Abstract
Moving towards species-relevant chemical safety assessments and away from animal testing requires access to reliable data to develop and build confidence in new approaches. The Integrated Chemical Environment (ICE) provides tools and curated data centered around chemical safety assessment. This article describes updates to ICE, including improved accessibility and interpretability of in vitro data via mechanistic target mapping and enhanced interactive tools for in vitro to in vivo extrapolation (IVIVE). Mapping of in vitro assay targets to toxicity endpoints of regulatory importance uses literature-based mode-of-action information and controlled terminology from existing knowledge organization systems to support data interoperability with external resources. The most recent ICE update includes Tox21 high-throughput screening data curated using analytical chemistry data and assay-specific parameters to eliminate potential artifacts or unreliable activity. Also included are physicochemical/ADME parameters for over 800,000 chemicals predicted by quantitative structure-activity relationship models. These parameters are used by the new ICE IVIVE tool in combination with the U.S. Environmental Protection Agency’s httk R package to estimate in vivo exposures corresponding to in vitro bioactivity concentrations from stored or user-defined assay data. These new ICE features allow users to explore the applications of an expanded data space and facilitate building confidence in non-animal approaches.
Keywords: Non-animal methods, IVIVE, regulatory toxicology
Introduction
As the momentum grows toward adoption of alternatives to animal use for chemical safety testing, there is a commensurate need for curated data to support method validation and establish scientific confidence in new approaches (Prior et al., 2019). Data describing the biological activity of a chemical are used for applications such as developing regulatory exposure limits, developing and evaluating new test methods, screening or prioritizing chemicals to identify potential adverse outcomes, and developing predictive models that can reduce the need for in vivo or in vitro testing. Finding appropriate and reliable data to support these efforts can be a challenge. In the absence of biological context, protocol information, and other metadata, the suitability of a data set for a particular purpose is not always clear. Even when the specific methods used to generate the data are named, it can be difficult for those with less familiarity with in vitro mechanistic approaches to determine how the methods may relate to a given in vivo adverse outcome.
Launched in 2017, the Integrated Chemical Environment (ICE) provides a free access point to data and tools for assessing and interpreting chemical bioactivity data (Bell et al., 2017). ICE was developed by the National Toxicology Program Interagency Center for the Evaluation of Alternative Toxicological Methods (NICEATM) to provide curated chemical bioactivity and property data for users evaluating alternatives to animal toxicity tests, such as method developers, chemical producers, and risk assessors. A primary goal of ICE is to increase access to reliable data to support the development, evaluation, and use of in vitro and in silico methods for characterizing the potential health impacts of chemicals.
In addition to data, users developing or evaluating new testing approaches also need easy-to-use, open-source computational tools. Recent updates to ICE focus on meeting the needs of users who are interested in exploring data and computational tools to relate in vitro bioactivity to in vivo testing results. New resources include an in vitro to in vivo extrapolation (IVIVE) tool that leverages the U.S. Environmental Protection Agency (EPA) high-throughput toxicokinetic (httk) R package (Pearce et al., 2017) and curated high-throughput screening (cHTS) data. Additionally, the mechanistic assays are annotated to toxicity endpoints using mode-of-action information according to terms from the NCI Metathesaurus (NCIm, https://ncim.nci.nih.gov/ncimbrowser/). Based on feedback from the scientific community, changes have also been made to ICE to improve the user experience, and additional help documentation has been added to make the resource easier to use.
This article will describe data available in ICE, data organization around regulatory toxicity endpoints, the curation of the ICE cHTS data, mapping of mechanistic assays to toxicity endpoints, and improvements to the ICE Search and IVIVE tools.
ICE data
Scope of ICE data
ICE was established to provide access to diverse types of relevant and reliable data for users developing and evaluating non-animal methods to assess chemical bioactivity. Potential ICE users include regulators, regulated industry, and test method developers. These users have diverse data needs, including in vivo assay data for regulatory endpoints and data from non-animal approaches that provide mechanistic information that may be relevant to the same regulatory endpoints. Users may also be looking for data about chemicals, including physicochemical properties and modeling parameters.
Table 1 summarizes data available in ICE. The organization of this table by regulatory endpoints reflects ICE’s emphasis on providing data in a manner useful for regulatory application. In vivo data in ICE include data from methods that adhere to, or closely resemble, accepted regulatory guidelines. Also included are data that do not have guideline-level documentation, including select in vivo data from public resources such as the National Library of Medicine’s Hazardous Substances Data Bank (HSDB, now part of PubChem, https://pubchem.ncbi.nlm.nih.gov/) (Fonger et al., 2014) and the Organisation for Economic Co-operation and Development’s eChemPortal (https://www.echemportal.org/echemportal/). ICE provides additional curation and review to increase the usefulness of these data, as described below. ICE also includes chemical lists compiled from public sources such as test method evaluations, test guidelines, and collaborative modeling projects. In vivo data in ICE are primarily rodent (rat), but include human, rabbit and guinea pig. In vitro data in ICE have been generated both from targeted assays designed to inform on a specific regulatory endpoint, such as estrogen receptor binding, and non-targeted assays that are designed to provide insight on mechanistic interactions with a biological system, such as oxidative stress.
Table 1:
Assay data in ICE
| Endpoint | Data Type (number of unique chemicals) | Example Assays/Models |
|---|---|---|
| Oral Systemic Toxicity | In vivo (10,335) | acute oral toxicity assay |
| In silico (838,911) | CATMoS1 | |
| Dermal Systemic Toxicity | In vivo (278) | acute dermal toxicity assay |
| Inhalation Systemic Toxicity | In vivo (225) | acute inhalation toxicity assay |
| Endocrine-Androgen | In vivo (140) | Hershberger (agonist/antagonist) |
| In vitro (164) | androgen receptor binding and transactivation (agonist/antagonist) | |
| In silico (838,911) | androgen receptor pathway model (agonist/antagonist), CoMPARA1 (agonist/antagonist) | |
| Endocrine-Estrogen | In vivo (118) | uterotrophic |
| In vitro (54) | estrogen receptor potency category, TG455 | |
| In silico (838,911) | estrogen receptor pathway model (agonist), CERAPP1 (agonist/antagonist) | |
| Eye Irritation/Corrosion | In vivo (183) | acute eye irritation |
| In vitro (117) | Vitrigel | |
| Skin Irritation/Corrosion | In vivo (120) | acute skin irritation/corrosion, 4h human patch test |
| In vitro (193) | reconstructed human epidermis irritation and corrosion, Vitrolife-Skin, Corrositex, TER, etc. | |
| Skin Sensitization | In vivo (572) | human potency assays, murine local lymph node assay |
| In vitro (121) | KeratinoSens, human cell line activation test, direct peptide reactivity assay | |
| cHTS2 | In vitro (9213) | high-throughput screening data from Tox21 and ToxCast |
| Physicochemical Properties (OPERA1 predictions) | In silico (838,911) | LogP, Henry’s Law, pKa, boiling point, etc. |
Predictions are generated using OPERA v2.5 (https://github.com/NIEHS/OPERA) from the predictive modeling projects CATMoS, the Collaborative Estrogen Receptor Activity Prediction Project (CERAPP; Mansouri et al., 2016), and the Collaborative Modeling Project for Androgen Receptor Activity (CoMPARA; Mansouri et al., 2020).
cHTS data currently in ICE are from EPA invitrodb v3.2 (accessed August 2019). Other endpoints listed in this table do not include cHTS data.
