Sirt2, a predominant cytoplasmic α-tubulin deacetylase, promotes the growth of hepatocellular carcinoma; indeed, HBV replication increases Sirt2 expression, and overexpression of Sirt2 is associated with hepatic fibrosis and epithelial-to-mesenchymal transition. Increased amounts of Sirt2 isoforms 1, 2, and 5 upon HBV replication might further upregulate HBV replication, leading to a vicious cycle of virus replication/disease progression. However, we show here that catalytically inactive nuclear Sirt2.5 antagonizes the effects of Sirt2.1 and Sirt2.2 on HBV replication, thereby inhibiting cccDNA level, transcription of cccDNA, and subsequent synthesis of replicative intermediate DNA. More Sirt2.5 was recruited to cccDNA than Sirt2.1, thereby increasing epigenetic modification by depositing transcriptional repressive markers, possibly through direct and/or indirect association with histone lysine methyltransferases, such as SETDB1, SUV39H1, EZH2, and/or PR-Set7, which represses HBV transcription. Thus, Sirt2.5 might provide a functional cure for HBV by silencing the transcription of HBV.
KEYWORDS: epigenetic modifications, histone lysine methyltransferases, Sirt2 isoform 5, cccDNA, hepatitis B virus
ABSTRACT
Sirtuin 2 (Sirt2), an NAD+-dependent protein deacetylase, deacetylates tubulin, AKT, and other proteins. Previously, we showed that Sirt2 isoform 1 (Sirt2.1) increased replication of hepatitis B virus (HBV). Here, we show that HBV replication upregulates the expression of Sirt2 primary and alternatively spliced transcripts and their respective isoforms, 1, 2, and 5. Since Sirt2 isoform 5 (Sirt2.5) is a catalytically inactive nuclear protein with a spliced-out nuclear export signal (NES), we speculated that its different localization affects its activity. The overexpression of Sirt2.5 reduced expression of HBV mRNAs, replicative intermediate DNAs, and covalently closed circular DNA (cccDNA), an activity opposite that of Sirt2.1 and Sirt2.2. Unlike the Sirt2.1-AKT interaction, the Sirt2.5-AKT interaction was weakened by HBV replication. Unlike Sirt2.1, Sirt2.5 activated the AKT/GSK-3β/β-catenin signaling pathway very weakly and independently of HBV replication. When the NES and an N-terminal truncated catalytic domain were added to the Sirt2.5 construct, it localized in the cytoplasm and increased HBV replication (like Sirt2.1 and Sirt2.2). Chromatin immunoprecipitation assays revealed that more Sirt2.5 was recruited to cccDNA than Sirt2.1. The recruitment of histone lysine methyltransferases (HKMTs), such as SETDB1, SUV39H1, EZH2, and PR-Set7, and their respective transcriptional repressive markers, H3K9me3, H3K27me3, and H4K20me1, to cccDNA also increased in Sirt2.5-overexpressing cells. Among these, the Sirt2.5–PR-Set7 and –SETDB1 interactions increased upon HBV replication. These results demonstrate that Sirt2.5 reduces cccDNA levels and viral transcription through epigenetic modification of cccDNA via direct and/or indirect association with HKMTs, thereby exhibiting anti-HBV activity.
IMPORTANCE Sirt2, a predominant cytoplasmic α-tubulin deacetylase, promotes the growth of hepatocellular carcinoma; indeed, HBV replication increases Sirt2 expression, and overexpression of Sirt2 is associated with hepatic fibrosis and epithelial-to-mesenchymal transition. Increased amounts of Sirt2 isoforms 1, 2, and 5 upon HBV replication might further upregulate HBV replication, leading to a vicious cycle of virus replication/disease progression. However, we show here that catalytically inactive nuclear Sirt2.5 antagonizes the effects of Sirt2.1 and Sirt2.2 on HBV replication, thereby inhibiting cccDNA level, transcription of cccDNA, and subsequent synthesis of replicative intermediate DNA. More Sirt2.5 was recruited to cccDNA than Sirt2.1, thereby increasing epigenetic modification by depositing transcriptional repressive markers, possibly through direct and/or indirect association with histone lysine methyltransferases, such as SETDB1, SUV39H1, EZH2, and/or PR-Set7, which represses HBV transcription. Thus, Sirt2.5 might provide a functional cure for HBV by silencing the transcription of HBV.
