SUMMARY
Basement membranes (BMs) are supramolecular matrices built on laminin and type IV collagen networks that provide tissues structural and signaling support. BM complexity, however, has hindered an understanding of its formation, dynamics, and regulation. Using genome editing, we tagged 29 BM matrix components and receptors in C. elegans with mNeonGreen. Here, we report a common template that initiates BM formation, which rapidly diversifies during tissue differentiation. Through photobleaching studies, we show that BMs are not static—surprisingly, many matrix proteins move within the laminin and collagen scaffoldings. Finally, quantitative imaging, conditional knockdown, and optical highlighting indicate that papilin, a poorly studied glycoprotein, is the most abundant component in the gonadal BM, where it facilitates type IV collagen removal during BM expansion and tissue growth. Together, this work introduces methods for holistic investigation of BM regulation, and reveals that BMs are highly dynamic and capable of rapid change to support tissues.
Keywords: Basement membrane, extracellular matrix, type IV collagen, laminin, organ growth, papilin, endogenous tagging, C. elegans, basement membrane dynamics
Graphical Abstract

eTOC blurb
Basement membranes (BMs) are complex extracellular structures that are difficult to study in vivo. Keeley et al. use CRISPR/Cas9 genome editing to fluorescently tag 29 BM components in C. elegans and discover that BMs are not static—many matrix proteins move within a stable laminin and collagen IV-based scaffold.
INTRODUCTION
Basement membranes (BMs) are thin, dense sheets of extracellular matrix (ECM) that surround most tissues (Yurchenco, 2011). Comparative phylogenetic studies have suggested that BMs are the most ancient ECM and arose at the time of animal multicellularity (Fidler et al., 2018). BMs are supramolecular assemblies composed of a core set of proteins that includes laminin networks, cross-linked type IV collagen grids, the glycoprotein nidogen, and often the heparan sulfate proteoglycans perlecan, agrin, and type XVIII collagen. Matricellular proteins such as fibulins and SPARC, proteases, protease inhibitors, and growth factors are also BM residents (Yurchenco, 2011). Numerous receptors, including integrin and dystroglycan, interact with BM matrix components to mediate laminin polymerization in embryonic cells, an event necessary to seed initial BM formation (Yurchenco, 2015). BMs have numerous essential structural roles in animals—they bear load to protect tissues from mechanical forces, filter blood in the kidney, help form the blood-brain barrier and neuromuscular junctions, and mediate tissue shaping (Morrissey and Sherwood, 2015; Rogers and Nishimune, 2017). BMs also have crucial signaling activities that polarize epithelia, regulate tissue growth, guide cell migration, and promote cell survival (Bunt et al., 2010; Li et al., 2017; Ma et al., 2017; Rasmussen et al., 2012). To achieve distinct functions, BMs vary significantly in composition, thickness, and mechanical properties (Chang and Chaudhuri, 2019). More than 20 distinct human diseases are associated with genetic disruption of BM components, highlighting the diverse and important roles of BMs (Nyström et al., 2017). The misregulation of BM component expression is also a key driver of cancer metastasis, and defective BM turnover leads to tissue decline in diabetes mellitus, fibrosis, and aging (Candiello et al., 2010; Halfter et al., 2017; Naba et al., 2014).
Despite the fundamental importance of BMs to development, homeostasis, and disease, mechanisms regulating their structure, growth, and turnover are poorly understood. BMs interact dynamically with tissues and thus require in vivo models to elucidate their regulation and function. Further, as each BM is a unique assemblage of matrix components, experimental approaches that address the complete make-up of BMs in vivo are needed. Because of BM compositional complexity, most studies have focused on only a handful of components. Mass-spectrometry approaches combined with bioinformatics are beginning to characterize the complete ECM-associated with tissues (Schiller et al., 2015), but limitations include challenges with protein contamination, solubility, identification, and confirmation of matrix proteins as bona fide BM inhabitants (Randles et al., 2017).
There is also a gap in our understanding of BM component dynamics. Previous work on BM turnover using pulse-chase experiments suggests that BMs turn over on a scale of weeks (Decaris et al., 2014; Trier et al., 1990). While these studies indicate that BM matrix components are stable and have long half-lives, they have not revealed whether matrix components are fixed in place once deposited within BMs. Live cell imaging in Drosophila with GFP-tagged type IV collagen, perlecan, nidogen, and laminin has revealed dynamic mechanisms of matrix deposition during egg chamber formation and initial BM assembly (Isabella and Horne-Badovinac, 2016; Matsubayashi et al., 2017). However, endogenously tagged BM matrix proteins are limited in Drosophila to only type IV collagen and perlecan (Ramos-Lewis et al., 2018) and BM-encased organs in larvae and adults lie deep within tissues beyond the reach of light microscopy, hindering analysis of matrix dynamics after deposition (Cetera et al., 2016).
Caenorhabitis elegans is a powerful model in which to examine BM biology. CRISPR/Cas9-mediated homologous recombination has facilitated the generation of endogenously tagged laminin, type IV collagen, nidogen, integrin, and dystroglycan genes with genetically encoded fluorophores (Jayadev et al., 2019; Matsuo et al., 2019; Naegeli et al., 2017; Walser et al., 2017). C. elegans is also small and translucent, which allows live imaging of BMs from the embryo through adulthood. In addition, unlike vertebrates where BM gene families have expanded, C. elegans has single genes encoding most major BM components, simplifying genetic and compositional studies (Clay and Sherwood, 2015). Finally, RNAi-mediated knockdown allows for conditional loss of BM components, facilitating later functional studies of BM components required for embryonic viability (Morrissey et al., 2016).
To comprehensively examine BMs, we used genome engineering to introduce mNeonGreen (mNG) into the genomic loci encoding 17 BM matrix components and 12 receptors. We show that a similar template of matrix components and receptors is associated with initial BM formation throughout the embryo shortly after gastrulation, but then BMs diversify dramatically on tissues as development proceeds. By performing fluorescent recovery after photobleaching (FRAP), we demonstrate that BMs consist of a stable scaffolding of collagen and laminin, within which the matrix proteins nidogen, fibulin, agrin, spondin, and peroxidasin move, thus revealing an unexpected dynamic property of BMs. Finally, we address a poorly understood aspect of BM biology—BM expansion during organ growth. Using quantitative live imaging and conditional knockdown, we reveal that the glycoprotein papilin is the most abundant matrix component within the rapidly growing gonadal BM, and that loss of papilin results in smaller gonads and type IV collagen accumulation into a fibrotic network during BM growth. Through optical highlighting of type IV collagen, we show that papilin promotes collagen removal during BM expansion, and that papilin functions in part by limiting BM access of collagen modifying ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) proteins. Together, this collection of endogenously tagged BM components provides a new resource to investigate BM formation, diversity, and regulation, and reveals that BMs are not static after deposition, but rather have highly mobile components that may confer dynamic properties to BMs.
RESULTS
Endogenous, Fluorescently Tagged Basement Membrane (BM) Proteins
The properties and regulation of BMs remain elusive in part because of the challenge of studying BM composition and dynamics in vivo. C. elegans is a useful model to examine BMs due to its optical clarity, amenability to genome editing, and conserved BM gene families that have not undergone the expansion found in vertebrates (Clay and Sherwood, 2015). To better understand BM regulation, we used CRISPR/Cas9-mediated homologous recombination to knock-in genetically encoded fluorescent proteins into 29 genes encoding conserved matrix components and receptors in C. elegans (Figures 1 and S1 and S2; Table S1) (Dickinson and Goldstein, 2016; Gotenstein et al., 2010; Hrus et al., 2007; Kramer, 2005; Topf and Chiquet-Ehrismann, 2011; Unsoeld et al., 2013). These genes encode: 1) the core basement membrane components laminin (αA/LAM-3, αB/EPI-1, β/LAM-1, γ/LAM-2), type IV collagen (α1/EMB-9 and α2/LET-2), perlecan/UNC-52, agrin/AGR-1, nidogen/NID-1, collagen XVIII/CLE-1); 2) the matricellular proteins SPARC/OST-1, fibulin/FBL-1, spondin/SPON-1, papilin/MIG-6, hemicentin/HIM-4; 3) the peroxidasin enzymes (PXN-1 and PXN-2); and 4) BM-associated receptors, including integrins (α/INA-1, α/PAT-2, and β/PAT-3), teneurin/TEN-1, dystroglycan (DGN-1 and DGN-2), syndecan/SDN-1, glypicans (GPN-1 and LON-2), LAR phosphatase/PTP-3, and discoidin domain receptors (DDR-1 and DDR-2) (Figure 1; Table S1). All genes were tagged with mNG to allow for direct comparisons of protein levels by fluorescence intensity. Select genes were tagged with the red fluorophore mKate2 (Figures S1 and S2; Table S1) (Heppert et al., 2016). Most fluorophores were attached at the C-terminus with an 18 amino acid non-polar flexible linker, although some were inserted N-terminally or internally to visualize multiple splice variants or avoid post-translational cleavage (Figures S1 and S2; Table S1; see below).
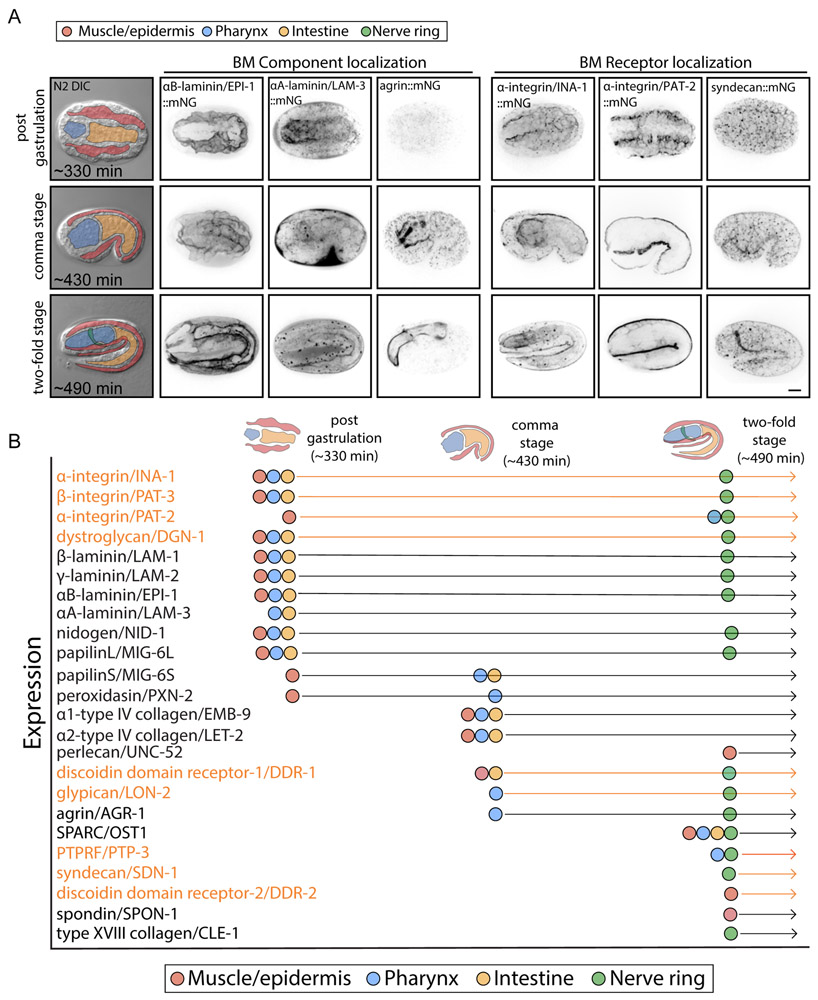
Figure 1. Basement Membrane (BM) Matrix Components and Receptors in C. elegans.
(A) A schematic of the embryonic and larval stages of the C. elegans life cycle labeled with developmental times post-fertilization (embryonic stages) or post-hatching (larval stages) at 20°C with the pharynx and gonad highlighted. (B) A schematic of BM composition showing the major families of conserved BM matrix components and receptors in C. elegans that were endogenously tagged using genome editing. See Methods for details on molecule illustrations.
To determine the validity of our tagging approach, we assessed knock-in strain viability and tagged BM protein localization (Table S1). For the majority of genes, insertion of the fluorophore resulted in superficially wild-type homozygous viable animals. However, α2-type IV collagen/let-2::mNG and SPARC/ost-1::mNG animals were non-viable as homozygotes and β-laminin/lam-1::mNG worms grew slowly. These strains were thus maintained as heterozygotes. The vast majority of proteins localized to BM. However, in four cases (papilin/mig-6 N-terminal, perlecan/unc-52 C- and N-terminal, and peroxidasin-2/pxn-2 C-terminal), the mNG signal localized diffusely in the extracellular space and was cleaved from the protein (Figures S1 and S3). Alternative tagging of papilin C-terminal, peroxidasin-2/pxn-2 N-terminal, and perlecan internal localized tightly to BM (Table S1; Figure S1). Further, Western blotting confirmed full-length papilinL/MIG-6L::mNG protein (Figure S3). We used these tagged strains for subsequent studies. These results demonstrate that endogenously tagging BM components with genetically encoded fluorophores is a viable approach for study of BMs in vivo.
Basement Membrane Emergence During Development
Studies in mice, C. elegans, and Drosophila embryos have indicated that laminin network formation is a requirement for initiation of BM formation (Huang, 2003; Matsubayashi et al., 2017; Smyth et al., 1999; Urbano et al., 2009). Cell culture work has also suggested that integrin and dystroglycan receptors mediate laminin polymerization (Yurchenco and Patton, 2009). In vivo studies have shown that the embryonic type IV collagen network is deposited after laminin in mice, C. elegans, and Drosophila (Graham et al., 1997; Matsubayashi et al., 2017; Pöschl et al., 2004; Urbano et al., 2009). The timing of the appearance of other BM components and matricellular proteins within embryonic BMs is less clear.
