Abstract
Glioma is a common type of tumor in human central nervous system, and it is characterized with high mobility and mortality. The prognosis of patients with advanced glioma remains poor. Thus, it is necessary to develop novel therapeutic approaches for the treatment of this disease. Circular RNAs are a group of noncoding RNAs which have been detected in eukaryotic cells. They are tissue-specific and characterized with a more stable structure compared with linear RNAs. Recently, studies have revealed that certain circular RNAs are involved in biological processes such as gene regulation; however, the functions of most circular RNAs remain unknown and require further investigation. Furthermore, circular RNAs can act as “sponges” of its target microRNA, consequently suppressing their activity. Additionally, impaired expression of circular RNAs is reported in different diseases including cancer. In our study, low expression of circular RNA Scm like with 4 Mbt domains 2 was detected in glioma samples. Furthermore, reduced circRNA Scm like with 4 Mbt domains 2 expression was observed in human glioma cell lines compared to normal astrocyte cells. Additionally, overexpression of circRNA Scm like with 4 Mbt domains 2 suppressed the growth and metastasis of glioma cells in vitro. Moreover, microRNA-182-5p could be a downstream molecule of circRNA Scm like with 4 Mbt domains 2. The influenced of microRNA-182-5p-induced proliferation, migration, and invasion of glioma cells could be abrogated by overexpressed circRNA Scm like with 4 Mbt domains 2. In addition, metastasis suppressor 1 was predicted as a novel target of microRNA-182-5p, and its expression was restored by circRNA Scm like with 4 Mbt domains 2. In summary, our findings provided novel insight into the roles of circRNA Scm like with 4 Mbt domains 2 in glioma. More importantly, circRNA Scm like with 4 Mbt domains 2/microRNA-182-5p/metastasis suppressor 1 axis could be a putative therapeutic target for the treatment of patients with glioma.
Keywords: glioma, circular RNA SFMBT2, cell growth, miR-182-5p, MTSS1
Introduction
Glioma is a type of aggressive and prevalent malignancy in human brain, whose prognosis is unfavorable. It accounts for ∼80% of malignant neoplasia and ∼40% of primary tumors in central nervous system.1 Although the recent treatments have been remarkably improved using modern approaches such as targeted biologic therapy and adjuvant radiotherapy, the overall survival rate of patients with glioma still remains poor.2-4 Therefore, the development of novel therapeutic strategies against glioma is urgent.
Circular RNAs (circRNAs) are a novel class of noncoding RNAs. Unlike linear RNAs, they form a continuous loop and are characterized with a more stable structure.5,6 Due to the absence of free 5′- or 3′- ends in circRNAs, they are resistant to exonuclease-induced degradation and could be more stable compared with linear RNAs.7 Furthermore, the expression of circRNAs is tissue-specific.6 Although some circRNAs have been reported as potential gene regulators, the regulatory functions of most circRNAs remain largely unknown.8 Recent studies have revealed that circRNAs could be able to function as microRNA (miRNA) “sponges” which inhibit the activity of miRNAs competitively.9 In addition, circRNAs are involved in the pathogenesis of numerous types of diseases, such as nervous system disorders and cancer.10-12 Previous study has revealed the regulatory functions of a novel circRNA Scm like with 4 Mbt domains 2 (circ_SFMBT2) in gastric cancer, and it could act as a sponge for microRNA-182-5p (miR-182-5p) to control the growth of cancer cells13; however, the underlying mechanisms and putative downstream molecules of circ_SFMBT2 in glioma have not been completely elucidated.
MicroRNAs are noncoding RNAs with the length of ∼22 nt, which are novel downstream targets of other noncoding RNAs such as long non-coding RNAs (lncRNAs) and circRNAs.14,15 Previous studies have indicated that miRNAs function through binding to the 3′-untranslated region (3′-UTR) of corresponding messenger RNAs (mRNAs).16,17 Impaired expression levels of miRNAs have been observed in patients with cancer, which consequently contributing to tumor progression.18 Furthermore, miR-93 was capable of promoting the proliferation of glioma cells by regulating phosphatidylinositol 3 kinase/protein kinase B pathway.16 Elevated expression of H19 was observed in colorectal cancer, which may enhance cell proliferation through miR-675.14 Additionally, miR-140 and miR-152 interact with certain lncRNAs in glioma19,20; however, the detailed functions of miRNAs in glioma remain elusive and require further investigation. Among the abovementioned miRNAs, miR-182-5p is involved in the pathogenesis of gastric and bladder cancer.13,21 Moreover, miR-182 could regulate the growth and metastasis of numerous types of tumors through metastasis suppressor 1 (MTSS1)22-24; MTSS1 was first identified as a metastasis suppressor missing in metastatic bladder carcinoma cell lines, and it is associated with cancer progression or tumor metastasis in a variety of organ sites through interacting with the actin cytoskeleton.23,24
In the present study, the influences of circ_SFMBT2-modulated signaling pathway on the proliferation, migration, and invasion of glioma cells were elucidated. Our findings revealed that circ_SFMBT2 was downregulated in glioma tissues and cells. Overexpression of circ_SFMBT2 was able to suppress cell growth and in vitro invasion. In addition, miR-182-5p could be a putative downstream target of circ_SFMBT2. Furthermore, overexpressed circ_SFMBT2 abrogated the effects of miR-182-5p/MTSS1 signaling on promoting the growth and metastasis of glioma cells. In summary, our article explained the essential roles of circ_SFMBT2 during the development of glioma, which could provide novel insight for the treatment of patients with glioma.
Material and Methods
Clinical Specimen
A total of 28 matched tumor and noncancerous samples were obtained from patients with glioma, who underwent surgical resection at the First Affiliated Hospital of Jinzhou Medical University between May 2016 and June 2018. None of these patients had received other types of treatment prior to the surgery. The protocol was designed in accordance with the Helsinki declaration and was approved by the institutional review board of the First Affiliated Hospital of Jinzhou Medical University (approval no. JYD160203). Written informed consents were signed obtained from all the patients. Metastasis was detected in 9 cases, and 20 patients were diagnosed with grade I or II glioma. The clinicopathological features of participants are presented in Table 1. All the samples were snap-frozen immediately in liquid nitrogen, and then stored at −80 °C for further use.