To be included in ICE, bioactivity data must:
Provide information on the biological/toxicological effect of a chemical on a whole organism, a cell-based system, or molecular pathways.
Define the chemical, experimental, or computational model setup, and result.
Adhere to community standards for the specific assay or computational model; for example, an assay described in an accepted test guideline must follow that guideline.
Be derived from an assay/computational model that is deemed appropriate for the endpoint it claims to inform on, based on validation studies, performance metrics, or other publicly available data.
Be available to be openly shared and distributed without restrictions.
There are some data in ICE that may not meet all the above criteria. For example, individual chemical data without an associated Chemical Abstracts Service Registry Number (CASRN) or where the chemical identity cannot be reasonably determined are not included in ICE. However, bioactivity data from chemical mixtures may be included, provided at least one component of the mixture has a CASRN, as is common in agrochemical formulations. Data sets developed as part of large-scale computational efforts at NICEATM often involve aggregation of data from public repositories as mentioned above. Verification that all inclusion criteria are met for these data sets is not always possible. However, before inclusion in ICE these datasets are curated to review and address formatting and data quality issues. One example of this curation of data from the public repositories is the acute oral toxicity data that were curated for use by the CATMoS (Collaborative Acute Toxicity Modeling Suite) modeling consortium (Kleinstreuer et al., 2018b).
Information on a chemical’s physical and pharmacological properties is also needed to establish a chemical’s suitability for testing in a specific assay or to use the chemical in a predictive model. For example, information on protein binding is a critical variable in describing how a chemical is processed by a biological system used in IVIVE. To address such needs, ICE contains in silico predictions for physicochemical properties, as well as absorption, distribution, metabolism, and excretion (ADME) properties (Table 1). These are generated using the Open Structure-activity/property Relationship App (OPERA, https://github.com/NIEHS/OPERA )(Mansouri et al., 2018), a free and open-source/open-data suite of quantitative structure-activity relationship (QSAR) models that provides predictions of physicochemical properties, environmental fate parameters, and regulatory toxicology endpoints. Availability of OPERA predictions not only provides users with access to predicted property values on a large number of structures but has also supported further development of ICE tools. For example, the OPERA predictions of ADME properties, such as partition coefficients and hepatic clearance rates, are key input parameters for the IVIVE tool described below.
ICE Chemical Quick Lists
ICE provides curated chemical lists that can be used with any of its tools. ICE Chemical Quick Lists are useful for evaluating specific types of toxicities represented in the ICE database (Table 2). ICE has two types of quick lists: reference chemical lists and non-reference chemical lists. Reference chemical lists are comprised of chemicals that cause a specified, well-characterized biological effect and therefore can be used to assess the performance of an assay designed to measure that effect. Non-reference chemical lists have less restrictive criteria for inclusion than reference chemical lists, and may include chemicals with uncharacterized or ambiguous biological effects.
Table 2:
ICE Chemical Quick Lists
| Chemical Quick Lists | Description1 | Reference Chemical List |
|---|---|---|
| AR In Vitro Agonist | 37 chemicals with androgen receptor agonist activity characterized in in vitro assays (Kleinstreuer et al., 2016a) | Yes |
| AR In Vitro Antagonist | 28 chemicals with androgen receptor antagonist activity characterized in in vitro assays (Kleinstreuer et al., 2016a) | Yes |
| AR In Vivo Agonists | 26 chemicals with androgen receptor agonist activity characterized in in vivo assays (Browne et al., 2018) | No |
| AR In Vivo Antagonists | 23 chemicals with androgen receptor antagonist activity characterized in in vivo assays (Browne et al., 2018) | No |
| EPA IRIS Carcinogenicity Classifications | 225 chemicals classified for weight of evidence of carcinogenicity according to the EPA Guidelines for Carcinogen Risk Assessment (https://www.epa.gov/risk/guidelines-carcinogen-risk-assessment) | No |
| ER In Vitro Agonist | 40 chemicals with estrogen receptor agonist activity characterized in vitro assays (Browne et al., 2015). 118 chemicals with estrogenic activity characterized in guideline-like rodent uterotrophic assays (Kleinstreuer et al., 2016b) | Yes |
| ER In Vivo Agonist | 43 chemicals with estrogenic activity characterized in guideline-like rodent uterotrophic assays (Kleinstreuer et al., 2016b) | Yes |
| Eye IrritationCorrosion | 123 chemicals recommended by ICCVAM for evaluating in vitro assays for eye irritation (https://ntp.niehs.nih.gov/go/40177) | Yes |
| IARC Classifications | 864 chemicals classified in IARC monographs evaluating carcinogenicity to humans. (https://monographs.iarc.fr/monographs-available/) | No |
| NTP Cancer Bioassay Chemicals |
542 chemicals from the NTP Technical Reports characterizing the toxicologic potential of agents in test animals. (https://ntp.niehs.nih.gov/publications/reports/index.html) |
No |
| RoC Classifications |
229 chemicals classified according to RoC listing criteria for human carcinogens. (https://ntp.niehs.nih.gov/whatwestudy/assessments/cancer/index.html) | No |
| Skin Corrosion | 32 chemicals recommended for assessing proficiency of conducting in vitro assays for skin corrosion (https://ntp.niehs.nih.gov/go/40193) | Yes |
| Steroidogenesis - Androgen | 36 chemicals with androgen synthesis effects characterized in the H295R steroidogenesis assay. (Pinto et al., 2018) | No |
| Steroidogenesis - Estrogen | 35 chemicals with estrogen synthesis effects characterized in the H295R steroidogenesis assay. (Pinto et al., 2018) | No |
| Thyroid | 34 chemicals with effects on thyroid activity characterized in up to three in vivo assays. (Wegner et al., 2016) | No |
| Tox21 | 9000+ chemicals tested in Tox21 Program. (https://tripod.nih.gov/tox21/assays/) | No |
Numbers and references are current as of February 2020
Users can download each of these lists with supporting bioactivity information from ICE on the Quick Lists page (https://ice.ntp.niehs.nih.gov/ChemicalQuickLists). These lists can be useful for evaluating data in ICE or for use in development of new approaches. Effort has been taken to have a reference chemical list available for all toxicity endpoints for which there is a non-animal method available in ICE.
On the other hand, the ICE non-reference chemical lists were developed to provide a list of chemicals to aid in exploratory queries relevant to a specific toxicity for which ICE has data but a “reference” list has not been established or is not available (for example Tox21 chemical list or the various cancer quick lists). Non-reference chemical lists are often compiled from review studies, which often do not meet the criteria allowing data extracted from them to serve as reference lists. The limitations of these sources may include substantial deviations from established test guidelines or results reported that did not signal a clear activity. Other non-reference lists represent useful lists of chemicals that do not relate to a specific bioactivity, such as the Tox21 chemical inventory quick list.
