Viruses hijack cellular proteins, termed viral receptors, to assist their entry into host cells. While viral receptors experience negative selection to maintain their normal functions, they also undergo positive selection due to an everlasting evolutionary arms race between viruses and hosts. A complete picture on how viral receptors evolve under two conflicting forces is still lacking. In this study, we systematically analyzed the evolution of 96 viral receptors in primates and human populations. We found around half of viral receptors underwent adaptive evolution and exhibit significantly elevated rates of adaptation compared to control genes in primates. We also found signals of past natural selection for 58 viral receptors in human populations. Interestingly, 49 viral receptors experienced different selection pressures in different human populations, indicating that viruses represent an important driver of local adaptation in humans. Our results suggest that host-virus arms races drive accelerated adaptive evolution in viral receptors.
KEYWORDS: adaptive evolution, phylogenetics, primates, viral receptors
ABSTRACT
Viral receptors are the cell surface proteins that are hijacked by viruses to initialize their infections. Viral receptors are subject to two conflicting directional forces, namely, negative selection due to functional constraints and positive selection due to host-virus arms races. It remains largely obscure whether negative pleiotropy limits the rate of adaptation in viral receptors. Here, we perform evolutionary analyses of 96 viral receptor genes in primates and find that 41 out of 96 viral receptors experienced adaptive evolution. Many positively selected residues in viral receptors are located at the virus-receptor interfaces. Compared with control proteins, viral receptors exhibit significantly elevated rate of adaptation. Further analyses of genetic polymorphisms in human populations reveal signals of positive selection and balancing selection for 53 and 5 viral receptors, respectively. Moreover, we find that 49 viral receptors experienced different selection pressures in different human populations, indicating that viruses represent an important driver of local adaptation in humans. Our findings suggest that diverse viruses, many of which have not been known to infect nonhuman primates, have maintained antagonistic associations with primates for millions of years, and the host-virus conflicts drive accelerated adaptive evolution in viral receptors.
IMPORTANCE Viruses hijack cellular proteins, termed viral receptors, to assist their entry into host cells. While viral receptors experience negative selection to maintain their normal functions, they also undergo positive selection due to an everlasting evolutionary arms race between viruses and hosts. A complete picture on how viral receptors evolve under two conflicting forces is still lacking. In this study, we systematically analyzed the evolution of 96 viral receptors in primates and human populations. We found around half of viral receptors underwent adaptive evolution and exhibit significantly elevated rates of adaptation compared to control genes in primates. We also found signals of past natural selection for 58 viral receptors in human populations. Interestingly, 49 viral receptors experienced different selection pressures in different human populations, indicating that viruses represent an important driver of local adaptation in humans. Our results suggest that host-virus arms races drive accelerated adaptive evolution in viral receptors.
INTRODUCTION
Delivering genetic material into host cells is an essential step in the life cycle of viruses. With diverse strategies, the entry into host cells by viruses is mainly mediated by the so-called viral receptors (1). Viral receptors are proteins with “normal” cellular functions on the surface of host cells but are hijacked by viruses to assist their infections (2). For instance, transferrin receptor (TFRC) protein is a housekeeping protein that regulates the import of iron into cells and plays a crucial role in iron homeostasis (3, 4). TFRC protein is used by at least three viruses (mouse mammary tumor virus, arenavirus, and parvovirus) to act as their receptor (3, 4). The relationship between viruses and viral receptors is not a simple one-to-one relationship (5). In fact, a virus can use several different proteins as its receptors to initialize its infection, and sometimes a protein can be hijacked by multiple viruses to act as their receptor (5).
In theory, the evolution of viral receptors is subject to two opposing directional evolutionary forces. On one hand, viral receptors experience negative selection due to functional constraints. On the other hand, viral receptors undergo perpetual evolutionary arms races with viruses: viral receptors escape binding by viral proteins to resist viral infections, and viral proteins, in turn, evolve to restore their binding with viral receptors (6, 7). The resistance and counterresistance cycle, a classic form of Red Queen dynamics (8), can drive rapid adaptive evolution of virus receptors (7, 9). However, if virus-driven selection occurs in sites that are crucial for the normal biological functions of viral receptors, it can result in a reduction of fitness. The negative pleiotropy might limit the rate of adaptation in viral receptors.
It remains largely obscure how viral receptor proteins evolved under two opposing selective pressures, although sporadic evolutionary analyses of viral receptors have been available. Several viral receptor proteins, such as TFRC, cluster of differentiation 4 (CD4; human immunodeficiency virus [HIV] receptor), sodium taurocholate cotransporting polypeptide (NTCP; hepatitis B virus [HBV] receptor), Niemann-Pick C1 (NPC1; filovirus receptor), and angiotensin-converting enzyme 2 (ACE2; the receptors of several coronaviruses), have been found to undergo positive selection, and some positively selected sites are located at the interface between viruses and virus receptors (3, 4, 10–12). However, a comprehensive picture of the evolution of viral receptors is still lacking. Systematic analyses of the evolution of viral receptors have significant implications in understanding the evolutionary arms races between viruses and hosts, the host specificity of viruses, and the origin of viral infectious diseases.
Here, we systematically analyzed the evolutionary pattern of 96 viral receptor genes in primates. First, we investigated the selective pressures acting on viral receptors across 20 primate species. Next, we compared the rate of adaptation between viral receptor genes and control genes. Finally, we employed population genetics approaches to detect signals of past selection in three different human populations (Asian, European, and African) and explored the association between polymorphisms in viral receptors and phenotypes of medical relevance.
RESULTS
Adaptive evolution is pervasive in primate viral receptors.
We systematically collected information on mammalian viral receptors by reviewing the literature (see Table S1 in the supplemental material) and the ViralZone database (13, 14). We assembled a data set of 96 viral receptors involving more than 107 viruses (Table S1). As expected, Gene Ontology (GO) analyses show that viral receptors are overrepresented in biological processes (“viral entry into host cell” [GO: 0046718; false discovery rate, or FDR, 5.49 × 10−121], “interaction with host” [GO: 0051701; FDR = 8.16 × 10−110], “viral life cycle” [GO: 0019058; FDR = 1.53 × 10−108], and “symbiont process” [GO: 0044403; FDR = 2.05 × 10−74]), molecular function (“virus receptor activity” [GO: 0001618; FDR = 2.59 × 10−126] and “hijacked molecular function” [GO: 0104005; FDR = 1.30 × 10−126]), and cellular component (“cell surface” [GO: 0009986; FDR = 1.68 × 10−44], “plasma membrane” [GO: 0005886; FDR = 1.46 × 10−44], and “cell periphery” [GO: 0071944; FDR = 6.61 × 10−44]) (Table 1 and Table S2). Besides the virus-related categories with extremely significant FDR values, it is interesting that molecular function categories, such as “molecular transducer activity” (GO: 0060089; FDR = 2.83 × 10−24) and “signaling receptor activity” (GO: 0038023; FDR = 5.47 × 10−24), are enriched in viral receptor genes (Table S2), suggesting that viral receptors mainly function as signaling receptors and transducers in host cells.
TABLE 1.