Table 1.
Clinicopathological Parameters of Patients With Glioma Enrolled in This Study.a
| Parameters | n | SFMBT2 expression | P value | |
|---|---|---|---|---|
| Low (14) | High (14) | |||
| Gender | .415 | |||
| Male | 16 | 9 | 7 | |
| Female | 12 | 5 | 7 | |
| Age (years) | .484 | |||
| >60 | 15 | 8 | 7 | |
| ≤60 | 13 | 6 | 7 | |
| Tumor size (cm) | .443 | |||
| >5 | 14 | 8 | 6 | |
| ≤5 | 14 | 6 | 8 | |
| Tumor grade | .018b | |||
| I-II | 20 | 8 | 12 | |
| III-IV | 8 | 6 | 2 | |
| Smoking | .439 | |||
| Yes | 12 | 7 | 5 | |
| No | 16 | 7 | 9 | |
| Liver metastasis | .016b | |||
| Yes | 9 | 7 | 2 | |
| No | 19 | 7 | 12 | |
a Differences among variable were analyzed using the χ2 test.
b The values have statistically significant differences.
Cell Culture
Human glioma cell lines (D54, A172, and U251) as well as normal human astrocyte cells (A735) were all obtained from the American Type Culture Collection. Dulbecco’s Modified Eagle’s Medium (DMEM) was used to culture the cells, which also contained 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 µg/mL streptomycin (HyClone; GE Healthcare Life Science). All the cells were maintained at 37 °C in a humidified 5% CO2 atmosphere.
Cell Transfection
To establish the circ_SFMBT2 overexpression model, wild-type (WT; o/e-circ_SFMBT2) or mutant (o/e-NC) circ_SFMBT2 fragment was integrated into PLCDH-cir vector (Ribobio). The lentiviral vector was generated by Hanbio. Transfection was performed according to the manufacturer’s protocols. Glioma cells were selected using 0.5 µg/mL puromycin (Sigma-Aldrich) for 2 weeks. To generate the circ_SFMBT2 knockdown model, small interfering RNA (siRNA) sequences targeting circ_SFMBT2 (si-SFMBT2; GTCGGTGACTAAGCAATCAAA) and negative control (NC) were designed and synthesized by Genepharm Co Ltd. The mimic or inhibitor of miR-182-5p and the corresponding NC were purchased from Genepharm Co Ltd. The mimics, inhibitors (100 pM), and siRNA (50 nM) were transfected into glioma cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc) according to the manufacturer’s protocols. At 8 hours posttransfection, the culture medium was replenished with fresh DMEM containing 10% FBS. The transfection efficiency was confirmed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
Reverse Transcription-Quantitative Polymerase Chain Reaction
Reverse transcription-quantitative polymerase chain reaction was used to determine the levels of circ_SFMBT2, miR-182-5p, and MTSS1 in the experimental groups. MiRNA was extracted by miRNeasy Mini Kit (Qiagen), and miR-21 expression was determined using the TaqMan MicroRNA Assay kit (cat. no. 4427975; Applied Biosystems; Thermo Fisher Scientific, Inc). Quantitative polymerase chain reaction was subsequently performed using an Applied Biosystem 7500 PCR instrument (Applied Biosystems; Thermo Fisher Scientific, Inc). The following thermocycling conditions were used for qPCR: 95 °C for 10 minutes, followed by 40 cycles at 95 °C for 15 seconds and 60 °C for 1 minute. U6 small nuclear RNA was used as the internal control.
Total RNA from clinical specimens or cells was extracted by TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc) according to the manufacturer’s protocols. The concentration of extracted RNA was evaluated using NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Inc). Complementary DNA was synthesized by a PrimeScript RT kit (Takara Biotechnology Co, Ltd), and qPCR was performed using SYBR Green PCR Master Mix (TaKaRa Biotechnology Co, Ltd) according to the manufacturer’s protocols. Endogenous glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as controls to normalize the expression of mRNA.
The sequences of forward and reverse primer were as follows: circ_SFMBT2, 5′-GCGTCGGTGACTAAGCAATC-3′ and 5′-CCAATCCCACATAGCGAAGG-3′; miR-182-5p, 5′-TGCGGTTTGGCAATGGTAGAAC-3′ and 5′-CCAGTGCAGGGTCCGAGGT-3′; MTSS1, 5′-TCAAGAACAGATGGAAGAATGG-3′ and 5′-TGCGGTAGCGGTAATGTG-3′; GAPDH, 5′-GCAAGAGCACAAGAGGAAGA-3′ and 5′-ACTGTGAGGAGGGGAGATTC-3′ and U6, 5′-CTCGCTTCGGCAGCACATA-3′ and 5′-AACGATTCACGAATTTGCGT-3′. Polymerase chain reaction program used for the thermocycler was 95 °C for 5 minutes, followed by 45 cycles of 95 °C for 15 seconds, 60 °C for 20 seconds, and 72 °C for 10 seconds. Relative expression levels were calculated using 2−ΔΔCq method.25
Western Blotting
Total protein was extracted from tissues/cells by radioimmunoprecipitation assay buffer (Beyotime Institute of Biotechnology). The concentration of extracted protein was determined using bicinchoninic acid assay (Beyotime Institute of Biotechnology). Same amount (40 μg) of protein samples were separated using sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE). The protein was transferred onto a nitrocellulose membrane (EMD Millipore). Then, the membranes were blocked using tris-buffered saline containing 5% skimmed milk at room temperature for 2 hours, followed by overnight incubation using corresponding primary antibodies: MTSS1 (1:500; cat. no. sc-101204; Santa Cruz Biotechnology Inc) or GAPDH (1:1,000; cat. no. sc-47724; Santa Cruz Biotechnology Inc) at 4 °C. Next day, the membranes were further incubated in horseradish peroxidase–conjugated secondary antibody (1:5,000; cat. no. sc-2371; Santa Cruz Biotechnology Inc) at room temperature for 2 hours. The signals were visualized with an enhanced chemiluminescence protein detection kit (Pierce Biotechnology; Thermo Fisher Scientific, Inc), and then the blots were quantified using Image J (NIH).
