Abstract
ecent advances in deep learning for medical image segmentation demonstrate expert-level accuracy.ecent advances in deep learning for medical image segmentation demonstrate expert-level accuracy.R However, application of these models in clinically realistic environments can result in poor generalization and decreased accuracy, mainly due to the domain shift across different hospitals, scanner vendors, imaging protocols, and patient populations etc. Common transfer learning and domain adaptation techniques are proposed to address this bottleneck. However, these solutions require data (and annotations) from the target domain to retrain the model, and is therefore restrictive in practice for widespread model deployment. Ideally, we wish to have a trained (locked) model that can work uniformly well across unseen domains without further training. In this paper, we propose a deep stacked transformation approach for domain generalization. Specifically, a series of n stacked transformations are applied to each image during network training. The underlying assumption is that the “expected” domain shift for a specific medical imaging modality could be simulated by applying extensive data augmentation on a single source domain, and consequently, a deep model trained on the augmented “big” data (BigAug) could generalize well on unseen domains. We exploit four surprisingly effective, but previously understudied, image-based characteristics for data augmentation to overcome the domain generalization problem. We train and evaluate the BigAug model (with n = 9 transformations) on three different 3D segmentation tasks (prostate gland, left atrial, left ventricle) covering two medical imaging modalities (MRI and ultrasound) involving eight publicly available challenge datasets. The results show that when training on relatively small dataset (n=10~32 volumes, depending on the size of the available datasets) from a single source domain: (i) BigAug models degrade an average of 11% (Dice score change) from source to unseen domain, substantially better than conventional augmentation (degrading 39%) and CycleGAN-based domain adaptation method (degrading 25%), (ii) BigAug is better than “shallower” stacked transforms (i.e. those with fewer transforms) on unseen domains and demonstrates modest improvement to conventional augmentation on the source domain, (iii) after training with BigAug on one source domain, performance on an unseen domain is similar to training a model from scratch on that domain when using the same number of training samples. When training on large datasets (n=465 volumes) with BigAug, (iv) application to unseen domains reaches the performance of state-of-the-art fully supervised models that are trained and tested on their source domains. These findings establish a strong benchmark for the study of domain generalization in medical imaging, and can be generalized to the design of highly robust deep segmentation models for clinical deployment.
Keywords: Domain generalization, Data augmentation, Deep learning, Medical image segmentation
I. Introduction
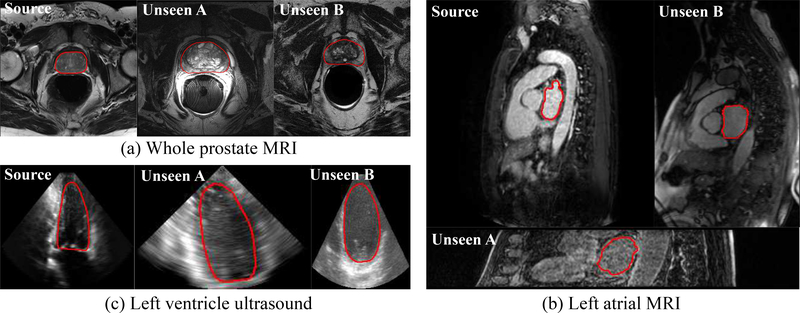
Successful clinical deployment of deep learning-based artificial intelligence (AI) models for medical imaging tasks requires a trained model to maintain a high level of accuracy when applied to unseen domains (i.e., different hospitals, scanner vendors, imaging protocols, patient populations, etc.) [1], as illustrated in Fig. 1. Ideally, highly generalizable models in medical imaging could be achieved when training datasets include a large quantity of high-quality images from multiple centers with diverse imaging vendors/protocols. Unfortunately, in current practice, datasets are often limited by the lack of annotations and difficulty in data sharing among centers [2]. These limitations have led to scenarios where small training datasets which lack diversity fail to maintain their performance on data from “unseen” domains. For example, the error rate of a deep model for retinal image analysis was 5.5% on images from the same vendor used in training dataset, but decreased to 46.6% on images from another vendor [3]. This issue of poor generalizability has become one of the major roadblocks for deploying deep learning models into clinical practice [4].
Fig. 1.
Medical image segmentation in the source and unseen domains (i.e., a specific medical imaging modality across different vendors, imaging protocols, and patient populations, etc.) for (a) whole prostate MRI, (b) left atrial MRI, and (c) left ventricle ultrasound. The illustrated images are processed with intensity normalization.
Given the limited quantity and quality of medical imaging data, it is infeasible to employ naive strategies that aggregate data from multiple source domains and impractical to train separate high-quality domain-specific (e.g., vendor specific) models. Two popular solutions have been proposed to improve the generalizability of deep learning models using data trained from a single source domain. The first, transfer learning, is the process of fine-tuning a portion of a pre-trained network (usually the last few layers [5] or shared convolutional filters [6]). Transfer learning is able to overcome some of the aforementioned issues by only requiring a small amount of annotated data in the unseen domain; however, it is limited in use due to the lack of pre-trained models developed on a large amount of medical imaging data. A second solution, domain adaptation [7], aims to generalize to a known target domain whose annotations are unknown during model training. Generative adversarial network (GAN) [8] and its variants (e.g., CycleGAN [9]) are frequently integrated into domain adaptation methods, by either learning domain-invariant features (seen in MRI [10], ultrasound [11], histopathology [12]), or translating image style between the source and target domains (used in X-ray [13], [14] and ultrasound [15]). Additionally, these methods have been used to model the imaging physics (e.g., estimating the T1–w pulse sequence) of the target MRI imaging domain, and by applying the model to create target data specific training features, an augmented deep model can be trained [16].
Despite their promising performance, the assumption of a known target domain requires specific image samples need to be collected (or even labeled) and a new model needs to be retrained before deployment. It is not feasible to obtain a pair of source and target domain images to implement the adversarial domain adaptation for every new application. Therefore, model deployment using these typs of techniques is impractical in diverse patient populations (e.g., multiple clinical centers) or unpredictable scenarios (e.g., emergency care or rural area use of ultrasound).
Domain generalization, which indicates settings that one has no access to any data from the unseen target domains, has the potential to overcome these issues. Particularly, in the field of medical imaging, we are usually faced with the difficult situation that the training dataset is derived from a single center and acquired on one vendor system with a specific protocol. Some non-deep-learning models have been shown to be robust to center-specific or vendor-specific variability. For example, by combining mesh-based computational atlas with Gaussian appearance model [17] or by Bayesian transfer learning [18], 2D brain MRI segmentation can be generalized to unseen domains at certain accuracy. Inclusion of a deep learning in the classical probabilistic generative model is also proposed to improve 2D brain MRI segmentation on unseen domains [19]. In 2D computer vision applications with deep learning, researchers recently made progress in this highly challenge setting [20], [21], [22]. Their approaches, essentially, are various complexities of data augmentations to expand the data distribution coverage (with higher variations). Specifically, additional training data samples are generated in image domain [20], semantic space [22], or by adversarial learning [21], respectively.
