Abstract
Background
Prediction of brain invasion pre-operatively rather than postoperatively would contribute to the selection of surgical techniques, predicting meningioma grading and prognosis. Here, we aimed to predict the risk of brain invasion in meningioma pre-operatively using a nomogram by incorporating radiomic and clinical features.
Methods
In this case-control study, 1728 patients from Beijing Tiantan Hospital (training cohort: n = 1070) and Lanzhou University Second Hospital (external validation cohort: n = 658) were diagnosed with meningiomas by histopathology. Radiomic features were extracted from the T1-weighted post-contrast and T2-weighted magnetic resonance imaging. The least absolute shrinkage and selection operator was used to select the most informative features of different modalities. The support vector machine algorithm was used to predict the risk of brain invasion. Furthermore, a nomogram was constructed by incorporating radiomics signature and clinical risk factors, and decision curve analysis was used to validate the clinical usefulness of the nomogram.
Findings
Sixteen features were significantly correlated with brain invasion. The clinicoradiomic model derived from the fusing MRI sequences and sex resulted in the best discrimination ability for risk prediction of brain invasion, with areas under the curves (AUCs) of 0•857 (95% CI, 0•831–0•887) and 0•819 (95% CI, 0•775–0•863) and sensitivities of 72•8% and 90•1% in the training and validation cohorts, respectively.
Interpretation
Our clinicoradiomic model showed good performance and high sensitivity for risk prediction of brain invasion in meningioma, and can be applied in patients with meningiomas.
Funding
This work was supported by the National Natural Science Foundation of China (81772006, 81922040); the Youth Innovation Promotion Association CAS (grant numbers 2019136); special fund project for doctoral training program of Lanzhou University Second Hospital (grant numbers YJS-BD-33).
Keywords: Meningioma, Brain invasion, Radiomics, Magnetic resonance images
1. Introduction
Meningiomas are the most common primary intracranial tumours in adults, accounting for 36•7% of all intracranial tumours [1]. Brain invasion in meningiomas was defined as the presence of meningioma tissue within the adjacent brain without a separating connective tissue layer in 1997 [2]. In the latest 2016 edition of the World Health Organization (WHO) classification of central nervous system (CNS) tumours [3], microscopic examination of brain invasion was added as an independent grading criterion for the diagnosis of WHO grade II atypical meningioma. Brain invasion is getting the highest clinical attention [4], and the main reasons are as follows: firstly, microsurgical resection is the most widely used treatment for the vast majority of meningiomas [5]. The choice of surgical technique is closely related to brain invasion, such as application of intraoperative navigation, expansion of surgical excision range, etc. Secondly, brain tumours now explicitly list brain invasion as a criterion of atypia [3]. Grading of meningiomas only depends on histopathological criteria, and higher grades are associated with worse prognosis, including higher rates of tumour recurrence and worse survival [6], but brain invasion may not be detected by histopathology due to a lack of brain tissue samples [5,6]. The authors reported 85% of samples as ‘unassessable’ pathologically [7]. Therefore, clinical pathological assessment of brain invasion is limited. Finally, brain invasion has gained distinct changes in clinical behaviour, and is a risk factor for preoperative seizures and postoperative haemorrhage [7,10]. From the above, we observed that brain invasion directly impacted therapeutic decisions, histopathologic grading and prognosis [7,8,11].
Histopathological examination is the only standard for the diagnosis of brain invasion in meningiomas [12]. However, some studies have reported that the frequency of brain invasion is significantly different from that diagnosed in neuropathological tissue samples [7,8]. A large portion of invasive tissue might not be detected during microscopic analyses without adjacent CNS tissue. Some grade I meningiomas (14•7% in our study) show brain invasion, which would affect the treatment selection and prognosis of patients when pathological examination was missed. Compared to focusing on local fine structures, imaging studies of brain invasion can analyse the entire tumour. In a recent study [7], Alborz et al. used routine preoperative MRI to predict brain invasion. Several imaging characteristics, such as irregular tumour shape, heterogeneous contrast enhancement, and peritumour oedema were identified as predictors of brain invasion. However, current image-guided brain invasion testing is non-specific and highly subjective [1,7], which depends on the experience of radiologists.
To overcome these problems, radiomics analysis is a reasonable tool that has rapidly emerged in the field of medical imaging analysis in recent years. Compared with subjective evaluation, radiomics analysis is more stable and objective, quantifying high-dimensional tumour features that cannot be observed by the naked eye, such as texture, intensity, and shape features [13,14]. It is also a non-invasive way to provide a quantitative method for the evaluation of tumour heterogeneity. Recent studies have shown several applications of radiomics in meningiomas, such as different diagnoses, prediction of the grade and histological subtype, and prediction recurrence-free survival in meningioma [5,[15], [16], [17]]. Moreover, a recent study showed that radiomics might predict preoperative cavernous sinus invasion by pituitary adenomas [18]. These studies show the value of radiomics in medical imaging, which can also be a potential method for the prediction of brain invasion in meningiomas on MRI.
To the best of our knowledge, until now there is no reported study predicting brain invasion in meningiomas based on the radiomic or texture features analysis. Therefore, the aim of our study was first to extract the radiomic features that are correlated with brain invasion from two MRI modalities (T1-weighted post-contrast [T1C] and T2-weighted [T2]), respectively. Second, to fuse these two modalities to generate a radiomic signature. Third, to establish a nomogram combining radiomic signatures and clinical risk factors to predict brain invasion in meningioma patients undergoing MRI.
2. Materials and methods
2.1. Study population
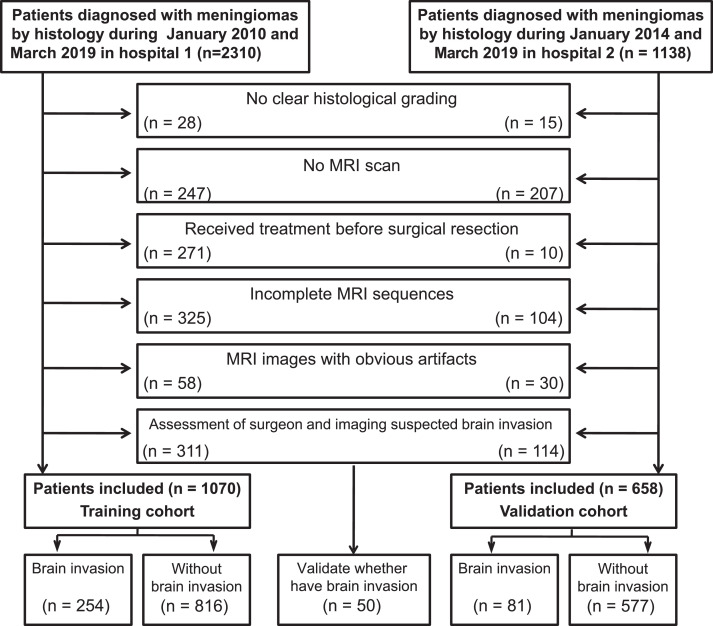
Our study was a case-control study [19]. Ethical approval was obtained from the Institutional Review Board of Lanzhou University Second Hospital and Beijing Tiantan Hospital, and informed consent of patients was waived. In the study, all patients with meningiomas who underwent surgery were enroled according to the inclusion and exclusion criteria. The inclusion criteria were: (a), all patients were diagnosed with meningioma and clearly graded by histology (including WHO grade I, II, and III meningiomas); (b), MR images, including T1C and T2 sequences, were performed within one week before surgical resection; and (c), the images of each patient were of good quality and without artefacts. The exclusion criteria were: (a), before surgical resection, patients received treatment such as radiotherapy, chemoradiotherapy or surgery; (b), patients without a clear histological grading; (c), patients with incomplete MRI sequences; (d), patient images with artefacts that impacted assessment; (e), assessment of surgeon and imaging suspected brain invasion. These exclusion criteria were chosen because some patients may have MRI examinations in other hospitals and lack of MR data in our institute, and some patients only have enhanced MR without plain scan (plain scan was done in other hospital), resulting in lack of sequence, etc.
