Abstract
RAS was identified as a human oncogene in the early 1980’s and found to be mutated in nearly 30% of all human cancers. More importantly, however, RAS plays a central role in driving tumor development and maintenance. Despite decades of effort, there remain no FDA approved drugs that directly inhibit RAS. The prevalence of RAS mutations in cancer and the lack of effective anti-RAS therapies stem from RAS’ core role in growth factor signaling, unique structural features, and biochemistry. However, recent advances have brought promising new drugs to clinical trials and shone a ray of hope in the field. Here, we will exposit the details of RAS biology that illustrate its key role in cell signaling and shed light on the difficulties in therapeutically targeting RAS. Furthermore, past and current efforts to develop RAS inhibitors will be discussed in depth.
Keywords: RAS, Cancer, Small molecule inhibitor, MAPK, Growth factor signaling, GTPase, Monobody, Autophagy
Introduction
Cancer is the second leading cause of death in the United States, responsible for over half a million deaths per year at a cost of nearly $150 billion and likely to increase (National Cancer Institute, 2019; Murphy et al., 2017). Nearly 30% of all cancers harbor an oncogenic mutation in a single set of genes encoding the RAS proteins (Baines et al., 2011). Moreover, RAS is the most frequently mutated oncogene in 3 of the top 4 most lethal cancers: pancreatic adenocarcinoma, colorectal cancer, and lung cancer (Baines, et al., 2011). Perhaps even more astonishing is the fact that there are currently no FDA approved treatments to directly target RAS despite over 40 years of research. How does RAS function? What accounts for its prominent role in cancer? How has it evaded pharmacological targeting for decades? These questions will be explored in this chapter.
1. RAS Biology
RAS proteins encompass a superfamily of small guanosine triphosphatases (GTPases) that function in myriad cellular processes from cell proliferation to vesicular transport (Cox and Der, 2010). For the purposes of this chapter, RAS refers to the “classical” members of the oncogenic RAS subfamily of GTPases: HRAS, NRAS, and KRAS. HRAS and KRAS were initially identified as viral oncogenes in rats during the 1960’s and ‘70’s (Harvey, 1964; Kirsten and Mayer, 1967; Scolnick et al., 1973), and later recognized to be endogenous human oncogenes in the 1980’s (Der et al., 1982; Parada et al., 1982; Santos et al., 1982). NRAS was subsequently identified as a transforming gene in several human tumor lines including a neuroblastoma line (Hall et al., 1983; Shimizu et al., 1983). RAS is a ubiquitously expressed key signaling hub for initiating cellular processes in response to extracellular signals. The structure, function, and biological as well as pathological roles of RAS will be discussed below.
1.1. RAS Structure
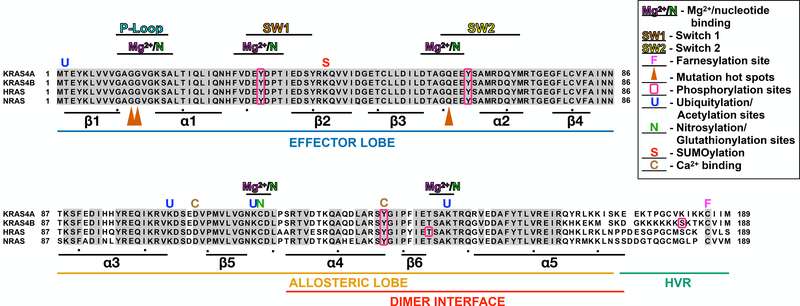
The four RAS proteins (HRAS, NRAS, and KRAS4A/4B) are 188–189 amino acid membrane associated GTPases encoded by three separate genes, with KRAS encoding two distinct proteins due to alternative splicing (Cox and Der, 2010). KRAS4B is the primary KRAS isoform and will be referred to as KRAS hereafter (Cox and Der, 2010). RAS isoforms are 85% identical and consist of a central G-domain that hydrolyzes GTP (residues 1–172) and a hypervariable region (HVR; residues 173–188/9) that includes the lipidation site(s) for membrane targeting (O’Bryan, 2019) (Fig. 1). The G-domain consists of an effector lobe (residues 1–86) and an allosteric lobe (residues 87–172) both of which are involved in nucleotide binding. The effector lobe encodes the Switch 1 (SW1) and Switch 2 (SW2) regions that mediate RAS-protein interactions, and is invariant between RAS isoforms (Gorfe et al., 2008). The allosteric lobe is ~86% identical between isoforms and regulated by numerous post-translational modifications (PTMs) and ionic interactions. Further, this region facilitates interactions with the plasma membrane (PM) and promote dimerization/nanoclustering of RAS to regulate its function (Fig. 1).
Figure 1. RAS structure.
Alignment of RAS isoforms. Secondary structure is indicated by the location of alpha helices and beta strands. PTMs to specific residues are indicated as follows: farnesylation, (F); ubiquitylation and acetylation, (U); nitrosylation and glutationylation, (N); SUMOylation, (S); and Ca2+ binding, (C); phosphorylation, boxed residues (O); mutation hotspots,orange arrowheads (▲). Additionally, the dimer interface (α4-β6-α5) in highlighted.
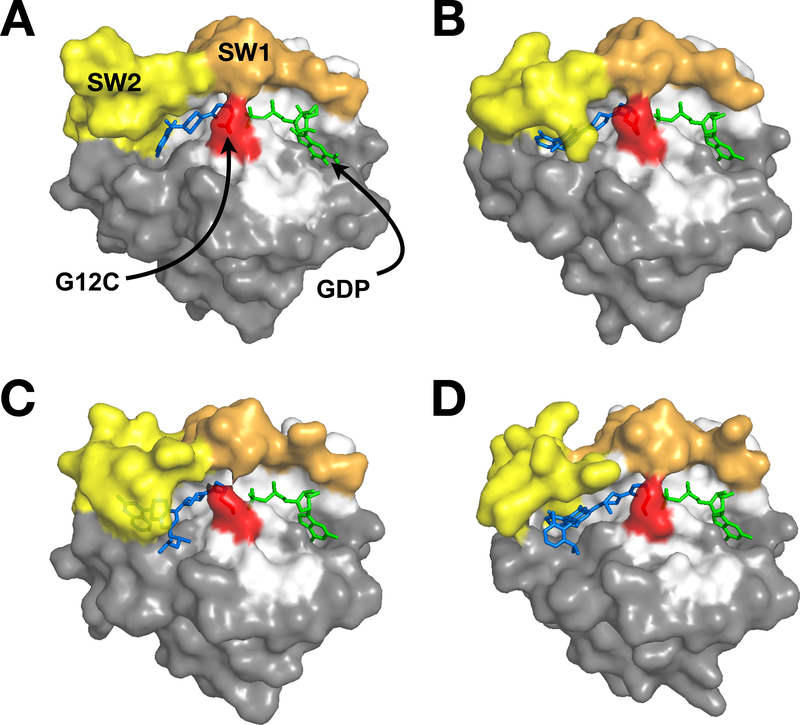
The secondary structure of RAS consists of 6 β-strands (β1- β6) that form a central β-sheet and 5 surrounding α-helices (α1- α5) spanning the G-domain (Fig. 1) (Pai et al., 1990). The nucleotide binding pocket is formed on one end by the SW1, SW2, and the P-loop in the effector lobe, and on the other end by the residues spanning β5-α4 and β6-α5 in the allosteric lobe (Fig. 2). The switch domains and P-loop coordinate with Mg2+ and surround the phosphates of GTP/GDP, while residues in the allosteric lobe interact with the base portion of the nucleotide (Fig. 2; see PDB:5P21) (Pai, et al., 1990).
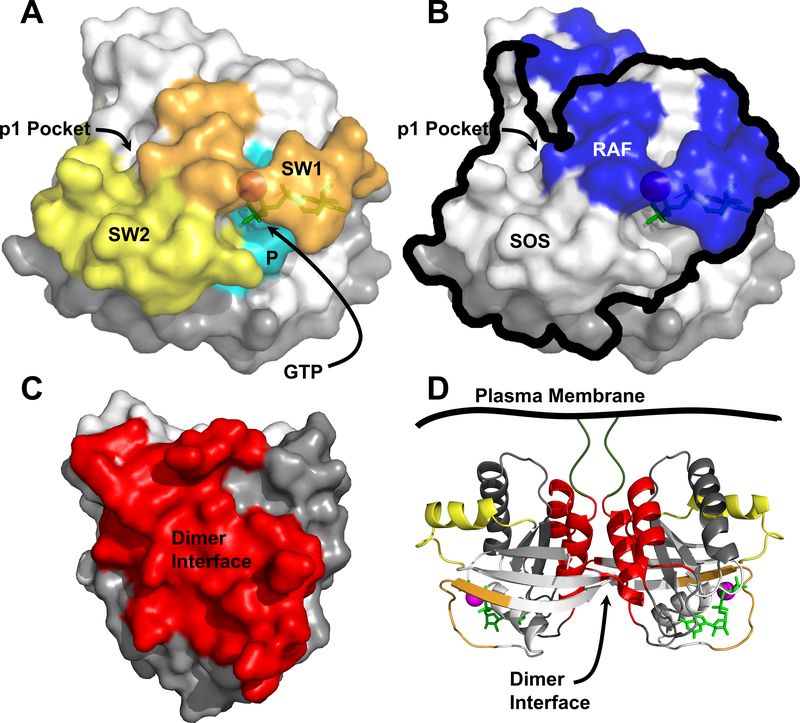
Figure 2. Three-dimensional visualization of RAS features.
All structures were generated using PDB: 5P21. A) Switch 1 (SW1; orange), Switch 2 (SW2; yellow), and the P-Loop (P; cyan) are shown in relation to the nucleotide binding pocket occupied by GTP and Mg2+ (green and magenta, respectively) and the p1 pocket. Effector lobe is shown as white and allosteric lobe, grey. B) RAS is shown in the same orientation as A) to illustrate the overlap of the RAF (blue) and SOS (black outline) interaction surfaces with the switch regions. The interfaces for RAF and SOS were extrapolated from Fetics et al and Boriack-Sjodin et al respectively (Boriack-Sjodin, et al., 1998; Fetics et al., 2015). C) The α4-α5 dimer interface is indicated in red. D) A RAS dimer is depicted with secondary structure highlighted, with the dimer interface colored red, SW1 and SW2 are orange and yellow, respectively.
Coupling of RAS to downstream effectors is primarily determined by conformational changes in SW1 and SW2 regions that are regulated by GTP vs GDP binding. Nucleotide loading requires Mg2+ as a cofactor to stabilize binding (Boriack-Sjodin et al., 1998; Simanshu et al., 2017). When loaded with GDP, RAS is in an “off” state. Spontaneous release of GDP by RAS is an intrinsically slow process [half-life of ~4hrs (Smith et al., 2013)] that is facilitated by guanine-nucleotide exchange factors (GEFs). GEFs destabilize nucleotide binding to promote formation of nucleotide-free RAS (Haney and Broach, 1994; Uejima et al., 2010). Due to the approximately 3-fold higher concentration of GTP vs GDP in the cytoplasm of cells [~0.5mM vs 0.15mM, respectively (Traut, 1994)] coupled with the picomolar affinity of RAS for guanine nucleotides (John et al., 1990), RAS rapidly binds GTP (Vetter and Wittinghofer, 2001). RAS is a relatively poor GTPase, with intrinsically slow GTP hydrolysis (30min) (Manne et al., 1985; Rudack et al., 2012). However, GTPase accelerating proteins (GAPs) enhance the GTPase activity of RAS by ~100-fold leading to inactivation (Adari, 1988).
In the GTP-bound state, the switch regions form an interaction surface with multiple conformations (Nakhaeizadeh et al., 2016). SW1 has two possible states, state 1 and state 2 (Johnson and Mattos, 2013). In the absence of other proteins, RAS-GTP slowly interconverts between these two states (Geyer et al., 1996). However, GAPs facilitate transition to state 1 in which an arginine finger mechanism promotes GTP hydrolysis (Geyer, et al., 1996; Kotting et al., 2006; te Heesen et al., 2007). State 2 is associated with effector binding which initiates downstream signaling cascades (discussed in section 1.3.) (Geyer, et al., 1996), and intrinsic GTP hydrolysis (Spoerner et al., 2010). When SW1 is in state 2, SW2 assumes either a catalytically inactive “T” state which promotes continued RAS signaling or a catalytically active “R” state that favors GTP hydrolysis and termination of signaling (Johnson and Mattos, 2013).
RAS binding of GTP is necessary for coupling to effectors but not sufficient to activate effectors such as RAF. Emerging evidence suggests that RAS must dimerize or form higher order complexes in order to properly signal (Fig. 2). Radiation inactivation experiments suggested that HRAS functioned as a dimer or potential trimer (Santos et al., 1988). Furthermore, monomeric RAS was insufficient to activate RAF in solution; however, artificial dimerization of RAS resulted in activation of RAF kinase activity both in vitro and in cells (Inouye et al., 2000). Consistent with these observations, RAS stimulates dimerization of RAF in cells (Rushworth et al., 2006; Weber et al., 2001), which is necessary for RAF kinase activation. Finally, RAS proteins form transient 6–7 member nanoclusters on the PM that are essential for RAS recruitment and activation of effectors (Plowman et al., 2005; Prior et al., 2003; Zhou and Hancock, 2015).
Analysis of RAS crystal structures provided further support for a role of dimerization in RAS function (Guldenhaupt et al., 2012; Kovrigina et al., 2015; Muratcioglu et al., 2015; Spencer-Smith et al., 2017). Guldenhaupt et al observed RAS dimers in 5 of 7 RAS crystal structures (Guldenhaupt, et al., 2012). Furthermore, a combination of molecular modeling, and biophysical analyses of NRAS anchored on artificial membranes suggested that NRAS dimerized through residues in the α4 and α5 helices (Guldenhaupt, et al., 2012). However, analysis by Spencer-Smith et al extended these observations, noting that these α4-α5 RAS dimers were only observed in active state RAS structures (Spencer-Smith, et al., 2017), supporting the notion that dimerization is necessary for RAS signaling. Interestingly, this same region had previously been implicated in RAS nanoclustering as well (Abankwa et al., 2008). RAS dimerization has also been observed through a β-sheet to β-sheet interaction as well as through a surface formed by the α3 and α4 helices that inhibit or promote RAS function, respectively (Muratcioglu, et al., 2015). Both α3-α4 and α4-α5 dimers were predicted by molecular modeling (Prakash et al., 2017). Superresolution microscopy studies established that KRAS dimers form at endogenous expression levels, and are required for RAS function (Nan et al., 2015). Furthermore, KRAS dimerization mediated the oncogenic activity and sensitivity to MEK inhibitors (Ambrogio et al., 2018).
Despite these findings, the importance of dimerization in RAS function remains the subject of significant debate. The isolated G-domain of RAS lacks the intrinsic ability to dimerize in solution (Ito et al., 1997; Kovrigina, et al., 2015; Kraulis et al., 1994; O’Connor and Kovrigin, 2008). Furthermore, the dimers observed by superresolution were mediated by the isolated C-terminal HVRs (Nan, et al., 2015) which were also sufficient to drive nanoclustering (Janosi and Gorfe, 2010; Janosi et al., 2012). Finally, recent work by Groves and colleagues failed to observe dimers of natively processed KRAS on supported lipid bilayers suggesting that RAS does not intrinsically dimerize but leaving open the possibility that other proteins may facilitate RAS dimerization (Chung et al., 2018).
The recent isolation of several tool biologics that target the α4-α5 (Spencer-Smith, et al., 2017) and the α3-α4 dimerization interfaces (Bery et al., 2019) provided additional support for the role of dimerization in RAS function. These reagents represent the first experimental tools that allow for perturbation of RAS dimerization and function. Indeed, they reveal the importance of these dimerization interfaces in oncogenic RAS signaling and transformation (Bery, et al., 2019; Khan et al., 2020; Spencer-Smith, et al., 2017). These reagents, along with a naturally occurring RAS dimerization inhibitor, will be discussed in more detail in section 2.2.2.
1.2. The RAS Lifecycle
1.2.1. RAS Prenylation and Membrane Association
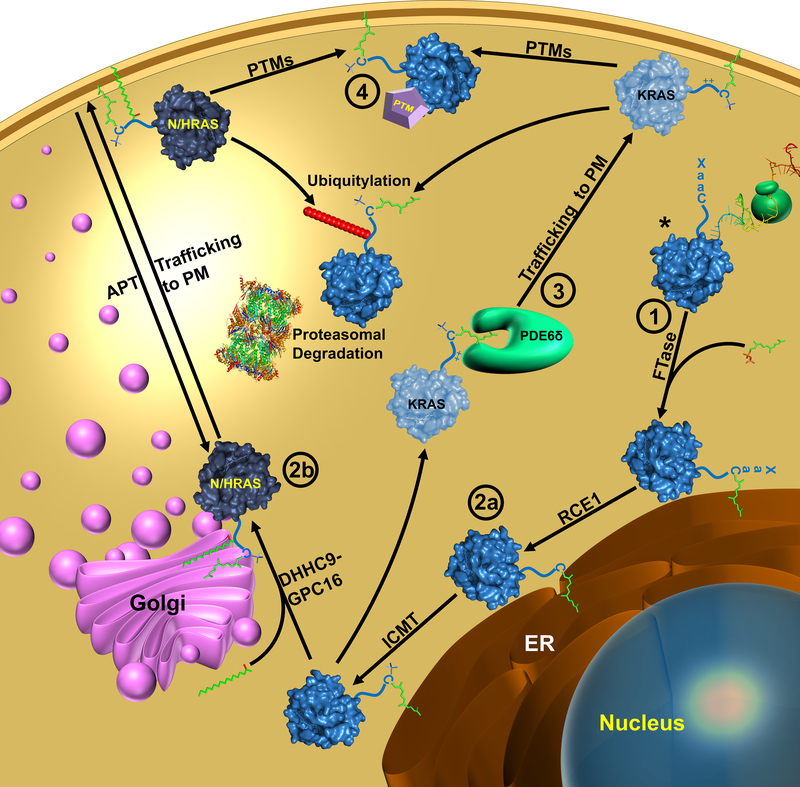
RAS is translated in the cytoplasm as an inactive soluble protein (Kikuchi and Williams, 1994; Willumsen et al., 1984). RAS must associate with the membrane to be active (Kikuchi and Williams, 1994), and the initial step in transporting RAS to the membrane is prenylation (Casey et al., 1989). Farnesyl transferase (FTase) recognizes the carboy-terminal CaaX (Cysteine-aliphatic-aliphatic-any amino acid) motif on RAS and catalyzes the covalent attachment of a farnesyl group to the Cys of the CaaX motif (Reiss et al., 1990). Farnesylated RAS then associates with the endoplasmic reticulum (ER), although the details of trafficking from the cytosol to the ER are unknown (Wright and Philips, 2006). In the ER, the carboxy-terminal tripeptide (aaX) is removed by RAS-converting enzyme-1 (RCE1) (Boyartchuk et al., 1997; Hollander et al., 2000). Proteolytically processed RAS is then modified by isoprenylcysteine carboxymethyltransferase (ICMT), which adds a methyl group to the new carboxyl-terminus of RAS (Dai et al., 1998) (Fig. 3).
Figure 3. RAS lifecycle.
RAS is translated in the cytoplasm (*) where the Cys of the CaaX motif is prenylated by FTase after which it associates with the endoplasmic reticulum (ER). In the ER, RAS is modified RCE1 followed by ICMT to remove the aaX of the Caax motif and methylation of the C-terminal Cys, respectively. KRAS is trafficked to the PM by PDEδ while N- and HRAS are transferred to the Golgi and palmitoylated by PAT. N- and HRAS are shuttled to the PM from the Golgi by classical vesicular trafficking mechanisms. RAS function is modulated by PTMs and targeted degradation. Numbers indicate points where RAS has been targeted for indirect inhibition: ① Prenylation inhibitors are discussed in section 2.1.1.; ② Inhibition of RAS processing in section 2.1.2.; ③ Inhibition of KRAS trafficking in section 2.1.3.; ④ Targeting RAS PTMs in section 2.1.4.
RAS processing and trafficking post-carboxymethylation differs between isoforms. In the case of NRAS, HRAS, and KRAS4a, the proteins are trafficked through the Golgi and on to the PM, whereas KRAS4b is trafficked directly to the PM in a microtubule-dependent manner (Choy et al., 1999; Thissen et al., 1997) (Fig. 3). These divergent pathways are mediated by “second signals” in the HVR N-terminal to the CaaX motif (Ahearn et al., 2011a). For NRAS, HRAS, and KRAS4a Cys located in the HVR (two for HRAS and one for NRAS and KRAS4a) serve as sites for palmitoylation (Hancock et al., 1989) (Fig. 3). Following carboxymethylation, NRAS, HRAS, and KRAS4a are transported to the Golgi where the palmitoyl acyltransferase (PAT) complex, DHHC9-GPC16, attaches palmitate to Cys within the HVR (Swarthout et al., 2005). These RAS isoforms are then shuttled to the membrane through classical vesicular trafficking mechanisms (Apolloni et al., 2000; Choy, et al., 1999). This is a reversible process in which acyl-protein thioesterase (APT) catalyzes a deacylation reaction that removes palmitate from the RAS C-terminus (Duncan and Gilman, 1998) (Fig. 3). This cycle of palmitoylation/depalmitoylation promotes HRAS and NRAS shuttling between the PM and the Golgi and is thought to maintain specific intracellular compartmentalization that facilitates RAS signaling (Rocks et al., 2005). Interestingly, this cycle is regulated by another PTM: peptidyl-prolyl isomerization. Specifically, FK506-binding protein 12 (FKBP12) binds palmitoylated HRAS and catalyzes cis-trans isomerization of Pro179, which enhances depalmitoylation of HRAS (Ahearn et al., 2011b).
In contrast, KRAS4b is not further lipidated post-prenylation (Fig. 3). Instead, a polybasic region within the HVR serves as the second signal for KRAS4b membrane attachment (Hancock et al., 1990). Phosphodiesterase 6 delta subunit (PDEδ) is a guanine-nucleotide dissociation inhibitor (GDI)-like solubilizing factor that binds to the farnesylated C-terminus of KRAS4b (as well as other RAS GTPases), chaperones it to the PM, and prevents random redistribution to other membranes in the cell for all RAS isoforms (Chandra et al., 2011) (Fig. 3). Once at the PM, KRAS release from PDEδ is catalyzed by the G protein Arl2 (Ismail et al., 2011). The positively charged polybasic region of KRAS4b forms an electrostatic interaction with negatively charged headgroups on the cytosolic face of the PM that, together with the farnesyl modification, are sufficient for KRAS4b to associate with the PM (Ahearn, et al., 2011a).
1.2.2. RAS Degradation
RAS is modulated by additional PTMs which are discussed below. However, RAS delivery to the membrane and subsequent function in signaling is a dynamically regulated process involving the constitutive translation and degradation of RAS as evidenced by its relatively short half-life [~9hrs (Kim et al., 2009)]. To date, reports of RAS degradation have been exclusively through ubiquitin-mediated mechanisms (Fig. 3). In one instance, RAS degradation is mediated by the same cellular machinery that regulates Wnt/β-catenin signaling. Specifically, RAS is phosphorylated on Thr144 and 148 (pThr144/148) by glycogen synthase kinase 3β (GSK3β), which is recognized by the F-box protein β-transducin repeat-containing protein (β-TrCP) (Jeong et al., 2012). β-TrCP acts as a recognition subunit for the E3 ubiquitin ligase Skp1-cullin-F-box (SCF) protein, which polyubiquitylates RAS and targets it for proteolytic degradation by the proteasome (Jeong, et al., 2012). A further level of regulation is enacted by the Smad ubiquitylation regulatory factor 2 (SMURF2). SMURF2 forms an E2:E3 complex with the E2 ubiquitin-conjugating enzyme UBCH5 that polyubiquitylates β-TrCP (Shukla et al., 2014). This leads to β-TrCP degradation and a loss of β-TrCP-mediated RAS degradation (Shukla, et al., 2014).
The E3 ligase, neural precursor cell expressed developmentally downregulated protein 4–1 (Nedd4–1), has also been implicated in RAS degradation (Zeng et al., 2014b). Interestingly, Nedd4–1 regulation of RAS functioned as a negative feedback loop to attenuate RAS signaling. RAS signaling enhanced expression of Nedd4–1, which ubiquitylated RAS targeting it for lysosomal degradation (Zeng, et al., 2014b). Thus, RAS degradation is initiated and regulated by multiple ubiquitylation systems.
1.3. RAS Signaling
1.3.1. Upstream Signaling to RAS
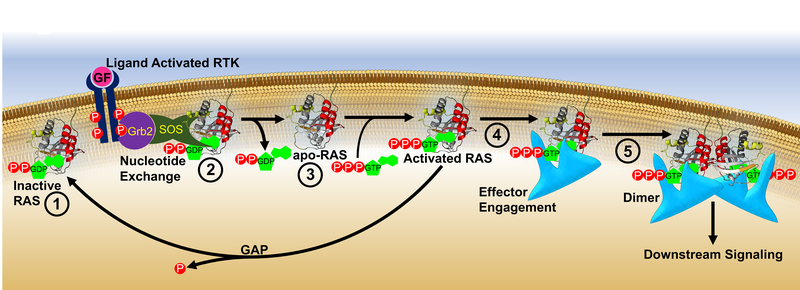
Receptor tyrosine kinases (RTKs) such as epidermal growth factor receptor (EGFR), fibroblast growth factor receptors (FGFR), and vascular endothelial growth factor receptors (VEGFR) among many others (Popovic and Wilson, 2010), are activated by growth factors that induce receptor dimerization and trans-autophosphorylation of Tyr in their cytoplasmic carboxy-termini (Fig. 4). These phospho-Tyr (pTyr) sites recruit effectors containing Src-homology (SH2) or protein tyrosine binding (PTB) domains (Pawson, 1997) that mediate assembly of a variety of signaling complexes. Of particular importance is the adaptor protein growth factor receptor-bound protein-2 (GRB2) which consists of a central Src homology 2 (SH2) domain and two flanking SH3 domains that constitutively interact with the Pro-rich region of the RAS GEF Son-of-Sevenless (SOS) (Li et al., 1993). Upon RTK activation, the GRB2 SH2 domain binds pTyr sites in the RTK C-terminus resulting in translocation of SOS to the PM where it facilitates the exchange of GDP for GTP on RAS resulting in RAS activation and stimulation of subsequent downstream signaling pathways (Buday and Downward, 1993; Chardin et al., 1993; Egan et al., 1993; Gale et al., 1993; Li, et al., 1993; McCormick, 1993; Olivier et al., 1993; Rozakis-Adcock et al., 1993; Simon et al., 1993) (Figs. 2 and 4).
Figure 4. RAS activation and direct inhibitors of RAS.
From left to right: ① RAS is inactive when bound to GDP. ② RTK activation results in recruitment of GRB2/SOS complex to the PM where SOS promotes GDP release from RAS and formation of transient nucleotide-free state (apo-RAS; ③). ④ Binding of GTP leads to RAS recruitment of effectors. ⑤ Subsequent dimerization of the RAS/effector complex activates effectors initiating signaling cascades. RAS inactivation occurs through GTP hydrolysis catalyzed by GAPs. Numbers indicate points of direct RAS inhibition: ① G12C inhibitors (section 2.2.1.2.); ② Inhibitors of SOS-RAS (sections 2.2.1.3., 2.2.1.4., 2.2.2.1., and 2.2.2.5.); ③ Inhibitors of apo-RAS (section 2.2.2.1.) ④ Inhibitors of effector interactions (sections 2.2.1.3., 2.2.1.5., 2.2.2.1., 2.2.2.3., 2.2.2.4., 2.2.2.5., 2.2.2.6., and 2.2.2.7.); ⑤ Anti-RAS Biologics (see sections 2.2.2.6., and 2.2.2.7.). Coloring of RAS molecule is the same as in Fig. 2.
1.3.2. Downstream RAS Signaling
RAS stimulates a number of prominent effectors such as RAF, PI3K, RALGDS, TIAM1, RASSF, and many others (Rajalingam et al., 2007; Repasky et al., 2004). Engagement of these targets involves RAS-GTP interaction with a RAS binding domain (RBD) in each target. Three distinct RBD sequences have been identified: (1) the RBD of RAF and TIAM 1, (2) the RBD from PI3K, and (3) the RAS association (RA) domains of RALGDS and AF6 (Herrmann, 2003).