To better understand how BMs are assembled during embryogenesis, we used our collection of endogenously tagged BM components to view BM emergence. C. elegans embryonic development takes 14 h from fertilization to hatching at 20°C (Figures 1A) (Hall, 2017). The primordial intestine, muscle, epidermis, and pharynx (contractile feeding organ) begin to assemble during gastrulation (150 - 330 min) (Figure 2) (Hall, 2017). Following gastrulation, the embryo elongates, progressing through comma (430 min) and two-fold stages (490 min; Figure 2) during which the nerve ring encircles the pharynx prior to hatching (840 min). γ-laminin, nidogen, and papilin were the first detectable BM components and were deposited around all tissues by the end of gastrulation (Figures 2 and S4). C. elegans has two α-laminins (LAM-3 and EPI-1) that pair with the shared β (LAM-1) and γ (LAM-2) chains (Huang, 2003; Kao et al., 2006). Both α-laminin chains were present in the primordial gut and pharynx, but only the α-laminin EPI-1 was in the muscle and epidermal BM. Additionally, the two isoforms of papilin were distributed differently—papilinL was in all BMs, while papilinS was restricted to the BM surrounding the body wall muscle (Figures 2 and S4). The integrins and dystroglycan were broadly distributed on all tissues at gastrulation. C. elegans has a single dystroglycan receptor, DGN-1 and two integrin heterodimers composed of either the α-integrins INA-1 or PAT-2 with the single β-integrin PAT-3 (Clay and Sherwood, 2015). The α-integrin PAT-2 localized strongly to muscle, while INA-1 was found along gut and pharynx surfaces. The β-integrin PAT-3 localized to all three tissues (Figures 2 and S5). Other BM-associated receptors present early appeared to be localized intracellularly (discoidin domain receptors 1 and 2/DDR-1 and DDR-2) or to cell-cell membrane interfaces (syndecan/SDN-1 and LAR phosphatase/PTP-3), where they might mediate intercellular communication (Figures 2 and S4). Together, these observations indicate that at BM emergence, a common extracellular laminin, nidogen, and papilin scaffold forms on all tissues while integrin and dystroglycan receptors localize to all BM-associated cell surfaces. Notably, even at initial BM formation, heterogeneities in laminin, papilin, and integrin isoforms distinguish nascent BMs.
Figure 2. BM emergence during development.
(A) The left column shows differential interference contrast (DIC) images of embryonic development at 20°C at the post-gastrulation (330 min post fertilization), comma (430 min), and two-fold stages (490 min). Color overlays highlight embryonic muscle and epidermis (red), pharynx (blue), intestine (yellow), and the nerve ring when it arises at the two-fold stage (green). The muscle and epidermis are colored together as they were difficult to differentiate; however, clear associations with muscle are noted in the text. Inverted grayscale maximum intensity projections show fluorescence localization of representative BM components (columns 2-4)— the α-laminin subunits (EPI-1 and LAM-3) and agrin/AGR-1. EPI-1 localizes to the BM of all tissues starting from the end of gastrulation, while LAM-3 shows a similar pattern, but is absent from muscle/epidermis and the nerve ring. Agrin is restricted to the pharynx BM starting at the comma stage. Maximum intensity projections (with single z-slice shown for PAT-2 at two-fold stage) of BM receptor localization (columns 5-7) of the α-integrins (INA-1 and PAT-2) and syndecan/SDN-1. INA-1 localizes to cell-BM interface of all tissues, while PAT-2 appears at the muscle-BM interface at gastrulation then later at the pharynx and nerve ring BM. Scale bar is 10μm. (B) A graphic summary of matrix component (black) and matrix receptor (orange) localization during embryonic development (n ≥ 5 animals examined for each stage and each BM component). Arrows represent developmental progress and dots represent the stage of emergence in each tissue.
As embryonic development proceeded to the comma stage, type IV collagen appeared in all three BMs (it was also present at this stage and later stages in large intracellular vesicles within body wall muscles, see (Morrissey et al., 2016)), perlecan localized to the body wall muscle BM, and agrin became visible in the pharynx BM (Figures 2 and S4). Dystroglycan and the integrin receptors maintained their localization, while DDR-1 localized to BMs in the muscle and intestine and the glypican LON-2 was present along the pharynx BM (Figures 2 and S5). By the two-fold stage, the nerve ring develops and many BM-associated receptors are enriched there, including the integrins, LAR phosphatase, glypican, and syndecan (Figures 2 and S5). The nerve ring BM also had high levels of all laminin chains, agrin, nidogen, type IV collagen, SPARC, papilin, and type XVIII collagen (Figures 2 and S5). Finally, peroxidasins (PXN-1 and PXN-2), hemicentin, and fibulin were only detected within BMs after embryonic development (see Figure 3). We conclude that differences in BM composition increase significantly over developmental time as tissues differentiate.
Figure 3. Quantification of BM composition and regional localization.
(A) Representative inverted grayscale maximum intensity confocal projections of whole animal γ-laminin::mNG (left) and α1-type IV collagen::mNG (right) at the L1 stage. (B) Normalized mean fluorescence intensity displayed as waffle plots compare the composition of the gonadal and pharynx BMs per unit area. Each square is normalized to the mean fluorescence intensity of the least abundant component, peroxidasin-1 (n ≥ 9 animals imaged for each matrix component). (C) Heatmaps of maximum intensity projections from confocal z-stacks of the L1 pharynx enlarged to emphasize regional differences (see schematic) in protein distribution— type IV collagen is in a gradient from posterior-to-anterior, laminin and nidogen are evenly distributed (but concentrated around nerve ring, orange arrowheads), papilinS is evenly distributed (signal from the epidermis makes it appear higher in the posterior bulb), agrin is enriched in the anterior region (arrow), type XVIII collagen is at high levels in the posterior bulb (white arrow, and also in nerve ring BM, orange arrowhead), fibulin is enriched in the anterior region (arrow), spondin is enriched in the metacorpus (arrow), and peroxidasin-1 and perlecan are present at low uniform levels (n = 10 animals examined for each). Scale bar is 10μm.
Quantitative Analysis and Comparisons of BM Composition Between Tissues
BMs are diverse in composition and changes in BMs influence cell and tissue function in normal and disease states (Naba et al., 2014; Randles et al., 2017). Biochemical, mass spectrometry, and fixed tissue microscopy studies have characterized important aspects of BM structure and composition, however, these approaches cannot easily determine the relative amounts of multiple BM components at defined locations and none can be used in living animals.
We next used our endogenously tagged BM matrix components to quantify and compare BM composition surrounding two different organs: the L1 larval stage pharynx and gonad (Figure 3). The pharynx is a rigid muscular pump that continuously beats to grind food. It is fully functional at the L1 stage, and electron microscopy studies have indicated the pharynx has a thick BM (Huang et al., 2003; Mango, 2007). In contrast, the L1 gonad is a nascent organ that envelops the germ cell and somatic gonad precursors. It grows rapidly during larval development and is encased by a thinner BM (Huang et al., 2003). Measurements along the posterior bulb of the pharynx and central dorsal region of the gonad showed that the core BM components laminin (γ-laminin::mNG), type IV collagen (α1-type IV collagen::mNG), and nidogen were present in both organs, as well as papilinS and type XVIII collagen (Figure 3B). The α1 chain of type IV collagen (EMB-9) is present in two copies per type IV collagen molecule, while the γ-chain of laminin (LAM-2) is one of three chains of laminin (Kramer, 2005). For simplicity, unless denoted, we refer to levels and presence of EMB-9/α1-type IV collagen::mNG as type IV collagen and LAM-2/γ-laminin::mNG as laminin in the text. PapilinS was the most abundant component in both BMs. Spondin, fibulin, agrin, perlecan, and peroxidasin-1 were present in the pharynx BM, but not the gonad (Figure 3B). Direct comparisons of BM component levels revealed that every matrix protein that was present in both tissues was higher in the pharynx, except for papilinS (Figure 3B). The most dramatic difference was in type IV collagen levels, which were ~3-fold higher in the pharynx BM (Jayadev et al., 2019). Given the size of type IV collagen (~400 nm) (Yurchenco, 2011), it may account for the greater thickness of the pharynx BM. Most matrix components in the pharynx BM were also present in gradients or enriched in specific sub-regions, indicating specialization within the pharynx BM (Figure 3C). Collectively, these results show that BMs vary in quantitative composition; that the make-up of the pharynx BM, which surrounds a functional tissue, is more complex than that of the immature gonad; and that BM composition can change across micron-scale distances.
A Stable Collagen and Laminin Scaffold Supports Mobile BM Proteins
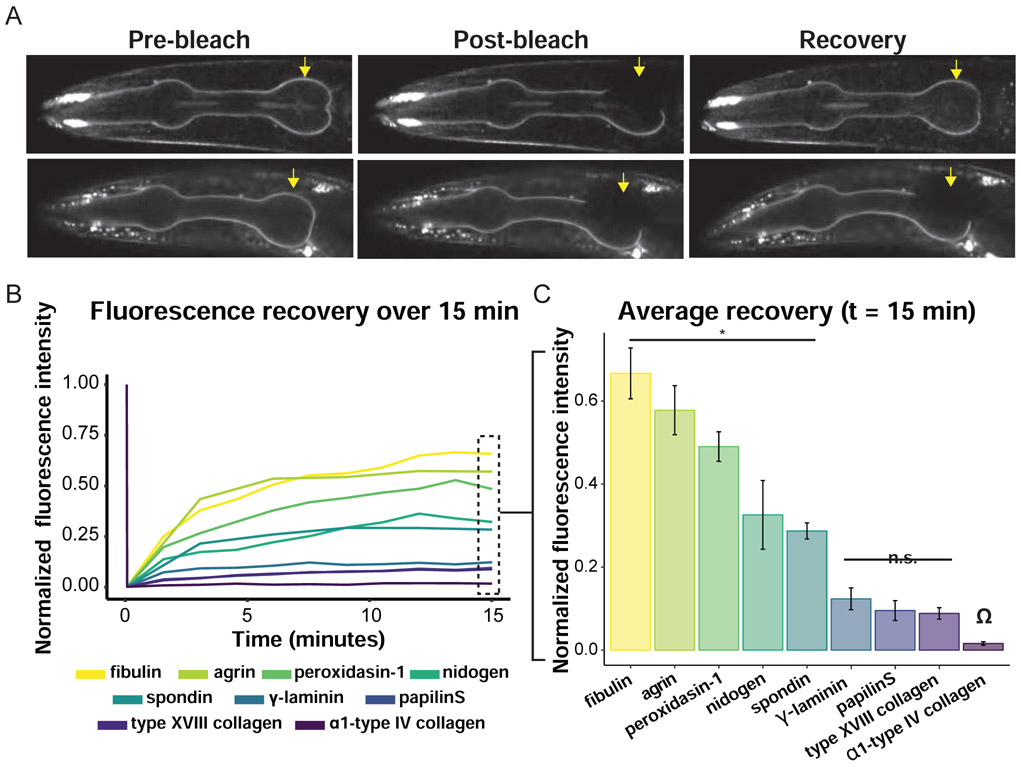
Previous work in vertebrates suggests that BMs turn over on a scale of weeks (Decaris et al., 2014; Trier et al., 1990). Whether matrix components are stably integrated within BMs or have dynamic associations after BMs are deposited, however, is unknown. To directly address the stability of individual BM matrix components, we performed FRAP experiments in the posterior bulb of the pharynx BM of L4-staged larvae (Figure 4A). We chose the pharynx as it contains most BM matrix components we tagged (perlecan levels, however, were too low to assess), it does not grow significantly during the L4 larval stage, and the different matrix component levels do not increase at this time. We hypothesized that fluorescence recovery would measure protein degradation and replacement rates if BM components are stably associated within BM or reveal more rapid dynamic exchange rates if their associations are transient. To determine if any matrix components show transient connections, we analyzed fluorescence recovery over a 15-min period, imaging every 90 sec. Fibulin, agrin, spondin, nidogen, and peroxidasin-1 recovered between ~30-65% of their original fluorescence intensity in only 15 min. In contrast, laminin, type XVIII collagen, papilinS, and type IV collagen exhibited ~10% or less recovery over this time period (Figures 4 and S6). Longer imaging revealed that type XVIII collagen and papilinS recovered <20% of their original intensity in 1 h and laminin and type IV collagen recovered ~30% of their original intensity in 5.5 h, confirming a more stable association with BM (Figure S6). These results indicate that laminin, collagen, and papilinS have a relatively stable association with BM after deposition, while all other components have dynamic associations.
Figure 4. FRAP of BM components reveal stable and dynamic matrix components.
(A) Confocal images (single z-slices) of the L4 pharynx show differences in recovery rate after photobleaching for mNG::fibulin and α1-type IV collagen::mNG over 15 min. Yellow arrows indicate the bleached half of the pharynx and dashed white line indicates the unbleached control region. Scale bar is 10μm. (B) Line graph showing the normalized fluorescence recovery of nine basement membrane components present in the L4 pharynx over 15 min (n = 5 animals for each). (C) Bar graph displays the mean recovery fraction for each protein at the end of 15 min. Error bars denote standard error. All means were compared to the α1-type IV collagen as denoted by the Ω symbol. Black bars indicate two groups: * mNG::fibulin, agrin-1::mNG, peroxidasin-1::mNG, nidogen::mNG, and spondin::mNG exhibited faster recovery than α1-type IV collagen (n = 5 animals for each, * p < 0.005, Dunnett’s test); however, no significant (n.s.) differences were observed for γ-laminin::mNG, papilinS::mNG, and α1-type XVIII collagen::mNG.
