Abstract
Novel severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) became pandemic by the end of March 2020. In contrast to the 2002–2003 SARS-CoV outbreak, which had a higher pathogenicity and lead to higher mortality rates, SARSCoV-2 infection appears to be much more contagious. Moreover, many SARS-CoV-2 infected patients are reported to develop low-titer neutralizing antibody and usually suffer prolonged illness, suggesting a more effective SARS-CoV-2 immune surveillance evasion than SARS-CoV. This paper summarizes the current state of art about the differences and similarities between the pathogenesis of the two coronaviruses, focusing on receptor binding domain, host cell entry and protease activation. Such differences may provide insight into possible intervention strategies to fight the pandemic.
Keywords: Sars-CoV-2, Furin, Receptor binding domain, Sars-CoV, Coronavirus
Introduction
At the end of December 2019, Chinese public health officials announced to the World Health Organization (WHO) that a novel virus caused in Wuhan a disease with symptoms similar to pneumonia [1]. They recognized that the virus was from the coronavirus family and was formally named severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). SARS-CoV-2 rapidly became pandemic by the end of March 2020, forcing much of the world to adopt lockdown strategies and putting health care systems under pressure while major concern about global health and economic stability arose. In contrast to the 2002–2003 SARS-CoV outbreak, which had a higher pathogenicity and lead to higher mortality rates, SARS-CoV-2 infection appears to be much more contagious, rapidly spreading to all continents. Compared to SARS-CoV, SARS-CoV-2 infection is characterized by a wider clinical spectrum, including asymptomatic infection, mild upper respiratory tract illness, severe viral pneumonia with respiratory failure and death [1, 2]. In contrast to SARS-CoV, many SARS-CoV-2-infected patients are reported to develop low-titer neutralizing antibody and usually suffer prolonged illness, suggesting a more effective SARS-CoV-2 immune surveillance evasion than SARS-CoV [3, 4]. Since the high transmission rate and viral immune escape may be involved in the SARS-CoV-2 widespread, both potentially representing a target for interventional strategies, it is of utmost importance to elucidate the molecular mechanisms which are involved in these atypical pathogenetic features.
Coronaviruses structure and replication
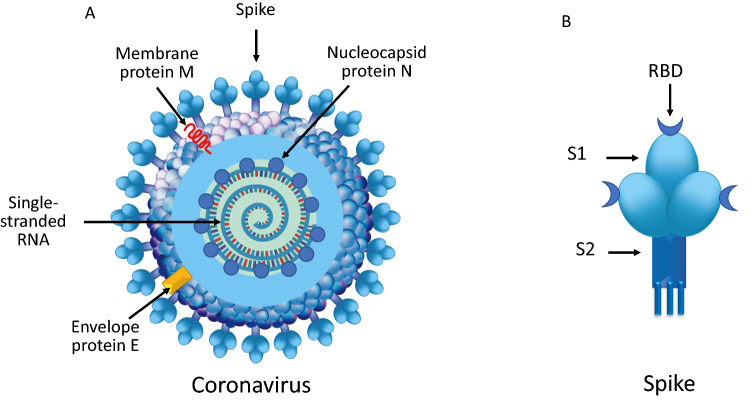
Human coronaviruses (hCoVs) are enveloped viruses with a positive-sense, single-stranded RNA genome [5]. HCoVs genome size is one of the largest among RNA viruses, ranging from 26.4 to 31.7 kilobases. Viral particles and envelope average diameters are around 125 nm and 85 nm, respectively. On electron microscopy, hCoVs show a characteristic club-shaped spikes that projects from their surface, creating an image reminiscent of the solar corona, from which their name originates [6]. The viral envelope consists of a lipid bilayer, in which the membrane (M), envelope (E) and spike (S) structural proteins are anchored (Fig. 1a) [5–7]. Inside the envelope, viral genome is enclosed, i.e., a ribonucleoprotein (RNP) core, comprising the nucleocapsid protein (N) that acts as a scaffold around the 29,900 nucleotides of RNA. The M and E proteins play a central role in forming the viral envelope and providing the structural integrity [7]. The surface spike (S) belongs to a class I fusion proteins which mediate the receptor binding and the fusion between virus and host cell membranes [8]. The S protein is composed by the S1 subunit, which forms the head of the spike and hosts the receptor-binding domain (RBD), and by the S2 subunit, the stem which anchors the spike to the viral envelope and, following protease activation, enables host cell fusion (Fig. 1b) [8, 9]. After cell entry, viral genome is released into the cell cytoplasm, host ribosomes begin to translate the first reading frame from the viral genome and then via the neo-formed RNA-dependent polymerases, the numerous sub-genomic RNAs are transcribed and then translated [10, 11]. Following genomic RNA replication, the viral structural proteins E and M move along the secretory pathway into the Golgi compartment and maturation of structural proteins occurs. M proteins direct most protein interactions required for assembly of viruses, whilst E proteins are involved in several other aspects of the virus’ life cycle, including envelope formation and budding [7, 11]. In addition to the 4 main structural proteins, hCoVs possess 16 non-structural proteins which assemble to form a multi-protein replicase–transcriptase complex (RTC). RTC promotes viral RNA replication, favors viral survival through inhibition of innate immunity responses, and enhances virulence power [7, 12]. Progeny viruses are released from the host cell by exocytosis through secretory vesicles. In humans, hCoVs infections can affect the respiratory, gastrointestinal, liver and central nervous systems [11, 12]. SARS-CoV and the novel SARS-CoV-2 share 79.5% sequence identity [5, 13–15] and this explains why not only similarities, but also differences can be detected in the epidemiology and clinical features in the disorders they cause [12, 14, 16]. Structural–functional analysis has identified differences in the mechanisms involved in host cell infection which could partially explain the dissimilarity in efficiency and speed of virus transmission between SARS-CoV and SARS-CoV-2.
