HIV-1 Env protein is exposed to the inhibition not only by humoral response, but also by host restriction factors, including serine incorporator 5 (SERINC5) and interferon-inducible transmembrane 3 (IFITM3). This study investigates how HIV-1 envelope glycoprotein (Env) manages to overcome the pressures from all these different host inhibition mechanisms over the long course of viral infection. HIV-1 Env preserves the resistance to SERINC5 but becomes sensitive to IFITM3 when infection progresses into the chronic stage. Our study also supports the possibility of using CD4 mimetic compounds to sensitize HIV-1 Env to the inhibition by SERINC5 as a potential therapeutic strategy.
KEYWORDS: HIV, IFITM3, SERINC5, envelope glycoprotein
ABSTRACT
Infection of human immunodeficiency virus type 1 (HIV-1) is subject to restriction by cellular factors. Serine incorporator 5 (SERINC5) and interferon-inducible transmembrane 3 (IFITM3) proteins represent two of these restriction factors, which inhibit HIV-1 entry into target cells. Both proteins impede fusion of the viral membrane with the cellular membrane and the formation of a viral fusion pore, and both are countered by the HIV-1 envelope glycoprotein (Env). Given the immense and lasting pressure which Env endures from host adaptive immune responses, it is important to understand whether and how HIV-1 Env is able to maintain the resistance to SERINC5 and IFITM3 throughout the course of infection. We have thus examined a panel of HIV-1 Env clones that were isolated at different stages of viral infection—transmission, acute, and chronic. While HIV-1 Env clones from the transmission stage are resistant to both SERINC5 and IFITM3, as infection progresses into the acute and chronic stages, the resistance to IFITM3 but not to SERINC5 is gradually lost. We further discovered a significant correlation between the resistance of HIV-1 Env to soluble CD4 inhibition and the resistance to SERINC5 but not to IFITM3. Interestingly, the miniprotein CD4 mimetic M48U1 sensitizes HIV-1 Env to the inhibition by SERINC5 but not IFITM3. Together, these data indicate that SERINC5 and IFITM3 exert differential inhibitory pressures on HIV-1 Env over different stages of HIV-1 infection and that HIV-1 Env uses varied strategies to resist these two restriction factors.
IMPORTANCE HIV-1 Env protein is exposed to the inhibition not only by humoral response, but also by host restriction factors, including serine incorporator 5 (SERINC5) and interferon-inducible transmembrane 3 (IFITM3). This study investigates how HIV-1 envelope glycoprotein (Env) manages to overcome the pressures from all these different host inhibition mechanisms over the long course of viral infection. HIV-1 Env preserves the resistance to SERINC5 but becomes sensitive to IFITM3 when infection progresses into the chronic stage. Our study also supports the possibility of using CD4 mimetic compounds to sensitize HIV-1 Env to the inhibition by SERINC5 as a potential therapeutic strategy.
INTRODUCTION
The HIV-1 envelope glycoprotein (Env) is not only under the selection pressure of adaptive immunity; it is also the target of innate immunity. A group of cellular factors, often interferon-induced, have been reported to inhibit HIV-1 Env-mediated virus entry (1). These include interferon-induced transmembrane (IFITM) proteins (2, 3), the 90K protein (4–6), serine incorporator 5 (SERINC5) (7, 8), membrane-associated RING-CH (MARCH) proteins (9–11), endoplasmic reticulum class 1 α-mannosidase (ERManI) (12), and guanylate binding protein 5 (GBP5) (13–15). Among these HIV-1 Env inhibitors, inhibition by IFITM and SERINC5 proteins has been shown to be overcome by Env mutations (16–21).
SERINC5 was originally discovered as the cellular restriction factor that is antagonized by HIV-1 Nef accessory protein (7, 8). In the absence of Nef, SERINC5 is incorporated into HIV-1 particles and impairs HIV-1 infectivity by inhibiting the expansion of a viral fusion pore (7, 8, 21). The ability of Nef to antagonize SERINC5 appears to be important for HIV-1 pathogenesis, since this ability of Nef is lost or severely attenuated in viruses from elite controllers (22). Nef is not the only mechanism used by HIV-1 to counter SERINC5. Our group and others have found that HIV-1 Env is able to resist SERINC5 restriction (20, 21). The V3 loop of Env has been further identified as one determinant of this function of Env (20). Similarly, IFITM3 can also inhibit HIV-1 entry by impairing the hemifusion of the viral membrane and the formation of a viral fusion pore (3, 23, 24). One difference is that IFITM3 is able to exert its inhibition either in the target cells or in the virus particles, while SERINC5 only inhibits when present in HIV-1 particles (7, 8, 25). Nonetheless, IFITM3 is also countered by HIV-1 Env, and the Env determinant of this resistance was mapped to the V3 loop (19).
In spite of these similarities between IFITM3 and SERINC5 in their anti-HIV-1 activity and HIV-1 countering mechanisms, these proteins are structurally and functionally very different. SERINC5 has 10 transmembrane domains, located on cellular membrane and implicated in lipid modification (26, 27), whereas IFITM3 has only 132 amino acids, with one transmembrane domain and an intramembrane domain and predominantly located in late endosomes (28, 29). It remains unclear how IFITM3 and SERINC5, once incorporated into HIV-1 particles, act on viral Env and whether Env reacts differently to resist these two inhibitors. It has been reported that transmitted founder (T/F) HIV-1 strains resist IFITM3 inhibition, but this resistance diminishes with the progression of HIV-1 infection as a result of the need of HIV-1 Env to change and escape from the inhibition by neutralizing antibodies (18). However, it is not entirely known how the susceptibility of HIV-1 Env to SERINC5 inhibition changes over the course of HIV-1 infection.
To answer these questions, we have examined a panel of primary HIV-1 Env clones for their susceptibility to IFITM3 and SERINC5 inhibition. These Env clones were derived from either T/F HIV-1 strains, acute or chronic infections. While HIV-1 Env becomes more sensitive to IFITM3 inhibition as the infection progresses to the chronic stage, the Env clones of all stages of infection are resistant to SERINC5 restriction.
RESULTS
HIV-1 Env clones of both acute and chronic infections manifest resistance to SERINC5 inhibition.
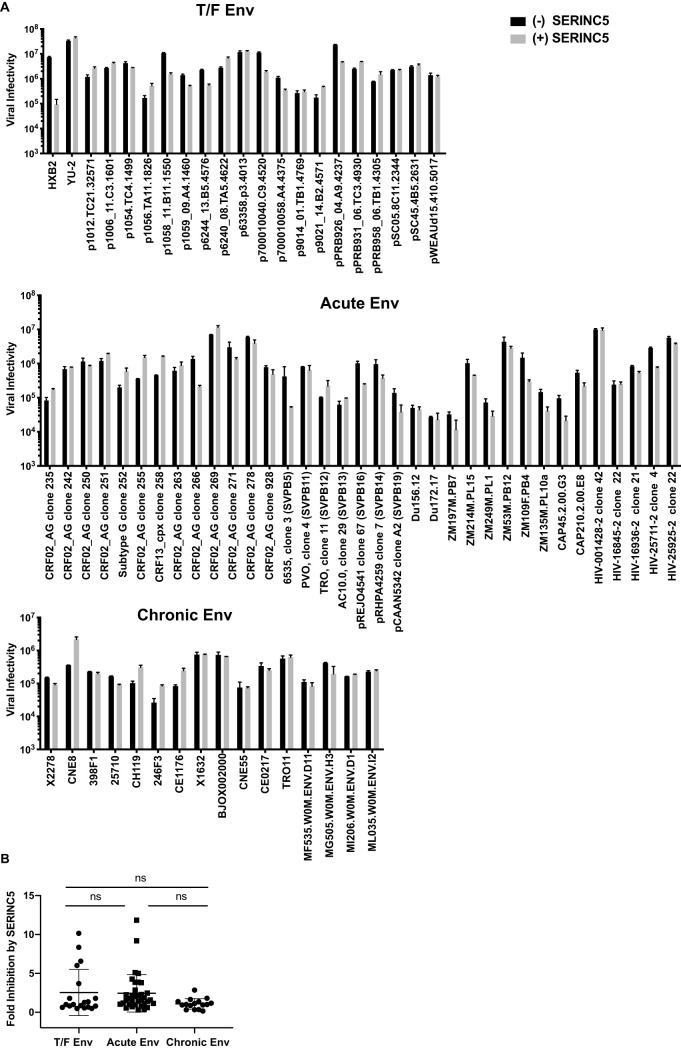
We asked to what extent primary HIV-1 Env resists the inhibition by IFITM3 and SERINC5 and whether the level of resistance persists with the progression of HIV-1 infection. To answer these questions, we examined 70 HIV-1 Env clones for inhibition by IFITM3 and by SERINC5. Among these Env clones, 19 isolates were derived from T/F HIV-1, 35 from acute infections, and 16 from chronic infection. We used the Nef-deleted and Env-deleted HIV-1 clone (NL4-3 ΔNefΔEnv) to produce virus particles that were pseudotyped with these primary Env proteins by cotransfecting HEK293T cells. IFITM3 or SERINC5 DNA was included in the cotransfection experiments to test their inhibition of the pseudotyped HIV-1 particles. The Nef-deleted HIV-1 was used in order to accurately measure the susceptibility of Env clones to SERINC5 and IFITM3 inhibition without the interference of Nef, which is able to antagonize SERINC5. The results shown in Fig. 1A showed that the lab-adapted HXB2 Env-mediated infection was inhibited by SERINC5 by up to 60-fold, whereas the primary YU-2 Env was resistant; this is in agreement with previous publications (20, 21).