1.3.2.1. RAS-RAF-MEK-ERK signaling
The RAS/RAF/MAPK pathway is mainly involved in transducing signaling from the extracellular milieu to the cell nucleus and is arguably the best characterized signal transduction pathway in cell biology. RAS-GTP recruits the RAF Ser/Thr kinase which subsequently stimulates the MEK-ERK kinase cascade. However, mutant RAS proteins result in constitutive activation of downstream effectors even in the absence of growth factor stimulation, conferring a proliferative advantage to tumors (Molina and Adjei, 2006; Simanshu, et al., 2017).
Active RAS promotes RAF recruitment to the cell membrane through the interaction of RAS SW1 with the RBD of RAF (Fig. 2), which induces a conformational change in RAF that exposes activating Ser/Thr and Tyr phosphorylation sites and dimerization of RAF protomers (Hibino et al., 2011; Rajakulendran et al., 2009). Activated RAF phosphorylates and activates the MEK1/2 dual-specificity protein kinase, which phosphorylates and activates the ERK1/2 mitogen-activated protein kinase (MAPK). ERK1 and ERK2 phosphorylate and activate a variety of nuclear transcription factors and kinases, including Elk-1, c-Ets1, c-Ets2, p90RSK1, MNK1, and MNK2 resulting in the induced expression of genes that promote cell-cycle progression (Liebmann, 2001; Schreck and Rapp, 2006). However, ERK is also involved in negative feedback regulation of the pathway which has important consequences for bypassing resistance to various targeted therapies of this pathway (Neel and Bivona, 2017).
1.3.2.2. RAS-PI3K Signaling
Phosphoinositide 3-kinases (PI3Ks) represent a conserved family of eight lipid kinases that phosphorylate phosphoinositides (PIs) at the 3-position of their inositol ring (Engelman et al., 2006). PI3Ks are subdivided into three major classes based on their structures and substrate specificity and several members are implicated in cancer. PIK3CA has received the most attention due to its mutational activation in a number of tumors including breast, colon, and liver (Engelman, et al., 2006). PIK3CA, hereafter referred to only as PI3K, consists of a regulatory p85 subunit and a catalytic p110 subunit. The p85 subunit links RTK activation with regulation of PI3K activity. However, PI3K is also a RAS effector as the p110 catalytic domain contains an RBD that directly binds RAS (Sheridan and Downward, 2013; Sjolander et al., 1991). All RAS isoforms do not activate the PI3K pathway equally and HRAS is a more potent activator than KRAS (Yan et al., 1998). Active PI3K converts phosphatidylinositol (4,5)-bisphosphate (PIP2) into phosphatidylinositol (3,4,5)-trisphosphate (PIP3). PIP3 recruits phosphatidyl inositol-dependent kinase 1 (PDK1) and AKT (protein kinase B) to the PM, where PDK1 phosphorylates and activates AKT at Thr308 (Downward, 2003). The mTORC2 complex phosphorylates Ser473 resulting in full activation of AKT. The PI3K-AKT axis promotes cell proliferation and survival by multiple mechanisms including inhibition of Bcl-2 family members BAD and BAX, activation of MDM2, and inhibition of FOXO family of transcription factors (Engelman, et al., 2006). Thus, RAS-PI3K signaling favors tumorigenesis by supporting cell proliferation and opposing apoptosis. Indeed, disrupting RAS-PI3K interaction through mutations of the RAS binding domain of PIK3CA led to a substantial decrease in RAS-induced tumorigenesis highlighting the importance of p110α in RAS-driven oncogenesis in vivo (Gupta et al., 2007). Despite significant efforts in developing PI3K inhibitors for anti-cancer therapy (Yang et al., 2019; Zhao et al., 2017), clinical trials with these inhibitors as monotherapy has shown limited clinical efficacy. These disappointing results are due in part to amplification, mutation of PIK3CA and PIK3CB, and extensive cross-talk with other signaling pathways leading to activation of compensatory pathways in response to PI3K inhibition (Huw et al., 2013; Nakanishi et al., 2016; Yang, et al., 2019).
1.3.2.3. RAS-RALGEF-RAL Axis
RAL (RAS Like) is a family of RAS-like GTPases consisting of two members, RALA and RALB. Both are ubiquitously expressed in humans and share 80% homology with RAS (Gentry et al., 2014; Repasky, et al., 2004). RAL exchange factors, i.e., RAL–GEFs, serve as a direct link between RAS and RAL activation. Four distinct RAL–GEFs (RALGDS, RGL, RGL2/RLF and RGL3) contain a common C-terminal RAS association (RA) domain and are RAS effectors. GTP-bound RAS relocates these RALGEFs to the PM where they promote the exchange of GDP for GTP on RALA and RALB. Despite their similarity in sequence and effectors, RALA and RALB appear to perform different and in some cases antagonistic functions. RALA is required for anchorage-independent proliferation of transformed cells, while RALB is required for the survival of transformed, but not normal, cells. Once activated, RAL signaling regulates many cellular processes including endocytosis, exocytosis, actin cytoskeletal dynamics, and transcription (Chien and White, 2003).
1.3.2.4. RAS-TIAM1 Signaling
T lymphoma invasion and metastasis protein 1 (TIAM1) is a GEF containing a RAS binding domain and is involved in RAS activation of RAC, CDC42, and to a lesser extent RHOA (Mertens et al., 2003). TIAM1−/− mice are resistant to the development of RAS-driven skin tumors and fibroblasts derived from these knockouts do not form foci when transfected with oncogenic RAS (Malliri et al., 2002). TIAM1 was identified in a retroviral insertional mutagenesis screen for genes conferring invasiveness to otherwise non-invasive murine T-lymphoma cells (Habets et al., 1994). Various post translational modifications including myristylation, phosphorylation, and phosphoinositol binding at the N-terminus of TIAM1 modulate its intracellular localization and activation (Mertens, et al., 2003). Membrane localization of TIAM1 is crucial for its ability to induce RAC-mediated membrane ruffles and activation of JNK. TIAM1 has been implicated in neurotrophin-induced Schwann cell migration (Yamauchi et al., 2005). In addition to its role in RAS signaling, TIAM1 is also involved in RAP1 mediated cell spreading (Arthur et al., 2004). TIAM1 has been reported to be both pro- and anti-apoptotic as well as pro- and anti-tumorigenic.
1.3.2.5. RAS-RASSF
Although potent activators of proliferation, RAS proteins stimulate apoptotic and senescence pathways as well (Donninger et al., 2016). RASSF proteins provide an important link between RAS activation and stimulation of these pathways. RASSFs are a family of ten proteins characterized by a RAS association (RA) domain at either the N-terminus or C-terminus (Zinatizadeh et al., 2019). Given the lack of intrinsic catalytic activity, RASSF proteins function as scaffolding molecules that link RAS to a wide range of signaling pathways including Hippo, BAX, and p53. Although several of the RASSF family members bind to MST kinases (mammalian homologs of Drosophila Hippo), RASSF1A appears to drive apoptosis through activation of MST kinases (Donninger, et al., 2016). Despite binding MST kinases, other RASSF family members do not appear to activate the Hippo pathway but rather link to apoptotic/senescence pathways through activation of alternative pathways including p53 and BAX. Thus, RASSF genes act as RAS-regulated tumor suppressors and the epigenetic inactivation of various RASSF genes in human tumors plays key role in RAS-driven transformation and metastasis (Akino et al., 2005).
1.3.3. Modulation of RAS Function by Post-Translational Modifications
PTMs regulate the structure, function, localization, and abundance of proteins. Common PTMs include phosphorylation, lipidation, acetylation, nitrosylation, ubiquitylation, glycosylation, SUMOylation, methylation, hydroxylation, and formation of disulfide bridges among others (Bürkle, 2001). Given the key role of RAS in cell biology, it is not surprising that RAS is regulated by many of these modifications as described below, with the exception of PTMs required for the RAS lifecycle discussed in Section 1.2 (Fig. 1).
1.3.3.1. RAS Phosphorylation
There are multiple phosphorylation sites on RAS, with some conserved between isoforms, and others isoform specific (Khan, et al., 2020) (Fig. 1). Src phosphorylates all RAS isoforms on Tyr32 and Tyr64 in the SW1 and SW2 regions, respectively (Bunda et al., 2014; Kano et al., 2019). Phosphorylation of Tyr32 on NRAS and HRAS, or both residues on KRAS, attenuates RAF binding thereby negatively regulating RAS signaling (Bunda, et al., 2014; Kano, et al., 2019). In the case of NRAS and HRAS, pTyr32 also concomitantly enhanced GAP association and GTP hydrolysis (Bunda, et al., 2014). Conversely, pTyr32/64 on KRAS inhibited both GAP and GEF interaction while enhancing the intrinsic GDP/GTP exchange rate (Kano, et al., 2019). The consequence was the formation of a “dark” GTP-loaded RAS (i.e. RAS in the “on” state but incapable of effector interaction). Importantly, the phosphatase SHP2 dephosphorylated Tyr32 on HRAS and NRAS (Bunda et al., 2015), and Tyr32/64 on KRAS (Kano, et al., 2019), resulting in derepression of RAS-RAF interaction and promotion of downstream signaling. The critical role of SHP2 phosphatase activity on RAS was underscored by work demonstrating that deletion of Ptpn11 (the gene encoding SHP2) nearly completely blocked the formation of pancreatic ductal adenocarcinoma (PDAC) and non-small-cell lung cancer (NSCLC) in pancreas-specific and lung-specific KRAS-driven mouse models, respectively (Ruess et al., 2018).
Tyr137 is another pan-RAS phosphorylation site. Unlike pTyr32/64, pTyr137 enhanced RAS-RAF interaction without any effect on GTPase activity (Ting et al., 2015). Interestingly, phosphorylation of Tyr137 appeared to be part of a positive feedback loop. RAS activation of its effector RAS- and RAB-interacting protein 1 (RIN1) resulted in activation of its own downstream effector, Abelson tyrosine-protein kinase (ABL), which in turn phosphorylated RAS on Tyr137 resulting in enhanced signaling through RAF (Ting, et al., 2015).
Protein kinase C (PKC) mediates phosphorylation of the KRAS C-terminus on Ser181 (Ballester et al., 1987; Bivona et al., 2006). This phosphorylation dissociated KRAS from the PM through a farnesyl-electrostatic switch mechanism resulting in relocalization of KRAS to internal membranes including the mitochondrial outer membrane and ER (Bivona, et al., 2006). The effects of pSer181 on KRAS signaling were not determined because of the confounding effects of PKC agonists on signaling pathways, and the fact that the phosphomimetic mutant KRAS(G12V/S181E) was highly cytotoxic to cells through a Bcl-XL-mediated apoptotic mechanism (Bivona, et al., 2006). Inositol trisphosphate (InsP3) receptors (IP3Rs) on the ER were later identified as novel KRAS effectors that were inhibited in their ability to promote mitochondrial respiration and inhibition of autophagy (Sung et al., 2013). GTP-loaded KRAS-pSer181 attenuated Bcl-XL sensitization of IP3R to InsP3. Consequently, the ER-to-mitochondria Ca2+ transfer required for IP3R-mediated mitochondrial respiration was inhibited, leading to apoptosis (Sung, et al., 2013).
Paradoxically, pSer181 is required for normal KRAS activity. Alvarez-Moya et al demonstrated a reciprocal relationship between KRAS-calmodulin (CaM) interaction and Ser181 phosphorylation that modulates KRAS activity (Alvarez-Moya et al., 2010). CaM inhibited phosphorylation of Ser181 by PKC, and conversely pSer181 inhibited CaM-KRAS interaction. Interestingly, while CaM-KRAS interaction reduced KRAS activation, if CaM and PKC were co-inhibited, KRAS activation and downstream signaling were still attenuated indicating that pSer181 is necessary for RAS activation (Alvarez-Moya, et al., 2010). Mechanistically, phosphorylation of Ser181 decreased GAP activity on KRAS promoting RAS-RAF interaction and downstream signaling (Alvarez-Moya, et al., 2010).
KRAS is also a substrate of cyclic GMP-dependent protein kinase (PKG). PKG phosphorylated KRAS on Ser181 downstream of AMP-activated protein kinase (AMPK) and required endothelial nitric oxide synthase signaling. As with PKC-mediated phosphorylation of Ser181, PKC promoted KRAS loss from the PM over time (Cho et al., 2016). In an intriguing twist on the work discussed above, however, the authors found that pSer181 transiently and acutely enhanced KRAS association with the PM, KRAS nanoclustering, and activation of both ERK and AKT, followed by loss of KRAS from the PM by endocytic internalization of KRAS (Cho, et al., 2016). Overall, the discovery of RAS phosphorylation has led to several new approaches to therapeutically inhibit RAS, which are discussed in section 2.1.4.
1.3.3.2. RAS Acetylation
RAS is also as a substrate for acetylation (Fig. 1), although the function of this modification remains uncertain. RAS acetylation on Lys104 and Lys147 was observed using recombinant RAS incubated with different lysine-acetyltransferases in vitro and with RAS immunoprecipitated from cell lysates (Knyphausen et al., 2016; Yang et al., 2012). Substitution of Lys104 on KRAS(G12V) with non-acetylatable residues had no effect on transforming activity or SOS-mediated nucleotide exchange (Yang, et al., 2012). However, KRAS(G12V/K104Q) (mimicking acetylation) rendered RAS resistant to SOS, suggesting that acetylation inactivated RAS (Yang, et al., 2012; Yin et al., 2017). Nevertheless, this amino acid substitution likely did not reliably reproduce the effects of acetylation. Using genetic code expansion to generate KRAS acetylated on Lys104 [KRAS(K104Ac) or Lys147 [KRAS(K147Ac)], Knyphausen et. al found that these modifications did not affect SOS-mediated nucleotide exchange on acetylated KRAS (Knyphausen, et al., 2016). Furthermore, K104Q RAS mutation reduced proliferation and cell survival of transformed cells in one study (Yang, et al., 2012), but did not affect the growth of wild-type RAS-expressing cells or the transformation potential of oncogenic KRAS(G12V) in later work (Yin, et al., 2017). GAP activity was similarly attenuated in KRAS(K104Q) (Yin, et al., 2017). The authors attributed the lack of effect of Gln substitution on cell growth to a counterbalancing inhibition of GAP activity (Yin, et al., 2017).
Inhibition or knockdown of HDAC6 and SIRT2 increased RAS acetylation, reduced proliferation of colorectal cancer cells and non-small-cell lung carcinoma (NSCLC) cells, and reduced the viability of KRAS(G12V), but not KRAS(G12V/K104A), transformed cells (Yang et al., 2013). These results suggested that HDAC6 and SIRT2 are RAS deacetylases and that Lys104 acetylation negatively regulates RAS activity and tumorigenicity. Thus, HDAC6 and SIRT2 inhibitors could be used as therapeutics to target oncogenic RAS. However, neither HDAC6 or SIRT2 directly deacetylated RAS(K104Ac) or RAS(K147Ac) (Knyphausen, et al., 2016). Thus, further work is needed to determine whether deacetylase inhibitors could be used to attenuate oncogenic RAS activity.
Recent studies suggested that N-terminal acetylation of KRAS is important for structural stability and function. Mass spectrometry of endogenous KRAS isolated from pancreatic cancer and colorectal cancer cells, or expressed in HEK293 cells, revealed loss of the initiator Met and N-acetylation of the exposed Thr residue (Buser et al., 2001; Dharmaiah et al., 2019; Ntai et al., 2018). Structural studies showed that KRAS(2–169) lacking the initiator Met and N-acetylation was Mg2+-free and adopted the inactive state 1 conformation of SW1 (Dharmaiah, et al., 2019). In comparison, N-acetylation of KRAS(2–169) resulted in Mg2+ binding and stabilization of the switch regions through interaction of the N-acetylated Thr with the central β-sheet (Dharmaiah, et al., 2019). This work suggests that removal of the N-terminal Met and subsequent N-acetylation of the remaining Thr residue is a critical processing event necessary for proper RAS folding and function.
1.3.3.3. RAS Nitrosylation
Nitric oxide (NO) was initially shown to stimulate RAS nucleotide exchange through modification of Cys118 (Lander et al., 1996; Lander et al., 1995a) (Fig. 1). Furthermore, RAS was critical for stress signaling, including NO, as evidenced by diminished NF-κB signaling in cells expressing dominant negative RAS compared to wild-type (Lander et al., 1995b). Structural and biochemical studies later determined that nitrosylation of Cys118 did not directly affect the structure of RAS, nucleotide exchange rates, or effector binding, but rather a RAS thiyl-radical intermediate in the biochemical process of Cys118 nitrosylation enhanced nucleotide dissociation (Heo and Campbell, 2004; Williams et al., 2003). Importantly, urethane-induced lung tumorigenesis was impaired in transgenic mice expressing KRAS(C118S), suggesting that activation of wild-type RAS by nitrosylation may contribute to oncogenesis (Huang et al., 2014).
A physiological role for RAS nitrosylation has been elucidated in the brain. While NO has neurodegenerative effects, it also participates in neurogenesis both in physiological processes and following injury [reviewed in (Contestabile and Ciani, 2004)]. NO-induced neurogenesis involved MAPK signaling, although the specific mechanism remained unidentified (Carreira et al., 2010). Cultured neuron stem cells (NCS) displayed upregulated MAPK signaling and enhanced proliferation in response to NO through RAS Cys118 nitrosylation (Santos et al., 2018). In a mouse model of brain seizure, NO was necessary for NCS proliferation post-injury (Carreira, et al., 2010). In vivo, RAS nitrosylation was observed in wild-type, but not mice lacking nitric oxide synthase suggesting that RAS nitrosylation is required for NCS proliferation post-injury (Santos, et al., 2018). Considering that the tumor microenvironment is under oxidative stress [conditions favorable to the formation of reactive oxygen species such as NO (Grek and Tew, 2010)] and that nitrosylation of wild-type RAS can contribute to tumorigenesis (Huang, et al., 2014), targeting RAS nitrosylation may be a viable therapeutic strategy in a wide variety of tumors.
1.3.3.4. RAS Glutathionylation
Glutathione is a tripeptide composed of Cys, Glu, and Gly that is synthesized by cells and helps to maintain redox homeostasis (Mieyal et al., 2008). Cys residues on proteins are susceptible to modification by glutathione – a process called glutathionylation (Grek et al., 2013). This reaction can either occur non-enzymatically on oxidized Cys or can be catalyzed by a number of enzymes (Xiong et al., 2011). As described in the previous section, Cys118 on RAS is sensitive to oxidation. Angiotensin II-mediated reactive oxygen species production leads to glutathionylation of RAS on Cys118, increased RAS activity, PI3K signaling, and protein synthesis contributing to vascular hypertrophy (Adachi et al., 2004). In contrast, in vitro measurements of RAS intrinsic nucleotide exchange, SOS-mediated nucleotide exchange, and GAP-mediated GTP rates hydrolysis were all unaffected by Cys118 glutathionylation (Hobbs et al., 2013a). In addition, glutathionylated RAS was resistant to redox-mediated nucleotide dissociation, indicating that Cys radicals formed in the step prior to RAS glutathionylation can activate RAS by inducing nucleotide exchange (similar to radicals formed by nitrosylation – see preceding section), but that the subsequent glutathionylation step actually neutralizes Cys radical-mediated RAS activation (Hobbs, et al., 2013a).
1.3.3.5. RAS Ubiquitylation and SUMOylation
Ubiquitylation of RAS can lead to RAS degradation (section 1.2.2.) or directly modulate RAS activity (Figs. 1 and 3). For example, similar to β-TrCP, leucine zipper-like transcription regulator 1 (LZTR1) bound RAS and acted as a recognition subunit for another E3 ligase, cullin 3, leading to RAS ubiquitylation (Bigenzahn et al., 2018). However, unlike β-TrCP, LZTR1-mediated RAS ubiquitylation at Lys170 did not induce RAS degradation, but rather disrupted RAS association with the membrane to inhibit RAS signaling (Steklov et al., 2018). Similarly, mono- and diubiquitylation of HRAS and NRAS enhanced endosomal partitioning of these RAS isoforms and attenuated MAPK signaling (Jura et al., 2006). The Rabex-5 E3 ligase also mediated RAS ubiquitylation leading to endosomal localization of RAS and attenuated signaling (Xu et al., 2010; Yan et al., 2010).
In contrast, RAS ubiquitylation may actually enhance RAS signaling. Sasaki et al identified Lys147 as a ubiquitylation site on KRAS and HRAS that enhanced GTP-loading (Sasaki et al., 2011). Indeed, ubiquitylated KRAS more effectively immunoprecipitated RAF and PI3K, and enhanced their in vitro kinase activities (Sasaki, et al., 2011). Further, ubiquitylation of HRAS Lys117 enhanced GDP/GTP exchange while KRAS Lys147 ubiquitylation impaired GAP-mediated GTPase activity (Baker et al., 2013; Hobbs et al., 2013b). These data suggest that inhibiting ubiquitylation of specific RAS sites may serve to attenuate RAS function.
Similar to ubiquitylation, SUMOylation involves the covalent attachment of the small ubiquitin-like modifier (SUMO) to lysine residues on proteins through an E1, E2, E3 ligase system (Sarge and Park-Sarge, 2011). Unlike ubiquitylation, SUMOylation does not target proteins for degradation, but rather modifies protein function. SUMOylation is involved in a number of pathological processes including neurodegenerative disease, heart disease, and cancer (Sarge and Park-Sarge, 2011). Lys42 on all RAS isoforms is a target of SUMOylation (Choi et al., 2018a). Lys42 was SUMOylated with the SUMO3 protein through the E3 ligase PIASγ, and mutation of Lys42 to Arg reduced MAPK signaling (Choi, et al., 2018a; Choi et al., 2018b). Treatment of transformed pancreatic cells with an inhibitor of SUMO E2 inhibited cell migration, suppressed expression of a mesenchymal cell marker, and concomitantly induced the epithelial cell marker zonula occludens-1 (ZO-1) (Choi, et al., 2018b). These effects correlated with a reduction in RAS SUMOylation suggesting that SUMO modification of Lys42 on RAS is an important event in tumorigenesis.
1.3.4. RAS Regulation by Allosteric Interactions
1.3.4.1. RAS Ionic Interactions
As discussed above, Mg2+ is a necessary co-factor for RAS and is coordinated by several residues within the nucleotide binding pocket (Ser15, Thr35, and Asp57) along with the nucleotide (Fig. 2) (Milburn et al., 1990; Pai et al., 1989; Pai, et al., 1990). Structures of SOS complexed with RAS showed that Mg2+ was displaced and nucleotides were excluded from the nucleotide binding pocket, indicating the importance of Mg2+ for nucleotide binding (Boriack-Sjodin, et al., 1998). Indeed, the intrinsic GDP/GTP exchange rate of RAS was increased 10-fold in low [Mg2+] compared to physiological concentrations (Hall and Self, 1986). Chelation of Mg2+ with EDTA is a common methodology in RAS biology for inducing nucleotide release from RAS [e.g. (Maurer et al., 2012)]. In addition to stabilizing nucleotide binding, Mg2+ is also important for GAP-mediated GTPase activity (Rudack, et al., 2012).
Ca2+ also allosterically regulates RAS function. Ca2+ binds RAS in the allosteric lobe by coordinating with Asp107 and Tyr137 (Buhrman et al., 2010). Ca2+ binding induced long-range conformational changes that resulted in ordering of SW2 and shifted Gln61 into the active site (Buhrman, et al., 2010). The authors proposed that RAS-Ca2+ interaction was critical for RAF-mediated GTPase activity that required Gln61, suggesting a reason that Gln61 is a mutational hot spot (Buhrman, et al., 2010). Earlier work supported this hypothesis by correlating structural features of RAS Gln61 mutants with the reduced ability of RAF to catalyze RAS GTPase activity (Buhrman et al., 2007).
1.3.4.2. RAS Interaction with the Plasma Membrane
RAS isoforms differ in how their C-termini are modified and interact with membranes (section 1.2.1.). These differences promote RAS segregation into distinct PM microdomains and depend on GDP versus GTP loading (Parker and Mattos, 2015). For example, HRAS-GDP localized to cholesterol-rich lipid rafts, but HRAS-GTP translocated to disordered membrane domains in galectin-1-stabilized clusters (Prior, et al., 2003). In contrast, inactive KRAS clustered in non-raft regions of the PM, and clustered in a distinct non-raft domain when GTP-loaded (Prior, et al., 2003; Weise et al., 2011). KRAS segregation may also be supported by galectin-3, which enhanced KRAS-mediated MAKP and PI3K signaling (Elad-Sfadia et al., 2004). Importantly, KRAS-GTP and HRAS-GTP were segregated into separate non-raft compartments, suggesting the potential for distinct signaling pathways to be activated by this compartmentalization of RAS (Parker and Mattos, 2015; Prior, et al., 2003). NRAS displays a third membrane partitioning scheme, concentrating at the interface between lipid rafts and non-ordered membrane when bound to GDP, but moving into rafts when activated (Kapoor et al., 2012; Parker and Mattos, 2015).
In addition, the activation state changes how RAS interacts with the membrane in an isoform-specific manner. GTP-loaded HRAS and NRAS reoriented to interact with the membrane through their α4 and α5 helices while activated KRAS remained more flexible (Gorfe et al., 2007; Kapoor, et al., 2012). Thus, effector interactions with HRAS and NRAS may be facilitated by membrane interactions (Gorfe, 2010). Activated KRAS interacted with the membrane in an orientation that occluded the effector-binding region. This orientation was reversed by addition of RAF or RALGDS RBDs, while the common G12V mutation enhanced an effector binding-domain exposed configuration (Mazhab-Jafari et al., 2015). These studies underscore the importance of RAS-membrane interactions in the distinct signaling profiles of the RAS isoforms and potentially point to isoform-specific therapeutic targets on the RAS molecule.
2. Therapeutic Targeting of RAS
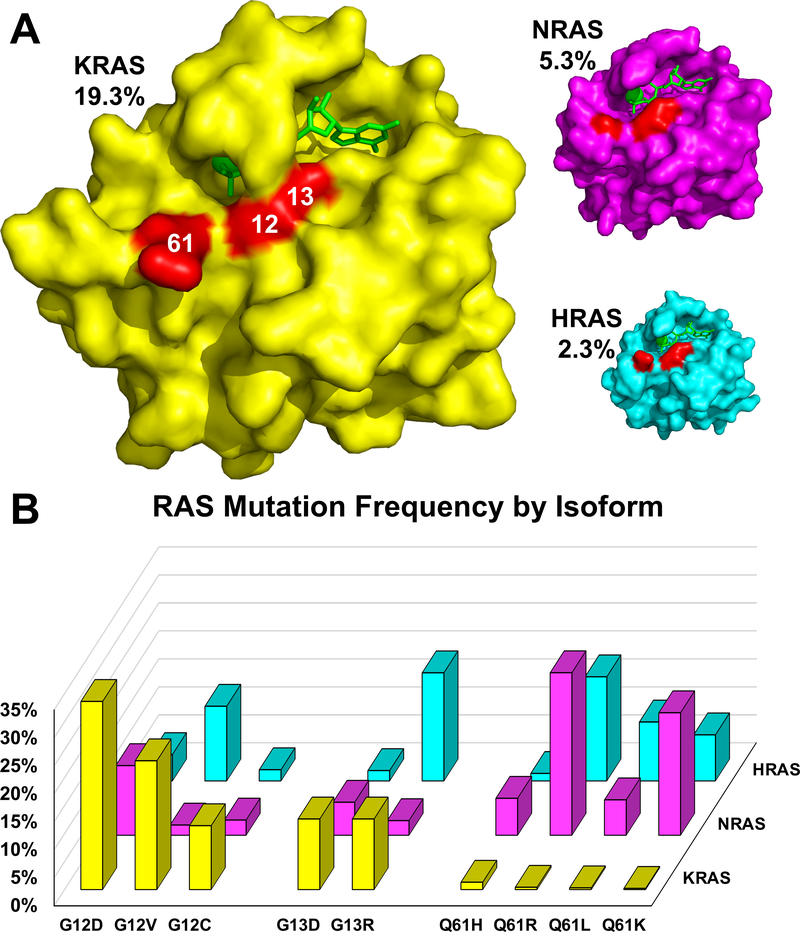
Human tumors are frequently (~27%) beset by RAS mutations and the annual number of deaths due to RAS mutant cancers worldwide is on par with deaths caused jointly by malaria and tuberculosis (Simanshu, et al., 2017). Furthermore, the importance of RAS in tumor maintenance underscores the necessity for RAS targeted therapies. The incidence of RAS mutations varies by tumor type with some cancers (e.g. pancreatic cancer) having a RAS mutation in >90% of tumors. Mutations in RAS primarily occur at codons 12,13, and 61, shifting RAS to the GTP-bound state due to impairment in both intrinsic and GAP-stimulated GTP hydrolysis (Gibbs et al., 1984; Scheffzek et al., 1997; Simanshu, et al., 2017) (Fig. 1). KRAS is most frequently mutated in solid tumors followed by NRAS and HRAS (Prior et al., 2012) (Fig. 5). KRAS mutations occur mostly at codon 12 (80%) while codon 61 mutations predominate in NRAS (60%). HRAS on the other hand displays a roughly equal distribution of mutations at codons 12, 13 and 61 (Fig. 5) (Khan, et al., 2020; O’Bryan, 2019).
