Abstract
The generation of induced pluripotent stem cells through somatic cell reprogramming requires a global reorganization of cellular functions. This reorganization occurs in a multi-phased manner and involves a gradual revision of both the epigenome and transcriptome. Recent studies have shown that the large-scale transcriptional changes observed during reprogramming also apply to long non-coding RNAs (lncRNAs), a type of traditionally neglected RNA species that are increasingly viewed as critical regulators of cellular function. Deeper understanding of lncRNAs in reprogramming may not only help to improve this process but also have implications for studying cell plasticity in other contexts, such as development, aging, and cancer. In this review, we summarize the current progress made in profiling and analyzing the role of lncRNAs in various phases of somatic cell reprogramming, with emphasis on the re-establishment of the pluripotency gene network and X chromosome reactivation.
Keywords: Mesenchymal-to-epithelial transition, X chromosome reactivation, Apoptosis, Proliferation
Introduction
High-throughput sequencing techniques have demonstrated that mammalian genomes are ubiquitously transcribed [1]. The transcription of large numbers of non-coding RNAs (ncRNAs) [2] might help to explain interspecies differences despite limited variations in the number and sequence of coding genes. ncRNAs are classified based on their length into small RNAs (< 200 nucleotides), e.g., microRNAs (miRNAs) and piwi-interacting RNAs (piRNAs), and long ncRNAs (lncRNAs, > 200 nucleotides) [3], [4].
lncRNAs are typically transcribed by RNA polymerase II, although this process occurs with different modalities and origins. They include circular RNAs (circRNAs) [5], enhancer RNAs (eRNAs) [6], antisense transcripts [7], and long intergenic ncRNAs (lincRNAs) [8], [9]. These lncRNAs recruit epigenetic regulators to chromatin and serve as scaffolds to stabilize protein complexes or as decoys for proteins and miRNAs [10], [11], [12]. These diverse regulatory modes are due to the complementary base pairing of lncRNAs with both DNA and other RNAs, the ability of lncRNAs to interact with proteins, and the localization of lncRNAs in different cellular compartments (e.g., cytoplasm, nucleus, or mitochondrion) [13], [14]. Interestingly, a large fraction of lncRNAs also associate with ribosomes and, paradoxically, some previously annotated lncRNAs produce small peptides with biological functions [15], [16]. The latter raises relevant questions regarding the true nature of some lncRNAs, but the general significance of these findings requires further investigation.
Although the functional relevance of lncRNAs has not yet been systematically explored, some are known to regulate key aspects of normal and pathological cellular functions such as proliferation [17], metabolism [18], [19], and epithelial cytoarchitecture [20]. The recent discovery that lncRNAs display cell-specific and development-specific expression patterns has also suggested a pivotal role of lncRNAs in cell fate determination [21], [22], [23]. In support of this idea, suppression of several embryonic stem cell (ESC)-specific lincRNAs influences pluripotency maintenance/exit and early lineage commitment [21]. Similarly, the heart-associated lncRNA Braveheart serves as a key player in the activation of the core vascular gene network during mouse cardiac cell fate specification [22]. In addition, lincRNA Yin Yang 1 (linc-YY1), which is highly conserved in humans, regulates myogenesis through dislodgement of the YY1/polycomb repressive complex on target promoters, resulting in the activation of muscle gene expression [23].
Here, we summarize the current knowledge of lncRNA profiling and functions in an extreme scenario of cell fate transition, somatic cell reprogramming.
lncRNAs are new players in somatic cell reprogramming
Mammalian somatic cells can be reprogrammed to an ESC state and this has revolutionized stem cell research [24], [25]. Originally, the reprogramming factor cocktail contained SOX2, KLF4, OCT4 (encoded by Pou5f1 or POU5F1 gene in mice or humans, respectively), and c-MYC (SKOM), but other combinations of exogenous factors are also effective [26], [27]. More recently, mouse reprogramming has been achieved using only chemicals [28]. Remarkably, induced pluripotent stem cells (iPSCs) provide a priori unlimited number of individual-specific stem cells that can be used for in vitro disease modeling, drug screening, and potential cell-based therapies [29], [30]. Although producing iPSCs from different cell sources and mammalian species is in general no longer an issue [31], [32], [33], [34], the underlying mechanisms remain unclear and clarifying them is important for improving iPSC quality [35].
Reprogramming occurs through a stepwise process that involves reorganization of most cell functions, culminating in the reactivation of the pluripotency gene program [36], [37]. This conversion is characterized by several roadblocks and checkpoints. For example, reprogramming cells must overcome the senescence/apoptosis barrier to acquire the ability to proliferate indefinitely [38], [39], switch their metabolism from oxidative phosphorylation to glycolysis [40], and undergo a mesenchymal-to-epithelial transition (MET) [41], [42]. By the end of these events, under standard conditions, only a small percentage of the original population activates the endogenous pluripotency gene circuitry.
Since the first demonstration of somatic cell reprogramming, multiple regulatory factors have been identified. Among these, miRNAs play essential roles in creating or removing reprogramming roadblocks [43], [44]. For example, components of miRNA cluster 302–367 suppress Tgfbr2 to neutralize the pro-mesenchymal effects of TGFβ cytokines secreted by somatic cells or present in serum, facilitating the MET [45], [46]. miRNA cluster 302–367 also targets genes encoding chromatin regulators (e.g., BAF170) that prevent the activation of pluripotency genes in the late phase of reprogramming [46]. Conceivably, lncRNAs, which are present in much higher numbers than miRNAs and are in principle more versatile players in cell regulation, could be important regulators of reprogramming too. In this regard, lncRNAs also experience phase-dependent changes during reprogramming, and their regulation shares the same epigenetic mechanisms that drive mRNA changes in reprogramming [47], [48]. These findings suggest a coordinated role between protein-coding and ncRNAs in reorganizing cellular functions. Studying lncRNAs is, therefore, important to understand reprogramming as a whole (Table 1). Such knowledge may also contribute to clarifying the role of lncRNAs in other contexts such as development, aging, and cancer.