Knowledge organization of ICE data
A major goal of the ICE update was to make ICE data more accessible to those with limited experience with the assays and data types represented in the database. Knowledge organization systems (KOS) such as controlled vocabularies, thesauri, and ontologies can improve the utility of data in accordance with FAIR principles of findability, accessibility, interoperability, and reproducibility (Harrow et al., 2019; Wilkinson et al., 2016). ICE data are annotated using KOS to organize the assays based on regulatory toxicology endpoints of interest and mechanistic target information. Use of the KOS facilitates expansion of the toxicity endpoint parent terms beyond typical in vivo assays to include in vitro assays with relevant biological targets. For example, Fig. 1 illustrates the mapping of an assay measuring the phosphorylation of a histone protein to the toxicity endpoint using controlled terminology and parentchild relationships. This organization allows users to easily identify and select assays associated with the toxicity endpoint of interest, without requiring deep familiarity with each technology or data source. The current ICE update therefore increases visibility of non-animal methods that can inform on regulatory endpoints. For example, the suite of in vitro assays probing the adverse outcome pathway for skin sensitization (OECD, 2014) has been included, as well as the ToxCast/Tox21 estrogen and androgen receptor pathway models (Judson et al., 2015; Kleinstreuer et al., 2016a).
Fig. 1. Example Mapping to KOS.
High-throughput screening assays from the Tox21 program are mechanistic in nature and do not intuitively link to toxicity endpoints. Annotation from the mechanistic targets of the Tox21 assay (Histone modification) to the parent toxicity endpoint (Cancer) is via the mode of action (KCC2). Identifiers refer to the NCImetathesaurus codes.
Many in vitro and in silico methods are mechanistic in nature and provide insight on biochemical activity, molecular signaling, or metabolite levels that are conceptually distant from in vivo regulatory endpoints like acute lethality. However, these mechanistic assays can still provide useful information relevant to in vivo endpoints, such as assays probing key events in an adverse outcome pathway framework (Ankley et al., 2010; Villeneuve et al., 2014a, 2014b). Mechanistic targets are used in ICE to help integrate in vitro assays with the regulatory endpoint-based framework of ICE data as shown in Fig. 1. Currently, over 40 mechanistic targets have been integrated into ICE (Supplemental File 1) defined largely by modes of action cited in peer-reviewed literature that are mapped to or associated with toxicity endpoints.
Curation of mechanistic targets to toxicity endpoint
Mapping of the assay annotations to the mechanistic targets is based on review of the assays by domain experts and those familiar with the technical aspects of the assays. Assays are described using available information from the assay and notations from the ICE curation team. A NICEATM team member with expertise in a given toxicity endpoint defines a set of modes of action based on the current literature. Assays in ICE are then mapped to the defined modes of action. Mechanistic target terms like “vascularization” and “epigenetic process” along with the mode of action are annotated to the assay. A data scientist reviews the new mechanistic targets and the mechanistic target/mode-of-action relationship alongside the existing mappings in ICE to identify any issues with internal logic, and selects the appropriate controlled vocabulary terms. ICE relies on terms available in the National Cancer Institute Metathesaurus (NCIm, https://ncim.nci.nih.gov/ncimbrowser/), as they cover multiple KOS relevant to the toxicology covered in ICE. The data scientist brings forward proposed mechanistic target-controlled vocabulary mapping and any parent/child relationships to a group of toxicologists and individuals familiar with the in vitro assays for discussion. Once terms and relationships have been agreed upon by the group, the ICE KOS terms updated appropriately.
Current mappings are available in Supplemental Table 1. Mapping of the ICE mechanistic target terms to terms in the NCIm creates a connection to widely used and established terminologies with controlled identifiers, allowing the annotations found in ICE to be accessed and the ICE data linked to other resources and terms. At this time, not all assays have been mapped to a mechanistic target. The mappings are actively maintained to keep up with current literature and to promote greater interoperability with related data resources, such as those from the EPA’s Chemistry Dashboard (https://comptox.epa.gov/dashboard; Williams et al., 2017), and we intend to add more terms in future ICE releases.
Data processing, curation, and mapping
One feature of ICE that sets it apart from resources such as HSDB or EPA’s Chemistry Dashboard is the ability to query and merge bioactivity data and chemical property data for a variety of endpoints (Table 1). Another important ICE feature is NICEATM’s data curation, which improves the interoperability of the data and facilitates analyses, setting ICE apart from other databases that simply serve as repositories for data. ICE data have standardized units, common identifiers, and values that are available in a tabular format, so users do not have to perform separate queries or processing for assay values. Curation is done by both subject matter experts and data scientists to ensure that values are technically accurate and retain relevant metadata such as sex, timing of the measurement, and cell viability. Such metadata are important for putting the data as originally published into the appropriate biological context.
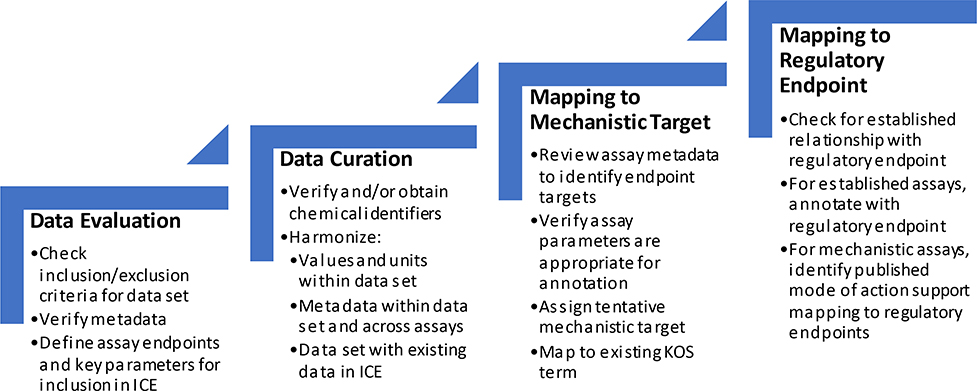
Fig. 2 presents the steps in the ICE data curation process. This process verifies that:
Fig. 2.
ICE data go through a four-step curation process. This process involves technical domain experts familiar with the biology and regulatory applications of the assay. Data scientists also review the data to ensure harmonization across ICE data sets and with established identifiers to promote the interoperability of ICE data.
Chemical identifiers are consistent and are in a standard format.
The intended assay targets and the endpoints needed to describe chemical activity are clearly defined.
Assay response values, units, and other relevant metadata are harmonized within and across assays to promote data interoperability.
These steps help to ensure that ICE queries capture all related data and that queries from other databases using shared identifiers can be integrated into in-house workflows and analysis pipelines without the need for extensive processing on the user side.