Data augmentation has proven to be among the most important regularization techniques related to deep learning’s generalization performance [23]. It helps prevent models from overfitting to the training data and generalize better on the testing data. However, majority of published work has focused on non-medical imaging data and default augmentation settings are either derived from the same source in training and validation or do not consider domain source at all [24], [23], [25], [26]. In specific applications of medical image segmentation, image rotation and GAN-based augmentations have been shown to improve the performance in 2D data for both CT and MRI [27], as they can extrapolate and interpolate the manifold of data, respectively. Recently, we proposed a reinforcement learning-based searching approach for selecting necessary data augmentations in 3D medical image segmentation tasks [28]. However, implementing data augmentation methods, even optimal on the source domain, does not guarantee the generalizability on data from unseen domains. Furthermore, while a large amount of medical imaging data is acquired in 3D, the majority of published work considers 2D data augmentation approaches due to augmentation in large 3D volumetric data being computationally expensive. The impact of 3D data augmentation on domain generalization in medical image segmentation tasks is largely unknown.
Medical images acquired by the same imaging modality, e.g., T2 MRI, across different vendors (GE, Philips, Siemens, etc.), scanning protocols (flip angle, repetition time, etc.), and patient populations are visually different in three aspects: image quality, image appearance, and spatial configuration (Refer to Fig. 1 (a), (b) for such examples). Some imaging modalities also have vendor-specific differences such as ultrasound (Fig. 1 (c)) and OCT, whereas CT (for the same phase) generally has more consistent image characteristics.
Motivated by the observed heterogeneity in medical imaging data, we propose a deep stacked transformation data augmentation approach (called BigAug) for generalizing 3D medical image segmentation models to unseen domains. Our main hypothesis is that the domain shift properties of medical imaging data can be simulated by applying a wide variety of data augmentation techniques on a (single) source domain, and consequently, a deep neural networks trained on augmented (or “big”) data that incorporates domain shift simulations would result in improved generalization on unseen domains. As far as we know, we are the first to investigate data augmentation for unseen domain generalization in medical imaging deep learning.
BigAug is designed to have individual images undergo nine stacked image transformations within each training iteration, in order to substantially augment the diversity of the data seen by the neural network during training. Each technique is controlled by two parameters which determine the probability and magnitude of the image transformation. The BigAug technique is integrated and demonstrated on a 3D Anisotropic Hybrid Network (AH-Net) [29] architecture. In the following experiments, we
systematically analyze the effect of each augmentation technique on the model’s generalization ability, revealing the major differences of medical (MRI and ultrasound) images caused by domain shift, and showing that augmentations used in BigAug are able to model these changes.
demonstrate BigAug to be uniformly effective for both MRI and ultrasound, with 11.6% average reduction in Dice coefficient on unseen domains, compared with 39.3% and 25.6% reduction using a standard method (random cropping only) and a CycleGAN-based domain adaptation method, respectively.
demonstrate BigAug to outperform “shallower” stacked transforms (with less transforms) on unseen domains, and modest improvement compared to the standard augmentation method on the source domain;
show that when trained with datasets of the same size, the model with BigAug can achieve a similar performance on the unseen domain as compared with a model trained from scratch on that unseen domain.
show that BigAug is a key component for achieving good generalization and state-of-the-art segmentation accuracy on several unseen (public) datasets when the model is trained with a larger training dataset.
II. Methods
We consider the problem of unseen domain generalization, where we are provided with data XS and annotations YS from a single source domain without any data and annotations from unseen domains. The goal is to train a model fS from the source domain and make it perform uniformly well across unseen domains. In our setting, both XS and YS are 3D volumes, and fS is a 3D segmentation network.
A. Deep Stacked Transformations
Our BigAug is a sequence of n stacked transformations τ(.), as formulated in Eq. 1, where each transformation is an image processing function, and each function is associated with two parameters: 1) the probability p to apply the function and 2) the magnitude m of the function. Given training data xs and associated annotation ys, augmented data and corresponding annotation could be generated after n transformations through Eq. 1.
| (1) |
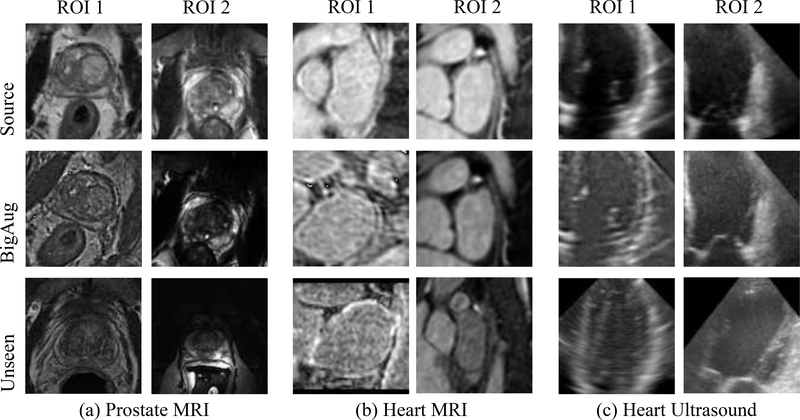
Image processing functions are mainly used to alter the three aspects (image quality, appearance, and spatial configuration) of medical images. Here, transformations are applied in each mini-batch during training to account for the contribution of domain-specific shifts in medical images. Our basic hypothesis is that augmenting image sets during the training can result in models that are robust over potential variations in unseen domains. Potentially it could be more effective and efficient than performing data processing/synthesis at inference stage (e.g., using CycleGAN to translate target image to source-like appearance). Fig. 2 shows some examples of BigAug results in different tasks in both MRI and ultrasound. It can be observed that the domain shift is composed of the combination of multiple factors, which can be simulated by BigAug. Note that the transformations in BigAug (Eq. 1) are without mandatory order. In our work, they are in the order as described below.
Fig. 2.
Examples of deep stacked transformations (BigAug) results on (a) whole prostate MRI, (b) left atrial MRI, and (c) left ventricle ultrasound. 1st row: ROIs randomly cropped in volumes from source domains; 2nd row: corresponding cropped ROIs after BigAug; 3rd row: ROIs randomly cropped in volumes from unseen domains. The image pairs of 2nd–3rd rows have better visual similarity than 1st–3rd rows.
Image Quality
sharpness, blurriness, and noise level are often associated with different image qualities in medical imaging. Blurriness caused by MR/ultrasound motion artifacts can affect the interpretability of images and the performance of segmentation algorithms. In our work, Gaussian filtering is used to blur the image as to simulate unseen more blurry images, with a magnitude (defined by the standard deviation of a Gaussian kernel) ranging between [0.25, 1.5]. On the other hand, to compensate blurriness as to simulate unseen sharper images, the image sharpening technique known as unsharp masking is utilized. Unsharp masking is done by applying the filter inverse to the blur,
| (2) |
where Iblurred and Ifilteredblurred are blurred images by applying Gaussian filtering on image I and image Iblurred, respectively, and α is the magnitude (strength of sharpening effect) ranging between [10, 30]. Noises are commonly observed in medical images, e.g., in Fig. 2 (b), the first unseen image has more noises than source images. To make our model robust to the noise, Gaussian noises were added with magnitude (std. of the Gaussian distribution) ranging between [0.1, 1.0] to the image. The three image quality transformations are mainly based on Gaussian function/filter, as a Gaussian distribution is commonly used to represent real-valued variables with unknown distributions. There exist many other specific functions/filters, such as speckle and Poisson noise, median and median filter, etc., which may improve the performance for special imaging modalities. The image quality-based transforms do not apply to annotations YS.
