Abstract
Whole-genome sequencing of 217 animals from three Danish commercial pig breeds (Duroc, Landrace [LL], and Yorkshire [YY]) was performed. Twenty-six million single-nucleotide polymorphisms (SNPs) and 8 million insertions or deletions (indels) were uncovered. Among the SNPs, 493,099 variants were located in coding sequences, and 29,430 were predicted to have a high functional impact such as gain or loss of stop codon. Using the whole-genome sequence dataset as the reference, the imputation accuracy for pigs genotyped with high-density SNP chips was examined. The overall average imputation accuracy for all biallelic variants (SNP and indel) was 0.69, while it was 0.83 for variants with minor allele frequency > 0.1. This study provides whole-genome reference data to impute SNP chip-genotyped animals for further studies to fine map quantitative trait loci as well as improving the prediction accuracy in genomic selection. Signatures of selection were identified both through analyses of fixation and differentiation to reveal selective sweeps that may have had prominent roles during breed development or subsequent divergent selection. However, the fixation indices did not indicate a strong divergence among these three breeds. In LL and YY, the integrated haplotype score identified genomic regions under recent selection. These regions contained genes for olfactory receptors and oxidoreductases. Olfactory receptor genes that might have played a major role in the domestication were previously reported to have been under selection in several species including cattle and swine.
Keywords: commercial pig, imputation, population structure, whole-genome sequencing
Introduction
Farm animals have been subject to strong artificial selection leading to remarkable phenotypic changes in body shape, physiology, and behavior. Identifying the genetic changes underlying these developments provides new insights into mechanisms by which genetic selection shapes phenotypic diversity (Rubin et al., 2010). Identification of genomic regions subject to selection in livestock may assist the discovery and validation of genomic regions involved in the manifestation of economically important traits. Commercial pig breeds have undergone intense artificial selection leading to dramatically improved performance. A comparison of the genomic features among breeds can give an in-depth understanding of how genomic variation has been shaped and advantageous characteristics have evolved through strong artificial selection. A comparison of breed-specific variants could reveal the genetic diversity of the different populations.
Advances in genomics have expanded our ability to study the genetics of organisms. Technological development and application of next-generation sequencing and high-throughput genotyping platforms have transformed the study of farm animal genetics. Numerous genome-wide studies have been undertaken, whereby samples from subpopulations are genotyped to investigate genomic differences between them. Previously, the majority of population genetic studies of farm animals were based on common variants found on genotyping arrays (Deniskova et al., 2018; Gautason et al., 2019; Huang et al., 2019). However, whole-genome sequencing (WGS) supports the studies of low frequency and rare variants. Rare variants are on average younger than common variants (Mathieson and McVean, 2014). Thus, rare variants are more powerful for distinguishing closely related populations and more informative with respect to recent demographic history. The large number of polymorphisms spread across the genome can reveal the fine genetic structure of the population and may provide evidence of adaptive selection across the genome (Barendse et al., 2009). This applies in particular to complex traits, where genetic and environmental factors combine to produce the phenotype.
The use of WGS is becoming more common in swine genomic research. The WGS of 10 unrelated wild boars from different geographical areas helped to reveal the porcine demography and evolution (Groenen et al., 2012). The sequencing of 69 Chinese pigs from various breeds and wild boars provided a suitable dataset to reveal the unique haplotype on the X chromosome that may respond to the adaptation of cold and hot environments (Ai et al., 2015). The WGS strategy has also been applied to study local pig breeds (Molnár et al., 2014; Choi et al., 2015). Furthermore, such as in humans and cattle, genotype imputation with a panel of WGS individuals could yield a WGS level marker set for a large pig population at affordable prices. This strategy has been applied for Quantitative traits loci (QTL) studies on lumbar number (Yan et al., 2017) and hematological traits (Yan et al., 2018), and it has been shown that the imputed WGS population have a higher genomic prediction accuracy compared with array-type genotyping (Song et al., 2019).
Pig producers in Denmark use a terminal crossbreeding system with three breeds. F1 sows are crosses of Yorkshire (YY) and Landrace (LL) that are mated to purebred Duroc (DD) boars to produce pigs for slaughter. Genetic evaluation is usually done within each of these breeds based on recorded phenotypes. Breeding goals differ between breeds: DD pigs are selected for growth, leanness, and feed efficiency, whereas YY and LL are selected for maternal traits. In-depth knowledge of the population structure and possible admixture events among these three breeds are important to maintain pure breeding programs. Strong directional selection and breeding in closed nucleus herds may have resulted in genetic differentiation among these three Danish pig breeds. This could be studied by signatures of selection and distribution of DNA polymorphisms, especially concerning functional annotation categories. There are several approaches for searching signatures of selection (Qanbari and Simianer, 2014). The fixation index (FST; Wright, 1949) has been widely used in human, animal, and plant populations. Recently, the integrated haplotype score (iHS) has been employed to examine selection signatures (Voight et al., 2006). The inheritance of identical haplotypes from a common ancestor creates long tracts of homozygous genotypes known as runs of homozygosity (ROH), which can be used to assess both recent and past inbreeding within a population (Ceballos et al., 2018).
The objective of this study was to investigate genetic diversity, population structure, and signatures of selection in three Danish commercial pig breeds as well as divergence of the three breeds using the observed polymorphisms at the whole-sequence level. We also examined the imputation accuracy for high-density (HD) single-nucleotide polymorphism (SNP) chip genotypes to the whole-genome sequence variants using a multi-breed reference population.
Materials and Methods
Institutional Animal Care and Use Committee approval was not obtained because this study did not involve animal handling and sample collection. The blood samples for DNA extraction for WGS were taken from an existing blood repository and the HD genotypes were obtained from the MetaPig project (http://www.metapig.eu).
Selection of animals for WGS
For each breed, the selection of animals for sequencing was done as follows. First, based on pedigree information, the genetic contribution of each animal in the pedigree to the target population was computed, which consisted of genotyped animals born in 2010, 2011, and 2012. An individual’s contribution to the target population was computed recursively as half the sum of all its offsprings’ contributions + 1 if the individual was itself a member of the target population. Second, animals were ranked based on their genetic contributions. The top-ranked animals with available blood samples were selected for sequencing following a sequential procedure based on the highest rank, but rejecting animals with relationships greater than or equal to 0.5 to animals selected for sequencing. The number of animals for WGS in each breed was 89 DD, 61 LL, and 67 YY.
DNA sequence alignment and variant calling
Genomic DNA was extracted from the blood samples stored at −20 °C using the QIAsymphony DNA Mini Kit (Qiagen). Whole-genome resequencing was performed on an Illumina HiSeq 2000 platform by AROS (now Eurofins), Aarhus, Denmark, and on BGISEQ PE100 at BGI, Copenhagen, Denmark.
AROS
About 1.2 μg genomic DNA was randomly fragmented by Covaris. The genomic library was prepared according to the manufacturer’s protocol (Illumina, True Seq DNA preparation guide) using the TruSeq DNA sample preparation kit. The paired-end library was sequenced on an Illumina HiSeq 2000 in 2 × 100 cycles using the v3 chemistry.
