Abstract
The aim of this study is to evaluate the antimicrobial resistance patterns and molecular frequency of blaGES-2 and blaoxa-48 genes in Pseudomonas aeruginosa and Acinetobacter baumannii strains isolated from burn wound infection in Tehran, Iran. In this study, 50 isolates of A. baumannii and 48 isolates of P. aeruginosa were collected from the Burn Unit of Shahid Motahari Hospital at Tehran, Iran. Antibiotic susceptibility tests of all isolates were carried out using the disc diffusion method, and the production of extended-spectrum β-lactamases (ESBLs) in isolates was surveyed by the double disc synergy method and based on CLSI (2019 AST M100) criteria. Finally, the frequency of blaGES-2 and blaoxa-48 genes was surveyed by PCR. Antibiotic susceptibility tests showed that 48/48 (100%) of P. aeruginosa isolates and 49/50 (98%) of A. baumannii isolates were resistant to ceftriaxone and cefotaxime, respectively. Ceftazidime exhibited the lowest (26/48; 54.1%) resistance rates against P. aeruginosa isolates. The production of ESBLs was seen in 8/48 (16.6%) and 3/50 (6%) of P. aeruginosa and A. baumannii isolates, respectively. On the basis of conventional PCR and sequencing, the frequencies of the blaGES-2 gene among P. aeruginosa and A. baumannii was 87.5% and 58%, respectively. Moreover, blaoxa-48 gene was detected in 70.83% and 92% of P. aeruginosa and A. baumannii isolates, respectively. Results suggest that antibiotic-resistant A. baumannii and P. aeruginosa strains isolated from burn patients are frequently found; therefore, it is absolutely necessary to implement continuous screening and follow-up programmes for detecting antimicrobial resistance.
Keywords: Acinetobacter baumannii, antibiotic resistance, bla, blaoxa-48, Pseudomonas aeruginosa
Introduction
Hospital infections are known as one of the most critical problems in health and treatment systems [1]. The factors causing such infections lead to a pervasive spread of such diseases in different hospital wards, and to a high mortality rate. Moreover, these factors are the reasons for the limited treatment efficacy and high costs [2]. Burning is one of the common and destructive injuries that requires immediate care to prevent its side effects. One of the most important concerns about burn patients is bacterial infections [3]. Pseudomonas aeruginosa, Acinetobacter baumannii and methicillin-resistant Staphylococcus aureus are the most common pathogens found in infections in burn patients [[4], [5], [6], [7]]. Some of the most noticeable infections induced by these agents are bacteraemia, ventilator-associated pneumonia, urinary tract infections, meningitis and wound infection among hospitalized patients, especially in the intensive care unit [8]. The intrinsic resistance of P. aeruginosa and A. baumannii against different groups of antibiotics and their ability to apply novel resistance mechanisms are the main problems when controlling infections at health-care centres [9,10]. Extended-spectrum β-lactamases (ESBLs) of GES type were detected for the first time in a clinical isolate of Klebsiella pneumoniae [11], and then in Enterobacteriaceae, P. aeruginosa and A. baumannii isolates [12,13]. Carbapenems are widely used to treat infections caused by P. aeruginosa and A. baumannii [14,15]. The ESBL genes stimulate resistance against extended-spectrum cephalosporins and cause many problems in and obstacles to the treatment of infections caused by P. aeruginosa and A. baumannii [16,17]. Currently, several GES-type β-lactamases, including GES-1, GES-2, GES-4, GES-5, GES-6, GES-8, GES-9, GES-11 and GES-19, have been identified in Enterobacteriaceae and among P. aeruginosa and A. baumannii around the world [18,19]. Changes in the amino acid position 170 in the case of GES-1, GES-2 and GES-5 genes can induce resistance against carbapenems [20]. The emergence of the OXA (oxacillinase) group of β-lactamases (Class D) resulted in several problems in controlling and treating opportunistic infections. The blaoxa48 gene is widespread in K. pneumoniae and plays a number of critical roles such as biofilm formation and resistance to carbapenems [21,22]. The blaOXA-48 gene is frequently recognized in Escherichia coli and K. pneumoniae [23]. However, only two studies have reported OXA-48 in A. baumannii and, so far, the blaOXA-48 gene has not been detected in P. aeruginosa isolates [24,25]. Screening for blaoxa48 in patients is essential to prevent the nosocomial outbreak before hospital admission, and identifying blaoxa48 and its variant with a short turnaround time promotes the time to active treatment [26]. Therefore, the objective of the present research is to evaluate the antimicrobial resistance patterns and molecular frequency of blaoxa-48 and blaGES-2 genes among P. aeruginosa and A. baumannii strains isolated from burn wound infection in Tehran, Iran.
