Abstract
Background
High recurrence and chemoresistance drive the high mortality in hepatocellular carcinoma (HCC). Although cancer stem cells are considered to be the source of recurrent and chemoresistant tumors, they remain poorly defined in HCC. Tonicity-responsive enhancer binding protein (TonEBP) is elevated in almost all HCC tumors and associated with recurrence and death. We aimed to identify function of TonEBP in stemness and chemoresistance of liver cancer.
Methods
Tumors obtained from 280 HCC patients were analyzed by immunohistochemical analyses. Stemness and chemoresistance of liver CSCs (LCSCs) were investigated using cell culture. Tumor-initiating activity was measured by implanting LCSCs into BALB/c nude mice.
Findings
Expression of TonEBP is higher in LCSCs in HCC cell lines and correlated with markers of LCSCs whose expression is significantly associated with poor prognosis of HCC patients. TonEBP mediates ATM-mediated activation of NF-κB, which stimulates the promoter of a key stem cell transcription factor SOX2. As expected, TonEBP is required for the tumorigenesis and self-renewal of LSCSs. Cisplatin induces the recruitment of the ERCC1/XPF dimer to the chromatin in a TonEBP-dependent manner leading to DNA repair and cisplatin resistance. The cisplatin-induced inflammation in LSCSs is also dependent on the TonEBP-ERCC1/XPF complex, and leads to enhanced stemness via the ATM-NF-κB-SOX2 pathway. In HCC patients, tumor expression of ERCC1/XPF predicts recurrence and death in a TonEBP-dependent manner.
Interpretation
TonEBP promotes stemness and cisplatin resistance of HCC via ATM-NF-κB. TonEBP is a key regulator of LCSCs and a promising therapeutic target for HCC and its recurrence.
Keywords: Hepatocellular carcinoma recurrence, SOX2, ERCC1/XPF, NF-κB
Research in context.
Evidence before this study
Hepatocellular carcinoma (HCC) is a common cancer with high mortality rate due to extremely high recurrence rate and lack of effective chemotherapeutic agents. We previously showed that expression of TonEBP in tumors and surrounding areas predicted recurrence and death, but underlying mechanisms remain undefined. Cancer stem cells (CSCs) are essential in tumor recurrence due to their tumor-initiating capacity and inherent chemoresistance. Although several surface markers of CSCs have been reported in HCC, molecular pathways involved are not known.
Added value of this study
Here, we uncover molecular pathways responsible for the recurrence and chemoresistance in HCC. TonEBP relays local inflammatory signals to the ATM-NF-κB pathway leading to SOX2 expression and HCC stemness indicating the importance of inflammatory microenvironment. Surprisingly, treatment with cisplatin activates this pathway in a manner dependent on the interaction between TonEBP and the ERCC1/XPF dimer, which is involved in a variety of DNA repair processes. High expression of ERCC1/XPF in tumors predicts recurrence and death in a manner dependent on the level TonEBP expression with high levels of significance. Thus, inflammatory signals and DNA damages caused by cisplatin activate surviving CSCs, both in a manner dependent on TonEBP.
Implications of all the available evidence
The ATM-NF-κB pathway is an attractive therapeutic target for HCC and its recurrence. Inhibiting the dual TonEBP actions might be an effective strategy for this.
Alt-text: Unlabelled box
1. Introduction
Most cancer patients die not from the primary tumor but from a reconstituted tumor. The 2nd or 3rd most likely leading cause of cancer-associated death is liver cancer, in which hepatocellular carcinoma (HCC) is the major histological subtype accounting for 70–85% of cases of primary liver cancer [1]. Surgical resection has been considered as a primary treatment option of liver cancer due to inherent resistance to chemotherapeutic agents [2]. However, recurrence is nearly 70% even after complete resection leading to a very high mortality. Despite numerous efforts, molecular pathways involved in recurrence as well as chemoresistance of HCC remain poorly understood [2, 3].
Cancer stem cells (CSCs) or tumor-initiating cells, a small subset of cells within a solid tumor, are considered to be the source of recurrent and chemoresistant tumors [4]. Identification of CSCs arises from the finding that tumor cells are heterogeneous [5, 6]. CSCs display elevated ability to cope with cellular stresses including resistance to chemotherapeutic agents and highly activated DNA damage response (DDR) [7]. CD44, CD133, EpCAM, or ALDH are established as CSC markers for various cancers [8], [9], [10], [11]. Expression of these markers is associated with recurrence and death. Recently, markers for liver stem cells (LCSCs) have been proposed [12, 13]. However, their functional role in HCC patients are not fully defined and mechanistic basis for tumor initiation leading to HCC is not known.
The transcription factors NF-κB are highly activated in tumor cells and CSCs [14] due to DNA damages caused by exposure to environmental toxins or chemotherapeutic agents. NF-κB reprograms transcription network required for the cancer stemness [15] and the cellular process involved in DDR [16]. Although ataxia–telangiectasia-mutated (ATM) protein kinase is responsible for the activation of NF-κB, upstream signals are not known.
Tonicity-responsive enhancer binding protein (TonEBP), also known as NFAT5, was discovered as a DNA binding transcriptional enhancer [17]. Numerous studies have revealed, however, that TonEBP has many other functions [18]. TonEBP is a central component of the NF-κB enhanceosome assembled on promoters of inflammatory genes [19]. TonEBP expression is stimulated by inflammation leading to elevated expression of pro-inflammatory genes and chronic inflammatory diseases [[20], [21], [22], [23], [24]]. In addition, TonEBP is recruited to the sites of DNA damage as an upstream regulator of DDR [25]. The same study also revealed that TonEBP interactome includes 250 proteins associated with DDR suggesting that TonEBP has more functions related to DDR. Of great interest, we recently reported that TonEBP promoted hepatocellular carcinogenesis and recurrence [26]. Expression of TonEBP is higher in tumors compared to surrounding non-tumor regions in greater than 90% of HCC patients regardless of the etiology of HCC. Although higher TonEBP expression in tumors predicted recurrence and death in the HCC patients, underlying mechanism has not been elucidated. In this study, we discover that TonEBP is a key regulator for the central signaling pathways in LCSCs: TonEBP is the upstream regulator of ATM in two separate pathways. TonEBP stimulates ATM in response to inflammatory signals leading to activation of NF-κB, which in turn enhances stemness and chemoresistance. In the other pathway, TonEBP recruits the ERCC1/XPF dimer, a prominent DNA repair protein complex, to the chromatin. The ERCC1/XPF stimulates the ATM-NF-κB axis in a TonEBP-dependent manner as well as DNA repair in response to cisplatin treatment. Thus, TonEBP regulates both of the two key properties of LCSCs – self-renewal and chemoresistance.
2. Materials and methods
2.1. Human HCC samples and clinical information
This research was approved by the Institutional Review Board at the Ulsan University Hospital (UUH 2015-12-018). A total of 280 patients who underwent hepatic resection for HCC from January 2008 to February 2017 at the Ulsan University Hospital, University of Ulsan, College of Medicine, Ulsan, Korea, were included in the study. Written consent form was received from each patient. All patients were HCC treatment-naïve before surgery. The patients were predominantly males (84.6%) with average age of 56.6 years. The median follow-up period was 38.0 months (range = 1–117 months). Postoperative recurrence was observed in 150 cases (53.6%). During postoperative follow-up period, metastasis and death were observed in 63 (22.5%) and 92 (32.9%) of cases, respectively. Data were expressed as mean +/- standard deviation or median (range). For statistical significance, Student t-test and chi-squared test were used for comparisons of variables between groups. The cumulative relapse and survival rates were evaluated by the Kaplan–Meier method, and differences were determined by the log-rank test. A multivariate analysis was carried out to identify the independent predictor for recurrence, metastasis and survival using the Cox regression hazard model. All data were analyzed using the statistical package SPSS for Windows (version 21.0; SPSS Inc.). In all cases, a two-tailed P-value less than 0.05 was considered statistically significant. Additional protocols and procedures are described below.
2.2. Tissue array
Human HCC patient tissue samples were collected as previously described [26]. H&E of each patient was analyzed by histologist, and hepatic tumor regions of 280 HCC patients in formalin-fixed paraffin-embedded tissue were marked and extracted for tissue array. Then, extracted tissues were arranged, molded by using tissue microarray cassette, and solidified for tissue array analysis. The arrays were processed simultaneously for immunohistochemistry.
2.3. Oncosphere formation assay
Cells were seeded on ultra-low attachment culture dishes (Corning) in serum-free medium. DMEM/F12 serum-free medium (Invitrogen) contained 2 mM l-glutamine, 1% sodium pyruvate (Invitrogen), 100 μg/ml penicillin G, and 100 U/ml streptomycin supplemented with 20 ng/ml epithelial growth factor (Invitrogen), 10 ng/ml fibroblast growth factor-2 (Invitrogen), N2 (Invitrogen), and B27 (Invitrogen). Cells were cultured for one to two weeks.
2.4. Animals
All the methods involving live mice were carried out in accordance with the approved guidelines. All experimental protocols were approved by the Institutional Animal Care and Use Committee of the Ulsan National Institute of Science and Technology (UNISTACUC-12-15-A).
To induce HCC, we administrated a single intraperitoneal injection of 25 mg/kg diethylnitrosamine (DEN) (N0756; Sigma) to 2-week-old C57BL/6 mice and euthanised them at 9 months of age [25].