Once a data set has been curated for the items outlined above (Fig.2), subject matter experts work with data scientists to annotate the assays and facilitate mapping to regulatory toxicity endpoints and appropriate KOS. NCIm incorporates controlled vocabularies and ontologies such as the Gene Ontology (Ashburner et al., 2000), Unified Medical Language System (Bodenreider, 2004), and National Cancer Institute Thesaurus (Sioutos et al., 2007), among others. Use of NCIm allows ICE to use the NCIm identifier to map the ICE term to multiple relevant KOS in environmental health sciences for describing health effects and biological processes (Fig. 1, CUI IDs) These mappings are based on technical details of the assay protocol, current and past U.S. and international regulatory guidelines, and available peer-reviewed literature. The mapping process includes review by data scientists, curators, and domain experts familiar with the relevant biology and regulatory statutes. In general, consensus is reached after discussion to review the annotations for mechanistic targets. If consensus cannot be reached, the mapping to the KOS is put on hold for the assay pending additional information. Currently, mapping of ICE data to KOS terms is limited to regulatory endpoints and mechanistic targets, with the initial efforts focused on cHTS assays. Ongoing activities include building out the annotation of the assay and model details with information about the testing platform, such as adherence to test guidelines and the type of model. Adding these metadata, mapped to the appropriate KOS, for the experimental details will improve the search and filtering capabilities within ICE, as well as providing users with additional information with which to evaluate the bioactivity data. Refining the annotation to the toxicity endpoint and the terms for the mechanistic assays will be a continuous process, as will expanding coverage to cover tasks such as identifying orthologous assays.
Curation of cHTS data
The curated high throughput screening (cHTS) data in ICE are compiled from public data released by the U.S. government interagency Tox21collaboration (Tice et al., 2013) (https://tripod.nih.gov/tox21/assays/) and EPA’s ToxCast high-throughput screening (HTS) program (Dix et al., 2007; Kavlock et al., 2012)(https://doi.org/10.23645/epacomptox.6062623). These HTS programs produce data on thousands of compounds for thousands of assays spanning multiple molecular targets and testing platforms. Therefore, one processing approach cannot be applied to all ToxCast and Tox21 assay data to be included in ICE. A curation workflow was applied to these HTS datasets for integration into ICE resulting in the cHTS inventory. Curation efforts sought to maximize the confidence in bioactivity calls and flag any chemical activity with high uncertainty. One step in this process involves removal of chemicals and/or assays in cases where there is reasonable certainty that the reported activity concentrations do not accurately reflect the bioactivity of the chemical within the expected variability of the assay, based on considerations such as erroneous concentration-response patterns and activity outside the tested concentration range (Richard et al., 2016). Another step involves removal of data for chemicals which have not passed purity and concentration confirmation.
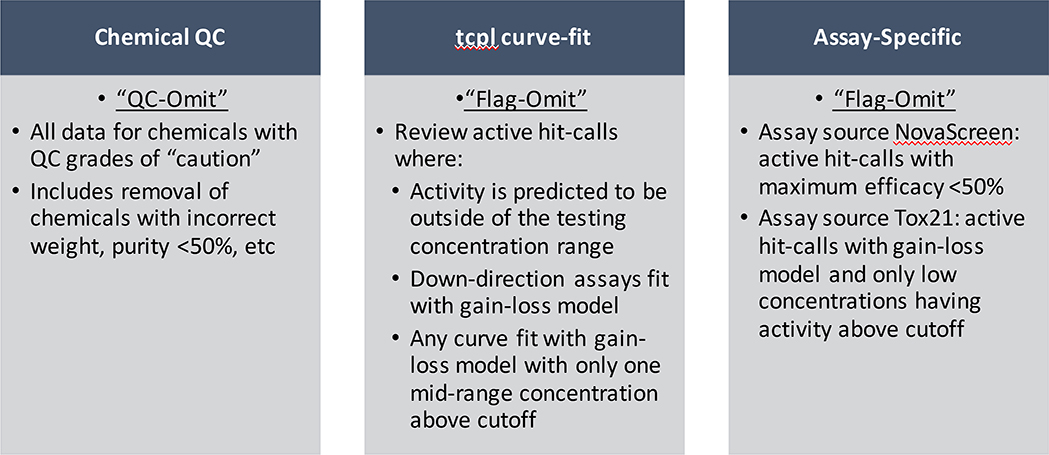
Integration of Tox21 and ToxCast data sets to form the cHTS data set in ICE begins with retrieval of output from a custom analysis algorithm, the ToxCast Pipeline (tcpl), an R package developed by EPA specifically for concentration-response analysis for HTS data (Filer et al., 2016). The EPA tcpl pipeline provides a representative curve for each assay-chemical pair, so technical and biological replicate information is not included in the ICE database, and would be challenging to integrate with other assay information contained in ICE. Work is ongoing to create linkages to other resources that house the technical replicate information for users who want that level of detail (Williams et al., 2017). Following retrieval of the representative curve, NICEATM curation processing then applies chemical- and assay-focused filtering steps as summarized in Fig. 3. Analytical chemistry quality control (QC) methods have been, or are being, developed for all unique chemicals in the Tox21 library, providing information on the purity and identity of each sample (Richard et al., 2016). Results of these analyses are used in the ICE cHTS curation process ensure that only chemicals with confirmed purity and concentration are reported. Any chemicals failing purity or concentration QC are flagged in ICE and the bioactivity data are not reported. (Supplemental File 2). NICEATM also uses information provided from tcpl outputs about the curve-fitting and specific assay platforms to develop custom flags. For example, custom criteria developed for ICE curation include flagging bioactivity calls where the measure of activity (AC50) is above the tested concentration range and assay-specific filtering (https://ice.ntp.niehs.nih.gov/DATASETDESCRIPTION). These chemical- and assay-based flags increase the confidence in the cHTS bioactivity calls for ICE users. Thus, cHTS endpoints can receive one of four calls: Active, Inactive, QC-omit (for cases of failed chemical QC) and Flag-omit (for cases where active calls were omitted due to filtering based on custom NICEATM-defined flags). A numeric activity value (i.e., AC50, the concentration at 50% the maximal response) is only given for “active” calls. ICE Search queries that include cHTS data will return the curated outputs in both the results table and download files.
Fig. 3. Curation criteria for cHTS data in ICE.
Three types of metadata are considered when curating the Tox21 and ToxCast HTS data for integration into ICE: chemical-based criteria from analytical chemistry quality control analysis, curve-fit information based on the tcpl algorithm curve-fitting output for all active hit-calls, and assay-specific criteria set by domain experts familiar with the assays.
Ease of access and interpretation of the diverse cHTS data in ICE are facilitated by mapping to mechanistic target groups and modes of action. For each mechanistic target or mode of action selected, the user is provided with a summary of the number of times a chemical is active, inactive, etc., based on all the HTS assays that are annotated to that mechanistic target or mode of action. Detailed information on the activity concentrations is available by downloading the cHTS data from the ICE Data Sets page (https://ice.ntp.niehs.nih.gov/DATASETDESCRIPTION).