BGI
About 1 μg genomic DNA was randomly fragmented by Covaris. The fragmented genomic DNA was selected by the Agencourt AMPure XP to an average size of 200 to 400 bp. Fragments were end-repaired and then 3′ adenylated. Adaptors were ligated to the ends of these 3′ adenylated fragments, and these products were subjected to amplification. The Polymerase chain reaction (PCR) products were purified by the Agencourt AMPure XP. The double-stranded PCR products were heat-denatured and circularized by the splint oligo sequence. The single-strand circle DNA was formatted as the final library. The paired-end 100 bp reads were obtained by combinatorial Probe-Anchor Synthesis using BGISEQ-500.
For each individual, paired-end read trimming was performed using trim-fastq from the PoPoolation package (Kofler et al., 2011). Reads were trimmed from the 3′ end by removing “N” characters. Next, quality trimming to a minimum base Phred quality of 20 using a modified Mott algorithm as implemented by Phred (described here: http://www.phrap.org/phredphrap/phred.html). Reads were removed if they: were shorter than 40 bp after trimming, had a mean quality score <20, or contained more than 3 “N” characters. Filtered reads were aligned to the porcine reference genome build 11 (Warr et al., 2019) by the Burrows-Wheeler Aligner (BWA version 0.7.17; Li, 2013, preprint), employing the “bwa-mem” algorithm for paired-end reads, and reads that became unpaired when the filtering step removed their mate. SAMtools version 1.8 (Li et al., 2009) was used for sorting, merging, and marking potential PCR duplicates. From here till the Variant Call Format (VCF) file, the reads were processed using the Genome Analysis Toolkit (GATK version 3.8; Poplin et al., 2018). Reads were realigned using RealignerTargetCreator and IndelRealigner; base quality was recalibrated using BaseRecalibrator; and information was summarized for each sample using HaplotypeCaller. The resulting Genomic Variant Call Format (GVCFs) were combined to a VCF file using CombineGVCFs using default parameter values. Variants were called using GenotypeGVCFs using default parameters.
The following quality control parameters were used for genotype calling by GATK (Poplin et al., 2018): heterozygosity=0.001, indel_heterozygosity=1.25E-4 heterozygosity_stdev=0.01, standard_min_confidence_threshold_for_calling=10.0, standard min confidence threshold for emitting=30.0, max alternate alleles=6, max_num_PL values=100, and min mapping quality score=20. Software defaults were used for all other parameters.
The statistic of the WGS marker sets in each breed
The minor allele frequency (MAF) and detailed genotype count reports were calculated by the --freq function and --freqx functions in PLINK v1.9 (Purcell et al., 2007). To estimate the extent of linkage disequilibrium (LD), we used mapthin (https://www.staff.ncl.ac.uk/richard.howey/mapthin/) to reduce the WGS marker set to 100 SNP per Mb. Then, we used the --r2 function in PLINK to calculate the pair-wise squared correlations in each breed, setting --ld-window-r2 to 0 and maximal pair-wise distance of 10 Mb to output all the r2 value for all markers within 10 Mb region for each marker. To plot the LD decay in each breed, we grouped the distance between two markers into bins of 100 kb and calculated the mean of r2 in each bin.
Annotation of genetic variants and distribution of high-impact functional variants
To annotate the variants called from our WGS dataset, Ensembl variant effect predictor (VEP ver. 99) (McLaren et al., 2016) was used to annotate variants with information in the VEP database (the merged cache file Sscrofa11.1 ver. 99). The default parameter of --distance 5 Kb of VEP was applied to define the upstream and downstream variants. We also applied --sift b option to the prediction of the SIFT score (Kumar et al., 2009) of missense variants.
Population structure
A principal component analysis (PCA) with 217 sequenced animals from three breeds was performed using the --pca function in PLINK software (Purcell et al., 2007) for all three breeds. We plotted the first and second principal components to show the clustering of individuals. The ancestry components of all WGS pigs (217 animals) were estimated using ADMIXTURE (Alexander et al., 2009) with K = 2 to 4. The best K value was chosen based on the cross-validation value and the separation of breeds.
FST estimation
The animals with admixture (ancestry < 0.99 to a breed) were removed from the rest of the analysis (FST, iHS, and ROH). The remaining animal numbers were 87 DD, 36 LL, and 33 YY. We used PLINK software (Purcell et al., 2007) to thin the markers based on pair-wise LD estimates. Firstly, we used the function --indep-pairwise to generate a set of SNPs in 20-kb windows with all pair-wise r2 < 0.2 within each breed. After LD pruning, the number of markers included in FST and ROH analyses was 266,914 (DD), 245,217 (LL), and 218,185 (YY). Then, we explored the differentiation of each marker between the following combinations of breeds: 1) DD vs. LL, 2) DD vs. YY, and 3) LL vs. YY using PLINK --fst function.
Runs of homozygosity
PLINK software (Purcell et al., 2007) was used to detect ROH, using the same marker sets as were used for the FST estimation (the reduced dataset in animals and markers). The function --homozyg-kb was used to detect ROH with a minimal length of 1 Mb, keeping the rest of the parameters at default values.
Integrated haplotype score
Integrated haplotype scores in each breed (the animals with ancestry assignment to one breed ≥ 0.99) were estimated to identify the selective pressure within each breed using the WGS marker set. The WGS genotype files for each breed were phased using Beagle 5 (Browning and Browning, 2007). Then, the haplotypes of each chromosome were analyzed using the R package REHH (Voight et al., 2006) to calculate the iHS value. In contrast to neutral evolution, selection tends to produce clusters of markers with outlier values of the iHS statistic, which can be used to identify regions under selection. Due to a lack of information, alleles are not identified as “ancestral” or “derived.” Though neglecting ancestry status reduces the strength of the signal, conspicuous values remain and identification of candidate regions under selection remains possible (Gautier et al., 2017). To determine the candidate regions under selection within a breed, we scanned through the chromosomes in sliding windows with the following parameters of the function cal_candidate_regions in the REHH package: threshold=8.5, pval=TRUE, window_size=1E6, overlap=1E5, and min_n_extr_mrk = 2. Here, a Bonferroni correction was used to set the significance threshold of −log10(P) = 8.5.
Enrichment analysis
The genes under selection in the candidate genomic regions returned from the iHS were searched based on the Sscrofa11.1 (https://www.ensembl.org/Sus_scrofa/Info/Index) pig genome assembly. Bedtools’ (Quinlan and Hall, 2010) function intersect was used to extract genes overlapping selective sweeps (candidate genomic regions calculated by REHH using the function calc_candidate_regions). The R packages clusterProfiler (Yu et al., 2012) and AnnotationHub (https://bioconductor.org/packages/release/bioc/html/AnnotationHub.html) were used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis.
Imputation from HD SNP array to WGS variants
A total of 487 three-way terminal crosses produced in the Danish commercial pig production system were genotyped using the Affymetrix Axiom PigHD SNPs chip (Axiom_PigHDv1, 658 k). The animals were part of the “MetaPig–Modulation of the pig gut metagenome to increase feed efficiency” project (http://www.metapig.eu). To determine marker positions in the Sscrofa11.1 assembly of the pig genome (Groenen et al., 2012), the probe sequences of the PigHD SNPs were mapped to the new assembly using BWA (Li, 2013). Markers with a unique map position and CIGAR value equal to “50M” were retained for further analyses. For the WGS marker set, non-autosomal markers and indels with a position coinciding with pigHD SNPs were removed. To phase the pigHD and WGS marker set, we used Eagle (Loh et al., 2016) with default parameter values. The WGS data set included 26,581,741 markers on 18 autosomes. Finally, we used Minimac3 (Das et al., 2016) to impute the pigHD marker set to the WGS level.