Cell Counting Kit 8 Assay
Cells were harvested 24 hours following transfection. A total of 2 × 104 cells were seeded onto a 96-well plate. The proliferative activity of the cells was examined by Cell Counting Kit 8 (CCK-8) assay (Dojindo Molecular Technologies, Inc) at day 1, 2, 3, and 4 postinoculation. Generally, CCK-8 solution (10 µL) was added into each well at the corresponding time points. Following the incubation at 37 °C for additional 2 hours, the absorbance at 450 nm was evaluated using a microplate reader (Bio-Rad Laboratories, Inc).
Transwell Migration and Invasion Assay
The migration and invasion of cells were examined using a Transwell assay. For cell migration assay, a total of 2 × 105 cells were diluted using FBS-free medium and placed onto the upper chamber (BD Biosciences). For cell invasion assay, cells were added into the upper chamber which was precoated with Matrigel (Sigma-Aldrich). Then, 500 µL of culture medium containing 10% FBS was added into the lower chamber. Following overnight incubation, nonmigratory/-invasive cells were removed using a cotton swab. The migrated/invaded cells in the lower chamber were fixed using 4% paraformaldehyde and stained with 0.5% crystal violet. The numbers of cells were counted within 5 randomly selected fields by an inverted light microscope (magnification ×200, Olympus Corporation).
Northern Blotting
Total RNA was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc) according to the manufacturer’s protocols. Equal amount of RNA (30 μg) was loaded into a 15% Tris/Borate/EDTA (TBE)-urea gel and separated by a 15% urea-PAGE gel, and then transferred onto positively charged nylon membranes (GE Healthcare Life Sciences) and cross-linked with ultraviolet irradiation. The blots were hybridized with digoxignin (DIG)-labeled probe for miR-182-5p (Exiqon) at 42 °C overnight. Moreover, the membranes were washed in low-stringency buffer (2 × SSC containing 0.1% SDS). Then, the levels of miRNAs were determined using a DIG Luminescent Detection Kit (Roche). U6 was used as a loading control.
Bioinformatic Prediction and Luciferase Activity Assay
Targetscan and miRanda were employed to predict the novel targets of circ_SFMBT2 or miR-182-5p. MiR-182-5p was predicted as the novel downstream molecule of circ_SFMBT2 through the bioinformatics database Targetscan version 6.2 April 2012 Release (available at: www.targetscan.org). Metastasis suppressor 1 was identified as the potential direct target of miR-182-5p via miRanda version August 2010 Release (available at: www.microrna.org). Wild-type fragment of the 3′-UTR of circ_SFMBT2/MTSS1 with potential binding sites of miR-182-5p were obtained from Shanghai GenePharma Co, Ltd. They were integrated into pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega Corporation) according to the manufacturer’s protocols. circ_SFMBT2/MTSS1-3′-UTR-MUT reporter vector which contained the mutant miR-182-5p binding site was also generated using QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene). Furthermore, the corresponding vectors were used to cotransfect with miR-NC or miR-182-5p mimics into D54 cells. Subsequently, luciferase activity was examined 48 hours after transfection using Dual Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocols, and firefly luciferase activity was normalized to Renilla luciferase.
Statistical Analysis
All the data were presented as means ± standard deviation, and then analyzed by SPSS 17.0 (SPSS, Inc). The significance of differences between/among groups was compared using Student t test or 1-way analysis of variance (ANOVA). A student-Newman-Keuls test was carried out after ANOVA. The relationship between RNA expression levels was investigated using Spearman correlation analysis. P < .05 was considered to indicate a statistically significant difference.
Results
The Level of CircRNA Circ_SFMBT2 Is Decreased in Glioma Tissues/Cells
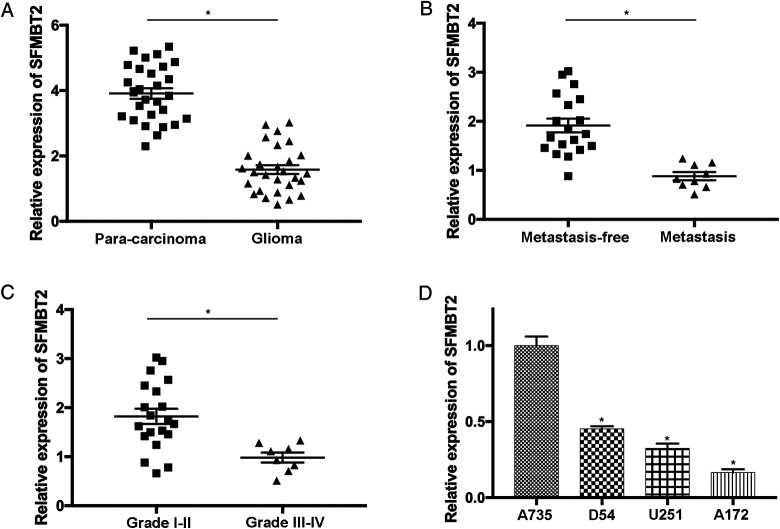
The expression of circ_SFMBT2 was determined in 28 glioma samples and paired noncancerous tissues using RT-qPCR. The results revealed significantly low expression of circ_SFMBT2 in glioma samples compared to the paracarcinoma tissues (P < .05; Figure 1A). Moreover, the association between circ_SFMBT2 level and the development of glioma was also studied. The data suggested that the expression of circ_SFMBT2 was notably reduced in patients with glioma with metastasis compared with the ones without metastasis (P < .05; Figure 1B). In addition, the expression level of circ_SFMBT2 was remarkably decreased in patients with advanced glioma (P < .05; Figure 1C), indicating that low expression of circ_SFMBT2 could be related to the development of this disease. By comparing to normal human astrocytes, significant reduction in circ_SFMBT2 was detected in glioma cells (P < .05; Figure 1D). In summary, the expression level of circ_SFMBT2 was notably reduced in glioma, which may result in the progression of tumor.
