Abstract
Members of the Roseobacter group are known for their different ecologically relevant metabolic traits and high abundance in many marine environments. This includes traits like carbon monoxide oxidation, sulfur oxidation, nitrogen oxidation, DMSP demethylation, denitrification, and production of bioactive compounds. Nevertheless, their role in the marine biogeochemical cycles remains to be elucidated. Roseobacter ponti DSM 106830T, also designated strain MM-7T (=KCTC 52469T =NBRC 112431T), is a novel type strain of the Roseobacter group, which was proposed as new Roseobacter species. It was isolated from seawater of the Yellow Sea in South Korea. We report the complete genome sequence of R. ponti DSM 106830T, which belongs to the family Rhodobacteraceae. The genome of R. ponti DSM 106830T comprises a single circular chromosome (3,861,689 bp) with a GC content of 60.52% and an additional circular plasmid (p1) of 100,942 bp with a GC content of 61.51%. The genome encodes 3,812 putative genes, including 3 rRNA, 42 tRNA, 1 tmRNA, and 3 ncRNA. The genome information was used to perform a phylogenetic analysis, which confirmed that the strain represents a new species. Moreover, the genome sequence enabled the investigation of the metabolic capabilities and versatility of R. ponti DSM 106830T. Finally, it provided insight into the high niche adaptation potential of Roseobacter group members.
Keywords: Roseobacter ponti DSM 106830T, genome, phylogeny, metabolism, aerobic anoxygenic phototrophy
Significance
The phylogeny of the Roseobacter group is complex and has recently been reconsidered. Members of the Roseobacter group are highly likely to play a key role in the ocean due to their inhabitation of various ecological niches and their general abundance. However, their role in the ocean as part of the biogeochemical cycling is not yet fully understood. Therefore, complete genomes of Roseobacter group members are required to tackle these questions. In particular, the here presented genome of the new type strain Roseobacter ponti DSM 106830T helps to improve the phylogenetic resolution by representing a potential missing link in the Roseobacter group. Furthermore, we provide insights into the group genomic equipment which reveals high adaptational and functional properties.
Introduction
The marine ecosystem is highly dynamic and bacterial diversity in the oceans is stunning. Bacterioplankton is dominated by a few marine bacterial clades, including the gammaproteobacterial SAR86 clade and the alphaproteobacterial SAR11, SAR116, and Roseobacter clades (Rappe et al. 2000; Suzuki et al. 2001; Kirchman 2008). Recently, the latter one has been described as Roseobacter group (Freese et al. 2017; Simon et al. 2017; Sonnenschein et al. 2018). These microorganisms are flexible in their metabolic potential, such as heterotrophy, photoheterotrophy, or autotrophy, lifestyle, such as free-living, particle-associated, or eukaryote-associated (Luo and Moran 2014). Roseobacter group members are widely distributed and in some marine ecosystems they constitute 15–20% of the bacterial community (Selje et al. 2004; Suzuki, Preston, et al. 2001; Moran et al. 2007). They possess different mechanisms for energy generation, such as utilization of organic and inorganic compounds including sulfur oxidation and catabolism of dimethylsulfoniopropionate (DMSP) to dimethyl sulfide (DMS), which is a climate-relevant gas (Wagner-Döbler and Biebl 2006). Luo and Moran (2014) compared Roseobacter group members based on genes mediating biogeochemical cycling including, Roseobacter denitrificans and Roseobacter litoralis which are capable of phototrophy (Shiba 1991). Both also produce a pink pigment, bacteriochlorophyll a (BchlA) and other bioactive secondary metabolites encoded by non-ribosomal peptide synthetases (NRPS) or polyketide synthase (PKS) gene clusters (Martens et al. 2007). Although various aspects of the Roseobacter group have been studied in recent years, complete genome sequences of isolates are lacking or limited to very few members. Thus, in depth biochemical and genomic characterization to elucidate ecological significance and evolutionary processes shaping the genomes of Roseobacter group members are still incomplete.