Comparison with pigHD and dbSNP database
The Affymetrix Axiom PigHD SNPs chip (Axiom_PigHDv1, 658 k) from the previous step was used to check the overlap of the WGS marker set. To check the overlap with the dbSNP database, the vcf file was downloaded from European Variation Archive (EVA) (https://www.ebi.ac.uk/eva/).
Results
General description of the variants
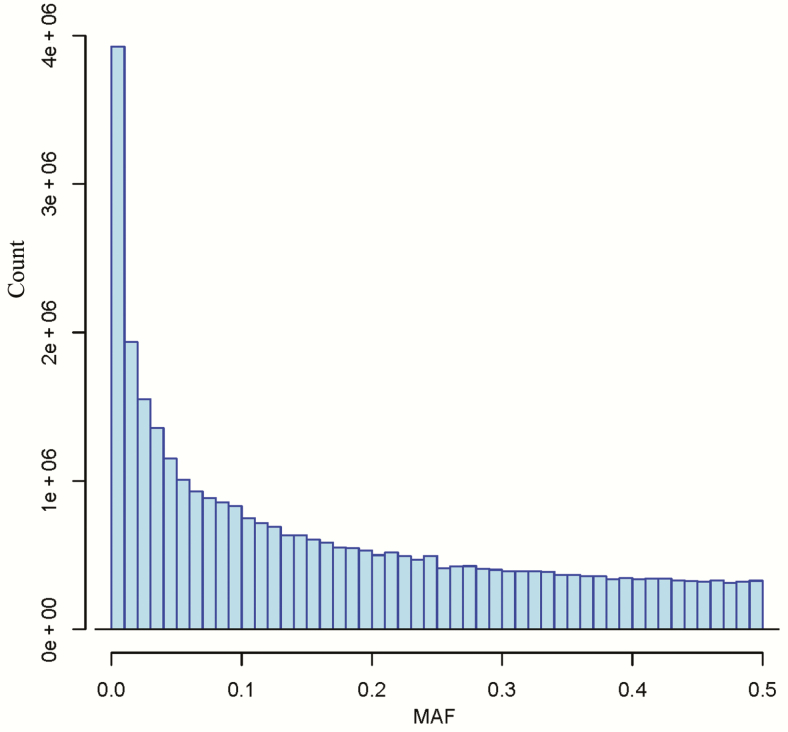
Thirty-four million variants across the pig genome were identified. Approximately, 32.21 million variants were located on the 18 autosomes. On 18 autosomes across the three breeds, there were 3.92 million variants with MAF ≤ 1% (rare), 5.99 million with MAF > 1%, but ≤5% (low frequency), and 22.29 million variants with MAF > 5% (common). A histogram of the distribution of MAFs across three breeds is presented in Figure 1.
Figure 1.
The distribution of MAF.
The WGS marker set was compared with pigHD and dbSNP marker sets. We successfully updated the location of 594,217 pigHD markers. A total of 534,536 variants overlapped with the pigHD marker set, and 21,197,692 markers were also present in dbSNP. Moreover, we had 11,471,879 new variants that were not in dbSNP and 43,946,701 dbSNP variants that were not segregating in these seqeunced animals.
Variant distribution and LD in each breed
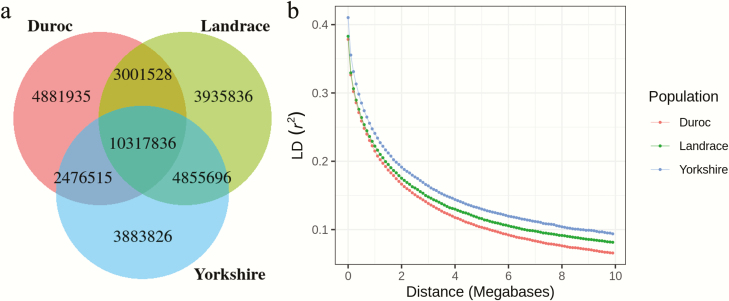
The number of genomic variants is similar in the three breeds. DD had 20,677,814 variants across the genome, while LL had 22,110,896 variants and YY had 21,533,874 variants. Almost half of the variants (10,317,836) segregated in all three breeds (Figure 2a). Moreover, DD had more variants (4,881,935) not found in LL or YY, and LL and YY had more variants (4,855,696) shared only between them (Figure 2a). To check whether a variant is shared or unique among these three breeds, we counted pair-wise common and breed-specific SNPs, insertions, and deletions (Table 1). The numbers are similar among three breeds; however, DD showed a higher number of breed-specific insertions though despite that DD had the lowest number of variants. The decay of LD with increasing distance in each of the breeds was estimated (Figure 2b). The result showed that DD exhibited the fastest decline of LD with physical distance, whereas YY had the longest.
Figure 2.
Counts of biallelic variants and LD decay in three breeds. (a) Venn diagram showing counts of shared and breed-specific variants in each breed and (b) decay of LD in each breed, the r2 is the mean of the r2 in 100 kb windows.
Table 1.
The number of variants (in millions) shared between breeds and breed-specific biallelic variants1
| Number of biallelic shared between pairs of breeds1 | Number of breed-specific variants | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPs | Deletions | Insertions | SNPs | Deletions | Insertions | |||||||
| DD | YY | LL | DD | YY | LL | DD | YY | LL | ||||
| DD | 16.21 | 9.88 | 10,31 | 1.86 | 0.78 | 0.83 | 2,55 | 1.11 | 1,24 | 3.94 | 0.71 | 0.91 |
| YY | 17.01 | 11.83 | 2.10 | 1.00 | 2.38 | 1.24 | 3.21 | 0.79 | 0.73 | |||
| LL | 17.51 | 2.08 | 2.48 | 3.28 | 0.70 | 0.71 | ||||||
1Diagonals are total biallelic variants within a breed; off-diagonals are shared variants.
Genetic relationships between the three breeds
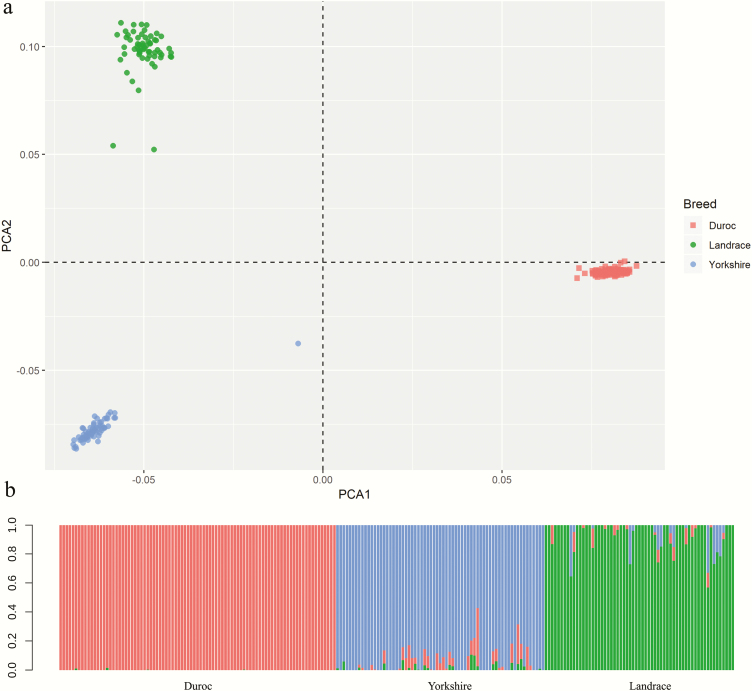
To assess the genetic structure of the three pig breeds, a PCA was performed using the WGS marker set. The first three components accounted for 42.56% (PC1), 29.69% (PC2), 3.31% (PC3), and 2.93% (PC4) of total variation (Figure 3a and Supplementary Figure S1). The first component separated the DD from LL and YY. The second component separated LL and YY (Figure 3a). There were three animals (two LL and one YY), which were highly admixed (Figure 3a). The Admixture analysis clearly distinguished three breeds as expected based on their origin with some admixed individuals (Figure 3b). Admixture analysis with K = 2, K = 3, and K = 4 (Supplementary Figure S2) was performed. The cross-validation error for K = 2 was 0.36, K = 3 was 0.28, and K = 4 was 0.28. Besides, the population assignment proportions of YY showed two components, except for K = 4 (Supplementary Figure S2). We concluded K = 3 was the optimal choice. In DD, we only detected a very small amount of admixture from LL in a few DD individuals. However, both in LL and YY, the analysis identified some individuals with admixture from the other two breeds.