Image Appearance
The appearance difference of medical imaging is related to the statistical characteristics of image intensities, such as variations of brightness and contrast, and intensity perturbation, which result from different scanners and scanning protocols. Refer to the 1st and 3rd rows in Fig. 2 for the image appearance differences in MRI and ultrasound. To adjust the brightness of the image, we randomly shift the intensity level with magnitude ranging between [−0.1, 0.1] for the image. To control the contrast of the image, we apply gamma correction with magnitude (gamma value) ranging between [0.5, 1.0] or [1.0, 4.5], where magnitude = 1 gives the original image and smaller/larger value makes image lighter/darker, respectively. Gamma correction is used in a highly competitive brain MRI segmentation algorithm [30], and contributes to the robust segmentation performance across multiple hospitals [31]. To perturbing image intensities, we multiply a scale factor and add a shift factor for the image, both with magnitude ranging between [−0.1, 0.1]. Such a method is a component in the state-of-the-art brain MRI segmentation algorithm [32]. The image appearance transforms are not applied to annotations YS.
Spatial Configuration
Spatial variations may include rotation (e.g., caused by different patient orientations during scanning), scaling (e.g., variation of organ/lesion size), and deformation (e.g., caused by organ motion or abnormality). Refer to the 1st and 3rd rows in Fig. 2 (a–c) for the spatial variations in MRI and ultrasound. These operations are computational expensive for large 3D volumetric data 1. A GPU-based acceleration approach [33] could be developed, but allocating the maximal capacity of GPU memory for model training only along with data augmentation on the fly are more desirable. In addition, since the whole 3D volume does not fit into the limited memory of the GPU, sub-volumes cropping are usually needed to fed into the model during the training. In this work, we develop an extremely efficient CPU-based spatial transform technique based on an open-source implementation2, which first calculates the 3D coordinate grid of sub-volume (with size of w×h×d voxels) to which the transformations (combining random 3D rotation, scaling, deformation, and cropping) are applied and then image interpolation is performed. We make further accelerations by only performing the interpolation within the minimal cuboid containing the 3D coordinate grid so that the computational time is independent from the input volume size (i.e., only depending on the cropping sub-volume size), and the spatial transform augmentation can be performed on the fly during training. The rotation and scaling are both performed along all three axes, and the magnitudes are controlled by rotation degree ranging between [−20°, 20°] and by scaling factor ranging between [0.4, 1.6], respectively. The deformation is achieved by sampling a grid of random offset vectors, which is smoothed by Gaussian smoothing filter (standard deviation ranging between [10, 13]) and rescaled by a random factor (ranging between [0, 1000]). The spatial transforms are applied to both data XS and annotations YS.
Note that instead of augmenting training images in such an explicit way, transformations (e.g., spatial) could be incorporated into the network learning process, through approaches like Spatial Transformer Networks [24]. However, the learned invariants are from the source domain, which may not generalize well on different unseen domains. The key idea of our BigAug is to use data augmentation to extrapolate the manifold of the source data, with the regularization of prior knowledge to handle the domain shift in medical imaging.
B. 3D Deep Segmentation
We use AH-Net [29] as the backbone of our 3D segmentation network. The AH-Net takes the advantage of both 2D and 3D deep segmentation networks by transferring deep features learned from large-scale 2D images to 3D encoder-decoder network. For the training, the inputs are sub-volumes cropped from the whole volume and outputs are the corresponding sub-volumes of segmentation masks with 1-channel annotations. To increase the variation of training data, sub-volumes are randomly cropped and equally distributed between the foreground and background. We use Dice loss [34] as the loss function which naturally balances the positive and negative voxel distribution. In testing, sliding window with overlapping is applied to the whole 3D volume to generate the final 3D segmentation.
III. Experiments
A. Experimental Design
3D medical imaging mainly includes CT, MRI, PET, ultrasound, and OCT. Therefore, we would like to evaluate the proposed method with various data from public resources, including Medical Segmentation Decathlon (MSD)1, Grand Challenges in Biomedical Image Analysis2, and recent MICCAI challenges.
Due to data availability and restrictions (only one public PET segmentation challenge is available from a single center3; an ideal public OCT dataset (containing three vendors)4 is available but can only be used for the challenge), and also to include sufficient image variabilities (CT imaging is fairly standardized to Hounsfield scale so the domain shift is usually less of a concern), we decided to use MRI and ultrasound to illustrate the capabilities of the proposed method. Prostate MRI and Heart MRI datasets from the MSD challenge are selected as the source domain data of our Task 1 and Task 2, respectively, because: 1) MSD [35] is a recent large scale annotated medical image dataset which represents the state-of-the-art dataset with high quality; 2) moreover, more than two other Prostate MRI and Heart MRI public datasets with annotations can be found, serving as multiple unseen domains. In addition, the CETUS Heart ultrasound dataset1 is selected as our Task 3, since it contains image data from the three major ultrasound vendors (i.e., GE, Philips, Siemens).
We first validate our method on three tasks as follows: Task 1: whole prostate segmentation in MRI volumes, Task 2: left atrial segmentation in MRI volumes, and Task 3: left ventricle segmentation in ultrasound volumes. Each model is trained and validated on a single source domain dataset with the same BigAug configuration, and applied/tested to 2–3 unseen domain sets. Second, we investigate the variation in model performances when the models are trained with a single augmentation transformation or a combination of several best-performing transformations. Third, we train deep models from scratch for whole prostate (WP), peripheral zone (PZ), and transition zone (TZ) segmentation in prostate MRI with different numbers of training data from a target domain, and compare their performances with the BigAug augmented model. Finally, models for whole prostate segmentation in MRI are trained on a self-collected big data with and without BigAug, and applied to four unseen domains.
B. Datasets: Source vs. Unseen Domain
Task 1
Four publicly available 3D prostate MRI datasets are used: MSD-Prostate (MSD-P)2, PROMISE123 [36], NCI-ISBI134, and ProstateX5 [37]. MSD-P serves as the single source domain, and others are different unseen domains.
Task 2
Three publicly available 3D heart MRI datasets are used: MSD-Heart (MSD-H)2, 2018 ASC6, and MM-WHS7 [38]. MSD-H serves as the single source domain, and others are different unseen domains.
Task 3
One publicly available 3D ultrasound dataset, CETUS1 is used, where data is equally acquired from three different ultrasound vendors (i.e., GE, Philips, Siemens, 10 volumes each). We used heuristics to identify vendor association, but we acknowledge that our split strategy may include wrong associations8. Vendor A is used as the single source domain, and Vendor B and C serve as unseen domains.