FIG 1.
Inhibition of HIV-1 Env clones by SERINC5. HEK293T cells were cotransfected with NL4-3 ΔNefΔEnv proviral DNA, different HIV-1 Env clones, and 100 ng of SERINC5 cDNA. 48 h after transfection, pseudotyped HIV-1 was used to infect TZM-bl cells. (A) Luciferase data of one representative experiment are presented. (B) Fold inhibition by SERINC5 was determined by calculating the ratio of infectivity of the SERINC5-free viruses to those of the SERINC5-bearing viruses. Fold inhibition by SERINC5 for each Env clone from three independent transfections is shown. The data of all Env clones are presented. The mean value of fold inhibition for each Env group is indicated by the whisker. The standard deviations are also presented. ns, not significant.
We then tested the primary Env clones (Fig. 1A) and summarized their folds of inhibition by SERINC5 in Fig. 1B and Tables 1 to 3. The mean fold of inhibition was 2.5 for T/F Env clones, 2.6 for acute Env clones, and 1.8 for chronic Env clones, which are not statistically different from each other and show similar levels of resistance to SERINC5 as the YU-2 Env. Out of the 19 T/F Env clones, 4 showed more than 5-fold inhibition, 2 out of the 35 acute clones were inhibited by more than 5-fold, and none of the 16 chronic Envs were inhibited by more than 5-fold. Therefore, resistance to SERINC5 is preserved by most of the HIV-1 Env clones over the course of infection, from transmission until the chronic stage.
TABLE 1.
Response of T/F Env clones to the inhibition by SERINC5, IFITM3, CD4 MAb (RPA-T4), sCD4, and CCR5 antagonist TAK-779
| T/F Enva | Subtype | Fold inhibition by SERINC5b | Fold inhibition by IFITM3c | IC50 values for:d,e |
||
|---|---|---|---|---|---|---|
| CD4 MAb (μg/ml) | sCD4 (nM) | TAK-779 (μM) | ||||
| p1012.TC21.32571 | B | 0.48 ± 0.01 | 8.09 ± 0.23 | 0.09 | 331 | 0.15 |
| p1006_11.C3.1601 | B | 0.80 ± 0.20 | 7.01 ± 0.80 | 0.05 | 241 | 0.06 |
| p1054.TC4.1499 | B | 1.8 ± 0.53 | 3.21 ± 0.02 | 0.11 | 113 | 0.05 |
| p1056.TA11.1826 | B | 0.6 ± 0.42 | 3.78 ± 0.70 | 0.06 | 635 | 0.4 |
| p1058_11.B11.1550* | B | 10.14 ± 1.30 | 9.22 ± 1.30 | 0.13 | 298 | 0.02 |
| p1059_09.A4.1460 | B | 3.7 ± 0.09 | 4.05 ± 0.022 | 0.13 | 201 | 0.09 |
| p6244_13.B5.4576 | B | 6.0 ± 0.06 | 5.4 ± 1.04 | 0.05 | 254 | 0.09 |
| p6240_08.TA5.4622 | B | 0.56 ± 0.01 | 2.2 ± 0.04 | 0.06 | 478 | 0.14 |
| p63358.p3.4013 | B | 1.25 ± 0.002 | 2.6 ± 0.04 | 0.05 | 538 | 0.03 |
| p700010040.C9.4520 | B | 8.34 ± 0.027 | 1.53 ± 0.01 | 0.17 | 97 | 0.03 |
| p700010058.A4.4375 | B | 0.85 ± 0.02 | 3.20 ± 0.37 | 0.06 | 413 | 0.3 |
| p9014_01.TB1.4769 | B | 0.99 ± 0.32 | 4.06 ± 0.35 | 0.06 | >1,000 | 0.54 |
| p9021_14.B2.4571 | B | 0.48 ± 0.20 | 1.82 ± 0.28 | 0.05 | 378 | 0.04 |
| pPRB926_04.A9.4237 | B | 6.56 ± 0.77 | 1.13 ± 0.04 | 0.18 | 93 | 0.03 |
| pPRB931_06.TC3.4930 | B | 0.67 ± 0.15 | 1.65 ± 0.16 | 0.12 | >1,000 | 0.16 |
| pPRB958_06.TB1.4305 | B | 0.81 ± 0.31 | 3.57 ± 0.02 | 0.1 | 141 | 0.12 |
| pSC05.8C11.2344 | B | 1.35 ± 0.33 | 1.14 ± 0.16 | ND | ND | ND |
| pSC45.4B5.2631 | B | 0.99 ± 0.10 | 2.96 ± 0.37 | 0.09 | 268 | 0.28 |
| pWEAUd15.410.5017* | B | 1.8 ± 0.06 | 2.40 ± 0.26 | ND | ND | ND |
All the Env clones are R5 tropic except the two clones marked by *, which are dual tropic.
Fold inhibition by SERINC5 was determined by calculating the ratio of infectivity of the SERINC5-free virus to that of the SERINC5-bearing virus.
Fold inhibition by IFITM3 was determined by calculating the ratio of infectivity of the IFITM3-free virus to that of the IFITM3-bearing virus.
The IC50 values were obtained from the study by Keele et al. (30).
ND, not determined.
TABLE 2.
