Abstract
Complex nervous systems have a modular architecture, whereby reiterative groups of neurons (“modules”) that share certain structural and functional properties are integrated into large neural circuits. Neurons develop from proliferating progenitor cells that, based on their location and time of appearance, are defined by certain genetic programs. Given that genes expressed by a given progenitor play a fundamental role in determining the properties of its lineage (i.e., the neurons descended from that progenitor), one efficient developmental strategy would be to have lineages give rise to the structural modules of the mature nervous system. It is clear that this strategy plays an important role in neural development of many invertebrate animals, notably insects, where the availability of genetic techniques has made it possible to analyze the precise relationship between neuronal origin and differentiation since several decades. Similar techniques, developed more recently in the vertebrate field, reveal that functional modules of the mammalian cerebral cortex are also likely products of developmentally defined lineages. We will review studies that relate cell lineage to circuitry and function from a comparative developmental perspective, aiming at enhancing our understanding of neural progenitors and their lineages, and translating findings acquired in different model systems into a common conceptual framework.
Keywords: mouse, zebrafish, Xenopus, Drosophila, neural progenitor, proliferation, cell lineage, cell fate, circuitry
Introduction
Highly ordered connections of neurons represent the structural basis of the complex brain circuitry controlling animal behavior. Neurons are highly diverse, and a continual effort in the neuroscience community lies in further discerning individual neuronal classes based on a number of molecular, structural, and/or physiological criteria (Darmanis et al. 2015; Tasic et al., 2016; Zhang et al., 2014). Aside from deepening our insight into the diversity of neural cell types, much effort is also directed to identify communalities between groups of neurons that form units of function. Thus, complex nervous systems of vertebrates and invertebrates alike can be subdivided into modules, defined as groups of neurons that share certain structural and functional properties. Modules, such as the anatomically distinct nuclei of the vertebrate brain stem, may differ from each other, each nucleus being defined by a specific population of afferent or efferent fibers. In other cases, modules can be reiterative, consisting of numerically and morphologically similar neuronal clusters, as for example the glomeruli of the olfactory system in vertebrate or insect brains, or the columns of the mammalian cerebral cortex (Chen and Shepherd, 2005; Harzsch and Krieger, 2018; Kaas, 2012).
An important question concerns the role of development during the formation of neuronal modules. Neurons develop from progenitor cells that, based on their location and time of appearance within the neural primordium, are defined by certain genetic programs. It stands to reason that the set of genes expressed by a given progenitor may play a fundamental role in determining the structural and functional phenotype of the neurons descended from that progenitor. Is it possible to relate a brain module, as defined structurally and functionally in the mature brain, to a specific genetically distinct neural progenitor?
In the insect brain, the question can be answered in the positive, as repeatedly reviewed in the recent literature (Boyan and Williams, 2011; Jiang and Reichert, 2014; Lin and Lee, 2012; Spindler and Hartenstein, 2010), and summarized below. Most parts of the insect brain develop from a relatively small number of stem cell-like progenitors, called neuroblasts, which appear early in the embryo, and which each produce an invariant lineage of neurons and/or glia. Insect neural lineages represent structural modules, whereby neurons of a lineage stay in close contact, projecting their axons in one or two compact tracts, and typically forming terminal branches in common brain neuropil compartments (Hartenstein et al., 2008; Ito and Awasaki, 2008; Larsen et al., 2009). We know a lot about the relationship between lineage and brain module in the insect brain because genetic or cellular techniques that label lineages (i.e., “clones”) have been available since several decades (del Valle Rodriguez et al., 2011; Garcia-Bellido and Merriam, 1971; Griffin et al., 2014; Lee and Luo, 2001). For the mammalian brain, clonal techniques that allow one to reconstruct the lineages of individual progenitors at high temporal and spatial resolution have recently been introduced (Bribian et al., 2016; Gao et al., 2014; Zong et al., 2005). As a result of these studies there is a renewed interest in the question of how the progeny of defined neural progenitors relates to modules of the brain. A growing body of evidence supports the notion that functional properties of neurons are dependent on their lineage relationships (He et al., 2015; Li et al., 2012; Ohtsuki et al., 2012; Tarusawa et al., 2016; Yu et al., 2009; 2012).
In the present review we will address a number of questions that focus on the relationship between cell lineage and modular brain architecture in different model systems. How do progenitor cells in different vertebrates and invertebrates divide over the course of neural development? At what time point can we define, in vertebrates, progenitors that give rise to structurally or functionally defined brain modules? How do such progenitors compare to invertebrate neuroblasts? Answers to these questions will contribute to our understanding of how genetic information is able to shape neural circuits and, thereby, animal behavior.
Neural progenitor proliferation and lineages: Drosophila and other invertebrate systems
Neuroblast specification and internalization
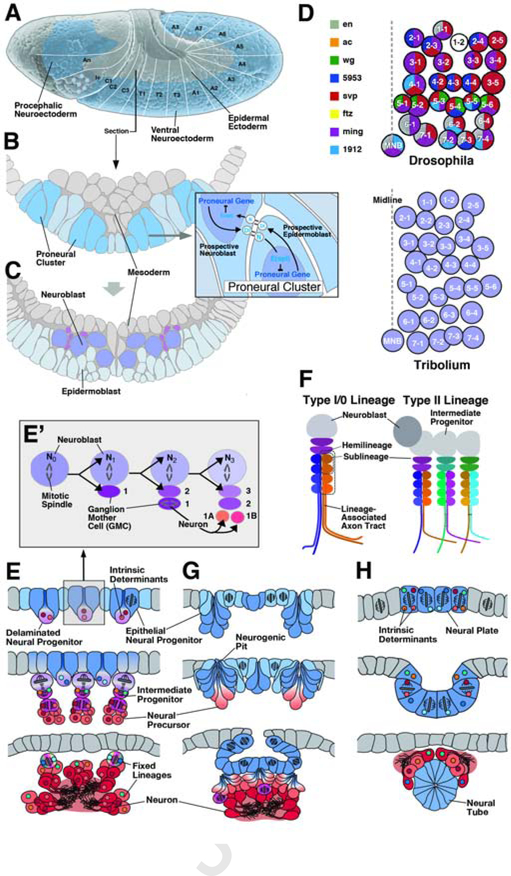
At an early stage of embryonic development, concomitant or even preceding gastrulation, a restricted domain within the ectodermal layer, the neuroectoderm, is endowed with the capability to generate neural progenitor cells. This step involves the function of maternal factors and conserved signaling pathways, including Wnt and BMP, which delineate the size and position of the neuroectoderm within the embryo (reviewed in Hartenstein and Stollewerk, 2015; Fig.1A). Subsequently, during the phase called neurulation, neural progenitors are specified within the neurectoderm and then become internalized. In Drosophila and other insects, neural progenitors (neuroblasts) delaminate to form a lose layer of cells in between the ectoderm and mesoderm (Fig.1B, C). Those cells of the neurectoderm that stay within the neuroectoderm maintain the potential to produce more neuroblasts at a later timepoint; once the full complement of neuroblast has become internalized, cells left behind at the surface give rise to part of the epidermis (epidermoblasts). The pattern of neuroblasts is invariant: each neuroblast forms a uniquely identifiable cell that appears at the same time and position in every individual of a given species (Fig.1D, top). Even comparing different insect species, such as Drosophila, grasshopper, and flour beetle, neuroblast patterns are almost identical (Bate, 1976; Biffar and Stollewerk, 2014; Thomas et al., 1984; Fig.1D).
Figure 1.
Structure and origin of neural lineages in Drosophila. (A) Lateral surface view of Drosophila embryo at a stage shortly after gastrulation. Large ectodermal domain forming part of the head (procephalic neuroectoderm; origin of the brain) and the trunk (ventral neuroectoderm; origin of ventral nerve cord) gives rise to neural lineages. White lines and lettering delineate segmental units (An antennal segment; Ic intercalary segment; C2–3 gnathal segments; T1-T3 thoracic segments; A1–8 abdominal segments). (B, C) Cross sections of ventral neuroectoderm before (B) and after (C) neuroblast segregation. Expression of bHLH genes of the Achaete-Scute-Complex divide the neuroectoderm into an orthogonal pattern of proneural clusters (dark lilac). Notch-Delta signaling within proneural clusters (inset) selects one cell that delaminates as a neuroblast (C). (D) Top: Pattern of neuroblasts of one Drosophila abdominal hemisegment. Each neuroblast is a unique cell (1–1 to 7–4), defined by its time of delamination, position, and combinatorial expression of transcription factors and signaling molecules (see key to color code on the left of panel; after Doe, 1992, with permission). The neuroblast pattern is highly conserved among different insect taxa, as exemplified by the coleopteran Tribolium castaneum (bottom of panel; after Biffar and Stollewerk, 2014, with permission). (E, E’) Schematic cross-sections of insect embryo at sequential stages of development, illustrating characteristics of neural progenitors in insects. Top panel depicts epithelial neuroectoderm (blue) from which neural progenitors (lilac) originate. Insect neural progenitors (neuroblasts) delaminate and initiate a series of determinate, asymmetric divisions, giving rise to intermediate progenitors (magenta) called ganglion mother cells (GMCs). GMCs typically undergo one more mitosis into two neuronal (or glial) precursors (orange) which subsequently differentiate into neurons (red). Inset (E’) shows details of neuroblast division. Freshly delaminated neuroblast (N0) divides asymmetrically into apical second generation neuroblast (N1,2…) and basal ganglion mother cell (GMC1,2…). Ganglion mother cells undergo one terminal mitosis into molecularly different neurons (1A, 1B). (F) Structure of neural lineages in insects. Canonical proliferation pattern, delineated in panel (E), results in a type I lineage. Neurons born as “A” daughter cells and “B” daughter cells of ganglion mother cells represent the A and B hemilineages. Neurons born sequentially in a defined time window define a sublineage. Certain subsets of neuroblasts of the brain generate type II lineages, whereby neuroblasts produce intermediate progenitors, which, in turn, each behave like a type I neuroblasts, generating neurons via ganglion mother cells. (G, H) Generation and proliferation of neural progenitors in chelicerates (spiders; G) and urochordates (sea squirts; H). In these phyla, parts of the neuroectderm invaginate as multiple small “neurogenic pits” (in chelicerates, as well as other arthropod taxa) or single large neural tube (in urochordates, as well as the closely related vertebrates). Prior to the birth of neural precursors or intermediate progenitors, the neuroepithelium (blue) undergoes a phase of abundant symmetric divisions.
The balance between how many neurectodermal cells delaminate at any given moment as neuroblasts, as well as the precise pattern of these cells, is determined by cell-cell signaling within the neuroectoderm. In the center of this complex genetic mechanism resides the Notch signaling pathway, which allocates the expression of proneural genes (bHLH transcription factors enabling cells to adopt a neural fate) to the delaminating neuroblasts (inset in Fig.1B; for review see Hartenstein and Wodarz, 2013; Huang et al., 2014; Kageyama et al., 2009; Quan and Hassan, 2005). Loss of Notch activity results in a large excess of delaminating neuroblasts at an early time point; activation of the pathway has the opposite effect. At later stages in neural development, the Notch pathway exerts additional effects. Well known among these are the differentiation between the two hemilineages generated by sequential division of neuroblast progeny, called ganglion mother cells (GMCs), and the promotion of self renewal in type 2 neuroblasts (Kumar et al., 2009; Lin et al., 2012; Truman et al., 2010; see below).
Neuroblasts appear in two broad regions of the embryo. The ventral neuroectoderm, stretching out along the trunk ectoderm and numbering approximately 3500 cells, produces the neuroblasts of the ventral nerve cord (Doe, 1992; Hartenstein and Campos-Ortega, 1984; 1985; Hartenstein et al., 1987; Younossi-Hartenstein et al., 1996). The head (procephalic) neurectoderm, comprised of several hundred cells, located in the anterior-dorsal part of the ectoderm, gives rise to neuroblasts that form the brain (Fig.1A). Neuroblasts are organized segmentally, with each segment comprising a set of approximately 60 neuroblasts (30 on each side; Fig.1D). Overall, the ventral neurectoderm generates approximately 750 neuroblasts, and the procephalic neurectoderm 200. Neuroblasts do not delaminate all at once, but form several groups (“neuroblast waves”) which move in sequentially following a tightly controlled pattern.
Neuroblasts produce fixed lineages by asymmetric division
First defined by Whitman (1878, 1887) and described in detail by Wheeler (1891) in embryos of grasshoppers based on histological sections, neuroblasts appear as large pale cells which “bud off” smaller, more strongly labeled cells, the neurons and glia of the embryonic nervous system. The idea that a neuroblast produces a readily identifiable cluster of neurons through a series of asymmetric divisions was introduced through in vitro and in vivo studies (Poulson, 1950; Seecof et al.,1973). The fixed nature of insect neural lineages, i.e., the concept that neuroblasts organize into stereotypic arrays and undergo an invariant number of mitotic divisions, producing neurons with precisely predetermined fates, was demonstrated by later studies (Booker and Truman, 1987; Doe and Goodman, 1985; Goodman and Spitzer, 1979; Taghert et al., 1984).
Neuroblasts have two proliferative phases. The first takes place in the embryo, where most neuroblasts undergo 5–8 divisions, producing 10–16 neurons per lineage (Hartenstein et al., 1987; Larsen et al., 2009). Division is asymmetric, following what has been termed a “stem cell mode” of mitosis. Here, as a result of the positioning and orientation of the mitotic spindle in the dividing cell, the two daughter cells are overtly different in size and location. In contrast, symmetric division refers to a process where the two daughter cells are of equal size, and obtain the same position in relation to the layer they form part of or are located next to. Note that in many cases, there is a third scenario where cell division can appear symmetric at the structural level, but where molecular factors (detectable only by special staining protocols) are distributed unevenly among the progeny. We will in the following refer to this type of division as “molecularly asymmetric”, to distinguish it from the (structurally) asymmetric type.
The asymmetric division of a neuroblast yields one daughter cell that is large and remains in contact with the overlying ectoderm, and a second one that is small, and obtains a position at the basal surface of its larger sibling (Fig.1E, E’). The large daughter cell resumes the proliferative fate of the mother neuroblast, and the other, small daughter cell differentiates as a neuron or glial cell, either following one more division, or directly. Recent works (Baumgardt et al., 2014) showed that the first one of these two modes of division (called “type I”) prevails only during the initial phase of neuroblast mitotic activity. During these type I divisions, the small daughter cell, which is called “ganglion mother cell”, undergoes one more molecularly asymmetric division, generating an “A” daughter neuron and a “B” daughter neuron (Truman et al. 2010; Fig.1E’, F). The cluster of sequentially produced “A” neurons and “B” neurons form their own “A” hemilineage and “B” hemilineage, respectively. After a certain number of divisions (typically 3–4), the neuroblast switches to a “type 0” mode, where the small daughter cell differentiates directly into a neuron (Baumgardt et al., 2014). Within a lineage or hemilineage, neurons are further divided according to birth date into smaller groups, called sublineages (Fig.1F).