2.5. Xenograft transplantation of CSCs
For subcutaneous injection models, different dilutions (101, 102, 103, or 104) of control and treated cells from oncosphere formation assay were implanted into mice (female BALB/c nude mice), aged 4 to 6 weeks, with Matrigel Basement Membrane Matrix (BD biosciences) into two sides of the same nude mouse at the posterior dorsal flank region (n = 6–8). Tumor-initiating frequency was calculated by limiting dilution assay according to the protocol available from web (http://bioinf.wehi.edu.au/software/elda/). The mice were maintained under standard conditions according to the institutional guidelines for animal care.
2.6. Immunohistochemistry (IHC) and histology analysis
Liver tissues were fixed overnight in 4% paraformaldehyde, embedded in paraffin, and cut into 5 μm sections. Paraffin sections were deparaffinized and dehydrated. Antigen was retrieved by citrate and peroxidase or EDTA and peroxidase in appropriate time for each antigen. Anti-ERCC1 antibodies (CST), anti-XPF antibodies (Abcam), anti-EpCAM antibodies (Millipore) and anti-CD44 antibodies (Abcam) were used for IHC using optimized conditions. Signal intensity from immunohistochemical analyses were measured and sub-grouped using image software (Fiji-Image J).
2.7. Cell line
Hep3B from American Type Culture Collection (ATCC) was maintained in modified Eagle's medium (Hyclone) supplemented with 10% fetal bovine serum (FBS; Thermo) with penicillin-streptomycin (Hyclone). PLC/PRF/5 and Huh7 cells from ATCC were maintained in Roswell Park Memorial Institute 1640 supplemented with 2.05 mM l-glutamine, 25 mM HEPES, 10% FBS, and penicillin-streptomycin. U2OS cells was maintained in Dulbecco's Modified Eagle Medium supplemented with 10% FBS with penicillin-streptomycin. Cells were maintained at 37 °C in incubator with 5% CO2. Cells were transfected with siRNA (3 nM) using lipofectamine RNAiMAX (Invitrogen) following the manufacturer's instructions. Control shRNA and TonEBP shRNA were used for generating lentivirus-mediated knockdown in HCC cell lines. Positive cells were selected with puromycin for 5 days.
Cell viability was determined by counting the viable cells or measuring the reduction of 3-(4,5-Dimethyl-2-thiazolyl)−2,5-diphenyl-2H-tetrazolium bromide (MTT, Sigma). To examine cell death, cells were cultured under described conditions in 96-well plates (Corning) and cell death was examined Caspase-Glo® (Promega) assays.
2.8. Quantitative real-time PCR (RT-qPCR)
Total RNA was isolated using the TRIzol reagent (Invitrogen) followed by chloroform and ethanol precipitation. cDNA was synthesized by M-MLV reverse transcriptase (Promega) according to the manufacturer's instructions. After reverse transcription, Q-PCR was performed using SYBR Green I Master and LightCycler 480 II (Roche). Measured cycle threshold values were normalized with GAPDH and they were expressed as fold-over control samples. All RT-qPCR reactions were duplicated.
| Name | Forward | Reverse |
|---|---|---|
| SOX2 | GCCGAGTGGAAACTTTTGTCG | GGCAGCGTGTACTTATCCTTCT |
| Oct4 | CTGGGTTGATCCTCGGACCT | CCATCGGAGTTGCTCTCCA |
| Nanog | TTTGTGGGCCTGAAGAAAACT | AGGGCTGTCCTGAATAAGCAG |
| c-Myc | GGCTCCTGGCAAAAGGTCA | CTGCGTAGTTGTGCTGATGT |
| KLF4 | ACGATCGTGGCCCCGGAAAAGGACC | CAACAACCGAAAATGCACCAGCCCCAG |
| Nestin | GCAAAGGAGCCTACTCCAAG | AGATGGAGCAGGCAAGAGAT |
| CD90 | AGGACGAGGGCACCTACAC | GCCCTCACACTTGACCAGTT |
| CD133 | AAGGCATATGAATCCAAAATTGA | CCACCAGAGGCATCAGAATAA |
| EpCAM | AGAACCTACTGGATCATCATTGAACTAA | CGCGTTGTGATCTCCTTCTG |
| E-cad | TGCCCAGAAAATGAAAAAGG | GTGTATGTGGCAATGCGTTC |
| N-cad | ACAGTGGCCACC TACAAAGG | CCGAGATGGGGTTGATAATG |
| FN1 | CAGTGGGAGACCTCGAGAAG | TCCCTCGGAACATCAGAAAC |
| Vm | GAGAACTTTGCCGTTGAAGC | GCTTCCTGTAGGTGGCAATC |
| TNFα | CTCTTCTCCTTCCTGATCGTGGCA | GTTGGATGTTCGTCCTCCTCACA |
| IL-8 | AAGGAACCATCTCACTGTGTGTAAAC | ATCAGGAAGGCTGCCAAGAG |
| CCL2 | GGCTGAGACTAACCCAGAAAC | GAATGAAGGTGGCTGCTATGA |
2.9. Immunoblotting
Cell lysis for protein extraction was performed as previously described [19, 26]. Briefly, Cells were washed two times with cold PBS and lysed in RIPA buffer (0.01 M Tris, pH 7.4, 0.15 M NaCl, 0.001 M EDTA, 0.001 M EGTA, 1% Triton-X 100, 0.002 M PMSF, and protease inhibitor (Roche). After centrifugation of lysate, supernatant was used for immunoblot assay. Protein concentration was measured by BCA system (Pierce). Equal amounts of protein from each sample were separated by SDS-PAGE and immunoblotted using specific primary antibodies. HRP-conjugated secondary antibodies were used for detection. The antigen-antibody binding was detected by enhanced chemiluminescence Western blotting detection reagents (GE healthcare life sciences). Anti-SOX2 antibodies (CST); anti-phospho-ATM antibodies (CST); anti-ATM antibodies (CST); anti-phospho-p65 antibodies (Santa Cruz); anti-p65 antibodies (CST); anti-phsohpo-STAT3 antibodies (CST); anti-STAT3 antibodies (CST).
2.10. Flow cytometry and cell sorting
Cells were stained using antibodies according to the manufacturer's instructions. Labeled cells were detected using FACSCalibur (BD Immunocytometry Systems). For sorting of CD133 and CD90 double positive cells, cocktail PE-conjugated anti-human CD133 (Miltenyi) and FITC-conjugated anti-human CD90 antibodies (Miltenyi) were incubated with cells. Then cells were sorted with FACS Aria III (BD Immunocytometry Systems). For sorting of CD133 positive cells, magnetic-activated cell sorting (MACS) technology based on CD133 (Miltenyi) were performed.
2.11. Oncosphere formation assay
Cells were seeded on ultra-low attachment culture dishes (Corning) in serum-free medium. DMEM/F12 serum-free medium (Invitrogen) contained 2 mM l-glutamine, 1% sodium pyruvate (Invitrogen), 100 μg/ml penicillin G, and 100 U/ml streptomycin supplemented with 20 ng/ml epithelial growth factor (Invitrogen), 10 ng/ml fibroblast growth factor-2 (Invitrogen), N2 (Invitrogen), and B27 (Invitrogen). Cells were cultured for one to two weeks. For limiting dilution assay, different number of cells were plated in 96-well multiwall plate and the number of wells not forming oncosphere was counted and stem cell frequency was calculated with limiting dilution assay according to the protocol available from web (http://bioinf.wehi.edu.au/software/elda/).
2.12. TCGA analysis
Upper Quantile normalized FPKM (FPKM-UQ) of RNA-seq and matched clinical information of HCC cohorts was extracted from metadata of TCGA of Genomics Data Commons portal (https://gdc-portal.nci.nih.gov). With R studio, the transcript level of TonEBP and SOX2 was analyzed in HCC patients expressing SOX2 (n = 271). Correlation analysis between TonEBP and SOX2 were generated using R-derived RNA-seq and plotted with Excel.
2.13. Annexin-V/propidium iodide (PI) double staining assay
Apoptotic cells were determined with an FITC-Annexin V Apoptosis Detection Kit (BD Biosciences, San Diego, CA, USA) according to the manufacturer's protocol. Briefly, the cells were washed and incubated for 15 min at room temperature in the dark containing FITC-Annexin V. Afterwards, apoptotic cells were analyzed by flow cytometry (BD Biosciences, San Diego, CA, USA).
2.14. Immunofluorescence, microscopy, and image analysis
Cells were plated in LabTek chamber slides (Thermo Fisher Scientific) and incubated before fixation with 4% paraformaldehyde for 20 min at 25 °C before permeabilization (15 min) with PBS containing 0.2% Triton X-100. Antibody incubation was performed as recommended, followed by mounting. Edu (Thermo Fisher Scientific) labeling is performed as manufacturer's instructions (Click-iT™ EdU Alexa Fluor™ 594 Imaging Kit).
2.15. Nuclear and cytoplasmic fractionation
Cells were harvested by using scrapper and centrifuged. The cell pellet was washed by suspension with PBS. The cell nucleus and cytoplasm were separated by using Nuclear and Cytoplasmic extraction kit (Pierce) according to manufacturer's instruction.
2.16. Tight chromatin-bound fractionation
Tight chromatin-bound fraction was isolated as previously described [27]. In brief, cell pellets were sequentially washed in CEBN buffer (10 mM HEPES [pH 7.8], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 0.2% NP-40, 1X protease inhibitor cocktail (Invitrogen), and 1X phosphatase Inhibitor cocktail), CEB buffer (CEBN buffer without NP-40), soluble nuclear buffer (3 mM EDTA, 0.2 mM EGTA, protease and phosphatase inhibitor), and salt buffers with 0.45 M NaCl buffer.
