Summary
A major class of small bacterial RNAs (sRNAs) regulate translation and mRNA stability by pairing with target mRNAs, dependent upon the RNA chaperone Hfq. Hfq, related to the Lsm/Sm families of splicing proteins, binds the sRNAs and stabilizes them in vivo and stimulates pairing with mRNAs in vitro. Although Hfq is abundant, the sRNAs, when induced, are similarly abundant. Therefore, Hfq may be limiting for sRNA function. We find that, when overexpressed, a number of sRNAs competed with endogenous sRNAs for binding to Hfq. This correlated with lower accumulation of the sRNAs (presumably a reflection of the loss of Hfq binding), and lower activity of the sRNAs in regulating gene expression. Hfq was limiting for both positive and negative regulation by the sRNAs. In addition, deletion of the gene for an expressed and particularly effective competitor sRNA improved the regulation of genes by other sRNAs, suggesting that Hfq is limiting during normal growth conditions. These results support the existence of a hierarchy of sRNA competition for Hfq, modulating the function of some sRNAs.
Introduction
Approximately 100 small RNAs (sRNAs) have been identified in Escherichia coli (E. coli) by genome-wide searches using a variety of approaches (reviewed in Sharma and Vogel, 2009; Waters and Storz, 2009; Backofen and Hess, 2010). Many of these sRNAs regulate gene expression either positively or negatively via partial base pairing with target genes (Gottesman, 2004; Waters and Storz, 2009). Hfq, an RNA-binding protein, is required for full action of this family of sRNAs in E. coli (reviewed in Brennan and Link, 2007). In most cases of positive regulation, sRNAs bind to and remodel secondary structures in the 5′ untranslated region (UTR) of a target gene to free the ribosome binding site (RBS), allowing translation. In negative regulation, on the other hand, sRNAs generally bind close to the RBS, resulting in decreased gene expression by blocking translation and/or decreasing the stability of the target mRNA.
Hfq, found in about 50% of bacterial species (Sun et al., 2002), was originally described as a host factor for the replication of phage Qβ in E. coli. It forms a homohexameric doughnut-like structure with similarity to Sm/Lsm proteins in eukaryotes (reviewed in Brennan and Link, 2007). Hfq binds to both sRNAs and mRNAs in vivo, as judged by immunoprecipitation experiments (Zhang et al., 2003; Sittka et al., 2008). Many Hfq-binding sRNAs are stabilized by Hfq in vivo; without Hfq, at least some of these sRNAs are degraded by RNase E. In addition, Hfq helps to promote pairing of sRNA and target mRNA. Thus, in the absence of Hfq, the accumulation of many sRNAs is low, generally resulting in loss of regulation of the target genes.
While sRNAs can be induced to high levels under stress conditions, the amount of Hfq per cell does not increase when an sRNA is overproduced (Zhang et al., 1998). This raises the possibility that Hfq may become limiting for sRNA action under some stress conditions. For instance, a single E. coli cell has been estimated to contain up to 10 000 Hfq hexamers, but one sRNA, OxyS, reaches a level of 4500 molecules per cell when induced by oxidative stress (Altuvia et al., 1997). Given the number of sRNAs that can bind Hfq, as well as binding of Hfq to mRNAs, it seems likely that Hfq could be limiting under some conditions. If Hfq is limiting, sRNAs will compete for Hfq binding and an induced sRNA might interfere with regulation by other sRNAs. This mechanism was proposed for OxyS-negative regulation of the sigma factor RpoS (Zhang et al., 1998). Overexpression of OxyS decreased the expression of RpoS; however, no base pairing between OxyS and the rpoS mRNA could be detected. Because Hfq and sRNAs are necessary for rpoS translation, Zhang et al. suggested that OxyS might compete for Hfq (Zhang et al., 1998). We observed a similar phenomenon (Mandin and Gottesman, 2010). Using an sRNA library and a PBAD–rpoS–lacZ translational fusion, we found that a few sRNAs, including OxyS, negatively regulated rpoS expression, and this effect was Hfq-dependent. These observations led to the hypothesis that induced OxyS interferes with the action of DsrA, which is a positive regulator of RpoS, by sequestering Hfq. One expected consequence of sequestration of Hfq would be that DsrA is rapidly degraded, providing a reason why RpoS is negatively regulated. Consistent with this model, DsrA and OxyS compete with each other for binding to Hfq in vitro (Sledjeski et al., 2001).
There are a number of expectations if competition is taking place: (i) if the stability and therefore the accumulation of an sRNA requires binding to Hfq, induction of one sRNA might reduce the accumulation of other sRNAs, (ii) induction of one sRNA should reduce the ability of other sRNAs to gain access to Hfq, as measured by lower levels of immunoprecipitation with Hfq, (iii) induction of one sRNA might interfere with the ability of other sRNAs to carry out regulation, while competing with them for Hfq binding, and (iv) deletion of the genes for a competing sRNA might improve the regulation by other sRNAs.
In this study, we investigated sRNA competition for Hfq. We have focused our attention on those sRNAs that are relatively abundant under normal growth conditions to allow monitoring of these sRNAs without specific induction treatments. We monitored total accumulation and Hfq binding to these sRNAs, as well as the activity of target genes for these sRNAs after induction of alternative sRNAs. We find that sRNAs can compete with each other for Hfq, limiting the extent of gene regulation by some sRNAs. Individual sRNAs show significant differences in their ability to compete. Thus, Hfq competition provides another regulatory mechanism that Hfq-dependent sRNAs can use in addition to regulation by base pairing. This conclusion is consistent with recent studies also showing competition between selected sRNAs for Hfq (Hussein and Lim, 2011; Olejniczak, 2011).
Results
Accumulation of sRNAs depends on Hfq
Many sRNAs in E. coli require Hfq for their function. Many research groups have shown that sRNAs are less stable and do not accumulate in the absence of Hfq (for example, see Massé et al., 2003; Basineni et al., 2009; Figueroa-Bossi et al., 2009; Moon and Gottesman, 2009; Papenfort et al., 2009). We have confirmed this with a set of seven sRNAs to be used in this work, under our growth conditions. The level of a given sRNA in an hfq mutant represents the expectation if Hfq were totally unavailable due to competition. RNA samples were collected from a wild type (NM22540, a derivative of MG1655) and an hfq deletion mutant at both exponential phase (OD600 = 0.5) and early stationary phase (OD600 = 1.5) at three different temperatures (32°C, 37°C and 42°C), and the levels of each sRNA were then compared (Fig. S1, Table 1). The levels of these sRNAs in hfq+ cells varied with temperature and growth phase (Fig. S1). As expected, DsrA was more abundant at low temperature than at 42°C (Repoila and Gottesman, 2001). Spot42 was also more abundant at lower temperatures than at 42°C; CyaR was more abundant at higher temperatures. These differences were most evident during exponential growth.
Table 1.
Comparison of the amount of sRNAs in the wild type and the hfq-deleted strain.
| Growth phase | Exponential phase (OD600 = 0.5) | Stationary phase (OD600 = 1.5) | |||||
|---|---|---|---|---|---|---|---|
| Temperature (°C) | 32 | 37 | 42 | 32 | 37 | 42 | |
| ArcZ | Long form | 0.2 (± 0) | 1.0 (± 0.2) | 2.8 (± 2.3) | 0.3 (± 0.2) | 0.7 (± 0.2) | 1.7 (± 0.7) |
| Short forma | > 100 | > 100 | > 100 | > 100 | > 100 | > 100 | |
| DsrA | Long form | 3.9 (± 1.8) | 4.7 (± 2.1) | 4.2 (± 2.3) | 4.1 (± 2.3) | 1.8 (± 1.1) | 2.1 (± 0.7) |
| Short form | 27 (± 19) | 22 (± 7.1) | 38 (± 14) | 243 (± 102) | 30 (± 14) | 24 (± 9) | |
| ChiX | 11 (± 3.5) | 15 (± 4.7) | 19 (± 7.8) | 11 (± 4.4) | 18 (± 8.4) | 23 (± 9.3) | |
| CyaR | 2.0 (± 1.3) | 1.9 (± 0.6) | 6.4 (± 3.4) | 5.8 (± 2.0) | 4.5 (± 1.8) | 11.3 (± 2.5) | |
| GcvB | 1.4 (± 0.6) | 1.6 (± 0.2) | 2.5 (± 0.9) | 1.5 (± 0.9) | 1.1 (± 0.5) | 2.0 (± 0.6) | |
| MgrR | 3.3 (± 0.7) | 3.5 (± 0.7) | 5.5 (± 3) | 3.5 (± 0.7) | 4.3 (± 2) | 5.4 (± 0.6) | |
| Spot 42 | 9.5 (± 4.8) | 3.8 (± 2) | 1.2 (± 0.2) | 1.6 (± 0.6) | 2.1 (± 1.2) | 1.5 (± 0.3) | |
For ArcZ, no short-form RNA was detectable above background in the Δhfq strain at any temperature; this has been arbitrarily shown here as > 100.
