Abstract
Background
This study was performed to explore the clinical and prognostic significance of APOB mRNA expression, DNA methylation and APOB mutation in patients with low-grade glioma (LGG).
Methods
Bioinformatic analysis was conducted using genomic, clinical and survival data from The Cancer Genome Atlas (TCGA) and Chinese Glioma Genome Atlas (CGGA) databases. Serum APOB protein levels were measured via immunoturbidimetry in 150 patients with LGG and 100 healthy controls from Hubei General Hospital.
Results
There was a negative association between the levels of APOB mRNA and DNA methylation (r=−0.355, P<0.0001) in patients with LGG from the TCGA database. Additionally, LGG patients with low levels of APOB mRNA exhibited better overall survival (OS) than those with high levels of APOB mRNA (HR=0.637, P=0.0085). The survival time of LGG patients with APOB hypermethylation was markedly longer than that of patients with APOB hypomethylation (HR=0.423, P=0.0185). The prognostic significance of APOB mRNA and DNA methylation was also validated with the CGGA cohort, and a similar conclusion was reached. APOB gene mutations were observed in 3% of patients with LGG from the TCGA database, and no association was detected between APOB mutations and OS (P=0.164). Furthermore, the levels of APOB protein were much lower in patients with LGG than in normal individuals (P=0.0022), and the expression of APOB protein was markedly different among groups when stratified by histological type (P<0.0001) and histological-molecular classification (P<0.0001).
Conclusion
APOB mRNA expression is negatively regulated by DNA methylation in patients with LGG. Low expression or hypermethylation of APOB might predict relatively favorable survival in patients with LGG.
Keywords: low-grade glioma, APOB, prognosis, methylation, bioinformatic analysis
Introduction
Glioma is one of the most common and aggressive tumors in the brain,1 and 15,000–17,000 patients are newly diagnosed each year in the United States,2 causing a potential threat to the health of human beings. Glioma is generally divided into low-grade gliomas (LGG) and glioblastoma multiforme (GBM). GBM is the most aggressive subtype among gliomas, with a median overall survival (OS) of 12–16 months.3 LGG is less aggressive, and patients with LGG possess highly diverse clinical outcomes.4 Hence, it is highly significant to precisely predict the prognosis of patients with LGG with novel and accessible biomarkers.
Epigenetics has become a research focus, and aberrant DNA methylation has been viewed as a hallmark of glioma.5–8 Chen et al9 reported that patients with glioma with MGMT promoter methylation exhibited better progression-free survival (especially for patients receiving alkylating agents) than those with MGMT unmethylated promoters. Moreover, Feldheim et al10 pointed out that methylation of the MGMT promoter had been viewed as an independent prognostic factor in the treatment of glioma and was correlated with a better response to chemotherapy. Mathur et al proposed that the MGMT promoter methylation level could serve as a potential biomarker for treatment decisions by predicting hypermutation risk at recurrence in patients with LGG.11 Hence, identifying novel genes regulated by DNA promoter methylation will lead to better insights into the molecular mechanisms of LGG and will thus make great contributions to the discovery of novel biomarkers in the prediction of prognosis.
Apolipoprotein B (APOB) has long been of interest due to its close association with cardiovascular diseases.12–14 However, recent studies have suggested that APOB also plays an important role in the occurrence and progression of various tumors, including hepatocellular carcinoma,15 kidney renal clear cell carcinoma,16 nonesophageal squamous cell carcinoma,17 and breast cancer.18 However, no study has specifically investigated the prognostic role of APOB in patients with glioma. Importantly, based on the bioinformatic analysis, we found that APOB mRNA expression was negatively regulated by DNA promoter methylation in patients with LGG. Whether APOB DNA methylation can affect the OS of patients with LGG is currently unknown. Thus, in this study, we initially characterized the association between APOB mRNA expression and DNA methylation. Then, we investigated the prognostic role of APOB mRNA, its DNA methylation and APOB mutation among patients with LGG in the TCGA database. Subsequently, we validated the prognostic role of APOB mRNA and its DNA methylation with the CGGA database. Finally, we compared the levels of serum APOB protein in patients with LGG and healthy individuals, investigated the diagnostic significance of serum APOB protein and explored the association between the levels of serum APOB protein and clinicopathological features.
Materials and Methods
Bioinformatic Analysis
RNA-seq data and clinical information of TCGA-LGG were downloaded from UCSC Xena (https://xenabrowser.net/),19 which is an online website to analyze data from the TCGA database. This database included 532 patients with LGG (diffuse LGG and intermediate-grade glioma, including WHO grades II and III20) with complete RNA-seq information. Among LGG patients with available genomic data, 531 cases had intact OS information. Moreover, DNA methylation data of APOB (measured by 450K BeadChip) were obtained through the UCSC Xena website. The DNA methylation value of APOB was defined as the average value for all CpG sites in the APOB DNA promoter region. A total of 532 LGG tissues were measured by 450K BeadChip. The unit of DNA methylation measurement is β value (Figure S1), which is calculated by the intensity of methylation (M for signal A) and unmethylation (U for signal B) alleles, and the ratio of fluorescence signal β = max (m, 0)/[max (M, 0) + max (U, 0) + 100]. Therefore, the β value ranges from 0 (completely unmethylated) to 1 (completely methylated). Among them, 528 patients with glioma with APOB DNA methylation data had intact OS data.
To verify the prognostic role of APOB mRNA and DNA methylation, the CGGA database21 (http://www.cgga.org.cn/) was also searched. The CGGA database is an easily accessible web application for data analysis to investigate brain tumors datasets more than 2000 specimens from Chinese cohorts (mainly from Beijing Tiantan Hospital). This database includes the mRNA sequencing (N=1018), DNA methylation (N=159), mRNA microarray (N=301), whole-exome sequencing (N=286) and microRNA microarray (N=198) and matched clinical information. A total of 605 patients with LGG with follow-up information and APOB mRNA data were included in this study. Additionally, 108 cases of patients with LGG with APOB methylation data were further applied to validate the clinical and prognostic significance of APOB methylation.
Collection of Serum APOB Protein in Patients with LGG
The concentrations of serum APOB protein from 150 patients with LGG and 100 healthy individuals [(1) no chronic disease or malignant tumor; (2) no acute infection; (3) no hyperlipemia] were retrospectively collected from Hubei General Hospital from January 2016 to December 2019. All patients with glioma were definitively diagnosed by pathological examination combined with radiological findings (brain CT or MRI), and serum APOB protein levels were detected before surgical operation. APOB protein was detected via immunoturbidimetry in this hospital. In addition, other clinical information, such as age, gender, maximum tumor diameter (MTD), IDH1-mutation status, histological type and histological-molecular classification (oligodendroglioma, astrocytoma IDH-wt, astrocytoma IDH1-mutant), was also searched using the Electronic Medical Record (EMR). All participants involved in this study were required to provide written informed consent. This study protocol was verified and successfully approved by the Institutional Review Board of Hubei General Hospital (No. 2018-CK61).