Materials and methods
Ethics statement
The study protocol was approved by the Ethics Committee of Islamic Azad University, Ahar Branch (IR.IAU-AHAR.REC.1398.105).
Bacterial isolates and species identification
In the current study, from May 2018 until the end of July 2019, 98 clinical isolates comprising 50 isolates of A. baumannii and 48 isolates of P. aeruginosa were collected from those patients hospitalized at the Burn Ward of Shahid Motahari Hospital in Tehran, Iran. Briefly, the surface layer of the burn wound was cleaned and washed with normal saline, and swab samples were collected. Samples were transferred to the medical laboratory using Stuart transport medium. In the next step, swab samples were inoculated into several bacterial growth media including blood agar, MacConkey agar and Tryptic Soy Broth, then were incubated at 37°C for 24 hours. Strains were identified as A. baumannii and P. aeruginosa using standard biochemical tests including Gram stain, pigment production on Mueller–Hinton agar (Merck, Darmstadt, Germany), catalase and oxidase test, growth on triple sugar iron agar and Kligler iron agar, oxidation–fermentation, citrate test, sulphide indole motility, Methyl Red, Voges–Proskauer tests, motility and growth at 42°C. Following a definitive diagnosis, A. baumannii and P. aeruginosa isolates were inoculated into trypticase soy broth (Merck) supplemented with 20% glycerol and were preserved at –70°C until further processing [27].
Antibiotic susceptibility testing
The susceptibility of A. baumannii and P. aeruginosa to piperacillin/tazobactam (10/100 μg), imipenem (10 μg), meropenem (10 μg), ceftazidime (30 μg), ceftriaxone (30 μg), cefotaxime (30 μg), cefepime (30 μg), aztreonam (30 μg), amikacin (30 μg), gentamicin (10 μg) and ciprofloxacin (5 μg) was determined using a Kirby–Bauer disc diffusion method (DDM) on Mueller–Hinton agar. Pseudomonas aeruginosa (ATCC 27853) was used as a control for DDM. The finding of the DDM method was then interpreted based on the CLSI (CLSI 2019 AST M100) criteria.
Based on the US Centers for Disease Control and Prevention and the European Centre for Disease Prevention and Control, multidrug-resistant (MDR) isolates were identified, and P. aeruginosa and A. baumannii isolates were selected as MDR, which were resistant to at least one antimicrobial among at least three or more antibiotic groups.
Phenotypic detection of ESBL production
To evaluate the production of ESBL by isolates, this study used the double-disc synergy test (DDST) according to the CLSI (2019 M100) criteria (with either 30 μg cefotaxime or 30 μg ceftazidime alone, or with either 30 μg cefotaxime or 30 μg ceftazidime plus 10 μg clavulanic acid), and the test was performed on Mueller–Hinton agar plates. If the inhibition zone produced by the combined effects of either cefotaxime or ceftazidime plus clavulanic acid was ≥5 mm larger than that produced by either cefotaxime or ceftazidime alone, the test would be determined positive.
DNA extraction and PCR surveying
Genomic DNA of A. baumannii and P. aeruginosa isolates was extracted using a High Pure PCR Template Preparation Kit (Roche, Mannheim, Germany) in line with the manufacturer's guidelines and was preserved at –80°C. The presence of blaoxa-48 and blaGES-2 genes was screened by PCR. The PCR was performed on a 25-μL reaction mixture containing 3 μL of 10 × PCR buffer without MgCl2, 2.5 mM MgCl2, 0.5 μL of 10 mM of each deoxynucleoside triphosphate (dNTPs), 1 μL of forward primer (10 pmol) and 1 μL of forward primer (10 pmol), 1 unit of Taq polymerase (Cinnagene, Tehran, Iran), 4 μL of template DNA, and sterile distilled water up to 25 μL.
The primer sequences used for PCR are listed in Table 1. Amplification reactions were carried out on a 9700 Gene Amp thermocycler (Applied Biosystems, Foster City, CA, USA). PCR conditions were as follows: one cycle of 95°C for 4 min, 35 cycles of denaturation at 95°C for 45 s, annealing at 52°C to 55°C (according to the primers) for each gene for 1 min and elongation at 72°C for 45 s with a final extension at 72°C for 10 min following the last cycle. PCR products were transferred to 1.5% agarose gel, stained with DNA safe stain (SinaClon Co., Tehran, Iran), visualized by a UV transilluminator, and screened in the presence of blaoxa-48 and blaGES-2 genes. Finally, amplicons representing each studied gene were confirmed based on sequencing analysis by ABI 3730X capillary sequencer (Pishgam; Macrogen, Seoul, Korea). Acinetobacter baumannii ATCC 19606 and distilled water were used as positive and negative controls, respectively.