Figure 1.
The expression level of circ_SFMBT2 is significantly reduced in glioma tissues and cells. (A) The level of circ_SFMBT2 was evaluated in 28 glioma samples and paired noncancerous controls using reverse transcription-quantitative polymerase chain reaction. (B) The expression of circ_SFMBT2 was determined in patients with glioma with or without metastasis. (C) circ_SFMBT2 expression was examined in patients with glioma with different grades. (D) The level of circ_SFMBT2 in normal human astrocyte (A735) and glioma cells (D54, A172 and U251) was analyzed. *P < .05. SFMBT2, circular RNA Scm like with 4 Mbt domains 2.
Overexpressed Circ_SFMBT2 Suppresses the Growth and Metastasis of Glioma Cells
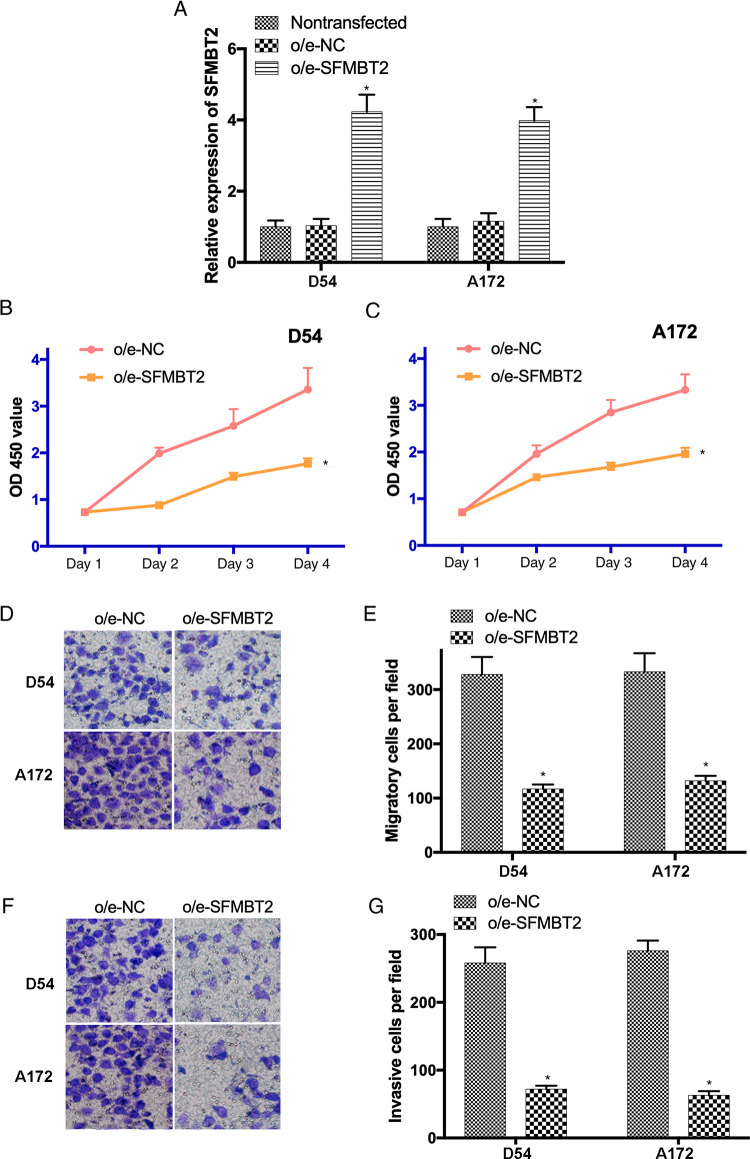
In order to investigate the effects of circ_SFMBT2 on the biological behavior of glioma cells, circ_SFMBT2 was ectopically overexpressed in D54 and A172 cells using lentiviral vectors. The transfection efficiency was evaluated by RT-qPCR (Figure 2A). In addition, CCK8 assay revealed that the proliferative activity of D54 and A172 cells transfected with o/e-circ_SFMBT2 was notably suppressed compared to the o/e-NC group (P < .05; Figure 2B and C). Furthermore, Transwell assay indicated that the migration and invasion of o/e-circ_SFMBT2-transfected cells was remarkably inhibited (P < .05; Figure 2D-G). Our findings suggested that the growth and metastasis of glioma cells could be suppressed by the overexpression of circ_SFMBT2.
Figure 2.
The growth and metastasis of D54 and A172 cells are inhibited by overexpressed SFMBT2. (A) Transfection efficiency of o/e-SFMBT2 was examined by reverse transcription-quantitative polymerase chain reaction. (B and C) The proliferation of transfected glioma cells was determined by Cell Counting Kit-8 assay. (D and E) The migrating ability of transfected D54 and A172 cells was evaluated using a Transwell assay (magnification ×200). (F and G) The invasive activity of glioma cells after the transfected with o/e-SFMBT2 or o/e-NC was examined (magnification ×200). *P < .05. NC indicates negative control. SFMBT2, circular RNA Scm like with 4 Mbt domains 2.
