Abstract
The cytochrome P450 enzyme CYP-sb21 from the rare actinomycete Sebekia benihana is capable of hydroxylating the immunosuppressive drug molecule cyclosporine A (CsA) primarily at the 4th N-methyl leucine (MeLeu4), giving rise to γ-hydroxy-N-methyl-l-Leu4-CsA (CsA-4-OH). This oxidative modification of CsA leads to dramatically reduced immunosuppressive activity while retaining the hair growth-promoting side-effect, thus demonstrating great application potential in both pharmaceutical and cosmetic industries. However, this P450 enzyme also hydroxylates CsA at the unwanted position of the 9th N-methyl leucine (MeLeu9), indicating that the regioselectivity needs to be improved for the development of CsA-4-OH into a commercial hair growth stimulator. Herein, we report the crystal structure of CYP-sb21 in its substrate-free form at 1.85 Å. Together with sequence and 3D structure comparisons, Autodock-based substrate docking, molecular dynamics (MD) simulation, and site-directed mutagenesis, we identified a number of key residues including R294, E264, and M179 that can improve catalytic efficiency or change the regioselectivity of CYP-sb21 towards CsA, setting the stage for better enzymatic preparation of CsA-4-OH. This study also provides new insights into the substrate recognition and binding mechanism of P450 enzymes that accommodate bulky substrates.
Keywords: Cytochrome P450 monooxygenase, Crystal structure, Cyclosporine A, Regioselectivity, Hair growth stimulator
1. Introduction
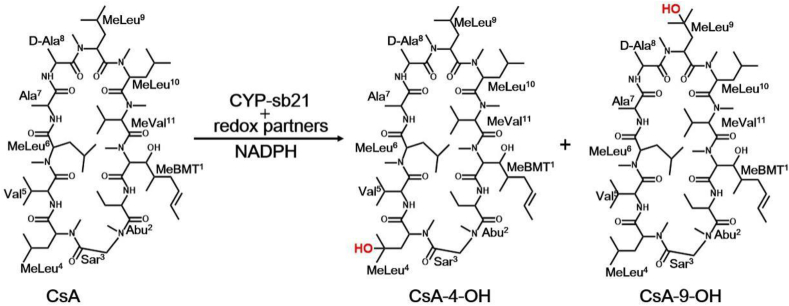
Cyclosporin A (CsA) is a cyclic peptide comprised of eleven amino acids including ten l-amino acids and a single d-amino acid (Fig. 1) [1]. Unlike ribosomal peptides, CsA is a natural product assembled by a nonribosomal peptide synthetase (NRPS), cyclosporin synthetase [2]. CsA was first isolated in 1971 from the fungus Tolypocladium inflatum and came into medical use in 1983 [3]. Today, CsA has been one of the most widely used immunosuppressants to treat and prevent the graft-versus-host diseases in bone marrow transplantation and to prevent rejection of kidney, heart, and liver transplants [[4], [5], [6]].
Fig. 1.
Alternative CsA hydroxylation reactions catalyzed by CYP-21b.
Interestingly, the hair growth stimulating side-effect of CsA has also attracted significant attention due to its application potential in both pharmaceutical and cosmetic industries. To date, significant efforts have been made for generation of the CsA derivatives that lose the immunosuppressive potency but retain the hair-growth-promoting activity. Among a wide spectrum of CsA derivatives with alternative amino acid substitutions, γ-hydroxy-N-methyl-L-Leu4-CsA (CsA-4-OH) and γ-hydroxy-N-methyl-L-Leu9-CsA (CsA-9-OH) were both found to show the desired decoupled activities, with the former favored due to its higher hair-restoring activity, which is 100-fold more effective than the existing commercial product minoxidil in a mice skin graft experiment. Moreover, CsA-4-OH displayed a 10-fold lower immunosuppressive activity than CsA-9-OH [[7], [8], [9], [10]]. Despite of these promising results, it is challenging to realize industrial production of CsA-4-OH for meaningful clinic tests by chemical methods, owing to the complex process, the low yield, and the prohibitive high cost. Therefore, the enzymes that can efficiently catalyze the hydroxylation at the specific site of CsA have long been pursued.
Cytochrome P450 (CYP) enzymes belong to a superfamily of heme-thiolate monooxygenases that exist in all kingdoms of life [11,12]. P450 enzymes can catalyze a vast variety of reactions such as C-, N-, and S-hydroxylation, epoxidation, dealkylation, desaturation, aromatic coupling, cyclopropanation, and nitration [[12], [13], [14]]. Compared with chemical catalysts, P450 enzymes have the significant advantage of being able to regio- and stereo-specifically activate and modify the inert C–H bond in various substrates under mild conditions [[12], [13], [14]].
CYP-sb21 (CYP107Z14) from the rare actinomycete Sebekia benihana was previously reported to be capable of introducing a hydroxyl group at the γ position in either the 4th N-methyl leucine (MeLeu [4]) or the 9th N-methyl leucine (MeLeu [9]) of CsA with the former regioselectivity preferred [7,16,17] (Fig. 1). Through domain swapping, it was revealed that the 2nd domain (136–225 amino acids) of CYP-sb21 plays a critical role in its regioselectivity [[7], [16], [17]]. However, the structural basis for substrate recognition and regioselectivity of CYP-sb21 has yet to be elucidated, with which a specific P450 CsA-4-OH synthase (rather than a dual hydroxylation enzyme) is expected to be engineered.
Thus, in this study, we solved the substrate-free structure of CYP-sb21 for the first time. Together with substrate docking, MD simulation, and comparative analysis of CYP-sb21 and the two other functionally related P450 enzymes, we identified a putative CsA binding pocket, in which nine amino acids were selected as mutation sites for tuning the substrate binding and regioselectivity of CYP-sb21. As a result, we obtained a number of mutants with the improved MeLeu4-hydroxylating activity, increased catalytic efficiency, or new oxidative products.
2. Materials and methods
2.1. Construction of recombinant plasmids
The vectors pET-28b-cyp-sb21, pET28b-sefdx, and pET28b-sefdR for expression of CYP-sb21, Synechococcus elongatus PCC 7942 ferredoxin (seFdx), and Synechococcus elongatus PCC 7942 ferredoxin reductase (seFdR), respectively, were previously constructed in our laboratories [17]. All mutants listed in Table S3 were generated by applying mutagenic PCR to pET28b-cyp-sb21 according to QuikChange protocol.
