Abstract
Natural killer (NK) cells recognize and eliminate infected and malignant cells. Their life histories are poorly understood, particularly in humans, due to lack of informative models and endogenous clonal markers. Here we apply transplantation of barcoded rhesus macaque hematopoietic cells to interrogate the landscape of NK cell production, expansion and life histories at a clonal level long-term and following proliferative challenge. We identify oligoclonal populations of rhesus CD56−CD16+ NK cells that are characterized by marked expansions and contractions over time, yet remained long-term clonally uncoupled from other hematopoietic lineages, including CD56+CD16− NK cells. Individual or groups of CD56−CD16+ expanded clones segregated with surface expression of specific killer immunoglobulin-like receptors (KIRs). These clonally distinct NK cell subpopulation patterns persisted for over 4 years, including following transient in vivo anti-CD16 mediated depletion and subsequent regeneration. Profound and sustained IL-15-mediated depletion was required to generate new oligoclonal CD56−CD16+ NK cells. Together, our results indicate that linear NK cell production from multipotent hematopoietic progenitors or less mature CD56+CD16− cells is negligible during homeostasis and moderate proliferative stress. In such settings, peripheral compartmentalized self-renewal can maintain the composition of distinct, differentiated NK cell subpopulations.
One Sentence Summary:
Macaque clonal barcoding reveals NK oligoclonal expansion and persistence independent of production from HSPC or immature NK
Introduction
Natural killer (NK) cells are defined as lymphocytes capable of target cell killing and cytokine production independent of somatic RAG-mediated antigen receptor recombination. Thereby distinguished from adaptive T and B cells, this fact precludes clonal tracking of NK cells based on receptor gene structure. In humans, NK cells have been phenotypically defined by the expression of CD56 and/or CD16, in the absence of T, B, and myeloid markers(1). More recently, a range of markers have been used to define functionally distinct human NK subsets(2–4). Models of human NK cell ontogeny have been proposed based on comparative phenotyping of NK cells in bone marrow(BM), lymph nodes(LN) and blood; kinetics of recovery following transplantation; and in vitro culture(5, 6). Precursor cells derived from multipotent hematopoietic stem and progenitor cells(HSPC) have been hypothesized to migrate from the BM to LN, generating CD56brightCD16− NK cells, followed by continuous maturation and release to maintain the predominant circulating CD56dimCD16+ NK cell subset. Deuterium labeling and Ki-67 staining studies have estimated a half-life of 14 days for circulating human NK cells and a proliferation rate of 4–5% per day, interpreted as evidence for ongoing release of immature progenitors. Nonetheless, almost instantaneous appearance of labeled CD56dim NK cells in the blood, consistent with proliferation of circulating cells, was also noted(7, 8)
Murine NK cells were shown to respond to a specific viral or hapten exposure, conferring protective immunity upon adoptive transfer and re-challenge, uncovering unexpected memory capabilities of NK cells similar to cytotoxic effector T cells(9). In addition, cytokine exposure has been shown to result in persistence of NK cell responses to viruses or tumors, even following adoptive transfer(10). In humans, subsets of CD56dimCD16+ NK cells expand upon CMV infection and may represent human analogs of murine adaptive NK cells(11–14). In contrast to CD56bright and canonical CD56dim NK cells, human “adaptive” NK cells variegatedly lack expression of several signaling proteins, and generally express KIR and NKG2C(13, 14). Epigenetically, adaptive NK cells approximate CD8+ effector T cells(14), with decreased methylation at the IFNG CNS1 locus(15), suggesting shared developmental pathways. Epigenetically imprinted “clonal” expansions in humans have been inferred by the presence of NK cells expressing distinct combinations of KIR following CMV infection or re-activation(11, 14). Mechanisms propagating functional NK cell diversity and memory in the absence of somatic receptor rearrangements have not been elucidated.
There are marked differences between the phenotype and function of mouse versus human NK cells (16, 17). Human NK cells are scantly produced in immunodeficient mice and robust in vitro clonal assays are lacking, thus processes maintaining their homeostasis and possible memory are poorly characterized in humans. Relative to rodents, rhesus macaque(RM) NK cells are well studied, evolutionarily close to humans, and share phenotypic and functional characteristics with human NK: a dominant blood CD56−CD16+ NK cell population functionally resembling human CD56dimCD16+ NK cells, and a CD56+CD16− NK cell population that is dominant in LN but less frequent in blood, resembling human CD56brightCD16− NK cells(18). Functional evidence for antigen-specific NK cell memory in RM following SIV/HIV vaccination has been reported(19). While human and rhesus KIR are evolutionary divergent, rhesus Mamu-KIR3D receptors are extremely diverse, bind MHC class I molecules analogous to human lineage II KIR, and show variegated distribution demonstrated via staining with the limited number of available anti-KIR monoclonal antibodies and/or MHC class I tetramer binding(20–22).
We developed a robust system of genetic “barcoding” to interrogate in vivo hematopoiesis following autologous RM HSPC transplantation, allowing tracking of clonal output from thousands of HSPC and their progeny. In our initial report of hematopoiesis early following engraftment, we were surprised to find that the majority of blood NK cells were clonally distinct from T, B, and myeloid cells derived from multipotent HSPC(23). The minor CD56+CD16− NK cell blood population was clonally related to the other lineages, however, at these early time points, the major blood population of CD56−CD16+ NK cells accounted for the divergence in clonal ontogeny. We interpreted these surprising results to be consistent with a distinct developmental pathway for CD56−CD16+ NK cells.
Here, we have dissected clonal patterns in NK cells for over 4 years, allowing a detailed long-term kinetic analysis of NK cell clonal populations in steady-state and upon proliferative challenge. Our results provide direct evidence for long-term clonal expansions, persistence and self-renewal of mature NK cells independent of progenitor cells, highlighting a previously unappreciated compartmentalization among NK cell subsets and forming the basis for a new model of NK cell dynamics.
Results
Low diversity of CD56−CD16+ NK cells and distinct clonal architecture by long-term clonal mapping
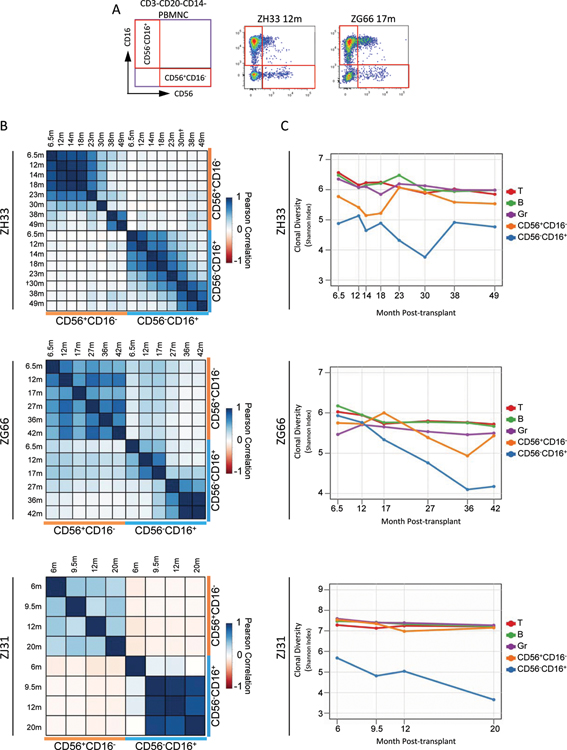
We tracked clonal contributions to NK cells for periods of more than four years following transplantation of RM with genetically-barcoded autologous CD34+ HSPC(Fig S1A), asking whether highly lineage-biased NK cells observed during the early post-engraftment period persisted(23) , and dissecting the characteristics and origins of NK cells in vivo at a clonal level. Each unique barcode marks a single HSPC and its clonal progeny. The majority of circulating RM PB NK cells are CD56−CD16+(77.3±3.2%, SEM), sharing properties with human CD56dimCD16+NK cells. A smaller and individually variable subset of rhesus CD16+NK cells express low levels of CD56. The minor PB NK cell population in rhesus is CD56+CD16−(5.8±1.1%), corresponding to human CD56brightCD16− cells(Fig 1A, Fig S2A–C)(18).
Figure 1: Clonal composition of peripheral blood NK cells.