MicroRNA-182-5p Is the Putative Downstream Molecule of Circ_SFMBT2 in Glioma
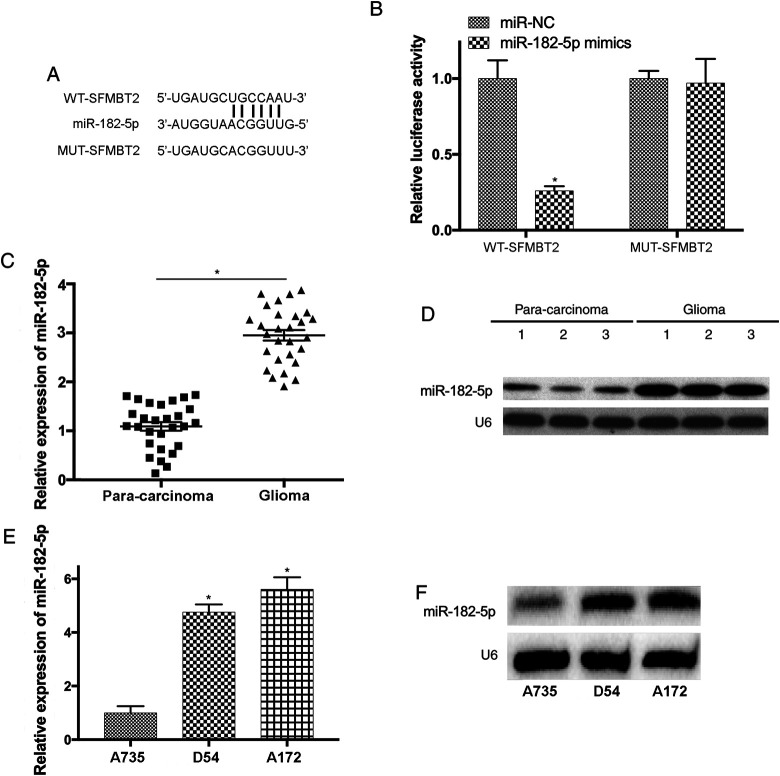
To study whether circ_SFMBT2 is a potential tumor suppressor in glioma and its functions through targeting downstream miRNAs, the complementary binding sites between miR-192-5p and circ_SFMBT2 were predicted by bioinformatics analysis (Figure 3A). The association between circ_SFMBT2 and miR-182-5p was also examined using luciferase activity assay. Luciferase reporters containing WT (circ_SFMBT2-WT) and mutant (circ_SFMBT2-MUT) sequence of predicted miR-182-5p binding sites were generated. Our results suggested that overexpressed miR-182-5p significantly reduced the activity of circ_SFMBT2-WT luciferase plasmid by comparing to the control (P < .05; Figure 3B). In addition, the data of RT-qPCR and northern blotting indicated that miR-182-5p was upregulated in glioma samples (P < .05; Figure 3C and D). Moreover, upregulation of miR-182-5p was detected in D54 and A172 cells (P < .05; Figure 3E and F).
Figure 3.
MiR-182-5p is a novel downstream molecule of SFMBT2 in glioma. (A) The complementary binding sites between miR-182-5p and SFMBT2 transcript were predicted. (B) Overexpressed miR-182-5p lead to significantly decreased luciferase activity of SFMBT2-WT, but not in SFMBT2-MUT. (C and D) The level of miR-182-5p was notably elevated in glioma samples. (E and F) The expression of miR-182-5p in D54 and A172 cells was upregulated. *P < .05. MiR indicates microRNA; miR-182-5p, microRNA-182-5p; MUT, mutant; SFMBT2, circular RNA Scm like with 4 Mbt domains 2; WT, wild-type.
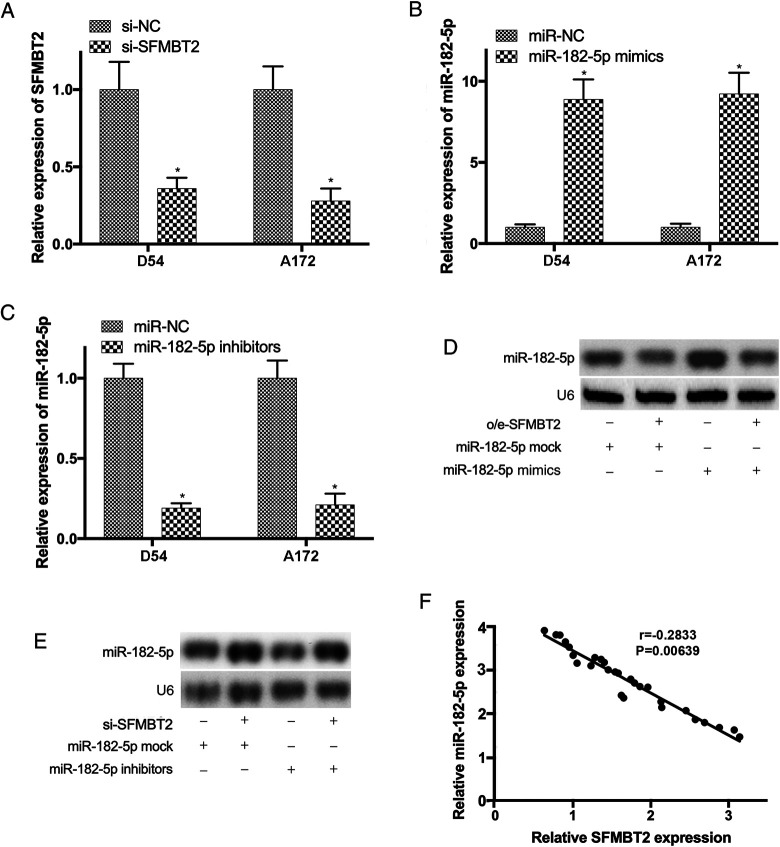
To further elucidate the effects of circ_SFMBT2 on the expression levels of miR-182-5p, D54, and A172 cells were cotransfected with o/e-circ_SFMBT2 and miR-182-5p mimics, or with sh-circ_SFMBT2 and miR-182-5p inhibitors, respectively. The transfection efficiencies were determined by RT-qPCR (Figure 4A-C). Northern blotting also revealed that the upregulation of miR-182-5p in D54 cells transfected with miR-182-5p mimics was abrogated by o/e-circ_SFMBT2 (Figure 4D). Vice versa, low expression of miR-182-5p in D54 cells transfected with miR-182-5p inhibitors was reversed following cotransfection with sh-circ_SFMBT2 (Figure 4E). Moreover, the association between circ_SFMBT2 and miR-182-5p was further investigated using Spearman correlation analysis, and the results revealed that the levels of miR-182-5p and circ_SFMBT2 were negatively correlated in glioma samples (Figure 4F).
