Abstract
The Common or European quail (Coturnix coturnix) is a Galliform bird of ecological importance for research in the field of animal migration. The Common quail is also a game bird, thus, of great interest for recreational activities and conservation management. Here, we generated a high quality de novo transcriptome for the Common quail for which no reference genome is to date publicly available. The transcriptome was obtained from a population of Common quail originated from captive founders raised under laboratory conditions. Paired-end RNA-Sequencing reads were obtained from extracted total RNA of brain tissue punches (preoptic-hypothalamic region) of 23 quails, which yielded to 5.5–11.2 million reads per individual bird for a total of 236 million reads. After assembly optimization, we used a stringent filtering analysis pipeline to remove redundant and low confidence transcripts. The final transcriptome consisted of 22,293 transcripts of which 21,551 (97%) were provided with annotation data. Our data offers a high quality pipeline for compiling transcriptomes of complex non-genomic species. Our data also provide a robust reference for gene expression studies in this species or other related Galliform species, including the Japanese quail.
Keywords: RNA-Sequencing, transcriptome, De novo assembly, annotation, Gene Ontology (GO) analysis, Common quail, Avian migration
Specifications Table
| Subject | Biology |
| Specific subject area | Transcriptomics and neurophysiology |
| Type of data | Transcriptome assembly, raw sequences |
| How data were acquired | High-throughput RNA-Sequencing performed on a HiSeq V4 platform at the Vienna Biocenter (Vienna, Austria) |
| Data format | Raw, Analysed |
| Parameters for data collection | All experimental birds were raised in captivity on a 16:8 hrs light:dark cycle. Upon young adulthood experimental quails were exposed to a gradual decline in day length (i.e. 30 min/week until the photoperiod reached 12:12 hrs light:dark cycle) to simulate autumn migration followed by a non-migratory stage in the non-breeding grounds. For the present RNA-Sequencing experiment, the assembly de novo was generated by pooling together reads of 23 individual birds (11 birds were sampled during the migratory phase and 12 birds were sampled during the non-migratory phase). |
| Description of data collection | Total RNA was extracted from brain tissue punches (preoptic-hypothalamic region). A paired-end 125 basepair Next Generation Sequencing library was prepared. Sequencing reads were analysed to obtain a de novo transcriptome assembly of the Common quail (Coturnix coturnix). |
| Data source location | Birds used in this experiment originated from a captive colony of breeding Common quails maintained at the Konrad Lorenz Institute of Ethology (Vetmeduni Vienna, Austria). All experimental procedures were carried out in laboratory conditions at the Konrad Lorenz Institute of Ethology. |
| Data accessibility | Raw RNA-Sequencing data and assembly file are available from the Sequence Read Archive (SRA) on NCBI, study accession number PRJNA630138 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA630138/). The associated annotation data are available as Supplementary Material. |
Value of the Data
-
•
In this article, we provide the first published de novo transcriptome for the migratory Galliform, the Common quail (Coturnix coturnix). These data provide an important reference to enhance our understanding of the molecular and physiological pathways associated with animal migration, especially in bird species.
-
•
The Common quail transcriptome data provided here can be used as a reference for differential expression analysis of RNA-Sequencing data.
-
•
These data will be fruitful for behavioural neuroendocrinologists, ecologists, and conservation biologists. These data are likely to provide important insights also to veterinarians and biomedical scientists working in the field of nutrition and focusing on studying gene expression pathways linked to rapid changes in body mass and fat stores, including metabolic disorders such as obesity.
1. Data description
Data reported in this article originate from RNA-Sequencing of brain tissue punches (hypothalamus) from 23 Common quails (Coturnix coturnix). Paired-end sequences obtained from each individual bird were subsequently assembled together to obtain the transcriptome assembly of the Common quail.
1.1. Quality check of RNA-Sequencing libraries
RNA-Sequencing libraries generated from hypothalamic samples of Common quail yielded between 5.5 and 11.2 million paired-end reads (on average 9.1 million reads) per individual bird for a total of 233.5 million paired-end reads. Quality filtering removed between 5.1% and 9.1% reads per sample, which produced a subset of high quality RNA-Sequencing paired-end reads containing between 4.3 and 9.5 million paired-end reads per individual (on average 7.7 million reads).
