Abstract
Serratia marcescens, rod-shaped Gram-negative bacteria is classified as an opportunistic pathogen in the family Enterobacteriaceae. It causes a wide variety of infections in humans, including urinary, respiratory, ocular lens and ear infections, osteomyelitis, endocarditis, meningitis and septicemia. Unfortunately, over the past decade, antibiotic resistance has become a serious health care issue; the effective means to control and dissemination of S. marcescens resistance is the need of hour. The whole genome sequencing of S. marcescens FGI94 strain contains 4434 functional proteins, among which 690 (15.56%) proteins were classified under hypothetical. In the present study, we applied the power of various bioinformatics tools on the basis of protein family comparison, motifs, functional properties of amino acids and genome context to assign the possible functions for the HPs. The pseudo sequences (protein sequence that contain ≤100 amino acid residues) are eliminated from the study. Although we have successfully predicted the function for 483 proteins, we were able to infer the high level of confidence only for 108 proteins. The predicted HPs were classified into various classes such as enzymes, transporters, binding proteins, cell division, cell regulatory and other proteins. The outcome of the study could be helpful to understand the molecular mechanism in bacterial pathogenesis and also provide an insight into the identification of potential targets for drug and vaccine development.
Keywords: Serratia marcescens, hypothetical proteins, functional annotation, potential targets
INTRODUCTION
Enterobacteriaceae family consists of more than 250 published bacterial species and Serratia is one of the most clinically important genera of the family (Alnajar and Gupta, 2017). The bacteria belonging to the Enterobacteriaceae are most commonly encountered organisms which are isolated from water, soil and clinical specimens. Serratia species is frequently found associated with animals and plants either detrimental or beneficial. Among the genus, Serratia marcescens (S. marcescens), Gram-negative bacteria have been recognized as a crucial cause of healthcare associated infections (nosocomial infections) in humans (Parente et al., 2016). Several strains of S. marcescens have been reported till date, and the complete genome sequence of Serratia strain FGI94 has been sequenced and its 16S rRNA has shown 99% highest nucleic acid identity with the pathogenic strain Serratia rubidaea JCM1240 (Aylward et al., 2013). In recent times, the majority of the healthcare associated urinary, respiratory, eye infections, osteomyelitis, wound infections, endocarditis and pulmonary infections were reported by S. marcescens (Padmavathi et al., 2014). Resistance to antimicrobial agents through intrinsic and acquired process is a notable feature in S. marcescens. A wide variety of gene cassettes containing resistance were identified in the chromosome and plasmids of S. marcescens. Certain strains of S. marcescens have shown resistance to antibiotics (Gentamicin, Cephalosporins, Fluoroquinolone, Cefotaxime and Ceftazidime) which already complicates the antibiotic therapy (Iguchi et al., 2014).
According to the World Health Organization (WHO) report 2018, Serratia spp. were classified under class-1 priority. The categorization was based on the antimicrobial resistance (AMR) mechanism of the pathogens (WHO Priority Pathogens List for R&D of New Antibiotics, 2018). AMR has emerged as a serious threat to the public health authorities at global level, particularly in intensive care and surgical units. Resistance leads to the long-term medication, higher medical cost, prolonged hospitalization, and on the severity, leading to death (Vazirianzadeh et al., 2013). The rapid emergence of antibiotic resistance is occurring worldwide as at least 2 million people acquire resistance each year and around 23,000 deaths were documented by CDC Report (CDC—Antibiotic Resistant Threats Report, 2013). According to the WHO report, AMR is one of the three major challenging problems of the human race (WHO Antimicrobial resistance, 2018). Global distribution of resistance plasmids among several bacterial species is alarming the entire world on AMR (Finley et al., 2013).
The recent technological innovations in the Next Generation Sequencing (NGS) generates larger amount of genomic data from wide range of bacterial species. However, very limited bacterial species have their complete proteome information, but sound knowledge of the microbial proteome is essential to understand the disease pathogenesis, virulent determinants, and their survival and propagation (Singh et al., 2015). For instance, in most bacterial species, nearly 30–40% of genes within in the genomes are marked as unknown or hypothetical (Hoskeri et al., 2010). Proteins with unknown function or conserved putative proteins, which are showing limited connection with functionally annotated proteins, are termed as Hypothetical Proteins (HPs). HPs are translated portions of nucleic acid sequences based on their sequence similarity, but for experimental existence functional and biochemical characterization has to be evaluated (Shahbaaz et al., 2013). Moreover, HPs also includes the low identity proteins, imprecisely described and vague functional proteins (Hoskeri et al., 2010). In general, HPs are classified into two major classes: (i) Uncharacterized Protein Families (UPFs), and (ii) Domains of Unknown Function (DUFs). The ones whose protein structures are available but not been functionally characterized or linked to any known functional gene, are referred as UPFs. Whereas the ones whose experimental existence of protein is available although not related to any known functional or structural domain, are represented as DUFs (Varma et al., 2015). Annotating the HPs have many advantages such as, deriving new structure-functional relationship, novel protein structures, and also helps in decoding additional pathways for better understanding of the pathogens. Functionally annotated HPs may serve as potential biomarkers for the screening and diagnostics of diseases. They can also be used as a pharmacological target to design and discover novel drugs (Singh et al., 2015). Function of the protein can be predicted using various strategies, such as phylogenetic profiling, mass spectrometry identification, SAGE analysis, lethal analysis, etc., (Varma et al., 2015; Gasperskaja and Kučinskas, 2017). In order to minimize the time and investment cost, various computational tools were developed to aid the functional annotation process (Hawkins and Kihara, 2007).
In the current study, we have used the power of computational tools to annotate the possible functions for the HPs in S. marcescens. Functional predictions of HPs from various bacterial genomes (V. cholera, N. gonorrhoeae, C. difficile, S. aureus, M. tuberculosis, H. influenza) through computational approaches were widely used in successful proteome annotation (Shahbaaz et al., 2013; Singh et al., 2015; Costa et al., 2018). Deciphering the function of entire gene coding regions in the genome is essential to merge the gaps in proteome to fully understand the pathogenicity and genome plasticity of S. marcescens.
MATERIALS AND METHODS
Sequence Retrieval
The entire protein sequences of S. marcescens FGI94 were identified by searching in NCBI (http://www.ncbi.nlm.nih.gov/) database. S. marcescens genome possesses 4434 protein-coding genes out of which 690 proteins were marked as HPs. All the HPs sequences were retrieved for the biological functional assignment using various functional annotation servers. In order to minimize the misinterpretations in functional annotation pipeline, pseudo genes (proteins having less than 100 amino acid residues) were excluded in this study. The various tools used in the functional predictions of HPs in S. marcescens are tabulated in Table 1.
Table 1. .
List of in silico servers and tools used for the function annotation of HPs
Physicochemical Characterization
Physicochemical characterization of the HPs was performed using ExPASy proteomics tools (Gasteiger et al., 2005). Parameters such as molecular weight (Mw), theoretical isoelectric point (pI), grand average of hydropathicity (GRAVY), aliphatic and instability index were computed for the 483 HPs in S. marcescens.
Protein Localization
Localization of the HPs was predicted using various tools including PSORTb (Yu et al., 2010), PSLpred (Bhasin et al., 2005) and CELLO (Yu et al., 2004). The signal peptide was predicted using SignalP server (Petersen et al., 2011). SecretomeP (Bendtsen et al., 2005) was used to identify the presence of HPs in non-classical secretory pathway. Transmembrane information about the HPs was predicted using TMHMM (Krogh et al., 2001) and HMMTOP (Tusnády and Simon, 1998) servers.