Table 1.
Primers used for detection of blaoxa-48 and blaGES-2 genes
| Primer | Gene sequence | Size (bp) |
|---|---|---|
| GES 2-F GES 2-R |
GTTTTGCAATGTGCTCAACG TGCCATAGCAATAGGCGTAG |
371 |
| OXA 48-F OXA 48-R |
TTGGTGGCATCGATTATCGG GAGCACTTCTTTTGTGATGGC |
744 |
Statistical analysis
The results of this study were analysed using the statistical package SPSS v.23.0 (SPSS Inc., Chicago, IL, USA) and descriptive statistic tests.
Results
Number of specimens and distribution of bacteria
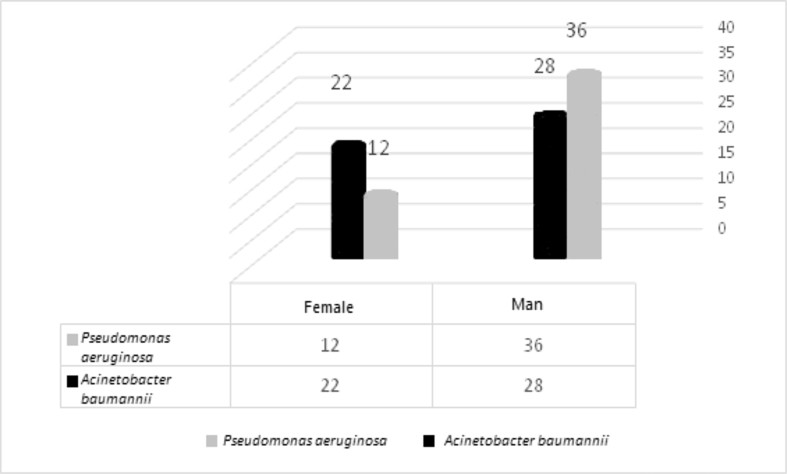
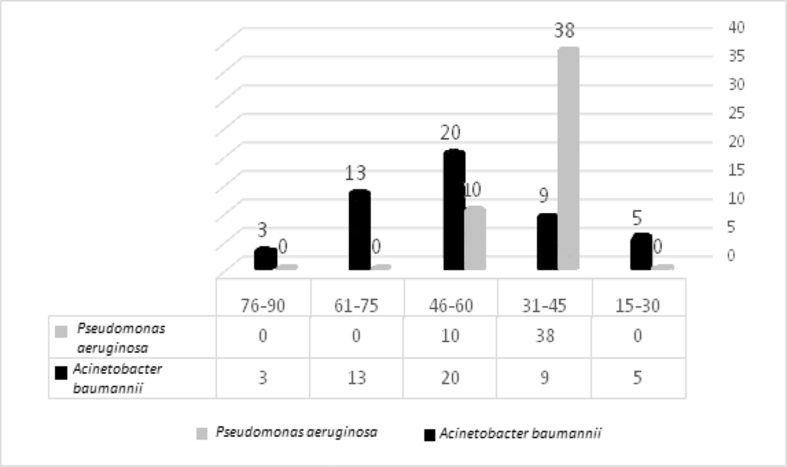
In this cross-sectional study, a total of 220 swab samples were collected from May 2018 until the end of July 2019. Of these samples, 98 cultures (44.5%) (50 (22.7%) A. baumannii and 48 (21.8%) P. aeruginosa) were determined to be positive for A. baumannii and P. aeruginosa. Of the isolates, 36/48 (75%) and 28/50 (56%) of the P. aeruginosa and A. baumannii isolates were male, respectively (Fig. 1) and the mean age was 44 years (range 15 days to 90 years). Pseudomonas aeruginosa (38/48; 79.2%) had the highest proportion in the 31–45-year age group. In contrast, A. baumannii (20/50; 40%) had the highest proportion in the 46–60-year age group. The frequency of P. aeruginosa and A. baumannii isolates is shown in Fig. 2 by different age groups.
Fig. 1.
Frequency and distribution of Pseudomonas aeruginosa and Acinetobacter baumannii isolates among the male and female gender.
Fig. 2.
The frequency of Pseudomonas aeruginosa and Acinetobacter baumannii isolates by different age groups.
