Abstract
New treatments for the diseases caused by apicomplexans are needed. Recently, we determined that tartrolon E (trtE), a secondary metabolite derived from a shipworm symbiotic bacterium, has broad-spectrum anti-apicomplexan parasite activity. TrtE inhibits apicomplexans at nM concentrations in vitro, including Cryptosporidium parvum, Toxoplasma gondii, Sarcocystis neurona, Plasmodium falciparum, Babesia spp. and Theileria equi. To investigate the mechanism of action of trtE against apicomplexan parasites, we examined changes in the transcriptome of trtE-treated T. gondii parasites. RNA-Seq data revealed that the gene, TGGT1_272370, which is broadly conserved in the coccidia, is significantly upregulated within 4 h of treatment. Using bioinformatics and proteome data available on ToxoDB, we determined that the protein product of this tartrolon E responsive gene (trg) has multiple transmembrane domains, a phosphorylation site, and localizes to the plasma membrane. Deletion of trg in a luciferase-expressing T. gondii strain by CRISPR/Cas9 resulted in a 68% increase in parasite resistance to trtE treatment, supporting a role for the trg protein product in the response of T. gondii to trtE treatment. Trg is conserved in the coccidia, but not in more distantly related apicomplexans, indicating that this response to trtE may be unique to the coccidians, and other mechanisms may be operating in other trtE-sensitive apicomplexans. Uncovering the mechanisms by which trtE inhibits apicomplexans may identify shared pathways critical to apicomplexan parasite survival and advance the search for new treatments.
Keywords: Toxoplasma, Anti-apicomplexan, Drug discovery, CRISPR/Cas9, Natural products, Tartrolon E
Graphical abstract

Highlights
-
•
Tartrolon E is a broad-spectrum anti-apicomplexan compound.
-
•
T. gondii responds to tartrolon E treatment by upregulating expression of the coccidian gene, trg.
-
•
The gene product of trg has multiple transmembrane domains and is phosphorylated and thus may play a role in signaling.
-
•
Deletion of trg results in an increase in parasite resistance to tartrolon E treatment.
1. Introduction
Apicomplexan parasites are the cause of significant diseases of both humans and domesticated animals. Despite the current therapies available to treat these parasitic infections, the challenge of overcoming the rapid development of parasite drug resistance underscores the need for new treatments.
Recently we described the broad-spectrum anti-apicomplexan activity of tartrolon E (trtE), a secondary metabolite of symbiotic bacteria of shipworms (O'Connor et al., 2020). Tartrolons are a class of macrolide dimers, or pseudodimers consisting of polyketide chains joined as diesters that commonly bind to boron. These compounds are produced by both marine and terrestrial bacteria and exhibit antibacterial (Elshahawi et al., 2013; Irschik et al., 1995; Schummer et al., 1994) and insecticidal activity (Lewer et al., 2003). We demonstrated that trtE inhibits many different apicomplexans at low nM concentrations in vitro, including Cryptosporidium parvum, Toxoplasma gondii, Sarcocystis neurona, Plasmodium falciparum, Babesia spp. and Theileria equi. In vivo efficacy of trtE has also been demonstrated, as the treatment of C. parvum-infected neonatal mice with trtE significantly reduced intestinal infection (O'Connor et al., 2020).
No other compound with this broad activity against apicomplexans has been reported, which makes the identification of the mechanism of action of trtE particularly important. To gain insights into the compound's anti-parasitic mechanism, we conducted an RNA-Seq analysis of trtE treated T. gondii. This analysis identified a conserved gene of unknown function that was upregulated during trtE treatment. When this gene was disrupted, T. gondii parasites became significantly more resistant to trtE, supporting a role for this gene product in the parasite's response to trtE treatment. We name this gene trg for trtE responsive gene, and present bioinformatic analyses characterizing the gene and protein in T. gondii and related species.
2. Methods
2.1. Parasites
T. gondii strains ME49 and RH were maintained by serial passage in human foreskin fibroblasts (HFF, ATCC SCRC-1041), cultured in DMEM supplemented with 15% fetal bovine serum as previously described (Roos et al., 1995).
2.2. RNA-seq of trtE-treated T. gondii RH parasites
Total RNA from T. gondii RH parasites treated with 24.2 nM trtE or 0.1% DMSO control was collected 4, 8, and 12 h post-treatment for RNA-Seq analysis. The details of the experimental analysis and results of the transcriptome are available in the GEO database (GSE140197). Briefly, the integrity and quantity of the total RNA samples were determined using a Fragment Analyzer™ (AATI, Ames, IA, USA). The RNA samples that satisfied the input requirement were used as input for the Illumina TruSeq Stranded Total RNA with Ribo-Zero® Gold sample preparation kit (Illumina, San Diego, CA, USA) following the manufacturer's instructions. The quality and molar concentration of the libraries were determined using the Fragment Analyzer. The libraries were pooled and sequenced using HiSeq 2500 High Output V4 chemistry (Illumina). The 75 million to 95 million reads per sample obtained were analyzed using the TopHat-Cufflinks pipeline. The expression level was estimated with Cuffdiff and represented as fragments per kilobase of exons per million mapped fragments (FPKM) to the T. gondii GT1 genome available on ToxoDB (Gajria et al., 2008) (http://toxodb.org). Genes were considered deferentially expressed when both the p-value and q-value, estimated based using a negative binomial model, were below a significance value of 0.05. The RNA-Seq experiment was conducted once.
2.3. Reverse transcriptase quantitative PCR
To confirm RNA-Seq results, HFF cells were infected with 2 × 106 T. gondii RH strain parasites per well in six-well plates. After an additional 24 h of incubation, infected cells were treated with 24.2 nM trtE, 10 μM pyrimethamine, or 0.1% DMSO carrier control in triplicate. RNA was extracted from treated, infected cells using an RNeasy® Kit (Qiagen, Germantown, MD, USA) at 1, 2, and 4 h post treatment. Synthesis of cDNA for each RNA sample was accomplished using iScript™ cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA) using 1 μg of RNA template. Quantitative PCR was performed on the CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad) using SsoAdvanced™ Universal SYBR® Green Supermix (Bio-Rad). The amplification conditions were: 98 °C for 30 s, 40 cycles of denaturation at 98 °C for 30 s, and annealing/extension at 60 °C for 30 s. A melting curve, representing a 1 s hold at every 0.5 °C between 65° and 95 °C, was generated to confirm a single peak for each primer pair. Primer sequences and product sizes for each pair are provided in Table S1. Relative expression of TGGT1_272370 (hereafter referred to as trg, GenBank accession no: XM_018781656) and bradyzoite antigen 1 (bag1; GenBank accession no: XM_002365075) was determined using the 2−ΔΔCt method (Livak and Schmittgen, 2001) using expression of the endogenous parasite actin gene (act 1; GenBank accession no: XM_002369622) for normalization.