Numbers are ratio between WT (NM22540) versus hfq− (NM22562). Northern blot signals were determined using biotinylated probes and quantified as described in Experimental procedures. Numbers represent the averages of three independent experiments; Fig. S1 shows a representative experiment.
As expected, the levels of the sRNAs were significantly lower when hfq was deleted (Fig. S1; Table 1). However, the extent of the loss in the hfq mutant varied with different sRNAs. For instance, the accumulation of MgrR and the long form of DsrA decreased three- to fivefold in the absence of Hfq compared with the wild type during exponential growth at all temperatures. CyaR and Spot42 decreased approximately 10-fold under the conditions at which they were best expressed (Fig. S1, Table 1). ChiX showed the most dramatic change, more than 15-fold under some conditions, while there was little change in the level of GcvB throughout all growth conditions. ArcZ is processed from a 120 nt form to an abundant 56 nt form, and, as previously observed, none of the processed form was detected in an hfq mutant (Sittka et al., 2009; Mandin and Gottesman, 2010). The loss of the processed form in the hfq mutant might reflect Hfq-dependent processing of ArcZ, instability of the processed form, or both. DsrA, in which a proportion of the sRNA is in a shorter form, showed a similar pattern; the long form decreased only modestly in the hfq mutant, but essentially no short form was detected.
These results suggest very different degrees of dependence upon Hfq for stability of different sRNAs. There is some correlation between the effect of deleting hfq and the half-lives measured in the presence of Hfq. For instance, GcvB has been reported to be unstable (half-life of 2 min) and its accumulation is not much changed in an hfq mutant, while ChiX, with a long half-life, is dramatically decreased in accumulation in the hfq mutant (compare Table 1 and Vogel et al., 2003). Why Hfq does not stabilize some Hfq-binding sRNAs remains to be determined.
Less Hfq lowers sRNA accumulation
The accumulation of many Hfq-binding sRNAs was dramatically changed in the absence of Hfq (Fig. S1, Table 1). However, competition for Hfq will not lead to total absence of Hfq but instead may lower the available pool. Therefore, we examined the effect of a mutation known to decrease Hfq levels on the levels of Hfq-binding sRNAs. Sharma et al. reported that DksA positively regulates Hfq expression in Shigella (Sharma and Payne, 2006). In the Shigella dksA mutant, the levels of both the hfq mRNA and protein decreased by 50% compared with the wild type. We therefore examined the effect of the dksA mutation on sRNA accumulation and function as well as competition for Hfq in E. coli.
The levels of Hfq in both the wild type and the dksA mutant were measured to confirm the phenotype in E. coli; protein samples were collected at mid-log phase (OD600 = 0.5) and stationary phase (OD600 = 1.5) at 30°C, and Western blots were performed with Hfq antiserum. As observed in Shigella, in the absence of DksA, the level of Hfq decreased to approximately 50% of that in wild-type cells (Fig. 1, bottom panel); a similar reduction was seen at 37°C.
Fig. 1.

Effect of a dksA mutant on levels of sRNAs. Wild type (MG1655) and an isogenic dksA deletion derivative (CF9240) were grown at 30°C and samples were taken for Western blot for Hfq and Northern analysis for the same sRNAs as in Fig. S1. The levels of the sRNAs were compared between the wild type and dksA mutant for the three experiments and the fold difference (FD) was obtained as the ratio of wild type to ΔdksA; these values are listed below each blot. For ArcZ and DsrA, the levels of the full-length long (L) and processed short (S) transcripts were analysed separately. Normalization was to levels of SsrA.
sRNA samples were analysed from the wild-type and dksA mutant strains (Fig. 1; a ratio of > 1 indicates less sRNA in the ΔdksA mutant). Accumulation of both ChiX and MgrR was decreased in the dksA mutant, compared with wild-type cells (Fig. 1). In the dksA mutant, CyaR levels were significantly less only in stationary phase (when it is most abundant), while Spot42, more abundant in exponential phase than in stationary (Fig. S1), showed moderately lower accumulation only in exponential phase (Fig. 1). ArcZ and DsrA long and short forms were analysed separately. In both cases, the effects of the dksA mutant were moderate. Note that there is enough Hfq, at least in stationary-phase cells, for significant accumulation of the short form of ArcZ in the dksA mutant (Fig. 1) in contrast to the situation in an hfq mutant (Fig. S1).
Competition by OxyS overexpression and rescue by increased Hfq
The results with a dksA mutant suggested that if a highly induced sRNA competes with other sRNAs for Hfq, we would expect a decrease in the total accumulation of these other sRNAs. We revisited the negative regulation of RpoS by OxyS as an initial test for competition (Zhang et al., 1998). RpoS is positively regulated by three sRNAs and Hfq (Majdalani et al., 1998; 2001; Sledjeski et al., 2001; Mandin and Gottesman, 2010). If Hfq is limiting for sRNA action when OxyS is overexpressed, we would expect a decrease in the accumulation of RpoS, as previously seen, as well as a decrease in levels of DsrA and other sRNAs. In addition, overexpression of Hfq should reduce or reverse sRNA competition.
This was examined in the experiments shown in Fig. 2. A plasmid carrying OxyS under the control of the Pbad promoter was used; cells also carried either a compatible low-copy vector control or a compatible low-copy plasmid expressing Hfq, also from a PBAD-inducible promoter. Hfq and/or OxyS expression was induced, and samples analysed for OxyS RNA and Hfq protein levels (Fig. S2). In addition to OxyS, the levels of five different Hfq-dependent sRNAs, including DsrA, were examined; all were chosen because their levels were high enough to be easily detected in cells growing under these conditions (LB, mid-log, 30°C; see Fig. S1). RpoS levels were also monitored.
Fig. 2.

OxyS induction effect on sRNA levels and rescue by extra Hfq. Strain KM153 (hfq+ ΔoxyS) containing either the vector pNM12 or pGFK1014, a plasmid expressing OxyS, and either the vector pBAD33 or pKMT4, a plasmid expressing Hfq, was grown in LB containing ampicillin and chloramphenicol at 30°C. Cells were induced with 0.2% arabinose at OD600 = 0.2–0.3 for 1 h.
A. Samples were taken at OD600 = 0.5, and processed as previously for Western blots for Hfq or RpoS, or Northern blots probed for the sRNAs.
B. The graphs represent the averages from three independent experiments as for (A), quantified as described in Experimental procedures and as shown in Fig. S2 for Hfq and OxyS. For OxyS, a standard curve was generated with a known amount of purified OxyS from in vitro transcription (Fig. S2). If the cell extracts undergo loss of sRNAs during preparation of samples, our estimates for the number of molecules may be low, but relative numbers in different samples should be similarly decreased. For all other graphs, the level of the protein (Hfq or RpoS) or sRNA in cells carrying the two vectors was set to 1, and the relative amounts expressed by comparison to this number. Note that for ArcZ the short form of the sRNA was plotted; for DsrA the short form and long form were combined and plotted. Normalization was to levels of SsrA (not shown). Brackets marked with an asterisk (*) indicate numbers considered significantly different (P < 0.05; NS: not significant, determined by t-est).