Materials and Methods
Isolation, Growth Conditions, and Genomic DNA Extraction
Roseobacter ponti DSM 106830T obtained from the “Deutsche Sammlung für Mikroorganismen und Zellkulturen” (DSMZ; Braunschweig, Germany) was originally isolated from seawater of the Yellow Sea in South Korea (Jung et al. 2017). A single colony from an active culture plate of R. ponti DSM 106830T was passed for 2 days in Medium 514 (DSMZ Medium 514 Bacto Marine Broth Difco 2216, Braunschweig, Germany) at 30 °C and 180 rpm (Infors AG, Bottmingen, Schweiz). Cells were pelleted at 10.020× for 15 min and washed with sterile water twice. Genomic DNA was extracted by using the MasterPure complete DNA purification kit as recommended by the manufacturer (Epicentre, Madison, WI). The quality of the DNA was checked via agarose gel electrophoresis (0.8%, 50 min, 100 V) and the concentration was determined photometrically using a NanoDrop ND-1000 Spectrophotometer (Thermo Scientific, Schwerte, Germany).
Genome Sequencing, Assembly, and Annotation
Illumina paired-end sequencing libraries were prepared using the Nextera XT DNA Sample Preparation kit (Illumina, San Diego, CA). Quality and size of the libraries were analyzed using Agilent Bioanalyzer 2100 and the Agilent High Sensitivity DNA kit as recommended by the manufacturer (Agilent Technologies, Waldbronn, Germany). Concentration of the libraries was determined using the Qubit dsDNA HS Assay kit as recommended by the manufacturer (Life Technologies GmbH, Darmstadt, Germany). Sequencing was performed using a MiSeq system and reagent kit v3 with 600 cycles as recommended by the manufacturer (Illumina, San Diego, CA). For Oxford Nanopore sequencing, 1.5 µg high molecular weight DNA was used for library preparation employing the Ligation Sequencing kit 1D (SQK-LSK109) and the Native Barcode Expansion kit (EXP-NBD103, Barcode 2) as recommended by the manufacturer (Oxford Nanopore Technologies, Oxford, UK). Sequencing was performed for 72 h using a MinION device Mk1B and a SpotON Flow Cell R9.4.1 as recommended by the manufacturer (Oxford Nanopore Technologies) using MinKNOW software v18.12.6 for sequencing and Guppy v3.0.3 (https://community.nanoporetech.com, last accessed April 29, 2019) for demultiplexing. Default parameters were used for all software unless otherwise specified. Reads were quality-filtered using fastp version 0.20.0 (Chen et al. 2018) and remaining sequencing adapters were removed with cutadapt v2.5 (Martin 2011). Unicycler version 0.4.8 (Wick et al. 2017) was used for a de novo hybrid assembly in normal mode. The quality of the assembly was assessed with CheckM v1.1.2 (Parks et al. 2015) and validated with Bandage 2.1 (Wick et al. 2015). Genome annotation was performed employing the Prokaryotic Genome Annotation Pipeline (PGAP) (Tatusova et al. 2016).
Phylogenetic Analysis
The Genome Taxonomy Database Toolkit (GTDB-Tk) v1.0.1 (Chaumeil et al. 2019), was used to provide an initial taxonomic placement. Afterwards, the genome sequence was uploaded to the Type (Strain) Genome Server (TYGS) (https://tygs.dsmz.de), for a whole-genome-based taxonomic analysis (Meier-Kolthoff and Göker 2019). Analysis was performed for the 16 closest relatives (supplementary table S1, Supplementary Material online) at 16S rRNA gene level and with whole-genome sequences as described by TYGS using default parameters (as of January 28, 2020). In addition, an extended 16S rRNA gene analysis including sequences of not fully genome-sequenced type strains was performed by employing TYGS (Meier-Kolthoff and Göker 2019). The fast homology search tool AAI-profiler (Medlar et al. 2018) was also used to assess the phylogeny by using the deduced proteome of R. ponti DSM 106830T.