To investigate the dose-dependent expression of trg during trtE treatment, HFF cells were infected with 2 × 106 T. gondii RH parasites in six-well plates. Twenty-four hours post-infection, infected cells were treated with 60.5, 24.2, 12.1, 6.1, 1.2 nM trtE or 0.1% DMSO control for 4 h before RNA extraction. Three technical replicates were performed on each sample. Data analysis was accomplished using GraphPad Prism 8.00 Software (GraphPad Software, San Diego, CA, USA).
2.4. Bioinformatics analysis of trg predicted protein product
The complete amino acid sequence of the ME49 strain Trg protein (GenBank accession no: XP_018636692) was retrieved from the National Center for Biotechnology Information (NCBI) database. The presence of multiple post-translational modification sites were predicted including phosphorylation sites using NetPhos 3.1 server (Blom et al., 1999) (https://services.healthtech.dtu.dk/service.php?NetPhos-3.1) with kinase-specific predictions (Blom et al., 2004), and palmitoylation sites using CSS-Palm 4.0 (Ren et al., 2008) (http://csspalm.biocuckoo.org/online.php). The TMHMM server v.2.0 (Krogh et al., 2001) (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0) was used to estimate transmembrane domains. Both WoLF PSORT (Horton et al., 2007) (https://wolfpsort.hgc.jp; using “Animal” as the organism type) and data from hyperplexed Localization of Organelle Proteins by Isotopic Tagging (hyperLOPIT) experiments using extracellular T. gondii tachyzoites (https://proteome.shinyapps.io/toxolopittzex/) accessed through ToxoDB (Gajria et al., 2008) (http://toxodb.org) were used to predict Trg localization. Identification of short linear motifs in the Trg sequence was accomplished using the eukaryotic linear motif (ELM) resource (Gouw et al., 2018) (http://elm.eu.org) filtering for predicted cell compartment and T. gondii taxonomic context. Protein sequences of Trg homologs in other coccidia were identified by searching NCBI Database resources (NCBI Resource Coordinators, 2016) using the basic local alignment search tool (Altschul et al., 1990), ToxoDB (Gajria et al., 2008) (http://toxodb.org), and Ensembl Genomes (Howe et al., 2019) (http://www.ensemblgenomes.org), aligned using the Clustal W (Thompson et al., 1994) algorithm of the AlignX module of Vector NTI (Life Technologies, Rockville, MD, USA) and evaluated for identity and similarity to Trg using the Sequence Manipulation Suite (Stothard, 2000) (http://www.bioinformatics.org/sms2/ident_sim.html). Domain families for protein sequences of Trg and homologs were identified using the NCBI conserved domain database (Marchler-Bauer et al., 2015) (https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml).
2.5. CRISPR/Cas9-mediated deletion of trg
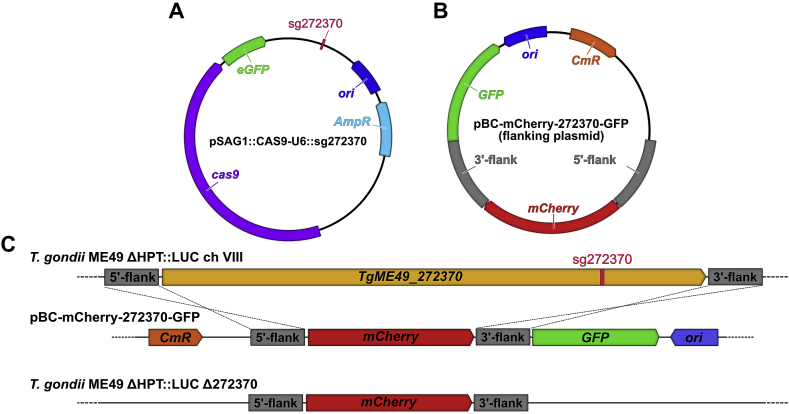
A clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 method (CRISPR/Cas9) was designed to knock out trg in the T. gondii ME49 ΔHPT:LUC strain (Tobin and Knoll, 2012). A guide sequence (5′-AGTGGATGCGGAGCCTGCTG-3′) targeting trg was identified using the Eukaryotic Pathogen CRISPR guide RNA/DNA Design Tool (Tarleton and Peng, 2015) (http://grna.ctegd.uga.edu) based on the efficiency score against the T. gondii ME49 genome available on ToxoDB (Gajria et al., 2008) (http://toxodb.org). The pSAG1:CAS9-U6:sg272370 (Fig. 1A) was prepared by replacing the single guide (sgRNA) sequence of the pSAG1:CAS9-U6 plasmid (Shen et al., 2014) with the identified guide sequence targeting trg using primers listed in Table S1 and the Q5® Site-Directed Mutagenesis Kit (New England Biolabs, Ipswich, MA, USA). A second plasmid provided the regions of sequence identity for homologous recombination and the mCherry gene substitution for trg. This plasmid, pBC-GFP-272370-mCherry (Fig. 1B, designated the “flanking plasmid”), was generated by Gibson assembly® (New England Biolabs) of PCR amplicons of 1 kb upstream and 1 kb downstream flanking sequences of trg, the mCherry expression cassette (pSAG1-mCherry-5′ DHFR 3′ UTR) amplified from the pBC-GFP-mCherry plasmid and the linearized pBC-GFP-mCherry plasmid (containing a GFP expression cassette, pTUB1-eGFP-SAG1 3′ UTR). Plasmids were produced in transfected E. coli D5α (New England Biolabs) and isolated using the QIAGEN Plasmid Maxi Kit (QIAGEN).T. gondii ME49 ΔHPT:LUC tachyzoites were transfected with the pSAG1:CAS9-U6:sg272370 and flanking plasmids by electroporation (Soldati and Boothroyd, 1993).
Fig. 1.