A. FACS analysis of CD3−CD20−CD14− PBMNC for ZH33 at 12m and ZG66 at 17m post-transplantation, with sorting gates for CD56+CD16− and CD56−CD16+ NK subpopulations shown. B. Pairwise Pearson correlation coefficients between fractional abundances of all barcodes for CD56+CD16− and CD56−CD16+ NK from 6m through most recent follow-up (r-values, p-values and 95% confidence intervals(CI) in Table S1). C. Shannon diversity indices for all barcode contributions to T, B, Granulocytes(Gr), CD56+CD16− NK, and CD56−CD16+ NK over time. Table S3 gives the number of barcodes above threshold for each sample. “†”: The 30m ZH33 CD56−CD16+ sample was additionally sorted to be CD8α+NKG2+.
The overall degree of relatedness between clonal contributions can be demonstrated via Pearson correlations between two samples for all barcodes. There was minimal overall correlation between contributions from all clones to CD56−CD16+ versus CD56+CD16− NK cells at all time points for the three animals(Fig 1B, r values/p values Table S1). We also noted markedly lower clonal diversity over time for CD56−CD16+ NK cells compared to other lineages, including CD56+CD16− NK cells, as assessed by Shannon diversity index, taking into account both overall clone number and skewing of contributions(Fig 1C). Diversity remained relatively stable and higher in other cell types for over three years(Fig 1C and(24)).
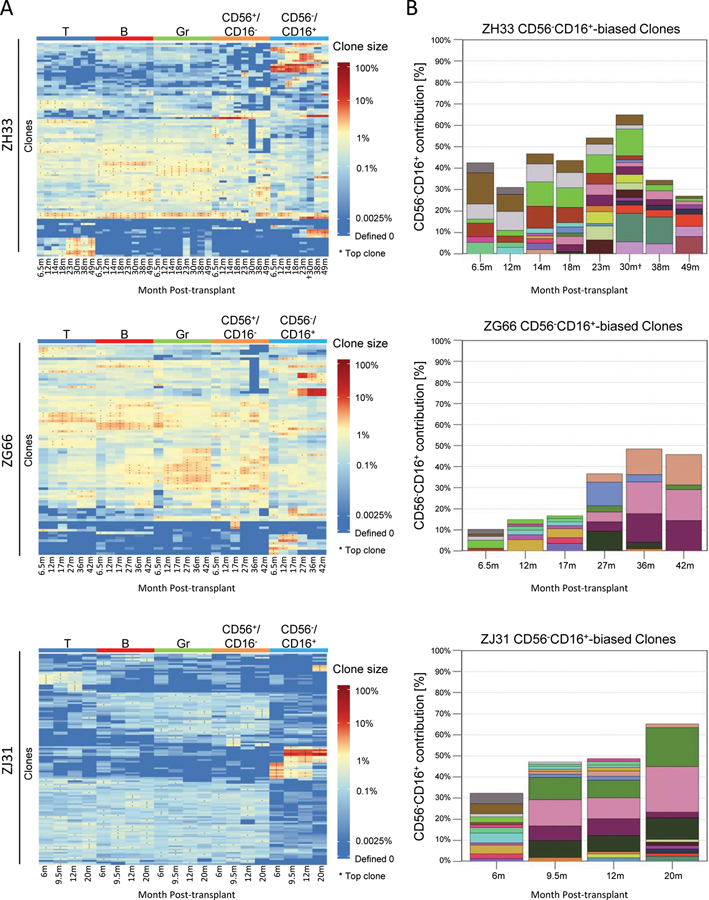
A focus on high contributing clones offers information on those most responsible for hematopoiesis, reducing the impact of sampling bias. Contributions from the ten highest contributing clones in each lineage at each time point are shown in a heat map for each animal, clustering clones by overall similarity of contributions across all samples. In all three animals, very high contributing CD56−CD16+ clones showed distinct contribution patterns in contrast to multipotent clones producing CD56+CD16−NK, granulocytes(Gr), T, and B cells, even as late as 49 months post-transplant(Fig 2A, Table S2). This pattern was most striking in ZJ31, with very high contributing CD56−CD16+ clones virtually undetectable in the other lineages. Of note, similar highly expanded T cell clones appeared in animal ZH33 much later following transplantation than expanded NK clones.
Figure 2: Clonal bias of CD56−CD16+ NK subpopulations.
A. Heat maps of the natural log fractional abundance of the highest contributing clones defined as all barcodes present as a top 10 highest contributing barcode in at least one of the samples, mapped over all samples for each animal. Corresponding % contributions in Table S2. Each row corresponds to one unique barcode; * indicates the barcode is one of the top 10 for that sample. Heat maps are organized by unsupervised hierarchical clustering of Euclidean distances between barcodes’ log fractional abundances, with relative contribution shown via color gradient. B. Highly biased CD56−CD16+ NK clone contributions over time. A barcode is defined as highly biased at a time point if it meets the following conditions: (i) >1% contribution to CD56−CD16+ cells, and (ii) > 10-fold abundant in contribution to CD56−CD16+ cells compared to T, B, Gr and CD56+CD16− cells. y-axes show the fraction of all barcode reads in CD56−CD16+ cells contributed by the “highly biased” clones. Each color represents the contribution of a single clone; the same color at different time points indicates the same clone. 30m ZH33 CD56−CD16+ sample was additionally sorted to be CD8α+NKG2+.
We further characterized highly biased NK clones, defined as a clone size >1%, and contributing to CD56−CD16+ NK cells at a ratio > 10:1 compared to fractional contribution to any other cell type, including CD56+CD16−NK, T, B or Gr. Figure S3 provides histograms validating a >10:1 ratio as clearly delineating a separate population of highly biased CD56−CD16+ clones. When using the same criteria for comparisons with other lineages, only the T cell compartment in ZH33 showed appreciable numbers of high contributing biased clones, developing late after transplant(Fig 2A, Fig S3). The highly biased CD56−CD16+ NK cell clones were few in number; a median of 8(range 4–15) per total 667(range 471–1503) clones detected in this cell type per time point (Table S3). However, these rare clones comprised a major fraction of blood CD56−CD16+ NK cells in all three animals(Fig 2B), indicating very marked expansion of these highly biased clones, with single clones contributing up to 21%.
Both CD8α and NKG2 represent additional markers expressed on virtually all CD56+CD16− as well as CD56−CD16+ RM NK cells(18). Antibodies that react only with human NKG2A bind to both NKG2A and NKG2C isoforms on RM NK cells(18, 25–27), and are thus referred to as NKG2. Sorting of CD3−CD20−CD14−CD8α+NKG2+ cells for CD56+ and CD16+ confirmed highly biased clones as contributing to distinct subsets of bona fide CD56+CD16− or CD56−CD16+ NK cells, respectively(Fig S2D, E).
Overall, these biased clonal patterns are difficult to explain via continuous ongoing production of terminally-differentiated short-lived rhesus CD56−CD16+ NK cells from HSPC via maturation through CD56+CD16− NK cells. Rather, NK cell clonal patterns could result from ongoing production via an independent pool of long-lived NK cell progenitors for these highly biased populations, or homotypic clonal expansions of the distinct CD56−CD16+ NK cells themselves, analogous to memory T cells(28).
