Abstract
Multienzyme complex has attracted increased attention in biofuel technology. They offer solutions to effective degradation of complex plant material into fermentable sugars. Microorganisms, especially bacteria and fungi, are well studied for their ability to produce enzymes complex unlike yeast. Yeast strain isolated from mushroom farm was studied for simultaneous production of cellulase, xylanase and ligninase enzymes using lignocellulose waste as substrates. A response surface methodology (RSM) involving Box-Behnken design (BBD) was used to investigate interaction between variables (moisture content, inoculum size, initial pH, incubation time) that affect enzyme production. Crude filtrate was partially purified and characterised. Yeast strain identified as Saccharomyces cerevisiae SCPW 17 was finally studied. Evaluation of lignocellulose waste for enzyme complex production revealed corn cob to be most effective substrate for cellulase, xylanase and ligninase production with enzyme activity of 17.63 ± 1.45 U/gds, 29.35 ± 1.67 U/gds and 150.75 ± 2.01 μmol/min respectively. Time course study showed maximum enzyme complex production was obtained by day 6 with cellulase activity of 12.5 U/gds, xylanase 48.3 U/gds and ligninase 90.8 μmol/min. Using RSM involving BBD, maximum enzyme activity was found to be 19.51 ± 0.32 U/gds, 56.86 ± 0.38 U/gds, 408.17 ± 1.04 μmol/min for cellulaase, xylanase and ligninase respectively. The developed models were highly significant at probability level of P = 0.0001 and multiple correlation co-efficient (R2) was 0.9563 for cellulase, 0.9532 for xylanase and 0.9780 for ligninase. Enzyme complex was stable at varying pH and temperature conditions. Saccharomyces cerevisiae (SCPW 17) studied produced enzyme complex which can be used for bioconversion of biomass to value-added chemicals.
Keywords: Bioinformatics, Biotechnology, Cell biology, Genetics, Microbiology, Molecular biology, Multienzyme complex, Cellulase, Xylanase, Ligninase, Corncorb, Saccharomyces cerevisiae
Bioinformatics; Biotechnology; Cell biology; Genetics; Microbiology; Molecular biology; Multienzyme complex, Cellulase, Xylanase, Ligninase, Corncorb, Saccharomyces cerevisiae
1. Introduction
Producing renewable fuels and chemicals from lignocellulosic biomass (which are largely abundant in nature) along the biochemical conversion route requires the hydrolysis of the polysaccharide components of biomass, cellulose and hemicellulose, into their various sugar constituents. The use of enzymes to catalyse the degradation of cellulose to glucose and hemicellulose to free sugars has long been considered the most viable strategy to provide cost-effective bioconversion of biomass to value-added chemicals. Micro-organisms possess several mechanisms for lignocellulose deconstruction. Fungi and bacteria especially have the free enzyme system and the most common mechanisms [1]. Furthermore, cellulosomes and xylanosomes are complex protein structures for the degradation of biomass, and are largely produced by anaerobic bacteria and aerobic fungi. Whereas, the potential to produce cellulase or xylanase or both is not commonly found among yeast strains and very scant literatures are available. Importantly, the economy of biofuel and chemical production from lignocellulosic biomass requires the use of cellulose, hemicellulose and lignin in order to obtain an economically feasible biomass conversion [2].
Several approaches are being proposed – use of enzyme degradation of lignocellulose biomass, the use of natural strains with multienzyme complex [3], enzyme cocktail (mixtures of various enzymes from different sources) or metabolically engineered microbes like Saccharomyces cerevisiae, Escherichia coli or Yarowia lipolytica to convert biomass hydrolysates into target products [4] Enzyme market is very promising in developed countries, like the US, Western Europe, Japan, and Canada. In these countries the demand for industrial enzymes is stable, unlike in developing economies, Asia-Pacific, Eastern Europe, Africa and the Middle East regions that are recently emerging as fast-growing markets for industrial enzymes [5].
Hydrolases are the most widely used class of enzymes in the industry [6]. Enzymes are very well-established products in biotechnology. In the world market, enzymes for industrial applications are expected to be worth $6·3 billion by 2021 [7]. The cost of enzymes for biofuel applications alone is envisaged to total $1·0 billion in 2020, with a CAGR of 10·4%. Again, by 2020, the US and European markets for biofuel enzymes are expected to be worth $355·7 million and $325·2 million, respectively, both with a CAGR >10% (BBC research 2015). Lignocellulose-degrading enzymes are applied in different fields, including technical use, food manufacturing, paper bleaching, biofuels, textile industry, and as tools for research and development [8].
High demand in recent years in the enzyme market highlights a need for development of novel microbial strains capable of producing high enzyme titres, including multienzyme complex, enzyme consortiums and cocktails combining different enzymes from various sources. This will help to reduce cost whilst achieving complete deconstruction of lignocellulose as well as building a balanced enzymatic mixture for industry processes. A promising approach is to source natural strains from unexploited environments that abound in Nigeria. Biodiversity environments in Nigeria provides great opportunity for isolation and characterization of novel yeast strains with enzymes producing potentials that can be used to completely deconstruct lignocellulose biomass. The mushroom farm environment in Nigeria is a typical example for the isolation of novel microbial strains capable of producing lignocellulases with high titer. This is more so when Nigeria has in high volumes lignocellulosic materials arising from various sources such as cassava peel, cassava pulp, grasses, plantain/banana waste, yam and cassava decay [9, 10]. Success in obtaining a novel yeast from the local environment will not only have the advantage of producing products from these waste materials, it will also help to provide a cleaner environment [9, 10].
The high moisture content from the lignocellulose materials, nutrients and favourable temperature for growth of the mushroom allows microbial activity to thrive. It is expected that microbes in this habitat possess the ability to produce lignocellulases that breakdown lignocellulasic biomass which characterize a mushroom farm – hence this study. To the best of the authors' knowledge, no studies have been carried out on the production of combined multienzyme – cellulase, xylanase and ligninase from yeasts from a mushroom farm. Studies reported in literature include production of xylanase using psychrophilic yeast Cryptococcus adeliae and Cryptococcus flavus isolate I-11 [11,12,13]. Gomes et al. [14], reported cellulolytic and xylanolytic activities from yeast strains isolated from bromelied tanks of Viresea minarum, with major genera of Cryptococcus, Fellomyces, Myriangiale and Ocultifer species. They also reported that only very few of the strains could elaborate cellulolytic and xylanolytic activities simultaneously. Again, Thongekkew et al. [15], reported of all the yeast strains isolated from various sources some strains displayed cellulase activity whilst others displayed xylanase activity. For production of multienzyme complex, Qadir et al. [16], studied co-culture of two Saccharomyces cerevisae strains, MK-157and MK-118 to produce endoglucanase, β-glucosidase and xylanase.
In this paper, we present the results of a novel study where simultaneous production of cellulase, xylanase and ligninase was achieved by a single yeast strain (Saccharomyces cerevisiae SCPW 17) isolated from a mushroom farm using lignocellulosic waste as substrates. Several factors such as cultivation conditions and source of nutrient will affect the production of these enzymes by microorganisms. Optimization of the process parameters for enhanced production of these enzymes was also studied and statistically tested using response surface (RSM) method [17, 18, 19]. RSM is a statistical tool for determining optimum conditions of a process response variables and evaluation of the correlations between a group of controlled experiments and observing the results of one or more selected variables [17, 18, 19]. It also brings to bear an alternative method of optimization where the interactions between all the variables are taken into consideration and estimate of their combined effect is expounded. We therefore extended the study to include variables such as moisture content, initial pH, inoculum size, and incubation time as the independent variables while the dependent variables (response) were cellulose, xylanase and ligninase production.
2. Materials and methods
2.1. Soil sample collection
Soil samples were collected from a mushroom farm in Yala Local Government Area of Cross River State, Nigeria. The samples were aseptically collected and placed in sterile sample bottles, properly labelled, and transported on ice packs to the Microbiology Laboratory of the University of Nigeria, Nsukka within 24 h of collection.