Table 1.
lncRNAs and their functions in somatic cell reprogramming
| Name | Function or mechanism | Refs. |
|---|---|---|
| lincRNA-RoR | Inhibiting p53 translation and acting as a miR-145 sponge to promote reprogramming efficiency | [51], [58] |
| lincRNA-p21 | Activating p21 expression and inhibiting expression of pluripotency genes by recruiting SETDB1 or DNMT1 to derail reprogramming | [52], [53] |
| LNCPRESS1 | Acting as a decoy for histone deacetylase SIRT6 | [56] |
| lincRNA-1463 | Inhibiting pre-iPSC to iPSC conversion | [52] |
| lincRNA-1526 | Inhibiting pre-iPSC to iPSC conversion | [52] |
| lincRNA-1307 | Promoting pre-iPSC to iPSC conversion | [52] |
| Zeb2-NAT | Maintaining Zeb2 expression and inhibiting the MET | [66] |
| Gas5 | Maintaining expression of Tet1 and pluripotency genes, and protecting NODAL mRNA from microRNA-mediated degradation | [70], [71] |
| Snhg14 | Binding to the promoter of Sox2 to enhance its expression, promoting reprogramming, and maintaining pluripotency in iPSCs | [75] |
| Peblr20 | Promoting reprogramming by activating endogenous Pou5f1 in a trans manner, and recruiting TET2 to the enhancer region of Pou5f1 to activate eRNAs | [76] |
| Ladr49 and Ladr83 | Inhibiting expression of muscle-related genes | [47] |
| Ladr86 and Ladr91 | Promoting expression of mitochondria-associated genes and blocking the metabolic switch | [47] |
| Xist | Impairing XCR, promoting MET, and inhibiting pre-iPSC to iPSC conversion | [87], [93] |
Note: iPSC, induced pluripotent stem cell; XCR, X chromosome reactivation; MET, mesenchymal-to-epithelial transition; lncRNA, long non-coding RNA; lincRNA-RoR, long intergenic non-coding RNA-regulator of reprogramming; SETDB1, SET domain bifurcated histone lysine methyltransferase 1; DNMT1, DNA methyltransferase 1; LNCPRESS1, lncRNA p53-regulated and ESC specific 1; SIRT6, sirtuin 6; Zeb2-NAT, zinc finger E-box binding homeobox 2-natural antisense transcript; Gas5, growth arrest specific 5; TET2, ten-eleven translocation 2; NODAL, nodal growth differentiation factor; Snhg14, Sox2 promoter-interacting lncRNA 14 (also known as Spilr14); Sox2, SRY-box transcription factor 2; Peblr20, Pou5f1 enhancer-binding lncRNA 20; eRNA, enhancer RNA; Ladr49, long non-coding RNA activated during reprogramming 49; Xist, X-inactive specific transcript.
p53-regulated lncRNAs in reprogramming
Proliferation facilitates the appearance of stochastic events needed for directing chromatin reorganization into a pluripotent state during reprogramming [38]. However, somatic cells have a limited life span/proliferation capacity, and reprogramming is a stressful process involving activation of senescence/cell death pathways. Accordingly, suppressing p53, p16Ink4a, or p19Arf enhances reprogramming efficiency by accelerating proliferation and/or reducing apoptosis [39], [49], [50].
Interestingly, several p53-regulated lncRNAs have strong impacts on reprogramming efficiency (Figure 1). For instance, expression of lincRNA-RoR (regulator of reprogramming) [51] and lincRNA-p21 [52], [53] is induced by p53 [54], [55], which can influence reprogramming in positive and negative manner, respectively. Similarly, expression of p53-repressed LNCPRESS1 (lncRNA p53-regulated and ESC-associated 1) is robustly induced during reprogramming and activates the pluripotency network [56]. Given these circumstances, it would be useful to systematically profile lncRNAs during reprogramming in the presence or absence of p53 or other pro-senescence regulators.
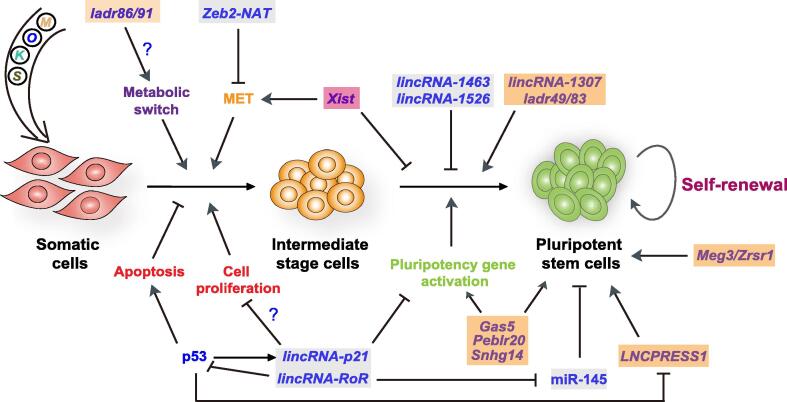
Figure 1.
lncRNAs regulating somatic cell reprogramming
Schematic depiction of lncRNAs regulating different stages of mouse or human reprogramming with exogenous factors. In the early phase of conversion from somatic cells to reprogramming intermediates, Xist is involved in the acquisition of an epithelial phenotype and ladr86/91 is involved in the metabolic switch from an aerobic to an anaerobic production of energy [47]. lincRNA-p21 is involved in the early as well as the late phases (reactivation of the pluripotency gene network) of reprogramming through suppression of cell proliferation and pluripotency gene activation, respectively. Similarly, lincRNA-RoR regulates the early phase of reprograming by suppressing p53 (a negative regulator of cell survival), and the late phase by acting as a sponge for miR-145, which targets multiple pluripotency gene transcripts. lincRNA-1463 and lincRNA-1526 negatively regulate the activation of pluripotency genes in reprogramming, whereas lincRNA-1307 and ladr49/83 are positive regulators. Gas5, Peblr20, Snhg14, and LNCPRESS1 positively regulate the activation of pluripotency genes in reprogramming and/or in ESCs. Expression of the imprinted lncRNAs Meg3/Zrsr1 is related to the acquisition of full pluripotency in mouse iPSCs. Repressive ncRNAs are marked in a gray box, promotive ncRNAs in a yellow box, and ncRNAs with a dual function in a red box. MET, mesenchymal-to-epithelial transition; ESC, embryonic stem cell; iPSC, induced pluripotent stem cell; lncRNA, long non-coding RNA; Xist, X-inactive specific transcript; ladr86, lncRNA activated during reprogramming 86; lincRNA-ROR, long intergenic non-coding RNA-regulator of reprogramming; LNCPRESS1, lncRNA p53-regulated and ESC associated 1; Meg3, maternally expressed 3; Zrsr1, lncRNA zinc finger (CCCH type), RNA binding motif and serine/arginine rich 1; Zeb2-NAT, zinc finger E-box binding homeobox 2-natural antisense transcript; Gas5, growth arrest specific 5; Peblr20, Pou5f1 enhancer-binding lncRNA 20; Snhg14, also known as Spilr14, Sox2 promoter-interacting lncRNA 14; S, SRY-box transcription factor 2 or SOX2; K, Kruppel like factor 4 or KLF4; O, OCT4; M, c-MYC.