2.17. Promoter and reporter assay
Human SOX2 promoter-containing plasmids were obtained from Addgene (pGL3-Sox2, Plasmid #101761) were inserted into pGL3 (Promega). κB binding sites in SOX2 promoter was mutated using cloned SOX2 promoter with primers and indicated as ΔκB. Cells were transfected luciferase plasmid. The Renilla luciferase reporter plasmid (pRL-TK, Promega) was used as a control for transfection efficiency. Luciferase activity after 6 h of stimulation was measured using the Dual-Luciferase Assay System (Promega) according to the manufacturer's instructions. Luciferase activity was normalized by activity of renilla luciferase.
2.18. Immunoprecipitation and chromatin immunoprecipitation (ChIP)
For preparation of lysates for IP, cells were washed three times with ice-cold PBS and lysed in RIPA buffer as described previously [18]. An appropriate antibody was added to lysates and incubated overnight at 4 °C, followed by incubation with Protein A/G Sepharose beads (GE Healthcare Sciences). After extensive washing with RIPA lysis buffer, complexes were eluted and analyzed by immunoblotting. Anti-ERCC1 antibodies (CST); anti-XPF antibodies (Abcam); anti-Myc antibodies (CST). ChIP was performed as previous studies [25]. In brief, cells were crosslinked with 1% formaldehyde followed by addition of 125 mM glycine. After washing, cells were sonicated and immunoprecipitated with anti-IgG, and anti-p65 (CST) antibodies at 4 °C overnight. After elution and reverse crosslinking the antibody/DNA complexes, DNA was purified by DNA purification kit (Qiagen) and analyzed by q-PCR using primer pairs covering specific region of the SOX2 promoter in duplicates.
2.19. TonEBP interaction analysis
We analyzed the candidate of TonEBP-interacting protein from the interactome data of previous report [25]. To isolate TonEBP-interacting proteins, HEK293 cells were transfected with the pcDNA5/FRT empty vector or pcDNA5/FRT-TonEBP-Flag plasmid. The cells were lysed in 1 ml of RIPA buffer supplemented with a protease inhibitor cocktail and 0.5 mM phenylmethylsulfonyl fluoride at 24 h post-transfection. Cell extracts were recovered by centrifugation at 12,000 rpm (Eppendorf, 5424R) for 15 min at 4 °C. The cleared supernatants were collected, and the cell lysates were mixed with 50 ml (50% slurry) of anti-Flag M2 resin (Sigma-Aldrich) and incubated at 4 °C for four hours. After centrifugation at 2500 rpm (Eppendorf, 5424R) for five minutes, the supernatants were discarded, and the collected resin samples were washed in 1 ml of RIPA lysis buffer five times. Flag-tagged TonEBP and bound proteins were eluted with excessive amounts of the 3 × Flag peptide (Sigma-Aldrich). Eluted proteins were subjected to immunoblotting. The molecular identities of the eluted proteins were determined by mass spectrometry.
2.20. Quantification and statistical analysis
Data are presented as means + S.D or means + S.E.M. Statistical significance (p < 0.05) was estimated by an unpaired t‐test for comparisons between two conditions. Two-way ANOVA was performed for multiple comparison. All statistical analyses were performed using Student's t-test. n = number of experimental replicates.
3. Results
3.1. TonEBP promotes self-renewal of liver cancer stem cells
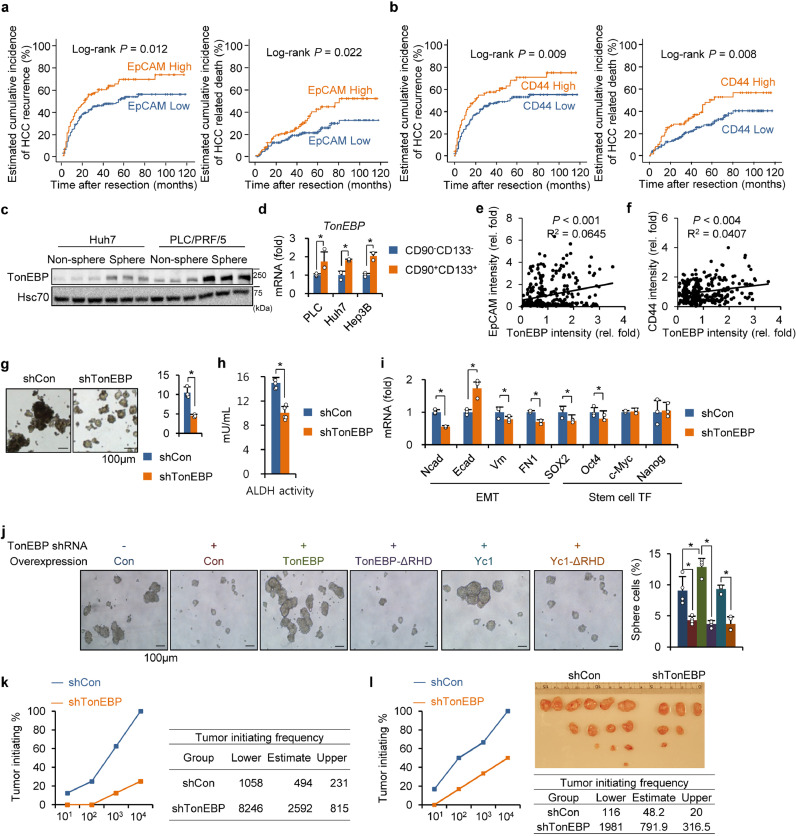
The motivation of this study was to understand how hepatic TonEBP expression leads to recurrence of HCC [26]. In order to gain a clue on molecular pathways involved in the recurrence, we decided to examine known markers of LCSCs (CD44, EpCAM, CD133, CD13, CD90, c-kit, and CK-19) in the cohort of HCC patients we had studied [26]. We found that high expression of EpCAM and CD44 was observed in 102 and 94 patients, respectively, out of 280 patients (Supplementary Fig. 1a,b). We did not pursue other markers (CD133, CD13, CD90, c-kit, and CK-19) because their expression was limited in our cohort, i.e., expression was detected in less than 10% of cases, making it difficult for meaningful analysis. Interestingly, the expression of EpCAM and CD44 correlated with each other with very high significance (Supplementary Fig. 1c and d). Kaplan-Meier analyses revealed that high expression of EpCAM and CD44 each correlated significantly with recurrence and death (Fig. 1a,b). These data provide evidence that LSCSs are involved in the recurrence of HCC in our cohort providing a basis for us to pursue cancer stem cells.
Fig. 1.
TonEBP is required for the self-renewal of cancer stem cells. (a,b) Kaplan–Meier plot of postoperative HCC recurrence and HCC related death in two groups of patients based on immunohistochemical signals in tissue microarrays prepared from 280 patients with HCC. (a) EpCAMlow (n = 178) versus EpCAMhigh (n = 102) and (b) CD44low (n = 186) versus CD44high(n = 94). (c) Huh 7 and PLC/PRF/5 cells were cultured for oncosphere formation. Cells in non-sphere and spheres were immunoblotted for TonEBP and Hsc70. (d) CD90−CD133− and CD90+CD133+ cells were obtained from PLC/PRF/5 (PLC), Huh7, and Hep3B cells. TonEBP mRNA was measured by RT-qPCR. Mean + SD. *p < 0.05. (e,f) Relative TonEBP versus (e) EpCAM or (f) CD44 abundance in tumors from the 280 patients with HCC. (g) TonEBP expression was stably reduced using TonEBP targeting lentivirus (shTonEBP) in PLC/PRF/5 cells, or not (shCon: control vector). Representative images from oncosphere formation assay (left). Percentage of sphere cells was obtained (right). Mean + SD. *p < 0.05 (h) CD90+CD133+ cells were isolated from the shTonEBP and shCon cells. ALDH activity was measured in cell lysates. Mean + SD, *p < 0.05. (i) RT-qPCR analyses of EMT genes and stem cell transcription factor (TF) genes in the spheres. Mean + SD. *p < 0.05. (j) The shCon (-) and shTonEBP cells (+) described above were transfected with an expression plasmid containing TonEBP, TonEBP-ΔRHD, Yc1, or Yc1-ΔRHD as indicated. Cells were cultured for oncosphere formation and representative images are shown (left). Percentages of sphere cells as Mean + SD (right). *p < 0.05. (k,l) Tumor initiation was measured from (k) Hep3B or (l) PLC/PRF/5 cells as described in Results and expressed as tumor initiating% (graph at left). From these data, tumorigenic cell frequency was calculated with limiting dilution assays according to the protocol available from web (http://bioinf.wehi.edu.au/software/elda/) and presented on the right as a table. Images of tumors formed are shown for PLC/PRF/5 cells.
Our previous study shows that expression of TonEBP is higher in tumors compared to adjacent non-tumor in HCC patients regardless of etiology [26]. Since expression of TonEBP in the tumor is significantly associated with recurrence [26], we set out to explore the role of TonEBP in LCSCs. LCSCs within the HCC cell lines in culture were enriched by oncosphere culture (Fig. 1c) or by selecting for surface markers CD90 and CD133 (Fig. 1d) [27], [28], [29]. Expression of TonEBP was higher in the oncospheres compared to non-sphere; likewise, CD90+CD133+ cells exhibited higher expression of TonEBP compared to their counterpart CD90−CD133− cells. In addition, expression of TonEBP showed weak but significant correlation with both EpCAM and CD44 in the tissue of microarrays of HCC patients (Fig. 1e, f). Furthermore, expression of cancer stem cell-related genes SOX2, Oct4, and Nanog was reduced in TonEBP haplodeficient animals in the DEN-induced HCC model [26] (Supplementary Fig. 1e). These observations suggest that TonEBP plays an important role in LCSCs.