CRISPR/Cas9 mediated deletion of trg. (A) The plasmid used to provide T. gondii-optimized expression of the Cas9 enzyme and sgRNA for targeting trg. (B) The plasmid containing a mCherry expression cassette (red, pSAG1-mCherry-DHFR 3′ UTR) surrounded by 1 kb areas of sequence identity to the flanking regions of trg (grey) and a GFP expression cassette (green, pTUB1-eGFP-SAG1 3′ UTR) used to provide a repair template for homologous recombination of disrupted trg locus. (C) Expression of Cas9 with sg272370 guide RNA (pink) targeted trg and induced a double-stranded break. Linearized pBC-mCherry-272370-GFP plasmid provided a template for double homologous recombination, which is designed to replace the entire trg gene with mCherry and lose GFP for sorting. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The desired mutant that successfully replaced trg with mCherry by homologous recombination must be mCherry+, and GFP− (Fig. 1C). Therefore, parasites that are were mCherry+ and GFP− were selected by cell sorting using BD FACSAria™ II (BD Biosciences, San Jose, CA, USA). The selected population was cloned by limiting dilution, as previously described (Pfefferkorn and Pfefferkorn, 1976). Eight individual clones were isolated.
2.6. Isolation of genomic DNA from T. gondii parasites
Total genomic DNA (gDNA) from parental and knockout parasites was isolated following methods described previously (Medina-Acosta and Cross, 1993). Briefly, parasites were harvested from infected flasks by syringe-lysis using a 25G needle and pelleted by centrifugation at 500×g for 10 min. The pellets were resuspended in 400 μl of TELT lysis buffer (2.5 M LiCl, 50 mM Tris-HCl pH 8.0, 4% (v/v) Triton X-100, 62.5 mM EDTA) supplemented with 10 μl of 10 mg/ml RNase A (Thermo Fisher Scientific, Waltham, MA, USA), mixed by gentle rotation until completely lysed, and combined with an equal volume of phenol/chloroform. The solutions were mixed as before and centrifuged for 5 min at 15,000×g to separate the nucleic acids in the aqueous phase from proteins and lipids in the interface and organic phase. For optimal recovery, we repeated the extraction on the spent phenol/chloroform using an equal volume of TELT lysis buffer. The nucleic acids from the pooled aqueous phases were isolated by ethanol precipitation (Sambrook and Russell, 2006) and dissolved in 10 mM Tris-HCl pH 8.0, 0.5 mM EDTA.
2.7. PCR test for trg
A total of 25 ng of gDNA from T. gondii ME49 ΔHPT:LUC parental strain and each of the eight ME49 ΔHPT:LUC Δtrg clones, was mixed with primers designed to amplify a 172 bp fragment of trg (Table S1) and Q5® Hot Start High-Fidelity 2X PCR Master Mix (New England Biolabs). The amplification conditions were: 98 °C for 30 s, 35 cycles of denaturation at 98 °C for 5 s, annealing at 68 °C, extension at 72 °C for 20 s, and a final extension at 72 °C for 2 min. Reactions were resolved on a 2% agarose gel using a 100 bp DNA ladder (New England Biolabs) for size estimation.
2.8. Southern blot analysis
Approximately 5 μg of gDNA from each strain was digested overnight with the restriction endonuclease Bsu36I (New England Biolabs) and resolved on a 0.8% agarose gel using a DIG-labeled DNA Molecular Weight Marker II (MilliporeSigma, St. Louis, MO, USA) for size estimation. After blotting and u. v. crosslinking to a nylon membrane (BrightStar™- Plus Positively Charged Nylon Membrane, Thermo Fisher Scientific), the membrane was hybridized to digoxigenin (DIG)-labeled mCherry gene probes generated by PCR using the PCR DIG Probe Synthesis Kit (MilliporeSigma), the primers shown in Table S1, and pBG-GFP-mCherry plasmid template. Hybridizations were performed overnight at 47 °C using DIG Easy Hyb™ hybridization buffer (MilliporeSigma), and final washes were made in 0.1X saline sodium citrate (SSC)/0.1% sodium dodecyl sulfate (SDS) solution for 15 min at 68 °C. Chemiluminescence of probe-target hybrids was detected by anti-DIG-alkaline phosphatase antibody (MilliporeSigma) and visualized using the ChemiDoc Touch™ Imaging System (Bio-Rad).
2.9. In vitro inhibitor response assays
HFF cells in 96-well plates were infected with 5 × 104 parental or trg knockout clones for 24 h before treatment with eight 2-fold dilutions of trtE (60.5–0.5 nM) or DMSO control with three technical replicates for each concentration. Treated parasites were incubated at 37 °C and 5% CO2 for 24 h before measuring luciferase (LUC) activity using the Bright-Glo™ Luciferase Assay System (Promega, Madison, WI, USA). Inhibition was calculated by comparing the LUC activity of trtE-treated and untreated parasites. Estimation of the EC50 value for trtE treatment using data from three independent experiments was accomplished using a four-parameter logistic regression, and comparisons between estimated EC50 values of the strains were made using an extra sum-of-squares F test using GraphPad Prism 8.00 Software (GraphPad Software).