INTRODUCTION
Hepatitis B virus (HBV) is a noncytopathic hepatotrophic DNA virus with a partially double-stranded relaxed circular (RC) DNA genome of 3.2 kb, which is converted to a covalently closed circular DNA (cccDNA) upon entry to the nucleus of a hepatocyte (1). This cccDNA, which is organized into a minichromosome by histones and nonhistone viral and cellular proteins, accumulates in the nucleus of transfected or infected cells (2–4). This minichromosome (a transcriptional template) transcribes four RNA species (3.5-, 2.4-, 2.1-, and 0.7-kb viral RNA transcripts), which are then transported to and translated in the cytoplasm to produce viral polymerase, HBc (core, C), viral HBs (surface, S), and HBx (X) proteins (5–7).
Although effective vaccines are available, HBV (which causes acute and chronic hepatitis) is a major health problem worldwide (5). Until now, two principal therapeutic strategies have been available to treat chronic hepatitis B: interferon alpha (IFN-α) and nucleos(t)ide analogues (8–10). These therapies reduce the viral load but cannot cure the disease due to the persistence of cccDNA. Since currently available therapeutics fail to eliminate or permanently silence cccDNA transcription, the virus can lay dormant and rebound after treatment (9–11).
Sirtuin 1 (Sirt1) represses HBV transcription by recruiting cccDNA during and after IFN-α treatment (12). cccDNA-recruited Sirt1 and histone deacetylase 1 (HDAC1) also inhibit the transcription of cccDNA in the absence of HBx (3). Transcription of HBV cccDNA is restricted by the cooperative action of Sirt3 and histone lysine methyltransferase (HKMT) suppressor of variation 3 to 9 homolog 1 (SUV39H1) and SET domain-containing 1A (SETD1A) (13). These HDAC1 and Sirt proteins belong to the HDAC superfamily, which comprises 18 human HDACs, namely, HDAC1 to -11 and Sirt1 to -7; these are grouped into four classes based on their homology with yeast proteins (14, 15). Sirt1 to -7 (class III HDACs) share a similar catalytic domain (CD) and use NAD+ as a cofactor for protein deacetylase catalytic activity; however, they differ with respect to substrate specificity and subcellular localization (15–17). Sirts also regulate aging, apoptosis, transcription, inflammation, and oxidative stress (18). Unlike Sirt1 and Sirt3 (19), the closely related Sirt2 (isoform 1; Sirt2.1) is expressed predominantly in the cytoplasm and increases HBV replication (3, 12, 13, 20). However, recruitment of Sirt2 to cccDNA, or the effect of Sirt2 on cccDNA transcription, has not been studied.
Sirt2 isoforms 2 and 5 (Sirt2.2 and Sirt2.5) in humans occur through alternative splicing of a Sirt2 primary transcript (21). Sirt2.1 and Sirt2.2 harbor a leucine-rich nuclear export signal (NES) and are localized mainly in the cytoplasm (20–22), where they regulate microtubule deacetylation, myelination in the central and peripheral nervous system, and gluconeogenesis (23–25). The third Sirt2 isoform, isoform 5, has spliced out exons 2 to 4, including an NES, and an N-terminal CD; therefore, it resides primarily in the nucleus (Fig. 1C) (20). This isoform also lacks deacetylase activity toward known Sirt2 substrates, possibly due to the spliced-out exon 2 to 4 region (21). Although Rack et al. (21) suggest an activity-independent nuclear function of Sirt2.5, its function has not been identified yet.