Approximately 1800 OxyS molecules per cell were present after 60 min of induction (Fig. 2B; OxyS panel, lane 2; Fig. S2); this increased to 2700 molecules when excess Hfq was present (OxyS panel, lane 4; Fig. S2). This increase suggests that OxyS stability is itself limited by availability of Hfq. Hfq levels were measured (Fig. S2) to be about 4000 Hfq hexamers per cell in the wild-type situation, increased to 6000–8000 hexamers per cell when the PBAD–hfq+ plasmid was induced, a modest overproduction of Hfq. Higher levels of Hfq from the plasmid were seen in the presence of the OxyS plasmid (Fig. 2B, Hfq panel, compare lanes 3 and 4); the reason for this is not known. The level of Hfq in the wild type (lane 1) was comparable to that seen by Kajitani et al. (1994) (5000–10 000 hexamers). RpoS levels during exponential phase growth are relatively low, and induction of OxyS had only a modest negative effect (Fig. 2B, RpoS panel, lane 2 versus lane 1). However, a twofold increase in Hfq resulted in a fourfold increase in RpoS levels in the absence of OxyS induction (RpoS panel, compare lane 3 with lane 1). Thus, Hfq is apparently limiting for RpoS translation, even in the absence of an overexpressed RNA. Overexpressing OxyS as well as Hfq modestly reduced this effect, suggesting that OxyS can still compete to some extent even with extra Hfq.
For the six endogenously encoded sRNAs examined, five decreased in levels as OxyS was induced (Fig. 2B: DsrA, ChiX, ArcZ, CyaR and MgrR, lanes 1 versus 2). In all cases, excess Hfq overcame this decrease (compare lanes 3 and 4), and, in fact, the sRNAs accumulated to a somewhat higher level than in wild-type cells (compare lane 3 to lane 1 for these panels). Thus, under normal growth conditions, Hfq levels were not sufficient to fully bind all sRNAs, and competition was greater under conditions of OxyS induction. Extra Hfq increased sRNA accumulation, with or without OxyS induction, presumably by binding more of the sRNAs and stabilizing them.
The only sRNA not significantly affected by OxyS overexpression was Spot 42. In parallel with this observation, Hfq overexpression did not significantly affect Spot 42 levels (Fig. 2B).
Competition for Hfq binding to sRNAs
The studies above measured accumulation of sRNAs as a proxy for the effect of Hfq binding on sRNA stability. From the results in Fig. 2, we would predict that induction of OxyS should also reduce the binding of other sRNAs to Hfq. This was examined by inducing OxyS, performing Hfq co-immunoprecipitation and measuring the sRNAs in the immunoprecipitate. To mimic the level of induction of OxyS by hydrogen peroxide, without the other complications of hydrogen peroxide treatment, we induced an arabinose-inducible promoter in place of the native oxyS promoter with arabinose for 1 h. Levels of OxyS were determined by comparison with an in vitro transcribed OxyS control. Induction of chromosomal PBAD–oxyS gave a level of OxyS within a factor of two of that seen after hydrogen peroxide induction, 600–1000 molecules per cell, less than that found previously (Zhang et al., 1998) and two- to fivefold less than that seen from the plasmid in Fig. 2 (Fig. S3). These induced cells were used for Hfq co-immunoprecipitation, and seven sRNAs were monitored by Northern blot (Fig. 3). The amounts of sRNA in the immunoprecipitate were normalized to levels without arabinose at the same time point. At 0 min (no accumulation of OxyS), this ratio is close to 1 for all the sRNAs (Fig. 3). When OxyS was induced for 60 min, the level of each of the sRNAs in the immunoprecipitate was significantly reduced. The least drastic effect was for immunoprecipitation of ChiX, possibly suggesting that OxyS is less effective in competing with ChiX for Hfq, compared with competition with other sRNAs (Fig. 3B). Because less OxyS was expressed here than in Fig. 2, there was less of a decrease in total RNA (Fig. 3A). However, the measurement of immunoprecipitated sRNAs was clearly a more sensitive measure of competition than accumulation of the sRNA. Immunoprecipitation also enriched for some sRNA species not detected in the total sRNA Northern blot (Fig. 3A); these may be degradation products forming during the immunoprecipitation procedure. These species were also less abundant after OxyS induction.
Fig. 3.

OxyS induction lowers sRNA binding to Hfq.
A. Strain KM94, harbouring PBAD–oxyS inserted into the host chromosome, was grown in LB at 30°C, induced at OD600 = 0.3 for 60 min with 0.2% arabinose, and samples processed as described in Experimental procedures. Control samples (lanes 1, 3, 5 and 7) were grown without arabinose. Co-IP: co-immunoprecipitate samples
B. Relative levels are expressed as the value in the immunoprecipitate after arabinose induction compared with levels without arabinose induction at 0 min and 60 min. Total input was normalized to levels of Elongation Factor TU (Ef-Tu). For immunoprecipitated samples, levels were normalized to levels of Ig visible on the gels. Graphs are the average of three experiments. The number of OxyS molecules was determined by comparison to the standard curve (Fig. S2). No significant difference was observed for the amount of Hfq with and without OxyS induction (NS: not significant). After OxyS induction, the amount of sRNAs in the immunoprecipitate was significantly lower than that observed without induction (P < 0.05). Asterisk indicates that the relative amount of ChiX in the immunoprecipitate after OxyS induction, while lower than without induction, was significantly higher than that for the other sRNAs (P < 0.05; determined by t-test). The values for both ArcZ and DsrA represent the levels of the short (processed) forms of these sRNAs.
Taken together, induction of OxyS, whether from a plasmid or from the chromosome at physiologically relevant levels, was sufficient to reduce levels of DsrA and other sRNAs, reduce binding of these sRNAs to Hfq, and this reduction in levels was overcome by increasing Hfq, consistent with competition for Hfq in vivo.
Competition for RpoS regulation by other sRNAs
OxyS was chosen for competition experiments here because it had previously been shown to negatively regulate rpoS without apparent base-pairing (Zhang et al., 1998). However, we would expect that other sRNAs, if overproduced, should also compete. In a previous study, downregulation of an rpoS–lacZ translational fusion by multiple sRNAs was observed and attributed to competition for Hfq (Mandin and Gottesman, 2010). The method used in that work may thus be appropriate for further investigating competition. In these experiments, each of 26 Hfq-binding sRNAs was expressed from a Plac promoter on a plasmid, and the activity of a translational fusion to a given target of interest was measured. We found that two of the plasmids, expressing DicF or IS118 (now known to encode a short ORF; K. Moon, unpublished), downregulated a Pbad–lacZ control fusion (Fig. S4). These plasmids were therefore omitted from further screening, leaving 24 Hfq-binding sRNAs that were screened in each experiment.
We first confirmed the previous results with an rpoS–lacZ fusion. Stimulation of expression by the three known positive regulators of rpoS translation was clearly seen (Fig. 4A). OxyS and five other sRNAs decreased expression of the PBAD–rpoS–lacZ fusion by at least twofold. Our results parallel those of Mandin and Gottesman; three sRNAs, MgrR, RydC, RyeB were just below the twofold cut-off in their work but just above it in this experiment (Mandin and Gottesman, 2010). The chromosome in these cells is wild type for all three of the positively acting sRNAs; previous work suggests that chromosomally encoded ArcZ and DsrA contribute the most to expression of the fusion under our growth conditions (Mandin and Gottesman, 2010). Thus, any competition seen presumably reflects competition with ArcZ and DsrA.
Fig. 4.

Competition for RpoS regulation by other sRNAs. Screening of the sRNA library with the PBAD–rpoS–lacZ translation fusion in (A) WT (PM1409), (B) ΔdksA (KM341). The cells were grown in LB containing ampicillin, 0.02% arabinose and 100 μM IPTG in microtitre plates at 37°C for 6 h. The effect of the overexpression of each sRNA on the rpoS–lacZ fusion was plotted as a function of the fold change compared with the basal activity of each strain containing a pBR-plac control vector (A: 40 units; B: 38 units). Fold changes greater than two were considered significant. Dark grey bars represent sRNAs for which effects were not considered significant; black and light grey bars indicate sRNAs having an activating or a repressing effect respectively.