In the late embryo, most neuroblasts enter a phase of mitotic quiescence that lasts for approximately 24 hours. Subsequently, they become active again, entering into their secondary, larval phase of neuron/glia production. Over the course of approximately three days, they undergo an estimated 75 rounds of mitosis, generating 150 neurons per lineage (Bello et al., 2008; Larsen et al., 2009). As in the embryo, most larval neuroblast divisions follow the type I pattern, giving birth to series of ganglion mother cells that undergo one more division to produce two hemilineages. It is not known whether, at a late larval stage, neuroblasts enter a phase of type 0 proliferation that omits ganglion mother cells. Secondary hemilineages (and, probably, primary hemilineages as well) typically have different fates, and form their own tract and projection domain. As a result, most lineages described for the ventral nerve cord emit two fibre bundles with different trajectories, corresponding to the two hemilineages (Harris et al., 2015; Truman et al., 2004; see below). In many lineages, particularly those of the brain, one hemilineage undergoes apoptotic cell death, leaving the lineage with only one hemilineage, and one fibre bundle/projection domain.
A subset of 8 neuroblasts, all localized close to the dorso-medial rim of a brain hemisphere, show a mode of proliferation that has been called type II. Here, neuroblasts divide asymmetrically, but yield an intermediate progenitor cell (IMP) which continues to undergo several rounds of asymmetric divisions before generating neurons, akin to what a regular (type I) neuroblast does (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008; Kang and Reichert, 2015; Fig.1F). As a result, lineages of type II neuroblasts are larger than those of type I neuroblasts, numbering 400–500 neurons per lineage. The dichotomous choice of type II neuroblast progeny to self renew or become IPM is also controlled by Notch activity; loss of N causes premature stop in self renewal and conversion to IMPs in these cells (Bowman et al., 2008; Wang et al., 2006).
Early neurogenesis in other invertebrates
The formation of neuronal modules from indvidual, genetically unique stem cells that divide in a fixed pattern, as described for Drosophila above, is a derived feature encountered in only a few invertebrate clades. Among arthropods, it exists in insects and crustaceans (Fig.1E). In other, more basally branching arthropods, like chelicerates and myriapods, the nervous system arises from groups (“pits”) of contiguous neuroectodermal cells that invaginate or ingress (Hartenstein and Stollewerk, 2015; Stollewerk, 2016; Fig.1G). Proliferation occurs mainly prior to invagination of the pits, and there is currently no evidence that individual pits are formed from single (epithelial) cells in a fixed lineage mechanism. Interestingly, regarding their overall number and spatial distribution, the pits bear a striking resemblance to the neuroblasts of insects. Furthermore, the cassette of proneural genes in conjunction with the Notch signaling pathway control the size of pits and its neuronal descendants (Stollewerk, 2016). It is possible that individual pits also form distinct neuronal modules, but this remains to be shown using markers for specific neuronal types. Another open question is how the conceptual framework of sequentially expressed cell fate determinants (transcription factors), as established for Drosophila (see below), translates to the more basal arthropod scenario.
Outside the arthropods, stem-cell like neural progenitors generating fixed lineages have been observed in hirudinea (leeches; Zhang and Weisblat, 2005), and may well exist in other annelids as well. A number of clades, among them rotifers, tardigrades, and nematodes are eutelic, which means that all cells of the body are generated in a fixed lineage pattern. For the nematode C. elegans, proliferation, cell movement and lineage relationships between cells have been reconstructed in detail (Hobert, 2010; Sulston et al., 1983; Wadsworth and Hedgecock, 1992). As described for Drosophila, neurogenesis in C. elegans is controlled by a great diversity of intrinsic determinants that are routed to specific neuronal precursors through asymmetric divisions. However, a mechanism by which the neuroectoderm forms “generic” neuroblasts that produce distinct neuronal modules does not exist. Instead, all the way towards a late stage of proliferation, individual cells generate quite distinct neural (or even non-neural) cell types. Neurons belonging to a functionally characterized class or circuit are not typically related by lineage (Hobert, 2010).
Interestingly, larval forms of urochordates (sea squirts), considered to be the sister group of vertebrates, also possess miniaturized nervous systems generated in a fixed lineage mechanism. The neurectoderm in these animals forms a dorsal neural plate that, similar to that of vertebrates described below, invaginates to form a neural tube (Fig.1H). Around the invagination process (“neurulation”) the neural plate possesses 40–60 cells arranged in a regular, symmetric pattern of orthogonal rows and columns (Nicol and Meinertzhagen, 1988a, b). Following neurulation, cells undergo another 1–4 rounds of structurally symmetric division, forming small clones of neural cells that add up to a total of approximately 350. About half of these cells remain undifferentiated epithelial (ependymal) cells; the remainder differentiates into different types of neurons. Some cases are documented where individual neural progenitors of the neural plate generate clones of like cells; examples are the pressure receptors (“coronet cells”) derived from progenitors a9.33 and a9.37 (Eakin and Kuda, 1971; Nicol and Meinertzhagen, 1991), or the ocellar photoreceptors that descend from the corresponding pair of progenitors (a9.33, a9.37) of the opposite side (Nicol and Meinertzhagen, 1991). However, it is not clear yet (and appears unlikely) if neural lineages of the urochordate tadpole represent structural or functional modules in general.
In summary, comparative embryological and genetic studies suggest that the last common ancestor of bilaterian animals (“Urbilateria”) possessed a proliferative neuroectoderm formed by neural progenitor cells that give rise to the nervous system. Becoming postmitotic, cells delaminate from the neuroepithelium and differentiate into neurons. With increasing complexity animals evolved mechanisms to increase neuron numbers. One mechanism has delaminating cells maintain their mitotic activity, thereby functioning as neural progenitors (neuroblasts, IMPs). Another device serving the same purpose is to invaginate the neuroectoderm, whereby either small domains (“pits”) move inside one by one, or, as seen in the case of chordates, the entire neuroectoderm folds inside the body to become a neural tube. In some instances, the proliferation pattern of neural progenitors within the neurectoderm becomes highly invariant, leading to the production of fixed lineages that form distinct structural/functional modules of the nervous system. Such a mechanism probably evolved multiple times independently; so far it has been described for subclades of arthropods and annelids. To what extent invariant lineages act as structural modules in vertebrates will be discussed below.
Neural progenitor proliferation and lineages: Vertebrates
Formation and growth of the neuroectoderm
The neuroectoderm of vertebrates forms a morphologically distinct domain, called neural plate, in the dorsal ectoderm of the embryo (Fig.2A). In the amphibian Xenopus laevis, direct cell counts yielded approximately 2900 cells (Hartenstein, 1989) for the neural plate, visible in the stage 13 embryo, which puts it in a similar range as Drosophila. For zebrafish, a similar number can be estimated for the neural plate of the two somite-stage embryo, based upon the analysis of cell proliferation by Kimmel and colleagues (Kane and Kimmel, 1993; Kimmel et al., 1994). Interestingly, the neural plate of the E7.5 mouse embryo, despite of the much larger number of neurons eventually produced, may start out with a similar number of cells: cell counts of the E7.5 embryo yielded approximately 8,000 cells for the ectoderm as a whole (Snow, 1977), of which the neuroectoderm can be assumed to account for less than half. As the neural plate invaginates to form the neural tube (Fig.2B, C), its neuroepithelial cells (“neuroepithelial progenitors”) continue to divide symmetrically, resulting in an overall increase in surface area (Fig.2D1). This expansion phase differs significantly among anamniotes (fishes, amphibians) and amniotes (birds, reptiles, mammals). The former hatch as motile larvae at an early, “premature” stage. The first neurons, called primary neurons, are born as early as the neural plate stage, following only a few rounds of mitoses of neuroepithelial progenitors (Hartenstein, 1989; Papan and Campos-Ortega, 1997; 1999; Fig.2D–G). In contrast, in mouse or chicken, the neural primordium increases at a least a thousand fold, to several hundred thousand cells or more, before neurons are born. Thus, in mouse, neuroepithelial progenitors continue to divide symmetrically until stage E10.5 (Fig.2H2) with a cell cycle length of approximately 4–8 hrs (Jiang and Nardelli, 2016; McShane et al., 2015), leaving time for >10 symmetric cell cycles.
Figure 2.
Early neurogenesis in the vertebrate embryo. (A) Lateral surface view of amphibian embryo after gastrulation. Dorsal neuroectoderm (neural plate; purple) gives rise to the brain (anterior neural plate, left) and spinal cord (posterior neural plate, right). (B-D) Cross section of neural plate (B) and neural tube (C). (D1, D2) depict enlarged subsections of neuroepithelium at an early phase (D1) and later phase (D2) of progenitor cell proliferation. The vertebrate neuroectoderm remains configured as a layer of neuroepithelial progenitors surrounding an inner lumen (ventricle). Progenitors initially divide symmetrically, resulting in an expansion of the neuroepithelium (D1). During a later phase, asymmetric (neurogenic) divisions generate primary neurons which delaminate from the ventricular layer (D2), whereas other daughter cells remain at the ventricular surface to continue dividing. (E-G) Small clones of primary neurons (E, F) and neuroepithelial progenitors (G) in Xenopus (E, G; from Hartenstein, 1989, with permission) and zebrafish (F; from Papan and Campos-Ortega, 1997, with permission) labeled by dye injection into individual cells at neural plate stage. (H1–4) Neural proliferation in mouse. Schematic cross sections of neuroepithelium at different stages of development. Early divisions of neural progenitors are symmetric, resulting in a great expansion of the neural tube (H2; prior to E10.5). Between E10.5 and E12.5, neurogenic mitotic activity commences (H3). Individual progenitors, now called apical radial glia (aRG), undergo a series of asymmetric divisions that give rise to 8–9 neural precursors and/or intermediate progenitors (H4; around E16.5). These cells remain in close contact and form an ontogenetic column (see also Fig.5C, D). Intermediate progenitors form the subventricular zone (SVZ). In mouse, these cells typically undergo only one more terminal division. Notch/Delta signaling promotes the fate of ventricular epithelial progenitors, as opposed to neural precursors or intermediate progenitors (inset in H2/3). (I1–2) In large mammals (e.g., primates) with folded cortex the subventricular zone splits into an inner and outer subventricular zone (iSVZ and oSVZ), respectively. Neural progenitors of the ventricular layer lose contact to the pial surface (“truncated radial glia”, tRG); progenitors of the oVZ (“basal radial glia”, bRG) continue to divide for an extended period, producing predominantly neurons of superficial cortical layers (from Nowakowski et al., 2016, with permission). (J1–4) Neurogenesis in the dorsal forebrain of different vertebrate clades (J1: reptiles, amphibians, fishes; J2: birds; J3: rodents; J4: primates; from Cardenas and Borrell, 2019, with permission). In anamniotes and reptiles, epithelial progenitors of the ventricular layer (apical radial glia; blue) directly give rise to postmitotic neural precursors (red) which form a basal neural layer (NL; “direct neurogenesis”). In birds, apical radial glia not only give rise to neural precursors, but also to mitotically active intermediate progenitors (magenta) which populate a subventricular zone (SVZ; “indirect neurogenesis”). This mode of indirect neurogenesis accounts for an increased number of neurons generated from the neuroepithelium. Indirect neurogenesis is even more pronounced in mammals, where neurons form a multilayered cortical plate (CP). Large mammals with a gryrated cortex show a dramatic expansion of intermediate neural progenitors, forming an inner and inner (iSVZ) and outer subventricular zone (oSVZ).
Neural proliferation
Neuronal birth (exit from the cell cycle) and cell cycle dynamics in vertebrates are interrelated in a complex pattern. It appears that neuronal birth is generally coupled to the occurrence of asymmetric divisions, which has been studied in detail for zebrafish and mouse. In the former, proliferation and cell fate could be studied directly by live imaging. Studies on spinal cord (Alexandre et al., 2010), forebrain (Dong et al., 2011) and retina (Baye and Link, 2007; Das et al., 2003; Poggi et al., 2005) over the period from 20h after fertilization to hatching (48–72h) demonstrated that individual neuroepithelial progenitors underwent 1–3 rounds of asymmetric divisions, whereby the mitotic spindle is directed perpendicular or at least at a significant tilt to the apical surface (Fig.2D). Accordingly, clones derived from individual neuroepithelial progenitors contained 2–8 cells, which represented a mixture of neurons and progenitors (Fig.2E–G). For spinal cord and forebrain it could be established that the daughter cell that ended up more apically exited the cell cycle and became a neuron, and the more basal cell continued to cycle as a neuroepithelial progenitor (Alexandre et al., 2010; Fig.2D2). This finding was surprising (and may differ from what happens in other vertebrates, including mouse), since it implies that the presumptive neuroepithelial progenitor has to reintegrate into the epithelium, and the cell fated as neuron has to actively exit (delaminate from) the epithelium.
In the developing telencephalon of the mouse (Florio and Huttner, 2014; Gao et al., 2013; Hartfuss et al., 2001; Jiang and Nardelli, 2016; Kriegstein et al., 2006; Noctor et al., 2004; Sun and Hevner, 2014), asymmetric mitotic activity sets in around E10.5 (Fig.2H3). At this stage, neuroepithelial progenitors start expressing some glial-specific proteins and other molecular markers (e.g., Pax 6; Asami et al., 2011), and are called “apical radial glia” (aRG). Clonal analysis performed on the dorsal telencephalon, which gives rise to the excitatory neurons of the cerebral cortex, indicates that there is a transitory phase between E10 and E12 where symmetric (“proliferative”) and asymmetric (“neurogenic”) divisions occur together. During symmetric division, an aRG generates two daughter aRGs; during asymmetric divisions, only one of the two aRG daughter cells maintains its proliferative fate, whereas the other one loses contact to the apical surface and either exits the cell cycle right away, or continues to divide symmetrically into several neurons. This second type of dividing progenitor (“intermediate progenitor”) no longer forms part of the neuroepithelium, but builds up its own proliferative layer, called the subventricular zone (Fig.2H4). The data for mouse suggest that symmetric versus neurogenic divisions of aRGs are mutually exclusive: once an aRG has entered the asymmetric, neurogenic phase, it continues with this mode of divisions for multiple rounds, generating a progeny of 8–9 cells (Gao et al., 2014). These cells remain in close contact, forming an “ontogenetic column”, a term and concept introduced several decades ago for primate cerebral cortex by Rakic and collaborators (Rakic, 1988; see below).