Figure 4.
The expression level of miR-182-5p was downregulated by SFMBT2. (A-C) The efficiencies of D54 and A172 cells transfected with sh-SFMBT2, miR-182-5p mimics, or inhibitors were confirmed. (D) The increase of miR-182-5p in glioma cells transfected with miR-182-5p mimics was reversed by o/e-SFMBT2. (E) The reduction of miR-182-5p in cells transfected with miR-182-5p inhibitors was abrogated by sh-SFMBT2. (F) The negative correlation between miR-182-5p and SFMBT2 in glioma tissues was revealed by Spearman correlation analysis (r = −0.2833; P = .00639). *P < .05. MiR indicates microRNA; miR-182-5p, microRNA-182-5p; SFMBT2, circular RNA Scm like with 4 Mbt domains 2.
MicroRNA-182-5p Serves Key Roles on the Growth and Metastasis of Glioma Cells
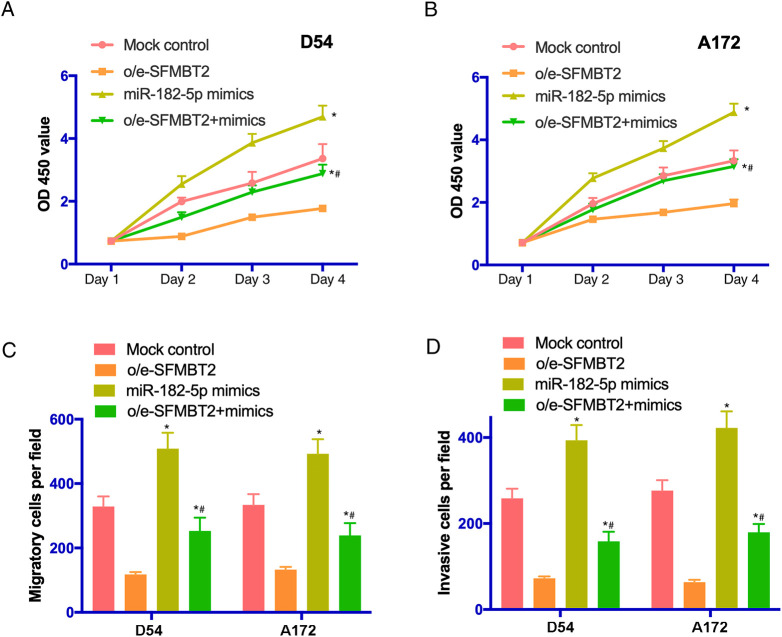
The influences of miR-182-5p on the proliferation, migration, and invasion of glioma cells were also studied. The results of CCK8 assay suggested that the proliferative activity of cells was notably enhanced following the transfection with miR-182-5p mimics (P < .05; Figure 5A and B). Furthermore, the migration of D54 and A172 cells was promoted in miR-96-5p mimics group (P < .05; Figure 5C). Additionally, the invasive ability of glioma cells was increased by miR-182-5p mimics (P < .05; Figure 5D). Taken all together, our findings indicated that miR-182-5p was capable of inducing the growth and metastasis of glioma cells.
Figure 5.
The proliferation, migration, and invasion of glioma cells were enhanced by miR-182-5p. (A and B) The proliferative activity of D54 and A172 cells transfected with mock control, o/e-SFMBT2, miR-182-5p mimics or cotransfected with o/e-SFMBT2 and miR-182-5p mimics was examined by Cell Counting Kit-8 assay. (C) The migrating ability of transfected glioma cells was evaluated using a Transwell assay. (D) The invasion of D54 and A172 cells was remarkably promoted following the treatment with miR-182-5p mimics. *P < .05 versus mock control. *# versus o/e-SFMBT2. MiR indicates microRNA; miR-182-5p, microRNA-182-5p; SFMBT2, circular RNA Scm like with 4 Mbt domains 2.
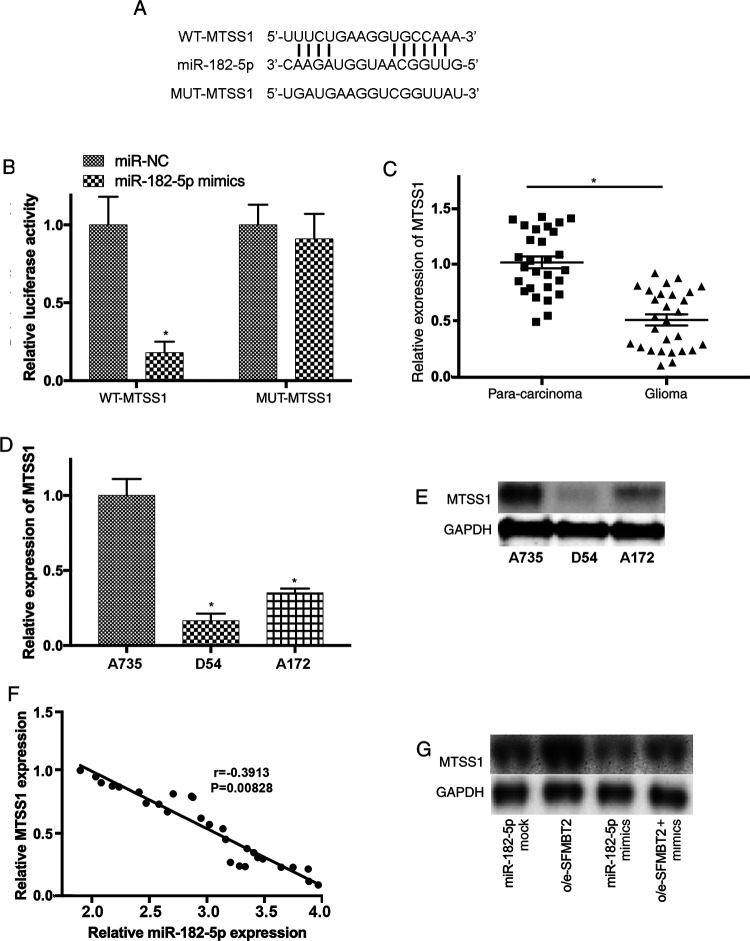
Metastasis Suppressor 1 Is a Novel Target of miR-182-5p
In order to investigate the potential downstream molecule of miR-182-5p, the complementary sequence between miR-182-5p and MTSS1 transcripts was predicted (Figure 6A). The association of MTSS1 and miR-182-5p was further elucidated using luciferase assay. Luciferase reporter vectors carrying the WT (MTSS1-WT) and mutant (MTSS1-MUT) sequence of predicted miR-182-5p binding sites were constructed. Our results revealed that overexpression of miR-182-5p remarkably reduce the activity of luciferase plasmids containing WT MTSS1 sequence but not in the one with mutant (Figure 6B). Furthermore, RT-qPCR indicated that the level of MTSS1 was downregulated in glioma samples (P < .05; Figure 6C). In addition, downregulated MTSS1 was detected in D54 and A172 cells (P < .05; Figure 6D and E). Moreover, the expression of MTSS1 and miR-182-5p was inversely correlated with glioma samples (Figure 6F).