If sRNAs compete for Hfq, we would expect anything else that limits Hfq levels to exacerbate competition. As shown earlier, a dksA mutant reduced the level of Hfq by 50%. We introduced the sRNA library into a dksA deletion mutant derivative of the PBAD–rpoS–lacZ fusion strain and compared the profile for sRNA competition to that for the wild-type strain. The basal level of rpoS was unchanged (40 units of β-galactosidase activity in WT versus 38 units in the ΔdksA strain), and ArcZ, DsrA and RprA were again detected as positive regulators (Fig. 4B). Negative regulation by all the sRNAs observed in the wild-type cells was seen, but the extent of negative regulation varied. Four additional sRNAs, FnrS, GcvB, RyhB and GlmZ, downregulated the fusion by twofold or more (Fig. 4B). These results are consistent with these sRNAs successfully competing when Hfq levels are lower, due to the dksA mutation, while they do not compete as well when Hfq is more abundant (wild-type cells). This may in part reflect the lower levels of DsrAandArcZ in the dksA mutant (Fig. 1). It is also possible that deletion of dksA affects the abundance of the competing sRNAs by affecting synthesis of either the sRNAs or their targets. Note that not all sRNAs competed; Spot42 and RybB had very little effect on rpoS translation in either the wild-type or dksA mutant cells. When hfq was deleted, the basal level of rpoS was twofold lower, and positive regulation by RprA and ArcZ was lost (Fig. S5A); multicopy DsrA was still able to act, as previously observed (Mandin and Gottesman, 2010). The negative regulation by other sRNAs was also reduced in the hfq mutant (Fig. S5A), consistent with the negative regulation by these sRNAs (Fig. 4) reflecting competition for Hfq.
To confirm these results, two of the sRNAs were examined further, testing effects on RpoS itself rather than the fusion, and directly measuring DsrA and ArcZ. ChiX was an effective competitor for rpoS, while Spot 42 did not compete in the microtitre assays (Fig. 4). These two sRNAs were each induced for 180 min from a plasmid under control of a Pbad promoter, to be more parallel to the tests with OxyS, and samples were analysed for DsrA, the short (processed) form of ArcZ (ArcZ56), and RpoS protein (Fig. 5; Fig. S6). Consistent with the results shown in Fig. 4, induction of ChiX significantly reduced the amount of RpoS (Fig. 5A), while Spot 42 did not (Fig. 5B). The levels of both DsrA and ArcZ were also affected by ChiX induction, with a somewhat greater effect on ArcZ (Fig. 5A). On the other hand, the induction of Spot42 had a much less dramatic effect on these two sRNAs. We note that less Spot42 accumulates (Fig. 5B; 1500 molecules of Spot 42 compared with 3000 molecules of ChiX at 120 min), but even at 30 min, when the level of ChiX is significantly less than the final level of Spot 42, there is a dramatic decrease in RpoS levels by ChiX. These observations suggest that different sRNAs have different abilities to compete for Hfq and consequently to affect Hfq titration, and imply tight binding of ChiX to Hfq.
Fig. 5.

ChiX but not Spot 42 interferes with RpoS and sRNA accumulation. Cells carrying vector (pNM12) or plasmids expressing ChiX (pGFK1035) (A) or Spot 42 (pGFK1034) (B) were grown in LB ampicillin at 30°C and induced with 0.2% arabinose at OD600 0.3. Both RNA and protein samples were taken at each time point as indicated for Northern blot for ChiX, DsrA, ArcZ and Western blot analysis of RpoS. The OD600 at 30 min and 60 min induction were approximately 0.5 and 0.8 respectively. SsrA levels were used as an internal control for the Northern blot. An example of a gel from this experiment is shown in Fig. S6. A standard curve was generated for ChiX and Spot42 by using in vitro transcribed RNA as described in Experimental procedures (see example for OxyS in Fig. S2). Values were determined by comparison to those curves. Averages from three experiments are expressed as the ratio of samples after sRNA induction to the same time points in the absence of sRNA induction (in cells carrying vector plasmid). Levels of the short (processed) form of ArcZ were used to generate the graph. The level of Hfq was consistent throughout the experiment with or without ChiX or Spot42 induction.
A. Strain KM255 (ΔchiX) containing either a vector control (pNM12) or a plasmid expressing ChiX (pGFK1035).
B. Strain KM349 (Δspf) containing either a vector control (pNM12) or a plasmid expressing Spot42 (pGFK1034).
Negatively regulated genes are also targets for Hfq competition
The studies discussed above confirmed that competition for Hfq can have significant effects on the positive regulation of rpoS. While positive regulation provides a particularly sensitive target for studying competition, if the model of Hfq titration is correct, we would expect a similar competition for sRNAs that negatively regulate targets. We would again expect competition to mimic the effect of an hfq mutant, in this case increasing expression.
To test this, four known negative targets of sRNAs were selected for study. In each case, the target gene is regulated by a relatively abundant sRNA that is expressed under normal growth conditions, allowing us to see regulation without special growth conditions. For each target, a translational fusion, under the control of the Pbad promoter, was constructed and tested with the library of sRNAs.
The first target tested was eptB, negatively regulated by MgrR, an sRNA that was an effective competitor for rpoS regulation (Moon and Gottesman, 2009 and Fig. 4). The sRNA library screening of the PBAD–eptB–lacZ fusion confirmed negative regulation by MgrR and demonstrated that eptB was also negatively regulated by ArcZ, even in the absence of MgrR (Fig. 6A and B). ArcZ acts directly on numerous targets, including positively regulating RpoS (Papenfort et al., 2009; Mandin and Gottesman, 2010); it can directly pair with eptB (Fig. S7 and K. Moon, unpubl. obs.). sRNAs ChiX and OmrA moderately upregulated eptB expression in the presence or absence of MgrR (Fig. 6). Two more sRNAs, MicC and RydC, acted as positive regulators only in a strain deleted for MgrR, suggesting that these sRNAs win the competition with ArcZ, but not with MgrR (Fig. 6B). In a strain deleted for hfq, both positive and negative regulation was lost (Fig. S5B), as expected.
Fig. 6.

sRNA competition for negative regulation of eptB. Cells harbouring a PBAD–eptB–lacZ translational fusion, which is negatively regulated by MgrR, were used for examining Hfq competition on a negatively regulated target. The cells were grown in LB containing ampicillin, 0.02% arabinose and 100 μM IPTG. The results were plotted as described in Fig. 4. The basal activity was that seen with the plac vector control plasmid for a given strain. Strains used were (A) WT (KM125); basal activity was 3.3 units; (B) ΔmgrR mutant (KM225); basal activity was 2.7 units.
dppA is a known target of GcvB. In a PBAD–dppA–lacZ strain, the negative regulation by GcvB was easily seen (Fig. 7A). When we screened the sRNA library in a strain carrying a PBAD–dppA–lacZstrain, only ChiX upregulated the expression of the dppA mRNA. This upregulation was reduced but was still retained if GcvB was deleted, suggesting that other sRNAs or Hfq itself may also downregulate dppA and be subject to competition (Fig. 7B). Candidates for other sRNA regulators are Spot42 and RyeB, which show negative regulation of dppA in the absence, but not in the presence of GcvB (Fig. 7B). In the strain deleted for GcvB, the expression of the fusion is higher, but two additional sRNAs, RyhB and MicC, can act as positive regulators for dppA (Fig. 7B); again, these may effectively compete with the weak regulation by Spot42 and RyeB but are not able to out-compete GcvB. In the absence of Hfq, the basal level of the fusion is significantly higher (154 units versus 74 units in the wild-type control), and all positive regulation is lost, consistent with positive regulation being due to Hfq titration (Fig. S5C). However, a number of sRNAs were still able to negatively regulate dppA, including GcvB. The other sRNAs may act indirectly (regulating GcvB) or act at some other level of dppA regulation.
Fig. 7.