Annotation of HTS assays to mechanistic targets
As described above (“Scope of ICE data”), ICE includes data from in vitro assays that are designed to provide mechanistic insight. While other efforts have been made to annotate the Tox21 and ToxCast data to gene or biochemical process (Richard et al., 2016), or KOS based on the assay platform (Cooper and Schürer, 2019), or processes specific to cancer (Chiu et al., 2018), annotation of assays specifically to link them to regulated toxicity endpoints has been lacking. ICE maps these in vitro assays to toxicity endpoints of regulatory interest through mechanistic targets.
ICE uses the EPA’s invitroDB database v3.2 (https://doi.org/10.23645/epacomptox.6062623, accessed October 2019) (EPA, 2019; Filer et al., 2016) to map cHTS assays to toxicity endpoints. Assays are annotated with fields such as “intended_target_family”, “intended_target_official_gene_symbol”, and “biological_process_target”. These annotations are applied for ICE integration in a manner that appropriately considers assay readouts unrelated to the main target of the assay. For example, background signal endpoints for internal controls are filtered out, and viability controls measuring cytotoxicity are annotated to cell viability or are filtered out as deemed appropriate by domain experts. Additional mapping of cHTS assays to mechanistic targets is also conducted and is based on NCIm terms as described above in “Curation of mechanistic targets to toxicity endpoint” relying heavily on expert-defined mode of action groups developed from peer-reviewed literature (Supplemental Table 1).
Using this approach gives users very clear statements of the association between the mechanistic target and the toxicity endpoint. For example, ICE could state that assays “may inform on” some mode of action “leading to” a toxicity endpoint “through a mechanistic target”. Specifically:
The assay: “APR_HepG2_MitoticArrest_1h_up”
May inform on: “KCC10: Cell Proliferation/Death/Energetics
Leading to: “Cancer”
Through the mechanistic target: “Cell Cycle”
That is a child of: “Cell Proliferation (CUI:C0596290)”
It is envisioned that ongoing and more specific annotation may allow assays to be associated with multiple modes of action. Currently, the endpoints in ICE for which mechanistic targets have mapped are limited to those that have peer-reviewed literature defining modes of action: acute lethality (Hamm et al., 2017; National Academies of Sciences, Engineering, and Medicine, 2015; Prieto et al., 2019), developmental and reproductive toxicity (van Gelder et al., 2010) and cancer based on the ten key characteristics of carcinogens (Smith et al., 2016). Mapping of in vitro assays to other toxicity endpoints ongoing, including skin sensitization, for which data described in the defined approach for skin sensitization testing (Casati et al., 2018; Kleinstreuer et al., 2018a) are available in ICE. The goal is to continue expanding and improving the mechanistic target mappings and assay annotation to toxicity endpoints, which aid in ease of access and computational modeling efforts.
ICE tools
ICE tools were developed in response to specific requests from NICEATM stakeholders. The ICE user interface enables users with little prior knowledge of chemical bioactivity testing to interact with and explore a wide variety of chemical testing data. The ICE Search tool allows users to integrate data from different models and testing systems to provide an overall view of a chemical’s activity for regulatory endpoints of interest. ICE also has tools that allows users to conduct simple IVIVE analyses, explore and compare the characteristics of one or two chemical lists.
The ICE Search and IVIVE tools have recently undergone major revisions in ICE 3.0 to increase their usability and are described in detail in the following sections. The ICE Chemical Characterization tool allows users to characterize and compare the physicochemical properties of one or two lists of chemicals to each other. Users can also compare specified chemicals to the property range of over 800,000 chemicals for which experimental or predicted data are available in the ICE database. This characterization can be useful for identifying features that might be driving assay performance across different groups of chemicals, investigating appropriate test substances based on technical limitations of the test system (such as highly volatile compounds), or checking for a bias in the range of properties represented in the list.
Search Tool
The ICE Search tool allows users to query and combine assay bioactivity data for over 10,000 chemicals and chemical property predictions for over 800,000 chemicals. The user builds an ICE Search query by specifying chemicals, assays, or both. Chemicals in a query can be provided using a combination of Chemical Quick Lists and user-specified CASRNs. ICE assays are organized in categories around toxicity endpoints of regulatory interest, with separate categories allowing queries specific to cHTS and physicochemical property data. Users can select multiple assays and endpoints across all categories. The organization of assays around toxicity endpoints of regulatory interest makes it easy to include data from both animal and non-animal tests relevant to that endpoint. The user therefore does not need to know what specific assay or test is relevant to. For example, if the user wishes to explore endocrine disruption, adding all endocrine assays to a query will by default include the relevant in vivo, in vitro, and in silico data, with the user having the option to include or exclude any category of assay or individual data source. Detailed information on the different data endpoints available from each assay can be found on the Data Sets page on the ICE website (https://ice.ntp.niehs.nih.gov/DATASETDESCRIPTION).
One of the challenges associated with identifying relevant data for a test chemical is that different salts of the chemical are often assayed. These salts may have the same core structure but different identifiers. In QSAR modeling, flat, desalted structures are used for modeling; that is, the chemical structure used for predictive purposes is often the two-dimensional representation of a chemical without the salt ion. ICE provides the user with the option to use this simplified representation of the chemical structure and add any chemicals with the same QSAR structure (QSAR Match) to their query, thus returning available information on the different salt forms. Query results indicate what “QSAR Match” chemicals were added to the dataset.
IVIVE Tool
IVIVE relates in vitro assay activity concentrations to in vivo exposures. This can support development of hypotheses on the exposure ranges that exert in vivo effects relevant to the bioactivity measured by in vitro assays (Bell et al., 2018). The ICE IVIVE tool brings together annotated in vitro assay data, QSAR model predictions of chemical ADME properties, and pharmacokinetic (PK) or physiologically based pharmacokinetic (PBPK) models of varied complexity to predict a daily equivalent administered dose (EAD) that would result in a plasma concentration equal to the activity concentration of any given in vitro assay. Using the ICE cHTS data, users can generate EAD predictions for over 9,000 chemicals. In addition, users can upload their own in vitro assay data to use in addition to or instead of cHTS data available in ICE. The PK/PBPK models of IVIVE tool are parameterized using values from the ICE database and not through the database included in the httk package. This currently limits the number of chemicals that can be run through the ICE browser tool, so inputs are limited to the 800,000 chemicals with predictions. For predictions of chemicals not currently these properties available in the ICE database, users are encouraged to use code available in GitHub for both the ICE IVIVE tool (https://github.com/NIEHS/ICE2.2_IVIVEpipeline) and the OPERA QSAR models (https://github.com/NIEHS/OPERA).
The ICE IVIVE tool uses PK and PBPK models (Table 3) to first estimate the plasma concentration of the chemical that would result from an in vivo exposure of 1 mg/kg at the specified dosing intervals. A linear extrapolation is then used to estimate the daily EADs resulting from a plasma concentration equivalent to the activity concentration of the in vitro assay. Currently, the ICE IVIVE tool provides three models with varied complexity and exposure routes: a one-compartment PK model, a three-compartment PK model, and a multi-compartment PBPK model for oral and intravenous routes (Table 3). Both the three-compartment PK and multi-compartment PBPK models come from the EPA httk package (Pearce et al., 2017), while the one-compartment model uses custom code based on published equations (Casey et al., 2018; Chang et al., 2014). The parameter values for executing the PK/PBPK models are either provided directly through the ICE database or calculated using embedded functions in the httk package using physicochemical property values from the ICE database.