Fig. 1.
a Coronaviruses structures. The membrane (M), envelope (E) and spike (S) structural proteins are anchored to the viral envelope which contains the ribonucleoprotein core, i.e., the nucleocapsid protein (N) which acts as a scaffold surrounding the single-stranded RNA. b The surface spike is composed by the S1 subunit, which harbors the receptor binding domain (RBD), and by the S2 subunit, the stem which anchors the spike to the viral envelope and, following protease activation, enables host cell fusion
SARS-CoV and SARS-CoV-2 host cell infection receptor recognition
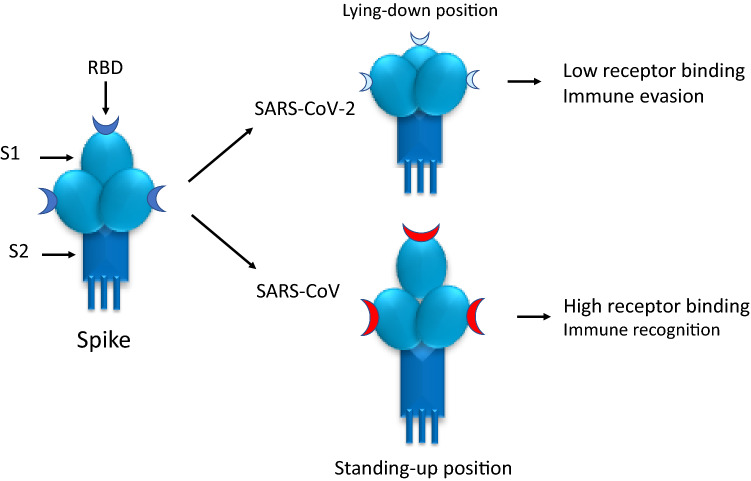
Receptor recognition is an important determinant of hCoVs infection and pathogenesis. The specific surface protein that provides the entry door in human cells for both SARS-CoV and SARS-CoV-2 is angiotensin-converting enzyme 2 (ACE2) [17–19]. The first difference between the two SARS-CoVs is that SARS-CoV-2 receptor-binding domain (RBD) has a higher ACE2-binding affinity, a characteristic which could lead to a more efficient cell entry [19]. However, ACE2-binding affinity of the entire SARS-CoV-2 S protein seems to be comparable to or even lower than that of SARS-CoV entire S protein. This observation suggests that SARS-CoV-2 RBD, even though more potent, is probably less exposed than SARS-CoV RBD [4]. There have been conflicting reports in the literature on the ACE2-binding affinities of the two SARS-CoVs spike proteins, probably because RBD constantly switches between a “standing-up” position and a “lying-down” position (Fig. 2a) [19, 20]. Evaluation by cryo-electron microscopy (Cryo-EM) of the crystal structure of the two SARS-CoVs RBD, complexed with ACE2 receptors, showed subtle, but functionally important differences [21]. SARS-CoV-2 RBD was mostly in the “lying-down” position, a state associated with ineffective receptor binding [21]. This observation was confirmed by flow cytometry in a different study [22]. In contrast to SARS-CoV-2 spike, Cryo-EM studies showed that in SARS-CoV spike protein, the RBD is mostly in the “standing-up” state [23]. As shown in an animal study, the lower accessibility of the SARS-CoV-2 RBD in “lying-down position”, a state associated with less effective receptor binding, may favor the immune evasion of SARS-CoV-2 as one of the conformational masking strategies. In mice, sera from SARS-CoV-infected animals poorly neutralized SARS-CoV-2 pseudovirus entry into host cells but they bound with high-affinity SARS-CoV RBD and potently neutralized SARS-CoV pseudovirus entry [4, 24, 25]. Conflicting reports on SARS-CoV-2 pathogenicity may also be related to genetic differences in the expression of the SARS-CoV-2 host cell entry factors among individuals and between populations [26, 27]. Indeed, to maintain its high infectivity while keeping its RBD less accessible, SARS-CoV-2 relies on a second strategy, i.e. host protease activation.