Domain Identification and Functional Annotation
Functions of the HPs were predicted using the web-based tools. NCBI-Blast (Altschul et al., 1990), SMART (Letunic et al., 2012), EBI-Interproscan (Hunter et al., 2011) and Motif (Kanehisa, 1997) were used to identify the functional domains/motifs present in the HPs. Family level classifications of the HPs were identified using Pfam (Finn et al., 2014), SCOP-Superfamily (Gough et al., 2001) and PANTHER (Thomas et al., 2006) families. Structural level classification of protein super families was predicted using CATH database (Orengo et al., 1997). Domain architecture of the HPs was predicted using SVMProt (Cai, 2003), CDART (Geer et al., 2002) and ProtoNet tools (Rappoport et al., 2012). Virulence activity of the HPs was predicted using Virulentpred (Garg and Gupta, 2008) and VCIM-pred (Saha and Raghava, 2006).
RESULTS AND DISCUSSION
Sequence Retrieval and Physicochemical Characterization
483 HPs were selected, which are having more than 100 amino acids residues for the physicochemical characterization and functional annotation. On the basis of amino acid sequences, predicted physicochemical properties of the 483 HPs were tabulated in Supplementary Table 1. Molecular weight is an important criterion in protein functional characterization. It can be seen that the proteins AGB82318.1 and AGB83651.1 have shown the lowest and highest Mw of 106 48.04 and 137 161.31 Da respectively. Isoelectric point (pI) is the pH at which the proteins have no net charge and the mobility based on charge will be zero. Prediction of pI is essential in the development of buffer system for purification and in isoelectric focusing. The predicted pI value of the HPs ranged from 3.53 to 11.83. Out of 483 HPs, 246 proteins were observed to be acidic and the predicted pI values ranged from 3.53 to 6.97. Similarly, 236 proteins have shown the pI values between 7.01 and 11.83, are considered basic and AGB83651.1 has shown neutral charge. Extinction co-efficient of the HPs were measured in water at 280 nm based on the concentration of Cystine, Tryptophan and Tyrosine amino acid residues in the protein sequences. Higher occupancy of these amino acid residues in the protein results in higher extinction co-efficient values. Small proteins with 100 plus amino acid residues containing minimal number of Cystine, Tryptophan and Tyrosine residues have shown very less extinction co-efficient values. Especially, AGB81485.1 and AGB84113.1 have not shown the extinction co-efficient values due to absence of Cystine, Tryptophan and Tyrosine amino acid residues. Computing the extinction co-efficient values helps in the quantitative analysis of protein-protein and protein-ligand interactions for the drug development process. Instability index of the HPs were found to be minimum value of –0.24 to the maximum of 119.78. Instability index value illustrates the stability of the proteins in test tube environment. The 228 HPs having the instability index values greater than 40 are classified as unstable and 225 proteins having the instability index less than 40 are classified as stable. Aliphatic amino acid residues present in the protein are directly proportional to the aliphatic index. Proteins with higher aliphatic index have shown higher thermal stability, especially in globular proteins. The computed aliphatic index of the 483 HPs falls between 38.03 and 157.52. The stability of the thermophilic proteins are mainly contributed by the aliphatic amino acids (A, V, I and L), which results in higher aliphatic index values to withstand the wide range of temperatures. GRAVY value represents the protein-water interactions and it illustrates the hydrophilic nature of the protein. The GRAVY values are computed based on the average sum of the hydrophilic and hydrophobic side chains in the amino acids. Least GRAVY value among the HPs is found to be –1.478 and the highest is 1.238.
Subcellular Localization
Proteins were characterized as drug and vaccine targets are mainly based on the subcellular localization. Proteins located in cytoplasmic matrix are capable to be potential drug targets, whereas both inner and outer membrane proteins can possibly act as potential vaccine targets. Crucial step in determining the function of the proteins is underlying on the knowledge of localization. Based on the knowledge of trained data sets, the HPs were predicted for their presence in any of these locations (cytoplasm, periplasm, extracellular, inner membrane and outer membrane) in the cell. We predicted the localization of 356 HPs out of 483 proteins subject for the study based on the concurrence hits. Among the predicted 356 HPs, 60% (214) of the proteins were shown to be present in Cytoplasm. Presence of HPs in inner membrane and outer membrane is 17.41% (62) and 2.52% (9) respectively. It is predicted that 14% (50) HPs are present in periplasm and 5.89% (21) in extra cellular matrix. However, locations of 127 proteins were not confirmed due to absence of concurrence results. Signal peptides are the key players in determining the transport of proteins to the target location. Hence, prediction of signal peptide is essential to know the transport system of the particular proteins and the cleavage sites. Apart from the certain cytoplasmic proteins, all other proteins have the signal peptides to facilitate the transport of proteins in and out of the membrane to reach the target cellular location or organelles. We predicted the presence of signal peptide sequences in 116 HPs out of 483. Similarly, 142 proteins were predicted to be involved in non-classical secretory pathway. Proteins which are secreted outside by the cells are secretomes, and these proteins are important to maintain the cell-cell communications, cell proliferation and pathogenesis. Membrane proteins are involved in various processes such as transport, signaling and energy transduction; hence half of the targets used for drug development are membrane proteins. The predicted 136 proteins have shown transmembrane helices from TMHMM server and 235 proteins from HMMTOP server. Prediction of membrane proteins is vital for the better understanding of drug targets to develop potent drug molecules. The details of the 483 HPs were shown in Supplementary Table 2.
Functional Annotation
Annotating the function of HPs is essential for the better understanding of the entire biological system towards the development of effective drugs. We have analyzed 483 HPs of S. marscences to predict their possible functions. Further, HPs were analyzed for the presence of functional domains and signature motifs in context with the biological function. The results of the domain and motif analysis are shown in Supplementary Table 3. Results of the sequence and structural features of the HPs were shown in the Supplementary Table 4. We have successfully annotated the function of 108 HPs with high confidence out of the 483 proteins. The list of 108 proteins with their functions assigned is illustrated in Table 2. Based on the analysis, only 22.36% of HPs functions were predicted and the remaining HPs have not shown concurrence results, which indicates that suitable experimental strategy has to be coupled for better functional assignment. Annotated proteins (Fig. 1) were majorly classified into the following six categories such as, enzymes, binding proteins, cell division proteins, cell regulatory proteins, transporters and the remaining proteins involved in different biological process.
Table 2. .