Antimicrobial susceptibility profile
The susceptibility profiles of A. baumannii and P. aeruginosa isolates to commonly used antimicrobials are shown in Table 2. The DDM results showed that 100% and 98% of P. aeruginosa and A. baumannii isolates exhibited resistance to ceftriaxone and cefotaxime, respectively. The P. aeruginosa strains showed a high degree of resistance to imipenem (95.83%), meropenem (93.75%), amikacin (93.75%), gentamicin (93.75%), ciprofloxacin (93.75%), cefepime (89.58%), aztreonam (81.25%) and piperacillin/tazobactam (81.25%), respectively. However, P. aeruginosa was found to have low levels of resistance to ceftazidime (54.17%). The A. baumannii strains showed a high level of resistance to ceftazidime (96%), imipenem (94%), meropenem (94%), cefepime (94%), amikacin (94%), ciprofloxacin (94%), piperacillin/tazobactam (90%), aztreonam (86%) and gentamicin (86%), respectively. In total, 98% (49/50) of A. baumannii and 100% (48/48) of P. aeruginosa were MDR.
Table 2.
Antibiotic susceptibility of the Acinetobacter baumannii and Pseudomonas aeruginosa by disc diffusion method
| Antibiotics |
Pseudomonas aeruginosa (n = 48) |
Acinetobacter baumannii (n = 50) |
Total (n = 98) |
|||
|---|---|---|---|---|---|---|
| Resistance | % | Resistance | % | Resistance | % | |
| Piperacillin + Tazobactam | 39 | 81.25 | 45 | 90.00 | 84 | 85.71 |
| Imipenem | 46 | 95.83 | 47 | 94.00 | 93 | 94.90 |
| Meropenem | 45 | 93.75 | 47 | 94.00 | 92 | 93.88 |
| Ceftazidime | 26 | 54.17 | 48 | 96.00 | 74 | 75.51 |
| Ceftriaxone | 48 | 100.00 | 49 | 98.00 | 97 | 98.98 |
| Cefotaxime | 48 | 100.00 | 49 | 98.00 | 97 | 98.98 |
| Cefepime | 43 | 89.58 | 47 | 94.00 | 90 | 91.84 |
| Aztreonam | 39 | 81.25 | 43 | 86.00 | 82 | 83.67 |
| Amikacin | 45 | 93.75 | 47 | 94.00 | 92 | 93.88 |
| Gentamicin | 45 | 93.75 | 43 | 86.00 | 88 | 89.80 |
| Ciprofloxacin | 45 | 93.75 | 47 | 94.00 | 92 | 93.88 |
DDST test result
The results of DDST showed that 16.6% (8/48) of P. aeruginosa strains and 6% (3/50) of A. baumannii strains for which the zone of inhibition for ceftazidime plus clavulanic acid was ≥5 mm larger than that for ceftazidime alone were positive in the DDST, respectively. Therefore, eight isolates of P. aeruginosa and three isolates of A. baumannii were classified as ESBL producers.
Screening for blaoxa-48 and blaGES-2 genes
The PCR was conducted to detect blaoxa-48 and blaGES-2 genes in P. aeruginosa and A. baumannii isolates using specific primers. PCR and sequencing showed that 87.5% (42/48) of P. aeruginosa and 58% (29/50) of A. baumannii were positive for blaGES-2 genes. In contrast, the frequencies of blaoxa-48 gene in P. aeruginosa and A. baumannii isolates were 70.8% (34/48) and 92% (46/50), respectively.
Discussion
This study evaluates the antimicrobial resistance patterns and molecular frequency of blaoxa-48 and blaGES-2 genes in a large number of P. aeruginosa and A. baumannii strains isolated from burn wound infections in Tehran, Iran. Both P. aeruginosa and A. baumannii are opportunistic and nosocomial pathogens that can induce several infections including otitis media, and respiratory tract, burn and wound infections with high mortality in patients, especially in immunocompromised individuals hospitalized in various wards of a hospital [[28], [29], [30]]. Antibiotic resistance among P. aeruginosa and A. baumannii has been accepted as a global public health problem around the world [31]. Based on a report by the WHO in 2017, carbapenem-resistant P. aeruginosa, carbapenem-resistant A. baumannii complex and carbapenem-resistant or ESBL-producing Enterobacteriaceae are critical priority pathogens [32,33]. Recently, the emergence of carbapenem (imipenem and meropenem) resistance among these bacteria has become a severe clinical problem, mainly in low- and middle-income countries [34]. Moreover, it is predictable that the unavailability of organized antibiotic resistance surveillance programmes in these countries will lead to unsuitable use between patients and health-care staff [1]. Among the antibiotics that were tested against A. baumannii and P. aeruginosa, ceftriaxone and cefotaxime had the highest resistance rate. Moreover, P. aeruginosa strains showed a high level of resistance to imipenem, meropenem, amikacin, gentamicin, ciprofloxacin, cefepime, aztreonam and piperacillin/tazobactam. Similarly, a high resistance rate was reported by Shariati et al., who claimed that 95% of P. aeruginosa strains were resistant