Given the association of TonEBP and LCSCs, we directly investigated the role of TonEBP in LCSCs. Lentiviral knockdown of TonEBP significantly reduced oncosphere formation (Fig. 1g), stem cell frequency (Supplementary Fig. 1f), and ALDH activity (Fig. 1h), while overexpression of TonEBP enhanced oncosphere formation (Supplementary Fig. 1 g, h). In addition, decreased expression of genes related to the cancer stemness and LCSCs markers was observed in the TonEBP-deficient oncosphere population (Fig. 1i and Supplementary Fig. 1i) while the opposite was observed in the TonEBP-overexpressed oncospheres (Supplementary Fig. 1j). These results demonstrate that TonEBP drives the self-renewal of LCSCs. Next, we asked which domain of TonEBP is responsible for the oncosphere formation. Rel-homology domain (RHD) consists of about 270 amino acid residues near the N-terminus of TonEBP. RHD is responsible for DNA binding [17], and interactions with NF-κB and proteins involved in DDR [19, 25]. In order to define the role of RHD in oncosphere formation, various constructs were transfected to the cells whose TonEBP was stably knocked down: TonEBP, TonEBP-ΔRHD (RHD deleted), Yc1 (N-terminal one third of TonEBP including RHD), and Yc1-ΔRHD. As shown in Fig. 1j, TonEBP and Yc1 restored the oncosphere formation while TonEBP-ΔRHD and Yc1-ΔRHD did not indicating that RHD is essential in the self-renewal of LCSCs. These results suggest that protein-protein interactions are critical for the TonEBP's ability to drive the self-renewal of LCSCs.
Ability to form reconstituted tumor is the key feature of CSCs. We next assessed the function of TonEBP in tumor initiation. The oncospheres from the TonEBP knocked down or control cells described above were trypsin digested to form single cell suspensions. 104, 103, 102 or 101 cells were subcutaneously injected into BALB/c nude mice and tumors were examined 3 months later. Both in Hep3B (Fig. 1k) and PLC/PRF/5 cells (Fig. 1l) tumor initiating capacity and tumor initiating frequency were dramatically reduced in the TonEBP-deficient cells demonstrating that TonEBP is required for reconstituted tumor formation. Taken together with the roles of TonEBP in the expression of markers for LCSCs and self-renewal of LCSCs described above, these results demonstrate that TonEBP promotes self-renewal of LCSCs.
3.2. TonEBP promotes cancer stemness via ATM-NF-κB-SOX2
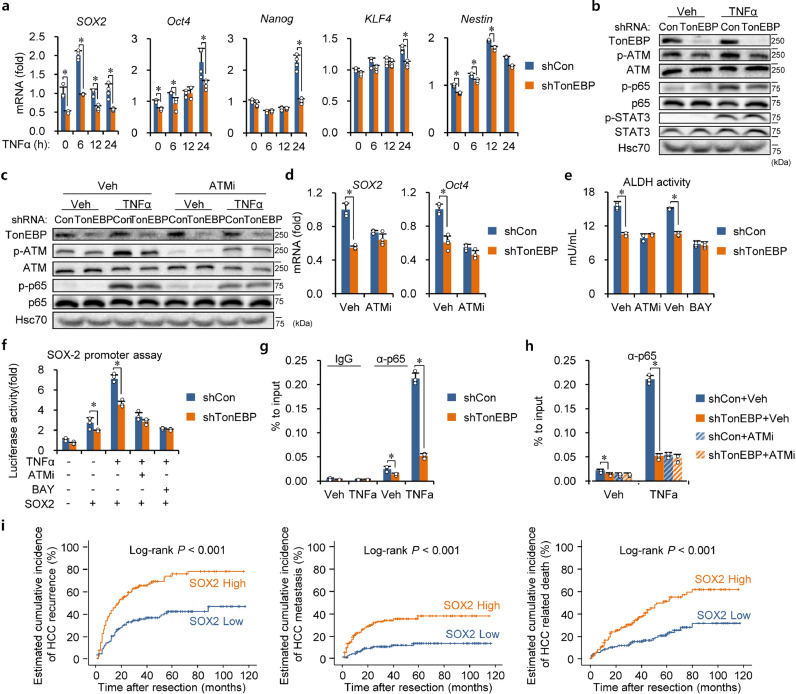
Inflammation has been recognized as a major contributing factor for recurrent tumors [30]. In fact, inflamed tumor microenvironment is critical for self-renewal of CSCs [14, 30]. We previously observed that hepatic TonEBP deficiency mitigated inflammation and liver injury [26]. To explore the hypothesis that TonEBP promotes cancer stemness in response to inflamed environment, we examined effects of TNFα on expression of stem cells transcription factors in HCC cells. While mRNA for Oct4, Nanog, KLF4 and Nestin was elevated in a TonEBP-dependent manner, their increase was preceded by that of SOX2 mRNA which peaked at 6 h of treatment (Fig. 2a). As expected, there is a clear increase in the protein level of SOX2, also in a TonEBP-dependent manner (Supplementary Fig. 2a). Thus, TonEBP has prominent influence on SOX2 expression in LCSCs.
Fig. 2.
TonEBP promotes cancer stemness via ATM-NF-κB-SOX2. (a) Stem cell transcription factors were analyzed in shCon and shTonEBP PLC/PRF/5 cells after treatment with TNFα (10 ng/ml) for up to 24 h, as indicated, using RT-qPCR. Mean + SD. *p < 0.05. (b) Immunoblot analyses of p-ATM, ATM, p-p65, p65, p-STAT, STAT3 and TonEBP after 2 h treatment with TNFα or vehicle (Veh). (c) Cells were pretreated with Veh or ATMi followed by a 24 h treatment with TNFα. mRNA of SOX2 and Oct4 were measured. Mean + SD. *p < 0.05. (d) PLC/PRF/5 cells were transfected with pGL3 (-, bottom) or SOX2 promoter reporter in pGL3 (+). Pretreatment with ATMi or BAY followed by treatment with TNFα (6 h) were performed as indicated. Luciferase activity is shown in mean + SD. *p < 0.05. (e) CD90+CD133+ cells were treated with ATMi or BAY (5 μM of BAY 11–7082) for 24 h. ALDH activity in mean + SD. *p < 0.05. (f) PLC/PRF/5 cells were transfected with pGL3 (-, bottom) or SOX2 promoter reporter in pGL3 (+). Pretreatment with ATMi or BAY followed by treatment with TNFα (6 h) were performed as indicated. Luciferase activity is shown in mean + SD. *p < 0.05. (g) ChIP was performed on cells treated with Veh or TNFα (2 h) using anti-p65 IgG (α-p65) or normal IgG (IgG). DNA fragments containing the κB site in the SOX2 promoter was quantified using qPCR. Mean + SD. *p < 0.05. (h) ChIP was performed using anti-p65 IgG on cells pretreated with Veh or ATMi followed by a 2 h treatment with Veh or TNFα. Mean + SD. *p < 0.05. (i) Kaplan-Meier plot of postoperative recurrence, metastasis, and HCC related death in two groups of patients: SOX2low (n = 137) vs. SOX2high (n = 143).
We next investigated how TonEBP regulates the expression of SOX2. Since SOX2 expression in CSCs is driven by NF-κB [31, 32], we examined NF-κB and its upstream regulator ATM [33]. As expected, phosphorylation (activation) of ATM and p65 was stimulated by TNFα (Fig. 2b) in RHD- dependent manner (Supplementary Fig. 2b). Since there is no evidence of direct interaction between TonEBP and ATM, we conclude that TonEBP indirectly activates ATM and subsequent signaling. Interestingly, the activation of ATM and p65 was TonEBP-dependent as it was blocked by TonEBP knockdown. ATM inhibitor (ATMi, KU55933) blocked not only the phosphorylation of p65 (Fig. 2c) but also expression of SOX2 and Oct4 mRNA (Fig. 2d) and ALDH activity (Fig. 2e). Our interpretation is that suppression of SOX2 to leads to overall reduction in stemness. Next, we examined the SOX2 promoter. The promoter was stimulated by TNFα in a TonEBP-dependent manner, while it was inhibited by ATMi and BAY 11–7082 (BAY, an inhibitor of IKK which blocks the activation of NF-κB) (Fig. 2f). The effects of BAY were supported by the observation that the stimulation of the promoter by TNFα was dependent on the presence of NF-κB binding site (Supplementary Fig. 2c). ChIP analyses revealed that binding of the p65 subunit of NF-κB to the SOX2 promoter was also stimulated by TNFα in a TonEBP-dependent (Fig. 2g) and ATM-dependent manner (Fig. 2h). Furthermore, TonEBP-mediated stimulation of SOX2 expression and cancer stemness was dependent on RHD (Supplementary Fig. 2d–f), and ATM is required for the function of TonEBP in stimulation of SOX2 expression (Supplementary Fig. 2g). These data demonstrate that TNFα stimulates the SOX2 promoter via the ATM-NF-κB pathway.
We asked prognostic association of SOX2. When the 280 HCC patients were stratified into two groups based on SOX2 immunohistochemical signals (Supplementary Fig. 2h), higher expression of SOX2 displayed strong association with HCC recurrence, HCC metastasis, and HCC related death (Fig. 2i). Furthermore, the SOX2 expression correlated with TonEBP expression with high significance in our cohort of HCC patients (Supplementary Fig. 2i, j) and in analysis of the RNA-seq data set from TCGA (Supplementary Fig. 2k) confirming the role of TonEBP in driving SOX2 expression. Taken together, these results demonstrate that TonEBP promotes HCC cancer stemness by driving SOX2 expression.