dppA, a negative target of GcvB, is subject to sRNA competition. Isogenic strains carrying a PBAD–dppA–lacZ fusion were used to observe sRNA competition, as for Fig. 4. Cells were grown and assayed as for Fig. 6, but 0.0005% arabinose was used to induce the fusion; for each strain, the basal activity was that seen with the plac control plasmid.
A. WT (KM 86); basal activity was 74 units.
B. ΔgcvB (KM339); basal activity was 116 units.
The third target examined, ompX, is negatively regulated by CyaR and MicA (Johansen et al., 2008; Papenfort et al., 2008; De Lay and Gottesman, 2009; Gogol et al., 2011) (Fig. 8). In a wild-type strain, no sRNAs showed positive regulation of this fusion above the twofold cut-off (Fig. 8A). Deleting either micA or cyaR from the chromosome did not uncover a better competitor (Fig. 8B and C). When either CyaR or MicA was deleted, negative regulation by Spot 42 was seen, although it is not known if this is direct (Fig. 8B and C).
Fig. 8.

ompX, a negative target of CyaR, was not subject to sRNA competition. Isogenic strains carrying a translational fusion from the second transcriptional start site of ompX fused to the araBAD promoter were examined for competition by overexpressed sRNAs. The ompX–lacZfusion was induced with 0.0005% arabinose in LB and specific activity was obtained as for Fig. 4. (A) Wild type (KM331); basal activity was 149 units; (B) ΔmicA (KM342); basal activity was 165 units; (C) ΔcyaR (KM340); basal activity was 130 units.
Another CyaR target, yqaE, was also tested. As for ompX, no strong positive regulators were detected, and, in the absence of CyaR, Spot 42 was able to negatively regulate this gene (Fig. S8). The finding that two different CyaR targets do not show competition may suggest that CyaR is not easily competed, or that these two targets have particularly strong pairing (see Discussion).
ChiX acts as a competitor at chromosomally encoded levels
In the experiments above, ChiX was consistently observed as a strong Hfq competitor (Figs 4–7), and levels of ChiX are significant in cells under our growth conditions (Fig. S1). In fact, chromosomally encoded ChiX has been shown to strongly negatively regulate the gene for an outer membrane porin, ybfM (chiM) (Figueroa-Bossi et al., 2009; Overgaard et al., 2009; Rasmussen et al., 2009). A reporter for ybfM was tested; although the basal level of β-galactosidase in a chiX+ strain was too low to measure, overexpression of three sRNAs, MicC, OmrA and RydC, reproducibly increased expression (Table S1), consistent with some competition for Hfq and therefore relief of ChiX repression. When the chromosomal copy of ChiX was deleted, the basal level of ybfM was significantly higher, and negative regulation by two sRNAs, ChiX, sRNAs, ChiX, as expected, and RprA, not previously described, was seen (Table S1). However, no positive regulators were detected. Because RprA is not well expressed under these growth conditions, and chiX is deleted, the lack of positive regulators is consistent with the likely absence of any direct pairing sRNAs for ybfM under these growth conditions, and therefore the absence of an sRNA to compete with.
It seemed possible that the chromosomally encoded levels of ChiX would be sufficient for it to compete for Hfq in a physiologically significant fashion. This was examined in a number of ways. The effect of the sRNA library on dppA was tested in the absence of ChiX. In this strain, multiple sRNAs were now able to positively regulate (compete), including those seen before (ChiX, MicC and RyhB), as well as FnrS, CyaR, RydC, RybB and DsrA (Fig. S9; compare with Fig. 7A). One interpretation of this is that chromosomally encoded ChiX competes for Hfq as effectively as some of the overproduced sRNAs. In the absence of both GcvB and ChiX, the pattern of regulation was similar to that seen in the absence of GcvB (compare Fig. S9B with Fig. 7B).
The ability of ChiX to compete was also evaluated by a somewhat more sensitive assay, quantitative RT-PCR, for both PBAD–eptB–lacZ and Pbad–dppA–lacZ fusions, in strains carrying deletions of gcvB, mgrR or chiX and appropriate combinations of deletions. RNA samples were isolated and analysed from cells growing under the same conditions as for the sRNA library screening. Cells harbouring a PBAD–lacZ–lacZ fusion were a negative control and did not show any changes in the different backgrounds, as expected (Fig. 9A). eptB is directly regulated by MgrR, but not by GcvB. As expected, the level of eptB–lacZ did not change in the absence of GcvB, while it increased dramatically in the absence of MgrR (Fig. 9B). The level of dppA–lacZ increased significantly in the absence of GcvB, again as expected. However, an increase was also seen in the mgrR deletion strain (Fig. 9C). Multicopy MgrR had no effect on the dppA–lacZ fusion (Fig. 7), making it unlikely that MgrR is a direct regulator of dppA–lacZ; this effect might be indirect (for instance, changing the levels of other sRNA regulators). Deletion of ChiX decreased the levels of eptB– and dppA–lacZ fusions by 50%. For both eptB and dppA, a double mutant of chiX and the regulating sRNA showed expression similar to that for the deletion of the direct regulator (ΔmgrR ΔchiX for Fig. 9B, ΔgcvB ΔchiX for Fig. 9C). This result strongly suggests that ChiX, under normal growth conditions, titrates Hfq, limiting the ability of MgrR and GcvB to negatively regulate their targets and thus modulating Hfq-dependent regulation of many genes.
Fig. 9.
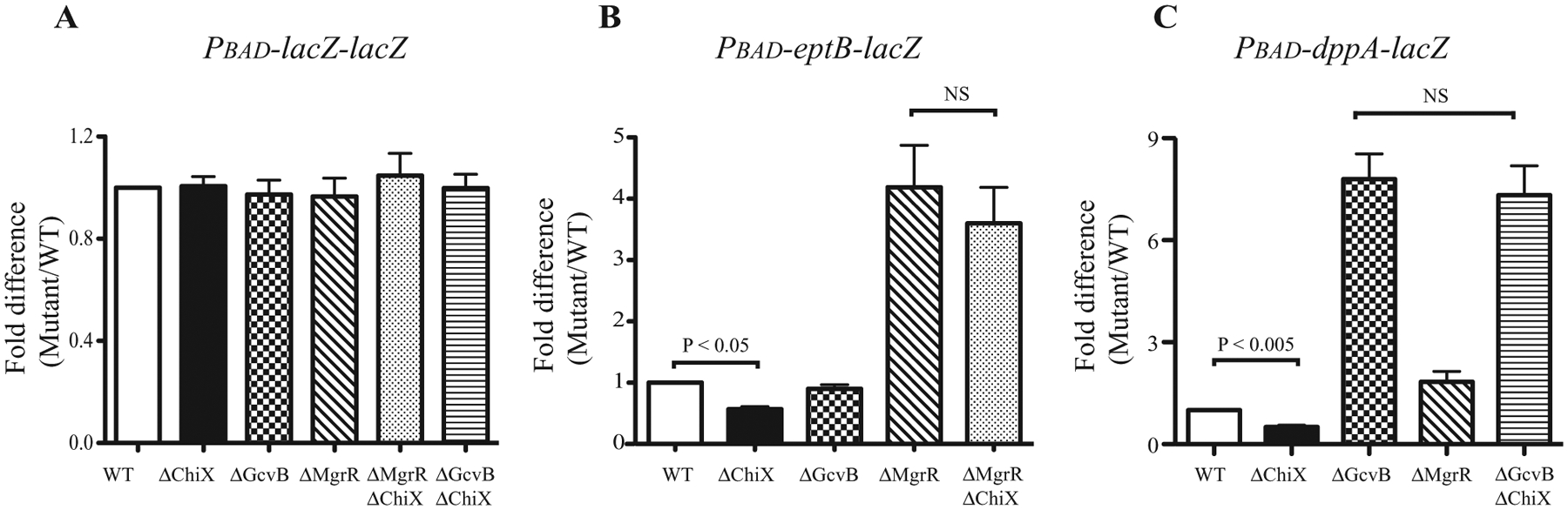
Chromosomally encoded ChiX affects the level of eptB and dppA. Isogenic derivatives of cells harbouring different lac fusions were grown in LB with arabinose (0.02% for both lacZ–lacZ and eptB–lacZ and 0.0005% for dppA–lacZ) at 37°C for 6 h, RNA isolated and cDNAs prepared for qRT-PCR as described in Experimental procedures. The normalized mRNA level of the WT strain for each set was designated as 1.0, and mutants compared with this value. For each set, a ΔgcvB derivative, an ΔmgrR derivative and a ΔchiX derivative were used.