to imipenem, meropenem and gentamicin [3]. Results of a previously conducted study in Mofid Children's Hospital revealed that the resistance rates of P. aeruginosa isolates, collected from 2013 to 2018, to imipenem, meropenem, gentamicin, amikacin, ciprofloxacin and piperacillin/tazobactam were 50.4%, 70.3%, 58.1%, 43.2%, 16.7% and 40.5%, respectively [1]. On the other hand, Farhan et al. reported low-level resistance [35]. According to the results of a published study, it can be concluded that two independent risk factors that include (a) prolonged hospitalization at the intensive care unit (>29 days) and (b) the existence of P. aeruginosa in the bacteriological specimens taken before treatment can lead to the acquisition of carbapenem resistance [31]. The present study also revealed that ceftazidime, in comparison with other antibiotics, showed the lowest resistance rate against P. aeruginosa isolates. Acinetobacter baumannii strains had a high level of resistance to all tested antibiotics, the results of which are in agreement with those obtained by Boral et al. [36], Azimi et al. [1], Romanin et al. [37], Rossi et al. [14], Kumar et al. [38] and Ardehali et al. [39]. In the current study, 98% of A. baumannii and 100% of P. aeruginosa were MDR. Similar findings were shown by Shariati et al. [3], Farhan et al. [35] and Ahmad and Ali [40]. Moreover, these results were in contrast with the results of Dutta et al. [41]. On the other hand, ESBL production was seen in 8/48 (16.6%) and 3/50 (6%) of P. aeruginosa and A. baumannii isolates, respectively. In recent years, the increase of MDR strains of A. baumannii and P. aeruginosa isolates has led to widespread and extensive use of carbapenems. Consequently, the degree of carbapenem resistance in A. baumannii and P. aeruginosa is now increasing around the world [42,43]. An increase in the health-care costs, prolonged hospitalization, reduced success rate of infection treatments, and an increase in morbidity and mortality rates are a number of the unfortunate consequences that result from MDR bacterial infections [44,45]. On the basis of conventional PCR and sequencing, the frequencies of blaGES-2 and blaoxa-48 genes among P. aeruginosa isolates were 87.5% and 70.83%, respectively. Moreover, blaGES-2 and blaoxa-48 genes were detected in 58% and 92% of A. baumannii isolates, respectively. These results were in contrast with those of Boral et al. [36] and Cheikh et al. [42] who reported that none of the A. baumannii isolates contained the blaoxa-48 gene. Romanin et al. reported that none of the A. baumannii isolates contained blaoxa-48 and blaGES-2 genes [37]. These results revealed that the frequency of blaoxa-48 could vary from country to country.
In conclusion, the results of the study showed that the prevalence of P. aeruginosa and A. baumannii resistant to multiple antibiotics dramatically increased, and the finding suggests that antibiotic-resistant A. baumannii and P. aeruginosa strains are frequently isolated from burn patients. Moreover, the results suggest that the use of antibiotics, especially carbapenems, must be carefully controlled in patients who are colonized by or infected with A. baumannii and P. aeruginosa. Finally, it is recommended that combination therapy including imipenem plus meropenem, aztreonam plus aminoglycosides, aztreonam plus colistin, ceftolozane plus tazobactam, ceftazidime/avibactam, piperacillin/tazobactam plus amikacin or piperacillin/tazobactam plus colistin, or meropenem/ceftazidime plus colistin could exert the highest synergistic effect against MDR and carbapenem-resistant A. baumannii and P. aeruginosa isolates, compared with each separate isolate.
Author contributions
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data. They played an active role in drafting the article or revising it critically to achieve important intellectual content, gave the final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Conflict of interest
All of the authors declare that there are no commercial, personal, political, nor any other potentially conflicting interests related to the submitted manuscript.
Funding
This research did not receive any specific grant from funding agencies in public, commercial and not-for-profit sectors.
Acknowledgements
We would like to thank the Department of Microbiology, Islamic Azad University, Ahar Branch, Ahar, Iran for their kind cooperation. The authors received no specific funding for this work.