2.2. Strain isolation and inoculum preparation
For initial isolation, appropriate dilutions of soil samples were made and aliquots inoculated onto agar plates, with media composition (g/L): NaNO3 - 2.0; KH2PO4 - 1.0; MgSO4.7H2O - 0.5; KCl 0.5; Proteose peptone - 2.0; Agar - 20, and 0.5% carboxymethyl cellulose (CMC) and xylan was added respectively. For ligninase, basal medium (LBM), comprised (g/L): KH2PO4 – 1.0; Yeast Extract - 0.01; C4HI2N2O6- 0.5; CuSO4.5H2O - 0.001; MgSO4.7H2O - 0.5; Fe2(SO4)3- 0.001; CaCl2.2H2O - 0.01 and MnSO4.H2O- 0.001. The LBM was supplemented with 0.25 % w/v lignin and 1.6 % w/v agar and autoclaved. This was supplemented with 10 mL of a separately sterilized 20 % w/v aqueous glucose solution. A 100 μg/ml chloramphenicol was added, adjusted to pH 6.0 and incubated for 3–5 days at 28±2 °C to isolate yeast colonies [20]. The morphologically different yeast strains were selected and transferred onto YPDA slants and maintained at 4 °C for further studies.
2.3. Qualitative (plate) screening of yeast isolates for lignocellulase production
The yeast isolates were screened qualitatively for their ability to produce cellulase, xylanase and ligninase simultaneously using CMC, xylan as well as lignin agar plates to detect their individual cellulase, xylanase and ligninase activity respectively. Cellulase and xylanase activity was carried out according to the method of Srilakshmi et al. [21], with slight modifications. After 3 days of incubating the plates at 28±2 °C, all the plates were flooded with 15 mL of 0.1% Congo red staining solution for 15 min and distained with 1M solution of NaCl. Isolates with halo zone from the point of spot inoculation of the culture outwards denotes substrate (CMC or Xylan) degradation as a result of the enzymes produced by these yeast isolates. A yellow-opaque zone around a colony indicates degradation of the substrate and red colour shows non-degraded substrate. To test for lignin degradation after 3–5 days incubation in the dark, the LBM plates were flooded with a 1 % w/v aqueous solution of FeCl3 and K3 (Fe(CN)6) prepared freshly before use. Phenols in undegraded lignin will stain blue-green, with clear zones around colonies indicating oxidation of phenolic components [22]. “The zone of clearance was measured and colonies with wider diameter of clearance were selected for further studies”.
2.4. Quantitative screening of yeast isolates for lignocellulase production
2.4.1. Submerged fermentation (SmF)
The yeast isolates with wider diameter were also assayed by quantitative method for their cellulose, xylanase and ligninase activity under submerged fermentation conditions. The isolates were grown in medium containing the following (g/L): NaNO3-2.0; KH2PO4-1.0; MgSO4.7H2O-0.5; KCl-0.5; Proteose peptone-2.0; CMC-5.0 for cellulase assay, Xylan-5.0 for xylanase assay and Lignin for ligninase was added respectively. Then 50 ml of the medium (pH 6.0) in a 250 mL Erlenmeyer flask was autoclaved at 121 °C for 15 min, cooled, inoculated with 1.0 mL (1.42 × 107 CFU/ml) of 24 h old culture and incubated at 28±2 °C. Samples were analyzed after every 24 h for 7 days. Extraction of crude enzyme from fermentation broth was done by centrifugation at 7,000 × g for 30 min and the clear supernatant (crude enzyme) was used for cellulase and xylanase assay using DNSA (dinitrosalicylic acid) method [23]. Isolates with high cellulase activity were also assayed for their xylanolytic ability and vice versa as well as ligninase. The isolate with most efficient cellulase, xylanase and ligninase producing ability was characterized by the amplification of the internal transcribed spacer (ITS1 and ITS2) regions as a means of identification. Ribosomal RNA (r-RNA) sequencing method and Sanger sequencing of the amplicons was done at the Core DNA Services Laboratory 188 of the University of Calgary.
2.4.2. Solid state fermentation (SSF)
For the initial study different local agro-waste materials such as corn cob (CC), sugar cane bagasse (SCB), opete bagasse (OB), rice husk (RH), ugu pod (UP), were assessed for their potential as substrate for cellulase, xylanase and ligninase production under solid state fermentation (SSF). Fermentation was carried out in 250 ml Erlenmeyer flasks that contain 10g of dry weight of each of the above named substrates having particle size of 425 μm. To determine the effect of particle size on enzyme production. The Endecotte machine was used to separate these particles into different sizes (300 μm, 425 μm, 710 μm and 800 μm). This was moistened at 80% level using the mineral salt medium solution (g/L: NaNO3-2.0; KH2PO4-1.0; MgSO4.7H2O-0.5; KCl-0.5; bambara meal-2.0) as the moistening agent. The initial pH of the nutrient solution was kept at 6.0 before sterilization. The flasks were sterilized at 121 °C for 30 min to kill all autochthonous microbes present in the sample, allowed to cool and inoculated with 1.0 mL of isolate suspension (containing 107 CFU/g of dry substrate). The contents of the flasks were thoroughly mixed, incubated at 28±2 °C for the 10 days and samples were taken every 24 h for analysis [24]. The initial moisture content was determined gravimetrically and all liquid added to the flasks was taken into consideration in calculating the moisture content. All the experiments were carried out in triplicate.
2.5. Box-Behnken design (BBD) for optimization of fermentation parameters using response surface methodology (RSM)
Response Surface Methodology (RSM) involving Box-Behnken design (BBD) was used to determine the interactions between and amongst all the variables and their combined effect on cellulase, xylanase and ligninase production. A Box-Behnken factorial design with four factors and three levels was used to optimize the cultural condition. The independent variables (factors) used for the analysis were moisture ratio (A), pH (B), inoculum size (C) and incubation time (D). The effect of these variables on cellulase, xylanase and ligninase production as well as the most suitable combination that gave the maximum enzyme yield was also analyzed. Cellulase, xylanase and ligninase activity extracted were taken as the dependent variable or response (Y). Each of four independent variables was studied at three different levels designated as low, middle and high level of each variable coded as −1, 0 and +1 respectively (Tables 1 and 2). A BBD with a total of 27 experimental runs shows the coded and actual values for each of the enzymes in Tables 3, 4, and 5. The variables were taken at a central code value considered as zero. The incubation temperature was kept constant at 28±2 °C throughout the entire experiments. Thereafter, the contents of all the flaks were analyzed for cellulase, xylanase and ligninase activity after 6 days of incubation. The second order polynomial equation below was adopted to study the effects of the independent variables on the response.
Table 1.
Experimental range, coded values and level of independent variables for the optimization of cellulase and xylanase production.
| Factors | Symbol | Coded Level |
||
|---|---|---|---|---|
| -1 | 0 | +1 | ||
| Moisture Content (%) | X1 | 75 | 80 | 85 |
| pH | X2 | 5 | 6 | 7 |
| Inoculum Size (ml) | X3 | 1.5 | 2.0 | 2.5 |
| Incubation time (days) | X4 | 5 | 6 | 7 |
Table 2.
Experimental range, coded values and level of independent variables for the optimization of ligninase production.
| Factors | Symbol | Coded Level |
||
|---|---|---|---|---|
| -1 | 0 | +1 | ||
| Moisture Content (%) | X1 | 50 | 60 | 70 |
| pH | X2 | 4.0 | 4.5 | 5.0 |
| Inoculum Size (ml) | X3 | 2.0 | 3.0 | 4.0 |
| Incubation time (days) | X4 | 5 | 6 | 7 |
Table 3.
Box–Behnken design matrix with experimental values of cellulase and xylanase activity optimization.