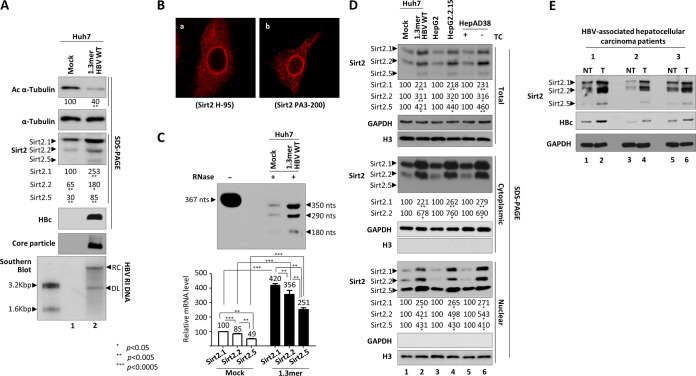
FIG 1.
Expression of three isoforms of endogenous Sirt2 transcripts and proteins increases upon HBV replication. (A) Expression of endogenous Sirt2 protein increases in HBV-replicating cells. Huh7 cells were mock transfected (lane 1) or transfected with 4 μg of 1.3mer HBV WT (ayw) (lane 2). Lysates were prepared at 72 h posttransfection. Prepared lysates were subjected to SDS-PAGE and immunoblotting to detect proteins or 1% native agarose gel electrophoresis followed by immunoblotting to detect core particles. Endogenous Sirt2, HBc, acetylated α-tubulin, and α-tubulin proteins were detected using rabbit polyclonal anti-Sirt2 PA3-200 (1:1,000), rabbit polyclonal anti-HBc (1:1,000) (65), mouse monoclonal anti-acetylated α-tubulin (1:1,000; T 6793; Sigma-Aldrich), and mouse monoclonal anti-α-tubulin (1:1,000; sc-8035; Santa Cruz) antibodies, respectively. Relative levels of acetylated α-tubulin and three isoforms of Sirt2 proteins were measured using ImageJ 1.46r. Tubulin was used as a loading control. To measure viral DNA synthesis, Southern blot analysis was performed. HBV DNA isolated from core particles was separated by agarose gel electrophoresis, transferred to a nylon membrane, and subjected to autoradiography after hybridization to 32P-labeled random-primed probe specific for the full-length HBV. Shown are HBV replicative intermediate, double-stranded linear, and partially double-stranded relaxed circular DNAs (HBV RI DNA, DL, and RC, respectively). (B) Comparison of anti-Sirt2 H-95 and anti-Sirt2 PA3-200 antibodies to show localization of Sirt2.5 in the nucleus and Sirt2.1 and Sirt2.2 in the cytoplasm. Endogenous Sirt2 levels in Huh7 cells were detected by anti-Sirt2 H-95 (1:300) (a) and PA3-200 (1:300) (b) antibodies. Digital images of stained cells were captured under a confocal microscope (LSM710; Zeiss, Germany). Data are representative of three independent experiments. (C) RPA to detect three transcripts of Sirt2 isoforms in HBV replicating cells. An in vitro-transcribed radiolabeled antisense RNA probe (367 nt) was hybridized overnight at 50°C with total RNA from mock-transfected and 4 μg of 1.3mer HBV WT-transfected Huh7 cells. Following RNase A/T1 (EN0551; Thermo Fisher Scientific) digestion, protected RNAs (Sirt2.1, 350 nt; Sirt2.2, 290 nt; and Sirt2.5, 180 nt) were electrophoresed in a 5% polyacrylamide–8 M urea gel and visualized by autoradiography. (D) HBV replication increases expression of Sirt2.1, Sirt2.2, and Sirt2.5 proteins. Transiently mock- and 1.3mer HBV WT-transfected (4 μg) Huh7 cells (lanes 1 and 2), HepG2 and HBV replicating stable HepG2.2.15 cells (lanes 3 and 4), and tetracycline-treated and -removed HepAD38 cells (lanes 5 and 6) were cultured for 72 h. Total lysates and cytoplasmic and nuclear fractions were prepared from whole cells by differential centrifugation (2, 69) and subjected to SDS-PAGE and immunoblotting with PA3-200 anti-Sirt2 (1:1,000) antibody. The purity of the cytoplasmic and nuclear fractions was examined by anti-GAPDH (1:5,000; sc-32233; Santa Cruz) and anti-histone H3 (1:5,000; ab1791; Abcam) antibodies, respectively. (E) Expression of Sirt2 in paired tumor and nontumor liver biopsy specimens from HBV-associated HCC patients. Protein lysates from biopsy specimens were prepared in a weight per volume ratio in mPER buffer. SDS-PAGE and immunoblotting were performed as described above. NT, adjacent nontumor; T, tumor. Data are presented as the means and standard deviations (SD) from three independent experiments. Statistical significance was evaluated using Student's t test. *, P < 0.05; **, P < 0.005; and ***, P < 0.0005; each relative to the control.