References
- 1.Azimi T., Maham S., Fallah F., Azimi L., Gholinejad Z. Evaluating the antimicrobial resistance patterns among major bacterial pathogens isolated from clinical specimens taken from patients in Mofid Children’s Hospital, Tehran, Iran: 2013–2018. Infect Drug Resist. 2019;12:2089–2102. doi: 10.2147/IDR.S215329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pormohammad A., Nasiri M.J., Azimi T. Prevalence of antibiotic resistance in Escherichia coli strains simultaneously isolated from humans, animals, food, and the environment: a systematic review and meta-analysis. Infect Drug Resist. 2019;12:1181–1197. doi: 10.2147/IDR.S201324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shariati A., Asadian E., Fallah F., Azimi T., Hashemi A., Sharahi J.Y. Evaluation of Nano-curcumin effects on expression levels of virulence genes and biofilm production of multidrug-resistant Pseudomonas aeruginosa isolated from burn wound infection in Tehran, Iran. Infect Drug Resist. 2019;12:2223–2235. doi: 10.2147/IDR.S213200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lan Y., Li W., Jiao Y., Guo R., Zhang Y., Xue W. Therapeutic efficacy of antibiotic-loaded gelatin microsphere/silk fibroin scaffolds in infected full-thickness burns. Acta Biomat. 2014;10:3167–3176. doi: 10.1016/j.actbio.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 5.Armin S., Karimi A., Fallah F., Rafiei Tabatabaii S., Hoseini Alfatemi S.M., Khiabanirad P. Antimicrobial resistance patterns of Acinetobacter baumannii, Pseudomonas aeruginosa and Staphylococcus aureus isolated from patients with nosocomial infections admitted to tehran hospitals. Arch Pediatr Infect Dis. 2015;3(4):e32554. [Google Scholar]
- 6.Norbury W., Herndon D.N., Tanksley J., Jeschke M.G., Finnerty C.C. Infection in burns. Surg Infect. 2016;17:250–255. doi: 10.1089/sur.2013.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pormohammad A., Mehdinejadiani K., Gholizadeh P., Nasiri M.J., Mohtavinejad N., Dadashi M. Global prevalence of colistin resistance in clinical isolates of Acinetobacter baumannii: a systematic review and meta-analysis. 2020;139:103887. doi: 10.1016/j.micpath.2019.103887. [DOI] [PubMed] [Google Scholar]
- 8.Noori M., Karimi A., Fallah F., Hashemi A., Alimehr S., Goudarzi H. High prevalence of metallo-beta-lactamase producing Acinetobacter baumannii isolated from two hospitals of Tehran, Iran. Arch Pediatr Infect Dis. 2014;2(3):e15439. [Google Scholar]
- 9.Pachori P., Gothalwal R., Gandhi P. Emergence of antibiotic resistant Pseudomonas aeruginosa in intensive care unit; a critical review. Genes Dis. 2019;6:109–119. doi: 10.1016/j.gendis.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bassetti M., Vena A., Croxatto A., Righi E., Guery B. How to manage Pseudomonas aeruginosa infections. Drugs Context. 2018;7:212527. doi: 10.7573/dic.212527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poirel L., Le Thomas I., Naas T., Karim A., Nordmann P. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2000;44:622–632. doi: 10.1128/aac.44.3.622-632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laudy A.E., Róg P., Smolińska-Król K., Ćmiel M., Słoczyńska A., Patzer J. Prevalence of ESBL-producing Pseudomonas aeruginosa isolates in Warsaw, Poland, detected by various phenotypic and genotypic methods. PloS One. 2017;12 doi: 10.1371/journal.pone.0180121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cantón R., Novais A., Valverde A., Machado E., Peixe L., Baquero F. Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Europe. Clin Microbiol Infect. 2008;14:144–153. doi: 10.1111/j.1469-0691.2007.01850.x. [DOI] [PubMed] [Google Scholar]
- 14.Rossi I., Royer S., Ferreira M.L., Campos P.A., Fuga B., Melo G.N. Incidence of infections caused by carbapenem-resistant Acinetobacter baumannii. Am J Infect Control. 2019;47:1431–1435. doi: 10.1016/j.ajic.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Walters M.S., Grass J.E., Bulens S.N., Hancock E.B., Phipps E.C., Muleta D. Carbapenem-resistant Pseudomonas aeruginosa at US emerging infections program sites, 2015. Emerg Infect Dis. 2019;25:1281–1288. doi: 10.3201/eid2507.181200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaikh S., Fatima J., Shakil S., Rizvi S.M.D., Kamal M.A. Antibiotic resistance and extended spectrum beta-lactamases: types, epidemiology and treatment. Saudi J Biol Sci. 2015;22:90–101. doi: 10.1016/j.sjbs.