, we could not determine whether the benefits of carrying PCSK9 variants go beyond reductions in apoB levels and could be due to lower plasma PCSK9 levels. Nevertheless, our results are in accordance with 2 other studies from our group showing that circulating PCSK9 levels are associated with bioprosthetic aortic valve degeneration (26,27). Our ex vivo data confirmed that PCSK9 is more abundant in the explanted aortic valve from patients with CAVS compared with control patients. However, evaluating the demographical and clinical variables between control and CAVS patients, we found that age and dyslipidemia significantly differed, being possible confounders. However, the used specimens were 6 per group, and regression analyses adjusted for age and/or dyslipidemia need to be interpreted with caution. Nevertheless, our in vitro experiments provided evidence that VIC are indeed able to produce and to secrete PCSK9. The proteins secreted by VIC are entrapped within the extracellular matrix and do not reach the blood stream. However, at the present time, we are not able to differentiate between the PCSK9 infiltrate from the blood stream and the PCSK9 locally produced by the VICs. Indeed, in vitro, we found that VICs submitted to a procalcifying milieu had an increase in PCSK9 levels, and we showed that a direct block of extracellular VIC-released PCSK9 was able to significantly reduce calcium accumulation. We believe that these results are important to generate further studies focused on the molecular mechanisms by which PCSK9 modulates VIC calcium accumulation and thus aortic valve detrimental processes.
Conclusions
We performed a large genetic association study as well as a series of experiments showing that variation in PCSK9 is linked with low cholesterol levels and protection against CAVS and that PCSK9 expression was higher in explanted aortic valves from patients with versus without CAVS. In human VICs, PCSK9 expression and secretion were induced by an exposure to a pro-osteogenic milieu and a PCSK9 neutralizing antibody reduced VIC calcium levels by more than 50%, thereby providing experimental support to our genetic findings.
Although our results highlight the potential role of PCSK9 in the etiology of CAVS, the ultimate proof of causality and clinical benefit would be a clinical trial showing that PCSK9 inhibition is linked with a reduction in valvular outcomes. However, to our knowledge, no such trials are currently planned. An ongoing trial is currently testing the hypothesis that PCSK9 inhibition will reduce aortic valve macrocalcification (measured by computed tomography) and microcalcification (measured by 18F-NaF positron-emission tomography/computed tomography) in patients with mild-to-moderate CAVS (PCSK9 Inhibitors in the Progression of Aortic Stenosis; NCT03051360). Interestingly, an exploratory analysis of the FOURIER trial recently revealed that PCSK9 inhibition with evolocumab could decrease CAVS incidence in patients with cardiovascular disease (28).
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: In the absence of a pharmacological treatment delaying CAVS progression and outcomes, PCSK9 inhibition could provide benefit to patients with CAVS and our results provide a rationale for a large clinical randomized trial to investigate the effect of PCSK9 inhibition in patients with CAVS.
TRANSLATIONAL OUTLOOK: Our results highlight the potential role of PCSK9 in the etiology of CAVS and support the notion that targeting apoB-containing lipoprotein particles such as LDL is key for optimal CAD risk reduction and potentially CAVS.
Acknowledgments
The authors thank all study participants as well as Anna Guarino and Barbara Micheli for the collection and processing of the control specimens (Cardiovascular Tissue Bank, Milan, Italy).