Metabolism and Secondary Metabolites
To investigate the metabolic potential of R. ponti DSM 106830T, BlastKOALA version 2.2 (Kanehisa et al. 2016) was used to get an overview of pathways. Putative secondary metabolites and putative phage regions were identified with AntiSMASH 5.1.0 (Blin et al. 2019) and PHASTER (Arndt et al. 2016), respectively.
Results and Discussion
Genomic Features
The genome of R. ponti DSM 106830T was sequenced using both, long-read (Oxford Nanopore) and short-read technology (Illumina). After quality-filtering, 201,474 long-reads with a mean length of 7,642 bp (Oxford Nanopore) and 2,980,230 paired-end Illumina reads (2 × 300 bp) were obtained. Unicycler version 0.4.8 (Wick et al. 2017) was used for a de novo hybrid assembly. This resulted in two circular contigs representing a chromosome and a plasmid with a total average coverage of 559-fold (chromosome) and 764-fold (plasmid). The assembly was manually validated with Bandage version 2.1 (Wick et al. 2015). CheckM detected a completeness of 99.25% and a contamination rate of 0.48%. The genome comprises one circular chromosome (3,861,689 bp) and one circular plasmid (100,942 bp) with a GC-content of 62.92% and 61.51%, respectively. Genome features are summarized in table 1.
Table 1.
Genomic Features of Roseobacter ponti DSM 106830T
| Features | Chromosome | Plasmid (p1) |
|---|---|---|
| Genome size (bp) | 3,861,689 | 100,942 |
| GC content (%) | 60.52 | 61.51 |
| Gene number | 3,715 | 97 |
| rRNA genes | 3 | 0 |
| tRNA genes | 42 | 0 |
| ncRNA | 2 | 0 |
| tm-RNA genes | 1 | 0 |
| Regulatory RNA | 8 | 0 |
| CRISPR | 0 | 0 |
| Phage | 1 | 0 |
In total, 52.1% of genes were annotated by BlastKOALA and classified into 23 functional categories according to the KEGG Orthology. Among all categories, the environmental information processing (11.8%) and carbohydrate metabolism (11.6%) were the most abundant.
Phylogeny of R. ponti DSM 106830T
GTDB-Tk (Chaumeil et al. 2019) revealed that this strain is novel and placed R. ponti DSM 106830T taxonomically into the family Rhodobacteracea, based on average nucleotide identity (N/A) and relative evolutionary divergence values (∼0.97). Phylogenetic assignment at genus level was not possible. To refine the phylogenetic position of R. ponti DSM 106830T a phylogenetic tree based on 16S rRNA gene sequences and whole-genome sequence was constructed. The 16S rRNA gene-based tree grouped R. ponti DSM 106830T together with R. litoralis OCh 149, R. denitrificans OCh 114, and R. denitrificans DSM 7001 into the genus Roseobacter (supplementary fig. S1A and S1C, Supplementary Material online). The phylogenetic tree based on Genome Blast Distance Phylogeny takes the whole-genome sequence into account and resulted in a different clustering (supplementary fig. S1B, Supplementary Material online). Briefly, R. ponti DSM 106830T was most closely assigned to Sulfitobacter marinus DSM 23422 based on GBDP calculations for the 16 closest relatives (supplementary table S1A and S1B, Supplementary Material online). The tree emphasizes the challenge to make valid phylogenetical classifications of new isolates from the Roseobacter group. The whole-genome tree supports the classification of R. ponti DSM 106830T into the Roseobacter group, but due to deep branching and low bootstrap values genus assignments are not supported (supplementary fig. S1B, Supplementary Material online). Additionally, proteome AAI-profile analysis further validates this finding with AAI values <80%. AAI-profiler assigned strain R. ponti DSM 106830T inconclusively to a Rhodobacteracea bacterium, though the closest sequence neighbors belong to both genera Roseobacter and Sulfitobacter. Consequently, new isolates including the here presented R. ponti DSM 106830T of the Roseobacter group should not be solely classified on the basis of the 16S rRNA gene as they share >89% 16S rRNA gene identity (Buchan et al. 2005), which can lead to incorrect phylogenetic assignments. Recently, several genus reassignments of the Roseobacter group are ongoing (Wirth and Whitman 2018). Based on the here presented data, we suggest R. ponti DSM 106830T as a missing link between the Sulfitobacter and Roseobacter genus rather than a new species within the Roseobacter genus.