3. Results
3.1. T. gondii responds to trtE treatment by upregulating expression of trg
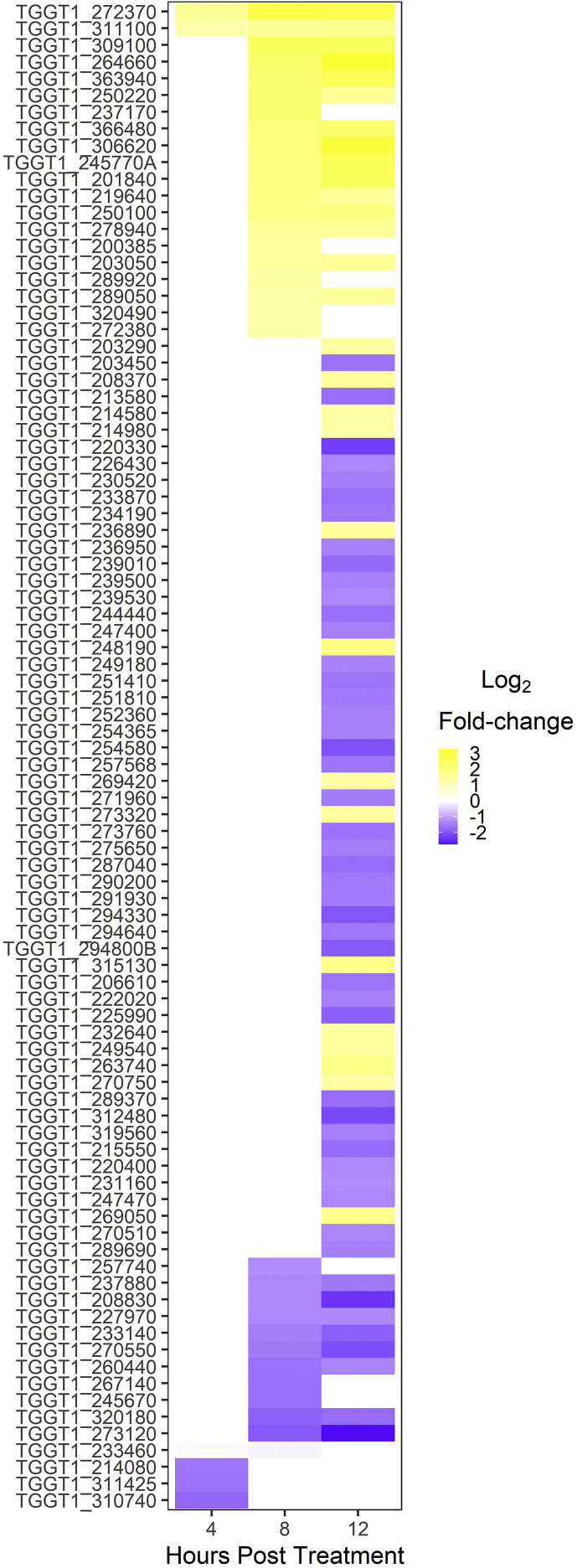
To identify proteins that may be targeted by trtE, we conducted a preliminary RNA-Seq experiment to examine the transcriptome of trtE treated T. gondii parasites. Of the 90 significantly and differentially expressed T. gondii genes over the 12-hr. treatment period (Fig. S1, Table S2), only trg (Fig. 2A) and TGGT1_311100 (Fig. S1) were upregulated within 4 h of treatment and remained upregulated throughout the entire treatment period.
Fig. 2.
T. gondii responds to trtE treatment by upregulating trg in a rapid dose-dependent manner. (A) Changes in transcript abundance of trg in T. gondii parasites treated with 24.2 nM trtE or DMSO carrier control. (B) Expression of trg quantified by RT-qPCR of RNA extracted from T. gondii parasites treated with trtE (24.2 nM), known inhibitory compound pyrimethamine (Pyr, 10 μM), or 0.1% DMSO carrier control at 1, 2, 4 h post treatment. Three technical replicates were performed. Treatment of parasites with trtE significantly increased the expression of trg at 4 h post treatment (p ≤ 0.0001). (C) Expression of trg quantified by RT-qPCR of RNA extracted from T. gondii parasites treated with various concentrations of trtE or 0.1% DMSO carrier control. RNA was isolated from each condition 4 h post treatment and RT-qPCR samples were run in triplicate. Fold-change in gene expression for RT-qPCR experiments was calculated using the 2−ΔΔCt method and normalized by the expression of parasite actin. The relationship between expression of trg and concentration of the trtE treatment was determined to be linear (R2 = 0.7776, p ≤ 0.0001).
To confirm RNA-Seq results, we treated T. gondii ME49 strain with trtE or DMSO for 1–4 h and measured the expression of trg and TGGT1_311100 homologs by RT-qPCR. Expression of trg but not TgME49_311100 (Fig. S2), was upregulated within 4 h of trtE-treatment (Fig. 2B). Treatment with the known T. gondii inhibitor pyrimethamine did not affect trg transcript abundance. The expression of bag1 was decreased after 4 h of trtE treatment (Fig. S2). Additionally, we observed a linear dose-dependent expression of trg 4 h after the treatment of T. gondii parasites with trtE (Fig. 2C).
3.2. Bioinformatic analyses of trg predicted protein sequence
The ME49 strain trg gene is 12,805 bp in length comprised of 9 exons, of which 7107 bp encode a protein of 2368 aa. To gain insight into the function of the protein, the presence of predicted post-translational modifications and functional motifs was investigated. NetPhos 3.1 (Blom et al., 1999) was used to predict 137 potential sites with a probability score higher than 75%. Of these, three positions (T393, S502, S530) were identified as protein kinase A (PKA) family-specific sites and six positions (S134, S174, T443, S1076, T2172, T2300) were identified as protein kinase C (PKC) family-specific sites; the rest were unspecified. The predicted phosphorylation sites were distributed as 75.2% (103/137) serine, 22.6% (31/137) threonine, and 2.2% (3/137) tyrosine. Predicted palmitoylation sites identified by CSS-Palm 4.0 (Ren et al., 2008) clustered around two areas (C635, C636, C641, C646; C1637, C1647, C1684) of Trg. While bioinformatic analyses predicted multiple phosphorylation and palmitoylation sites, only one residue (T2262) has experimental evidence of phosphorylation (Treeck et al., 2011). Trg was not identified in the palmitoylated proteins discovered by DIA-MS (Foe et al., 2015) or ABE-MS (Caballero et al., 2016). To examine the predicted localization of Trg, we used both computational prediction tools and reports of experimental proteome data of tagged proteins. TMHMM v2.0 (Krogh et al., 2001) predicted five transmembrane domains in the protein sequence (positions 610–632, 748–770, 1618–1640, 1653–1675, and 2192–2214). WoLF PSORT (Horton et al., 2007) analysis of the protein sequence predicted the uncharacterized protein to be localized to the plasma membrane with 81.25% (26/32) nearest neighbors associated with the plasma membrane, 12.5% (4/32) neighbors associated with the nucleus, and 6.25% (2/32) neighbors associated with the cytoplasm. The hyperLOPIT proteomic data for T. gondii tachyzoites accessed through ToxoDB (Gajria et al., 2008) also localized Trg to the plasma membrane.
To look for known functional domains in Trg, we examined the protein sequence for linear motifs. Filtering the results of the Eukaryotic Linear Motifs (ELM) search (Gouw et al., 2018) for motifs common to T. gondii and plasma membrane proteins identified four (1809–1812, 1814–1817, 1862–1865, 1919–1922, 2129–2132) tyrosine-based endocytic sorting signals (ELM accession no: ELME000120) in Trg. These Yxxphi motifs are found in the cytosolic tail of membrane proteins and act as a sorting signal that directs traffic within the endosomal and the secretory pathways.