lincRNA-RoR
lincRNA-RoR (2603 nucleotides) is located on chromosome 18q21.31. Its expression is induced by p53 [54] and facilitates human reprogramming by suppressing the p53-mediated transcriptional response to oxidative stress and DNA damage [51]. However, overexpression or knockdown of lincRNA-RoR does not influence cell growth at the early stages of reprogramming. In cancer cells, lincRNA-RoR suppresses p53 translation through heterogeneous nuclear ribonucleoprotein I (hnRNP I), a classical RNA-binding protein (RBP) involved in splicing [54]. In this regard, a fraction of hnRNP I is localized in the cytoplasm, where it promotes the translation of p53 through binding to internal ribosome entry sites. Apart from suppressing the p53 pathway, lincRNA-RoR sequesters pro-differentiation miRNAs, including miR-145, to maintain the expression of the core pluripotency transcription factors OCT4, SOX2, and NANOG in human ESCs [57], [58]. Although these two latter mechanisms have not yet been tested in reprogramming, it is likely that they also contribute to the effects of lincRNA-RoR in this process. Notably, lincRNA-RoR is more highly expressed in human iPSCs than in ESCs [51], which may be a consequence of a selective advantage conferred during reprogramming.
lincRNA-p21
lincRNA-p21 (3121 nucleotides, located on chromosome 6p21.2) resides 5 kb upstream of p21. It was originally discovered as an executioner of the p53-mediated apoptotic response [55]. Expression of lincRNA-p21 is induced by p53 and impairs mouse reprogramming, but different cis or trans modes of action have been proposed [52], [53]. The cis-acting model is based on the observation that conditional excision of the lincRNA-p21 promoter and first exon reduces the expression of p21 mRNA, resulting in enhanced proliferation and reprogramming efficiency based on alkaline phosphatase activity (an early marker of reprogramming) [53]. However, it is important to note that removal of the lincRNA-p21 locus leads to the loss of multiple enhancers that control the transcription of nearby genes, including p21 itself, independently of lincRNA-p21 [59]. In fact, removal of the lincRNA-p21 locus alters the expression of nearby genes even in tissues with no detectable lincRNA-p21 transcript. Conversely, the trans-acting model of lincRNA-p21 in reprogramming proposes that lincRNA-p21 impairs reprogramming independently of proliferation by binding to pluripotency loci (e.g., Nanog) and blocking their reactivation [52]. In this model, lincRNA-p21 represses pluripotency loci by recruiting the histone 3 lysine 9 (H3K9) methyltransferase SETDB1 or the maintenance DNA methyltransferase DNMT1. Binding of lincRNA-p21 to these epigenetic regulators is mediated by the classical RBP hnRNP-K. Hence, suppressing hnRNP-K also results in enhanced reprogramming efficiency, although it is unlikely that it acts exclusively through lincRNA-p21. The idea that expression of p53-induced lncRNAs such as lincRNA-p21 prevents reprogramming independent of proliferation or apoptosis is attractive, as this may have implications for the control of cell fate when stem cells are under stress or in aging and in p53-negative cancers.
In a screen for lncRNAs modulating the conversion of pre-iPSCs to iPSCs, lincRNA-p21 was identified as a negative regulator of reprogramming [52]. Pre-iPSCs are stable but incompletely reprogrammed clones, and their conversion to iPSCs is commonly used as a proxy for the late phase of reprogramming [60]. The same screen also identified lincRNA-1463 and lincRNA-1526 as negative regulators of the conversion of pre-iPSCs to iPSCs. However, it is unclear whether expression of these two lncRNAs is also regulated by p53.
LNCPRESS1
Another screening study conducted on differentiating human ESCs led to the discovery of LNCPRESS1 (832 nucleotides, located on chromosome 7q22.1). LNCPRESS1 is a p53-repressed lncRNA that positively regulates the pluripotency gene network in human ESCs [56]. LNCPRESS1 acts as a decoy for the histone deacetylase SIRT6, thereby enriching the activating H3K56/K9 acetylation marks at pluripotency loci. Interestingly, expression of LNCPRESS1 is strongly induced during reprogramming [56], suggesting a potential role of LNCPRESS1 in improving reprogramming efficiency.
Other lncRNAs regulating reprogramming
In addition to lncRNAs regulated by p53, several other lncRNAs have been identified which can regulate somatic cell reprogramming through MET, metabolic switch, or reactivation of the pluripotency gene network (Figure 1). ESCs are epithelial-like, therefore, reprogramming of mesenchymal-like cells such as fibroblasts to iPSCs unavoidably involves the acquisition of an epithelial phenotype. This occurs in the early phase of reprogramming through MET [41], [42], [61]. Since pluripotent cells and somatic cells have different metabolic profiles, reprogramming, logically, also requires metabolic remodeling. iPSCs exhibit a metabolic shift from oxidative phosphorylation to glycolysis, which is associated with a decreased number and complexity of mitochondria. Importantly, blocking MET or the metabolic switch derails reprogramming [41], [42], [62], but not every reprogramming intermediate that successfully passes through these checkpoints achieves full activation of the pluripotency gene network. Likewise, even when somatic cells have been successfully reprogrammed to iPSCs, their DNA methylation patterns often do not faithfully reflect those of ESCs. For example, iPSCs can show aberrant DNA methylation of imprinted regions, which alters the expression of lncRNAs encoded by those regions [63], [64], [65].