Roseobacter ponti DSM 106830T Metabolic Versatility
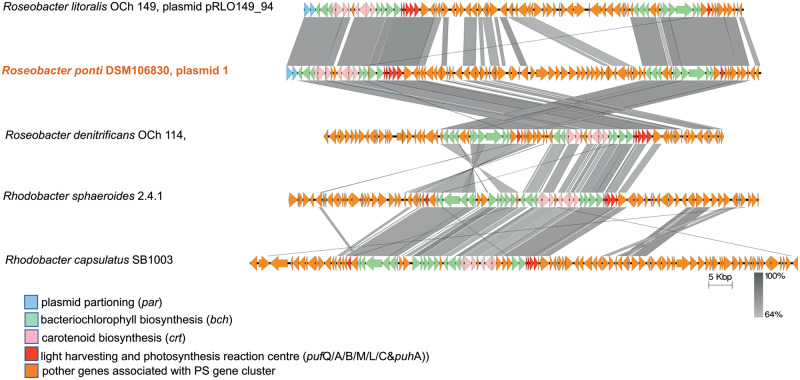
Roseobacter group members are equipped with a diverse toolkit of metabolic capabilities, which partly explains their success in colonizing a broad range of different marine habitats (Buchan et al. 2005). Some metabolic properties of R. ponti DSM 106830T were studied by Jung et al. (2017). The analysis of the genome sequence of R. ponti DSM 106830T revealed specific traits and metabolic adaptations to ecological niches. Jung and coworkers screened the strain for presence of genes encoding photosynthetic reaction center proteins (pufL, pufM, and puhA), enabling aerobic anoxygenic photoheterotrophy (AAP), by PCR and detected puhA but not pufL and pufM. However, the whole-genome sequence confirmed that all three genes responsible for AAP are present (G3256_19015, G3256_18695, and G3256_18700). In addition, the genome harbors putative genes important for a functional photosynthetic gene cluster (PGC) and the production of BchlA (Zheng et al. 2011). These included the bch genes important for BchlA biosynthetic pathways (G3256_18600, G3256_18655, G3256_18670, G3256_18675, G3256_18980), the puf operon involved in formation of the reaction center (G3256_18680–G3256_18705), puhA participating in reaction center assembly (G3256_19015), and crt genes (G3256_1860, G3256_18620, G3256_18640, G3256_18645, G3256_18650) responsible for carotenoid biosynthesis and regulation (Pradella et al. 2004; Zheng et al. 2011; Petersen et al. 2012; Chi et al. 2015). The puf operon (pufQBALMC(X)) analysis of Petersen et al. (2012) to was extended to determine the closest related synteny to the R. ponti DSM 106830Tpuf operon (fig. 1). The puf operon comprises six genes encoding a cytochrome subunit (pufQ), light harvesting proteins (pufA and pufB), the photosynthetic reaction center subunits L and M (pufL and pufM, respectively) the photosynthetic reaction center cytochrome C (pufC or pufX in Rhodobacter) (Kortlüke et al. 1997; Zheng et al. 2011). Roseobacter ponti DSM 106830T contains pufC, which is also present in R. denitrificans OCh 114 and R. litoralis OCh 149 (fig. 1) (Kortlüke et al. 1997). The analysis showed that the puf operon of R. ponti DSM 106830T is most similar to a puf operon of R. litoralis OCh 149 (fig. 1). In both organisms, the operon is encoded by plasmids (Pradella et al. 2004) and share PGC genes. Plasmid genes that are unrelated to the puf operon or PGC are genetically not conserved. Remarkably, in other members of the Roseobacter group, such as Rhodobacter encapsulates and R. denitrificans, the puf operon is encoded by the chromosome (Kortlüke et al. 1997; Petersen et al. 2012), indicating an evolutionary adaption to a specific ecological niche of some Roseobacter group members via plasmid acquisition.