List of 108 proteins function assigned using the in silico analysis and its general classification
| S. No. | Accession number | Protein function |
|---|---|---|
| ENZYMES | ||
| 1. | AGB80450.1 | O-ACETYLTRANSFERASE |
| 2. | AGB80553.1 | THIOESTERASE FAMILY PROTEIN |
| 3. | AGB80565.1 | ACYL-COA N-ACYLTRANSFERASES (NAT) |
| 4. | AGB80880.1 | HNH ENDONUCLEASE |
| 5. | AGB80945.1 | L-RHAMNOSEMUTAROTASE |
| 6. | AGB81206.1 | ENDONUCLEASE/EXONUCLEASE/PHOSPHATASE FAMILY PROTEIN |
| 7. | AGB81233.1 | L,D-TRANSPEPTIDASE CATALYTIC DOMAIN-LIKE |
| 8. | AGB81255.1 | L,D-TRANSPEPTIDASE CATALYTIC DOMAIN-LIKE |
| 9. | AGB81307.1 | ACYL CARRIER PROTEIN PHOSPHODIESTERASE |
| 10. | AGB81573.1 | PECTIN LYASE FOLD/VIRULENCE FACTOR |
| 11. | AGB81931.1 | L,D-TRANSPEPTIDASE CATALYTIC DOMAIN-LIKE |
| 12. | AGB82203.1 | 2-METHYL ACONITATECIS-TRANS ISOMERASEPRPF |
| 13. | AGB82249.1 | GLUCONATE 2-DEHYDROGENASE, MEMBRANE-BOUND, GAMMA SUBUNIT |
| 14. | AGB82301.1 | KINASE/PYROPHOSPHORYLASE |
| 15. | AGB82307.1 | THIOESTERASE |
| 16. | AGB82315.1 | L,D-TRANSPEPTIDASE CATALYTIC DOMAIN-LIKE |
| 17. | AGB82349.1 | COPPER/ZINC SUPEROXIDE DISMUTASE (SODC) |
| 18. | AGB82393.1 | GTP CYCLOHYDROLASE I FOLE2PEROXIDASE |
| 19. | AGB82584.1 | GRPB FAMILY PROTEIN |
| 20. | AGB82609.1 | ATPASE LIKE |
| 21. | AGB82744.1 | RIBONUCLEASE H-LIKE |
| 22. | AGB82969.1 | THIOESTERASE/THIOL ESTER DEHYDRASE-ISOMERASE |
| 23. | AGB83112.1 | ENDONUCLEASE/EXONUCLEASE/PHOSPHATASE FAMILY PROTEIN |
| 24. | AGB83389.1 | ELONGATION FACTOR P HYDROXYLASE |
| 25. | AGB83394.1 | SMR DOMAIN-LIKE |
| 26. | AGB83655.1 | P-LOOP CONTAINING NUCLEOTIDE TRIPHOSPHATE HYDROLASES |
| 27. | AGB83673.1 | DNA ENDONUCLEASE SMRA |
| 28. | AGB83960.1 | SPRT FAMILY ZINC-DEPENDENT METALLOPROTEASE |
| 29. | AGB84006.1 | CYSTEINE DIOXYGENASE TYPE I |
| 30. | AGB84093.1 | STREPTOMYCIN 3''-ADENYLYLTRANSFERASE |
| 31. | AGB84302.1 | 4-HYDROXY-2-KETOVALERATE ALDOLASE |
| 32. | AGB84572.1 | OXIDOREDUCTASEMOLYBDOPTERIN-BINDING DOMAIN |
| 33. | AGB84650.1 | GLYCOSIDE HYDROLASE/DEACETYLASE |
| 34. | AGB84659.1 | POLYSACCHARIDE DEACETYLASE FAMILY PROTEIN |
| TRANSPORTER | ||
| 35. | AGB80902.1 | PORINS |
| 36. | AGB81989.1 | TYPE VI SECRETION PROTEIN |
| 37. | AGB82622.1 | MULTIDRUG RESISTANCE EFFLUX TRANSPORTER EMRE |
| 38. | AGB82815.1 | PORINS |
| 39. | AGB82887.1 | MANGANESE EFFLEX PUMP MNTP |
| 40. | AGB83037.1 | TYPE VI SECRETION PROTEIN |
| 41. | AGB83342.1 | PTS ASCORBATE TRANSPORTER SUBUNIT IIC |
| 42. | AGB83371.1 | TRIPARTITE TRICARBOXYLATE TRANSPORTER PERMEASE |
| 43. | AGB84290.1 | LPS EXPORT ABC TRANSPORTER PERIPLASMIC PROTEIN LPTC |
| BINDING PROTEIN | ||
| 44. | AGB80434.1 | PK BETA-BARREL DOMAIN-LIKE |
| 45. | AGB80485.1 | PHET/TILS DOMAIN |
| 46. | AGB82209.1 | LIGB-LIKE |
| 47. | AGB82889.1 | POLYPHENOL OXIDOREDUCTASE |
| 48. | AGB83171.1 | NAD(P)-BINDING ROSSMANN-FOLD DOMAINS |
| 49. | AGB83384.1 | COPPER-BINDING PROTEIN |
| 50. | AGB83476.1 | CHITIN-BINDING PROTEIN CBP21 |
| 51. | AGB83721.1 | GLYOXALASE/BLEOMYCIN RESISTANCE PROTEIN/ DIOXYGENASE SUPERFAMILY |
| 52. | AGB84196.1 | LIGB-LIKE |
| 53. | AGB84203.1 | CYTH-LIKE PHOSPHATASES |
| 54. | AGB84358.1 | MMCQ/YJBR FAMILY DNA-BINDING PROTEIN |
| CELL DIVISION PROTEIN | ||
| 55. | AGB81074.1 | CELL DIVISION PROTEIN ZAPD |
| 56. | AGB81957.1 | MACRODOMAINTER PROTEIN MATP |
| 57. | AGB83358.1 | SPORULATION RELATED DOMAIN |
| 58. | AGB83861.1 | CELL DIVISION PROTEIN ZAPA |
| 59. | AGB84389.1 | CAPSULE ASSEMBLY WZI FAMILY PROTEIN |
| 60. | AGB84499.1 | SPORULATION RELATED DOMAIN |
| CELL REGULATORY PROCESS PROTEIN | ||
| 61. | AGB80600.1 | DNA RECOMBINATION PROTEIN RMUC |
| 62. | AGB80822.1 | REGULATOR OF RIBONUCLEASE ACTIVITY B |
| 63. | AGB80981.1 | CREA PROTEIN |
| 64. | AGB81139.1 | RECOMBINATION REGULATOR RECX |
| 65. | AGB82748.1 | SPOVR FAMILY PROTEIN |
| 66. | AGB82870.1 | DNA DAMAGE-INDUCIBLE PROTEIN YEBG |
| 67. | AGB83410.1 | ANTITOXIN HICB |
| 68. | AGB83544.1 | TYPE II TOXIN-ANTITOXIN SYSTEM RELE/PARE FAMILY TOXIN |
| 69. | AGB84710.1 | DER GTPASE-ACTIVATING PROTEIN YIHI |
| OTHER PROTEINS | ||
| 70. | AGB80442.1 | DCRB PROTEIN PRECURSOR |
| 71. | AGB80489.1 | HD-DOMAIN/PDEASE-LIKE |
| 72. | AGB80549.1 | CUPIN FOLD METALLOPROTEIN, WBUC FAMILY |
| 73. | AGB80602.1 | SCP-LIKE |
| 74. | AGB80728.1 | TRANSLOCATION/ASSEMBLY MODULE TAMB |
| 75. | AGB80756.1 | RIBOSOME MATURATION FACTOR |
| 76. | AGB80987.1 | PEROXIDE STRESS PROTEIN YAAA |
| 77. | AGB81382.1 | DSREFH-LIKE |
| 78. | AGB81401.1 | PHEROMONE SHUTDOWN, TRAB |
| 79. | AGB81427.1 | CUPIN DOMAIN |
| 80. | AGB81476.1 | YBFN-LIKE LIPOPROTEIN |
| 81. | AGB81581.1 | INNER MEMBRANE PROTEIN YEDI |
| 82. | AGB81636.1 | SH3 DOMAIN-CONTAINING PROTEIN |
| 83. | AGB81695.1 | ETHD DOMAIN |
| 84. | AGB81860.1 | PUTATIVE INNER MEMBRANE PROTEIN OF ENTEROBACTERIACEAE |
| 85. | AGB81892.1 | ATP-DEPENDENT CLP PROTEASE ADAPTOR PROTEIN CLPS |
| 86. | AGB82063.1 | BRCT DOMAIN |
| 87. | AGB82081.1 | MSYB PROTEIN |
| 88. | AGB82090.1 | YCEI-LIKE DOMAIN |
| 89. | AGB82146.1 | CUPIN PROTEIN |
| 90. | AGB82367.1 | CUPIN DOMAIN |
| 91. | AGB82486.1 | VIRULENCE FACTOR |
| 92. | AGB82487.1 | VIRULENCE FACTOR SRFB |
| 93. | AGB82504.1 | OUTER MEMBRANE LIPOPROTEIN-SORTING PROTEIN |
| 94. | AGB82704.1 | PUTATIVE DSDNA MIMIC |
| 95. | AGB82764.1 | OUTER MEMBRANE LIPOPROTEIN SLP FAMILY |
| 96. | AGB82925.1 | GALACTOSE-BINDING DOMAIN-LIKE |
| 97. | AGB83073.1 | BESTROPHIN HOMOLOG 22 |
| 98. | AGB83302.1 | YFAZ PRECURSORPF04453 |
| 99. | AGB83335.1 | YFBU DOMAIN |
| 100. | AGB83445.1 | FLXA-LIKE PROTEIN |
| 101. | AGB83581.1 | TETRATRICOPEPTIDE REPEAT-LIKE DOMAIN |
| 102. | AGB83698.1 | DTW DOMAIN-CONTAINING PROTEIN |
| 103. | AGB83707.1 | OUTER MEMBRANE LIPOPROTEIN RCSF |
| 104. | AGB83712.1 | YAEQ PROTEIN |
| 105. | AGB84085.1 | DOXX FAMILY MEMBRANE PROTEIN |
| 106. | AGB84181.1 | ANTIBIOTIC BIOSYNTHESIS MONOOXYGENASE |
| 107. | AGB84506.1 | TYPE IV PILUS ASSEMBLY PROTEIN PILM |
| 108. | AGB84710.1 | DER GTPASE-ACTIVATING PROTEIN YIHI |
Fig. 1.