Footnotes
This study was funded by the European Research Area Network on Cardiovascular Disease (ERA-CVD) Joint Transnational Call 2018 (PICASSO JTC2018-042), which is a European Research Area Network (ERA-Net) comprising 24 partners from 19 countries/regions that has been granted for funding through the current EU Framework Programme for Research and Innovation ‘Horizon 2020,’ (Drs. Poggio, Arsenault, Capoulade, and Mass) by the Italian Ministry of Health (GR-2018-12366423) (Dr. Poggio), by the Fondation de l’IUCPQ (Dr. Arsenault), by the Fondazione Gigi & Pupa Ferrari ONLUS (FPF-14) Dr. Poggio, Merck (Dr. Arsenault), and Pfizer (Dr. Arsenault). The EPIC-Norfolk Study is funded by Cancer Research UK grant number 14136 and the Medical Research Council grant number G1000143 (Dr. Wareham). The COFRASA (Aortic Stenosis in Elderly: Determinant of Progression; NCT00338676) and GENERAC (Genetic of Aortic Valve Stenosis-Clinical and Therapeutic Implications; NCT00647088) studies are supported by grants from the Assistance Publique-Hôpitaux de Paris (PHRC National 2005 and 2010, and PHRC régional 2007) (Dr. Messika-Zeitoun). Dr. Capoulade is supported by a “Connect Talent” research chair from Région Pays de la Loire and Nantes Métropole. Dr. Mass is supported by the German Research Foundation (Excellence Cluster ImmunoSensation), the Fritz Thyssen Foundation and Daimler and Benz Foundation. Drs. Clavel, Thériault, and Arsenault hold junior scholar awards from the Fonds de Recherche du Québec: Santé (FRQS). Ms. Chen was funded by a studentship from the McGill University Health Centre Foundation. Dr. Le Tourneau is supported by the Fédération Française de Cardiologie, a Fondation Coeur et Recherche and an Inserm Translational Research grant. Dr. Pibarot holds the Canada Research Chair in Valvular Heart Disease and his research program is supported by a Foundation Scheme Grant from the Canadian Institutes of Health Research (CIHR). Dr. Smith was supported by grants from the Swedish Heart-Lung Foundation (2016-0134 and 2016-0315), the Swedish Research Council (2017-02554), the European Research Council (ERC-STG-2015-679242), the Crafoord Foundation, Skåne University Hospital, the Scania county, governmental funding of clinical research within the Swedish National Health Service, a generous donation from the Knut and Alice Wallenberg foundation to the Wallenberg Center for Molecular Medicine in Lund, and funding from the Swedish Research Council (Linnaeus grant Dnr 349-2006-237, Strategic Research Area Exodiab Dnr 2009-1039) and Swedish Foundation for Strategic Research (Dnr IRC15-0067) to the Lund University Diabetes Center. Dr. Mathieu holds a FRQS Research Chair on the Pathobiology of Calcific Aortic Valve Disease. Prof. Bossé holds a Canada Research Chair in Genomics of Heart and Lung Diseases. Dr. Thanassoulis is supported by R01 HL128550 from the National Institutes of Health/National Heart, Lung, and Blood Institute; and has received research research funding from Servier and Ionis Pharmaceuticals; has been a consultant for Amgen, Sanofi/Regerenon, Boehringer Ingelheim, and Ionis Pharmaceuticals, Novartis and HLS Therapeutics. Dr. Clavel has received funding from Medtronic; and her institution has a core laboratory contract with Edwards Lifesciences for which she is not directly compensated. Dr. Le Tourneau has received funding from Abbott/St. Jude. Dr. Messika-Zeitoun has received funding from Edwards Lifesciences. Dr. Pibarot has received funding from Edwards Lifesciences and Medtronic. Dr. Mathieu has been a consultant for Casebia Therapeutics. Dr. Arsenault has received research funding from Pfizer, Merck, and Ionis Pharmaceuticals; and has been a consultant for Novartis. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the JACC: Basic to Translational Scienceauthor instructions page.
Appendix
For an expanded Methods section and supplemental figures and tables, please see the online version of this paper.
Contributor Information
Paolo Poggio, Email: paolo.poggio@cardiologicomonzino.it.
Benoit J. Arsenault, Email: benoit.arsenault@criucpq.ulaval.ca.