Statistical Analysis
All the data were analyzed using the SPSS 22.0 program (SPSS Inc., Chicago, IL, USA) or GraphPad Prism 5.0 (GraphPad Inc., La Jolla, CA, USA). Patients with LGG were divided into low and high AOPB mRNA expression groups according to the median value of APOB mRNA. Similarly, LGG patients were divided into hypermethylation and hypomethylation groups based on the median value of APOB DNA methylation. The Kolmogorov–Smirnov (K-S) method was initially applied to determine the normality assumption of continuous variables. Independent t-tests were applied to examine the differences in APOB expression, DNA methylation, and APOB protein between groups. For three groups, analysis of variance (ANOVA) was employed to compare differences. The association between APOB mRNA expression along with DNA methylation and the clinicopathological features in patients with LGG was detected by the χ2 test or Fisher’s exact test.22 Correlation analysis was conducted to evaluate the Pearson correlation coefficient between APOB mRNA expression and APOB DNA methylation level. The Log rank test was used to assess the significance of the difference between two survival curves. All the variables from TCGA-LGG dataset were selected for the univariate Cox analysis and variables with P<0.05 were further selected for the multivariate Cox regression. Multivariate Cox regression was performed to explore the independent risk factors in the prognosis of patients with LGG. P <0.05 at both tails was regarded as statistically significant.
Results
APOB Expression is Negatively Regulated by DNA Methylation
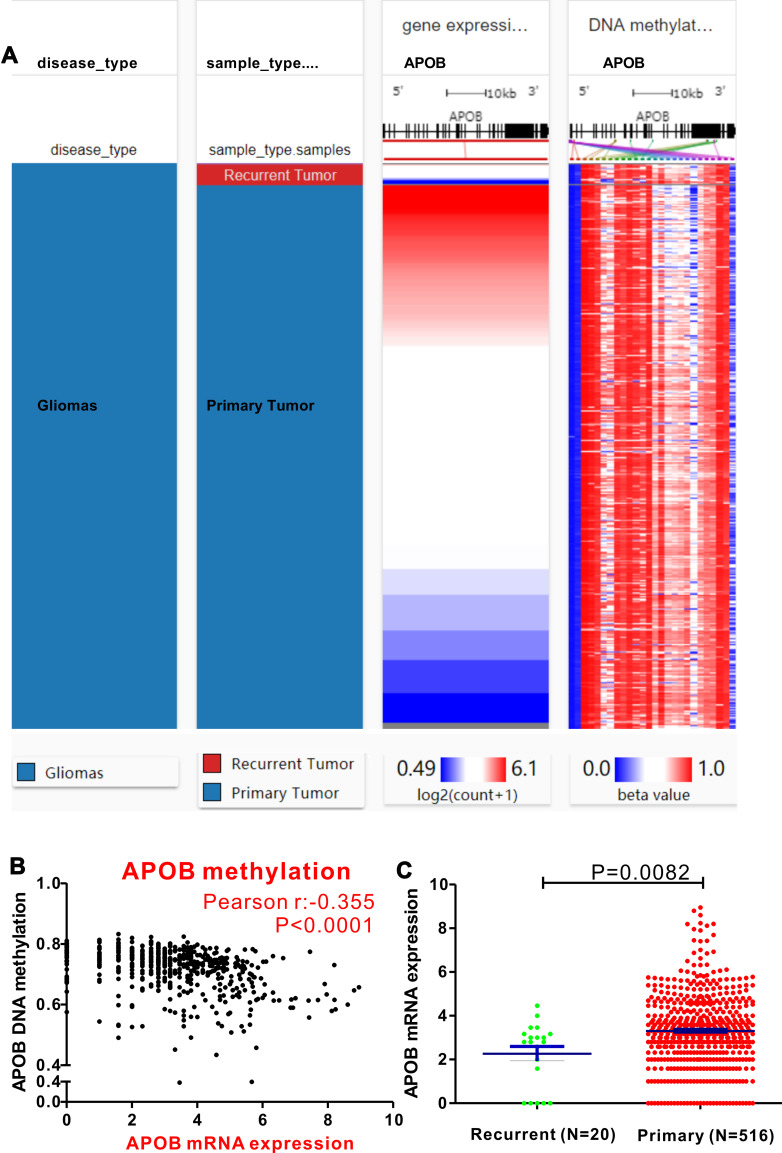
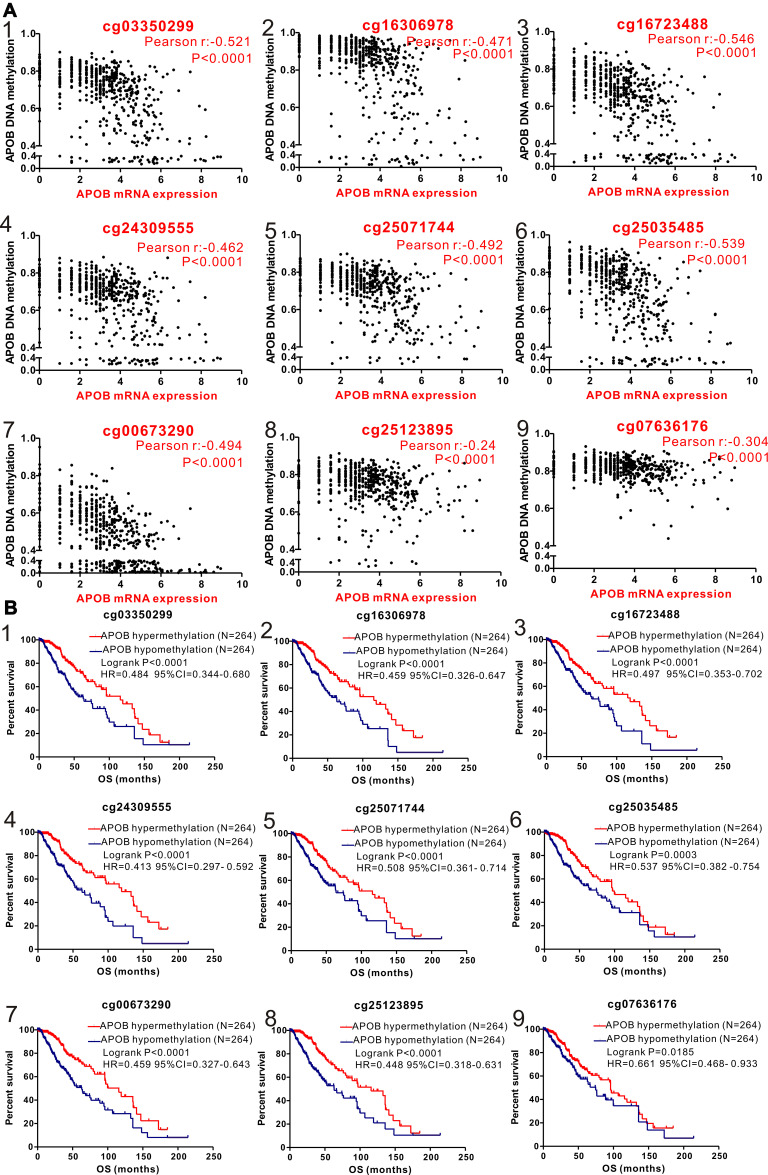
First, a comparable heatmap that included the sample type, APOB mRNA expression and APOB DNA methylation in TCGA-LGG was generated through the UCSC Xena website. As illustrated in Figure 1, APOB mRNA expression was negatively regulated by APOB DNA methylation (Pearson r=−0.355, P<0.0001, Figure 1B). Moreover, we found that the levels of APOB mRNA were significantly higher in patients with primary LGG than in those with recurrent LGG [3.305 ± 0.076 vs 2.265 ± 0.329 Log2(count+1), P=0.0082, Figure 1C]. Furthermore, Pearson correlation analyses were also undertaken to investigate the associations between APOB mRNA expression and DNA methylation of the specific CpG sites (Table S1). Both the heatmap and the correlation analyses revealed that APOB mRNA expression was negatively correlated with the nine CpG sites of APOB DNA methylation (Figure 2A). Additionally, nine CpG sites (|Pearson r|>0.2) of APOB DNA hypermethylation were all well correlated with favorable OS in patients with LGG (Figure 2B).