NK cell clonal dynamics over time
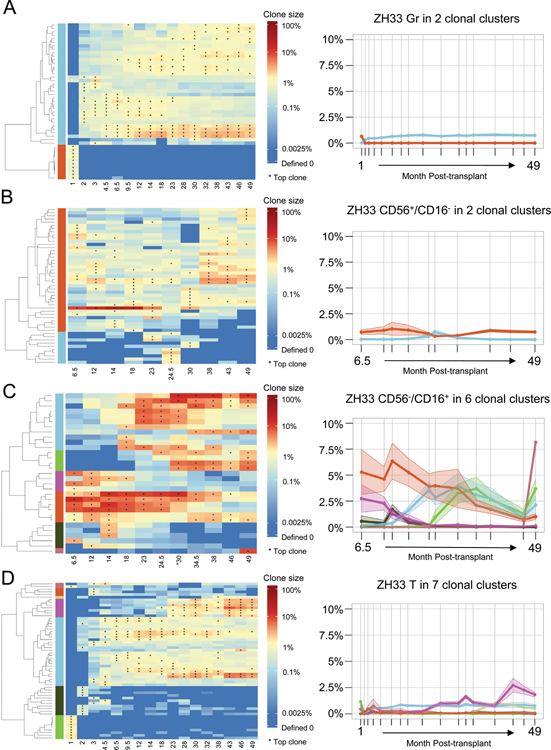
To study the clonal dynamics of NK subpopulations, clones were grouped hierarchically into clusters within each cell type based on overall similarities of contributions over time, and the mean fractional contributions of each cluster over time were plotted. In ZH33 Gr, there were two clusters, one representing short-term engrafting progenitors that disappeared by 2 months, and the second representing very stable ongoing production from long-term clones beginning at 2 months and persisting to 49 months(Fig 3A left panel). Each clone contributed an average of less than 1%(Fig 3A right panel). In CD56+CD16− cells, the major cluster shows very stable output from clones contributing an average of less than 1–2% per clone(Fig 3B). These clones were primarily multi-lineage, also producing T, B and myeloid cells(Fig 2A). Similar patterns emerged in additional animals(Fig S4).
Figure 3: Cluster tracking of barcodes with similar kinetic behavior over time.
Heat maps (left panels) show the log fractional contribution to hematopoiesis of the highest contributing clones (top 10 in each sample, as defined in Figure 2A) organized by unsupervised hierarchical clustering in each cell type over time for animal ZH33. The hierarchical tree in each cell type was cut at a level to retrieve clear clusters via visual inspection of the dendrograms and heat maps. The clusters so defined are designated by color bars on the left of the heat maps. The fractional abundances of each clone in these clusters are averaged for all clones in the cluster, and plotted over time in the right panels, with the shaded ribbons around each line representing the SEM of the average fractional abundance. Line colors match the colors of the clusters in the corresponding heat map. A. Gr, B. CD56+CD16− NK C. CD56−CD16+ NK D. T cells.
The CD56−CD16+ NK cell patterns were strikingly different in all three animals, both in kinetics and in magnitude of individual contributions(Fig 3C, S4). There were clear expansions and contractions of groups of clones over time, with some clusters comprised of clones with average contributions of 6–8%. Plots of the changes over time for ZH33 clones shown individually are shown in Fig S5. Notably, a similar pattern of expanding and contracting clusters was observed in T cells later post-transplant in animal ZH33(Fig 3D).
NK cell clones segregate with specific cell surface receptor phenotypes
KIRs are diverse and rapidly evolving receptors mediating activating and inhibitory interactions with target cells. NK cell expansions occurring coincident with CMV reactivation post-transplantation in humans are frequently KIR+, with clonal-like variegated patterns of specific KIRs expressed on subsets of expanding NK cells(11, 12). Expression of highly polymorphic and polygenic KIR genes has been described as stochastic and is presumed to be maintained epigenetically. Investigation of NK responses in vivo at a clonal level has been previously impossible, with no ability to link different KIR repertories to specific clonal expansions.
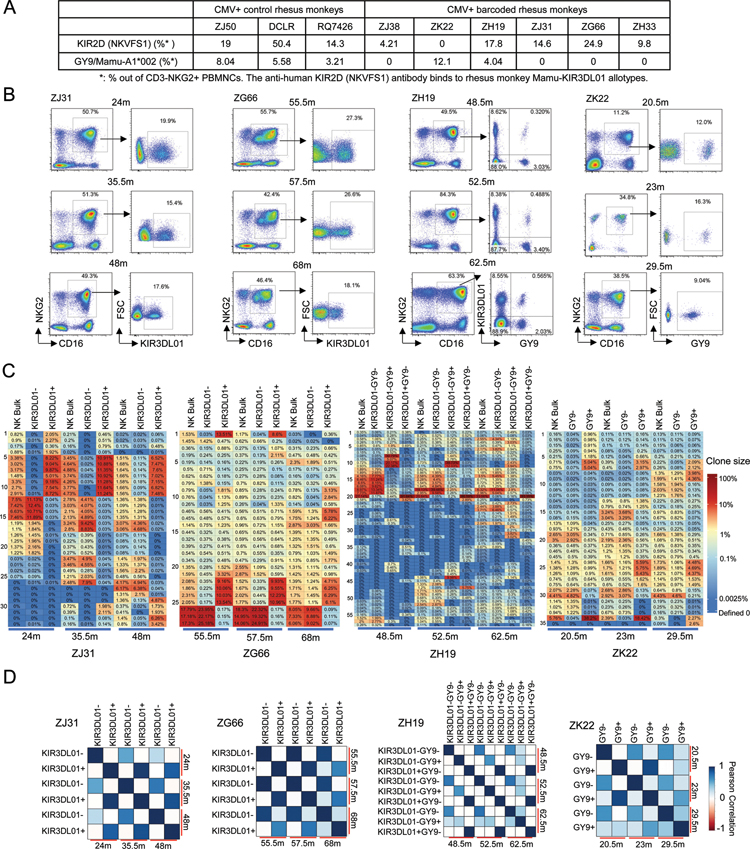
Many specific antibodies have been developed to detect diverse human KIRs. However, almost all human KIR antibodies lack cross-reactivity with RM KIRs, given the highly diverse and rapidly evolving nature of these loci in primates, and few rhesus-specific KIR antibodies have been developed(20, 29). An anti-human KIR2D (clone NKVFS1) antibody has been shown to recognize rhesus KIR3DL01(30, 31). An alternate approach to identifying expression of specific rhesus KIR has been staining with tetrameric peptide-MHC-I(pMHC-I) complexes shown to bind to KIR and segregate macaque NK cell populations with specific patterns of KIR expression(22, 32).
We stained CD3−NKG2+ NK cells from 9 RMs with anti-human KIR2D and the rhesus pMHC-I tetramer SIV-Gag GY9/Mamu-A1*002. Positive and negative subpopulations after staining with these reagents were distinct, and varied between animals(Fig 4A–B), reminiscent of observed variegated, persistent KIR expression patterns on human NK cells(11). We sorted subpopulations of CD3−CD14−CD20−CD16+NKG2+ NK cells from 4 barcoded monkeys for KIR2D and pMHC-I tetramer staining and performed barcode analysis on each distinct subpopulation of sufficient size for barcode retrieval(Fig 4B). The highly biased and expanded CD16+NKG2+ NK clones showed clear segregation between sorted subpopulations(Fig 4C–D). For instance, in monkeys ZJ31 and ZG66, different sets of expanded clones completely segregated into the same KIR3DL01+ or KIR3DL01− subpopulations. The clones present in the KIR3DL01− fraction likely expressed other KIRs not reactive with available reagents. In ZH19, three major NK subpopulations were sorted based on both KIR3D01 and GY9/Mamu-A1*002 staining, and each population contained a set of specific clones. Within the KIR3DL01− NK subset, specific GY9+ clones were enriched and contributed major fractions to this subpopulation, although within the bulk population these clones were relatively small contributors, for instance #8, 9, 10, and 11 at 48.5m, and #44 at 52.5m(Fig 4C). ZK22 had no detectable KIR3DL01+ NK cells, but clear populations of GY9/Mamu-A1*002+cells showing barcode segregation. In all four animals, clonal segregation linked to KIR expression was stable long-term, documented from nine months-two years(Fig 4D).
Figure 4: Clonal segregation of CD16+NKG2+ NK cells based on KIR3DL01 expression and/or SIV Gag GY9/Mamu-A1*002 tetramer staining.
A. NK surface receptor subpopulations of CD3−NKG2+ NK cells from RM PB (n=9) detected by SIV Gag GY9/Mamu-A1*002 tetramer and anti-KIR2D(NKVFS1) staining. B. FACS plots of SIV Gag GY9/Mamu-A1*002 tetramer and anti-KIR2D staining of PB CD3−CD16+NKG2+ NK cells from barcoded macaques. C. Top 10 barcoded clone heatmaps for bulk CD16+NKG2+ NK cells and sorted NK subpopulations based on anit-KIR2D and tetramer staining for the time points and animals shown in panel B. Distinct clonal distribution within different NK subpopulations distinguished by presence and/or absence of staining are shown. D. Pearson correlation coefficients comparing pairwise fractional contributions between NK KIR subpopulations from 4 monkeys same as in panel C. The color scale for r values is shown on the right(r-values, p-values and 95% CI in Table S1).