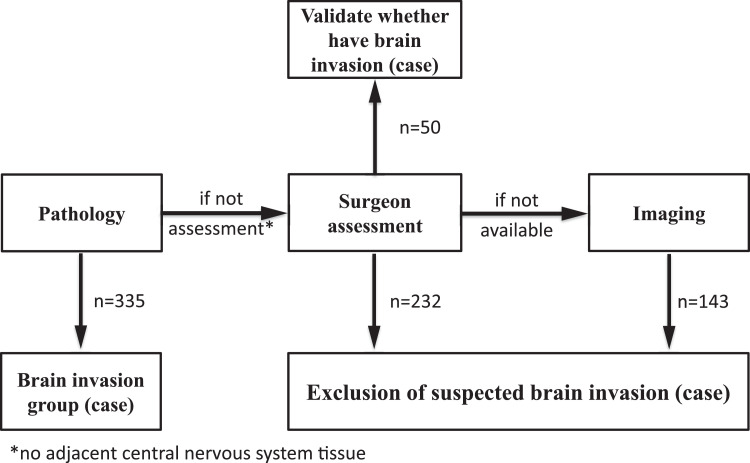
Tumour resection was performed with the aid of a microscope for all patients, and brain invasion was diagnosed according to the pathological records [12]. Brain invasion may not have been determined in some cases due to the lack of adjacent brain tissue. the samples (excluding incomplete MRI sequences, brain invasion, etc.) including 1127 cases in the training cohort and 691 cases in the validation cohort were evaluated for brain invasion by the surgeon and imaging records [9]. Surgical records described obvious adhesion between the surface of the tumour and the brain. If surgical records were not available, diagnostic imaging reports were used for evaluation. T2 showed that the tumour boundary was blurred, irregular tumour shape, and significant peritumour oedema. Thus 311 cases in the training cohort and 114 cases in the validation cohort with suspected brain invasion were excluded. The basic characteristics (such as age, sex, histological subtype, and the WHO grade etc.) of these cases are consistent with that in corresponding patients included in training cohort and validation cohort.
Finally, a total of 1070 patients from Beijing Tiantan Hospital were used as the training cohort from January 2010 to March 2019, and 658 patients of the Lanzhou University Second Hospital were identified for the external validation cohort from January 2014 to March 2019. Thus, the study included 1728 patients (training cohort: 276 males and 794 females, with a mean age of 51•89 ± 10•12 years; validation cohort: 137 males and 521 females, with a mean age of 52•5 ± 9•99 years). The patient recruitment flowchart is shown in Fig. 1. Data concerning the following five conventional clinical variables were obtained from the electronic medical records: age, sex, WHO grade, histological subtype, and Ki-67 expression level, which were blindly and independently conducted by two radiologists (P L and J Z) to ensure the accuracy of the extracted data. In this retrospective study, there was not a difference in the data recording methods during the study period.
Fig. 1.
Inclusion and exclusion criteria.
An assessment of the brain invasion and suspected brain invasion groups’ flowchart is shown in Fig. 2. Thus, 335 patients (training cohort: n = 254; validation cohort: n = 81) were enroled in the invasion group, while 1393 patients (training cohort: n = 816; validation cohort: n = 577) were enroled in the non-invasion group. For suspected brain invasion, we randomly selected 50 cases (based on the surgeon assessment) and applied a clinicoradiomic model to validate whether have brain invasion.
Fig. 2.
Assessment of brain invasion and suspected brain invasion groups flowchart.
2.2. Image acquisition, segmentation, and normalization
MRI examinations were completed one week before the operation. The MRIs combined two hospitals with 3.0-T scanners (Siemens Verio; Siemens Trio Tim; GE Discovery MR750; Philips Achieva) and a 1.5-T scanner (Siemens Magnetom Aera). The MRI protocols of each hospital included T2 and T1C images, and the detailed parameters for each scanner are illustrated in Table.S1.
To ensure the validity and accuracy of the data, two radiologists (readers 1 and 2, with 12 and 15 years of experience in brain MRI interpretation, respectively) independently performed manual segmentation of the MR images with an open-source ITK-SNAP software (www.itksnap.org), and without prior knowledge of the operative and pathological records. A more senior radiologist with 19 years of experience conformed the segmentation. Reader 1 (P L) segmented all training cases, reader 2 (J Z) segmented all validation cases. When the two radiologists were unsure, reader 3 (Z Y Z) with 19 years of experience confirmed segmentation. The regions of interest (ROIs) of images were manually delineated on each slice of the tumour without the surrounding brain tissue, and oedema on both axial T1C and T2 images. For T2 images, it was drawn separately with T1C as visual guidance. The volume of interest (VOI) was generated by fusing the segmented tissues on each slice.
After manual segmentation, T1C and T2 images were standardized using z-score normalization to obtain a standard normal distribution of the image intensities. To evaluate the reproducibility and robustness of feature extraction, after two months, 50 patients in the training cohort were randomly selected and segmented again by Reader 1 and Reader 2 to build the re-segmentation set, and 50 patients were randomly selected from each MR scanner to build the different MR scanners set for calculating the intra-/interclass correlation coefficients (ICCs), respectively.
2.3. Feature extraction and selection
The pyradiomic platform was used to extract standardized radiomic features from the medical imaging data [20], and feature extraction followed the Image Biomarker Standardization Initiative (IBSI) guideline in this study [21]. A total of 1595 radiomic features were extracted from the VOI of each modality of MR images: 14 shape features, 306 first-order features, and 1275 textural features.T1C features were extracted from the VOI of T1C images, while T2 features were extracted from the VOI of T2 images. The Supplementary Methods list the details of the radiomic features.
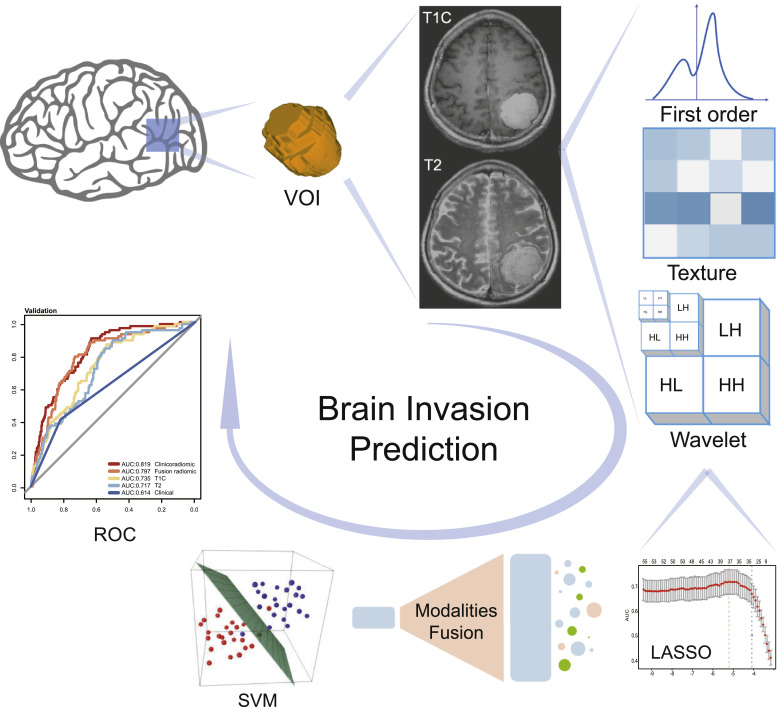
For both T1C and T2 features, the least absolute shrinkage and selection operator (LASSO) regression with three-fold cross-validation was used to select the radiomic features highly correlated with brain invasion. Features with a coefficient lower than 0.25, or a P value greater than 0.05, were removed accordingly. To determine whether the selected features are correlated with each other, we used Pearson correlation analysis to estimate the correlation between these features from T1C and T2 images. In addition, the correlation between the radiomics features and the Ki-67 expression level was evaluated by Pearson correlation coefficient analysis. A flowchart of the study is shown in Fig. 3.