1.2. Trinity parameter optimization of de novo transcriptome assembly
1.2.1. Quality assessment and read representation
We compiled three independent de novo assemblies using different parameters within the Trinity package. As can be seen in Table 1, overall, there were relatively minor differences among the three compiled transcriptomes. The total number of assembled bases ranged between 344,461,191 to 441,854,562 among the three de novo assemblies with a median transcript length between 360 and 380 (Table 1a). A similar number of reads aligned back to the transcriptome in the Trinity run with the default setting and in the Trinity run with target K-mer coverage set to 30 (84.1% and 83.7%, respectively), whereas re-alignment rate was in comparison lower (78.8%) in the Trinity run with K-mer minimum coverage set to 2 (Table 1c). As shown in Table 1c, the largest number of reads that aligned concordantly back to the transcriptome exactly one time was obtained in the Trinity run with minimum K-mer set to 2 (31,659,608 paired reads out of 176,376,164 total paired reads, 18.0%).
Table 1.
Summary Trinity statistics of the three compiled de novo assemblies of the Common quail transcriptome. Data are provided for all transcripts (a) or, for the longest isoform per unigene (b). Summary read mapping results from bowtie2 is provided in (c). T indicates the target K-mer coverage and minimum K-mer indicates the minimum number of K-mers to be included in the initial Inchworm contigs.
| T = 50 | T = 30 | minimum K-mer = 2 | |
|---|---|---|---|
| (a) All transcript contigs | |||
| Contig N50 | 1,329 | 1,355 | 1,589 |
| Median contig length | 370 | 380 | 360 |
| Average contig | 744.1 | 759.98 | 781.53 |
| Total assembled bases | 441,854,562 | 441,167,562 | 344,461,191 |
| (b) Longest isoform per gene | |||
| Contig N50 | 594 | 605 | 637 |
| Median contig length | 328 | 328 | 304 |
| Average contig | 522.25 | 527.68 | 524.44 |
| Total assembled bases | 225,940,673 | 217,544,496 | 155,153,297 |
| (c) Bowtie2 | |||
| failed alignments | 4320,019 (2.45%) | 4516,006 (2.56%) | 5653,533 (3.21%) |
| aligned concordantly 1 time | 23,815,747 (13.50%) | 24,265,641 (13.76%) | 31,659,608 (17.95%) |
| aligned concordantly >1 times | 148,240,398 (84.05%) | 147,594,517 (83.68%) | 139,063,023 (78.84%) |
1.2.2. Assessment of assembly completeness
Most of the avian core genes were successfully recovered in all the three Trinity assemblies (Table 2). Specifically, out of a total of 4,915 of the single-copy orthologs between 81% and 84% were recovered completely, and between 7% and 10% were recovered partially. Only between 8% and 11% of the 4,915 single-copy orthologs were classified as missing among the three compiled assemblies. These data indicate very good coverage and high quality of the assemblies of the protein-coding transcriptomes for the Common quail.
Table 2.
Summary of the complete, duplicated, fragmented, and missing orthologs from Benchmarking Universal Single-Copy Orthologs (BUSCO) search against the 4,915 single-copy orthologs for Aves within the three compiled de novo assembly. T indicates the target K-mer coverage and minimum K-mer indicates the minimum number of K-mers to be included in the initial Inchworm contigs.
| Type of BUSCO ortholog | T = 50 | T = 30 | minimum K-mer = 2 |
|---|---|---|---|
| Complete | 4,130 (84.03%) | 3,989 (81.16%) | 4,042 (82.24%) |
| (a) Complete and single-copy | 1,394 (28.36%) | 1,335 (27.16%) | 1,426 (29.01%) |
| (b) Complete and duplicated | 2,736 (55.67%) | 2,654 (54.00%) | 2,616 (53.22%) |
| Fragmented | 370 (7.53%) | 481 (9.79%) | 348 (7.08%) |
| Missing | 415 (8.44%) | 445 (9.05%) | 525 (10.68%) |
1.2.3. Removal of redundant transcripts and assembly selection
Depending on the assembly parameters used in the Trinity package, the TransDecoder filtering of redundant transcripts resulted in the reduction of the number of assembled transcripts by 9–11 fold. As can be seen in Table 3, the largest number of transDecoder ORFs was obtained in the assembly with target K-mer coverage set to 30 (54,532 ORFs), intermediate values were obtained in the assembly with default settings (52,272 ORFs), while the lowest value of ORFs was detected in the assembly with minimum K-mer set to 2 (50,345 ORFs).