Zeb2-NAT
Zinc finger E-box binding homeobox 2-natural antisense transcript (Zeb2-NAT, 430 nucleotides, located on chromosome 2q22.3) is a natural antisense transcript for the transcription factor ZEB2 [66]. ZEB2 is a master regulator of the epithelial-to-mesenchymal transition (EMT) that represses the expression of many epithelial genes [67]. Zeb2-NAT is involved in maintaining Zeb2 expression by preventing the splicing of the Zeb2 5′-untranslated region (5′-UTR) [66]. Interestingly, Zeb2-NAT is highly expressed in fibroblasts from aged mice [68], which are known to be less amenable to reprogramming [69]. The reprogramming efficiency of these aged fibroblasts is enhanced when expression of Zeb2-NAT is reduced. Given that aging promotes the accumulation of DNA damage, thereby activating p53, it is plausible that expression of Zeb2-NAT is also induced by p53 in aged fibroblasts and during reprogramming. However, this speculation has not yet been tested.
Gas5
Expression of the lncRNA growth arrest specific 5 (Gas5, 656 nucleotides, located on chromosome 1q25.1) is controlled by pluripotency transcription factors. Gas5 plays a pivotal role in reprogramming and self-renewal of mouse ESCs by maintaining the expression of genes encoding key pluripotency factors and ten-eleven translocation 1 (Tet1) [70]. The effect of Gas5 on Tet1 expression suggests that Gas5 regulates active DNA demethylation in reprogramming. Its human ortholog, GAS5, also controls human ESC pluripotency by protecting NODAL mRNA from miRNA-mediated degradation [71], a mechanism that may also participate in human reprogramming. GAS5 is also a well-known regulator of cell proliferation and apoptosis in various cell contexts, including breast cancer, lung cancer, and differentiating mouse ESCs [70], [72], [73].
Ladr lncRNAs
Single-cell transcriptomic analysis of mouse reprogramming has unveiled numerous lncRNAs that are activated or repressed during this process [47]. Among them, knockdown of two lncRNAs activated during reprogramming 49 and 83 (Ladr49 and Ladr83), showed modest effects on reprogramming efficiency, but led to the upregulation of muscle-related genes in reprogramming intermediate cells. Interestingly, these two lncRNAs have been previously shown to physically associate with polycomb repressive complex 2 (PRC2) [21], [74]. On the other hand, depletion of Ladr86 and Ladr91, which show upregulated expression in reprogramming, represses mitochondria-associated genes, suggesting a role in the metabolic switch during reprogramming.
Promoter/enhancer-interacting lncRNAs
A recently devised approach chromatin-RNA in situ reverse-transcription sequencing (CRIST-seq) took advantage of the specificity of the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (Cas9) system for DNA to profile pluripotency-specific lncRNAs that interact with the promoter of the core pluripotency genes Sox2 and Pou5f1 [75]. Among the identified lncRNAs interacting with the Sox2 promoter, 59 of them were differentially expressed in reprogramming. Notably, overexpression of one of these differentially expressed lncRNAs, Sox2 promoter-interacting lncRNA 14 (Spilr14, also known as Snhg14), led to significant enhancement of reprogramming efficiency, whereas its knockdown caused loss of pluripotency in iPSCs. Another lncRNA, Pou5f1 enhancer-binding lncRNA 20 (Peblr20), was identified utilizing a strategy combining RNA reverse transcription-associated capture sequencing (RAT-seq) and RNA sequencing [76]. Peblr20 is expressed at higher levels in iPSCs than in mouse embryonic fibroblasts and promotes reprogramming by activating endogenous Pou5f1 in trans through the recruitment of TET2 to the enhancer region, which enhances the expression of eRNAs.
Imprinted lncRNAs
Maternally expressed 3 (Meg3, also known as Gtl2) is localized at the imprinted Dlk1–Dio3 gene cluster on mouse chromosome 12qF1, which also encodes numerous miRNAs [77], [78]. ncRNAs harbored in this region are maternally expressed in mammals. Interestingly, they are strongly repressed in most mouse iPSCs compared to ESCs, which is responsible for the failure to support the development of all-iPSC mice [63], [79]. This inability can be reversed through reactivation of the imprinted Dlk1–Dio3 locus in reprogramming using the histone deacetylase inhibitor valproic acid or ascorbic acid (vitamin C, Vc) [63], [80], [81]. Vc facilitates the conservation of imprinting at this locus by interfering with reprogramming factor-induced loss of H3K4 methylation, which prevents the recruitment of DNA methyltransferase 3A (DNMT3A). Importantly, MEG3, which is encoded in the DLK1–DIO3 locus, is also frequently silenced in human iPSCs [82]. Similarly, hypomethylation of the maternally expressed imprinted lncRNA zinc finger (CCCH type), RNA binding motif, and serine/arginine rich 1 (Zrsr1) is associated with reduced pluripotency in mouse iPSCs, which could not be rescued by treating reprogramming cells with Vc or PD0325901 and CHIR99021 (inhibitors of MEK1/2 and GSK3, respectively) [64]. It remains to be clarified how these imprinted lncRNAs regulate the reprogramming process.
X chromosome reactivation during reprogramming
In the late stage of reprogramming, another important process involving lncRNAs is X chromosome reactivation (XCR) (Figure 2). During the development of mice and other mammals, one of the two X chromosomes in females is randomly silenced in somatic cells shortly after implantation to maintain the dosage equivalence between the sexes [83], [84]. X chromosome inactivation (XCI) is initiated by the expression of X-inactive specific transcript (Xist), a 17,918-nucleotide lncRNA localized in the X chromosome inactivation center (XIC) [85]. Xist coats the X chromosome, which gradually removes RNA polymerase II and active histone marks such as H3K4 trimethylation. This is followed by a sequential gain of diverse repressive marks including H3K27 trimethylation (H3K27me3), macroH2A.1 histone (macroH2A), and DNA methylation on the inactivated X chromosome (Xi). Consequently, Xi is silenced throughout life with remarkable stability. Despite possessing a stable nature, Xi can be reactivated in specific contexts such as primordial germ cell differentiation, somatic cell nuclear transfer (SCNT), and somatic cell reprogramming [86]. Due to its ease and reproducibility, reprogramming provides an unprecedented tool for characterizing the events involved in XCR [87], [88].