2.2. Protein expression, purification and concentration measurement
Escherichia coli BL21(DE3) was transformed by a recombinant plasmid, and a single colony of the transformant was inoculated into 750 mL LB media containing kanamycin (50 mg/L, Sangon, China), thiamine (1 mM, Sigma, USA), and 10% glycerol (m/v), and then cultured at 37 °C until OD600 reached 0.6–1.0 (~6–8 h). Next, 0.2 mM isopropyl β-d-thiogalactoside (IPTG, Sigma, USA) was added to induce protein expression, and 1 mM δ-aminolevulinic acid (5-ALA, Sigma, USA) was supplemented to supply the heme synthetic precursor. The cells were shaking incubated at 160 rpm for 16–20 h at 20 °C for better protein folding. All proteins first were purified with Ni-NTA affinity chromatography as described before [18]. To further improve protein purity, wild-type or mutant CYP-sb21, or seFdR were load onto a HiLoad 16/600 Supdex 200 pg column (GE Healthcare, USA), and seFdx was load onto a HiLoad 16/600 Supdex 75 pg column (GE Healthcare, USA). These columns were equilibrated with buffer A (50 mM NaH2PO4, 300 mM NaCl, 10% (w/v) glycerol, pH 8.0), and the protein peaks were collected, combined, and concentrated for enzyme reactions. After wild-type CYP-sb21 was diluted with buffer B (50 mM Tris, 10% (w/v) glycerol, pH 8.0) and loaded onto a Mono Q 5/50 GL column (GE Healthcare, USA), it was eluted by a linear gradient of buffer B and buffer C (50 mM Tris, 1 M NaCl pH 8.0), and the resulting peak was collected for crystallization. All samples were snap frozen in liquid nitrogen and stored at - 80 °C for later use. The functional concentrations of CYP-sb21 and its mutants were calculated from the CO-bound reduced difference spectra using an extinction coefficient (ε450–490) of 91,000 M−1 cm−1 [19]. The concentrations of redox partner proteins and CYP-sb21 prepared for crystallization were determined by SimpliNano (GE Healthcare, USA).
2.3. In vitro enzyme assays for CsA hydroxylation by CYP-sb21
A standard in vitro reaction mixture in 100 μL reaction buffer (50 mM NaH2PO4, 10% glycerol, pH 7.4) contained 10 μM wild-type or mutant CYP-sb21, 10 μM seFdx, 10 μM seFdR, 20 or 50 μM CsA (diluted from a 50 mM stock solution in methanol), and 1 mM NADPH; and 5 U glucose dehydrogenase (GDH) and 20 mM glucose were supplemented as an NADPH regeneration system. The reaction was quenched by adding 100 μL methanol after the incubation at 30 °C for 16 h. The samples were centrifuged at 20,000 ×g to remove the precipitated proteins, and the supernatants were analyzed by high-performance liquid chromatography (HPLC) with an SB-C18 column and the HPLC method the same as previously reported [17].
2.4. Liquid chromatography-high resolution mass spectrometry (LC-HRMS) analysis
The LC-HRMS analysis was performed as reported [18] on a Waters symmetry column (4.6 × 150 mm, RP18) using the negative-mode electrospray ionization with a linear gradient of 10–100% acetonitrile in ddH2O with 0.1% formic acid over 20 min, and followed by 100% acetonitrile for 5 min at a flow rate of 0.5 mL/min. The high resolution mass spectra were recorded on a Dionex Ultimate 3000 coupled to a Bruker Maxis Q-TOF.
2.5. Docking and MD simulation
CsA was docked into the structure of CYP-sb21 in the zwitterionic form using AutoDock 4.2 [20]. Regents and water molecules were removed before docking. All side chains were set as rigid body and grid spacing was set to 1 Å. Other parameters remained as their default values. The top 10 lowest energy docking poses of CsA from 2,500,000 searching results were chosen and they all showed relevant enzyme-substrate interactions. MD simulations were carried out using Gromacs 4.5 [21] and the Amber ff99SB force field [22]. In the simulation, the CYP-sb21 with CsA was solvated by adding 10.0 Å TIP3P water in a rectangular box. After 50,000 steps of energy minimization and 10 ns of equilibrium MD, 50 ns of production run was performed at constant pressure (1 atm) and temperature (298 K), with the snapshots saved every 50 ps which were then used in data analysis.
2.6. Crystallization and structure determination
The crystal of substrate-free CYP-sb21 was obtained at 16 °C by hanging drop vapor diffusion and the crystal screen droplets consisted of a 1:1 (v/v) protein at 20 mg/mL and the well solution of 100 mM Bis-Tris, pH 6.5, 200 mM magnesium chloride hexahydrate, and 25% PEG3350. Crystals appeared after 3 d and were ready for data collection in two weeks. The crystals were flash-frozen in liquid nitrogen. The diffraction data were collected at 100 K under the synchrotron radiation at beamline BL19U1 of the Shanghai Synchrotron Radiation Facility (SSRF). The data sets were integrated and scaled with the HKL3000 package [23]. The structure was determined by molecular replacement with the structure of saAcmM (PDB ID: 5NWS) as the initial search model with the program Phaser. The programs Refmac5 and Coot9 were used for the refinement and model building [[24], [25], [26], [27]]. Ramachandran plots were generated with Coot9. The statistics for data processing and structure refinement are shown in Table 1. The coordinate was deposited to Protein Data Bank with the PDB ID code of 6M4S. Figures were prepared using PYMOL (http://www.pymol.org).
Table 1.