3.3. TonEBP promotes cisplatin resistance and DDR in LCSCs
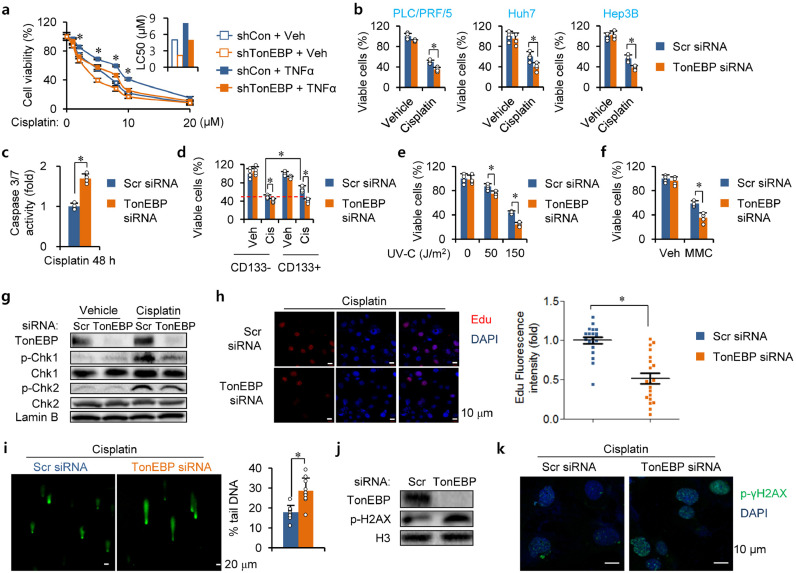
CSCs contribute to the resistance of cancers to chemotherapy [4–6]. This is due, at least in part, to their elevated DDR [4]. Since the data discussed above show that TonEBP promotes self-renewal of LCSCs, we next asked whether TonEBP is involved in the chemoresistance. We used cisplatin which damages DNA mainly by forming various DNA crosslinks and induces cell death [34]. We first examined cell survival in response to cisplatin treatment (Fig. 3a). Cisplatin killed PCL/PRF/5 cells in a dose-dependent manner. Interestingly, the cell death increased after TonEBP knockdown leading to a decrease in LD50 indicating that TonEBP was protective. On the other hand, the cell death was blocked by TNFα. The protective effect of TNFα is likely due to increased stemness as discussed above.
Fig. 3.
TonEBP is required for cisplatin resistance and DNA damage response. (a) PLC/PRF/5 cells transfected with indicated shRNA were treated with TNFα or Veh in combination with various concentrations of cisplatin for 48 h. Cell viability was analyzed by MTT assay (left) and LC50 values were calculated (right). Mean + SD. *p < 0.05. (b) HCC cell lines were transfected with siRNA indicated, followed by treatment with cisplatin (5 μM) or vehicle (dimethylformaldehyde 0.025%) for 48 h before counting viable cells. (c) Activity of caspase 3 and 7 (3/7) was measured from siRNA transfected PLC/PRF/5 cells after a 48 h treatment with cisplatin. (d) CD133+ and CD133− cells were isolated from siRNA transfected cells. Viable cells were counted after incubation with cisplatin (Cis) or Veh for 48 h. Mean + SD, two-way ANOVA, *p < 0.05. (e) siRNA transfected cells were treated with various doses of UV as indicated. Viable cells were counted 48 h later. (f) Cells were treated with Veh or 5 μM MMC for 48 h and viable cells were counted. (g) Nuclear extracts from cells treated for 4 h with Veh or Cisplatin were immunoblotted. (h) Representative images of EdU incorporation in PLC/PRF/5 cells after a 12 h treatment with cisplatin (left). Fluorescence intensity of EdU per cell is shown on the right. (i) CD133+ cells were isolated from siRNA transfected cells and then treated with cisplatin. Alkaline comet assay was performed and representative images are shown (left).% of intensity in tail is shown (right). (j,k) p-γH2AX was detected in the siRNA transfected CD133+ cells which had been treated with cisplatin using (j) immunoblotting and (k) immunohistochemistry.
Since TonEBP is protective from cisplatin-induced cell death independent of TNFα (Fig. 3a), we examined this more closely. The effect of RNAi-mediated transient knockdown of TonEBP was observed in the three HCC cell lines based on both live cell counting and MTT assay (Fig. 3b, Supplementary Fig. 3a). As expected, activity of caspases 3/7 (Fig. 3c and Supplementary Fig. 3b) and Annexin V-positive (apoptotic) cells (Supplementary Fig. 3c) were elevated by cisplatin treatment also in a TonEBP-dependent manner. Reversely, TonEBP overexpression was protective in 3 different HCC cell lines (Supplementary Fig. 3d). Cell populations enriched for LCSCs by selecting for CD133 (CD133+ cells) were less sensitive to cisplatin compared to CD133− (Fig. 3d, Supplementary Fig. 3e). Of note, TonEBP-dependent cisplatin resistance was greater in the CD133+ cells consistent with elevated DDR in LCSCs. Since TonEBP interacts with many proteins in DDR [25], it is likely that the TonEBP-mediated cisplatin resistance is due to DDR.
Cisplatin targets DNA by forming DNA adducts, both intrastrand and interstrand crosslinks [34]. We asked which crosslink was responsible for the TonEBP-dependent cell death. UV was used to produce intrastrand crosslinks and mitomycin C (MMC) to produce interstrand crosslinks. TonEBP deficiency increased cell death in response to both agents (Fig. 3e, f and Supplementary Fig. 3f, g) indicating that TonEBP protects from both DNA adducts produced by cisplatin. Next we examined DNA damages and DDR in response to cisplatin treatment. Cisplatin treatment resulted in activation (phosphorylation) of both checkpoint kinase 1 (Chk1) and 2 (Chk2) (Fig. 3g) consistent with single-strand and double-strand DNA breaks. As expected, abundant DNA repair activity was detected by EdU (5-ethynyl-2′-deoxyuridine) labeling (Fig. 3h). Cisplatin-induced DNA damages measured by alkaline comet assay (Fig. 3i) and H2AX phosphorylation (Fig. 3j, k) were elevated in the TonEBP-deficient cells. Thus, the TonEBP-deficient cells displayed reduced DNA damage signals and DNA repair activity despite elevated DNA damages providing strong evidence that TonEBP mediates DDR as well as resistance in response to cisplatin treatment.
3.4. TonEBP interacts with ERCC1/XPF via RHD domain
In order to address molecular basis of the TonEBP-mediated DNA repair activity described above, we examined the 250 DDR proteins that interacted with TonEBP [25]. Becuase nucleotide exchange repair (NER) is a major repair pathway for DNA damage caused by cisplatin [33], we looked for proteins involved in NER and found three proteins - MMS19, ERCC1 and XPF. We became interested in ERCC1 and XPF because the ERCC1/XPF heterodimer repairs intrastrand and intrastrand crosslinks caused by cisplatin [34]. Expression of ERCC1/XPF is associated with cisplatin resistance and poor outcome after anti-cancer therapy in a variety of cancer types [35, 36]. However, little is known about the role of ERCC1/XPF in liver cancer.
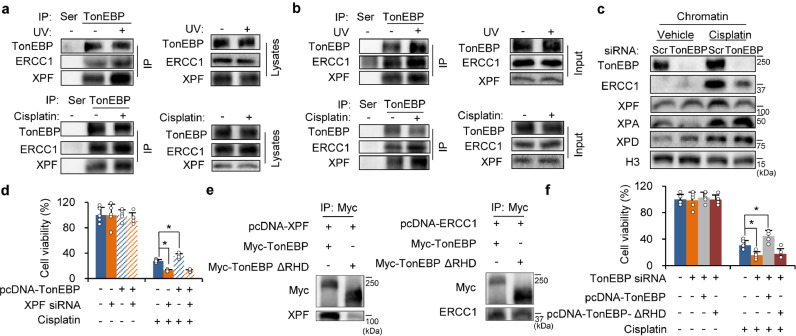
We confirmed that ERCC1 and XPF were co-immunoprecipitated by TonEBP from cell lysates (Fig. 4a). In addition, the interactions were stimulated by UV or cisplatin treatment without changes in their expression levels. The interactions and their stimulation by UV or cisplatin were also observed in chromatin fractions (Fig. 4b). The cisplatin-responsive chromatin binding of ERCC1/XPF was reduced in TonEBP deficient cells without changes in their upstream signaling proteins; XPA and XPD (Fig. 4c, Supplementary Fig. 4a). mRNA levels for the genes encoding these proteins were not affected by TonEBP (Supplementary Fig. 4b) indicating that the TonEBP-mediated DNA repair activity was independent of transcription regulation. As expected, TonEBP-mediated chemoresistance was blocked by XPF knockdown (Fig. 4d) demonstrating that ERCC1/XPF mediates the TonEBP-dependent cisplatin resistance.
Fig. 4.
TonEBP interacts with ERCC1/XPF through RHD. (a) PLC/PRF/5 cell lysates were prepared 2 h after treatment without (-) or with UV (+, 150 J/m2) (upper panel) or after a 4 h treatment with vehicle (-) or cisplatin (+, 5 μM). Immunoprecipitation was performed using normal rabbit serum (Ser) or anti-TonEBP serum (TonEBP). Immunoprecipitates (left) and cell lysates (right) were immunoblotted. (b) Chromatin fractions were prepared and analyzed after cells were treated as above. (c) Chromatin fractions were prepared from siRNA transfected PLC/PRF/5 cells after a 4 h treatment with Veh or cisplatin. Immunoblotting was performed for proteins indicated. H3, Histone H3 (a marker for nuclear protein). (d) PLC/PRF/5 cells were transfected with pcDNA (-) or pcDNA-TonEBP followed by transfection with siRNA indicated. The double transfected cells were treated for 48 h with cisplatin or vehicle (-). Viable cells were measured by MTT assay. Mean + SD. *p < 0.05. (e) Lysates of HEK293 cells transfected with various combinations of expression vectors as indicated were immunoprecipitated with anti-Myc antibodies, followed by immunoblotting. (f) PLC/PRF/5 cells were transfected with scr or TonEBP siRNA followed by a second transfection with empty expression vector (pcDNA) or expression vector for TonEBP or TonEBP ΔRHD as indicated. Cell viability was measured after a 48 h treatment with cisplatin.