We postulated that the rapid recovery seen in many of the BM components was not through protein degradation and replacement, as BM proteins have long half-lives (Decaris et al., 2014). Consistent with this, Western blot analysis confirmed that fibulin, nidogen, agrin, and spondin were present predominantly as full-length mNG fusion proteins (Figure S3). Most matrix proteins in the pharynx BM (with the exception of agrin) are secreted from distant tissues and recruited to the pharynx BM (Clay and Sherwood, 2015; Gotenstein et al., 2010; Kramer, 2005). We hypothesized that the dynamic BM proteins might be replaced by rapidly exchanging with components in the extracellular fluid, might move within the BM itself, or a combination of both. To test these possibilities, we photobleached the entire posterior bulb of the pharynx then examined fluorescence recovery every 10 sec to assess the initiation of fluorescence recovery (Figure 5). If a matrix protein was predominantly rapidly exchanging with the extracellular fluid, we predicted that it would recover evenly across the bleached region. Alternatively, if the matrix protein moved within the BM, we predicted it would recover first at the edge of the bleached region. Strikingly, all dynamic components—fibulin, nidogen, agrin, spondin, and peroxidasin-1—recovered more quickly near the edge of the bleached region (Figures 5A and S7A and S7B; Video S1), and moved 5 μm into the unbleached region at rates that closely matched the level of their fluorescence recovery in 15 min: fibulin (0.17 ± 0.04 μm/sec), peroxidasin-1 (0.19 ± 0.02 μm/sec), agrin (0.08 ± 0.02 μm/sec), nidogen (0.06 ± 0.02 μm/sec), and spondin (0.09 ± 0.01 μm/sec; n = 5, mean ± SD). We also examined movement of the bleached fibulin and observed that the unbleached fibulin adjacent to the site of photobleaching declined in fluorescence intensity prior to interior regions (presumably as the bleached fibulin moved into this region, Figure 5A), as would be expected if fibulin moved bi-directionally within the BM. Bleaching the anterior pharynx of agrin::mNG worms revealed that agrin also moved here (n = 5/5 animals, Video S2). To further test if matrix movement is a general BM property, we examined the gonadal BM, which contains nidogen (but not other dynamic components; Figure 3B). Similar to the pharynx BM, nidogen was mobile while laminin did not move (Figure S7C; Video S3). As a different test to estimate the contribution of matrix components within the extracellular fluid to the recovery of dynamic BM components after photobleaching, we photobleached mNG::fibulin in the entire pharynx BM. We examined fibulin as it does not localize strongly to the adjoining gut BM. Thus, the fluorescence recovery should be predominantly from extracellular sources rather than movement within the BM. We found that only 9.5 ± 3.5% of mNG::fibulin recovered when the entire pharynx was photobleached compared to 68.7 ± 8.2% recovery when only the posterior bulb was photobleached (mean ± SD, 15 min recovery, n = 5), suggesting that exchange with the extracellular fluid occurs, but is a minor component of dynamic fibulin. Lastly, we determined whether the dynamic recovery of BM components was active or passive, focusing on fibulin, the most dynamic pharynx BM component. We first treated worms with dicyclohexylcarbodiimide (DCCD), an ATP synthase inhibitor, and found that while it did not affect the levels of BM fibulin (STAR Methods), there was a dramatic reduction in recovery rate after photobleaching (Figure 5C). We hypothesized that a possible ATP driven regulator of BM movement might be muscle contractions, as these could distribute energy into BMs that surround tissues. Notably, in our FRAP experiments above we immobilized worms with polystyrene beads, which hinders worm movement, but allows for muscle contractions. Strikingly, we found that muscle paralysis induced by levamisole (sustained contraction), levamisole and tricaine (contraction and relaxation), or 2,3-Butanedione monoxime (BDM, muscle relaxation alone) dramatically reduced the percent fluorescence recovery of mNG::fibulin and the movement of mNG::fibulin (Figure 5C, Video S4; 0.17 ± 0.04 μm/sec (control) versus 0.08 ± 0.02 μm/sec levamisole, 0.08 ± 0.03 μm/sec levamisole & tricaine, 0.11 ± 0.05 μm/sec BDM, n = 5 animals examined each, p < 0.01 for each compared to control, Student’s t-test). These experiments indicate that the dynamic matrix components move non-directionally within the BM and that muscle contractions contribute to dynamic matrix mobility.
Figure 5. The matrix component fibulin moves within the BM.
(A) (Left) A 10-min time-lapse (imaged every 10 s) of mNG::fibulin FRAP. Images show a single confocal z-slice of the pharynx and an inset of the photobleached region with a heatmap prior to photobleaching (prebleach), after photobleaching (0 min), and at 4- and 10-min recovery timepoints. Gray boxes represent the control region distant from the bleached area, green boxes the unbleached region of the BM outside the edge of the bleached area, blue boxes the edge of the bleached area, and purple boxes the midregion of the photobleached area. (Right) A line graph of normalized fluorescence recovery reveals that recovery first occurs at the edge of the bleached region (blue) before recovering in the middle (magenta). The graph also shows greater loss of fluorescence in the BM closer to the photobleached region (green) than the control region (gray) (n = 5/5 animals). (B) (Left) A representative 30-min time-lapse (imaged every 2 min) of type XVIII collagen::mNG FRAP and (Right) a line graph of recovery. Dim fluorescence signal recovers uniformly across the bleached region independent of proximity to the edge of the bleached region (n = 5/5 animals examined). (C) (Left) Confocal images (single z-slices) of the L4 pharynx show differences in recovery rate of mNG::fibulin after photobleaching between control (polystyrene bead immobilization allowing muscle contractions) and tricaine/levamisole and BDM muscle paralysis treatments. Yellow arrows indicate the bleached half of the pharynx and dashed white box indicates the unbleached control region. (Right) Bar graph displays the mean recovery fraction at the end of the 15-min interval. Error bars denote standard error. All means were compared to the mNG::fibulin control (n = 5 animals for each, ** p < 0.001, *** p < 0.0001, Student’s t-test). Scale bar is 10μm.
Finally, we examined how the more stable components recovered after photobleaching. Type XVIII collagen was the only stable BM protein that had detectable recovery within a 30-min window (~10%), allowing for spatiotemporal FRAP analysis. In contrast to the more rapidly recovering components, we found that type XVIII collagen was recruited evenly throughout the photobleached region (Figure 5B; Video S1), suggesting it is replaced by extracellular sources. Further, long term static imaging of recovering photobleached regions of type IV collagen, laminin, and papilinS did not reveal movement within the BM, suggesting they are also replaced by extracellular sources (Figure S7D). Taken together, these observations indicate that BMs are built on a stable laminin and collagen scaffold that includes type XVIII collagen and papilin within which other matrix proteins dynamically associate.
Papilin/MIG-6 Promotes Removal of Type IV Collagen During BM Expansion
BMs must expand during organ growth and in neoplastic diseases. Mechanisms that promote BM enlargement, and especially expansion of the stable collagen and laminin scaffolding, are unknown. We next used our collection of endogenously tagged BM components to examine BM enlargement in the gonad, which has a BM-encased surface area that increases more than 90-fold from hatching to early adulthood (Jayadev et al., 2019).
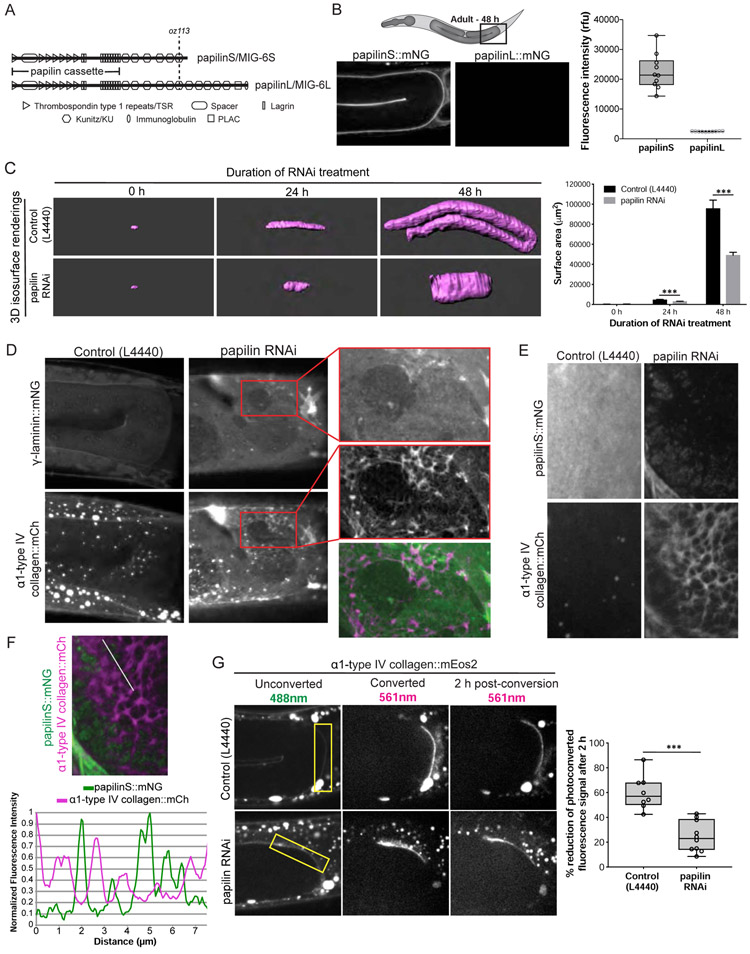
We focused on papilinS, which was present at levels exceeding all other matrix components combined at the initiation of gonadal BM expansion (Figure 3B). Papilin is a conserved proteoglycan that encodes at least two isoforms: one long (papilinL/MIG-6L) and one short (papilinS/MIG-6S; Figure 6A). We found that papilinL, which is secreted by the distal tip cells (DTCs) as they migrate, is present in the BM during gonad growth, but at dramatically lower levels than papilinS (~50-fold lower, Figure 6B). Genetic loss of papilinS is embryonic or early larval lethal (Kawano et al., 2009). We thus depleted both isoforms during gonad growth by feeding RNAi to L1-staged worms and analyzed laminin and collagen in gonadal BM of early adult animals (48 h post-hatching). Reduction of papilin perturbed gonad growth and BM expansion (Figure 6C, more than 50% reduction in surface area compared to control animals), but did not result in BM dissolution or ruptures (n = 0/20 animals with ruptured gonads observed), as occurs after RNAi-mediated loss of laminin or collagen (Jayadev et al., 2019). Instead, we found that papilin loss led to increased and fibrotic α1-type IV collagen::mNG compared to its smooth appearance in control animals (1.83-fold increase, n = 12 animals, p < 0.0001, Student’s t-test). Notably, we saw the same fibrotic phenotype with α1-type IV collagen::mCherry (Figure 6D)(Ihara et al., 2011), which is tagged in a different location within α1-type IV collagen (Figure S1; n = 10/10 animals). In contrast, the smooth laminin network was unperturbed after papilin loss (Figure 6D), similar to controls (n = 10/10). To assess if papilin was uniquely required during BM expansion, we initiated RNAi feeding at the L3 stage (24 h post-hatching, near the end of growth), 48 h adult (after growth), and 72 h adult (after growth), and found that type IV collagen was patterned normally 48 h after RNAi treatment in all cases (n = 8/8 animals each), suggesting that papilin is not required for maintenance of collagen. We also examined animals harboring a mutant of papilin (mig-6(oz113)) that truncates the C-terminal domain of papilinL, leaving only the papilinS product (Figure 6A). These animals did not produce the type IV collagen fibrosis phenotype (n = 0/10 animals), suggesting that domains unique to papilinL are not required for collagen remodeling during BM expansion.
Figure 6. Papilin promotes type IV collagen network remodeling during BM growth.
(A) A diagram of papilin/MIG-6 shows the protein domains of papilinS and papilinL isoforms and the papilinL truncation mutant (mig-6(oz113)). (B) (Top left) A diagram of the C. elegans gonad (gray) where levels of papilinS::mNG and papilinL::mNG fluorescence were compared (black box). (Bottom left) Single confocal z-slices of papilinS::mNG and papilinL::mNG at the young adult stage and (Right) quantification of fluorescence (boxplot, n = 10 animals for each). (C) (Left) 3D isosurface renderings of gonadal α1-type IV collagen::mCh following 0, 24, or 48 h of RNAi treatment in control and papilin RNAi animals. (Right) Quantification of gonadal surface area; bar graphs show mean surface area and error bars represent standard deviation (n = 3-6 animals examined for each, *** p<0.0001, Student’s t-test). (D) Confocal maximum intensity projections show co-localization of γ-laminin::mNG and α1-type IV collagen::mCh in the turn region of the adult gonad in an untreated control and a papilin RNAi knockdown animal. Reduction of papilin disrupted gonad growth and led to a fibrous increase in type IV collagen. The laminin network in the same animal was normal (magnified insets with overlay, n = 10 animals). (E) Maximum intensity projections show co-localization of the even distribution of papilinS::mNG and α1-type IV collagen::mCh within control animals BM compared to the patchy localization of papilinS and fibrous α1-type IV collagen::mCh after RNAi mediated loss of papilin. (F) Overlay of residual papilinS::mNG and α1-type IV collagen::mCh maximum intensity projection images after RNAi knockdown of papilin. A line plot indicated by the white line shows that areas with low papilin have high type IV collagen and vice versa (n = 5/5 animals). (G) (Left) Single confocal z-slices of α1-type IV collagen::mEos2 prior to, immediately after, and 2 h after photoconversion in a control and papilin RNAi knockdown animal. Optically highlighted α1-type IV collagen::mEos2 is indicated by yellow boxes. (Right) Quantification of the percentage reduction in mean photoconverted (red) mEos2 signal after 2 h (boxplot, n = 8-10 animals for each, *** p < 0.0001, Student’s t-test). Scale bars are 10μm except panel C, which is 25μm.
To further understand the role of papilin during BM growth, we analyzed RNAi-mediated loss of papilinS protein and the patterning of type IV collagen. We found that after L1 RNAi-initiated loss of papilin, the papilinS protein declined dramatically, but was still present in a patchy, punctate pattern in the gonads of 48 h young adult animals (Figure 6E). The localization of this residual papilinS correlated with areas where the type IV collagen was smoothly patterned, whereas regions of papilin absence was where collagen levels were increased and fibrotic (Figure 6E and F). To examine how papilin regulates type IV collagen during BM growth, we generated an α1-type IV collagen::mEos2 knock-in strain (Figure S1). mEos2 is a highly stable photoconvertible fluorophore that converts from green to red with ultraviolet light (McKinney et al., 2009). Optical highlighting of α1-type IV collagen::mEos2 within the BM of the gonad in 48 h young adults revealed a ~60% loss of photoconverted collagen signal after 2 h in control animals, but only a ~20% reduction in papilin RNAi treated animals (Figure 6G). Taken together, these results indicate that papilin plays a localized role in mediating type IV collagen removal during BM expansion, which likely accounts for the buildup of BM collagen after papilin loss.
Papilin Limits BM Association of Collagen Modifying ADAMTS Proteases & Peroxidasin
Papilins are structurally similar to ADAMTS matrix-associated metalloproteinases, and share a papilin cassette (thrombospondin type-1 repeats (TSR) with a spacer sequence rich in cysteine residues); but lack a catalytic domain (Figure 6A). Drosophila papilin can bind to and inhibit a vertebrate procollagen N-protease ADAMTS in vitro (Kramerova et al., 2000). Studies with dominant papilin mig-6 alleles, have suggested they may regulate the localization or the activity of ADAMTS protease MIG-17 to control the path of DTC migration (Kawano et al., 2009).