Fig. 2.
Receptor-binding domain (RBD) of the S protein may constantly switch between a “lying-down” and a “standing-up” position. In SARS-CoV-2, RBD is mostly in the “lying-down” position, a state associated with not only ineffective receptor binding, but also immune evasion. In SARS-CoV, RBD is mostly in “standing-up” position, a state associated with not only high effective receptor binding, but also immune recognition
Proteolytic activation
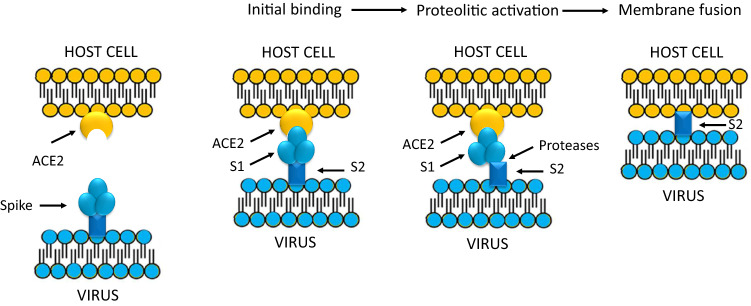
After initial receptor binding, hCoVs need to fuse their envelope with the host cell membrane to deliver the viral nucleocapsid into the target cells (Fig. 2b) [28–31]. To fuse membranes, SARS-CoV spike needs to be proteolytically activated at the S1/S2 level. The S2 subunit, cleaved from S1 subunit by host cell proteases, facilitates membrane fusion, bringing the virion into the host cells (Fig. 3) [28]. The major proteases involved in the two SARS-CoVs entry are the cell surface transmembrane protease serine 2 (TMPRSS2) and the lysosomal proteases cathepsins [28, 29, 32]. SARS-CoV S proteins contains cleavage sites for both TMPRSS2 and cathepsins. In experimental SARS-CoV infection, inhibition of both proteases is required to block SARS-CoV entry in cell cultures, however, only TMPRSS2 activity seems to be essential for inhibiting viral replication and spread [29, 32]. An important difference between the two SARS-CoVs is that SARS-CoV-2 S proteins contains also a furin-like cleavage site, absent in the SARS-CoV S protein [4, 29, 32, 33]. Cleavage of S protein by furin at the S1/S2 site is an essential process for cell–cell fusion and SARS-CoV-2 entry into human lung cells [29, 32–34]. Furin pre-activation provides a gain-of-function for a more efficient spreading, enhancing SARS-CoV-2 entry into cells with relatively low expressions of TMPRSS2 and/or lysosomal cathepsins [4, 29, 32–34]. However, protease cleavage of CoVs spikes leads to major structural rearrangement of the S2 subunit [35]. This process, which is irreversible, may reduce entry efficiency in some types of cells with high expressions of TMPRSS2 and cathepsins, as shown in in vitro studies performed in different cell line cultures [36]. Examining the whole native SARS-CoV-2 architecture by transmission electron microscopy, it has been shown that many S molecules had already undergone the structural changes associated with less efficiency [4, 37]. As shown in in vitro experiments, the reduced viral entry capacity induced by furin pre-activation may be more relevant for “not fresh” virus particles [4]. Indeed, conformational modifications of the S molecules, which may slowly occur spontaneously, can be facilitated by a variety of environmental factors, such as physical force, high temperature or chemicals [4]. Like SARS-CoV, also SARS-CoV-2 spread depends on TMPRSS2 activity, but in vitro studies showed that a TMPRSS2 inhibitor, camostat mesylate [38], only partially blocked SARS-2-S-driven entry into human epithelial cell line cells. This finding suggests that that furin-mediated precleavage at the S1/S2 site in infected cells could promote subsequent TMPRSS2-dependent entry into target cells [32]. However, being a clinically proven and commercial serine protease inhibitor, camostat mesylate might be helpful for clinicians at intensive care unit treating severely ill COVID-19 patients [39]. Finally, SARS-CoV-2 also infect endothelial cells and, during COVID-19, one of the serine-proteases activated is thrombin. Part of SARS-CoV-2 pathogenesis is caused by enzymatic activation of the clotting cascade by thrombin activation at the endothelial surface of capillaries with a significant risk of thrombotic complications, ranging from microvascular thrombosis, to venous thromboembolic, disseminated intravascular coagulation and endothelial plasma leakage and thus alveolar obstruction [40]. The growing awareness and mechanistic understanding of the prothrombotic state of COVID-19 patients is driving efforts to more stringent diagnostic screening and to the early institution of antithrombotic drugs for both prevention and treatment of thrombotic complications [40, 41].