Top 20 GO biological process terms enriched for viral receptor genes
| Function | GO code | No. of genes | No. of receptors | Fold enrichment | FDR |
|---|---|---|---|---|---|
| Viral entry into host cell | 0046718 | 94 | 67 | 100 | 5.49 × 10−121 |
| Entry into other organism involved in symbiotic interaction | 0051828 | 102 | 67 | 100 | 1.73 × 10−119 |
| Entry into cell of other organism involved in symbiotic interaction | 0051806 | 102 | 67 | 100 | 1.15 × 10−119 |
| Entry into host | 0044409 | 102 | 67 | 100 | 8.63 × 10−120 |
| Entry into host cell | 0030260 | 102 | 67 | 100 | 6.91 × 10−120 |
| Interaction with host | 0051701 | 158 | 67 | 92.95 | 8.16 × 10−110 |
| Viral life cycle | 0019058 | 167 | 67 | 87.94 | 1.53 × 10−108 |
| Viral process | 0016032 | 589 | 68 | 25.31 | 1.12 × 10−77 |
| Symbiont process | 0044403 | 664 | 68 | 22.45 | 2.05 × 10−74 |
| Interspecies interaction between organisms | 0044419 | 709 | 68 | 21.02 | 1.21 × 10−72 |
| Multiorganism process | 0051704 | 2355 | 71 | 6.61 | 4.10 × 10−43 |
| Biological adhesion | 0022610 | 912 | 44 | 10.57 | 1.69 × 10−30 |
| Cell adhesion | 0007155 | 906 | 41 | 9.92 | 5.83 × 10−27 |
| Localization | 0051179 | 5550 | 75 | 2.96 | 8.31 × 10−23 |
| Immune system process | 0002376 | 2630 | 52 | 4.33 | 2.40 × 10−19 |
| Movement of cell or subcellular component | 0006928 | 1465 | 38 | 5.69 | 2.07 × 10−16 |
| Locomotion | 0040011 | 1244 | 35 | 6.17 | 7.10 × 10−16 |
| Response to stimulus | 0050896 | 8264 | 80 | 2.12 | 9.73 × 10−16 |
| Cell surface receptor signaling pathway | 0007166 | 2360 | 45 | 4.18 | 2.77 × 10−15 |
| Cell-cell adhesion | 0098609 | 481 | 24 | 10.94 | 2.90 × 10−15 |
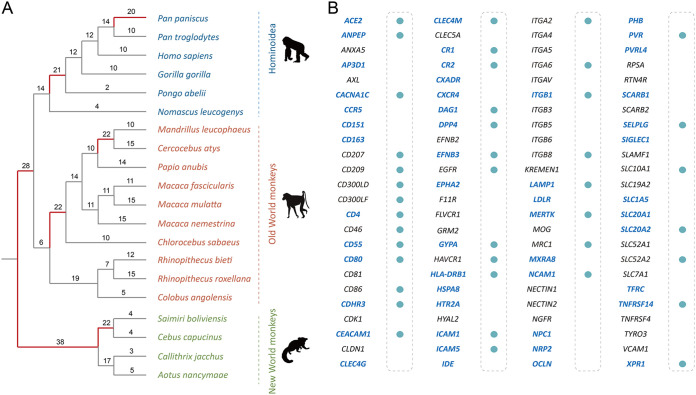
Because most of the viral receptors identified to date involve viruses that infect humans, we used primates as our focal organisms, including four New World monkeys, ten Old World monkeys, and six Hominoidea species (Fig. 1A). The selection pressure acting on a gene could be measured by the ratio of the number of nonsynonymous substitutions per nonsynonymous site (dN) to the number of synonymous substitutions per synonymous site (dS). We estimated dN/dS values for each viral receptor gene and found the overall dN/dS ratio varies from 0.02 to 1.80. The median dN/dS (0.25) of viral receptors seems to be higher than the genome-wide median dN/dS (0.18) previously estimated for primates, although these dN/dS values are estimated from data sets with different primate species (15).
FIG 1.
Adaptive evolution of viral receptor genes. (A) The phylogenetic tree represents the evolutionary relationship of 20 primates, including four New World monkeys, ten Old World monkeys, and six Hominoidea species. The numbers of genes subject to positive selection on a particular branch are labeled. (B) The filled circles represent the viral receptor genes that have residues subject to positive selection inferred in the PAML analysis. The genes that have signatures of positive selection in the BUSTED analysis are highlighted in blue and boldface.
The dN/dS statistic is a conservative test of positive selection (16). We used the branch-site unrestricted statistical test for episodic diversification (BUSTED) to test whether a gene underwent positive selection for at least one branch or one site (17) and found that over half (50/96, 52.1%) of viral receptor genes experienced episodic selection across the primate phylogeny. We next used the branch model to detect specific lineages subject to positive selection. The proportion of positively selected lineages ranges from 0/38 to 23/38 (Fig. 1A and Fig. S1). Moreover, we used the codon model to detect positively selected sites in viral receptor genes. We found evidence of positive selection for 41 out of 96 (42.7%) viral receptor genes, that is, there were positively selected sites (Fig. 1B). The proportion of positively selected sites within viral receptor genes varies from 0% to 18%. Taken together, all these lines of evidence suggest that positive selection pervasively occurred in viral receptor genes during the evolutionary course of primates.
Many positively selected sites overlap virus-receptor interfaces.
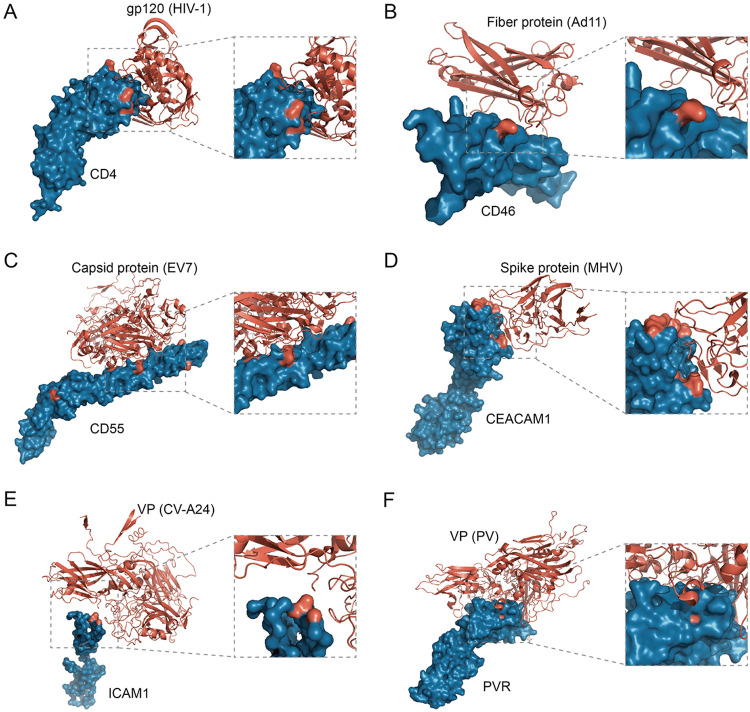
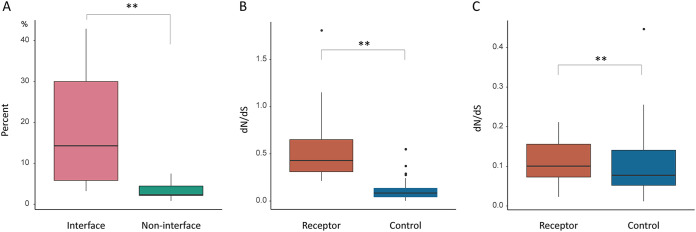
We explored the relationship between positively selected sites detected in viral receptors and receptor-virus interacting interfaces. We found many positively selected sites overlap virus-receptor interacting interfaces. (i) T-cell surface glycoprotein CD4 is the primary receptor of HIV-1, and the positively selected sites, P73, N77, A80, and D113, are located in the virus-receptor interfaces (Fig. 2A) (18). (ii) The only positively selected residue detected in NTCP, K157, is a key binding site of HBV, and mutation at this site can effectively inhibit HBV infection (10, 19). (iii) For the adenovirus receptor, membrane cofactor protein CD46, a positively selected site, R103, was mapped to the virus-receptor interaction region (Fig. 2B) (20). (iv) Enterovirus interacts with complement decay-accelerating factor CD55 to invade host cells, and at least nine positively selected sites, V124, V155, R170, D175, V176, G178, I206, Q230, and H263, overlap the virus binding sites (Fig. 2C) (21). (v) Carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) mediates the entry into host cells by mouse hepatitis virus (MHV), a beta-coronavirus. Among the positively selected sites detected in primate CEACAM1 genes, sites F63, Y68, G75, G85, T86, Q88, and S127 are located in the virus-receptor interaction region (Fig. 2D) (22). (vi) Intercellular adhesion molecule 1 (ICAM1) is the receptor of coxsackievirus A21 and rhinovirus (23). The positively selected sites P28, K29, and P70 are the binding sites of rhinovirus, and K29 is also one of the binding sites of coxsackievirus (Fig. 2E). (vii) Nectin-like protein 5, also known as poliovirus receptor (PVR), acts as the receptor of poliovirus, and the residue Q80 at the interaction surface was subject to positive selection (Fig. 2F) (24). Moreover, for these seven viral receptors, we found the proportion of positively selected residues lying at the virus-host interaction interface is significantly higher than the proportion of positively selected residues in other regions (P = 0.007 by Mann-Whitney U test) (Fig. 3A). It should be noted that only a few crystal structures of viral receptors in complex with viral proteins have been available to date. Nevertheless, our results suggest the molecular arms races between viral receptors and viral proteins drive adaptive evolution in many, if not all, viral receptors.