Data collection and refinement statistics for CYP-sb21.
| CYP-sb21 | |
|---|---|
| Data collection | |
| Beamline | BL19U1 |
| Wavelength | 0.979 |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 41.636 91.092 52.975 |
| a, b, g (°) | 90.000 94.649 90.000 |
| Resolution (Å) | 50.0–1.85 (1.88-1.85) |
| Unique reflections | 33433 (1679) |
| Rmerge | 0.052 (0.411) |
| Rpim | 0.027(0.224) |
| I/σ | 23.1 (3.36) |
| CC1/2 | 0.991 (0.902) |
| Completeness (%) | 99.4 (99.2) |
| Redundancy | 6.7 (6.4) |
| Refinement | |
| Resolution (Å) | 1.85 |
| No. reflections | 33433 |
| Rwork/Rfree | 0.174/0.225 |
| No. atoms | |
| Protein | 3046 |
| Heme | 43 |
| Water | 261 |
| B-factors | |
| Protein | 23 |
| Heme | 14 |
| Water | 33 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.019 |
| Bond angles (°) | 1.983 |
Highest-resolution shell is shown in parentheses
3. Results
3.1. Overall features of the CYP-sb21 structure
We determined the crystal structure of the substrate-free CYP-sb21 at 1.85 Å resolution (PDB ID: 6M4S) with one molecule in the asymmetric unit. The molecular replacement was conducted using the structure of saAcmM (PDB ID: 5NWS) as the initial search model. The CYP-sb21 crystals belong to space group P21, with unit-cell parameters a = 41.6 Å, b = 91.1 Å, c = 53.0 Å, α = 90.0°, β = 94.6°, and γ = 90.0°. Detailed crystallographic data statistics for CYP-sb21 are shown in Table 1. However, we were unable to obtain the structure of CYP-sb21 in complex with CsA despite of our greatest efforts on both co-crystallization and substrate-soaking attempts. The unsuccess was likely due to the very poor aqueous solubility of CsA and the low substrate binding affinity.
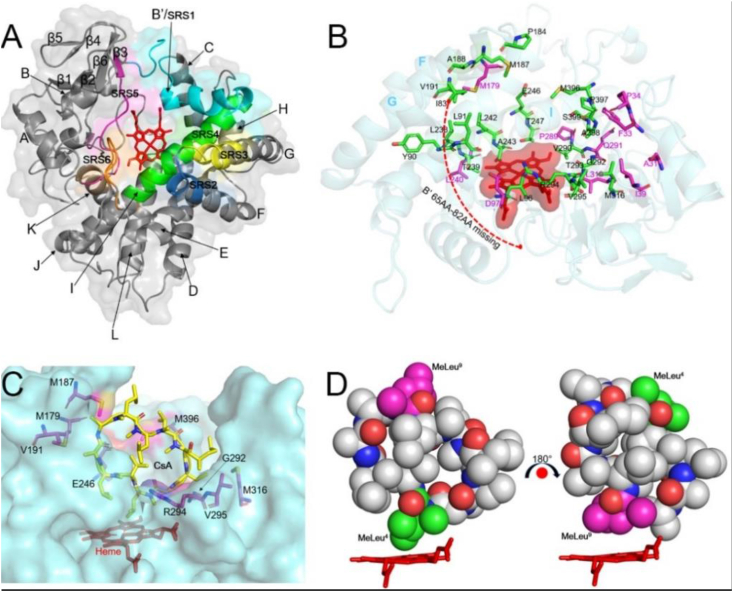
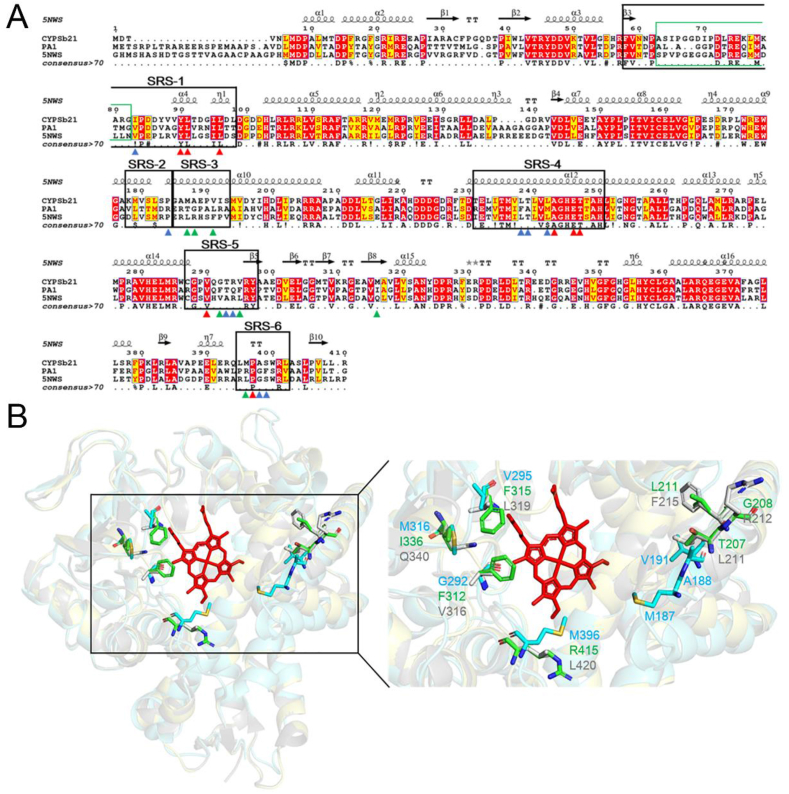
The three-dimensional structure of CYP-sb21 adopts a canonical P450 fold, consisting of mainly α-helix (A-L) and β-sheet (β1-β6) rich regions (Fig. 2A). The six substrate recognition sites (SRSs) of CYP-sb21 including SRS1 (59–100 aa), SRS2 (178–184 aa), SRS3 (185–192 aa), SRS4 (230–250 aa), SRS5 (287–297 aa), and SRS6 (395–402 aa) were defined according to the previously reported method [[24], [25], [26], [27]] (Fig. 2, Fig. 3A). The B’ helix and BC-loop regions that are well-known to play key roles in substrate entry and products egress [29,30], in our structure are flexible with the 65–82 residues not being observed due to missing electron density (Fig. 2A and B). Collectively, our structure reflects an open conformation [18,31]] and the missing BC-loop appears to be a common characteristic for the P450 enzymes that accommodate bulky substrates [[18], [31]].
Fig. 2.