Macaques in this study were rhCMV+ at the time of transplantation. We documented reactivation of latent rhCMV immediately post-transplant (Fig S6A). Although antibodies to markers for PLZF, FcεRγ, EAT-2, or SYK distinctly stained RM lymphocyte subsets, down-regulation of these signaling molecules, previously linked to human adaptive NK cells in the context of CMV infection(14), was not observed in CD16+CD56− NK cells of CMV+ compared to CMV− macaques(Fig S6B). We also examined methylation of the CNS1 locus upstream of the IFNG promoter (Fig S6C–D). Methylation at this locus strongly inversely correlates with human adaptive NK cell expansion following CMV infection(15). Although methylation at CNS1 was decreased in CD56−CD16+ as compared to CD56+CD16− NK cells and B cells(Fig S6E–F), the methylation levels were consistent with CNS1 methylation in human CD56bright versus canonical CD56dim NK cells, respectively. Thus, in our macaques we did not identify NK cells directly analogous to human adaptive NK cells.
Altogether, these results are consistent with expansion and maintenance of distinct CD16+ NK cell clones identified by KIR expression patterns. The relationship between distinct KIR expression patterns and specific barcoded clones persisted for up to two years, consistent with a model of peripheral homotypic expansion and maintenance of phenotypically and epigenetically distinct NK cell proliferating clones.
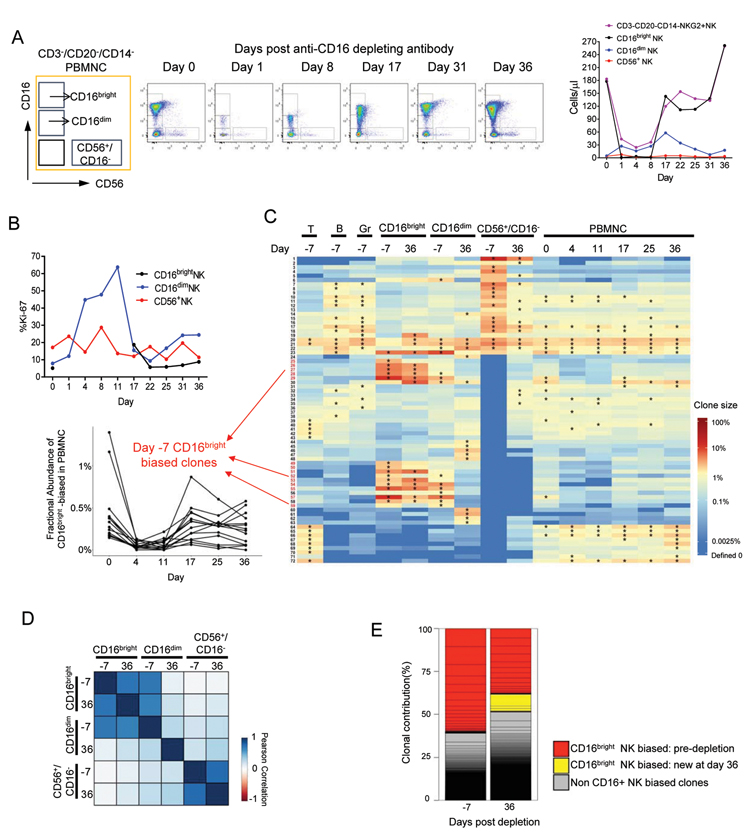
In vivo depletion via anti-CD16 associated with regeneration of biased CD56−CD16+ NK clones
To gain insights to peripheral NK cell homeostasis, we asked whether regeneration of circulating CD56−CD16+ NK cells following in vivo depletion would be supported via proliferation of pre-existing biased dominant clones, appearance of a new group of dominant clones, or maturation of a polyclonal population from multipotent HSPC clones via CD56+CD16− NK cells. To this end, we administered the anti-CD16 antibody 3G8 to macaque ZH33 32 months post-transplant, previously reported to deplete greater than 90% of RM NK cells in vivo with a single dose(33). By day 1, 99% of CD16bright NK cells disappeared from blood, with negligible impact on circulating CD56+CD16− NK cells(Fig 5A), however a population of CD16dim NK cells persisted. There was no impact on the numbers or barcode composition of T, B, or myeloid cells(Fig S7A–C). To confirm true depletion of CD56−CD16+ cells, versus trogocytosis or CD16 epitope blockade, we demonstrated nearly equivalent depletion of CD3−CD20−CD14−NKG2+ cells(Fig 5A). Highly biased CD56−CD16+ clones (#25–29, #49–55,57,59) were profoundly depleted from PBMC on days 4 and 11(Fig 5C). The number and fraction of CD56−CD16dim NK cells increased by day 1 and peaked at day 17. CD56−CD16bright NK cells began to reappear by day 17 and normalized in number by day 36.
Figure 5: In vivo depletion of CD16+ cells.
FACS plots and NK subset absolute concentrations (A) and Ki-67+ % of PB NK subsets (B) before and after 50mg/kg anti-CD16-depleting antibody treatment of ZH33 32m post-transplant. C. Top 15 clones heat map shows the log fractional abundance of the in T, B, Gr, CD16bright, CD16dim, CD56+ and total PBMC samples over time before and after antibody. Depletion and regeneration of individual Day −7 CD56−CD16+ highly-biased NK clones(numbered in red; defined in Fig 2) is shown to the left. The fractional contributions of these highly biased clones decreased on day 4 vs day 0(p=0.002, paired t-test). D. Pearson correlation between clonal contributions before and after anti-CD16(r-values, p-values, 95% CI in Table S1). E. Stacked bar plots of relative clonal contributions to CD56−CD16bright NK cells from CD16+ highly-biased clones versus multi-lineage or clones biased towards other lineages before and after anti-CD16. NK-biased clones defined as > 10-fold abundant in contribution to CD56−CD16+ cells compared to T, B, Gr and CD56+CD16− cells regardless of clone size. Grey bars: non CD16bright NK biased clones’ contributions; CD16bright NK biased clone contributions are shown in colors reflecting time point of appearance, each individual clone’s contribution is delineated by lines, and stacked to create bars.
At baseline, as previously reported for human NK cell subsets(8), fewer CD56−CD16+ compared to CD56+CD16− NK cells were actively cycling, as determined by Ki-67 staining(Fig 5B). Ki-67 was expressed in 8.8–19.8% of the specific KIR+ subsets(Fig S7F), consistent with the hypothesis that these fully mature cells can self-renew. During regeneration, there was a marked increase in cycling CD16dim cells, peaking just before CD56−CD16bright cells reappeared at day 17, consistent with regeneration of CD16bright from CD16dim NK cells. Notably, despite a higher baseline, there was no consistent increase in cycling of CD56+CD16− cells.
In terms of cellular barcodes, overall clonal contributions to CD56−CD16bright NK cells comparing baseline day −7 to day 36 were strongly correlated(r=0.74 Fig 5D). The majority of highly-biased individual NK clones present pre-depletion appeared again after recovery(Fig 5C). The overall fraction of clonal contributions to CD56−CD16bright NK cells derived from biased clones compared to multipotent clones decreased somewhat following recovery but remained almost 50%(Fig 5E). Many CD56−CD16dim clones were shared with CD56−CD16bright clones at baseline and their overall clonal patterns were highly correlated(Fig 5D). However, some biased dominant CD56−CD16dim clones decreased contributions to the CD56−CD16dim NK population by day 36, perhaps reflecting rapid proliferation and partial depletion of these clones via maturation during regeneration of the CD16brightCD56− compartment by certain clones within the CD56−CD16dim compartment.
In contrast, we found only limited recruitment of multi-lineage, unbiased clones to regenerate CD56−CD16+ NK cells following this short-term depletion. Only three large new highly biased clones appeared following recovery(Fig 5C, #6, 30, 58). Overall clonal correlations between CD56−CD16+ NK and other lineages, including CD56+CD16− NK cells, remained very low(Fig 5D). Even under this proliferative stress, our data indicate that regeneration of CD56−CD16+ cells from circulating CD56+CD16− precursors is unlikely. Rather, clonal patterns remained generally stable, suggesting self-renewal and high proliferative potential of CD56−CD16+ NK cells. These NK cells may have regenerated from circulating CD56−CD16dim clonally-restricted NK cells surviving antibody depletion, given the induction of rapid cycling in this subset prior to refilling of the CD56−CD16+ compartment.