Fig. 1.
Synteny comparison of Roseobacter ponti DSM 106830T plasmid 1 to closest relatives. References include Roseobacter litoralis OCh 149, plasmid pRLO149_94 (CP002624.1), and cutted regions of Roseobacter denitrificans OCh 114 chromosome (CP000362.1), Rhodobacter sphaeroides 2.4.1, chromosome 1 (CP000143.2), and Rhodobacter capsulatus SB1003, chromosome (CP001312.1). The comparison was performed with Easyfig (Sullivan et al. 2011) using BlastN percent identities. Synteny between related regions is indicated by vertical gray-shaded areas and black lines. The legend indicates the biological categories of genes involved in the photosynthesis cluster.
The genome analysis by BlastKOALA resulted in a variety of pathway, such as genes involved in biogeochemical cycling (Luo and Moran 2014) including the dissimilatory nitrite reductase (nirK), dimethylsulfoniopropionate demethylase (dmdA), sulfur oxidation protein complex (soxB), and large subunit of carbon monoxide dehydrogenase (coxL). In comparison to R. denitrificans OCh 114 nasA, nirB, napA, narG, and nirS involved in the nitrogen metabolism are absent in the genome of R. ponti DSM 106830T. It is indicated that R. ponti DSM 106830T is capable of degrading DMSP and performing carbon monoxide oxidation.
Additionally, the search for gene clusters involved in secondary metabolite synthesis identified six putative clusters of which five were encoded by the chromosome and one by the plasmid. Interestingly, the plasmid harbors one putative terpene cluster which encodes the synthesis of the carotenoid spheroidenone, which is the main light-harvesting carotenoid of Roseobacter group members (fig. 1) (Wagner-Döbler and Biebl 2006). Pigment gene clusters are a typical feature of AAP bacteria and cell color can range from yellow/orange over brown or pink/red to purple (Zheng et al. 2011). In addition to the photosynthetic apparatus including the puf operon, the carotenoids are encoded on the plasmid. Carotenoids are, amongst others, protective against harmful radicals, such as oxygen and radiation (Chi et al. 2015) and could be advantageous from an evolutionary point of view.
Roseobacter ponti DSM 106830T Horizontal Gene Transfer
The Roseobacter group members occur in a wide variety of different ecological niches in the marine oceans, indicating a high adaptation potential (Wagner-Döbler and Biebl 2006; Brinkhoff et al. 2008). Evolutionary driving forces for genetic diversification by horizontal gene transfer (HGT) and mechanisms of DNA exchange include phages (transduction), plasmids (conjugation), and virus-like particles or gene transfer agents (GTA) (Wall et al. 1975; Pal et al. 2005; Zhan et al. 2016). To investigate the evolutionary potential of R. ponti DSM 106830T the genome was screened for indicators of HGT. This revealed one putative chromosomal phage region (17.4 kb, 2,122,003–2,139,468). PHASTER classified the completeness as questionable and the typical insertion sites attL/attR were not identified. It is likely that this element is rather a GTA, a virus-like particle which is proposed to originate from ancient prophage remnants (Lang and Beatty 2000). Since gene equipment and content of GTAs are similar to phages, detection by PHASTER is expected. Previous genomic studies showed that nearly all genomes of the Roseobacter group possess GTAs (Lang and Beatty 2000; Newton et al. 2010; Huang et al. 2011). In detail, the detected region comprises 19 putative phage-associated genes of which three were annotated as GTA.