Figure 1.
The epigenetic mechanism underlying aberrant expression of APOB in low-grade glioma (LGG) revealed by bioinformatic analysis. (A) Heatmap of the association between expression of APOB mRNA and the methylation of APOB DNA CpG sites. (B) A negative correlation is observed between APOB mRNA and its DNA methylation (r=−0.355). (C) The levels of APOB mRNA are significantly higher in patients primary LGG than those with recurrent LGG (P=0.0082).
Figure 2.
Correlation analysis of the relationship between APOB mRNA expression and methylation of APOB DNA CpG sites (A) and association between methylation of APOB DNA CpG sites and overall survival (OS) in patients with LGG (B) in TCGA database. (1) cg03350299; (2) cg16306978; (3) cg16723488; (4) cg24309555; (5) cg25071744; (6) cg25035485; (7) cg00673290; (8) cg25123895; (9) cg07636176.
Prognostic Significance of APOB and DNA Methylation in the TCGA Database
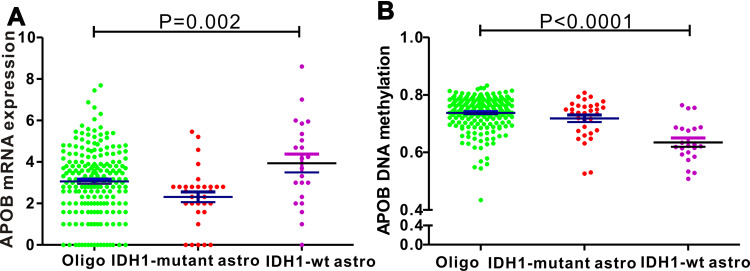
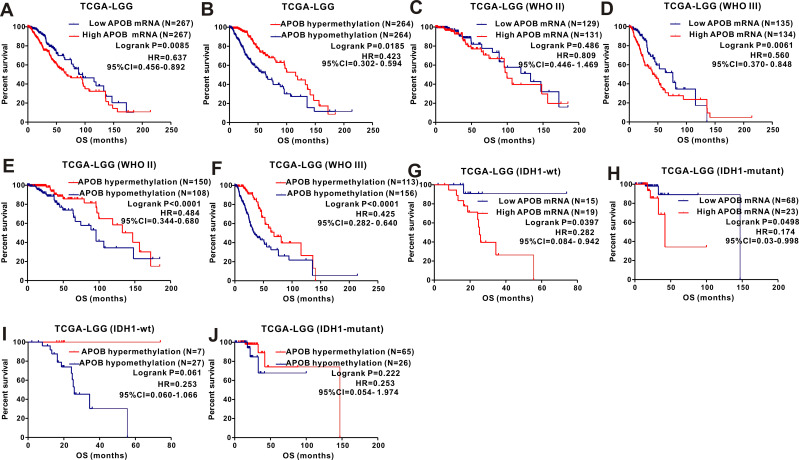
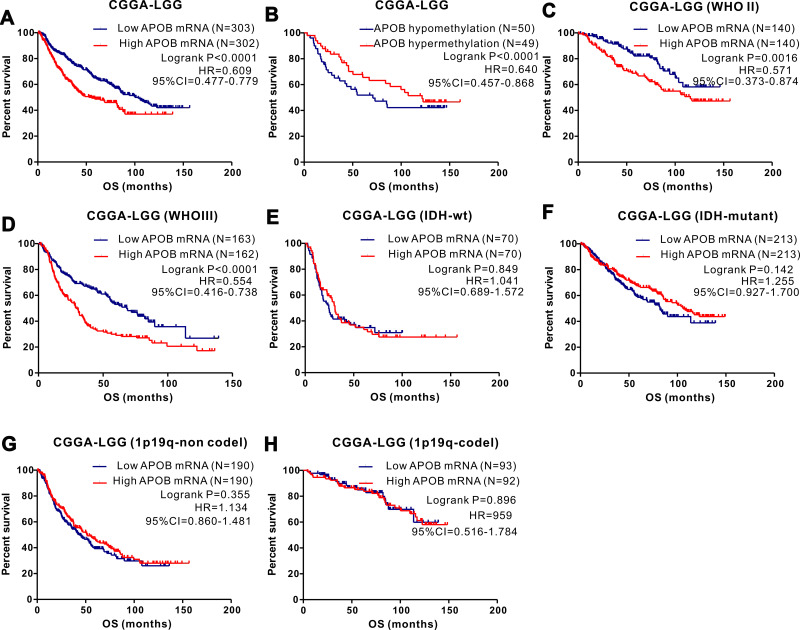
We analyzed the association between APOB mRNA expression and clinicopathological features among 532 patients with LGG. Patients with LGG were divided into two groups by the median value of APOB mRNA expression. As revealed in Table 1, we found that IDH1-mutation (P=0.001), cancer status (P=0.048), survival (P=0.001) and methylation status (P<0.001) could significantly affect the expression levels of APOB mRNA. For the integrated histological-molecular classification, we noticed that the expression of APOB mRNA (Figure 3A) and APOB DNA methylation (Figure 3B) were quite different among patients with LGG with oligodendroglioma, IDH1-wildtype and IDH1-mutation. Then, Kaplan-Meier curves were plotted to explore the prognostic significance of APOB mRNA in 531 patients with LGG. As shown in Figure 4A, patients with low APOB mRNA expression possessed significantly more favorable OS than those with high APOB mRNA expression (HR=0.637, 95% CI=0.456–0.892, P=0.0085). Moreover, this strong association also existed in patients with LGG with WHO III (Figure 4D), IDH1-wildtype (Figure 4G) and IDH1-mutation (Figure 4H), while no association existed in patients with WHO II (Figure 4C).