A. Overall structure of CYP-sb21. The conserved secondary structure components (α-helices and β-sheets) are labelled; and the regions of six substrate recognition sites (SRSs) are highlighted in cyan (SRS1, 59–100 aa), blue (SRS2, 178–184 aa), yellow (SRS3, 185–192 aa), green (SRS4, 230–250 aa), magenta (SRS5, 287–297 aa) and orange (SRS6, 395–402 aa). B. The substrate binding pocket for Autodock and MD analyses. The amino acids shown as sticks in green represent the key residues analyzed by both Autodock and MD. The sticks in magenta denote the key amino acids only analyzed by MD. The central heme prosthetic group is shown as stick and surface in red. The unstructured B′ region (56–82 aa) is indicated by a red dashed line. The key helices are labelled by blue capital letters. C. The selected mutation sites. The amino acids that were subject to mutagenesis analysis are shown as sticks in magenta. D. The two productive binding modes of CsA as spheres. MeLeu4 and MeLeu9 are colored in green and magenta, respectively.
Fig. 3.
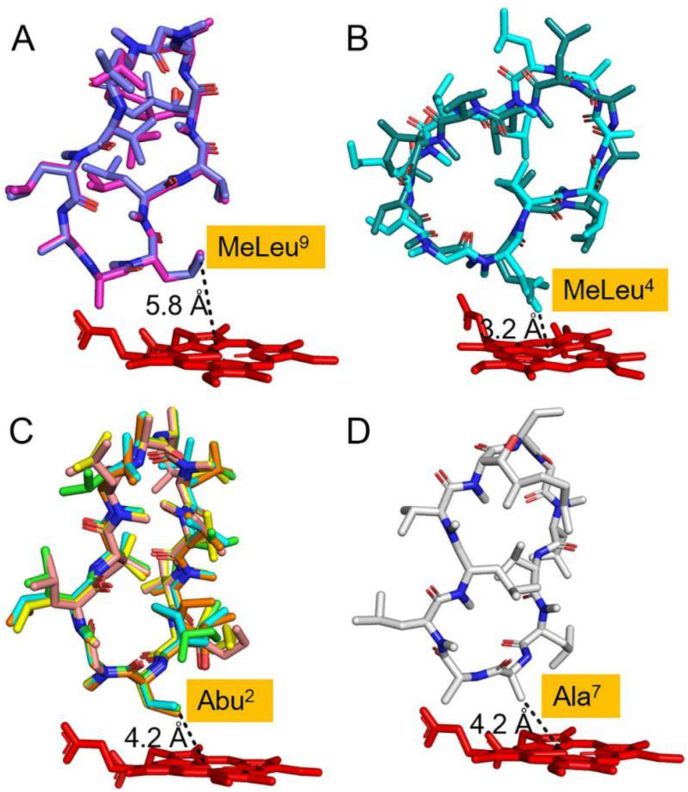
Top ten Autodock poses divided into four groups of conformations. A. Conformation 1 (pose 2 and 6), MeLeu9 nearest the heme-iron center. B. Conformation 2 (pose 8 and 9), MeLeu4 nearest the heme-iron center. C. Conformation 3 (Pose 1, 3, 4, 7 and 10), Abu2nearest the heme-iron center. D. Conformation 4 (Pose 5), Ala7 nearest the heme-iron center. The distances between the nearest amino acids to the heme-iron center are labelled by black dashed lines.
3.2. Substrate docking and MD simulation for CYP-sb21
Due to lack of the crystal structure for the CYP-sb21/CsA complex, the program AutoDock was used to explore potential CsA binding poses. The structure of CYP-sb21 was used as template for creating the starting coordinates. The top 10 lowest energy docking solutions from 2,500,000 searching results were chosen for further analysis. We revealed that the top 10 poses could be divided into four groups of conformations and the key sites nearest the heme-iron reactive center were MeLeu9 (5.8 Å), MeLeu4(3.2 Å), Abu2 (4.2 Å), and Ala7 (4.2 Å) (Fig. 3). Since CYP-sb21 experimentally hydroxylates CsA at MeLeu4 or MeLeu9, the conformations 1 and 2 apparently reflect the actual poses during the catalytic process. Importantly, CsA is roughly a centrosymmetric molecule with MeLeu9 and MeLeu4 located at opposite sides of the symmetry center (Fig. 1, Fig. 2D). Thus, we reason that CYP-sb21 may be unable to well distinguish conformations 1 and 2, thereby giving rise to the two products CsA-4-OH and CsA-9-OH at an accumulative conversion rate of 86% (Fig. 5A) and a 1.5-to-1 product ratio (Fig. 5B). The distance between MeLeu4 and heme-iron in conformation 2 is 3.2 Å, while the distance between MeLeu9 and heme-iron in conformation 1 is 5.8 Å (Fig. 3A and B). This difference in distance may explain why CsA-4-OH is the major product over CsA-9-OH. Although we also obtained two other conformations 3 and 4 (Fig. 3C and D), the hydroxylation products of Abu2 and Ala7 have not been observed either in vitro or in vivo [7,28] for unknown reasons.
Fig. 5.