Proliferation of intermediate progenitors in the developing mouse telencephalon is limited to a single mitosis before exiting the cell cycle; in larger species of mammals, including primates, an extended phase of proliferation of intermediate progenitors is thought to account for the large increase in cortical thickness and cell number (Betizeau et al., 2013; Fietz et al., 2010; Martinez-Cerdeno et al., 2012). Correspondingly, the ontogenetic column in these larger mammals can be expected to be much bigger than the 8–9 cells established for mouse. Aside from the increased number of divisions of intermediate progenitors, another layer of complexity is added to the process of corticogenesis in larger mammals which, typically, have a folded (gyrencephalic) cerebral cortex: the subventricular zone splits into two, an inner and outer subventricular zone (iSVZ, oSVZ). The oSVZ is populated by former apical radial glial cells that have lost contact to the ventricular surface, but still project a long process towards the basal (pial) surface (“basal radial glia”; Fig.2I). At the same time, the apical radial glia of the ventricular layer becomes “truncated”, losing contact to the pial surface (Fig.2I). Subsequently, radial glia of the oSVZ proliferate actively, increasing their own number and generating cortical neurons predominantly destined to populate the superficial cortical layers. Formation of the oSVZ is thought to be essential for the tangential spread of cortical neurons underlying the formation of gyri (Kriegstein et al., 2006; Nowakowski et al., 2016; Reillo et al., 2011).
The evolution of corticogenesis among vertebrates is depicted in a way, illustrated in Fig.2J (from Cardenas and Borrell, 2019), that assumes as a starting point a scenario where postmitotic neural precursor cells move out of the ventricular layer and differentiate as neurons (“direct neurogenesis”; Fig.2J1). A subventricular zone, consisting of proliferating intermediate progenitors, is absent. This mode of neurogenesis in general, and of corticogenesis in particular, is encountered in fishes, amphibians, and reptiles. It is thought that neurons populating the simple telencephalic cortex in these animals show characteristics of the deep layer neurons of the mammalian neocortex (Cardenas and Borrell, 2019). Beginnning with birds one encounters subventricular intermediate progenitors that form neurons after a few rounds of division (“indirect neurogenesis”; Fig.2J2). The same mode of cortex formation persists in many mammalian taxa, including rodents (Fig.2J3). Here, apical radial glia and intermediate progenitors undergo a prolonged phase of proliferation, giving rise to neurons that assemble into multiple layers. In larger mammals there appears a second stratum of intermediate progenitors, constituting the outer subventricular zone, which allows for the generation of much larger numbers of neurons of preferentially outer layer identity, and the folding of the cortex into gyri and sulci (Fig.2J4).
Proneural genes and Notch signaling in vertebrate neurogenesis
The pattern of asymmetric divisions in vertebrate neurogenesis, that is, the timing and spatial distribution of cells that withdraw from the cell cycle and differentiate, is controlled by similar molecular mechanisms as those discovered in Drosophila. Homologs of the bHLH proneural genes, as well as components of the Notch signaling pathway, appear in a complex pattern in the proliferating neuroepithelium of the brain and spinal cord. Proneural genes are among the first in a long cascade of determinants that provide cells with the potential to express particular neural or glial fates (Bertrand et al., 2002; Castro and Guillemot, 2011; Chitnis, 1999; Huang et al., 2014; Kageyama et al., 1995). Notch signaling becomes active in the course of asymmetric divisions, when the fate switch of either remaining a neuroepithelial progenitor (radial glia) or becoming a postmitotic neuron/glia is executed (Dong et al., 2012; Gaiano and Fishell, 2002; Yoon and Gaiano, 2005; Yoon et al., 2008). As a result of the asymmetric division, Notch activity becomes concentrated in one of the daughters, which remains the epithelial progenitor (Fig.2H2/3, inset). Lowering of Notch activity in the sibling cell (the postmitotic neuron, or the delaminated intermediate progenitor) is accompanied by neural differentiation. Inhibition of the Notch pathway at any stage in neurogenesis results in an increased number of neurons produced at that stage, and concomitant loss of neuroepithelial progenitors, with the effect that later formed neurons are decreased in number.
Neural progenitor proliferation and the specification of neuronal fate
Drosophila neuroblasts express intrinsic determinants of cell fate in spatially and temporally restricted manner
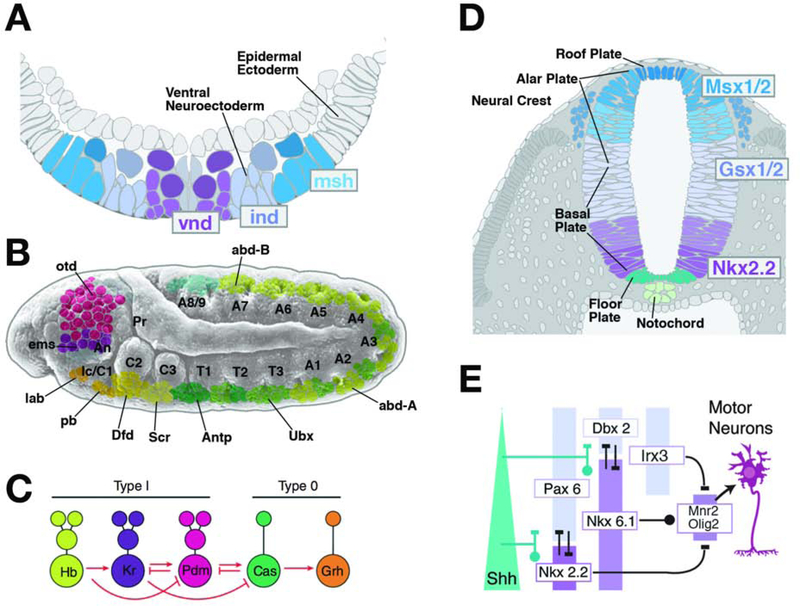
Vertebrates and invertebrates alike express systems of intrinsic determinants that influence profoundly the development of neural progenitors forming at different positions within the neuroectoderm. These determinants (which molecularly, for the most part, represent transcriptional regulators) subdivide the neuroectoderm into a “mosaic” with many different domains, each one characterized by a unique genetic identity. In Drosophila, beginning at the blastoderm stage, members of different classes of homeobox transcription factors are expressed within the neuroectoderm in discrete transverse and longitudinal zones (Akam, 1987; Harding et al., 1985; Urbach and Technau, 2004; Fig.3A, B). Expression continues in the neuroblasts that delaminate from within these zones, and the progeny (lineage) of the neuroblasts. For example, neuroblasts delaminating from within a medial neurectodermal domain that expresses the homeobox gene ventral nerve cord defective (vnd), or an anterior domain defined by orthodenticle (otd), will continue to express these and many other genes once inside the embryo (Hirth et al., 1995; Zhao et al., 2007; Younossi-Hartenstein et al., 1997; Fig.3A, B). In this manner, the neuroectodermal mosaic of expression domains of transcription factors results in a pattern of genetically uniquely specified neuroblasts. With the advance of molecular techniques it was possible to identify a host of transcription factors whose differential expression provides each neuroblast and their progeny with a unique genetic code (Doe, 1992; Skeath and Thor, 2003; Urbach and Technau, 2004). The early expressed code of transcription factors specifies lineage identity; modifying this code by genetic means transforms the fate of the entire lineage (Sen et al., 2014; see below).
Figure 3.
Neural proliferation and specification of neural fate. (A, B) Transcriptional regulators determining neural fate subdivide the neuroectoderm into discrete domains. (A) depicts schematic cross section of Drosophila ventral neuroectoderm following neuroblast delamination; the genes vnd, ind and msh are expressed in longitudinal columns of neuroectoderm and resulting neuroblasts. (B) shows expression pattern of Hox genes (lab, pb, Dfd, Scr, Antp, Ubx, abdA, abdB) and head gap genes (otd, ems) in discrete domains of neuroectoderm and neuroblast layer along the antero-posterior axis. (C) A cassette of sequentially expressed transcription factors, conisting of Hunchback (Hb), Kruppel (Kr), Nubbin (Nub), Castor (Cas) and Grainyhead (Grh), specifies the fate of embryonic Drosophila neuroblasts. Direct interactions between these factors orchestrate the temporal dynamics of their expression (from Allan and Thor, 2015, with permission). (D, E) The expression pattern of transcriptional regulators, many of them homologous to those discovered in Drosophila (see conserved color coding between panels A and D), divide the neural tube into longitudinal columns. The vnd homolog Nkx2.2, as well as Nkx6.1, activated by high levels of the signal Sonic hedgehog (Shh), are expressed in nested columns adjacent to the floor plate (E). Other transcription factors (e.g., Pax6, Dbx2, Irx3, Msx1/2, Gsx1/2) are activated by a dorsally originating BMP signal. Inhibitory interactions among the transcription factors delineate subdomains, as exemplified by the MN domain that gives rise to motor neurons and oligodendrocytes (E).
The phenotypic differences among different sublineages that are born during specific time intervals are also likely based on genetic factors activated sequentially as neuroblasts undergo asymmetric cell divisions. For example, during their primary phase of proliferation, Drosophila neuroblasts sequentially express the factors Hunchback (Hb), Kruppel (Kr), Nubbin (Nub), Castor (Cas) and Grainyhead (Grh; Brody and Odenwald, 2005; Pearson and Doe, 2004; Fig.3C). The secondary phase of proliferation that takes place in the larva is divided into an early period, characterized by the expression of the transcription factor Chinmo, from a late period where neuroblast switch to the expression of the Broad protein (Doe, 2006; Syed et al., 2017). Numerous additional transcription factors, as well as RNA-binding proteins have been identified which are expressed in distinct temporal patterns during the larval (secondary) phase of neuroblast proliferation (Doe, 2017; Sullivan et al., 2019; Syed et al., 2017). It is thought that the transcriptional code manifested during a certain time window of neuroblast proliferation endows the sublineage produced during this phase with its own characteristic structural and functional characteristics (Kao and Lee, 2010; Kohwi and Doe, 2013; Li et al., 2013).
Intrinsic determinants of neural fate in vertebrates
As described for Drosophila, the vertebrate neuroectoderm (i.e., the neural plate followed by the neural tube) is divided by the expression of intrinsic determinants of specific neural fates into distinct transverse and longitudinal domains. Expression domains are delineated by global and local signaling events, involving, among others, the Shh, BMP, Wnt pathways. By the time when neurogenic divisions start to occur (e.g., around stage E10 in mouse embryos), these signals have specified within the neural tube a stable pattern of expression domains of numerous transcription factors, many of which have homologies to factors expressed in a similar pattern in Drosophila (Briscoe et al., 2000; Dessaud et al., 2008; Graham et al., 1989; Jessell, 2000; Fig.3D). For example, the homeobox genes Nkx2.2 (homolog of the Drosophila gene vnd; see above) and Nkx6.1 are expressed close to the ventral midline under the influence of high levels of the Shh morphogen (Briscoe et al., 1999; Sander et al., 2000; Fig.3D, E). Further laterally, other transcription factors, including Irx3, Gsh1/2 (homologs of Drosophila ind), and Msx1/2 (homologs of Drosophila msh) are activated under the input of BMP signaling (Illes et al., 2009; Ramos and Robert, 2005; Winterbottom et al., 2010). Inhibitory interactions between these factors delineate small columnar expression domains with different progenitor fates. For example, domain MN, generating progenitors of motor neurons and oligodendrocytes, is defined by high levels of Nkx6.1 and low levels of Nkx2.2 and Irx3 (Fig.3E). In vitro culture of individual Nkx6.1-positive progenitors taken at E9.5 or E10 produced clones of less than 20 to several hundred cells that, in many cases, maintained expression of individual intrinsic markers (Agalliu and Schieren, 2009). This finding and many other studies indicate that at the time point when progenitors enter the phase of neurogenic divisions, the expression of intrinsic determinants sets the fate of the lineage these cells give rise to.
Research on the anterior neural tube, in particular the forebrain vesicle that gives rise to the cerebral cortex, also documented the role of early expressed transcriptional regulators in later neural fate. Morphogen gradients (e.g. BMP, EGF, FGF, and Wnt), followed by regional expression of various transcription factors (e.g. COUP-TF1, Emx2), pattern the anterior neuroectoderm into a “mosaic” where contiguous aRGCs express distinct combinations of fate determinants, and genes specific to distinct pools of aRGCs have been linked to the types of neurons these progenitors give rise to (Azzarelli et al., 2015; Rubenstein et al., 1999; Takahashi and Liu, 2006). There is also mounting evidence to suggest that cortical neurons with different structural fates (e.g., projections to different cortical or subcortical target regions) are generated in a sequential manner under the control of transcription factors regulated by intrinsic programs within the progenitors (Britanova et al., 2008; Chen et al., 2005a, b; Hanashima et al., 2004; Molyneaux et al., 2005; 2007; Okano and Temple, 2009; Shen et al., 2006). Similar to the Drosophila Hb-Kr-Nub-Cas-Grh cassette, these factors control the fate of neuron populations born during these different time periods. For example, the factors Fezf1 and Fezf2 are expressed at an early stage in progenitors and remain on in deep layer corticofugal neurons (Chen et al., 2005a; Eckler and Chen, 2014; Greig et al., 2013; Molyneaux et al., 2005). By contrast, Cut-like homeobox 1 and 2 (Cux1 and Cux2), as well as Satb2, appear at later stages and specify cells of superficial layers, including the callosal neurons (Britanova et al., 2008; Weiss and Nieto, 2019).
Lineages as structural and functional units: Drosophila
Neuroblasts generate lineages with discrete anatomical features
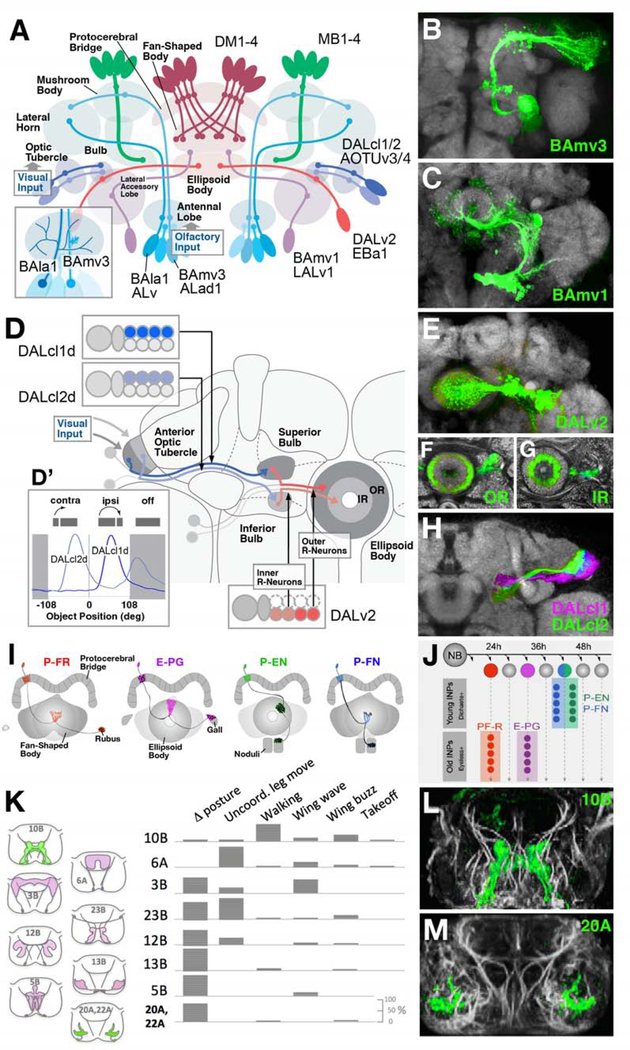
Neural lineages in Drosophila form highly stereotyped anatomical units, such that neurons which derive from a single neuroblast remain in close proximity, with their axons and dendrites forming coherent bundles that innervate discrete neuropil domains. Most lineages have been mapped anatomically at a high level of resolution (Cardona et al., 2010; Ito et al., 2013; Lovick et al., 2013; Pereanu and Hartenstein, 2006; Truman et al., 2004; Wong et al., 2013; Yu et al., 2013). One can picture the fly brain as a mosaic of structural/functional modules (the lineages) which, in large part, are genetically specified at an early stage of development (Fig.4A). Some of the well-studied examples of neural lineages in the Drosophila brain briefly discussed in the following include those which form the olfactory center, called antennal lobe, the central complex, and the ventral nerve cord.