Table 1.
Correlation Between APOB mRNA Expression/Methylation and Clinicopathologic Features of 532 Patients with LGG in TCGA Database
| Clinical Features | APOB Expression | P value | APOB Methylation | P value | |||
|---|---|---|---|---|---|---|---|
| Low | High | Low | High | ||||
| Age | ≤50 | 196 | 178 | 0.066 | 172 | 202 | 0.004 |
| >50 | 69 | 89 | 94 | 64 | |||
| Gender | Female | 117 | 124 | 0.596 | 127 | 114 | 0.258 |
| Male | 148 | 143 | 139 | 152 | |||
| Race | White/American | 243 | 248 | 0.425 | 242 | 249 | 0.094 |
| Black/Africa american | 9 | 13 | 16 | 6 | |||
| Asian | 6 | 3 | 5 | 4 | |||
| Histological Type | Astrocytoma | 100 | 97 | 0.507 | 126 | 71 | <0.001 |
| Oligodendroglioma | 104 | 97 | 81 | 120 | |||
| Mixed glioma | 61 | 73 | 59 | 75 | |||
| Family history of cancer | Yes | 59 | 79 | 0.155 | 73 | 65 | 0.433 |
| No | 112 | 110 | 108 | 114 | |||
| IDH1-mutation | Mutant | 68 | 23 | 0.001 | 26 | 65 | <0.001 |
| Wildtype | 15 | 19 | 27 | 7 | |||
| Laterality | Left | 137 | 121 | 131 | 127 | ||
| Midline | 5 | 2 | 3 | 4 | |||
| Right | 119 | 143 | 129 | 133 | |||
| WHO grade | II | 129 | 132 | 0.828 | 108 | 153 | <0.001 |
| III | 136 | 134 | 157 | 113 | |||
| Cancer status | With tumor | 113 | 140 | 0.048 | 134 | 119 | 0.011 |
| Tumor free | 93 | 78 | 69 | 102 | |||
| KPS | ≤80 | 10 | 19 | 0.448 | 15 | 14 | 0.649 |
| >80 | 17 | 22 | 18 | 21 | |||
| Sample type | pLGG | 253 | 261 | 0.146 | 253 | 261 | 0.055 |
| rLGG | 12 | 6 | 13 | 5 | |||
| Radiotherapy | Yes | 138 | 149 | 0.604 | 159 | 128 | <0.001 |
| No | 89 | 87 | 66 | 110 | |||
| OS | Alive | 214 | 178 | <0.001 | 173 | 219 | <0.001 |
| Dead | 48 | 88 | 92 | 44 | |||
| MTD | ≤2cm | 71 | 131 | 0.087 | 93 | 109 | 0.153 |
| >2cm | 3 | 16 | 12 | 7 | |||
| Seizure history | Yes | 150 | 164 | 0.512 | 161 | 153 | 0.652 |
| No | 93 | 90 | 88 | 95 | |||
| APOB mRNA expression | Low | - | - | - | 97 | 168 | <0.001 |
| High | - | - | 169 | 98 | |||
| APOB Methylation |
Low | 97 | 169 | <0.001 | - | - | - |
| High | 168 | 98 | - | - | - | ||
Abbreviations: KPS, Kamofsky performance status; WHO, World health organization; OS, overall survival; MTD, maximum tumor diameter; pLGG, primary low-grade glioma; rLGG, recurrent low-grade glioma.
Figure 3.
Differences in APOB mRNA expression and its DNA methylation among three histological-molecular types. (A) Patients with IDH1-wildtype astrocytoma possess highest levels of APOB mRNA, while the lowest expression is observed in oligodendroglioma. (B) Levels of APOB DNA methylation are highest in oligodendroglioma and lowest in IDH1-wildtype astrocytoma.
Figure 4.
The prognostic significance of APOB as well as its methylation in patients with LGG from the TCGA database and subgroup analyses. Low levels of APOB mRNA (A) or its hypermethylation (B) predicts favorable OS in LGG patients. Subgroup analyses are performed stratified by WHO grade (C–F), IDH1-mutation and 1p19q (G–J).
Based on 532 cases of LGG with intact clinical information and APOB methylation data, the χ2 test or Fisher’s exact test was applied to detect the correlation between APOB DNA methylation and clinical parameters. As exhibited in Table 1, the APOB DNA methylation status was different when stratified by age (≤50 vs >50, P=0.004), histological type (astrocytoma vs oligodendroglioma vs mixed glioma, P<0.0001), IDH1 mutation (wildtype vs mutant, P<0.001), WHO grade (II vs III, P<0.001), cancer status (with tumor vs tumor free, P=0.011), radiotherapy (yes vs no, P<0.001), survival (living vs dead, P<0.001) and APOB mRNA (high vs low, P<0.001). Then, the Log rank test of the OS curves (Figure 5B) showed that patients with APOB DNA hypermethylation were associated with more favorable OS than those with APOB DNA hypomethylation (HR=0.423, 95% CI=0.302–0.594, P=0.0185). When stratified by WHO grade and IDH1 mutation, we found that this trend still existed in WHO II (Figure 4E) and III (Figure 4F) groups, but disappeared in groups with IDH1-wildtype (Figure 4I) and IDH1 mutation (Figure 4J). Finally, Cox regression analyses were performed to identify the independent risk factors in the prognosis of patients with LGG (Table 2), and the results revealed that hypermethylation of APOB was an independent prognostic factor among patients with LGG (HR=0.05, 95% CI=0.004–0.332, P=0.013).
Figure 5.