Appendix
References
- 1.Otto C.M. Calcific aortic stenosis--time to look more closely at the valve. N Engl J Med. 2008;359:1395–1398. doi: 10.1056/NEJMe0807001. [DOI] [PubMed] [Google Scholar]
- 2.Rossebo A.B., Pedersen T.R., Boman K. Intensive lipid lowering with simvastatin and ezetimibe in aortic stenosis. N Engl J Med. 2008;359:1343–1356. doi: 10.1056/NEJMoa0804602. [DOI] [PubMed] [Google Scholar]
- 3.Chan K.L., Teo K., Dumesnil J.G., Ni A., Tam J. Effect of lipid lowering with rosuvastatin on progression of aortic stenosis: results of the aortic stenosis progression observation: measuring effects of rosuvastatin (ASTRONOMER) trial. Circulation. 2010;121:306–314. doi: 10.1161/CIRCULATIONAHA.109.900027. [DOI] [PubMed] [Google Scholar]
- 4.Cowell S.J., Newby D.E., Burton J. Aortic valve calcification on computed tomography predicts the severity of aortic stenosis. Clin Radiol. 2003;58:712–716. doi: 10.1016/s0009-9260(03)00184-3. [DOI] [PubMed] [Google Scholar]
- 5.Arsenault B.J., Boekholdt S.M., Mora S. Impact of high-dose atorvastatin therapy and clinical risk factors on incident aortic valve stenosis in patients with cardiovascular disease (from TNT, IDEAL, and SPARCL) Am J Cardiol. 2014;113:1378–1382. doi: 10.1016/j.amjcard.2014.01.414. [DOI] [PubMed] [Google Scholar]
- 6.Perrot N., Boekholdt S.M., Mathieu P., Wareham N.J., Khaw K.-T., Arsenault B.J. Life's simple 7 and calcific aortic valve stenosis incidence in apparently healthy men and women. Int J Cardiol. 2018;269:226–228. doi: 10.1016/j.ijcard.2018.07.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lagace T.A., Curtis D.E., Garuti R. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J Clin Invest. 2006;116:2995–3005. doi: 10.1172/JCI29383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen J.C., Boerwinkle E., Mosley T.H., Jr., Hobbs H.H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 9.Benn M., Nordestgaard B.G., Grande P., Schnohr P., Tybjaerg-Hansen A. PCSK9 R46L, low-density lipoprotein cholesterol levels, and risk of ischemic heart disease: 3 independent studies and meta-analyses. J Am Coll Cardiol. 2010;55:2833–2842. doi: 10.1016/j.jacc.2010.02.044. [DOI] [PubMed] [Google Scholar]
- 10.Sabatine M.S., Giugliano R.P., Keech A.C. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med. 2017;376:1713–1722. doi: 10.1056/NEJMoa1615664. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz G.G., Steg P.G., Szarek M. Alirocumab and cardiovascular outcomes after acute coronary syndrome. N Engl J Med. 2018;379:2097–2107. doi: 10.1056/NEJMoa1801174. [DOI] [PubMed] [Google Scholar]
- 12.Smith J.G., Luk K., Schulz C.A. Association of low-density lipoprotein cholesterol-related genetic variants with aortic valve calcium and incident aortic stenosis. JAMA. 2014;312:1764–1771. doi: 10.1001/jama.2014.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langsted A.G., Nordestgaard B.G., Benn M., Tybjærg-Hansen A.R., Kamstrup P.R. PCSK9 R46L loss-of-function mutation reduces lipoprotein(a), LDL cholesterol, and risk of aortic valve stenosis. J Clin Endocrinol Metab. 2016;101:3281–3287. doi: 10.1210/jc.2016-1206. [DOI] [PubMed] [Google Scholar]
- 14.Willer C., Schmidt E., Sengupta S. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Day N., Oakes S., Luben R. EPIC-Norfolk: study design and characteristics of the cohort. European Prospective Investigation of Cancer. Br J Cancer. 1999;80(Suppl 1):95–103. [PubMed] [Google Scholar]