Fig. 3.
Flowchart of the process of radiomics. The tumours were segmented on T1-weighted post-contrast (T1C) and T2-weighted (T2) MRI to form the volume of interest (VOI). The least absolute shrinkage and selection operator (LASSO) was used to select the features. The support vector machine (SVM) algorithm was then used to fit the predictive model. The optimal model was selected with the best performance for brain invasion prediction by model comparison.
For clinical factors, the correlation between clinical factors and brain invasion were tested via the Chi-square test and a Student's t-test with the P-value set to 0.05. Features with P-value more than 0.05 and postoperative factors were excluded from the model. There were a priori variables to be excluded regardless of the statistical significance, such as WHO grade and Ki-67 expression level (postoperative factors).
2.4. Fusion of modalities
According to the principles and characteristics of T1C and T2 modalities, T1C, which is derived from the injection of contrast medium, represents the blood supply and whether the blood-brain barrier has been breached, while T2, which is correlated with water content, mainly reflects tissue oedema, and is sensitive to oedema around the tumour. Hence, in order to increase the discrimination ability of the radiomic model, we fused the two modalities by combining the selected radiomic features together, reflecting the factors that influence brain invasion from different perspectives.
2.5. Radiomic signature building
After fusing modalities, based on the selected radiomic features, the support vector machine (SVM) algorithm was used to build a radiomic model with a radial basis kernel for risk prediction of brain invasion. The T1C and T2 models were built based on T1C and T2 features, respectively, while the fusion model was built based on T1C and T2 fusion features (all 16 radiomic features).
The maximum area under the curve (AUC) in the training cohort with three-fold cross-validation determined the final regularization parameter. Then, the radiomic model predicted a radiomic signature showing the likelihood of brain invasion for each patient.
2.6. Nomogram development and validation
After analysing the clinical characteristics, the Chi-square test was used to compare the differences in sex and WHO grade, while the Student's t-test was used to compare the differences in age and Ki-67 expression level between the invasion and non-invasion groups in the training and validation cohorts. Generally, P values < 0•05 (two-sided) were considered statistically significant.
Integrated discrimination improvement (IDI) was conducted to quantify the performance improvement. After the inclusion of radiomic signature, the P-values showed whether the improvement in reclassification was statistically significant.
After the selection of clinical characteristics and the model comparison, a nomogram incorporating the radiomic signature and correlated clinical risk factors was developed in the training cohort and validated in the validation cohort, using multivariate logistic regression coefficients. This method provides a more understandable and convenient tool for clinicians and patients.
The discrimination ability of the radiomic-clinical nomogram was assessed by the calibration curves for the training and validation cohorts, and the agreement between the observed outcomes and predicted risks of brain invasion was assessed using the Hosmer-Lemeshow (H-L) test. Decision curve analysis (DCA) was used to quantify the net benefits at different threshold probabilities to assess the clinical usefulness of the nomogram [22].
2.7. Imaging analysis by the radiologists
To compare the diagnostic performance of the radiomics signature with visual assessment, MRIs (T1C and T2 sequences) of all 1728 cases were independently assessed by two radiologists (J Z and Z Y Z). Both radiologists had no prior knowledge of the histopathological results and all personal information was de-identified prior to analysis. Image analysis was based on clinical experience. Diagnostic accuracy, sensitivity, and specificity were calculated and compared with the histopathological results.
2.8. Statistical analysis
In this study, all the statistical analyses were achieved with Python 3.7.1 (https://www.python.org), R software (version 3.4.1; http://www.Rproject.org) and IBM SPSS 22.0 for Windows (IBM Corp, Armonk, NY, USA). Python was used to extract and select the radiomic features as well as build the prediction models. R software was used to evaluate the prediction models. SPSS was used to compare the variables between different cohorts. Logistic regression with the LASSO penalty and the SVM model were implemented using Python 3.7.1 in the Scikit-learn package (version 0.21.3). The data were adjusted for multiple testing (Bonferroni correction) in all data analysis. Pearson's Chi-squared test was used to explore the difference between the training and validation cohorts. Descriptive statistics for continuous variables are expressed as mean ± standard deviation. The Chi-square test was used to compare the difference in sex and WHO grade, while a Student's t-test was used to compare the difference in age and Ki-67 expression level between brain invasion and non-invasion in the training and validation cohorts. Generally, P values of less than 0•05 (two-sided) were considered statistically significant. The intra-/inter-class correlation coefficients (ICCs) were used to assess the agreement of extracted features by two radiologists and different MR scanners, respectively. Kappa test analyses were conducted to determine the inter-observer agreement.
2.9. Role of the funding source
This study has received funding from the National Natural Science Foundation of China, the Youth Innovation Promotion Association CAS, and Special fund project for doctoral training program of Lanzhou University Second Hospital. The funders (J L Z) had role in study design and data interpretation.
3. Results
3.1. Clinical characteristics
Considering the larger sample size of 1070 cases from Beijing Tiantan Hospital, this group was set as the training cohort, while the 658 cases from Lanzhou University Second Hospital were set as the validation cohort. The clinical characteristics of the patients are shown in Table 1. In these two cohorts, sex and the pathological WHO grade were found to be significantly different statistically (P < 0•001 for all), and age was not significantly different (P > •05) between the invasion and non-invasion groups. For meningiomas with brain invasion, the mean Ki-67 expression level was 5•7 ± 4•8, 7•3 ± 5•6, which were significantly higher than that of without brain invasion with a mean value of 3•8 ± 2•4, 3•7 ± 3•3 (P < 0•001) in the training and validation cohorts, respectively.
Table 1.
Patient clinical characteristics in the training and validation cohorts.
| Characteristics | Training cohort |
P value | Validation cohort |
P value | ||
|---|---|---|---|---|---|---|
| Invasion | Non-invasion | Invasion | Non-invasion | |||
| Sex | <0.001* | <0.001* | ||||
| Male | 99 | 177 | 33 | 105 | ||
| Female | 155 | 639 | 48 | 472 | ||
| Age (years, mean ± SD) | 51.89±13.53 | 51.79±9.92 | 0.900 | 52.5 ± 10.5 | 52.2 ± 10.0 | 0.823 |
| WHO grade | <0.001* | <0.001* | ||||
| I grade | 0 | 786 | 0 | 545 | ||
| II grade | 248 | 27 | 74 | 30 | ||
| Ⅲ grade | 6 | 3 | 7 | 2 | ||
| Ki-67 expression level | 5.7 ± 4.8 | 3.8 ± 2.4 | <0.001* | 7.3 ± 5.6 | 3.7 ± 3.3 | <0.001* |
Note: The chi-square test was used to compare the difference in sex and WHO grade, while a Student's t-test was used to compare the difference in age and Ki-67 expression level.
P <.05. SD, standard deviation.
The distribution of different subtypes of meningiomas between the brain invasion and non-invasion groups is shown in Supplementary Table S2. The frequency of brain invasion in WHO grade I transitional meningioma (9•25%; 4•55%) and WHO grade II atypical meningioma (2•71%; 4•56%) was much higher than other subtypes in the training and validation cohorts, respectively.
3.2. Radiomic features correlated with brain invasion
The ICCs were calculated to evaluate agreement of features extracted by two radiologists and different MR scanners, respectively, and all values > 0•75, reflecting good agreement. In total, 3190 radiomic features were extracted from axial T1C and T2 sequences from each patient. Amongst them, eight T1C features and eight T2 features were selected, and most of them (14/16) were extracted from the filter-filtered images and were more relevant with brain invasion. These 16 features included 2 shape features, 4 first-order features, and 10 texture features, which can be seen in Table 2.