The prognostic role of APOB as well as its methylation in LGG patients from the CGGA database and subgroup analyses. Low expression (A) or hypermethylation (B) of APOB is associated with better OS in LGG patients. The association between low APOB mRNA and favorable OS existed in patients with WHO II group (C) and WHO III group (D), while not existed in I DH1-widetype (E) and mutation group (F), 1p19q non-codel (G) and codel groups (H).
Table 2.
Cox Regression Analyses of Potential Risk Factors for OS in Patients with LGG from TCGA Database
| Univariate Regression | Multivariate Regression | |||||
|---|---|---|---|---|---|---|
| P value | HR | 95% CI | P value | HR | 95% CI | |
| Age (≤50 vs >50) | <0.001 | 3.535 | 2.471–5.057 | 0.423 | 2.553 | 0.257–25.343 |
| Gender (Female vs Male) | 0.495 | 1.126 | 0.801–1.582 | – | – | – |
|
Race White/American Black/Africa American Asian |
0.605 | – | – | – | – | – |
| 0.317 | 1.477 | 0.688–3.170 | – | – | – | |
| 0.961 | 0.003 | 0.001–606.14 | – | – | – | |
| Histological Astrocytoma | 0.009 | – | – | 0.585 | ||
| Type Oligodendroglioma | 0.002 | 0.546 | 0.370–0.806 | 0.311 | 0.244 | 0.016–3.726 |
| Mixed | 0.137 | 0.716 | 0.461–1.112 | 0.841 | 0.831 | 0.139–4.977 |
| Laterality Left | 0.343 | – | – | – | – | – |
| Midline | 0.154 | 1.290 | 0.909–1.832 | – | – | – |
| Right | 0.644 | 1.321 | 0.405–4.307 | – | – | – |
| IDH1 (wildtype vs mutant) | 0.001 | 0.181 | 0.068–0.485 | 0.789 | 1.245 | 0.250–6.207 |
| KPS (>80 vs ≤80) | 0.369 | 0.741 | 0.386–1.425 | – | – | – |
| Radiotherapy (Yes vs No) | 0.001 | 0.440 | 0.276–0.701 | 0.400 | 0.380 | 0.040–3.620 |
| Family history of cancer | ||||||
| (Yes vs No) | 0.039 | 1.575 | 1.023–2.425 | 0.539 | 1.948 | 0.232–16.381 |
| Grade (III vs II) | <0.001 | 3.217 | 2.221–4.659 | 0.008 | 9.876 | 3.154–22.758 |
| Seizure history (Yes vs No) | 0.508 | 0.888 | 0.624–1.263 | – | – | – |
| Cancer status (With tumor vs without tumor) | <0.001 | 24.072 | 5.938–97.585 | 0.939 | 13.41 | 0.012–89.131 |
| MTD (>2cm vs ≤2) | 0.743 | 0.789 | 0.191–3.259 | – | – | – |
| Sample type (pLGG vs rLGG) | 0.706 | 1.104 | 0.592–2.058 | – | – | – |
| APOB mRNA (High vs Low) | 0.007 | 1.627 | 1.144–2.313 | 0.286 | 4.807 | 0.269–85.770 |
| Methylation (High vs Low) | <0.001 | 0.402 | 0.281–0.577 | 0.013 | 0.05 | 0.004–0.332 |
Abbreviations: OS, overall survival; KPS, Kamofsky performance status; WHO, World health organization; MTD, maximum tumor diameter; pLGG, primary low-grade glioma; rLGG, recurrent low-grade glioma.
Validation of the Prognostic Significance of APOB and DNA Methylation in the CGGA Database
To verify the results in the TCGA database, we further investigated the expression and prognostic role of APOB mRNA, as well as APOB DNA methylation, in the CGGA database. We analyzed the potential factors affecting the expression of APOB mRNA among 605 patients with LGG (Figure S2A). LGG patients with IDH1-wildtype expressed more APOB mRNA than those with IDH1-mutant (Figure S2A5). Moreover, we also studied the potential factors affecting APOB methylation status (Figure S2B), and gender (Figure S2B1) as well as WHO grade (Figure S2B4) could significantly affect APOB DNA methylation.
To assess the prognostic significance of APOB for LGG in the CGGA, Kaplan-Meier analyses were applied to evaluate the association between APOB mRNA expression and the clinical survival data. As shown in Figure 5A, patients with LGG with low APOB mRNA levels possessed markedly better OS than those with high APOB mRNA levels (HR=0.609, 95% CI=0.477–0.779, P<0.0001). We also determined that low expression of APOB mRNA was significantly correlated with relatively favorable OS both in patients with WHO II (Figure 5C) and WHO III (Figure 5D), while this association disappeared when stratified by IDH1 mutation (Figure 5E and F) and 1p19q (Figure 5G and H). Additionally, based on 99 patients with LGG with intact survival information along with APOB DNA methylation data, Kaplan–Meier survival analyses (Figure 5B) revealed that patients with APOB hypermethylation exhibited markedly better OS than those with APOB hypomethylation (HR=0.640, 95% CI=0.457–0.868, P<0.0001). Due to the limited number of patients with LGG with methylation data, we could not further conduct survival analyses stratified by WHO grade, IDH1-mutation and 1p19q.
Association Between APOB Mutations and Prognosis in LGG Patients
The CbioPortal website23 (https://www.cbioportal.org/) was searched to investigate APOB mutations in patients with LGG. As shown in Figure S3A, the APOB mutation rate was 3.0% (N=16), and missense mutation was the most common type among patients with LGG. In addition, we further explored the association between APOB mutation status and prognosis. As exhibited in Figure S3B, the log rank test shows that there was no significant difference in OS between patients with LGG without APOB mutations and those with APOB mutations (P=0.164).
Correlation Between Serum APOB Protein and Clinical Features
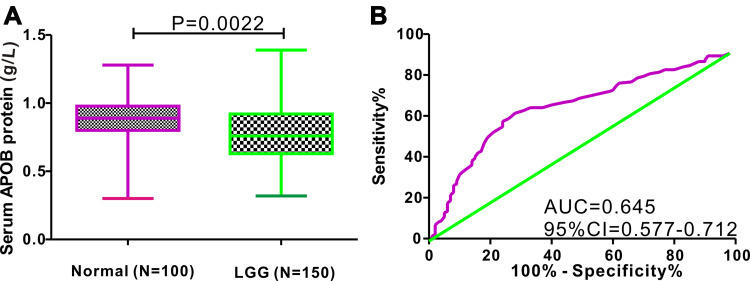
A total of 150 patients with LGG from the Department of Neurosurgery and 100 healthy controls from the health examination center were retrospectively included in this study. A t-test was used to compare the difference in serum APOB protein between patients with LGG and healthy individuals, and the results showed that the levels of serum APOB protein were relatively higher in healthy individuals than in patients with LGG (0.869 ± 0.017 vs 0.788 ± 0.018 g/L, P=0.0022; Figure 6A). Next, ROC analysis was performed to evaluate the diagnostic significance of serum APOB to differentiate patients with LGG from healthy controls (Figure 6B). The predictive accuracy of serum APOB as reflected by area under curve (AUC) for identifying LGG from healthy controls was 0.645 (95% CI=0.577–0.712). With the optimal cutoff value of 0.815 g/L, the sensitivity and specificity of serum APOB in differentiating glioma from healthy controls were 61.33% and 72%, respectively. Furthermore, we also explored the association between serum APOB protein and clinicopathological characteristics among patients with LGG. As shown in Figure S4, we observed that the levels of serum APOB were markedly different between oligodendroglioma and astrocytoma (P<0.0001).