Figure 5. RAS mutation frequency.
(A) Structures of wild-type KRAS (PDB:6GOD; yellow), NRAS (PDB:5UHV; magenta), and HRAS (PDB:5P21; cyan) loaded with GppNHp were aligned and sized relative to their mutation frequency in cancer (indicated as percentages). Mutation hotspots are indicated in red and labeled with codon number. (B) The frequency of specific missense mutations is graphed as the percentage of occurrences relative to the total number of mutations within each isoform. Data were compiled from the Catalogue of Somatic Mutations (COSMIC), v86 (Forbes et al., 2017).
Although decades of effort have failed to produce a single FDA-approved anti-RAS therapeutic, the recent development of compounds that pharmacologically inhibit oncogenic KRAS(G12C) provide hope that clinically viable treatments for RAS-driven cancers are on the horizon (Canon et al., 2019; Fell et al., 2018; Hallin et al., 2020; Janes et al., 2018; Ostrem et al., 2013). The following sections will discuss prior efforts to pharmacologically target oncogenic RAS, the reasons for their inadequacy, and recent findings on compounds and treatment modalities that may yet prove effective.
2.1. Indirect Inhibitors of RAS
A characteristic feature of RAS proteins is the lack of deep pockets other than the nucleotide binding pocket (Fig. 2). However, the high affinity of RAS for nucleotides (picomolar affinity) coupled with the abundance of guanine nucleotides in the cell has led to the belief that targeting the nucleotide pocket may be folly. These properties initially led many to believe that RAS may be “undruggable”. Thus, efforts focused on developing approaches to indirectly target RAS. These endeavors have included inhibiting RAS processing and trafficking, modulating RAS PTMs, and blocking RAS-mediated signaling pathways (Fig. 3 and Table 1).
Table 1.
Indirect Inhibitors of RAS
| Name | Potency | Type | Binding Site | Mechanism of Action | Model Tested | Clinical Trials | Selected References |
|---|---|---|---|---|---|---|---|
| Peptidomimetic FTase Inhibitors (FTI) | IC50 = 0.5–830nM | FTI | Catalytic domain of FTase | ↓ RAS farnesylation and membrane attachment | Cell culture; Mouse models of pancreatic, lung, and breast cancer | (James, et al., 1993; Kohl, et al., 1994; Reiss, et al., 1990) | |
| Natural Products-Based FTIs and Derivatives | IC50 = 12nm-13μM | FTI | Catalytic domain of FTase | ↓ RAS farnesylation and membrane attachment | Cell culture; Mouse fibrosarcoma model | (Gibbs, et al., 1993; Hara, et al., 1993) | |
| Lonafarnib (SCH-66336) | IC50 = 1.9nM | FTase inhibitor | FTase | ↓ RAS farnesylation and membrane attachment | Cell culture; Mouse models; Cancer clinical trials; Progeria clinical trials | Used in nearly 40 clinical trials for various cancers, progeria, and Hep D. Progeria: NCT00425607 Hep D.: NCT03719313 | (Desjardins, et al., 2011; Eskens, et al., 2001; Gordon, et al., 2018; Kieran, et al., 2007; Kim, et al., 2005; Njoroge, et al., 1998; Ravoet, et al., 2008; Yurdaydin, et al., 2018) |
| Tipifarnib (R115777) | IC50 = 7.9nM | FTase inhibitor | FTase | ↓ RAS farnesylation and membrane attachment | Cell culture; Mouse models; Clinical trials; Currently in Phase II clinical trials for head and neck tumors. | Used in over 80 trials for various cancers; Currently: NCT02383927, NCT03496766 | (Alsina, et al., 2004; Burnett, et al., 2012; Crul, et al., 2002; End, et al., 2001; Rao, et al., 2004) |
| BMS-214662 | IC50 = 100nM (HRAS); >2.5μM (KRAS) | FTase inhibitor | Catalytic domain of FTase | ↓ RAS farnesylation and membrane attachment | Cell culture; Mouse models of lung, colon, bladder, gastric, and pancreatic cancer; Phase I clinical trials – mostly solid tumors | NCT00006242 NCT00006213 NCT00005973 NCT00022529 NCT00006018 NCT00004877 | (Rose, et al., 2001) |
| L-744832 | IC50 = 2–20μM | FTase inhibitor | FTase | ↓ RAS farnesylation and membrane attachment | Cell culture; | (Sepp-Lorenzino, et al., 1995) | |
| DPI-1 and DPI-2 | IC50 = 340nM (DPI-1); 207nM (DPI-2) | Dual prenylation inhibitor (DPI) | FTase and GGTase | ↓ RAS family prenylation and membrane attachment | Cell culture; Mouse model of pancreatic cancer | (Lobell, et al., 2001) | |
| GGTI-2418 | GGTase inhibitor | GGTase | ↓ RALA, RALB, CDC42, and RAC geranylgeranylation and membrane attachment | Phase I trial for advanced solid tumors | NCT03900442 | (Karasic, et al., 2019) | |
| L-778,123 | IC50 = 100nm-162μM | DPI | FTase and GGTase | ↓ RAS family prenylation and membrane attachment | Cell culture; Phase I trial for solid and liquid tumors | NCT00003430 NCT00004057 | (Hahn, et al., 2002; Lobell, et al., 2002; Morgan, et al., 2012) |
| Bisphospohates/ Zoledronic acid | FPPS inhibitor | FPPS | ↓ farnesyl and geranylgeranyl lipid synthesis, RAS lipidation, and membrane localization | Cell culture; Mouse models and human trials of pancreatic, prostate, lung, and breast cancer; Clinical trials | Used in 100s of clinical trials, primarily for bone-related maladies | (Salzano, et al., 2016; Xu, et al., 2019) | |
| NSC1011 and derivatives | IC50 ≈ 5 – 10μM | RCE1 inhibitor | RCE1 | ↓ RAS membrane localization | Cell culture | (Mohammed, et al., 2016) | |
| Cysmethynil and derivatives | IC50 ≈ 2 – 25μM | ICMT inhibitor | ICMT | ↓ RAS membrane localization | Cell culture | (Ramanujulu, et al., 2013) | |
| Salirasib | Kd = 2 –10μM | Farnesylcysteine mimetic | Galectins | ↓ Activated RAS membrane localization | Cell culture; Mouse models of neurofibromatosis, colon cancer, and pancreatic cancer; Phase I and II trials for solid tumors | NCT00531401 | (Borthakur, et al., 2007; Bustinza-Linares, et al., 2010; Furuse, et al., 2018; Riely, et al., 2011; Rotblat, et al., 2008) |
| Palmostatin B | IC50 = 670nM | APT inhibitor | APT | ↑Random H/NRAS membrane distribution | Cell culture | (Dekker, et al., 2010) | |
| Deltarasin | Kd = 41nM | PDEδ inhibitor | Prenyl-binding pocket | ↓ KRAS/PDEδ interaction and RAS trafficking to the plasma membrane | Cell culture; Mouse pancreatic cancer model | (Zimmermann, et al., 2013) | |
| Deltazinone 1 | Kd = 8nM | PDEδ inhibitor | Prenyl-binding pocket | ↓ KRAS/PDEδ interaction and RAS trafficking to the plasma membrane | Cell culture | (Papke, et al., 2016) | |
| Deltasonamide 1 and 2 | Kd = 203pM (1) Kd = 385pM (2) |
PDEδ inhibitor | Prenyl-binding pocket | ↓ KRAS/PDEδ interaction and RAS trafficking to the plasma membrane | Cell culture | (Martin-Gago, et al., 2017) | |
| Deltaflexin-1 and -2 | IC50 ≅ 10μM | PDEδ inhibitor | Prenyl-binding pocket | ↓ KRAS/PDEδ interaction and RAS trafficking to the plasma membrane | Cell culture | (Siddiqui, et al., 2020) | |
| NSC87877 | IC50 ≅ 300nM | SHP1/2 inhibitior | Catalytic domain | ↑pY32 / ↓effector binding | Cell culture; Mouse leukemia model | (Chen, et al., 2006; Perez-Fernandez, et al., 2019; Tsutsumi, et al., 2018) | |
| II-B08 | IC50 = 5.5μM | SHP2 inhibitor | Catalytic domain | ↑pY32 / ↓effector binding | Cell culture; Mouse glioma model | (Bunda, et al., 2015; Tsutsumi, et al., 2018; Zhang, et al., 2010) | |
| 11a-1 | IC50 = 200nM | SHP2 inhibitor | Catalytic domain | ↑pY32 / ↓effector binding | 2/3D cell culture | (Kano, et al., 2019; Tsutsumi, et al., 2018; Zeng, et al., 2014a) | |
| PHPS1 | IC50 = 2.1μM; Ki = 0.73μM | SHP2 inhibitor | Catalytic domain | ↑pY32 / ↓effector binding | Ex vivo 3D-transdifferentiation assay | (Hellmuth, et al., 2008; Ruess, et al., 2018; Zhao et al., 2019) | |
| GS493 | IC50 = 71nM | SHP2 inhibitor | Catalytic domain | ↑pY32 / ↓effector binding | Cell culture; Mouse pancreatic and lung cancer models | (Grosskopf, et al., 2015; Ruess, et al., 2018; Tsutsumi, et al., 2018) | |
| cmpd #57 | SHP2 inhibitor | SHP2 | ↑pY32 / ↓effector binding | Cell culture | (Mainardi, et al., 2018) | ||
| SHP099 | IC50 = 71nM | SHP2 inhibitor | N-terminal/C-terminal/Phosphatase domain interface | ↑pY32 / ↓effector binding | Cell culture; Mouse pancreatic, lung, ovarian, and breast cancer models | (Chen, et al., 2016; Jiang, et al., 2019; Kano, et al., 2019; Pandey, et al., 2019; Wong, et al., 2018; Zhao, et al., 2019) | |
| TNO155 | SHP2 inhibitor | SHP2 | ↑pY32 / ↓effector binding | Phase I clinical trial dose finding study for patients with advanced solid tumors | NCT03114319 | ||
| JAB-3068 | SHP2 inhibitor | SHP2 | ↑pY32 / ↓effector binding | Phase I clinical trial dose finding study for patients with advanced solid tumors | NCT03565003 | ||
| RMC-4550 | EC50 = 49nM | SHP2 inhibitor | SHP2 allosteric site | ↓ Nucleotide exchange | Cell culture; Mouse xenografts | (Nichols, et al., 2018) | |
| RMC-4630 | SHP2 inhibitor | SHP2 | ↑pY32 / ↓effector binding | Phase I clinical trial dose finding study for patients with advanced relapsed or refractory solid tumors | NCT03634982 | ||
| Bryostatin-1 | PKC agonist/inhibitor | PKC | ↑pS181 on RAS/inhibition of RAS phosphorylation | Cell culture; Mouse xenografts; 30+ Phase I and II trials for solid and liquid tumors | NCT00031694 NCT00006389 | (Barcelo, et al., 2014; Bivona, et al., 2006; Lam, et al., 2010; Mohammad, et al., 1998) | |
| Prostratin | PKC agonist | PKC | Cell culture; Mouse transformed fibroblast and pancreatic cancer models | (Wang, et al., 2015) | |||
| Edelfosine | PKC inhibitor | PKC | Inhibition of RAS phosphorylation | Mouse xenografts | (Barcelo, et al., 2014) |
2.1.1. Inhibition of RAS Prenylation
RAS association with the PM is dependent on prenylation of the carboxy-terminal CaaX domain by FTase (Fig. 3). The seminal paper describing farnesylation of RAS by FTase demonstrated that peptides as short as the last 4 amino acids of all RAS isoforms (i.e. the CaaX motifs) were sufficient to inhibit FTase-mediated prenylation of RAS (Reiss, et al., 1990). Thus, were born the first FTase inhibitors (FTIs) (Table 1). Although the tetrapeptide FTIs displayed poor cellular uptake and were degraded intracellularly (Basso et al., 2006), improved peptidomimetics were developed. In one example, a methyl ester derivative of the tetrapeptide, CVIM (the KRAS4b C-terminus), inhibited delivery of RAS to the membrane and activation of MAPK signaling (Lerner et al., 1995). In addition, a number of non-peptide mimetics of the CaaX motif proved to be “true” FTIs in that they were not modified or inactivated by FTase (Nigam et al., 1993; Qian et al., 1994; Vogt et al., 1995), and restored normal growth patterns to RAS-transformed cells (James et al., 1993). A number of these compounds even showed preclinical efficacy in RAS-driven models of pancreatic cancer (Kohl et al., 1994), lung cancer (Sun et al., 1995), and breast cancer (Kohl et al., 1995).
Non-peptidomimetic FTIs were also developed. These compounds were discovered through screening of small molecules or targeting FTase-CaaX interaction, FTase-farnesyl pyrophosphate (FPP) interaction, or both. Antagonists of FTase-FPP interaction showed some efficacy in inhibiting growth of RAS-transformed cells (Gibbs et al., 1993; Hara et al., 1993). Similarly, bisubstrate FTIs (inhibitors mimicking both CaaX and FPP interaction with FTase) also substantially decreased growth of RAS-transformed cells, and restored contact inhibition and normal cytoskeletal morphology (Manne et al., 1995). However, two of the most successful FTIs were small molecule CaaX-competitive inhibitors of FTase (Basso, et al., 2006). Specifically, the compounds SCH66336 (lonafarnib) and R115777 (tipifarnib) displayed single-digit nanomolar IC50 concentrations with respect to FTase inhibition (End et al., 2001; Njoroge et al., 1998).
Certain properties of FTIs presaged the eventual disappointing results in clinical trials. First, in many cases, growth inhibition of oncogenic KRAS transformed cells and tumor cell lines with FTIs required higher doses than for HRAS inhibition, or were simply ineffective against KRAS transformed cells (End, et al., 2001; Lerner, et al., 1995; Rose et al., 2001). This was due in part to the higher affinity of FTase for the KRAS4b C-terminus versus the other RAS isoforms (Cox and Der, 1997; Reiss, et al., 1990). However, a more potent explanation resided in the discovery that KRAS and NRAS can be alternatively prenylated. In addition to FTase, two other prenylating enzymes are expressed by mammalian cells: geranylgeranyl prenyltransferase-1 and −2 (GGTase-1 and GGTase-2). FTase recognizes CaaX domains terminating in Ser, Met, Glu, and Ala, while GGTase-1 typically binds CaaX domains ending in Leu, and GGTase-2 recognizes C-termini displaying two Cys of the terminal four residues (Basso, et al., 2006). Unlike FTase, GGTases catalyze the covalent attachment of a 20-carbon geranylgeranyl moiety to the reactive C-terminal thiol group on substrate proteins (Sebti and Hamilton, 2000). A number of studies in the mid-90’s demonstrated that KRAS4b and NRAS could be prenylated by GGTase-1 when FTase was inhibited (James et al., 1995; Rowell et al., 1997; Whyte et al., 1997; Zhang et al., 1997). As a result, FTI treatment of KRAS4b-mutant cells leads to geranylgeranylation of KRAS4b, unaltered KRAS membrane association, and continued KRAS function. Given the predominance of KRAS mutations in human solid tumors, FTI clinical trials have met with disappointing results thus far.
A second unexpected characteristic of FTIs was that the sensitivity of cancer cells to FTIs was not dependent on the mutational status of RAS. In a large panel of tumor cell lines from diverse origins 70% of the lines were growth inhibited by the FTI, L-744832, but there was no correlation to either the tumor origin or the presence of mutated RAS (Sepp-Lorenzino et al., 1995). Indeed, some of the resistant cells harbored oncogenic RAS. Similarly, the FTI, BMS-214662, was potently cytotoxic to a number of the cancer cells, but without any correlation to the presence of oncogenic RAS (Rose, et al., 2001). These unanticipated results may have been due to whether or not the lines were dependent on RAS signaling, as opposed to the presence of oncogenic RAS per se (Cox and Der, 1997). That being said, a number of other proteins are farnesylated, including proteins involved in structural properties of the nucleus, apoptosis, growth and cell cycle progression, mitosis, protein folding, and stress response (Basso, et al., 2006). Reduced farnesylation of these additional FTase substrates likely accounted for the effects of FTIs on cancer cells, but suggested a number of potential non-RAS effects that may contribute to toxicity.
Nevertheless, a handful of FTIs entered clinical trials although most were not carried beyond Phase I due to significant side effects and a lack of response (Appels et al., 2005; Basso, et al., 2006; Crul et al., 2001). However, SCH66336 (lonafarnib) and R115777 (tipifarnib) have now been tested in over 100 clinical trials (Khan, et al., 2020) (Table 1). Clinical trials in cancer patients were directed at both solid and liquid tumors, and in monotherapy or combination therapy (Basso, et al., 2006). In general, treatment of solid tumors with either agent regardless of whether or not they were combined with other chemotherapies were ineffective. Some cases of partial response or stabilized disease were observed with lonafarnib monotherapy of NSCLC (Adjei et al., 2000; Eskens et al., 2001). Similarly, a case of partial response to tipifarnib was also observed in NSCLC (Crul et al., 2002), and cases of partial response and stable disease were seen in urothelial tract transitional cell carcinoma and breast cancer (Johnston et al., 2003; Rosenberg et al., 2005). However, Phase II and III trials in lung, pancreatic, and colorectal cancer failed to produce any response to tipifarnib monotherapy (Adjei et al., 2003; Cohen et al., 2003; Heymach et al., 2004; Rao et al., 2004). Comparably, lonafarnib treatment of pediatric brain tumors appeared to have positive results in a Phase I study (Kieran et al., 2007), but a dose-finding study in malignant glioma reported severe adverse events (Desjardins et al., 2011), and further trials utilizing lonafarnib to treat brain tumors have not been reported.
Combining either lonafarnib or tipifarnib with chemotherapeutics did not significantly improve the outcomes of solid tumor clinical trials. For example, combining tipifarnib with gemcitabine to treat pancreatic cancer did not improve survival (Van Cutsem et al., 2004), and combining tamoxifen with tipifarnib to treat breast cancer only yielded a handful of partial responses or stable disease cases (Lebowitz et al., 2005). Lonafarnib combined with paclitaxel to treat NSCLC was well tolerated and gave some promising results in Phase II trials (Kim et al., 2005), however, subsequent Phase III trials were terminated due to insufficient activity (Basso, et al., 2006).
In contrast to studies on solid tumors, liquid tumors showed more promising initial results. In a Phase I trial treating patients with refractory and relapsed acute leukemias with tipifarnib 10 of 35 patients responded including 2 complete remissions (Karp et al., 2001). However, a later Phase II/III trial combining tipifarnib with cytarabine showed no effects on response or survival in patients with acute myeloid leukemia (AML) (Burnett et al., 2012). Although positive responses were also observed in a number of other blood malignancies (Alsina et al., 2004; Cortes et al., 2003; Kurzrock et al., 2004; Kurzrock et al., 2003), tipifarnib was rejected for approval in the treatment of AML due to an insufficient complete response rate (Basso, et al., 2006). Studies in which blood cancer patients were treated with lonafarnib showed early promising results (Borthakur et al., 2006; Cortes et al., 2007), but the most recent Phase II trial reported only limited activity in patients with myelodysplastic syndrome or secondary AML (Ravoet et al., 2008).
Despite the bleak results from clinical trials, FTIs may yet show clinical benefit. Tipifarnib is currently in Phase II trials to treat squamous head and neck cancer in patients with mutant HRAS (which is not alternatively prenylated) (ClinicalTrials.gov Identifier: NCT02383927). Additionally, conditions that arise from other proteins dependent on prenylation may benefit from FTIs. One such case is hepatitis D, which is caused by infections with the hepatitis delta virus (HDV) (Hughes et al., 2011). Production of HDV viral particles is dependent on prenylation of the large delta hepatitis antigen (Bordier et al., 2002), and therefore clinical trials have been carried out to test the effectiveness of lonafarnib in treating HDV infections including ongoing Phase II trials (ClinicalTrials.gov Identifier: NCT03719313). Published reports indicate that 4 weeks of lonafarnib treatment is effective in lowering viral titers in a manner that correlates to serum concentrations of the drug (Koh et al., 2015). More recently, 6 of 12 HDV patients receiving lonafarnib displayed negativity for HDV RNA when treated for a longer, 12-week course, indicating the clinical viability of lonafarnib in treating HDV (Yurdaydin et al., 2018).
Lamin A is another major target of FTase. Mutation of LMNA, the gene encoding lamin A, gives rise to multiple diseases (Burke and Stewart, 2002; Reiss, et al., 1990) including Hutchinson-Gilford Progeria Syndrome (HGPS) (De Sandre-Giovannoli et al., 2003; Eriksson et al., 2003). HGPS is a rare condition that results in premature ageing characterized by early hair loss, a thin nose and lips, prominent eyes and ears, and an average life span of 13 years (Merideth et al., 2008). HGPS mutations in LMNA result in a dominant negative (DN) form of lamin A (Eriksson, et al., 2003). Normally, after lamin A is farnesylated, the last 15 residues of lamin A are cleaved off, removing the farnesylated portion of the protein and producing a mature lamin protein that helps form the nuclear envelope (Reddy and Comai, 2012). In HGPS, lamin A mutations create a version of the protein that cannot be cleaved, and the persistently farnesylated protein intercalates in the nuclear envelope causing cellular damage (Casasola et al., 2016). HGPS patients treated with lonafarnib in two Phase II clinical trials exhibited reduced mortality rates and improvements in weight gain, cardiovascular measures, skeletal rigidity, and hearing (Gordon et al., 2012; Gordon et al., 2018). Although not a cure, these trials are a start to improving morbidity and mortality in HGPS patients.
Since KRAS and NRAS are alternatively prenylated (James, et al., 1995; Rowell, et al., 1997; Whyte, et al., 1997; Zhang, et al., 1997), inhibition of both FTase and GGTase has been suggested as an alternate treatment strategy. Treatment of tumor cells with combined FTIs and GGTIs or dual prenylation inhibitors (DPIs) that block both FTase and GGTase was more effective at reducing KRAS prenylation and inducing apoptosis than either FTIs or GGTIs alone (Lobell et al., 2001). Animals infused with either combined FTIs and GGTIs or DPIs for 72hrs at concentrations high enough to prevent KRAS prenylation died within two weeks (Lobell, et al., 2001). Although 24hr infusions were tolerated, head to head comparison of an FTI against a DPI showed that the two treatments had comparable outcomes, but neither treatment showed a statistically significant increase in tumor cell apoptosis (Lobell, et al., 2001). Thus, the dose-limiting toxicity of combined FTIs and GGTIs or DPIs would likely limit the therapeutic benefit of these treatment strategies.
Despite these concerns, there were reasons to believe that GGTI-treatment alone could be more effective in treating cancers. A number of RAS-superfamily proteins that are downstream of RAS or essential for RAS-driven tumorigenesis and metastasis are exclusively geranylgeranylated including RALA, RALB, CDC42, and RAC (Berndt et al., 2011). Therefore, clinical trials were initiated using the inhibitor, GGTI-2418. Thus far, no objective responses have been observed and one trial has been terminated due to lack of efficacy (Karasic et al., 2019) (see also ClinicalTrials.gov Identifier: NCT03900442 and https://drugs.ncats.io/drug/M67G28K74K). The DPI, L-778,123, was also tested in clinical trials. Inhibition of prenylation of both FTI and GGTI substrates could be observed in blood cells, but inhibition of KRAS prenylation was not (Britten et al., 2001; Lobell et al., 2002). However, a Phase I study with nine NSCLC or head and neck cancer patients given L-778,123 in combination with radiotherapy found one partial and five complete responses, indicating the potential efficacy of L-778,123 (Hahn et al., 2002). A Phase I dosing study conducted in pancreatic cancer patients concomitantly given L-778,123 and radiotherapy reported “acceptable toxicity” levels and one partial response (Martin et al., 2004). More recently, L-778,123 treatment of leukemia cell lines in vitro suggested that this compound could be effective in treating AML (Onono et al., 2008), especially in combination with chemotherapeutics (Morgan et al., 2012). However, no further clinical work with this inhibitor has been reported.
Finally, inhibition of FPP synthase (FPPS) has been tested as a way to prevent RAS prenylation by limiting the amount of FPP and geranylgeranyl pyrophosphate (GGPP) substrate available for the FTase and GGTase reactions. Bisphosphonates, such as zoledronate, are a class of drugs that act as synthetic analogs of pyrophosphate and inhibit FPPS (Gong et al., 2011). The aminobisphosphonate, zoledronic acid (ZOL), induced tumor cell apoptosis, inhibited tumor growth in a number of in vitro models (Marra et al., 2012), as well as inhibited RAS prenylation and signaling (Santini et al., 2006; Xu et al., 2019). However, ZOL is rapidly cleared from the blood through excretion and uptake by bone tissue (Caraglia et al., 2010). New nanoparticle formulations have been developed to facilitate targeted delivery and prevent clearance of ZOL that have shown effectiveness in mice harboring glioblastoma and prostate adenocarcinoma xenografts (Marra, et al., 2012; Marra et al., 2011; Porru et al., 2014; Salzano et al., 2016). Clinical trials utilizing ZOL to treat RAS-driven cancers have yet to be reported (Table 1).
2.1.2. Targeting Post-Prenylation RAS Processing
Disrupting proteolytic cleavage of prenylated RAS by RCE1 or blocking methylation of cleaved RAS by ICMT would be predicted to retard RAS trafficking to the PM and inhibit RAS signaling. However, results have been mixed. RCE1 inactivation worsened KRAS-driven myeloproliferative disease whereas ICMT inactivation reduced tumor development (Wahlstrom et al., 2007; Wahlstrom et al., 2008). In contrast, ICMT inactivation accelerated neoplastic progression in a KRAS-driven PDAC mouse model (Court et al., 2013).
Despite these results, small molecule inhibitors of RCE1 and ICMT have been developed (Table 1). While some small molecules disrupted RAS membrane localization in HCT-116 cells (Mohammed et al., 2016), the most effective in vitro inhibitors were the substrate mimetics that were not cell permeable (Hampton et al., 2018). Although additional work is required to develop RCE1 inhibitors with more drug-like properties, it remains unclear whether such compounds would be effective at treating malignancies given the transgenic animal studies referenced above.
N-Acetyl-S-farnesyl-l-cysteine based inhibitors compete with farnesylated proteins for ICMT binding and inhibit RAS carboxymethylation in cells (Volker et al., 1991). Similarly, cysmethynil competitively inhibited ICMT, resulting in defective RAS trafficking, reduced signaling, and autophagic cell death [(Ramanujulu et al., 2013) and reviewed in (Yang et al., 2017)]. However, current inhibitors are too limited in their efficacy, safety, and solubility profiles for clinical use (Yang, et al., 2017).
The farnesylcysteine mimetic Salirasib inhibits RAS by blocking interaction with galectins, which facilitate nanoclustering of activated RAS (Table 1) (Elad-Sfadia, et al., 2004; Prior, et al., 2003; Rotblat et al., 2008; Weise, et al., 2011). As such, Salirasib selectively targets activated RAS, disrupting RAS association with the PM, inhibiting RAS signaling, and attenuating growth of RAS-transformed cells and cancer cell lines in vitro and in vivo [reviewed in (Rotblat, et al., 2008)]. Phase I clinical trials of Salirasib indicated minimal toxicity (Badar et al., 2015; Borthakur et al., 2007; Bustinza-Linares et al., 2010; Furuse et al., 2018; Laheru et al., 2012). However, a Phase II trial of Salirasib in mutant KRAS lung adenocarcinoma failed to show any response [(Riely et al., 2011); ClinicalTrials.gov Identifier: NCT00531401]. No further clinical trials have been reported at this time.