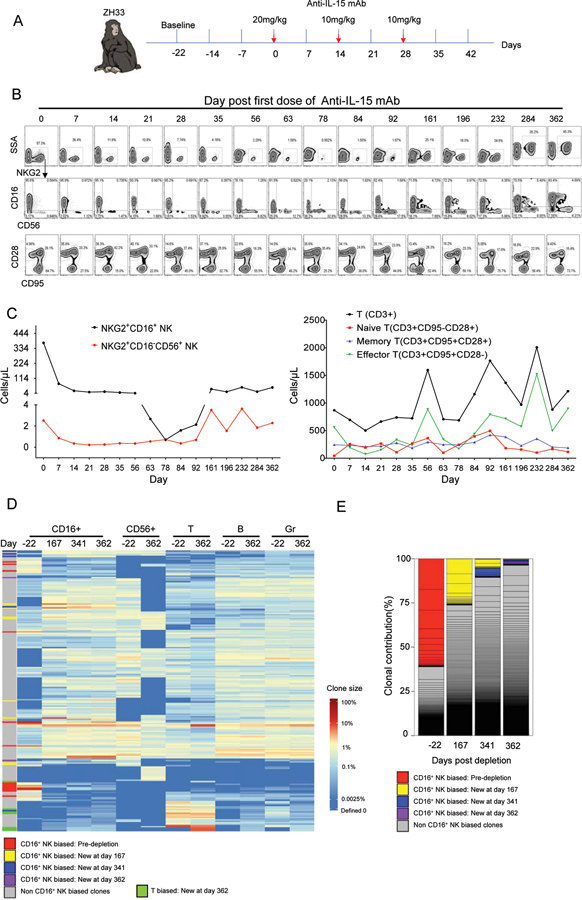
Prolonged profound in vivo NK cell depletion via anti-IL15
Next, we aimed to examine NK cell dynamics upon long-lasting in vivo depletion of NK cells. 18 months following full recovery from anti-CD16 antibody treatment and clonal stabilization, we administered three biweekly doses of an anti-IL15 neutralizing antibody to macaque ZH33. This treatment is reported to result in profound and very prolonged depletion of all NK subsets within RM blood and tissues(34)(Fig 6A). This antibody resulted in greater than 540-fold depletion of PB NKG2+CD56−CD16+NK cells and more than 10-fold depletion of NKG2+CD16−CD56+ NK cells. In contrast to anti-CD16 antibody effects, both CD16bright and CD16dim cells were depleted by anti-IL15. Recovery of NK cell numbers did not begin until more than three months post-treatment, not reaching a normal range until greater than 12 months later(Fig 6B, C). As reported previously, effector T cells were also depleted by this antibody(34), but to a lesser degree than NK cells and only short-term(Fig 6B–C). The majority of the residual CD16+ cells in PB during the NK depletion were NKG2−CD8α− cells expressing CD11c and HLA-DR: these cells likely represented atypical monocytes(Fig S7D).
Figure 6: In vivo depletion of NK cells with anti-IL15.
A. Schematic of anti–IL-15 treatment. ZH33 received 20 mg/kg of anti–IL-15 mAb on day 0 and 10 mg/kg on days 14 and 28. B. Upper and middle rows show NK subsets based on NKG2, CD16 and CD56 expression in CD3−CD14−CD20− PBMNC; bottom row shows CD95 and CD28 expression on CD3+ T cells. C. Absolute PB concentration of NK and T cell subpopulations. D. Heat map of top 50 most abundant clones in CD16+ NK, CD56+ NK, T, B, Mono and Gr samples over time. Depleted and newly arising clones of different bias types are designated on the left of the heat map(defined per criteria in Figure 2 legend). Newly biased T clones are defined as non NK-biased clones contributing to T cells at a ratio of 10:1 compared to fractional contribution to any other cell type, including CD56+CD16–NK, CD56-CD16+NK, B or Gr, at day 362. CD56+CD16− NK samples from days 167 and 341 were not obtained. E. Stacked bar plots of relative contributions to CD56−CD16+ NK from CD56−CD16+ NK-biased clones versus multi-lineage or clones biased towards other lineages before and after anti-IL15, using same criteria defined in 5E. Each individual clone’s contribution is delineated by lines, and stacked to create the bars, with colors designating clone type. NK-biased clones defined as > 10-fold abundant in contribution to CD56−CD16+ cells compared to T, B, Gr and CD56+CD16− cells regardless of clone size.
The highly expanded and biased NK clones present at baseline(day −22) were profoundly depleted following the anti-IL15 antibody treatment, and in contrast to the effect of anti-CD16 NK cell depletion, these clones almost completely disappeared upon NK cell recovery(Fig 6D, designated by red bars on left of heat map and in Fig 6E). Instead, new clonal expansions arose in the CD16+ NK compartment at day 167 following treatment, the earliest time point sufficient NK cells could be collected for clonal analysis. These expanded clones were present specifically in CD56−CD16+ cells, and in contrast to results following the less profound and shorter-term depletion induced by the anti-CD16 antibody, were not the same expanded clones present at baseline(compare clones labeled with red bars to those with yellow, blue and purple bars in Fig 6D–E). While some of these new expanded NK cell clones were biased(Fig 6D–E, yellow, blue and purple bars), the majority did contribute at detectable but relatively lower levels to T, B, Gr and CD56+NK cells(Fig 6D). In addition, expanded clones not fitting criteria for CD16-bias began contributing to this compartment post recovery(Fig 6D and E, grey bars). New sets of expanded T cell clones also appeared (Fig 6D, green bar). These results suggest that following profound and lasting depletion of all NK cells, including biased and expanded CD56−CD16+ clones, new NK cells could not be regenerated peripherally from long-lived self-renewing NK cells, and instead had to be produced from long-term engrafted HSPC also contributing to other cell lineages.
Discussion
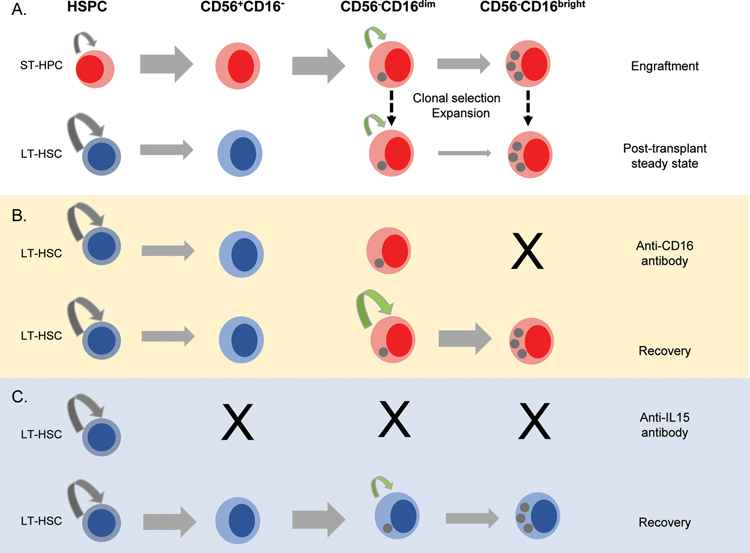
Maintenance of distinct cell populations is a function of survival, self-renewal and differentiation. The prevailing human model posits that CD56bright NK cells are produced from BM precursors and continuously generate CD56dim NK cells in secondary lymphoid tissues(5, 36). A CD34+CD38+CD123−CD45RA+CD7+CD10+CD127− NK cell progenitor was recently identified in human hematopoietic tissues, and could differentiate to CD56bright and rarer CD56dim NK cells in vitro and in xenografted mice(37). We recently utilized the macaque clonal tracking approach to identify sites of NK cell production(43). CD56+CD16− NK cells were produced in the marrow from HSPC, in contrast, oligoclonally expanded CD56−CD16+ NK cells were homogeneously distributed between blood, nodes and multiple marrow locations, suggesting clonal expansions occur outside the marrow and nodes, even if the final steps in maturation occur in nodes(44). As evidenced by Ki-67 staining, human blood CD56bright NK cells were reported to cycle more rapidly than CD56dim NK cells(8). In the present study, we also observed higher steady-state turnover of rhesus CD56+CD16− compared to CD56−CD16+ NK cells. But in contrast to predictions from a continuous lineage differentiation model, the barcode composition of CD56+CD16− and CD56−CD16+ NK cells remained distinct under homeostatic conditions over several years. Barcodes displayed unique, longitudinally-stable profiles even within small populations of differentiated KIR-expressing CD56−CD16+ NK cells. Moreover, transient depletion of CD56−CD16+ NK cells did not increase CD56+CD16− NK cell proliferation, but rather that of CD56−CD16dim NK cells, which displayed the same clonal make-up as the CD56−CD16bright NK cell population. These findings do not fit the paradigm of continuous differentiation from HSPC and CD56bright NK cells during steady-state maintenance of mature NK cells, and support a model of compartmentalized NK cell homeostasis(Fig 7).