Fig. 3.
After initial binding of the ACE2 receptor, SARS-CoV spike is proteolytically activated and enzymatically cleaved at the S1/S2 level. S1 than dissociates from S2 and the truncated 2 subunit of the Spike protein facilitates fusion of viral and cellular membranes [28, 38]
Conclusions
Both SARS-CoV and SARS-CoV-2 use human ACE2 as entry receptor and human proteases as entry activators. In vitro studies have identified strategies that SARS-CoV-2 adopts to infect human cells that potentially contribute to wide spread of the virus and to immune evasion. These cell entry mechanisms may represent substantial target for host immune surveillance and provide insight into possible intervention strategies to fight the pandemic induced by this novel agent.
Acknowledgements
Open access funding provided by Università degli Studi di Roma La Sapienza within the CRUI-CARE Agreement.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y, Peng F, Wang R, et al. The deadly coronaviruses: the 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J Autoimmun. 2020;109:102434. doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shang J, Wan Y, Luo C, et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neuman BW, Adair BD, Yoshioka C, Quispe JD, Orca G, Kuhn P, et al. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J Virol. 2006;80:7918–7928. doi: 10.1128/JVI.00645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldsmith CS, Tatti KM, Ksiazek TG, et al. Ultrastructural characterization of SARS coronavirus. Emerg Infect Dis. 2004;10:320–326. doi: 10.3201/eid1002.030913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. In: Maier HJ, Bickerton E, Britton P, editors. Coronaviruses. Methods in molecular biology, vol 1282. Berlin: Springer; 2015. pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heald-Sargent T, Gallagher T. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses. 2012;4:557–580. doi: 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Follis KE, York J, Nunberg JH. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell-cell fusion but does not affect virion entry. Virology. 2006;350:358–369. doi: 10.1016/j.virol.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simmons G, Zmora P, Gierer S, et al. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai MM, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Astuti I, Ysrafil Y. severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab Syndr. 2020;14:407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neuman BW, Buchmeier MJ. Supramolecular architecture of the coronavirus particle. Adv Virus Res. 2016;96:1–27. doi: 10.1016/bs.aivir.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tortorici MA, Veesler D. Structural insights into coronavirus entry. Adv Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang LF, Shi Z, Zhang S, Field H, et al. Review of bats and SARS. Emerg Infect Dis. 2006;12:1834–1840. doi: 10.3201/eid1212.060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Zhao S, Teng T, et al. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12:244. doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W, Moore MJ, Vasilieva N, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan R, Zhang Y, Li Y, Xia L, et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan Y, Cao D, Zhang Y, et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gui M, Song W, Zhou H, et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia S, Liu M, Wang C, et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gheblawi M, Wang K, Viveiros A, et al. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ortiz-Fernandez L, Sawalha AH. Genetic variability in the expression of SARS-CoV-2 host cell entry factors across populations. bioRxiv. 2020 doi: 10.1101/2020.04.06.027698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Belouzard S, Millet JK, Licitra BN, et al. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rabaan AA, Al-Ahmed SH, Haque S, et al. SARS-CoV-2, SARS-CoV, and MERS-COV: a comparative overview. Infez Med. 2020;28:174–184. [PubMed] [Google Scholar]
- 30.Hasöksüz M, Kiliç S, Saraç F. Coronaviruses and SARS-COV-2. Turk J Med Sci. 2020;50:549–556. doi: 10.3906/sag-2004-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, Zheng X, Tong Q, et al. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J Med Virol. 2020;92:491–494. doi: 10.1002/jmv.25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann M, Kleine-Weber H, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coutard B, Valle C, de Lamballerie X, et al. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmann M, Kleine-Weber H, Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020;78(4):779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li F, Berardi M, Li W, et al. Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. J Virol. 2006;80:6794–6800. doi: 10.1128/JVI.02744-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walls AC, Park YJ, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu C, Yang Y, Gao Y, et al. Viral architecture of SARS-CoV-2 with post-fusion spike revealed by Cryo-EM. bioRxiv. 10.1101/2020.03.02.972927.
- 38.Millet JK, Whittaker GR. Host cell proteases: citical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawase M, Shirato K, van der Hoek L, Taguchi F, Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J Virol. 2012;86:6537–6545. doi: 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKee DL, Sternberg A, Stange U, Laufer S, Naujokat C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol Res. 2020;157:104859. doi: 10.1016/j.phrs.2020.104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McFadyen JD, Stevens H, Peter K. The emerging threat of (micro)thrombosis in COVID-19 and its therapeutic implications. Circ Res. 2020;10:1161. doi: 10.1161/CIRCRESAHA.120.317447. [DOI] [PMC free article] [PubMed] [Google Scholar]