HKMTs such as SETD1A (also known as SET1A) or SUV39H1 and SETDB1 (SET domain bifurcated 1) activates or silence, respectively, HBV cccDNA transcription (13, 26–28). Some HKMTs act on chromatin through cooperative action with Sirts; this activity involves the deacetylation of both histone and nonhistone proteins (13, 29, 30). For example, cccDNA recruited Sirt3 deacetylate histone H3 lysine 9 (H3K9), thereby increasing the recruitment of SUV39H1 to cccDNA; this leads to methylation of H3K9 (H3K9me1) to yield trimethyl-H3K9 (H3K9me3), which represses transcription of cccDNA (13). Although Sirt2 can bind to HKMT PR-Set7 (also known as SET8 or KMT5A) to deacetylate H4K16ac and PR-Set7 at acetyl-K90 to regulate monomethylation on H4K20 (H4K20me1) (30), it has never reported whether either PR-Set7 or Sirt2 acts on HBV cccDNA.
Accumulating evidence indicates that epigenetic mechanisms affect the persistence of HBV (3, 13, 26–28, 31–34); therefore, an epigenetic therapy may be an attractive option to treat chronic hepatitis B infection (28). A novel strategy that targets epigenetic modifications was used successfully to treat leukemia (35). However, use of general epigenetic modifiers to control cccDNA would be risky due to potential harmful effects on cell homeostasis (9, 10).
Here, we examined the roles of catalytically inactive Sirt2.5 (21) on the HBV life cycle. Although overexpression of Sirt2.1 enhanced HBV replication (20), we found that the overexpression of Sirt2.5 inhibited the synthesis of HBV RNA and DNA. A Sirt2.5 mutant in which the NES and N-terminal truncated CD were restored localized in the cytoplasm and increased HBV replication, much like Sirt2.1 and Sirt2.2, demonstrating that the N-terminal 40 amino acids are not essential for activity of Sirt2.1 and Sirt2.2. Furthermore, we demonstrated that overexpression of Sirt2.5 reduces cccDNA levels, that Sirt2.5 recruited onto cccDNA to a greater extent than Sirt2.1, and that Sirt2.5 induces epigenetic modifications through the Sirt2.5-SETDB1 and –PR-Set7 interactions. Overexpression of Sirt2.5 in HBV-infected cells increased recruitment of SUV39H1 and EZH2 (enhancer of zeste homolog 2) without interacting with them directly. Together with these four HKMTs, the deposition of respective transcriptional repressive markers, such as H3K9me3, H3K27me3, and H4K20me1, increased, possibly through cooperative actions. Taken together, these results indicate that Sirt2.5 exhibits anti-HBV activity by silencing transcription of cccDNA. This unprecedented functional role of catalytically inactive nuclear Sirt2.5 may open up new avenues to achieving a functional cure for HBV infection.
RESULTS
HBV replication increases expression of endogenous Sirt2.1 to Sirt2.5 transcript and protein.