Table 3.
Summary results from TransDecoder for each type of predicted open reading frame. T indicates the target K-mer coverage and minimum K-mer indicates the minimum number of K-mers to be included in the initial Inchworm contigs.
| Type of coding sequence | T = 50 | T = 30 | minimum K-mer = 2 |
|---|---|---|---|
| Complete (both start and stop codons) | 33,362 (63.82%) | 33,218 (60.91%) | 30,495 (60.57%) |
| 5prime partial alignments | 10,393 (19.88%) | 11,571 (21.22%) | 11,328 (22.50%) |
| 3prime partial alignments | 5,140 (9.83%) | 6,022 (11.04%) | 4,925 (9.78%) |
| Internal alignments | 3,377 (6.46%) | 3,721 (6.82%) | 3,597 (7.14%) |
| total | 52,272 | 54,532 | 50,345 |
Despite the relatively minor differences across the three compiled Trinity assemblies, data gathered by Bowtie2 and TransDecoder suggested that the transcriptome in which target K-mer coverage was set to 30 yielded to a reasonable trade-off between read representation and specificity. Consequently, we selected the latter de novo assembly for all the further RNA-Sequencing analyses in the pipeline.
1.2.4. Clustering of non-redundant transcripts
The non-redundant transcripts in the selected Trinity assembly were clustered together using CD-Hit-EST according to a stringent similarity threshold of 0.95. We thus obtained a total number of 22,293 clusters (41% out of the TransDecoder transcript sets).
1.3. Assembly functional annotation
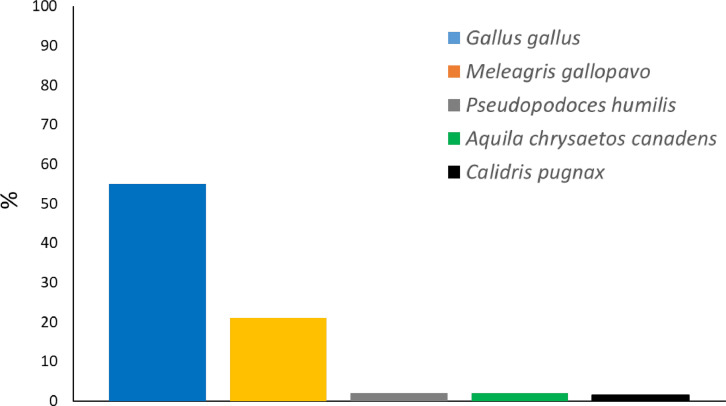
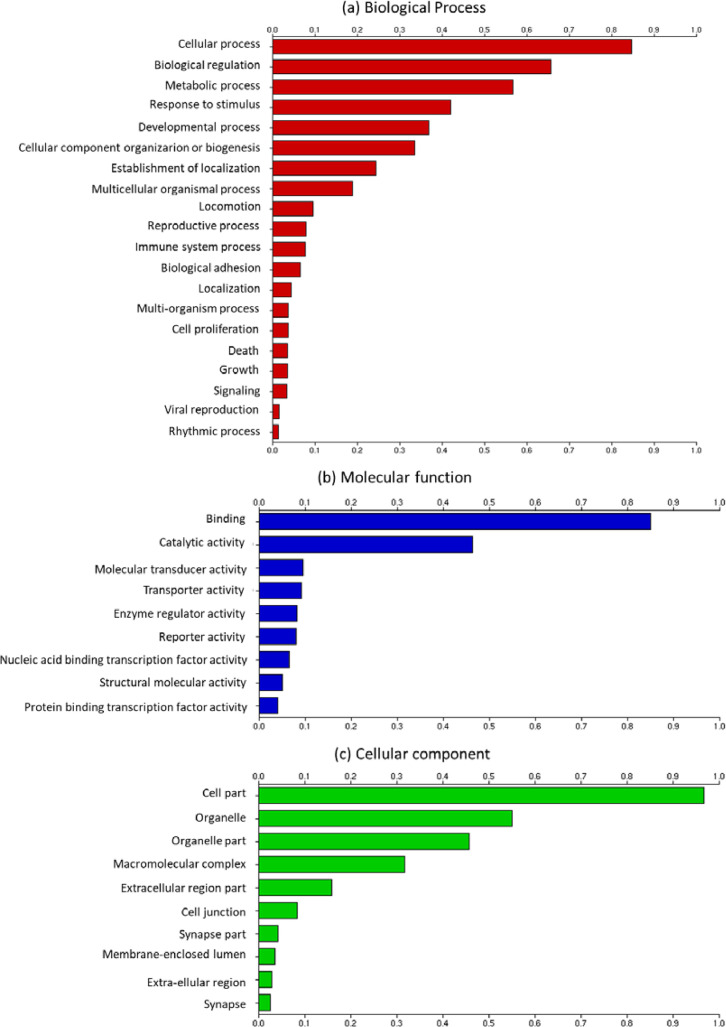
We were able to annotate the majority of transcripts of the final assembly (97%, 21,551 out of 22,293 transcripts). Out of 21,551 total hits detected, 20,462 hits (95%) belonged to the class Aves. Out of 20,462 total hits belonging to Aves, 11,850 hits (58%) were in common with the chicken (Gallus gallus). The taxonomic distribution for the top five most represented bird species in our annotated final transcriptome is shown in Fig. 1. Several Gene Ontology (GO) terms with a relatively enriched representation of cellular, molecular, and biological processes were also identified (Fig. 2). The distribution of the number of transcripts associated with the 15 most frequently occurring EggNOG identifiers is provided in Table 4.
Fig. 1.
Taxonomic distribution of five most represented species (out of 21,551 hits) detected using FunctionAnnotator.
Fig. 2.
Frequency distribution of Gene Ontology (GO) terms for transcripts of Common quail identified in FunctionAnnotator for each primary category: (a) biological process, (b) molecular function, and (c) cellular component.
Table 4.
Number of non-redundant clustered transcripts assigned to the 15 most occurring EggNOG identifiers.
| EggNOG Identifier | Description | N transcripts |
|---|---|---|
| COG5048 | Zinc finger protein | 344 |