All datasets have annotations provided by the data source, except for the ProstateX where no prostate segmentation is available and the annotations of both peripheral zone (PZ) and transition zone (TZ) are provided by our radiologist collaborators. One patient’s study in ProstateX was excluded due to prior surgical procedure to resect a large portion of the TZ (transurethral resection of the prostate), which deformed the appearance of the prostate. Table I briefly summarizes the used datasets.
TABLE I.
Datasets Used In Our Experiment.
| Task | 1. MRI - whole prostate | 2. MRI - left atrial | 3. Ultrasound - left ventricle | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Domain | Source | Unseen | Source | Unseen | Source | Unseen | ||||
| Dataset | MSD-P | PROMISE12 | NCI-ISBI13 | ProstateX | MSD-H | ASC | MM-WHS | CETUS-A | CETUS-B | CETUS-C |
| # Data | 26/6 | 50 | 60 | 98 | 16/4 | 100 | 20 | 8/2 | 10 | 10 |
In addition to the benchmark datasets, a large MRI dataset including 465 patients is used in the final experiment. Our collaborated radiologists collected 465 MRI data (denoted as MultiCenter) from multiple medical centers worldwide, representing multiple MRI vendors (i.e. GE, Philips, Siemens), and various center-specific MRI protocols. The whole prostate boundaries were manually traced in three planes on T2-weighted MRI by a radiologist with over 10 years of experience in interpretation of prostate MRIs. A second radiologist with < 1 year experience in reading prostate MRI was trained under supervision of the expert and performed segmentation in the same fashion using the same segmentation software. The segmentations from the expert radiologist were considered ground truth. This ‘MultiCenter’ dataset serves as a large source of training data, and MSD-P, PROMISE12, NCI-ISBI13, and ProstateX are four unseen domains.
C. Implementation
This work is implemented using NVIDIA Transfer Learning Toolkit for Medical Imaging1. We first resample all the data in source domain into a fixed resolution of 1.0mm ×1.0mm × 1.0mm. Then, image intensities I are normalized to [0, 1] by (I − min)/(max − min), where min=0, max=2048 for all MRIs2 except for ASC dataset, and min=0, max=255 for ultrasound and ASC which range from [0, 255]. In BigAug, the probability to apply each transformation is set to 0.5; transformations are in the order as described in Section II-A. Performances of models are not sensitive to different orders based on our preliminary experiments (prostate MRI segmentation) – the generated images might have slight differences if changing the order of transformations; however, considering the comprehensive changes after BigAug and the overall large amount of generated training samples for training, these differences tend to result in minor differences on the network performance. Image intensities are not renormalized after BigAug, as renormalization results in lower performance in empirical experiments. The cropped sub-volumes are of the following sizes: 96×96×32 (w × h × d ) for Task 1, and 96 × 96 × 96 for Task 2 and Task 3. ResNet50’s weights pretrained on ImageNet are used to initialize the encoder part of AH-Net. We use ADAM to optimize the network with the initial learning rate of 0.0001. Task 1 and Task 2 are trained on 4 GPUs on the NVIDIA DGX cluster, and Task 3 is trained in 1 NVIDIA Titan XP GPU, all using SGD and with a mini-batch size of 4 ROIs per GPU. Since randomness exists in the whole training process, each model is trained for 300 epochs on the source domain for three times, and the model with the best performance on the validation set of the source domain is selected to be applied to unseen domains.
In model inference, the testing data is resampled into 1.0mm×1.0mm×1.0mm and normalized to [0, 1], and the stride of sliding window is (w−16)×(h−16)×(d−16). For Task 1, since MSD-P has 2-class annotations for PZ and TZ, we first train a 2-class model and then combine the output into 1-class as the whole prostate after inference; and only T2-weighted image is used, as most unseen data only has T2.
D. Experimental Results and Analysis
1). BigAug vs. Standard Model vs. Domain Adaptation (CycleGAN)
For each segmentation task, we trained models on the source data, including a baseline model with random cropping only, nine models each with a single augmentation/transformation, and a BigAug model. The train/validation splits in the source data are shown in Table I. Additionally, we implemented a popular domain adaptation method – CycleGAN [9], [13], [14], which first transfers the unseen testing image to the appearance similar to the source domain, and then applies the baseline model on the transferred image. Such a method has been shown to be no worse than traditional inference-time image transformation approach, such as histogram matching [14]. More specifically, we split each dataset with a ratio of 4:1 for the training and validation of CycleGAN. 2D image slices are extracted at certain amount of interval from 3D volumes, in order to balance the slice numbers in source and target sets. All the image slices are resized to 256×256, and rescaled to [0, 255]. We train the CycleGAN model for 200 epochs.
We report the Dice coefficient for the segmentation on the validation sets in the source domains and on the testing sets in unseen domains in Table II. The Dice coefficient is a standard metric to report the segmentation performance. All of the public challenge datasets utilized in this study use Dice as one of or even the single evaluation metric (i.e., ASC, MM-WHS). While distance-based metrics, such as Hausdorff distance, are also important, we only use Dice to keep simplicity for interpreting results and facilitate comparison with supervised methods on all public datasets. The numbers of testing images for each unseen dataset are listed in Table I. The baseline model degrades dramatically on unseen domains, from 89.1% to 49.8% on average. The major findings are:
TABLE II.
The Effect of BigAug and Various Augmentation Methods on Unseen Domain Generalization (Measured with Dice Scores). Source Columns Indicate The Dataset Used for Training, and its Dice Scores are Validation Dice Scores (Using a Split) for Comparisons. Unseen Columns List Dice Results When Applied to Unseen Datasets (of The Model Trained on the Source). Here Baseline Refers to a Random Crop With no Further Augmentation. Top4 Stands for the Combination of Four Best Performing Augmentations (Sharpening, Brightness, Contrast, Scaling). Supervised Indicates the State-of-the-Art Literature Results, When a Model is Trained and Tested on the Same Dataset.