Response of acute Env clones to the inhibition by SERINC5 and IFITM3
| Acute Enva | Subtype | Fold inhibition by SERINC5b | Fold inhibition by IFITM3c |
|---|---|---|---|
| CRF02_AG clone 235 | A/G | 1.13 ± 0.55 | 1.57 ± 0.25 |
| CRF02_AG clone 242 | A/G | 2.08 ± 0.64 | 6.83 ± 1.19 |
| CRF02_AG clone 250 | A/G | 9.18 ± 0.32 | 4.75 ± 1.54 |
| CRF02_AG clone 251 | A/G | 0.54 ± 0.11 | 2.28 ± 0.51 |
| Subtype G clone 252 | A/G | 0.41 ± 0.09 | 5.8 ± 0.66 |
| CRF02_AG clone 255 | A/G | 1.74 ± 0.56 | 3.41 ± 0.88 |
| CRF13_cpx clone 258 | A/G | 0.77 ± 0.11 | 1.7 ± 0.46 |
| CRF02_AG clone 263 | A/G | 2.24 ± 0.66 | 8.06 ± 2.49 |
| CRF02_AG clone 266 | A/G | 1.43 ± 0.24 | 4.57 ± 0.06 |
| CRF02_AG clone 269 | A/G | 2.06 ± 0.60 | 2.78 ± 0.19 |
| CRF02_AG clone 271 | A/G | 1.11 ± 0.18 | 11.61 ± 0.75 |
| CRF02_AG clone 278 | A/G | 1.37 ± 0.40 | 14.7 ± 1.84 |
| CRF02_AG clone 928 | A/G | 1.91 ± 0.07 | 10.67 ± 0.002 |
| 6535, clone 3 (SVPB5) | B | 11.83 ± 1.98 | 4.69 ± 0.54 |
| PVO, clone 4 (SVPB11) | B | 0.96 ± 0-0.04 | 1.98 ± 0.13 |
| TRO, clone 11 (SVP. B12) | B | 0.37 ± 0.03 | 5.66 ± 0.63 |
| AC10.0, clone 29 (SVPB13) | B | 0.60 ± 0.15 | 6.50 ± 1.51 |
| pREJO4541 clone 67 (SVPB16) | B | 3.86 ± 0.56 | 4.82 ± 0.67 |
| pRHPA4259 clone 7 (SVPB14) | B | 2.177 ± 0.49 | 15.56 ± 1.0 |
| pCAAN5342 clone A2 (SVPB19) | B | 0.71 ± 0.018 | 3.98 ± 0.40 |
| Du156.12 | C | 1.17 ± 0.33 | 1.26 ± 0.20 |
| Du172.17 | C | 1.40 ± 0.55 | 1.05 ± 0.005 |
| ZM197M.PB7 | C | 4.26 ± 2.33 | 0.98 ± 0.09 |
| ZM214M.PL15 | C | 2.28 ± 0.50 | 2.97 ± 0.39 |
| ZM249M.PL1 | C | 2.84 ± 1.30 | 1.21 ± 0.03 |
| ZM53M.PB12 | C | 1.58 ± 0.54 | 1.22 ± 0.14 |
| ZM109F.PB4 | C | 5.10 ± 1.71 | 12.30 ± 0.63 |
| ZM135M.PL10a | C | 3.87 ± 1.40 | 4.53 ± 0.64 |
| CAP45.2.00.G3 | C | 5.00 ± 1.92 | 1.55 ± 0.17 |
| CAP210.2.00.E8 | C | 2.46 ± 0.10 | 5.85 ± 0.14 |
| HIV-001428-2 clone 42 | C | 1.04 ± 0.06 | 8.19 ± 0.52 |
| HIV-16845-2 clone 22 | C | 0.95 ± 0.08 | 9.13 ± 0.39 |
| HIV-16936-2 clone 21 | C | 1.57 ± 0.15 | 10.96 ± 0.70 |
| HIV-25711-2 clone 4 | C | 3.80 ± 0.27 | 7.81 ± 0.04 |
| HIV-25925-2 clone 22 | C | 1.52 ± 0.03 | 15.64 ± 4.07 |
All the Env clones are R5 tropic.
Fold inhibition by SERINC5 was determined by calculating the ratio of infectivity of the SERINC5-free virus to that of the SERINC5-bearing virus.
Fold inhibition by IFITM3 was determined by calculating the ratio of infectivity of the IFITM3-free virus to that of the IFITM3-bearing virus.
TABLE 3.
Response of chronic Env clones to the inhibition by SERINC5 and IFITM3
| Chronic Enva | Subtype | Fold inhibition by SERINC5b | Fold inhibition by IFITM3c |
|---|---|---|---|
| X2278 | B | 1.64 ± 0.13 | 6.89 ± 1.72 |
| CNE8 | A/E | 0.67 ± 0.02 | 3.27 ± 0.40 |
| 398F1 | A | 1.159 ± 0.07 | 6.4 ± 1.42 |
| 25710 | C | 1.8 ± 0.0.07 | 9.11 ± 1.35 |
| CH119 | B/C | 0.33 ± 0.03 | 4.2 ± 0.57 |
| 246F3 | A/C | 0.31 ± 0.08 | 0.96 ± 0.27 |
| CE1176 | C | 0.34 ± 0.02 | 13.368 ± 5.03 |
| X1632 | G | 0.993 ± 0.10 | 12.238 ± 1.15 |
| BJOX002000 | B/C | 1.12 ± 0.17 | 7.93 ± 1.37 |
| CNE55 | A/E | 1.02 ± 0.27 | 5.3 ± 2.7 |
| CE0217 | C | 1.37 ± 0.33 | 11.7 ± 1.33 |
| TRO11 | B | 0.93 ± 0.24 | 4.13 ± 0.8 |
| MF535.W0M.ENV.D11 | D/A | 1.35 ± 0.11 | 2.22 ± 0.2 |
| MG505.W0M.ENV.H3 | A | 2.8 ± 1.4 | 15.26 ± 3.5 |
| MI206.W0M.ENV.D1 | A | 0.86 ± 0.006 | 19.2 ± 0.7 |
| ML035.W0M.ENV.I2 | D/A | 0.93 ± 0.08 | 19.6 ± 2.18 |
All the Env clones are R5 tropic.
Fold inhibition by SERINC5 was determined by calculating the ratio of infectivity of the SERINC5-free virus to that of the SERINC5-bearing virus.
Fold inhibition by IFITM3 was determined by calculating the ratio of infectivity of the IFITM3-free virus to that of the IFITM3-bearing virus.
HIV-1 Env clones present distinct profiles of susceptibility to SERINC5 and IFITM3 restriction.
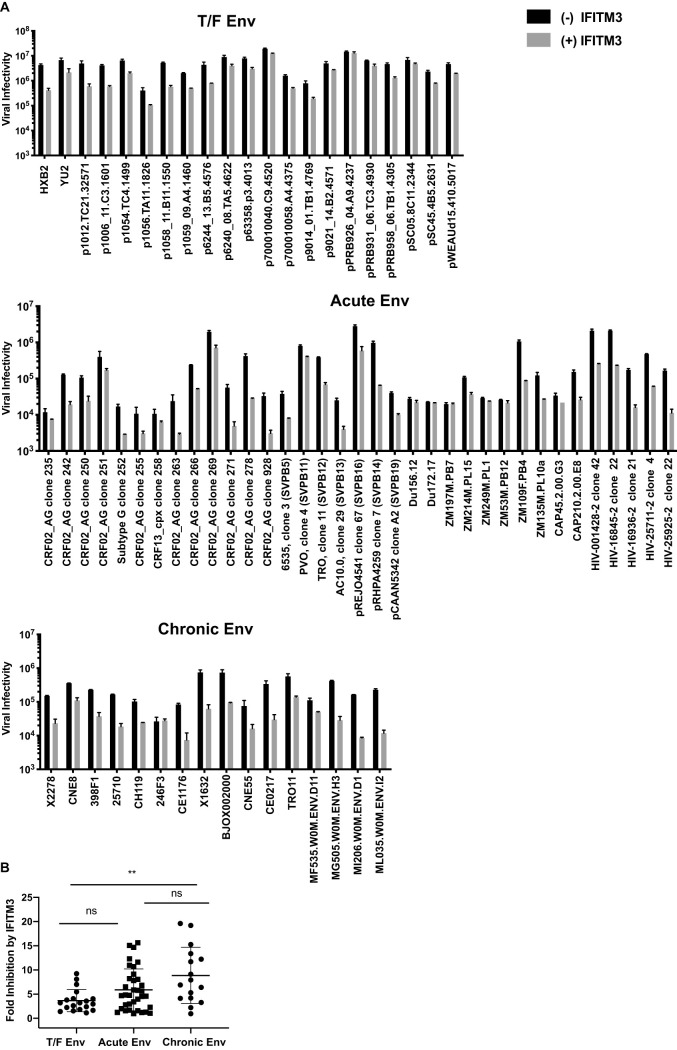
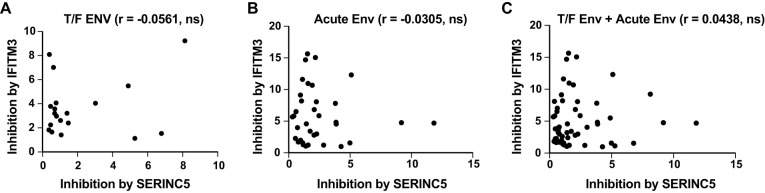
We next measured IFITM3 inhibition of these primary Env clones. As controls, the HXB2 Env was inhibited by 10-fold, while the YU-2 Env was relatively resistant to IFITM3 inhibition (Fig. 2A). The mean fold of inhibition by IFITM3 was 3.6 for the T/F Env clones, whereas the value increased to 5.8 for the acute Env clones and to 8.17 for the chronic Env clones, both of which are higher than that of the T/F Env (Fig. 2B; Tables 1 to 3). Therefore, as opposed to the persistent resistance of HIV-1 Env clones to SERINC5 across different stages of infection, the T/F Env clones are relatively resistant to IFITM3 restriction, but this resistance property is lost as the infection advances, which supports the findings by Foster et al. (18). Importantly, we observed no significant correlations between the restriction by IFITM3 and SERINC5 in either of the Env groups (Fig. 3), thus indicating that the same Env is not necessarily sensitive or resistant to both IFITM3 and SERINC5.