Figure 3.
PCA plot and ADMIXTURE analysis to show the genetic relationship between three pig breeds (all 217 animals). (a) PCA plot of three breeds, indicating genetic variation along the first two eigenvectors. The dots indicate DD (salmon), YY (blue), and LL (green). (b) Population assignment proportions per individual based on results from ADMIXTURE analysis (K = 3). In the plot, each vertical bar represents a single individual and the different colors reflect the genetic contribution from each of the three components.
Runs of homozygosity
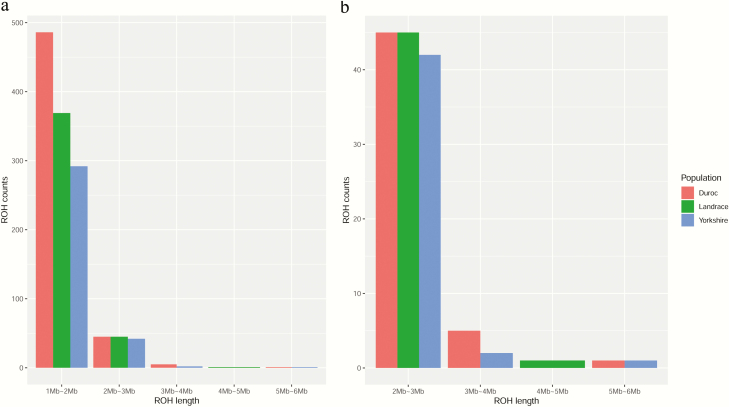
ROH in the three breeds with a length ≥1 Mb were detected (Figure 4a). DD had the largest number of short (1 to 2 Mb) ROH (Figure 4a). For longer ROH (2 to 3 Mb), the three breeds had similar numbers of ROH (Figure 4a and b). Overall, the three breeds had similar numbers of ROH with lengths ranging from 2 to 6 Mb.
Figure 4.
The distribution of ROH in three breeds. DD: red, LL: green, and YY: blue. (a) The length distribution of ROH longer than 1 Mb in three breeds and (b) the length distribution of ROH larger than 2 Mb in three breeds.
Population differentiation (FST) and integrated haplotype Score (iHS)
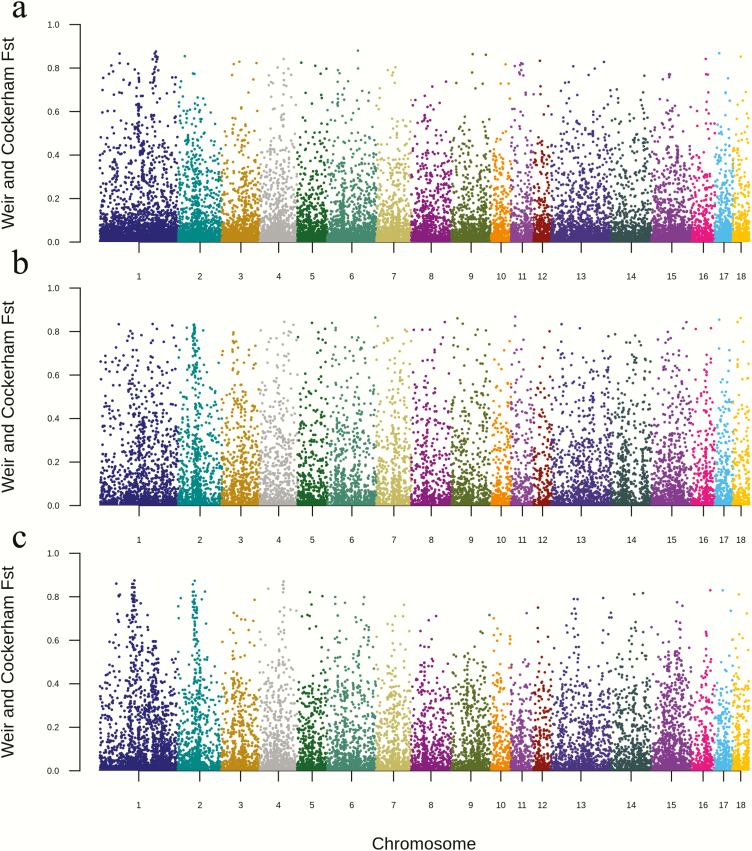
The simplest, and most common, statistic used to detect genomic divergence due to selection in a structured population is the FST. The FST analyses were performed as pair-wise comparisons of DD vs. LL (Figure 5a and Supplementary Table S1), DD vs. YY (Figure 5b and Supplementary Table S1), and LL vs. YY (Figure 5c and Supplementary Table S1). The average FST value of DD vs. LL was 0.046, DD vs. YY was 0.077, and LL vs. YY was 0.043. Moreover, Manhattan plots (Figure 5) showed that markers with high fixation values (FST > 0.8) were distributed uniformly across the genome, and without any clear pattern. In DD vs. LL, there were 36 variants with FST > 0.8 (Supplementary Table S1). In DD vs. YY, there were 49 variants with FST > 0.8 (Supplementary Table S1). In LL vs. YY, 41 variants had FST > 0.8 (Supplementary Table S1). Three variants showed FST > 0.8 in both DD vs. LL and DD vs. YY. They were 1:184624047 (an intronic variant of FBXO34), 17:15354511 (an intergenic variant close to ENSSSCG00000043546), and 18:26729134 (an intergenic variant close to KCND2). The low FST of the three comparisons indicated a low fixation of alleles in these three breeds.
Figure 5.
The Manhattan plot of the FST value of three pair-wise comparisons. (a) The Manhattan plot of FST between DD and LL; (b) the Manhattan plot of FST between DD and YY; and (c) the Manhattan plot of the FST between LL and YY.