| Task 1. MRI - whole prostate | Task 2. MRI- left atrial | Task 3. Ultrasound - left ventricle | All Tasks | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Source | Unseen | Source | Unseen | Source | Unseen | Source | Unseen | |||||
| MSD-P | PROMISE | NCI-ISBI | Prostate X | MSD-H | ASC | MM-WHS | CETUS-A | CETUS-B | CETUS-C | Average | Average | |
| Baseline | 89.6 | 60.4 | 58.0 | 76.8 | 91.9 | 4.4 | 72.9 | 85.8 | 51.7 | 39.2 | 89.1 | 49.8 |
| Shaipening | 90.6 | 65.5 | 82.8 | 84.0 | 91.5 | 5.7 | 78.9 | 83.7 | 59.5 | 78.5 | 88.6 | 62.9 |
| Blurring | 86.1 | 63.9 | 67.0 | 79.9 | 90.9 | 3.3 | 76.9 | 90.5 | 73.4 | 72.4 | 89.2 | 61.1 |
| Noise | 91.1 | 59.3 | 67.4 | 81.4 | 91.4 | 8.3 | 78.0 | 87.3 | 66.8 | 62.2 | 90.0 | 59.0 |
| Brightness | 89.7 | 63.3 | 66.9 | 83.0 | 91.3 | 12.2 | 80.2 | 85.5 | 63.6 | 83.1 | 88.8 | 63.6 |
| Contrast | 91.1 | 72.7 | 60.7 | 86.1 | 91.3 | 12.7 | 78.6 | 88.4 | 58.4 | 85.5 | 90.3 | 63.6 |
| Perturb | 90.1 | 63.4 | 69.5 | 81.5 | 91.7 | 6.6 | 77.3 | 88.5 | 63.6 | 83.1 | 90.1 | 55.7 |
| Rotation | 87.4 | 59.0 | 57.9 | 75.1 | 91.2 | 5.2 | 72.1 | 78.0 | 60.4 | 62.6 | 85.5 | 54.7 |
| Scaling | 90.8 | 59.3 | 60.8 | 78.8 | 91.3 | 7.4 | 75.3 | 91.0 | 84.1 | 68.2 | 91.0 | 61.3 |
| Deform | 89.7 | 61.4 | 61.5 | 81.2 | 91.6 | 7.8 | 69.2 | 86.3 | 62.4 | 31.4 | 89.2 | 51.1 |
| Top4 | 91.0 | 73.5 | 83.0 | 86.5 | 91.6 | 45.4 | 79.4 | 90.9 | 81.9 | 80.5 | 91.2 | 74.9 |
| CycleGAN | - | 74.7 | 76.4 | 81.2 | - | 18.0 | 76.2 | - | 65.3 | 66.6 | - | 63.5 |
| BigAug (ours) | 91.3 | 80.2 | 85.4 | 86.5 | 91.4 | 65.5 | 80.0 | 92.1 | 84.9 | 81.3 | 91.6 | 80.0 |
| Supervised | - | 91.4 [39] | 89.3 [40] | 91.9* | - | 94.2 [41] | 88.6# | - | 92.5* | 92.5* | - | 91.5 |
Indicates Inter-Observer Variability.
On average, across all tasks on unseen domains, BigAug (Dice=80.0%) performs substantially better than any one of the tested augmentations, and significantly better than the baseline model (49.8%) and CycleGAN (63.5%). Using only simple random crop (baseline) does not generalize well on unseen datasets with Dice dropping as much as 40%, which supports the importance of data augmentation in general. It is surprising that the BigAug based domain generalization is even better than CycelGAN based domain adaptation which has seen the target domain.
The major imaging differences caused by domain shift of MRI are image quality and appearance, in which sharpening is the most important one, followed by contrast, brightness, and intensity perturbation. Refer to Fig. 1 for some examples. Fig. 1(a) demonstrate that contrast and sharpening are the major differences with unseen A (PROMISE12) and unseen B (NCI-ISBI13), respectively, compared to the source image (MSD-P). Note that the spatial transforms seem to be less important for prostate MRI, but they contribute to transform heart MRIs where the shape, size, and orientation of heart can be very different (refer to Fig. 1 (b) and Fig. 2 (b)). This is likely due to the prostate is relatively static while the heart is a moving/beating object.
The imaging differences caused by domain shift of different Ultrasound vendors are more comprehensive, which could be related to the spatial transform, image appearance and quality, in which 3D scaling is the most important one, followed by brightness, blurring, and contrast. Refer to Fig. 1(c) for some examples: compared to the source image (CETUS-A), scaling and contrast are the major differences with unseen A (CETUS-B) and unseen B (CETUS-C), respectively. Spatial transformations substantially contribute to heart ultrasound segmentation task, partially because the heart is a deformable object and different angles between the ultrasound probe and heart can result in images with different rotation degrees. In addition, the size of training dataset CETUS-A is small, not covering enough geometric variations.
For a specific unseen domain (e.g., ASC in Task 2), all settings with a single augmentation perform poorly (Dice lower than 12.7%) including CycleGAN (18.0%) which cannot synthesize spatial difference, but BigAug could significantly boost the segmentation performance (Dice = 65.5%). This is due to the very different characteristics in the objects in the unseen domain with a mix of changes in the morphology and image quality & appearance. Thus, comprehensive transforms are required to represent such large changes.
Overall, for both MRI and ultrasound, the top 4 augmentations are contrast (Dice = 63.6%), brightness (63.6%), sharpening (62.9%), and 3D scaling (61.3%), each of which is comparable with CycleGAN (63.5%).
BigAug performance is ~10% worse compared to those with fully supervised methods, as they have advantages of training and testing on the same domain and more training data. This gap can be reduced by using a larger source dataset (as shown in later Section III-D4), in which case the BigAug performance is comparable to the supervised methods.
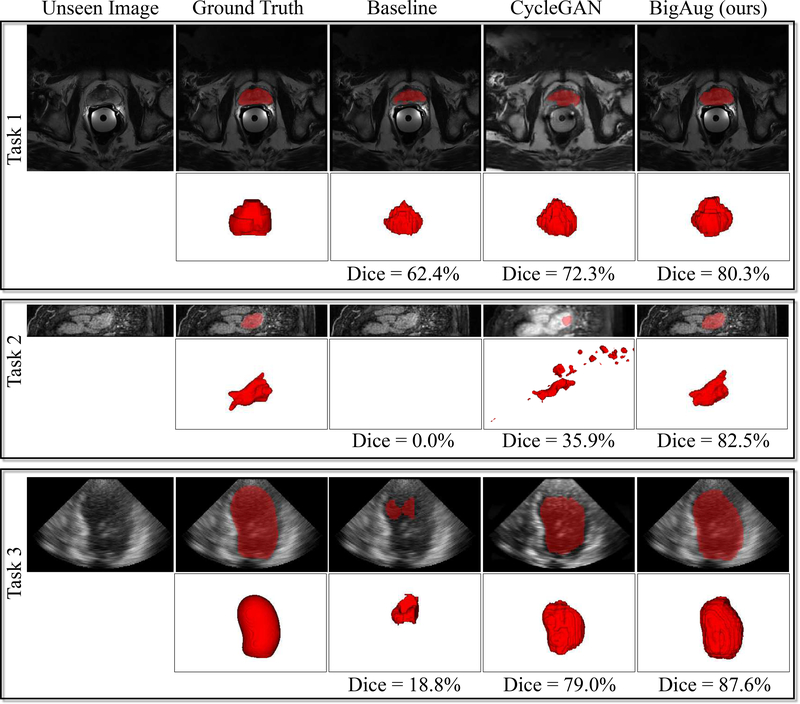
Examples of unseen domain segmentation produced by baseline model, CycleGAN-based domain adaptation, and our BigAug domain generalization are shown in Fig. 3. The baseline and BigAug models are trained only on individual source domains, while CycleGAN requires images from target/unseen domain to train an additional generative model.
Fig. 3.