Figure 4.
Lineage-based architecture of the Drosophila brain. (A) Schematic of the Drosophila brain, illustrating representative lineages (BAla1/ALv, BAmv3/ALad1, BAmv1/LALv1, DALv2/EBa1, DALcl1/2/AOTUv3, MB1–4, DM1–4) and the compartments innervated by them. BAla1 and BAmv3 both form projection neurons connecting the antennal lobe (olfactory center) with the mushroom body; however, they differ in dendritic geometry (inset at lower left), with BAla1 forming wide-spread multiglomerular branches, and BAmv3 narrow, uni- or bi-glomerular branches. (B, C) Z-projections of frontal confocal sections of adult fly brain, illustrating GFP-labeled clones of lineages BAmv3/ALad1 and BAmv1/LALv1, respectively. (D-H) Lineage-based composition of the anterior visual pathway (AVP), which conducts visual information from the optic lobe via anterior optic tubercle and bulb to the ellipsoid body, as shown schematically in (D). Two hemilineages, DALcl1/AOTUv3 d and DALcl2/AOTUv4 d [inset at upper left of (D); GFP-labeled in confocal image shown in (H)] form parallel pathways between discrete subdomains of the tubercle and bulb. Based on Calcium-imaging of these neuron populations, DALcl1 d neurons react in a retinotopic manner to small stimuli in the ipsilateral visual field; DALcl2 d neurons do not show any retinotopy, and are active when stimulated contralaterally, as well as after cessation of the stimulus [inset at bottom left of (D)]. Lineage DALv2/EBa1 generates neurons that continue the parallel visual pathways from bulb to ellipsoid body (inset at bottom right of D). Early born (outer ring) neurons connect the superior bulb to the periphery of the ellipsoid body (OR); later born (inner ring) neurons project from inferior bulb to ellipsoid body center [IR; panels (F, G) and inset at bottom right of (D)]. (I, J) Sublineages of the type II neuroblasts DM1–4 form discrete classes of columnar neurons of the central complex (P-FR, E-PG, P-EN, P-FN), born during different time intervals from different intermediate progenitors (INPs; panel J). Early INP offspring are specified by expression of Dichaete; late offspring by Eyeless (from Sullivan et al., 2019; with permission). (K-M) Hemilineages form spatially and functionally discrete populations of interneurons in the ventral nerve cord. (K, Left) Schematic frontal sections of ventral nerve cord with domains innervated by lineages indicated (10B, 6A, 3B, 23B, 12B, 13B, 5B, 20A/22A) rendered in colors. (K, right) Types of behaviors preferentially elicited by stimulating hemilineage indicated. (L, M) Z-projections of frontal confocal sections of ventral nerve cord, showing GFP-labeled hemilineages 10B (elicits walking and wing beat) and 20A (involved in leg posture; from Harris et al., 2015, with permission).
The fly olfactory system is formed by olfactory receptor neurons (ORNs) whose somata and dendrites are located in sensilla that are subjected to chemical stimuli from the external world, and whose axons terminate in the antennal lobe, the primary olfactory processing compartment in the central brain. Each ORN expresses a single odorant receptor gene (out of approximately 50), and neurons which share a given receptor converge onto a single glomerulus in the antennal lobe (Wilson, 2013). Information is received from ORNs by an anatomically stereotyped population of olfactory projection neurons, which exhibit dendrites in the antennal lobe and axonal outputs in the mushroom body calyx and/or the lateral horn, two higher-order brain structures (Fig.4A). Projection neurons are members of four paired lineages, generated from neuroblasts BAlc/ALl1, BAmv3/ALad1, BAla1/ALv, and lastly, BAlp4/ALlv1 (Das et al., 2008; 2010; 2013; Lai et al., 2008; Lin et al., 2010; 2012; Yu et al., 2010). (Note that for the adult Drosophila brain, two different designations (e.g., “BAlc” and “ALl1”) exist for many lineages; the first is based on the analysis of Wong et al., 2013, which uses the nomenclature introduced for brain lineages identified by their characteristic axon tracts that remain detectable throughout development (Cardona et al., 2009; Hartenstein et al., 2015; Lovick et al., 2013; Pereanu and Hartenstein, 2006). The second system of lineage designations was launched by Ito et al. (2013) and Yu et al. (2013) specifically for the lineages (“clonal units”) of the adult brain, based on their relationship to neuropil compartments). Antennal lobe projection neurons belonging to different lineages form discrete classes in terms of dendritic arborization and axonal projection. For example, the lineage BAmv3/ALad1 includes uni-glomerular neurons, whose dendrites are restricted to a single glomerulus, and project via the medial antennal lobe tract to the mushroom body calyx and the lateral horn (Fig.4A, inset; B). Lineage BAla1/ALv generates neurons with widely branching multiglomerular dendrites, and axons that exclusively target the lateral horn via a different tract, the medio-lateral antennal lobe tract (Fig.4A, inset).
Experimental evidence supports the idea that intrinsic genetic programs act at an early stage to establish the type of neuron a given neuroblast will generate. For example, the homeobox gene orthodenticle (otd) is normally expressed in a pair of adjacent progenitors, those of lineage BAmv3/ALad1 and its neighbor, BAmv1/LALv1, whose neurons innervate the central complex (Fig.4A–C). If otd is specifically eliminated in BAmv1/LALv1, the fate of the entire lineage (in the order of 100 neurons) of this neuroblast is homeotically transformed into that of the neighboring BAmv3/ALad1; Sen et al., 2014). Thus, in these mutant brains, dendritic and axonal arborization, as well as transmitter phenotype of BAmv1/LALv1 neurons are absent, and replaced by supernumerary axons/dendrites of the BAmv3/ALad1 class.
Lineages or hemilineages can be divided into smaller building blocks
The above discussed studies clearly demonstrate that neurons forming part of lineages share certain fundamental functional and anatomical characters. In addition, within a given lineage, smaller sets of neurons defined by different birth date (sublineages) frequently differ in detail. For example, the lineage BAmv3/ALad1 introduced above includes uni-glomerular neurons, whose dendrites are restricted to a single glomerulus. Within this overall pattern, neurons born at different time points target different glomeruli. In other words, BAmv3/ALad1, as a whole lineage, innervates the neuropil volume constituting the antennal lobe; individual neurons of BAmv3/ALad1 subdivide this volume into smaller units, the glomeruli (Berck et al., 2017; Jefferis et al., 2001; 2004; Ramaekers et al., 2005; Yu et al., 2010).
A similar “tiling” behavior has been demonstrated for neurons of lineage DALv2/EBa1, which forms the ring (R-) neurons of the central complex (Fig.4D–G). The central complex, a higher order brain center important for visually-guided behaviors, has become a heavily studied brain structure with the lineage concept in mind (Boyan and Liu, 2014; Boyan and Reichert, 2011; Boyan and Williams, 2011; Boyan et al., 2017; Omoto et al., 2017; 2018; Lovick et al., 2017; Sullivan et al., 2019; Yang et al., 2013). It consists of four compartments, the protocerebral bridge (PB), fan-shaped body (FB), noduli (NO), and ellipsoid body (EB; Fig.4A). Lineage DALv2 R-neurons have a circular axon that branches within the torus-shaped ellipsoid body (Fig.4D–G). Individual R-neurons differ in regard to the position of their axon within the ellipsoid body. Early born R-neurons have axons located closer to the periphery (outer ring neurons, OR), and receive input from DALcl1/AOTUv3 d in the superior bulb (Fig.4D, F); later born R neurons are targeted by DALcl2/AOTUv4 d in the inferior bulb, and extend axons towards the center of the ellipsoid body (inner ring neurons, IR; Omoto et al., 2017; 2018; Fig.4D, G).
Four type II lineages, DM1–4, generate the large number of so-called columnar neurons that interconnect the neuropils of the central complex in a topologically highly ordered manner. More than 10 subclasses of columnar neurons have been distinguished (Hanesch et al., 1989; Wolff and Rubin, 2015). Among these are the P-EG neurons (connect the protocerebral bridge with the ellipsoid body and the gall, a major output domain of the central complex), the P-FR neurons (protocerebral bridege, fan-shaped body, rubus), P-EN neurons (protocerebral bridge, ellipsoid body, noduli) and P-FN neurons (protocerebral bridge, fan-shaped body, noduli; Fig.4I). The four classes represent sublineages of the DM1–4 Type II neuroblasts. P-FR and E-PG are the descendants of intermediate progenitors born around 24h and 32h after larval hatching, respectively; P-EN and P-FN are formed by one progenitor born at around 42h (Sullivan et al., 2019; Fig.4J). As explained above (see Fig.1F), intermediate progenitors, just like regular Type I neuroblasts, generate series of neurons in a strict temporal order, and different transcription factors, expressed during different time windows, specify neuronal fate. P-FR and E-PG are born during late divisions of their respective progenitors, defined by the expression of Eyeless (Ey); by contrast P-EN and P-FN descend from early progenitor divisions, which express Dichaete (D; Fig.4J). Ey is absolutely required to promote the P-FR anf E-PG, and at the same time inhibit P-EN and P-FN (Sullivan et al., 2019). Loss of ey results in the absence of E-PG neurons and ectopic P-EN cells.
Hemilineages and sublineages as functional modules
The significance of clonal relationships in the construction of functional circuitry has been assessed in different regions of the Drosophila CNS. Two lineages forming part of a pathway (the anterior visual pathway or AVP) that conducts visual input from the optic lobe to the central complex illustrate the principle that many lineages form functional neuronal classes defined by discrete response properties. The AVP begins with neurons located in the medulla of the optic lobe; from here, visual information reaches the anterior optic tubercle (AOTU), followed by the bulb (BU) and, finally, the ellipsoid body (EB) of the central complex (Fig.4D). Two discrete hemilineages, DALcl1/AOTUv3 d and DALcl2/AOTUv4 d, produce the neurons that connect the anterior optic tubercle with the bulb (Fig.4D, H). Neuroanatomical analysis and two-photon calcium imaging revealed that DALcl1 d and DALcl2 d form two parallel channels with different functional properties. DALcl1 d neurons possess small, retinotopically ordered receptive fields, and preferentially respond to bright objects presented to the ipsilateral eye. In contrast, DALcl2 d neurons possess, large overlapping receptive fields; they are excited by bright objects presented to the contralateral eye, and inhibited as the stimulus moves into the ipsilateral field (Fig.4D’). In a similar manner, the previously mentioned sublineages of neuroblasts DM1–4, including PF-R, E-PG, P-EN, and P-FN, also subserve distinct functions. For example, E-PGs are required in navigation behavior (Giraldo et al., 2018; Green et al., 2017); elimination of these cells, resulting from a loss of ey-expression, abolishes the flies’ ability to orient relative to bright focal stimuli (“virtual sun”), leaving other aspects of motor control intact (Sullivan et al., 2019).
Another prominent example of hemilineages representing functional modules is that of the ventral nerve cord (VNC), the analog of the vertebrate spinal cord. Like the brain, the VNC is comprised of neurons that for the most part arise during the secondary phase of neuroblast proliferation. Secondary neuroblasts and their lineages have been mapped from the larval to adult stage (Shepherd et al., 2016; Truman et al., 2004). As they form the majority of VNC interneurons, interacting with sensory afferents from the body and motorneuron efferents controling the muscles, Harris et al. (2015) hypothesized that lineally organized neuronal ensembles would also be functionally organized to somehow coordinate locomotor behaviors. Utilizing a novel suite of genetic tools, they were able to stably and specifically label individual hemilineages in the fly VNC and document their anatomical characteristics (Fig.4K). In addition to simple labeling, they expressed a heat-sensitive ion channel in specific hemilineages and assessed the behavioral consequence of targeted activation of these neurons. Interestingly, hemilineage-specific activation led to a stereotyped behavioral response, the type and complexity of which correlated with the projection location and spatial distribution of hemilineage neurites (Harris et al., 2015; Fig.4K–M).
Lineages as structural and functional modules: Vertebrates
What role, if any, do developmentally defined lineages play in defining anatomical modules in the mature brain of vertebrates? Neural progenitors of vertebrates remain part of the neurectoderm throughout their proliferatory history. For a long period, their mode of division is symmetric, resulting in an exponential growth of the size of the neurectoderm (see above). The number of neural progenitors is not invariant and, at least in the mammalian system, very large. The first question that arises when studying clones of cells derived from individual neural progenitors is at what stage the given progenitor was labeled. Early induction, during the phase of symmetric division of neural progenitors, must invariably lead to large, heterogenous clones distributed widely over the brain (McCarthy et al., 2001; Price and Thurlow, 1988; Walsh and Cepko, 1988; 1992). But at what time point in the life of a neural progenitor does it make sense to expect this cell to constitute a discrete founder of a structural or functional module, akin to a neuroblast in the fly? Recent work focusing on mouse cerebral cortex development has brought us closer to address this question (Gao et al., 2014). Thus, as described above, clones of cells generated during the neurogenic phase of proliferation form radial columns of contiguous cells, called “ontogenetic columns”. Experimental data support the idea that neurons belonging to such ontogenetic columns become indeed anatomically and functionally closely connected in the mature cortex. Before reviewing these data, a brief discussion of the term and concept of a “cortical column” will be helpful.