Figure 6.
MTSS1 is a potential target of miR-182-5p in glioma. (A) The complementary sequences of miR-182-5p on the transcripts of MTSS1 were predicted. (B) The association between MTSS1 and miR-182-5p was further confirmed using luciferase reporter assay. (C) The level of MTSS1 was notably decreased in glioma samples. (D and E) The expression of MTSS1 was downregulated in D54 and A172 cells. (F) The expression levels of MTSS1 and miR-182-5p were inversely correlated with glioma samples (r = −0.3913; P = .00828). (G) MTSS1 expression was significantly reduced by miR-182-5p mimics and upregulated by overexpression of SFMBT2 in D54 cells. MiR indicates microRNA; miR-182-5p, microRNA-182-5p; MTSS1, metastasis suppressor 1; SFMBT2, circular RNA Scm like with 4 Mbt domains 2.
Our previous findings suggested that MTSS1 could inhibit the migration and invasion of glioma cells by targeting CTTN.26 In order to further investigate MTSS1-modulated signaling pathways associated with the progression of glioma, the expression level of MTSS1 was examined in D54 cells following the transfection with mock control, o/e-circ_SFMBT2, miR-182-5p mimics or cotransfected with o/e-circ_SFMBT2, and miR-182-5p mimics. Significantly low expression of MTSS1 was observed after the treatment with miR-182-5p mimics, while its expression level was restored by overexpression of circ_SFMBT2 in glioma cells (Figure 6G). These findings suggested that MTSS1 could be a novel target of miR-182-5p in glioma. More importantly, circ_SFMBT2/miR-182-5p/MTSS1 axis could serve essential roles during the development of glioma.
Discussion
In the present study, low expression of circRNA SFMBT2 was detected in glioma tissues and cells. Further experiments were performed to investigate the downstream molecules of circ_SFMBT2 in glioma. The results indicated that circ_SFMBT2 binds to miR-182-5p, whose expression was remarkably increased in glioma tissues and cells. Moreover, the level of miR-182-5p was downregulated by circ_SFMBT2 in glioma cells, and their expression levels were inversely correlated in glioma samples. Additionally, reduction in miR-182-5p lead to suppressed growth and metastasis of glioma. However, controversial findings have been reported in previous study as circ_SFMBT2 promoted the growth of gastric cancer cells by targeting miR-182-5p,13 suggesting that circ_SFMBT2 may have both oncogenic and tumor-suppressor functions.
Moreover, MTSS1 was identified as the downstream target of miR-182-5p. Further study also indicated that MTSS1 was downregulated in glioma tissues and cells, and the level of MTSS1 could be reduced by miR-182-5p mimics and restored by overexpressed circ_SFMBT2, respectively. The expression levels of MTSS1 have been previously evaluated in different types of tissues including prostate, spleen, and thymus, and low expression of MTSS1 was detected in gastric, breast, and bladder cancer,27-29 which could lead to unfavorable survival rate in the patients. In consistence with our findings, miR-182 is able to regulate the growth and metastasis of numerous types of cancer cells including esophageal, hepatocellular, and prostate cancer.22-24 In summary, the findings of present study suggested the substantial roles of circ_SFMBT2/miR-182-5p/MTSS1 axis on the development of glioma.
Taken all together, our data revealed that circ_SFMBT2 was a putative tumor suppressor in glioma that could induce the upregulation of MTSS1 by suppressing miR-182-5p, consequently inhibiting the progression of glioma. However, there were some limitations in this study. For example, the levels of proliferation-associated molecules such as BrdU, proliferating cell nuclea antigen (PCNA), and Ki-67 could be evaluated to confirm the existing findings of CCK-8 assay. In addition, MTSS1 knockdown model could be generated to evaluate the effects of MTSS1 on circ_SFMBT2-regulated cell growth and migration. These findings indicated the essential effects of circ_SFMBT2 on tumorigenesis and elucidated the potential mechanisms underlying the proliferation, migration, and invasion of glioma cells, which could provide important evidences on the regulatory functions of circ_SFMBT2 during the progression of glioma. More importantly, circ_SFMBT2/miR-182-5p/MTSS1 axis could be a novel therapeutic target for the treatment of patients with glioma.