Table 3.
PK/PBPK Models provided: IVIVE models in ICE IVIVE Tool
| Model Name | Model Source | Plasma levels calculateda | Routes | Parameters |
|---|---|---|---|---|
| 1C | NICEATM | Steady state | NA | NA |
| Solve_3Comp | httk v1.10.1 | Maximal | Oral, IV | Exposure intervals, Simulation length |
| Solve_pbtk | httk v1.10.1 | Maximal | Oral, IV | Exposure intervals, Simulation length |
The one-compartment (1C) model calculates the steady-state plasma concentration at the 50th and 90th percentile chemical concentration using physiological and pharmacokinetic parameters of a Monte Carlo simulated population. Other models calculate the maximal concentration at the 50th percentile based on average physiological and pharmacokinetic parameter values.
The output of the IVIVE tool includes a table summarizing the model inputs as well as the predictions of plasma concentration of the chemical at 1 mg/kg at the specified dosing intervals and EAD values. Interactive tabular and graphic representations of the EAD values allows users to explore results in detail and apply filters to examine specific chemicals or assays. Additionally, users can choose to overlay a selection of in vivo data from the ICE database over the EAD plots, allowing a comparison of experimental results and predicted values. Efforts are currently underway to use the KOS mapping of in vitro assay to better guide users on selecting what in vivo data for comparison, with predicted results, might benefit their search. An expanded availability of in vivo data is also in development through links to other resources such as CEBS (https://manticore.niehs.nih.gov/cebssearch) and the Chemistry Dashboard (https://comptox.epa.gov/dashboard).
Discussion
The updates to ICE since its launch in 2017 improve the user experience and are intended to support FAIR principles through additional data curation and mapping to KOS. NICEATM’s interactions with the chemical safety assessment and regulatory communities have helped us to develop ICE to prioritize commonly requested resources. These resources include the curated data sets and chemical lists focused on specific bioactivity or regulatory endpoints, and the ability to put the in vitro assays into an in vivo context using IVIVE.
Supplementary Material
Highlights:
ICE provides curated data and tools to support chemical safety testing
In vitro data are mapped to mechanistic targets and regulatory endpoints
Tox21 data are curated using analytical chemistry and assay-specific information
In silico predictions are available for physchem and ADME properties
IVIVE tool uses ICE or user data to estimate in vivo exposure levels
Acknowledgements
We would like to acknowledge Charles Schmitt and Nisha Sipes for their helpful feedback and discussions as well as David Fargo and Frank Day for maintenance and upkeep of the ICE servers. This project was funded in whole or in part with federal funds from the NIEHS, NIH under Contract No. HHSN273201500010C.
NICEATM supports the Interagency Coordinating Committee on the Validation of Alternative Methods (ICCVAM), a committee of 16 U.S. federal agencies that require, use, generate or disseminate toxicological and safety testing information. The three strategic goals of NICEATM, in support of ICCVAM, are to connect end users with developers of new approach methodologies, to foster the use of and establish confidence in these methods, and encourage adoption of these methods by federal agencies and regulated industries (ICCVAM, 2018). The curation discussed herein directly impacts all three of these goals by removing the common roadblocks of lack of familiarity with mechanistic testing platforms and access to reliable reference data, thereby making data from new approach methodologies easier to navigate and relate to regulatory endpoints for users of all backgrounds. Adding in the annotation and mapping to the KOS in conjunction with the online IVIVE tool address some commonly expressed and previously unmet needs of stakeholders. Continued development of ICE will provide more end user support, building out additional features and availability of online tools, and expanding datasets. This will aid in making computational toxicology methods more broadly accessible, which we hope will increase users’ comfort and understanding of new approaches to chemical bioactivity testing.
Abbreviations:
- ADME
absorption, distribution, metabolism, and excretion
- CASRN
Chemical Abstracts Service Registry Number
- cHTS
curated high-throughput screening
- EAD
equivalent administered dose
- EPA
U.S. Environmental Protection Agency
- HSDB
Hazardous Substances Data Bank
- HTS
high-throughput screening
- httk
high-throughput toxicokinetic
- ICCVAM
Interagency Coordinating Committee on the Validation of Alternative Methods
- ICE
Integrated Chemical Environment
- IVIVE
in vitro to in vivo extrapolation
- KOS
knowledge organization systems
- NCIm
NCI Metathesaurus
- NICEATM
National Toxicology Program Interagency Center for the Evaluation of Alternative Toxicological Methods
- OPERA
Open Structure-activity/property Relationship App
- PBPK
physiologically based pharmacokinetic
- PK
pharamacokinetic
- QSAR
quantitative structure-activity relationship
- QC
quality control
- tcpl
ToxCast Pipeline
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ankley GT, Bennett RS, Erickson RJ, Hoff DJ, Hornung MW, Johnson RD, Mount DR, Nichols JW, Russom CL, Schmieder PK, Serrrano JA, Tietge JE, Villeneuve DL, 2010. Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem. 29, 730–741. 10.1002/etc.34 [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, 2000. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SM, Chang X, Wambaugh JF, Allen DG, Bartels M, Brouwer KLR, Casey WM, Choksi N, Ferguson SS, Fraczkiewicz G, Jarabek AM, Ke A, Lumen A, Lynn SG, Paini A, Price PS, Ring C, Simon TW, Sipes NS, Sprankle CS, Strickland J, Troutman J, Wetmore BA, Kleinstreuer NC, 2018. In vitro to in vivo extrapolation for high throughput prioritization and decision making. Toxicol. In Vitro 47, 213–227. 10.1016/j.tiv.2017.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SM, Phillips J, Sedykh A, Tandon A, Sprankle C, Morefield SQ, Shapiro A, Allen D, Shah R, Maull EA, Casey WM, Kleinstreuer NC, 2017. An Integrated Chemical Environment to Support 21st-Century Toxicology. Environ. Health Perspect. 125, 054501 10.1289/EHP1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenreider O, 2004. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 32, D267–D270. 10.1093/nar/gkh061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne P, Judson RS, Casey WM, Kleinstreuer NC, Thomas RS, 2015. Screening Chemicals for Estrogen Receptor Bioactivity Using a Computational Model. Environ. Sci. Technol. 10.1021/acs.est.5b02641 [DOI] [PubMed] [Google Scholar]