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abdar M.H., Taheri-Kalani M., Taheri K., Emadi B., Hasanzadeh A., Sedighi A. Prevalence of extended-spectrum beta-lactamase genes in Acinetobacter baumannii strains isolated from nosocomial infections in Tehran, Iran. GMS Hyg Infect Control. 2019;14:Doc02. doi: 10.3205/dgkh000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogaerts P., Naas T., El Garch F., Cuzon G., Deplano A., Delaire T. GES extended-spectrum β-lactamases in Acinetobacter baumannii isolates in Belgium. Antimicrob Agents Chemother. 2010;54:4872–4878. doi: 10.1128/AAC.00871-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poirel L., Weldhagen G.F., Naas T., De Champs C., Dove M.G., Nordmann P. GES-2, a class A β-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob Agents Chemother. 2001;45:2598–2603. doi: 10.1128/AAC.45.9.2598-2603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart N.K., Smith C.A., Frase H., Black D.J., Vakulenko S.B. Kinetic and structural requirements for carbapenemase activity in GES-type β-lactamases. Biochemistry. 2014;54:588–597. doi: 10.1021/bi501052t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh S.K., Gupta M.J.M.P. blaOXA-48 carrying clonal colistin resistant-carbapenem resistant Klebsiella pneumoniae in neonate intensive care unit, India. Microb Pathog. 2016;100:75–77. doi: 10.1016/j.micpath.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Ramos-Vivas J., Chapartegui-González I., Fernández-Martínez M., González-Rico C., Fortún J., Escudero R. Biofilm formation by multidrug resistant Enterobacteriaceae strains isolated from solid organ transplant recipients. Sci Rep. 2019;9:1–10. doi: 10.1038/s41598-019-45060-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakthavatchalam Y.D., Anandan S., Veeraraghavan B.J.J. Laboratory detection and clinical implication of oxacillinase-48 like carbapenemase: the hidden threat. J Global Infect Dis. 2016;8:41–50. doi: 10.4103/0974-777X.176149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathlouthi N., Areig Z., Al Bayssari C., Bakour S., Ali El Salabi A., Ben Gwierif S. Emergence of carbapenem-resistant Pseudomonas aeruginosa and Acinetobacter baumannii clinical isolates collected from some Libyan hospitals. 2015;21:335–341. doi: 10.1089/mdr.2014.0235. [DOI] [PubMed] [Google Scholar]
- 25.Aruhomukama D., Najjuka C.F., Kajumbula H., Okee M., Mboowa G., Sserwadda I. Bla VIM-and bla OXA-mediated carbapenem resistance among Acinetobacter baumannii and Pseudomonas aeruginosa isolates from the Mulago hospital intensive care unit in Kampala, Uganda. BMC Infect Dis. 2019;19:1–8. doi: 10.1186/s12879-019-4510-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arana D., Saez D., García-Hierro P., Bautista V., Fernández-Romero S., De la Cal M.Á. Concurrent interspecies and clonal dissemination of OXA-48 carbapenemase. Clin Microbiol Infect. 2015;21:148.e1–148.e4. doi: 10.1016/j.cmi.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 27.Pormohammad A., Lashkarbolouki S., Azimi T., Gholizadeh P., Bostanghadiri N., Safari H. vol. 32. a prospective study; Iran: 2019. p. 100594. (Clinical characteristics and molecular epidemiology of children with meningitis in Tehran). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allegranzi B., Nejad S.B., Combescure C., Graafmans W., Attar H., Donaldson L. Burden of endemic health-care-associated infection in developing countries: systematic review and meta-analysis. Lancet. 2011;377(9761):228–241. doi: 10.1016/S0140-6736(10)61458-4. [DOI] [PubMed] [Google Scholar]
- 29.Lim C., Takahashi E., Hongsuwan M., Wuthiekanun V., Thamlikitkul V., Hinjoy S. Epidemiology and burden of multidrug-resistant bacterial infection in a developing country. Elife. 2016;5 doi: 10.7554/eLife.18082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shariati A., Azimi T., Ardebili A., Chirani A.S., Bahramian A., Pormohammad A. Insertional inactivation of oprD in carbapenem-resistant Pseudomonas aeruginosa strains isolated from burn patients in Tehran, Iran. New Microb New Infect. 2018;21:75–80. doi: 10.1016/j.nmni.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labaste F., Grossac J., Bounes F.V., Conil J.M., Ruiz S., Seguin T. Risk factors for acquisition of carbapenem-resistance during treatment with carbapenem in the intensive care unit: a prospective study. Eur J Clin Microbiol Infect Dis. 2019;38:1–9. doi: 10.1007/s10096-019-03644-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tillotson G. A crucial list of pathogens. Lancet Infect Dis. 2018;18:234–236. doi: 10.1016/S1473-3099(17)30754-5. [DOI] [PubMed] [Google Scholar]