Recently, we reported that HBV replication upregulates endogenous expression of Sirt2 mRNA and protein, leading to tubulin deacetylation (20). However, we did not discriminate expression of the three Sirt2 isoforms with respect to their effects on HBV replication (20). Therefore, we examined the expression of the three Sirt2 isoforms in mock- and 1.3mer HBV wild-type (WT)-transfected Huh7 cells (Fig. 1A). As expected, HBV replicative intermediate (RI) DNAs, such as RC and double-stranded linear (DL) DNAs, were detected in 1.3mer HBV WT-transfected Huh7 cells (Fig. 1A, bottom, lane 1 versus 2). Previously, we used an anti-Sirt2 H-95 antibody (1:1,000; Santa Cruz Biotechnology) to detect Sirt2 proteins; this antibody was raised against amino acids 1 to 95 of Sirt2 and as such can only detect the cytoplasmic isoforms 1 and 2 (Sirt2.1 and Sirt2.2), not the nuclear isoform 5 (Sirt2.5), which lacks amino acids 6 to 76 of Sirt2.1 (20). To detect all three isoforms of Sirt2 (Fig. 1A, third panel), we used an anti-Sirt2 PA3-200 antibody (1:1,000; Thermo Fisher Scientific), which was raised against amino acids 341 to 352 of Sirt2. When we examined endogenous levels of Sirt2 protein in mock- and HBV WT-transfected Huh7 cells (Fig. 1A, third panel), we found that expression of all three isoforms of Sirt2 (Sirt2.1, Sirt2.2, and Sirt2.5) in HBV-replicating cell was higher than that in mock control cells (Fig. 1A; third panel, lane 1 versus 2). Consistent with this, α-tubulin in HBV-replicating cells was deacetylated to a greater extent than that in mock control cells (20) (Fig. 1A, top, lane 1 versus 2).
To further verify the ability of anti-Sirt2 antibodies to detect all three Sirt2 isoforms, we performed immunofluorescence analysis using confocal microscopy (Fig. 1B). As expected, the H-95 antibody (1:300) detected mainly cytoplasmic Sirt2, most likely Sirt2.1 and Sirt2.2 (Fig. 1B, a), whereas the PA3-200 antibody (1:300) detected cytoplasmic and nuclear Sirt2, most likely Sirt2.1, Sirt2.2, and Sirt2.5 (Fig. 1B, b).
Since expression of all three isoforms of Sirt2 increased upon HBV replication, we next conducted RNase protection assays (RPA) to examine the expression of mRNA encoding Sirt2 isoforms in mock- and 1.3mer HBV WT-transfected Huh7 cells (Fig. 1C). Protected Sirt2.1, Sirt2.2, and Sirt2.5 mRNAs comprised 350, 290, and 180 nucleotides (nt), respectively. Consistent with Fig. 1A, expression of all three isoforms of Sirt2 mRNA was higher in HBV WT-transfected cells than in mock-transfected cells (Fig. 1C).
Since Sirt2.1 and Sirt2.2 are localized mainly in the cytoplasm and Sirt2.5 localizes in the nucleus (Fig. 1B) (20, 21), we further examined expression in cytoplasmic and nuclear fractions from HBV-replicating cells (Fig. 1D). Total, cytoplasmic, and nuclear fractions were obtained from HBV-replicating stable HepG2.2.15 (36) and HepAD38 (37) cells and from mock- or transiently transfected Huh7 cells with 1.3mer HBV WT (Fig. 1D). Consistent with Fig. 1A, HBV replication increased the expression of all three isoforms of Sirt2 protein (Fig. 1D, top). Consistent with the results shown in Fig. 1B and previous reports (20, 21), Sirt2.1 and Sirt2.2 localized predominantly in the cytoplasm, whereas Sirt2.5 accumulated in the nucleus (Fig. 1D). These results demonstrate that HBV replication enhances the expression of all three isoforms of Sirt2 mRNA and protein.