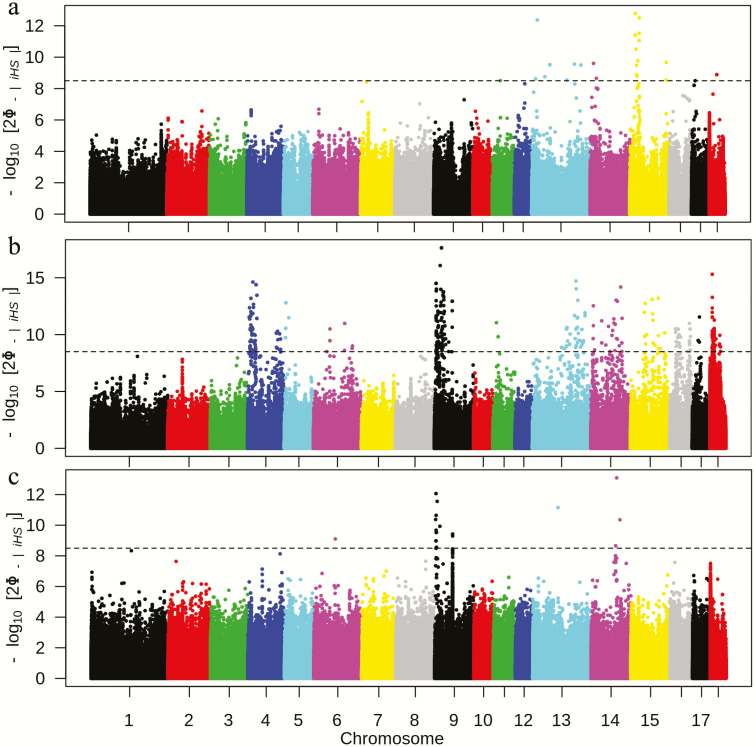
Integrated haplotype scores are shown in Figure 6 and Supplementary Table S2. Integrated haplotype score values for regions on chromosomes 4, 5, 6, 9, 11, 13, 14, 15, 16, 17, and 18 showed clear signals of genomic regions under selection. By contrast, a relatively smaller number of peaks in DD and YY were detected (Figure 6a and c and Supplementary Table S2). Chromosomes 11, 13, 14, 15, 17, and 18 in DD (and Supplementary Table S2) and chromosomes 9 in YY (and Supplementary Table S2) had regions under recent selection. The candidate region for LL and YY had one overlapping region on chromosome 9: 62600000 ~ 64400000. The iHS results showed that LL had more genomic regions under selection and reflected the fact that the three breeds are selected for different goals.
Figure 6.
The Manhattan plot of the –log10(P) value of iHS for each breed; the value of 8.5 was indicated as the threshold for significance (Bonferroni correction). (a) The Manhattan plot of –log10(P) values of iHS for DD; (b) the Manhattan plot of –log10(P) values of iHS for LL; and (c) the Manhattan plot of –log10(P) values of iHS for YY.
The genes within each candidate genomic region under selection were investigated for the enrichment of GO terms and KEGG terms. The genes in the candidate genomic region of DD did not show any enrichment (Table 2 and Supplementary Table S3). For LL, two enriched GO terms: “olfactory receptor activity” (GO:0004984, false discovery rate [FDR] < 0.01 for 42 genes; Supplementary Table S3) and “oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen” (GO:0016705, FDR < 0.01 for 15 genes; Supplementary Table S3) were discovered. For YY, three GO terms were enriched: “olfactory receptor activity” (GO:0004984, FDR < 0.01 and 21 genes; Supplementary Table S3), “oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen” (GO:0016709, FDR < 0.01 and 3 genes; Supplementary Table S3), and “NADP binding” (GO:0050661, FDR < 0.01 for 3 genes; Supplementary Table S3). The enrichment analysis of the KEGG pathway yielded one term enriched in YY, “Drug metabolism—cytochrome P450” (FDR < 0.05 for 5 genes; Table 2).
Table 2.
KEGG enrichment analysis
| Breed | KEGG | Adjusted P | Gene number | Genes |
|---|---|---|---|---|
| YY | Drug metabolism—cytochrome P450 | 2.44e-02 | 5 | FMO3, ENSSSCG00000022759, FMO2, FMO1, and FMO4 |
Annotation
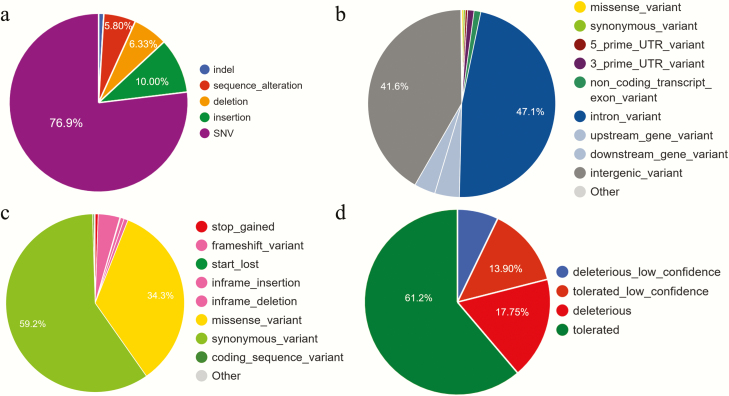
Variants across the genome were annotated with VEP. VEP annotated 99.52% (34,415,508 out of 34,581,909) of the variants, leaving out the variants located on the unplaced contigs. In total, 31,728 genes and 62,796 transcripts harbored at least one variant. About 77% of all the variants were single-nucleotide variants (Figure 7a). Similarly, based on the consequence type for each variant, intron variants were the most abundant variants followed by intergenic variants (Figure 7b). In this dataset, 29,430 variants were annotated as “high impact,” which included stop-gained, stop-lost, out-of-frame frameshift, start-lost, splice acceptor, and splice donor variants. Among variants in coding regions, synonymous variants were the most abundant type (493,099), followed by missense variants (161.833); see Figure 7c. Among the missense variants, most (99,046) had SIFT scores that indicated they were “tolerated” (Figure 7d), whereas 28,727 had scores that classified them as “deleterious.”
Figure 7.
Distribution of VEP annotation. (a) Variant class, (b) predicted consequences for each variant, (c) coding variants, and (d) distribution of four categories of SIFT scores among missense variants.
Imputation
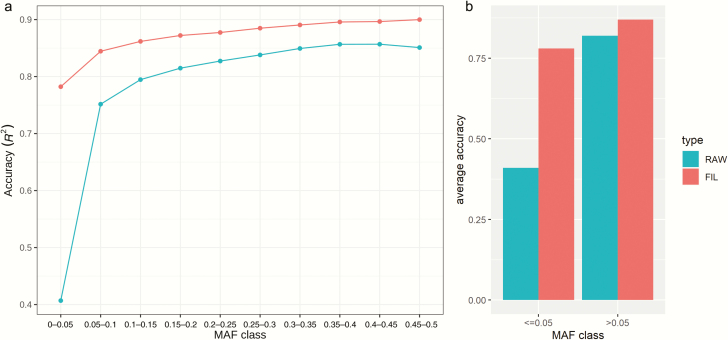
To examine the suitability of the WGS marker set as a reference for the imputation of SNP array genotypes, we used a three-way terminal-cross pig population with 487 animals genotyped using the pigHD chip (Affymetrix Axiom Pig HD SNPs, 658 K) to perform the imputation. The average imputation accuracy of all variants was 0.69. The imputation accuracy was above 0.83 for variants with MAF larger than 0.1 (Figure 8a). MAF is a key factor affecting imputation accuracy. For variants with MAF ≤ 0.05, the average accuracy was less than 0.50 (Figure 8b). The majority of the variants with MAF > 0.05 had an imputation accuracy above 0.8, with an average of 0.85 (Figure 8b).