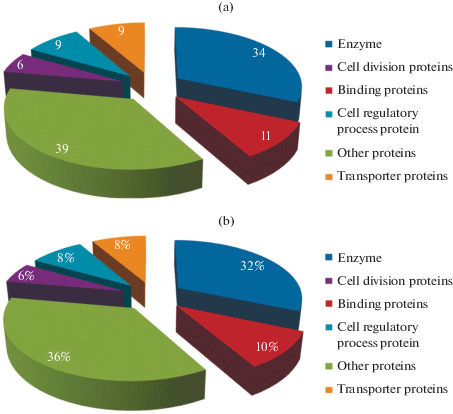
Distribution of predicted 108 proteins: (a) pie chart shows the number wise distribution of HPs. (b) Pie chart shows the percentage wise distribution of HPs.
Enzymes
Bacterial enzymes play a catalyst role in all the metabolic and catholic process, leading to the supply of essential nutrient for the growth and also responsible for the pathogenesis of the organism (Gurung et al., 2013). We have characterized 34 proteins which act as enzymes, out of which 9 were shown to be transferases. Transferases catalyze the transfer of a functional group (methyl group or glycosyl group) from one molecule (act as donor) to another molecule (act as acceptor). Six proteins were identified as endonucleases, which function in destroying the invaded foreign DNA (Van den Broek et al., 2005). Two proteins AGB81206.1 and AGB83112.1 were predicted as a member of the exonuclease-endonuclease-phosphate domain super family which plays a crucial role in the intracellular signaling activities in bacteria (Dlakic, 2000). Disrupting the intracellular signaling activities either arrests the physiological role or even kills the organism, therefore it is widely considered as a potential target for drug development (Kohanski et al., 2010). The protein AGB83394.1 is predicted to contain SMR domain like. It is observed that, SMR domain like protein is broadly classified into three sub-families. Family-1 closely relates to the C-terminal domain of the MutS, presumably found in Deltaproteobacteria, Firmicutes, Bacteroidetes and Epsilonproteobacteria phyla of bacteria and plants. The proteins under this family are responsible for the protection of cells from oxidative DNA damages. Family-2 closely relates to the C-terminal domain of MutS (eukaryotes), whereas Family-3 indicates the MutS that are found in E. coli. Overall, these three families of proteins are involved in endoculease activity and also responsible for the formation of the branched DNA structures (Fukui and Kuramitsu, 2011).
The protein AGB81307.1 is predicted to have acyl carrier protein phosphodiesterase and functions essentially in catalyzing the hydrolytic cleavage of the 4′ phosphopantetheine residue from acyl carrier protein phosphodiesterase with the generation of apo acyl carrier protein phosphodiesterase. It also plays a significant role in the regulation of fatty acid synthesis (Fischi and Kennedy, 1990). The fatty acid metabolic regulation in bacteria is essential to maintain the lipid homeostasis in the growth and stationary phases as well as in various physical and nutrient states (Fujita et al., 2007). AGB82203.1 is predicted as 2-methyl aconitate cis/trans isomerase PrpF. These proteins catalyze the inter-conversion of 2-methy CAA and 2-methyl TAA in the 2-methylcitric acid cycles (Du et al., 2017). We predicted AGB83389.1 as an elongation factor P. The factor P is required for the synthesis of proteins containing polyproline motifs and functions in ribosomal stalling (Hersch et al., 2013). AGB83655.1 is predicted as P-loop containing nucleoside triphosphate hydrolase superfamily. These families of proteins function as kinases in many pathways and also work as motor to drive reactions through conformational changes (Leipe et al., 2004). The protein AGB84572.1 is found to be oxidoreductase molybdopterin-binding domain and these proteins are reported to be involved in the H2 metabolism and bioenergetic pathways. The protein involved in these pathways help in the production of ATP molecules by using oxidation-reduction reactions and thereby providing energies to the bacteria for the normal cellular activities (Li et al., 2009). The observed function of the HPs helps to understand the crucial role of new proteins in bacterial growth and can be targeted as a potential targets for drug discovery.
Binding Proteins
We identified 11 HPs under binding proteins. AGB80434.1 is classified as PK beta barrel domain like protein which is reported to be involved in chromosome formation and prerequisite for the initiation of chromophore maturation. It is well understood that PK beta barrel domain like protein play a significant role in wide variety of cellular processes including DNA replication, DNA repair and horizontal gene transfer (Stepanenko et al., 2013). Thus, such proteins are important for the survival of the pathogens in the environment. We have characterized AGB82209.1 and AGB84196.1 as Leptospira immunoglobulin like protein B and it is observed that these proteins plays a major role in binding with fibrinogen, collagen, laminin and elastin and inhibit fibrin clot formation (Lin et al., 2011). The molecular mechanism of protein binding plays a critical role in the coagulation cascade and platelet aggregation, tissue regeneration, and immune responses proves to be a potential target for many pathogens (Choy et al., 2011).
Cell Division Proteins
Cell division in bacteria is closely linked to bacterial multiplication. Knowledge on the mechanism of cell division is essential to explore novel targets in drug development. The AGB81074.1 and AGB83861.1 are predicted to be cell division protein (ZapD). It is understood that the members of the ZapD family of proteins share a common role in cell division machinery and mechanism (Durand-Heredia et al., 2012). The protein AGB84389.1 is predicted to a capsule assembly Wzi family protein. This protein is classified as an outer membrane protein which is involved in extracellular capsule formation in many pathogenic bacteria (Bushell et al., 2013), and can be used as novel drug target.