FIG 2.
Positively selected sites overlap the virus-receptor interacting interfaces. In the crystal structures of the virus-receptor complex, viruses and receptors are labeled in red and blue, respectively. The positively selected residues in viral receptor proteins are labeled in red. (A) CD4 protein bound to the envelope glycoprotein gp120 of human immunodeficiency virus type 1 (HIV-1) (PDB entry 1RZJ); (B) CD46 protein bound to the fiber protein of adenovirus virus type 11 (Ad11) (PDB entry 2O39); (C) CD55 protein bound to the capsid protein of echovirus 7 (EV7) (PDB entry 3IYP); (D) CEACAM1 protein bound to the N-terminal domain of spike glycoprotein of mouse hepatitis coronavirus (MHV) (PDB entry 3R4D); (E) ICAM1 protein bound to structural protein VP1-3 of coxsackievirus A24 (CV-A24) (PDB entry 6EIT); (F) PVR protein bound to structural protein VP1-4 of poliovirus (PV) (PDB entry 3EPC).
FIG 3.
Analyses of selection pressure in receptor genes. (A) The comparison of the proportion of positively selected sites at the virus-host interacting interfaces with that in other locations for seven viral receptors. The pink box represents the proportion of positively selected sites at the interfaces, and the green box represents the proportion of positively selected sites at other locations. (B and C) Hierarchical comparison of selective pressures between receptor and control genes. The receptor genes were divided into two parts, viral receptor genes with top 50% dN/dS values (B) and the other viral receptor genes (C), based on their dN/dS values. The difference in selection pressure between viral receptor genes and the corresponding control genes then was compared.
Rate of adaptation is elevated in primate viral receptors.
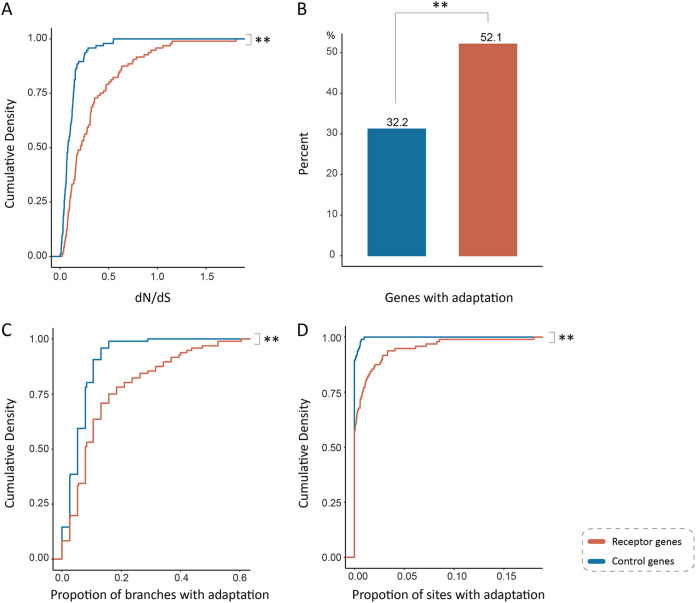
For each viral receptor, we chose a control gene that shares similar GO biological process categories with the viral receptor (see Materials and Methods for details) (Table S3). We then compared the rate of adaptive evolution between viral receptor genes and control genes. We found the mean dN/dS of viral receptor genes is significantly higher than that of the control genes (0.32 versus 0.11; P = 6.8 × 10−9 by Mann-Whitney U test) (Fig. 3B and 4A). The proportion of viral receptor genes that underwent episodic adaptive evolution is significantly greater than that of control genes (52.1% versus 31.2%; P < 0.001 by χ2 test) (Fig. 4B). More lineages in viral receptors are found to be under adaptive evolution than in control genes (13.69% versus 5.96%; P = 5.2 × 10−7 by Mann-Whitney U test) (Fig. 4C). The proportion of positively selected residues in viral receptor genes is also significantly higher than that in control genes (0.97% versus 0.05%; P = 3.7 × 10−4 by Mann-Whitney U test), indicating ∼95% of positively selected sites are associated with host-virus conflicts (Fig. 4D). Taken together, these results strongly suggest that the rate of adaptive evolution is significantly elevated in viral receptor genes, and adaptive evolution in viral receptors is mainly driven by viruses.
FIG 4.
Elevated adaptation rate in primate viral receptors. (A) The comparison of dN/dS ratio between viral receptor genes and control genes. (B) The comparison of proportion of the genes subject to positive selection detected by BUSTED analyses between viral receptor genes and control genes. (C) Comparison of the proportion of the branches subject to positive selection between viral receptor genes and control genes. (D) Comparison of the proportion of the sites subject to positive selection between viral receptor genes and control genes. Viral receptor genes and control genes are labeled in orange and blue, respectively.
We further analyzed the difference in GO enrichment between viral receptors and control genes. Not unexpectedly, similar to viral receptor genes, control genes are enriched in GO categories: biological process (“immune system process” [GO: 0002376], “cell surface receptor signaling pathway” [GO: 0007166], “localization” [GO: 0051179], “response to stimulus” [GO: 0050896]), molecular function (“signaling receptor binding” [GO: 0005102], “cell adhesion molecule binding” [GO: 0050839], and “protein binding” [GO: 0005515]), and cellular component (“plasma membrane part” [GO: 0044459], “cell periphery” [GO: 0071944], and “plasma membrane” [GO: 0005886]) (Table S2). The main difference in GO enrichment between viral receptors and control genes appears to be virus-related GO terms. It follows that viruses are likely the driving force for the elevated adaptive evolution of viral receptor genes.