- 16.Arsenault B.J., Boekholdt S.M., Dube M.P. Lipoprotein(a) levels, genotype, and incident aortic valve stenosis: a prospective Mendelian randomization study and replication in a case-control cohort. Circ Cardiovasc Genet. 2014;7:304–310. doi: 10.1161/CIRCGENETICS.113.000400. [DOI] [PubMed] [Google Scholar]
- 17.Gurdasani D., Sjouke B., Tsimikas S. Lipoprotein(a) and risk of coronary, cerebrovascular, and peripheral artery disease: the EPIC-Norfolk prospective population study. Arterioscler Thromb Vasc Biol. 2012;32:3058–3065. doi: 10.1161/ATVBAHA.112.255521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sudlow C., Gallacher J., Allen N. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12 doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poggio P., Branchetti E., Grau J.B. Osteopontin-CD44v6 interaction mediates calcium deposition via phospho-Akt in valve interstitial cells from patients with noncalcified aortic valve sclerosis. Arterioscler Thromb Vasc Biol. 2014;34:2086–2094. doi: 10.1161/ATVBAHA.113.303017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao A.S., Lindholm D., Rivas M.A., Knowles J.W., Montgomery S.B., Ingelsson E. Large-scale phenome-wide association study of variants demonstrates protection against ischemic stroke. Circ Genom Precis Med. 2018;11 doi: 10.1161/CIRCGEN.118.002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verbeek R., Boyer M., Boekholdt S.M. Carriers of the PCSK9 R46L variant are characterized by an antiatherogenic lipoprotein profile assessed by nuclear magnetic resonance spectroscopy—brief report. Arterioscler Thromb Vasc Biol. 2017;37:43–48. doi: 10.1161/ATVBAHA.116.307995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sliz E.V., Kettunen J.O., Holmes M.Y. Metabolomic consequences of genetic inhibition of PCSK9 compared with statin treatment. Circulation. 2018;138:2499–2512. doi: 10.1161/CIRCULATIONAHA.118.034942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poggio P., Songia P., Cavallotti L. PCSK9 involvement in aortic valve calcification. J Am Coll Cardiol. 2018;72:3225–3227. doi: 10.1016/j.jacc.2018.09.063. [DOI] [PubMed] [Google Scholar]
- 24.O'Donoghue M.L., Fazio S., Giugliano R.P. Lipoprotein(a), PCSK9 inhibition and cardiovascular risk: insights from the FOURIER trial. Circulation. 2019;139:1483–1492. doi: 10.1161/CIRCULATIONAHA.118.037184. [DOI] [PubMed] [Google Scholar]
- 25.Stiekema L.C.A., Stroes E.S.G., Verweij S.L. Persistent arterial wall inflammation in patients with elevated lipoprotein(a) despite strong low-density lipoprotein cholesterol reduction by proprotein convertase subtilisin/kexin type 9 antibody treatment. Eur Heart J. 2019;40:2775–2781. doi: 10.1093/eurheartj/ehy862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salaun E., Mahjoub H., Dahou A. Hemodynamic deterioration of surgically implanted bioprosthetic aortic valves. J Am Coll Cardiol. 2018;72:241–251. doi: 10.1016/j.jacc.2018.04.064. [DOI] [PubMed] [Google Scholar]
- 27.Nsaibia M.J., Mahmut A., Mahjoub H. Association between plasma lipoprotein levels and bioprosthetic valve structural degeneration. Heart. 2016;102:1915. doi: 10.1136/heartjnl-2016-309541. [DOI] [PubMed] [Google Scholar]
- 28.Bergmark B.A., O'Donoghue M.L., Murphy S.A. An exploratory analysis of proprotein convertase subtilisin/kexin type 9 inhibition and aortic stenosis in the FOURIER trial. JAMA Cardiol. 2020;6:709–713. doi: 10.1001/jamacardio.2020.0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.