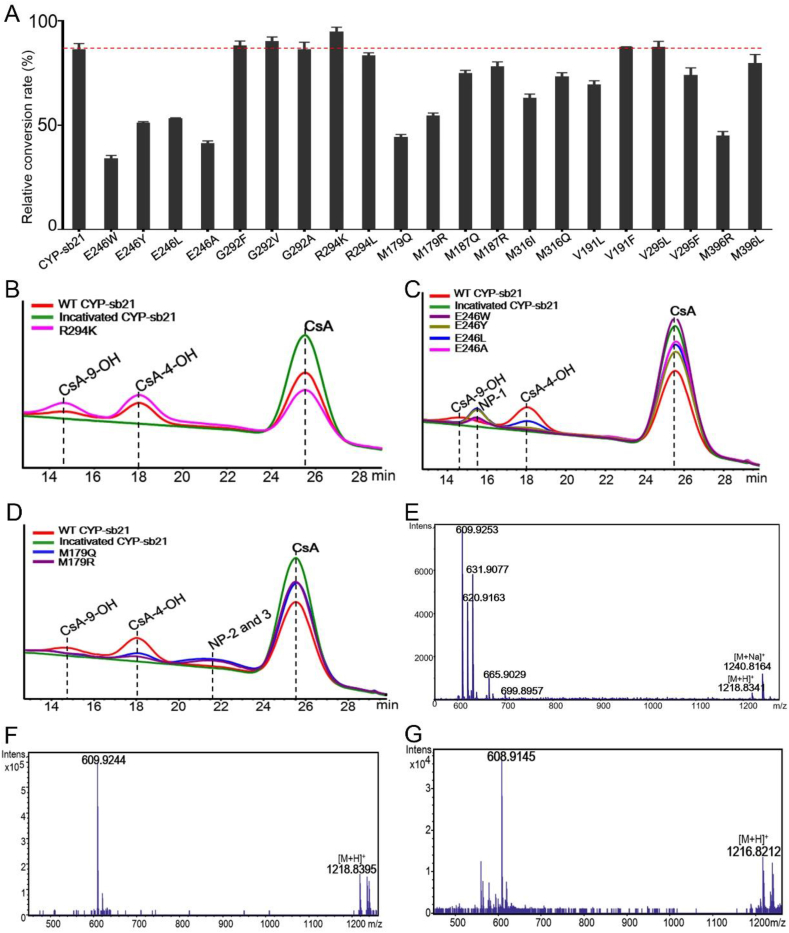
CsA-hydroxylating activities of different CYP-sb21 mutants. A. Substrate conversion rates of wild-type and mutant CYP-21b. A standard in vitro reaction mixture contained 10 μM CYP-21b, 10 μM ferredoxin, 10 μM ferredoxin reductase, 20 μM CsA, and 1 mM NADPH; and 5 U glucose dehydrogenase (GDH) and 20 mM glucose as the NADPH regeneration system. B. HPLC analysis (210 nm) of the mutant R294K with improved activity against 50 μM CsA. C. HPLC analysis (210 nm) of E246W, E246Y, E246L, and E246A that produce CsA-4-OH, CsA-9-OH, and a new product NP-1 from 50 μM CsA. D. HPLC analysis (210 nm) of M179Q and M179R that produce CsA-4-OH, CsA-9-OH, and two co-eluted new products NP-2 and NP-3 from 50 μM CsA. E. The high resolution mass spectrum of NP-1. F. The high resolution mass spectrum of NP-2. G. The high resolution mass spectrum of NP-3.
Next, we carefully analyzed the four docking modes in conformations 1 and 2 using LigPlot+ (v.1.4.5) [32], and revealed 24 amino acids that form the substrate binding pocket of CYP-sb21 (Fig. 2B, Fig. S1, Table S1). Among these residues, Gly292 and Arg294 interact with CsA through hydrogen bonds, which likely play essential roles in specific substrate binding (Fig. S1). To validate this docking mode, we chose pose 8 which is supposed to site-specifically hydroxylate MeLeu [4] as the starting configuration for MD simulation. The binding energies of the important residues within 6 Å of CsA were calculated using the g_energy tool of the MD software Gromacs [21], and the top 10 amino acids in substrate binding pocket were selected according to their energy values (Table S1).
3.3. Identification of the key amino acids that could influence the regioselectivity of CYP-sb21
To further identify the key amino acids that could influence regioselectivity of CYP-sb21, we performed multiple protein sequence alignment and 3D structure comparison for CYP-sb21, CYP-pa1, and saAcmM (i.e., CYP-X). CYP-pa1, another CsA hydroxylase from Pseudonocardia autotrophica, was reported to show the opposite regioselectivity (i.e., MeLeu9 rather than MeLeu4 is the preferred hydroxylation site) when compared with CYP-sb21 [7]. saAcmM, which was identified from S. antibioticus, catalyzes a specific hydroxylation of bicyclic chromopeptide lactones [31]. The sequence identities and similarities between CYP-sb21 and CYP-pa1, CYP-sb21 and saAcmM, and CYP-pa1 and saAcmM are 55% and 67%, 52% and 64%, and 48% and 58%, respectively (Fig. 4A). The root-mean-square deviation (RMSD) between CYP-sb21 and the modelled structure of CYP-pa1, and between CYP-sb21 and saAcmM are 0.73 and 0.74, respectively. These results indicate that the three enzymes are analogous to one another. Their substrate binding pockets are conservative with only 7 out of 24 amino acids being non-conservative (Fig. 4B, Table S1). We hypothesized that these differential residues might play important roles in the determination of substrate specificity, catalytic activity, and regioselectivity of CsA. Thus, from the substrate binding pocket we selected the 7 amino acids (E246, M179, M187, V191, V295, M316 and M396) that are non-conservative in all three P450 enzymes and the other two amino acids (G292 and R294) that may play key roles in substrate binding based on Autodock and MD analyses (Fig. 2C) as the targets for mutagenesis analysis.
Fig. 4.
Comparison of CYP-sb21, CYP-pa1 and saAcmM (CYP-X). A. Protein sequence alignment of CYP-sb21, CYP-pa1, and saAcmM. The 24 key amino acids in the CsA binding pocket (as described in Fig. 1B) indicated by red, blue, and green triangles denote all identical, two out of three identical, and all different amino acids among the three P450 enzymes, respectively. The regions of six substrate recognition sites (SRSs) are marked by black rectangles. The green rectangle represents the missing region both in CYP-sb21 and saAcmM three-dimensional structures. Sequence analysis was performed using Expresso through the T-COFFEE online service, and the figure was prepared by ESPript 3.0. The η symbol represents a 310-helix. α-Helices and β-strands are indicated as helices and black arrows, respectively; strict β-turns as TT letters and strict α-turns as TTT. B. The 3D structures of CYP-sb21 (PDB ID: 6M4S), CYP-pa1 (simulated by SWISS-MODEL), and saAcmM (PDB ID: 5NWS) shown as cartoon in aquamarine, yelloworange, and gray, respectively. The 7 non-conservative residues among the three P450 enzymes are shown as sticks in aquamarine (CYP-sb21), green (CYP-pa1), and gray (saAcmM).