Cell Regulatory Process Protein
Gene regulation in prokaryotes and eukaryotes includes wide range of mechanisms for the production of desired gene product. This regulatory process is a complex network that controls the expression of the various transcriptional units in the bacteria, presumably maintains the microbial pathogenesis, growth and survival. The AGB80600.1 protein was predicted to be DNA recombination protein RmuC. Although the function of this protein is not clearly understood, it shows high level of sequence similarities with myosins and structural maintenance of chromosome proteins (Gaudermann et al., 2006). Protein AGB80981.1 is a CreA (DNA binding protein), which function in the repression and control of alc regulon expression, that are necessary for the ethanol utilization pathways (Panozzo et al., 1998). Protein AGB81139.1 is a recombination regulatory (RecA) protein and functions in DNA pairing, strand exchange and recombinational DNA repair (Cox, 1991). RecA are multifunctional proteins that are found in all forms of living organisms. In the form of recombinase, the protein exhibits DNA dependent ATPase activity and also plays a role in the regulatory system that controls the induction of the SOS response. As in the case of nucleoprotein filament, the pair plays a role in DNA strand exchange (Gruenig et al., 2008). We found AGB82748.1 protein in association with stage V sporulation protein R related protein. Beall and Moran (1994) in their work described the involvement of SpovR from Bacillus subtilis in spore cortex formation (Beall and Moran, 1994). Proteins AGB83410.1 and AGB83544.1 are antitoxin systems (ReiI/ParE) composed in all bacterial genome and play a significant role in the formation of persistence cells, involvement in biofilm formation and in the pathogenesis of the organisms (Wen et al., 2014). We identified AGB84710.1 as Der GTPase activating protein YihI; the families of these proteins are more conserved in Eubacteria and involved in the cell survival and bacterial growth (Verstraeten et al., 2011). The findings are helpful to conclude that by inhibiting these proteins bang the normal cellular process and thereby reduce the bacterial pathogenecity.
Transporters
Bacteria contains various transport proteins for the import and export of substances such as nutrients, ions, metabolites, amino acids, etc., through the cell membrane, to exclude the unwanted by-products, and modify their cytoplasmic content of protons and salts needed for growth and development of the micro-organisms. We predicted AGB81989.1 and AGB83037.1 as type VI secretion system and these proteins play a major role in virulence, antibacterial activity and also participate in metal ion uptake conferring an advantage during bacteria-bacteria competition (Gallique et al., 2017). AGB82622.1 belongs to multi-drug resistance efflux transporter. EmrE belongs to the SMR family of small multidrug transporters (Ninio et al., 2004). Recent data suggest that over expression of SMR causes bacteria to become resistant to wide range of antiseptics and antibiotics such as ethidium bromide, methyl viologen and intercalating dyes (Ma and Chang, 2004). AGB82887.1 is predicted to be Manganese Efflux Pump MntP. Metals such as Manganese, Copper and Zinc are essential for all micro-organisms and appropriate maintenance of metal homeostasis is important to prevent toxicity and mismetallation for both eukaryotes and bacteria (Procheronet et al., 2013). The identification of such pumps in various bacteria such as E. coli (MntP) (Martin et al., 2015), Streptococcus pneumoniae (MntE) (Turner et al., 2015) and Neisseria spp. (MntX) (Veyrier et al., 2011) play an important role in removing excess manganese before levels become toxic. The protein AGB83371.1 is found to be tripartite tricarboxylate transporter permease. This kind of protein is most abundant in beta-proteobacteria and plays a significant role in the transport of carboxylate (Rosa et al., 2018). We identified AGB84290.1 as Lipopolysaccharides export ABC transporter periplasmic protein LptC. It is found that the lipopolysaccharide acts as hydrophilic barrier to wide range of hydrophobic antibiotics and plays a counterpart in the pathogenicity of the organisms (Hicks and Jia, 2018).
Other Proteins
We categorized 39 hypothetical proteins function in different biological process under one category named other proteins. The protein AGB80728.1 was predicted as Translocation and Assembly Module (TAM), which plays a major role in outer membrane biogenesis and virulence mechanisms in bacterial kingdom (Josts et al., 2017). AGB8163.1 is found to be SH3 domain and it has been identified in various bacteria and plays a critical role in the targeting domains involved in bacterial cell wall recognition and metal binding (Kamitori and Yoshida, 2015). We identified AGB82063.1 as BRCT domain, and it is recognized as a C-terminal region of BRCA1 gene which participates in the signal transduction and in protein targeting motif in the DNA damage response system (Woods et al., 2012). Further, structural analysis of BRCT domain reveals that it may appear either in singleton (single BRCT) or tandem pair (double BRCT) and it also has a phosphate binding site in which the phosphate molecules are bound either in the DNA end or in the phosphopeptide (Sheng et al., 2011). We found the protein AGB82090.1 belongs to Ycel protein and to the subgroup of lipocalin superfamily, and plays a role in isoprenoid quinine metabolism, transport and storage (Handa et al., 2005).
The proteins AGB81427.1, AGB82146.1 and AGB82367.1 are predicted to be the cupin superfamily, presumably involved cell wall synthesis, reactions involving sugar modification and also play a role in oxalate metabolism. We predicted AGB82764.1 as outer membrane lipoprotein Slp family which is encoded by the gene XAC1113 (Ferreira et al., 2016). Bacterial Lipoproteins are a class of membrane-anchored proteins and play important role in bacterial physiology and pathogenesis (Zuckert, 2014). De Souza et al., 2004 reported Slp involvement in the biofilm formation. The protein AGB83581.1 is found to be tetratricopeptide repeat like domain. This kind of proteins are found in wide variety of prokaryotic and eukaryotic organisms and plays an vital role in cell processes and associated with virulence mechanisms of bacterial pathogens (Cerveny et al., 2013). AGB83707.1 is predicted to be outer membrane lipoprotein RcsF, presumably involved in signal transduction pathway (Shiba et al., 2012). We identified AGB84181.1 protein as antibiotic biosynthesis mono-oxygenases. Increased production of reactive oxygen species (ROS) causes oxidative damages to the cells. The primary ROS generated within mitochondria are limited to flavoproteins and flavins that activate the molecular oxygens. The generated monooxygenases do variety of functions in all living organisms and catalyze a wide range of reactions such as participating in respiratory chain within the cytoplasmic membrane and protecting themselves against external reactive oxygen species (Khan et al., 2017). The protein AGB84506.1 type IV pilli are bacterial cell surface proteins which plays a major role in the twitching motility, bacteriophage infection, virulence, biofilm formation, natural transformation and bacteriophage infections (van Schaik et al., 2005). Proteins AGB81476.1, AGB82504.1, AGB82764.1 and AGB83707.1 are classified as bacterial lipoproteins. Bacterial lipoproteins possess fatty acids at their amino-termini that facilitate the hydrophilic protein to anchor onto bacterial cell membranes (Buddelmeijer, 2005). These lipoproteins are universally distributed in the bacterial kingdom and accounts to about 1–3% of the total genome. These lipoproteins play pivotal roles in many physiological and cellular processes such as cell wall metabolism, antibiotic resistance, nutrient uptake, cell division, signal transduction and virulence. Thus, the identified HPs are predicted to play an important role in the survival and pathogenesis of the pathogens and the identified functions of HPs may provide clue as a therapeutic target for the drug design and developments.
Virulence Proteins
Virulence factors are essential to invade the bacteria to colonize, causes disease and to overcome the defenses of the host. Therefore, understanding the molecular mechanisms of microbial virulence plays a central role in the pathogenesis of the bacteria. So, we used VICMpred and VirulentPred, an SVM based method to predict the bacterial virulence factor among the 483 HPs. The predicted results are shown in Supplementary Table 5. We found that 29 HPs as virulent factors, 226 as a cellular process, 15 as information and storage and 120 as metabolism molecules. All these proteins can be used as potential targets for drug design and development.