Anatomically and functionally defined columns in the mammalian cortex
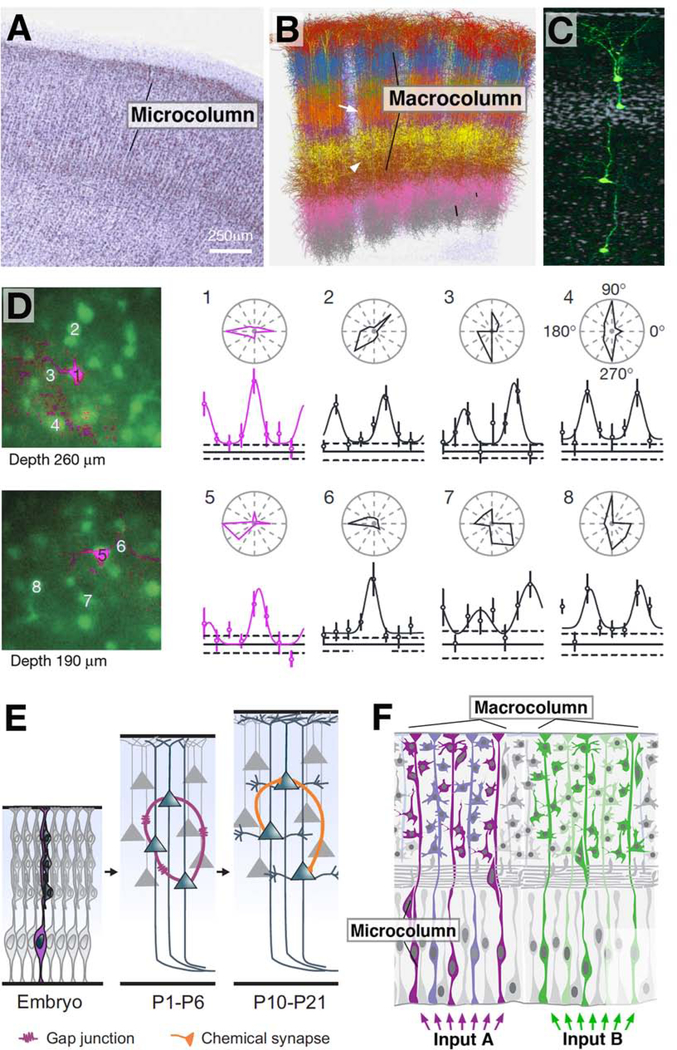
Quite different entities within the mammalian cerebral cortex have been called columns. In the anatomical sense, the term can refer to “microcolumns”, which are visible on histological stains of mammalian brain slices as radially oriented, linear arrays of 50–80μm diameter. Quantitative studies of human brain estimated that microcolumns contain an average of 11 neurons (Jones, 2000; Rockland, 2010; Fig.5A). On the other hand, much larger groupings of cortical neurons are referred to as columns, including the ocular dominance columns in the visual cortex, or the whisker-specific barrels in the somatosensory cortex (Feldmeyer et al., 2013; Lang et al., 2011; Lübke and Feldmeyer, 2007; Fig.5B). These “macrocolumns”, which are almost ten times larger in diameter than the microcolumns, often correspond to physiologically detectable modules in the cortex. As early as the 1950’s, Vernon Mountcastle suggested that the mammalian cortex exhibits a columnar organization; these so-called “cortical columns” were proposed to behave as the elementary computational unit of the brain (Mountcastle et al., 1955). Evidence that cortical columns operate as functional units were supported by the studies of Hubel and Wiesel in the cat visual cortex, who observed that neurons located within a given column were similarly tuned, that is, responded to the same or similar stimulus (Hubel and Wiesel, 1962; 1968).
Figure 5.
Significance of cell lineage in the mammalian cerebral cortex. (A) Nissl-stained frontal section of the human cerebral cortex, showing columnar arrangement of neuronal cell bodies (from Jones, 2000, with permission). (B) Digital 3D reconstruction of five neighboring barrel columns in rat somatosensory cortex. Branched neurite trees of different types of neurons are rendered in different colors (after Egger et al., 2014; with permission by Dr. Marcel Oberlaender). Macrocolumns are spatially well separated in cortical layer IV (arrow), but not in deep or superficial layers (arrowhead). (C) Neurons derived from one apical radial glia progenitor at the onset of neurogenic divisions form a coherent ontogenetic column (GFP-labeled; from Gao et al., 2014, with permission). (D) Sibling neurons forming part of one ontogenetic column have direction preference. Shown at the left are tangential confocal sections of the visual cortex at two different depths. Sibling neurons appear in magenta, general neurons in green. To the right are polar plots of orientation tuning of neurons #1–8. Note similar tuning of siblings #1 and 5 (from Li et al., 2012, with permission). (E) Sibling neurons are strongly electrically coupled by gap junctions (purple) during the first postnatal weeks (P1-P6). At a later stage, the same neurons form preferentially chemical synapses (orange) among themselves (from Gao et al., 2013, with permission). (F) Schematic representation of relationship between microcolumn and macrocolumn. Numerous adjacent microcolumns, representing developmentally based ontogenetic columns, are bundled into larger units (macrocolumns) by shared thalamic input (e.g., afferents from a single vibrissa) or other connections.
Anatomically, macrocolumns are not as clearly defined as microcolumns. Typically, adjacent macrocolumns are not rigidly separated, but interdigitate, or blend into each other. Dendritic arborizations of a neuron located within a given, functionally defined macrocolumn cross column boundaries into neigboring territories (Fig.5B). Also axonal input may cross column boundaries. Significantly, as in the case of the well studied somatosensory columns in the rodent or primate cortex, one type of input (e.g., thalamocortical afferents) may be highly localized and confined to a column (Fig.5B, arrow), whereas a second type maybe much more divergent, crossing column boundaries (Lang et al., 2010; Lübke and Feldmeyer, 2007; Fig.5B, arrowheads). These complications have led to numerous ongoing debates concerning the question “what is a column” [see, for discussion, previous reviews by Herculano-Houzel et al. (2008), Rakic (2008), Rockland (2010), among others].
Microcolumns as developmentally defined ontogenetic columns
The studies of mammalian brain development discussed in the previous section demonstrated that progenitors of the neocortex located in the ventricular zone divide asymmetrically and “bud” off cells which migrate apically or vertically along long processes made by the radial glial cells to their final destination where they differentiate, eventually distributing into the six cortical layers (Rakic 1971, 1972). Neurons which derive from a single progenitor appear to form vertical columns, called ontogenetic column, which could be retained even into the adult (Rakic, 1988). This concept is referred to as the radial unit hypothesis; it posits that a radial glial cell (neural progenitor) not only sequentially generates neurons, but also serves to guide neurons out of the VZ along long processes. Neurons migrate radially and minimally laterally, forming vertical columns extending the depth of the cortex and its six layers. Neurons end up in cortical layers in an inside-out fashion, with cells born first lying in deeper layers, and neurons born later in more superficial layers (reviewed in McConnell, 1988; Rakic, 1974; 1978, 1988).
Early attempts of experimental clonal analysis, mainly based on retroviral labeling of random, sparse assemblies of individual progenitor cells contributed numerous new insights into the gradual acquisition of cell fate, but were not able to provide support for the concept of ontogenetic columns or the radial unit hypothesis. These studies (e.g., Luskin et al., 1988; Price and Thurlow, 1988; Walsh and Cepko, 1992; Tan et al., 1998) yielded clones of different cell numbers and distributions; in many cases, neurons were distributed widely along the horizontal plane, casting doubt on the idea that progeny of an individual radial glial progenitor should remain together. However, early studies lacked in spatial and temporal resolution, and did not conceptually distinguish between pyramidal neurons and local interneurons that, as we know now, originate from completely different locations and follow different proliferatory and migratory patterns (Fig.2H1; see also below). Only more recently developed techniques, among them MADM (Mosaic Analysis with Double Markers; Hippenmeyer et al., 2010; Zong et al., 2005; Gao et al., 2014), were able to precisely define the time point and type of progenitor in which the genetic label was activated, and visualized for the first time the ontogenetic column in mouse cortex. These studies confirmed the earlier radial unit hypothesis, showing that radial glial progenitors from E10.5 onward generated clones of neurons that initially migrated along the glial process of origin, and then stayed together as a radial column (Fig5.C, D).
Ontogenetic columns as functional units
When combining single progenitor lineage tracing with physiological characterization of clonally-related sister neurons the functional role of lineage, at least for excitatory cortical neurons, become apparent. Li et al. (2012) turned to the mouse visual cortex; their method allowed them to identify GFP-labelled, clonally-related sister neuron pairs. Injection of calcium indicator dye near the labeled pair allowed them to optically record the activity of this pair, along with unlabeled, presumably non-sister neighboring neurons, using two-photon excitation microscopy. By presenting the mice with a wide-field grating visual stimulus with different orientations, they found that clonally-related sister neurons prefer similarly oriented visual stimuli, a correlation that was not observed between clonally-related and unrelated neurons (Li et al., 2012; Fig.5E). Similarly, Kondo et al. (2016) noted that neurons with similar orientation preference were clustered vertically in a volume that corresponded to part of a microcolumn.
The similar tuning properties of sister cortical neurons makes sense considering the results of other studies, showing that clonally-related neurons exhibit preferential chemical synaptic interactions (Yu et al., 2009), a property dependent upon transient electrical coupling via gap junctions during early development (Li et al., 2012; Yu et al., 2012). Lineage-specific electrical communication enhances the synchronous firing of these neurons, which is thought to underlie the preferential synaptic wiring observed later in development (Fig.5F). Transcriptional regulation of cell adhesion genes, or birthdate-dependent migration and “inside-out” patterning, may also contribute to connectivity between clonally-related neurons (He et al., 2015; Lv et al., 2019; Tarusawa et al., 2016). Thus, a framework for the role of lineage in sculpting vertebrate brain circuits has emerged; clonally-related sister neurons from a given progenitor talk to each other early and often, growing radially into a column to process information as a group.
Does lineage play a role in sculpting every neural circuit? In the case of GABAergic interneurons of the cortex, the answer seems debatable. Most cortical interneurons derive from the medial ganglion eminence and preoptic area, which migrate tangentially to populate multiple laminae of the cortex (reviewed in Batista-Brito and Fishell, 2009; Fig.2H1). Brown et al. (2011) and Ciceri et al. (2013) suggested that clonally-related interneurons tend to form non-random, spatially-discrete clusters, with both intralaminar and interlaminar distributions. Clonally related interneurons are electrically coupled, but show no preferential connections via chemical synapses (Zhang et al., 2017; Fig.5G). However, related interneurons do establish chemical synapses to shared sets of pyramidal neurons (Fig.5H). Other studies challenged the assertion of clustering, suggesting that there does not seem to be a clear relationship between lineage and the spatial distribution of cortical interneurons (Harwell et al., 2015; Mayer et al., 2015). The discrepancy may lie in the methodology; the former utilized selective and stable fluorescent labeling of individual progenitors and their progeny, whereas the latter utilized retrovirus-based barcoded libraries as a readout. Although follow up rebuttals from each group acknowledge certain shared conclusions and advantages and/or disadvantages of their respective approaches (Mayer et al., 2016; Sultan et al., 2016), the debate seemingly continues.
Conclusion: Ontogenetic modules in vertebrates and Drosophila
The picture that emerges in vertebrate brain development is that cortical macrocolumns are composite structures that contain hundreds of ontogenetically specified microcolumns (Fig.5I). The defining feature of a macrocolumn is functional and anatomical, but not lineage-related. Considering the example of the macrocolumns outlined by the barrels in the rodent somatosensory cortex, it is the pool of trigeminal and thalamic afferents innervated by one whisker that determines the size/cell number of one cortical barrel, and not a predetermined set of lineages within the cortex (Jhaveri et al., 1991; Killackey and Belford, 1979; Schlaggar and O’Leary, 1993; 1994; Senft and Woolsey, 1991; Fig.4E). Likewise, the size of visual cortical columns depends on afferents from the geniculate nucleus (Hubermann, 2007; Penn and Shatz, 1999). The macrocolumn (in sensory cortical domains) could be considered then as a functional module, comprising the circuitry required to process the sensory input from a discrete set of spatially or functionally related afferents. Towards this end, afferents invading the developing cortex recruit a relatively large number of small, contiguous ontogenetic modules to become part of the macrocolumn. Ontogenetic columns may provide “conveniently sized” packages of highly interconnected neurons, which may subserve discrete subroutines within the overall operation of the macrocolumn.
In contrast, ontogenetic modules formed by lineages in the Drosophila brain are much larger, relative to overall brain size, compared to vertebrate microcolumns. The entire (central) brain of Drosophila is formed by lineages/sublineages that number in the hundreds, rather than the millions or even billions that stand for microcolumns of the mammalian cortex. Drosophila lineages/sublineages are also much more different from each other anatomically and functionally than the relatively homogenous microcolumns, as discussed for DALcl1 and DALcl2 (see section 3 above) which innervate largely non-overlapping dendritic and axonal territories, and include neurons that are tuned to completely different visual stimuli (Omoto et al., 2017). It should be noted that modular structures that are not controlled by lineage descent also exist in invertebrates; a notable example is the Drosophila compound eye, which is formed by strictly invariant units (ommatidia) whose cellular components are assembled by a cascade of cell-cell interactions (Treisman, 2013).
The comparison between the vertebrate and insect mode of building ontogenetic modules reveals several fundamental differences, one of them being the time point in development at which a genetic switch seals the fate of neurons generated by a given progenitor cell, molding these neurons into a coherent structural/functional module. In Drosophila, this time point is early, occurring right with the birth of the progenitor (neuroblast) from the neurectoderm, thus giving the progenitor many divisions to produce a relatively large module. In vertebrate (considering mouse cortical development) the corresponding time point is much later. Initially, neural progenitors within the neurectoderm undergo many rounds of symmetric divisions, in which progeny does not appear to be bound together by intrinsically expressed genetic factors. Only following the onset of asymmetric, neurogenic divisions does the formation of ontogenetic modules set in, and that stage the progenitor produces only a small number of neurons, from 8 or so in mouse to possibly a few hundreds in larger mammals, where intermediate progenitors multiply the progeny of an asymmetrically dividing progenitor (Gao et al., 2013). Once more detail has come to light, it will be informative to compare the principles of how brain circuits are wired, and how they function, under the conditions exemplified by mammalian cortex and Drosophila central brain.
Highlights.