Because trtE is broadly effective against multiple apicomplexan parasites, we looked for proteins that shared sequence identity with Trg among the apicomplexa. However, after searching various databases (NCBI, ToxoDB.org, and Ensembl Genomes), we identified homologs with similar predicted transmembrane structures in the coccidia only (Table 1, Fig. 3, and Fig. S3). No homologs of Trg were found in other more distantly related apicomplexans.
Table 1.
Identity and similarity of Trg homologs to TgTrg
| Organism | GeneID | Accession No. | Identity (%) [No. aa] | Similarity (%) [No. aa] | Coverage (%) |
|---|---|---|---|---|---|
| H. hammondi H.H.34 | HHA_272370 | KEP66945.1 | 73.31 [1736] | 77.53 [100] | 84 |
| B. besnoiti Ger 1 | BESB_032870 | XP_029215099.1 | 35.43 [958] | 47.08 [315] | 30 |
| N. caninum Liverpool | NCLIV_034940 | CEL67703.1 | 32.07 [813] | 42.96 [276] | 48 |
| C. suis Wien I | CSUI_002351 | PHJ23796.1 | 20.33 [615] | 31.93 [351] | 23 |
| C. cayetanensis NF1_C8 | LOC34621828 | XP_026189647 | 17.09 [443] | 25.27 [212] | 11 |
| E. tenella Houghton | ETH_00032795 | CDJ37990.1 | 16.08 [411] | 23.79 [197] | 12 |
| S. neurona SN3 | SN3_00202415 | NAa | 10.19 [269] | 18.55 [221] | 14 |
Not available in the NCBI database.
Fig. 3.
Schematic of identified homologs of Trg in coccidian parasites. Protein homologs to Trg (TGME49_272370) with predicted transmembrane domains (green) in order of descending sequence similarity. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Deletion of trg results in an increase in parasite resistance to trtE-treatment
To determine if Trg played a direct role in the response of T. gondii parasites to treatment with trtE, we generated trg deletion mutants using CRISPR/CAS9. After the selection of putative knockout parasites by FACS, and two rounds of cloning by limiting dilution, the presence of trg was assessed by PCR in eight ME49 ΔHPT:LUC Δtrg isolates (Fig. 4A); and screened for erroneous insertion events by Southern blot analysis (Fig. 4B). We were unable to detect trg in ME49ΔHPT:LUC Δtrg clones by PCR (Fig. 4A), indicating disruption of the gene. No erroneous insertion events of the mCherry expression cassette into the chromosome of ME49ΔHPT:LUC Δtrg clones were detected by Southern blot (Fig. 4B). There were no apparent differences in growth rates between wt and the ME49 ΔHPT:LUC Δtrg mutants, as was previously observed (Sidik et al., 2016).
Fig. 4.
Deletion of trg increases parasite resistance to trtE treatment. (A) PCR detecting trg in gDNA isolated from T. gondii ME49ΔHPT:LUC parental line (P) and ME49 ΔHPT:LUC Δtrg clones. The anticipated 172 bp amplicon is detected in the parental line (white triangle) but was absent in the Δtrg clones. (B) Southern blot analysis of gDNA from the parental line and Δtrg clones digested with Bsu36I using mCherry probe to detect insertion events. The single anticipated 7.6 kb fragment present in all Δtrg clones (black triangle) was absent from the parental sample. (C) Parental (ME49 ΔHPT:LUC) and three Δtrg gene deletion mutants infected host cells for 24 h before being treated with varying concentrations of trtE. The growth of each strain was evaluated 24 h post treatment by LUC expression. Samples were run in triplicate, and three independent experiments performed. Estimation of the EC50 for each strain was accomplished using a four-parametric logistic regression in GraphPad Prism. The curve generated for the parental strain (R2 = 0.9797) estimated the EC50 to be 3.027 ng/ml (2.309–3.968; 95% CI). One curve adequately fit all the data from the three Δtrg clones tested (R2 = 0.9215, p = 0.16) with an estimated average EC50 of 5.088 ng/ml (4.144–6.247; 95% CI), which was significantly different from the EC50 of the parental line by extra sum-of-squares F test (p = 0.0004).
We compared the susceptibility of the parental strain and three of the eight ME49 ΔHPT:LUC Δtrg clones (1, 3, and 4) to trtE-treatment by quantifying the EC50 of the compound for each clone. ME49 ΔHPT:LUC Δtrg parasites were, on average 68% more resistant to trtE treatment than the ME49 ΔHPT:LUC parental line (Fig. 4C) with an estimated EC50 of 5.088 ng/ml (4.144–6.247 ng/ml; 95% CI) for the 3 deletion mutants clones as compared to an EC50 of 3.027 ng/ml (2.309–3.968 ng/ml; 95% CI) for the parental line.
4. Discussion
This study has identified a component of the response of T. gondii parasites to the pan-anti-apicomplexan compound, trtE. Because trtE-treated T. gondii parasites die within 1 h of treatment (O'Connor et al., 2020), genes that were differentially expressed after 4 h of treatment were targeted in this study. RNA-Seq of trtE-treated T. gondii parasites identified five genes in addition to trg (TGGT1_311100, TGGT1_233460, TGGT1_214080, TGGT1_311425, and TGGT1_310740) that were differentially expressed after 4 h of treatment (Fig. S1 and Table S2). Two of these genes have characterized protein products (GeneID:protein; TgME49_214080:toxofilin, and TgME49_233460:SRS29B:SAG1). Toxofilin is an actin-binding protein (Poupel et al., 2000) that has been demonstrated to dissemble host actin and facilitate rapid parasite invasion of host cells (Delorme-Walker et al., 2012). The parasite surface protein SAG1 has been shown to play an active role in attachment and invasion of T. gondii tachyzoites (Mineo and Kasper, 1994) and anti-SAG1 antibodies partially block invasion (Mineo et al., 1993). The expression of both genes was downregulated 4 h post trtE-treatment suggesting trtE may affect tachyzoite invasion into host cells. Pretreatment of tachyzoites with trtE prevents the parasite from establishing infection, but it was not determined if growth inhibition was pre or post-invasion (O'Connor et al., 2020). Of the remaining genes differentially expressed within 4 h of treatment, only trg and TGGT1_311100 were significantly upregulated throughout the entire RNA-Seq experiment; these genes were selected for further study.