FIG 2.
Inhibition of HIV-1 Env clones by IFITM3. HEK293T cells were cotransfected with NL4-3 ΔNefΔEnv proviral DNA, different HIV-1 Env clones (as indicated), and 100 ng of IFITM3 DNA. The infectivity of viruses was determined by infecting TZM-bl cells. (A) Luciferase data of one representative experiment are shown. (B) Fold inhibition by IFITM3 was determined by calculating the ratio of infectivity of the IFITM3-free virus to those of the IFITM3-bearing virus. Fold inhibition by IFITM3 from three independent transfections are presented. The mean value of fold inhibition for each Env group is indicated by the whisker. The standard deviations are also presented. *, P < 0.05; ns, not significant.
FIG 3.
Lack of correlation between SERINC5 and IFITM3 inhibition. (A to C) Correlation analysis between SERINC5 inhibition and IFITM3 inhibition for HIV-1 T/F Env (A), HIV-1 acute Env (B), and T/F Env and acute Env together (C). Correlation was assessed using the Spearman rank test. ns, not significant.
SERINC5-resistant HIV-1 Envs tend to be refractory to soluble CD4 inhibition.
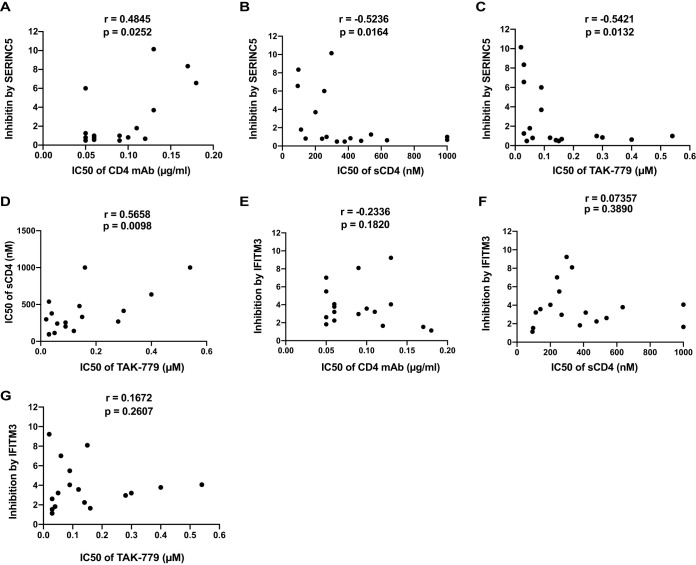
HIV-1 Env sequentially engages CD4 and CCR5 before triggering the fusion of the viral membrane with the cellular membrane. We thus asked whether the efficiency of using CD4 and/or CCR5 by Env correlates with the susceptibility to restriction by SERINC5 and IFITM3. Answering this question is facilitated by the data that are available for T/F Env clones in regard to their sensitivity to agents that inhibit CD4 or CCR5 (Table 1) (30). We first ran the correlation analysis between the 50% inhibitory concentration (IC50) values of T/F Env against a CD4 monoclonal antibody (MAb), RPA-T4, and the folds of inhibition by SERINC5. A significant positive correlation was detected (Fig. 4A), which indicates that the SERINC5-resistant T/F Env clones (with lower folds of inhibition) tend to require greater levels of CD4 (with lower IC50 of CD4 MAb) for entry into target cells. This is likely because these SERINC5-resistant T/F Env clones may have lower affinity for soluble CD4 (sCD4), given the significant negative correlation between the folds of inhibition by SERINC5 and the IC50 values for sCD4 against the T/F Env clones (Fig. 4B). We then examined the correlation of SERINC5 inhibition and the response to the CCR5 inhibitor TAK-779 and observed a significant negative correlation (Fig. 4C). This suggests that SERINC5-resistant Envs tend to be more independent of the CCR5 levels present in the target cell. Interestingly, the T/F Env clones which are more resistant to sCD4 (with higher IC50 of sCD4) tend to be more independent on CCR5 (higher IC50 of TAK-779) (Fig. 4D). However, when we ran the same analysis for the inhibition by IFITM3, no significant correlation was observed with the responses to any of these three agents (Fig. 4E to G), suggesting that SERINC5 and IFITM3 target Envs sampling different conformations.
FIG 4.
Correlation between the inhibition of HIV-1 Env clones by SERINC5 and IFITM3 with the responses of HIV-1 Env clones to CD4 MAb, sCD4, and CCR5 inhibitor TAK-799. (A to C) Correlation analyses were performed between the folds of SERINC5 inhibition and the IC50 values of T/F Env clones for CD4 MAb (A), sCD4 (B), and TAK-779 (C). The IC50 values of these agents were obtained from the study by Keele et al. (30). (D) Correlation between the IC50 values of sCD4 and TAK-779. (E to G) Correlation between the folds of inhibition by IFITM3 and the IC50 values for CD4 MAb (E), sCD4 (F), and TAK-779 (G). Correlation were assessed using the Spearman rank test.