Table 2.
Radiomics features extracted from T1C and T2 that were significantly relevant with brain invasion.
| T1C | T2 |
|---|---|
| original_shape_Maximum2DDiameterSlice original_shape_Maximum3DDiameter wavelet-LHH_glcm_Imc2 wavelet-HHL_glcm_Imc2 exponential_firstorder_10Percentile exponential_glcm_InverseVariance squareroot_firstorder_10Percentile squareroot_glcm_MCC |
wavelet-LLL_firstorder_Median lbp-3D-m2_glrlm_ShortRunHighGrayLevelEmphasis logarithm_glrlm_LongRunHighGrayLevelEmphasis square_ngtdm_Busyness squareroot_firstorder_90Percentile squareroot_glrlm_LongRunHighGrayLevelEmphasis squareroot_glszm_GrayLevelVariance squareroot_ngtdm_Busyness |
The Pearson correlation analysis showed that the features extracted from T1C were consistent with some features extracted from T2, P value < 0•0005 (Supplementary Fig.S1). For example, T2_wavelet-LLL_first order_ Median feature was correlated with T1C_ original_ shape_ Maximum2DDiameterSlice, T1C_ original_ shape_ Maximum3D Diameter and T1C_ wavelet-LHH_glcm_Imc2 features in both the training and validation cohorts. The correlation between these features showed that the two groups of features remained highly similar and stable in both the training and validation cohorts. The Ki-67 expression level was significantly correlated with radiomic features, with a P-value of 0.0003 in the validation cohort.
3.3. Fusion of modalities and model building
Considering the different principles of T1C and T2 modalities, their radiomic features may correspond to different information. To analyse this information jointly, modality fusion was conducted at the feature level. By combining the selected radiomic features together, the radiomics signature can reflect the influencing factors of brain invasion from different perspectives. Univariate analysis showed that sex, histopathological WHO grade and Ki-67 expression level were significantly different between patients with brain invasion and non-invasion, P < 0•001. However, pathological grade and Ki-67 expression levels results were obtained after surgery. Thus, the radiomics signature and sex were selected for the clinicoradiomic model building.
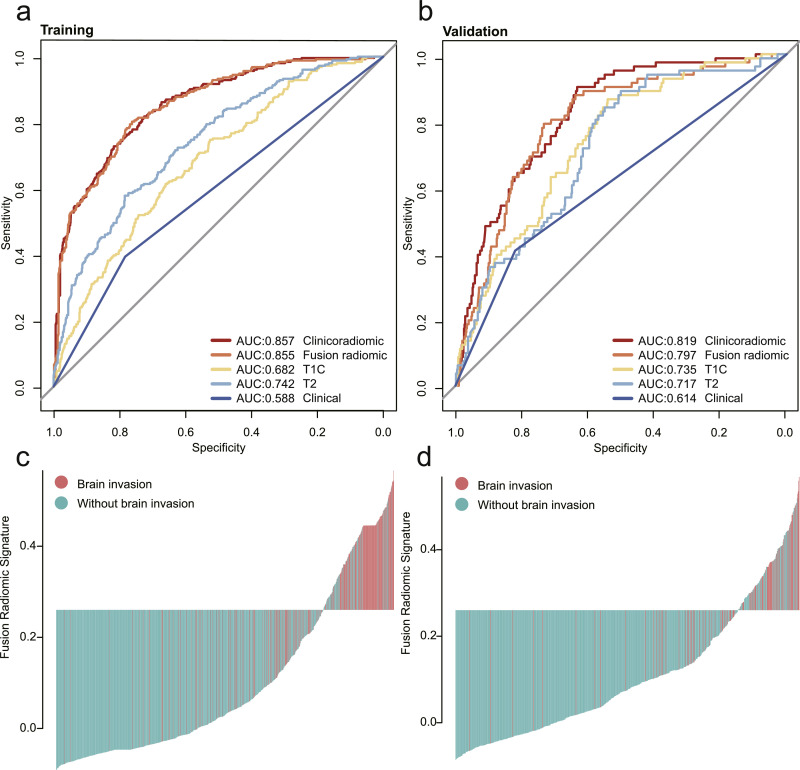
The discrimination ability of these models was first assessed in the training cohort and then externally validated in the independent validation cohort. The performances of T1C, T2, the radiomic (T1C and T2 combined), and the clinicoradiomic models are shown in Table 3. The clinicoradiomic model demonstrated the best performance, resulting in an AUC of 0•857 (95% CI, 0•831–0•883) and 0•819 (95% CI, 0•775–0•863), with a sensitivity of 72•8% (95% CI, 67•3%−78•4%) and 90•1% (95% CI, 83•47%−97%) for brain invasion prediction in the training and validation cohorts, respectively. In addition, the clinicoradiomic model was applied to surgically suspected brain invasion, and 48 out of 50 cases were diagnosed with brain invasion. Supplementary results listed the calculation formula of the clinicoradiomic model. The receiver operating characteristic (ROC) curves for T1C, T2, the radiomic, and the clinicoradiomic model are plotted in Fig. 4.
Table 3.
Performance of models for brain invasion prediction.
| Cohort | Model | AUC | ACC (%) | SEN (%) | SPE (%) | PPV | NPV |
|---|---|---|---|---|---|---|---|
| Training set | T1C | 0.682(0.645–0.719) | 64.77(61.85–67.70) | 61.42(55.37–67.52) | 65.81(62.55–69.06) | 0.359(0.313–0.404) | 0.846(0.817–0.874) |
| T2 | 0.742(0.708–0.776) | 73.46(70.90–76.06) | 57.87(52.03–64.0) | 78.31(75.54–81.05) | 0.454(0.400–0.509) | 0.857(0.832–0.882) | |
| Radiomic | 0.855(0.829–0.882) | 76.92(74.40–79.50) | 80.32(75.56–85.25) | 75.86(72.90–78.84) | 0.509(0.461–0.558) | 0.925(0.906–0.945) | |
| Clinicoradiomic | 0.857(0.831–0.883) | 79.35(76.93–81.76) | 72.83(67.34–78.40) | 81.37(78.65–84.05) | 0.549(0.494–0.603) | 0.906(0.885–0.927) | |
| Validation set | T1C | 0.735(0.682–0.789) | 57.90(54.18–61.58) | 86.42(78.84–93.95) | 53.90(49.87–57.90) | 0.208(0.164–0.251) | 0.966(0.946–0.986) |
| T2 | 0.717(0.661–0.772) | 54.71(51.07–58.43) | 88.89(82.02–95.69) | 49.91(46.0–53.90) | 0.199(0.159–0.240) | 0.970(0.950–0.990) | |
| Radiomic | 0.796(0.747–0.845) | 73.25(69.86–76.59) | 79.01(70.19–87.95) | 72.44(68.77–76.04) | 0.287(0.228–0.345) | 0.961(0.943–0.979) | |
| Clinicoradiomic | 0.819(0.775–0.863) | 65.96(62.38–69.60) | 90.12(83.39–96.97) | 62.56(58.65–66.52) | 0.253(0.204–0.302) | 0.978(0.963–0.994) |
Note: Radiomic, combination of T1C and T2; Clinicoradiomic, fusion of radiomic signature and sex information; T1C, contrast-enhanced T1-weighted imaging; T2, T2-weighted imaging; ACC, balanced accuracy; AUC, area under receiver operating characteristic curve; SEN, sensitivity; SPE, specificity; PPV, positive predictive value; NPV, negative predictive value.
Fig. 4.