- Browne P, Kleinstreuer NC, Ceger P, Deisenroth C, Baker N, Markey K, Thomas RS, Judson RJ, Casey W, 2018. Development of a curated Hershberger database. Reprod. Toxicol. 81, 259–271. 10.1016/j.reprotox.2018.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati S, Aschberger K, Barroso J, Casey W, Delgado I, Kim TS, Kleinstreuer N, Kojima H, Lee JK, Lowit A, Park HK, Régimbald-Krnel MJ, Strickland J, Whelan M, Yang Y, Zuang V, 2018. Standardisation of defined approaches for skin sensitisation testing to support regulatory use and international adoption: position of the International Cooperation on Alternative Test Methods. Arch Toxicol 92, 611–617. 10.1007/s00204-017-2097-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey WM, Chang X, Allen DG, Ceger PC, Choksi NY, Hsieh J-H, Wetmore BA, Ferguson SS, DeVito MJ, Sprankle CS, Kleinstreuer NC, 2018. Evaluation and Optimization of Pharmacokinetic Models for in Vitro to in Vivo Extrapolation of Estrogenic Activity for Environmental Chemicals. Environ. Health Perspect. 126, 97001 10.1289/EHP1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang X, Kleinstreuer N, Ceger P, Hsieh J-H, Allen D, Casey W, 2014. Application of Reverse Dosimetry to Compare In Vitro and In Vivo Estrogen Receptor Activity. Appl In Vitro Toxicol 1, 33–44. 10.1089/aivt.2014.0005 [DOI] [Google Scholar]
- Chiu WA, Guyton KZ, Martin MT, Reif DM, Rusyn I, 2018. Use of high-throughput in vitro toxicity screening data in cancer hazard evaluations by IARC Monograph Working Groups. ALTEX 35, 51–64. 10.14573/altex.1703231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DJ, Schürer S, 2019. Improving the Utility of the Tox21 Dataset by Deep Metadata Annotations and Constructing Reusable Benchmarked Chemical Reference Signatures. Molecules 24, 1604 10.3390/molecules24081604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix DJ, Houck KA, Martin MT, Richard AM, Setzer RW, Kavlock RJ, 2007. The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol. Sci. 95, 5–12. 10.1093/toxsci/kfl103 [DOI] [PubMed] [Google Scholar]
- OECD, 219. eChemPortal. https://www.echemportal.org/echemportal/ (accessed Dec 2019)
- EPA’s National Center for Computational Toxicology, 2019. ToxCast Database (invitroDB). 10.23645/EPACOMPTOX.6062623.V4 (accessed Dec 2019) [DOI]
- Filer DL, Kothiya P, Setzer RW, Judson RS, Martin MT, 2016. tcpl: The ToxCast Pipeline for High-Throughput Screening Data. Bioinformatics 33, 618–620. 10.1093/bioinformatics/btw680 [DOI] [PubMed] [Google Scholar]
- Fonger GC, Hakkinen P, Jordan S, Publicker S, 2014. The National Library of Medicine’s (NLM) Hazardous Substances Data Bank (HSDB): background, recent enhancements and future plans. Toxicology 325, 209–216. 10.1016/j.tox.2014.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J, Sullivan K, Clippinger AJ, Strickland J, Bell S, Bhhatarai B, Blaauboer B, Casey W, Dorman D, Forsby A, Garcia-Reyero N, Gehen S, Graepel R, Hotchkiss J, Lowit A, Matheson J, Reaves E, Scarano L, Sprankle C, Tunkel J, Wilson D, Xia M, Zhu H, Allen D, 2017. Alternative approaches for identifying acute systemic toxicity: Moving from research to regulatory testing. Toxicol In Vitro 41, 245–259. 10.1016/j.tiv.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow I, Balakrishnan R, Jimenez-Ruiz E, Jupp S, Lomax J, Reed J, Romacker M, Senger C, Splendiani A, Wilson J, Woollard P, 2019. Ontology mapping for semantically enabled applications. Drug Discovery Today 24, 2068–2075. 10.1016/j.drudis.2019.05.020 [DOI] [PubMed] [Google Scholar]
- ICVAM, 2018. A Strategic Roadmap for Establishing New Approaches to Evaluate the Safety of Chemicals and Medical Products in the United States. National Toxicology Program (NTP) 10.22427/NTP-ICCVAM-ROADMAP2018 [DOI] [Google Scholar]
- Judson RS, Magpantay FM, Chickarmane V, Haskell C, Tania N, Taylor J, Xia M, Huang R, Rotroff DM, Filer DL, Houck KA, Martin MT, Sipes N, Richard AM, Mansouri K, Setzer RW, Knudsen TB, Crofton KM, Thomas RS, 2015. Integrated Model of Chemical Perturbations of a Biological Pathway Using 18 In Vitro High-Throughput Screening Assays for the Estrogen Receptor. Toxicol. Sci. 148, 137–154. 10.1093/toxsci/kfv168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavlock R, Chandler K, Houck K, Hunter S, Judson R, Kleinstreuer N, Knudsen T, Martin M, Padilla S, Reif D, Richard A, Rotroff D, Sipes N, Dix D, 2012. Update on EPA’s ToxCast Program: Providing High Throughput Decision Support Tools for Chemical Risk Management. Chemical Research in Toxicology 25, 1287–1302. 10.1021/tx3000939 [DOI] [PubMed] [Google Scholar]
- Kleinstreuer NC, Ceger P, Watt ED, Martin M, Houck K, Browne P, Thomas RS, Casey WM, Dix DJ, Allen D, Sakamuru S, Xia M, Huang R, Judson R, 2016a. Development and Validation of a Computational Model for Androgen Receptor Activity. Chem. Res. Toxicol. 10.1021/acs.chemrestox.6b00347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstreuer NC, Ceger PC, Allen DG, Strickland J, Chang X, Hamm JT, Casey WM, 2016b. A Curated Database of Rodent Uterotrophic Bioactivity. Environ. Health Perspect. 124, 556–562. 10.1289/ehp.1510183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstreuer NC, Hoffmann S, Alépée N, Allen D, Ashikaga T, Casey W, Clouet E, Cluzel M, Desprez B, Gellatly N, Göbel C, Kern PS, Klaric M, Kühnl J, Martinozzi-Teissier S, Mewes K, Miyazawa M, Strickland J, van Vliet E, Zang Q, Petersohn D, 2018a. Non-animal methods to predict skin sensitization (II): an assessment of defined approaches. Crit. Rev. Toxicol. 48, 359–374. 10.1080/10408444.2018.1429386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstreuer NC, Karmaus AL, Mansouri K, Allen DG, Fitzpatrick JM, Patlewicz G, 2018b. Predictive models for acute oral systemic toxicity: A workshop to bridge the gap from research to regulation. Computational Toxicology 8, 21–24. 10.1016/j.comtox.2018.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri K, Abdelaziz A, Rybacka A, Roncaglioni A, Tropsha A, Varnek A, Zakharov A, Worth A, Richard AM, Grulke CM, Trisciuzzi D, Fourches D, Horvath D, Benfenati E, Muratov E, Wedebye EB, Grisoni F, Mangiatordi GF, Incisivo GM, Hong H, Ng HW, Tetko IV, Balabin I, Kancherla J, Shen J, Burton J, Nicklaus M, Cassotti M, Nikolov NG, Nicolotti O, Andersson PL, Zang Q, Politi R, Beger RD, Todeschini R, Huang R, Farag S, Rosenberg SA, Slavov S, Hu X, Judson RS, 2016. CERAPP: Collaborative Estrogen Receptor Activity Prediction Project. Environ. Health Perspect. 124, 1023–1033. 10.1289/ehp.1510267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri K, Grulke CM, Judson RS, Williams AJ, 2018. OPERA models for predicting physicochemical properties and environmental fate endpoints. J Cheminform 10, 10 10.1186/s13321-018-0263-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri K, Kleinstreuer N, Abdelaziz AM, Alberga D, Alves VM, Andersson PL, Andrade CH, Bai F, Balabin I, Ballabio D, Benfenati E, Bhhatarai B, Boyer S, Chen J, Consonni V, Farag S, Fourches D, García-Sosa AT, Gramatica P, Grisoni F, Grulke CM, Hong H, Horvath D, Hu X, Huang R, Jeliazkova N, Li J, Li X, Liu H, Manganelli S, Mangiatordi GF, Maran U, Marcou G, Martin T, Muratov E, Nguyen D-T, Nicolotti O, Nikolov NG, Norinder U, Papa E, Petitjean M, Piir G, Pogodin P, Poroikov V, Qiao X, Richard AM, Roncaglioni A, Ruiz P, Rupakheti C, Sakkiah S, Sangion A, Schramm K-W, Selvaraj C, Shah I, Sild S, Sun L, Taboureau O, Tang Y, Tetko IV, Todeschini R, Tong W, Trisciuzzi D, Tropsha A, Van Den Driessche G, Varnek A, Wang Z, Wedebye EB, Williams AJ, Xie H, Zakharov AV, Zheng Z, Judson RS, 2020. CoMPARA: Collaborative Modeling Project for Androgen Receptor Activity. Environ. Health Perspect. 128, 27002 10.1289/EHP5580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Academies of Sciences, Engineering, and Medicine, 2015. Application of Modern Toxicology Approaches for Predicting Acute Toxicity for Chemical Defense. The National Academies Press, Washington, DC: 10.17226/21775 [DOI] [PubMed] [Google Scholar]
- National Library of Medicine, 2019. PubChem https://pubchem.ncbi.nlm.nih.gov/ (accessed Dec 2019).