In conclusion, the analysis of the R. ponti DSM 106830T genome sequence shows that phylogenetic classifications of new Roseobacter group members should not be performed by 16S rRNA gene but on the whole-genome comparisons. We suggest R. ponti DSM 106830T as a missing link between the genera Sulfitobacter and Roseobacter rather than as a new species in the genus Roseobacter. Notably, the plasmid p1 of R. ponti DSM 106830T encodes the AAB operon, which was described for only six Rhodobacteracea members including R. litoralis OCh 149 (Pradella et al. 2004; Brinkmann et al. 2018). Additionally, a putative GTA was detected in the chromosome. Both indicate the adaptive capabilities to a specific ecological niche and the oligotrophic marine environment by HGT via plasmids and GTAs. Finally, the genome analysis confirms previous studies (Jung et al. 2017) and is the foundation for future physiological analyses of R. ponti. Additionally, the data provided here will be valuable for studies targeting PGC within the Roseobacter group.
Supplementary Material
Acknowledgments
We thank Sarah Teresa Schüßler and Melanie Heinemann for technical assistance. This study was partly supported by the Deutsche Forschungsgemeinschaft (DFG) as part of the collaborative research center TRR51. The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Data deposition: The whole-genome sequence project has been deposited at DDBJ/ENA/GenBank under the accession numbers CP048788 (chromosome) and CP048789 (plasmid). The NCBI BioProject and BioSample IDs are PRJNA605959 and SAMN14082710, respectively. The raw reads have been deposited at NCBI SRA database under the accession numbers SRR11069767 (paired-end Illumina) and SRR11069766 (Oxford Nanopore).
Literature Cited
- Arndt D, et al. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44(W1):W16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K, et al. 2019. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acid Res. 47:81–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkhoff T, Giebel H-A, Simon M.. 2008. Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch Microbiol. 189(6):531–539. [DOI] [PubMed] [Google Scholar]
- Brinkmann H, Göker M, Koblížek M, Wagner-Döbler I, Petersen J.. 2018. Horizontal operon transfer, plasmids, and the evolution of photosynthesis in Rhodobacteraceae. ISME J. 12(8):1994–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan A, González JM, Moran MA.. 2005. Overview of the marine Roseobacter lineage. Appl Environ Microbiol. 71(10):5665–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH.. 2019. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics btz848:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J.. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34(17):i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SC, et al. 2015. Assembly of functional photosystem complexes in Rhodobacter sphaeroides incorporating carotenoids from the spirilloxanthin pathway. Biochim Biophys Acta. 1847(2):189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freese HM, Methner A, Overmann J.. 2017. Adaptation of surface-associated bacteria to the open ocean: a genomically distinct subpopulation of Phaeobacter gallaeciensis colonizes pacific mesozooplankton. Front Microbiol. 8:1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Zhang Y, Chen F, Jiao N.. 2011. Complete genome sequence of a marine roseophage provides evidence into the evolution of gene transfer agents in alphaproteobacteria. Virol J. 8(1):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y-T, Park S, Lee J-S, Yoon J-H.. 2017. Roseobacter ponti sp. nov., isolated from seawater. Int J Syst Evol Microbiol. 67(7):2189–2194. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Morishima K.. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 428(4):726–731. [DOI] [PubMed] [Google Scholar]
- Kirchman DL. 2008. Microbial ecology of the oceans. Second ed New Jersey: Wiley-Blackwell. [Google Scholar]
- Kortlüke C, Breese K, Gad'on N, Labahn A, Drews G.. 1997. Structure of the puf operon of the obligately aerobic, bacteriochlorophyll a-containing bacterium Roseobacter denitrificans OCh114 and its expression in a Rhodobacter capsulatus puf puc deletion mutant. J Bacteriol. 179(17):5247–5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AS, Beatty JT.. 2000. Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc Natl Acad Sci USA. 97(2):859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Moran MA.. 2014. Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev. 78(4):573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens T, et al. 2007. Bacteria of the Roseobacter clade show potential for secondary metabolite production. Microb Ecol. 54(1):31–42. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 17(1):10–17. [Google Scholar]