Natural selection acts on virus receptors in human populations.
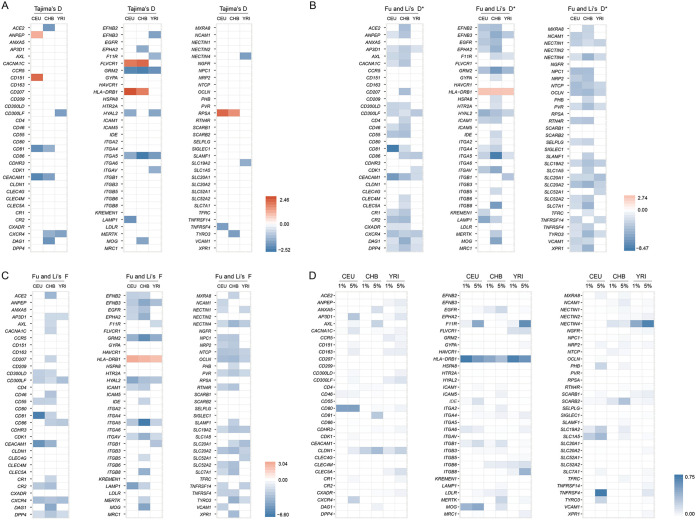
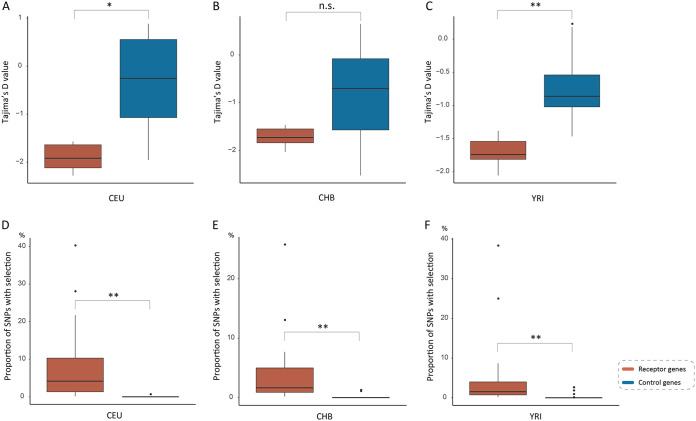
To explore how viral receptor genes evolved in human populations, we employed population genetics approaches to detect signals of past natural selection that occurred in virus receptor genes with polymorphism data from three human populations of different ancestries, namely, Utah residents with Northern and Western European ancestry (CEU), Han Chinese (CHB), and Yoruba in Ibadan, Nigeria (YRI), which are available from the pilot 3 phase of the 1000 Genomes Project. We found 7, 10, and 10 viral receptor genes with significantly negative Tajima’s D values in CEU, CHB, and YRI populations, respectively (Fig. 5A and Table S4). Moreover, a total of 15, 19, and 30 viral receptor genes was found to have extremely high integrated haplotype homozygosity (|iHS|) values in CEU, CHB, and YRI populations, respectively (Fig. 5D and 6, and Table S6). Taken together, at least 53 viral receptor genes might have undergone directional selection in different human populations. Moreover, 5, 3, and 1 viral receptor genes appear to be subject to balancing selection (with significantly positive Tajima’s D) in CEU, CHB, and YRI populations, respectively (Fig. 5A and Table S4). Therefore, we found evidence of natural selection in 58 viral receptor genes involving diverse viruses, such as coronaviruses, enteroviruses, retroviruses, hepatitis B virus, and others. Moreover, the selection pressure (measured by Tajima’s D and the proportion of single-nucleotide polymorphisms [SNPs] with top 1% genome-wide outlying |iHS| scores within the gene) experienced by the viral receptor genes subject to selective sweep is significantly stronger than that of the corresponding control genes in all three human populations (with the exception of Tajima’s D in the CHB population, but the proportion of SNPs with top 1% genome-wide outlying |iHS| scores is significantly higher in the viral receptor genes than in the control genes in the CHB populations) (Fig. 7). These results suggest that natural selection in the viral receptor genes in human populations was also driven mainly by viruses.
FIG 5.
Signals of natural selection in viral receptors in human populations. (A) The heatmap of Tajima’s D in three populations (CEU, CHB, and YRI). Only values of statistical significance are shown. Significantly negative and positive values are labeled in blue and red, respectively. (B) The heatmap of Fu and Li’s D* in three populations (CEU, CHB, and YRI). Only values of statistical significance are shown. Significantly negative and positive values are labeled in blue and red, respectively. (C) The heatmap of Fu and Li’s F* in three populations (CEU, CHB, and YRI). Only values of statistical significance are shown. Significantly negative and positive values are labeled in blue and red, respectively. (D) The heatmap of the proportion of SNPs with top 1% and top 5% genome-wide outlying |iHS| scores within the gene.
FIG 6.
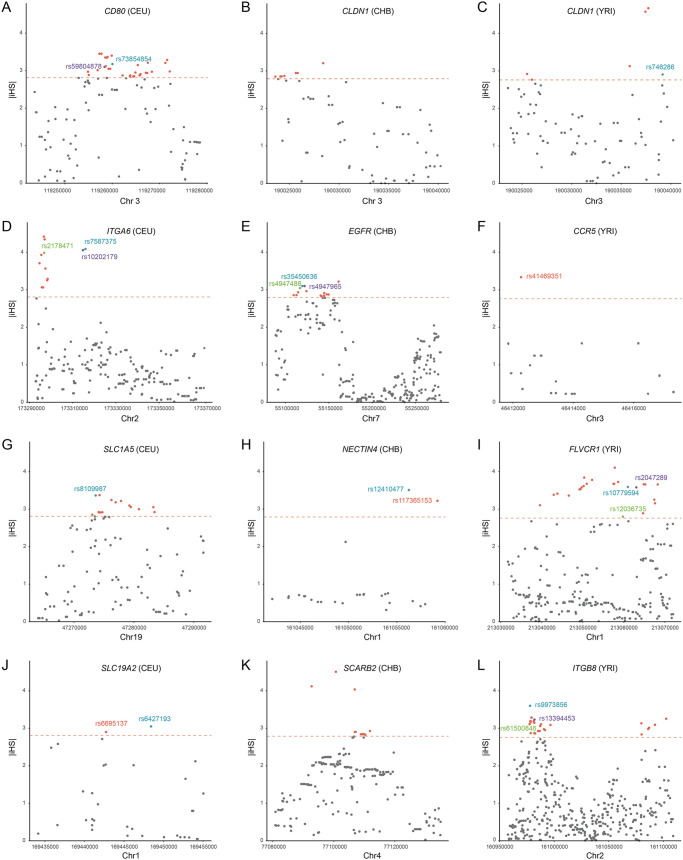
Selection and population differentiation of representative viral receptors. The |iHS| scores of SNPs within 11 representative receptor genes (CCR5, CD80, CLDN1, EGFR, FLVCR1, ITGA6, ITGB8, NECTIN4, SCARB2, SLC19A2, and SLC1A5) are shown. The dashed line represents the top 1% of the |iHS| scores at the genome-wide level. The outlying SNPs with Fst of greater than 0.15 between different populations are labeled in different colors.
FIG 7.