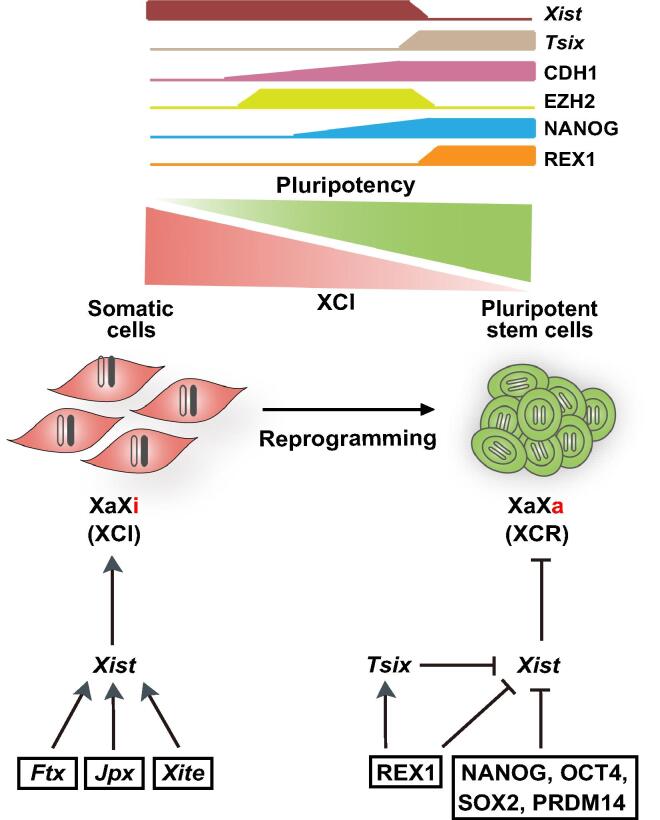
Figure 2.
X chromosome reactivation in somatic cell reprogramming
Both X chromosomes are active in mammalian female blastocysts and inactivation of one of them is an epigenetic hallmark of embryonic development. Xist is transcribed from XIC and is the main responsible for carrying out XCI during development. The Xi is reactivated during reprogramming of somatic cells to iPSCs through repression of Xist. In ESCs and iPSCs, Xist is regulated by different pluripotency factors including NANOG, OCT4, PRDM14, and REX1, along with Tsix. XCR, X chromosome reactivation; XCI, X chromosome inactivation; XIC, X-chromosome inactivation center; Xa, active X chromosome; Xi, inactive X chromosome; Tsix, Xist antisense RNA; CDH1, cadherin 1 (E-cadherin); EZH2, enhancer of zeste 2 polycomb repressive complex 2 subunit; NANOG, nanog homeobox; Ftx, five prime to Xist; Jpx, just proximal to Xist; Xite, X-inactivation intergenic transcription element; PRDM14, PR/SET domain 14.
A relevant question in the field of reprogramming is the order of the epigenetic events leading to XCR. High-resolution single-cell time course analyses of reprogramming using immunofluorescence and RNA fluorescence in situ hybridization (FISH) [87], [89] demonstrate that XCR occurs in the following order: (1) recruitment of H3K27 methyltransferase EZH2 to Xi (XiEZH2+) in E-cadherin (CDH1) positive (CDH1+) cells, (2) activation of NANOG in XiEZH2+ cells, (3) loss of EZH2, H3K27me3, Xist coating, and macroH2A1 on Xi in NANOG+ cells, and (4) removal of DNA methylation on Xi and activation of the transcribed antisense to Xist (Tsix). Therefore, except for DNA demethylation, the events of XCR in reprogramming follow the inverse order of the events of XCI in development. Notably, the removal of DNA methylation during XCR in reprogramming is TET-independent and hence passive, and the expression of Tsix is paradoxically dispensable.
Conspicuously, despite the initiation of XCR in NANOG+ cells, full reactivation of the core pluripotency circuitry precedes XCR in reprogramming. Accordingly, XCR serves as one of the crucial standards for bona fide mouse iPSC identification [90]. This is consistent with the observation that the pluripotency factor PRDM14 represses Xist expression [91]. Although ectopic expression of Xist impairs XCR in reprogramming, its depletion does not affect XCR or reprogramming efficiency [87]. This is inconsistent with the observations in SCNT, the efficiency of which is greatly improved following Xist elimination [92]. A possible explanation could be that Xist plays opposite roles at different stages of somatic cell reprogramming, as SCNT has no obvious phases. In support of this idea, Xist depletion impairs the MET in the early phase of reprogramming but significantly improves the conversion of pre-iPSCs to iPSCs [93]. Interestingly, the reprogramming booster Vc [81] promotes XCR in reprogramming by preventing the relocalization of macroH2A onto Xi [93]. This is likely mediated by the boosting effect of Vc on H3K27me3 demethylases [94], [95]. It remains to be determined whether Vc also acts through unrelated mechanisms, for example by decreasing the levels of N6-methyladenosine (m6A) deposition on Xist, as Vc is also a cofactor for another two dioxygenases, the alpha-ketoglutarate-dependent dioxygenase fat mass and obesity-associated protein (FTO) and AlkB homolog 5 (ALKBH5), which are responsible for erasing this epitranscriptomic mark [96], [97], [98].
Notably, the aforementioned mechanisms have been investigated in mouse cell reprogramming, but it is unclear whether the same principles apply to human cells. In this regard, XCR is either absent or unstable in female donors when human iPSCs are generated using standard culture conditions [99], but is present in reprogramming to naïve pluripotency [100]. Further studies are needed to understand the differences and similarities between XCR in mouse and human cell reprogramming.
Perspectives
It is becoming increasingly evident that lncRNAs are critical players in cell fate regulation. Few lncRNAs, mostly related to p53 or cell senescence, have been well studied thus far, but the repertoire of lncRNAs that regulate pluripotency/reprogramming is likely large. Systematic profiling is needed for functional characterization of these lncRNAs. This could be facilitated by developing highly efficient/deterministic reprogramming systems [101], as low-efficiency protocols are prone to cell heterogeneity and the cell reprogramming kinetics are asynchronous, which complicates the interpretations. In addition, specialized high-throughput RNA sequencing approaches such as global nuclear run-on sequencing (GRO-seq) and sequencing with RNase R treatment are necessary to enrich for certain lncRNAs such as eRNAs and circRNAs [102], [103], respectively, since they are rarely detected using conventional RNA sequencing methodologies. In this regard, several circRNAs exhibit human iPSC/ESC-specific expression [104], [105]. Among them, circBIRC6 and circCORO1C positively regulate pluripotency maintenance and reprogramming [104]. circBIRC6 acts as a sponge for differentiation-mediated miRNAs, whereas the mechanism underlying circCORO1C regulation is unknown. An important consideration is that functional lncRNA screens with short hairpin RNAs (shRNAs) or small interfering RNAs (siRNAs) have inevitable limitations because many lncRNAs are localized in the nucleus, where shRNAs/siRNAs cannot effectively silence transcripts. As a solution, CRISPR/Cas9-based screening systems assisted by comprehensive computer prediction algorithms could be used to increase on-target efficiency and minimize off-target effects. This approach requires eliminating the whole DNA fragment (rather than introducing a frame shift for coding genes), potentially in the promoter region, to suppress expression/functionality of lncRNAs without affecting the nearby genes [106]. CRISPR/Cas13 and CRISPR interference are potential alternatives, as they can be used to edit RNA or repress gene expression without altering the genome [107], [108]. Collectively, the characterization of lncRNAs in reprogramming will not only help to uncover new layers of regulation for the diverse pathways modulating reprogramming but may also have implications in other types of cell fate transitions.