Generalization to unseen domains for three different 3D medical image segmentation tasks. Baseline standard deep models have the low performances on unseen MRI and ultrasound images from different clinical centers, scanner vendors, etc. CycleGAN based domain adaptation method help improve the segmentation performances. BigAug training generates robust models which significantly improve the segmentation performances on unseen domains. Segmentation masks (red) overlayed on unseen or CycleGAN synthesized images are illustrated.
To further demonstrate the general effectiveness of BigAug, a paired cross-domain evaluation is performed among vendors, i.e., picking one for training and one for testing from CETUS-A, -B, and -C at each time. Results in Table III show that BigAug can generalize substantially better than baseline model regardless of training on which ultrasound vendors. However, the absolute accuracies on unseen vendors can be different depending on different source-unseen pairs.
TABLE III.
Results of Paired Cross-Domain Evaluation Among Vendors on the CETUS Heart Ultrasound Dataset. Results are Presented As Dice Scores of Baseline / Bigaug.
| Test╲Train | CETUS-A | CETUS-B | CETUS-C |
|---|---|---|---|
| CETUS-A | 85.8 / 92.1 | 51.7 / 84.9 | 39.2 / 81.3 |
| CETUS-B | 70.5 / 79.4 | 92.0/ 91.5 | 39.2 / 82.0 |
| CETUS-C | 55.8 / 73.6 | 54.8 / 74.7 | 92.8 / 93.2 |
2). BigAug vs. Shallower Stacked Transformations
Individual augmentation transforms may perform slightly better on some isolated cases (e.g., brightness augmentation for MM-WHS in Task 2 in Table II), but on average only BigAug consistently shows good generalization.
To investigate the optimal augmentation configuration for domain generalization, i.e., how many and which transformations should be used, we combine the four best performing transformations as “Top4” (i.e., sharpness, brightness, contrast, and scaling). “Top4” are competitive but “shallower” stacked transformations, which cover at least one aspect across image quality, appearance and spatial transform. The results are shown in Table II. Overall, the shallower competitor (top4) achieves a Dice of 74.9%, which is substantially higher than the baseline model (49.8%), but lower than BigAug (80.0%) which uses all transformations. This also applies to each individual task – except for one case, i.e., brightness augmentation for MM-WHS in Task 2. This could be explained by a more diverse data distribution, which helps better prevent overfitting while improving generalization. For Task 1 and Task 3, BigAug is better than the baseline and top4 for both source and unseen testing sets, which indicates the effect of BigAug on small sized data (e.g., 10 training data for CETUS).
Besides the significant improvement (30.2%) on unseen domains, BigAug could also slightly improve (2.5%) the performance on source domains, from 89.1% to 91.6% on average (note sometimes can be slightly worse, e.g., Task 2). This is an important benefit of BigAug, i.e., it retains performance on the source domain. Therefore, using all the presented transformations is recommended in general.
3). BigAug vs. Training From Scratch on Target Domain
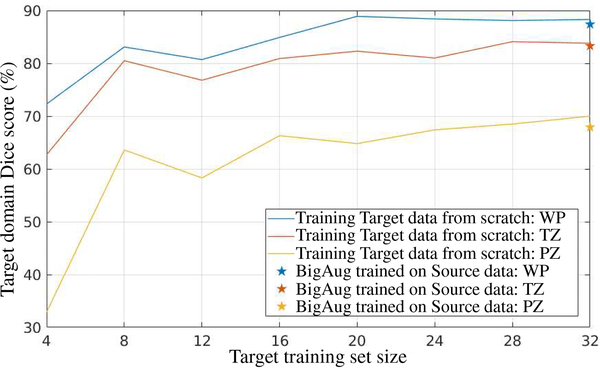
Another important finding is that models trained with BigAug on the source domain have comparable, or slightly lower performance than a model that is trained from scratch on target domain using the same amount of data.
In this section, we trained models to segment WP, TZ, and PZ in prostate MRI images. A BigAug model is trained on all 32 (train/validation: 25/7) volumes in MSD-P dataset and then applied to 66 volumes in ProstateX dataset. On the other hand, another set of BigAug models are trained from scratch on different training set sizes of 4, 8, 12, 16, 20, 24, 28, and 32 (train:validation: 3:1) volumes in ProstateX dataset and tested on the same 66 volumes in ProstateX dataset as well. Results are shown in Fig. 4. When trained on 32 volumes on source domain (MSD-P), BigAug achieves a Dice of 87.4%, 83.3%, and 67.9% on unseen domain (ProstateX). Such a performance is close to the model that is trained from scratch on 32 volumes (88.3%, 83.8%, and 70.0%) from the target domain (ProstateX).
Fig. 4.
Comparison between BigAug model trained on source domain and training from scratch on target domain for segmentation of whole prostate (WP), transition zone (TZ), and peripheral zone (PZ) in prostate MRI. Source domain: MSD-P; Target domain: ProstateX.
4). BigAug with Larger Data
Thus far, we have demonstrated that BigAug is able to significantly improve the 3D medical image segmentation performance on unseen domains when the training data size is relatively small, i.e., < 32 volumes. In this section, we experimentally demonstrate that BigAug is still able to boost the performances on the unseen domain when a larger dataset could be used for model training, and it helps achieve equivalent performances of state-of-the-art fully supervised methods.
We train models with and without BigAug on the ‘MultiCenter’ dataset (randomly split into 4:1 for training and validation) and apply the trained models to all other four prostate MRI datasets, which serve as four different unseen domains. During the training, there were a few differences compared to the implementation described in section III-C. Instead of normalizing intensities to [0, 1], we used zero mean and unit std. for the normalization due to wide variation in intensities of data from different vendors; we crop larger ROIs (128 × 128 × 32) for both training and inference; we train the model to segment the whole prostate directly – the average performance on unseen domains is actually the same as training a PZ & TZ segmentation model.
Results are shown in Table IV. The major findings are:
TABLE IV.
The Effect of Bigaug With Big Data (465 MRI Volumes From Multiple Medial Centers, MRI Vendors, and Protocols) for the Task of Whole Prostate Segmentation in MRI Volumes. Note That the State-of-the-Art Methods Marked With
| Source | Unseen | ||||||
|---|---|---|---|---|---|---|---|
| train | val | MSD-P | PROMISE | NCI-ISBI | ProstateX | Average | |
| Baseline | 95.6 | 89.9 | 87.8 | 82.9 | 88.8 | 90.6 | 87.5 |
| BigAug (ours) | 94.1 | 91.8 | 89.1 | 88.1 | 89.4 | 91.9 | 89.6 |
| State-of-the-art | - | - | - | 91.4* [39] | 89.3* [40] | 91.9* | 90.9 |
are Trained And Tested on the Same Domain or Inter-Observer Variability (91.9%). No Evaluation of the Whole Prostate Segmentation is Available in MSD Challenge (http://medicaldecathlon.com/results.html)
BigAug is still able to promote the segmentation accuracy (preventing overfitting the source domain) when a larger training dataset (covering large variations) is available. It achieves 91.8% and 89.6% Dice scores on source and unseen domains, respectively, which are 1.9% and 2.1% higher than the baseline model, respectively. It is particularly helpful on challenging cases, e.g., PROMISE12 dataset, where some MRI images are with very low image quality, brightness, and contrast.