Comparison of population genetic metrics between the viral receptor genes subject to selective sweep and the corresponding control genes in three different human populations. (A, B, and C) Comparison of Tajima’s D between the viral receptor genes subject to selective sweep and the corresponding control genes in three human populations. (D, E, and F) Comparison of the proportion of SNPs with top 1% genome-wide outlying |iHS| scores between the viral receptor genes subject to selective sweep and the corresponding control genes in three human populations.
Interestingly, we found some viral receptor genes underwent different selection pressures in different human populations, a pattern consistent with local adaptation (Fig. 5 and Table S5). To further explore the possibility of local adaptation, we used the fixation index (Fst) to assess population differentiation for viral receptor genes that are subject to directional selection (with high |iHS|). Among the 42 receptor genes with significantly high |iHS| values, 28 exhibit strong pattern of population differentiation (with high Fst values [>0.15]; Tables S6 and S7). Therefore, our results indicate that viruses represent an important driver of local adaptation in humans.
Case studies of important virus receptors.
Different coronaviruses use different receptors, and at least four receptors, ACE2 (for severe acute respiratory syndrome coronavirus [SARS-CoV], SARS-CoV-2, and human coronavirus NL63), CEACAM1 (for MHV), alanine aminopeptidase (ANPEP; for porcine transmissible gastroenteritis coronavirus and porcine respiratory coronavirus), and dipeptidyl peptidase 4 (DPP4; for Middle East respiratory syndrome coronavirus), have been identified (25). In the CHB population, the genetic diversity (π) of the ACE2 gene is extremely low (π = 9.4 × 10−5), and the neutrality tests indicate a selective sweep occurred (Tajima’s D < 0; Fu and Li’s D* < 0; Fu and Li’s F* < 0) (Fig. 5A to C and Table S4). In contrast, no evidence of natural selection was found for the ACE2 gene in the CEU or YRI populations. The CHB population exhibits strong genetic differentiation with the CEU and YRI populations (for CHB versus CEU, Fst = 0.36; for CHB versus YRI, Fst = 0.33), indicating that the ACE2 gene has experienced local adaptation in the CHB population (Table S5). These results indicate that ACE2-utilizing coronaviruses (other viruses or environmental factors cannot be formally excluded) infected the CHB population in the past and drive the evolution of the ACE2 gene in the CHB population. In both CEU and CHB populations, the CEACAM1 gene displays low genetic diversity and have significantly negative D, D*, and F* values, suggesting that directional selection acted on the CEACAM1 gene in both populations (Fig. 5A to C and Table S4). The ANPEP gene displayed significantly positive D values in the CEU population and extremely high |iHS| scores in the YRI population (Fig. 5 and Table S6). Weak evidence of selection was also found for the DPP4 gene in all three populations. Consistent with population genetics analyses in human populations, strong evidence of positive selection was found for all four coronavirus receptor genes in primates. Therefore, our results suggest various coronaviruses have undergone a perpetual evolutionary arms race with primates (including humans), and coronaviruses might not be new for primates or humans.
Both metabotropic glutamate receptor 2 (GRM2) and neural cell adhesion molecule 1 (NCAM1) have been reported to act as receptors for rabies virus (26, 27). For the GRM2 gene, D, D*, and F* values were significantly lower than 0 in the CEU, CHB, and YRI populations (Fig. 5A to C and Table S4). For the NCAM1 gene, the |iHS| values of its SNPs are outliers in CEU, CHB, and YRI populations (Fig. 5D and Table S6). These results indicate rabies virus or related viruses are an important selective agent for their receptor genes across different human populations.
Hepatitis B virus seriously threatens public health worldwide. We found an SNP (rs36115704) in the NTCP gene (the HBV receptor) that displays a significantly high |iHS| score in the CHB population, but no signal of selection was found in the CEU or YRI population. Moreover, the SNP had an extreme Fst value (0.23) for CHB versus YRI (Fig. 5D and Table S7). It follows that HBV may have long been circulating and represents an ancient serious infectious disease in the Chinese population.
We found many retrovirus receptor genes are subject to natural selection in human populations. CD4, C-C chemokine receptor type 5 (CCR5), and C-X-C chemokine receptor type 4 (CXCR4) act as the receptors or the coreceptors of HIV (28, 29). The CD4 gene has two SNPs with extremely high |iHS| scores in the CHB population (Table S6). For the CCR5 gene, the SNP rs41469351 had a significantly high |iHS| score in the YRI population, and for this SNP, the YRI population shows strong genetic differentiation with the other two populations (Fst = 0.34 for YRI versus CEU; Fst = 0.35 for YRI versus CHB) (Fig. 6 and Table S7). For the CXCR4 gene, significantly negative Tajima’s D values were found in both the CHB and YRI populations (Fig. 5A and Table S4). In the YRI population, the SNPs rs11311779, rs71337118, and rs550519394 of the AP-3 complex subunit delta-1 (AP3D1; the receptor of bovine leukemia virus) gene display extremely high |iHS| scores (Table S6). Feline leukemia virus subgroup C receptor-related protein 1 (FLVCR1; the receptor of feline leukemia virus) displays significantly positive Tajima’s D values in the CEU and CHB populations, suggesting balancing selection acted on the FLVCR1 gene in these populations. Several SNPs of the FLVCR1 gene were found to have extremely high |iHS| values in the YRI population (Fig. 5D and 6). Moreover, signals of natural selection were also detected for hyaluronidase-2 (HYAL2; the receptor of Jaagsiekte sheep retrovirus) in CEU and YRI populations, thiamine transporter 1 (also known as solute carrier family 19 member 2 [SLC19A2]; receptor of feline leukemia virus and murine leukemia virus) in CEU and YRI populations, and sodium/glucose cotransporter 1 (SLC1A5; receptor for feline endogenous virus RD114 and baboon M7 endogenous virus) in the CEU population (Tables S4 and S6). Taken together, our results indicate a complex evolutionary history of the interaction between various retroviruses and human populations.
DISCUSSION
Viral receptors are cell surface proteins that perform normal cellular functions but are hijacked by the viruses to assist their infections (2). Thus, it is expected that viral receptors evolved under two conflicting evolutionary forces, namely, negative selection to maintain their own functions and positive selection due to the evolutionary chase of viruses. Thus, one allele that escapes binding by viruses might affect normal host cellular functions and, thus, is selected against. This negative pleiotropy is expected to limit the rate of adaptation. In this study, we systematically analyzed the evolutionary patterns of 96 viral receptor genes related to more than 100 viruses in primates. We found positive selection pervasively occurred in viral receptor genes during the course of primate evolution. Many positively selected residues are mapped to virus-host interfaces. Moreover, the rate of adaptive evolution in viral receptors is significantly elevated compared to that in control genes. Therefore, our results suggest that the evolution of virus receptor genes can take the paths that minimize negatively pleiotropic effects, and the host-virus arms races did drive accelerated adaptive evolution of viral receptors in primates (Fig. 1 and 4).