A. PBAD–lacZ–lacZ derivatives grown in 0.02% arabinose: WT: (PM1410); ΔchiX (KM359); ΔgcvB (KM360), ΔmgrR (KM361), ΔmgrR ΔchiX (KM390), ΔgcvB ΔchiX (KM391).
B. PBAD–eptB–lacZ derivatives grown in 0.02% arabinose; WT: KM125; ΔchiX (KM363); ΔgcvB (KM364), ΔmgrR (KM225), ΔmgrR ΔchiX (KM381).
C. PBAD–dppA–lacZ derivatives grown in 0.0005% arabinose; WT: KM86; ΔchiX (KM352); ΔgcvB (KM339), ΔmgrR (KM366), ΔgcvB ΔchiX (KM353).
Error bar shows standard deviation among four different trials. NS indicates no significant difference observed among samples; all the samples in (A) showed no significant differences.
Discussion
Trans-encoded Hfq-dependent sRNAs regulate various genes via base pairing. Our results and the results of others (Hussein and Lim, 2011) suggest that Hfq is a limiting factor for sRNA regulation. We have shown that the limitation of the available Hfq pool for sRNAs causes competition among them, affecting the stability and activity of the sRNAs and, consequently, changing the outcome of gene regulation. Under conditions of overexpression of one sRNA, accumulation of other sRNAs decreases, binding of these other sRNAs to Hfq is reduced, and the regulation of target genes is perturbed. Even without overexpression, we find that an abundant sRNA affects the regulation of targets, apparently by limiting Hfq availability.
The idea of competition for Hfq is not new. Zhang and co-workers suggested in 1998 that OxyS negatively regulates RpoS by competing for Hfq (Zhang et al., 1998). Our experiments confirm this, demonstrating that both the accumulation of sRNAs important for RpoS translation (DsrA and ArcZ) and binding of these sRNAs to Hfq is reduced when OxyS is overexpressed, and the competition is overcome by increasing levels of Hfq (Figs 2 and 3). Thus in this physiologically relevant example, OxyS is able to downregulate RpoS by interfering with positive regulation. Overexpression of ArcZ has been observed to lead to widespread changes in mRNA profiles, consistent with titration of Hfq (Papenfort et al, 2009).
Hfq is a limiting factor for sRNA accumulation
Hfq plays multiple roles in stimulating sRNA-dependent function (Brennan and Link, 2007). One role that is relatively easy to monitor is that binding to Hfq stabilizes sRNAs, protecting them from RNase E. If Hfq becomes limiting, we expected this to be reflected in lower levels of sRNA accumulation. In an initial test, the effect of deleting Hfq on sRNA accumulation was found to differ significantly for different sRNAs. Those not significantly affected by deletion of hfq may be stable even in the absence of Hfq, or may be unstable even when bound to Hfq. ChiX is very stable in wild-type strains (Vogel et al., 2003), and its accumulation is drastically reduced in an hfq mutant (Fig. S1, Table 1). GcvB is unstable even in the presence of Hfq (Vogel et al., 2003) and deletion of hfq had minimal effects on its accumulation (Fig. S1, Table 1), consistent with GcvB either binding Hfq poorly, transiently, or in a manner that does not stabilize the sRNA.
This experiment, carried out at three temperatures and in both exponential and stationary phase, leads to additional conclusions. ArcZ has previously been reported to be processed; the appearance of the processed form is Hfq-dependent (Sittka et al., 2009; Mandin and Gottesman, 2010). At low temperature (32°C), the full-length RNA was more abundant in the absence of Hfq than in the hfq+ strain (Fig. S1; Table 1). This is consistent with Hfq-dependent processing, although it does not rule out instability of the processed form in the absence of Hfq as well. Possibly at 32°C the pathway for degradation of the full-length RNA is slow enough to see accumulation in the hfq mutant. The dsrA promoter is known to be less active at high temperature (42°C) (Repoila and Gottesman, 2001; 2003); this temperature effect was less striking in stationary phase (Fig. S1). CyaR is positively regulated by cyclic AMP and CRP, while Spot 42 is negatively regulated (Møller et al., 2002; Johansen et al., 2008; Papenfort et al., 2008; De Lay and Gottesman, 2009). In our experiments, the accumulation of CyaR and Spot 42 was reciprocal, with CyaR most abundant at high temperature or in stationary phase and Spot 42 low under those conditions. MgrR was highly dependent upon Hfq for accumulation in exponential phase, but not in stationary-phase cells, particularly at 37°C. Possibly the degradation machinery is limiting under these conditions.
Different sRNAs compete to different extents
We have used a library of plasmids, each expressing an sRNA, to examine the hierarchy of sRNAs for competition, for both positive and negative regulation. The results support rather different behaviours for different sRNAs and/or sRNA/mRNA pairs. Some of these differences may reflect differences in accumulation and/or stability of the sRNAs, but others seem likely to reflect differences in Hfq binding affinity and possibly in mode of Hfq binding (see, for instance, Olejniczak, 2011).
ChiX represents an example of an sRNA that competes particularly well. ChiX overexpression disrupted positive regulation of RpoS (Figs 4 and 5), and negative regulation of eptB and dppA (Figs 6 and 7). Overproduction of OxyS had much less effect on binding of ChiX to Hfq, compared with other sRNAs (Fig. 3). Finally, deletion of chiX was sufficient to improve GcvB-dependent regulation of dppA and MgrR-dependent regulation of eptB (Fig. 9), strongly suggesting that ChiX helps to modulate sRNA regulation under normal growth conditions. It seems likely this reflects strong binding of ChiX to Hfq. This is consistent with evidence from others suggesting that ChiX may be unusual in its interaction with Hfq (Overgaard et al., 2009). ChiX is reported to act catalytically, stimulating the degradation of multiple mRNA target molecules/sRNA (Overgaard et al., 2009). If pairing usually leads to dissociation of Hfq and the destruction of both sRNA and mRNA (Massé et al., 2003; De Lay and Gottesman, 2011; Hussein and Lim, 2011), possibly ChiX binding by Hfq is qualitatively or quantitatively different. It may not be displaced from Hfq after pairing (or by competition). Spot 42, on the other hand, represents an sRNA that seems to be unable to compete effectively (Figs 4, 5 and 7). Levels of Spot 42 did not decrease significantly when OxyS was overproduced (Fig. 2), and decreased only modestly in a dksA mutant (Fig. 1). Spot 42 clearly binds to Hfq, and immunoprecipitation with Hfq was significantly reduced when OxyS was overproduced (Fig. 3). The simplest conclusion is that Spot 42, even when overproduced, does not bind tightly enough to compete with DsrA, ArcZ and MgrR, for instance. We also note that Spot 42 has many mRNA targets (Beisel and Storz, 2011). If pairing with a mRNA helps to remove an sRNA from Hfq (Massé et al., 2003; Hussein and Lim, 2011 and see below), it is possible that Spot 42 binding to Hfq is particularly transient. In fact, for two of the substrates we tested, Spot 42 proved to have some ability to negatively regulate them, possibly directly (Figs 7 and 8), although it is striking that this regulation could only be detected when the primary regulators were deleted from the chromosome, again suggesting that Spot 42 competes poorly with other sRNAs.
Other sRNAs fell between these two extremes. MicC, RydC and OmrA were able to compete modestly with ChiX (Table S1), and also competed for regulation of eptB, but only in the absence of MgrR, when ArcZ may be regulating it (Fig. 6B). Similarly, MicC and RyhB competed for regulation of dppA, but again only when the primary regulator, GcvB is deleted; direct regulation in this case may be by Spot 42 or RyeB (Fig. 7). Others (RprA, DsrA, MicA, MicF, among others) were never able to compete sufficiently to cause a twofold change in expression.