In the case of HRAS and NRAS, palmitoylation is another post-prenylation processing step that could be targeted to reduce RAS localization to the PM, and thereby inhibit RAS signaling. As discussed in section 1.2.2.1., the DHHC9-GPC16 protein complex catalyzes the transfer of a palmitate moiety onto one or two of the cysteine residues (depending on the isoform) in the RAS HVR N-terminal to the farnesylated cysteine (Swarthout, et al., 2005). There are a number of other DHHC (Asp-His-His-Cys)-containing proteins that function as PATs [reviewed in (Lobo, 2013)], and some also palmitoylate RAS proteins and/or are considered oncogenes (Ducker et al., 2004; Lobo, 2013). To date, no specific or potent PAT inhibitors have been developed (Lobo, 2013). The existing inhibitors tend to be highly cytotoxic and there are a number of issues that make screening for selective DHHC protein inhibitors difficult (e.g. high level of homology in the active site of PATs; inability to purify active PAT), limiting the drive to develop this class of compounds (Lobo, 2013). Counterintuitively, blocking depalmitoylation by inhibiting APT has also been studied as an approach to inhibit RAS (Dekker et al., 2010). The concept behind this work centered around the observation that a palmitoylation/depalmitoylation cycle is needed to maintain proper localization of RAS proteins (Rocks, et al., 2005). Surprisingly, the inhibitor palmostatin B reduced depalmitoylation and induced a random HRAS and NRAS membrane distribution in cells (Dekker, et al., 2010). These effects correlated with a phenotypic reversion of HRAS(G12V) transformed cells treated with palmostatin B (Dekker, et al., 2010).
2.1.3. Inhibition of RAS Trafficking to the Membrane
After prenylation and processing in the ER, the next step at which RAS could be blocked is trafficking to the PM. In the case of HRAS and NRAS this would entail targeting classical secretory vesicle trafficking, which would necessarily be non-specific. Moreover, there is evidence that HRAS has alternative trafficking pathways (Zheng et al., 2007). Conversely, KRAS is chaperoned to the membrane through its interaction with the δ subunit of phosphodiesterase 6 (PDEδ) (Chandra, et al., 2011). Similar to the palmitoylation/depalmitoylation cycle of NRAS and HRAS, cyclic KRAS-PDEδ and KRAS-membrane interaction is thought to prevent the random distribution of KRAS in endomembranes and facilitate its trafficking to the PM (Schmick et al., 2014). Therefore, PDEδ inhibitors were predicted to reduce KRAS enrichment in the PM and decrease KRAS signaling. High-throughput screening followed by structure-guided design was used to develop the compound deltarasin which bound to the prenyl-binding pocket of PDEδ with nanomolar affinity and displayed good solubility and membrane permeability (Zimmermann et al., 2013). Deltarasin rapidly and effectively blocked KRAS-PDEδ interaction in this system, displaying an “in cell” affinity surprisingly similar to the affinity of deltarasin for isolated PDEδ (41nM) (Zimmermann, et al., 2013). Deltarasin treatment of PDAC cell lines resulted in KRAS redistribution to endomembranes, inhibited proliferation, induced cell death, reduced ERK1/2 phosphorylation, and significantly attenuated tumor growth in a PDAC mouse xenograft model (Zimmermann, et al., 2013).
Unfortunately, deltarasin bound to other proteins at the effective concentrations leading to non-specific cytotoxicity (Papke et al., 2016). A novel class of PDEδ inhibitors was therefore developed, leading to a new compound, deltazinone 1, which inhibited PDEδ-KRAS interaction, RAS signaling, and growth of PDAC cells, but without the general cytotoxicity of deltarasin (Papke, et al., 2016). Although these compounds bound to PDEδ with nanomolar affinity, micromolar concentrations were required for growth inhibition in cells (Martin-Gago et al., 2017). Deltasonamide 1 and 2 represent a new generation of molecules that bind PDEδ with picomolar affinity (Martin-Gago, et al., 2017). Like their predecessors, these compounds inhibited KRAS-PDEδ interaction, affected KRAS membrane redistribution, and inhibited PDAC cell growth; however, low membrane partitioning coefficients limited the availability of these compounds in the cytosol (Martin-Gago, et al., 2017). Therefore, further development is required to improve the membrane solubility of these compounds.
A new class of PDEδ inhibitor has been developed with enhanced cell penetration properties. While some PDEδ inhibitors are “ejected” from PDEδ by Arl-2 due to the same mechanism that unloads KRAS from PDEδ (see section 1.2.1), deltaflexin-1 and −2 were designed to include moieties to prevent unloading of the inhibitors from PDEδ and enhance cell penetration (Siddiqui et al., 2020). Deltaflexin-1 and −2 bound to PDEδ with KDs of 3.6μM and 2.9μM, respectively, reduced FRET between KRAS(G12V) and PDEδ, and reduced nanoclustering of KRAS(G12V) with an IC50 of ~1.7μM (Siddiqui, et al., 2020). Concordantly, Deltaflexin-1 inhibited the proliferation of cells expressing mutant KRAS (IC50 ≅ 10μM), but not HRAS or wild-type RAS. Both compounds inhibited tumorsphere formation, indicating the potential for these compounds to reduced tumor growth in vivo (Table 1) (Siddiqui, et al., 2020).
2.1.4. Targeting Other RAS Post-Translational Modifications
2.1.4.1. SHP2 Inhibition
RAS phosphorylation has been described as having both inhibitory and activating effects on RAS (see section 1.3.3.1.). Therefore, compounds that both agonize and antagonize RAS kinases and phosphatases have been studied for their effects on the ability of RAS to promote oncogenesis. The best studied of the compounds targeting RAS phosphorylation are those that inhibit SHP2 (discussed in section 1.3.3.1.). SHP2 is one of two non-receptor protein Tyr phosphatases discovered in the mid-1990s and determined to play a role in RAS signaling (Deb et al., 1998; Feng, 1993; Freeman et al., 1992; Noguchi et al., 1994). The two N-terminal SH2 domains negatively regulate the phosphatase domain (Hof et al., 1998). SHP2 inhibitors were developed (e.g. NSC87877) that bound the catalytic cleft of both SHP1 and SHP2, inhibited their activity, and attenuated activation of RAS and MAPK (Chen et al., 2006). NSC87877 reduced proliferation of leukemia cell lines and enhanced survival of mice with HL-60 xenografts when co-treated with a PKC activator, indicating that oncogenic RAS is susceptible to SHP2 inhibition in vivo (Perez-Fernandez et al., 2019). II-B08 bound the catalytic domain of SHP2 and inhibited EGF-stimulated MAPK activation in HEK293 and NIH 3T3 cells (Zhang et al., 2010). Importantly, II-B08 also inhibited proliferation and growth of astrocytes derived from an HRAS(G12V)-driven glioblastoma mouse model and glioblastoma cell lines, as well as reduced tumor progression and size in a spontaneous transgenic glioma mouse model (Bunda, et al., 2015). Similarly, 11a-1 bound the SHP2 active site, inhibited MAPK activation, and reduced proliferation of cancer cell lines (Zeng et al., 2014a). 11a-1 also inhibited EGF-induced MAPK activation, increased markers of apoptosis in PDAC cell lines and PDAC patient derived xenograft (PDX) cells, and reduced growth and viability of PDAC cells grown in 3D tissue culture (Kano, et al., 2019). PHPS1 also targeted the SHP2 catalytic domain and blocked MAPK activation in tumor cell lines (Hellmuth et al., 2008). Chemical optimization of PHPS1 led to GS493, which inhibited epithelial-to-mesenchymal transition of PDAC cells and reduced growth of NSCLC cells in soft agar and mouse xenografts (Grosskopf et al., 2015). Importantly, while GS493 alone only modestly reduced tumor progression in established mouse PDAC and NSCLC models, co-treatment with MEK inhibitors reduced ERK activation, decreased tumor proliferation, and induced tumor regression (Ruess, et al., 2018). Analogously, the SHP2 inhibitor, compound #57, failed to affect proliferation in NSCLC cells alone, but acted synergistically with MEK inhibitors to attenuate cell growth (Mainardi et al., 2018). These results suggested that SHP2 dephosphorylation of RAS is involved in mechanisms of resistance to MEK inhibitors. However, caution must be used in interpreting data from studies with NSC-87877, IIB-08, 11a-1, and GS493, as these inhibitors have off-target effects (Tsutsumi et al., 2018).
The most well studied of the SHP2 inhibitors, SHP099, binds an allosteric site and stabilizes the auto-inhibited conformation of SHP2 (Chen et al., 2016; Garcia Fortanet et al., 2016). Cell lines expressing oncogenic RTKs or possessing RTK amplification were sensitive to SHP099, but lines with RAS or BRAF mutations were resistant (Chen, et al., 2016). SHP099 also inhibited an esophageal cancer mouse xenograft model and nearly eliminated circulating RTK-driven AML cells in an orthotopic mouse model (Chen, et al., 2016). Concordantly, SHP099 reduced circulating tumor cell numbers and splenomegaly in a mouse bone marrow transplant model of resistant AML expressing mutant FLT3 (Pandey et al., 2019).
In contrast to the resistance of RAS-driven hematopoietic cancer and colorectal cancer cell lines to SHP099, SHP099 was effective against a variety of cancer cells harboring mutant KRAS, especially when combined with MEK inhibition. Specifically, SHP099 reduced the viability of PDAC organoids and significantly inhibited tumor growth in PDAC xenografts in mice (Kano, et al., 2019). In addition, KRAS mutant PDAC PDX organoids were sensitized to the MEK inhibitor trametinib by SHP099 (Ruess, et al., 2018). Similarly, while KRAS mutant lung cancer cell proliferation was not inhibited by SHP099 alone, combination treatment with MEK inhibitors synergistically inhibited cell growth of lung, pancreatic, and colorectal cancer cell lines (Jiang et al., 2019; Mainardi, et al., 2018). However, SHP099 treatment alone was sufficient to stop NSCLC mouse xenograft tumor growth (Mainardi, et al., 2018). This discrepancy was found to be due to SHP2 inhibition-induced senescence in growth factor-limited conditions which was enhanced by MEK inhibitors (Mainardi, et al., 2018). Comparable results were observed with combined trametinib and SHP099 treatment in KRAS-mutant pancreas, lung, ovarian, and gastric cancers, and triple-negative breast cancers expressing wild type RAS (Fedele et al., 2018; Wong et al., 2018).
Sensitivity to SHP099 may be dependent on the specific RAS mutation. While cancer cells with KRAS(G12) mutations responded to SHP2/MEK co-inhibition, KRAS(G13D), KRAS(Q61K), and NRAS(Q61R) mutant cells were insensitive (Ahmed et al., 2019). Interestingly, a subset of BRAF mutant cells were sensitive to SHP099 co-treatment with BRAF inhibitors suggesting that SHP2 activity may be a general mechanism of resistance for cells with mutations or amplifications in the RTK-RAS-RAF signaling pathway.
RMC-4550, an allosteric inhibitor similar to SHP009, inhibited KRAS(G12C)-mutant, NF1 mutant, or class 3 BRAF mutant but was ineffective against tumors driven by other KRAS mutations (Nichols et al., 2018). Importantly, RMC-4550 inhibited SHP2 association with GRB2 and the RTK-proximal scaffolding protein GAB1, but cells expressing a constitutively active form of SOS were insensitive to RMC-4550-mediated reduction of MAPK signaling (Nichols, et al., 2018). These results suggested that SHP2 may regulate SOS-mediated activation of RAS (Nichols, et al., 2018) as opposed to directly regulating RAS dephosphorylation.
2.1.4.2. Kinase Agonists and Antagonists
As discussed earlier, PKC phosphorylation of RAS was found to promote apoptosis (Ballester, et al., 1987; Bivona, et al., 2006). Therefore, PKC activation has been studied as a means to treat RAS-driven cancers. Treatment of mice bearing PDAC xenograft tumors with Bryostatin-1, an acute PKC agonist, resulted in significant inhibition of tumor growth (Mohammad et al., 1998). However, in addition to the fact that PKC has many cellular targets (Nestler EJ, Lippincott-Raven, 1999), Bryostatin-1 also inhibits PKC under prolonged exposure (Kortmansky and Schwartz, 2003). Given the daily administration of Bryostatin-1 (Mohammad, et al., 1998), it remained unclear whether agonism or antagonism of RAS phosphorylation was involved in the anti-tumor properties of Bryostatin-1 in this context. Reduced growth of Bryostatin-1 treated KRAS(G12V) fibroblasts, but not KRAS(G12V/S181A) fibroblasts, was observed in vitro and in vivo, thus providing evidence that Bryostatin-1 acts mechanistically by promoting Ser181 phosphorylation (Bivona, et al., 2006). In a Phase II study combining paclitaxel and Bryostatin-1 to treat advanced pancreatic cancer, no objective responses were observed and treatment was discontinued due to progressive disease and significant adverse effects (ClinicalTrials.gov Identifier: NCT00031694) (Lam et al., 2010). Similarly, a Phase II study for treating patients with metastatic or unresectable stomach cancer that combined cisplatin with Bryostatin-1 was also discontinued due to lack of response (Table 1) (ClinicalTrials.gov Identifier: NCT00006389).
Consistent with the above results, the atypical PKC activator, prostratin, inhibited KRAS-CaM interaction in KRAS mutant PDAC cells and transformed cells (Wang et al., 2015). Prostratin also decreased the viability of KRAS(G12V) transformed fibroblasts and pancreatic cancer cells, but not KRAS(G12V/S181D) transformed fibroblasts (Wang, et al., 2015). Importantly, the number and volume of tumors formed by KRAS(G12V)-mutant fibroblasts and PDAC xenografts was reduced by prostratin (Wang, et al., 2015). Moreover, orthotopic mouse KRAS mutant PDAC tumor models displayed reduced tumor size, tumor burden, and metastasis in response to prostratin (Wang, et al., 2015). Established tumor growth was also inhibited by prostratin in both subcutaneous and orthotopic xenograft models (Wang, et al., 2015). Similarly, inhibition of PKG-mediated KRAS Ser181 phosphorylation attenuated proliferation in NSCLC cells (Cho, et al., 2016). These results suggested that phosphorylation of Ser181 was antagonistic to RAS function.
In contrast to the above studies, KRAS(G12V)-transformed xenograft tumor growth was enhanced by expression of KRAS(G12V/S181D) (i.e. a phosphomimetic mutant), and nearly abolished in xenografts expressing KRAS(G12V/S181A) (Barcelo et al., 2014). Similar inhibition of tumor grow was observed in xenografts with the PKC inhibitors, Edelfosine and Bryostatin-1 (Barcelo, et al., 2014). While these studies indicate the potential of targeting protein kinases in RAS-driven cancers, the mechanism by which they act on mutant RAS remains unclear: some of the studies indicate a pro-tumorigenic role for RAS-phosphorylation while others suggest it is anti-tumorigenic. Furthermore, the ultimate practicality of this approach remains in doubt as approval of drugs targeting PKC in the clinic has remained elusive (Mochly-Rosen et al., 2012).
2.1.4.2. Co-Targeting Autophagy and MAPK Signaling
Although inhibitors targeting signaling pathways downstream of RAS are relevant to treating RAS-driven and RAS-dependent cancers, it is outside the scope of this chapter. There are a number of excellent reviews that discuss such approaches [e.g. (Baines, et al., 2011; Liu et al., 2018)]. Nevertheless, there are a handful of recent papers that have shown success in treating oncogenic RAS-dependent cancers by co-treating with inhibitors of MAPK signaling and autophagy inhibitors.
In a physiological context, autophagy is a cellular quality control process that recycles misfolded or aggregated cytoplasmic proteins and damaged organelles (Hansen and Johansen, 2011). In addition, autophagy is a stress response to starvation that provides cells with nutrients through digestion of cellular organelles, or if starvation persists, initiation of a distinct form of cell death called autophagic cell death (Galluzzi et al., 2018; Kroemer and Levine, 2008). Importantly, due to their nutrient and oxygen deprived microenvironment, tumor cells utilize autophagy (Degenhardt et al., 2006), and autophagy is increasingly recognized to play a role in acquisition of drug resistance in cancer (Sui et al., 2013; Zambrano and Yeh, 2016).
Pancreatic cancers depend on autophagy for tumor growth (Yang et al., 2011). Multiple KRAS-mutant PDAC cell lines displayed increased levels of autophagic flux in response to inhibition of the RAS-MAPK pathway (Bryant et al., 2019; Kinsey et al., 2019). Although treatment of PDACs with inhibitors of the MAPK pathway have proven ineffective clinically (Liu, et al., 2018), treatment of PDAC cells and tumor xenografts with a combination of an autophagy inhibitor and an inhibitor to a member of the RAS-RAF-MAPK pathway resulted in a synergistic inhibition of PDAC cells in culture as well as tumors in mice (Bryant, et al., 2019; Kinsey, et al., 2019). This synergistic inhibition was not limited to PDAC but was also seen in NRAS-mutant melanomas and BRAF-mutant colorectal cancers. Similar results were observed in KRAS-mutant colorectal cancer cells and PDAC cells by siRNA co-depletion of CRAF, BRAF, and the autophagy E1 ligase ATG7 with minimal effects on wild-type cells (Lee et al., 2019). Perhaps most striking, compassionate treatment of a PDAC patient refractory to standard-of-care treatment with trametinib (a MEK inhibitor) and hydroxychloroquine (an autophagy inhibitor) resulted in reduced cancer pain, reduced CA19–9 levels, a significant reduction in the primary tumor, and near complete resolution of the metastatic liver lesions (Kinsey, et al., 2019). Thus, combinatorial inhibition of the MAPK and autophagy pathways may provide a novel therapeutic approach for an historically intractable disease. Indeed, several clinical trials are currently in progress to assess this approach in PDAC as well as other RAS or RAF mutant cancers (ClinicalTrials.gov Identifier: NCT03979651; NCT04214418; NCT04132505; NCT03754179; NCT02257424).
2.2. Direct Inhibitors of RAS
Targeting RAS directly by small molecules has been challenging due to a lack of amenable binding pockets. Design of inhibitors targeting the nucleotide pocket has not been considered possible due to the picomolar affinity of RAS for nucleotides coupled with the millimolar levels of guanine nucleotides in cellulo (Cox et al., 2014). To circumvent such challenges, alternative approaches have been employed. Virtual screening of small molecules, peptides targeting RAS-effector or RAS-GEF interactions, medicinal chemistry to target specific mutated residues, and generation of RAS biologics (e.g. antibodies, Monobodies, and DARPins) have identified numerous lead compounds. However, their efficacy has been limited to either cell lines or mouse models and most have not advanced to clinical evaluation. Nevertheless, a few promising leads have been developed, and some have shown early clinical promise, renewing hope that oncogenic RAS will become a more tractable therapeutic target in the near future (Table 2).
Table 2.
Direct RAS Inhibitors
| Name | Potency | Type | Binding Site | Mechanism of Action | Model Tested | Clinical Trials | Selected References |
|---|---|---|---|---|---|---|---|
| Sulindac | Small molecule | Unknown | ↓ GEF nucleotide exchange, GAP GTPase, RAF interaction | Cell culture; Rat breast cancer model; Clinical trials | Nearly 40 clinical trials in various cancers | (Gurpinar, et al., 2014; Herrmann, et al., 1998; Thompson, et al., 1995; Thompson, et al., 1997) | |
| 643000 and 117028 | Small molecule | P3 pocket? | ↓ RAS-GTP | Cell culture | (Grant, et al., 2011) | ||
| DCAI | EC50 = 15.8μM | Small molecule | P1 pocket | ↓ SOS interaction | Cell culture | (Maurer, et al., 2012) | |
| Cmpd 12 | KD = 190μM | Small molecule | P1 pocket | ↓ Nucleotide exchange | In vitro | (Sun, et al., 2012) | |
| Kobe0065 and Kobe 2602 | IC50 = 10 – 20μM | Small molecule | P1 pocket - state 1 of SW1 | ↓ SOS-mediated nucleotide exchange and RAF interaction | Cell culture; Mouse colon carcinoma model | (Prakash, et al., 2015a; Shima, et al., 2013) | |
| BI-2852 | EC50 = 5.8 – 6.7μM | Small molecule | Pocket between SW1/SW2 | ↓ GEF, GAP, and effector interactions | Cell culture | (Kessler, et al., 2019) | |
| Cmpd 11 | IC50 = 5μM | Small molecule | P1 pocket | ↓ RAF interaction | Cell culture | (McCarthy, et al., 2019) | |
| Cmpd2 | EC50 = 2.7μM | Small molecule | Membrane/P1 pocket | ↓ Effector engagement | Cell culture | (Fang, et al., 2018; Jansen, et al., 2017) | |
| SCH-53239 SCH-53870 SCH-54292 | IC50 ≈ 10 – 20μM | Small molecule | Pocket adjacent SW2 | ↓ Nucleotide exchange | Cell culture | (Peri, et al., 2005; Taveras, et al., 1997) | |
| Cmpd4 | > 100μM needed for RAS inhibition | Small molecule | SOS | ↑ Nucleotide exchange → negative feedback | Cell culture | (Burns, et al., 2014; Howes, et al., 2018) | |
| cmpd17 cmpd19 | > 50μM needed for RAS inhibition | Small molecule | SOS | ↑ Nucleotide exchange → negative feedback | In vitro | (Abbott, et al., 2018) | |
| BAY-293 | IC50 ≈ 1 – 3μM | Small molecule | SOS | ↓ SOS interaction | Cell culture | (Hillig, et al., 2019) | |
| HBS3 | Kd = 28 – 158μM | Peptide | SOS1 interaction site | ↓ SOS interaction and nucleotide exchange | Cell culture | (Patgiri, et al., 2011) | |
| SAH-SOS1A | IC50 = 5 – 15μM | Peptide | SOS1 interaction site | ↓ SOS interaction and nucleotide exchange | Cell culture | (Leshchiner, et al., 2015) | |
| RASGRF1-TAT | Activity observed at 500nM | Peptide | RASGRF1 interaction site | ↓ Nucleotide exchange | Cell culture | (Sacco, et al., 2012) | |
| MCP1 and Derivatives | IC50 = 17.9μM | Small molecule | Unknown | ↓ RAF interaction | Cell culture; Mouse tumor models | (Gonzalez-Perez, et al., 2010; Skobeleva, et al., 2007) | |
| 3144 | IC50= 3.8μM | Small molecule | SW1/SW2 | ↓ Effector interaction | Cell culture; Mouse breast and pancreatic cancer models | (Welsch, et al., 2017) | |
| cyclorasin 9A5 | IC50 = 0.12μM | Cyclic peptide | RAS-GTP SW1 loop | ↓ RAF interaction | Cell culture | (Upadhyaya, et al., 2015) | |
| Cmpd 12 | EC50 = 0.32μM | Small molecule | SII-P/C12 of RAS(G12C) | Disrupts SW1/2 conformation; traps KRAS in a GDP-bound state; ↓ interaction with effectors and activators | Cell culture | (Ostrem, et al., 2013) | |
| ARS-853 | IC50 ≈ 2μM | Small molecule | SII-P/C12 of RAS(G12C) | Disrupts SW1/2 conformation; traps KRAS in a GDP-bound state; ↓ interaction with effectors and activators | Cell culture | (Patricelli, et al., 2016) | |
| ARS-1620 | IC50 = 120nM | Small molecule | SII-P/C12 of RAS(G12C) | Disrupts SW1/2 conformation; traps KRAS in a GDP-bound state; ↓ interaction with effectors and activators | Cell culture; Mouse pancreatic and lung cancer models | (Janes, et al., 2018) | |
| MRTX849 | IC50 ≅ 10nM | Small molecule | SII-P/C12 of RAS(G12C) | Disrupts SW1/2 conformation; traps KRAS in a GDP-bound state; ↓ interaction with effectors and activators | Cell culture; Mouse pancreatic and lung cancer models; Phase I/II clinical trial for patients with advanced solid tumors with KRAS G12C mutation | NCT03785249; Positive results in lung and colon adenocarcinoma patients | (Hallin, et al., 2020) |
| AMG 510 | IC50 = 4 – 32nM | Small molecule | SII-P/C12 of RAS(G12C) | Disrupts SW1/2 conformation; traps KRAS in a GDP-bound state; ↓ interaction with effectors and activators | Cell culture: Mouse pancreatic and lung cancer models: Phase I/II clinical trial for patients with advanced solid tumors with KRAS G12C mutation | NCT03600883; Positive results in lung cancer patients | (Canon, et al., 2019) |
| 2C07 | βME50 = 1.10 – 2.53mM | Small molecule | SII-G | Stabilizes GDP state; ↓ RAS/SOS interaction and nucleotide exchange and PI3K interaction | In vitro | (Gentile, et al., 2017) | |
| Y13–259 | Monoclonal antibody | HRAS SW2 | Sequestration of RAS in intracellular aggregates | Cell culture | (Cardinale, et al., 1998; Furth, et al., 1982; Mulcahy, et al., 1985) | ||
| anti-p21ser | Monoclonal antibody | Viral KRAS(G12S) residues 5–16 | ↓ GTP loading | Cell culture | (Clark, et al., 1985; Feramisco, et al., 1985) | ||
| iDab#6 | Kd = 26 – 180nM | Intrabody | SW1/SW2 of RAS-GTP | ↓ Effector interaction | Cell culture; Mouse fibrosarcoma, colorectal cancer, and lung cancer models | (Quevedo, et al., 2018; Tanaka, et al., 2003; Tanaka and Rabbitts, 2010; Tanaka, et al., 2007) | |
| Abd-7 | IC50 = 8 – 10μM | Small molecule | P1 pocket | ↓ PI3K, RAF, and RALGDS interaction | Cell culture | (Bery, et al., 2018; Quevedo, et al., 2018) | |
| RT11(-i) | IC50 = 5 – 13μM | Chimeric cell-penetrating antibody | RAS-GTP | ↓ PI3K, RAF, and RALGDS interaction | Cell culture; Mouse pancreatic, fibrosarcoma, and colorectal cancer models | (Kang, et al., 2018; Shin, et al., 2017; Shin, et al., 2020) | |
| inRas37 | IC50 = 1.5 – 8μM | (Shin, et al., 2020) | |||||
| R11.1.6 | Kd = 4 – 40nM | High affinity scaffold based on sso7d | RAS-GTP SW2 | ↓ GTP hydrolysis and RAF association | Cell culture | (Kauke, et al., 2017) | |
| K27 and K55 | IC50 = 2.4 (K27); 167nM (K55) | Designed Ankyrin Repeat Proteins (DARPins) | K27: RAS-GDP SW1 K55: RAS-GTP SW1/SW2 and prevented RAF interaction |
K27: ↓ SOS interaction K55: ↓ RAF interaction |
Cell culture | (Guillard, et al., 2017) | |
| K13 and K19 | K13: Kd = 30nM K19: Kd = 10nM |
DARPins | KRAS(G12V) α3-α4 dimerization interface |
↓ RAS dimerization, SOS nucleotide exchange, and RAF, PI3K, and RALGDS interaction | Cell culture | (Bery, et al., 2019) | |
| NS1 | Kd = 15 nM (HRAS) Kd = 65 nM (KRAS) |
Monobody | α4-α5 dimerization interface | ↓ H/KRAS dimerization and signaling | Cell culture; Mouse pancreatic, endometrial, and lung cancer models | (Khan, et al., 2019; Spencer-Smith, et al., 2017; Spencer-Smith, et al., 2019) |
2.2.1. Small Molecule Inhibitors of RAS
2.2.1.1. Nonsteroidal Anti-Inflammatory Drugs
Nonsteroidal anti-inflammatory drugs (NSAIDs) were recognized in the late 1980’s to early 1990’s to have anti-tumorigenic activity (Ip et al., 1985; Pepin et al., 1992; Rao et al., 1995). The oxidative metabolite of the NSAID Sulindac inhibited chemically-induced mammary tumors that were prone to HRAS mutations (Table 2) (Thompson et al., 1995; Thompson et al., 1997). Sulindac directly bound HRAS and prevented its interaction with RAF, the GEF CDC25, and p120GAP and inhibited proliferation, MAPK activation, and RAF kinase activity in HRAS-transformed fibroblasts (Herrmann et al., 1998). Clinically, NSAIDs were not found to have potent anti-cancer effects, possibly due to dose-limiting side effects (Gurpinar et al., 2014).
2.2.1.2. The p1 Pocket
Four novel pockets (p1–4) on RAS were discovered using a combination of computational methods and molecular dynamics simulations (Grant et al., 2011). Virtual screening identified lead compounds (643000 and 117028) that bound to the p3 pocket. Although some activity was observed in vitro, further development has not been published. However, subsequent work identified the p1 pocket as a viable target of compound binding (Fig. 2 and Table 2) (McCarthy et al., 2019).
The first reports of p1 pocket binding compounds came independently from two groups. DCAI and cmpd12 bound a pocket adjacent to SW1/SW2 (Maurer, et al., 2012; Sun et al., 2012), later recognized as the p1 pocket (Prakash et al., 2015a; Prakash et al., 2015b). Both compounds inhibited nucleotide exchange (Sun, et al., 2012), and in the case of DCAI, nucleotide release was attenuated by blocking RAS-SOS interaction (Maurer, et al., 2012). Cmpd12 displayed a KD for KRAS of 190μM, but only 78% inhibition of nucleotide exchange was observed at 1mM (Sun, et al., 2012). In cells, DCAI prevented GTP-loading of RAS, and inhibited EGF-induced RAS-GTP (Maurer, et al., 2012). However, inhibition of RAS signaling was not reported in either study and no further studies utilizing these compound have been published.