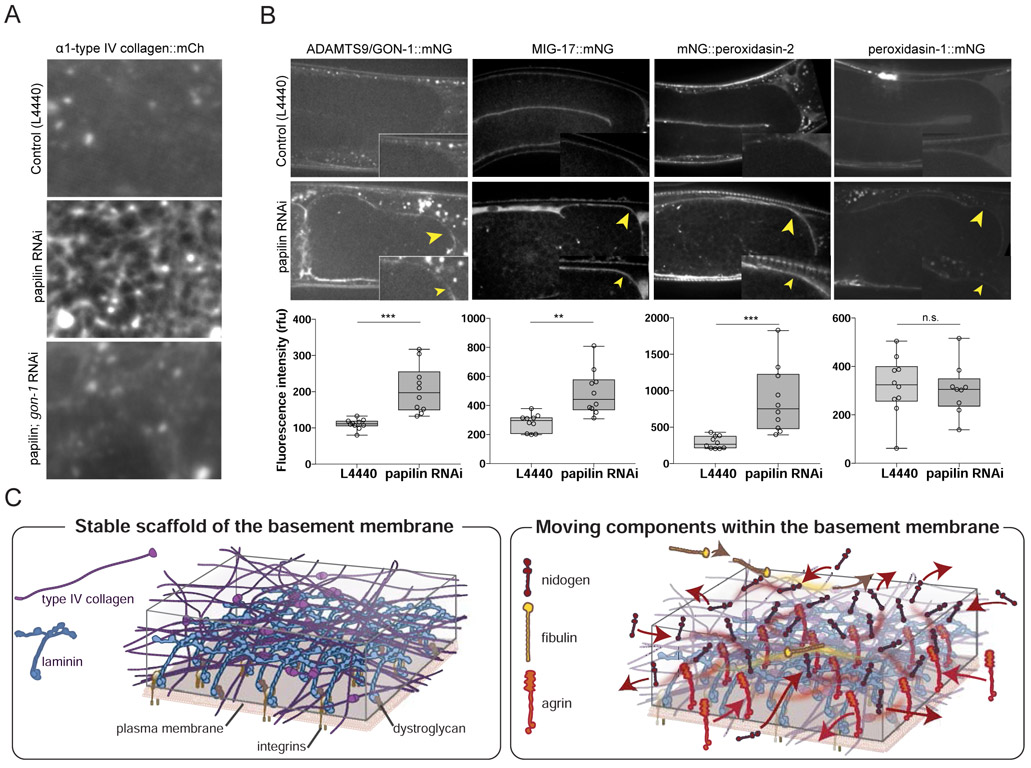
As our data indicated that papilin promotes type IV collagen removal, we first depleted the five C. elegans ADAMTS proteases—adt-1, adt-2, adt-3, gon-1, mig-17—by RNAi at the L1 stage to determine if loss of any phenocopied papilin loss. Reduction of these ADAMTS proteases, however, did not lead to a buildup of type IV collagen (n ≥ 10 animals for each), suggesting that papilin does not promote ADAMTS proteases activity to regulate collagen removal. We next examined whether papilin might inhibit an ADAMTS protease, and co-depleted papilin in combination with each of the five ADAMTS proteases to see if loss of any suppressed the cobweb collagen buildup. Strikingly, reduction of gon-1 in combination with papilin restored smooth collagen patterning (n = 21/23 rescued, versus 0/16 papilin RNAi alone, n ≥ 10 animals for each ADAMTS; Figure 7A). To examine the localization of GON-1, an ortholog to vertebrate ADAMTS9 (Clay and Sherwood, 2015), we generated a knock-in strain (GON-1::mNG; Figure S2) and found that reduction of papilin led to a significant increase in ADAMTS9/GON-1 associated with the gonadal BM (Figure 7B). MIG-17::mNG levels were similarly increased in the BM after papilin loss (Figure 7B). We also examined two additional type IV collagen modifying enzymes—peroxidasin-1/PXN-1 and peroxidasin-2/PXN-2. Peroxidasin-2 mediates covalent sulfilimine bond formation between type IV collagen molecules, while peroxidasin-1 antagonizes peroxidasin-2 activity (Gotenstein et al., 2010). Loss of papilin led to increased levels of peroxidasin-2 within the BM, but not peroxidasin-1 (Figure 7B). RNAi-mediated reduction of peroxidasin-2 in combination with papilin, however, did not suppress the collagen phenotype resulting from papilin loss (n = 15/15 animals had fibrotic collagen). Collectively, these observations indicate that papilin plays a localized role in limiting BM access (and perhaps activity) of ADAMTS9/GON-1 to promote collagen removal, and exerts a similar influence on the localization of the ADAMTS protease MIG-17 and the peroxidasin-2 enzyme. Thus, papilin might play a broad role in collagen remodeling during BM growth.
Figure 7. Papilin limits ADAMTS protease and peroxidasin gonadal BM association.
(A) Confocal sum projections of α1-type IV collagen::mCh fluorescence in the gonadal BM. RNAi knockdown of papilin led to an increased fibrous type IV collagen network (n = 16/16) that was rescued by loss of gon-1 (n = 21/23). (B) (Top) Single confocal slices through the gonad arm shows GON-1::mNG, MIG-17::mNG, mNG::peroxidasin-2, and peroxidasin-1::mNG in control or papilin RNAi treated animals. Loss of papilin resulted in increased BM association of GON-1, MIG-17, and peroxidasin-2, but not peroxidasin-1. (Bottom) Quantification of BM fluorescence intensity, (boxplot, control n = 10 and papilin RNAi n = 10 for each, n.s., not significant, ** p < 0.001 or *** p < 0.0001, Student’s t-test). Scale bars are 10μm. (C) A model for BM dynamics. (Left) Laminin (and possibly type IV collagen (Jayadev et al., 2019)) associate with the cell membranes through integrin and dystroglycan receptors forming a stable scaffolding. (Right) Nidogen, fibulin, and agrin (spondin and peroxidasin-1 not shown) move dynamically within the laminin and type IV collagen network.
DISCUSSION
A major challenge in BM biology is examining the dynamics and regulation of these dense, varied, and complex matrices in vivo (Hohenester and Yurchenco, 2013). Live antibody labeling of laminin and type IV collagen in ex vivo cultures in mice (Harunaga et al., 2014; Pflicke and Sixt, 2009) and a type I collagen-GFP that localizes adjacent to BMs in zebrafish (van den Berg et al., 2019), have provided insights into BM dynamics during cell migrations and glandular morphogenesis. However, no BM components have been tagged with a genetically encoded fluorophore in a vertebrate to directly follow BM dynamics. In Drosophila, GFP-tagged transgenes expressing laminin and nidogen, and GFP inserted via exon traps into type IV collagen and perlecan genes, have revealed their regulation and functions during BM deposition, signaling, tissue shaping, and wound healing (Isabella and Horne-Badovinac, 2016; Ma et al., 2017; Matsubayashi et al., 2017; Ramos-Lewis et al., 2018). Drosophila studies, however, have not endogenously tagged most BM components and live imaging of BMs after embryogenesis has only been achieved using dissected tissues (Cetera et al., 2016). To overcome these limitations, we extended our previous studies of endogenous mNG tagged γ-laminin, dystroglycan, and α- and β-integrins (Jayadev et al., 2019; Naegeli et al., 2017) to complete a comprehensive assembly of endogenously tagged BM components with 17 tagged BM matrix components and 12 tagged receptors. In addition, this evolving collection includes type IV collagen tagged with the photoconvertible fluorophore mEos2 and two BM-localized ADAMTS proteases—ADAMTS9/GON-1 (this study) and MIG-17 (Ji et al., 2019). Importantly, most of these BM components have strong loss-of-function phenotypes, including embryonic or larval lethality (laminin, type IV collagen, spondin, papilin, teneurin, perlecan, SPARC, and integrin), and sterility or low brood size (fibulin, hemicentin, dystroglycan, and ADAMTS9) (Kawano et al., 2009; Kramer, 2005; Kramerova et al., 2000; Muriel et al., 2005; Trzebiatowska et al., 2008; Vogel and Hedgecock, 2001; Woo et al., 2008). The normal health of the genome-edited strains, the tight association of the tagged proteins with BMs, and the presence of full-length protein products provide strong evidence that the tagged proteins show endogenous regulation and localization.
Although BMs in adult tissues are remarkably diverse, it is unknown when during development this diversity arises. Work on the initial formation of BMs in mice, C. elegans, and Drosophila embryos has established that laminin deposition occurs early and is required for BM assembly (Hohenester and Yurchenco, 2013; Kao et al., 2006; Matsubayashi et al., 2017; Urbano et al., 2009). In C. elegans, laminin is deposited and BMs start forming near the end of gastrulation (Huang et al., 2003). We found that in addition to laminin, nidogen, and papilin were present on all BMs at this time, suggesting these components form an initiating BM template. Notably, at this nascent phase, distinct forms of laminin and papilin had tissue-specific BM localization. Further, the collagen cross-linking enzyme peroxidasin-2 and the integrin receptor PAT-2/PAT-3 were restricted to muscle BMs. Thus, BMs have a common early template, but tissue-specific differences are also present. As development proceeded, BM complexity and heterogeneity between tissues increased, and gradients and regionally increased concentrations of matrix components appeared. In the L1-stage pharynx BM, type IV collagen was found in a gradient, while components like type XVIII collagen and spondin were strongly enriched in specific pharynx sub-regions. A gradient of GFP-tagged type IV collagen has also been observed in the Drosophila egg chamber BM, where it translates into a stiffness gradient that elongates the egg chamber (Crest et al., 2017). Local differences and gradients in BM components are likely common, but overlooked features of BMs that have important roles during organ formation and function.
How BMs increase in size during organ growth in development, regeneration, and tumor expansion is a crucial, but unknown aspect of BM regulation. Using our ability to compare fluorescence and therefore BM protein levels, we found that papilinS was present at higher levels than all other BM components combined in the L1-stage gonad—an organ where the BM will enlarge more than 90-fold during subsequent development (Jayadev et al., 2019). We found that loss of papilin limited BM expansion and perturbed gonad growth, suggesting an important role for papilin in BM enlargement. RNAi-mediated depletion of papilin during gonad growth revealed that in BM areas where papilinS protein was no longer detectable, the collagen scaffold was increased in levels and fibrous. In contrast, laminin retained its uniform localization after papilin loss. Optical highlighting of a photoconvertible type IV collagen showed that the collagen was more stably associated with the BM after papilin reduction, indicating that papilin promotes type IV collagen removal from BMs during gonad growth, which may account for its function in facilitating BM expansion and tissue growth. Papilins have structural similarities to ADAMTS secreted matrix-associated metalloproteinases, but they lack a proteolytic catalytic site. In vitro studies have shown that Drosophila papilin binds to and inhibits the activity of a vertebrate procollagen N-proteinase ADAMTS (Kramerova et al., 2000). Our studies suggest that in addition to directly inhibiting ADAMTS proteases, papilin also limits the BM association of ADAMTS proteins. Loss of papilin led to a significant increase in the BM association of the ADAMTS proteinases GON-1 and MIG-17, as well as the type IV collagen cross-linking enzyme peroxidasin-2. Supporting a possible functional role for papilin’s exclusion of these enzymes, we found that reduction of gon-1 in combination with papilin partially rescued the normal patterning of type IV collagen on the gonad BM. Although it might seem counterintuitive that loss of an ADAMTS protease reduces collagen buildup and fibrotic appearance in the gonadal BM, a number of human ADAMTS proteins are pro-fibrotic and associated with increased ECM deposition (Perrucci et al., 2018). Aggrecan and versican are the only characterized substrates for ADAMTS9 (Kelwick et al., 2015), the vertebrate GON-1 ortholog; however, these proteoglycans are not present in C. elegans. Thus, GON-1 (and ADAMTS9) likely has an additional substrate(s) that regulates type IV collagen removal from BMs.
No studies have yet assessed the dynamics of BM components within BMs after deposition. By performing FRAP experiments on the mNG-tagged matrix proteins, we found that laminin and type IV collagen, the two central scaffolding components of BM, are stable and have slow recovery times, on the order of many hours. Papilin and type XVIII collagen also displayed slow recovery after photobleaching. These slow recovery rates may reflect the normal degradation and replacement rates of these proteins during development. All other matrix components that were assessed—fibulin, nidogen, agrin, peroxidasin-1, and spondin—showed rapid recovery of bleached regions on the order of minutes. We considered that these BM components might be rapidly exchanging between the BM and the extracellular fluid. However, fluorescence recovery did not occur evenly along the bleached region of BM. Instead, recovery occurred first at the edge of the photobleached region, indicating that BM components predominantly recover by moving within the more stable scaffolding components. We also found that this movement was non-directional and in the case of the mobile component fibulin, was enhanced by muscle contractions. Together, these observations provide a new model for BM dynamics, where a relatively stable scaffolding of laminin and type IV collagen hosts more dynamic regulatory components that move within this scaffolding (Figure 7C; Video S5). We suspect that the more dynamic components have numerous weak interactions with other BM components, which maintain their association with the BM, but also allow for their movement. Consistent with this, nidogen has many BM binding partners, including laminin, type IV collagen, and perlecan (Yurchenco, 2011). Fibulins also associate with many other BM proteins (Kobayashi et al., 2007). The mobile matrix components might allow BMs to dynamically respond to mechanical changes in tissues, particularly as the mobility of fibulin is affected by the mechanical activity of muscle contraction. Further, as growth factors can bind to agrin and fibulin (Bányai et al., 2010; Fresco et al., 2016), mobile matrix components may serve as conduits for growth factor trafficking.
We have shown here that we can examine BM emergence, growth, composition, and dynamics with our comprehensive collection of endogenously tagged BM components. We envision future studies engineering disease-associated mutations into these strains to probe how these mutations affect BM component trafficking, localization, and dynamics. In addition, studies can now be conducted in combination with FRAP and photoconversion to identify genes mediating the turnover of the stable collagen and laminin scaffoldings—networks that accumulate in fibrosis, diabetes, aging, and tissue decline (Candiello et al., 2010; Mak and Mei, 2017; Tsilibary, 2003). Thus, the use of these endogenously tagged BM components is poised to further deepen our understanding of this ancient and fascinating animal ECM.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David R. Sherwood (david.sherwood@duke.edu).
Materials Availability
All worm strains will be available from the Caenorhabditis Genomics Center (CGC, cgc.umn.edu).
Plasmids generated in this study are available upon request.
Data and Code Availability
The published article includes all data generated or analyzed during this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Worm handling and strains
Worms were grown under standard conditions and N2 Bristol strain was wild type. L1 animals were obtained by bleach synchronization. All worms used in this study were hermaphrodites. L4 animal staging was determined by DIC or fluorescence based on gonad and vulval morphology, with gonads having reflected, but not completing secondary migration. All alleles and strains used in this study are listed in Key Resources and annotated according to wormbase guidelines: https://wormbase.org/about/userguide/nomenclature#ecgkh5m87dlji1bf923a064--10).