Hfq competition affected the downregulation of both eptB and dppA but was not seen for ompX regulation (Fig. 8). ompX is negatively regulated by CyaR and MicA; it is the chromosomally encoded levels of these sRNAs that would be subject to competition. Whether the failure to see competition reflects something about ompX mRNA itself, about the sRNAs pairing with it and how they bind Hfq as compared with ChiX, for instance, or whether the interaction of CyaR and MicA with this target is particularly effective is not yet known (see Fig. S7 for predicted pairing). We speculate that each sRNA’s abundance and Hfq binding affinity plays a crucial role in determining the hierarchy of sRNAs in the system, although competitive affinity studies will be needed to establish some of the relevant values.
Hfq is an abundant protein. Nonetheless, our work and work reported by others suggest that it is normally limiting for many of its functions. Thus, increasing Hfq levels modestly increased accumulation of sRNAs, significantly increased RpoS levels, and decreased competition (Fig. 2). Reducing Hfq levels by twofold, via a deletion of dksA, increased the ability of sRNAs to compete, suggesting more stringent competition for Hfq (Fig. 4B). Hussein and Lim (2011) found that increasing Hfq levels increased both positive regulation (RpoS synthesis) and negative regulation (silencing of target genes), consistent with our findings.
What is the mechanism of competition for Hfq?
For sRNAs to compete for Hfq, Hfq must be limiting, and binding to one sRNA must be transient enough to allow another sRNA to displace it before it is able to act. In a recent study, Fender et al. propose a model for Hfq binding and exchange of sRNAs in which an incoming sRNA competes with an sRNA bound to Hfq in a concentration-dependent fashion (Fender et al., 2010). In vitro experiments suggested a slow off-rate for sRNAs bound to Hfq in the absence of such a second competing RNA. However, their data suggest that in vivo one would expect sRNAs to move on and off Hfq rapidly. The competition we see is consistent with this. Hfq binding to sRNAs were dramatically reduced when another sRNA was induced (Fig. 3).
In another study, Hussein and Lim also reported that Hfq is a limiting factor for sRNA-dependent regulation (Hussein and Lim, 2011). In their experiments, the ability of an overproduced sRNA to regulate a direct target mRNA, also overproduced, was improved by increasing the level of Hfq, and overproducing either an sRNA or a mRNA interfered with regulation of other sRNA : mRNA pairs. They observed that this competition was lessened if the sRNA and its partner mRNA were both overproduced, suggesting that after pairing, Hfq sequestration is lost; we did not investigate this in our system. They suggest that the availability of the target mRNA for a bound sRNA eases competition, by allowing pairing and subsequent release from Hfq and degradation of the two RNAs. In this model, an excess of sRNA over target, probably the case in our overproduction experiments, would be expected to exacerbate competition. This would also assume that each sRNA is used for pairing once, a condition that may not hold for tight binding sRNAs such as ChiX.
Our results agree with and extend these observations, establishing evidence for differential competition by different sRNAs and evidence for a role for competition under physiologically relevant conditions. Some sRNAs, such as ChiX, may compete so well that they are never induced to very high levels, but instead are regulated in a unique fashion, by regulated destruction (Figueroa-Bossi et al., 2009; Overgaard et al., 2009). Others may compete only when induced. Others are apparently unable to compete effectively, although they still bind Hfq sufficiently well to regulate their targets.
This discussion assumes similar requirements for binding of the sRNAs we have investigated. While most sRNAs are believed to bind to the distal face of Hfq, others may use the proximal face (reviewed in Vogel and Luisi, 2011), and competition presumably would not occur between sRNAs binding at different sites (Olejniczak, 2011). Similarly, mRNAs may also compete for binding, certainly with other mRNAs and possibly with sRNAs. We have not investigated these issues here.
Overall, our study provides evidence that Hfq levels are such that competition for Hfq modulates the effectiveness of the sRNA. Under specific induction conditions, many, but not all, sRNAs will shut down or diminish the effects of other, uninduced sRNAs, by competing for Hfq, thus providing an additional and novel level of connectivity to the various sRNA regulons in the cell. While this work and others cited here have been carried out in E. coli, competition for Hfq will certainly occur in other bacteria. In addition, competition among RNAs in eukaryotes has recently been proposed, in which the microRNAs serve as connections and modulators (Salmena et al., 2011); the work in E. coli may serve as a model for what might be expected in eukaryotic systems.
Experimental procedures
Bacterial strains and plasmids
Strains and plasmids used in this study are listed in Table S2. All E. coli strains in this work are derived from MG1655. The recipient strain for standard cloning procedures was DH5α. Strains carrying lacZ fusions were constructed in PM1205 (Mandin and Gottesman, 2009), which contains the PBAD–catsacB segment upstream of lacZ at the chromosomal lacZ site. To construct a lacZ translational fusion of a gene of interest, primers were designed to give a PCR product with 40 nt homologous to the PBAD promoter followed by the specific gene from +1 to codon 9 followed by lac homology (40 nt). Primers are listed in Table S3. The PCR products were introduced into the PM1205 chromosome using the red recombination system (Yu et al., 2000; Mandin and Gottesman, 2009). For the sRNA library, the plasmids were those described by Mandin and Gottesman (2010). The ΔdksA mutants were provided by Dr M. Cashel (NICHD, NIH).
Media and growth conditions
All strains were grown in LB. Antibiotic concentrations (in micrograms per millilitre) were as follows: ampicillin on plates, 50; ampicillin in liquid cultures, 100; kanamycin, 25; chloramphenicol, 25, tetracycline, 25. Each plasmid was freshly transformed into the strain of interest, and a colony was cultured in LB with the appropriate antibiotic overnight at 30°C. Overnight cultures were subcultured into fresh medium at OD600 0.05 (about 1:500 dilution) and grown to OD600 ≈ 0.3. Cells were induced with arabinose for the designated time. For co-immnunoprecipitation experiments, 2 OD600 of cells were collected before and after arabinose induction. Cells were washed with cold DEPC-PBS and then frozen at −80°C for the next steps. Unless otherwise indicated, cells were grown at 30°C to increase the expression of DsrA.
RNA isolation and Northern blot analysis
To examine the effect of OxyS expressed from an arabinose-inducible promoter, cells were treated with 0.2% arabinose at OD600 = 0.3. Cells were collected and total RNA extracted at different time points by the hot-phenol method as previously described (Massé et al., 2003) and detailed in Moon and Gottesman (2009). Detection was performed with the corresponding biotinylated probes in Table S3 (‘Biotinylated probes used for Northern blots’). Bands were quantified by imaging using a LAS-4000 mini camera (FujiFilm Medical Systems USA) before the blots were saturated, and analysed with MultiGauge software (Fuji); all samples were normalized to SsrA loading controls.
For quantification of sRNA molecules per cell, we used RNAs transcribed in vitro to establish a standard curve (see Fig. S2). In vitro transcribed RNA was eluted from the gel, precipitated with ethanol, resuspended in buffer and quantified using a Nanodrop (Thermo-Fisher) in triplicate; the average value was used for the standard curve.
In vitro transcription
To produce specific sRNAs in vitro, we used a template harbouring the T7 promoter followed by the sRNA sequence. For OxyS in vitro transcription, the plasmid template pSP64-OxyS was linearized with a restriction enzyme and purified using a QIAGEN PCR purification kit. The primer templates used for DsrA, ChiX and Spot42 are listed in Table S3. sRNAs were synthesized using MEGAscript T7 (ambion) Kit followed by TBE 5%-urea gel purification to remove remaining NTPs and other non-specific fragments.
Protein sample preparation and Western blot analysis
Protein samples were collected in parallel with RNA samples. One millilitre of cells were collected at each time point and processed as described in Ranquet and Gottesman (2007), with details described in Supporting information.
Co-immunoprecipitation
Cell samples were resuspended with 400 μl of lysis buffer and were sonicated for 5 min at medium intensity followed by centrifugation at 1400 r.p.m. for 10 min at 4°C. The supernatant of each sample was processed based on the method described by Sun et al. (2006) and adapted as described in Supporting information.
sRNA library screening
The library was introduced into a specific background strain by TSS transformation and spotted on an LB plate containing ampicillin as described previously (Mandin and Gottesman, 2010); details are given in Supporting information.