| Coded values |
Response (Cellulase activity U/gds) |
Response (Xylanase activity U/gds) |
||||||
|---|---|---|---|---|---|---|---|---|
| Run | X1 | X2 | X3 | X4 | Observed value | Predicted value | Observed value | Predicted value |
| 1 | -1 | -1 | 0 | 0 | 14.67 | 14.48 | 35.07 | 37.86 |
| 2 | -1 | +1 | 0 | 0 | 14.16 | 13.07 | 39.19 | 42.35 |
| 3 | +1 | -1 | 0 | 0 | 8.88 | 9.34 | 44.43 | 45.55 |
| 4 | +1 | +1 | 0 | 0 | 11.70 | 11.26 | 41.78 | 43.27 |
| 5 | 0 | 0 | -1 | -1 | 6.98 | 5.67 | 13.55 | 10.72 |
| 6 | 0 | 0 | -1 | +1 | 6.22 | 6.22 | 31.83 | 32.46 |
| 7 | 0 | 0 | +1 | -1 | 9.89 | 9.26 | 22.08 | 25.73 |
| 8 | 0 | 0 | +1 | +1 | 16.48 | 17.16 | 45.70 | 52.81 |
| 9 | -1 | 0 | 0 | -1 | 6.46 | 7.99 | 15.37 | 16.02 |
| 10 | -1 | 0 | 0 | +1 | 13.11 | 13.98 | 45.49 | 44.17 |
| 11 | +1 | 0 | 0 | -1 | 7.04 | 7.04 | 24.58 | 24.05 |
| 12 | +1 | 0 | 0 | +1 | 10.17 | 8.74 | 47.23 | 44.73 |
| 13 | 0 | -1 | -1 | 0 | 7.37 | 7.78 | 31.76 | 31.45 |
| 14 | 0 | -1 | +1 | 0 | 16.83 | 16.71 | 51.09 | 48.33 |
| 15 | 0 | +1 | -1 | 0 | 9.48 | 9.70 | 30.84 | 31.75 |
| 16 | 0 | +1 | +1 | 0 | 15.60 | 15.29 | 51.79 | 50.25 |
| 17 | -1 | 0 | -1 | 0 | 10.51 | 10.03 | 33.70 | 33.07 |
| 18 | -1 | 0 | +1 | 0 | 19.51 | 18.88 | 51.98 | 47.33 |
| 19 | +1 | 0 | -1 | 0 | 6.98 | 8.14 | 31.72 | 33.94 |
| 20 | +1 | 0 | +1 | 0 | 12.81 | 13.82 | 56.86 | 55.06 |
| 21 | 0 | -1 | 0 | -1 | 6.34 | 6.38 | 18.37 | 18.70 |
| 22 | 0 | -1 | 0 | +1 | 11.78 | 11.17 | 42.03 | 40.87 |
| 23 | 0 | +1 | 0 | -1 | 6.07 | 7.20 | 18.83 | 17.56 |
| 24 | 0 | +1 | 0 | +1 | 10.37 | 10.86 | 46.97 | 44.22 |
| 25 | 0 | 0 | 0 | 0 | 11.39 | 11.10 | 44.89 | 41.12 |
| 26 | 0 | 0 | 0 | 0 | 11.03 | 11.10 | 40.01 | 41.12 |
| 27 | 0 | 0 | 0 | 0 | 10.88 | 11.10 | 38.47 | 41.12 |
Table 4.
Box–Behnken design matrix of ligninase activity optimization.
| Coded values |
Response |
|||||
|---|---|---|---|---|---|---|
| Run | X1 | X2 | X3 | X4 | Ligninase activity (μmol/min) |
|
| Observed | Predicted | |||||
| 1 | -1 | -1 | 0 | 0 | 352.258 | 341.326 |
| 2 | -1 | +1 | 0 | 0 | 377.419 | 381.416 |
| 3 | +1 | -1 | 0 | 0 | 366.237 | 362.635 |
| 4 | +1 | +1 | 0 | 0 | 360.645 | 372.132 |
| 5 | 0 | 0 | -1 | -1 | 232.043 | 237.581 |
| 6 | 0 | 0 | -1 | +1 | 341.075 | 330.071 |
| 7 | 0 | 0 | +1 | -1 | 203.548 | 215.107 |
| 8 | 0 | 0 | +1 | +1 | 338.280 | 333.298 |
| 9 | -1 | 0 | 0 | -1 | 218.065 | 206.907 |
| 10 | -1 | 0 | 0 | +1 | 313.548 | 318.053 |
| 11 | +1 | 0 | 0 | -1 | 220.860 | 218.725 |
| 12 | +1 | 0 | 0 | +1 | 304.731 | 318.260 |
| 13 | 0 | -1 | -1 | 0 | 346.667 | 364.030 |
| 14 | 0 | -1 | +1 | 0 | 357.850 | 366.988 |
| 15 | 0 | +1 | -1 | 0 | 408.172 | 401.405 |
| 16 | 0 | +1 | +1 | 0 | 394.194 | 379.201 |
| 17 | -1 | 0 | -1 | 0 | 355.699 | 361.311 |
| 18 | -1 | 0 | +1 | 0 | 346.022 | 353.838 |
| 19 | +1 | 0 | -1 | 0 | 380.215 | 369.474 |
| 20 | +1 | 0 | +1 | 0 | 366.237 | 357.700 |
| 21 | 0 | -1 | 0 | -1 | 223.656 | 217.234 |
| 22 | 0 | -1 | 0 | +1 | 329.140 | 323.596 |
| 23 | 0 | +1 | 0 | -1 | 240.430 | 243.049 |
| 24 | 0 | +1 | 0 | +1 | 343.871 | 347.368 |
| 25 | 0 | 0 | 0 | 0 | 360.645 | 360.828 |
| 26 | 0 | 0 | 0 | 0 | 353.011 | 360.828 |
| 27 | 0 | 0 | 0 | 0 | 368.828 | 360.828 |
Table 5.
Analysis of variance (ANOVA) test for Box–Behnken design Cellulase.
| Source | DF | Adj SS | Adj MS | F-Value | P-Value |
|---|---|---|---|---|---|
| Model | 14 | 334.134 | 23.867 | 18.78 | 0.000 |
| Linear | 4 | 248.202 | 62.051 | 48.81 | 0.000 |
| A-Moisture content | 1 | 36.192 | 36.192 | 28.47 | 0.000 |
| B-pH | 1 | 0.190 | 0.190 | 0.15 | 0.706 |
| C-Inoculum | 1 | 158.268 | 158.268 | 124.50 | 0.000 |
| D-Time | 1 | 53.552 | 53.552 | 42.13 | 0.000 |
| Square | 4 | 60.931 | 15.233 | 11.98 | 0.000 |
| A∗A | 1 | 2.193 | 2.193 | 1.73 | 0.214 |
| B∗B | 1 | 0.472 | 0.472 | 0.37 | 0.554 |
| C∗C | 1 | 5.057 | 5.057 | 3.98 | 0.069 |
| D∗D | 1 | 33.200 | 33.200 | 26.12 | 0.000 |
| 2-Way Interaction | 6 | 25.001 | 4.167 | 3.28 | 0.038 |
| A∗B | 1 | 2.772 | 2.772 | 2.18 | 0.165 |
| A∗C | 1 | 2.512 | 2.512 | 1.98 | 0.185 |
| A∗D | 1 | 3.098 | 3.098 | 2.44 | 0.144 |
| B∗C | 1 | 2.789 | 2.789 | 2.19 | 0.164 |
| B∗D | 1 | 0.325 | 0.325 | 0.26 | 0.622 |
| C∗D | 1 | 13.506 | 13.506 | 10.62 | 0.007 |
| Error | 12 | 15.254 | 1.271 | ||
| Lack-of-Fit | 10 | 15.117 | 1.512 | 22.00 | 0.044 |
| Pure Error | 2 | 0.137 | 0.069 | ||
| Total | 26 | 349.389 |
| Model Summary | |||
|---|---|---|---|
| S | R-Sq | R-Sq (adj) | R-Sq (pred) |
| 1.12747 | 95.63% | 90.54% | 74.99% |
An empirical second order polynomial equation was adopted to find the effects of independent variables to the response.
| Y = β0 + β1A + β2B + β3C + β4D + β1,1A2 + β2,2B2 + β3,3C2 + β4,4D2 + β1,2AB + β1,3AC + β1,4AD + β2,3BC + β2,4BD + β3,4CD |
Where:
Y represents cellulase, xylanase and ligninase activity (response),
β0 is the constant term;
β1, β2, β3 and β4are the coefficient of linear terms;
β1,1, β2,2, β3,3 and β4,4are the coefficient of quadratic terms;
β1,2, β1,3, β1,4, β2,3, β2,4 and β3,4 are the coefficient of cross product terms respectively. The goodness of fit of the polynomial equation was expressed by coefficient of determination R2 and its statistical significance level was checked by F-test.
2.6. Analytical methods for enzyme assays
2.6.1. Enzyme extraction
Enzyme extraction was carried out at 24 h intervals during the incubation period. The crude enzyme was extracted by adding 50 mM citrate buffer (pH 4.8) at 5 mL of buffer/g of fermented substrate and agitated at 200 rpm for 30 min. The solid material was separated by passing the slurry through a muslin cloth and Whatman glass microfiber filter paper. The filtrate was collected, centrifuged at 1500 ×g for 20 min to obtain a clear supernatant and was used to analyze for cellulase and xylanase activity by 3,5-dinitrosalicylic acid (DNS) method described by Miller [23] while ligninase assay was done according to the method of Tien and Kirk [25] with modifications.