METHOD DETAILS
CRISPR Strain Construction
We used CRISPR/Cas9-mediated genome editing with a self-excising hygromycin selection cassette (SEC) to generate endogenous BM tags with modifications from previous methods ((Dickinson and Goldstein, 2016; Jayadev et al., 2019); Table S1; Figures S1 and S2). Briefly, we generated new SEC plasmids by removing the 3xFlag tag downstream of the SEC cassette sequence and attached an 18 amino acid flexible linker (flexlink) (6x glycine-alanine-serine) in frame and directly upstream of the fluorophore. For most genes the fluorophore was inserted just upstream of the stop codon to generate a C-terminally tagged protein. However, in cases where there were multiple isoforms that shared an N-terminus but varied at the C-terminus, we generated an N-terminal knock-in. For N-terminal knock-ins, we ran a signal peptide prediction (SignalP-5.0) to determine signal peptide cleavage sites and inserted the fluorophore immediately downstream of the site. For all N-terminal knock-ins, we used a 2aa upstream linker and 4aa downstream linker strategy (bothlink) (Morin et al., 2001), except for papilin/mig-6, for which we used the flexlink cassette with 18aa amino acid linker being present on the N-terminus. The papilinS isoform-specific tag prevented splicing of the papilinL isoform, resulting in homozygous animals that exhibit sterility and gonad phenotypes consistent with loss of papilinL (Kawano et al., 2009). We thus reported values for heterozygous (papilinS::mNG/+) animals in all analyses. In the specific cases of emb-9 and let-2, we used the bothlink cassette to insert the mNG in the same locus as the endogenous vkg::GFP in D. melonagaster (Morin et al. 2001). To generate the internally tagged perlecan/unc-52 we inserted a synthetic exon (Morin et al. 2001) between two exons near the C-terminus of the gene (Figures S1 and S2; Table S1). For all constructs 2-3kb of DNA centered on the insertion site was amplified from N2 genomic DNA and cloned into an in intermediate vector (TOPO) to be used as homology arms. Homology arms were mutated to introduce as many silent point mutations as possible adjacent to the Cas9 cut site. No mutations were made for epi-1, let-2, pxn-2, unc-52 C-terminal, or unc-52 exon as the insertion split the PAM site. For ost-1 and ten-1 only one silent mutation could be introduced, for all other constructs 5-13 mutations were introduced between the two sgRNA sites chosen. Mutated homology regions were inserted into the appropriate repair plasmid using Gibson assembly. We generated 1 or 2 guide RNA (sgRNA) plasmids for each target by inserting the respective sgRNA sequences into the pDD122 plasmid. The sgRNA sequences used are listed (Figures S1 and S2; Table S1).
For each strain, we injected a mixture of 50ng/μl Cas9-sgRNA plasmids (25ng/μl of each guide plasmid in cases where two sgRNAs were available), 50-100ng/μl repair template plasmid, and 2.5ng/μl pCFJ90 (myo-2p>mCherry as a co-injection marker) into the gonads of ~10-40 young adult N2 animals. Injected animals were singled and allowed to lay eggs for 3-4 days at 20-23°C in the absence of selection. 500μl of 2mg/ml hygromycin solution was added to each plate, and the plates were returned to 20°C for 4-5 days. Candidate knock-in animals were dominant roller [sqt-1(e1350)] worms that survived hygromycin treatment and lacked red fluorescent extrachromosomal array markers. To excise the SEC, we heat-shocked plates containing ~6 L3/L4 rollers each at 34°C in a water bath for 4 h, then grew the animals at 20°C for 3-4 days. Adult wild-type animals (worms that lost both copies of the SEC) were singled and successful genome editing was verified by visualizing fluorescence and PCR genotyping. Sequencing of the fluorophore insertion site was also performed in all cases to verify proper insertion.
CRISPR strain viability analysis
We assessed genome-edited strains for viability at the plate level for non-N2 visible phenotypes (e.g., uncoordinated (Unc), lethal (Let), dumpy (Dpy), and sterile (Ste). To assess viability, edited worms were compared to N2 at 20°C. Five L4 animals were plated on 200μl of OP50 and allowed to grow until starvation. One day prior to starvation, five L4 animals were picked to new plates for a second generation. This was repeated again for a third generation. If no differences were seen, the worms were considered to have N2 viability.
Fusion protein localization and cleavage analysis
To assess the validity of our basement membrane component protein fusions, homozygous viable strains were screened for fluorescence localization using confocal microscopy. Localization of fluorescence in each strain was examined for BM localization and compared to a GFP secreted into the extracellular space. Strains that exhibited a diffuse localization pattern like secreted GFP were analyzed for proteolysis. Amino acid sequences were analyzed using ProP 1.0 (http://www.cbs.dtu.dk/services/ProP/) that predicts arginine-lysine cleavage by a proprotein convertase. Proteins that were found to exhibit diffuse fluorescence and possess a cleavage site indicating that the fluorescent protein may be cleaved were removed from analysis and new endogenous tags were generated. Cleavage of the original tag and presence of a full-length protein product from the secondary CRISPR tag were confirmed by Western blot analysis. We did not detect DGN-2::mNG or GPN-1::mNG fluorescence, suggesting they might be present at low levels or in response to environmental conditions not tested.
Fusion protein western blot analysis
Worms were collected in sterile water and washed several times before being pelleted and snap-frozen in liquid nitrogen. Worms were thawed in lysis buffer (2% SDS, 10% glycerol, 50 μm Tris 6.8), heated for 5 min at 95°C, then vortexed. Samples were mixed with 4x Laemmli sample buffer (Biorad), heated for 5 min at 95°C, then loaded into a Criterion TGX 10% gel (Biorad) that was run at 100V. The contents of the gel were transferred to nitrocellulose membrane (Biorad) at 20V for 30 min. Membranes were blocked in 5% milk diluted in Tris-buffered saline-Tween 20 (TBS-T) for 1 h before incubation with the anti-mNeonGreen at 1:1000 (32F6, Chromotek) diluted in 2.5% milk/TBS-T. After washing, horseradish peroxidase (HRP)-conjugated secondary antibodies (Jackson-ImmunoResearch) were diluted in 5% milk/TBS-T. SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) was used for detection.
RNA interference
RNAi constructs were obtained from the Vidal and Ahringer libraries and all RNAi experiments were performed using the feeding method (Source Bioscience; (Kamath et al., 2003; Rual et al., 2004)). RNAi bacterial cultures were grown in selective media (1:1000 ampicilin) for 12-14 h at 37°C. 1mM Isopropyl b-D-1-thiogalactopyranoside (IPTG) was added to induce dsRNA expression and cultured for an additional hour. RNAi plates were prepared by spreading a 1:1 mixture of 1M IPTG and 100mg/ml ampicillin (5μl each) on NGM agar plates and left at room temperature overnight following seeding with induced RNAi culture to allow for further induction and drying. For co-depletion experiments, the relevant induced bacterial cultures were mixed at a 1:1 ratio before they were seeded on plates. Most experiments were performed using synchronized L1 worms, which were placed on RNAi plates and allowed to feed for 24-72 h at 20°C. The L4440 empty vector was used as a negative control. For depletion of papilin later in development, synchronized L1 worms were fed L4440 bacteria until the L3, 48 h adult, or 72 h adult stages before they were transferred to papilin RNAi plates for 48 h.
ATP depletion and muscle paralysis
To compare worms immobilized with polystyrene beads alone (muscle contractions, control), L4 hermaphrodites were bathed in 20 uM dicyclohexylcarbodiimide (DCCD) in M9 for 1 h (ATP synthase inhibitor) (Hong and Pedersen, 2008), 0.02% levamisole in M9 for 30 min (muscle hypercontraction), 0.2% tricaine and 0.02% levamisole (muscle contraction and relaxant) in M9 for 30 min, or 75mM 2,3-Butanedione monoxime (BDM, muscle relaxant) in M9 for 1 h (Petzold et al., 2011), then transferred to 5% noble agar pads with polystyrene microbeads. The cover slip was partially sealed with VALAP, flooded with relevant buffer or drug solution to prevent the agar from drying, and then fully sealed with VALAP. FRAP analysis was then carried out as outlined below. Importantly, examination of mNG::fibulin levels after all treatments indicated that fibulin levels were not different from control worms (n = 5 for each, p > 0.05, Student’s t-test).
Imaging
Confocal images were acquired on a Zeiss AxioImager microscope equipped with a Yokogawa CSU-10 spinning disc confocal controlled by Micromanager (Edelstein et al., 2010) or Metamorph software using a Zeiss 40x Plan-APOCHROMAT 1.4NA oil immersion objective or 100x Plan-APOCHROMAT 1.4NA oil immersion objective and a Hamamatsu ORCA R2 CCD camera, Orca-Fusion sCMOS camera, or ImageEM EMCCD camera.
For still images of embryonic stages, embryos were collected from OP50 plates, washed with M9, then placed on 5% agar pads with 0.01M sodium azide. Embryo stages were identified under the microscope and acquired at 40x magnification. Exposure times for confocal imaging varied depending on fluorescence intensity (range: 500ms to 1500ms). Images were displayed either as a single z-slice or as confocal maximum intensity z-projections of confocal slices through the embryo.
For images of larval and adult stages, worms were mounted on 5% agar pads containing 0.01M sodium azide. Images were displayed as single z-slice, maximum intensity, or sum intensity projections.
For fluorescence recovery after photobleaching (FRAP) experiments, photobleaching was performed using an iLas2 targeted laser system from BioVision equipped with an Omicron Lux 60mW 405nm continuous wave laser and controlled with MetaMorph software. Laser power and duration of photobleach were the same for all images within a group of experiments. For all FRAP experiments, worms were immobilized using undiluted 100nm polystyrene bead solution (CV 5%; Polysciences cat. #64010). Duration of experiments and imaging intervals were determined empirically to limit photobleaching. For FRAP experiments where the entire pharynx was photobleached, a slightly higher laser power for a shorter time was used to limit the duration of laser exposure and movement of animals. In all cases the entire surface of the pharynx or gonad was photobleached. To assess the effects of our FRAP conditions on animal health, animals were rescued after posterior pharynx photobleaching and all developed normally (n = 30/30 animals). Further, we never observed pharyngeal morphological defects (n > 100 animals) after photobleaching. Finally, we stained photobleached BMs with mitoTracker Red, which in addition to mitochondria also enriches in intact BMs (Sherwood et al., 2005). BMs stained normally immediately after photobleaching (n = 8/8 animals).
Photoconversion of α1-type IV collagen::mEos2 was achieved with the iLas2 targeted laser system using the following parameters: 0% 405nm laser power, 40 iterations, thickness 3. Photoconversion was limited to the turn region of the gonad in 48 h post-hatching young adult control and papilin RNAi treated animals immobilized on 5% noble agar pads containing 0.01 M sodium azide. Immediately after optical highlighting, animals were recovered, washed in M9, and allowed to feed for 2 h on OP50, then mounted again to determine remaining fluorescence levels of photoconverted α1-type IV collagen::mEos2.
To determine gonad surface area, 3D isosurfaces were constructed as previously described (Jayadev et al., 2019) using Imaris software (Bitplane). At 0 h and 24 h treatment timepoints, isosurfaces of the entire developing gonad were generated, but at the 48 h timepoint only images of a single gonad arm could be obtained (because of its large size), and the surface area of the gonad was determined by doubling the surface area of the individual gonad arm.
Parameters for experiments are described in the figure legends. All image and video processing and analysis were performed in Fiji 2.0 (Schindelin et al., 2012). Figures were constructed using Adobe Illustrator (CC 2017), and graphs were exported from R or Microsoft Excel.
Generation of model BM proteins and receptors for schematics
For Video S5 three-dimensional models of the BM proteins were generated using structural models from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB, https://www.rcsb.org/) and previously published studies. For collagen IV (chain: 1cag, head: 1m3d) and laminin (alpha: 2y38 (Hussain et al., 2011), 4yeq (Moran et al., 2015), beta: 4aqs (Carafoli et al., 2012), gamma: 5mc) model assembly was based on (Hohenester and Yurchenco, 2013) and references therein. For nidogen (G1: laminin-like G domain - 1pz7 (Stetefeld et al., 2004), G2: 1h4u (Hopf et al., 2001), G3: 1npe (Takagi et al., 2003), and EGF domains (from 1gl4 (Kvansakul et al., 2001)) and the laminin binding site as in 1klo (Stetefeld et al., 1996), the model assembly was based on (Lössl et al., 2014). For agrin (Follistatin-like: 6maa (McCoy et al., 2019), laminin-like-EGF, laminin G: 1pz7 (Stetefeld et al., 2004), and EGF domains), the model based on (Hrus et al., 2007). For fibulin (anaphylatoxin-like: 4hwj (Bajic et al., 2013) and EGF domains), the model assembly based on (Hesselson and Kimble, 2006) For integrins (active form extrapolated from inactive extracellular model 1jv2 (Xiong et al., 2001)). All pdb domains are represented by their solvent-accessible surface models as obtained from Chimera software. The entire protein models were assembled manually. The exact representation of the entire proteins is therefore somewhat artistic (their isoforms / unbound and bound conformations / precise folding may not be accurate). For other BM proteins and receptors that were not modeled in 3D, protein schematic sizes were based off of the number of amino acids in each protein from Wormbase (https://wormbase.org/) and shapes were based on the results of conserved domain search using InterPro (https://www.ebi.ac.uk/interpro/) as well as structural information from Uniprot (https://www.uniprot.org/) to determine which key domains should be represented. The ratio and dynamics of matrix components shown in the BM schematics (Figures 1 and 7C and Video S5) are based on the matrix ratios found in the L1 pharynx (Figure 3) and the dynamics (revealed by FRAP) of each molecule and its mode of movement (Figures 4 and 5 and S6 and S7).