Cell lineages are developmentally-genetically and anatomically-functionally defined modules of the brain
Progenitors of lineages (neuroblasts) in insects are formed at an early stage
Insect neuroblasts produce a relative small number of large, diverse lineages
Progenitors of lineages (radial glia) in vertebrates are formed late after a phase of symmetric amplifying divisions
Vertebrate neural progenitors generate a large number of small lineages
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agalliu D, Schieren I, 2009. Heterogeneity in the developmental potential of motor neuron progenitors revealed by clonal analysis of single cells in vitro. Neural Dev. 4:2. doi: 10.1186/1749-8104-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akam M, 1987. The molecular basis for metameric pattern in the Drosophila embryo. Development 101,1–22. [PubMed] [Google Scholar]
- Alexandre P, Reugels AM, Barker D, Blanc E, Clarke JD, 2010.Neurons derive from the more apical daughter in asymmetric divisions in the zebrafish neural tube. Nat. Neurosci. 13, 673–9. [DOI] [PubMed] [Google Scholar]
- Asami M, Pilz GA, Ninkovic J, Godinho L, Schroeder T, Huttner WB, Gotz M, 2011. The role of Pax6 in regulating the orientation andmode of cell division of progenitors in the mouse cerebral cortex. Development 138, 5067–5078. 10.1242/dev.074591 (dev.074591 [pii]). [DOI] [PubMed] [Google Scholar]
- Azzarelli R, Hardwick LJ, Philpott A, 2015. Emergence of neuronal diversity from patterning of telencephalic progenitors. Wiley Interdiscip. Rev. Dev. Biol. 4, 197–214. doi: 10.1002/wdev.174. [DOI] [PubMed] [Google Scholar]
- Bate CM, 1976. Embryogenesis of an insect nervous system. I. A map of the thoracic and abdominal neuroblasts in Locusta migratoria. J. Embryol. Exp. Morphol. 35, 107–23. [PubMed] [Google Scholar]
- Batista-Brito R, Fishell G, 2009. The developmental integration of cortical interneurons into a functional network. Curr. Top. Dev. Biol. 87, 81–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgardt M, Karlsson D, Salmani BY, Bivik C, MacDonald RB, Gunnar E, Thor S, 2014. Global programmed switch in neural daughter cell proliferation mode triggered by a temporal gene cascade. Dev. Cell. 30, 192–208. doi: 10.1016/j.devcel.2014.06.021. [DOI] [PubMed] [Google Scholar]
- Baye LM, Link BA, 2007. Interkinetic nuclear migration and the selection of neurogenic cell divisions during vertebrate retinogenesis. J. Neurosci. 27, 10143–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello BC, Izergina N, Caussinus E, Reichert H, 2008. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 3:5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berck ME, Khandelwal A, Claus L, Hernandez-Nunez L, Si G, Tabone CJ, Li F, Truman JW, Fetter RD, Louis M, Samuel AD, Cardona A, 2017. The wiring diagram of a glomerular olfactory system. Elife 13;5. pii: e14859. doi: 10.7554/eLife.14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F, 2002. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517–30. [DOI] [PubMed] [Google Scholar]
- Betizeau M, Cortay V, Patti D, Pfister S, Gautier E, Bellemin-Menard A, Afanassieff M, Huissoud C, Douglas RJ, Kennedy H, Dehay C, 2013. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron 80, 442–457. 10.1016/j.neuron.2013.09.032(S0896-6273(13)00863-5 [pii]). [DOI] [PubMed] [Google Scholar]
- Biffar L, Stollewerk A, 2014. Conservation and evolutionary modifications of neuroblast expression patterns in insects. Dev. Biol. 388, 103–16. doi: 10.1016/j.ydbio.2014.01.028. [DOI] [PubMed] [Google Scholar]
- Booker R, Truman JW, 1987. Postembryonic neurogenesis in the CNS of the tobacco hornworm, Manduca sexta. I. Neuroblast arrays and the fate of their progeny during metamorphosis. J. Comp. Neurol. 255, 548–559. doi: 10.1002/cne.902550407. [DOI] [PubMed] [Google Scholar]
- Boone JQ, Doe CQ, 2008. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev. Neurobiol. 68, 1185–95. doi: 10.1002/dneu.20648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA, 2008. The tumor suppressors Brat and Numb regulate transitamplifying neuroblast lineages in Drosophila. Dev. Cell 14, 535–46. doi: 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyan G, Liu Y, 2014., Timelines in the insect brain: fates of identified neural stem cells generating the central complex in the grasshopper Schistocerca gregaria. Dev. Genes Evol. 224, 37–51. doi: 10.1007/s00427-013-0462-8. [DOI] [PubMed] [Google Scholar]
- Boyan G, Liu Y, Khalsa SK, Hartenstein V, 2017. A conserved plan for wiring up the fan-shaped body in the grasshopper and Drosophila. Dev. Genes Evol. 227, 253–269. doi: 10.1007/s00427-017-0587-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyan GS, Reichert H, 2011. Mechanisms for complexity in the brain: generating the insect central complex. Trends Neurosci. 34, 247–57. doi: 10.1016/j.tins.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Boyan G, Williams L, 2011. Embryonic development of the insect central complex: insights from lineages in the grasshopper and Drosophila. Arthropod Struct. Dev. 40, 334–48. doi: 10.1016/j.asd.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Bribián A, Figueres-Oñate M, Martín-López E, López-Mascaraque L, 2016. Decoding astrocyte heterogeneity: New tools for clonal analysis. Neuroscience 323, 10–9. doi: 10.1016/j.neuroscience.2015.04.036. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Sussel L, Serup P, Hartigan-O’Connor D, Jessell TM, Rubenstein JL, Ericson J, 1999. Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature 398, 622–7. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell TM, Ericson J, 2000. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 101, 435–45. [DOI] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, Sestan N, Molnár Z, Tarabykin V, 2008. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron 57, 378–92. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Brody T, Odenwald WF, 2005. Regulation of temporal identities during Drosophila neuroblast lineage development. Curr. Opin. Cell Biol. 17, 672–5. [DOI] [PubMed] [Google Scholar]
- Brown KN, Chen S, Han Z, Lu CH, Tan X, Zhang XJ, Ding L, Lopez-Cruz A, Saur D, Anderson SA, et al. , 2011. Clonal production and organization of inhibitory interneurons in the neocortex. Science 334, 480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas A, Borrell V, 2019. Molecular and cellular evolution of corticogenesis in amniotes. Cell Mol. Life Sci. 2019 Sep 28. doi: 10.1007/s00018-019-03315-x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona A, Saalfeld S, Arganda I, Pereanu W, Schindelin J, Hartenstein V, 2010. Identifying neuronal lineages of Drosophila by sequence analysis of axon tracts. J. Neurosci. 30, 7538–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro DS, Guillemot F, 2011. Old and new functions of proneural factors revealed by the genome-wide characterization of their transcriptional targets. Cell Cycle 10, 4026–31. doi: 10.4161/cc.10.23.18578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Schaevitz LR, McConnell SK, 2005a. FezI regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc. Natl. Acad. Sci. U S A. 102, 17184–17189. doi: 10.1073/pnas.05087332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Rasin MR, Kwan KY, Sestan N, 2005b. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proc. Natl. Acad. Sci. U S A. 102, 17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WR, Shepherd GM, 2005. The olfactory glomerulus: a cortical module with specific functions. J. Neurocytol. 34, 353–60. [DOI] [PubMed] [Google Scholar]
- Ciceri G, Dehorter N, Sols I, Huang ZJ, Maravall M, Marín O, 2013. Lineage-specific laminar organization of cortical GABAergic interneurons. Nat. Neurosci.16, 1199–210. [DOI] [PubMed] [Google Scholar]
- Chitnis AB, 1999. Control of neurogenesis--lessons from frogs, fish and flies. Curr. Opin. Neurobiol. 9, 18–25. [DOI] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, Quake SR, 2015. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. U S A. 112, 7285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Sen S, Lichtneckert R, Okada R, Ito K, Rodrigues V, Reichert H, 2008. Drosophila olfactory local interneurons and projection neurons derive from a common neuroblast lineage specified by the empty spiracles gene. Neural Dev. 3:33. doi: 10.1186/1749-8104-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Reichert H, Rodrigues V, 2010. Notch regulates the generation of diverse cell types from the lateral lineage of Drosophila antennal lobe. J. Neurogenet. 24, 42–53. doi: 10.3109/01677060903582202. [DOI] [PubMed] [Google Scholar]
- Das A, Gupta T, Davla S, Prieto-Godino LL, Diegelmann S, Reddy OV, Raghavan KV, Reichert H, Lovick J, Hartenstein V, 2013. Neuroblast lineage-specific origin of the neurons of the Drosophila larval olfactory system. Dev. Biol. 373, 322–37. doi: 10.1016/j.ydbio.2012.11.003. Epub 2012 Nov 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T, Payer B, Cayouette M, Harris WA, 2003. In vivo time-lapse imaging of cell divisions during neurogenesis in the developing zebrafish retina. Neuron 37, 597–609. [DOI] [PubMed] [Google Scholar]
- del Valle Rodríguez A, Didiano D, Desplan C, 2011. Power tools for gene expression and clonal analysis in Drosophila. Nat. Methods 9, 47–55. doi: 10.1038/nmeth.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaud E, McMahon AP, Briscoe J, 2008. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development 135, 2489–503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- Doe CQ,1992. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development 116, 855–63. [DOI] [PubMed] [Google Scholar]
- Doe CQ, Goodman CS, 1985. Early events in insect neurogenesis. I. Development and segmental differences in the pattern of neuronal precursor cells. Dev. Biol. 111, 193–205. doi: 10.1016/0012-1606(85)90445-2. [DOI] [PubMed] [Google Scholar]
- Doe CQ, 2006. Chinmo and neuroblast temporal identity. Cell 127, 254–6. [DOI] [PubMed] [Google Scholar]
- Doe CQ, 2017. Temporal Patterning in the Drosophila CNS. Annu. Rev. Cell Dev. Biol. 33, 219–240. doi: 10.1146/annurev-cellbio-111315-125210. [DOI] [PubMed] [Google Scholar]
- Dong Z, Wagle M, Guo S, 2011. Time-lapse Live Imaging of Clonally Related Neural Progenitor Cells in the Developing Zebrafish Forebrain. J. Vis. Exp 50: 2594. doi: 10.3791/2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Yang N, Yeo SY, Chitnis A, Guo S, 2012. Intralineage directional Notch signaling regulates self-renewal and differentiation of asymmetrically dividing radial glia. Neuron 74, 65–78. doi: 10.1016/j.neuron.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eakin RM, Kuda A,1971. Ultrastructure of sensory receptors in Ascidian tadpoles. Z. Zellforsch. Mikrosk. Anat.112, 287–312. [DOI] [PubMed] [Google Scholar]
- Eckler MJ, Chen B, 2014. Fez family transcription factors: controlling neurogenesis and cell fate in the developing mammalian nervous system. Bioessays 36, 788–97. doi: 10.1002/bies.201400039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmeyer D, Brecht M, Helmchen F, Petersen CC, Poulet JF, Staiger JF, Luhmann HJ, Schwarz C, 2013. Barrel cortex function. Prog. Neurobiol. 103:3–27. doi: 10.1016/j.pneurobio.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Fietz SA, Kelava I, Vogt J,Wilsch-Brauninger M, Stenzel D, Fish JL, Corbeil D, Riehn A, Distler W, Nitsch R, Huttner WB, 2010. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat. Neurosci. 13, 690–699. 10.1038/nn.2553 (nn.2553 [pii]). [DOI] [PubMed] [Google Scholar]
- Florio M, Huttner WB, 2014. Neural progenitors, neurogenesis and the evolution of the neocortex. Development 141, 2182–2194. 10.1242/dev.090571 (141/11/2182 [pii]). [DOI] [PubMed] [Google Scholar]
- Gaiano N, Fishell G, 2002. The role of notch in promoting glial and neural stem cell fates. Annu. Rev. Neurosci. 25, 471–490. [DOI] [PubMed] [Google Scholar]
- Gao P, Sultan KT, Zhang XJ, Shi SH, 2013. Lineage-dependent circuit assembly in the neocortex. Development 140, 2645–55. doi: 10.1242/dev.087668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Postiglione MP, Krieger TG, Hernandez L, Wang C, Han Z, Streicher C, Papusheva E, Insolera R, Chugh K, Kodish O, Huang K, Simons BD, Luo L, Hippenmeyer S, Shi SH, 2014. Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell 159, 775–88. doi: 10.1016/j.cell.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bellido A, Merriam JR, 1971. Genetic analysis of cell heredity in imaginal discs of Drosophila melanogaster. Proc. Natl. Acad. Sci. U S A. 68, 2222–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo YM, Leitch KJ, Ros IG, Warren TL, Weir PT, Dickinson MH, 2018. Sun Navigation Requires Compass Neurons in Drosophila. Curr. Biol. 28, 2845–2852.e4. doi: 10.1016/j.cub.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CS, Spitzer NC, 1979. Embryonic development of identified neurones: differentiation from neuroblast to neurone. Nature 280, 208–214. doi: 10.1038/280208a0. [DOI] [PubMed] [Google Scholar]
- Graham A, Papalopulu N, Krumlauf R, 1989.The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell 57, 367–78. [DOI] [PubMed] [Google Scholar]
- Green J, Adachi A, Shah KK, Hirokawa JD, Magani PS, Maimon G, 2017. A neural circuit architecture for angular integration in Drosophila. Nature 546, 101–106. doi: 10.1038/nature22343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD, 2013. Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci 14, 755–69. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin R, Binari R, Perrimon N, 2014. Genetic odyssey to generate marked clones in Drosophila mosaics. Proc. Natl. Acad. Sci. U S A. 111, 4756–63. doi: 10.1073/pnas.1403218111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanashima C, Li SC, Shen L, Lai E, Fishell G, 2004. Foxg1 suppresses early cortical cell fate. Science 303, 56–9. [DOI] [PubMed] [Google Scholar]
- Hanesch U, Fischbach K-F, and Heisenberg M,1989. Neuronal architecture of the central complex in Drosophila melanogaster. Cell Tissue Res. 257, 343–366. [Google Scholar]
- Harding K, Wedeen C, McGinnis W, Levine M, 1985. Spatially regulated expression of homeotic genes in Drosophila. Science 229, 1236–42. [DOI] [PubMed] [Google Scholar]
- Harris RM, Pfeiffer BD, Rubin GM, Truman JW, 2015. Neuron hemilineages provide the functional ground plan for the Drosophila ventral nervous system. Elife 20;4. doi: 10.7554/eLife.04493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V, Campos-Ortega JA, 1984. Early neurogenesis in wildtype Drosophila melanogaster. Wilhelm Roux’s Arch. Dev. Biol.193, 308–325. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Campos-Ortega JA,1985. Fate mapping in wildtype Drosophila melanogaster. I. The pattern of embryonic cell divisions. Wilhelm Roux’s Arch. Dev. Biol. 194, 181–195. [Google Scholar]