Abbreviations
- ANOVA
analysis of variance
- CCK-8
Cell Counting Kit 8
- circRNA
circular RNAs
- circ_SFMBT2
circRNA Scm like with 4 Mbt domains 2
- DMEM
Dulbecco’s Modified Eagle’s Medium
- FBS
fetal bovine serum
- lncRNA
long non-coding RNA
- miR-182-5p
microRNA-182-5p
- miRNA
microRNA
- mRNA
messenger RNA
- MTSS1
metastasis suppressor 1
- NC
negative control
- PAGE
polyacrylamide gel electrophoresis
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- siRNA
small interfering RNA
- 3′-UTR
3′-untranslated region
Footnotes
Authors’ Note: W.G. initiated the present study. S.Z. and W.G. completed all the experiments and interpreted the data. Both authors drafted and approved the final manuscript. The data sets generated or analyzed during the present study are included in this published article. The present study was approved by the Ethics Committee of the First Affiliated Hospital of Jinzhou Medical University (Jinzhou, China). Written informed consents were obtained from all patients for the purpose of clinical use.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The present study was funded by the Natural Science Fund Program of Liaoning Province (grant no. 2019-ZD-0835).
ORCID iD: Wenshi Guo  https://orcid.org/0000-0002-4803-0626
https://orcid.org/0000-0002-4803-0626
References
- 1. Lai NS, Wu DG, Fang XG, Lin YC, Chen SS, Li ZB, Xu SS. Serum microRNA-210 as a potential noninvasive biomarker for the diagnosis and prognosis of glioma. Br J Cancer. 2015;112(7):1241–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sathornsumetee S, Rich JN. New treatment strategies for malignant gliomas. Expert Rev Anticancer Ther. 2006;6(7):1087–1104. [DOI] [PubMed] [Google Scholar]
- 3. Mansouri A, Mansouri S, Hachem LD, et al. The role of 5-aminolevulinic acid in enhancing surgery for high-grade glioma, its current boundaries, and future perspectives: a systematic review. Cancer. 2016;122(16):2469–2478. [DOI] [PubMed] [Google Scholar]
- 4. Delgado-López PD, Corrales-García EM. Survival in glioblastoma: a review on the treatment modalities. Clin Transl Oncol. 2016;18(11):1062–1071. [DOI] [PubMed] [Google Scholar]
- 5. Salzman J, Gawad C, Wang P, Lacayo N, Brown P. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;4 9:5333–5338. [DOI] [PubMed] [Google Scholar]
- 7. Vicens Q, Westhof E. Biogenesis of circular RNAs. Cell. 2014;159(1):13–14. [DOI] [PubMed] [Google Scholar]
- 8. Conn S, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. [DOI] [PubMed] [Google Scholar]
- 9. Hansen T, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. [DOI] [PubMed] [Google Scholar]
- 10. You X, Vlatkovic I, Babic A, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18(4):603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li J, Song Y. Circular RNA GLI2 promotes osteosarcoma cell proliferation, migration, and invasion by targeting miR-125b-5p. Tumour Biol. 2017;39(7):1010428317709991. [DOI] [PubMed] [Google Scholar]
- 12. Yao Z, Luo J, Hu K, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11(4):422–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sun H, et al. Circ-SFMBT2 promotes the proliferation of gastric cancer cells through sponging miR-182-5p to enhance CREB1 expression. Cancer Manag Res. 2018;10:5725–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tsang WP, Ng EK, Ng SS, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31(3):350–358. [DOI] [PubMed] [Google Scholar]
- 15. Zhao X, Sun J, Chen Y, et al. LncRNA PFAR promotes lung fibroblast activation and fibrosis by targeting miR-138 to regulate the YAP1-Twist axis. Mol Ther. 2018;26(9):2206–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ohta K, Hoshino H, Wang J, et al. MicroRNA-93 activates c-Met/PI3K/Akt pathway activity in hepatocellular carcinoma by directly inhibiting PTEN and CDKN1A. Oncotarget. 2015;6(5):3211–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cimmino A, Calin GA, Fabbri M, et al. Mir-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2015;6(5):3211–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liang Z, Feng Q, Xu L, Li S, Zhou L. CREPT regulated by miR-138 promotes breast cancer progression. Biochem Biophys Res Commun. 2017;493(1):263–269. [DOI] [PubMed] [Google Scholar]
- 19. Zhao H, Peng R, Liu Q, et al. The lncrna H19 interacts with miR-140 to modulate glioma growth by targeting iASPP. Arch Biochem Biophys. 2016;610:1–7. [DOI] [PubMed] [Google Scholar]
- 20. Chen L, Wang Y, He J, Zhang C, Chen J, Shi D. Long non-coding RNA H19 promotes proliferation and invasion in human glioma cells by downregulating miR-152. Oncol Res. 2018;26(9):1419–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xie F, Li Y, Wang M, et al. Circular RNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis. Mol Cancer. 2018;17(1):144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang Q, Ren Y, Cheng J, Qin J. Effect of miR-182 targeting MTSS1 on the proliferation and metastasis of esophageal cancer. Int J Clin Exp Pathol. 2016;9(11):10871–10877. [Google Scholar]
- 23. Wang J, Li J, Shen J, Wang C, Yang L, Zhang X, et al. MicroRNA-182 downregulates metastasis suppressor 1 and contributes to metastasis of hepatocellular carcinoma. BMC Cancer. 2012;12:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hirata H, et al. MicroRNA-182-5p promotes cell invasion and proliferation by downregulating FOXF2, RECK and MTSS1 genes in human prostate cancer. PLoS One. 2013;8(1):e55502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCq method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- 26. Zhang S, Qi Q. MTSS1 suppresses cell migration and invasion by targeting CTTN in glioblastoma. J Neurooncol. 2015;121(3):425–431. [DOI] [PubMed] [Google Scholar]
- 27. Lee YG, Macoska JA, Korench, uk S, Pienta K. A potential metastasis suppressor gene in bladder cancer. Neoplasia. 2002;4(4):291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu K, Wang G, Ding H, Chen Y, Yu G, Wang J. Downregulation of metastasis suppressor 1 (MTSS1) is associated with nodal metastasis and poor outcome in Chinese patients with gastric cancer. BMC Cancer. 2010;10:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parr C, Jian, g WG. Metastasis suppressor 1 (MTSS1) demonstrates prognostic value and anti-metastatic properties in breast cancer. Eur J Cancer. 2009;45:1673–1683. [DOI] [PubMed] [Google Scholar]