A larger sized dataset is a key to the success of deep segmentation models. Compared to training on 32 volumes, training on 465 volumes increases 5.6% Dice score, from 84.0% to 89.6% on unseen domains on average.
BigAug trained with larger MultiCenter dataset produces competitive performance on unseen domains, only 1.3% lower than state-of-the-art methods on average. But this does not mean that our BigAug model is slightly inferior to state-of-the-art methods. On the contrary, note that the Dice scores of state-of-the-art methods are actually evaluated on their “source” domains or obtained by human experts, while these domains are unseen to our model trained using BigAug. Also note that since the ground truth of these public data have different annotation style (i.e., definition of boundary location between zones) compared with our training data, it is not surprising to observe a modest decrease in performance. Among the four different unseen domains, BigAug obtains a Dice score that is no worse than the two of the compared state-of-the-art methods.
Last, and perhaps most importantly, our BigAug model achieves a similar performance compared with intra-reader variability between two licensed radiologists (relative novice versus expert) on the unseen domain. Specifically, it achieves a Dice score of 91.9% on the unseen ProstateX dataset. In contrast, the Dice score between a novice versus expert radiologist annotations on ProstateX is also 91.9%.
IV. Conclusion
In this paper, we propose a deep stacked data augmentation (BigAug) training approach for generalizing deep-learning based medical image segmentation models to unseen domains. We exploit and extensively evaluate four important characteristics of BigAug on three different 3D segmentation tasks (prostate, left atrial, left ventricle) involving two medical imaging modalities (MRI and ultrasound). The experiments utilize eight public challenge datasets and establish a strong benchmark for the study of domain generalization in medical imaging. The empirical evaluation, performance analysis, and conclusive insights can be generalized to the design of really practical, highly robust, and competitive deep segmentation models for other medical imaging tasks.
Acknowledgments
Manuscript received XX YY, 2019. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This project was supposed in part by the Intramural Research Program of the NIH. Asterisk indicates corresponding authors.
This work was performed when L. Zhang was with Nvidia Corporation.
Footnotes
For example, a typical MR scan consisting of hundreds of 512×512 slices requires about 1 minute to perform all three spatial transform operations, then training 100 scans to converge (usually requiring 300 epochs in our work) needs about 500 hours.
We recognize different vendors by visually observing the CETUS image appearance, as the vendor information is not provided. Patients 1,2,8,9,13 are from vendor A, 3,4,12,14,15 vendor B, and 5,6,7,10,11 vendor C.
This normalization works better than normalizing to zero mean and unit std. in this experiment in a preliminary comparison.
Contributor Information
L. Zhang, PAII Inc., Bethesda, MD 20817, USA
X. Wang, Nvidia Corporation, Bethesda, MD 20814, USA.
D. Yang, Nvidia Corporation, Bethesda, MD 20814, USA.
H. Roth, Nvidia Corporation, Bethesda, MD 20814, USA.
A. Myronenko, Nvidia Corporation, Bethesda, MD 20814, USA.
D. Xu, Nvidia Corporation, Bethesda, MD 20814, USA.
Z. Xu, Nvidia Corporation, Bethesda, MD 20814, USA.
T. Sanford, National Institutes of Health Clinical Center, Bethesda, MD 20892, USA.
B. Turkbey, National Institutes of Health Clinical Center, Bethesda, MD 20892, USA.
B. J. Wood, National Institutes of Health Clinical Center, Bethesda, MD 20892, USA.
S. Harmon, Clinical Research Directorate, Frederick National Laboratory for Cancer Research sponsored by the National Cancer Institute, Bethesda, MD 20892.
References
- [1].Abràmoff MD, Lavin PT, Birch M, Shah N, and Folk JC, “Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices,” npj Digital Medicine, vol. 1, no. 1, p. 39, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hosny A, Parmar C, Quackenbush J, Schwartz LH, and Aerts HJ, “Artificial intelligence in radiology,” Nature Reviews Cancer, vol. 18, no. 8, p. 500, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].De Fauw J, Ledsam JR, Romera-Paredes B, Nikolov S, Tomasev N, Blackwell S, Askham H, Glorot X, ODonoghue B, Visentin D et al. , “Clinically applicable deep learning for diagnosis and referral in retinal disease,” Nature Medicine, vol. 24, no. 9, p. 1342, 2018. [DOI] [PubMed] [Google Scholar]
- [4].Yasaka K and Abe O, “Deep learning and artificial intelligence in radiology: Current applications and future directions,” PLoS Medicine, vol. 15, no. 11, p. e1002707, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ghafoorian M, Mehrtash A, Kapur T, Karssemeijer N, Marchiori E, Pesteie M, Guttmann CR, de Leeuw F-E, Tempany CM, van Ginneken B et al. , “Transfer learning for domain adaptation in MRI: Application in brain lesion segmentation,” in MICCAI. Springer, 2017, pp. 516–524. [Google Scholar]
- [6].Karani N, Chaitanya K, Baumgartner C, and Konukoglu E, “A lifelong learning approach to brain MR segmentation across scanners and protocols,” in MICCAI, 2018, pp. 476–484. [Google Scholar]
- [7].Tzeng E, Hoffman J, Saenko K, and Darrell T, “Adversarial discriminative domain adaptation,” in CVPR, vol. 1, no. 2, 2017, p. 4. [Google Scholar]
- [8].Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, and Bengio Y, “Generative adversarial nets,” in NIPS, 2014, pp. 2672–2680. [Google Scholar]
- [9].Zhu J-Y, Park T, Isola P, and Efros AA, “Unpaired image-to-image translation using cycle-consistent adversarial networks,” in ICCV, 2017, pp. 2223–2232. [Google Scholar]
- [10].Kamnitsas K, Baumgartner C, Ledig C, Newcombe V, Simpson J, Kane A, Menon D, Nori A, Criminisi A, Rueckert D et al. , “Unsupervised domain adaptation in brain lesion segmentation with adversarial networks,” in IPMI. Springer, 2017, pp. 597–609. [Google Scholar]
- [11].Degel MA, Navab N, and Albarqouni S, “Domain and geometry agnostic CNNs for left atrium segmentation in 3D ultrasound,” in MICCAI, 2018, pp. 630–637. [Google Scholar]