Comparison of the receiver operating characteristic (ROC) curves of different models. (a, b) ROC curves of the different models in the training and validation cohorts. The clinicoradiomic model demonstrated the best discriminating ability amongst these models, with an AUC of 0.857 in the training cohort and an AUC of 0.819 in the validation cohort. (c, d) Radiomic signature histogram of the training and validation cohorts. The red bar shows the sample with brain invasion, and the blue bar shows the sample without brain invasion. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Model comparison
The DeLong test was used to compare the AUC estimates of the discrimination performance between different predictive models. The integrated discrimination improvement (IDI) index was measured to evaluate the incremental predictive utility of different predictive models. The clinicoradiomic model improved by 2•41% compared with the radiomic model in integrated discrimination (P = 0•0142) in the validation cohort (Table 4). In addition, comparisons between different models in the training cohort are shown in Table S3.
Table 4.
Comparison between different models in validation cohort.
| Initial model | Model introducing new factor | Performance improvement (IDI) |
|---|---|---|
| Combination of T1C and T2 | Clinicoradiomic | 2.41% P = 0.0142 |
| T1C | Combination of T1C and T2 | 4.77% P = 0.0016 |
| T2 | Combination of T1C and T2 | 6.34% P < 0.0001 |
Compared with the T1C and T2 models, the performance of combination of T1C and T2 model improved by 4.77% and 6.34% in discrimination ability, respectively. Compared with combination of T1C and T2 model, the performance of clinicoradiomic model improved by 2.42% in discrimination ability. IDI: Integrated discrimination improvement; Clinicoradiomic, fusion of radiomic signature and sex information.
3.5. Radiomic-Clinical nomogram performance assessment
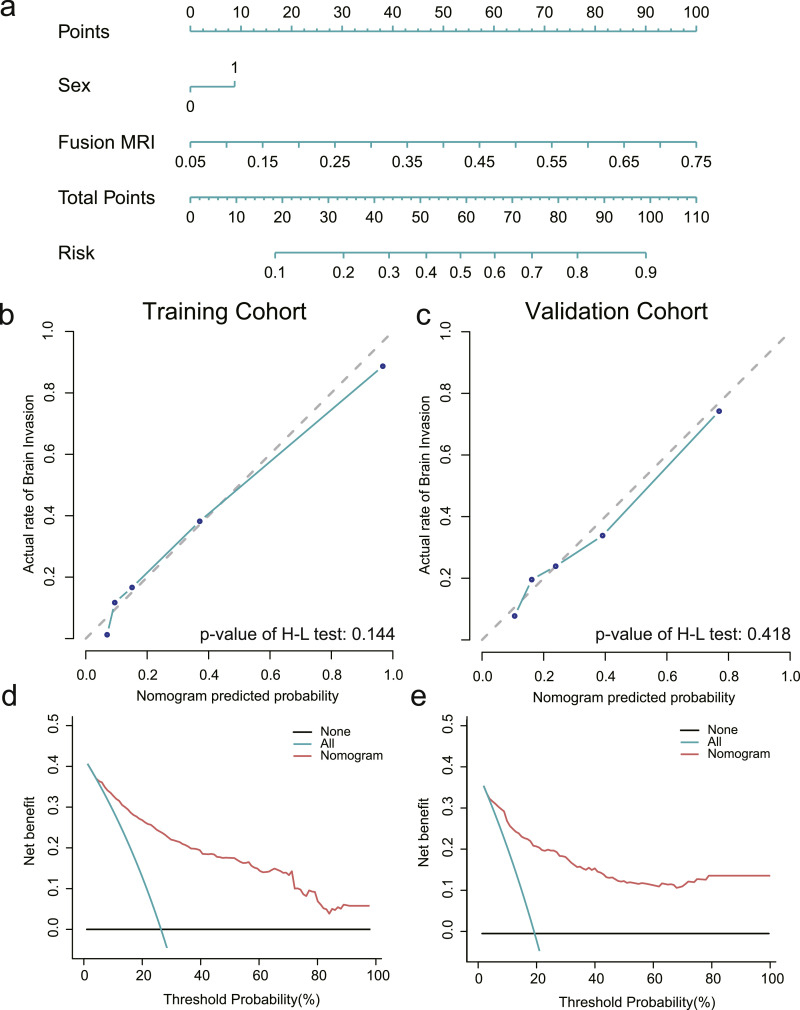
The clinicoradiomic model indicated the best performance and defined the radiomic nomogram. The H-L test together with the calibration curve was applied to measure the consistency between actual brain invasion outcomes and the probability of brain invasion predicted by the clinicoradiomic model. The actual brain invasion outcomes were consistent with the predicted probability of brain invasion in both the training and validation cohorts, with P-values of 0•144 and 0•418, respectively, as shown in Fig. 5b-c,
Fig. 5.
Establishment and performance of the clinicoradiomic model. (a) The clinicoradiomic model was conducted to develop a nomogram. (b, c) Calibration curves of the clinicoradiomic nomogram for the training and validation cohorts. The x-axis represents the probability of brain invasion measured using the clinicoradiomic model, and the y-axis represents the actual rate of brain invasion. The solid line represents the discrimination ability of the nomogram, while the diagonal dotted line represents an ideal evaluation by a perfect model. The P-value of the Hosmer-Lemeshow test was 0.144 and 0.418 in the training and validation cohorts, respectively. A closer fit to the diagonal dotted line represents a better evaluation. (d, e) Decision curve analysis for the clinicoradiomic model. The x-axis shows the threshold probability, and the y-axis measures the net benefit. The blue line represents all patients with brain invasion, while the black line represents all patients without brain invasion. The red line represents the clinicoradiomic model. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Based on clinical applications, the decision curve assessed the discrimination ability of the clinicoradiomic model, therefore, establishing its clinical utility. A net benefit was provided by the clinicoradiomic model in the DCA over the brain invasion scheme or non-invasion scheme at a threshold probability >20% (Fig. 5d-e). This result showed that the clinicoradiomic data were clinically available.
3.6. Imaging analysis by the radiologists
The diagnostic performance of the MRI visual assessment was evaluated. The accuracy of the MRI assessments were 64•4% and 65•7%, as shown in Table S4. Inter-observer agreement showed a kappa value of 0•734 (95% CI 0•595–0•873).
4. Discussion
Until now, this is the preliminary study to develop a clinicoradiomic model to predict the risk of brain invasion in meningiomas based on MRI with a big scale data. The performance of this fusing model was validated via discrimination, calibration curve and DCA in an external validation cohort. Sixteen radiomic features showed high correlation with brain invasion and maintained stable amongst multiple centres. A multi-sequence (combining T1C and T2) model of radiomics demonstrated better predictive power in both training (AUC: 0•855) and validation (AUC: 0•796) cohorts. Meantime, the nomogram, based on radiomic features and sex information, showed the best discrimination ability in both training (AUC: 0•856) and validation (AUC: 0•818) cohorts and can discriminate about 90% of cases of brain invasion in meningiomas.
In this study, the incidence of brain invasion in high grade meningiomas (50%) is consistent with other reports (22–77%) [12,23,24], and amongst tumours of all WHO grades, the incidence of brain invasion (19%) reflected much higher than the previous study (4–11%) [7,12]. The main reasons are as follows. First, according to the current WHO classification, neurosurgeons pay great attention to brain invasion and select more sampling of different surface sites to explicitly provide adequate information to the neuropathologist. Therefore, the detection rate of brain invasion has probably improved. Second, patients preference for hospital choice, and a large amount of data excluded for various reasons may cause data deviation. Moreover, amongst the subtypes of meningiomas, the frequency of brain invasion in transitional and atypical meningioma is higher than in other subtypes, which indicates that these two subtypes are more aggressive than other subtypes.