2.6.2. Enzyme assays
Cellulase and xylanase activity was determined according to the method of Bailey et al. [26] with slight modifications. This was done by mixing 0.5 ml of 1% (w/v) carboxymethylcellulose (CMC) and xylan prepared in 50 mM citrate buffer (pH 4.8) with 0.5 ml of the appropriately diluted enzyme respectively. The enzyme-substrate mixture was incubated at 50 °C for 10 min. Thereafter, 1.5 mL of the 3,5-dinitrosalicylic acid (DNS) reagent was added to terminate the reaction. Enzyme blanks and a control that contained all the reagents were also run concurrently but in enzyme blank, the reaction was terminated prior to the addition of enzyme extract whereas in control, distilled water was added instead. Thereafter, the tubes were placed in boiling water for 10 min, cooled to room temperature in water for stabilization. The released reducing sugars were determined spectrophotometrically at 540 nm using of 3,5-dinitrosalicylic acid (DNS) method with glucose and xylose calibration curve used as standards for cellulase and xylanase activity respectively [26]. One unit of cellulose and xylanas is defined as the amount of enzyme that liberates 1 μmol of glucose and xylose equivalents per minute, respectively under the standard assay conditions.
While the ligninase peroxide (LiP) assay was performed using 2.6 mL of the reaction mixture containing 1.0 mL citrate buffer (pH 4.5), 1.0 mL of 4.0mM veratryl alcohol (3,4-dimethoxybenzylalcohol), 500 μL of 1mM H2O2 and 100μL of the crude enzyme. A blank containing all other reagents except the sample was used. Absorbance was read after a 10 min reaction interval at 427 nm (Molar absorbtivity, ϵ313 = 9300 M−1 cm−1). One unit (U) of LiP activity was defined as the amount of enzyme which converted 1μmole of veratryl alcohol to veratraldehyde under standard conditions. The experiments were carried out in triplicate and results reported as mean ± standard deviation.
2.6.3. Ammonium sulphate precipitation
Protein precipitation by salting out technique using ammonium sulphate (NH4(SO4)2) was carried out with constant gentle stirring [27]. The crude enzyme was partially purified from the culture supernatant, for the purpose, various ammonium sulphate concentrations, i.e. 40, 50, 60, 70 and 80 % were used for the precipitation of enzyme. This was left overnight and the precipitate was collected by centrifugation at 10,000 ×g for 10 min. The precipitate obtained was dissolved in phosphate buffer (50 mM, pH 8.0) and dialyzed against the same buffer for 24 h. The precipitates were collected and analyzed for enzyme complex activity.
2.6.4. Effect of pH on enzyme complex activity and stability
The effect of pH on crude filterate containing enzyme complex (cellulase, xylanase and ligninase) activity was measured in the pH range of 3–9, using the appropriate buffers at concentration of 100 mM (3.0–6.0, sodium citrate; 4.0–6.0, sodium acetate; 6.0–8.0 sodium phosphate; 7.0–9.0, Tris) under standard assay conditions. To study stability as a function of pH, 100 μL of the partially purified enzyme was mixed with 100 μL of the buffer solutions and incubated at 28 °C for 1 h then aliquots of the mixture were taken to measure the residual enzyme complex activity (%) under standard assay conditions.
2.6.5. Effect of temperature on enzyme activity and stability
The influence of temperature on activity of enzyme complex was studied by incubating the reaction mixture at different temperatures (30, 40, 50, 60, 70). The relative enzyme activity was recorded at 1 h interval during a period of 6 h. The activity of the enzyme was considered as 100% under standard assay conditions.
2.6.6. Protein content of enzyme
The protein content of the enzyme was determined according to Lowry et al. [28], using bovine serum albumin (BSA) as standard. The enzyme sample was properly mixed and 0.5 mL of it transferred to a 10 mL glass tube. 0.7 mL of Lowry solution was added, mixed and incubated for 20 min at room temperature (28 °C) in the dark. After 20 min of incubation, 0.1 mL of dilute Folin reagent was added to each tube and mixed. This was incubated at room temperature in the dark. The absorbance was measured at 550nm against a blank. Standard curve was prepared using BSA (1 mg/mL) dissolved in distilled water. All readings were taken in triplicates.
3. Results and discussion
3.1. Screening and identification of lignocellulose producing yeast
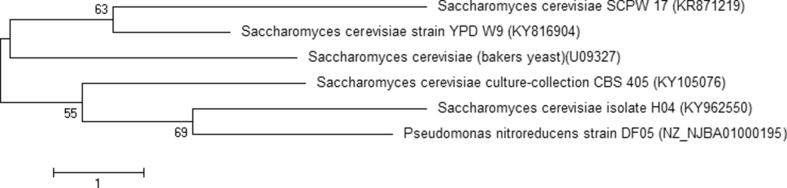
Yeast isolated from mushroom farm capable of simultaneously producing cellulase, xylanase and ligninase were screened qualitatively, ten yeast colonies were picked on the basis of the clear zones around the colonies. Secondary screening on liquid medium was also carried out to select strains with the potential to produce the desired enzyme complex. The yeast strain coded X10 which showed maximum enzyme complex production was selected. This selected yeast was therefore subjected to further investigations. Gomes et al. [14], reported isolation of yeast strains, with cellulolytic and xylanolytic activities with major genera including Cryptococcus, Fellomyces, Myriangiale and Ocultifer. Just few of these strains could elaborate cellulolytic and xylanolytic activities, simultaneously. Thongekkaew et al. [15], also described the ability of 61 yeast strains isolated from different environment to produce xylanase and cellulose activity. The yeast strain under study was identified on the basis of the amplification of its internal transcribed spacer (ITS1 and ITS2) regions as Saccharomyces cerevisiae SCPW 17. The phylogenetic tree displaying the strain relatedness is shown in Figure 1. It is important to note that in recent times, novel microbial strains are being sort and developed especially strains with unique phenotypes that can be used for industry applications. The environments from which microorganisms are isolated play critical role in the phenotypes they express. A genotype-by-environment interaction enables organisms to acclimatize to environmental variation [29, 30]. Many organisms are able to adapt to an environment and this influences the phenotype they express [31]. Saccharomyces cerevisiae like other microbes produce extracellular enzymes, and the yeasts strains isolated from the mushroom soil showed varying degrees of lignocellulase enzyme activity more importantly ligninase. This could have been conferred mainly because the genes responsible for expressing hydrolytic enzymes are present in the strain and the environment as well as nutrient present influenced the strains ability to express the enzymes. Hence, the reasons for carrying out this study.
Figure 1.
Phylogenic tree showing relationship of Saccharomyces cerevisiae strain SCPW 17 among the Saccharomyces genus.
3.2. Evaluation of different substrates for enzyme complex production under solid state fermentation (SSF)
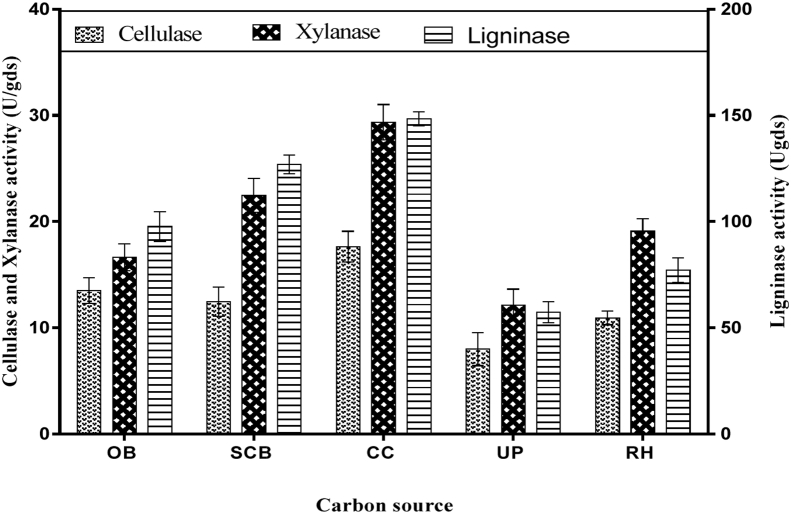
Different agro-residues were screened for their ability to be used as substrate for the production of cellulase, xylanase and ligninase enzyme complex. Lignocellulosic wastes used include, sugar cane bagasse (SCB), opete bagasse (OB), corn cob (CC), ugu pod (UP), and rice husk (RH). Our results showed that corn cob was the most effective substrate for cellulase, xylanase and ligninase production with enzyme activity of 17.63 ± 1.45 U/gds 29.35 ± 1.67 U/gds and 150.75 ± 2.01 μmol/min respectively. SCB was the next best substrate (12.45 ± 1.37 U/gds cellulase; 22.48 ± 1.58 U/gds xylanase and ligninase128.14 ± 1.73 μmol/min) as shown in (Figure 2). The lignocellulose biomass were used directly without any pre-treatment as substrate by solid state fermentation for enzyme complex production, which is typical of what is found in the environment as in the case of mushroom farm. This part of the experiment is very important as pre-treatment would incur additional costs in industrial processes. It is important to mention that the composition of lignocellulose can vary and therefore will affect their accessibility to the yeast for the enzyme production. This in turn could affect to varying degrees the production of enzyme complex. This notwithstanding, the yeast still exhibited the ability to utilize the other substrates to produce the required enzyme complex.