| COG0666 | Ankyrin Repeat | 173 |
| COG4886 | Leucine Rich Repeat | 170 |
| COG0515 | Serine Threonine protein kinase | 156 |
| ENOG410XRW9 | Receptor | 145 |
| COG5069 | Microtubule associated monoxygenase, Calponin and LIM domain containing | 113 |
| COG5059 | Kinesin family member | 76 |
| COG5022 | Myosin heavy chain | 75 |
| ENOG410XQHI | Cadherins are calcium dependent Cell adhesion proteins (By similarity) | 75 |
| COG5599 | Protein tyrosine phosphatase | 66 |
| COG0553 | Helicase | 64 |
| COG1226 | PotAssium voltage-gated channel | 63 |
| COG2940 | Histone-lysine N-methyltransferase | 60 |
| COG1100 | GTP-binding Protein | 57 |
| COG5021 | Ubiquitin protein ligase | 57 |
2. Experimental design, materials, and methods
2.1. Experimental animals
The birds used in this article were obtained from a breeding captive colony of outbred Common quails kept at the Konrad Lorenz Institute of Ethology (Vetmeduni Vienna, Austria). Our stock birds originated from wild founders captured during the spring migration/breeding season on the southern Italian coast near Palermo in 2008, 2009, and 2010 (Istituto Sperimentale Zootecnico per la Sicilia, Palermo, Italia). As previously shown [1], microsatellite and mtDNA screening excluded the presence of admixture with the Japanese quail in our study population. Experimental birds were reared under a 16:8 hrs light:dark cycle since hatching until they were about 7 weeks of age (46–59 days-old). Food and water were provided ad libitum throughout the course of the experiment. The indoor rooms were under constant climate control at 20–24 °C.
2.2. Experimental manipulation of the migratory phenotype
From about 7 weeks of age until about 14 weeks of age, all experimental birds were exposed to a gradual decline in day length (30 min/week) until the photoperiod reached 12:12 hrs light:dark. This photoperiod schedule was then maintained constant until the end of the experiment. This photoperiod manipulation aimed to simulate autumn migration followed by a non-migratory life history stage in the non-breeding grounds. After 8 weeks since the start of the photoperiod decline (average age of the birds was about 15 weeks, 102–114 days-old), half of the birds were randomly chosen and allocated to the migratory sampling group (October 2017), whereas the remaining birds were allocated to the non-migratory group (December 2017). For both groups, sampling procedures were performed at a standardised time of the day. For the RNA-Sequencing experiment presented here, we selected 11 birds sampled during the migratory phase (female: 6, male: 5) and 12 birds sampled during the non-migratory phase (female: 7, male: 5) – all the birds used here were sampled at night time (i.e. 21:00 h, three hours after lights were switched off).