Nonetheless, the p1 pocket has remained an attractive site to target on RAS. The inhibitors Kobe0065 and Kobe2602 were discovered by a virtual screen targeting the p1 pocket on GTP-loaded HRAS(P40D) (Prakash, et al., 2015a; Shima et al., 2013). The compounds inhibited the interaction of HRAS(G12V) with RAF, attenuated downstream MAPK signaling, and reduced levels of phosphorylated AKT and GTP-loaded RALA (signaling proteins downstream of PI3K and RALGDS discussed above). While the Kobe compounds inhibited SOS-mediated nucleotide exchange in vitro, in cells they failed to alter the levels of RAS-GTP indicating that unlike previous p1 binders, the Kobe compounds specifically inhibited the interaction of HRAS(G12V) with RAF (Shima, et al., 2013). Importantly, the Kobe compounds inhibited the growth of HRAS(G12V)-transformed cells as well as cancer cell lines harboring an oncogenic K, H-, or NRAS mutation and reduced growth of colorectal cancer cell xenografts by 50% when given orally (Shima, et al., 2013). Despite these promising results, further studies with the Kobe compounds have not been reported.
More potent p1 pocket inhibitors have been recently described. BI-2852 was discovered using NMR-based fragment screening and optimized by structure-based design (Kessler et al., 2019). BI-2852 bound similarly to KRAS WT, (G12D) and (G12C) mutants regardless of GTP vs GDP. Further, BI-2852 effectively inhibited the interaction with SOS, CRAF, and PI3K resulting in decreased nucleotide exchange, MAPK and AKT activation, and proliferation. Interestingly, BI-2852 also inhibited the drop in KRAS(G12C)-GTP levels caused by ARS-1620 treatment, indicating that BI-2852 decreasing intrinsic GTPase activity or reduces residual interaction of KRAS(G12C) with GAPs.
Another p1 binding compound, Cmpd11, bound RAS with nanomolar affinity and inhibited RAS-RAF interaction (>10-fold reduction with 1μM cmpd11), as SOS-mediated nucleotide exchange and release was only mildly affected. Treatment of KRAS mutant lung tumor cells with cmpd11 reduced pERK levels and cell proliferation (McCarthy, et al., 2019). Testing of the compounds in vivo was not reported.
A unique inhibitor that simultaneously binds the KRAS p1 pocket and the membrane was recently described using a novel screening assay that measured the BRAF activation by fully processed KRAS(G12V) bound to liposomes (Jansen et al., 2017). Similar to the above p1 pocket binders, the initial hits were indole compounds, and medicinal chemistry generated additional compounds including cmpd2 that inhibited KRAS(G12V) activation in the screening assay with a submicromolar IC50. Cmpd2 inhibited the growth of KRAS(G12D)-expressing pancreatic cancer cells, and dose-dependently reduced AKT and ERK activation. Importantly, cmpd2 preferentially engaged prenylated, membrane bound KRAS vs soluble KRAS (Jansen, et al., 2017). NMR measurements of paramagnetic relaxation enhancement on nanodisc-tethered KRAS(G12V) revealed that cmpd2 simultaneously engaged KRAS(G12V) and membrane. This dual interaction favored an orientation of KRAS relative to the membrane that occluded the effector binding domain and prevented RAF engagement (Fang et al., 2018). However, the in vivo efficacy of these compounds remains to be evaluated.
2.2.1.3. Modulation of SOS-Mediated Nucleotide Exchange
Targeting RAS:SOS interaction has also been used as an approach to develop direct RAS inhibitors (Table 2). Compounds SCH-53239, SCH-53870, and SCH-54292 bound RAS in a pocket adjacent to the SW2 region (Taveras et al., 1997). These compounds inhibited intrinsic nucleotide exchange of NRAS(G12V) and reduced nerve growth factor-induced neurite outgrowth of PC12 cells (Taveras, et al., 1997). Peri et al used molecular modeling to develop compounds that inhibited CDC25-stimulated nucleotide exchange on HRAS (Peri et al., 2005). Furthermore, these compounds downregulated pERK in fibroblasts, inhibited RAS-dependent yeast growth, and proliferation of KRAS(G12R)-transformed fibroblasts (Peri, et al., 2005). Further work has not been reported for these compounds.
A number of compounds bind SOS in a hydrophobic pocket in the CDC25 domain adjacent to where RAS binds (Abbott et al., 2018; Burns et al., 2018; Burns et al., 2014; Hillig et al., 2019; Hodges et al., 2018). These compounds were initially derived from the same screen that discovered cmpd12 (Burns, et al., 2014; Sun, et al., 2012), and synthesis of derivatives resulted in cmpd4 which displayed improved activation of SOS-mediated nucleotide exchange on KRAS(G12D) in vitro and enhanced RAS-GTP levels in HeLa cells (Burns, et al., 2014). While many of these small molecules enhanced SOS-mediated nucleotide exchange, they paradoxically inhibited RAS signaling due to negative feedback phosphorylation of SOS by ERK which disrupted SOS-GRB2 interaction, SOS membrane localization, and consequently reduced RAS activation (Abbott, et al., 2018; Burns, et al., 2018; Burns, et al., 2014; Hodges, et al., 2018; Howes et al., 2018). More potent compounds were developed that inhibited ERK [cmpd17 and cmpd19, see (Abbott, et al., 2018)], however, their effects on cell growth in vitro or tumor growth in vivo were not studied.
In contrast, BAY-293 binds to the same pocket on SOS1 but inhibits SOS1-KRAS interaction and nucleotide exchange activity (Hillig, et al., 2019). This compound was generated by chemical linking hits from an NMR-based fragment screen with optimized hits from a high throughput screen and displayed enhanced binding affinity for SOS1 and inhibition of SOS1-KRAS interaction (Hillig, et al., 2019). In wild-type cells, BAY-293 inhibited RAS activation, MAPK signaling, and cell proliferation. In contrast, BAY-293 was no more effective than the inactive enantiomer at inhibiting KRAS(G12C) cells. However, when combined with a RAS(G12C) inhibitor (see below), BAY-293 synergistically inhibited cells indicating that cotreatment could improve the efficacy of KRAS(G12C)-targeted inhibitors (Hillig, et al., 2019). While promising, further study is needed to determine the clinical viability of SOS-binding small molecules.
Multiple groups have developed peptides that inhibit RAS:SOS interaction. The HBS3 peptide was based on the RAS-contacting αH helix of SOS (Patgiri et al., 2011). A hydrogen bond surrogate (HBS) approach was used to produce stabilized α-helices with the relevant SOS residues oriented to interact with RAS, and HBS3 was optimized for water solubility and helical structure (Patgiri, et al., 2011). HBS3 bound to apoRAS and RAS-GDP, and inhibited SOS-mediated nucleotide exchange in vitro (Patgiri, et al., 2011). HBS3 treatment reduced RAS-GTP levels and MAPK activation in response to EGF stimulation (Patgiri, et al., 2011). An independent group used a similar “stapled” peptide approach to develop stabilized α-helix SOS (SAH-SOS)-based peptides (Leshchiner et al., 2015). The resulting SAH-SOS1A peptide inhibited nucleotide binding by both wild-type and mutant KRAS, disrupted KRAS-SOS interaction, and was cell permeable (Leshchiner, et al., 2015). SAH-SOS1A treatment of KRAS mutant cancer cells reduced their viability which correlated with reduced MAPK activity (Leshchiner, et al., 2015). Similarly, a cell permeable RASGRF1 peptide inhibited RASGRF1-catalyzed nucleotide exchange in vitro, MAPK activation in cells, growth of KRAS-transformed fibroblasts, and IGF-stimulated migration and invasion of urothelial-carcinoma-derived 5637 cells at 500nM (Sacco et al., 2012). Unfortunately, none of these peptides were tested in mammalian animal models, and therefore further study is required.
2.2.1.4. Inhibitors of RAS/RAF interaction
A number of compounds with distinct or unknown binding sites block RAS/RAF interaction (Table 2). MCP1 and its derivatives inhibited RAS signaling and RAF activation, reduced RAS/RAF interaction in mammalian cells, induced reversion and inhibited the invasiveness of HRAS(G12V) transformed fibroblasts and NRAS(Q61K) HT-1080 fibrosarcoma cells, and inhibited growth of KRAS(G12S) A549 lung carcinoma cells and KRAS(G12D) pancreatic adenocarcinoma cells (Gonzalez-Perez et al., 2010; Kato-Stankiewicz et al., 2002). The MCP1 derivative, MCP110, synergistically induced apoptosis and inhibited growth of multiple RAS mutant human cell lines and Kaposi’s sarcoma-associated herpesvirus cell models when combined with MAPK inhibitors or microtubule-targeting drugs (Skobeleva et al., 2007). In mice, MCP110 displayed reasonable tolerability, bioavailability, and persistence following oral, intravenous, and intraperitoneal administration and moderately inhibited lung adenocarcinoma xenograft growth a monotherapy (Skobeleva, et al., 2007). When combined with paclitaxel, MCP110 drastically reduced the growth rate of mouse colorectal carcinoma xenografts (Skobeleva, et al., 2007). Unfortunately, further preclinical evaluation has not been reported.
The small molecule 3144 was discovered by a computational docking screen focused on critical residues in the RAF binding region (SW1/SW2) on KRAS(G12D) (Welsch et al., 2017). Although the authors were unable to obtain co-crystal structures of 3144 with KRAS, several biophysical approaches demonstrated direct interaction with KRAS. Indeed, mutations of predicted SW1/SW2 interacting residues reduced binding. 3144 treatment inhibited ERK and AKT activity and stimulated apoptosis in several RAS mutant cancer cell lines in vitro. Administration of 3144 either orally, or by a combination of i.v. and i.p. injections, reduced the growth of KRAS-mutant xenograft tumors and NRAS mutant PDX tumors (Welsch, et al., 2017). Further work on this promising drug candidate has not been reported.
Cyclorasin cyclic peptides were discovered in a combinatorial peptide library screen, inhibited RAS-RAF interaction, and were cell permeable (Upadhyaya et al., 2015). One modified version, cyclorasin 9A5, displayed robust cellular uptake and potent antiproliferative activity against RAS mutant cancer cells. Cyclorasin 9A5 selectively bound GTP-loaded KRAS(G12V) near the SW1 loop, potentially overlapping with the p1 pocket as determined by NMR (Grant, et al., 2011; Upadhyaya, et al., 2015). Treatment with cyclorasin 9A5 reduced RAS:BRAF interaction as well as AKT and ERK activation in various KRAS mutant cell lines (Upadhyaya, et al., 2015). As with many other small molecules recently developed, further translational work is needed to explore the full potential of this compound.
2.2.1.5. The Switch 2 Pocket
An important milestone in direct RAS inhibition was achieved recently by the groundbreaking discovery of compounds that covalently bind to KRAS(G12C) at the Cys12 residue in a unique binding pocket called the SW2 pocket (SII-P) (Ostrem, et al., 2013). Surprisingly, these compounds only reacted with GDP- and not GTP-loaded RAS yet effectively inhibited KRAS(G12C) mutant cells. This was due to the fact that RAS(G12C) retains sufficient intrinsic GTPase activity that it continues to cycle in cells thereby allowing for reaction with the inhibitor once GDP loaded. Newer iterations of these compounds have been developed, e.g. ARS-853 (Patricelli et al., 2016) and ARS-1620 (Janes, et al., 2018), with enhanced binding and pharmacological properties compared to the initial compounds and are available to the research community as tool compounds.
Recently, two additional KRAS(G12C) inhibitors, AMG510 (Canon, et al., 2019) and MRTX849 (Hallin, et al., 2020), have advanced to clinical trials (ClinicalTrials.gov Identifiers: NCT03600883 and NCT03785249, respectively). Like the earlier KRAS(G12C) inhibitors, these compounds lock KRAS(G12C) in the inactive GDP-bound state (Canon, et al., 2019; Fell, et al., 2018; Hallin, et al., 2020; Janes, et al., 2018; Ostrem, et al., 2013; Patricelli, et al., 2016). Structurally, one of the salient features of AMG510 is the improvement in binding to KRAS(G12C) by interaction with a cryptic groove in KRAS(G12C) to enhance potency and selectivity approximately 10-fold as compared to ARS-1620 (Canon, et al., 2019) (Fig. 6).
Figure 6. Comparative structures of KRAS(G12C) inhibitors.
(A) Shokat cmpd 16 (PDB: M422); (B) ARS1620 (PDB:5V9U); (C) MRTX cmpd12 (PDB: 6N2K); (D) AMG510 (PDB: 6OIM). Coloring is the same as described in Fig. 2A
AMG510 and MRTX849 treatment of KRAS(G12C) mutant cancer lines resulted in accumulation of KRAS in the inactive state, and pronounced inhibition of KRAS-dependent signaling, cancer cell growth, and regression of tumors in xenograft models (Canon, et al., 2019; Hallin, et al., 2020). Initial clinical results with both AMG510 and MRTX849 have been highly encouraging: significant tumor inhibition was observed in both lung and colon adenocarcinoma cancer patients harboring KRAS(G12C) mutations (Canon, et al., 2019; Hallin, et al., 2020). However, caution is warranted as cancer cells are notorious for acquiring resistance to monotherapies, particularly those targeting oncogenic driver enzymes (van Maldegem and Downward, 2020), and blocking the RAS pathway will inevitably lead to emergence of drug resistance (Molina-Arcas et al., 2019). For KRAS(G12C) inhibitors it might be doubly disadvantageous as higher levels of downstream effectors like RAF might associate significantly with GTP-bound active RAS and reduce its cycling to the GDP state thereby limiting reaction with KRAS(G12C) inhibitors (McCormick, 2020). Given this concern, both Amgen and Mirati have explored various combination strategies including co-inhibition of EGFR, PI3K, SHP2, AKT, mTOR, or MEK which improved responses in lung and colon tumor xenograft models (Canon, et al., 2019; Hallin, et al., 2020; van Maldegem and Downward, 2020). Given that KRAS mutations lead to an overall immune-suppressive tumor microenvironment (Coelho et al., 2017; Cullis et al., 2018), AMG510 synergized with immunotherapy (anti-PD1) to provide enhanced anti-tumor response in a preclinical colorectal tumor model (Canon, et al., 2019). Thus, specific inhibition of KRAS within tumor cells can alter the immune-suppressive tumor micro-environment and can synergize with immune therapies. Thus, many are waiting with great expectations as these compounds progress through clinical trials.
In an extension of the work to target G12C mutants, Genitle et al employed disulfide tethering of a non-natural cysteine on KRAS(M27C) to discover compounds targeting an expanded pocket around SW2 (Gentile et al., 2017). The compound 2C07 bound an extended groove around the SII-P pocket which the group referred to as the SW2 groove (SII-G). Importantly, 2C07 bound both GTP- and GDP-loaded K- and HRAS(M72C) (Gentile, et al., 2017). Surprisingly, 2C07 did not affect the ability of GTP-bound HRAS(M72C) to interact with the RAF-RBD despite significant alterations to SW1. However, 2C07 inhibited the interaction of HRAS(M72C) with SOS to reduce nucleotide exchange on HRAS(G12V/M72C). Furthermore, targeting SII-G with a derivative of 2C07 inhibited HRAS(G12V/M72C) activation of PI3K (Gentile, et al., 2017). It is important to note that 2C07 is only active on M72C mutants (the residue to which it is tethered) and the above assays were performed in vitro. However, 2C07 represents a lead compound for further development of inhibitors targeting SII-G on RAS.
2.2.2. Biologics-Based RAS Inhibitors
A number of RAS inhibitors have been generated based on antibodies and synthetic protein scaffolds (Khan, et al., 2020). While the applicability of these biological RAS inhibitors as therapeutics may be limited, they will nevertheless assist in defining new drug targets on RAS.
2.2.2.1. RAS Antibodies
Antibodies against RAS were the first developed RAS inhibitors. The rat monoclonal antibody Y13–259 was produced against viral HRAS and cross-reacted with both viral and endogenous RAS from multiple rodent cell lines (Furth et al., 1982). Y13–259 bound near the SW2 region and was proposed to inhibit effector interactions (Furth, et al., 1982; Lacal and Aaronson, 1986). When injected into NIH 3T3 fibroblasts, Y13–259 inhibited cell division (Mulcahy et al., 1985). Expression of a single-chain variable fragment (scFv) of Y13–259 sequestered endogenous RAS in aggregates and induced cell-cycle arrest (Cardinale et al., 1998). The anti-p21ser antibody was generated against a synthetic peptide consisting of amino acids 5–16 of viral KRAS(G12S) (Clark et al., 1985). Microinjection of the antibody reverted viral KRAS-transformed cells to a normal phenotype (Clark, et al., 1985; Feramisco et al., 1985). While not useful as therapeutics, these tools have indicated where RAS could be targeted by small molecules.
Interestingly, anti-p21ser inhibited GTP loading and failed to bind nucleotide-loaded RAS suggesting that anti-p21ser bound to apoRAS (Clark, et al., 1985). These results raise the possibility of targeting nucleotide binding as an approach to inhibit RAS despite the high affinity of RAS for nucleotide. Indeed, recent studies support this possibility. A number of RAS mutants (Q61L, G13D, and A146T) possess high intrinsic nucleotide dissociation rates suggesting that they spontaneously release GTP and reside in a nucleotide-free state more frequently than wild type RAS or other RAS mutants (Smith, et al., 2013). In addition, our group discovered a role for apoRAS in regulating the PI3K Class 2β isoform (PIK3C2B) (Wong et al., 2012). PIK3C2B activity was inhibited by apoRAS in vitro suggesting a potential role for apoRAS in cell signaling. Furthermore, PIK3C2B interacted preferentially with dominant-negative RAS vs WT or oncogenic versions in cells suggesting that PIK3C2B was targeting apoRAS. Together, these findings suggest that it indeed may be possible to target apoRAS as an approach to inhibit RAS function in cells. Some success with such an approach has been found with heterotrimeric G-proteins. BIM-46187 inhibited Gq by targeting the nucleotide-free state (Schmitz et al., 2014). Whether similar success is possible with RAS remains to be seen.
2.2.2.2. RAS Intrabodies
Intracellular antibodies (intrabodies) are single chain variable fragments of antibodies (scFvs) composed of the variable light and heavy chains of antibodies linked together (Cattaneo and Biocca, 1999; Rondon and Marasco, 1997). Individually, the variable chains are considered the smallest immunoglobulin-based recognition units (aka Domain antibodies or Dabs) (Ward et al., 1989). Intracellular Dab #6 (iDab#6) was isolated as a specific HRAS(G12V) affinity probe but also displayed cross-reactivity to N- and KRAS (Table 2) (Tanaka et al., 2003). iDab#6 bound the SW1/SW2 region of RAS-GTP, prevented effector binding, inhibited HRAS(G12V)-mediated transformation and activation of ERK and AKT, and blocked the growth of mutant-RAS colorectal cancer and fibrosarcoma cell lines in culture (Tanaka, et al., 2003; Tanaka and Rabbitts, 2010; Tanaka et al., 2007). When tested in vivo, iDab#6 blocked the ability of HT-1080 fibrosarcoma cells [NRAS(Q61K)] and DLD-1 colorectal cancer cells [KRAS(G13D)] to form tumors in athymic nude mice (Tanaka, et al., 2007). Furthermore, iDab#6 inhibited spontaneous lung tumor formation in a genetic mouse model (Tanaka and Rabbitts, 2010).
Using iDab#6 as a tool for drug discovery, Quevedo et al utilized a surface plasmon resonance (SPR)-based screen to identify compounds that bound HRAS(G12V) but not when complexed with iDab#6 (Bery et al., 2018; Quevedo et al., 2018). Crystallography of the lead compound showed that it bound in the p1 pocket and allowed for development of the derivative Abd-7 which inhibited interaction of mutant H-, K-, and NRAS with the RBDs from PI3Kα, PI3Kγ, CRAF, and RALGDS (Quevedo, et al., 2018). Moreover, Abd-7 (20μM) inhibited EGF-stimulated ERK and AKT activation and proliferation of RAS-mutant tumor cells (Quevedo, et al., 2018). Future work will be required to determine the efficacy of this compound in vivo.
Intrabodies against RAS that are self-internalizing have also been developed (Table 2). Shin et al isolated a chimeric antibody that replaced the variable heavy chain of the cell penetrating antibody, TMab4, with a variable heavy chain targeting KRAS(G12D) to yield the antibody, RT11 (Shin et al., 2017). RT11 targeted a diverse set of oncogenic RAS mutants in all three RAS isoforms, and blocked RAS interaction with CRAF, RALGDS, and PI3Kα both in vitro and in cells resulting in the concomitant inhibition of these signaling pathways (Shin, et al., 2017). Mutational analysis indicated that RT11 bound the SW1/SW2 regions (Shin, et al., 2017). Furthermore, RT11 inhibited the growth of a diverse set of human cancer cell lines expressing mutant K- and NRAS and showed no cytotoxicity towards wild-type RAS-expressing cells (Shin, et al., 2017). Addition of an RDG peptide that specifically bound tumor associated integrins conferred tumor specificity to the antibody. The resulting chimeric antibody, RT11-i, exhibited improved anti-proliferative activity on cultured cancer cells, reduced growth of KRAS and NRAS mutant tumors in mice, and synergized with anti-tumor agents including cetuximab (Shin, et al., 2017) and gemcitabine (Kang et al., 2018) to inhibit the growth of colorectal and pancreatic tumors in mice.
Recently, a second generation of RT11-i was isolated with enhanced internalization and endosomal escape, improved affinity of the VH chain for RAS, improved tumor affinity of the RGD peptide, and a modified constant chain that conferred resistance to Fcγ receptor–mediated clearance (Shin et al., 2020). The resulting antibody, inRas37, had improved binding to mutant RAS isoforms (~2-fold), and like RT11-i, colocalized on the inner membrane with KRAS(G12V) and KRAS(G13D) in tumor cells, but remained cytosolic in wild-type RAS-expressing cells (Shin, et al., 2020). inRas37, like RT11-I, disrupted mutant RAS-RAF interaction in cultured cells, reduced oncogenic RAS-induced MAPK and AKT activation, and inhibited the viability of mutant KRAS-expressing cells with single digit micromolar IC50’s. inRas37 displayed significantly enhanced anti-tumor activity over RT11-i, reducing growth of LoVo tumor xenografts in mice by 75% compared to 45% for RT11-i (Shin, et al., 2020). In mice with preestablished tumors, inRas37 inhibited growth of RAS mutant tumors but not tumors with wild type RAS or tumors resistant to RAS knockdown due to additional activating mutations in PI3K and/or β-catenin (Shin, et al., 2020). Combination of inRas37 with inhibitors of PI3K or β-catenin synergized to reduce viability of the resistant cell lines in spheroid cultures and mouse xenografts (Shin, et al., 2020). Additional studies will be required for translation to the clinic (Table 2).
2.2.2.3. sso7d-Based RAS Inhibitors
Proteins based on the sso7d protein scaffold represent another family of engineered proteins used to target RAS. R11.1.6 is a sso7d protein that favored binding to GppNHp-loaded KRas(G12D) over wild-type RAS depending on the isoform of wild-type RAS (Table 2)(Kauke et al., 2017). KRAS(G12D) interacted with R11.1.6 through the SW1/SW2 region, with one residue extending into the p1 pocket (Kauke, et al., 2017). Intriguingly, the interaction inhibited GTP hydrolysis, but comparison of crystallographic structures indicated that R11.1.6 sterically competed with RAF for binding to KRAS(G12D). In cells, R11.1.6 colocalized with KRAS(G12D) at the membrane, pulled down both KRAS(G12D) and wild-type KRAS from cell lysates, reduced co-immunoprecipitation of KRAS with BRAF, and inhibited MAPK signaling (Kauke, et al., 2017). Although it is unlikely this inhibitor could be translated to the clinic due to delivery issues, it provides a useful tool for the study of RAS.
2.2.2.4. RAS-Targeting Monobodies
Monobodies (Mbs) are synthetic binding proteins based on the fibronectin type III (FN3) domain (Koide et al., 2012; Sha et al., 2017). These designer proteins have been widely used to target many proteins given their ability to selectively bind regions important to the function of the target protein (Sha, et al., 2017). Indeed, the NS1 Mb bound with nanomolar affinity to both H- and KRAS but did not bind NRAS or related small GTPases such as RRAS2 (Table 2) (Spencer-Smith, et al., 2017). NS1 inhibited oncogenic HRAS and KRAS signaling both in vitro and in vivo but did not impair signaling from NRAS or downstream oncogenic kinases such BRAF(V600E) or MEK(DD) (Khan et al., 2019; Spencer-Smith, et al., 2017). Although NS1 inhibited oncogenic HRAS(G12V) activation of RAF, it did not affect HRAS:RAF interaction. Structural studies revealed that NS1 bound the α4-α5 interface in the allosteric lobe, a region implicated in RAS dimerization. Indeed, NS1 inhibited HRAS and KRAS dimerization and nanoclustering (Spencer-Smith, et al., 2017). However, mutations of residues proposed to mediate dimerization, i.e., Asp154 or Arg161, (Guldenhaupt, et al., 2012), did not affect HRAS(G12V) signaling suggesting that RAS may utilize an alternative interface for dimerization, such as the α3-α4 interface (Bery, et al., 2019; Spencer-Smith, et al., 2017). Nevertheless, the importance of the α4-α5 interface in RAS function was illustrated by the ability of NS1 to inhibit KRAS-dependent tumor growth in xenograft models (Khan, et al., 2019).
Additional studies revealed that NS1 inhibited RAS at multiple levels (Spencer-Smith et al., 2019). While NS1 did not affect HRAS interaction with RAF, KRAS:RAF interaction was inhibited by NS1. This effect was mediated by the HVR region. Exchanging the HRAS HVR for the KRAS HVR resulted in a chimeric oncogenic HRAS protein that bound RAF to same extent as KRAS. Furthermore, NS1 reduced interaction of RAF with this HRAS chimera. In contrast, exchanging the KRAS HVR for the HRAS HVR resulted in an oncogenic KRAS protein in which interaction with RAF was unaffected by NS1. These results suggest that NS1 may alter the KRAS HVR interaction with the PM thereby reducing KRAS levels at the PM (Spencer-Smith, et al., 2017). Furthermore, NS1 inhibited GTP loading of WT RAS in cells following growth factor stimulation despite having no effect on SOS-mediated nucleotide exchange on the isolated G-domain in vitro. These results suggested that NS1 binding of the α4-α5 interface may alter the orientation of RAS and interfere with GEF binding. Together, the results with NS1 suggest that targeting the α4-α5 interface may provide a path to inhibit RAS in vivo.
2.2.2.5. DARPin-Based RAS Inhibitiors
Designed ankyrin repeat proteins (DARPins) are another engineered protein scaffold used to target diverse proteins including RAS (Binz et al., 2003). DARPins K27 and K55 were selected for binding KRAS(G12V), and both inhibited SOS-mediated nucleotide exchange and RAS-RAF interaction (Table 2)(Guillard et al., 2017). Neither DARPin was selective for any RAS isoform or mutation, but they were specific for the nucleotide-bound state of RAS: K27 favored RAS-GDP (KD = 3.9nM) and K55 RAS-GTP (KD = 167nM) (Guillard, et al., 2017). K27 bound RAS-GDP to inhibit nucleotide exchange while K55 bound to GTP-loaded RAS. Both DARPins prevented interaction with multiple effectors, reduced ERK and AKT activation, and inhibited anchorage-independent growth of RAS-mutant human tumor cells (Guillard, et al., 2017).
Recently, two additional DARPins, K13 and K19, that targeted KRAS(G12V) independent of the nucleotide status with KD’s of 30 and 10nM, respectively (Bery, et al., 2019). These DARPins bound the α3-α4 helices in the allosteric lobe of KRAS(G12V) and inhibited KRAS dimerization, suggesting an alternative dimerization interface to the α4-α5 interface described in section 1.1. K13 and K19 were selective to KRAS vs NRAS and HRAS. As with K27, K13 and K19 inhibited nucleotide exchange, but not effector interactions. In cells, both DARPins inhibited KRAS interaction with multiple effectors resulting in reduced MEK, ERK and AKT activation without affecting wild type or mutant HRAS and NRAS signaling (Bery, et al., 2019). As with many protein-based inhibitors, the RAS DARPins are not clinically viable in their current forms, but nevertheless represent important tools for research and drug discovery.