Figure 6.
Difference in serum APOB protein between LGG patients and healthy controls, and ROC analysis. (A) Serum APOB protein levels are higher in healthy individuals than that in patients with LGG (P=0.0022). (B) Serum APOB protein shows an acceptable diagnostic performance for identifying LGG (AUC=0.645).
Discussion
In the present study, we identified a novel gene (APOB) in LGG that is commonly suppressed by DNA CpG methylation. Then, our study revealed that both APOB mRNA and methylation were novel prognostic biomarkers in LGG through integrative database analysis. It is noteworthy that two large cohorts (532 patients with LGG from the TCGA database and 605 patients from the CGGA database) were incorporated into this analysis. Furthermore, we analyzed the differential expression of APOB at the protein level and found that serum APOB protein concentrations were much lower in patients with LGG than in healthy individuals. Taken together, APOB is downregulated by DNA methylation in LGG, and hypermethylation of APOB serves as an independent prognostic factor in patients with LGG.
Recent studies demonstrated that APOB was implicated in the progression and recurrence of some kinds of malignant tumors. A recent study reported18 that APOB was an independent risk factor for intraocular metastasis in patients with breast cancer, and APOB could be utilized as a serum biomarker to identify intraocular metastasis from non-intraocular metastasis. Besides, a comprehensive network analysis revealed that APOB might be a tumor suppressor gene, and aberrant expression of APOB was markedly associated with unfavorable clinical outcome in patients with liver cancer.15 Furthermore, the Cancer Genome Atlas Research Network analyzed 196 cases of liver cancer by DNA methylation and discovered that APOB was downregulated by DNA methylation.24 Currently, no related studies have specifically explored the role of APOB in glioma. To the best of our knowledge, this is the first study to discover the negative association between APOB mRNA expression and DNA CpG methylation in patients with LGG and systematically investigate the prognostic role of APOB mRNA, DNA methylation and APOB gene mutations in patients with LGG. In this study, we not only assessed the prognostic significance of APOB mRNA expression and DNA CpG methylation in the TCGA database but also further confirmed this prognostic significance with the CGGA database. Encouragingly, the results from both databases were almost consistent.
Aberrant DNA methylation leads to the abnormal expression of genes, one of the main factors in the occurrence and progression of glioma.25–27 Our study revealed that the levels of APOB DNA methylation were higher in patients with oligodendroglioma than in those with IDH-wild type astrocytoma. Furthermore, Cox regression analysis demonstrated that APOB hypermethylation was an independent factor of relatively favorable survival in patients with LGG. The OS time of patients with glioma with APOB hypermethylation was significantly longer than that of patients with APOB hypomethylation, as reflected by Kaplan–Meier survival curves. In particular, APOB hypermethylation was markedly associated with better OS in patients with WHO grade II or III. However, this trend disappeared when stratified by IDH1-mutation, probably due to the small number of patients in the IDH1-wildtype group from the TCGA database. The clinical outcomes of LGG mainly depend on the aggressiveness of individual tumors. Quite a few patients experience tumor recurrence after radical surgery. Chemotherapy or radiotherapy after radical surgery may benefit patients with LGG,28,29 but their role is still fairly controversial. Hence, novel prognostic indicators may provide a more accurate risk assessment for patients with glioma, which could guide personalized treatment. Several studies30–32 have reported that DNA promoter methylation could be used as a promising predictive biomarker in glioma. Our study suggests that APOB hypermethylation may serve as a novel prognostic indicator for patients with LGG.
APOB protein, encoded by the APOB gene, plays a critical role in the formation of lipoprotein complexes of low-density lipoprotein and high-density lipoprotein.33 APOB protein is an established risk factor for metabolic diseases and cardiovascular diseases.34 However, some researchers have begun to focus on the interplay between serum APOB protein and cancer. Li et al reported that the levels of serum APOB protein were significantly higher in post-chemotherapeutic patients with breast cancer than in prechemotherapeutic patients.35 Moreover, a population-based cohort study revealed that serum APOB protein was positively correlated with colorectal cancer and lung cancer among male patients and inversely associated with female breast cancer.36 Li et al37 conducted a retrospective clinical study incorporating 1044 patients with breast cancer and could not find any association between circulating APOB levels and OS. In addition, a recent study38 assessed the serum APOB protein in relation to clinical features and prognosis and found that high APOB levels were significantly associated with improved OS and cancer-specific survival in patients with colorectal cancer but were not statistically associated with TNM stage. In this study, we specifically explored the status of serum APOB protein in patients with LGG and normal individuals and associations with clinicopathological features. Our study demonstrated that the levels of APOB protein were significantly lower in patients with LGG than in normal controls, and the expression of APOB protein was significantly associated with histological type and histological-molecular classification. More importantly, this study revealed that the serum APOB concentration could identify LGG from healthy controls with acceptable diagnostic performance (AUC=0.645), indicating that circulating APOB protein might serve as a potential diagnostic biomarker for patients with LGG.
APOB protein is a dietary lipid transporter that plays a vital role in lipid metabolism.36,39 Recent studies have demonstrated that lipid metabolism reprogramming is implicated in the involvement of cell growth, angiogenesis, signal transduction and progression of several cancers,40,41 including glioma.42,43 Moreover, Chimienti et al44 reported that APOA inhibited the secretion of IL-6 by interfering with lipopolysaccharide signaling in a human astrocytoma cell line. Hashemi et al45 discovered that APOA1 was a key oncogene in malignant astrocytoma tumors due to its functional consequences in tumor growth, angiogenesis and migration. As described previously, APOB is mainly responsible for the transportation of lipids, and methylation of APOB leads to the accumulation of extracellular lipids. Extracellular lipid loading contributes to glioma angiogenesis mediated by the hypoxic paracrine signaling pathway.46 Based on the above studies, we can infer that APOB is a novel therapeutic target for the treatment of glioma from the perspective of lipid metabolism reprogramming.