2.3. Brain collection
Birds were sacrificed using an overdose of sodium pentobarbital 200 mg/ml via intraperitoneal administration. Birds were then decapitated, and brains removed after 6 min postmortem on average (4–9 min). The brains were immediately placed on dry ice and stored at −80 °C for further dissections. Punch dissections were performed as previously described in the related Japanese quail [2]. Briefly, the brains were placed ventral side up into a frozen brain holder matrix (Hugo Sachs Elektronik, Germany) with a 1 mm graduated scale. Two blades were positioned ∼ 4 mm from the rostral pole and ∼2 from the cerebellum to obtain a 2mm-tick coronal section. The cutting plane was adjusted to match as closely as possible the plane of the chicken brain atlas ([3] – coronal brain sections interaural 2.08–2.56 mm). Then, one single punch per individual brain was obtained from the basal hypothalamus spanning the third ventricle, and immediately stored at −80 °C until laboratory analysis.
2.4. RNA isolation
Total RNA was extracted using the Rneasy Microarray Tissue Mini Kit (Qiagen, Hilden, Germany), with a DNase digestion step to remove DNA contaminants using RNase-Free DNase Set (Qiagen, Hilden, Germany) following previously validated protocol in the Japanese quail [2]. Both purity and integrity of RNA were assessed respectively using a Nanodrop spectrophotometer (ThermScientific, Waltham, MA, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Hypothalamic RNA concentrations averaged 86.3 0± 5.80 ng/μl, ratio 260/280: 2.01 ± 0.01, Agilent RIN scores averaged 9.03 ± 0.06 (mean ± se for all).
2.5. cDNA library construction and RNA-Sequencing
For each individual hypothalamic sample, 500 ng of high-quality RNA (RIN ≥ 8.3 for all) was used to construct 23 barcoded sequencing libraries using standard Lexogen Sense mRNA-sequencing Library Preparation kit (Lexogen, Vienna, Austria) according to the manufacturer's instructions. The obtained cDNA libraries were then sequenced paired-end in one lane with a sequencing run length of 125 on a HiSeq V4 platform at the Vienna Biocenter (Vienna, Austria).
2.6. Transcriptome data analysis
2.6.1. Standard quality assessment of sequencing reads
The sequencing reads (BAM format) were pre-processed following standard guidelines. After conversion to fastq format, we used the software Trimmomatic v0.38 [4] to remove Illumina adapter sequences (up to two mismatches were allowed when recognizing adaptor sequences) and to filter out low quality reads (average Phred quality score < 30 in a sliding window of 8 base pairs across entire reads and reads below a minimum length of 50). Quality control of processed reads was confirmed and visualized using FastQC v0.11.7 (https://www.bioinformatics.babraham.ac.uk/index.html).
2.6.2. Trinity de novo assembly and parameter optimization
A reference genome for the Common quail is currently not available. Thus, we assembled the transcriptome de novo using the quality filtered paired-end sequencing reads. Assembly de novo strategies can outperform reference genome transcriptome mapping strategies for species with complex genomes [5], [6], [7]. The program Trinity v2.7.0 was used for the de-novo assembly of the pre-processed reads [8]. Briefly, Trinity assembly pipeline consists of three consecutive modules: Inchworm, Chrysalis, and Butterfly. In the first stage, all overlapping K-mers are extracted from the RNA-Seq reads. The Inchworm module then assembles initial contigs by extending sequences with abundant K-mers. In the following stage, Chrysalis clusters overlapping Inchworm contigs, builds de Bruijn graphs for each cluster, and partitions reads among clusters. Finally, Butterfly resolves all probable sequences from each graph component predicting alternatively spliced and paralogous transcripts independently for each cluster. In order to assess software performance with our data we used three independent strategies to compile the transcriptome assembly. Apart from assembly using default parameters (K-mer size set to 25 and target K-mer coverage set to 50, –normalize_max_read_cov), a second assembly was obtained using default parameters but with the target K-mer coverage set to 30. The third assembly was obtained by using all default parameters except for the K-mer minimum coverage (–min_kmer_cov) which was set at 2 in order to exclude all singletons obtained in the initial Inchworm contigs. We compared the three assemblies using standardised metrics following recommendations in the Trinity software (https://github.com/trinityrnaseq/trinityrnaseq/wiki). The de novo assemblies were performed on the Department of Integrative Biology and Evolution server with 4 processors, 80 cores and 378 GB RAM.