We tried to compare the Sirt2 protein levels in biopsied tumor and adjacent nontumor liver tissues from three HBV-associated hepatocellular carcinoma (HCC) patients (Fig. 1E). HBV DNAs from HCC patients 1, 2, and 3 were identified as 6,400,000, 287,000, and 939,000 IU/ml, respectively. Immunoblot analysis revealed that although three isoforms of Sirt2 were expressed in both tumor and adjacent nontumor liver tissues, HBV-associated HCC tumors expressed higher levels of Sirt2 than the paired adjacent nontumor (Fig. 1E, top, lane 1 versus 2, 3 versus 4, and 5 versus 6). When HBc expression levels from biopsied tumor and adjacent nontumor liver tissues were compared, higher levels of HBc were detected in tumor than adjacent nontumor, which showed expression patterns similar to those of Sirt2 (Fig. 1E, second panel).
Overexpression of Sirt2.5 reduces levels of HBV transcripts and cccDNA.
Recently, we reported that the overexpression of Sirt2.1 increases HBV transcription (20). Therefore, we hypothesized that nuclear Sirt2.5 regulates HBV transcription, since different localizations imply different activities. To examine the effect of Sirt2.5 overexpression on HBV promoter activity, we conducted a luciferase reporter assay in Huh7 and HepG2 cells (Fig. 2A). Consistent with a previous report, we found that the promoter activity of all HBV enhancers and promoters (enhancer I/X promoter [EnhI/Xp], enhancer II/Core promoter [EnhII/Cp], preS1p, and preS2p) was upregulated upon overexpression of Sirt2.1 (Fig. 2A) (20). However, upon overexpression of Sirt2.5, the promoter activity of all HBV enhancers and promoters was downregulated (Fig. 2A).
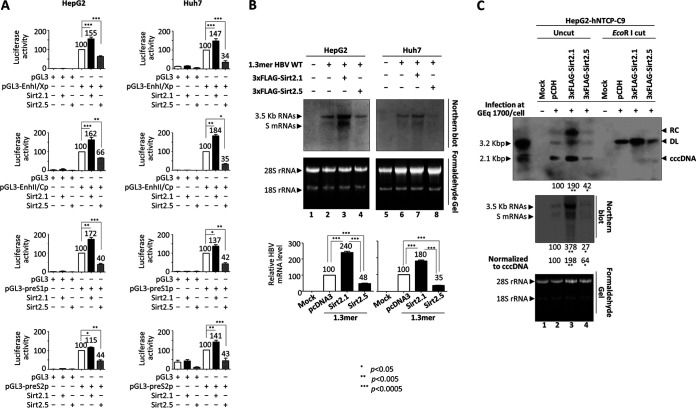
FIG 2.
Overexpression of Sirt2.5 reduces HBV viral transcription and cccDNA levels. (A) Luciferase reporter assay shows reduced HBV enhancer and promoter activity upon overexpression of Sirt2.5. HepG2 (first row) and Huh7 (second row) cells were transiently transfected with 2 μg of the indicated luciferase reporter vectors in the presence of 3×FLAG-Sirt2.1 (2 μg) or 3×FLAG-Sirt2.5 (2 μg). At 72 h posttransfection, lysates were prepared (E153A; Promega) and luciferase activity was measured. Luciferase activity relative to the respective control luciferase reporter vectors is presented. (B) Northern blotting to show the reduced levels of HBV transcripts upon overexpression of Sirt2.5. HepG2 (lanes 1 to 4) and Huh7 (lanes 5 to 8) cells were transiently mock transfected (lanes 1 and 5) or transfected with 4 μg of 1.3mer HBV WT (ayw) (lanes 2 and 6), 4 μg of 1.3mer HBV WT (ayw) plus 4 μg of 3×FLAG-Sirt2.1 construct (lanes 3 and 7), or 4 μg of 1.3mer HBV WT (ayw) plus 4 μg of 3×FLAG-Sirt2.5 construct (lanes 4 and 8). The amount of transfected DNA was adjusted using pcDNA3. At 72 h posttransfection, total RNA was extracted for Northern blotting; 20 μg of total RNA was separated by 1.2% formaldehyde gel electrophoresis, transferred to nylon membranes, hybridized, and subjected to autoradiography as described in the legend of Fig. 1A for Southern blotting. The 3.5-kb pgRNA and the 2.1- and 2.4-kb mRNAs encoding the S protein are indicated. Ribosomal RNAs (28S and 18S rRNAs) are included as a loading control. (C) Overexpression of Sirt2.5 reduced the amounts of HBV cccDNA and viral mRNA in HBV-infected cells. HepG2-hNTCP-C9 cells transduced with pCDH (lane 2), 