2.2.2.6. DIRAS3: An Endogenous RAS Inhibitor
DIRAS3 is a 26 kDa RAS family GTPase sharing 50–60% homology with the classical RAS family members. Like RAS, DIRAS3 is prenylated at a C-terminal CaaX site, binds GTP with high affinity, exhibits weak GTPase activity, and requires GTP-loading and membrane association for its biological function (Luo et al., 2003). In contrast, the biological function of DIRAS3 is to inhibit cell growth suggesting its role as a tumor suppressor. Interestingly, DIRAS3 is downregulated in a number of cancers, and re-expression induces cytotoxicity, inhibits the growth of tumor xenografts in mice, and reduces MAPK and PI3K signaling (Sutton et al., 2019). Moreover, low DIRAS3 expression in PDAC patients predicts poor survival (Sutton, et al., 2019). A recent study suggested that DIRAS3 functions as an endogenous inhibitor of RAS (Sutton, et al., 2019). DIRAS3 interacted with KRAS at the PM and reduced RAS nanoclustering. DIRAS3 bound the α5 helix of KRAS. Similar to NS1 Mb, DIRAS3 interacted with HRAS and KRAS but not NRAS (Spencer-Smith, et al., 2017; Sutton, et al., 2019). DIRAS3 inhibited oncogenic HRAS and KRAS mediated signaling and transformation; however, a DIRAS3 N-terminal deletion mutant (ΔNT DIRAS3) did not inhibit KRAS function despite retaining comparable binding affinity to and subcellular localization with KRAS as WT DIRAS3 (Sutton, et al., 2019). Thus, DIRAS3 may represent a naturally occurring RAS inhibitor. Whether inducing DIRAS3 re-expression in tumors could be a valid therapeutic avenue remains to be seen. Nevertheless, these data underscore the value of targeting the RAS allosteric lobe with Monobodies and DARPins as an approach to inhibit RAS. However, the challenge with these and other anti-RAS biologics is how to effectively deliver such therapeutic to tumor cells.
Conclusions/Future Directions
RAS plays an indispensable role in normal cell signaling and mammalian physiology, but is frequently mutated as part of the tumorigenic processes. Thus, RAS has been a major focus of cancer research in the ~40 years since it was first recognized as an oncogene. Although there remain no FDA approved anti-RAS therapeutics, the recent development of highly specific drugs that directly target KRAS(G12C) has provided significant hope for success. These drugs have shown encouraging preclinical as well as early Phase 1 clinical results (Canon, et al., 2019; Hallin, et al., 2020). Thus, success in therapeutically inhibiting RAS appears in sight. Nevertheless, these drugs only inhibit a small portion of RAS mutant cancers (<15%), and continued persistence will be needed to develop drugs that inhibit a wider range of mutant RAS proteins. In addition, resistance to these inhibitors is all but certain, thus necessitating the implementation of combination therapies to inhibit potential bypass resistance pathways and/or engage the immune system in the hopes of establishing more durable therapeutic responses. Given the newfound appreciation that is indeed possible to directly inhibit RAS with clinically viable small molecules, there has been a resurgence in interest in pharmacologically targeting RAS. Thus, there is great optimism that we will eventually succeed in developing viable anti-RAS therapeutics for cancer patients, with the light at the end of the tunnel indeed growing brighter.
Acknowledgements
The authors wish to apologize in advance for any omissions of references to our colleagues’ work. J.P.O. was supported in part by a Merit Review Award (1I01BX002095) from the United States (US) Department of Veterans Affairs Biomedical Laboratory Research and Development Service, NIH awards (CA212608 and CA138313), and start-up funds from the Hollings Cancer Center at MUSC. The contents of this article do not represent the views of the US. Department of Veterans Affairs or the United States Government.
Footnotes
Disclosures: The authors have nothing to disclose.
Bibliography
- National Cancer Institute. (2019). Cancer Statistics. Available from: https://www.cancer.gov/about-cancer/understanding/statistics.
- Abankwa D, Hanzal-Bayer M, Ariotti N, Plowman SJ, Gorfe AA, Parton RG, McCammon JA, and Hancock JF (2008). EMBO J 27, 727–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott JR, Hodges TR, Daniels RN, Patel PA, Kennedy JP, Howes JE, Akan DT, Burns MC, Sai J, Sobolik T, Beesetty Y, Lee T, Rossanese OW, Phan J, Waterson AG, and Fesik SW (2018). J Med Chem 61, 6002–6017. [DOI] [PubMed] [Google Scholar]
- Adachi T, Pimentel DR, Heibeck T, Hou X, Lee YJ, Jiang B, Ido Y, and Cohen RA (2004). J Biol Chem 279, 29857–62. [DOI] [PubMed] [Google Scholar]
- Adari H (1988). Science 240, 518. [DOI] [PubMed] [Google Scholar]
- Adjei AA, Erlichman C, Davis JN, Cutler DL, Sloan JA, Marks RS, Hanson LJ, Svingen PA, Atherton P, Bishop WR, Kirschmeier P, and Kaufmann SH (2000). Cancer Res 60, 1871–7. [PubMed] [Google Scholar]
- Adjei AA, Mauer A, Bruzek L, Marks RS, Hillman S, Geyer S, Hanson LJ, Wright JJ, Erlichman C, Kaufmann SH, and Vokes EE (2003). J Clin Oncol 21, 1760–6. [DOI] [PubMed] [Google Scholar]
- Ahearn IM, Haigis K, Bar-Sagi D, and Philips MR (2011a). Nat Rev Mol Cell Biol 13, 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahearn IM, Tsai FD, Court H, Zhou M, Jennings BC, Ahmed M, Fehrenbacher N, Linder ME, and Philips MR (2011b). Mol Cell 41, 173–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed TA, Adamopoulos C, Karoulia Z, Wu X, Sachidanandam R, Aaronson SA, and Poulikakos PI (2019). Cell Rep 26, 65–78 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akino K, Toyota M, Suzuki H, Mita H, Sasaki Y, Ohe-Toyota M, Issa JP, Hinoda Y, Imai K, and Tokino T (2005). Gastroenterology 129, 156–69. [DOI] [PubMed] [Google Scholar]
- Alsina M, Fonseca R, Wilson EF, Belle AN, Gerbino E, Price-Troska T, Overton RM, Ahmann G, Bruzek LM, Adjei AA, Kaufmann SH, Wright JJ, Sullivan D, Djulbegovic B, Cantor AB, Greipp PR, Dalton WS, and Sebti SM (2004). Blood 103, 3271–7. [DOI] [PubMed] [Google Scholar]
- Alvarez-Moya B, Lopez-Alcala C, Drosten M, Bachs O, and Agell N (2010). Oncogene 29, 5911–22. [DOI] [PubMed] [Google Scholar]
- Ambrogio C, Kohler J, Zhou ZW, Wang H, Paranal R, Li J, Capelletti M, Caffarra C, Li S, Lv Q, Gondi S, Hunter JC, Lu J, Chiarle R, Santamaria D, Westover KD, and Janne PA (2018). Cell 172, 857–868 e15. [DOI] [PubMed] [Google Scholar]
- Apolloni A, Prior IA, Lindsay M, Parton RG, and Hancock JF (2000). Mol Cell Biol 20, 2475–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appels NM, Beijnen JH, and Schellens JH (2005). Oncologist 10, 565–78. [DOI] [PubMed] [Google Scholar]
- Arthur WT, Quilliam LA, and Cooper JA (2004). J Cell Biol 167, 111–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badar T, Cortes JE, Ravandi F, O’Brien S, Verstovsek S, Garcia-Manero G, Kantarjian H, and Borthakur G (2015). Clin Lymphoma Myeloma Leuk 15, 433–438 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines AT, Xu D, and Der CJ (2011). Future Med Chem 3, 1787–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R, Wilkerson EM, Sumita K, Isom DG, Sasaki AT, Dohlman HG, and Campbell SL (2013). J Biol Chem 288, 36856–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballester R, Furth ME, and Rosen OM (1987). J Biol Chem 262, 2688–95. [PubMed] [Google Scholar]
- Barcelo C, Paco N, Morell M, Alvarez-Moya B, Bota-Rabassedas N, Jaumot M, Vilardell F, Capella G, and Agell N (2014). Cancer Res 74, 1190–9. [DOI] [PubMed] [Google Scholar]
- Basso AD, Kirschmeier P, and Bishop WR (2006). J Lipid Res 47, 15–31. [DOI] [PubMed] [Google Scholar]
- Berndt N, Hamilton AD, and Sebti SM (2011). Nat Rev Cancer 11, 775–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bery N, Cruz-Migoni A, Bataille CJ, Quevedo CE, Tulmin H, Miller A, Russell A, Phillips SE, Carr SB, and Rabbitts TH (2018). Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bery N, Legg S, Debreczeni J, Breed J, Embrey K, Stubbs C, Kolasinska-Zwierz P, Barrett N, Marwood R, Watson J, Tart J, Overman R, Miller A, Phillips C, Minter R, and Rabbitts TH (2019). Nat Commun 10, 2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigenzahn JW, Collu GM, Kartnig F, Pieraks M, Vladimer GI, Heinz LX, Sedlyarov V, Schischlik F, Fauster A, Rebsamen M, Parapatics K, Blomen VA, Muller AC, Winter GE, Kralovics R, Brummelkamp TR, Mlodzik M, and Superti-Furga G (2018). Science 362, 1171–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz HK, Stumpp MT, Forrer P, Amstutz P, and Pluckthun A (2003). J Mol Biol 332, 489–503. [DOI] [PubMed] [Google Scholar]
- Bivona TG, Quatela SE, Bodemann BO, Ahearn IM, Soskis MJ, Mor A, Miura J, Wiener HH, Wright L, Saba SG, Yim D, Fein A, Perez de Castro I, Li C, Thompson CB, Cox AD, and Philips MR (2006). Mol Cell 21, 481–93. [DOI] [PubMed] [Google Scholar]
- Bordier BB, Marion PL, Ohashi K, Kay MA, Greenberg HB, Casey JL, and Glenn JS (2002). J Virol 76, 10465–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, and Kuriyan J (1998). Nature 394, 337–43. [DOI] [PubMed] [Google Scholar]
- Borthakur G, Kantarjian H, Daley G, Talpaz M, O’Brien S, Garcia-Manero G, Giles F, Faderl S, Sugrue M, and Cortes J (2006). Cancer 106, 346–52. [DOI] [PubMed] [Google Scholar]
- Borthakur G, Ravandi F, O’Brien S, Beran M, Williams B, Goldsweig H, and Giles F (2007). Blood 110, 1600–1600. [Google Scholar]
- Boyartchuk VL, Ashby MN, and Rine J (1997). Science 275, 1796–800. [DOI] [PubMed] [Google Scholar]
- Britten CD, Rowinsky EK, Soignet S, Patnaik A, Yao SL, Deutsch P, Lee Y, Lobell RB, Mazina KE, McCreery H, Pezzuli S, and Spriggs D (2001). Clin Cancer Res 7, 3894–903. [PubMed] [Google Scholar]
- Bryant KL, Stalnecker CA, Zeitouni D, Klomp JE, Peng S, Tikunov AP, Gunda V, Pierobon M, Waters AM, George SD, Tomar G, Papke B, Hobbs GA, Yan L, Hayes TK, Diehl JN, Goode GD, Chaika NV, Wang Y, Zhang GF, Witkiewicz AK, Knudsen ES, Petricoin EF 3rd, Singh PK, Macdonald JM, Tran NL, Lyssiotis CA, Ying H, Kimmelman AC, Cox AD, and Der CJ (2019). Nat Med 25, 628–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buday L, and Downward J (1993). Cell 73, 611–20. [DOI] [PubMed] [Google Scholar]
- Buhrman G, Holzapfel G, Fetics S, and Mattos C (2010). Proc Natl Acad Sci U S A 107, 4931–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhrman G, Wink G, and Mattos C (2007). Structure 15, 1618–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunda S, Burrell K, Heir P, Zeng L, Alamsahebpour A, Kano Y, Raught B, Zhang ZY, Zadeh G, and Ohh M (2015). Nat Commun 6, 8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunda S, Heir P, Srikumar T, Cook JD, Burrell K, Kano Y, Lee JE, Zadeh G, Raught B, and Ohh M (2014). Proc Natl Acad Sci U S A 111, E3785–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B, and Stewart CL (2002). Nat Rev Mol Cell Biol 3, 575–85. [DOI] [PubMed] [Google Scholar]
- Bürkle A (2001). In “Encyclopedia of Genetics”, pp. 1533, Elsevier BV. [Google Scholar]
- Burnett AK, Russell NH, Culligan D, Cavanagh J, Kell J, Wheatley K, Virchis A, Hills RK, Milligan D, and Institute, A. M. L. W. G. o. t. U. N. C. R. (2012). Br J Haematol 158, 519–22. [DOI] [PubMed] [Google Scholar]
- Burns MC, Howes JE, Sun Q, Little AJ, Camper DV, Abbott JR, Phan J, Lee T, Waterson AG, Rossanese OW, and Fesik SW (2018). Anal Biochem 548, 44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MC, Sun Q, Daniels RN, Camper D, Kennedy JP, Phan J, Olejniczak ET, Lee T, Waterson AG, Rossanese OW, and Fesik SW (2014). Proc Natl Acad Sci U S A 111, 3401–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buser CA, Dinsmore CJ, Fernandes C, Greenberg I, Hamilton K, Mosser SD, Walsh ES, Williams TM, and Koblan KS (2001). Anal Biochem 290, 126–37. [DOI] [PubMed] [Google Scholar]
- Bustinza-Linares E, Kurzrock R, and Tsimberidou AM (2010). Future Oncol 6, 885–91. [DOI] [PubMed] [Google Scholar]
- Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, Gaida K, Holt T, Knutson CG, Koppada N, Lanman BA, Werner J, Rapaport AS, San Miguel T, Ortiz R, Osgood T, Sun JR, Zhu X, McCarter JD, Volak LP, Houk BE, Fakih MG, O’Neil BH, Price TJ, Falchook GS, Desai J, Kuo J, Govindan R, Hong DS, Ouyang W, Henary H, Arvedson T, Cee VJ, and Lipford JR (2019). Nature 575, 217–223. [DOI] [PubMed] [Google Scholar]
- Caraglia M, Marra M, Naviglio S, Botti G, Addeo R, and Abbruzzese A (2010). Expert Opin Pharmacother 11, 141–54. [DOI] [PubMed] [Google Scholar]
- Cardinale A, Lener M, Messina S, Cattaneo A, and Biocca S (1998). FEBS Lett 439, 197–202. [DOI] [PubMed] [Google Scholar]
- Carreira BP, Morte MI, Inacio A, Costa G, Rosmaninho-Salgado J, Agasse F, Carmo A, Couceiro P, Brundin P, Ambrosio AF, Carvalho CM, and Araujo IM (2010). Stem Cells 28, 1219–30. [DOI] [PubMed] [Google Scholar]
- Casasola A, Scalzo D, Nandakumar V, Halow J, Recillas-Targa F, Groudine M, and Rincon-Arano H (2016). Nucleus 7, 84–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey PJ, Solski PA, Der CJ, and Buss JE (1989). Proc Natl Acad Sci U S A 86, 8323–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo A, and Biocca S (1999). Trends Biotechnol 17, 115–21. [DOI] [PubMed] [Google Scholar]
- Chandra A, Grecco HE, Pisupati V, Perera D, Cassidy L, Skoulidis F, Ismail SA, Hedberg C, Hanzal-Bayer M, Venkitaraman AR, Wittinghofer A, and Bastiaens PI (2011). Nat Cell Biol 14, 148–58. [DOI] [PubMed] [Google Scholar]
- Chardin P, Camonis JH, Gale NW, van Aelst L, Schlessinger J, Wigler MH, and Bar-Sagi D (1993). Science 260, 1338–43. [DOI] [PubMed] [Google Scholar]
- Chen L, Sung SS, Yip ML, Lawrence HR, Ren Y, Guida WC, Sebti SM, Lawrence NJ, and Wu J (2006). Mol Pharmacol 70, 562–70. [DOI] [PubMed] [Google Scholar]
- Chen YN, LaMarche MJ, Chan HM, Fekkes P, Garcia-Fortanet J, Acker MG, Antonakos B, Chen CH, Chen Z, Cooke VG, Dobson JR, Deng Z, Fei F, Firestone B, Fodor M, Fridrich C, Gao H, Grunenfelder D, Hao HX, Jacob J, Ho S, Hsiao K, Kang ZB, Karki R, Kato M, Larrow J, La Bonte LR, Lenoir F, Liu G, Liu S, Majumdar D, Meyer MJ, Palermo M, Perez L, Pu M, Price E, Quinn C, Shakya S, Shultz MD, Slisz J, Venkatesan K, Wang P, Warmuth M, Williams S, Yang G, Yuan J, Zhang JH, Zhu P, Ramsey T, Keen NJ, Sellers WR, Stams T, and Fortin PD (2016). Nature 535, 148–52. [DOI] [PubMed] [Google Scholar]
- Chien Y, and White MA (2003). EMBO Rep 4, 800–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KJ, Casteel DE, Prakash P, Tan L, van der Hoeven D, Salim AA, Kim C, Capon RJ, Lacey E, Cunha SR, Gorfe AA, and Hancock JF (2016). Mol Cell Biol 36, 3086–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi BH, Chen C, Philips M, and Dai W (2018a). Oncotarget 9, 4440–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi BH, Philips MR, Chen Y, Lu L, and Dai W (2018b). J Biol Chem 293, 17574–17581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov IE, and Philips MR (1999). Cell 98, 69–80. [DOI] [PubMed] [Google Scholar]
- Chung JK, Lee YK, Denson JP, Gillette WK, Alvarez S, Stephen AG, and Groves JT (2018). Biophys J 114, 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R, Wong G, Arnheim N, Nitecki D, and McCormick F (1985). Proc Natl Acad Sci U S A 82, 5280–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho MA, de Carne Trecesson S, Rana S, Zecchin D, Moore C, Molina-Arcas M, East P, Spencer-Dene B, Nye E, Barnouin K, Snijders AP, Lai WS, Blackshear PJ, and Downward J (2017). Immunity 47, 1083–1099 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SJ, Ho L, Ranganathan S, Abbruzzese JL, Alpaugh RK, Beard M, Lewis NL, McLaughlin S, Rogatko A, Perez-Ruixo JJ, Thistle AM, Verhaeghe T, Wang H, Weiner LM, Wright JJ, Hudes GR, and Meropol NJ (2003). J Clin Oncol 21, 1301–6. [DOI] [PubMed] [Google Scholar]
- Contestabile A, and Ciani E (2004). Neurochem Int 45, 903–14. [DOI] [PubMed] [Google Scholar]
- Cortes J, Albitar M, Thomas D, Giles F, Kurzrock R, Thibault A, Rackoff W, Koller C, O’Brien S, Garcia-Manero G, Talpaz M, and Kantarjian H (2003). Blood 101, 1692–7. [DOI] [PubMed] [Google Scholar]
- Cortes J, Jabbour E, Daley GQ, O’Brien S, Verstovsek S, Ferrajoli A, Koller C, Zhu Y, Statkevich P, and Kantarjian H (2007). Cancer 110, 1295–302. [DOI] [PubMed] [Google Scholar]
- Court H, Amoyel M, Hackman M, Lee KE, Xu R, Miller G, Bar-Sagi D, Bach EA, Bergo MO, and Philips MR (2013). J Clin Invest 123, 4681–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, and Der CJ (1997). Biochim Biophys Acta 1333, F51–71. [DOI] [PubMed] [Google Scholar]
- Cox AD, and Der CJ (2010). Small GTPases 1, 2–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Fesik SW, Kimmelman AC, Luo J, and Der CJ (2014). Nat Rev Drug Discov 13, 828–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crul M, de Klerk GJ, Beijnen JH, and Schellens JH (2001). Anticancer Drugs 12, 163–84. [DOI] [PubMed] [Google Scholar]
- Crul M, de Klerk GJ, Swart M, van’t Veer LJ, de Jong D, Boerrigter L, Palmer PA, Bol CJ, Tan H, de Gast GC, Beijnen JH, and Schellens JH (2002). J Clin Oncol 20, 2726–35. [DOI] [PubMed] [Google Scholar]
- Cullis J, Das S, and Bar-Sagi D (2018). Cold Spring Harb Perspect Med 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q, Choy E, Chiu V, Romano J, Slivka SR, Steitz SA, Michaelis S, and Philips MR (1998). J Biol Chem 273, 15030–4. [DOI] [PubMed] [Google Scholar]
- De Sandre-Giovannoli A, Bernard R, Cau P, Navarro C, Amiel J, Boccaccio I, Lyonnet S, Stewart CL, Munnich A, Le Merrer M, and Levy N (2003). Science 300, 2055. [DOI] [PubMed] [Google Scholar]
- Deb TB, Wong L, Salomon DS, Zhou G, Dixon JE, Gutkind JS, Thompson SA, and Johnson GR (1998). J Biol Chem 273, 16643–6. [DOI] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, and White E (2006). Cancer Cell 10, 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker FJ, Rocks O, Vartak N, Menninger S, Hedberg C, Balamurugan R, Wetzel S, Renner S, Gerauer M, Scholermann B, Rusch M, Kramer JW, Rauh D, Coates GW, Brunsveld L, Bastiaens PI, and Waldmann H (2010). Nat Chem Biol 6, 449–56. [DOI] [PubMed] [Google Scholar]
- Der CJ, Krontiris TG, and Cooper GM (1982). Proc Natl Acad Sci U S A 79, 3637–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins A, Reardon DA, Peters KB, Threatt S, Coan AD, Herndon JE 2nd, Friedman AH, Friedman HS, and Vredenburgh JJ (2011). J Neurooncol 105, 601–6. [DOI] [PubMed] [Google Scholar]
- Dharmaiah S, Tran TH, Messing S, Agamasu C, Gillette WK, Yan W, Waybright T, Alexander P, Esposito D, Nissley DV, McCormick F, Stephen AG, and Simanshu DK (2019). Sci Rep 9, 10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donninger H, Schmidt ML, Mezzanotte J, Barnoud T, and Clark GJ (2016). Semin Cell Dev Biol 58, 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J (2003). Nat Rev Cancer 3, 11–22. [DOI] [PubMed] [Google Scholar]
- Ducker CE, Stettler EM, French KJ, Upson JJ, and Smith CD (2004). Oncogene 23, 9230–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JA, and Gilman AG (1998). J Biol Chem 273, 15830–7. [DOI] [PubMed] [Google Scholar]
- Egan SE, Giddings BW, Brooks MW, Buday L, Sizeland AM, and Weinberg RA (1993). Nature 363, 45–51. [DOI] [PubMed] [Google Scholar]
- Elad-Sfadia G, Haklai R, Balan E, and Kloog Y (2004). J Biol Chem 279, 34922–30. [DOI] [PubMed] [Google Scholar]
- End DW, Smets G, Todd AV, Applegate TL, Fuery CJ, Angibaud P, Venet M, Sanz G, Poignet H, Skrzat S, Devine A, Wouters W, and Bowden C (2001). Cancer Res 61, 131–7. [PubMed] [Google Scholar]
- Engelman JA, Luo J, and Cantley LC (2006). Nat Rev Genet 7, 606–19. [DOI] [PubMed] [Google Scholar]
- Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Dutra A, Pak E, Durkin S, Csoka AB, Boehnke M, Glover TW, and Collins FS (2003). Nature 423, 293–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskens FA, Awada A, Cutler DL, de Jonge MJ, Luyten GP, Faber MN, Statkevich P, Sparreboom A, Verweij J, Hanauske AR, Piccart M, European Organization for R, and Treatment of Cancer Early Clinical Studies, G. (2001). J Clin Oncol 19, 1167–75. [DOI] [PubMed] [Google Scholar]
- Fang Z, Marshall CB, Nishikawa T, Gossert AD, Jansen JM, Jahnke W, and Ikura M (2018). Cell Chem Biol 25, 1327–1336 e4. [DOI] [PubMed] [Google Scholar]
- Fedele C, Ran H, Diskin B, Wei W, Jen J, Geer MJ, Araki K, Ozerdem U, Simeone DM, Miller G, Neel BG, and Tang KH (2018). Cancer Discov 8, 1237–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell JB, Fischer JP, Baer BR, Ballard J, Blake JF, Bouhana K, Brandhuber BJ, Briere DM, Burgess LE, Burkard MR, Chiang H, Chicarelli MJ, Davidson K, Gaudino JJ, Hallin J, Hanson L, Hee K, Hicken EJ, Hinklin RJ, Marx MA, Mejia MJ, Olson P, Savechenkov P, Sudhakar N, Tang TP, Vigers GP, Zecca H, and Christensen JG (2018). ACS Med Chem Lett 9, 1230–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G (1993). Science 259, 1607. [DOI] [PubMed] [Google Scholar]
- Feramisco JR, Clark R, Wong G, Arnheim N, Milley R, and McCormick F (1985). Nature 314, 639–42. [DOI] [PubMed] [Google Scholar]
- Fetics SK, Guterres H, Kearney BM, Buhrman G, Ma B, Nussinov R, and Mattos C (2015). Structure 23, 505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, Stefancsik R, Harsha B, Kok CY, Jia M, Jubb H, Sondka Z, Thompson S, De T, and Campbell PJ (2017). Nucleic Acids Res 45, D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman RM Jr., Plutzky J, and Neel BG (1992). Proc Natl Acad Sci U S A 89, 11239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth ME, Davis LJ, Fleurdelys B, and Scolnick EM (1982). J Virol 43, 294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse J, Kurata T, Okano N, Fujisaka Y, Naruge D, Shimizu T, Kitamura H, Iwasa T, Nagashima F, and Nakagawa K (2018). Cancer Chemother Pharmacol 82, 511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale NW, Kaplan S, Lowenstein EJ, Schlessinger J, and Bar-Sagi D (1993). Nature 363, 88–92. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, Alnemri ES, Altucci L, Amelio I, Andrews DW, Annicchiarico-Petruzzelli M, Antonov AV, Arama E, Baehrecke EH, Barlev NA, Bazan NG, Bernassola F, Bertrand MJM, Bianchi K, Blagosklonny MV, Blomgren K, Borner C, Boya P, Brenner C, Campanella M, Candi E, Carmona-Gutierrez D, Cecconi F, Chan FK, Chandel NS, Cheng EH, Chipuk JE, Cidlowski JA, Ciechanover A, Cohen GM, Conrad M, Cubillos-Ruiz JR, Czabotar PE, D’Angiolella V, Dawson TM, Dawson VL, De Laurenzi V, De Maria R, Debatin KM, DeBerardinis RJ, Deshmukh M, Di Daniele N, Di Virgilio F, Dixit VM, Dixon SJ, Duckett CS, Dynlacht BD, El-Deiry WS, Elrod JW, Fimia GM, Fulda S, Garcia-Saez AJ, Garg AD, Garrido C, Gavathiotis E, Golstein P, Gottlieb E, Green DR, Greene LA, Gronemeyer H, Gross A, Hajnoczky G, Hardwick JM, Harris IS, Hengartner MO, Hetz C, Ichijo H, Jaattela M, Joseph B, Jost PJ, Juin PP, Kaiser WJ, Karin M, Kaufmann T, Kepp O, Kimchi A, Kitsis RN, Klionsky DJ, Knight RA, Kumar S, Lee SW, Lemasters JJ, Levine B, Linkermann A, Lipton SA, Lockshin RA, Lopez-Otin C, Lowe SW, Luedde T, Lugli E, MacFarlane M, Madeo F, Malewicz M, Malorni W, Manic G, et al. (2018). Cell Death Differ 25, 486–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Fortanet J, Chen CH, Chen YN, Chen Z, Deng Z, Firestone B, Fekkes P, Fodor M, Fortin PD, Fridrich C, Grunenfelder D, Ho S, Kang ZB, Karki R, Kato M, Keen N, LaBonte LR, Larrow J, Lenoir F, Liu G, Liu S, Lombardo F, Majumdar D, Meyer MJ, Palermo M, Perez L, Pu M, Ramsey T, Sellers WR, Shultz MD, Stams T, Towler C, Wang P, Williams SL, Zhang JH, and LaMarche MJ (2016). J Med Chem 59, 7773–82. [DOI] [PubMed] [Google Scholar]