Figure 8.
The statistics of the imputation accuracy (r2 reported by Minimac3). (a) The imputation accuracy across all MAF classes. The blue line shows the imputation accuracy of all variants in different MAF classes. The red line shows the imputation accuracy of variants with imputation accuracy, r2 > 0.4 for different MAF classes. (b) The imputation accuracy across for variants with MAF ≤ 0.05 and MAF > 0.05. The blue bars show the imputation accuracy of all variants (RAW). The red bars show the imputation accuracy of variants with r2 > 0.4 (FIL).
Discussion
As the price of next-generation sequencing decreases and analysis pipelines continue to improve, large-scale sequencing of livestock populations becomes affordable and practicable. Here, 217 animals from three Danish commercial pig breeds were sequenced to investigate the population structure, detect selection signatures, and produce a reference to impute SNP chip genotypes to WGS variants. The variant calling procedure detected 34 million variants across the whole genome. A study with 69 Chinese pigs from 11 diverse breeds and 3 wild boar populations identified more than 40 million variants (Ai et al., 2015). The number of variants in our marker set is within the range observed from this report, considering we had three commercial breeds. Similarly, in cattle, a WGS study with 234 bulls identified 28.3 million variants (Daetwyler et al., 2014). Another WGS study in cattle with 65 animals reported between 17.7 and 22.0 million variants, depending on the method used and sampling scheme (Baes et al., 2014).
A whole-genome marker set is a good resource for genome-wide association studies (GWAS) (Yan et al., 2017; Cai et al., 2019a) and population genetics studies (Francioli et al., 2014). Besides, the annotation of variants by VEP revealed more than 200,000 amino acid altering variants, 29,000 with high-impact predicted consequences. A previous study has shown that variant annotation is a good resource for post-GWAS analysis (Cai et al., 2019c). Moreover, the uniform distribution of WGS variants without ascertainment bias toward common variants and high marker density gives us the ability to investigate the genome and population in a less biased manner than when using ascertained marker sets included in SNP arrays.
One application of a WGS marker set is imputation. Imputation has been used for GWAS in other species, for example, cattle (Cai et al., 2018, 2019a, 2019b). However, the use of WGS variant sets for imputation in pigs is still limited. In this study, we investigated the imputation accuracy in a challenging setup by imputing from a pig HD marker genotyped in a crossbred population. This imputation setup was challenging for the following reasons: 1) the test population pigs were not closely related to the WGS reference panel; 2) the test population consisted of crossbred animals, whereas the reference population was drawn from three purebred populations; and 3) the relatively small size of the WGS reference population. A previous study indicated that factors affecting the imputation accuracy included reference group size, degree of relationship between validation and reference groups, LD, MAF, among others (van Binsbergen et al., 2014). Even with the experimental limitations, an imputation accuracy comparable to cattle imputation with similar reference population sizes was evident (Daetwyler et al., 2014). In this cattle study, the imputation accuracy was also low for markers with MAF < 0.1, and the overall average of imputation accuracy was below 0.8 (Daetwyler et al., 2014). We achieved higher imputation accuracy compared to other pig studies (Yan et al., 2018; van den Berg et al., 2019). One reason for the higher imputation accuracy could be improved genome assembly (Sscrofa11.1) and reliability of variant calling of the WGS animals since the quality in the assembly and variants in the reference panel affect the imputation accuracy (van Binsbergen et al., 2014). Other reasons could be improved determination of marker positions in the new reference genome and the animals in the test set were crossbred, having higher LD than purebred animals. The imputation accuracy also indicated proper handling of the HD and WGS marker set during the imputation, including the accuracy of lift-over of genome location to the new assembly and appropriateness of quality control procedures adopted.
The PCA analysis and ADMIXTURE showed that the three breeds were clustered by breed (Figure 3). DD, compared with LL and YY, had less admixture from the other two populations (Figure 3b). Moreover, the higher number of private variants in DD suggests that DD is less admixed with the other two populations. A higher number of ROH in DD compared with LL and YY was observed. Moreover, the difference in ROH counts was more pronounced for short ROH (1 to 2 Mb, Figure 4a). These two characteristics of ROH indicated stronger inbreeding in the past (background relationship) in DD. The level of inbreeding could affect LD patterns in the genome (Gaut and Long, 2003). The extent of LD in these three breeds (Figure 2b) was similar to the LD observed in five pure pig lines (Veroneze et al., 2014) since r2 dropped below 0.2 after approximately 1 Mb. Moreover, the LD has a shorter range in DD than in LL and YY, which supported that inbreeding happened in early generations. A previous study of LD decay in the Danish pig populations is in accordance with these findings (Wang et al., 2013).
The mean FST of three pair-wise combinations did not indicate strong differentiation among the breeds; only 4% to 7% of the genetic variation was due to drift. FST < 0.05 indicated little genetic differentiation. FST ranging from 0.05 to 0.15 means moderate genetic difference (Hartl et al., 1997). Therefore, DD vs. LL and LL vs. YY exhibited little genetic difference; DD and YY had a moderate genetic differentiation. Besides, Manhattan plots of FST (Figure 5) revealed that fixed loci were occurred across the whole genome without showing any clear hot spots. We also applied an iHS scan to identify genomic regions that had undergone a recent selection. Within genomic regions recently under selection, we found that LL and YY had enrichment for genes involved in the olfactory receptor activity and oxidoreductase activity. Genes related to oxidoreductase activity may act as an antioxidant extending lifespan in fly and mice (Mitsui et al., 2002; Umeda-Kameyama et al., 2007). In the study of long-lived rodents, an enrichment of oxidoreductase activity was found in the selected genes (Sahm et al., 2018). The pig genome was found to have undergone expansion for olfactory receptor genes (Groenen et al., 2012). Pigs have the largest repertoire of functional olfactory receptor genes (Groenen et al., 2012). There are 1,301 porcine olfactory receptor genes. In addition, there are 343 partial olfactory receptor genes in the pig genome (Groenen et al., 2012). Previous studies in other pig populations or cattle populations also identified selection signature in olfactory receptor genes (Groenen, 2016; Bahbahani et al., 2018; Muñoz et al., 2019). The selective sweep in the olfactory receptor genes suggests that olfactory loci play a major role in the domestication of species (Ramey et al., 2013). Olfactory receptors have also been found to be under selection in humans (Nielsen et al, 2005). In summary, our results indicated that the genes involved in the olfactory receptor activity, oxidoreductase activity, and P450 genes have been under recent selection in LL and YY.
In conclusion, the DNA from 217 pigs from three commercial breeds was sequenced. From this dataset, 34 million variants across the genome were discovered, which provides a good reference for imputation. The variant annotation showed potential functional variants in our marker set, which could be utilized for post-GWAS analysis. The genomic differences among the breeds were revealed by this marker set. Moreover, the iHS identified olfactory receptors and oxidoreductases genes to have been targets of recent selection.
Supplementary Material
Acknowledgments
We acknowledge Søren Svendsen and Thu Hong Le, Center for Quantitative Genetics and Genomics, Aarhus University, Denmark, for their help in arranging the blood samples for whole-genome sequencing. “MetaPig–Modulation of the pig gut metagenome to increase feed efficiency” project is acknowledged for sharing PigHD genotype data. This research was supported in part by the Center for Genomic Selection in Animals and Plants (GenSAP) funded by Innovation Fund Denmark, grant number 0603-00519B.