Figure 7: Model for NK cell homeostasis and regeneration.
A. Long-term repopulating stem cells(LT-HSPC), short-term repopulating progenitors(ST-HSPC), or NK-restricted progenitors may be lentivirally-barcoded. Each may give rise to mature CD16+NK, containing barcodes corresponding to the parental progenitors, red for those derived from ST-HSPC and blue for those derived from LT-HSPC. A few of these mature NK cells expand to large clonal populations maintaining receptor expression and barcodes of the originating NK and persisting for months-years. ST-HSPC disappear, explaining lack of overlap of ST-HSC-derived NK with other hematopoietic lineages, including CD56+CD16− immature NK. B. Upon anti-CD16 antibody depletion of CD56−CD16brightNK, CD56−CD16dimNK proliferate and regenerate the mature NK pool without significant contributions from CD56+CD16−NK, thus the populations remain clonally-distinct. C. Following prolonged depletion of all NK with anti-IL-15, many expanded clones disappear, to be replaced by a different set of expanded NK clones upon return of IL-15. Some of the new expanded NK clones have barcodes overlapping multiple lineages because they develop from mature NK that have arisen from LT-HSPC after anti-IL-15 treatment.
The persistence of differentiated, oligoclonal expanded NK cell clones, even in the setting of proliferative stress, also argues against the notion of mature rhesus CD16+ or human canonical CD56dim NK cells as senescent(38). This conclusion is also supported by results of clinical NK cell infusions and murine adoptive transfer studies, documenting in vivo persistence of mature NK cells for several months without dependence on regeneration from marrow HSPC(39, 40). We recently reported evidence from two clinical syndromes suggesting that human CD56dim NK cells can be maintained independently of ongoing production from HSPC, suggesting longevity and self-renewal(41, 42). Hypothetically, high levels of homeostatic cytokines, such as IL-7 and IL-15, or even inflammatory cytokines, may be required to break the steady-state barrier between these subsets to rejuvenate the CD56−CD16+NK cell pool. Very severe and prolonged depletion of the entire NK cell pool, using IL-15 depleting reagents, was required to disrupt the clonal make-up of this CD56−CD16+ NK cell compartment and eventually recruit new, distinctly NK clones into the CD56−CD16+ NK compartment. The lack of overlap between initial populations of expanded and biased CD56−CD16+ clones arising and persisting post-transplantation and long-term multipotent HSPC clones may result from rapid initial recovery of NK cells from short-term progenitors unable to contribute to other lineages long-term, with acquisition of homotypic self-renewal in a subset of these initially-produced NK cells, allowing clonal persistence without clonal overlap with HSPC or CD56+CD16− NK cells(Fig 7). Identification of the precise engrafting CD34+ short-term multipotent progenitor or NK-committed precursor serving as the source of these cells is the subject of future barcoding studies.
Our observations raise important questions as to how dynamics among NK cell subpopulations is orchestrated. The segregation of specific expanded clones with distinct surface KIR expression in our model, with many individual clones being uniformly positive or negative for a given KIR, provides evidence that a single original cell which proliferated to generate this clone had already undergone the process of KIR receptor acquisition and epigenetic stabilization and was likely a fully mature NK cell. This striking result provides clear evidence for the stability of specific clonal KIR expression patterns through multiple cell divisions, for time periods up to several years. Specific KIR segregating with individual or groups of barcoded NK clones may be directly responsible for signaling expansion or may simply be stably co-expressed with other receptors driving expansion and persistence of a relatively small number of contributing clones. Whether response to a specific stimulus or stochastic factors are responsible for NK clonal expansion and persistence is unclear. Waxing and waning groups of expanded NK clones over time suggests an environmentally-driven process. Recent reports that CMV infection can induce the differentiation of epigenetically unique and stable human adaptive or memory-like CD56dim NK cell populations implies that rhesus CMV reactivation post-transplantation might drive oligoclonal expansions(13, 14), and can be tested via analysis of NK clonal patterns in CMV negative macaques, and following de novo CMV infection. However, we could not demonstrate that RM CD56−CD16+ NK cells expressed the signature of signaling molecules and transcription factors previously linked to human adaptive NK cells arising in the context of CMV infection. Regrettably, no antibodies reactive with macaque CD57 or NKG2C exist, markers also associated with human adaptive or memory-like NK cell subsets following CMV and other viral infections, although a recent report utilized RNA probes and flow cytometry to distinguish NKG2C versus NKG2A-expressing macaque NK cells(45, 46). Therefore, it remains unclear whether the rhesus macaque models human adaptive NK cell differentiation and homeostasis.
In conclusion, the tracking of individual NK cell clonal histories in macaques provides direct evidence for the peripheral expansion and stable persistence of differentiated mature NK clones, and associates these clones with stable expression of specific KIRs, offering insights into the development and maintenance of mature circulating NK cells. Under homeostatic conditions, our findings suggest a barrier between HSPC-dependent CD56bright and the more stably maintained CD56dim NK cell populations, providing a model for understanding NK differentiation and homeostasis(Fig 7). In addition, these findings have translational implications, since development of NK cell therapies requires an understanding of how, where and why specific NK cells are generated(47).
Materials and Methods
Barcoded autologous transplantation model and experimental design
Animal studies were approved by the NHLBI Animal Care and Use Committee. RM CD34+ HSPC transductions with barcoded lentiviral libraries and autologous transplantation following 1000 rad total body irradiation were performed as described (23, 48), with individual animal data summarized in Fig S1A. The transplanted product contained virtually no barcoded mature cells, Thus, barcoded hematopoiesis following engraftment was generated from CD34+ HSPC, not from residual NK cells present in the initial graft (Fig S1B).
Barcode libraries with a diversity 10 times greater than that necessary to be more than 95% certain that each individual barcode marked a single CD34+ cell were used, at a transduction efficiency resulting in single barcode per HSPC (23, 49). DNA samples from hematopoietic lineage cells were used to amplify the barcode region via PCR followed by next generation sequencing to recover the barcodes for lineage clonal relationship analysis. Output was processed using custom Python code to identify barcoded clones contributing above sequencing error and sampling thresholds (23, 24). Each clone’s fractional contribution to a sample could then be calculated. Custom R code is available on GitHub at http://github.com/d93espinoza/barcodetrackR.
Cell purification and FACS analysis
Blood, LN and BM cells were isolated on a density gradient and stained with various antibody panels for FACS analyses and sorting. Antibodies are listed in Table S5. Cellular subsets were FACS sorted to >98% purity using gating as shown in Figure S1C. Intracellular Ki-67 staining was performed utilizing the eBioscience Foxp3/Transcription Factor Staining Buffer. Cells were sorted on a BD FACSAriaII Sorter. All FACS data was analyzed using FlowJo. GY9/Mamu-A1*002 monomers, graciously provided by Prof. David A. Price (Cardiff University), were tetramerized with either PE or APC streptavidin and used for staining one million PBMNC with 0.6 µg tetramer for 15min at room temperature, then staining with other surface marker antibodies for 30min at 4°C.
Barcode retrieval
Cell DNA was extracted with the DNeasy Kit(Qiagen). 100–500ng DNA underwent 28-cycle PCR using Phusion High-Fidelity DNA Polymerase (ThermoFisher). A universal reverse primer and a unique forward primer(Table S4) were used to multiplex sample sequencing. Following gel-purification, 10 to 24 multiplexed samples were then pooled for sequencing on an Illumina HiSeq 2500 or 3000. Python code used for extraction from FASTQ files is located at: https://github.com/d93espinoza/barcode_extracter.