- Gentile DR, Rathinaswamy MK, Jenkins ML, Moss SM, Siempelkamp BD, Renslo AR, Burke JE, and Shokat KM (2017). Cell Chem Biol 24, 1455–1466 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry LR, Martin TD, Reiner DJ, and Der CJ (2014). Biochim Biophys Acta 1843, 2976–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer M, Schweins T, Herrmann C, Prisner T, Wittinghofer A, and Kalbitzer HR (1996). Biochemistry 35, 10308–20. [DOI] [PubMed] [Google Scholar]
- Gibbs JB, Pompliano DL, Mosser SD, Rands E, Lingham RB, Singh SB, Scolnick EM, Kohl NE, and Oliff A (1993). J Biol Chem 268, 7617–20. [PubMed] [Google Scholar]
- Gibbs JB, Sigal IS, Poe M, and Scolnick EM (1984). Proc Natl Acad Sci U S A 81, 5704–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L, Altman RB, and Klein TE (2011). Pharmacogenet Genomics 21, 50–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Perez V, Reiner DJ, Alan JK, Mitchell C, Edwards LJ, Khazak V, Der CJ, and Cox AD (2010). J Mol Signal 5, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon LB, Kleinman ME, Miller DT, Neuberg DS, Giobbie-Hurder A, Gerhard-Herman M, Smoot LB, Gordon CM, Cleveland R, Snyder BD, Fligor B, Bishop WR, Statkevich P, Regen A, Sonis A, Riley S, Ploski C, Correia A, Quinn N, Ullrich NJ, Nazarian A, Liang MG, Huh SY, Schwartzman A, and Kieran MW (2012). Proc Natl Acad Sci U S A 109, 16666–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon LB, Shappell H, Massaro J, D’Agostino RB Sr., Brazier J, Campbell SE, Kleinman ME, and Kieran MW (2018). JAMA 319, 1687–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfe AA (2010). Curr Med Chem 17, 1–9. [DOI] [PubMed] [Google Scholar]
- Gorfe AA, Grant BJ, and McCammon JA (2008). Structure 16, 885–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfe AA, Hanzal-Bayer M, Abankwa D, Hancock JF, and McCammon JA (2007). J Med Chem 50, 674–84. [DOI] [PubMed] [Google Scholar]
- Grant BJ, Lukman S, Hocker HJ, Sayyah J, Brown JH, McCammon JA, and Gorfe AA (2011). PLoS One 6, e25711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grek CL, and Tew KD (2010). Current Opinion in Pharmacology 10, 362–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grek CL, Zhang J, Manevich Y, Townsend DM, and Tew KD (2013). J Biol Chem 288, 26497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskopf S, Eckert C, Arkona C, Radetzki S, Bohm K, Heinemann U, Wolber G, von Kries JP, Birchmeier W, and Rademann J (2015). ChemMedChem 10, 815–26. [DOI] [PubMed] [Google Scholar]
- Guillard S, Kolasinska-Zwierz P, Debreczeni J, Breed J, Zhang J, Bery N, Marwood R, Tart J, Overman R, Stocki P, Mistry B, Phillips C, Rabbitts T, Jackson R, and Minter R (2017). Nat Commun 8, 16111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldenhaupt J, Rudack T, Bachler P, Mann D, Triola G, Waldmann H, Kotting C, and Gerwert K (2012). Biophys J 103, 1585–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, Nye E, Stamp G, Alitalo K, and Downward J (2007). Cell 129, 957–68. [DOI] [PubMed] [Google Scholar]
- Gurpinar E, Grizzle WE, and Piazza GA (2014). Clin Cancer Res 20, 1104–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habets GG, Scholtes EH, Zuydgeest D, van der Kammen RA, Stam JC, Berns A, and Collard JG (1994). Cell 77, 537–49. [DOI] [PubMed] [Google Scholar]
- Hahn SM, Bernhard EJ, Regine W, Mohiuddin M, Haller DG, Stevenson JP, Smith D, Pramanik B, Tepper J, DeLaney TF, Kiel KD, Morrison B, Deutsch P, Muschel RJ, and McKenna WG (2002). Clin Cancer Res 8, 1065–72. [PubMed] [Google Scholar]
- Hall A, Marshall CJ, Spurr NK, and Weiss RA (1983). Nature 303, 396–400. [DOI] [PubMed] [Google Scholar]
- Hall A, and Self AJ (1986). J Biol Chem 261, 10963–5. [PubMed] [Google Scholar]
- Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, Sudhakar N, Bowcut V, Baer BR, Ballard JA, Burkard MR, Fell JB, Fischer JP, Vigers GP, Xue Y, Gatto S, Fernandez-Banet J, Pavlicek A, Velastagui K, Chao RC, Barton J, Pierobon M, Baldelli E, Patricoin EF 3rd, Cassidy DP, Marx MA, Rybkin II, Johnson ML, Ou SI, Lito P, Papadopoulos KP, Janne PA, Olson P, and Christensen JG (2020). Cancer Discov 10, 54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton SE, Dore TM, and Schmidt WK (2018). Crit Rev Biochem Mol Biol 53, 157–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JF, Magee AI, Childs JE, and Marshall CJ (1989). Cell 57, 1167–77. [DOI] [PubMed] [Google Scholar]
- Hancock JF, Paterson H, and Marshall CJ (1990). Cell 63, 133–9. [DOI] [PubMed] [Google Scholar]
- Haney SA, and Broach JR (1994). J Biol Chem 269, 16541–8. [PubMed] [Google Scholar]
- Hansen TE, and Johansen T (2011). BMC Biol 9, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M, Akasaka K, Akinaga S, Okabe M, Nakano H, Gomez R, Wood D, Uh M, and Tamanoi F (1993). Proc Natl Acad Sci U S A 90, 2281–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey JJ (1964). Nature 204, 1104–5. [DOI] [PubMed] [Google Scholar]
- Hellmuth K, Grosskopf S, Lum CT, Wurtele M, Roder N, von Kries JP, Rosario M, Rademann J, and Birchmeier W (2008). Proc Natl Acad Sci U S A 105, 7275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo J, and Campbell SL (2004). Biochemistry 43, 2314–22. [DOI] [PubMed] [Google Scholar]
- Herrmann C (2003). Current Opinion in Structural Biology 13, 122–129. [DOI] [PubMed] [Google Scholar]
- Herrmann C, Block C, Geisen C, Haas K, Weber C, Winde G, Moroy T, and Muller O (1998). Oncogene 17, 1769–76. [DOI] [PubMed] [Google Scholar]
- Heymach JV, Johnson DH, Khuri FR, Safran H, Schlabach LL, Yunus F, DeVore RF 3rd, De Porre PM, Richards HM, Jia X, Zhang S, and Johnson BE (2004). Ann Oncol 15, 1187–93. [DOI] [PubMed] [Google Scholar]
- Hibino K, Shibata T, Yanagida T, and Sako Y (2011). J Biol Chem 286, 36460–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillig RC, Sautier B, Schroeder J, Moosmayer D, Hilpmann A, Stegmann CM, Werbeck ND, Briem H, Boemer U, Weiske J, Badock V, Mastouri J, Petersen K, Siemeister G, Kahmann JD, Wegener D, Bohnke N, Eis K, Graham K, Wortmann L, von Nussbaum F, and Bader B (2019). Proc Natl Acad Sci U S A 116, 2551–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs GA, Bonini MG, Gunawardena HP, Chen X, and Campbell SL (2013a). Free Radic Biol Med 57, 221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs GA, Gunawardena HP, Baker R, and Campbell SL (2013b). Small GTPases 4, 186–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges TR, Abbott JR, Little AJ, Sarkar D, Salovich JM, Howes JE, Akan DT, Sai J, Arnold AL, Browning C, Burns MC, Sobolik T, Sun Q, Beesetty Y, Coker JA, Scharn D, Stadtmueller H, Rossanese OW, Phan J, Waterson AG, McConnell DB, and Fesik SW (2018). J Med Chem 61, 8875–8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, and Shoelson SE (1998). Cell 92, 441–50. [DOI] [PubMed] [Google Scholar]
- Hollander I, Frommer E, and Mallon R (2000). Anal Biochem 286, 129–37. [DOI] [PubMed] [Google Scholar]
- Howes JE, Akan DT, Burns MC, Rossanese OW, Waterson AG, and Fesik SW (2018). Mol Cancer Ther 17, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Huang L, Carney J, Cardona DM, and Counter CM (2014). Nat Commun 5, 5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SA, Wedemeyer H, and Harrison PM (2011). The Lancet 378, 73–85. [DOI] [PubMed] [Google Scholar]
- Huw LY, O’Brien C, Pandita A, Mohan S, Spoerke JM, Lu S, Wang Y, Hampton GM, Wilson TR, and Lackner MR (2013). Oncogenesis 2, e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye K, Mizutani S, Koide H, and Kaziro Y (2000). J Biol Chem 275, 3737–40. [DOI] [PubMed] [Google Scholar]
- Ip C, Carter CA, and Ip MM (1985). Cancer Res 45, 1997–2001. [PubMed] [Google Scholar]
- Ismail SA, Chen YX, Rusinova A, Chandra A, Bierbaum M, Gremer L, Triola G, Waldmann H, Bastiaens PI, and Wittinghofer A (2011). Nat Chem Biol 7, 942–9. [DOI] [PubMed] [Google Scholar]
- Ito Y, Yamasaki K, Iwahara J, Terada T, Kamiya A, Shirouzu M, Muto Y, Kawai G, Yokoyama S, Laue ED, Walchli M, Shibata T, Nishimura S, and Miyazawa T (1997). Biochemistry 36, 9109–19. [DOI] [PubMed] [Google Scholar]
- James GL, Goldstein JL, and Brown MS (1995). J Biol Chem 270, 6221–6. [DOI] [PubMed] [Google Scholar]
- James GL, Goldstein JL, Brown MS, Rawson TE, Somers TC, McDowell RS, Crowley CW, Lucas BK, Levinson AD, and Marsters JC Jr. (1993). Science 260, 1937–42. [DOI] [PubMed] [Google Scholar]
- Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, Chen Y, Babbar A, Firdaus SJ, Darjania L, Feng J, Chen JH, Li S, Li S, Long YO, Thach C, Liu Y, Zarieh A, Ely T, Kucharski JM, Kessler LV, Wu T, Yu K, Wang Y, Yao Y, Deng X, Zarrinkar PP, Brehmer D, Dhanak D, Lorenzi MV, Hu-Lowe D, Patricelli MP, Ren P, and Liu Y (2018). Cell 172, 578–589 e17. [DOI] [PubMed] [Google Scholar]
- Janosi L, and Gorfe AA (2010). Biophys J 99, 3666–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janosi L, Li Z, Hancock JF, and Gorfe AA (2012). Proc Natl Acad Sci U S A 109, 8097–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen JM, Wartchow C, Jahnke W, Fong S, Tsang T, Pfister K, Zavorotinskaya T, Bussiere D, Cheng JM, Crawford K, Dai Y, Dove J, Fang E, Feng Y, Florent JM, Fuller J, Gossert AD, Hekmat-Nejad M, Henry C, Klopp J, Lenahan WP, Lingel A, Ma S, Meyer A, Mishina Y, Narberes J, Pardee G, Ramurthy S, Rieffel S, Stuart D, Subramanian S, Tandeske L, Widger S, Widmer A, Winterhalter A, Zaror I, and Hardy S (2017). PLoS One 12, e0174706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong WJ, Yoon J, Park JC, Lee SH, Lee SH, Kaduwal S, Kim H, Yoon JB, and Choi KY (2012). Sci Signal 5, ra30. [DOI] [PubMed] [Google Scholar]
- Jiang L, Xu W, Chen Y, and Zhang Y (2019). Artif Cells Nanomed Biotechnol 47, 3231–3238. [DOI] [PubMed] [Google Scholar]
- John J, Sohmen R, Feuerstein J, Linke R, Wittinghofer A, and Goody RS (1990). Biochemistry 29, 6058–65. [DOI] [PubMed] [Google Scholar]
- Johnson CW, and Mattos C (2013). Enzymes 33 Pt A, 41–67. [DOI] [PubMed] [Google Scholar]
- Johnston SR, Hickish T, Ellis P, Houston S, Kelland L, Dowsett M, Salter J, Michiels B, Perez-Ruixo JJ, Palmer P, and Howes A (2003). J Clin Oncol 21, 2492–9. [DOI] [PubMed] [Google Scholar]
- Jura N, Scotto-Lavino E, Sobczyk A, and Bar-Sagi D (2006). Mol Cell 21, 679–87. [DOI] [PubMed] [Google Scholar]
- Kang YW, Lee JE, Jung KH, Son MK, Shin SM, Kim SJ, Fang Z, Yan HH, Park JH, Han B, Cheon MJ, Woo MG, Lim JH, Kim YS, and Hong SS (2018). Cancer Lett 438, 174–186. [DOI] [PubMed] [Google Scholar]
- Kano Y, Gebregiworgis T, Marshall CB, Radulovich N, Poon BPK, St-Germain J, Cook JD, Valencia-Sama I, Grant BMM, Herrera SG, Miao J, Raught B, Irwin MS, Lee JE, Yeh JJ, Zhang ZY, Tsao MS, Ikura M, and Ohh M (2019). Nat Commun 10, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor S, Weise K, Erlkamp M, Triola G, Waldmann H, and Winter R (2012). Eur Biophys J 41, 801–13. [DOI] [PubMed] [Google Scholar]
- Karasic TB, Chiorean EG, Sebti SM, and O’Dwyer PJ (2019). Target Oncol 14, 613–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp JE, Lancet JE, Kaufmann SH, End DW, Wright JJ, Bol K, Horak I, Tidwell ML, Liesveld J, Kottke TJ, Ange D, Buddharaju L, Gojo I, Highsmith WE, Belly RT, Hohl RJ, Rybak ME, Thibault A, and Rosenblatt J (2001). Blood 97, 3361–9. [DOI] [PubMed] [Google Scholar]
- Kato-Stankiewicz J, Hakimi I, Zhi G, Zhang J, Serebriiskii I, Guo L, Edamatsu H, Koide H, Menon S, Eckl R, Sakamuri S, Lu Y, Chen QZ, Agarwal S, Baumbach WR, Golemis EA, Tamanoi F, and Khazak V (2002). Proc Natl Acad Sci U S A 99, 14398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauke MJ, Traxlmayr MW, Parker JA, Kiefer JD, Knihtila R, McGee J, Verdine G, Mattos C, and Wittrup KD (2017). Sci Rep 7, 5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler D, Gmachl M, Mantoulidis A, Martin LJ, Zoephel A, Mayer M, Gollner A, Covini D, Fischer S, Gerstberger T, Gmaschitz T, Goodwin C, Greb P, Haring D, Hela W, Hoffmann J, Karolyi-Oezguer J, Knesl P, Kornigg S, Koegl M, Kousek R, Lamarre L, Moser F, Munico-Martinez S, Peinsipp C, Phan J, Rinnenthal J, Sai J, Salamon C, Scherbantin Y, Schipany K, Schnitzer R, Schrenk A, Sharps B, Siszler G, Sun Q, Waterson A, Wolkerstorfer B, Zeeb M, Pearson M, Fesik SW, and McConnell DB (2019). Proc Natl Acad Sci U S A 116, 15823–15829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I, Rhett JM, and O’Bryan JP (2020). Biochim Biophys Acta Mol Cell Res 1867, 118570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I, Spencer-Smith R, and O’Bryan JP (2019). Oncogene 38, 2984–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieran MW, Packer RJ, Onar A, Blaney SM, Phillips P, Pollack IF, Geyer JR, Gururangan S, Banerjee A, Goldman S, Turner CD, Belasco JB, Broniscer A, Zhu Y, Frank E, Kirschmeier P, Statkevich P, Yver A, Boyett JM, and Kun LE (2007). J Clin Oncol 25, 3137–43. [DOI] [PubMed] [Google Scholar]
- Kikuchi A, and Williams LT (1994). J Biol Chem 269, 20054–9. [PubMed] [Google Scholar]
- Kim ES, Kies MS, Fossella FV, Glisson BS, Zaknoen S, Statkevich P, Munden RF, Summey C, Pisters KM, Papadimitrakopoulou V, Tighiouart M, Rogatko A, and Khuri FR (2005). Cancer 104, 561–9. [DOI] [PubMed] [Google Scholar]
- Kim SE, Yoon JY, Jeong WJ, Jeon SH, Park Y, Yoon JB, Park YN, Kim H, and Choi KY (2009). J Cell Sci 122, 842–8. [DOI] [PubMed] [Google Scholar]
- Kinsey CG, Camolotto SA, Boespflug AM, Guillen KP, Foth M, Truong A, Schuman SS, Shea JE, Seipp MT, Yap JT, Burrell LD, Lum DH, Whisenant JR, Gilcrease GW 3rd, Cavalieri CC, Rehbein KM, Cutler SL, Affolter KE, Welm AL, Welm BE, Scaife CL, Snyder EL, and McMahon M (2019). Nat Med 25, 620–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsten WH, and Mayer LA (1967). J Natl Cancer Inst 39, 311–35. [PubMed] [Google Scholar]
- Knyphausen P, Lang F, Baldus L, Extra A, and Lammers M (2016). Biol Chem 397, 1071–85. [DOI] [PubMed] [Google Scholar]
- Koh C, Canini L, Dahari H, Zhao X, Uprichard SL, Haynes-Williams V, Winters MA, Subramanya G, Cooper SL, Pinto P, Wolff EF, Bishop R, Ai Thanda Han M, Cotler SJ, Kleiner DE, Keskin O, Idilman R, Yurdaydin C, Glenn JS, and Heller T (2015). The Lancet Infectious Diseases 15, 1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl NE, Omer CA, Conner MW, Anthony NJ, Davide JP, deSolms SJ, Giuliani EA, Gomez RP, Graham SL, Hamilton K, and et al. (1995). Nat Med 1, 792–7. [DOI] [PubMed] [Google Scholar]
- Kohl NE, Wilson FR, Mosser SD, Giuliani E, deSolms SJ, Conner MW, Anthony NJ, Holtz WJ, Gomez RP, Lee TJ, and et al. (1994). Proc Natl Acad Sci U S A 91, 9141–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide A, Wojcik J, Gilbreth RN, Hoey RJ, and Koide S (2012). J Mol Biol 415, 393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortmansky J, and Schwartz GK (2003). Cancer Investigation 21, 924–936. [DOI] [PubMed] [Google Scholar]
- Kotting C, Blessenohl M, Suveyzdis Y, Goody RS, Wittinghofer A, and Gerwert K (2006). Proc Natl Acad Sci U S A 103, 13911–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovrigina EA, Galiakhmetov AR, and Kovrigin EL (2015). Biophys J 109, 1000–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis PJ, Domaille PJ, Campbell-Burk SL, Van Aken T, and Laue ED (1994). Biochemistry 33, 3515–31. [DOI] [PubMed] [Google Scholar]
- Kroemer G, and Levine B (2008). Nat Rev Mol Cell Biol 9, 1004–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzrock R, Albitar M, Cortes JE, Estey EH, Faderl SH, Garcia-Manero G, Thomas DA, Giles FJ, Ryback ME, Thibault A, De Porre P, and Kantarjian HM (2004). J Clin Oncol 22, 1287–92. [DOI] [PubMed] [Google Scholar]
- Kurzrock R, Kantarjian HM, Cortes JE, Singhania N, Thomas DA, Wilson EF, Wright JJ, Freireich EJ, Talpaz M, and Sebti SM (2003). Blood 102, 4527–34. [DOI] [PubMed] [Google Scholar]
- Lacal JC, and Aaronson SA (1986). Mol Cell Biol 6, 1002–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laheru D, Shah P, Rajeshkumar NV, McAllister F, Taylor G, Goldsweig H, Le DT, Donehower R, Jimeno A, Linden S, Zhao M, Song D, Rudek MA, and Hidalgo M (2012). Invest New Drugs 30, 2391–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam AP, Sparano JA, Vinciguerra V, Ocean AJ, Christos P, Hochster H, Camacho F, Goel S, Mani S, and Kaubisch A (2010). Am J Clin Oncol 33, 121–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander HM, Milbank AJ, Tauras JM, Hajjar DP, Hempstead BL, Schwartz GD, Kraemer RT, Mirza UA, Chait BT, Burk SC, and Quilliam LA (1996). Nature 381, 380–1. [DOI] [PubMed] [Google Scholar]
- Lander HM, Ogiste JS, Pearce SFA, Levi R, and Novogrodsky A (1995a). Journal of Biological Chemistry 270, 7017–7020. [DOI] [PubMed] [Google Scholar]
- Lander HM, Ogiste JS, Teng KK, and Novogrodsky A (1995b). J Biol Chem 270, 21195–8. [DOI] [PubMed] [Google Scholar]
- Lebowitz PF, Eng-Wong J, Widemann BC, Balis FM, Jayaprakash N, Chow C, Clark G, Gantz SB, Venzon D, and Zujewski J (2005). Clin Cancer Res 11, 1247–52. [PubMed] [Google Scholar]
- Lee CS, Lee LC, Yuan TL, Chakka S, Fellmann C, Lowe SW, Caplen NJ, McCormick F, and Luo J (2019). Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner EC, Qian Y, Blaskovich MA, Fossum RD, Vogt A, Sun J, Cox AD, Der CJ, Hamilton AD, and Sebti SM (1995). J Biol Chem 270, 26802–6. [DOI] [PubMed] [Google Scholar]
- Leshchiner ES, Parkhitko A, Bird GH, Luccarelli J, Bellairs JA, Escudero S, Opoku-Nsiah K, Godes M, Perrimon N, and Walensky LD (2015). Proc Natl Acad Sci U S A 112, 1761–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, and Schlessinger J (1993). Nature 363, 85–8. [DOI] [PubMed] [Google Scholar]
- Liebmann C (2001). Cell Signal 13, 777–85. [DOI] [PubMed] [Google Scholar]
- Liu F, Yang X, Geng M, and Huang M (2018). Acta Pharm Sin B 8, 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell RB, Liu D, Buser CA, Davide JP, DePuy E, Hamilton K, Koblan KS, Lee Y, Mosser S, Motzel SL, Abbruzzese JL, Fuchs CS, Rowinsky EK, Rubin EH, Sharma S, Deutsch PJ, Mazina KE, Morrison BW, Wildonger L, Yao SL, and Kohl NE (2002). Mol Cancer Ther 1, 747–58. [PubMed] [Google Scholar]
- Lobell RB, Omer CA, Abrams MT, Bhimnathwala HG, Brucker MJ, Buser CA, Davide JP, deSolms SJ, Dinsmore CJ, Ellis-Hutchings MS, Kral AM, Liu D, Lumma WC, Machotka SV, Rands E, Williams TM, Graham SL, Hartman GD, Oliff AI, Heimbrook DC, and Kohl NE (2001). Cancer Res 61, 8758–68. [PubMed] [Google Scholar]
- Lobo S (2013). In “Drug Discovery”, IntechOpen. [Google Scholar]
- Luo RZ, Fang X, Marquez R, Liu SY, Mills GB, Liao WS, Yu Y, and Bast RC (2003). Oncogene 22, 2897–909. [DOI] [PubMed] [Google Scholar]
- Mainardi S, Mulero-Sanchez A, Prahallad A, Germano G, Bosma A, Krimpenfort P, Lieftink C, Steinberg JD, de Wit N, Goncalves-Ribeiro S, Nadal E, Bardelli A, Villanueva A, and Bernards R (2018). Nat Med 24, 961–967. [DOI] [PubMed] [Google Scholar]
- Malliri A, van der Kammen RA, Clark K, van der Valk M, Michiels F, and Collard JG (2002). Nature 417, 867–71. [DOI] [PubMed] [Google Scholar]
- Manne V, Bekesi E, and Kung HF (1985). Proc Natl Acad Sci U S A 82, 376–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manne V, Yan N, Carboni JM, Tuomari AV, Ricca CS, Brown JG, Andahazy ML, Schmidt RJ, Patel D, Zahler R, and et al. (1995). Oncogene 10, 1763–79. [PubMed] [Google Scholar]
- Marra M, Salzano G, Leonetti C, Porru M, Franco R, Zappavigna S, Liguori G, Botti G, Chieffi P, Lamberti M, Vitale G, Abbruzzese A, La Rotonda MI, De Rosa G, and Caraglia M (2012). Biotechnol Adv 30, 302–9. [DOI] [PubMed] [Google Scholar]
- Marra M, Salzano G, Leonetti C, Tassone P, Scarsella M, Zappavigna S, Calimeri T, Franco R, Liguori G, Cigliana G, Ascani R, La Rotonda MI, Abbruzzese A, Tagliaferri P, Caraglia M, and De Rosa G (2011). Nanomedicine 7, 955–64. [DOI] [PubMed] [Google Scholar]
- Martin NE, Brunner TB, Kiel KD, DeLaney TF, Regine WF, Mohiuddin M, Rosato EF, Haller DG, Stevenson JP, Smith D, Pramanik B, Tepper J, Tanaka WK, Morrison B, Deutsch P, Gupta AK, Muschel RJ, McKenna WG, Bernhard EJ, and Hahn SM (2004). Clin Cancer Res 10, 5447–54. [DOI] [PubMed] [Google Scholar]
- Martin-Gago P, Fansa EK, Klein CH, Murarka S, Janning P, Schurmann M, Metz M, Ismail S, Schultz-Fademrecht C, Baumann M, Bastiaens PI, Wittinghofer A, and Waldmann H (2017). Angew Chem Int Ed Engl 56, 2423–2428. [DOI] [PubMed] [Google Scholar]
- Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, Fauber BP, Pan B, Malek S, Stokoe D, Ludlam MJ, Bowman KK, Wu J, Giannetti AM, Starovasnik MA, Mellman I, Jackson PK, Rudolph J, Wang W, and Fang G (2012). Proc Natl Acad Sci U S A 109, 5299–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazhab-Jafari MT, Marshall CB, Smith MJ, Gasmi-Seabrook GM, Stathopulos PB, Inagaki F, Kay LE, Neel BG, and Ikura M (2015). Proc Natl Acad Sci U S A 112, 6625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MJ, Pagba CV, Prakash P, Naji AK, van der Hoeven D, Liang H, Gupta AK, Zhou Y, Cho KJ, Hancock JF, and Gorfe AA (2019). ACS Omega 4, 2921–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick F (1993). Nature 363, 15–6. [DOI] [PubMed] [Google Scholar]
- McCormick F (2020). Cancer Cell 37, 3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merideth MA, Gordon LB, Clauss S, Sachdev V, Smith AC, Perry MB, Brewer CC, Zalewski C, Kim HJ, Solomon B, Brooks BP, Gerber LH, Turner ML, Domingo DL, Hart TC, Graf J, Reynolds JC, Gropman A, Yanovski JA, Gerhard-Herman M, Collins FS, Nabel EG, Cannon RO 3rd, Gahl WA, and Introne WJ (2008). N Engl J Med 358, 592–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens AE, Roovers RC, and Collard JG (2003). FEBS Letters 546, 11–16. [DOI] [PubMed] [Google Scholar]
- Mieyal JJ, Gallogly MM, Qanungo S, Sabens EA, and Shelton MD (2008). Antioxid Redox Signal 10, 1941–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn MV, Tong L, deVos AM, Brunger A, Yamaizumi Z, Nishimura S, and Kim SH (1990). Science 247, 939–45. [DOI] [PubMed] [Google Scholar]
- Mochly-Rosen D, Das K, and Grimes KV (2012). Nat Rev Drug Discov 11, 937–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad RM, Dugan MC, Mohamed AN, Almatchy VP, Flake TM, Dergham ST, Shields AF, Al-Katib AA, Vaitkevicius VK, and Sarkar FH (1998). Pancreas 16, 19–25. [DOI] [PubMed] [Google Scholar]
- Mohammed I, Hampton SE, Ashall L, Hildebrandt ER, Kutlik RA, Manandhar SP, Floyd BJ, Smith HE, Dozier JK, Distefano MD, Schmidt WK, and Dore TM (2016). Bioorg Med Chem 24, 160–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina JR, and Adjei AA (2006). J Thorac Oncol 1, 7–9. [PubMed] [Google Scholar]
- Molina-Arcas M, Moore C, Rana S, van Maldegem F, Mugarza E, Romero-Clavijo P, Herbert E, Horswell S, Li LS, Janes MR, Hancock DC, and Downward J (2019). Sci Transl Med 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MA, Onono FO, Spielmann HP, Subramanian T, Scherr M, Venturini L, Dallmann I, Ganser A, and Reuter CW (2012). J Mol Med (Berl) 90, 149–61. [DOI] [PubMed] [Google Scholar]
- Mulcahy LS, Smith MR, and Stacey DW (1985). Nature (London) 313, 241–243. [DOI] [PubMed] [Google Scholar]
- Muratcioglu S, Chavan TS, Freed BC, Jang H, Khavrutskii L, Freed RN, Dyba MA, Stefanisko K, Tarasov SG, Gursoy A, Keskin O, Tarasova NI, Gaponenko V, and Nussinov R (2015). Structure 23, 1325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S, Xu J, Kochanek K, and Arias E (2017). NCHS Data Brief 328. [Google Scholar]
- Nakanishi Y, Walter K, Spoerke JM, O’Brien C, Huw LY, Hampton GM, and Lackner MR (2016). Cancer Res 76, 1193–203. [DOI] [PubMed] [Google Scholar]