CONCLUSIONS
In general, about 30–50% of the sequenced genome are referred as hypothetical proteins, and the rapid accumulation of the genome data containing HPs is one of the emerging challenges in modern biology. HPs hamper the identification of novel drug targets which can specifically act on pathogens to combat the pathogenicity. In the case of bacterial pathogens, these HPs play a crucial role in the identification of potential drug targets and also enhance our understanding of their virulence capacity and pathogenicity. In this study, we have characterized the functions for the 108 HPs from S. marcescens with a high level of confidence using various bioinformatics approaches. Various types of enzymes, transporters, cell division, binding proteins were characterized which play an essential role in the growth, survival virulence and pathogenesis of S. marcescens. Characterization of proteins on the basis of physiochemical properties and subcellular localization helps in differentiating the vital drug targets from vaccine targets. In addition, we also identified 18 virulence proteins that are predicted to play a crucial role in the pathogenesis of the organisms. Hence, this study may facilitate future studies on the predicted HPs as novel therapeutic targets for the drug and vaccine development.
Supplementary material
ACKNOWLEDGMENTS
DP thankfully acknowledges Indian Council of Medical Research [no. ISRM/11/(17)/2017] for Senior Research Fellowship. JJ greatly acknowledges DST [EMR/2016/000498], ICMR [BIC/12(07)/2015] and BRNS [35/14/02/2018-BRNS/35009] for providing financial assistance through research projects. PS gratefully acknowledges BRNS for providing financial assistance. The authors thankfully acknowledge the DST-FIST [SR/FST/LSI-667/2016(C)], DST-PURSE [SR/PURSE Phase 2/38 (G)] and MHRD-RUSA 2.0 [F.24/51/2014-U, Policy (TNMulti-Gen), Dept. of Edn. Govt. of India] for the financial support and infrastructure facilities.
COMPLIANCE WITH ETHICAL STANDARDS
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Electronic supplementary material
Supplementary materials are available for this article at 10.1134/S1062359020300019 and are accessible for authorized users.
REFERENCES
- 1.Alnajar S., Gupta R.S. Phylogenomics and comparative genomic studies delineate six main clades within the family Enterobacteriaceae and support the reclassification of several polyphyletic members of the family. Infect. Genet. Evol. 2017;54:108–127. doi: 10.1016/j.meegid.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Aylward F.O., Tremmel D.M., Starrett G.J., Bruce D.C., Chain P., Chen A., Davenport K.W., Detter C., Han C.S., Han J., Huntemann M. Complete genome of Serratia sp. strain FGI 94, a strain associated with leaf-cutter ant fungus gardens. Genome Announce. 2013;1:e0023912. doi: 10.1128/genomeA.00239-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beall B., Moran C.P. Cloning and characterization of spoVR, a gene from Bacillus subtilis involved in spore cortex formation. J. Bacteriol. 1994;176:2003–2012. doi: 10.1128/jb.176.7.2003-2012.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendtsen J.D., Kiemer L., Fausbøll A., Brunak S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005;5:58. doi: 10.1186/1471-2180-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bharat Siva Varma P., Adimulam Y.B., Kodukula S. In silico functional annotation of a hypothetical protein from Staphylococcus aureus. J. Infect. Publ. Health. 2015;8:526–532. doi: 10.1016/j.jiph.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Bhasin M., Garg A., Raghava G.P.S. PSLpred: prediction of subcellular localization of bacterial proteins. Bioinformatics. 2005;21:2522–2524. doi: 10.1093/bioinformatics/bti309. [DOI] [PubMed] [Google Scholar]
- 8.Buddelmeijer N. The molecular mechanism of bacterial lipoprotein modification-how, when and why. FEMS Microbiol. Rev. 2015;39:246–261. doi: 10.1093/femsre/fuu006. [DOI] [PubMed] [Google Scholar]
- 9.Bushell S.R., Mainprize I.L., Wear M.A., Lou H., Whitefield C., Maismith J.H. Wzi is an outer membrane lectin that underpins group 1 capsule assembly in Escherichia coli. Structure. 2013;21:844–853. doi: 10.1016/j.str.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai C.Z. SVM-Prot: web-based support vector machine software for functional classification of a protein from its primary sequence. Nucleic Acids Res. 2003;31:3692–3697. doi: 10.1093/nar/gkg600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.CDC, Antibiotic resistant threats report, 2013. https://www.cdc.gov/drugresistance/biggest_threats.html.
- 12.Cerveny L., Straskova A., Dankova V., Hartiova A., Ceckova M., Staud F., Stulik J. Tetratricopeptide repeat motifs in the world of bacterial pathogens: role in virulence mechanisms. Infect. Immun. 2013;81:629–635. doi: 10.1128/IAI.01035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choy H.A., Kelley M.M., Croda J., Matsunaga J., Babbitt J.T., Ko A.I., Picardeau M., Haake D.A. The multifunctional LigB adhesin binds homeostatic proteins with potential roles in cutaneous infection by pathogenic Leptospira interrogans. PLoS One. 2011;6:e16879. doi: 10.1371/journal.pone.0016879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox M.M. The RecA protein as a recombinational repair system. Mol. Microbiol. 1991;5:1295–1299. doi: 10.1111/j.1365-2958.1991.tb00775.x. [DOI] [PubMed] [Google Scholar]
- 15.da Costa W.L.O., Araújo C.L.A., Dias L.M., Pereira L.C.S., Alves J.T.C., Araújo F.A., Folador E.L., Henriques I., Silva A., Folador A.R.C. Functional annotation of hypothetical proteins from the Exiguobacterium antarcticum strain B7 reveals proteins involved in adaptation to extreme environments, including high arsenic resistance. PLoS One. 2018;13:e0198965. doi: 10.1371/journal.pone.0198965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Souza A.A., Takita M.A., Coletta-Filho H.D., Caldana C., Yanai G.M., Muto N.H., de Oliveira R.C., Nunes L.R., Machado M.A. Gene expression profile of the plant pathogen Xylella fastidiosa during biofilm formation in vitro. FEMS Microbiol. Lett. 2004;237:341–353. doi: 10.1016/j.femsle.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 17.Dlakic M. Functionally unrelated signaling proteins contain a fold similar to Mg2+ dependent endoculeases. Trends Biochem. Sci. 2000;25:272–273. doi: 10.1016/s0968-0004(00)01582-6. [DOI] [PubMed] [Google Scholar]
- 18.Du C., Cao S., Shi X., Nie X., Zheng J., Deng Y., Ruan L., Peng D., Sun M. Genetic and biochemical characterization of a gene operon for trans-aconitic acid, a novel nematicide from Bacillus thuringiensis. J. Biol. Chem. 2017;292:3517–3530. doi: 10.1074/jbc.M116.762666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durand-Heredia J., Rivkin E., Fan G., Morales J., Janakiraman A. Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J. Bacteriol. 2012;194:3189–3198. doi: 10.1128/JB.00176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferreira R.M., Moreira L.M., Ferro J.A., Soares M.R.R., Laia M.L., Varani A.M., de Oliveira J.C.F., Ferro M.I.T. Unravelling potential virulence factor candidates in Xanthomonas citri subsp. citri by secretome analysis. PeerJ. 2016;4:e1734. doi: 10.7717/peerj.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finley R.L., Collignon P., Larsson D.J., McEwen S.A., Li X.Z., Gaze W.H., Reid-Smith R., Timinouni M., Graham D.W., Topp E. The scourge of antibiotic resistance: the important role of the environment. Clin. Infect. Dis. 2013;57:704–710. doi: 10.1093/cid/cit355. [DOI] [PubMed] [Google Scholar]