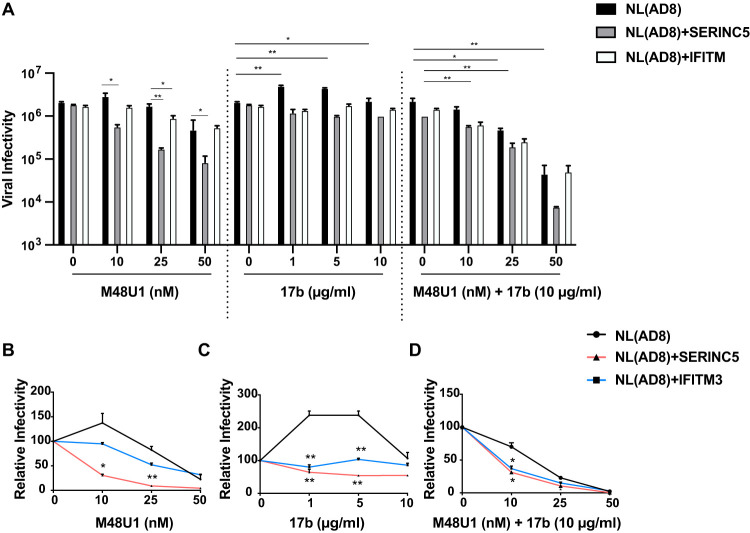
Given the correlation of Env resistance to SERINC5 with CD4 binding, we tested whether there is an opportunity to sensitize the SERINC5-resistant HIV-1 Env to SERINC5 inhibition by using the miniprotein CD4 mimetic (CD4mc) M48U1 which binds to the CD4-binding pocket in gp120 (31). It is possible that the quaternary architecture of primary Envs may resist engagement with proteins such as sCD4, but because of their smaller size, CD4mc might bypass such constraints. We used the NL(AD8) virus, which carries the Env sequence of the primary AD8 strain and is completely resistant to both SERINC5 and IFITM3 (19, 20). At the 10-nM and 25-nM concentrations, M48U1 did not notably inhibit NL(AD8) (Fig. 5A and B). However, the NL(AD8) virus became markedly inhibited by SERINC5 upon exposure to M48U1, known to stabilize the “open” CD4-bound conformation (32) (Fig. 5A and B). A moderate inhibition by IFITM3 was also observed (Fig. 5B). We further tested the response of NL(AD8) to the anticoreceptor binding site 17b antibody, a CD4-induced (CD4i) antibody (33). The infectivity of NL(AD8) increased by more than 2-fold upon exposure to 1 and 5 μg/ml 17b (Fig. 5C). This phenotype was previously reported and shown to be determined primarily by the configuration of the V1, V2, and V3 variable loops (34–37). If a conformation of high free energy is required to activate the trimer, the binding by 17b could stabilize a lower-energy conformation which favors viral entry. In the case of 17b, it has been suggested that suboptimal occupation of the binding site (i.e., binding to one subunit of the trimer) might induce conformational changes in the unoccupied subunits, thus facilitating entry (37). However, occupation of the other subunits abrogates this activity, consistent with the lack of enhancement of viral entry at higher 17b concentrations (10 μg/ml; Fig. 5C). Interestingly, the 17b-infectivity-enhancing effect was abrogated by SERINC5 or IFITM3 (Fig. 5C), suggesting that these restriction factors stabilize Env in conformation(s) that more readily expose the coreceptor binding site. In agreement with the capacity of SERINC5 and IFITM3 to stabilize more “open” Env conformation(s), HIV-1 carrying either SERINC5 or IFITM3 became more sensitive to the inhibition by 17b (10 μg/ml) and M48U1 (10 nM) compared to the control HIV-1 (Fig. 5D). This sensitizing effect was less evident when 25 nM or 50 nM M48U1 was used, because at these concentrations, M48U1 together with 17b already strongly diminished HIV-1 infectivity even in the absence of SERINC5 or IFITM3 (Fig. 5D).
FIG 5.
Effect of CD4 mimetic M48U1 and 17b antibody on the inhibition of HIV-1 Env by SERINC5 and IFITM3. (A) The NL(AD8) viruses that carried either SERINC5 or IFITM3 were incubated with different concentrations of M48U1 or the 17b antibody or the combination of M48U1 and 17b antibody (10 μg/ml) before infecting the TZM-bl cells. Luciferase data of one representative experiment are shown. (B to D) Responses of the control NL(AD8) virus or NL(AD8) bearing SERINC5 or IFITM3 to the inhibition by M48U1 (B), 17b (C), or the combination of M48U1 and 17b (D). The level of viral infectivity without M48U1 or 17b treatment was arbitrarily set as 100. The results were calculated from three independent infection experiments. *, P < 0.05; **, P < 0.01.
Taken together, these data suggest that SERINC5 and IFITM 3 stabilize more “open” Env conformations that result in the exposure of certain CD4i-epitopes such as the coreceptor binding site. This provides an opportunity to sensitize HIV-1 to the inhibition by SERINC5 by targeting the gp120 Phe43 cavity using small CD4mc (38, 39).
The combination effect of SERINC5 and IFITM3 on HIV-1 infection.
Since HIV-1 is exposed to both SERINC5 and IFITM3 during the natural course of infection, we tested whether these two restriction factors together elicit stronger inhibition in combination than alone. We first examined the T/F Env clones. The mean fold of inhibition by SERINC5 and IFITM3 was higher than that by either SERINC5 or IFITM3 but only statistically significant for IFITM3 (Fig. 6A). This might be because the mean fold of inhibition by SERINC5 is already higher than that by IFITM3. We next examined the chronic Env clones and did not observe significantly stronger inhibition by the combination of SERINC5 and IFITM3 compared to IFITM3 alone (Fig. 6B). Taken together, these data suggest that the combination of IFITM3 and SERINC5 does not tend to inhibit HIV-1 more than the stronger inhibitor between IFITM3 and SERINC5.
FIG 6.
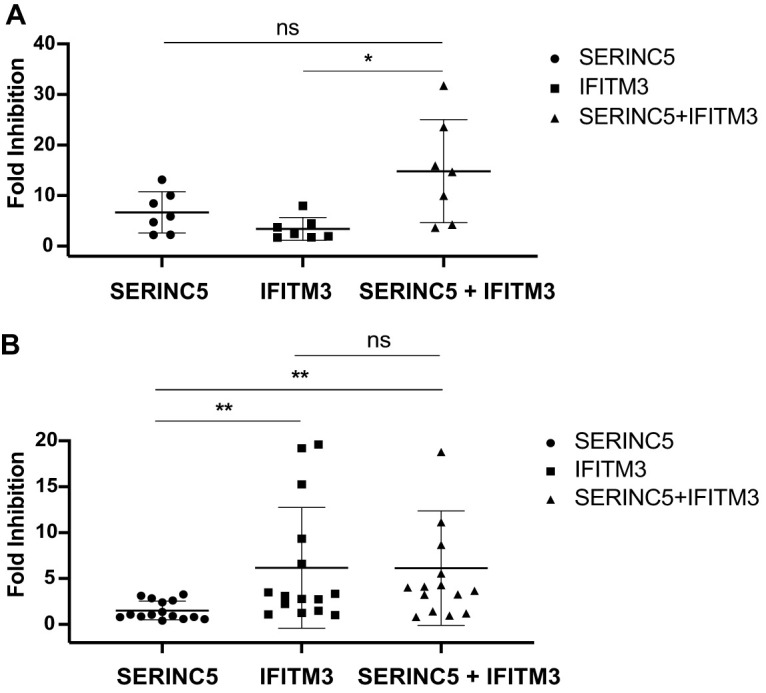
Inhibition of HIV-1 Env by SERINC5 and IFITM3 together. (A) The NL4-3ΔNefΔEnv viruses were pseudotyped with the T/F Env clones. Viruses that carried either SERINC5, IFITM3, or both of these two proteins were used to infect TZM-bl cells. Folds of inhibition were calculated from the data of three independent experiments. (B) Inhibition of chronic Env clones by either SERINC5 or IFITM3 alone or by these two proteins together. Folds of inhibition were calculated from data of three independent experiments. The mean value of fold inhibition for each Env group is indicated by the whisker. The standard deviations are also presented. *, P < 0.05; ns, not significant.
DISCUSSION
In this study, we report the preserved resistance to SERINC5 by HIV-1 Env across different stages of HIV-1 infection, from HIV-1 transmission to acute infection and subsequent chronic infection. In contrast, while resisting IFITM3 inhibition during transmission, HIV-1 Env gradually loses this resistance property as the infection progresses to the chronic stage, which is in agreement with the findings by Foster et al. (18). These different responses of Env to SERINC5 and IFITM3 over the course of HIV-1 infection may be attributed to the fact that expression of IFITM3 is induced by interferon, while SERINC5 is constitutively expressed (40). Therefore, SERINC5 poses a constant inhibitory pressure on HIV-1. In contrast, as interferon response tapers off, the IFITM3 level also goes down, in particular, as HIV-1 infection advances to the chronic stage. With the need to evade the inhibition by neutralizing antibodies, Env constantly changes and thus may lose the resistance to IFITM3 (18).