Figure 2.
Cellulase, xylanase and ligninase activity of different carbon sources by solid state fermentation using Saccharomyces cerevisiae SCPW 17 with bambara meal as nitrogen source. Key: OB-opete bagasse; SCB-sugar cane bagasse; CC-corn cob; UP-ugu pod; RH-rice husk.
Reports are available in the literature on the production of cellulases and xylanase from filamentous fungi and bacteria [32, 33, 34] using crude LC substrates. However, production of cellulase, xylanase is not very common with yeast, not to mention ligninase. Gomes et al. [14], reported cellulolytic and xylolytic activity in some yeast strains, while Qadir et al. [16], reported cellulase and xylanase activity in coculture of Saccharomyces cerevisiae MK-157 and Candida tropicalis MK-118. None of these microorganisms are efficient at cellulase, xylanase and ligninase activities simultaneously as observed in the present study. Multienzyme complex (MEC) has become appealing in biofuel technology as they possibly offer solutions to the more effective degradation of complex plant material into fermentable sugars, microorganisms involved in carbon cycling produce an array of carbohydrate degrading enzymes, including cellulase and xylanases. A number of microorganisms produce these as free extracellular enzymes, while others produce a multi-enzyme complex (MEC) such as the cellulosome in which several enzymes such as cellulases and xylanases are combined within a complex. The Saccharomyces cerevisiae SCPW 17 isolated from mushroom farm soil produced cellulase, xylanase and ligninase enzymes simultaneously in all test substrate although corn cob was the best and was used for further studies.
3.3. Production profile of cellulase, xylanase and ligninase
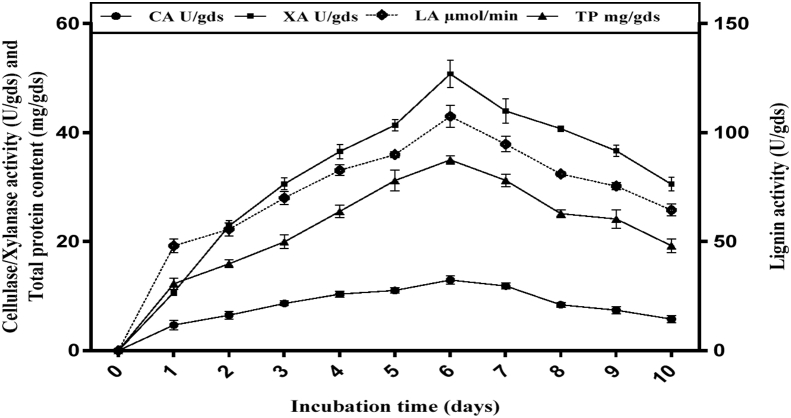
SSF is an attractive method for the production of value-added products making use of inexpensive agricultural residues such as CC. So far, simultaneous production of cellulase, xylanase and ligninase under SSF of CC by yeasts has not been reported. The time course study of enzyme production by Saccharomyces cerevisiae SCPW 17 grown on corn corb at 28 °C in SSF, is shown in Figure 3. Maximum enzyme complex production was obtained by day 6 with cellulase activity of 12.95 ± 0.76 U/gds, xylanase 50.79 ± 2.48 U/gds and ligninase 107.46 ± 5.07 U/gds. The accumulation of these extracellular enzymes by Saccharomyces cerevisiae SCPW 17 started after 24 h and increased steadily up to 144 h where the peak production was observed. The peak production of enzymes for most microorganisms occurs during the exponential growth phase of the microbe and this has direct effect on the cost of the production process because a fast growing strain in optimally rich medium is desirable and gives high enzyme yields. This agrees with the report of Mahalakshmi and Jayalakshmi [35] that the highest lignocellulase yield is achieved at the log stage of the growth curve and decreases with decreases in cell concentration. Several authors have reported the use of corn cob for lignocellulase production with varying degrees of enzyme activity [35, 36]. Essentially enzyme productions depend on the chemical composition of the substrate, accessibility and physiochemical association between its components [37].
Figure 3.
Time course study of enzyme production by Saccharomyces cerevisiae SCPW 17 grown on corn corb at 28 °C in SSF. Key: CA - cellulase activity; XA - xylanase activity; LA - ligninase activity; TP - total protein content of enzyme; U/gds - unit per gram of dry substrate; - micromole per min; mg/gds - milligram per gram of dry substrate.
3.4. Optimization of culture conditions for lignocellulase production
3.4.1. Box-Behnken design (BBD) for optimization of fermentation parameters using response surface methodology (RSM)
Response Surface Methodology (RSM) with BBD was applied to determine the maximum cellulase, xylanase and ligninase yields and to study the interactive effect of moisture to substrate ratio, initial pH, inoculum size and incubation time on enzyme production as well as to derive a statistical model for their effects. The results of the BBD experiments along with the mean observed and predicted responses for cellulase, xylanase and ligninase revealed that these data are in reasonable agreement as shown in Tables 3 and 4 respectively. The response Eq. (1) represents a suitable model for cellulase recovery while Eq. (2) represents that of xylanase recovery and Eq. (3) shows ligninase recovery.
| P = 11.100–1.737A + 0.126B + 3.632C + 2.112D + 0.641A2 + 0.297B2 + 0.974C2 - 2.495D2 + 0.832AB - 0.792AC - 0.880AD - 0.835BC - 0.285BD + 1.837CD | (1) |
| P = 41.12 + 2.15A + 0.55B + 8.84C + 12.21D + 1.52A2 - 0.39B2 - 0.29C2 - 10.40D2 - 1.69AB + 1.72AC - 1.87AD + 0.41BC + 1.12BD + 1.34CD | (2) |
| P = 360.83 + 3.01 A + 12.40 B - 4.81 C + 52.67 D - 6.89 A2 + 10.44 B2 + 6.64 C2- 88.45 D2 - 7.65 AB - 1.08 AC - 2.90 AD - 6.29 BC - 0.51 BD + 6.43 CD | (3) |
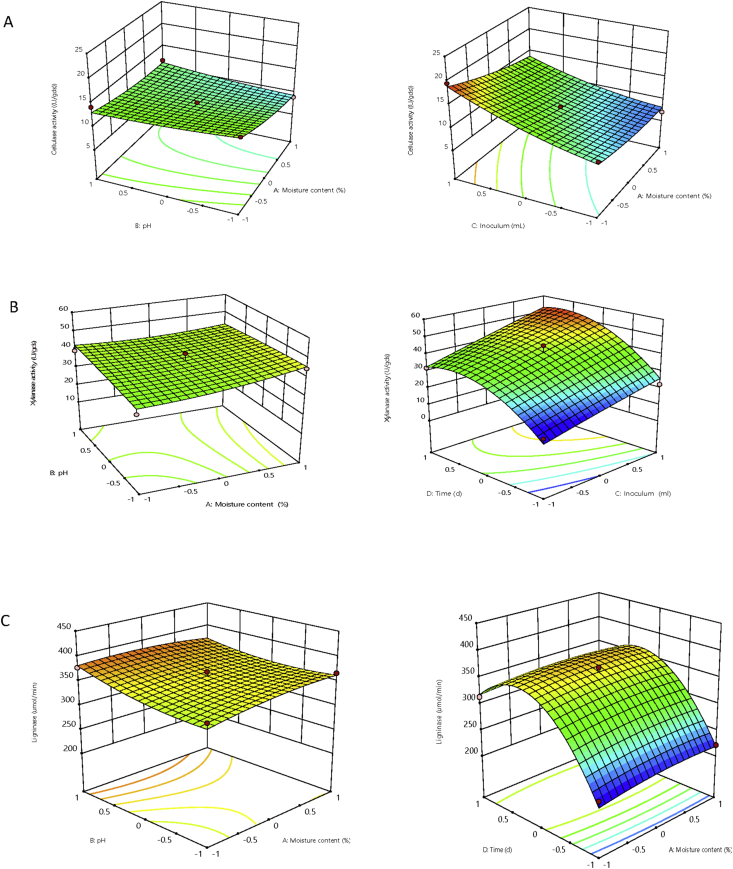
Where, P is the square root of predicted response, and A, B, C and D are the un-coded (real) values of moisture ratio, initial pH, inoculum size and incubation time respectively. From the RSM results obtained, run number 18 produced the highest cellulase (19.51 U/gds) while run 20 produced the highest xylanase (56.86 U/gds) activity which represent 40.36% and 23.29% increase from the earlier result obtained (13.90 U/gds and 46.12 U/gds) respectively. Run 15 gave the highest ligninase activity (408.172 U/gds). This shows the importance of optimizing the production parameters. Three-dimensional response surface plots were constructed from the developed models (Figure 4a, b, c) and they exhibit the individual and interactive effect of the process variables (see supplementary material, Figures 1a,b, 2a,b, 3a,b). ANOVA was used to find out the adequacy and fitness of the developed model and the results are shown in Tables 5, 6, and 7. It was found that the developed models were highly significant at probability level of P = 0.000. The Fisher's values (F-values) were calculated and found to be (18.78 for cellulase, 17.47 for xylanase and 38.03 for ligninase) with very low probability value (P = 0.000), which exhibit a high degree of adequacy of quadratic models [38]. The regression equation obtained indicated the R2 value of 0.9563 for cellulase, 0.9532 for xylanase and 0.9780 for ligninase respectively. These values have satisfactorily demonstrated that the quadratic model was highly significant and could explain about 95% of the variability in the enzyme yield. Similarly different authors have reported analysis of variance having high coefficient of determination (R2) of 0.9978–0.9070 and P < 0.05 of significant level [39, 40, 41, 42].