We find signatures of positive selection in primates in regions known to be critical in the interaction between nonprimate hosts and viruses, suggesting related viruses are antagonizing these cellular factors in primates. The case of the receptor of mouse hepatitis virus CEACAM1 is of special interest. Positively selected sites identified in CEACAM1, F63, Y68, G75, G85, T86, Q88, and S127, are mapped to the virus-receptor interaction region (Fig. 2) (22). It follows that unknown coronaviruses that use CEACAM1 as their receptor might undergo an evolutionary arms race with primates for millions of years. Positively selected residues in ICAM1, the receptor of coxsackievirus and rhinovirus (both of them belong to the Enterovirus genus of the Picornaviridae family), also overlap the virus-receptor interaction region, suggesting enteroviruses have infected primates for millions of years. The only residue, K157, under positive selection in NTCP protein, the receptor of HBV, is crucial for HBV entry, consistent with the fact that HBV have been isolated from many primates. However, only a few crystal structures of receptors in complex with viral proteins have been resolved, limiting our interpretation of the significance of positively selected sites. On the other hand, our analyses provide valuable candidate sites of functional importance, which merit further experimental characterization. Nevertheless, our results indicate that pathogenic viruses represent important selective agents for the adaptive evolution of viral receptor genes in primates (10, 19).
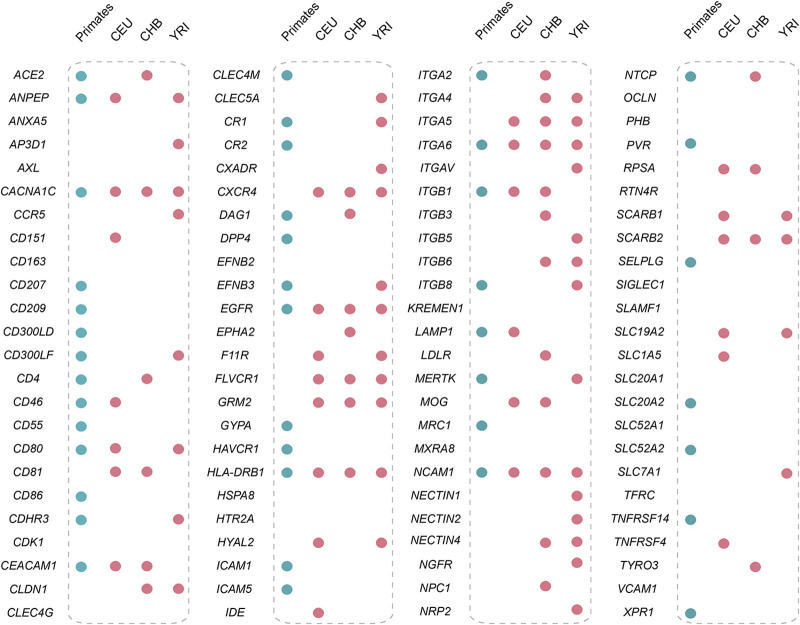
Population genetic analyses show that natural selection also acted on many viral receptor genes in human populations (Fig. 8). Our results suggest that diverse viruses, such as coronaviruses, enteroviruses, retroviruses, and many others, infected humans in the distant past and have shaped the evolution of viral receptor genes. The possibility that the positive selection acting on viral receptor genes is driven by factors other than viral infections cannot be formally excluded. However, when exploring the relationship between SNPs subject to natural selection and phenotypes of medical relevance, we found that a total of 27 SNPs with outlying |iHS| scores from 14 receptor genes are associated with phenotypes of medical relevance (Table 2). Moreover, most adaptive evolution in viral receptor genes appears to be driven by host-virus arms races in primates. Therefore, viruses are likely the most important factor driving the adaptive evolution of virial receptor genes in humans. We hypothesized that the host-virus conflicts drive the spread of some disease risk variants in human populations. We also found that a viral receptor gene might experience different selective pressures in different human populations, a pattern consistent with local adaptation. Given that some viruses are restricted in certain geographic regions (30), viruses may represent an important driver of local adaptation in human populations.
FIG 8.
Selection of viral receptors at macroevolutionary and microevolutionary levels. The blue-filled circles indicate the receptor genes subject to positive selection in primates (inferred by the PAML analyses), and the red-filled circles represent the presence of past selection in human populations.
TABLE 2.
Association between SNPs with selection signals and human phenotypes
| Gene | SNP | Population | |iHS| | Reference allele | Alternate allele | Risk allele | P value | Traita |
|---|---|---|---|---|---|---|---|---|
| AP3D1 | rs75483641 | CEU | 2.0193 | C | T | T | 8 × 10−10 | Male-pattern baldness |
| AXL | rs4802111 | CEU | −2.0706 | C | T | C | 1 × 10−21 | Heel bone mineral density |
| CHB | −2.5279 | C | T | C | 1 × 10−21 | Heel bone mineral density | ||
| CACNA1C | rs216026 | CEU | 2.8285 | T | G | T | 8 × 10−6 | Fractional exhaled nitric oxide |
| rs11062222 | CEU | −2.2341 | A | G | A | 9 × 10−6 | Short-term memory | |
| rs7301013 | CEU | −1.9981 | A | G | A | 3 × 10−12 | Heel bone mineral density | |
| CD300LF | rs9906320 | CEU | 2.0893 | G | A | A | 3 × 10−9 | Plateletcrit |
| CD80 | rs6804441 | CEU | 2.4862 | A | G | A | 3 × 10−16 | Systemic lupus erythematosus |
| HLA-DRB1 | rs9256938 | CEU | −3.1420 | C | A | A | 2 × 10−12 | Blood protein levels |
| YRI | −3.4035 | C | A | A | 2 × 10−12 | Blood protein levels | ||
| rs34075049 | YRI | −3.5202 | G | A | A | 9 × 10−6 | Neurofibrillary tangles | |
| ITGA4 | rs12988934 | CHB | 2.1631 | C | T | T | 2 × 10−14 | White blood cell types |
| rs1375493 | YRI | 2.0981 | G | A | A | 1 × 10−10 | Lymphocyte percentage of white cells | |
| rs10209150 | YRI | −2.0076 | A | G | G | 6 × 10−6 | Neurofibrillary tangles | |
| ITGB8 | rs10231365 | YRI | 2.8091 | C | T | T | 9 × 10−9 | Waist circumference adjusted for BMI |
| rs4721902 | YRI | 2.5428 | T | C | T | 3 × 10−8 | Waist-to-hip ratio adjusted for BMI | |
| rs2214442 | YRI | 2.5235 | A | G | A | 2 × 10−7 | Waist circumference adjusted for BMI | |
| rs3823974 | YRI | −2.4516 | T | C | C | 3 × 10−21 | Body fat distribution | |
| LDLR | rs688 | CEU | 2.0982 | C | T | C | 1 × 10−25 | Total cholesterol levels |
| 5 × 10−30 | Low-density lipoprotein cholesterol levels | |||||||
| rs5927 | CHB | −2.6008 | A | G | A | 9 × 10−6 | Cortisol levels | |
| MERTK | rs4374383 | CHB | −2.6794 | A | G | A | 1 × 10−9 | Hepatitis C-induced liver fibrosis |
| MOG | rs2252711 | CEU | 2.7605 | T | C | T | 4 × 10−7 | Pulmonary function |
| NCAM1 | rs2186709 | CHB | 4.5103 | A | G | G | 2 × 10−24 | Age of smoking initiation |
| rs7110863 | CHB | −3.9668 | A | G | G | 3 × 10−11 | Smoking cessation | |
| rs7111153 | CHB | −3.7995 | T | C | T | 4 × 10−18 | Self-reported math ability | |
| rs17115088 | CHB | 2.7324 | C | A | C | 2 × 10−42 | Heel bone mineral density | |
| SLC19A2 | rs1983546 | CEU | −2.0123 | A | G | G | 1 × 10−15 | QT interval |
| SLC7A1 | rs9508495 | CEU | 2.0207 | C | T | T | 1 × 10−9 | Systolic blood pressure |
| rs3803266 | CEU | 2.0069 | G | C | G | 7 × 10−10 | Medication use |
BMI, body mass index.