While Env protein is able to overcome the inhibition by SERINC5, it does not prevent incorporation of SERINC5 into HIV-1 particles (20), which gives SERINC5 the opportunity to act on Env and, as a result, sensitize the virus to neutralizing antibodies and Env-targeting compounds (20, 21). This property of SERINC5 necessitates its removal from HIV-1 particles by Nef. We noticed that the chronic Env clones tend to be more resistant to SERINC5 than the T/F and acute Env clones. This may partly result from the accumulated polymorphisms in viral Nef protein over the long course of chronic infection, which impair the ability of Nef to counter SERINC5 (41). Partial loss of SERINC5 antagonism by Nef could lay more pressure on HIV-1 Env to resist SERINC5. Our observation that chronic Env becomes sensitive to IFITM3 while maintaining resistance to SERINC5 suggests that HIV-1 Env has different strategies to evade the inhibition by these two restriction factors.
Our data suggest a correlation between the susceptibility of HIV-1 Env to SERINC5 and the affinity of Env to CD4. We observed that SERINC5-resistant Envs are more resistant to sCD4 inhibition (Fig. 4), which suggests a low affinity of SERINC5-resistant Envs for CD4. Our results also suggest that SERINC5 and IFITM3 stabilize Env in more “open” conformation(s), resulting in the exposure of the coreceptor binding site and potential neutralization by otherwise nonneutralizing antibodies. To prevent this from happening, primary HIV-1 Envs assume a “closed” conformation, thus effectively concealing epitopes recognized by nonneutralizing antibodies as well as antibodies that mediate antibody-dependent cellular toxicity (ADCC) (42, 43). We speculate that by doing so, HIV-1 Env also happens to gain resistance to SERINC5. In support of this speculation, cell surface expression of CD4 renders the SERINC5-resistant HIV-1 Env prone to SERINC5 inhibition, through induction of an “open” conformation of Env as a result of interaction with CD4 (44). Furthermore, our study showed that CD4 mimetic transforms the SERINC5-resistant Env to a sensitive one, likely through its ability to “open up” Env trimers, which has been shown to enhance antibody access and consequently promotes ADCC (1, 32, 45). It is thus not surprising that HIV-1 has evolved multiple strategies to downregulate CD4 in the infected cells, including Vpu and Nef (46–48), because premature interaction of CD4 with Env trimer exposes Env not only to antibodies (49) but also to restriction factors, including SERINC5. At the same time, agents like CD4mc are expected to sensitize HIV-1 Env to the attack by both antibodies and SERINC5, which might have therapeutic potential.
The conclusions of this study are based on the analysis of a relatively large group of HIV-1 Env clones from different stages of infection and thus may not apply to each and every Env clone that has been tested here or remains to be tested. For example, some Env clones can have low IC50 values of sCD4, i.e., high affinity to CD4, yet exhibit resistance to SERINC5. This variation among the Env proteins of different HIV-1 strains indicates that more factors than the affinity to CD4 modulate Env susceptibility to SERINC5 restriction.
In summary, our results indicate a constant inhibitory pressure on HIV-1 Env imposed by SERINC5 over the course of HIV-1 infection. Adopting a “closed” Env conformation may have allowed HIV-1 to evade not only the humoral response but also the restriction by SERINC5, even though this mechanism may not be effective against IFITM3. Along this line, “opening up” Env trimers with CD4mc could expose HIV-1 to attack by both antibodies and SERINC5 restriction.
MATERIALS AND METHODS
Plasmid DNA.
pNL4-3 DNA was obtained from the NIH AIDS Reagent Program. pNL(AD8) DNA was kindly provided by Eric O. Freed (50). pBJ6-SERINC5-HA was obtained from EURIPRED (reference number 100107). The pQCXIP retroviral expression vector was purchased from Clontech (catalog number 631516). N-terminal Flag-tagged QCXIP-IFITM3 was generated as previously described (19). NL4-3ΔNefΔEnv was generated by inserting stop codons to amino acids positions 31/32 in Nef and amino acid positions 39/40 in Env. HIV-1 Env-expressing clones tested in this paper were obtained from the NIH AIDS Reagents Program catalog number 11663 (30), catalog number 11227 (51), catalog number 11326 (52–54), catalog number 11672 (55), catalog number 11673 (55), catalog number 12670 (56), and catalog number 11674 (57). Of note, each Env clone was isolated from a different individual.
Virus production.
HIV-1 was produced by transfecting human embryonic kidney cell line HEK293T with HIV-1 proviral DNA. Viruses in the supernatants were clarified by centrifugation in a CS-6R centrifuge (Beckman Coulter) at 3,000 rpm for 25 min at 4°C. The amounts of viruses were determined by measuring viral reverse transcriptase (RT) activity. To produce NL4-3ΔNefΔEnv viruses carrying different HIV-1 Envs, 200 ng of NL4-3ΔNefΔEnv proviral DNA was cotransfected with 25 ng of HIV-1 Env-expressing plasmid DNA.
To investigate the effect of SERINC5 or IFITM3 on HIV-1 infectivity, 200 ng of HIV-1 proviral DNA was cotransfected with 100 ng of SERINC5 DNA or 100 ng of IFITM3 DNA into HEK293T cells that were seeded in 6-well plates. Viruses thus produced were used to infect the TZM-bl indicator cells as described below. The amount of 100 ng of SERINC5 or IFITM3 DNA used in the cotransfection experiments was informed by the results of titration experiments as described in our previous studies (19, 20).
Measuring viral infectivity.
Viral infectivity was measured by infecting TZM-bl indicator cells, which contain an HIV-1 long terminal repeat (LTR)-luciferase expression cassette. These cells were obtained from the NIH AIDS Reagent Program (catalog number 8129). TZM-bl cells were first seeded into 24-well plates (40,000 cells per well) before being infected with HIV-1. At 48 h after viral infection, the TZM-bl cells were lysed in passive lysis buffer. Cell lysates were mixed with luciferase substrate, and luciferase activity was measured using a luminometer. The levels of luciferase activity were normalized by the relative quantities of viral reverse transcriptase activity, and the results represent the infectivity of the virus particles. The amount of HIV-1 used in TZM-bl infection is in the range of 2,000 cpm (count per minute) of viral reverse transcriptase activity.
M48U1 and 17b inhibition assay.
Viruses were incubated with different concentrations of the CD4 peptide mimetic M48U1 compound (45) or the 17b nonneutralizing antibody (43) for 1 h at 37°C and then used to infect TZM-bl cells to assess their infectivity levels. After 48 h, infected TZM-bl cells were lysed, and the levels of luciferase activity were measured.
Correlation analysis.
The data published by Keele et al. (30) were adopted to examine the potential correlation between the SERINC5 or IFITM3 sensitivity and the responses of HIV-1 T/F Env clones to the inhibition by CD4 monoclonal antibody (MAb) RPA-T4 (555344, BD PharMingen), sCD4 (514-CD, R&D Systems), or CCR5 inhibitor TAK779 (4983, NIH AIDS Reagents Program).
Statistics.
P values were calculated with Student’s t test. The R values and P values of correlation graphs were calculated using correlation Spearman in GraphPad Prism.
ACKNOWLEDGMENTS
We thank Eric Freed for providing the AD8-1 proviral DNA and the NIH AIDS Reagent Program for providing the Env clones. We are grateful to Myles McLean for critical readings of the manuscript.
This study was supported by funding from the Canadian Institutes of Health Research to C.L. and A.F. (MOP-133479, PJT-166048). A.F. is the recipient of a Canada Research Chair on Retroviral Entry (RCHS0235 950-232424).