Using qRT-PCR, we confirmed that the trg transcript (but not TGGT1_311100) was significantly upregulated in a rapid, dose-dependent manner in response to trtE treatment. This observation appears to be specific to trtE-treatment and not related to a general inhibition of the parasites since treatment with the anti-Toxoplasma compound pyrimethamine did not induce an increase in trg transcripts. Stress can cause T. gondii parasites to form bradyzoites in vitro (Mayoral et al., 2020). However, trtE treatment reduced expression of the bradyzoite-specific gene bag1 after 4 h of treatment, suggesting that the increase in expression of trg is not related to stress-associated bradyzoite formation. This observation is corroborated by studies showing that trg expression is not upregulated during bradyzoite formation (Behnke et al., 2008). In general, it appears that trg is not stage-specific as transcripts are detected during the merozoite (Behnke et al., 2014), oocyst, tachyzoite, and bradyzoite stages (Fritz et al., 2012). In a CRISPR genome-wide loss of function screen (Sidik et al., 2016), trg was found to be dispensable and, consistent with this report, we did not observe any obvious differences in growth rate between the parental and the trg knock out clones.
Bioinformatic analysis of the Trg sequence did not provide definitive clues as to the function of the protein. Trg has multiple transmembrane domains; it likely localizes to the plasma membrane and is a substrate for phosphorylation. The predictions for localization and endosomal targeting, common to membrane-bound proteins, are supported by both mass spectrometry data that identified the protein in the membrane fraction of T. gondii RH parasites (Dybas et al., 2008), and hyperLOPIT proteome data of extracellular tachyzoites (https://proteome.shinyapps.io/toxolopittzex/). There is experimental evidence that one of the predicted threonine phosphorylation sites (T2262) of Trg is phosphorylated (Treeck et al., 2011). Altogether these observations suggest the protein may play a role in a signaling cascade.
The trg gene is conserved amongst coccidia, and homologs were identified in Besnoitia besnoiti, Cyclospora cayetanensis, Cytoisopora suis, Eimeria tenella, Hammondia hammondi, N. caninum, and S. neurona (Table 1, Fig S3). Of the coccidia, we have confirmed the activity of trtE against S. neurona (O'Connor et al., 2020). Homologs to Trg in other trtE-sensitive apicomplexans may have sufficiently diverged such that we could not identify them by protein sequence identity, especially as there are no identified functional motifs in Trg. Alternatively, this response to trtE may be unique to the coccidians, and other mechanisms may be operating in Plasmodium, the piroplasms, and Cryptosporidium.
We anticipate that trtE has potassium ionophore activity since it is structurally similar to the known potassium ionophores trtB (Surup et al., 2018) and boromycin (Moreira et al., 2016; Pache and Zähner, 1969). Ionophores are a promising source of treatments for drug-resistant parasites (Kevin Ii et al., 2009). For example, monensin, a sodium ionophore isolated from the bacterium Streptomyces cinnamonensis, has activity against Plasmodium spp., C. parvum, and T. gondii (Couzinet et al., 2000; Gumila et al., 1997; McDonald et al., 1990). Using a forward genetic screen to identify mutant T. gondii parasites resistant monensin, Garrison and Arrizabalaga found that the disruption of a mitochondrial DNA repair enzyme (TgMSH-1) contributed to parasite resistance to both monensin and the potassium ionophore salinomycin (Garrison and Arrizabalaga, 2009). A study of the transcriptomic response of T. gondii parasites to monensin and salinomycin identified significant upregulation of histones and other genes associated with cell cycle arrest (Lavine and Arrizabalaga, 2011). However, the transcriptomic profile of trtE-treated T. gondii (. Table S2 and. Fig S1) was not in any way similar to that of monensin/salinomycin-treated parasites, suggesting that trtE has a different mechanism of action from these ionophores.
Using a combination of transcriptomic and genomic strategies, we identified a conserved coccidian gene that plays a role in the susceptibility of T. gondii parasites to the anti-apicomplexan compound, trtE. The function of the trg gene product remains entirely unclear, as does its relevance to the mechanism of action of trtE against non-coccidian apicomplexans. Further study into the function of Trg may clarify the specific mechanism of action of trtE against coccidans and identify a new drug target common to coccidian parasites of human and animal importance.
Declaration of competing interest
Please declare any financial or personal interests that might be potentially viewed to influence the work presented. Interests could include consultancies, honoraria, patent ownership or other. If there are none state ‘there are none’.
Acknowledgements
This work was supported by the National Center for Complimentary and Integrative Health, National Institutes of Health (grant number R21 AT009174) to RMO and support from the Department of Veterinary Microbiology and Pathology, Washington State University, College of Veterinary Medicine. GDB is a recipient of a Poncin Scholarship for Medical Research awarded by the Cora May Poncin Trust.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ijpddr.2020.07.003.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Alignment of Trg homologs. Protein sequences for all homologs of Trg identified in this study were aligned using Clustal W and annotated using the AlignX module of Vector NTI.
figs1RNA-Seq results. Differentially expressed genes of trtE-treated T. gondii RH parasites at 4, 8, and 12 h post treatment as determined by RNA-seq.