In our study, sex and histopathological grade were significantly different in the subgroup analysis. Male and histopathological high-grade meningiomas were prone to brain invasion, which was similar to the results of other studies. Recent studies have shown an increase in the proportion of men or histopathologically high-grade patients with brain invasion compared with non-invasive meningioma [7,25], especially according to the latest 2016 WHO classification of CNS tumour criteria. The mean age did not differ significantly in the continuous variable analysis. Ki-67 is commonly used as a proliferation marker in tumours [26,27]. In our study, the mean Ki-67 expression level of meningiomas with brain invasion was significantly higher than that in patients without brain invasion. This indicated that the tumour cells proliferated more actively in the brain invasion group, in agreement with published reports [28], [29], [30]. However, some authors have reported that brain invasion was not associated with Ki-67 [23,31]. It may be related to the sample size and the presence of more hot spots for counting Ki-67 cells in the central part of the specimen.
Accurate preoperative prediction of brain invasion may assist clinicians in making a personalised treatment plan to improve the quality of life because it is closely related to the selection of surgical techniques, predicting meningioma grading and prognosis. For example, the tumour-brain interface is not typically separated initially, but instead usually fully decompressed from the inside of the tumour, in order to reduce the damage of brain tissue with brain invasion. Neurophysiological testing and intraoperative navigation (with ultrasound or MRI) are needed during surgery to avoid damage to adjacent brain tissue, especially for tumours in functional areas. The surgical excision boundary for meningiomas with brain invasion is larger than that for non-invasive meningiomas. Neurosurgeons enlarge the extent of resection to remove the invaded tissue around the tumours, especially for tumours in non-functional areas. Moreover, patients with brain invasion tend to need preparation of adequate blood products for transfusion, especially for patients with rare blood types. Therefore, preoperative prediction of brain invasion is very important. At present, brain invasion can only be determined by histopathological examination [10,32], however, a considerable portion of invasive brain tissue may not be detected due to the lack of adjacent central nervous system tissue [7,32]. Imaging findings of brain invasion in meningiomas have been reported, but the image data are qualitative and subjective [33,34]. MR radiomics can reproducibly extract quantitative and objective data from different sequences, such as T1C, T2, and DWI, to predict grading, differential diagnosis, and prediction of recurrence in meningiomas [17,[34], [35], [36]], especially in the meningiomas grade. Hamerla et al. have reported comparison of machine learning classifiers for differentiation of grade 1 from higher gradings in meningioma, the results indicated that the combined model (T1WI, T1C, ADC mapping, Flair) has achieved an AUC of 0.97 [37]. Some literature results demonstrated that a radiomics prediction model performed well in predicting the meningiomas grade [38,39], which can provide visually imperceptible information about the tumour. Given this background, the radiomics model is a non-invasive, convenient, without the need for gene sequencing and tissue biopsy, and may be a potentially valuable approach to predict brain invasion in meningiomas. Moreover, in our study, the radiomics model (accuracy [ACC]: 79•35% in the training cohort) outperform the evaluation of brain invasion by two radiologists (ACC:64•5%, 65•7%).
Therefore, we explored the relationships between radiomic features and brain invasion. Of the 1595 radiomic features, eight features of a single sequence demonstrated high correlations with brain invasion and were stable across multiple centres. Amongst these features, most were textural features of images, which demonstrated microscopic description of the tumour including cellularity, peritumoural oedema, and compression of normal brain tissue by tumour. Some features cannot be easily identified by the human visual system or be interpreted to understand the specific meaning [34,35,40,41]. We observed that radiomics features including T1C_ original_ shape_ maximum 2D diameter slice, T1C_ original_ shape_ maximum 3D diameter, T2_lbp-3D-m2_glrlm_short run high grey level emphasis, etc. were associated significantly with brain invasion. Amongst them, T1C_ shape_ maximum 2D diameter slice and T1C_shape_maximum 3D diameter showed the size and shape of the tumour region. Compared with the non-invasion group, the values of these features were higher in brain invasion and showed the size and shape of meningiomas were associated with brain invasion. Chen et al. [42] previously reported that shape_ volume was associated with grade. It indicated that brain invasion, grade, and these features are related to each other. It has also been shown that these features were closely correlated to MRI characteristics, because irregular tumour shape was a risk factor for brain invasion [7]. Neighbouring grey-tone difference matrix (NGTDM) features, including busyness, contrast, and coarseness, may reflect microscopic heterogeneity within the tumours [1], which is associated with brain invasion. The heterogeneous distribution of cell density was quantified by these features in our study. After analysis of the correlation between radiomic features and the Ki-67 expression level, we found that there was a significant correlation between the two, which was consistent with other studies [1]. Thus, the radiomic feature, as a new tool, could predict brain invasion in meningiomas.
Moreover, we selected T1C and T2 sequences to develop the model. T1C imaging is usually used to define the boundaries of gross tumours, and to evaluate the extent of tumour invasion and blood supply [43]. T2 imaging is sensitive to water tissue and can be used to detect the presence of oedema [44]. Through Pearson correlation analysis, we found that the features extracted from T1C were consistent with some features extracted from T2 in the training and validation cohorts. Thus, we realized that this result revealed that the radiomic features of T1C also reflected some radiomic features of T2. For example, we assumed that a radiomic feature of T2 corresponded to oedema, which cannot be found in T1C, but it was related to other features from T1C. This indicated that different features from the two groups affected each other. It also proved that the single sequence was only sensitive to some features, and the combination of the two sequences could be more sensitive to detect more and minor features. At the same time, the correlation between the two groups of features remained highly similar in the training and validation cohorts, indicating that the selected features remained stable from different centres and different acquisition parameters. In this study, a multi-sequence (combining T1C and T2) model of radiomics features demonstrated better predictive power than the T1C or T2 models alone, in both the training and validation cohorts. These results show that multiple sequences could provide more information about tumours and improve the model's performance and accuracy of prediction of brain invasion in meningiomas.
Compared to radiomic models based on T1C, T2, and T1C/T2 images, incorporating sex information as a clinical risk factor of the nomogram demonstrated the best predictive performance. Our nomogram included preoperative risk factors without postoperative factors. This individualised nomogram was helpful for the preoperative prediction of brain invasion in meningiomas for both clinicians and radiologists. The results were more beneficial than the radiomics model and could be used in clinical applications for meningioma patients who underwent MRI scans.
Our study had several limitations. First, the retrospective study used pathological results as the gold standard. However, brain invasion may not be detected by histopathology due to a lack of brain tissue sample. In order to avoid false negatives, the samples (excluding incomplete MRI sequences, brain invasion, etc.) were assessed by surgeons and imaging, however, surgeon and imaging assessments are subjective. Second, since almost 50% of the patients were excluded for various reasons in the training and validation cohorts, there might be an inevitable selection bias, especially in 425 patients with suspected brain invasion excluded after assessment of surgeon and imaging, which may decrease the discrimination ability of our model. Third, the MRI scans were retrospectively collected from two centres with different device and acquisition parameters, and radiomic features were sensitive to parameters and other factors, we therefore standardised T1C and T2 images to obtain a standard normal distribution of the image intensities after feature selection. Fourth, in this large data study, two neuroradiologists spent plenty of time to manually delineate tumours, thus, efficient automatic segmentation was available for meningiomas in the future research. Finally, T1C and T2 sequences were selected for this study. On the T2 sequence, some cases had unclear boundaries. Although we referred to the T1C sequences for visual guidance to delineate, there were still deviations. In the future, multimodal studies such as FLAIR and DWI sequences could be incorporated into the model in order to further improve accuracy. In addition, although this is a case-control study, the main predictors enroled the clinicoradiomic model occurred before the end, so the authenticity of the study results is improved to a certain extent, further prospective studies are needed to confirm. Due to the limitation of case-control studies, the threshold of the algorithm needs to be adjusted according to the prevalence of brain invasion in different data sets when applied to the clinic [45].