REFERENCES
- 1.Beitari S, Wang Y, Liu SL, Liang C. 2019. HIV-1 envelope glycoprotein at the interface of host restriction and virus evasion. Viruses 11:311. doi: 10.3390/v11040311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu J, Pan Q, Rong L, He W, Liu SL, Liang C. 2011. The IFITM proteins inhibit HIV-1 infection. J Virol 85:2126–2137. doi: 10.1128/JVI.01531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lodermeyer V, Suhr K, Schrott N, Kolbe C, Sturzel CM, Krnavek D, Munch J, Dietz C, Waldmann T, Kirchhoff F, Goffinet C. 2013. 90K, an interferon-stimulated gene product, reduces the infectivity of HIV-1. Retrovirology 10:111. doi: 10.1186/1742-4690-10-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Q, Zhang X, Han Y, Wang X, Gao G. 2016. M2BP inhibits HIV-1 virion production in a vimentin filaments-dependent manner. Sci Rep 6:32736. doi: 10.1038/srep32736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lodermeyer V, Ssebyatika G, Passos V, Ponnurangam A, Malassa A, Ewald E, Sturzel CM, Kirchhoff F, Rotger M, Falk CS, Telenti A, Krey T, Goffinet C. 2018. The antiviral activity of the cellular glycoprotein LGALS3BP/90K is species specific. J Virol 92 doi: 10.1128/JVI.00226-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, Santoni FA, Pizzato M. 2015. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526:212–217. doi: 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Usami Y, Wu Y, Gottlinger HG. 2015. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526:218–223. doi: 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tada T, Zhang Y, Koyama T, Tobiume M, Tsunetsugu-Yokota Y, Yamaoka S, Fujita H, Tokunaga K. 2015. MARCH8 inhibits HIV-1 infection by reducing virion incorporation of envelope glycoproteins. Nat Med 21:1502–1507. doi: 10.1038/nm.3956. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Tada T, Ozono S, Yao W, Tanaka M, Yamaoka S, Kishigami S, Fujita H, Tokunaga K. 2019. Membrane-associated RING-CH (MARCH) 1 and 2 are MARCH family members that inhibit HIV-1 infection. J Biol Chem 294:3397–3405. doi: 10.1074/jbc.AC118.005907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Lu J, Liu X. 2018. MARCH2 is upregulated in HIV-1 infection and inhibits HIV-1 production through envelope protein translocation or degradation. Virology 518:293–300. doi: 10.1016/j.virol.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Zhou T, Frabutt DA, Moremen KW, Zheng YH. 2015. ERManI (endoplasmic reticulum class I alpha-mannosidase) is required for HIV-1 envelope glycoprotein degradation via endoplasmic reticulum-associated protein degradation pathway. J Biol Chem 290:22184–22192. doi: 10.1074/jbc.M115.675207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krapp C, Hotter D, Gawanbacht A, McLaren PJ, Kluge SF, Sturzel CM, Mack K, Reith E, Engelhart S, Ciuffi A, Hornung V, Sauter D, Telenti A, Kirchhoff F. 2016. Guanylate binding protein (GBP) 5 is an interferon-inducible inhibitor of HIV-1 infectivity. Cell Host Microbe 19:504–514. doi: 10.1016/j.chom.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 14.McLaren PJ, Gawanbacht A, Pyndiah N, Krapp C, Hotter D, Kluge SF, Gotz N, Heilmann J, Mack K, Sauter D, Thompson D, Perreaud J, Rausell A, Munoz M, Ciuffi A, Kirchhoff F, Telenti A. 2015. Identification of potential HIV restriction factors by combining evolutionary genomic signatures with functional analyses. Retrovirology 12:41. doi: 10.1186/s12977-015-0165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujiwara Y, Hizukuri Y, Yamashiro K, Makita N, Ohnishi K, Takeya M, Komohara Y, Hayashi Y. 2016. Guanylate-binding protein 5 is a marker of interferon-gamma-induced classically activated macrophages. Clin Transl Immunology 5:e111. doi: 10.1038/cti.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding S, Pan Q, Liu SL, Liang C. 2014. HIV-1 mutates to evade IFITM1 restriction. Virology 454–455:11–24. doi: 10.1016/j.virol.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jia R, Ding S, Pan Q, Liu SL, Qiao W, Liang C. 2015. The C-terminal sequence of IFITM1 regulates its anti-HIV-1 activity. PLoS One 10:e0118794. doi: 10.1371/journal.pone.0118794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster TL, Wilson H, Iyer SS, Coss K, Doores K, Smith S, Kellam P, Finzi A, Borrow P, Hahn BH, Neil SJD. 2016. Resistance of transmitted founder HIV-1 to IFITM-mediated restriction. Cell Host Microbe 20:429–442. doi: 10.1016/j.chom.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Pan Q, Ding S, Wang Z, Yu J, Finzi A, Liu SL, Liang C. 2017. The V3 loop of HIV-1 Env determines viral susceptibility to IFITM3 impairment of viral infectivity. J Virol 91:e02441-16. doi: 10.1128/JVI.02441-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beitari S, Ding S, Pan Q, Finzi A, Liang C. 2017. Effect of HIV-1 Env on SERINC5 antagonism. J Virol 91 doi: 10.1128/JVI.02214-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sood C, Marin M, Chande A, Pizzato M, Melikyan GB. 2017. SERINC5 protein inhibits HIV-1 fusion pore formation by promoting functional inactivation of envelope glycoproteins. J Biol Chem 292:6014–6026. doi: 10.1074/jbc.M117.777714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin SW, Alsahafi N, Kuang XT, Swann SA, Toyoda M, Gottlinger H, Walker BD, Ueno T, Finzi A, Brumme ZL, Brockman MA. 2019. Natural HIV-1 Nef polymorphisms impair SERINC5 downregulation activity. Cell Rep 29:1449–1457.e5. doi: 10.1016/j.celrep.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li K, Markosyan RM, Zheng YM, Golfetto O, Bungart B, Li M, Ding S, He Y, Liang C, Lee JC, Gratton E, Cohen FS, Liu SL. 2013. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog 9:e1003124. doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desai TM, Marin M, Chin CR, Savidis G, Brass AL, Melikyan GB. 2014. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog 10:e1004048. doi: 10.1371/journal.ppat.1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trautz B, Wiedemann H, Luchtenborg C, Pierini V, Kranich J, Glass B, Krausslich HG, Brocker T, Pizzato M, Ruggieri A, Brugger B, Fackler OT. 2017. The host-cell restriction factor SERINC5 restricts HIV-1 infectivity without altering the lipid composition and organization of viral particles. J Biol Chem 292:13702–13713. doi: 10.1074/jbc.M117.797332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inuzuka M, Hayakawa M, Ingi T. 2005. Serinc, an activity-regulated protein family, incorporates serine into membrane lipid synthesis. J Biol Chem 280:35776–35783. doi: 10.1074/jbc.M505712200. [DOI] [PubMed] [Google Scholar]