Since the interaction between TonEBP and ERCC1/XPF was confirmed by co-localization (Supplementary Fig. 4c), we performed co-immunoprecipitation experiments to characterize the interaction in more detail. Overexpressed TonEBP and ERCC1 or XPF were mutually pulled down by each other (Supplementary Fig. 4d). TonEBP interacts with other proteins such as NF-κB, YY1, SHPRH and USP1 via the rel-homology domain (RHD) [19, 25, 26]. We asked whether TonEBP interaction with ERCC1/XPF was also mediated by RHD. Deletion of RHD abolished the interaction of TonEBP with ERCC1/XPF (Fig. 4e and Supplementary Fig. 4e), and TonEBP's ability to prevent cell death from cisplatin treatment (Fig. 4f). These data show that TonEBP forms a complex with ERCC1/XPF to promote DNA repair and resistance to cisplatin.
3.5. TonEBP-ERCC1/XPF promotes DNA damage-induced ATM/inflammation activation
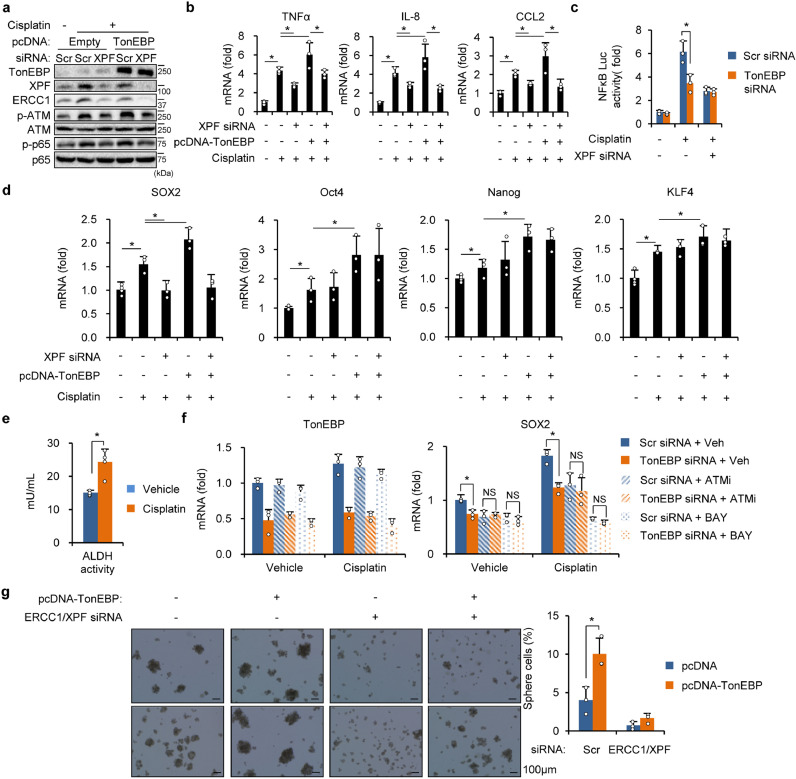
Cisplatin treatment induces inflammation in tumors and the kidney [37, 38]. Since TonEBP activates ATM-NF-κB leading to inflammation and also recruits ERCC1/XPF in response to cisplatin, we asked whether ERCC1/XPF activated ATM. We found that cisplatin stimulated ATM and NF-κB, and this stimulation was reduced in TonEBP-deficient cells (Supplementary Fig. 5a). In addition, TonEBP-dependent cisplatin resistance in cell viability was obliterated by treatment with inhibitors of ATM and NF-κB (Supplementary Fig. 5b). Reversely, overexpression of TonEBP potentiated the cisplatin-induced phosphorylation of ATM and NF-κB (Fig. 5a). These data demonstrate that cisplatin activate the ATM-NF-κB pathway in a TonEBP-dependent manner.
Fig. 5.
TonEBP-ERCC1/XPF mediates cisplatin-induced inflammation and stemness. (a) Cells were transfected with pcDNA (-) or pcDNA-TonEBP (+) followed by transfection with siRNA as indicated. The cells were then treated with cisplatin or vehicle (-) for 48 h before immunoblotting. (b) Cells were transfected with pcDNA or pcDNA-TonEBP followed by transfection with siRNA as indicated. Inflammatory genes were analyzed by RT-qPCR after a 6 h treatment with vehicle (-) or cisplatin. Mean + SD, two-way ANOVA, *p < 0.05. (c) siRNA transfected cells were transfected again with an NF-κB reporter construct. Luciferase activity was measured after a 2 h treatment with vehicle (-) or cisplatin. (d) Cells were transfected with various combinations of siRNA and pcDNA-TonEBP as indicated. Stem cell transcription factors were analyzed by RT-qPCR after a 6 h treatment with vehicle (-) or cisplatin. Mean + SD, two-way ANOVA, *p < 0.05. (e) ALDH activity was analyzed after a 24 h treatment of vehicle or cisplatin in CD133+ cells. (f) siRNA transfected cells were pretreated for 1 h with Veh, ATMi, or BAY followed by a 6 h treatment with vehicle or cisplatin. Expression of TonEBP and SOX2 was analyzed by RT-qPCR. (g) Cells were transfected with pcDNA (-) or pcDNA-TonEBP followed by a second transfection with scr siRNA (-) or ERCC1 siRNA + XPF siRNA as indicated. Cells were cultured for oncosphere formation and representative images are shown (left). Percentages of sphere cells in Mean + SD (right). *p < 0.05.
Next we examined the role of ERCC1/XPF. Knockdown of XPF also reduced the expression of ERCC1 (Fig. 5a), as reported previously [39]. Deficiency of ERCC1/XPF was associated with blockade of cisplatin-induced activation of ATM and NF-κB. While this activation was greater when TonEBP was overexpressed, it was blocked by XPF knockdown consistent with the role of TonEBP-ERCC1/XPF. As expected, cisplatin-stimulated expression of pro-inflammatory cytokine genes was blocked by knockdown of TonEBP or XPF to the same degrees (Supplementary Fig. 5c), potentiating that they were in the same pathway. Essentially the same observations were observed by treatment with UV (Supplementary Fig. 5d, e) and MMC (Supplementary Fig. 5f, g) indicating that both intrastrand and interstrand crosslinks were associated with the TonEBP-ERCC1/XPF pathway. Reversely, overexpression of TonEBP stimulated the expression of pro-inflammatory genes in association with XPF (Fig. 5b); both were blocked by knockdown of ERCC1/XPF. And TonEBP-mediated activation of cisplatin-responsive NF-κB activation was dependent on XPF (Fig. 5c). Finally, we found that ATMi blocked cisplatin-induced activation of p65 (Supplementary Fig. 5h) and expression of pro-inflammatory genes (Supplementary Fig. 5i) confirming the role of ATM in the activation of NF-κB. Taken together, these results show that DNA crosslinks produced by cisplatin induce inflammation by activation of the ATM-NF-κB pathway via the TonEBP-ERCC1/XPF complex
3.6. TonEBP-ERCC1/XPF promotes cancer stemness
Since TonEBP-ERCC1/XPF activated the ATM-NF-κB pathway, we asked whether TonEBP-ERCC1/XPF promoted the expression of SOX2 and cancer stemness. In addition, RHD, a responsible region of TonEBP to interact with ERCC1/XPF, is a mediator of TonEBP-dependent regulation of HCC stemness. We observed that cisplatin stimulated the expression of SOX2 and other stemness transcription factors (Fig. 5d) and elevated ALDH activity (Fig. 5e) indicating promotion of cancer stemness. Only the stimulation of SOX2 expression displayed clear dependence on XPF and TonEBP when cells are shortly exposed to cisplatin (Fig. 5d) consistent with a direct role of TonEBP-ERCC1/XPF. The cisplatin-induced SOX2 expression was sensitive to ATM inhibitor and NF-κB inhibitor, as expected (Fig. 5f).
Interestingly, LCSCs stemness measure by oncosphere formation was also dependent on TonEBP and ERCC1/XPF in the absence of cisplatin (Fig. 5g). Under this condition, ERCC1/XPF association with the chromatin was still dependent on TonEBP (Supplementary Fig. 6a) and oncosphere formation was dependent on ERCC and XPF (Supplementary Fig. 6b). These observations are likely due to inherent activity of ERCC1/XPF in CSCs due to high level of DNA damages [7]. Thus, TonEBP-ERCC1/XPF appears to be an essential feature of LCSCs responsible for driving cancer stemness even in the absence of external DNA damaging agents.