5. Conclusion
Preoperative identification of brain invasion would contribute to ameliorate clinical decision-making, predicting meningioma grading and prognosis. Though radiomic analysis, sixteen radiomic features showed high correlation with brain invasion, and maintained stable amongst multiple centres. The radiomics signature can effectively predict brain invasion in meningiomas based on MR imaging (combination of T1C and T2). The clinicoradiomic model incorporating the radiomic signatures and sex information showed great performance and high sensitivity in predicting brain invasion, and can be used in patients with meningioma.
Research in context
Evidence before this study
We searched publications with the following terms on PubMed and Web of Science: “(radiomics OR texture analysis) AND (predict OR prediction) AND (brain invasion OR brain non-invasion) AND meningiomas AND MRI”. The articles were not limited to English language publications and did not have date restrictions. There has been no research on the preoperative prediction of brain invasion in meningiomas.
Added value of this study
Microscopic examination of brain invasion has been added as an independent grading criterion for the diagnosis of WHO grade II atypical meningioma, according to the latest 2016 edition of the World Health Organization (WHO) Classification of Central Nervous System (CNS) tumours. Therefore, accurate prediction of brain invasion in meningiomas is urgently needed. To our knowledge, this is the first study to build a clinicoradiomic model based on radiomic features and clinical factors to predict brain invasion in meningiomas. This model demonstrated good performance and might be a potential non-invasive tool to assist in the clinical and surgical management, meningioma grading, and prediction of prognosis.
Implications of all the available evidence
The clinicoradiomic model, a new non-invasive way, incorporating the fusing radiomic features and sex information showed good performance for risk prediction of brain invasion in patients with meningioma. This model can be used in clinical applications for meningioma patients who undergo MRI scans to potentially improve their management and outcomes.
Funding sources
This study has received funding from National Natural Science Foundation of China (81772006, 81922040), and the funders had role in study design and data interpretation. The Youth Innovation Promotion Association CAS (grant numbers 2019136). Special fund project for doctoral training program of Lanzhou University Second Hospital (grant numbers YJS-BD-33).
Data sharing
Data are available from the corresponding author upon reasonable request.
Author contributions
Conception and design: Junting Zhang, Jie Tian, Junlin Zhou
Collection and assembly of the data: Jing Zhang, Kuan Yao, Panpan Liu, Zhenyu Liu, Tao Han, Zhiyong Zhao, Yuntai Cao, Guojin Zhang
Development of the methodology: Kuan Yao, Zhenyu Liu, Jie Tian
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of the manuscript: All authors
Declaration of Competing Interest
The authors declare no potential conflicts of interest.
Acknowledgements
We would like to thank Xiaobin Hu (from Lanzhou University) for the study design guidance and data interpretation.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102933.
Contributor Information
Junting Zhang, Email: zhangjunting2003@aliyun.com.
Jie Tian, Email: jie.tian@ia.ac.cn.
Junlin Zhou, Email: ery_zhoujl@lzu.edu.cn.
Appendix. Supplementary materials
Reference
- 1.Park Y.W., Oh J., You S.C., Han K., Ahn S.S., Choi Y.S. Radiomics and machine learning may accurately predict the grade and histological subtype in meningiomas using conventional and diffusion tensor imaging. Eur Radiol. 2019;29(8):4068–4076. doi: 10.1007/s00330-018-5830-3. [DOI] [PubMed] [Google Scholar]
- 2.Perry A., Stafford S.L., Scheithauer B.W., Suman V.J., Lohse C.M. Meningioma grading: an analysis of histologic parameters. Am J Surg Pathol. 1997;21(12):1455–1465. doi: 10.1097/00000478-199712000-00008. [DOI] [PubMed] [Google Scholar]
- 3.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 4.Jenkinson M.D., Javadpour M., Haylock B.J., Young B., Gillard H., Vinten J. The ROAM/EORTC-1308 trial: radiation versus Observation following surgical resection of Atypical Meningioma: study protocol for a randomised controlled trial. Trials. 2015;16:519. doi: 10.1186/s13063-015-1040-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niu L., Zhou X., Duan C., Zhao J., Sui Q., Liu X. Differentiation researches on the meningioma subtypes by radiomics from contrast-enhanced magnetic resonance imaging: a preliminary study. World Neurosurg. 2019;126:e646–e652. doi: 10.1016/j.wneu.2019.02.109. [DOI] [PubMed] [Google Scholar]
- 6.Rogers L., Barani I., Chamberlain M., Kaley T.J., McDermott M., Raizer J. Meningiomas: knowledge base, treatment outcomes, and uncertainties. A RANO review. J Neurosurg. 2015;122(1):4–23. doi: 10.3171/2014.7.JNS131644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adeli A., Hess K., Mawrin C., Streckert E.M.S., Stummer W., Paulus W. Prediction of brain invasion in patients with meningiomas using preoperative magnetic resonance imaging. Oncotarget. 2018;9(89):35974–35982. doi: 10.18632/oncotarget.26313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hess K., Spille D.C., Wagner A., Stummer W., Paulus W., Brokinkel B. Letter: brain invasion in meningiomas-sex-associated differences are not related to estrogen- and progesterone receptor expression. Neurosurgery. 2017;81(2):E25–e27. doi: 10.1093/neuros/nyx147. [DOI] [PubMed] [Google Scholar]
- 9.Zwirner K., Paulsen F., Schittenhelm J., Gepfner-Tuma I., Tabatabai G., Behling F. Integrative assessment of brain and bone invasion in meningioma patients. Radiation Oncology. 2019;14(1) doi: 10.1186/s13014-019-1341-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chernov M. Letter to the Editor: seizures and invasive meningiomas. J Neurosurg. 2016;125(6):1615–1616. doi: 10.3171/2016.8.JNS161962. [DOI] [PubMed] [Google Scholar]
- 11.Goldbrunner R., Minniti G., Preusser M., Jenkinson M.D., Sallabanda K., Houdart E. EANO guidelines for the diagnosis and treatment of meningiomas. Lancet Oncol. 2016;17(9):e383–e391. doi: 10.1016/S1470-2045(16)30321-7. [DOI] [PubMed] [Google Scholar]
- 12.Spille D.C., Hess K., Sauerland C., Sanai N., Stummer W., Paulus W. Brain invasion in meningiomas: incidence and correlations with clinical variables and prognosis. World Neurosurg. 2016;93:346–354. doi: 10.1016/j.wneu.2016.06.055. [DOI] [PubMed] [Google Scholar]
- 13.Lambin P., Rios-Velazquez E., Leijenaar R., Carvalho S., van Stiphout R.G., Granton P. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aerts H.J., Velazquez E.R., Leijenaar R.T., Parmar C., Grossmann P., Carvalho S. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olar A., Goodman L.D., Wani K.M., Boehling N.S., Sharma D.S., Mody R.R. A gene expression signature predicts recurrence-free survival in meningioma. Oncotarget. 2018;9(22):16087–16098. doi: 10.18632/oncotarget.24498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X., Lu Y., Xiong J., Wang D., She D., Kuai X. Presurgical differentiation between malignant haemangiopericytoma and angiomatous meningioma by a radiomics approach based on texture analysis. J Neuroradiol. 2019;46(5):281–287. doi: 10.1016/j.neurad.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 17.Zhu Y., Man C., Gong L., Dong D., Yu X., Wang S. A deep learning radiomics model for preoperative grading in meningioma. Eur J Radiol. 2019;116:128–134. doi: 10.1016/j.ejrad.2019.04.022. [DOI] [PubMed] [Google Scholar]