3.4. Mutagenesis analysis of CYP-sb21
To investigate the functionality of the 9 selected amino acids, we re-interrogated the Autodock poses and found that the bulky substrate CsA mainly interacts with these residues via hydrophobic contacts; and only G292 and R294 in some poses interact with CsA through hydrogen bonds (Fig. S1). Thus, we designed a number of mutants including G292A, G292F, G292V, R294L and R294K to determine the influence of the size, the hydrophobicity, and the charge of these two amino acids on the activity of CYP-sb21. E246 is the only residue that does not contact CsA in pose 8 and 9 that are presumably responsible for the MeLeu4 hydrxylation (Fig. S2). Thus, we mutated it into hydrophobic amino acids with differently sized side chains (E246W, E246Y, E246L, E246A), attempting to reduce by-products by changing the pocket's polarity and size. M179, which is located in FG helices region (Fig. 2B and C) that is important in substrate entry and product egress [33,34], demonstrated high energy change (−12.73 kJ/mol) in MD analysis (Table S1). Therefore, we mutated it into a couple of amino acids (M179Q, M179R) with a similar size of side chain and that may be more likely to form hydrogen bond with the substrate. The remaining five key amino acids which are non-conservative in sequence alignment were mutated into the amino acids in the same positions of the other two enzymes (Fig. 4, Table S1), including M187Q, M198R, M316I, M316Q, V191L, V191F, V295L, V295F, 396R, and M396L.
The HPLC analysis of the activities of mutants (supported by the cyanobacterial surrogate redox partner proteins seFdx and seFdR) [35,36] showed that the substrate conversion rates of G292 and R294, especially R294K (94.6%) were improved compared to that of wild-type CYP-sb21 (86.0%) (Fig. 5A and B). The CsA-4-OH ratio of R294K (65.0%) is similar to that of wild-type CYP-sb21 (69.0%). Although the conversion rates of E246 and M179 mutants were reduced, these mutants completely lost the ability to produce CsA-9-OH (Fig. 5C and D). The product ratios of CsA-4-OH of E246W, E246Y, E246L, E246A, M179Q and M179R were 5.6%, 17.0%, 42.7%, 8.2%, 60.4% and 40.0%, respectively. Interestingly, in addition to CsA-4-OH, the tested E246 and M179 mutants also generated to different extent the new hydroxylated product NP-1 and the co-eluted unknown hydroxylated and carbonylated products NP-2 and NP-3, respectively, on the basis of their mass spectra (Fig. 5C–G). These HPLC results indicated that we had identified the key amino acids that can influence activity or regioselectivity of CYP-sb21. In the future, one could apply random, saturation, or/and combinatorial mutagenesis towards these residues to screen out mutants with a more specific high-yield of CsA-4-OH or new oxidative products.
4. Discussion
Since the first cytochrome P450 structure of P450cam (i.e., CYP101A1) was solved in 1985 [37], a lot of P450 structures have been solved and the overall P450-fold has turned out to be quite conserved, which consists of mainly α-helix and β-sheet rich regions (Fig. 2A). It has been widely accepted that a P450 catalytic process undergoes through the conformational interchanges of F/G helices and/or B′ helix between the closed or open forms for substrates entry and product egress [29,30]. Compared with other P450 structures, the substrate-free structure of CYP-sb21 is quite similar to those of Rif16 and saAcmM, in all of which B′ helix is highly flexible and there is no electron density for this region, suggesting a widely open conformation. The protein sequence alignment of CYP-sb21, CYP-pa1, and saAcmM indicates that all these P450 enzymes have a very long B’ region as reported for Rif16 [[29], [30]] (Fig. S3). Interestingly, these P450 enzymes all accommodate bulky substrates.
Until March 2020, there have been a total number of 367 unique substrates/ligands co-crystalized with P450s in PDB (http://www.rcsb.org/). As shown in Fig. S4, most (81%) P450 substrates are small molecules with molecular weight (MW) of 0–500. The molecules with MW of 500–700 (15%) are almost all inhibitors [[38], [39], [40], [41]]. Among the molecules with MW of 700–1000 (not including the fatty acyl-acyl carrier protein substrate for P450BioI [42] since this substrate contains a protein moiety to provide a protein-protein interface), only erythromycin D, mycinamicin IV, and rifamycin L are native substrates. Only two substrates in complex with a P450 enzyme have their MWs greater than 1000, and both substrates are attached to a long linker that extends out of the active site [43]. Thus, the mechanism of a P450 enzyme recognizing bulky substrates such as CsA (MW 1202) remains to be elucidated. In the PDB database, rifamycin L (MW 754) represents the largest native substrate of a P450 enzyme. The rifamycin oxidase Rif16 also has a disordered B′ region and adopts an open conformation upon substrate binding [[35], [36]]. These results suggest that the long B’ region of P450s might have to be flexible in order to accept a bulky substrate, thereby leading to an open conformation all the time throughout the catalytic process.
We assume that the low solubility of CsA is one of the main reasons why we failed to obtain the CYP-21b/CsA complex structure. However, there are indeed some structures with CsA as an inhibitor in complex with cyclophilins of different species [[44], [45], [46], [47], [48]]; and a common feature is that 5–6 conservative hydrogen bonds are formed between the CsA binding pocket residues and the 1-, 2-, 3-, 10-, and 11-amino acids of CsA, while the remaining part of CsA is exposed to solvent (Fig. S5, Table S2). This binding mode again suggests that the B’ region should be flexible throughout the whole catalytic process in order to harbor a bulky substrate like CsA. Otherwise, the substrate would likely become an inhibitor with much higher binding affinity. The low substrate binding affinity may be another key reason why we could not obtain the complex of CsA with CYP-sb21, and why there is still no any available co-crystal structure of a P450 enzyme in complex with a small-molecule substrate with MW greater than 1000 in the PDB databank.
Protein rational design can effectively reduce the number of variants to be generated, which in turn significantly reduce the screening workload. However, rational design is usually limited to the enzymes whose structure-activity relationships are well-established [15,49]. Because we could not solve the co-crystallization conditions, AutoDock and MD analyses were therefore used to explore potential CsA binding poses. In our results, we found that the mutants of G292 and R294, especially R294K, improved the conversion efficiency of CYP-21b to a certain extent; the mutants of E264 completely abolished the formation CsA-9-OH; and the mutants of E264 and M179 produced new hydroxylation and carbonylation products. However, the single mutants of 5 non-conservative amino acids in the substrate binding pocket did not achieve the predominant regioselectivity at MeLeu [4]. Thus, we will subsequently use the semi-rational strategies for the identified key positions and screen out mutants with the specific high-yield of CsA-4-OH or any new meaningful products. Importantly, our research provides new insights into substrate recognition and binding mechanisms of P450 enzymes that accommodate bulky substrates, which is anticipated to facilitate the following engineering work for these challenging but useful P450 enzymes [[44], [45], [46], [47], [48]].