3.7. Tumor ERCC1 and XPF predicts poor prognosis of HCC in TonEBP dependent manner
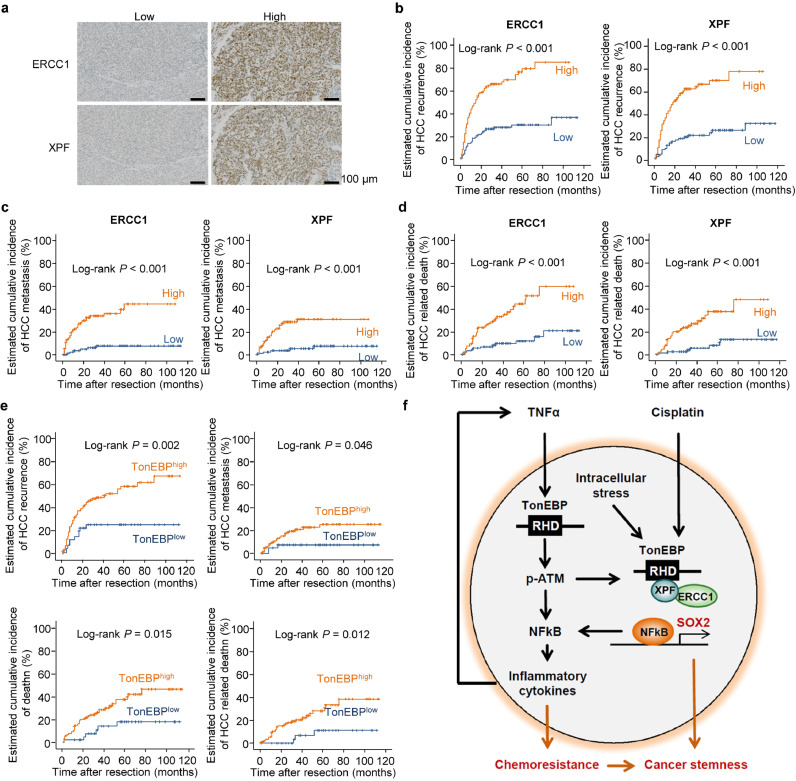
In order to examine the role of ERCC1/XPF in the recurrence of HCC, we examined hepatic tissues obtained from 280 HCC patients (Supplementary Table 1). ERCC1 stained negative in 89 patients while positive in 191 patients, of which 95 showed low signal and 96 showed high signal (Fig. 6a, Supplementary Table 2). Likewise, XPF stained negative in 103 patients while positive in 173 patients, of which 89 showed low signal and 88 showed high signal (Fig. 6a, Supplementary Table 2). For further analyses described below, those patients who displayed no immunohistochemical signal were excluded from analyses. Univariate analysis of the two layers of patients in each of ERCC1 and XPF showed highly significant association with recurrence, metastasis and death (Supplementary Table 2). Kaplan-Meier plot confirmed the higher recurrence, metastasis, and death in patients with high tumor ERCC1/XPF expression (Fig. 6b–d). This was analyzed further using multivariate analyses (Supplementary Table 3). As for recurrence, tumor size, microvascular invasion along with tumor ERCC1/XPF expression displayed strong association. Tumor size and tumor ERCC1/XPF expression showed a robust association with metastasis. Finally, microvascular invasion and tumor ERCC1/XPF were significantly associated with survival. Taken together, these data provide extremely strong evidence that hepatic ERCC1/XPF expression predicts post-operative recurrence, metastasis, and death in HCC patients.
Fig. 6.
Tumor ERCC1/XPF predicts poor post-operative prognosis in HCC patients in TonEBP-dependent manner. (a) Representative immunohistochemical images of ERCC1 and XPF in hepatic tumors from patients with HCC. (b–d) Kaplan–Meier plot of postoperative (b) recurrence, (c) metastasis, and (d) HCC-related death in four groups of patients: ERCC1low (n = 95) vs. ERCC1high(n = 96) and XPFlow (n = 89) vs. XPFhigh (n = 88). (e) Kaplan-Meier plot of postoperative recurrence, metastasis, death and HCC related death in two groups of patients: ERCC1high;XPFhigh;TonEBPlow (n = 41) vs. ERCC1high;XPFhigh;TonEBPhigh (n = 115). (f) Proposed model for the regulation of liver cancer stemness and chemoresistance by TonEBP.
We next examined TonEBP dependency in the ERCC1/XPF-mediated poor post-operative prognosis. For this, we analyzed 156 patients who displayed high signal for both ERCC1 and XPF. And patients were further stratified into 2 groups by previously reported TonEBP signals [26]. Of these patients, 41 showed low TonEBP signal while 115 showed high TonEBP signals (Supplementary Table 4). The high TonEBP group displayed higher recurrence, metastasis and death in univariate analysis (Supplementary Table 4) and Kaplan-Meier analysis (Fig. 6e) confirming the role of TonEBP-ERCC1/XPF in poor prognosis as predicted by its role in cancer stemness described above. These results provide strong evidence that ERCC1/XPF mediates the TonEBP's actions on poor post-operative outcome in HCC patients.
4. Discussion
The motivation of this study is to understand how high TonEBP in tumors leads to recurrence after resection of the tumors in HCC patients [26]. In a cohort of 280 HCC patients, we find clear evidence that EpCAM and CD44 are markers for LCSCs related to recurrence. Interestingly, expression of EpCAM and CD44 in HCC tumors correlates with that of TonEBP suggesting TonEBP's role. Indeed, studies using HCC cell lines has shown that TonEBP is involved in self-renewal and tumor-initiating activity of LCSCs via two separate pathways as summarized in Fig. 6f. TonEBP relays inflammatory signals to ATM leading to SOX2 expression and self-renewal of LCSCs. In the other pathway, TonEBP recruits the ERCC1/XPF dimer to the chromatin using protein-protein interactions. High level of DNA damages (both intrastrand and interstrand DNA crosslinks) due to intracellular stress within LCSCs activates ATM via the TonEBP-ERCC1/XPF complex on the chromatin. This pathway is responsible for the inflammation and cancer stemness induced by cisplatin treatment. Consistent with this model, tumor expression of ERCC1 and XPF predicts recurrence with extremely high significance in TonEBP-dependent manner. The TonEBP actions described here provide molecular basis for two common observations. One is that inflammatory microenvironment is critical for CSCs [14,30]. The other is that cisplatin-induced inflammatory microenvironment contributes CSCs to form chemoresistant and reconstituted tumor [40, 41]. The TonEBP's role in cisplatin resistance requires additional evidence using TonEBP overexpressing tumors in animals in addition to the TonEBP knockdown tumors described here. The TonEBP-dependence of the association of ERCC1 and XPF with recurrence in HCC patients (Fig. 6e) provides solid grounds for future investigation into underlying molecular pathways.
TonEBP is a pleiotropic transcriptional regulator – DNA binding transcriptional enhancer [17], transcriptional cofactor [19, 26] and epigenetic suppressor [24]. A recent study has uncovered non-transcriptional functions of TonEBP where TonEBP recognizes bulky DNA adducts and regulate ubiquitination of proliferating cell nuclear antigen through dynamic interactions with E3 ubiquitin ligase SHPRH and deubiquitinase USP1 [25]. This study adds more functions of TonEBP related to intracellular signal transduction and DDR as discussed above. Since the TonEBP interactome includes more than 450 proteins [25], other functions of TonEBP mediated by protein-protein interactions are likely to be uncovered in the future.
Although markers for LCSCs have been proposed based on clinical studies [12, 13], cellular pathways involved have not been defined. This study provides first clues related to EpCAM and CD44. Our results demonstrate that these two markers are related to TonEBP which drives SOX2 expression. Although enhanced drug efflux has been extensively characterized in chemoresistance, the role of DNA repair in HCC has not been clearly defined. Here we uncover that various DNA crosslinks activates the ATM-NF-κB-SOX2 pathway in a manner dependent on TonEBP and ERCC1/XPF. It appears that the process of repairing the DNA crosslinks is required for the activation of ATM.
The data presented here revealed underlying mechanism in HCC and suggest that targeting TonEBP is an attractive strategy to prevent recurrence of HCC and sensitizing HCC to chemotherapy. The recruitment of ERCC1/XPF to the chromatin by TonEBP involves protein-protein interaction between TonEBP and XPF. Breaking up this interaction might sensitize HCC cells to chemotherapy as well as blocking self-renewal of LCSCs.
Funding sources
This work was supported by the National Research Foundation grants (NRF- 2018R1A5A1024340, 2017R1E1A1A074673 and 2019R1A2C1089260) of Korea. This work was also supported by UNIST fund (1.190039.01). J.H.L. was supported by Global PhD Fellowship of Korea (NRF-2014H1A2A1019656). These funding sources primarily provided financial help for the purchase of reagents.
Declaration of Competing Interest
The authors declare no potential conflicts of interest.
Acknowledgments
We wish to thank the patients who generously provided tissues in our studies.
We thank Orlando D. Schärer (UNIST) to provide ERCC1/XPF cDNA constructs.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102926.
Contributor Information
Neung Hwa Park, Email: nhparkmd@gmail.com.
Hyug Moo Kwon, Email: hmkwon@unist.ac.kr.