figs2RT-qPCR verification of RNA-Seq results. Expression of trg, TGGT1_311100, and bag1 transcripts in RNA extracted from T. gondii parasites treated with trtE (24.2 nM, red) or DMSO carrier control (0.1%, black) at 1, 2, and 4 h post treatment. Fold-change in gene expression was calculated using the 2−ΔΔCt method with three replicates. Treatment of parasites with trtE significantly increased the expression of trg at all time points (p = 0.04, 0.01, and 0.004 at 1, 2, 4 h, respectively). Expression of TGGT1_311100 during trtE treatment was not significantly different from treatment with DMSO (p = 0.4, 0.5, and 0.7 at 1, 2, 4 h respectively). At 4 h post treatment with trtE, expression of bradyzoite-specific gene bag1 was significantly lower (p = 0.001) than DMSO-treated parasites. Three technical replicates were performed on all samples.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Behnke M.S., Radke J.B., Smith A.T., Sullivan W.J., White M.W. The transcription of bradyzoite genes in Toxoplasma gondii is controlled by autonomous promoter elements. Mol. Microbiol. 2008;68:1502–1518. doi: 10.1111/j.1365-2958.2008.06249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke M.S., Zhang T.P., Dubey J.P., Sibley L.D. Toxoplasma gondii merozoite gene expression analysis with comparison to the life cycle discloses a unique expression state during enteric development. BMC Genom. 2014;15 doi: 10.1186/1471-2164-15-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N., Gammeltoft S., Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4(6):1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Caballero M.C., Alonso A.M., Deng B., Attias M., De Souza W., Corvi M.M. Identification of new palmitoylated proteins in Toxoplasma gondii. Biochim. Biophys. Acta Protein Proteonomics. 2016;1864:400–408. doi: 10.1016/j.bbapap.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzinet S., Dubremetz J.F., Buzoni-Gatel D., Jeminet G., Prensier G. In vitro activity of the polyether ionophorous antibiotic monensin against the cyst form of Toxoplasma gondii. Parasitology. 2000;121:359–365. doi: 10.1017/S0031182099006605. [DOI] [PubMed] [Google Scholar]
- Delorme-Walker V., Abrivard M., Lagal V., Anderson K., Perazzi A., Gonzalez V., Page C., Chauvet J., Ochoa W., Volkmann N., Hanein D., Tardieux I. Toxofilin upregulates the host cortical actin cytoskeleton dynamics, facilitating Toxoplasma invasion. J. Cell Sci. 2012;125:4333–4342. doi: 10.1242/jcs.103648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybas J.M., Madrid-Aliste C.J., Che F.Y., Nieves E., Rykunov D., Angeletti R.H., Weiss L.M., Kim K., Fiser A. Computational analysis and experimental validation of gene predictions in Toxoplasma gondii. PloS One. 2008;3:e3899. doi: 10.1371/journal.pone.0003899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshahawi S.I., Trindade-Silva A.E., Hanora A., Han A.W., Flores M.S., Vizzoni V., Schrago C.G., Soares C.A., Concepcion G.P., Distel D.L., Schmidt E.W., Haygood M.G. Boronated tartrolon antibiotic produced by symbiotic cellulose-degrading bacteria in shipworm gills. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E295–E304. doi: 10.1073/pnas.1213892110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe I.T., Child M.A., Majmudar J.D., Krishnamurthy S., Van Der Linden W.A., Ward G.E., Martin B.R., Bogyo M. Global analysis of palmitoylated proteins in Toxoplasma gondii. Cell Host Microbe. 2015;18:501–511. doi: 10.1016/j.chom.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz H.M., Buchholz K.R., Chen X., Durbin-Johnson B., Rocke D.M., Conrad P.A., Boothroyd J.C. Transcriptomic analysis of toxoplasma development reveals many novel functions and structures specific to sporozoites and oocysts. PloS One. 2012;7 doi: 10.1371/journal.pone.0029998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajria B., Bahl A., Brestelli J., Dommer J., Fischer S., Gao X., Heiges M., Iodice J., Kissinger J.C., Mackey A.J., Pinney D.F., Roos D.S., Stoeckert C.J., Wang H., Brunk B.P. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkm981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E.M., Arrizabalaga G. Disruption of a mitochondrial MutS DNA repair enzyme homologue confers drug resistance in the parasite Toxoplasma gondii. Mol. Microbiol. 2009;72:425–441. doi: 10.1111/j.1365-2958.2009.06655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouw M., Michael S., Sámano-Sánchez H., Kumar M., Zeke A., Lang B., Bely B., Chemes L.B., Davey N.E., Deng Z., Diella F., Gürth C.-M., Huber A.-K., Kleinsorg S., Schlegel L.S., Palopoli N., Roey K.V., Altenberg B., Reményi A., Dinkel H., Gibson T.J. The eukaryotic linear motif resource - 2018 update. Nucleic Acids Res. 2018;46:D428–D434. doi: 10.1093/nar/gkx1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumila C., Ancelin M.L., Delort A.M., Jeminet G., Vial H.J. Characterization of the potent in vitro and in vivo antimalarial activities of ionophore compounds. Antimicrob. Agents Chemother. 1997;41:523–529. doi: 10.1128/aac.41.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P., Park K.-J., Obayashi T., Fujita N., Harada H., Adams-Collier C.J., Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:585–587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K.L., Contreras-Moreira B., De Silva N., Maslen G., Akanni W., Allen J., Alvarez-Jarreta J., Barba M., Bolser D.M., Cambell L., Carbajo M., Chakiachvili M., Christensen M., Cummins C., Cuzick A., Davis P., Fexova S., Gall A., George N., Gil L., Gupta P., Hammond-Kosack K.E., Haskell E., Hunt S.E., Jaiswal P., Janacek S.H., Kersey P.J., Langridge N., Maheswari U., Maurel T., Mcdowall M.D., Moore B., Muffato M., Naamati G., Naithani S., Olson A., Papatheodorou I., Patricio M., Paulini M., Pedro H., Perry E., Preece J., Rosello M., Russell M., Sitnik V., Staines D.M., Stein J., Tello-Ruiz M.K., Trevanion S.J., Urban M., Wei S., Ware D., Williams G., Yates A.D., Flicek P. Ensembl Genomes 2020-enabling non-vertebrate genomic research. Nucleic Acids Res. 2019;48:689–695. doi: 10.1093/nar/gkz890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irschik H., Schummer D., Gerth K., Höfle G., Reichenbach H. The tartrolons, new boron-containing antibiotics from a myxobacterium, Sorangium cellulosum. J. Antibiot. (Tokyo) 1995;48:26–30. doi: 10.7164/antibiotics.48.26. [DOI] [PubMed] [Google Scholar]