- 18.Niu J., Zhang S., Ma S., Diao J., Zhou W., Tian J. Preoperative prediction of cavernous sinus invasion by pituitary adenomas using a radiomics method based on magnetic resonance images. Eur Radiol. 2019;29(3):1625–1634. doi: 10.1007/s00330-018-5725-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nam J.G., Park S., Hwang E.J., Lee J.H., Jin K.N., Lim K.Y. Development and validation of deep learning-based automatic detection algorithm for malignant pulmonary nodules on chest radiographs. Radiology. 2019;290(1):218–228. doi: 10.1148/radiol.2018180237. [DOI] [PubMed] [Google Scholar]
- 20.van Griethuysen J.J.M., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zwanenburg A., Vallières M., Abdalah M.A., Aerts H., Andrearczyk V., Apte A. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295(2):328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fitzgerald M., Saville B.R., Lewis R.J. Decision curve analysis. JAMA. 2015;313(4):409–410. doi: 10.1001/jama.2015.37. [DOI] [PubMed] [Google Scholar]
- 23.Vranic A., Popovic M., Cor A., Prestor B., Pizem J. Mitotic count, brain invasion, and location are independent predictors of recurrence-free survival in primary atypical and malignant meningiomas: a study of 86 patients. Neurosurgery. 2010;67(4):1124–1132. doi: 10.1227/NEU.0b013e3181eb95b7. [DOI] [PubMed] [Google Scholar]
- 24.Pizem J., Velnar T., Prestor B., Mlakar J., Popovic M. Brain invasion assessability in meningiomas is related to meningioma size and grade, and can be improved by extensive sampling of the surgically removed meningioma specimen. Clin Neuropathol. 2014;33(5):354–363. doi: 10.5414/NP300750. [DOI] [PubMed] [Google Scholar]
- 25.Hess K., Spille D.C., Adeli A., Sporns P.B., Brokinkel C., Grauer O. Brain invasion and the risk of seizures in patients with meningioma. J Neurosurg. 2018;130(3):789–796. doi: 10.3171/2017.11.JNS172265. [DOI] [PubMed] [Google Scholar]
- 26.Ma J., Zhang Y., Chen L., Chen Y., Yang Y., Li D. Low expression of phosphatase and tensin homolog and high expression of Ki-67 as risk factors of prognosis in cranial meningiomas. World Neurosurg. 2020;136:e196–e203. doi: 10.1016/j.wneu.2019.12.108. [DOI] [PubMed] [Google Scholar]
- 27.Maier A.D., Bartek J., Jr., Eriksson F., Ugleholdt H., Juhler M., Broholm H. Clinical and histopathological predictors of outcome in malignant meningioma. Neurosurg Rev. 2020;43(2):643–653. doi: 10.1007/s10143-019-01093-5. [DOI] [PubMed] [Google Scholar]
- 28.Suwa T., Kawano N., Oka H., Ito H., Kameya T. Invasive meningioma: a tumour with high proliferating and "recurrence" potential. Acta Neurochir (Wien) 1995;136(3–4):127–131. doi: 10.1007/BF01410613. [DOI] [PubMed] [Google Scholar]
- 29.Suwa T., Kawano N., Kameya T., Ito H., Oka H., Yada K. Invasive meningiomas in relation to high proliferating potential. Noshuyo Byori. 1993;10(1):63–67. [PubMed] [Google Scholar]
- 30.Madsen C., Schroder H.D. Ki-67 immunoreactivity in meningiomas–determination of the proliferative potential of meningiomas using the monoclonal antibody Ki-67. Clin Neuropathol. 1997;16(3):137–142. [PubMed] [Google Scholar]
- 31.Backer-Grondahl T., Moen B.H., Arnli M.B., Torseth K., Torp S.H. Immunohistochemical characterization of brain-invasive meningiomas. Int J Clin Exp Pathol. 2014;7(10):7206–7219. [PMC free article] [PubMed] [Google Scholar]
- 32.Brokinkel B., Stummer W. Brain invasion in meningiomas: the rising importance of a uniform neuropathologic assessment after the release of the 2016 world health organization classification of central nervous system tumors. World Neurosurg. 2016;95:614–615. doi: 10.1016/j.wneu.2016.08.047. [DOI] [PubMed] [Google Scholar]
- 33.Hwang W.L., Marciscano A.E., Niemierko A., Kim D.W., Stemmer-Rachamimov A.O., Curry W.T. Imaging and extent of surgical resection predict risk of meningioma recurrence better than WHO histopathological grade. Neuro Oncol. 2016;18(6):863–872. doi: 10.1093/neuonc/nov285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Chen J.H., Chen T.Y., Lim S.W., Wu T.C., Kuo Y.T. Radiomics approach for prediction of recurrence in skull base meningiomas. Neuroradiology. 2019 doi: 10.1007/s00234-019-02259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanazawa T., Minami Y., Jinzaki M., Toda M., Yoshida K., Sasaki H. Preoperative prediction of solitary fibrous tumor/hemangiopericytoma and angiomatous meningioma using magnetic resonance imaging texture analysis. World Neurosurg. 2018;120:e1208–e1216. doi: 10.1016/j.wneu.2018.09.044. [DOI] [PubMed] [Google Scholar]
- 36.Lu Y., Liu L., Luan S., Xiong J., Geng D., Yin B. The diagnostic value of texture analysis in predicting WHO grades of meningiomas based on ADC maps: an attempt using decision tree and decision forest. Eur Radiol. 2019;29(3):1318–1328. doi: 10.1007/s00330-018-5632-7. [DOI] [PubMed] [Google Scholar]
- 37.Hamerla G., Meyer H.J., Schob S., Ginat D.T., Altman A., Lim T. Comparison of machine learning classifiers for differentiation of grade 1 from higher gradings in meningioma: a multicenter radiomics study. Magn Reson Imaging. 2019;63:244–249. doi: 10.1016/j.mri.2019.08.011. [DOI] [PubMed] [Google Scholar]
- 38.Laukamp K.R., Shakirin G., Baessler B., Thiele F., Zopfs D., Hokamp N.G. Accuracy of radiomics-based feature analysis on multiparametric magnetic resonance images for noninvasive meningioma grading. World Neurosurg. 2019 doi: 10.1016/j.wneu.2019.08.148. [DOI] [PubMed] [Google Scholar]
- 39.Chu H., Lin X., He J., Pang P., Fan B., Lei P. Value of MRI radiomics based on enhanced T1WI images in prediction of meningiomas grade. Acad Radiol. 2020 doi: 10.1016/j.acra.2020.03.034. [DOI] [PubMed] [Google Scholar]
- 40.Yan P.F., Yan L., Hu T.T., Xiao D.D., Zhang Z., Zhao H.Y. The potential value of preoperative MRI texture and shape analysis in grading meningiomas: a preliminary investigation. Transl Oncol. 2017;10(4):570–577. doi: 10.1016/j.tranon.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Q., Yao K., Liu Z., Li L., Zhao X., Wang S. Radiomics analysis of placenta on T2WI facilitates prediction of postpartum haemorrhage: a multicentre study. EBioMedicine. 2019;50:355–365. doi: 10.1016/j.ebiom.2019.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C., Guo X., Wang J., Guo W., Ma X., Xu J. The diagnostic value of radiomics-based machine learning in predicting the grade of meningiomas using conventional magnetic resonance imaging: a preliminary study. Front Oncol. 2019;9:1338. doi: 10.3389/fonc.2019.01338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou M., Scott J., Chaudhury B., Hall L., Goldgof D., Yeom K.W. Radiomics in brain tumor: image assessment, quantitative feature descriptors, and machine-learning approaches. AJNR Am J Neuroradiol. 2018;39(2):208–216. doi: 10.3174/ajnr.A5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q., Li Q., Mi R., Ye H., Zhang H., Chen B. Radiomics nomogram building from multiparametric MRI to predict grade in patients with glioma: a cohort study. J Magnetic Resonance Imag. JMRI. 2019;49(3):825–833. doi: 10.1002/jmri.26265. [DOI] [PubMed] [Google Scholar]
- 45.Park S.H., Han K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology. 2018;286(3):800–809. doi: 10.1148/radiol.2017171920. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.