CRediT authorship contribution statement
Fengwei Li and Shengying Li designed research; Fengwei Li, Li Ma, Xingwang Zhang, Jingfei Chen, Feifei Qi, Yinyue Huang, Zepeng Qu, Lishan Yao and Wei Zhang performed research; Fengwei Li, Xingwang Zhang, Jingfei Chen, Eung-Soo Kim and Shengying Li analyzed results; Fengwei Li, Eung-Soo Kim and Shengying Li wrote the manuscript; all authors read and approved the manuscript.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2019YFA0905100 to S.L.), the National Natural Science Foundation of China (31800664 to F.L., 31872729 to S.L. and 31600045 to L.M.), General Support from China Postdoctoral Science Foundation (Grant No. 2017M622293 to F.L.), Shandong Provincial Natural Science Foundation, China (ZR2019ZD22 to S.L. and ZR2016CQ05 to L.M.), the Applied Basic Research Programs of Science and Technology of Qingdao (17-1-1-60-jch to L.M.), and National Research Foundation of Korea (No. NRF-2017R1A2A2A05069859 to E.-S.K.). We thank the staff members in beamline BL19U1 of Shanghai Synchrotron Radiation Facility for help in data collection.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2020.07.004.
Contributor Information
Fengwei Li, Email: lifengwei@sdu.edu.cn.
Eung-Soo Kim, Email: eungsoo@inha.ac.kr.
Shengying Li, Email: lishengying@sdu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Borel J.F. History of the discovery of cyclosporin and of its early pharmacological development. Wien Klin Wochenschr. 2002;114:433–437. [PubMed] [Google Scholar]
- 2.Lawen A. Biosynthesis of cyclosporins and other natural peptidyl prolyl cis/trans isomerase inhibitors. Biochim Biophys Acta. 2015;1850:2111–2120. doi: 10.1016/j.bbagen.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Watts R. 2009. Oxford desk reference: rheumatology. [Google Scholar]
- 4.Starzl T.E., Klintmalm G.B.G., Porter K.A., Iwatsuki S., Schroter G.P.J. Liver-transplantation with use of cyclosporin-a and prednisone. N Engl J Med. 1981;305:266–269. doi: 10.1056/NEJM198107303050507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calne R.Y., Thiru S., Mcmaster P. Cyclosporin A in patients receiving renal allografts from cadaver donors. Lancet. 1998;312:1323–1327. doi: 10.1016/s0140-6736(78)91970-0. [DOI] [PubMed] [Google Scholar]
- 6.Kolata G. FDA speeds approval of cyclosporin. Science. 1983;221:1273. doi: 10.1126/science.221.4617.1273-a. [DOI] [PubMed] [Google Scholar]
- 7.Woo M.W., Lee B.R., Nah H.J., Choi S.S., Li S., Kim E.S. Domain characterization of cyclosporin regio-specific hydroxylases in rare actinomycetes. 2015;25:1634–1639. doi: 10.4014/jmb.1505.05034. [DOI] [PubMed] [Google Scholar]
- 8.Kim S.N.A.H., Lee C.W., Lee M.H., Kim J.H., Kim J.I., Kim S.J., Cho H.S., Lee H.S., Kim H.J. Patent DE-1392224; 2004. The use of nonimmunosuppressive [γ-hydroxyN-methyl-L-leucine4] cyclosporin derivatives for treating hair loss. WO02/092033 A1. [Google Scholar]
- 9.Kim S.N.A.H., Lee C.W., Lee M.H., Kim J.H., Kim J.I., Kim S.J., Cho H.S., Lee H.S., Kim H.J. Use of 3-position cyclosporine derivatives for hair growth. US patent. 2004;6:762. 164. [Google Scholar]
- 10.LG Household and Health Care. Korean Patent 1008652110000. 2007.
- 11.Henry H.L. Cytochrome P450 structure, mechanism, and biochemistry. J Am Chem Soc. 1996;118:10945. [Google Scholar]
- 12.Zhang X., Li S. Expansion of chemical space for natural products by uncommon P450 reactions. Nat Prod Rep. 2017;34:1061–1089. doi: 10.1039/c7np00028f. [DOI] [PubMed] [Google Scholar]
- 13.Hussain H.A., Ward J.M. Enhanced heterologous expression of two streptomyces griseolus cytochrome P450s and streptomyces coelicolor ferredoxin reductase as potentially efficient hydroxylation catalysts. Appl Environ Microbiol. 2003;69:373–382. doi: 10.1128/AEM.69.1.373-382.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaufrey F., Delort A.M., Jeminet G., Dauphin G. Bioconversion of monensin by a soil bacterium, Sebekia benihana. J Antibiot. 1990;43:1189–1191. doi: 10.7164/antibiotics.43.1189. [DOI] [PubMed] [Google Scholar]
- 15.Xu L.H., Du Y.L. Rational and semi-rational engineering of cytochrome P450s for biotechnological applications. Synth Syst Biotechnol. 2018;3:283–290. doi: 10.1016/j.synbio.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee M.J., Kim H.B., Yoon Y.J., Han K., Kim E.S. Identification of a cyclosporine-specific P450 hydroxylase gene through targeted cytochrome P450 complement (CYPome) disruption in Sebekia benihana. Appl Environ Microbiol. 2013;79:2253–2262. doi: 10.1128/AEM.03722-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma L., Du L., Chen H., Sun Y., Huang S., Zheng X. Reconstitution of the in vitro activity of the cyclosporine-specific P450 hydroxylase from Sebekia benihana and development of a heterologous whole-cell biotransformation system. Appl Environ Microbiol. 2015;81:6268–6275. doi: 10.1128/AEM.01353-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi F., Lei C., Li F., Zhang X., Wang J., Zhang W. Deciphering the late steps of rifamycin biosynthesis. Nat Commun. 2018;9:2342. doi: 10.1038/s41467-018-04772-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Omura T., Sato R. The carbon monoxide-binding pigment of liver microsomes. Ii. Solubilization, purification, and properties. J Biol Chem. 1964;239:2379–2385. [PubMed] [Google Scholar]