- 22.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., Sonnhammer E. L. L., Tate J., Punta M. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischi A.S., Kennedy E.P. Isolation and properties of acyl carrier protein phosphodiesterase of Escherichia coli. J. Bacteriol. 1990;172:5445–5449. doi: 10.1128/jb.172.9.5445-5449.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujita Y., Matsuoka H., Hirooka K. Regulation of fatty acid metabolism in bacteria. Mol. Microbiol. 2007;66:829–839. doi: 10.1111/j.1365-2958.2007.05947.x. [DOI] [PubMed] [Google Scholar]
- 25.Fukui, K. and Kuramitsu, S., Structure and function of the small MutS-related domain, Mol. Biol. Int., 2011, art. ID 691735, 10.4061/2011/691735 [DOI] [PMC free article] [PubMed]
- 26.Gallique M., Bouteiller M., Merieau A. The type VI secretion system: a dynamic system for bacterial communication. Front. Microbiol. 2017;8:1454. doi: 10.3389/fmicb.2017.01454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garg A., Gupta D. VirulentPred: a SVM based prediction method for virulent proteins in bacterial pathogens. BMC Bioinf. 2008;9:62. doi: 10.1186/1471-2105-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gasperskaja E., Kučinskas V. The most common technologies and tools for functional genome analysis. Acta Med. Litu. 2017;24:1–11. doi: 10.6001/actamedica.v24i1.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. The Proteomics Protocols Handbook. Totowa, NJ: Humana; 2005. [Google Scholar]
- 30.Gaudermann P., Vogl I., Zientz E., Moya A., Gross R., Dandekar T. Analysis of and function predictions for previously conserved hypothetical or putative proteins in Biochmannia floridanus. BMC Microbiol. 2006;6:1. doi: 10.1186/1471-2180-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geer L.Y., Domrachev M., Lipman D.J., Bryant S.H. CDART: protein homology by domain architecture. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gough J., Karplus K., Hughey R., Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 33.Gruenig M.C., Renzette N., Long E., Chitteni-Pattu S., Inman R.B., Cox M.M., Sandler S.J. RecA-mediated SOS induction requires an extended filament conformation but no ATP hydrolysis. Mol. Microbiol. 2008;69:1165–1179. doi: 10.1111/j.1365-2958.2008.06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gurung, N., Ray, S., Bose, S., and Rai, V., A broader view: microbial enzymes and their relevance industries, medicine, and beyond, Biomed. Res. Int., 2013, art. ID 329121. 10.1155/2013/329121 [DOI] [PMC free article] [PubMed]
- 35.Handa N., Terada T., Doi-Katayama Y., Hirota H., Tame J.R, Park S.Y., Kuramitsu S., Shirouzu M., Yokoyama S. Crystal structure of a novel polyisoprenoid-binding protein from Thermus thermophilus HB8. Protein Sci. 2005;14:1004–1010. doi: 10.1110/ps.041183305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hawkins T., Kihara D. Function prediction of uncharacterized proteins. J. Bioinf. Comput. Biol. 2007;5:1–30. doi: 10.1142/s0219720007002503. [DOI] [PubMed] [Google Scholar]
- 37.Hersch, S.J., Wang, M., Zou, S.B., Moon, K.M., Foster, L.J., Ibba, M., and Navarre, W.W., Divergent protein motifs direct elongation factor P-mediated translational regulation in Salmonella enteric and Escherichia coli,mBio, 2013, vol. 4. 10.1128/mBio.00180-13 [DOI] [PMC free article] [PubMed]
- 38.Hicks G., Jia Z. Structural basis for the lipoplysaccharide export activity of the bacterial lipopolysaccharide transport system. Int. J. Mol. Sci. 2018;19:E2680. doi: 10.3390/ijms19092680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoskeri J.H., Krishna V., Amruthavalli C. Functional annotation of conserved hypothetical proteins in Rickettsia massiliae MTU5. J. Comput. Sci. Syst. Biol. 2010;3:50–51. [Google Scholar]
- 40.Hunter S., Jones P., Mitchell A., Apweiler R., Attwood T.K., Bateman A., Bernard T., Binns D., Bork P., Burge S., de Castro E., Coggill P., Corbett M., Das U., Daugherty L. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iguchi A., Nagaya Y., Pradel E., Ooka T., Ogura Y., Katsura K., Kurokawa K., Oshima K., Hattori M., Parkhill J., Sebaihia M. Genome evolution and plasticity of Serratia marcescens, an important multidrug-resistant nosocomial pathogen. Genome Biol. Evol. 2014;6:2096–2110. doi: 10.1093/gbe/evu160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Josts I., Stubenrauch C.J., Vadlamani G., Mosbahi K., Walker D., Lithgow T., Grinter R. The structure of a conserved domain of TamB reveals a hydrophobic β Taco fold. Structure. 2017;25:1898–1906. doi: 10.1016/j.str.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamitori S., Yoshida H. SH Domains: Structure, Mechanisms and Applications. New York: Springer-Verlag; 2015. [Google Scholar]
- 44.Kanehisa M. Linking databases and organisms: GenomeNet resources in Japan. Trends Biochem. Sci. 1997;22:442–444. doi: 10.1016/s0968-0004(97)01130-4. [DOI] [PubMed] [Google Scholar]
- 45.Khan M.N., Mobin M., Abbas Z.K., AlMutairi K.A., Siddiqui Z.H. Role of nanomaterials in plants under challenging environments. Plant Physiol. Biochem. 2017;110:194–209. doi: 10.1016/j.plaphy.2016.05.038. [DOI] [PubMed] [Google Scholar]
- 46.Kohanski M.A., Dwyer D.J., Collins J.J. How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 2010;8:423–435. doi: 10.1038/nrmicro2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 48.Leipe D.D., Koonin E.V., Aravind L. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 2004;383:1–28. doi: 10.1016/j.jmb.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 49.Letunic I., Doerks T., Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X., Luo Q., Wofford N.Q., Keller K.L., Mcinerney M.J., Wall J.D., Krumholz L.R. A molybdopterin oxidoreductase is involved in H2 oxidation in Desulfovibrio desulfuricans G20. J. Bacteriol. 2009;191:2675–2682. doi: 10.1128/JB.01814-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Y.P., McDonough S.P., Sharma Y., Chang Y.F. Leptospira immunoglobulin-like protein B (LigB) binding to the C-terminal fibrinogen αC domain inhibits fibrin clot formation, platelet adhesion and aggregation. Mol. Microbiol. 2011;79:1063–1076. doi: 10.1111/j.1365-2958.2010.07510.x. [DOI] [PubMed] [Google Scholar]
- 52.Ma C., Chang G. Structure of the multidrug resistance efflux transporter EmrE from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2852–2857. doi: 10.1073/pnas.0400137101. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Martin J.E., Waters L.S., Storz G., Imlay J.A. The Escherichia coli small protein MntS and exporter MntP optimize the intracellular concentration of manganese. PLoS Genet. 2015;11:e1004977. doi: 10.1371/journal.pgen.1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ninio S., Elbaz Y., Schuldiner S. The membrane topology of EmrE—a small multidrug transporter from Escherichia coli. FEBS Lett. 2004;562:193–196. doi: 10.1016/S0014-5793(04)00240-6. [DOI] [PubMed] [Google Scholar]