However, there are two unavoidable limitations in our study. First, we could not determine the prognostic significance of serum APOB protein among 150 cases of patients with LGG because obtaining survival information of all the patients in the short run is not an easy task. Due to the limited number of normal brain tissues in both the TCGA and CGGA databases, we could not compare the difference in APOB mRNA or DNA methylation between LGG and normal brain tissues. Hence, in our future work, we will continue the inquiry about patient survival information and further investigate the potential biological role of APOB in LGG by a series of biomedical experiments.
Conclusion
The expression of APOB protein is downregulated in patients with LGG compared with normal individuals. APOB mRNA expression is negatively regulated by DNA methylation. APOB mRNA and APOB DNA methylation play an essential role in the prognosis of patients with LGG. In addition, APOB hypomethylation is an independent risk factor for unfavorable survival in patients with LGG. Collectively, APOB methylation may serve as a novel and promising prognostic biomarker for patients with LGG.
Funding Statement
This work was financially supported by the National Natural Science Fund of China (NSFC; Grant No. 81660211; 81960230).
Data Sharing Statement
All data in this study are available from the corresponding author upon reasonable request.
Disclosure
The authors confirm that there are no conflicts of interest.
References
- 1.Guan X, Vengoechea J, Zheng S, et al. Molecular subtypes of glioblastoma are relevant to lower grade glioma. PLoS One. 2014;9:e91216. doi: 10.1371/journal.pone.0091216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro Oncol. 2018;20:v1–v86. doi: 10.1093/neuonc/noy131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126 [DOI] [PubMed] [Google Scholar]
- 4.Huang SP, Chan YC, Huang SY, Lin YF. Overexpression of PSAT1 gene is a favorable prognostic marker in lower-grade gliomas and predicts a favorable outcome in patients with IDH1 mutations and chromosome 1p19q codeletion. Cancers. 2019;12:13. doi: 10.3390/cancers12010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malta TM, de Souza CF, Sabedot TS, et al. Glioma CpG island methylator phenotype (G-CIMP): biological and clinical implications. Neuro Oncol. 2018;20:608–620. doi: 10.1093/neuonc/nox183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gusyatiner O, Hegi ME. Glioma epigenetics: from subclassification to novel treatment options. Semin Cancer Biol. 2018;51:50–58. doi: 10.1016/j.semcancer.2017.11.010 [DOI] [PubMed] [Google Scholar]
- 7.Switzeny OJ, Christmann M, Renovanz M, Giese A, Sommer C, Kaina B. MGMT promoter methylation determined by HRM in comparison to MSP and pyrosequencing for predicting high-grade glioma response. Clin Epigenetics. 2016;8:49. doi: 10.1186/s13148-016-0204-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briand J, Nadaradjane A, Bougras-Cartron G, Olivier C, Vallette FM, Cartron PF. Diuron exposure and Akt overexpression promote glioma formation through DNA hypomethylation. Clin Epigenetics. 2019;11:159. doi: 10.1186/s13148-019-0759-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Yan Y, Zhou J, et al. Clinical prognostic value of isocitrate dehydrogenase mutation, O-6-methylguanine-DNA methyltransferase promoter methylation, and 1p19q co-deletion in glioma patients. Ann Transl Med. 2019;7:541. doi: 10.21037/atm.2019.09.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feldheim J, Kessler AF, Monoranu CM, Ernestus RI, Lohr M, Hagemann C. Changes of O(6)-Methylguanine DNA Methyltransferase (MGMT) promoter methylation in glioblastoma relapse-a meta-analysis type literature review. Cancers. 2019;11:1837. doi: 10.3390/cancers11121837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathur R, Zhang Y, Grimmer MR, et al. MGMT promoter methylation level in newly diagnosed low-grade glioma is a predictor of hypermutation at recurrence. Neuro Oncol. 2020. doi: 10.1093/neuonc/noaa059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batais MA, Almigbal TH, Shaik NA, Alharbi FK, Alharbi KK, Ali KI. Screening of common genetic variants in the APOB gene related to familial hypercholesterolemia in a Saudi population: a case-control study. Medicine. 2019;98:e14247. doi: 10.1097/MD.0000000000014247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohwada T, Sakamoto T, Kanno Y, et al. Apolipoprotein B correlates with intra-plaque necrotic core volume in stable coronary artery disease. PLoS One. 2019;14:e212539. doi: 10.1371/journal.pone.0212539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alves AC, Benito-Vicente A, Medeiros AM, Reeves K, Martin C, Bourbon M. Further evidence of novel APOB mutations as a cause of familial hypercholesterolaemia. Atherosclerosis. 2018;277:448–456. doi: 10.1016/j.atherosclerosis.2018.06.819 [DOI] [PubMed] [Google Scholar]
- 15.Lee G, Jeong YS, Kim DW, et al. Clinical significance of APOB inactivation in hepatocellular carcinoma. Exp Mol Med. 2018;50:147. doi: 10.1038/s12276-018-0174-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song J, Liu YD, Su J, Yuan D, Sun F, Zhu J. Systematic analysis of alternative splicing signature unveils prognostic predictor for kidney renal clear cell carcinoma. J Cell Physiol. 2019;234:22753–22764. doi: 10.1002/jcp.28840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S, Li X, Wen X, et al. Prognostic nomogram integrated baseline serum lipids for patients with non-esophageal squamous cell carcinoma. Ann Transl Med. 2019;7:548. doi: 10.21037/atm.2019.09.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu JX, Yuan Q, Min YL, et al. Apolipoprotein A1 and B as risk factors for development of intraocular metastasis in patients with breast cancer. Cancer Manag Res. 2019;11:2881–2888. doi: 10.2147/CMAR.S191352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehrer S, Rheinstein PH, Rosenzweig KE. Glioblastoma multiforme: fewer tumor copy number segments of the SGK1 gene are associated with poorer survival. Cancer Genomics Proteomics. 2018;15:273–278. doi: 10.21873/cgp.20085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brat DJ, Verhaak RG, Aldape KD, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia D, Li S, Li D, Xue H, Yang D, Liu Y. Mining TCGA database for genes of prognostic value in glioblastoma microenvironment. Aging. 2018;10:592–605. doi: 10.18632/aging.101415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hess AS, Hess JR. Understanding tests of the association of categorical variables: the Pearson chi-square test and Fisher’s exact test. Transfusion. 2017;57:877–879. doi: 10.1111/trf.14057 [DOI] [PubMed] [Google Scholar]