- 20.Morris G.M., Ruth H., William L., Sanner M.F., Belew R.K., Goodsell D.S. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2010;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hess B., Kutzner C., van der Spoel D., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theor Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 22.Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Mccoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 28.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 29.Modi S., Sutcliffe M.J., Primrose W.U., Lian L.Y., Roberts G.C. The catalytic mechanism of cytochrome P450 BM3 involves a 6 A movement of the bound substrate on reduction. Nat Struct Biol. 1996;3:414–417. doi: 10.1038/nsb0596-414. [DOI] [PubMed] [Google Scholar]
- 30.Shingo N., Huiying L., Hideaki S., Clinton N., Hiroshi O., Montellano P.R., Ortiz De. Crystal structures of epothilone D-bound, epothilone B-bound, and substrate-free forms of cytochrome P450epoK. J Biol Chem. 2003;278:44886–44893. doi: 10.1074/jbc.M308115200. [DOI] [PubMed] [Google Scholar]
- 31.Semsary S., Crnovcic I., Driller R., Vater J., Loll B., Keller U. Ketonization of proline residues in the peptide chains of actinomycins by a 4-oxoproline synthase. Chembiochem : Eur J Chem Biol. 2018;19:706–715. doi: 10.1002/cbic.201700666. [DOI] [PubMed] [Google Scholar]
- 32.Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 33.Lee Y-T, Wilson RF, Rupniewski I, Goodin DB. P450cam visits an open conformation in the absence of substrate. Biochemistry.49:3412-3419. [DOI] [PMC free article] [PubMed]
- 34.Poulos TL, Finzel BC, Howard AJ. Crystal structure of substrate-free Pseudomonas putida cytochrome P-450. Biochemistry.25:5314-5322. [DOI] [PubMed]
- 35.Zhang W, Du L, Li F, Zhang X, Qu Z, Han L, et al. Mechanistic insights into interactions between bacterial class I P450 enzymes and redox partners. ACS Catal.9992-10003.
- 36.Li S., Du L., Bernhardt R. Redox partners: function modulators of bacterial P450 enzymes. Trends Microbiol. 2020;28(6):445–454. doi: 10.1016/j.tim.2020.02.012. [DOI] [PubMed] [Google Scholar]
- 37.Poulos T.L., Finzel B.C., Gunsalus I.C., Wagner G.C., Kraut J. The 2.6-A crystal structure of Pseudomonas putida cytochrome P-450. J Biol Chem. 1985;260:16122–16130. [PubMed] [Google Scholar]
- 38.Sevrioukova I.F., Poulos T.L. Pyridine-substituted desoxyritonavir is a more potent inhibitor of cytochrome P450 3A4 than ritonavir. J Med Chem. 2013;56:3733–3741. doi: 10.1021/jm400288z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vieira D.F., Choi J.Y., Roush W.R., Podust L.M. Expanding the binding envelope of CYP51 inhibitors targeting Trypanosoma cruzi with 4-aminopyridyl-based sulfonamide derivatives. Chembiochem : Eur J Chem Biol. 2014;15:1111–1120. doi: 10.1002/cbic.201402027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hargrove T.Y., Garvey E.P., Hoekstra W.J., Yates C.M., Wawrzak Z., Rachakonda G. Crystal structure of the new investigational drug candidate VT-1598 in complex with Aspergillus fumigatus sterol 14alpha-demethylase provides insights into its broad-spectrum antifungal activity. Antimicrob Agents Chemother. 2017:61. doi: 10.1128/AAC.00570-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calvet C.M., Vieira D.F., Choi J.Y., Kellar D., Cameron M.D., Siqueira-Neto J.L. 4-Aminopyridyl-based CYP51 inhibitors as anti-Trypanosoma cruzi drug leads with improved pharmacokinetic profile and in vivo potency. J Med Chem. 2014;57:6989–7005. doi: 10.1021/jm500448u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cryle M.J., Schlichting I. Structural insights from a P450 Carrier Protein complex reveal how specificity is achieved in the P450(BioI) ACP complex. Proc Natl Acad Sci U S A. 2008;105:15696–15701. doi: 10.1073/pnas.0805983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunn A.R., Dmochowski I.J., Bilwes A.M., Gray H.B., Crane B.R. Probing the open state of cytochrome P450cam with ruthenium-linker substrates. Proc Natl Acad Sci U S A. 2001;98:12420–12425. doi: 10.1073/pnas.221297998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellis P.J., Carlow C.K., Ma D., Kuhn P. Crystal structure of the complex of brugia malayi cyclophilin and cyclosporin A. Biochemistry. 2000;39:592–598. doi: 10.1021/bi991730q. [DOI] [PubMed] [Google Scholar]
- 45.Ke H., Zhao Y., Luo F., Weissman I., Friedman J. Crystal structure of murine cyclophilin C complexed with immunosuppressive drug cyclosporin A. Proc Natl Acad Sci U S A. 1993;90:11850–11854. doi: 10.1073/pnas.90.24.11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kajitani K., Fujihashi M., Kobayashi Y., Shimizu S., Tsujimoto Y., Miki K. Crystal structure of human cyclophilin D in complex with its inhibitor, cyclosporin A at 0.96-A resolution. Proteins. 2008;70:1635–1639. doi: 10.1002/prot.21855. [DOI] [PubMed] [Google Scholar]
- 47.Peterson M.R., Hall D.R., Berriman M., Nunes J.A., Leonard G.A., Fairlamb A.H. The three-dimensional structure of a Plasmodium falciparum cyclophilin in complex with the potent anti-malarial cyclosporin A. J Mol Biol. 2000;298:123–133. doi: 10.1006/jmbi.2000.3633. [DOI] [PubMed] [Google Scholar]
- 48.Rasaiyaah J., Tan C.P., Fletcher A.J., Price A.J., Blondeau C., Hilditch L. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature. 2013;503:402–405. doi: 10.1038/nature12769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Z., Jiang Y., Guengerich F.P., Ma L., Li S., Zhang W. Engineering cytochrome P450 enzyme systems for biomedical and biotechnological applications. J Biol Chem. 2020;295:833–849. doi: 10.1074/jbc.REV119.008758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.