Appendix. Supplementary materials
Reference
- 1.Forner A., Bruix J. Biomarkers for early diagnosis of hepatocellular carcinoma. Lancet Oncol. 2012;13:750–751. doi: 10.1016/S1470-2045(12)70271-1. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag H.B. Hepatocellular carcinoma. New Engl J Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 3.Fan S.T., Lo C.M., Poon R.T., Yeung C., Liu C.L., Yuen W.K., Lam C.M., Ng K.K., Chan S.C. Continuous improvement of survival outcomes of resection of hepatocellular carcinoma: a 20-year experience. Ann Surg. 2011;253:745–758. doi: 10.1097/SLA.0b013e3182111195. [DOI] [PubMed] [Google Scholar]
- 4.Nassar D., Blanpain C. Cancer stem cells: basic concepts and therapeutic implications. Annu Rev Pathol: Mech Dis. 2016;11:47–76. doi: 10.1146/annurev-pathol-012615-044438. [DOI] [PubMed] [Google Scholar]
- 5.Clarke M.F. Clinical and therapeutic implications of cancer stem cells. N Engl J Med. 2019;380:2237–2245. doi: 10.1056/NEJMra1804280. [DOI] [PubMed] [Google Scholar]
- 6.Easwaran H., Tsai H.-.C., Baylin S.B. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell. 2014;54:716–727. doi: 10.1016/j.molcel.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bao S., Wu Q., McLendon R.E., Hao Y., Shi Q., Hjelmeland A.B., Dewhirst M.W., Bigner D.D., Rich J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 8.Wang Z., Tang Y., Xie L., Huang A., Xue C., Gu Z. The prognostic and clinical value of CD44 in colorectal cancer: a meta-analysis. Front Oncol. 2019;9:309. doi: 10.3389/fonc.2019.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glumac P.M., LeBeau A.M. The role of CD133 in cancer: a concise review. Clin Transl Med. 2018;7:18. doi: 10.1186/s40169-018-0198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamashita T., Ji J., Budhu A., Forgues M., Yang W., Wang H.Y., Jia H., Ye Q., Qin L.X., Wauthier E. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–1024. doi: 10.1053/j.gastro.2008.12.004. e1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu S., Liu C., Min X., Ji Y., Wang N., Liu D., Cai J., Li K. Prognostic value of cancer stem cell marker aldehyde dehydrogenase in ovarian cancer: a meta-analysis. PLoS One. 2013;8:e81050. doi: 10.1371/journal.pone.0081050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sukowati C.H., Rosso N., Crocè L.S., Tiribelli C. Hepatic cancer stem cells and drug resistance: relevance in targeted therapies for hepatocellular carcinoma. World J Hepatol. 2010;2:114. doi: 10.4254/wjh.v2.i3.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H., Choi G.H., Na D.C., Ahn E.Y., Kim G.I., Lee J.E., Cho J.Y., Yoo J.E., Choi J.S., Park Y.N. Human hepatocellular carcinomas with “Stemness”-related marker expression: keratin 19 expression and a poor prognosis. Hepatology. 2011;54:1707–1717. doi: 10.1002/hep.24559. [DOI] [PubMed] [Google Scholar]
- 14.Rinkenbaugh A.L., Baldwin A.S. The NF-κB pathway and cancer stem cells. Cells. 2016;5:16. doi: 10.3390/cells5020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grivennikov S.I., Karin M. Dangerous liaisons: STAT3 and NF-κB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 2010;21:11–19. doi: 10.1016/j.cytogfr.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Z.-.H., Shi Y., Tibbetts R.S., Miyamoto S. Molecular linkage between the kinase ATM and NF-κB signaling in response to genotoxic stimuli. Science. 2006;311:1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- 17.Miyakawa H., Woo S.K., Dahl S.C., Handler J.S., Kwon H.M. Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity. Proc Natl Acad Sci. 1999;96:2538–2542. doi: 10.1073/pnas.96.5.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi S.Y., Lee-Kwon W., Kwon H.M. The evolving role of TonEBP as an immunometabolic stress protein. Nat Rev Nephrol. 2020 doi: 10.1038/s41581-020-0261-1. [DOI] [PubMed] [Google Scholar]
- 19.Lee H.H., Sanada S., An S.M., Ye B.J., Lee J.H., Seo Y.-.K., Lee C., Lee-Kwon W., Küper C., Neuhofer W. LPS-induced NFκB enhanceosome requires TonEBP/NFAT5 without DNA binding. Sci Rep. 2016;6:24921. doi: 10.1038/srep24921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buxadé M., Lunazzi G., Minguillón J., Iborra S., Berga-Bolaños R., del Val M., Aramburu J., López-Rodríguez C. Gene expression induced by toll-like receptors in macrophages requires the transcription factor NFAT5. J Exp Med. 2012;209:379–393. doi: 10.1084/jem.20111569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi S.Y., Lim S.W., Salimi S., Yoo E.J., Lee-Kwon W., Lee H.H., Lee J.H., Mitchell B.D., Sanada S., Parsa A. Tonicity-responsive enhancer-binding protein mediates hyperglycemia-induced inflammation and vascular and renal injury. J Am Soc Nephrol. 2018;29:492–504. doi: 10.1681/ASN.2017070718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi S., You S., Kim D., Choi S.Y., Kwon H.M., Kim H.-.S., Hwang D., Park Y.-.J., Cho C.-.S., Kim W.-.U. Transcription factor NFAT5 promotes macrophage survival in rheumatoid arthritis. J Clin Investig. 2017;127:954–969. doi: 10.1172/JCI87880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halterman J.A., Kwon H.M., Leitinger N., Wamhoff B.R. NFAT5 expression in bone marrow-derived cells enhances atherosclerosis and drives macrophage migration. Front Physiol. 2012;3:313. doi: 10.3389/fphys.2012.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J.H., Lee H.H., Ye B.J., Lee-Kwon W., Choi S.Y., Kwon H.M. TonEBP suppresses adipogenesis and insulin sensitivity by blocking epigenetic transition of PPARγ2. Sci Rep. 2015;5:10937. doi: 10.1038/srep10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang H.J., Park H., Yoo E.J., Lee J.H., Choi S.Y., Lee-Kwon W., K-y Lee, Hur J.-.H., Seo J.K., Ra J.S. TonEBP Regulates PCNA polyubiquitination in response to DNA damage through interaction with SHPRH and USP1. iScience. 2019;19:177–190. doi: 10.1016/j.isci.2019.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee J.H., Suh J.H., Choi S.Y., Kang H.J., Lee H.H., Ye B.J., Lee G.R., Jung S.W., Kim C.J., Lee-Kwon W. Tonicity-responsive enhancer-binding protein promotes hepatocellular carcinogenesis, recurrence and metastasis. Gut. 2019;68:347–358. doi: 10.1136/gutjnl-2017-315348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Hagan Heather M., Wang W., Sen S., DeStefano Shields C., Lee Stella S., Zhang Yang W., Clements Eriko G., Cai Y., Van Neste L., Easwaran H. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell. 2011;20:606–619. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y., He L., Du Y., Zhu P., Huang G., Luo J., Yan X., Ye B., Li C., Xia P. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Qiu L., Li H., Fu S., Chen X., Lu L. Surface markers of liver cancer stem cells and innovative targeted-therapy strategies for HCC. Oncol Lett. 2018;15:2039–2048. doi: 10.3892/ol.2017.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taniguchi K., Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat Rev Immunol. 2018;18(5):309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 31.Liu M., Sakamaki T., Casimiro M.C., Willmarth N.E., Quong A.A., Ju X., Ojeifo J., Jiao X., Yeow W.-.S., Katiyar S. The canonical NF-κB pathway governs mammary tumorigenesis in transgenic mice and tumor stem cell expansion. Cancer Res. 2010;70:10464–10473. doi: 10.1158/0008-5472.CAN-10-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zakaria N., Mohd Yusoff N., Zakaria Z., Widera D., Yahaya B.H. Inhibition of NF-κB signaling reduces the stemness characteristics of lung cancer stem cells. Front Oncol. 2018:8. doi: 10.3389/fonc.2018.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X., Li F., Huang Q., Zhang Z., Zhou L., Deng Y., Zhou M., Fleenor D.E., Wang H., Kastan M.B. Self-inflicted DNA double-strand breaks sustain tumorigenicity and stemness of cancer cells. Cell Res. 2017;27:764. doi: 10.1038/cr.2017.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dasari S., Tchounwou P.B. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol. 2014;740:364–378. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olaussen K.A., Dunant A., Fouret P., Brambilla E., André F., Haddad V., Taranchon E., Filipits M., Pirker R., Popper H.H. DNA repair by ERCC1 in non–small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 36.Bellmunt J., Paz-Ares L., Cuello M., Cecere F., Albiol S., Guillem V., Gallardo E., Carles J., Mendez P., de la Cruz J. Gene expression of ERCC1 as a novel prognostic marker in advanced bladder cancer patients receiving cisplatin-based chemotherapy. Ann Oncol. 2007;18:522–528. doi: 10.1093/annonc/mdl435. [DOI] [PubMed] [Google Scholar]
- 37.Miller R.P., Tadagavadi R.K., Ramesh G., Reeves W.B. Mechanisms of Cisplatin nephrotoxicity. Toxins. 2010;2:2490–2518. doi: 10.3390/toxins2112490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edwardson D.W., Boudreau J., Mapletoft J., Lanner C., Kovala A.T., Parissenti A.M. Inflammatory cytokine production in tumor cells upon chemotherapy drug exposure or upon selection for drug resistance. PLoS One. 2017;12 doi: 10.1371/journal.pone.0183662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arora S., Kothandapani A., Tillison K., Kalman-Maltese V., Patrick S.M. Downregulation of XPF–ERCC1 enhances cisplatin efficacy in cancer cells. DNA Repair. 2010;9:745–753. doi: 10.1016/j.dnarep.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y.-.P., Yang C.-.J., Huang M.-.S., Yeh C.-.T., Wu A.T.H., Lee Y.-.C., Lai T.-.C., Lee C.-.H., Hsiao Y.-.W., Lu J. Cisplatin selects for multidrug-resistant CD133+ cells in lung adenocarcinoma by activating notch signaling. Cancer Res. 2013;73:406–416. doi: 10.1158/0008-5472.CAN-12-1733. [DOI] [PubMed] [Google Scholar]
- 41.Gilbert L.A., Hemann M.T. DNA damage-mediated induction of a chemoresistant niche. Cell. 2010;143:355–366. doi: 10.1016/j.cell.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.