- 55.Orengo C., Michie A., Jones S., Jones D., Swindells M., Thornton J. CATH—a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1109. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 56.Padmavathi A.R., Abinaya B., Pandian S.K. Phenol, 2,4-bis(1,1-dimethylethyl) of marine bacterial origin inhibits quorum sensing mediated biofilm formation in the uropathogen Serratia marcescens. Biofouling. 2014;30:1111–1122. doi: 10.1080/08927014.2014.972386. [DOI] [PubMed] [Google Scholar]
- 57.Panozzo C., Cornillot E., Felenbok B. The CreA repressor is the sole DNA-binding protein responsible for carbon catabolite repression of the alcA in Aspergillus nidulans via its binding to a couple of specific sites. J. Biol. Chem. 1998;273:6367–6372. doi: 10.1074/jbc.273.11.6367. [DOI] [PubMed] [Google Scholar]
- 58.Parente T.M.A.L., de Lima Rebouças E., dos Santos V.C.V., Barbosa F.C.B., Zanin I.C.J. Serratia marcescens resistance profile and its susceptibility to photodynamic antimicrobial chemotherapy. Photodiagn. Photodyn. Ther. 2016;14:185–190. doi: 10.1016/j.pdpdt.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 59.Petersen T.N., Brunak S., von Heijne G., Nielsen H., Signal P. 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 60.Procheron G., Garenaux A., Proulx J., Sabri M., Dozois C.M. Iron, copper, zinc, and manganese transport and regulation in pathogenic Enterobacteria: correlations, between strains, site of infection and the relative importance of the different metals transport systems for virulence. Front. Cell. Infect. Microb. 2013;3:90. doi: 10.3389/fcimb.2013.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rappoport N., Karsenty S., Stern A., Linial N., Linial M. ProtoNet 6.0: organizing 10 million protein sequences in a compact hierarchical family tree. Nucleic Acids Res. 2012;40:D313–D320. doi: 10.1093/nar/gkr1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosa L.T., Bianconi M.E., Thomas G.H., Kelly D.J. Tripartite ATP-independent periplasmic (TRAP) transporters and tripartite tricarboxylate transporters (TTT): from uptake to pathogenicity. Front. Cell. Infect. Microbiol. 2018;8:33. doi: 10.3389/fcimb.2018.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saha S., Raghava G.P.S. VICMpred: an SVM-based method for the prediction of functional proteins of gram-negative bacteria using amino acid patterns and composition. Genomics Proteomics Bioinf. 2006;4:42–47. doi: 10.1016/S1672-0229(06)60015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shahbaaz M., Imtaiyaz Hassan M., Ahmad F. Functional annotation of conserved hypothetical proteins from Haemophilus influenzae Rd KW20. PLoS One. 2013;8:e84263. doi: 10.1371/journal.pone.0084263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sheng Z.Z., Zhao Y.Q., Huang J.F. Functional evolution of BRCT domains from binding DNA to protein. Evol. Bioinf. Online. 2011;7:87–97. doi: 10.4137/EBO.S7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shiba Y., Miyagawa H., Nagahama H., Matsumoto K., Kondo D., Matsuoka S., Matsumoto K., Hara H. Exploring the relationship between lipoprotein mislocalization and activation of the Rcs signal transduction system in Escherichia coli. Microbiology. 2012;158:1238–1248. doi: 10.1099/mic.0.056945-0. [DOI] [PubMed] [Google Scholar]
- 67.Singh A., Singal B., Nath O., Singh I.K. Functional annotation and classification of the hypothetical proteins of Neisseria meningitidis H44/76. Am. J. Biosci. Bioeng. 2015;3:57–64. [Google Scholar]
- 68.Stepanenko O.V., Stepanenko O.V., Kuznetsova I.M., Verkhusha V.V., Turoverov K.K. Beta-barrel scaffold of fluorescent proteins: folding, stability and role in chromophore formation. Int. Rev. Cell. Mol. Biol. 2013;302:221–278. doi: 10.1016/B978-0-12-407699-0.00004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thomas P.D., Kejariwal A., Guo N., Mi H., Campbell M.J., Muruganujan A., Lazareva-Ulitsky B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–W650. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turner, A.G., Ong, C.L., Gillen, C.M., Davies, M.R., West, N.P., McEwan, A.G., and Walker, M.J., Manganese homeostasis in group A Streptococcus is critical for resistance to oxidative stress and virulence, mBio, 2015, vol. 6. 10.1128/mBio.00278-15 [DOI] [PMC free article] [PubMed]
- 71.Tusnády G.E., Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 72.van den Broek B., Noom M.C., Wuite G.J. DNA-tension dependence of restriction enzyme activity reveals mechanochemical properties of the reaction pathway. Nucleic Acids Res. 2005;33:2676–2684. doi: 10.1093/nar/gki565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Schaik E.J., Giltner C.L., Audette G.F., Keizer D.W., Bautista D.L., Slupsky C.M., Sykes B.D., Irvin R.T. DNA binding: a novel function of Pseudomonas aeruginosa type IV pili. J. Bacteriol. 2005;187:1455–1464. doi: 10.1128/JB.187.4.1455-1464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vazirianzadeh B., Dehghani R., Mehdinejad M., Sharififard M., Nasirabadi N. The first report of drug resistant bacteria isolated from the brown-banded cockroach, Supella longipalpa, in Ahvaz, South-Western Iran. J. Arthropod-Born Dis. 2013;8:53–59. [PMC free article] [PubMed] [Google Scholar]
- 75.Verstraeten N., Fauvart M., Versees W., Michiels J. The university conserved prokaryotic GTPases. Microbiol. Mol. Biol. Rev. 2011;75:507–542. doi: 10.1128/MMBR.00009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Veyrier F.J., Boneca I.G., Cellier M.F., Taha M.K. A novel metal transporter mediating manganese export (MntX) regulates the Mn toFe intracellular ration and Neisseria meningitidis virulence. PLoS Pathog. 2011;7:e1002261. doi: 10.1371/journal.ppat.1002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wen Y., Behieis E., Devreese B. Toxin-antitoxin systems: their role in persistence, biofilm formation and pathogenicity. Pathog. Dis. 2014;70:240–249. doi: 10.1111/2049-632X.12145. [DOI] [PubMed] [Google Scholar]
- 78.WHO priority pathogens list for R&D of new antibiotics, 2018a. https://www.who.int/medicines/publications/ WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf.
- 79.WHO, Antimicrobial Resistance (WHO Fact Sheet), Geneva: World Health Org., 2018b. http://www.who.int/en/news-room/factsheets/detail/antimicrobial-resistance. Accessed September 25, 2018.
- 80.Woods N.T., Mesquita R.D., Sweet M., Carvalho M.A., Li X., Nguyen H., Thomas C.E., Iversen Jr., Marsillac S., Karchin R., Koomen J., Monteiro A.N. Charting the landscape of tandem BRCT domain-mediated protein interactions. Sci. Signaling. 2012;5:rs6. doi: 10.1126/scisignal.2002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yu C.S., Lin C.J., Hwang J.K. Predicting subcellular localization of proteins for gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004;13:1402–1406. doi: 10.1110/ps.03479604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu N.Y., Wagner J.R., Laird M.R., Melli G., Rey S., Lo R., Dao P., Cenk Sahinalp S., Ester M., Foster L.J., Brinkman F.S.L. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zückert W.R. Secretion of bacterial lipoproteins: through the cytoplasmic membrane, the periplasm and beyond, Biochim. Biophys. Acta, Mol. Cell Res. 2014;1843:1509–1516. doi: 10.1016/j.bbamcr.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