- OECD, 2014. The Adverse Outcome Pathway for Skin Sensitisation Initiated by Covalent Binding to Proteins, OECD Series on Testing and Assessment. OECD Publishing, Paris: 10.1787/20777876 [DOI] [Google Scholar]
- Pearce R, Strope C, Setzer W, Sipes N, Wambaugh J, 2017. httk: R Package for High-Throughput Toxicokinetics. Journal of Statistical Software 79 10.18637/jss.v079.i04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto CL, Markey K, Dix D, Browne P, 2018. Identification of candidate reference chemicals for in vitro steroidogenesis assays. Toxicology in Vitro 47, 103–119. 10.1016/j.tiv.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto P, Graepel R, Gerloff K, Lamon L, Sachana M, Pistollato F, Gribaldo L, Bal-Price A, Worth A, 2019. Investigating cell type specific mechanisms contributing to acute oral toxicity. ALTEX 36, 39–64. 10.14573/altex.1805181 [DOI] [PubMed] [Google Scholar]
- Prior H, Casey W, Kimber I, Whelan M, Sewell F, 2019. Reflections on the progress towards nonanimal methods for acute toxicity testing of chemicals. Regulatory Toxicology and Pharmacology 102, 30–33. 10.1016/j.yrtph.2018.12.008 [DOI] [PubMed] [Google Scholar]
- Richard AM, Judson RS, Houck KA, Grulke CM, Volarath P, Thillainadarajah I, Yang C, Rathman J, Martin MT, Wambaugh JF, Knudsen TB, Kancherla J, Mansouri K, Patlewicz G, Williams AJ, Little SB, Crofton KM, Thomas RS, 2016. ToxCast Chemical Landscape: Paving the Road to 21st Century Toxicology. Chem. Res. Toxicol. 29, 1225–1251. 10.1021/acs.chemrestox.6b00135 [DOI] [PubMed] [Google Scholar]
- Sioutos N, Coronado S. de, Haber MW, Hartel FW, Shaiu W-L, Wright LW, 2007. NCI Thesaurus: A semantic model integrating cancer-related clinical and molecular information. Journal of Biomedical Informatics, Bio*Medical Informatics 40, 30–43. 10.1016/j.jbi.2006.02.013 [DOI] [PubMed] [Google Scholar]
- Smith MT, Guyton KZ, Gibbons CF, Fritz JM, Portier CJ, Rusyn I, DeMarini DM, Caldwell JC, Kavlock RJ, Lambert PF, Hecht SS, Bucher JR, Stewart BW, Baan RA, Cogliano VJ, Straif K, 2016. Key Characteristics of Carcinogens as a Basis for Organizing Data on Mechanisms of Carcinogenesis. Environ Health Perspect 124, 713–721. 10.1289/ehp.1509912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice RR, Austin CP, Kavlock RJ, Bucher JR, 2013. Improving the Human Hazard Characterization of Chemicals: A Tox21 Update. Environmental Health Perspectives 121, 756–765. 10.1289/ehp.1205784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelder MMHJ, van Rooij IALM, Miller RK, Zielhuis GA, de Jong-van den Berg LTW, Roeleveld N, 2010. Teratogenic mechanisms of medical drugs. Hum. Reprod. Update 16, 378–394. 10.1093/humupd/dmp052 [DOI] [PubMed] [Google Scholar]
- Villeneuve DL, Crump D, Garcia-Reyero N, Hecker M, Hutchinson TH, LaLone CA, Landesmann B, Lettieri T, Munn S, Nepelska M, Ottinger MA, Vergauwen L, Whelan M, 2014a. Adverse Outcome Pathway (AOP) Development I: Strategies and Principles. Toxicol. Sci. 142, 312–320. 10.1093/toxsci/kfu199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve DL, Crump D, Garcia-Reyero N, Hecker M, Hutchinson TH, LaLone CA, Landesmann B, Lettieri T, Munn S, Nepelska M, Ottinger MA, Vergauwen L, Whelan M, 2014b. Adverse Outcome Pathway Development II: Best Practices. Toxicol. Sci. 142, 321–330. 10.1093/toxsci/kfu200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner S, Browne P, Dix D, 2016. Identifying reference chemicals for thyroid bioactivity screening. Reproductive Toxicology 65, 402–413. 10.1016/j.reprotox.2016.08.016 [DOI] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg Ij.J., Appleton G, Axton M, Baak A, Blomberg N, Boiten J-W, Santos LB da S, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJG, Groth P, Goble C, Grethe JS, Heringa J, Hoen P.A.C. ‘t, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, Schaik R van, Sansone S-A, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, Lei J van der, Mulligen E van, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Mons B, 2016. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3, 1–9. 10.1038/sdata.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AJ, Grulke CM, Edwards J, McEachran AD, Mansouri K, Baker NC, Patlewicz G, Shah I, Wambaugh JF, Judson RS, Richard AM, 2017. The CompTox Chemistry Dashboard: a community data resource for environmental chemistry. Journal of Cheminformatics 9, 61 10.1186/s13321-017-0247-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.