- [12].Ren J, Hacihaliloglu I, Singer EA, Foran DJ, and Qi X, “Adversarial domain adaptation for classification of prostate histopathology whole-slide images,” in MICCAI, 2018, pp. 201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang Y, Miao S, Mansi T, and Liao R, “Task driven generative modeling for unsupervised domain adaptation: Application to X-ray image segmentation,” in MICCAI, 2018. [Google Scholar]
- [14].Chen C, Dou Q, Chen H, and Heng P-A, “Semantic-aware generative adversarial nets for unsupervised domain adaptation in chest X-ray segmentation,” in International Workshop on Machine Learning in Medical Imaging Springer, 2018, pp. 143–151. [Google Scholar]
- [15].Yang X, Dou H, Li R, Wang X, Bian C, Li S, Ni D, and Heng P-A, “Generalizing deep models for ultrasound image segmentation,” in MICCAI. Springer, 2018, pp. 497–505. [Google Scholar]
- [16].Jog A and Fischl B, “Pulse sequence resilient fast brain segmentation,” in MICCAI. Springer, 2018, pp. 654–662. [Google Scholar]
- [17].Puonti O, Iglesias JE, and Van Leemput K, “Fast and sequence-adaptive whole-brain segmentation using parametric Bayesian modeling,” NeuroImage, vol. 143, pp. 235–249, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kouw WM, Ørting SN, Petersen J, Pedersen KS, and de Bruijne M, “A cross-center smoothness prior for variational Bayesian brain tissue segmentation,” in IPMI. Springer, 2019, pp. 360–371. [Google Scholar]
- [19].Brudfors M, Balbastre Y, and Ashburner J, “Nonlinear markov random fields learned via backpropagation,” in IPMI. Springer, 2019, pp. 805–817. [Google Scholar]
- [20].Romera E, Bergasa LM, Alvarez JM, and Trivedi M, “Train here, deploy there: Robust segmentation in unseen domains,” in 2018 IEEE Intelligent Vehicles Symposium (IV) IEEE, 2018, pp. 1828–1833. [Google Scholar]
- [21].Volpi R, Namkoong H, Sener O, Duchi J, Murino V, and Savarese S, “Generalizing to unseen domains via adversarial data augmentation,” in NeurIPS, 2018. [Google Scholar]
- [22].Truong T-D, Duong CN, Luu K, Tran M-T, and Do M, “Beyond domain adaptation: Unseen domain encapsulation via universal nonvolume preserving models,” arXiv preprint arXiv:1812.03407, 2018. [Google Scholar]
- [23].Zhang C, Bengio S, Hardt M, Recht B, and Vinyals O, “Understanding deep learning requires rethinking generalization,” in ICLR, 2017. [Google Scholar]
- [24].Jaderberg M, Simonyan K, Zisserman A, and Kavukcuoglu K, “Spatial transformer networks,” in NIPS, 2015, pp. 2017–2025. [Google Scholar]
- [25].Perez L and Wang J, “The effectiveness of data augmentation in image classification using deep learning,” arXiv preprint arXiv:1712.04621, 2017. [Google Scholar]
- [26].Cubuk ED, Zoph B, Mane D, Vasudevan V, and Le QV, “Autoaugment: Learning augmentation strategies from data,” in CVPR, 2019, pp. 113–123. [Google Scholar]
- [27].Bowles C, Chen L, Guerrero R, Bentley P, Gunn R, Hammers A, Dickie DA, Hernández MV, Wardlaw J, and Rueckert D, “GAN augmentation: Augmenting training data using generative adversarial networks,” arXiv preprint arXiv:181010863, 2018. [Google Scholar]
- [28].Yang D, Roth H, Xu Z, Milletari F, Zhang L, and Xu D, “Searching learning strategy with reinforcement learning for 3D medical image segmentation,” in MICCAI. Springer, 2019, pp. 3–11. [Google Scholar]
- [29].Liu S, Xu D, Zhou SK, Pauly O, Grbic S, Mertelmeier T, Wicklein J, Jerebko A, Cai W, and Comaniciu D, “3D anisotropic hybrid network: Transferring convolutional features from 2D images to 3D anisotropic volumes,” in MICCAI. Springer, 2018, pp. 851–858. [Google Scholar]
- [30].Isensee F, Kickingereder P, Wick W, Bendszus M, and Maier-Hein KH, “Brain tumor segmentation and radiomics survival prediction: Contribution to the brats 2017 challenge,” in International MICCAI Brainlesion Workshop Springer, 2017, pp. 287–297. [Google Scholar]
- [31].Kickingereder P, Isensee F, Tursunova I, Petersen J, Neuberger U, Bonekamp D, Brugnara G, Schell M, Kessler T, Foltyn M et al. , “Automated quantitative tumour response assessment of MRI in neurooncology with artificial neural networks: a multicentre, retrospective study,” The Lancet Oncology, vol. 20, no. 5, pp. 728–740, 2019. [DOI] [PubMed] [Google Scholar]
- [32].Myronenko A, “3D MRI brain tumor segmentation using autoencoder regularization,” in International MICCAI Brainlesion Workshop Springer, 2018, pp. 311–320. [Google Scholar]
- [33].Rister B, Yi D, Shivakumar K, Nobashi T, and Rubin DL, “CT organ segmentation using GPU data augmentation, unsupervised labels and IOU loss,” arXiv preprint arXiv:1811.11226, 2018. [Google Scholar]
- [34].Milletari F, Navab N, and Ahmadi S-A, “V-Net: Fully convolutional neural networks for volumetric medical image segmentation,” in International Conference on 3D Vision (3DV) IEEE, 2016, pp. 565–571. [Google Scholar]
- [35].Simpson AL, Antonelli M, Bakas S, Bilello M, Farahani K, van Ginneken B, Kopp-Schneider A, Landman BA, Litjens G, Menze B et al. , “A large annotated medical image dataset for the development and evaluation of segmentation algorithms,” arXiv preprint arXiv:1902.09063, 2019. [Google Scholar]
- [36].Litjens G, Toth R, van de Ven W, Hoeks C, Kerkstra S, van Ginneken B, Vincent G, Guillard G, Birbeck N, Zhang J et al. , “Evaluation of prostate segmentation algorithms for MRI: the PROMISE12 challenge,” Medical Image Analysis, vol. 18, no. 2, pp. 359–373, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Litjens G, Debats O, Barentsz J, Karssemeijer N, and Huisman H, “Computer-aided detection of prostate cancer in MRI,” TMI, vol. 33, no. 5, pp. 1083–1092, 2014. [DOI] [PubMed] [Google Scholar]
- [38].Zhuang X and Shen J, “Multi-scale patch and multi-modality atlases for whole heart segmentation of MRI,” Medical Image Analysis, vol. 31, pp. 77–87, 2016. [DOI] [PubMed] [Google Scholar]
- [39].Zhu Q, Du B, and Yan P, “Boundary-weighted domain adaptive neural network for prostate MR image segmentation,” TMI, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jia H, Xia Y, Song Y, Zhang D, Huang H, Zhang Y, and Cai W, “3D APA-Net: 3D Adversarial Pyramid Anisotropic Convolutional Network for Prostate Segmentation in MR Images,” TMI, 2019. [DOI] [PubMed] [Google Scholar]
- [41].Xiong Z, Fedorov VV, Fu X, Cheng E, Macleod R, and Zhao J, “Fully automatic left atrium segmentation from late gadolinium enhanced magnetic resonance imaging using a dual fully convolutional neural network,” TMI, vol. 38, no. 2, pp. 515–524, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]