Competing interests
The authors have declared no competing interests.
Acknowledgements
We thank all members of the Esteban lab for their support. Research in the authors’ laboratories is supported by the National Key R&D Program of China (Grant Nos. 2016YFA0100701, 2016YFA0100102, 2018YFA0106903, and 2016YFA0100300); the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA16030502); the Youth Innovation Promotion Association of the Chinese Academy of Sciences (Grant No. 2015294), the National Natural Science Foundation of China (Grant Nos. 31671537, 31571524, and 31501192); the Natural Science Foundation of Guangdong Province (Grant No. 2018B030306042), the Guangdong Province Science and Technology Program (Grant Nos. 2014A030312001, 2016A050503037, 2016B030229007, and 2017B050506007), the Science and Technology Planning Project of Guangdong Province (Grant No. 2017B030314056), the Pearl River Science and Technology Nova Program of Guangzhou (Grant No. 201610010107), and the Guangzhou Science and Technology Program (Grant No. 201807010066), China. SK is supported by a President’s International Fellowship Initiative program from the Chinese Academy of Sciences and CW is supported by a Pearl River Overseas Young Talents Postdoctoral Fellowship.
Handled by Xiu-Jie Wang
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
Contributor Information
Miguel A. Esteban, Email: miguel@gibh.ac.cn.
Xichen Bao, Email: bao_xichen@gibh.ac.cn.
References
- 1.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hon C.C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J., Gough J. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mattick J.S., Makunin I.V. Non-coding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 4.Mattick J.S. The state of long non-coding RNA biology. Noncoding RNA. 2018;4:17. doi: 10.3390/ncrna4030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 6.Li W., Notani D., Rosenfeld M.G. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17:207–223. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- 7.Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 8.Kung J.T., Colognori D., Lee J.T. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 10.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamamura S., Imai-Sumida M., Tanaka Y., Dahiya R. Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci. 2018;75:467–484. doi: 10.1007/s00018-017-2626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon J.H., Abdelmohsen K., Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitt A.M., Chang H.Y. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim K.M., Noh J.H., Abdelmohsen K., Gorospe M. Mitochondrial noncoding RNA transport. BMB Rep. 2017;50:164–174. doi: 10.5483/BMBRep.2017.50.4.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruiz-Orera J., Messeguer X., Subirana J.A., Alba M.M. Long non-coding RNAs as a source of new peptides. Elife. 2014;3 doi: 10.7554/eLife.03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballantyne M.D., Pinel K., Dakin R., Vesey A.T., Diver L., Mackenzie R. Smooth muscle enriched long noncoding RNA (SMILR) regulates cell proliferation. Circulation. 2016;133:2050–2065. doi: 10.1161/CIRCULATIONAHA.115.021019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kornfeld J.W., Bruning J.C. Regulation of metabolism by long, non-coding RNAs. Front Genet. 2014;5:57. doi: 10.3389/fgene.2014.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X.Y., Lin J.D. Long Noncoding RNAs: a new regulatory code in metabolic control. Trends Biochem Sci. 2015;40:586–596. doi: 10.1016/j.tibs.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Q., Deng F., Qin Y., Zhao Z., Wu Z., Xing Z. Long non-coding RNA regulation of epithelial-mesenchymal transition in cancer metastasis. Cell Death Dis. 2016;7 doi: 10.1038/cddis.2016.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G. LincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klattenhoff C.A., Scheuermann J.C., Surface L.E., Bradley R.K., Fields P.A., Steinhauser M.L. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou L., Sun K., Zhao Y., Zhang S., Wang X., Li Y. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat Commun. 2015;6:10026. doi: 10.1038/ncomms10026. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 26.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 27.Buganim Y., Markoulaki S., van Wietmarschen N., Hoke H., Wu T., Ganz K. The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell. 2014;15:295–309. doi: 10.1016/j.stem.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou P., Li Y., Zhang X., Liu C., Guan J., Li H. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- 29.Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10:678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Cherry A.B., Daley G.Q. Reprogrammed cells for disease modeling and regenerative medicine. Annu Rev Med. 2013;64:277–290. doi: 10.1146/annurev-med-050311-163324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honda A., Choijookhuu N., Izu H., Kawano Y., Inokuchi M., Honsho K. Flexible adaptation of male germ cells from female iPSCs of endangered Tokudaia osimensis. Sci Adv. 2017;3 doi: 10.1126/sciadv.1602179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou T., Benda C., Duzinger S., Huang Y., Li X., Li Y. Generation of induced pluripotent stem cells from urine. J Am Soc Nephrol. 2011;22:1221–1228. doi: 10.1681/ASN.2011010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loh Y.H., Agarwal S., Park I.H., Urbach A., Huo H., Heffner G.C. Generation of induced pluripotent stem cells from human blood. Blood. 2009;113:5476–5479. doi: 10.1182/blood-2009-02-204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aasen T., Raya A., Barrero M.J., Garreta E., Consiglio A., Gonzalez F. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 35.Liang G., Zhang Y. Genetic and epigenetic variations in iPSCs: potential causes and implications for application. Cell Stem Cell. 2013;13:149–159. doi: 10.1016/j.stem.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plath K., Lowry W.E. Progress in understanding reprogramming to the induced pluripotent state. Nat Rev Genet. 2011;12:253–265. doi: 10.1038/nrg2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanna J., Saha K., Pando B., van Zon J., Lengner C.J., Creyghton M.P. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tapia N., Scholer H.R. p53 connects tumorigenesis and reprogramming to pluripotency. J Exp Med. 2010;207:2045–2048. doi: 10.1084/jem.20101866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu J., Ocampo A., Belmonte J.C.I. Cellular metabolism and induced pluripotency. Cell. 2016;166:1371–1385. doi: 10.1016/j.cell.