In vivo depletion of NK cells
The mouse anti-human CD16 monoclonal NK depleting antibody(3G8) and the anti-IL15 neutralizing antibody(M111) were obtained from the Nonhuman Primate Reagent Resource(http://www.nhpreagents.org) and use for NK depletion as described(33, 34).
Computational and statistical analyses
Data analysis, Pearson correlations, Euclidean distances, p values, and plot generation were performed using R (Foundation for Statistical Computing) and Prism (GraphPad Software). R code utilized is available online at http://github.com/d93espinoza/barcodetrackR.
Supplementary Material
10: Table S3: The number of barcodes above threshold for each sample from ZH33, ZG66 and ZJ31 in Figures 1 and 2.
8: Table S1: Pearson correlation coefficient r-values, p-values and 95% confidence interval (CI) for the correlation coefficient.
9: Table S2: Corresponding % contributions to the samples presented in Figure 2A.
11: Table S4: Primer sequence information.
12: Table S5: Antibody information.
1: Figure S1: Barcoded CD34+ HSPC monkey transplantation parameters, and hematopoietic lineage cells FACS sort gating strategy.
2: Figure S2: Phenotype of NK cell populations in rhesus macaque blood, marrow and lymph nodes.
3: Figure S3: Bias of all clones towards T, B, Gr, CD56+CD16− NK and CD56−CD16+ NK fractions.
4: Figure S4: Cluster tracking of barcodes with similar kinetic behavior over time for ZG66 and ZJ31.
5: Figure S5: Individual clonal tracking of CD56−CD16+ biased clones in ZH33.
6: Figure S6: Epigenetic profile and functionality of NKG2+CD56−CD16+ NK Cells.
7: Figure S7: The effect of in vivo depletion of CD16+ NK cells on other lineages and Ki-67 expression on RM PB NK subsets.
Acknowledgments:
We thank Naoya Uchida for the χHIV plasmid, Keyvan Keyvanfar, NHLBI FACS Core, DNA Sequencing Core, animal care staff, and the NIH Biowulf High-Performance Computing Resource.
Funding: Supported by NHLBI and NIAID Divisions of Intramural Research, the Scientific Research Training Program for Young Talents sponsored by Union Hospital, Tongji Medical College, Huazhong University of Science and Technology(DY); the European Research Council/European Union Seventh Framework Programme (FP/2007–2013, ERC 311335), Swedish Research Council, Swedish Foundation for Strategic Research, Swedish Cancer Foundation, Wallenberg Foundation, Stockholm County Council and Karolinska Institutet Center for Innovative Medicine (YTB, HS); and Leibnitz ScienceCampus Chronic Inflammation German Research Foundation Grants SFB TRR2141(B02), R03565/4–1 and Heisenberg Program R03565/1–1 (CR).
Footnotes
Competing interests: The authors declare they have no competing interests.
References
- 1.Caligiuri MA, Human natural killer cells. Blood 112, 461–469 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freud AG, Mundy-Bosse BL, Yu J, Caligiuri MA, The Broad Spectrum of Human Natural Killer Cell Diversity. Immunity 47, 820–833 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cichocki F, Schlums H, Theorell J, Tesi B, Miller JS, Ljunggren HG, Bryceson YT, Diversification and Functional Specialization of Human NK Cell Subsets. Curr. Top. Microbiol. Immunol 395, 63–94 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, Dekker CL, Mackey S, Maecker H, Swan GE, Davis MM, Norman PJ, Guethlein LA, Desai M, Parham P, Blish CA, Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci. Transl. Med 5, 208ra145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freud AG, Becknell B, Roychowdhury S, Mao HC, Ferketich AK, Nuovo GJ, Hughes TL, Marburger TB, Sung J, Baiocchi RA, Guimond M, Caligiuri MA, A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity 22, 295–304 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Freud AG, Yokohama A, Becknell B, Lee MT, Mao HC, Ferketich AK, Caligiuri MA, Evidence for discrete stages of human natural killer cell differentiation in vivo. J. Exp. Med 203, 1033–1043 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Wallace DL, de Lara CM, Ghattas H, Asquith B, Worth A, Griffin GE, Taylor GP, Tough DF, Beverley PC, Macallan DC, In vivo kinetics of human natural killer cells: the effects of ageing and acute and chronic viral infection. Immunology 121, 258–265 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lutz CT, Karapetyan A, Al-Attar A, Shelton BJ, Holt KJ, Tucker JH, Presnell SR, Human NK cells proliferate and die in vivo more rapidly than T cells in healthy young and elderly adults. J. Immunol 186, 4590–4598 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Sullivan TE, Sun JC, Lanier LL, Natural Killer Cell Memory. Immunity 43, 634–645 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper MA, Elliott JM, Keyel PA, Yang L, Carrero JA, Yokoyama WM, Cytokine-induced memory-like natural killer cells. Proc. Natl. Acad. Sci. U.S.A 106, 1915–1919 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beziat V, Liu LL, Malmberg JA, Ivarsson MA, Sohlberg E, Bjorklund AT, Retiere C, Sverremark-Ekstrom E, Traherne J, Ljungman P, Schaffer M, Price DA, Trowsdale J, Michaelsson J, Ljunggren HG, Malmberg KJ, NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 121, 2678–2688 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foley B, Cooley S, Verneris MR, Pitt M, Curtsinger J, Luo X, Lopez-Verges S, Lanier LL, Weisdorf D, Miller JS, Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood 119, 2665–2674 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M, Scott JM, Kamimura Y, Lanier LL, Kim S, Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity 42, 431–442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlums H, Cichocki F, Tesi B, Theorell J, Beziat V, Holmes TD, Han H, Chiang SC, Foley B, Mattsson K, Larsson S, Schaffer M, Malmberg KJ, Ljunggren HG, Miller JS, Bryceson YT, Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 42, 443–456 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luetke-Eversloh M, Hammer Q, Durek P, Nordstrom K, Gasparoni G, Pink M, Hamann A, Walter J, Chang HD, Dong J, Romagnani C, Human cytomegalovirus drives epigenetic imprinting of the IFNG locus in NKG2Chi natural killer cells. PLoS Pathog 10, e1004441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colucci F, Di Santo JP, Leibson PJ, Natural killer cell activation in mice and men: different triggers for similar weapons? Nat. Immunol 3, 807–813 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Sungur CM, Murphy WJ, Utilization of mouse models to decipher natural killer cell biology and potential clinical applications. Hematology Am. Soc. Hematol. Educ. Program 2013, 227–233 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Webster RL, Johnson RP, Delineation of multiple subpopulations of natural killer cells in rhesus macaques. Immunology 115, 206–214 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reeves RK, Li H, Jost S, Blass E, Li H, Schafer JL, Varner V, Manickam C, Eslamizar L, Altfeld M, von Andrian UH, Barouch DH, Antigen-specific NK cell memory in rhesus macaques. Nat. Immunol 16, 927–932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hermes M, Weil S, Groth A, Dressel R, Koch J, Walter L, Characterisation of mouse monoclonal antibodies against rhesus macaque killer immunoglobulin-like receptors KIR3D. Immunogenetics 64, 845–848 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreland AJ, Guethlein LA, Reeves RK, Broman KW, Johnson RP, Parham P, O’Connor DH, Bimber BN, Characterization of killer immunoglobulin-like receptor genetics and comprehensive genotyping by pyrosequencing in rhesus macaques. BMC Genomics 12, 295 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maloveste SM, Chen D, Gostick E, Vivian JP, Plishka RJ, Iyengar R, Kruthers RL, Buckler-White A, Brooks AG, Rossjohn J, Price DA, Lafont BA, Degenerate recognition of MHC class I molecules with Bw4 and Bw6 motifs by a killer cell Ig-like receptor 3DL expressed by macaque NK cells. J. Immunol 189, 4338–4348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu C, Li B, Lu R, Koelle SJ, Yang Y, Jares A, Krouse AE, Metzger M, Liang F, Lore K, Wu CO, Donahue RE, Chen IS, Weissman I, Dunbar CE, Clonal tracking of rhesus macaque hematopoiesis highlights a distinct lineage origin for natural killer cells. Cell Stem Cell 14, 486–499 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koelle SJ, Espinoza DA, Wu C, Xu J, Lu R, Li B, Donahue RE, Dunbar CE, Quantitative stability of hematopoietic stem and progenitor cell clonal output in rhesus macaques receiving transplants. Blood 129, 1448–1457 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mavilio D, Benjamin J, Kim D, Lombardo G, Daucher M, Kinter A, Nies-Kraske E, Marcenaro E, Moretta A, Fauci AS, Identification of NKG2A and NKp80 as specific natural killer cell markers in rhesus and pigtailed monkeys. Blood 106, 1718–1725 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaBonte ML, Choi EI, Letvin NL, Molecular determinants regulating the pairing of NKG2 molecules with CD94 for cell surface heterodimer expression. J. Immunol 172, 6902–6912 (2004). [DOI] [PubMed] [Google Scholar]