β-Galactosidase assay
The β-galactosidase activity of strains carrying lacZ fusions was assayed on a SpectraMax 250 (Molecular Devices) microtitre plate reader as described previously (Majdalani et al., 1998). Specific activities are represented as the Vmax divided by the OD600.
Real-time RT-PCR analysis
To determine more quantitatively the expression of each lacZ fusion gene in various background strains, real-time RT-PCR was performed on RNA obtained from wild-type and mutant strains carrying the control PBAD–lacZ–lacZ fusion or either PBAD–eptB–lacZ or PBAD–dppA–lacZ fusions grown in LB at 37°C as described above (see sRNA library screening). Real-time PCR was performed using an iQcycler (Bio-Rad) as described in Moon and Gottesman (2009). Expression of the SsrA gene was used as an internal standard, and SYBR Green Supermix was used as a signal reporter. Each sample was tested in quadruplicate and analysed as described in Moon and Gottesman (2009).
Supplementary Material
Acknowledgements
We thank members of the laboratory and G. Storz and Kumaran Ramamurthi for comments on the manuscript and discussions throughout this work. We thank R. Gourse for pointing out the effect of dksA on Hfq levels, M. Cashel for dksA mutants and A. Zhang for OxyS plasmids. J.H. Kim (LMP, NCI) is thanked for use of the iQcycler. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article.
References
- Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, and Storz G (1997) A small stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90: 43–53. [DOI] [PubMed] [Google Scholar]
- Backofen R, and Hess WR (2010) Computational predictions of sRNAs and their targets in bacteria. RNA Biol 7: 33–42. [DOI] [PubMed] [Google Scholar]
- Basineni SR, Madhugiri R, Kolmsee T, Hengge R, and Klug G (2009) The influence of Hfq and ribonucleases on the stability of the small non-coding RNA OxyS and its target rpoS in E. coli is growth phase dependent. RNA Biol 6: 584–594. [DOI] [PubMed] [Google Scholar]
- Beisel CL, and Storz G (2011) The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol Cell 41: 286–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan RG, and Link TM (2007) Hfq structure, function and ligand binding. Curr Opin Microbiol 10: 125–133. [DOI] [PubMed] [Google Scholar]
- De Lay N, and Gottesman S (2009) The Crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J Bacteriol 191: 461–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, and Gottesman S (2011) Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA 17: 1172–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fender A, Elf J, Hampel K, Zimmermann B, and Wagner EG (2010) RNAs actively cycle on the Sm-like protein Hfq. Genes Dev 24: 2621–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa-Bossi N, Valentini M, Malleret L, and Bossi L (2009) Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev 23: 1981–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogol EB, Rhodius VA, Papenfort K, Vogel J, and Gross CA (2011) Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of the global stress regulon. Proc Natl Acad Sci USA 108: 12875–12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S (2004) The small RNA regulators of Escherichia coli: roles and mechanisms. Annu Rev Microbiol 58: 273–301. [DOI] [PubMed] [Google Scholar]
- Hussein R, and Lim HL (2011) Disruption of small RNA signaling caused by competition for Hfq. Proc Natl Acad Sci USA 108: 1110–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen J, Eriksen M, Kallipolitis B, and Valentin-Hansen P (2008) Down-regulation of outer membrane proteins by noncoding RNAs: unraveling the cAMP-CRP- and σE-dependent CyaR-ompX regulatory case. J Mol Biol 383: 1–9. [DOI] [PubMed] [Google Scholar]
- Kajitani M, Kato A, Wada A, Inokuchi Y, and Ishihama A (1994) Regulation of the Escherichia coli hfq gene encoding the host factorfor phage Q beta. J Bacteriol 176: 531–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, and Gottesman S (1998) DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci USA 95: 12462–12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Chen S, Murrow J, St. John K, and Gottesman S (2001) Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol Microbiol 39: 1382–1394. [DOI] [PubMed] [Google Scholar]
- Mandin P, and Gottesman S (2009) A genetic approach for finding small RNA regulators of genes of interest identifies RybC as regulating the DpiA/DpiB two-component system. Mol Microbiol 72: 551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandin P, and Gottesman S (2010) Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J 29: 3094–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Escorcia FE, and Gottesman S (2003) Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17: 2374–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller T, Franch T, Udesen C, Gerdes K, and Valentin-Hansen P (2002) Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev 16: 1696–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K, and Gottesman S (2009) A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol Microbiol 74: 1314–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olejniczak M (2011) Despite similar binding to the Hfq protein regulatory RNAs widely differ in their competition performance. Biochemistry 50: 4427–4440. [DOI] [PubMed] [Google Scholar]
- Overgaard M, Johansen J, Moller-Jensen J, and Valentin-Hansen P (2009) Switching off small RNA regulation with trap-mRNA. Mol Microbiol 73: 790–800. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Pfeiffer V, Lucchini S, Sonawane A, Hinton JC, and Vogel J (2008) Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol Microbiol 68: 890–906. [DOI] [PubMed] [Google Scholar]
- Papenfort K, Said N, Welsink T, Lucchini S, Hinton JC, and Vogel J (2009) Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol 74: 139–158. [DOI] [PubMed] [Google Scholar]
- Ranquet C, and Gottesman S (2007) Translational regulation of the Escherichia coli stress factor RpoS: a role for SsrA and Lon. J Bacteriol 189: 4872–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen AA, Johansen J, Nielsen JS, Overgaard M, Kallipolitis B, and Valentin-Hansen P (2009) A conserved small RNA promotes silencing of the outer membrane protein YbfM. Mol Microbiol 72: 566–577. [DOI] [PubMed] [Google Scholar]
- Repoila F, and Gottesman S (2001) Signal transduction cascade for regulation of RpoS: temperature regulation of DsrA. J Bacteriol 183: 4012–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repoila F, and Gottesman S (2003) Temperature sensing by the dsrA promoter. J Bacteriol 185: 6609–6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, and Pandolfi PP (2011) A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell 146: 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma AK, and Payne SM (2006) Induction of expression of hfq by DksA is essential for Shigella flexneri virulence. Mol Microbiol. 62: 469–479. [DOI] [PubMed] [Google Scholar]
- Sharma CM, and Vogel J (2009) Experimental approaches for the discovery and characterization of regulatory small RNAs. Curr Opin Microbiol 12: 536–546. [DOI] [PubMed] [Google Scholar]
- Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, et al. (2008) Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet 4: e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittka A, Sharma CM, Rolle K, and Vogel J (2009) Deep sequencing of Salmonella RNA associated with heterologous Hfq proteins in vivo reveals small RNAs as a major target class and identifies RNA processing phenotypes. RNA Biol 6: 266–275. [DOI] [PubMed] [Google Scholar]
- Sledjeski DD, Whitman C, and Zhang A (2001) Hfq is necessary for regulation by the untranslated RNA DsrA. J Bacteriol 183: 1997–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun BK, Deaton AM, and Lee JT (2006) A transient heterochromatic state in Xist preempts × inactivation choice without RNA stabilization. Mol Cell 21: 617–628. [DOI] [PubMed] [Google Scholar]
- Sun X, Zhulin I, and Wartell RM (2002) Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res 30: 3663–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, and Luisi BF (2011) Hfq and its constellation of RNA. Nat Rev Microbiol 9: 578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Bartels V, Tang HH, Churakov G, Slagter-Jager JG, Huttenhofer A, and Wagner EGH (2003) RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res 31: 6435–6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, and Storz G (2009) Regulatory RNAs in bacteria. Cell 136: 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DG, Ellis HM, Lee EC, Jenkins NA, Copeland NG, and Court DL (2000) An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA 97: 5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, and Storz G (1998) The oxyS regulatory RNA represses rpoS translation by binding Hfq (HF-1) protein. EMBOJ 17: 6061–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, and Gottesman S (2003) Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 50: 1111–1124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