QUANTIFICATION AND STATISTICAL ANALYSIS
BM composition comparisons
To compare the composition of the gonadal and pharynx BMs in L1 animals, we used synchronized L1 animals except for perlecan::mNG, for which L1 animals were picked off NGM plates. We first visually screened each strain to determine if the tagged matrix component was present above background levels in the gonadal and pharynx BMs, excluding those that could not be detected. Stacks were acquired with step sizes of 374nm and fluorescence intensity values were generated by drawing an ~0.5μm wide, 4.5μm long line through a clearly defined section of a single confocal z-slice in the mid-plane of the posterior bulb of the pharynx and central dorsal region of the gonadal BM. At least 9 animals were imaged for each matrix component. Background levels were defined by drawing a square region outside the worm and determining the mean intensity. Multiple average background intensities were pooled, and the same background value was subtracted across all strains for consistency to allow for direct comparison. The α1-type chain of collagen (EMB-9) is present in two copies per type IV collagen molecule, and was quantified for compositional comparisons, while the γ-chain of laminin (LAM-2) is one of three different chains in laminin and was quantified for comparisons (Kramer, 2005). For simplicity, we used the levels of α1-type chain as type IV collagen. To generate waffle plots, the mean intensities of all matrix proteins present in the pharynx or gonad were sorted and all mean values were normalized by dividing by the lowest mean value (peroxidasin-1) and rounding to the nearest whole number. The rounded, normalized value was represented as number of squares in the waffle plot, with the lowest value represented as a single square. Color-coding was kept consistent between waffle plots of gonadal and pharynx BM composition to allow for direct comparison.
FRAP analysis
L4 animals were used for most analyses. Levels of perlecan::mNG in the pharynx were too low to accurately measure for FRAP analysis and SPARC::mNG was not analyzed as this strain was not homozygous viable. Background was subtracted as described above. Fluorescence intensity values were generated by drawing an ~0.5μm wide, 4.5μm long line through a clearly defined section of the mid-plane of a single z-slice of the pharynx or gonadal BM at the prebleach time point and measuring this region at each subsequent time point, manually correcting for animal movement. Fluorescence intensity was normalized by: 1) subtracting any residual post-bleach fluorescence from the fluorescence intensity for each time point, 2) dividing each time point by the fluorescence intensity of the pre-bleach time point, and 3) averaging the normalized values. For normalizing the fluorescence intensities in the control region, step 1 was omitted as no photobleaching occurred. For figure 4C the mean normalized recovery value at 15 min was of each protein compared to that of α1-type IV collagen/EMB-9 using Dunnett’s test (n = 5). To confirm that our experiments represented the steady-state fluorescence recovery of each protein and not addition of new protein to the BM on top of the existing photobleached protein, we measured a control region for each set of short and long term FRAP experiments to ensure that fluorescence in the BM did not increase significantly during the duration of the experiment (see Figure S6A for 15-min recovery controls; long term experiments: γ-laminin::mNG - prebleach control region average fluorescence intensity value 4598 rfu SD ± 1331 rfu; ~5.5 h recovery control region average fluorescence intensity value 4289 rfu SD ± 1724 rfu, p = 0.76, Student’s t-test. α1-type IV collagen::mNG prebleach control region average fluorescence intensity value 24298 rfu SD ± 3761 rfu; ~5.5 h recovery control region average fluorescence intensity value 24317 rfu SD ± 6068 rfu, p = 0.995, Student’s t-test).
Photoconversion analysis
Mean fluorescence intensity values of photoconverted α1-type IV collagen::mEos2 were generated by drawing an ~0.5μm wide, 2μm long line through the photoconverted BM region. Background signal was determined with a similar line in a region outside the BM (but within the worm) with no visible fluorescence. To obtain percentage reduction in mean photoconverted α1-type IV collagen::mEos2 signal, the difference between the background-corrected mean photoconverted signal intensities immediately after photoconversion and 2 h after photoconversion was divided by the background-corrected mean photoconverted signal intensity immediately after photoconversion.
Protein movement imaging
To determine mode of recovery of photobleached matrix components, the width of the pharynx was photobleached to limit fluorescence recovery coming from z-planes above or below the imaging plane. Most proteins were assessed for movement in the posterior bulb of the L4 pharynx, however, spondin::mNG recovery was examined in the isthmus and agrin::mNG was examined at the L1 stage where they were present at higher levels. Further, nidogen::mNG and γ-laminin::mNG movement was examined in the bend of L4 gonad arms. For BM components that showed dynamic recovery, imaging intervals were decreased to 6 or 10 s between frames to allow sensitive analysis of initial fluorescence recovery. For analysis of fluorescence recovery, single z-slice images were first registered using the StackReg function in Fiji, then rotated so that the anterior-posterior axis of the pharynx was horizontal with the anterior pharynx facing to the left. For mNG::fibulin, four ~4μm square boxes were drawn: one in the anterior pharynx far from the bleached region, one in an unbleached region adjacent to the bleached region, one at the bleached edge of the bleached region, one 5-10μm into the bleached region. For all other images, two 4μm square boxes were drawn, one at the bleached edge of the bleached region and the other 5-10μm into the bleached region. Background was subtracted as described above, and total fluorescence intensity in each box was measured at each time point, normalized to the pre-bleach intensity value in that region, and plotted as a line graph. The rate of matrix movement was determined by assessing the time fluorescently labeled matrix protein was first detected 5μm into the unbleached region and this time was divided into 5μm to determine rate in μm/sec. For all recovery experiments at least five animals were examined for each matrix component. To examine recovery in the anterior pharynx, the procorpus was photobleached in L1 agrin::mNG animals and assessed for recovery. Matrix proteins with that showed minimal fluorescence recovery in 30 min were photobleached and reimaged after 1 h (papilinS/MIG-6S) or were rescued back to a plate, and re-mounted and imaged after ~5.5 h (γ-laminin/LAM-2, α1-type IV collagen/EMB-9). To determine if papilinS, γ-laminin, and α1-type IV collagen move within the BM, background was subtracted as above and two 4μm square boxes were drawn, one at the bleached edge of the bleached region and the other 5-10 μm into the bleached region. The total fluorescence intensity in each box was measured, pooled, and the two measurements compared (n = 5-6 animals for each component). A higher fluorescent signal within the box nearest the bleach site would indicate movement, however, no significant differences in fluorescence intensity between the two boxes was observed in any of the components (Figure S7D).
To assess the rate of extracellular exchange of a dynamic matrix component with the BM, mNG::fibulin was bleached in the entire pharynx BM and the BM imaged (as outlined above in FRAP analysis) every 10 s for 15 min to assess recovery from extracellular fluid. mNG::fibulin is present at low levels in the gut BM, which is contiguous with the pharynx BM, and thus minimized the contribution of mNG::fibulin moving within the gut BM and into the pharynx BM after photobleaching. Secreted GFP was also photobleached with similar conditions and over 60% recovered adjacent to the BM in 1 min (n = 5 animals), indicating rapid recovery of the pool of fluorescent protein within the extracellular fluid.
Papilin isoform intensity comparison
Images were acquired using a spinning disc confocal as described above. Both papilinS::mNG and papilinL::mNG animals were acquired at the same imaging settings to allow for direct comparison of fluorescence intensities. We compared background subtracted mean fluorescence intensities using boxplots and Student’s t-test. Levels of papilinS::mNG were displayed as fold increase above papilinL::mNG levels by taking the mean fluorescence intensity of the papilinS::mNG animals divided by the papilinL::mNG animals. For each experiment (n ≥ 10 animals for each).
RNAi knockdown efficiency
To assess the efficiency of papilin RNAi, papilinL::mNG or papilinS::mNG animals from RNAi and control plates were imaged 48 h post plating. Mean gonadal BM fluorescence intensity was measured as described above. Background was subtracted and mean fluorescence intensities were compared using boxplots and Student’s t-tests. Percent knockdown was calculated as the difference between the background-corrected mean fluorescence intensities of animals on the RNAi plate and L4440 control plate divided by the mean fluorescence intensity of animals on the L4440 control plate for each isoform. For L1 platings, 88% reduction of papilinS and 61% reduction of papilinL was observed (n = 10 animals examined each). For the knockdown of papilin later in development, 61%, 69%, and 63% reduction of papilinS was observed with 24 h L3 stage, 48 h adult, and 72 h adult platings, respectively (n = 5-15 animals for each).
Scoring of papilin knockdown phenotype
Animals with an endogenous α1-type IV collagen::mNG or α1-type IV collagen::mCh transgene (emb-9p>emb-9::mCherry) were plated on RNAi and assessed for a fibrous type IV collagen network 48 h after plating. To assess the relationship between patterning of residual papilin and fibrous type IV collagen after papilin depletion, L1 stage papilinS::mNG; the α1-type IV collagen::mCh worms were placed on papilin RNAi and imaged after 48 h. Maximum intensity projections of the gonad were generated and an ~0.5 μm wide line was drawn through a region where the type IV collagen network had a fibrous appearance and patches of residual papilinS::mNG were present. Background was subtracted and fluorescence intensities were normalized and plotted on a line graph.
Quantification of ADAMTS localization following papilin knockdown
To quantify the localization of GON-1::mNG, MIG-17::mNG, peroxidasin-2::mNG, and peroxidasin-1::mNG following papilin RNAi knockdown, L1 animals were plated on papilin RNAi and were imaged at ~48 h post plating. Quantification of BM fluorescence intensity was assessed by generating a three-slice sum projection in each animal and drawing a 0.5 μm wide line along the BM at the turn of the gonad. Background was subtracted and mean fluorescence intensities were compared using boxplots and Student’s t-tests (n ≥ 10 animals for each).
Statistical Analysis
For quantification of fluorescence levels, sample size was validated a posteriori for variance and statistical significance. Statistical analysis was performed in R using the stats package (stats v3.6.1) unless otherwise specified. Distribution of data was assessed for normality using the Shapiro-Wilk test. For comparisons of mean fluorescence intensities between two populations, we used an unpaired, two-tailed Student’s t-test. To compare mean fluorescence recovery of multiple samples to a single control, we performed one-way ANOVA followed by a post-hoc Dunnet’s test (DescTools v0.99.19). Waffle plots and FRAP line graphs were prepared in R using the ggplot2 package and bloxplots were prepared in GraphPad Prism. In boxplots, box edges depict the 25th and 75th percentiles, the line in the box indicates the median value, and whiskers mark the minimum and maximum values. All other bar and line graphs were prepared in Microsoft Excel. Figure legends indicate sample sizes, statistical tests used, and p-values.
Supplementary Material
Table S1. Genome edited basement membrane (BM) proteins, Related to Figure 1.
Complete list of basement membrane (BM) matrix encoding genes and interacting receptors where CRISPR/Cas-9-mediated homologous recombination was used to knock in mNeonGreen (mNG) and other fluorophores. Table also includes human and mouse orthologs, sequences for sgRNAs, location where each gene was tagged, fluorophore(s) used for tagging, the linker used, results of viability analysis, and citations for previous localization reported in C. elegans.
Video S1. Fibulin moves within the pharynx BM. Related to Figure 5.
FRAP analysis of mNG::fibulin in the L4 stage pharynx posterior bulb with rapid sampling. (Top, left) After photobleaching of the posterior bulb, mNG::fibulin moves within the BM (n = 5/5 animals). (Top, right) A heatmap highlights the boundary (dashed line) of photobleaching and the movement of fibulin. (Bottom) FRAP analysis of type XVIII collagen::mNG with a 30-min time-lapse. After photobleaching type XVIII collagen::mNG showed uniform recovery in the posterior bulb (n = 5/5 animals). Images are single confocal z-slices and were acquired every 10 s for either 10 min (mNG::fibulin) or every 2 min for 30 min (type XVIII collagen::mNG). Scale bar is 10μm.
Video S2. Agrin moves within the anterior pharynx BM. Related to Figure 5.
(Left) FRAP analysis of agrin::mNG in the anterior region (left, bracket) of the L1-stage pharynx. (Right) A heatmap highlights the boundary (dashed line) of photobleaching and movement of agrin::mNG into the photobleached area. Images are single confocal z-slices and were acquired every 10 s for 10 min. Scale bar is 10μm.
Video S3. Nidogen moves within the gonadal BM. Related to Figure 5.
FRAP analysis of nidogen::mNG and γ-laminin::mNG in the bend of the gonad arm of an L4- stage animal. (Top, Left) After photobleaching of the bend in the gonad arm, nidogen::mNG moves within the BM (n = 10/10 animals). (Right) A heatmap highlights the boundary (yellow box) of photobleaching and movement of nidogen::mNG into the photobleached area. (Bottom) Similar photobleaching reveals that γ-laminin::mNG does not move within the BM (n = 10/10 animals). Images are single confocal z-slices and were acquired every 6 s for 10 min. Scale bar is 10μm.
Video S4. Fibulin movement is enhanced by muscle contractions. Related to Figure 5.
FRAP analysis of mNG::fibulin in the L4 stage pharynx posterior bulb in a worm immobilized with polystyrene beads, which allow muscle contractions. (Top, left) After photobleaching of the posterior bulb, mNG::fibulin moves rapidly within the BM (n = 5/5 animals). (Top, right) A heat map highlights the boundary of photobleaching and the movement of fibulin. (Bottom) FRAP analysis of mNG;;fibulin in worms whose muscles have been paralyzed with tricaine and levamisole reveals that fibulin movement is reduced (n = 5/5 animals). Images are single confocal z-slices and were acquired every 10 s for 15 min. Scale bar is 10μm.
Video S5. Video illustration of BM dynamics. Related to Figures 4 and 5.