3×FLAG-Sirt2.1 (lane 3), or 3×FLAG-Sirt2.5 (lane 4) were plated on collagen-coated 6-well plates, mock infected (lane 1) or infected with 1.7 × 103 GEq of HBV per cell (lanes 2 to 4), and incubated for 5 days. Total RNA then was extracted and Northern blotting was performed as described above (third panel). At 9 days postinfection at 100% confluence, cccDNA was extracted through Hirt protein-free DNA extraction procedure, with minor modifications (2, 38). Southern blotting of cccDNA without linearization (top) or following linearization with EcoRI (second panel) was performed. Data are presented as the means and SD from three (A and C) or four (B) independent experiments. Statistical significance was evaluated using Student's t test. *, P < 0.05; **, P < 0.005; ***, P < 0.0005; each relative to the control.
Since the activity of all HBV promoters and enhancers fell upon the overexpression of Sirt2.5 (Fig. 2A), we performed Northern blotting of HepG2 or Huh7 cells transfected with 1.3mer HBV WT or cotransfected with 1.3mer HBV WT plus Sirt2.1 or Sirt2.5 (Fig. 2B). Consistent with a previous report (20) and the data shown in Fig. 2A, the expression of pgRNA and subgenomic S mRNAs increased significantly upon overexpression of Sirt2.1 (Fig. 2B, lane 2 versus 3 and 6 versus 7). However, the expression of HBV pgRNA and subgenomic S mRNAs fell significantly upon overexpression of Sirt2.5 (Fig. 2B, lane 2 versus 4 and 6 versus 8), demonstrating that overexpression of Sirt2.5 inhibits HBV transcription.
To further verify the above-described findings, we transduced HepG2-hNTCP-C9 cells with a lentivirus encoding control pCDH, 3×FLAG-Sirt2.1, or 3×FLAG-Sirt2.5 to generate control, Sirt2.1-, or Sirt2.5-overexpressing cells. We then infected control, Sirt2.1-, or Sirt2.5-overexpressing HepG2-hNTCP-C9 cells with 1,700 genome equivalents (GEq) of HBV (Fig. 2C). As expected, the amount of HBV pgRNA and subgenomic S mRNAs in Sirt2.1-overexpressing HBV-infected HepG2-hNTCP-C9 cells increased (Fig. 2C, third panel, lane 2 versus 3), whereas that in Sirt2.5-overexpressing HBV-infected HepG2-hNTCP-C9 cells decreased (Fig. 2C, third panel, lane 2 versus 4).
Since the overexpression of Sirt2.1 increased pgRNA and S mRNAs and overexpression of Sirt2.5 decreased pgRNA and S mRNAs, we asked whether cccDNA levels are affected by overexpression of either Sirt2.1 or Sirt2.5. Since the activity of all HBV enhancers and promoters was upregulated or downregulated by overexpression of Sirt2.1 or Sirt2.5, respectively, we hypothesized that cccDNA levels were altered. Therefore, we examined the expression of HBV cccDNA in HBV-infected cells by Southern blotting (2, 38). To our surprise, Sirt2.1-overexpressing cells expressed higher levels of cccDNA than expected (Fig. 2C, first and second panels, lane 2 versus 3), whereas Sirt2.5-overexpressing cells expressed lower levels than expected (Fig. 2C, first and second panels, lane 2 versus 4). However, when we normalized HBV transcripts from cccDNA, we found that transcriptional activity was increased and decreased significantly by Sirt2.1 and Sirt2.5, respectively (Fig. 2C, third panel, lane 2 versus 3 versus 4). Thus, we conclude that Sirt2.1 and Sirt2.5 not only upregulate and downregulate, respectively, cccDNA levels significantly but also further upregulate and downregulate viral transcription, respectively.