- Nakhaeizadeh H, Amin E, Nakhaei-Rad S, Dvorsky R, and Ahmadian MR (2016). PLoS One 11, e0167145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Tamguney TM, Collisson EA, Lin LJ, Pitt C, Galeas J, Lewis S, Gray JW, McCormick F, and Chu S (2015). Proc Natl Acad Sci U S A 112, 7996–8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel DS, and Bivona TG (2017). NPJ Precis Oncol1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ GP (1999). In “Basic Neurochemistry: Molecular, Cellular and Medical Aspects” (A. B. Siegel GJ, Albers RW, et al., ed.). Lippincott-Raven; , Philadelphia. [Google Scholar]
- Nichols RJ, Haderk F, Stahlhut C, Schulze CJ, Hemmati G, Wildes D, Tzitzilonis C, Mordec K, Marquez A, Romero J, Hsieh T, Zaman A, Olivas V, McCoach C, Blakely CM, Wang Z, Kiss G, Koltun ES, Gill AL, Singh M, Goldsmith MA, Smith JAM, and Bivona TG (2018). Nat Cell Biol 20, 1064–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam M, Seong CM, Qian Y, Hamilton AD, and Sebti SM (1993). J Biol Chem 268, 20695–8. [PubMed] [Google Scholar]
- Njoroge FG, Taveras AG, Kelly J, Remiszewski S, Mallams AK, Wolin R, Afonso A, Cooper AB, Rane DF, Liu YT, Wong J, Vibulbhan B, Pinto P, Deskus J, Alvarez CS, del Rosario J, Connolly M, Wang J, Desai J, Rossman RR, Bishop WR, Patton R, Wang L, Kirschmeier P, Ganguly AK, and et al. (1998). J Med Chem 41, 4890–902. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Matozaki T, Horita K, Fujioka Y, and Kasuga M (1994). Mol Cell Biol 14, 6674–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntai I, Fornelli L, DeHart CJ, Hutton JE, Doubleday PF, LeDuc RD, van Nispen AJ, Fellers RT, Whiteley G, Boja ES, Rodriguez H, and Kelleher NL (2018). Proc Natl Acad Sci U S A 115, 4140–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Bryan JP (2019). Pharmacol Res 139, 503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor C, and Kovrigin EL (2008). Biochemistry 47, 10244–6. [DOI] [PubMed] [Google Scholar]
- Olivier JP, Raabe T, Henkemeyer M, Dickson B, Mbamalu G, Margolis B, Schlessinger J, Hafen E, and Pawson T (1993). Cell 73, 179–91. [DOI] [PubMed] [Google Scholar]
- Onono FO, Morgan MA, Ganser A, and Reuter CWM (2008). Blood 112, 2627–2627.18819192 [Google Scholar]
- Ostrem JM, Peters U, Sos ML, Wells JA, and Shokat KM (2013). Nature 503, 548–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai EF, Kabsch W, Krengel U, Holmes KC, John J, and Wittinghofer A (1989). Nature 341, 209–14. [DOI] [PubMed] [Google Scholar]
- Pai EF, Krengel U, Petsko GA, Goody RS, Kabsch W, and Wittinghofer A (1990). EMBO J 9, 2351–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R, Ramdas B, Wan C, Sandusky G, Mohseni M, Zhang C, and Kapur R (2019). J Clin Invest 129, 5468–5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papke B, Murarka S, Vogel HA, Martin-Gago P, Kovacevic M, Truxius DC, Fansa EK, Ismail S, Zimmermann G, Heinelt K, Schultz-Fademrecht C, Al Saabi A, Baumann M, Nussbaumer P, Wittinghofer A, Waldmann H, and Bastiaens PI (2016). Nat Commun 7, 11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada LF, Tabin CJ, Shih C, and Weinberg RA (1982). Nature 297, 474–8. [DOI] [PubMed] [Google Scholar]
- Parker JA, and Mattos C (2015). Molecular Cancer Research 13, 595–603. [DOI] [PubMed] [Google Scholar]
- Patgiri A, Yadav KK, Arora PS, and Bar-Sagi D (2011). Nat Chem Biol 7, 585–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, Chen Y, Kucharski JM, Feng J, Ely T, Chen JH, Firdaus SJ, Babbar A, Ren P, and Liu Y (2016). Cancer Discov 6, 316–29. [DOI] [PubMed] [Google Scholar]
- Pawson T (1997). Science 278, 2075. [DOI] [PubMed] [Google Scholar]
- Pepin P, Bouchard L, Nicole P, and Castonguay A (1992). Carcinogenesis 13, 341–8. [DOI] [PubMed] [Google Scholar]
- Perez-Fernandez A, Lopez-Ruano G, Prieto-Bermejo R, Ijurko C, Diez-Campelo M, Sanchez-Guijo F, and Hernandez-Hernandez A (2019). J Exp Clin Cancer Res 38, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri F, Airoldi C, Colombo S, Martegani E, van Neuren AS, Stein M, Marinzi C, and Nicotra F (2005). Chembiochem 6, 1839–48. [DOI] [PubMed] [Google Scholar]
- Plowman SJ, Muncke C, Parton RG, and Hancock JF (2005). Proc Natl Acad Sci U S A 102, 15500–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic N, and Wilson E (2010). In “Comprehensive Toxicology”, Elsevier. [Google Scholar]
- Porru M, Zappavigna S, Salzano G, Luce A, Stoppacciaro A, Balestrieri ML, Artuso S, Lusa S, De Rosa G, Leonetti C, and Caraglia M (2014). Oncotarget 5, 10446–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash P, Hancock JF, and Gorfe AA (2015a). Proteins 83, 898–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash P, Sayyed-Ahmad A, Cho KJ, Dolino DM, Chen W, Li H, Grant BJ, Hancock JF, and Gorfe AA (2017). Sci Rep 7, 40109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash P, Sayyed-Ahmad A, and Gorfe AA (2015b). PLoS Comput Biol 11, e1004469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior IA, Lewis PD, and Mattos C (2012). Cancer Res 72, 2457–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior IA, Muncke C, Parton RG, and Hancock JF (2003). J Cell Biol 160, 165–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Blaskovich MA, Saleem M, Seong CM, Wathen SP, Hamilton AD, and Sebti SM (1994). J Biol Chem 269, 12410–3. [PubMed] [Google Scholar]
- Quevedo CE, Cruz-Migoni A, Bery N, Miller A, Tanaka T, Petch D, Bataille CJR, Lee LYW, Fallon PS, Tulmin H, Ehebauer MT, Fernandez-Fuentes N, Russell AJ, Carr SB, Phillips SEV, and Rabbitts TH (2018). Nat Commun 9, 3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, and Therrien M (2009). Nature 461, 542–5. [DOI] [PubMed] [Google Scholar]
- Rajalingam K, Schreck R, Rapp UR, and Albert S (2007). Biochim Biophys Acta 1773, 1177–95. [DOI] [PubMed] [Google Scholar]
- Ramanujulu PM, Yang T, Yap SQ, Wong FC, Casey PJ, Wang M, and Go ML (2013). Eur J Med Chem 63, 378–86. [DOI] [PubMed] [Google Scholar]
- Rao CV, Rivenson A, Simi B, Zang E, Kelloff G, Steele V, and Reddy BS (1995). Cancer Res 55, 1464–72. [PubMed] [Google Scholar]
- Rao S, Cunningham D, de Gramont A, Scheithauer W, Smakal M, Humblet Y, Kourteva G, Iveson T, Andre T, Dostalova J, Illes A, Belly R, Perez-Ruixo JJ, Park YC, and Palmer PA (2004). J Clin Oncol 22, 3950–7. [DOI] [PubMed] [Google Scholar]
- Ravoet C, Mineur P, Robin V, Debusscher L, Bosly A, Andre M, El Housni H, Soree A, Bron D, and Martiat P (2008). Ann Hematol 87, 881–5. [DOI] [PubMed] [Google Scholar]
- Reddy S, and Comai L (2012). Exp Cell Res 318, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss Y, Goldstein JL, Seabra MC, Casey PJ, and Brown MS (1990). Cell 62, 81–8. [DOI] [PubMed] [Google Scholar]
- Repasky GA, Chenette EJ, and Der CJ (2004). Trends Cell Biol 14, 639–47. [DOI] [PubMed] [Google Scholar]
- Riely GJ, Johnson ML, Medina C, Rizvi NA, Miller VA, Kris MG, Pietanza MC, Azzoli CG, Krug LM, Pao W, and Ginsberg MS (2011). J Thorac Oncol 6, 1435–7. [DOI] [PubMed] [Google Scholar]
- Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, and Bastiaens PI (2005). Science 307, 1746–52. [DOI] [PubMed] [Google Scholar]
- Rondon IJ, and Marasco WA (1997). Annu Rev Microbiol 51, 257–83. [DOI] [PubMed] [Google Scholar]
- Rose WC, Lee FY, Fairchild CR, Lynch M, Monticello T, Kramer RA, and Manne V (2001). Cancer Res 61, 7507–17. [PubMed] [Google Scholar]
- Rosenberg JE, von der Maase H, Seigne JD, Mardiak J, Vaughn DJ, Moore M, Sahasrabudhe D, Palmer PA, Perez-Ruixo JJ, and Small EJ (2005). Cancer 103, 2035–41. [DOI] [PubMed] [Google Scholar]
- Rotblat B, Ehrlich M, Haklai R, and Kloog Y (2008). In “Small GTPases in Disease, Part B”, pp. 467–489. [Google Scholar]
- Rowell CA, Kowalczyk JJ, Lewis MD, and Garcia AM (1997). J Biol Chem 272, 14093–7. [DOI] [PubMed] [Google Scholar]
- Rozakis-Adcock M, Fernley R, Wade J, Pawson T, and Bowtell D (1993). Nature 363, 83–5. [DOI] [PubMed] [Google Scholar]
- Rudack T, Xia F, Schlitter J, Kotting C, and Gerwert K (2012). Proc Natl Acad Sci U S A 109, 15295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruess DA, Heynen GJ, Ciecielski KJ, Ai J, Berninger A, Kabacaoglu D, Gorgulu K, Dantes Z, Wormann SM, Diakopoulos KN, Karpathaki AF, Kowalska M, Kaya-Aksoy E, Song L, van der Laan EAZ, Lopez-Alberca MP, Nazare M, Reichert M, Saur D, Erkan MM, Hopt UT, Sainz B Jr., Birchmeier W, Schmid RM, Lesina M, and Algul H (2018). Nat Med 24, 954–960. [DOI] [PubMed] [Google Scholar]
- Rushworth LK, Hindley AD, O’Neill E, and Kolch W (2006). Mol Cell Biol 26, 2262–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco E, Metalli D, Spinelli M, Manzoni R, Samalikova M, Grandori R, Morrione A, Traversa S, Alberghina L, and Vanoni M (2012). Biotechnol Adv 30, 233–43. [DOI] [PubMed] [Google Scholar]
- Salzano G, Zappavigna S, Luce A, D’Onofrio N, Balestrieri ML, Grimaldi A, Lusa S, Ingrosso D, Artuso S, Porru M, Leonetti C, Caraglia M, and De Rosa G (2016). J Biomed Nanotechnol 12, 811–30. [DOI] [PubMed] [Google Scholar]
- Santini D, Caraglia M, Vincenzi B, Holen I, Scarpa S, Budillon A, and Tonini G (2006). Nat Clin Pract Oncol 3, 325–38. [DOI] [PubMed] [Google Scholar]
- Santos AI, Carreira BP, Izquierdo-Álvarez A, Ramos E, Lourenço AS, Filipa Santos D, Morte MI, Ribeiro LF, Marreiros A, Sánchez-López N, Marina A, Carvalho CM, Martínez-Ruiz A, and Araújo IM (2018). Antioxidants & Redox Signaling 28, 15–30. [DOI] [PubMed] [Google Scholar]
- Santos E, Nebreda AR, Bryan T, and Kempner ES (1988). J Biol Chem 263, 9853–8. [PubMed] [Google Scholar]
- Santos E, Tronick SR, Aaronson SA, Pulciani S, and Barbacid M (1982). Nature 298, 343–7. [DOI] [PubMed] [Google Scholar]
- Sarge KD, and Park-Sarge OK (2011). Int Rev Cell Mol Biol 288, 167–83. [DOI] [PubMed] [Google Scholar]
- Sasaki AT, Carracedo A, Locasale JW, Anastasiou D, Takeuchi K, Kahoud ER, Haviv S, Asara JM, Pandolfi PP, and Cantley LC (2011). Sci Signal 4, ra13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, and Wittinghofer A (1997). Science 277, 333–8. [DOI] [PubMed] [Google Scholar]
- Schmick M, Vartak N, Papke B, Kovacevic M, Truxius DC, Rossmannek L, and Bastiaens PIH (2014). Cell 157, 459–471. [DOI] [PubMed] [Google Scholar]
- Schmitz AL, Schrage R, Gaffal E, Charpentier TH, Wiest J, Hiltensperger G, Morschel J, Hennen S, Haussler D, Horn V, Wenzel D, Grundmann M, Bullesbach KM, Schroder R, Brewitz HH, Schmidt J, Gomeza J, Gales C, Fleischmann BK, Tuting T, Imhof D, Tietze D, Gutschow M, Holzgrabe U, Sondek J, Harden TK, Mohr K, and Kostenis E (2014). Chem Biol 21, 890–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreck R, and Rapp UR (2006). Int J Cancer 119, 2261–71. [DOI] [PubMed] [Google Scholar]
- Scolnick EM, Rands E, Williams D, and Parks WP (1973). J Virol 12, 458–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebti SM, and Hamilton AD (2000). Expert Opin Investig Drugs 9, 2767–82. [DOI] [PubMed] [Google Scholar]
- Sepp-Lorenzino L, Ma Z, Rands E, Kohl NE, Gibbs JB, Oliff A, and Rosen N (1995). Cancer Res 55, 5302–9. [PubMed] [Google Scholar]
- Sha F, Salzman G, Gupta A, and Koide S (2017). Protein Sci 26, 910–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C, and Downward J (2013). Enzymes 34 Pt. B, 107–36. [DOI] [PubMed] [Google Scholar]
- Shima F, Yoshikawa Y, Ye M, Araki M, Matsumoto S, Liao J, Hu L, Sugimoto T, Ijiri Y, Takeda A, Nishiyama Y, Sato C, Muraoka S, Tamura A, Osoda T, Tsuda K, Miyakawa T, Fukunishi H, Shimada J, Kumasaka T, Yamamoto M, and Kataoka T (2013). Proc Natl Acad Sci U S A 110, 8182–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Goldfarb M, Suard Y, Perucho M, Li Y, Kamata T, Feramisco J, Stavnezer E, Fogh J, and Wigler MH (1983). Proc Natl Acad Sci U S A 80, 2112–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SM, Choi DK, Jung K, Bae J, Kim JS, Park SW, Song KH, and Kim YS (2017). Nat Commun 8, 15090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SM, Kim JS, Park SW, Jun SY, Kweon HJ, Choi DK, Lee D, Cho YB, and Kim YS (2020). Sci Adv 6, eaay2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Allam US, Ahsan A, Chen G, Krishnamurthy PM, Marsh K, Rumschlag M, Shankar S, Whitehead C, Schipper M, Basrur V, Southworth DR, Chinnaiyan AM, Rehemtulla A, Beer DG, Lawrence TS, Nyati MK, and Ray D (2014). Neoplasia 16, 115–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui FA, Alam C, Rosenqvist P, Ora M, Sabt A, Manoharan GB, Bindu L, Okutachi S, Catillon M, Taylor T, Abdelhafez OM, Lonnberg H, Stephen AG, Papageorgiou AC, Virta P, and Abankwa D (2020). ACS Omega 5, 832–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanshu DK, Nissley DV, and McCormick F (2017). Cell 170, 17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MA, Dodson GS, and Rubin GM (1993). Cell 73, 169–77. [DOI] [PubMed] [Google Scholar]
- Sjolander A, Yamamoto K, Huber BE, and Lapetina EG (1991). Proc Natl Acad Sci U S A 88, 7908–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skobeleva N, Menon S, Weber L, Golemis EA, and Khazak V (2007). Mol Cancer Ther 6, 898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MJ, Neel BG, and Ikura M (2013). Proc Natl Acad Sci U S A 110, 4574–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer-Smith R, Koide A, Zhou Y, Eguchi RR, Sha F, Gajwani P, Santana D, Gupta A, Jacobs M, Herrero-Garcia E, Cobbert J, Lavoie H, Smith M, Rajakulendran T, Dowdell E, Okur MN, Dementieva I, Sicheri F, Therrien M, Hancock JF, Ikura M, Koide S, and O’Bryan JP (2017). Nat Chem Biol 13, 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer-Smith R, Li L, Prasad S, Koide A, Koide S, and O’Bryan JP (2019). Small GTPases 10, 378–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoerner M, Hozsa C, Poetzl JA, Reiss K, Ganser P, Geyer M, and Kalbitzer HR (2010). J Biol Chem 285, 39768–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steklov M, Pandolfi S, Baietti MF, Batiuk A, Carai P, Najm P, Zhang M, Jang H, Renzi F, Cai Y, Abbasi Asbagh L, Pastor T, De Troyer M, Simicek M, Radaelli E, Brems H, Legius E, Tavernier J, Gevaert K, Impens F, Messiaen L, Nussinov R, Heymans S, Eyckerman S, and Sablina AA (2018). Science 362, 1177–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, Han W, Lou F, Yang J, Zhang Q, Wang X, He C, and Pan H (2013). Cell Death Dis 4, e838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Qian Y, Hamilton AD, and Sebti SM (1995). Cancer Res 55, 4243–7. [PubMed] [Google Scholar]
- Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, Lee T, Rossanese OW, and Fesik SW (2012). Angew Chem Int Ed Engl 51, 6140–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung PJ, Tsai FD, Vais H, Court H, Yang J, Fehrenbacher N, Foskett JK, and Philips MR (2013). Proc Natl Acad Sci U S A 110, 20593–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MN, Lu Z, Li YC, Zhou Y, Huang T, Reger AS, Hurwitz AM, Palzkill T, Logsdon C, Liang X, Gray JW, Nan X, Hancock J, Wahl GM, and Bast RC Jr. (2019). Cell Rep 29, 3448–3459 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, and Linder ME (2005). J Biol Chem 280, 31141–8. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Lobato MN, and Rabbitts TH (2003). Journal of Molecular Biology 331, 1109–1120. [DOI] [PubMed] [Google Scholar]
- Tanaka T, and Rabbitts TH (2010). Oncogene 29, 6064–70. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Williams RL, and Rabbitts TH (2007). EMBO J 26, 3250–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taveras AG, Remiszewski SW, Doll RJ, Cesarz D, Huang EC, Kirschmeier P, Pramanik BN, Snow ME, Wang YS, del Rosario JD, Vibulbhan B, Bauer BB, Brown JE, Carr D, Catino J, Evans CA, Girijavallabhan V, Heimark L, James L, Liberles S, Nash C, Perkins L, Senior MM, Tsarbopoulos A, Webber SE, and et al. (1997). Bioorg Med Chem 5, 125–33. [DOI] [PubMed] [Google Scholar]
- te Heesen H, Gerwert K, and Schlitter J (2007). FEBS Lett 581, 5677–84. [DOI] [PubMed] [Google Scholar]
- Thissen JA, Gross JM, Subramanian K, Meyer T, and Casey PJ (1997). J Biol Chem 272, 30362–70. [DOI] [PubMed] [Google Scholar]
- Thompson HJ, Briggs S, Paranka NS, Piazza GA, Brendel K, Gross PH, Sperl GJ, Pamukcu R, and Ahnen DJ (1995). J Natl Cancer Inst 87, 1259–60. [PubMed] [Google Scholar]
- Thompson HJ, Jiang C, Lu J, Mehta RG, Piazza GA, Paranka NS, Pamukcu R, and Ahnen DJ (1997). Cancer Res 57, 267–71. [PubMed] [Google Scholar]
- Ting PY, Johnson CW, Fang C, Cao X, Graeber TG, Mattos C, and Colicelli J (2015). FASEB J 29, 3750–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut TW (1994). Mol Cell Biochem 140, 1–22. [DOI] [PubMed] [Google Scholar]
- Tsutsumi R, Ran H, and Neel BG (2018). FEBS Open Bio 8, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uejima T, Ihara K, Goh T, Ito E, Sunada M, Ueda T, Nakano A, and Wakatsuki S (2010). J Biol Chem 285, 36689–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya P, Qian Z, Selner NG, Clippinger SR, Wu Z, Briesewitz R, and Pei D (2015). Angew Chem Int Ed Engl 54, 7602–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cutsem E, van de Velde H, Karasek P, Oettle H, Vervenne WL, Szawlowski A, Schoffski P, Post S, Verslype C, Neumann H, Safran H, Humblet Y, Perez Ruixo J, Ma Y, and Von Hoff D (2004). J Clin Oncol 22, 1430–8. [DOI] [PubMed] [Google Scholar]
- van Maldegem F, and Downward J (2020). Immunity 52, 14–16. [DOI] [PubMed] [Google Scholar]
- Vetter IR, and Wittinghofer A (2001). Science 294, 1299–304. [DOI] [PubMed] [Google Scholar]
- Vogt A, Qian Y, Blaskovich MA, Fossum RD, Hamilton AD, and Sebti SM (1995). J Biol Chem 270, 660–4. [DOI] [PubMed] [Google Scholar]
- Volker C, Miller RA, McCleary WR, Rao A, Poenie M, Backer JM, and Stock JB (1991). J Biol Chem 266, 21515–22. [PubMed] [Google Scholar]
- Wahlstrom AM, Cutts BA, Karlsson C, Andersson KM, Liu M, Sjogren AK, Swolin B, Young SG, and Bergo MO (2007). Blood 109, 763–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlstrom AM, Cutts BA, Liu M, Lindskog A, Karlsson C, Sjogren AK, Andersson KM, Young SG, and Bergo MO (2008). Blood 112, 1357–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MT, Holderfield M, Galeas J, Delrosario R, To MD, Balmain A, and McCormick F (2015). Cell 163, 1237–1251. [DOI] [PubMed] [Google Scholar]
- Ward ES, Gussow D, Griffiths AD, Jones PT, and Winter G (1989). Nature 341, 544–6. [DOI] [PubMed] [Google Scholar]
- Weber CK, Slupsky JR, Kalmes HA, and Rapp UR (2001). Cancer Res 61, 3595–8. [PubMed] [Google Scholar]
- Weise K, Kapoor S, Denter C, Nikolaus J, Opitz N, Koch S, Triola G, Herrmann A, Waldmann H, and Winter R (2011). J Am Chem Soc 133, 880–7. [DOI] [PubMed] [Google Scholar]
- Welsch ME, Kaplan A, Chambers JM, Stokes ME, Bos PH, Zask A, Zhang Y, Sanchez-Martin M, Badgley MA, Huang CS, Tran TH, Akkiraju H, Brown LM, Nandakumar R, Cremers S, Yang WS, Tong L, Olive KP, Ferrando A, and Stockwell BR (2017). Cell 168, 878–889 e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte DB, Kirschmeier P, Hockenberry TN, Nunez-Oliva I, James L, Catino JJ, Bishop WR, and Pai JK (1997). J Biol Chem 272, 14459–64. [DOI] [PubMed] [Google Scholar]
- Williams JG, Pappu K, and Campbell SL (2003). Proc Natl Acad Sci U S A 100, 6376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willumsen BM, Norris K, Papageorge AG, Hubbert NL, and Lowy DR (1984). EMBO J 3, 2581–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong GS, Zhou J, Liu JB, Wu Z, Xu X, Li T, Xu D, Schumacher SE, Puschhof J, McFarland J, Zou C, Dulak A, Henderson L, Xu P, O’Day E, Rendak R, Liao WL, Cecchi F, Hembrough T, Schwartz S, Szeto C, Rustgi AK, Wong KK, Diehl JA, Jensen K, Graziano F, Ruzzo A, Fereshetian S, Mertins P, Carr SA, Beroukhim R, Nakamura K, Oki E, Watanabe M, Baba H, Imamura Y, Catenacci D, and Bass AJ (2018). Nat Med 24, 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KA, Russo A, Wang X, Chen YJ, Lavie A, and O’Bryan JP (2012). PLoS One 7, e45360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright LP, and Philips MR (2006). J Lipid Res 47, 883–91. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Uys JD, Tew KD, and Townsend DM (2011). Antioxid Redox Signal 15, 233–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Pan Q, and Ju W (2019). Anticancer Drugs 30, 821–827. [DOI] [PubMed] [Google Scholar]
- Xu L, Lubkov V, Taylor LJ, and Bar-Sagi D (2010). Curr Biol 20, 1372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi J, Miyamoto Y, Tanoue A, Shooter EM, and Chan JR (2005). Proc Natl Acad Sci U S A 102, 14889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Jahanshahi M, Horvath EA, Liu HY, and Pfleger CM (2010). Curr Biol 20, 1378–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Roy S, Apolloni A, Lane A, and Hancock JF (1998). J Biol Chem 273, 24052–6. [DOI] [PubMed] [Google Scholar]
- Yang J, Nie J, Ma X, Wei Y, Peng Y, and Wei X (2019). Mol Cancer 18, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MH, Laurent G, Bause AS, Spang R, German N, Haigis MC, and Haigis KM (2013). Mol Cancer Res 11, 1072–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MH, Nickerson S, Kim ET, Liot C, Laurent G, Spang R, Philips MR, Shan Y, Shaw DE, Bar-Sagi D, Haigis MC, and Haigis KM (2012). Proc Natl Acad Sci U S A 109, 10843–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’antonio G, Mautner J, Tonon G, Haigis M, Shirihai OS, Doglioni C, Bardeesy N, and Kimmelman AC (2011). Genes Dev 25, 717–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WS, Yeo SG, Yang S, Kim KH, Yoo BC, and Cho JY (2017). Amino Acids 49, 1469–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin G, Kistler S, George SD, Kuhlmann N, Garvey L, Huynh M, Bagni RK, Lammers M, Der CJ, and Campbell SL (2017). J Biol Chem 292, 4446–4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurdaydin C, Keskin O, Kalkan C, Karakaya F, Caliskan A, Karatayli E, Karatayli S, Bozdayi AM, Koh C, Heller T, Idilman R, and Glenn JS (2018). Hepatology 67, 1224–1236. [DOI] [PubMed] [Google Scholar]
- Zambrano J, and Yeh ES (2016). Breast Cancer (Auckl) 10, 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LF, Zhang RY, Yu ZH, Li S, Wu L, Gunawan AM, Lane BS, Mali RS, Li X, Chan RJ, Kapur R, Wells CD, and Zhang ZY (2014a). J Med Chem 57, 6594–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng T, Wang Q, Fu J, Lin Q, Bi J, Ding W, Qiao Y, Zhang S, Zhao W, Lin H, Wang M, Lu B, Deng X, Zhou D, Yin Z, and Wang HR (2014b). Cell Rep 7, 871–82. [DOI] [PubMed] [Google Scholar]
- Zhang FL, Kirschmeier P, Carr D, James L, Bond RW, Wang L, Patton R, Windsor WT, Syto R, Zhang R, and Bishop WR (1997). J Biol Chem 272, 10232–9. [DOI] [PubMed] [Google Scholar]
- Zhang X, He Y, Liu S, Yu Z, Jiang ZX, Yang Z, Dong Y, Nabinger SC, Wu L, Gunawan AM, Wang L, Chan RJ, and Zhang ZY (2010). J Med Chem 53, 2482–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Guo W, Wu Y, Yang C, Zhong L, Deng G, Zhu Y, Liu W, Gu Y, Lu Y, Kong L, Meng X, Xu Q, and Sun Y (2019). Acta Pharm Sin B 9, 304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Qiu Y, and Kong D (2017). Acta Pharm Sin B 7, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, McKay J, and Buss JE (2007). J Biol Chem 282, 25760–8. [DOI] [PubMed] [Google Scholar]
- Zhou Y, and Hancock JF (2015). Biochim Biophys Acta 1853, 841–9. [DOI] [PubMed] [Google Scholar]
- Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, Hoffmann M, Hahn SA, Triola G, Wittinghofer A, Bastiaens PI, and Waldmann H (2013). Nature 497, 638–42. [DOI] [PubMed] [Google Scholar]
- Zinatizadeh MR, Momeni SA, Zarandi PK, Chalbatani GM, Dana H, Mirzaei HR, Akbari ME, and Miri SR (2019). Genes Dis 6, 378–384. [DOI] [PMC free article] [PubMed] [Google Scholar]