- 33.Lee Y.-L., Lu M.C., Shao P.L., Lu P.L., Chen Y.H., Cheng S.H. Nationwide surveillance of antimicrobial resistance among clinically important Gram-negative bacteria, with an emphasis on carbapenems and colistin: results from the Surveillance of Multicenter Antimicrobial Resistance in Taiwan (SMART) in 2018. Int J Antimicrob Agents. 2019;54:318–328. doi: 10.1016/j.ijantimicag.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 34.Young K., Painter R.E., Raghoobar S.L., Hairston N.N., Racine F., Wisniewski D. In Vitro studies evaluating the activity of imipenem in combination with relebactam against Pseudomonas aeruginosa. BMC Microbiol. 2019;19:150. doi: 10.1186/s12866-019-1522-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farhan S.M., Ibrahim R.A., Mahran K.M., Hetta H.F., Abd El-Baky R.M. Antimicrobial resistance pattern and molecular genetic distribution of metallo-β-lactamases producing Pseudomonas aeruginosa isolated from hospitals in Minia, Egypt. Infect Drug Resist. 2019;12:2125–2133. doi: 10.2147/IDR.S198373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boral B., Unaldi Ö., Ergin A., Durmaz R., Eser Ö.K. A prospective multicenter study on the evaluation of antimicrobial resistance and molecular epidemiology of multidrug-resistant Acinetobacter baumannii infections in intensive care units with clinical and environmental features. Ann Clin Microbiol Antimicrob. 2019;18:1–9. doi: 10.1186/s12941-019-0319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romanin P., Palermo R.L., Cavalini J.F., Fávaro L.D.S., De Paula-Petroli S.B., Fernandes E.V. Multidrug- and extensively drug-resistant Acinetobacter baumannii in a tertiary hospital from Brazil: the importance of carbapenemase encoding genes and epidemic clonal complexes in a 10-year study. Microb Drug Resist. 2019;25:1365–1373. doi: 10.1089/mdr.2019.0002. [DOI] [PubMed] [Google Scholar]
- 38.Kumar S., Patil P.P., Singhal L., Ray P., Patil P.B., Gautam V. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates reveals the emergence of blaOXA-23 and blaNDM-1 encoding international clones in India. Infect Genet Evol. 2019;75:103986. doi: 10.1016/j.meegid.2019.103986. [DOI] [PubMed] [Google Scholar]
- 39.Ardehali S.H., Azimi T., Fallah F., Owrang M., Aghamohammadi N., Azimi L. Role of efflux pumps in reduced susceptibility to tigecycline in Acinetobacter baumannii. New Microb New Infect. 2019;30:100547. doi: 10.1016/j.nmni.2019.100547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmad S.S., Ali F.A. Detection of ESBL, AmpC and Metallo Beta-Lactamase mediated resistance in Gram-negative bacteria isolated from women with genital tract infection. Eur Sci J. 2014;10(9):193–209. [Google Scholar]
- 41.Dutta H., Nath R., Saikia L. Multi-drug resistance in clinical isolates of Gram-negative bacilli in a tertiary care hospital of Assam. Indian J Med Res. 2014;139:643–645. [PMC free article] [PubMed] [Google Scholar]
- 42.Cheikh H.B., Domingues S., Silveira E., Kadri Y., Rosário N., Mastouri M. Molecular characterization of carbapenemases of clinical Acinetobacter baumannii–calcoaceticus complex isolates from a University Hospital in Tunisia. 3 Biotech. 2018;8:1–8. doi: 10.1007/s13205-018-1310-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emaneini M., Kalantar-Neyestanaki D., Jabalameli L., Hashemi M., Beigverdi R., Jabalameli F. Molecular analysis and antimicrobial resistance pattern of distinct strains of Pseudomonas aeruginosa isolated from cystic fibrosis patients in Iran. Iranian J Microbiol. 2019;11:98–107. [PMC free article] [PubMed] [Google Scholar]
- 44.Mahmoudi S., Mahzari M., Banar M., Pourakbari B., Ashtiani M.T.H., Mohammadi M. Antimicrobial resistance patterns of Gram-negative bacteria isolated from bloodstream infections in an Iranian referral paediatric hospital: a 5.5-year study. J Glob Antimicrob Resist. 2017;11:17–22. doi: 10.1016/j.jgar.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 45.Bahramian A., Shariati A., Azimi T., Sharahi J.Y., Bostanghadiri N., Gachkar L. First report of New Delhi metallo-β-lactamase-6 (NDM-6) among Klebsiella pneumoniae ST147 strains isolated from dialysis patients in Iran. Infect Genet Evol. 2019;69:142–145. doi: 10.1016/j.meegid.2019.01.030. [DOI] [PubMed] [Google Scholar]