Glossary
Abbreviations
- BWA
Burrows-Wheeler Aligner
- DD
Duroc
- FDR
false discovery rate
- FST
fixation index
- GATK
Genome Analysis Toolkit
- GO
Gene Ontology
- GWAS
genome-wide association studies
- iHS
integrated haplotype score
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LD
linkage disequilibrium
- LL
Landrace
- MAF
minor allele frequency
- PCA
principal component analysis
- ROH
runs of homozygosity
- SNP
single-nucleotide polymorphisms
- VEP
variant effect predictor
- WGS
whole-genome sequencing
- YY
Yorkshire
Data Availability Statement
The data that support the findings of this study are available from SEGES Danish Pig Research Centre, 1609, Copenhagen V, Denmark (Whole genome sequence data) and Department of Veterinary and Animal Sciences, University of Copenhagen, 1870 Frederiksberg C, Copenhagen, Denmark (High density SNP array data). Restrictions apply to the availability of these data, which were used under license for this study. Data are available from the authors with the permission of SEGES Danish Pig Research Centre and University of Copenhagen.
Conflict of interest statement
The authors declare no conflict of interest.
Literature Cited
- Ai H., Fang X., Yang B., Huang Z., Chen H., Mao L., Zhang F., Zhang L., Cui L., He W.,. et al. 2015. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat. Genet. 47:217–225. doi: 10.1038/ng.3199 [DOI] [PubMed] [Google Scholar]
- Alexander D. H., Novembre J., and Lange K... 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655–1664. doi: 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baes C. F., Dolezal M. A., Koltes J. E., Bapst B., Fritz-Waters E., Jansen S., Flury C., Signer-Hasler H., Stricker C., Fernando R.,. et al. 2014. Evaluation of variant identification methods for whole genome sequencing data in dairy cattle. BMC Genomics 15:948. doi: 10.1186/1471-2164-15-948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahbahani H., Salim B., Almathen F., Al Enezi F., Mwacharo J. M., and Hanotte O... 2018. Signatures of positive selection in African Butana and Kenana dairy zebu cattle. PLoS One. 13(1):e0190446. doi: 10.1371/journal.pone.0190446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barendse W., Harrison B. E., Bunch R. J., Thomas M. B., and Turner L. B... 2009. Genome wide signatures of positive selection: the comparison of independent samples and the identification of regions associated to traits. BMC Genomics 10:178. doi: 10.1186/1471-2164-10-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg S., Vandenplas J., van Eeuwijk F. A., Bouwman A. C., Lopes M. S., and Veerkamp R. F... 2019. Imputation to whole-genome sequence using multiple pig populations and its use in genome-wide association studies. Genet. Sel. Evol. 51:2. doi: 10.1186/s12711-019-0445-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Binsbergen R., Bink M. C., Calus M. P., van Eeuwijk F. A., Hayes B. J., Hulsegge I., and Veerkamp R. F... 2014. Accuracy of imputation to whole-genome sequence data in Holstein Friesian cattle. Genet. Sel. Evol. 46:41. doi: 10.1186/1297-9686-46-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning S. R., and Browning B. L... 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81:1084–1097. doi: 10.1086/521987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z., Guldbrandtsen B., Lund M. S., and Sahana G... 2018. Prioritizing candidate genes post-GWAS using multiple sources of data for mastitis resistance in dairy cattle. BMC Genomics 19:656. doi: 10.1186/s12864-018-5050-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z., Guldbrandtsen B., Lund M. S., and Sahana G... 2019a. Dissecting closely linked association signals in combination with the mammalian phenotype database can identify candidate genes in dairy cattle. BMC Genet. 20:15. doi: 10.1186/s12863-019-0717-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z., Guldbrandtsen B., Lund M. S., and Sahana G... 2019b. Prioritizing candidate genes for fertility in dairy cows using gene-based analysis, functional annotation and differential gene expression. BMC Genomics 20:255. doi: 10.1186/s12864-019-5638-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z., Guldbrandtsen B., Lund M. S., and Sahana G... 2019c. Weighting sequence variants based on their annotation increases the power of genome-wide association studies in dairy cattle. Genet. Sel. Evol. 51:20. doi: 10.1186/s12711-019-0463-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos F. C., Joshi P. K., Clark D. W., Ramsay M., and Wilson J. F... 2018. Runs of homozygosity: windows into population history and trait architecture. Nat. Rev. Genet. 19:220–234. doi: 10.1038/nrg.2017.109 [DOI] [PubMed] [Google Scholar]
- Choi J. W., Chung W. H., Lee K. T., Cho E. S., Lee S. W., Choi B. H., Lee S. H., Lim W., Lim D., Lee Y. G.,. et al. 2015. Whole-genome resequencing analyses of five pig breeds, including Korean wild and native, and three European origin breeds. DNA Res. 22:259–267. doi: 10.1093/dnares/dsv011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., Capitan A., Pausch H., Stothard P., van Binsbergen R., Brøndum R. F., Liao X., Djari A., Rodriguez S. C., Grohs C.,. et al. 2014. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 46:858–865. doi: 10.1038/ng.3034 [DOI] [PubMed] [Google Scholar]
- Das S., Forer L., Schönherr S., Sidore C., Locke A. E., Kwong A., Vrieze S. I., Chew E. Y., Levy S., McGue M.,. et al. 2016. Next-generation genotype imputation service and methods. Nat. Genet. 48:1284–1287. doi: 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deniskova T. E., Dotsev A. V., Selionova M. I., Kunz E., Medugorac I., Reyer H., Wimmers K., Barbato M., Traspov A. A., Brem G.,. et al. 2018. Population structure and genetic diversity of 25 Russian sheep breeds based on whole-genome genotyping. Genet. Sel. Evol. 50:29. doi: 10.1186/s12711-018-0399-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francioli L. C., Menelaou A., Pulit S. L., Van Dijk F., Palamara P. F., Elbers C. C., Neerincx P. B., Ye K., Guryev V., and Kloosterman W. P... 2014. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 46(8):818. doi: 10.1038/ng.3021 [DOI] [PubMed] [Google Scholar]
- Gaut B. S., and Long A. D... 2003. The lowdown on linkage disequilibrium. Plant Cell. 15:1502–1506. doi: 10.1105/tpc.150730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautason E., Schönherz A. A., Sahana G., and Guldbrandtsen B... 2019. Relationship of Icelandic cattle with Northern and Western European cattle breeds, admixture and population structure. Acta Agric. Scand. A Anim. Sci. 69:1–14. doi: 10.1080/09064702.2019.1699951 [DOI] [Google Scholar]
- Gautier M., Klassmann A., and Vitalis R... 2017. REHH 2.0: a reimplementation of the R package REHH to detect positive selection from haplotype structure. Mol. Ecol. Resour. 17:78–90. doi: 10.1111/1755-0998.12634 [DOI] [PubMed] [Google Scholar]
- Groenen M. A. 2016. A decade of pig genome sequencing: a window on pig domestication and evolution. Genet. Sel. Evol. 48:23. doi: 10.1186/s12711-016-0204-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen M. A., Archibald A. L., Uenishi H., Tuggle C. K., Takeuchi Y., Rothschild M. F., Rogel-Gaillard C., Park C., Milan D., Megens H. J.,. et al. 2012. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491:393–398. doi: 10.1038/nature11622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl D. L., Clark A. G., and Clark A. G... 1997. Principles of population genetics. Sunderland (MA):Sinauer Associates. [Google Scholar]