Figure 4.
Response surface plots of effect of different variables on enzyme production (a) cellulase (b) xylanase (c) ligninase by S. cerevisae SCPW 17.
Table 6.
Analysis of variance (ANOVA) test for Box–Behnken design Xylanase.
| Source | DF | Adj SS | Adj MS | F-Value | P-Value |
|---|---|---|---|---|---|
| Model | 14 | 3599.01 | 257.07 | 17.47 | 0.000 |
| Linear | 4 | 2785.04 | 696.26 | 47.32 | 0.000 |
| A-Moisture content | 1 | 55.47 | 55.47 | 3.77 | 0.076 |
| B-pH | 1 | 3.69 | 3.69 | 0.25 | 0.626 |
| C-Inoculum | 1 | 938.10 | 938.10 | 63.75 | 0.000 |
| D-Time | 1 | 1787.79 | 1787.79 | 121.50 | 0.000 |
| Square | 4 | 763.99 | 191.00 | 12.98 | 0.000 |
| A∗A | 1 | 12.32 | 12.32 | 0.84 | 0.378 |
| B∗B | 1 | 0.80 | 0.80 | 0.05 | 0.820 |
| C∗C | 1 | 0.46 | 0.46 | 0.03 | 0.863 |
| D∗D | 1 | 576.99 | 576.99 | 39.21 | 0.000 |
| 2-Way Interaction | 6 | 49.98 | 8.33 | 0.57 | 0.750 |
| A∗B | 1 | 11.46 | 11.46 | 0.78 | 0.395 |
| A∗C | 1 | 11.76 | 11.76 | 0.80 | 0.389 |
| A∗D | 1 | 13.95 | 13.95 | 0.95 | 0.349 |
| B∗C | 1 | 0.66 | 0.66 | 0.04 | 0.836 |
| B∗D | 1 | 5.02 | 5.02 | 0.34 | 0.570 |
| C∗D | 1 | 7.13 | 7.13 | 0.48 | 0.500 |
| Error | 12 | 176.58 | 14.71 | ||
| Lack-of-Fit | 10 | 154.11 | 15.41 | 1.37 | 0.494 |
| Pure Error | 2 | 22.47 | 11.23 | ||
| Total | 26 | 3775.59 |
| Model Summary | |||
|---|---|---|---|
| S | R-Sq | R-Sq (adj) | R-Sq (pred) |
| 3.83597 | 92.32% | 89.87% | 75.15% |
Table 7.
Analysis of variance (ANOVA) test for Box–Behnken design Ligninase.
| Source | DF | Adj SS | Adj SS | F-Value | P-Value |
|---|---|---|---|---|---|
| Model | 14 | 92146.7 | 6581.9 | 38.03 | 0.000 |
| Linear | 4 | 35520.3 | 8880.1 | 51.31 | 0.000 |
| A-Moisture content | 1 | 108.5 | 108.5 | 0.63 | 0.444 |
| B-pH | 1 | 1844.2 | 1844.2 | 10.66 | 0.007 |
| C-Inoculum | 1 | 277.8 | 277.8 | 1.61 | 0.229 |
| D-Time | 1 | 33289.9 | 33289.9 | 192.34 | 0.000 |
| Square | 4 | 56029.6 | 14007.4 | 80.93 | 0.000 |
| A∗A | 1 | 253.0 | 253.0 | 1.46 | 0.250 |
| B∗B | 1 | 581.0 | 581.0 | 3.36 | 0.092 |
| C∗C | 1 | 235.2 | 235.2 | 1.36 | 0.266 |
| D∗D | 1 | 41728.6 | 41728.6 | 241.10 | 0.000 |
| 2 Way Interaction | 6 | 596.7 | 99.5 | 0.57 | 0.744 |
| A∗B | 1 | 234.0 | 234.0 | 1.35 | 0.268 |
| A∗C | 1 | 4.6 | 4.6 | 0.03 | 0.873 |
| A∗D | 1 | 33.7 | 33.7 | 0.19 | 0.667 |
| B∗C | 1 | 158.3 | 158.3 | 0.91 | 0.358 |
| B∗D | 1 | 1.0 | 1.0 | 0.01 | 0.939 |
| C∗D | 1 | 165.1 | 165.1 | 0.95 | 0.348 |
| Error | 12 | 2076.9 | 173.1 | ||
| Lack-of-Fit | 10 | 1951.8 | 195.2 | 3.12 | 0.267 |
| Pure Error | 2 | 125.1 | 62.6 | ||
| Total | 26 | 94223.6 |
| Model Summary | |||
|---|---|---|---|
| S | R-Sq | R-Sq (adj) | R-Sq (pred) |
| 13.1559 | 97.80% | 95.22% | 87.77% |
3.5. Characterization of enzyme complex
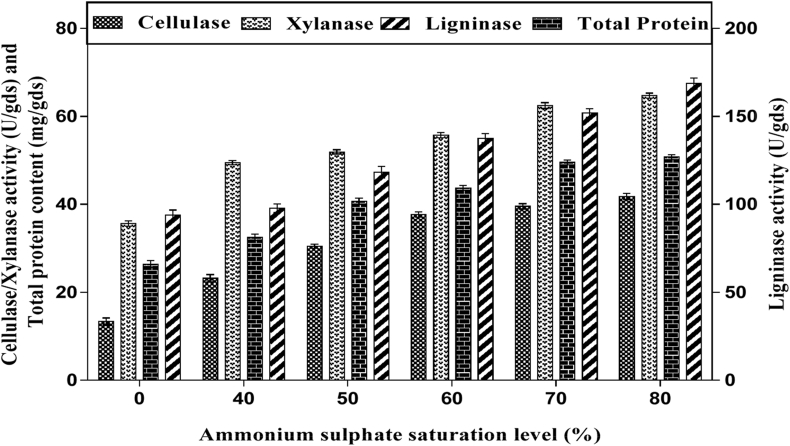
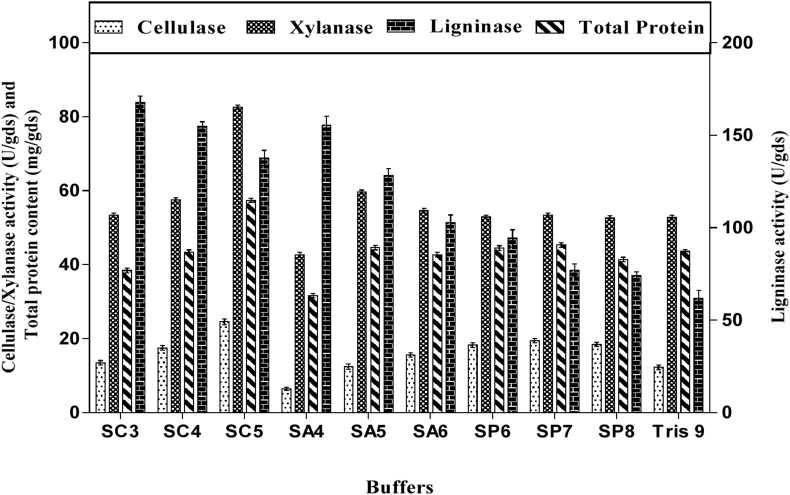
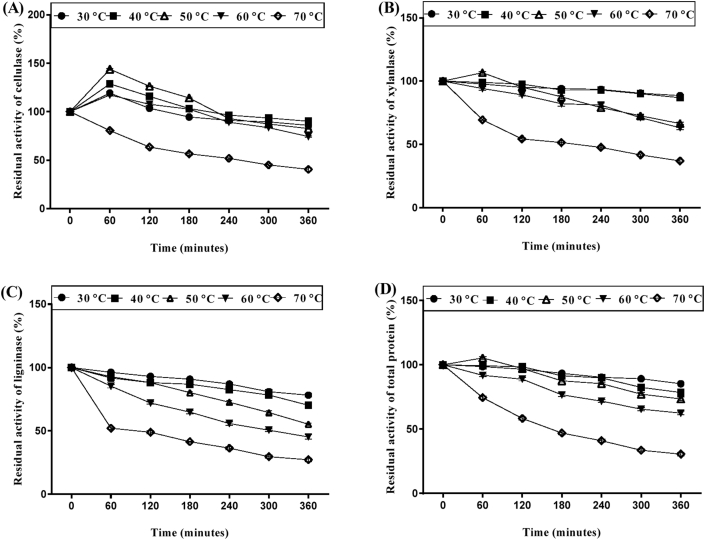
There are several reports on purification and characterization of enzymes from various microbial sources [43, 44] but for the first time we report the purification and characterization of culture filtrate with multienzyme complex from solid state fermentation of LC by Saccharomyces cerevisiae SCPW 17. The crude filtrate containing the enzyme complex (cellulase, xylanase and ligninase) was subjected to different ammonium sulphate concentrations (40, 50, 60, 70 and 80%) for precipitation (Figure 5). It was observed that enzyme activity increased with increase in (NH4)2SO4 concentration, hence 80% (NH4)2SO4 concentration showed better performance for the enzyme complex precipitation. The pH activity and stability of the partially purified enzyme complex was determined by measuring the enzyme activity at varying pH values ranging from 3–9 using different suitable buffers. It was observed that maximum cellulase and xylanase activity was established at pH 5.0, sodium citrate buffer, while ligninase was at pH 3.0 sodium citrate buffer (Figure 6). Stability of the enzyme is a very important factor in studying characteristics, effect of temperature on partially purified enzyme activity was recorded over a broad range of temperature (30–70 °C). In case of partially purified cellulase and xylanase were stable at temperatures 30 °C–60 °C for 4 h of incubation and retained over 80% activity. At higher temperature values enzyme stability was gradually declined (Figure 7). However with ligninase stability was maintained for a period of 4h at temperature range of 30–50 and 80% of the activity was retained. At 60 and 70 °C degrees enzyme stability gradually declined.
Figure 5.
Cellulase, xylanase, ligninase activity and total protein at different ammonium sulphate saturation levels, data is presented as mean ± SD of the three replicates.
Figure 6.
Effect of different buffers on partially purified cellulase, xylanase, ligninase from Saccharomyces cerevisiae SCPW 17, data is presented as mean ± SD of the three replicates. Keys: SC3-sodium citrate pH3; SC4-sodium citrate pH4; SC5-sodium citrate pH5; SA4-sodium acetate pH4; SA5-sodium acetate pH5; SA6-sodium acetate pH6; SP6- sodium phosphate pH6; SP7- sodium phosphate pH7; SP8- sodium phosphate pH8; Tris 9- tris buffer pH9.
Figure 7.
Thermostability of partially purified (A) cellulase, (B) xylanase (C) ligninase (D) total protein from Saccharomyces cerevisiae SCPW 17, data is presented as mean ± SD of the three replicates.
4. Conclusion