Laminin associates with the plasma membrane through binding to integrin and dystroglycan receptors forming a stable BM framework with type IV collagen. Nidogen, fibulin, and agrin (spondin and peroxidasin-1 not shown) move dynamically within the laminin and type IV collagen scaffolding. See Methods for details on molecule illustrations.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| RNAi feeding strain | Caenorhabditis Genetics Center | HT115 (DE3) |
| Vidal RNAi Library | Open Biosystems | ORF RNAi Collection V1.1 |
| Ahringer RNAi Library | Source Bioscience | C. elegans RNAi Collection (Ahringer) |
| Chemical, Peptides, and Recombinant Proteins | ||
| MitoTracker™ Red CMXRos | Molecular Probes | M7512 |
| Dicyclohexylcarbodiimide (DCCD) | Sigma | 633267 |
| Ethyl 3-aminobenzoate methanesulfonate (Tricaine) | Sigma | E10521 |
| Levamisole hydrochloride | Sigma | 1359302 |
| 2,3-Butanedione monoxime (BDM) | Sigma | B0753 |
| MitoTracker™ Red CMXRos | Molecular Probes | M7512 |
| 4x Laemmli Sample Buffer | BIO RAD | 1610747 |
| Anti-mNeongreen antibody | chromotek | 32F6 |
| HRP-conjugated secondary antibody | Jackson ImmunoResearch | 115-035-003 |
| Supersignal West Pico Chemiluminescent Substrate | ThermoFisher Scientific | 34579 |
| Experimental Models: Organisms/Strains | ||
| C. elegans wild-type strain | Caenorhabditis Genetics Center | WB-STRAIN:N2_(ancestral) |
| emb-9p::emb-9::mCh transgene; qyIs46 | Ihara et al. 2011 | NK364 |
| emb-9p::emb-9::mCh transgene; qyIs44 | Ihara et al. 2011 | NK362 |
| emb-9p::emb-9::mCh transgene with mig-6L mutant; qyIs46; mig-6(oz113)/nT1 | qyIs46 (Ihara et al. 2011) | NK2546 |
| emb-9p::emb-9::mCh transgene with lam-2::mNG CRISPR knock-in; qyIs44; qy20 | qyIs44 (Ihara et al., 2011), qy20 (This study) | NK2545 |
| emb-9p::emb-9::mCh transgene with mig-6S::mNG/+ CRISPR knock-in; qyIs46; qy72/+ | qyIs46 (Ihara et al., 2011), qy72 (This study) | NK2544 |
| emb-9::mNG CRISPR knock-in; qy24 | This study | NK2326 |
| emb-9::mEos2 CRISPR knock-in; qy89 | This study | NK2604 |
| let-2::LL::mNG CRISPR knock-in; qy61 | This study | NK2558 |
| lam-1::LL::mNG (9aa linker) CRISPR knock-in; qy63 | This study | NK2123 |
| lam-2::LL::mNG CRISPR knock-in; qy20 | This study | NK2335 |
| lam-2::LL::mNG CRISPR knock-in with plasma membrane mCherry; qy20;cpls55 | This study | NK2336 |
| lam-2::LL::mNG CRISPR knock-in; qy41 | Jayadev et al., 2019 | NK2446 |
| lam-3::LL::mNG CRISPR knock-in; qy28 | This study | NK2425 |
| epi-1::LL::mNG CRISPR knock-in; qy31 | This study | NK2404 |
| mNG::mig-6N terminal CRISPR knock-in; qy37 | This study | NK2442 |
| mig-6S::LL::mNG CRISPR knock-in; qy72/+ | This study | NK2556 |
| mig-6L::LL::mNG CRISPR knock-in; qy73 | This study | NK2557 |
| unc-52::mNG C terminal CRISPR knock-in; qy53 | This study | NK2500 |
| mNG::unc-52 N terminal CRISPR knock-in; qy75 | This study | NK2555 |
| unc-52::mNG synthetic exon CRISPR knock-in; qy80 | This study | NK2583 |
| nid-1::LL::mNG CRISPR knock-in; qy38 | This study | NK2443 |
| cle-1::LL::mNG CRISPR knock-in; qy22 | This study | NK2322 |
| mNG::fbl-1 N terminal CRISPR knock-in; qy62 | This study | NK2579 |
| agr-1::LL::mNG CRISPR knock-in; qy27 | This study | NK2353 |
| spon-1::LL::mNG CRISPR knock-in; qy30 | This study | NK2580 |
| pxn-1::LL::mNG CRISPR knock-in; qy40 | This study | NK2445 |
| mNG::pxn-2 N terminal CRISPR knock-in; qy76 | This study | NK2565 |
| pxn-2::LL::mNG C terminal CRISPR knock-in; qy39 | This study | NK2444 |
| ost-1::LL::mNG CRISPR knock-in; qy60 | This study | NK2559 |
| him-4::LL::mNG CRISPR knock-in; qy33 | This study | NK2422 |
| ina-1::LL::mNG CRISPR knock-in; qy23 | Jayadev et al., 2019 | NK2324 |
| ina-1::LL::mKate CRISPR knock-in; qy46 | This paper | NK2476 |
| pat-2::LL::mNG CRISPR knock-in; qy26 | Jayadev et al., 2019 | NK2330 |
| pat-2::LL::2XmNG CRISPR knock-in; qy49 | Jayadev et al., 2019 | NK2479 |
| pat-3::LL::mNG CRISPR knock-in; qy36 | Jayadev et al., 2019 | NK2436 |
| gpn-1::LL::mNG CRISPR knock-in; qy35 | This study | NK2581 |
| lon-2::LL::mNG CRISPR knock-in; qy55 | This study | NK2582 |
| dgn-1::LL::mNG CRISPR knock-in; qy18 | Naegeli et at. 2017 | NK2318 |
| ten-1::LL::mNG CRISPR knock-in; qy56 | This study | NK2502 |
| ddr-1::LL::mNG CRISPR knock-in; qy43 | This study | NK2456 |
| ddr-2::LL::mNG CRISPR knock-in; qy44 | This study | NK2457 |
| ptp-3::LL::mNG CRISPR knock-in; qy47 | This study | NK2477 |
| sdn-1::LL::mNG CRISPR knock-in; qy29 | This study | NK2413 |
| dgn-2::LL::mNG CRISPR knock-in; qy 82 | This study | NK2587 |
| gon-1::LL::mNG CRISPR knock-in; qy45 | This study | NK2590 |
| mig-17::mNG::3xFlag; shc19 | Ji et al. 2019 | FDU1506 |
| Oligonucleotides | ||
| Table S2 | ||
| Recombinant DNA | ||
| pDD122 (Peft-3::Cas9 + ttTi5605 sgRNA) | Addgene | 47550 |
| mNG-C1^SEC^linker | This study | mNG-SEC-LL |
| mNG-C1^SEC^bothlink | This study | mNG-SEC-bothlink |
| mEos2-C1^SEC | This study | mEos2-SEC |
| L4440 RNAi empty vector control | Addgene | 1654 |
| mig-6 RNAi | This study / Ahringer | mig-6 RNAi |
| Software and Algorithms | ||
| Fiji | Schindelin et al., 2012 | Image J 2.0.0-rc-69/1.52p, http://fiji.sc/ |
| Photoshop | Adobe | Version CC 2018 |
| Imaris | Bitplane | Version 9 |
| Illustrator | Adobe | Version CC 2018 |
| R | GNU Project | Open Source |
| μManager | Edelstein et al., 2010 | Version 1.4, https://micromanager.org/ |
| iLas | Biovision | V.1.2.3 |
| MetaMorph | Molecular Devices | 7.10.3.279 |
| Excel | Microsoft | Version 15.39 |
| GraphPad Prism | GraphPad | Version 7 |
Highlights:
In vivo visualization of 29 basement membrane (BM) proteins fused with mNeonGreen
Embryonic BMs share a common template but diversify greatly during development
Stable BM proteins form a scaffold that supports movement of dynamic BM components
Papilin promotes local BM collagen removal by limiting GON-1 protease localization
ACKNOWLEDGEMENTS
We thank Lisa Cameron of the Duke University LMCF for imaging advice, M. Clay for helpful discussions, and K. Gordon for comments on the manuscript. We thank A. Kawska (info@illuscientia) for her work on schematics and H Buelow for advice on sdn-1 tagging. E.H. is supported by 129351-PF-16-024-01-CSM from ACS, S.P by F31 HD097901, L.C.K. by F32GM103148, S.P. and D.R.S. by R35GM118049 and R21HD084290. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATIONS OF INTEREST
The authors declare no competing interests.
REFERENCES
- Bajic G, Yatime L, Klos A, and Andersen GR (2013). Human C3a and C3a desArg anaphylatoxins have conserved structures, in contrast to C5a and C5a desArg. Protein Sci. 22, 204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bányai L, Sonderegger P, and Patthy L (2010). Agrin binds BMP2, BMP4 and TGFbeta1. PLoS One 5, e10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg MCW, MacCarthy-Morrogh L, Carter D, Morris J, Ribeiro Bravo I, Feng Y, and Martin P (2019). Proteolytic and Opportunistic Breaching of the Basement Membrane Zone by Immune Cells during Tumor Initiation. Cell Rep. 27, 2837–2846.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunt S, Hooley C, Hu N, Scahill C, Weavers H, and Skaer H (2010). Hemocyte-secreted type IV collagen enhances BMP signaling to guide renal tubule morphogenesis in Drosophila. Dev. Cell 19, 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candiello J, Cole GJ, and Halfter W (2010). Age-dependent changes in the structure, composition and biophysical properties of a human basement membrane. Matrix Biol 29, 402–410. [DOI] [PubMed] [Google Scholar]
- Carafoli F, Hussain S-A, and Hohenester E (2012). Crystal structures of the network-forming short-arm tips of the laminin β1 and γ1 chains. PLoS One 7, e42473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetera M, Lewellyn L, and Horne-Badovinac S (2016). Cultivation and live imaging of drosophila ovaries. Methods Mol. Biol 1478, 215–226. [DOI] [PubMed] [Google Scholar]
- Chang J, and Chaudhuri O (2019). Beyond proteases: Basement membrane mechanics and cancer invasion. J. Cell Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay MR, and Sherwood DR (2015). Basement Membranes in the Worm: A Dynamic Scaffolding that Instructs Cellular Behaviors and Shapes Tissues. Curr Top Membr 76, 337–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crest J, Diz-Munoz A, Chen D-Y, Fletcher DA, and Bilder D (2017). Organ sculpting by patterned extracellular matrix stiffness. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaris ML, Gatmaitan M, FlorCruz S, Luo F, Li K, Holmes WE, Hellerstein MK, Turner SM, and Emson CL (2014). Proteomic analysis of altered extracellular matrix turnover in bleomycin-induced pulmonary fibrosis. Mol. Cell Proteomics 13, 1741–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, and Goldstein B (2016). CRISPR-Based Methods for Caenorhabditis elegans Genome Engineering. Genetics 202, 885–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, and Stuurman N (2010). Computer control of microscopes using μManager. Curr. Protoc. Mol. Biol Chapter 14, Unit14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler AL, Boudko SP, Rokas A, and Hudson BG (2018). The triple helix of collagens - an ancient protein structure that enabled animal multicellularity and tissue evolution. J. Cell Sci 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresco VM, Kern CB, Mohammadi M, and Twal WO (2016). Fibulin-1 Binds to Fibroblast Growth Factor 8 with High Affinity: EFFECTS ON EMBRYO SURVIVAL. J. Biol. Chem 291, 18730–18739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotenstein JR, Swale RE, Fukuda T, Wu Z, Giurumescu CA, Goncharov A, Jin Y, and Chisholm AD (2010). The C. elegans peroxidasin PXN-2 is essential for embryonic morphogenesis and inhibits adult axon regeneration. Development 137, 3603–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham PL, Johnson JJ, Wang S, Sibley MH, Gupta MC, and Kramer JM (1997). Type IV collagen is detectable in most, but not all, basement membranes of Caenorhabditis elegans and assembles on tissues that do not express it. J. Cell Biol 137, 1171–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter W, Moes S, Asgeirsson DO, Halfter K, Oertle P, Melo Herraiz E, Plodinec M, Jenoe P, and Henrich PB (2017). Diabetes-related changes in the protein composition and the biomechanical properties of human retinal vascular basement membranes. PLoS One 12, e0189857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DH (2017). Introduction to C. elegans Embryo Anatomy. WormAtlas. [Google Scholar]
- Harunaga JS, Doyle AD, and Yamada KM (2014). Local and global dynamics of the basement membrane during branching morphogenesis require protease activity and actomyosin contractility. Dev. Biol 394, 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppert JK, Dickinson DJ, Pani AM, Higgins CD, Steward A, Ahringer J, Kuhn JR, and Goldstein B (2016). Comparative assessment of fluorescent proteins for in vivo imaging in an animal model system. Mol. Biol. Cell 27, 3385–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselson D, and Kimble J (2006). Growth control by EGF repeats of the C. elegans Fibulin-1C isoform. J. Cell Biol 175, 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenester E, and Yurchenco PD (2013). Laminins in basement membrane assembly. Cell Adh Migr 7, 56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, and Pedersen PL (2008). ATP synthase and the actions of inhibitors utilized to study its roles in human health, disease, and other scientific areas. Microbiol. Mol. Biol. Rev 72, 590–641, Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf M, Gohring W, Ries A, Timpl R, and Hohenester E (2001). Crystal structure and mutational analysis of a perlecan-binding fragment of nidogen-1. Nat. Struct. Biol 8, 634–640. [DOI] [PubMed] [Google Scholar]
- Hrus A, Lau G, Hutter H, Schenk S, Ferralli J, Brown-Luedi M, Chiquet-Ehrismann R, and Canevascini S (2007). C. elegans agrin is expressed in pharynx, IL1 neurons and distal tip cells and does not genetically interact with genes involved in synaptogenesis or muscle function. PLoS One 2, e731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C -c. (2003). Laminin subunits and their role in C. elegans development. Development 130, 3343–3358. [DOI] [PubMed] [Google Scholar]
- Huang C-C, Hall DH, Hedgecock EM, Kao G, Karantza V, Vogel BE, Hutter H, Chisholm AD, Yurchenco PD, and Wadsworth WG (2003). Laminin alpha subunits and their role in C. elegans development. Development 130, 3343–3358. [DOI] [PubMed] [Google Scholar]
- Hussain S-A, Carafoli F, and Hohenester E (2011). Determinants of laminin polymerization revealed by the structure of the α5 chain amino-terminal region. EMBO Rep. 12, 276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara S, Hagedorn EJ, Morrissey MA, Chi Q, Motegi F, Kramer JM, and Sherwood DR (2011). Basement membrane sliding and targeted adhesion remodels tissue boundaries during uterine-vulval attachment in Caenorhabditis elegans. Nat. Cell Biol 13, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabella AJ, and Horne-Badovinac S (2016). Rab10-Mediated Secretion Synergizes with Tissue Movement to Build a Polarized Basement Membrane Architecture for Organ Morphogenesis. Dev. Cell 38, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayadev R, Chi Q, Keeley DP, Hastie EL, Kelley LC, and Sherwood DR (2019). α-Integrins dictate distinct modes of type IV collagen recruitment to basement membranes. J. Cell Biol 218, 3098–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji T, Wang K, Fan J, Huang J, Wang M, Dong X, Shi Y, Manning L, Zhang X, Shao Z, et al. (2019). ADAMTS-family protease MIG-17 regulates synaptic allometry by modifying the extracellular matrix and modulating glia morphology during growth. BioRxiv. [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- Kao G, Huang C, Hedgecock EM, Hall DH, and Wadsworth WG (2006). The role of the laminin beta subunit in laminin heterotrimer assembly and basement membrane function and development in C. elegans. Dev. Biol 290, 211–219. [DOI] [PubMed] [Google Scholar]
- Kawano T, Zheng H, Merz DC, Kohara Y, Tamai KK, Nishiwaki K, and Culotti JG (2009). C. elegans mig-6 encodes papilin isoforms that affect distinct aspects of DTC migration, and interacts genetically with mig-17 and collagen IV. Development 136, 1433–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelwick R, Desanlis I, Wheeler GN, and Edwards DR (2015). The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol. 16, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, Kostka G, Garbe JHO, Keene DR, Bachinger HP, Hanisch F-G, Markova D, Tsuda T, Timpl R, Chu M-L, et al. (2007). A comparative analysis of the fibulin protein family. Biochemical characterization, binding interactions, and tissue localization. J. Biol. Chem 282, 11805–11816. [DOI] [PubMed] [Google Scholar]