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 41.Li R., Liang J., Ni S., Zhou T., Qing X., Li H. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 42.Samavarchi-Tehrani P., Golipour A., David L., Sung H.K., Beyer T.A., Datti A. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 43.Bao X., Zhu X., Liao B., Benda C., Zhuang Q., Pei D. MicroRNAs in somatic cell reprogramming. Curr Opin Cell Biol. 2013;25:208–214. doi: 10.1016/j.ceb.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 44.Luginbuhl J., Sivaraman D.M., Shin J.W. The essentiality of non-coding RNAs in cell reprogramming. Noncoding RNA Res. 2017;2:74–82. doi: 10.1016/j.ncrna.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao B., Bao X., Liu L., Feng S., Zovoilis A., Liu W. MicroRNA cluster 302–367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J Biol Chem. 2011;286:17359–17364. doi: 10.1074/jbc.C111.235960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subramanyam D., Lamouille S., Judson R.L., Liu J.Y., Bucay N., Derynck R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim D.H., Marinov G.K., Pepke S., Singer Z.S., He P., Williams B. Single-cell transcriptome analysis reveals dynamic changes in lncRNA expression during reprogramming. Cell Stem Cell. 2015;16:88–101. doi: 10.1016/j.stem.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hussein S.M., Puri M.C., Tonge P.D., Benevento M., Corso A.J., Clancy J.L. Genome-wide characterization of the routes to pluripotency. Nature. 2014;516:198–206. doi: 10.1038/nature14046. [DOI] [PubMed] [Google Scholar]
- 49.Spike B.T., Wahl G.M. p53, stem cells, and reprogramming: tumor suppression beyond guarding the genome. Genes Cancer. 2011;2:404–419. doi: 10.1177/1947601911410224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonizzi G., Cicalese A., Insinga A., Pelicci P.G. The emerging role of p53 in stem cells. Trends Mol Med. 2012;18:6–12. doi: 10.1016/j.molmed.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 51.Loewer S., Cabili M.N., Guttman M., Loh Y.H., Thomas K., Park I.H. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bao X., Wu H., Zhu X., Guo X., Hutchins A.P., Luo Z. The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell Res. 2015;25:80–92. doi: 10.1038/cr.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dimitrova N., Zamudio J.R., Jong R.M., Soukup D., Resnick R., Sarma K. LincRNA-p21 activates p21 in cis to promote polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54:777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang A., Zhou N., Huang J., Liu Q., Fukuda K., Ma D. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013;23:340–350. doi: 10.1038/cr.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huarte M., Guttman M., Feldser D., Garber M., Koziol M.J., Kenzelmann-Broz D. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jain A.K., Xi Y., McCarthy R., Allton K., Akdemir K.C., Patel L.R. LncPRESS1 is a p53-regulated lncRNA that safeguards pluripotency by disrupting SIRT6-mediated de-acetylation of histone H3K56. Mol Cell. 2016;64:967–981. doi: 10.1016/j.molcel.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barta T., Peskova L., Collin J., Montaner D., Neganova I., Armstrong L. Inhibition of miR-145 enhances reprogramming of human dermal fibroblasts to induced pluripotent stem cells. Stem Cells. 2016;34:246–251. doi: 10.1002/stem.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y., Xu Z., Jiang J., Xu C., Kang J., Xiao L. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 59.Groff A.F., Sanchez-Gomez D.B., Soruco M.M.L., Gerhardinger C., Barutcu A.R., Li E. In vivo characterization of linc-p21 reveals functional cis-regulatory DNA elements. Cell Rep. 2016;16:2178–2186. doi: 10.1016/j.celrep.2016.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T.W., Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Esteban M.A., Bao X., Zhuang Q., Zhou T., Qin B., Pei D. The mesenchymal-to-epithelial transition in somatic cell reprogramming. Curr Opin Genet Dev. 2012;22:423–428. doi: 10.1016/j.gde.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 62.Xu X., Duan S., Yi F., Ocampo A., Liu G.H., Izpisua Belmonte J.C. Mitochondrial regulation in pluripotent stem cells. Cell Metab. 2013;18:325–332. doi: 10.1016/j.cmet.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 63.Stadtfeld M., Apostolou E., Akutsu H., Fukuda A., Follett P., Natesan S. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang G., Gao S., Hou X., Xu Z., Liu Y., Kang L. High-throughput sequencing reveals the disruption of methylation of imprinted gene in induced pluripotent stem cells. Cell Res. 2014;24:293–306. doi: 10.1038/cr.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Panopoulos A.D., Smith E.N., Arias A.D., Shepard P.J., Hishida Y., Modesto V. Aberrant DNA methylation in human iPSCs associates with MYC-binding motifs in a clone-specific manner independent of genetics. Cell Stem Cell. 2017;20:505–517. doi: 10.1016/j.stem.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Beltran M., Puig I., Pena C., Garcia J.M., Alvarez A.B., Pena R. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vandewalle C., Comijn J., De Craene B., Vermassen P., Bruyneel E., Andersen H. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005;33:6566–6578. doi: 10.1093/nar/gki965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bernardes de Jesus B., Marinho S.P., Barros S., Sousa-Franco A., Alves-Vale C., Carvalho T. Silencing of the lncRNA Zeb2-NAT facilitates reprogramming of aged fibroblasts and safeguards stem cell pluripotency. Nat Commun. 2018;9:94. doi: 10.1038/s41467-017-01921-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mahmoudi S., Brunet A. Aging and reprogramming: a two-way street. Curr Opin Cell Biol. 2012;24:744–756. doi: 10.1016/j.ceb.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tu J., Tian G., Cheung H.H., Wei W., Lee T.L. Gas5 is an essential lncRNA regulator for self-renewal and pluripotency of mouse embryonic stem cells and induced pluripotent stem cells. Stem Cell Res Ther. 2018;9:71. doi: 10.1186/s13287-018-0813-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu C., Zhang Y., Wang Q., Xu Z., Jiang J., Gao Y. Long non-coding RNA GAS5 controls human embryonic stem cell self-renewal by maintaining NODAL signalling. Nat Commun. 2016;7:13287. doi: 10.1038/ncomms13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mourtada-Maarabouni M., Pickard M.R., Hedge V.L., Farzaneh F., Williams G.T. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 73.Li W., Huang K., Wen F., Cui G., Guo H., Zhao S. Genetic variation of lncRNA GAS5 contributes to the development of lung cancer. Oncotarget. 