The continuous search for novel microbial strains capable of enzyme complex production for various industrial application is on the rise. Many studies in this quest to produce multi enzyme complex that can be used for hydrolysis of waste substrates such as lignocellulosic materials have been studied and reported. Lignocellulose waste materials are abundant and transforming them into useful products will be an added advantage to the economy. None of the report has so far produced microorganisms capable of producing combined cellulase, xylanase and ligninase activity. The yeast strain Saccharomyces cerevisiae SCPW 17 studied demonstrated the potential to simultaneously produce cellulase, xylanase and ligninase using low cost renewable agricultural waste (corn cob) by solid state fermentation, without any pre-treatment of substrate, this is important as pre-treatment would incur additional costs in industrial processes. Box benkan, Response surface methodology (RSM) employed for the improvement of cellulase, xylanase and ligninase production was successful and was found to be an appropriate method for optimizing the fermentation parameters to obtain maximum enzyme activity. The stability of the enzyme complex under a wide range of pH and at varied temperature also showed the potential of Saccharomyces cerevisiae SCPW 17 for applications in various biotechnology industries.
Declarations
Author contribution statement
Onyetugo C. Amadi, Egong J. Egong: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Tochukwu N. Nwagu, Gloria Okpala, Chukwudi O. Onwosi, Greg C. Chukwu, Bartholomew N. Okolo, Reginald C. Agu, Anene N. Moneke: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflicts of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Malgas S., Thoresen M., van Dyk J.S., Pletschke B.I. Time dependence of enzyme synergism during the degradation of model and natural lignocellulosic substrates. Enzym. Microb. Technol. 2017;103:1–11. doi: 10.1016/j.enzmictec.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Binod P., Sindhu R., Singhania R.R., Vikram S., Devi L., Nagalakshmi S., Kurien N., Sukumaran R.K., Pandey A. Bioethanol production from rice straw: an overview. Bioresour. Technol. 2010;101:4767–4774. doi: 10.1016/j.biortech.2009.10.079. [DOI] [PubMed] [Google Scholar]
- 3.van Dyk J.S., Sakka M., Sakka K., Pletschke B.I. Characterisation of the multi-enzyme complex xylanase activity from Bacillus licheniformis SVD1. J. Enzyme and Microb. Technol. 2010;47:174–177. [Google Scholar]
- 4.Sarkar N., Ghosh S.K., Bannerjee S., Aikat K. Bioethanol production from agricultural wastes: an overview. Renew. Energy. 2012;37:19–27. [Google Scholar]
- 5.Sarrouh B., Santos T.M., Miyoshi A., Dias R., Azevedo V. Up-to-date insight on industrial enzymes applications and global market. J. Bioprocess. Biotech. 2012;s1:1–10. [Google Scholar]
- 6.Novozymes . 2017. Enzymes at Work; p. 76.https://www.novozymes.com/en/-/media/Novozymes/en/aboutus/brochures/Documents/Enzymes_at_work.pdf+&cd= 1&hl=pt-BR&ct=clnk&gl=br 76. Available at. [Google Scholar]
- 7.BBC Research . BBC Research; 2017. Global Markets for Enzymes in Industrial Application - BIO030J. Wellesley, MA.https://www.bccresearch.com/market-research/biotechnology/enzymes-industrial-applications-reportbio030j.html Available at. [Google Scholar]
- 8.Howard R.L., Abotsi E., Jansen van Rensburg E.L., Howard S. Lignocellulose biotechnology: issues of bioconversion and enzyme production. Afr. J. Biotechnol. 2003;2:602–619. [Google Scholar]
- 9.Okolo B.N., Agu R.C. Use of waste materials from plant origin (yam) as a major raw material for bio-ethanol production – a practical approach. J. Solid Waste Technol. Manag. 2013;39(2):150–156. [Google Scholar]
- 10.Amadi O.C., Onyema N., Nwagu T.N., Moneke A.N., Okolo B.N., Agu R.C. Evaluating the potential of wild cocoyam “caladium bicolor” for ethanol production using indigenous fungal isolates. Procedia Environ. Sci. 2016;35:809–817. [Google Scholar]
- 11.Petrescu I., Lamotte-Brasseur J., Chessa J.P., Ntarima P., Claeyssens M., Devreese B., Gerday C. Xylanase from the psychrophilic yeast Cryptococcus adeliae. Extremophiles. 2000;4(3):137–144. doi: 10.1007/s007920070028. [DOI] [PubMed] [Google Scholar]
- 12.Parachin N.S., Siqueira S., de Faria F.P., Torres F.A.G., de Moraes L.M.P. Xylanases from Cryptococcus flavus isolate I-11: enzymatic profile, isolation and heterologous expression of CfXYN1 in Saccharomyces cerevisiae. J. Mol. Catal. B Enzym. 2009;59(1):52–57. [Google Scholar]
- 13.Otero D.M., Cadaval C.L., Teixeira L.M., Rosa C.A., Sanzo A.V.L., Kalil S.J. Screening of yeasts capable of producing cellulase-free xylanase. Afr. J. Biotechnol. 2015;14(23):1961–1969. [Google Scholar]
- 14.Gomes F.C.O., Safar S.V.B., Marques A.R., Medeiros A.O., Santos A.R.O., Carvalho C., Lachance M., Sampaio J.P., Rosa C.A. The diversity and extracellular enzymatic activities of yeasts isolated from water tanks of Vriesea minarum, an endangered bromeliad species in Brazil, and the description of Occultifur brasiliensis f.a., sp. nov. Antonie Leeuwenhoek. 2015;107:597–611. doi: 10.1007/s10482-014-0356-4. [DOI] [PubMed] [Google Scholar]
- 15.Thongekkaew J., Patangtasa W., Songklanakarin A.J. Cellulase and xylanase production from Candida easanensis using agricultural wastes as a substrate. J. Sci. Technol. 2014;36(6):607–613. [Google Scholar]
- 16.Qadir F., Shariq M., Ahmed A., Sohail M. Evaluation of a yeast co-culture for cellulase and xylanase production under solid state fermentation of sugarcane bagasse using multivariate approach. Ind. Crop. Prod. 2018;123:407–415. [Google Scholar]
- 17.Pal A., Khanum F. Production and extraction optimization of xylanase from Aspergillus niger DFR-5 through solid-state-fermentation. Bioresour. Technol. 2010;101(19):7563–7569. doi: 10.1016/j.biortech.2010.04.033. [DOI] [PubMed] [Google Scholar]