- 23.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wheeler DA, Roberts LR; Cancer GARN. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341. doi: 10.1016/j.cell.2017.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou D, Alver BM, Li S, et al. Distinctive epigenomes characterize glioma stem cells and their response to differentiation cues. Genome Biol. 2018;19:43. doi: 10.1186/s13059-018-1420-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Z, Tan H, Zhao W, et al. Integrative analysis of DNA methylation and gene expression profiles identifies MIR4435-2HG as an oncogenic lncRNA for glioma progression. Gene. 2019;715:144012. doi: 10.1016/j.gene.2019.144012 [DOI] [PubMed] [Google Scholar]
- 27.Huang GH, Du L, Li N, et al. Methylation-mediated miR-155-FAM133A axis contributes to the attenuated invasion and migration of IDH mutant gliomas. Cancer Lett. 2018;432:93–102. doi: 10.1016/j.canlet.2018.06.007 [DOI] [PubMed] [Google Scholar]
- 28.Baumert BG, Hegi ME, van den Bent MJ, et al. Temozolomide chemotherapy versus radiotherapy in high-risk low-grade glioma (EORTC 22033–26033): a randomised, open-label, phase 3 intergroup study. Lancet Oncol. 2016;17:1521–1532. doi: 10.1016/S1470-2045(16)30313-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryken TC, Parney I, Buatti J, Kalkanis SN, Olson JJ. The role of radiotherapy in the management of patients with diffuse low grade glioma: a systematic review and evidence-based clinical practice guideline. J Neurooncol. 2015;125:551–583. doi: 10.1007/s11060-015-1948-1 [DOI] [PubMed] [Google Scholar]
- 30.Bell EH, Zhang P, Fisher BJ, et al. Association of MGMT promoter methylation status with survival outcomes in patients with high-risk glioma treated with radiotherapy and temozolomide: an analysis from the NRG oncology/RTOG 0424 trial. JAMA Oncol. 2018;4:1405–1409. doi: 10.1001/jamaoncol.2018.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao P, Ostrom QT, Stetson L, Barnholtz-Sloan JS. Models of epigenetic age capture patterns of DNA methylation in glioma associated with molecular subtype, survival, and recurrence. Neuro Oncol. 2018;20:942–953. doi: 10.1093/neuonc/noy003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong X, Deng Q, Nie X, et al. Downregulation of HTATIP2 expression is associated with promoter methylation and poor prognosis in glioma. Exp Mol Pathol. 2015;98:192–199. doi: 10.1016/j.yexmp.2015.01.013 [DOI] [PubMed] [Google Scholar]
- 33.Pischon T, Girman CJ, Sacks FM, Rifai N, Stampfer MJ, Rimm EB. Non-high-density lipoprotein cholesterol and apolipoprotein B in the prediction of coronary heart disease in men. Circulation. 2005;112:3375–3383. doi: 10.1161/CIRCULATIONAHA.104.532499 [DOI] [PubMed] [Google Scholar]
- 34.Goff DCJ, Lloyd-Jones DM, Bennett G, et al. ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American college of cardiology/American heart association task force on practice guidelines. Circulation. 2013;2014(129):S49–S73. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Liu Z, Wu Y, et al. Status of lipid and lipoprotein in female breast cancer patients at initial diagnosis and during chemotherapy. Lipids Health Dis. 2018;17:91. doi: 10.1186/s12944-018-0745-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borgquist S, Butt T, Almgren P, et al. Apolipoproteins, lipids and risk of cancer. Int J Cancer. 2016;138:2648–2656. doi: 10.1002/ijc.30013 [DOI] [PubMed] [Google Scholar]
- 37.Li X, Tang H, Wang J, et al. The effect of preoperative serum triglycerides and high-density lipoprotein-cholesterol levels on the prognosis of breast cancer. Breast. 2017;32:1––6.. doi: 10.1016/j.breast.2016.11.024. [DOI] [PubMed] [Google Scholar]
- 38.Sirniö P, Väyrynen JP, Klintrup K, et al. Decreased serum apolipoprotein A1 levels are associated with poor survival and systemic inflammatory response in colorectal cancer. Sci Rep-Uk. 2017;7:5374. doi: 10.1038/s41598-017-05415-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cefalu AB, Pirruccello JP, Noto D, et al. A novel APOB mutation identified by exome sequencing cosegregates with steatosis, liver cancer, and hypocholesterolemia. Arterioscler Thromb Vasc Biol. 2013;33:2021–2025. doi: 10.1161/ATVBAHA.112.301101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Q, Luo Q, Halim A, Song G. Targeting lipid metabolism of cancer cells: a promising therapeutic strategy for cancer. Cancer Lett. 2017;401:39–45. doi: 10.1016/j.canlet.2017.05.002 [DOI] [PubMed] [Google Scholar]
- 41.Luo X, Cheng C, Tan Z, et al. Emerging roles of lipid metabolism in cancer metastasis. Mol Cancer. 2017;16:76. doi: 10.1186/s12943-017-0646-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou L, Wang Z, Hu C, et al. Integrated metabolomics and lipidomics analyses reveal metabolic reprogramming in human glioma with IDH1 mutation. J Proteome Res. 2019;18:960–969. doi: 10.1021/acs.jproteome.8b00663 [DOI] [PubMed] [Google Scholar]
- 43.Ahmad F, Patrick S, Sheikh T, et al. Telomerase reverse transcriptase (TERT) - enhancer of zeste homolog 2 (EZH2) network regulates lipid metabolism and DNA damage responses in glioblastoma. J Neurochem. 2017;143:671–683. doi: 10.1111/jnc.14152 [DOI] [PubMed] [Google Scholar]
- 44.Chimienti G, Mezzapesa A, Liuzzi GM, Latronico T, Pepe G. Apolipoprotein(a) inhibits lipopolysaccharide-induced IL-6 secretion in human astrocytoma cell line by interfering with lipopolysaccharide signaling. Inflamm Res. 2011;60:329–335. doi: 10.1007/s00011-010-0272-7 [DOI] [PubMed] [Google Scholar]
- 45.Hashemi M, Pooladi M, Razi AS. The investigation of changes in proteins expression (Apolipoprotein A1 and albumin) in malignant astrocytoma brain tumor. J Cancer Res Ther. 2014;10:107–111. doi: 10.4103/0973-1482.131413 [DOI] [PubMed] [Google Scholar]
- 46.Offer S, Menard JA, Perez JE, et al. Extracellular lipid loading augments hypoxic paracrine signaling and promotes glioma angiogenesis and macrophage infiltration. J Exp Clin Cancer Res. 2019;38:241. doi: 10.1186/s13046-019-1228-6 [DOI] [PMC free article] [PubMed] [Google Scholar]