- Hartenstein V, Rudloff E, Campos-Ortega JA, 1987. The pattern of proliferation of the neuroblasts in the wildtype embryo of Drosophila melanogaster. Wilhelm Roux’s Arch. Dev. Biol. 196, 473–485. [DOI] [PubMed] [Google Scholar]
- Hartenstein V,1989. Early neurogenesis in Xenopus laevis: The spatiotemporal pattern of proliferation and cell lineages in the embryonic spinal cord. Neuron 3, 399–411. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Spindler S, Pereanu W, Fung S, 2008. The development of the Drosophila larval brain. Adv. Exp. Med. Biol. 628, 1–31. doi: 10.1007/978-0-387-78261-4_1. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Wodarz A, 2013. Initial neurogenesis in Drosophila. Wiley Interdisc. Rev. Dev.Biol. 2, 701–21. doi: 10.1002/wdev.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V, Stollewerk A, 2015. The evolution of early neurogenesis. Dev. Cell 32, 390–407. doi: 10.1016/j.devcel.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V, Younossi-Hartenstein A, Lovick JK, Kong A, Omoto JJ, Ngo KT, Viktorin G, 2015. Lineage-associated tracts defining the anatomy of the Drosophila first instar larval brain. Dev. Biol. 406, 14–39. doi: 10.1016/j.ydbio.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartfuss E, Galli R, Heins N, Gotz M, 2001. Characterization of CNS precursor subtypes and radial glia. Dev. Biol. 229, 15–30. 10.1006/dbio.2000.9962. [DOI] [PubMed] [Google Scholar]
- Harwell CC, Fuentealba LC, Gonzalez-Cerrillo A, Parker PR, Gertz CC, Mazzola E, Garcia MT, Alvarez-Buylla A, Cepko CL, Kriegstein AR, 2015. Wide Dispersion and Diversity of Clonally Related Inhibitory Interneurons. Neuron 87, 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harzsch S, Krieger J, 2018. Crustacean olfactory systems: A comparative review and a crustacean perspective on olfaction in insects. Prog. Neurobiol. 161, 23–60. doi: 10.1016/j.pneurobio.2017.11.005. [DOI] [PubMed] [Google Scholar]
- He S, Li Z, Ge S, Yu YC, Shi SH, 2015. Inside-Out Radial Migration Facilitates Lineage-Dependent Neocortical Microcircuit Assembly. Neuron. 86, 1159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S, Collins CE, Wong P, Kaas JH, Lent R, 2008. The basic nonuniformity of the cerebral cortex. Proc. Natl. Acad. Sci. U S A. 105, 12593–8. doi: 10.1073/pnas.0805417105. Epub 2008. August 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S, Youn YH, Moon HM, Miyamichi K, Zong H, Wynshaw-Boris A, Luo L, 2010. Genetic mosaic dissection of Lis1 and Ndel1 in neuronal migration. Neuron 68, 695–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirth F, Therianos S, Loop T, Gehring WJ, Reichert H, Furukubo-Tokunaga K, 1995. Developmental defects in brain segmentation caused by mutations of the homeobox genes orthodenticle and empty spiracles in Drosophila. Neuron 15, 769–78. [DOI] [PubMed] [Google Scholar]
- Hobert O, 2010. Neurogenesis in the nematode Caenorhabditis elegans. WormBook 4:1–24. doi: 10.1895/wormbook.1.12.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Chan JA, Schuurmans C, 2014. Proneural bHLH genes in development and disease. Curr. Top. Dev. Biol. 110, 75–127. doi: 10.1016/B978-0-12-405943-6.00002-6 [DOI] [PubMed] [Google Scholar]
- Hubel DH, Wiesel TN, 1962. Receptive fields, binocular interaction and functional architecture in the cat’s visual cortex. J. Physiol. (Lond.) 160, 106–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubel DH, Wiesel TN, 1968. Receptive fields and functional architecture of monkey striate cortex. J. Physiol. (Lond.) 195, 215–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD, 2007. Mechanisms of eye-specific visual circuit development. Curr. Opin. Neurobiol. 17, 73–80. [DOI] [PubMed] [Google Scholar]
- Illes JC, Winterbottom E, Isaacs HV, 2009. Cloning and expression analysis of the anterior parahox genes, Gsh1 and Gsh2 from Xenopus tropicalis. Dev. Dyn. 238, 194–203. [DOI] [PubMed] [Google Scholar]
- Ito K, Awasaki T, 2008. Clonal unit architecture of the adult fly brain. Adv. Exp. Med. Biol. 628, 137–58. doi: 10.1007/978-0-387-78261-4_9. [DOI] [PubMed] [Google Scholar]
- Ito M, Masuda N, Shinomiya K, Endo K, Ito K, 2013. Systematic analysis of neural projections reveals clonal composition of the Drosophila brain. Curr. Biol. 23, 644–655. [DOI] [PubMed] [Google Scholar]
- Jefferis GS, Marin EC, Stocker RF, Luo L, 2001. Target neuron prespecification in the olfactory map of Drosophila. Nature 8;414, 204–8. [DOI] [PubMed] [Google Scholar]
- Jefferis GS, Vyas RM, Berdnik D, Ramaekers A, Stocker RF, Tanaka NK, Ito K, Luo L, 2004. Developmental origin of wiring specificity in the olfactory system of Drosophila. Development 131, 117–30. [DOI] [PubMed] [Google Scholar]
- Jessell TM, 2000. Neuronal specification in the spinal cord: inductive signals and trans-criptional codes. Nat. Rev. Genet. 1, 20–9. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Jhaveri S, Erzurumlu RS, Crossin K, 1991. Barrel construction in rodent neocortex: role of thalamic afferents versus extracellular matrix molecules. Proc. Natl. Acad. Sci. U S A. 88, 4489–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Nardelli J, 2016. Cellular and molecular introduction to brain development. Neurobiol. Dis. 92(Pt A):3–17. doi: 10.1016/j.nbd.2015.07.007. Epub 2015. July 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Reichert H, 2014. Drosophila neural stem cells in brain development and tumor formation. J. Neurogenet. 28, 181–9. doi: 10.3109/01677063.2014.898639. [DOI] [PubMed] [Google Scholar]
- Jones EG, 2000. Microcolumns in the cerebral cortex. Proc. Natl. Acad. Sci. USA 97, 5019–5021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas JH, 2012. Evolution of columns, modules, and domains in the neocortex of primates. Proc. Natl. Acad. Sci. U S A. 109 Suppl 1, 10655–60. doi: 10.1073/pnas.1201892109. Epub 2012 Jun 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R, Sasai Y, Akazawa C, Ishibashi M, Takebayashi K, Shimizu C, Tomita K, Nakanishi S, 1995. Regulation of mammalian neural development by helix-loop-helix transcription factors. Crit. Rev. Neurobiol. 9, 177–88. [PubMed] [Google Scholar]
- Kageyama R, Ohtsuka T, Shimojo H, Imayoshi I, 2009. Dynamic regulation of Notch signaling in neural progenitor cells. Curr. Opin. Cell Biol. 21, 733–740. doi: 10.1016/j.ceb.2009.08.009 [DOI] [PubMed] [Google Scholar]
- Kane DA, Kimmel CB, 1993. The zebrafish midblastula transition. Development 119, 447–56. [DOI] [PubMed] [Google Scholar]
- Kang KH, Reichert H, 2015. Control of neural stem cell self-renewal and differentiation in Drosophila. Cell Tiss. Res. 359, 33–45. doi: 10.1007/s00441-014-1914-9. [DOI] [PubMed] [Google Scholar]
- Kao CF, Lee T, 2010. Birth time/order-dependent neuron type specification. Curr. Opin. Neurobiol. 20, 14–21. doi: 10.1016/j.conb.2009.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killackey HP, Belford GR, 1979. The formation of afferent patterns in the somatosensory cortex of the neonatal rat. J. Comp. Neurol. 183, 285–303. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM, Kane DA, 1994. Cell cycles and clonal strings during formation of the zebrafish central nervous system. Development 120, 265–76. [DOI] [PubMed] [Google Scholar]
- Kohwi M, Doe CQ, 2013. Temporal fate specification and neural progenitor competence during development. Nat. Rev. Neurosci. 14, 823–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Yoshida T, Ohki K, 2016. Mixed functional microarchitectures for orientation selectivity in the mouse primary visual cortex. Nat. Commun.7, 13210. doi: 10.1038/ncomms13210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Noctor S, Martínez-Cerdeño V., 2006. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat. Rev. Neurosci. 7, 883–90. [DOI] [PubMed] [Google Scholar]
- Kumar A, Bello B, Reichert H, 2009. Lineage-specific cell death in postembryonic brain development of Drosophila. Development 136, 3433–42. doi: 10.1242/dev.037226. [DOI] [PubMed] [Google Scholar]
- Lai SL, Awasaki T, Ito K, Lee T, 2008. Clonal analysis of Drosophila antennal lobe neurons: diverse neuronal architectures in the lateral neuroblast lineage. Development 135, 2883–2893. doi: 10.1242/dev.024380. [DOI] [PubMed] [Google Scholar]
- Lang S, Dercksen VJ, Sakmann B, Oberlaender M, 2011. Simulation of signal flow in 3D reconstructions of an anatomically realistic neural network in rat vibrissal cortex. Neural Netw. 24, 998–1011. doi: 10.1016/j.neunet.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Larsen C, Shy D, Spindler SR, Fung S, Pereanu W, Younossi-Hartenstein A, Hartenstein V, 2009. Patterns of growth, axonal extension and axonal arborization of neuronal lineages in the developing Drosophila brain. Dev. Biol. 335, 289–304. doi: 10.1016/j.ydbio.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L, 2001. Mosaic analysis with a repressible cell marker (MARCM) for Drosophilaneural development. Trends Neurosci. 24, 251–4. [DOI] [PubMed] [Google Scholar]
- Li X, Chen Z, Desplan C, 2013. Temporal patterning of neural progenitors in Drosophila. Curr. Top. Dev. Biol.105, 69–96. doi: 10.1016/B978-0-12-396968-2.00003-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lu H, Cheng PL, Ge S, Xu H, Shi SH, Dan Y, 2012. Clonally related visual cortical neurons show similar stimulus feature selectivity. Nature 486, 118–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Lee T, 2012. Generating neuronal diversity in the Drosophila central nervous system. Dev. Dyn. 241, 57–68. doi: 10.1002/dvdy.22739. [DOI] [PubMed] [Google Scholar]
- Lin S, Lai SL, Yu HH, Chihara T, Luo L, Lee T, 2010. Lineage-specific effects of Notch/Numb signaling in post-embryonic development of the Drosophila brain. Development 137, 43–51. doi: 10.1242/dev.041699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Kao CF, Yu HH, Huang Y, Lee T, 2012. Lineage analysis of Drosophila lateral antennal lobe neurons reveals notch-dependent binary temporal fate decisions. PLoS Biol. 10(11):e1001425. doi: 10.1371/journal.pbio.1001425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovick JK, Ngo KT, Omoto JJ, Wong DC, Nguyen JD, Hartenstein V, 2013. Postembryonic lineages of the Drosophila brain: I. Development of the lineage-associated fiber tracts. Dev. Biol. 384, 228–57. doi: 10.1016/j.ydbio.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovick JK, Omoto JJ, Ngo KT, Hartenstein V, 2017. Development of the anterior visual input pathway to the Drosophila central complex. J. Comp. Neurol. 525, 3458–3475. doi: 10.1002/cne.24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lübke J, Feldmeyer D, 2007. Excitatory signal flow and connectivity in a cortical column: focus on barrel cortex. Brain Struct. Funct. 212, 3–17. [DOI] [PubMed] [Google Scholar]
- Luskin MB, Pearlman AL, and Sanes JR,1988. Cell lineage in the cerebral cortex of the mouse studied in vivo and in vitro with a recombinant retrovirus. Neuron 1, 635–647. [DOI] [PubMed] [Google Scholar]
- Lv X, Ren SQ, Zhang XJ, Shen Z, Ghosh T, Xianyu A, Gao P, Li Z, Lin S, Yu Y, Zhang Q, Groszer M, Shi SH., 2019. TBR2 coordinates neurogenesis expansion and precise microcircuit organization via Protocadherin 19 in the mammalian cortex. Nat. Commun.10, 3946. doi: 10.1038/s41467-019-11854-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Cerdeno V, Cunningham CL, Camacho J, Antczak JL, Prakash AN, Cziep ME, Walker AI, Noctor SC, 2012. Comparative analysis of the subventricular zone in rat, ferret andmacaque: evidence for an outer subventricular zone in rodents. PLoS One 7, e30178. 10.1371/journal.pone.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Jaglin XH, Cobbs LV, Bandler RC, Streicher C, Cepko CL, Hippenmeyer S, Fishell G, 2015. Clonally Related Forebrain Interneurons Disperse Broadly across Both Functional Areas and Structural Boundaries. Neuron. 87, 989–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Bandler RC, Fishell G, 2016. Lineage Is a Poor Predictor of Interneuron Positioning within the Forebrain. Neuron 92(1):45–51. doi: 10.1016/j.neuron.2016.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy M, Turnbull DH, Walsh CA, and Fishell G, 2001. Telencephalic neural progenitors appear to be restricted to regional and glial fates before the onset of neurogenesis. J. Neurosci. 21, 6772–6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SK, 1988. Development and decision-making in the mammalian cerebral cortex. Brain Res. 472, 1–23. [DOI] [PubMed] [Google Scholar]
- McShane SG, Molè MA, Savery D, Greene ND, Tam PP, Copp AJ, 2015. Cellular basis of neuroepithelial bending during mouse spinal neural tube closure. Dev. Biol. 404, 113–24. doi: 10.1016/j.ydbio.2015.06.003. Epub 2015 Jun 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD, 2005. FezI is required for the birth and specification of corticospinal motor neurons. Neuron 47, 817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD, 2007. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 8, 427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Mountcastle VB, Berman AL, Davies PW, 1955. Topographic organization and modality representation in first somatic area of cat’s cerebral cortex by method of single unit analysis. Am. J. Physiol. 183, 464. [Google Scholar]
- Nicol D, Meinertzhagen IA, 1988a. Development of the central nervous system of the larva of the ascidian, Ciona intestinalis L. I. The early lineages of the neural plate. Dev. Biol. 130, 721–36. [DOI] [PubMed] [Google Scholar]
- Nicol D, Meinertzhagen IA, 1988b. Development of the central nervous system of the larva of the ascidian, Ciona intestinalis L. II. Neural plate morphogenesis and cell lineages during neurulation. Dev. Biol.130, 737–66. [DOI] [PubMed] [Google Scholar]
- Nicol D, Meinertzhagen IA, 1991. Cell counts and maps in the larval central nervous system of the ascidian Ciona intestinalis (L.). J. Comp. Neurol. 309, 415–29. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR, 2004. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7, 136–144. 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Nowakowski TJ, Pollen AA, Sandoval-Espinosa C, Kriegstein AR, 2016. Transformation of the Radial Glia Scaffold Demarcates Two Stages of Human Cerebral Cortex Development. Neuron 91,1219–1227. doi: 10.1016/j.neuron.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki G, Nishiyama M, Yoshida T, Murakami T, Histed M, Lois C, Ohki K, 2012. Similarity of visual selectivity among clonally related neurons in visual cortex. Neuron, 75, 65–72. doi: 10.1016/j.neuron.2012.05.023. [DOI] [PubMed] [Google Scholar]
- Okano H, Temple S,2009. Cell types to order: temporal specification of CNS stem cells. Curr. Opin. Neurobiol. 19, 112–9. [DOI] [PubMed] [Google Scholar]