- 27.Pye VE, Rosa A, Bertelli C, Struwe WB, Maslen SL, Corey R, Liko I, Hassall M, Mattiuzzo G, Ballandras-Colas A, Nans A, Takeuchi Y, Stansfeld PJ, Skehel JM, Robinson CV, Pizzato M, Cherepanov P. 2020. A bipartite structural organization defines the SERINC family of HIV-1 restriction factors. Nat Struct Mol Biol 27:78–83. doi: 10.1038/s41594-019-0357-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ling S, Zhang C, Wang W, Cai X, Yu L, Wu F, Zhang L, Tian C. 2016. Combined approaches of EPR and NMR illustrate only one transmembrane helix in the human IFITM3. Sci Rep 6:24029. doi: 10.1038/srep24029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia R, Xu F, Qian J, Yao Y, Miao C, Zheng YM, Liu SL, Guo F, Geng Y, Qiao W, Liang C. 2014. Identification of an endocytic signal essential for the antiviral action of IFITM3. Cell Microbiol 16:1080–1093. doi: 10.1111/cmi.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grupping K, Selhorst P, Michiels J, Vereecken K, Heyndrickx L, Kessler P, Vanham G, Martin L, Arien KK. 2012. MiniCD4 protein resistance mutations affect binding to the HIV-1 gp120 CD4 binding site and decrease entry efficiency. Retrovirology 9:36. doi: 10.1186/1742-4690-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richard J, Veillette M, Brassard N, Iyer SS, Roger M, Martin L, Pazgier M, Schon A, Freire E, Routy JP, Smith AB 3rd, Park J, Jones DM, Courter JR, Melillo BN, Kaufmann DE, Hahn BH, Permar SR, Haynes BF, Madani N, Sodroski JG, Finzi A. 2015. CD4 mimetics sensitize HIV-1-infected cells to ADCC. Proc Natl Acad Sci U S A 112:E2687–94. doi: 10.1073/pnas.1506755112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol 69:5723–5733. doi: 10.1128/JVI.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sullivan N, Sun Y, Li J, Hofmann W, Sodroski J. 1995. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell line-passaged human immunodeficiency virus type 1 isolates. J Virol 69:4413–4422. doi: 10.1128/JVI.69.7.4413-4422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schutten M, Andeweg AC, Bosch ML, Osterhaus AD. 1995. Enhancement of infectivity of a non-syncytium inducing HIV-1 by sCD4 and by human antibodies that neutralize syncytium inducing HIV-1. Scand J Immunol 41:18–22. doi: 10.1111/j.1365-3083.1995.tb03528.x. [DOI] [PubMed] [Google Scholar]
- 36.Schutten M, Andeweg AC, Rimmelzwaan GF, Osterhaus AD. 1997. Modulation of primary human immunodeficiency virus type 1 envelope glycoprotein-mediated entry by human antibodies. J Gen Virol 78:999–1006. doi: 10.1099/0022-1317-78-5-999. [DOI] [PubMed] [Google Scholar]
- 37.Sullivan N, Sun Y, Binley J, Lee J, Barbas CF 3rd, Parren PW, Burton DR, Sodroski J. 1998. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J Virol 72:6332–6338. doi: 10.1128/JVI.72.8.6332-6338.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding S, Grenier MC, Tolbert WD, Vezina D, Sherburn R, Richard J, Prevost J, Chapleau JP, Gendron-Lepage G, Medjahed H, Abrams C, Sodroski J, Pazgier M, Smith AB 3rd, Finzi A. 2019. A new family of small-molecule CD4-mimetic compounds contacts highly conserved aspartic acid 368 of HIV-1 gp120 and mediates antibody-dependent cellular cytotoxicity. J Virol 93 doi: 10.1128/JVI.01325-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finzi A. 2019. Exposing HIV-1 Env: implications for therapeutic strategies. Clin Invest Med 42:E2–E6. doi: 10.25011/cim.v42i4.33109. [DOI] [PubMed] [Google Scholar]
- 40.Zutz A, Scholz C, Schneider S, Pierini V, Munchhoff M, Sutter K, Wittmann G, Dittmer U, Draenert R, Bogner JR, Fackler OT, Keppler OT. 2020. SERINC5 is an unconventional HIV restriction factor that is upregulated during myeloid cell differentiation. J Innate Immun doi: 10.1159/000504888:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sudderuddin H, Kinloch NN, Jin SW, Miller RL, Jones BR, Brumme CJ, Joy JB, Brockman MA, Brumme ZL. 2020. Longitudinal within-host evolution of HIV Nef-mediated CD4, HLA and SERINC5 downregulation activity: a case study. Retrovirology 17:3. doi: 10.1186/s12977-019-0510-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richard J, Prevost J, Alsahafi N, Ding S, Finzi A. 2018. Impact of HIV-1 envelope conformation on ADCC responses. Trends Microbiol 26:253–265. doi: 10.1016/j.tim.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 43.Alsahafi N, Bakouche N, Kazemi M, Richard J, Ding S, Bhattacharyya S, Das D, Anand SP, Prevost J, Tolbert WD, Lu H, Medjahed H, Gendron-Lepage G, Ortega Delgado GG, Kirk S, Melillo B, Mothes W, Sodroski J, Smith AB 3rd, Kaufmann DE, Wu X, Pazgier M, Rouiller I, Finzi A, Munro JB. 2019. An asymmetric opening of HIV-1 envelope mediates antibody-dependent cellular cytotoxicity. Cell Host Microbe 25:578–587.e5. doi: 10.1016/j.chom.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Shi J, Qiu X, Chai Q, Frabutt DA, Schwartz RC, Zheng YH. 2019. CD4 expression and Env conformation are critical for HIV-1 restriction by SERINC5. J Virol 93 doi: 10.1128/JVI.00544-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Acharya P, Luongo TS, Louder MK, McKee K, Yang Y, Kwon YD, Mascola JR, Kessler P, Martin L, Kwong PD. 2013. Structural basis for highly effective HIV-1 neutralization by CD4-mimetic miniproteins revealed by 1.5 A cocrystal structure of gp120 and M48U1. Structure 21:1018–1029. doi: 10.1016/j.str.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen MY, Maldarelli F, Karczewski MK, Willey RL, Strebel K. 1993. Human immunodeficiency virus type 1 Vpu protein induces degradation of CD4 in vitro: the cytoplasmic domain of CD4 contributes to Vpu sensitivity. J Virol 67:3877–3884. doi: 10.1128/JVI.67.7.3877-3884.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aiken C, Konner J, Landau NR, Lenburg ME, Trono D. 1994. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell 76:853–864. doi: 10.1016/0092-8674(94)90360-3. [DOI] [PubMed] [Google Scholar]
- 48.Garcia JV, Miller AD. 1991. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature 350:508–511. doi: 10.1038/350508a0. [DOI] [PubMed] [Google Scholar]
- 49.Ding S, Gasser R, Gendron-Lepage G, Medjahed H, Tolbert WD, Sodroski J, Pazgier M, Finzi A. 2019. CD4 incorporation into HIV-1 viral particles exposes envelope epitopes recognized by CD4-induced antibodies. J Virol 93 doi: 10.1128/JVI.01403-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Freed EO, Englund G, Martin MA. 1995. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J Virol 69:3949–3954. doi: 10.1128/JVI.69.6.3949-3954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, Mlisana K, Karim SA, Hong K, Greene KM, Bilska M, Zhou J, Allen S, Chomba E, Mulenga J, Vwalika C, Gao F, Zhang M, Korber BT, Hunter E, Hahn BH, Montefiori DC. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in southern Africa. J Virol 80:11776–11790. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 54.Williamson C, Morris L, Maughan MF, Ping LH, Dryga SA, Thomas R, Reap EA, Cilliers T, van Harmelen J, Pascual A, Ramjee G, Gray G, Johnston R, Karim SA, Swanstrom R. 2003. Characterization and selection of HIV-1 subtype C isolates for use in vaccine development. AIDS Res Hum Retroviruses 19:133–144. doi: 10.1089/088922203762688649. [DOI] [PubMed] [Google Scholar]
- 55.Kulkarni SS, Lapedes A, Tang H, Gnanakaran S, Daniels MG, Zhang M, Bhattacharya T, Li M, Polonis VR, McCutchan FE, Morris L, Ellenberger D, Butera ST, Bollinger RC, Korber BT, Paranjape RS, Montefiori DC. 2009. Highly complex neutralization determinants on a monophyletic lineage of newly transmitted subtype C HIV-1 Env clones from India. Virology 385:505–520. doi: 10.1016/j.virol.2008.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.deCamp A, Hraber P, Bailer RT, Seaman MS, Ochsenbauer C, Kappes J, Gottardo R, Edlefsen P, Self S, Tang H, Greene K, Gao H, Daniell X, Sarzotti-Kelsoe M, Gorny MK, Zolla-Pazner S, LaBranche CC, Mascola JR, Korber BT, Montefiori DC. 2014. Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 88:2489–2507. doi: 10.1128/JVI.02853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu X, Parast AB, Richardson BA, Nduati R, John-Stewart G, Mbori-Ngacha D, Rainwater SM, Overbaugh J. 2006. Neutralization escape variants of human immunodeficiency virus type 1 are transmitted from mother to infant. J Virol 80:835–844. doi: 10.1128/JVI.80.2.835-844.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]