Our analyses come with several caveats. (i) The possibility that there are potential viral receptors not identified yet in control genes cannot be fully excluded. However, GO analyses show that control genes are not enriched in GO terms related to viral infection, indicating that most control genes are not viral receptors. (ii) Some viral receptors might be not authentic ones and might only represent statistical noise in our study. (iii) Adaptive evolution in viral receptors might be driven by nonviral factors. However, the elevated rate of adaptive evolution in viral receptor genes and the overlaps between positively selected sites and the interaction interfaces suggest virus-host conflicts do contribute significantly to the adaptive evolution of viral receptors.
MATERIALS AND METHODS
Viral receptors and their orthologs in primates.
The viral receptor information was retrieved from the literature (see Table S1 in the supplemental material) and the ViralZone database (https://viralzone.expasy.org) (13, 14). We assembled a total of 96 viral receptors involving 107 viruses (for details, see Table S1). We used 20 primate species as our focal organisms, including four New World monkeys, ten Old World monkeys, and six Hominoidea species. To identify orthologs in primates for each viral receptor gene, we used the BLAST algorithm with viral receptor proteins from human as queries. The orthologous genes were aligned based on the codon evolution model using PRANK (31).
GO annotations and control gene mapping.
Gene Ontology annotations of 96 viral receptors were retrieved from the Gene Ontology website (https://geneontology.org) in August 2018 (32). A total of 238 viral receptor-related GO biological process categories with raw P values of <10−5 were retrieved, providing a framework for control gene mapping (33). A gene was treated as a control for the virus receptor if it fulfills the following criteria: (i) at least 50% of items were identical to the virus receptor and (ii) the number of GO items does not exceed 150% of the virus receptor (33). If there are multiple candidate control genes for each receptor, we randomly selected a control gene from candidate control genes.
Evolutionary analyses in primates.
To quantify selective pressures for viral receptors, maximum likelihood analysis of dN/dS values was performed using the Codeml program in the PAML 4.9 package (17, 34). The phylogeny of primates was used as the input tree. For the branch model analyses, we estimated dN/dS values for all branches of the primate phylogeny with the free ratio model (model = 1). For the site model analyses, we used two codon substitution models (M7 and M8) to identify residues under positive selection. M7 is a neutral model that assumes a beta distribution of dN/dS values. M8 is a positive selection model that matched M7, except that it allows a dN/dS of >1. The likelihood ratio test then was used to assess whether the data significantly fit to the positive selection model M8 or the neutral model M7. Empirical Bayes analysis was used to calculate the posterior probability of the site classes. The M8 model was also used to quantify the dN/dS for each receptor gene across 20 primate species. The BUSTED method, implemented in HyPhy, was also used to test whether receptor genes had experienced episodic positive selection across the primate phylogeny (17).
Population genetic analyses.
The human polymorphism data were retrieved from pilot phase 3 in the 1000 Genomes Project (http://www.1000genomes.org) (35). We analyzed three representative populations: Utah residents with Northern and Western European ancestry (CEU), Han Chinese (CHB), and Yoruba in Ibadan, Nigeria (YRI). We first estimated the nucleotide diversity (π) and the Watterson’s estimator (θw) for each viral receptor gene. The frequency-based methods (Tajima’s D, Fu and Li’s F*, and Fu and Li’s D*) were performed using DNASP v6 (36–38). To evaluate statistical significance, we performed coalescent simulations for each test with 1,000 replicates. The integrated haplotype (|iHS|) score was calculated by selscan v1.1.0a with default settings (39). We used the SNPs that fulfill the following conditions: (i) biallelic and (ii) minor allele frequency in the three populations of greater than 5%. After generating the unstandardized |iHS| scores, we normalized the scores separately in each population with 10 equally sized allele frequency bins. We calculated both the mean and weighted Weir and Cockerham's fixation index (Fst) to quantify the population differentiation. Three pairwise population comparisons (CEU versus CHB, CEU versus CHB, and CHB versus YRI) were performed using VCFtools (40, 41).
GWAS.
Information on virus receptor genes from the genome-wide association studies (GWAS) was retrieved from the GWAS catalog website (https://www.ebi.ac.uk/gwas) and the GWAS central website (https://www.gwascentral.org). We retrieved the traits and disease-risk alleles associated with the SNPs that display extremely high |iHS| values.
Statistical analysis.
Data on the proportion of selected genes were analyzed using chi-square test on SPSS software. All other data were analyzed using R and were subsequently analyzed with Wilcoxon-Mann-Whitney test. The threshold for significance for all tests was set to 0.05. Significance was indicated by one asterisk (P < 0.05), two asterisks (P < 0.01), and n.s. (not significant; P > 0.05).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31922001 and 31701091) and the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Grove J, Marsh M. 2011. The cell biology of receptor-mediated virus entry. J Cell Biol 195:1071–1082. doi: 10.1083/jcb.201108131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coffin JM. 2013. Virions at the gates: receptors and the host-virus arms race. PLoS Biol 11:e1001574. doi: 10.1371/journal.pbio.1001574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demogines A, Abraham J, Choe H, Farzan M, Sawyer SL. 2013. Dual host-virus arms races shape an essential housekeeping protein. PLoS Biol 11:e1001571. doi: 10.1371/journal.pbio.1001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaelber JT, Demogines A, Harbison CE, Allison AB, Goodman LB, Ortega AN, Sawyer SL, Parrish CR. 2012. Evolutionary reconstructions of the transferrin receptor of Caniforms supports canine parvovirus being a re-emerged and not a novel pathogen in dogs. PLoS Pathog 8:e1002666. doi: 10.1371/journal.ppat.1002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baranowski E, Ruiz-Jarabo CM, Domingo E. 2001. Evolution of cell recognition by viruses. Science 292:1102–1105. doi: 10.1126/science.1058613. [DOI] [PubMed] [Google Scholar]
- 6.Daugherty MD, Malik HS. 2012. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet 46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- 7.Sironi M, Cagliani R, Forni D, Clerici M. 2015. Evolutionary insights into host-pathogen interactions from mammalian sequence data. Nat Rev Genet 16:224–236. doi: 10.1038/nrg3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Valen L. 1973. A new evolutionary law. Evol Theory 1:1–30. [Google Scholar]