- Medlar AJ, Törönen P, Holm L.. 2018. AAI-profiler: fast proteome-wide exploratory analysis reveals taxonomic identity, misclassification and contamination. Nucleic Acid Res. 46(W1):W479–W485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Göker M.. 2019. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 10(1):2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran MA, et al. 2007. Ecological genomics of marine Roseobacters. Appl Environ Microbiol. 73(14):4559–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton RJ, et al. 2010. Genome characteristics of a generalist marine bacterial lineage. ISME J. 4(6):784–798. [DOI] [PubMed] [Google Scholar]
- Pal C, Papp B, Lercher MJ.. 2005. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat Genet. 37(12):1372–1375. [DOI] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW.. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25(7):1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, et al. 2012. Think pink: photosynthesis, plasmids and the Roseobacter clade. Environ Microbiol. 14(10):2661–2672. [DOI] [PubMed] [Google Scholar]
- Pradella S, et al. 2004. Genome organization and localization of the pufLM genes of the photosynthesis reaction center in phylogenetically diverse marine alphaproteobacteria. Appl Environ Microbiol. 70(6):3360–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappe MS, Vergin K, Giovannoni SJ.. 2000. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol Ecol. 33(3):219–232. [DOI] [PubMed] [Google Scholar]
- Selje N, Simon M, Brinkhoff T.. 2004. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 427(6973):445–448. [DOI] [PubMed] [Google Scholar]
- Shiba T. 1991. Roseobacter litoralis gen. nov., sp. nov., and Roseobacter denitrificans sp. nov., aerobic pink-pigmented bacteria which contain Bacteriochlorophyll a. Syst Appl Microbiol. 14(2):140–145. [Google Scholar]
- Simon M, et al. 2017. Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 11(6):1483–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenschein EC, et al. 2018. Phylogenetic distribution of roseobacticides in the Roseobacter group and their effect on microalgae. Environ Microbiol Rep. 10(3):383–393. [DOI] [PubMed] [Google Scholar]
- Sullivan MJ, Petty NK, Beatson SA.. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27(7):1009–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MT, Beja O, Taylor LT, DeLong EF.. 2001. Phylogenetic analysis of ribosomal RNA operons from uncultivated coastal marine bacterioplankton. Environ Microbiol. 3(5):323–331. [DOI] [PubMed] [Google Scholar]
- Suzuki MT, Preston CM, Chavez FP, DeLong EF.. 2001. Quantitative mapping of bacterioplankton populations in seawater: field tests across an upwelling plume in Monterey Bay. Aquat Microb Ecol. 24:117–127. [Google Scholar]
- Tatusova T, et al. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44(14):6614–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner-Döbler I, Biebl H.. 2006. Environmental biology of the marine Roseobacter lineage. Annu Rev Microbiol. 60(1):255–280. [DOI] [PubMed] [Google Scholar]
- Wall JD, Weaver PF, Gest H.. 1975. Gene transfer agents, bacteriophages, and bacteriocins of Rhodopseudomonas capsulata. Arch Microbiol. 105(1):217–224. [DOI] [PubMed] [Google Scholar]
- Wick RR, Judd LM, Gorrie CL, Holt KE.. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 13:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Schultz MB, Zobel J, Holt KE.. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31(20):3350–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth JS, Whitman WB.. 2018. Phylogenomic analyses of a clade within the Roseobacter group suggest taxonomic reassignments of species of the genera the proposal of six novel genera Aestuariivita, Citreicella, Loktanella, Nautella, Pelagibaca, Ruegeria, Thalassobius, Thiobacimonas and Tropicibacter, and the proposal of six novel genera. Int J Syst Evol Microbiol. 68(7):2393–2411. [DOI] [PubMed] [Google Scholar]
- Zhan Y, Huang S, Voget S, Simon M, Chen F.. 2016. A novel Roseobacter phage possesses features of podoviruses, siphoviruses, prophages and gene transfer agents. Sci Rep. 6:4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, et al. 2011. Diverse arrangement of photosynthetic gene clusters in aerobic anoxygenic phototrophic bacteria. PLoS One 6(9):e25050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.