2.6.3. Assessment of assembly quality, read content, and assembly completeness
First, we retrieved basic quality metrics statistics included in the Trinity package. These data were obtained for all transcript contigs and also for only the longest contig from each predicted gene. To further assess the quality of the three de novo assemblies we then examined the number of input RNA-Sequencing reads that were represented in each assembled transcriptome. Thus, we mapped cleaned reads back to their corresponding assemblies using Bowtie2 v2.3.1 in local read alignment to maximize the alignment score (–local) and by suppressing records for reads that failed to align (–no unal) [9]. In order to assess transcriptome completeness we also compared our assembled transcripts sets against a set of known highly conserved single-copy orthologs using BUSCO v 3.0.2 (Benchmarking Universal Single-Copy Orthologs). We used the predefined lineage Aves ortholog dataset (i.e. Aves_odb9, Eukaryota) containing 4,915 single-copy orthologs from the OrthoDB database [10]. We consequently calculated the number of complete (length is within two standard deviations of the mean length of the given BUSCO), duplicated (complete BUSCOs represented by more than one transcript), fragmented (partially recovered BUSCOs) and missing (not recovered) transcripts in each of our three Trinity assembly.
2.6.4. Removal of redundant transcripts and hierarchical clustering
We used TransDecoder v5.3.0 ([8]; https://github.com/TransDecoder) to first extract all predicted protein coding sequences and then to select the single-best open reading frame (ORF) for each transcript within our three de novo assemblies (–single_best_orf, default setting). Any transcripts with ORFs with less than 200 bp in length were removed before proceeding with further analyses in the pipeline.
2.6.5. Hierarchical clustering in non-redundant transcripts
Using the selected assembly de novo, the candidate coding regions identified using TransDecoder were further filtered using the software CD-Hit-Est v4.6.8 [11]. CD-Hit-Est clustered highly similar nucleotide sequences that meet a sequence identity threshold of 0.95 (default = 0.90).
2.6.7. Functional annotation
In order to obtain a comprehensive annotation of our final sets of transcripts we used FunctionAnnotator, which is especially well-suited for non-model organism annotation [12]. The uploaded final transcriptome of Common quail in FunctionAnnotator was used for homology searches against the NCBI-NR (Non-redundant) protein sequences database with a cut-off e-value of 1e−3. Further functional annotations to explore distribution and frequencies of KEGG (Kyoto Encyclopedia of Genes and Genomes; [13]) and eggNOG [14] was performed using Trinotate v3.2.0 [15].
Ethical statement
All animal procedures were performed in compliance with the Austria legislation with approval of the Ethics Committee of the University of Veterinary Medicine Vienna, and the Federal Ministry of Science, Research and Economy (BMWFW-68.205/0037-WF/V/3b/2017).
Funding information
The work was funded by a Marie Sklodowska-Curie Individual Fellowshipto to VM (#704582) and by intramural funds to LF. VM was additionally supported by a FWF Der Wissenschaftsfonds Lise Meitner Fellowship(#M2520-B29).
Author contribution
VM designed the study, performed the animal work, contributed in supervision of laboratory work, performed data analysis, and wrote the manuscript. LF gave input on study design and provided comments on earlier drafts of the manuscript. GP established the colony of captive Common quails from which the birds used in this experiment originated and contributed to the planning of the experiments. SS gave input on study design, supervised laboratory and data analyses, and provided comments on earlier drafts of the manuscript.
Declaration of Competing Interest
We declare no competing interests.