- Kramer JM (2005). Basement membranes. WormBook 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerova IA, Kawaguchi N, Fessler LI, Nelson RE, Chen Y, Kramerov AA, Kusche-Gullberg M, Kramer JM, Ackley BD, Sieron AL, et al. (2000). Papilin in development; a pericellular protein with a homology to the ADAMTS metalloproteinases. Development 127, 5475–5485. [DOI] [PubMed] [Google Scholar]
- Kvansakul M, Hopf M, Ries A, Timpl R, and Hohenester E (2001). Structural basis for the high-affinity interaction of nidogen-1 with immunoglobulin-like domain 3 of perlecan. EMBO J. 20, 5342–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Qi Y, McKee K, Liu J, Hsu J, and Yurchenco PD (2017). Integrin and dystroglycan compensate each other to mediate laminin-dependent basement membrane assembly and epiblast polarization. Matrix Biol 57–58, 272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lössl P, Kölbel K, Tänzler D, Nannemann D, Ihling CH, Keller MV, Schneider M, Zaucke F, Meiler J, and Sinz A (2014). Analysis of nidogen-1/laminin γ1 interaction by cross-linking, mass spectrometry, and computational modeling reveals multiple binding modes. PLoS One 9, e112886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Cao X, Dai J, and Pastor-Pareja JC (2017). Basement membrane manipulation in drosophila wing discs affects dpp retention but not growth mechanoregulation. Dev. Cell 42, 97–106.e4. [DOI] [PubMed] [Google Scholar]
- Mak KM, and Mei R (2017). Basement membrane type IV collagen and laminin: an overview of their biology and value as fibrosis biomarkers of liver disease. Anat Rec (Hoboken) 300, 1371–1390. [DOI] [PubMed] [Google Scholar]
- Mango SE (2007). The C. elegans pharynx: a model for organogenesis. WormBook 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y, Louani A, Dragu A, Sánchez-Sánchez BJ, Serna-Morales E, Yolland L, Gyoergy A, Vizcay G, Fleck RA, Heddleston JM, et al. (2017). A moving source of matrix components is essential for de novo basement membrane formation. Curr. Biol. 27, 3526–3534.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K, Koga A, and Ihara S (2019). Visualization of endogenous NID-1 and EMB-9 in C. elegans. MicroPublication Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy JC, Walker RG, Murray NH, and Thompson TB (2019). Crystal structure of the WFIKKN2 follistatin domain reveals insight into how it inhibits growth differentiation factor 8 (GDF8) and GDF11. J. Biol. Chem 294, 6333–6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, and Looger LL (2009). A bright and photostable photoconvertible fluorescent protein. Nat. Methods 6, 131–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran T, Gat Y, and Fass D (2015). Laminin L4 domain structure resembles adhesion modules in ephrin receptor and other transmembrane glycoproteins. FEBS J. 282, 2746–2757. [DOI] [PubMed] [Google Scholar]
- Morin X, Daneman R, Zavortink M, and Chia W (2001). A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA 98, 15050–15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey MA, and Sherwood DR (2015). An active role for basement membrane assembly and modification in tissue sculpting. J. Cell Sci. 128, 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey MA, Jayadev R, Miley GR, Blebea CA, Chi Q, Ihara S, and Sherwood DR (2016). SPARC promotes cell invasion in vivo by decreasing type IV collagen levels in the basement membrane. PLoS Genet. 12, e1005905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriel JM, Dong C, Hutter H, and Vogel BE (2005). Fibulin-1C and Fibulin-1D splice variants have distinct functions and assemble in a hemicentin-dependent manner. Development 132, 4223–4234. [DOI] [PubMed] [Google Scholar]
- Naba A, Clauser KR, Lamar JM, Carr SA, and Hynes RO (2014). Extracellular matrix signatures of human mammary carcinoma identify novel metastasis promoters. Elife 3, e01308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naegeli KM, Hastie E, Garde A, Wang Z, Keeley DP, Gordon KL, Pani AM, Kelley LC, Morrissey MA, Chi Q, et al. (2017). Cell Invasion In Vivo via Rapid Exocytosis of a Transient Lysosome-Derived Membrane Domain. Dev. Cell 43, 403–417.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyström A, Bornert O, and Kühl T (2017). Cell therapy for basement membrane-linked diseases. Matrix Biol 57–58, 124–139. [DOI] [PubMed] [Google Scholar]
- Perrucci GL, Rurali E, and Pompilio G (2018). Cardiac fibrosis in regenerative medicine: destroy to rebuild. J Thorac Dis 10, S2376–S2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold BC, Park S-J, Ponce P, Roozeboom C, Powell C, Goodman MB, and Pruitt BL (2011). Caenorhabditis elegans body mechanics are regulated by body wall muscle tone. Biophys. J 100, 1977–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflicke H, and Sixt M (2009). Preformed portals facilitate dendritic cell entry into afferent lymphatic vessels. J. Exp. Med 206, 2925–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöschl E, Schlötzer-Schrehardt U, Brachvogel B, Saito K, Ninomiya Y, and Mayer U (2004). Collagen IV is essential for basement membrane stability but dispensable for initiation of its assembly during early development. Development 131, 1619–1628. [DOI] [PubMed] [Google Scholar]
- Ramos-Lewis W, LaFever KS, and Page-McCaw A (2018). A scar-like lesion is apparent in basement membrane after wound repair in vivo. Matrix Biol 74, 101–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randles MJ, Humphries MJ, and Lennon R (2017). Proteomic definitions of basement membrane composition in health and disease. Matrix Biol 57–58, 12–28. [DOI] [PubMed] [Google Scholar]
- Rasmussen JP, Reddy SS, and Priess JR (2012). Laminin is required to orient epithelial polarity in the C. elegans pharynx. Development 139, 2050–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers RS, and Nishimune H (2017). The role of laminins in the organization and function of neuromuscular junctions. Matrix Biol 57–58, 86–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J-F, Ceron J, Koreth J, Hao T, Nicot A-S, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, et al. (2004). Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14, 2162–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller HB, Fernandez IE, Burgstaller G, Schaab C, Scheltema RA, Schwarzmayr T, Strom TM, Eickelberg O, and Mann M (2015). Time- and compartment-resolved proteome profiling of the extracellular niche in lung injury and repair. Mol. Syst. Biol 11, 819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood DR, Butler JA, Kramer JM, and Sternberg PW (2005). FOS-1 promotes basement-membrane removal during anchor-cell invasion in C. elegans. Cell 121, 951–962. [DOI] [PubMed] [Google Scholar]
- Smyth N, Vatansever HS, Murray P, Meyer M, Frie C, Paulsson M, and Edgar D (1999). Absence of basement membranes after targeting the LAMC1 gene results in embryonic lethality due to failure of endoderm differentiation. J. Cell Biol 144, 151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetefeld J, Mayer U, Timpl R, and Huber R (1996). Crystal structure of three consecutive laminin-type epidermal growth factor-like (LE) modules of laminin gamma1 chain harboring the nidogen binding site. J. Mol. Biol 257, 644–657. [DOI] [PubMed] [Google Scholar]
- Stetefeld J, Alexandrescu AT, Maciejewski MW, Jenny M, Rathgeb-Szabo K, Schulthess T, Landwehr R, Frank S, Ruegg MA, and Kammerer RA (2004). Modulation of agrin function by alternative splicing and Ca2+ binding. Structure 12, 503–515. [DOI] [PubMed] [Google Scholar]
- Takagi J, Yang Y, Liu J-H, Wang J-H, and Springer TA (2003). Complex between nidogen and laminin fragments reveals a paradigmatic beta-propeller interface. Nature 424, 969–974. [DOI] [PubMed] [Google Scholar]
- Topf U, and Chiquet-Ehrismann R (2011). Genetic interaction between Caenorhabditis elegans teneurin ten-1 and prolyl 4-hydroxylase phy-1 and their function in collagen IV-mediated basement membrane integrity during late elongation of the embryo. Mol. Biol. Cell 22, 3331–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trier JS, Allan CH, Abrahamson DR, and Hagen SJ (1990). Epithelial basement membrane of mouse jejunum. Evidence for laminin turnover along the entire crypt-villus axis. J. Clin. Invest 86, 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzebiatowska A, Topf U, Sauder U, Drabikowski K, and Chiquet-Ehrismann R (2008). Caenorhabditis elegans teneurin, ten-1, is required for gonadal and pharyngeal basement membrane integrity and acts redundantly with integrin ina-1 and dystroglycan dgn-1. Mol. Biol. Cell 19, 3898–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsilibary EC (2003). Microvascular basement membranes in diabetes mellitus. J. Pathol 200, 537–546. [DOI] [PubMed] [Google Scholar]
- Unsoeld T, Park J-O, and Hutter H (2013). Discoidin domain receptors guide axons along longitudinal tracts in C. elegans. Dev. Biol 374, 142–152. [DOI] [PubMed] [Google Scholar]
- Urbano JM, Torgler CN, Molnar C, Tepass U, Lopez-Varea A, Brown NH, de Celis JF, and Martin-Bermudo MD (2009). Drosophila laminins act as key regulators of basement membrane assembly and morphogenesis. Development 136, 4165–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel BE, and Hedgecock EM (2001). Hemicentin, a conserved extracellular member of the immunoglobulin superfamily, organizes epithelial and other cell attachments into oriented line-shaped junctions. Development 128, 883–894. [DOI] [PubMed] [Google Scholar]
- Walser M, Umbricht CA, Frohli E, Nanni P, and Hajnal A (2017). β-Integrin de-phosphorylation by the Density-Enhanced Phosphatase DEP-1 attenuates EGFR signaling in C. elegans. PLoS Genet. 13, e1006592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo W-M, Berry E, Hudson ML, Swale RE, Goncharov A, and Chisholm AD (2008). The C. elegans F-spondin family protein SPON-1 maintains cell adhesion in neural and non-neural tissues. Development 135, 2747–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, and Arnaout MA (2001). Crystal structure of the extracellular segment of integrin alpha Vbeta3. Science 294, 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenco PD (2011). Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb. Perspect. Biol 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenco PD (2015). Integrating Activities of Laminins that Drive Basement Membrane Assembly and Function. Curr Top Membr 76, 1–30. [DOI] [PubMed] [Google Scholar]
- Yurchenco PD, and Patton BL (2009). Developmental and pathogenic mechanisms of basement membrane assembly. Curr. Pharm. Des 15, 1277–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Genome edited basement membrane (BM) proteins, Related to Figure 1.
Complete list of basement membrane (BM) matrix encoding genes and interacting receptors where CRISPR/Cas-9-mediated homologous recombination was used to knock in mNeonGreen (mNG) and other fluorophores. Table also includes human and mouse orthologs, sequences for sgRNAs, location where each gene was tagged, fluorophore(s) used for tagging, the linker used, results of viability analysis, and citations for previous localization reported in C. elegans.
Video S1. Fibulin moves within the pharynx BM. Related to Figure 5.
FRAP analysis of mNG::fibulin in the L4 stage pharynx posterior bulb with rapid sampling. (Top, left) After photobleaching of the posterior bulb, mNG::fibulin moves within the BM (n = 5/5 animals). (Top, right) A heatmap highlights the boundary (dashed line) of photobleaching and the movement of fibulin. (Bottom) FRAP analysis of type XVIII collagen::mNG with a 30-min time-lapse. After photobleaching type XVIII collagen::mNG showed uniform recovery in the posterior bulb (n = 5/5 animals). Images are single confocal z-slices and were acquired every 10 s for either 10 min (mNG::fibulin) or every 2 min for 30 min (type XVIII collagen::mNG). Scale bar is 10μm.
Video S2. Agrin moves within the anterior pharynx BM. Related to Figure 5.
(Left) FRAP analysis of agrin::mNG in the anterior region (left, bracket) of the L1-stage pharynx. (Right) A heatmap highlights the boundary (dashed line) of photobleaching and movement of agrin::mNG into the photobleached area. Images are single confocal z-slices and were acquired every 10 s for 10 min. Scale bar is 10μm.
Video S3. Nidogen moves within the gonadal BM. Related to Figure 5.
FRAP analysis of nidogen::mNG and γ-laminin::mNG in the bend of the gonad arm of an L4- stage animal. (Top, Left) After photobleaching of the bend in the gonad arm, nidogen::mNG moves within the BM (n = 10/10 animals). (Right) A heatmap highlights the boundary (yellow box) of photobleaching and movement of nidogen::mNG into the photobleached area. (Bottom) Similar photobleaching reveals that γ-laminin::mNG does not move within the BM (n = 10/10 animals). Images are single confocal z-slices and were acquired every 6 s for 10 min. Scale bar is 10μm.
Video S4. Fibulin movement is enhanced by muscle contractions. Related to Figure 5.
FRAP analysis of mNG::fibulin in the L4 stage pharynx posterior bulb in a worm immobilized with polystyrene beads, which allow muscle contractions. (Top, left) After photobleaching of the posterior bulb, mNG::fibulin moves rapidly within the BM (n = 5/5 animals). (Top, right) A heat map highlights the boundary of photobleaching and the movement of fibulin. (Bottom) FRAP analysis of mNG;;fibulin in worms whose muscles have been paralyzed with tricaine and levamisole reveals that fibulin movement is reduced (n = 5/5 animals). Images are single confocal z-slices and were acquired every 10 s for 15 min. Scale bar is 10μm.
Video S5. Video illustration of BM dynamics. Related to Figures 4 and 5.
Laminin associates with the plasma membrane through binding to integrin and dystroglycan receptors forming a stable BM framework with type IV collagen. Nidogen, fibulin, and agrin (spondin and peroxidasin-1 not shown) move dynamically within the laminin and type IV collagen scaffolding. See Methods for details on molecule illustrations.
Data Availability Statement
The published article includes all data generated or analyzed during this study.