- Omoto JJ, Keleş MF, Nguyen BM, Bolanos C, Lovick JK, Frye MA, Hartenstein V, 2017. Visual Input to the Drosophila Central Complex by Developmentally and Functionally Distinct Neuronal Populations. Curr. Biol. 27, 1098–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto JJ, Nguyen BM, Kandimalla P, Lovick JK, Donlea JM, Hartenstein V, 2018. Neuronal Constituents and Putative Interactions Within the Drosophila Ellipsoid Body Neuropil. Front. Neural Circuits, 12:103. doi: 10.3389/fncir.2018.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papan C, Campos-Ortega JA, 1997. A clonal analysis of spinal cord development in the zebrafish. Dev. Genes Evol. 207, 71–81. [DOI] [PubMed] [Google Scholar]
- Papan C, Campos-Ortega JA, 1999. Region-specific cell clones in the developing spinal cord of the zebrafish. Dev. Genes Evol. 209, 135–44. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ, 2004. Specification of temporal identity in the developing nervous system. Annu. Rev. Cell Dev. Biol. 20, 619–47. [DOI] [PubMed] [Google Scholar]
- Penn AA, Shatz CJ, 1999. Brain waves and brain wiring: the role of endogenous and sensory-driven neural activity in development. Pediatr. Res. 45, 447–58. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Hartenstein V, 2006. Neural lineages of the Drosophila brain: a three-dimensional digital atlas of the pattern of lineage location and projection at the late larval stage. J. Neurosci. 26, 5534–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggi L, Vitorino M, Masai I, Harris WA, 2005. Influences on neural lineage and mode of division in the zebrafish retina in vivo. J. Cell Biol. 171, 991–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulson DF, 1950. Histogenesis, organogenesis and differentiation in the embryo of Drosophila melanogaster Meigen. Biology of Drosophila. Wiley New York, 168–274. [Google Scholar]
- Price J, Thurlow L, 1988. Cell lineage in the rat cerebral cortex: a study using retroviral-mediated gene transfer. Development 104, 473–82. [DOI] [PubMed] [Google Scholar]
- Quan XJ, Hassan BA, 2005. From skin to nerve: flies, vertebrates and the first helix. Cell. Mol. Life Sci. 62, 2036–49. doi: 10.1007/s00018-005-5124-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P, 1971. Guidance of neurons migrating to the fetal monkey neocortex. Brain Res. 33, 471–6. [DOI] [PubMed] [Google Scholar]
- Rakic P, 1972. Mode of cell migration to the superficial layers of fetal monkey neocortex. J. Comp. Neurol. 145, 61–83. [DOI] [PubMed] [Google Scholar]
- Rakic P, 1974. Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science 183, 425–7. [DOI] [PubMed] [Google Scholar]
- Rakic P, 1978. Neuronal migration and contact guidance in the primate telencephalon. Postgrad. Med. J. 54 Suppl 1, 25–40. [PubMed] [Google Scholar]
- Rakic P, 1988. Specification of cerebral cortical areas. Science 241, 170–176. [DOI] [PubMed] [Google Scholar]
- Rakic P, 2008. Confusing cortical columns. Proc. Natl. Acad. Sci. U S A 105, 12099–100. doi: 10.1073/pnas.0807271105. Epub [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaekers A, Magnenat E, Marin EC, Gendre N, Jefferis GS, Luo L, Stocker RF, 2005. Glomerular maps without cellular redundancy at successive levels of the Drosophila larval olfactory circuit. Curr. Biol. 15, 982–92. [DOI] [PubMed] [Google Scholar]
- Ramos C, Robert B, 2005. msh/Msx gene family in neural development. Trends Genet. 21, 624–32. [DOI] [PubMed] [Google Scholar]
- Reillo I, de Juan Romero C, García-Cabezas MÁ, Borrell V, 2011. A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb. Cortex 21, 1674–94. doi: 10.1093/cercor/bhq238 [DOI] [PubMed] [Google Scholar]
- Rockland KS, 2010. Five points on columns. Front. Neuroanat. 4:22. doi: 10.3389/fnana.2010.00022. eCollection 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL, Anderson S, Shi L, Miyashita-Lin E, Bulfone A, Hevner R, 1999. Genetic control of cortical regionalization and connectivity. Cereb. Cortex. 9, 524–532. doi: 10.1093/cercor/9.6.524. [DOI] [PubMed] [Google Scholar]
- Sander M, Paydar S, Ericson J, Briscoe J, Berber E, German M, Jessell TM, Rubenstein JL, 2000. Ventral neural patterning by Nkx homeobox genes: Nkx6.1 controls somatic motor neuron and ventral interneuron fates. Genes Dev. 14, 2134–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaggar BL, O’Leary DD, 1994. Early development of the somatotopic map and barrel patterning in rat somatosensory cortex. J. Comp. Neurol. 346, 80–96. [DOI] [PubMed] [Google Scholar]
- Schlaggar BL, O’Leary DD, 1993. Patterning of the barrel field in somatosensory cortex with implications for the specification of neocortical areas. Perspect. Dev. Neurobiol. 1, 81–91. [PubMed] [Google Scholar]
- Seecof RL, Donady JJ, Teplitz RL, 1973. Differentiation of Drosophila neuroblasts to form ganglion-like clusters of neurons in vitro. Cell Differ. 2, 143–149. [DOI] [PubMed] [Google Scholar]
- Sen S, Cao D, Choudhary R, Biagini S, Wang JW, Reichert H, VijayRaghavan K, 2014. Genetic transformation of structural and functional circuitry rewires the Drosophila brain. Elife 29;3. doi: 10.7554/eLife.04407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senft SL, Woolsey TA, 1991. Growth of thalamic afferents into mouse barrel cortex. Cereb. Cortex. 1, 308–35. [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S, 2006. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat. Neurosci. 9, 743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Shepherd D, Harris R, Williams DW, Truman JW, 2016. Postembryonic lineages of the Drosophila ventral nervous system: Neuroglian expression reveals the adult hemilineage associated fiber tracts in the adult thoracic neuromeres. J. Comp. Neurol. 524, 2677–95. doi: 10.1002/cne.23988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeath JB, Thor S, 2003. Genetic control of Drosophila nerve cord development. Curr. Opin. Neurobiol. 13(1):8–15. [DOI] [PubMed] [Google Scholar]
- Snow MHL, 1977. Gastrulation in the mouse: Growth and regionalization of the epiblast. J. Embryol. Exp. Morph. 42, 293–303. [Google Scholar]
- Spindler SR, Hartenstein V, 2010. The Drosophila neural lineages: a model system to study brain development and circuitry. Dev. Genes Evol. 220, 1–10. doi: 10.1007/s00427-010-0323-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stollewerk A, 2016. A flexible genetic toolkit for arthropod neurogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150044. doi: 10.1098/rstb.2015.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LF, Warren TL, Doe CQ, 2019. Temporal identity establishes columnar neuron morphology, connectivity, and function in a Drosophilanavigation circuit. Elife 8. pii: e43482. doi: 10.7554/eLife.43482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN, 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64–119. [DOI] [PubMed] [Google Scholar]
- Sultan KT, Han Z, Zhang XJ, Xianyu A, Li Z, Huang K, Shi SH, 2016. Clonally Related GABAergic Interneurons Do Not Randomly Disperse but Frequently Form Local Clusters in the Forebrain. Neuron 92, 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Hevner RF, 2014. Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat. Rev. Neurosci 15, 217–232. 10.1038/nrn3707 (nrn3707 [pii]). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed MH, Mark B, Doe CQ, 2017. Steroid hormone induction of temporal gene expression in Drosophila brain neuroblasts generates neuronal and glial diversity. Elife 10;6. pii: e26287. doi: 10.7554/eLife.26287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghert PH, Doe CQ, Goodman CS, 1984. Cell determination and regulation during development of neuroblasts and neurones in grasshopper embryo. Nature 307, 163–5. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Liu FC, 2006. Genetic patterning of the mammalian telencephalon by morphogenetic molecules and transcription factors. Birth Defects Res. C. Embryo Today. 78, 256–66. [DOI] [PubMed] [Google Scholar]
- Tan SS, Kalloniatis M, Sturm K, Tam PP, Reese BE, and Faulkner-Jones B, 1998. Separate progenitors for radial and tangential cell dispersion during development of the cerebral neocortex. Neuron 21, 295–304. [DOI] [PubMed] [Google Scholar]
- Tarusawa E, Sanbo M, Okayama A, Miyashita T, Kitsukawa T, Hirayama T, Hirabayashi T, Hasegawa S, Kaneko R, Toyoda S, Kobayashi T, Kato-Itoh M, Nakauchi H, Hirabayashi M, Yagi I, Yoshimura Y, 2016. Establishment of high reciprocal connectivity between clonal cortical neurons is regulated by the Dnmt3b DNA methyltransferase and clustered protocadherins. BMC Biol. 14(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, Bertagnolli D, Goldy J, Shapovalova N, Parry S, Lee C, Smith K, Bernard A, Madisen L, Sunkin SM, Hawrylycz M, Koch C, Zeng H, 2016. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 19, 335–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JB, Bastiani MJ, Bate M, Goodman CS, 1984. From grasshopper to Drosophila: a common plan for neuronal development. Nature 310, 203–7. [DOI] [PubMed] [Google Scholar]
- Treisman JE, 2013. Retinal differentiation in Drosophila. Wiley Interdiscip. Rev. Dev. Biol. 2, 545–57. doi: 10.1002/wdev.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman JW, Schuppe H, Shepherd D, Williams DW, 2004. Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development 131, 5167–5184. [DOI] [PubMed] [Google Scholar]
- Truman JW, Moats W, Altman J, Marin EC, Williams DW, 2010. Role of Notch signaling in establishing the hemilineages of secondary neurons in Drosophila melanogaster. Development 137, 53–61. doi: 10.1242/dev.041749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach R, Technau GM 2004. Neuroblast formation and patterning during early brain development in Drosophila. Bioessays 26, 739–51. [DOI] [PubMed] [Google Scholar]
- Wadsworth WG, Hedgecock EM, 1992. Guidance of neuroblast migrations and axonal projections in Caenorhabditis elegans. Curr. Opin. Neurobiol. 2, 36–41. [DOI] [PubMed] [Google Scholar]
- Walsh C, Cepko CL,1988. Clonally related cortical cells show several migration patterns. Science 241, 1342–1345. [DOI] [PubMed] [Google Scholar]
- Walsh C, Cepko CL,1992. Widespread dispersion of neuronal clones across functional regions of the cerebral cortex. Science 255, 434–440. [DOI] [PubMed] [Google Scholar]
- Wang H, Somers GW, Bashirullah A, Heberlein U, Yu F, Chia W, 2006. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 20, 3453–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Nieto M, 2019. The crux of Cux genes in neuronal function and plasticity. Brain Res, 1705, 32–42. doi: 10.1016/j.brainres.2018.02.044. [DOI] [PubMed] [Google Scholar]
- Wheeler WM, 1891. Neuroblasts in the arthropod embryo. J. Morphol. 4, 337–343. doi: 10.1002/jmor.1050040305. [DOI] [Google Scholar]
- Whitman CO, 1878. Memoirs: the embryology of Clepsine. Quart. J. Micr. Sci. 2, 215–315. [Google Scholar]
- Whitman CO, 1887. A contribution to the history of the germlayers in Clepsine. J. Morphol. 1, 105–182. doi: 10.1002/jmor.1050010107. [DOI] [Google Scholar]
- Wilson RI, 2013. Early olfactory processing in Drosophila: mechanisms and principles. Annu. Rev. Neurosci. 36, 217–41. doi: 10.1146/annurev-neuro-062111-150533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterbottom EF, Illes JC, Faas L, Isaacs HV, 2010. Conserved and novel roles for the Gsh2 transcription factor in primary neurogenesis. Development 137, 2623–31. doi: 10.1242/dev.047159. [DOI] [PubMed] [Google Scholar]
- Wolff T, Iyer NA, and Rubin GM, 2015. Neuroarchitecture and neuroanatomy of the Drosophila central complex: A GAL4-based dissection of protocerebral bridge neurons and circuits. J. Comp. Neurol. 523, 997–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DC, Lovick JK, Ngo KT, Borisuthirattana W, Omoto JJ, Hartenstein V, 2013. Postembryonic lineages of the Drosophila brain: II. Identification of lineage projection patterns based on MARCM clones. Dev. Biol. 384, 258–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Awasaki T, Yu HH, He Y, Ding P, Kao JC, Lee T, 2013. Diverse neuronal lineages make stereotyped contributions to the Drosophila locomotor control center, the central complex. J. Comp. Neurol. 521, 2645–2662. doi: 10.1002/cne.23339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K, Gaiano N, 2005. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat. Neurosci. 8, 709–715. [DOI] [PubMed] [Google Scholar]
- Yoon KJ, Koo BK, Im SK, Jeong HW, Ghim J, Kwon MC, Moon JS, Miyata T, Kong YY, 2008. Mind bomb 1-expressing intermediate progenitors generate Notch signaling to maintain radial glial cells. Neuron 58, 519–31. doi: 10.1016/j.neuron.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nassif C, Hartenstein V, 1996. Early neurogenesis of the Drosophila brain. J. Comp. Neur. 370, 313–329. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Green P, Liaw G, Rudolph K, Lengyel J, Hartenstein V, 1997. Control of early neurogenesis of the Drosophila brain by the head gap genes tll, otd, ems, and btd. Dev. Biol. 182, 270–283. [DOI] [PubMed] [Google Scholar]
- Yu HH, Kao CF, He Y, Ding P, Kao JC, Lee T, 2010. A complete developmental sequence of a Drosophila neuronal lineageas revealed by twin-spot MARCM. PLoS Biol. 8(8). pii: e1000461. doi: 10.1371/journal.pbio.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HH, Awasaki T, Schroeder MD, Long F, Yang JS, He Y, Ding P, Kao JC, Wu GY, Peng H, Myers G, Lee T, 2013. Clonal development and organization of the adult Drosophila central brain. Curr. Biol. 23, 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YC, Bultje RS, Wang X, Shi SH, 2009. Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature 458, 501–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YC, He S, Chen S, Fu Y, Brown KN, Yao XH, Ma J, Gao KP, Sosinsky GE, Huang K, Shi SH, 2012. Preferential electrical coupling regulates neocortical lineage-dependent microcircuit assembly. Nature 486, 113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SO, Weisblat DA, 2005. Applications of mRNA injections for analyzing cell lineage and asymmetric cell divisions during segmentation in the leech Helobdella robusta. Development 132, 2103–13. [DOI] [PubMed] [Google Scholar]
- Zhang XJ, Li Z, Han Z, Sultan KT, Huang K, Shi SH, 2017. Precise inhibitory microcircuit assembly of developmentally related neocortical interneurons in clusters. Nat. Commun. 8:16091. doi: 10.1038/ncomms16091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, Wu JQ, 2014. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Wheeler SR, Skeath JB, 2007. Genetic control of dorsoventral patterning and neuroblast specification in the Drosophila Central Nervous System. Int. J. Dev. Biol. 51,107–15. [DOI] [PubMed] [Google Scholar]
- Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L, 2005. Mosaic analysis with double markers in mice. Cell 121, 479–492. [DOI] [PubMed] [Google Scholar]