- 18.Kumar S.S., Haridas M., Sabu A. Process optimization for production of a fibrinolytic enzyme from newly isolated marine bacterium Pseudomonas aeruginosa KU1. Biocat. Agric. Biotechnol. 2018;14:33–39. [Google Scholar]
- 19.Limkar M.B., Pawar S.V., Rathod V.K. Statistical optimization of xylanase and alkaline protease co-production by Bacillus spp using Box-Behnken Design under submerged fermentation using wheat bran as a substrate. Biocatal. Agric. Biotech. 2019;17:455–464. [Google Scholar]
- 20.Neethu K., Rubeena M., Sajith S., Sreedevi S., Priji P., Unni K.N., Benjamin S. A novel strain of Trichoderma viride shows complete lignocellulolytic activities. Adv. Biosci. Biotechnol. 2012;3(8):1160. [Google Scholar]
- 21.Srilakshmi A., Dakshayani R., Saigopal D.V.R., Narasimha G. Influence of forest litter on soil enzyme activities. Ecol. Environ. Conserv. 2012;18:105–112. [Google Scholar]
- 22.Pointing S.B. Qualitative methods for the determination of lignocellulolytic enzyme production by tropical fungi. Fungal Divers. 1999;2:17–33. [Google Scholar]
- 23.Miller G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959;31(3):426–428. [Google Scholar]
- 24.Bansal N., Tewari R., Soni R., Soni S.K. Production of cellulases from Aspergillus niger NS-2 in solid state fermentation on agricultural and kitchen waste residues. Waste Manage. 2012;32(7):1341–1346. doi: 10.1016/j.wasman.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Tien M., Kirk T.K. Lignin-degrading enzyme from Phanerochaete chrysosporium: purification, characterization, and catalytic properties of a unique H2O2- requiring oxygenase. Proc. Natl. Acad. Sci. Unit. States Am. 1984;81:2280–2284. doi: 10.1073/pnas.81.8.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailey M.J., Biely P., Poutanen K. Inter laboratory testing of methods for assay of xylanase activity. J. Biotechnol. 1992;23(3):257–270. [Google Scholar]
- 27.Jagannathan V., Singh K., Damodaran M. Carbohydrate metabolism in citric acid fermentation. Purification and properties of aldolase from Aspergillus niger. Biochem. J. 1996;63(1):94–105. doi: 10.1042/bj0630094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 29.DeWitt T.J., Sih A., Wilson D.S. Costs and limits of phenotypic plasticity. Trends Ecol. Evol. 1998;13:77–81. doi: 10.1016/s0169-5347(97)01274-3. [DOI] [PubMed] [Google Scholar]
- 30.Sterns S. The evolutionary significance of phenotypic plasticity. Bioscience. 1989;39:436–445. [Google Scholar]
- 31.Miner B.G., Sultan S.E., Morgan S.G., Padilla D.K., Relyea R.A. Ecological consequences of phenotypic plasticity. Trends Ecol. Evol. 2005;20:685–692. doi: 10.1016/j.tree.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Andersen B., Poulsen R., Hansen G.H. Cellulolytic and xylanolytic activities of common indoor fungi. Int. Biodeterior. Biodegrad. 2016;107:111–116. [Google Scholar]
- 33.Do Vale L.H.F., Filho E.X.F., Miller R.N.G., Ricart C.A.O., de Sousa M.V. Cellulase systems in Trichoderma: an overview. Biotechnology and Biology of Biotechnol. Biol Trichoderma. 2014:229–245. Elsievier. [Google Scholar]
- 34.Gilbert H.J., Hazlewood G.P. Bacterial cellulases and xylanases. J. Gen. Microbiol. 1993;139:187–194. [Google Scholar]
- 35.Mahalakshmi N., Jayalakshmi S. Amylase, Cellulase and Xylanase production from a novel bacterial isolate Achromobacter xylosoxidans isolated from marine environment. Int. J. Adv Res Biolog. Sci. 2016;3:230–233. [Google Scholar]
- 36.Ghoshal G., Banerjee U.C., Shivhare U.S. Utilization of agrowaste and xylanase production in solid state fermentation. J. Biochem. Technol. 2015;6:1013–1024. [Google Scholar]
- 37.Rao R.S., Bhadra B., Shivaji S. Isolation and characterization of ethanolproducing yeasts from fruits and tree barks. Lett. Appl. Microbiol. 2008;47:9–24. doi: 10.1111/j.1472-765X.2008.02380.x. [DOI] [PubMed] [Google Scholar]
- 38.Maran J.P., Priya B. Ultrasound-assisted extraction of pectin from sisal waste. Carbohydr. Polym. 2015;115:732e738. doi: 10.1016/j.carbpol.2014.07.058. [DOI] [PubMed] [Google Scholar]
- 39.Coman G., Bahrim G. Optimization of xylanase production by Streptomyces sp P12- 137 using response surface methodology and central composite design. Ann. Microbiol. 2011;61:773–779. doi: 10.1007/s13213-010-0195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khusro A., Kaliyan B.K., Al-Dhabi N.A., ValanArasu M., Agastian P. Statistical optimization of thermo-alkali stable xylanase production from Bacillus tequilensis strain ARMATI. Electron. J. Biotechnol. 2016;22:16–25. [Google Scholar]
- 41.Perince S., Duran K. Optimization of enzymatic & ultrasonic bio-scouring of linen fabrics by aid of Box-Behnken Experimental Design. J. Clean. Prod. 2016;135:1179–1188. [Google Scholar]
- 42.Sanjivkumar M., Silambarasan T., Balagurunathan R., Immanuel G. Biosynthesis, molecular modeling and statistical optimization of xylanase from a mangrove associated actinobacterium Streptomyces variabilis (MAB3) using Box- Behnken design with its bioconversion efficacy. Int. J. Biol. Macromol. 2018;118:195–208. doi: 10.1016/j.ijbiomac.2018.06.063. [DOI] [PubMed] [Google Scholar]
- 43.Kamble R.D., Jadhav A.R. Isolation, purification, and characterization of xylanase produced by a new species of Bacillus in solid state fermentation. Int. J. Microbiol. 2012 doi: 10.1155/2012/683193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaur R., Tiwari S. Isolation, production, purification and characterization of an organic-solvent-thermostable alkalophilic cellulase from Bacillus vallismortis RG-07. BMC Biotechnol. 2015;15:19. doi: 10.1186/s12896-015-0129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.