Acknowledgments
We are most grateful to Antonio Console (Istituto Sperimentale Zootecnico per la Sicilia, Palermo) for providing us with the original stock of Common quails from which our experimental birds were produced. We thank Wolfgang Pegler and Chrystal Grabmayer for assistance with the animal husbandry, Roland Sasse for help with building the cages, Filipa Paiva Vilar Queirós (Erasmus MSc student) for help with the preparatory phase prior the experiment, Aryan Havrest (MSc student) for help with the animal experiment, Maria Kral for performing RNA-extraction and cDNA library preparation, Hannes Hofmann for technical assistance with the Department server, Pawel Herzyk and Jean Elbers for advice on the RNA-Sequencing data analyses.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2020.106041.
Appendix. Supplementary materials
References
- 1.Smith S., Fusani L., Boglarka B., Sanchez-Donoso I., Marasco V. Lack of introgression of Japanese quail in a captive population of common quail. Eur. J. Wildl. Res. 2018;64:1–9. doi: 10.1007/s10344-018-1209-7. [DOI] [Google Scholar]
- 2.Marasco V., Herzyk P., Robinson J., Spencer K.A. Pre- and post-natal stress programming developmental exposure to glucocorticoids causes long-term brain region specific changes to transcriptome in the precocial Japanese quail. J. Neuroendocrinol. 2016;28 doi: 10.1111/jne.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puelles L., Martinez-de-la-Torre M., Paxinos G., Watson C., Martinez S. Academic Press; Amsterdam: 2007. The Chick Brain in Stereotaxic Coordinates: an Atlas Featuring Neuromeric Subdivisions and Mammalian Homologies. [Google Scholar]
- 4.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ockendon N.F., O'Connell L.A., Bush S.J., Monzón-Sandoval J., Barnes H., Székely T., Hofmann H.A., Dorus S., Urrutia A.O. Optimization of next-generation sequencing transcriptome annotation for species lacking sequenced genomes. Mol. Ecol. Resour. 2016;16:446–458. doi: 10.1111/1755-0998.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Visser E.A., Wegrzyn J.L., Steenkmap E.T., Myburg A.A., Naidoo S. Combined de novo and genome guided assembly and annotation of the Pinus patula juvenile shoot transcriptome. BMC Genomics. 2015;16:1057. doi: 10.1186/s12864-015-2277-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ungaro A., Pech N., Martin J.-.F., McCairns R.J.S., Mévy J.-.P., Chappaz R., Gilles A. Challenges and advances for transcriptome assembly in non-model species. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0185020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M., Macmanes M.D., Ott M., Orvis J., Pochet N., Strozzi F., Weeks N., Westerman R., William T., Dewey C.N., Henschel R., Leduc R.D., Friedman N., Regev A. De novo transcript sequence reconstruction from RNA-seq using the Trinity plat- form for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langmead B., Salzberg S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. doi: doi.org/10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zdobnov E.M., Tegenfeldt F., Kuznetsov D., Waterhouse R.M., Simão F.A., Ioannidis P., Seppey M., Loetscher A., Kriventseva E.V. OrthoDB v9.1: cataloguing evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res. 2016;45:1–15. doi: 10.1093/nar/gkw1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinfor. - Matics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen T.-.W., Gan R.-.C., Fang Y.-.K., Chien K.-.Y., Liao W.-.C., Chen C.-.C., Wu T.H., Chang I.Y-F., Yang C., P-J Huang, Yeh Y.-.M., Chiu C.-.H., Huang T.-.W., Tang P. FastAnnotator: a versitle and efficient web tool for non-model organism annotation. Sci. Rep. 2017;10430 doi: 10.1038/s41598-017-10952-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powell S., Szklarczyk D., Trachana K., Roth A., Kuhn M., Muller J., Arnold R., Rattei T., Letunic I., Doerks T., Jensen L.J., Von Mering C., Bork P. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2012;40:284–289. doi: 10.1093/nar/gkr1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bryant D.M., Johnson K., DiTommaso T., Tickle T., Couger M.B., Payzin-Dogru D., Lee T.J., Leigh N.D., Kuo T.H., Davis F.G., Bateman J., Bryant S., Guzikowski A.R., Tsai S.L., Coyne S., Ye W.W., Freeman R.M., Peshkin L., Tabin C.J., Regev A., Haas B.J., Whited J.L. A tissue-mapped axolotl de novo transcriptome enables identification of limb regen- eration factors. Cell Rep. 2017;18:762–776. doi: 10.1016/j.celrep.2016.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.