- Huang M., Yang B., Chen H., Zhang H., Wu Z., Ai H., Ren J., and Huang L... 2019. The fine‐scale genetic structure and selection signals of Chinese indigenous pigs. Evol. Appl. 13(2):458–475. doi: 10.1111/eva.12887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R., Orozco-terWengel P., De Maio N., Pandey R. V., Nolte V., Futschik A., Kosiol C., and Schlötterer C... 2011. PoPoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS One. 6:e15925. doi: 10.1371/journal.pone.0015925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Henikoff S., and Ng P. C... 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4:1073–1081. doi: 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv, arXiv:1303.3997. doi: 10.1101/2020.03.24.006650, preprint [DOI] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079. doi: 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh P.-R., Palamara P. F., and Price A. L... 2016. Fast and accurate long-range phasing in a UK Biobank cohort. Nat. Genet. 48(7):811–816. doi: 10.1038/ng.3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I., and McVean G... 2014. Demography and the age of rare variants. PLoS Genet. 10:e1004528. doi: 10.1371/journal.pgen.1004528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W., Gil L., Hunt S. E., Riat H. S., Ritchie G. R., Thormann A., Flicek P., and Cunningham F... 2016. The Ensembl variant effect predictor. Genome Biol. 17:122. doi: 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui A., Hamuro J., Nakamura H., Kondo N., Hirabayashi Y., Ishizaki-Koizumi S., Hirakawa T., Inoue T., and Yodoi J... 2002. Overexpression of human thioredoxin in transgenic mice controls oxidative stress and life span. Antioxid. Redox Signal. 4:693–696. doi: 10.1089/15230860260220201 [DOI] [PubMed] [Google Scholar]
- Molnár J., Nagy T., Stéger V., Tóth G., Marincs F., and Barta E... 2014. Genome sequencing and analysis of Mangalica, a fatty local pig of Hungary. BMC Genomics 15:761. doi: 10.1186/1471-2164-15-761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz M., Bozzi R., García-Casco J., Núñez Y., Ribani A., Franci O., García F., Škrlep M., Schiavo G., Bovo S.,. et al. 2019. Genomic diversity, linkage disequilibrium and selection signatures in European local pig breeds assessed with a high density SNP chip. Sci. Rep. 9:13546. doi: 10.1038/s41598-019-49830-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Bustamante C., Clark A. G., Glanowski S., Sackton T. B., Hubisz M. J., Fledel-Alon A., Tanenbaum D. M., Civello D., White T. J., Sninsky J. J., Adams M. D., Cargill M.. 2005. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 3(6):e170. doi: 10.1371/journal.pbio.0030170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplin R., Ruano-Rubio V., DePristo M. A., Fennell T. J., Carneiro M. O., Van der Auwera G. A., Kling D. E., Gauthier L. D., Levy-Moonshine A., and Roazen D... 2018. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv, bioRxiv:201178. doi: 10.1101/201178 [DOI] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J.,. et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81:559–575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qanbari S., and Simianer H... 2014. Mapping signatures of positive selection in the genome of livestock. Livest. Sci. 166:133–143. doi: 10.1016/j.livsci.2014.05.003 [DOI] [Google Scholar]
- Quinlan A. R., and Hall I. M.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26(6):841–842. doi: 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey H. R., Decker J. E., McKay S. D., Rolf M. M., Schnabel R. D., and Taylor J. F... 2013. Detection of selective sweeps in cattle using genome-wide SNP data. BMC Genomics 14:382. doi: 10.1186/1471-2164-14-382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin C. J., Zody M. C., Eriksson J., Meadows J. R., Sherwood E., Webster M. T., Jiang L., Ingman M., Sharpe T., Ka S.,. et al. 2010. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 464:587–591. doi: 10.1038/nature08832 [DOI] [PubMed] [Google Scholar]
- Sahm A., Bens M., Szafranski K., Holtze S., Groth M., Görlach M., Calkhoven C., Müller C., Schwab M., Kraus J.,. et al. 2018. Long-lived rodents reveal signatures of positive selection in genes associated with lifespan. PLoS Genet. 14:e1007272. doi: 10.1371/journal.pgen.1007272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Ye S., Jiang Y., Zhang Z., Zhang Q., and Ding X... 2019. Using imputation-based whole-genome sequencing data to improve the accuracy of genomic prediction for combined populations in pigs. Genet. Sel. Evol. 51:58. doi: 10.1186/s12711-019-0500-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda-Kameyama Y., Tsuda M., Ohkura C., Matsuo T., Namba Y., Ohuchi Y., and Aigaki T... 2007. Thioredoxin suppresses Parkin-associated endothelin receptor-like receptor-induced neurotoxicity and extends longevity in Drosophila. J. Biol. Chem. 282:11180–11187. doi: 10.1074/jbc.M700937200 [DOI] [PubMed] [Google Scholar]
- Veroneze R., Bastiaansen J. W., Knol E. F., Guimarães S. E., Silva F. F., Harlizius B., Lopes M. S., and Lopes P. S... 2014. Linkage disequilibrium patterns and persistence of phase in purebred and crossbred pig (Sus scrofa) populations. BMC Genet. 15:126. doi: 10.1186/s12863-014-0126-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight B. F., Kudaravalli S., Wen X., and Pritchard J. K... 2006. A map of recent positive selection in the human genome. PLoS Biol. 4:e72. doi: 10.1371/journal.pbio.0040072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Sørensen P., Janss L., Ostersen T., and Edwards D... 2013. Genome-wide and local pattern of linkage disequilibrium and persistence of phase for 3 Danish pig breeds. BMC Genet. 14:115. doi: 10.1186/1471-2156-14-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr A., Affara N., Aken B., Beiki H., Bickhart D. M., Billis K., Chow W., Eory L., Finlayson H. A., and Flicek P... 2020. An improved pig reference genome sequence to enable pig genetics and genomics research. GigaScience 9(6):giaa051.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. 1949. The gebetical structure of populations. Ann. Eugen. 15(1):323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x [DOI] [PubMed] [Google Scholar]
- Yan G., Guo T., Xiao S., Zhang F., Xin W., Huang T., Xu W., Li Y., Zhang Z., and Huang L... 2018. Imputation-based whole-genome sequence association study reveals constant and novel loci for hematological traits in a large-scale swine F2 resource population. Front. Genet. 9:401. doi: 10.3389/fgene.2018.00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G., Qiao R., Zhang F., Xin W., Xiao S., Huang T., Zhang Z., and Huang L... 2017. Imputation-based whole-genome sequence association study rediscovered the missing QTL for lumbar number in Sutai pigs. Sci. Rep. 7(1):1–10. doi: 10.1038/s41598-017-00729-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L. G., Han Y., and He Q. Y... 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. doi: 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from SEGES Danish Pig Research Centre, 1609, Copenhagen V, Denmark (Whole genome sequence data) and Department of Veterinary and Animal Sciences, University of Copenhagen, 1870 Frederiksberg C, Copenhagen, Denmark (High density SNP array data). Restrictions apply to the availability of these data, which were used under license for this study. Data are available from the authors with the permission of SEGES Danish Pig Research Centre and University of Copenhagen.