- 27.LaBonte ML, McKay PF, Letvin NL, Evidence of NK cell dysfunction in SIV-infected rhesus monkeys: impairment of cytokine secretion and NKG2C/C2 expression. Eur. J. Immunol 36, 2424–2433 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Surh CD, Boyman O, Purton JF, Sprent J, Homeostasis of memory T cells. Immunol. Rev 211, 154–163 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Kruse PH, Rosner C, Walter L, Characterization of rhesus macaque KIR genotypes and haplotypes. Immunogenetics 62, 281–293 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Ries M, Reynolds MR, Bashkueva K, Crosno K, Capuano S 3rd, Prall TM, Wiseman R, O’Connor DH, Rakasz EG, Uno H, Lifson JD, Evans DT, KIR3DL01 upregulation on gut natural killer cells in response to SIV infection of KIR- and MHC class I-defined rhesus macaques. PLoS. Pathog 13, e1006506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schafer JL, Colantonio AD, Neidermyer WJ, Dudley DM, Connole M, O’Connor DH, Evans DT, KIR3DL01 recognition of Bw4 ligands in the rhesus macaque: maintenance of Bw4 specificity since the divergence of apes and Old World monkeys. J. Immunol 192, 1907–1917 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price DA, Brenchley JM, Ruff LE, Betts MR, Hill BJ, Roederer M, Koup RA, Migueles SA, Gostick E, Wooldridge L, Sewell AK, Connors M, Douek DC, Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J. Exp. Med 202, 1349–1361 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi EI, Wang R, Peterson L, Letvin NL, Reimann KA, Use of an anti-CD16 antibody for in vivo depletion of natural killer cells in rhesus macaques. Immunology 124, 215–222 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeGottardi MQ, Okoye AA, Vaidya M, Talla A, Konfe AL, Reyes MD, Clock JA, Duell DM, Legasse AW, Sabnis A, Park BS, Axthelm MK, Estes JD, Reiman KA, Sekaly RP, Picker LJ, Effect of Anti-IL-15 Administration on T Cell and NK Cell Homeostasis in Rhesus Macaques. J. Immunol 197, 1183–1198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cichocki F, Verneris MR, Cooley S, Bachanova V, Brunstein CG, Blazar BR, Wagner J, Schlums H, Bryceson YT, Weisdorf DJ, Miller JS, The Past, Present, and Future of NK Cells in Hematopoietic Cell Transplantation and Adoptive Transfer. Curr. Top. Microbiol. Immunol 395, 225–243 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freud AG, Yokohama A, Becknell B, Lee MT, Mao HC, Ferketich AK, Caligiuri MA, Evidence for discrete stages of human natural killer cell differentiation in vivo. J Exp Med 203, 1033–1043 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renoux VM, Zriwil A, Peitzsch C, Michaelsson J, Friberg D, Soneji S, Sitnicka E, Identification of a Human Natural Killer Cell Lineage-Restricted Progenitor in Fetal and Adult Tissues. Immunity 43, 394–407 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Bjorkstrom NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA, Bjorklund AT, Flodstrom-Tullberg M, Michaelsson J, Rottenberg ME, Guzman CA, Ljunggren HG, Malmberg KJ, Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK-cell differentiation uncoupled from NK-cell education. Blood 116, 3853–3864 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Szmania S, Lapteva N, Garg T, Greenway A, Lingo J, Nair B, Stone K, Woods E, Khan J, Stivers J, Panozzo S, Campana D, Bellamy WT, Robbins M, Epstein J, Yaccoby S, Waheed S, Gee A, Cottler-Fox M, Rooney C, Barlogie B, van Rhee F, Ex vivo-expanded natural killer cells demonstrate robust proliferation in vivo in high-risk relapsed multiple myeloma patients. J. Immunother 38, 24–36 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun JC, Beilke JN, Bezman NA, Lanier LL, Homeostatic proliferation generates long-lived natural killer cells that respond against viral infection. J. Exp. Med 208, 357–368 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corat MA, Schlums H, Wu C, Theorell J, Espinoza DA, Sellers SE, Townsley DM, Young NS, Bryceson YT, Dunbar CE, Winkler T, Acquired somatic mutations in PNH reveal long-term maintenance of adaptive NK cells independent of HSPCs. Blood 129, 1940–1946 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlums H, Jung M, Han H, Theorell J, Bigley V, Chiang SC, Allan DS, Davidson-Moncada JK, Dickinson RE, Holmes TD, Hsu AP, Townsley D, Winkler T, Wang W, Aukrust P, Nordoy I, Calvo KR, Holland SM, Collin M, Dunbar CE, Bryceson YT, Adaptive NK cells can persist in patients with GATA2 mutation depleted of stem and progenitor cells. Blood 129, 1927–1939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu C, Espinoza DA, Koelle SJ, Potter EL, Lu R, Li B, Yang D, Fan X, Donahue RE, Roederer M, Dunbar CE, Geographic clonal tracking in macaques provides insights into HSPC migration and differentiation. J. Exp. Med 215, 217–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu J, Freud AG, Caligiuri MA, Location and cellular stages of natural killer cell development. Trends. Immunol 34, 573–582 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez-Verges S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, Houchins JP, Miller S, Kang SM, Norris PJ, Nixon DF, Lanier LL, Expansion of a unique CD57(+)NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci U S A 108, 14725–14732 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ram DR, Manickam C, Hueber B, Itell HL, Permar SR, Varner V, Reeves RK, Tracking KLRC2 (NKG2C)+ memory-like NK cells in SIV+ and rhCMV+ rhesus macaques. PLoS Pathog 14, e1007104 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knorr DA, Bachanova V, Verneris MR, Miller JS, Clinical utility of natural killer cells in cancer therapy and transplantation. Semin. Immunol 26, 161–172 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donahue RE, Kuramoto K, Dunbar CE, Large animal models for stem and progenitor cell analysis. Curr Protoc Immunol Chapter 22, Unit 22A 21 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Lu R, Neff NF, Quake SR, Weissman IL, Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat. Biotechnol 29, 928–933 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
10: Table S3: The number of barcodes above threshold for each sample from ZH33, ZG66 and ZJ31 in Figures 1 and 2.
8: Table S1: Pearson correlation coefficient r-values, p-values and 95% confidence interval (CI) for the correlation coefficient.
9: Table S2: Corresponding % contributions to the samples presented in Figure 2A.
11: Table S4: Primer sequence information.
12: Table S5: Antibody information.
1: Figure S1: Barcoded CD34+ HSPC monkey transplantation parameters, and hematopoietic lineage cells FACS sort gating strategy.
2: Figure S2: Phenotype of NK cell populations in rhesus macaque blood, marrow and lymph nodes.
3: Figure S3: Bias of all clones towards T, B, Gr, CD56+CD16− NK and CD56−CD16+ NK fractions.
4: Figure S4: Cluster tracking of barcodes with similar kinetic behavior over time for ZG66 and ZJ31.
5: Figure S5: Individual clonal tracking of CD56−CD16+ biased clones in ZH33.
6: Figure S6: Epigenetic profile and functionality of NKG2+CD56−CD16+ NK Cells.
7: Figure S7: The effect of in vivo depletion of CD16+ NK cells on other lineages and Ki-67 expression on RM PB NK subsets.