- Kevin Ii D.A., Meujo D.A., Hamann M.T. Polyether ionophores: broad-spectrum and promising biologically active molecules for the control of drug-resistant bacteria and parasites. Expet Opin. Drug Discov. 2009;4:109–146. doi: 10.1517/17460440802661443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., Von Heijne G., Sonnhammer E.L.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lavine M.D., Arrizabalaga G. The antibiotic monensin causes cell cycle disruption of Toxoplasma gondii mediated through the DNA repair enzyme TgMSH-1. Antimicrob. Agents Chemother. 2011;55:745–755. doi: 10.1128/AAC.01092-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewer P., Chapin E.L., Graupner P.R., Gilbert J.R., Peacock C. Tartrolone C: a novel insecticidal macrodiolide produced by Streptomyces sp. CP1130. J. Nat. Prod. 2003;66:143–145. doi: 10.1021/np020451s. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., Geer R.C., He J., Gwadz M., Hurwitz D.I., Lanczycki C.J., Lu F., Marchler G.H., Song J.S., Thanki N., Wang Z., Yamashita R.A., Zhang D., Zheng C., Bryant S.H. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayoral J., Di Cristina M., Carruthers V.B., Weiss L.M. Methods in Molecular Biology. Humana Press Inc.; 2020. Toxoplasma gondii: bradyzoite differentiation in vitro and in vivo; pp. 269–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald V., Stables R., Warhurst D.C., Barer M.R., Blewett D.A., Chapman H.D., Connolly G.M., Chiodini P.L., McAdam K.P.W.J. In vitro cultivation of Cryptosporidium parvum and screening for anticryptosporidial drugs. Antimicrob. Agents Chemother. 1990;34:1498–1500. doi: 10.1128/AAC.34.8.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Acosta E., Cross G.A.M. Rapid isolation of DNA from trypanosomatid protozoa using a simple “mini-prep” procedure. Mol. Biochem. Parasitol. 1993;59:327–329. doi: 10.1016/0166-6851(93)90231-L. [DOI] [PubMed] [Google Scholar]
- Mineo J.R., Kasper L.H. Attachment of Toxoplasma gondii to host cells involves major surface protein, SAG-1 (P-30) Exp. Parasitol. 1994;79:11–20. doi: 10.1006/expr.1994.1054. [DOI] [PubMed] [Google Scholar]
- Mineo J.R., McLeod R., Mack D., Smith J., Khan I.A., Ely K.H., Kasper L.H. Antibodies to Toxoplasma gondii major surface protein (SAG-1, P30) inhibit infection of host cells and are produced in murine intestine after peroral infection. J. Immunol. 1993;150:3951–3964. [PubMed] [Google Scholar]
- Moreira W., Aziz D.B., Dick T. Boromycin kills mycobacterial persisters without detectable resistance. Front. Microbiol. 2016;7 doi: 10.3389/fmicb.2016.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators Database resources of the national center for Biotechnology information. Nucleic Acids Res. 2016;44:D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor R.M., Nepveux V.F.J., Abenoja J., Bowden G., Reis P., Beaushaw J., Relat R.M.B., Driskell I., Gimenez F., Riggs M.W., Schaefer D.A., Schmidt E.W., Lin Z., Distel D.L., Clardy J., Ramadhar T.R., Allred D.R., Fritz H.M., Rathod P., Chery L., White J. A symbiotic bacterium of shipworms produces a compound with broad spectrum anti-apicomplexan activity. PLoS Pathog. 2020;16(5) doi: 10.1371/journal.ppat.1008600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pache W., Zähner H. Metabolic products of microorganisms. 77. Studies on the mechanism of action of boromycin. Arch. für Mikrobiol. 1969;67:156–165. [PubMed] [Google Scholar]
- Pfefferkorn E.R., Pfefferkorn L.G. Toxoplasma gondii: isolation and preliminary characterization of temperature-sensitive mutants. Exp. Parasitol. 1976;39:365–376. doi: 10.1016/0014-4894(76)90040-0. [DOI] [PubMed] [Google Scholar]
- Poupel O., Boleti H., Axisa S., Couture-Tosi E., Tardieux I. Toxofilin, a novel actin-binding protein from Toxoplasma gondii, sequesters actin monomers and caps actin filaments. Mol. Biol. Cell. 2000;11:355–368. doi: 10.1091/mbc.11.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 2008;21:639–644. doi: 10.1093/protein/gzn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos D.S., Donald R.G.K., Morrissette N.S., Moulton A.L.C. Molecular tools for genetic dissection of the Protozoan parasite Toxoplasma gondii. Methods Cell Biol. 1995;45:27–63. doi: 10.1016/S0091-679X(08)61845-2. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. Standard ethanol precipitation of DNA in microcentrifuge tubes. CSH Protoc. 2006;2006(1) doi: 10.1101/pdb.prot4456. pdb.prot4456. Published 2006 Jun 1. [DOI] [PubMed] [Google Scholar]
- Schummer D., Irschik H., Reichenbach H., Höfle G. Antibiotics from gliding bacteria, LVII. Tartrolons: new boron‐containing macrodiolides from Sorangium cellulosum. Liebigs Ann. Chem. 1994:283–289. doi: 10.1002/jlac.199419940310. 1994. [DOI] [Google Scholar]
- Shen B., Brown K.M., Lee T.D., David Sibley L. Efficient gene disruption in diverse strains of Toxoplasma gondii Using CRISPR/CAS9. mBio. 2014;5 doi: 10.1128/mBio.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidik S.M., Huet D., Ganesan S.M., Huynh M.-H., Wang T., Nasamu A.S., Thiru P., Saeij J.P.J., Carruthers V.B., Niles J.C., Lourido S. A genome-wide CRISPR screen in toxoplasma identifies essential apicomplexan genes. Cell. 2016;166:1423–1435. doi: 10.1016/j.cell.2016.08.019. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati D., Boothroyd J.C. Transient transfection and expression in the obligate intracellular parasite Toxoplasma gondii. Science (80-. ) 1993;260:349–352. doi: 10.1126/science.8469986. [DOI] [PubMed] [Google Scholar]
- Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28 doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- Surup F., Chauhan D., Niggemann J., Bartok E., Herrmann J., Keck M., Zander W., Stadler M., Hornung V., Müller R. Activation of the NLRP3 inflammasome by hyaboron, a new asymmetric boron-containing macrodiolide from the myxobacterium Hyalangium minutum. ACS Chem. Biol. 2018;13:2981–2988. doi: 10.1021/acschembio.8b00659. [DOI] [PubMed] [Google Scholar]
- Tarleton R., Peng D. EuPaGDT: a web tool tailored to design CRISPR guide RNAs for eukaryotic pathogens. Microb. Genom. 2015;1 doi: 10.1099/mgen.0.000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins+ D.G., Gibson T.J. Nucleic Acids Research; 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin C.M., Knoll L.J. 2012. A Patatin-like Protein Protects Toxoplasma Gondii from Degradation in a Nitric Oxide-dependent Manner. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treeck M., Sanders J.L., Elias J.E., Boothroyd J.C. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites' boundaries. Cell Host Microbe. 2011;10:410–419. doi: 10.1016/j.chom.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of Trg homologs. Protein sequences for all homologs of Trg identified in this study were aligned using Clustal W and annotated using the AlignX module of Vector NTI.