- 9.Demogines A, Farzan M, Sawyer SL. 2012. Evidence for ACE2-utilizing coronaviruses (CoVs) related to severe acute respiratory syndrome CoV in bats. J Virol 86:6350–6353. doi: 10.1128/JVI.00311-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacquet S, Pons JB, De Bernardo A, Ngoubangoye B, Cosset FL, Régis C, Etienne L, Pontier D. 2018. Evolution of hepatitis B virus receptor NTCP reveals differential pathogenicity and species-specificities of hepadnaviruses in primates, rodents and bats. J Virol 93:e01738-18. doi: 10.1128/JVI.01738-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pontremoli C, Forni D, Cagliani R, Filippi G, De Gioia L, Pozzoli U, Clerici M, Sironi M. 2016. Positive selection drives evolution at the host-filovirus interaction surface. Mol Biol Evol 33:2836–2847. doi: 10.1093/molbev/msw158. [DOI] [PubMed] [Google Scholar]
- 12.Meyerson NR, Rowley PA, Swan CH, Le DT, Wilkerson GK, Sawyer SL. 2014. Positive selection of primate genes that promote HIV-1 replication. Virology 454-455:291–298. doi: 10.1016/j.virol.2014.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Zhu Z, Chen W, Cai Z, Xu B, Tan Z, Wu A, Ge X, Guo X, Tan Z, Xia Z, Zhu H, Jiang T, Peng Y. 2019. Cell membrane proteins with high N-glycosylation, high expression and multiple interaction partners are preferred by mammalian viruses as receptors. Bioinformatics 35:723–728. doi: 10.1093/bioinformatics/bty694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hulo C, de Castro E, Masson P, Bougueleret L, Bairoch A, Xenarios I, Le Mercier P. 2011. ViralZone: a knowledge resource to understand virus diversity. Nucleic Acids Res 39:D576–D582. doi: 10.1093/nar/gkq901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLaren PJ, Gawanbacht A, Pyndiah N, Krapp C, Hotter D, Kluge SF, Götz N, Heilmann J, Mack K, Sauter D, Thompson D, Perreaud J, Rausell A, Munoz M, Ciuffi A, Kirchhoff F, Telenti A. 2015. Identification of potential HIV restriction factors by combining evolutionary genomic signatures with functional analyses. Retrovirology 12:41. doi: 10.1186/s12977-015-0165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Z, Bielawski JP. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol Evol 15:496–503. doi: 10.1016/s0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murrell B, Weaver S, Smith MD, Wertheim JO, Murrell S, Aylward A, Eren K, Pollner T, Martin DP, Smith DM, Scheffler K, Kosakovsky Pond SL. 2015. Gene-wide identification of episodic selection. Mol Biol Evol 32:1365–1371. doi: 10.1093/molbev/msv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang CC, Venturi M, Majeed S, Moore MJ, Phogat S, Zhang MY, Dimitrov DS, Hendrickson WA, Robinson J, Sodroski J, Wyatt R, Choe H, Farzan M, Kwong PD. 2004. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci U S A 101:2706–2711. doi: 10.1073/pnas.0308527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeuchi JS, Fukano K, Iwamoto M, Tsukuda S, Suzuki R, Aizaki H, Muramatsu M, Wakita T, Sureau C, Watashi K. 2018. A single adaptive mutation in sodium taurocholate cotransporting polypeptide induced by hepadnaviruses determines virus species specificity. J Virol 93:e01432-18. doi: 10.1128/JVI.01432-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Persson BD, Reiter DM, Marttila M, Mei YF, Casasnovas JM, Arnberg N, Stehle T. 2007. Adenovirus type 11 binding alters the conformation of its receptor CD46. Nat Struct Mol Biol 14:164–166. doi: 10.1038/nsmb1190. [DOI] [PubMed] [Google Scholar]
- 21.Plevka P, Hafenstein S, Harris KG, Cifuente JO, Zhang Y, Bowman VD, Chipman PR, Bator CM, Lin F, Medof ME, Rossmann MG. 2010. Interaction of decay-accelerating factor with echovirus 7. J Virol 84:12665–12674. doi: 10.1128/JVI.00837-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng G, Sun D, Rajashankar KR, Qian Z, Holmes KV, Li F. 2011. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc Natl Acad Sci U S A 108:10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao C, Bator CM, Bowman VD, Rieder E, He Y, Hébert BT, Bella J, Baker TS, Wimmer E, Kuhn RJ, Rossmann MG. 2001. Interaction of coxsackievirus A21 with its cellular receptor, ICAM-1. J Virol 75:2444–2451. doi: 10.1128/JVI.75.5.2444-2451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang P, Mueller S, Morais MC, Bator CM, Bowman VD, Hafenstein S, Wimmer E, Rossmann MG. 2008. Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses. Proc Natl Acad Sci U S A 105:18284–18289. doi: 10.1073/pnas.0807848105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li F. 2015. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol 89:1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Wang Z, Liu R, Shuai L, Wang X, Luo J, Wang C, Chen W, Wang X, Ge J, He X, Wen Z, Bu Z. 2018. Metabotropic glutamate receptor subtype 2 is a cellular receptor for rabies virus. PLoS Pathog 14:e1007189. doi: 10.1371/journal.ppat.1007189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thoulouze MI, Lafage M, Schachner M, Hartmann U, Cremer H, Lafon M. 1998. The neural cell adhesion molecule is a receptor for rabies virus. J Virol 72:7181–7190. doi: 10.1128/JVI.72.9.7181-7190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chabot DJ, Chen H, Dimitrov DS, Broder CC. 2000. N-linked glycosylation of CXCR4 masks coreceptor function for CCR5-dependent human immunodeficiency virus type 1 isolates. J Virol 74:4404–4413. doi: 10.1128/jvi.74.9.4404-4413.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Endres MJ, Clapham PR, Marsh M, Ahuja M, Turner JD, McKnight A, Thomas JF, Stoebenau-Haggarty B, Choe S, Vance PJ, Wells TN, Power CA, Sutterwala SS, Doms RW, Landau NR, Hoxie JA. 1996. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell 87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 30.Holmes EC. 2008. Evolutionary history and phylogeography of human viruses. Annu Rev Microbiol 62:307–328. doi: 10.1146/annurev.micro.62.081307.162912. [DOI] [PubMed] [Google Scholar]
- 31.Löytynoja A, Goldman N. 2008. A model of evolution and structure for multiple sequence alignment. Philos Trans R Soc Lond B Biol Sci 363:3913–3919. doi: 10.1098/rstb.2008.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gene Ontology Consortium. 2015. Gene Ontology Consortium: going forward. Nucleic Acids Res 43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enard D, Cai L, Gwennap C, Petrov DA. 2016. Viruses are a dominant driver of protein adaptation in mammals. Elife 5:e12469. doi: 10.7554/eLife.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 35.Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MH, Konkel MK, Malhotra A, Stütz AM, Shi X, Casale FP, Chen J, Hormozdiari F, Dayama G, Chen K, Malig M, Chaisson MJP, Walter K, Meiers S, Kashin S, Garrison E, Auton A, Lam HYK, Mu XJ, Alkan C, Antaki D, Bae T, Cerveira E, Chines P, Chong Z, Clarke L, Dal E, Ding L, Emery S, Fan X, Gujral M, Kahveci F, Kidd JM, Kong Y, Lameijer EW, McCarthy S, Flicek P, Gibbs RA, Marth G, Mason CE, Menelaou A, Muzny DM, Nelson BJ, Noor A, Parrish NF, Pendleton M, Quitadamo A, Raeder B, Schadt EE, Romanovitch M, Schlattl A, Sebra R, Shabalin AA, Untergasser A, Walker JA, Wang M, Yu F, Zhang C, Zhang J, Zheng-Bradley X, Zhou W, Zichner T, Sebat J, Batzer MA, McCarroll SA, 1000 Genomes Project Consortium, Mills RE, Gerstein MB, Bashir A, Stegle O, Devine SE, Lee C, Eichler EE, Korbel JO. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu YX, Li WH. 1993. Statistical tests of neutrality of mutations. Genetics 133:693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tajima F. 1993. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics 135:599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 39.Szpiech ZA, Hernandez RD. 2014. Selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol Biol Evol 31:2824–2827. doi: 10.1093/molbev/msu211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, 1000 Genomes Project Analysis Group. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.