2017;8:91025–91029. doi: 10.18632/oncotarget.19955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao J., Ohsumi T.K., Kung J.T., Ogawa Y., Grau D.J., Sarma K. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang S., Wang Y., Jia L., Wen X., Du Z., Wang C. Profiling the long noncoding RNA interaction network in the regulatory elements of target genes by chromatin in situ reverse transcription sequencing. Genome Res. 2019;29:1521–1532. doi: 10.1101/gr.244996.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang C., Jia L., Wang Y., Du Z., Zhou L., Wen X. Genome-wide interaction target profiling reveals a novel Peblr20-eRNA activation pathway to control stem cell pluripotency. Theranostics. 2020;10:353–370. doi: 10.7150/thno.39093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.da Rocha S.T., Edwards C.A., Ito M., Ogata T., Ferguson-Smith A.C. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24:306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 78.Kagami M., Sekita Y., Nishimura G., Irie M., Kato F., Okada M. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet. 2008;40:237–242. doi: 10.1038/ng.2007.56. [DOI] [PubMed] [Google Scholar]
- 79.Liu L., Luo G.Z., Yang W., Zhao X., Zheng Q., Lv Z. Activation of the imprinted Dlk1-Dio3 region correlates with pluripotency levels of mouse stem cells. J Biol Chem. 2010;285:19483–19490. doi: 10.1074/jbc.M110.131995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stadtfeld M., Apostolou E., Ferrari F., Choi J., Walsh R.M., Chen T. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat Genet. 2012;44:398–405. doi: 10.1038/ng.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Esteban M.A., Wang T., Qin B., Yang J., Qin D., Cai J. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 82.Nishino K., Toyoda M., Yamazaki-Inoue M., Fukawatase Y., Chikazawa E., Sakaguchi H. DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee J.T. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Froberg J.E., Yang L., Lee J.T. Guided by RNAs: X-inactivation as a model for lncRNA function. J Mol Biol. 2013;425:3698–3706. doi: 10.1016/j.jmb.2013.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Plath K., Mlynarczyk-Evans S., Nusinow D.A., Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet. 2002;36:233–278. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- 86.Ohhata T., Wutz A. Reactivation of the inactive X chromosome in development and reprogramming. Cell Mol Life Sci. 2013;70:2443–2461. doi: 10.1007/s00018-012-1174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pasque V., Tchieu J., Karnik R., Uyeda M., Sadhu Dimashkie A., Case D. X chromosome reactivation dynamics reveal stages of reprogramming to pluripotency. Cell. 2014;159:1681–1697. doi: 10.1016/j.cell.2014.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Papp B., Plath K. Epigenetics of reprogramming to induced pluripotency. Cell. 2013;152:1324–1343. doi: 10.1016/j.cell.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pasque V., Plath K. X chromosome reactivation in reprogramming and in development. Curr Opin Cell Biol. 2015;37:75–83. doi: 10.1016/j.ceb.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., Arnold K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 91.Payer B., Rosenberg M., Yamaji M., Yabuta Y., Koyanagi-Aoi M., Hayashi K. Tsix RNA and the germline factor, PRDM14, link X reactivation and stem cell reprogramming. Mol Cell. 2013;52:805–818. doi: 10.1016/j.molcel.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Inoue K., Kohda T., Sugimoto M., Sado T., Ogonuki N., Matoba S. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science. 2010;330:496–499. doi: 10.1126/science.1194174. [DOI] [PubMed] [Google Scholar]
- 93.Chen Q., Gao S., He W., Kou X., Zhao Y., Wang H. Xist repression shows time-dependent effects on the reprogramming of female somatic cells to induced pluripotent stem cells. Stem Cells. 2014;32:2642–2656. doi: 10.1002/stem.1775. [DOI] [PubMed] [Google Scholar]
- 94.Kamikawa Y.F., Donohoe M.E. Histone demethylation maintains Prdm14 and Tsix expression and represses Xist in embryonic stem cells. PLoS One. 2015;10 doi: 10.1371/journal.pone.0125626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang T., Chen K., Zeng X., Yang J., Wu Y., Shi X. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011;9:575–587. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 96.Patil D.P., Chen C.-K., Pickering B.F., Chow A., Jackson C., Guttman M. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kuiper C., Vissers M.C. Ascorbate as a co-factor for Fe- and 2-oxoglutarate dependent dioxygenases: physiological activity in tumor growth and progression. Front Oncol. 2014;4:359. doi: 10.3389/fonc.2014.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barakat T.S., Ghazvini M., de Hoon B., Li T., Eussen B., Douben H. Stable X chromosome reactivation in female human induced pluripotent stem cells. Stem Cell Rep. 2015;4:199–208. doi: 10.1016/j.stemcr.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sahakyan A., Kim R., Chronis C., Sabri S., Bonora G., Theunissen T.W. Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell. 2017;20:87–101. doi: 10.1016/j.stem.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mor N., Rais Y., Sheban D., Peles S., Aguilera-Castrejon A., Zviran A. Neutralizing Gatad2a-Chd4-Mbd3/NuRD complex facilitates deterministic induction of naive pluripotency. Cell Stem Cell. 2018;23:1–14. doi: 10.1016/j.stem.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Core L.J., Waterfall J.J., Lis J.T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Suzuki H., Zuo Y., Wang J., Zhang M.Q., Malhotra A., Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34 doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu C.Y., Li T.C., Wu Y.Y., Yeh C.H., Chiang W., Chuang C.Y. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun. 2017;8:1149. doi: 10.1038/s41467-017-01216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lei W., Feng T., Fang X., Yu Y., Yang J., Zhao Z.A. Signature of circular RNAs in human induced pluripotent stem cells and derived cardiomyocytes. Stem Cell Res Ther. 2018;9:56. doi: 10.1186/s13287-018-0793-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pulido-Quetglas C., Aparicio-Prat E., Arnan C., Polidori T., Hermoso T., Palumbo E. Scalable design of paired CRISPR guide RNAs for genomic deletion. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu S.J., Horlbeck M.A., Cho S.W., Birk H.S., Malatesta M., He D. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355:eaah7111. doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]