Overexpression of Sirt2.5 inhibits HBV DNA synthesis.
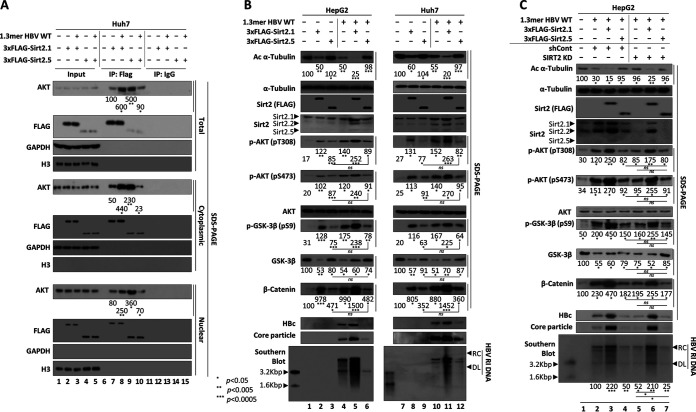
Since the levels of cccDNA and HBV RNAs in Sirt2.5-overexpressing HBV-infected HepG2-hNTCP-C9 cells decreased (Fig. 2C), we reasoned that HBV DNA synthesis is also affected by Sirt2.5 (Fig. 3). To test this, we transiently transfected HepG2 (Fig. 3A, lanes 1 to 4) or Huh7 (Fig. 3A, lanes 5 to 8) cells with 1.3mer HBV WT or cotransfected them with 1.3mer HBV WT plus 3×FLAG-Sirt2.1 or 3×FLAG-Sirt2.5. Consistent with our previous report (20) and with the above-described results (Fig. 1A, top, lane 2), transfection of the 1.3mer HBV WT resulted in α-tubulin deacetylation (Fig. 3A, top, lane 1 versus 2 and 5 versus 6). Upon overexpression of Sirt2.1, HBV replication (as shown by levels of HBc protein, core particle formation, and DNA synthesis) increased (Fig. 3A, fifth, sixth, and seventh panels, lane 2 versus 3 and lane 6 versus 7), as did α-tubulin deacetylation (Fig. 3A, top, lane 2 versus 3 and lane 6 versus 7) (20). When Sirt2.5 was overexpressed, HBV replication was inhibited significantly (Fig. 3A, fifth, sixth, and seventh panels, lane 2 versus 4 and lane 6 versus 8). Since Sirt2.5 lacks deacetylase activity (21) and inhibits HBV replication, α-tubulin was deacetylated only minutely (Fig. 3A, top, lanes 4 and 8).
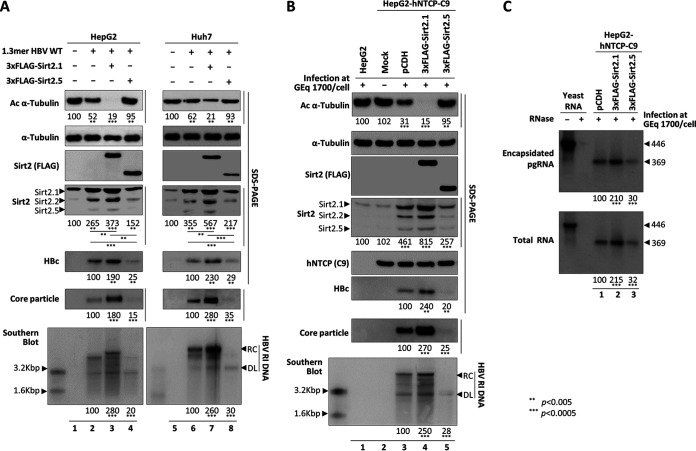
FIG 3.
Overexpression of Sirt2.5 decreases HBV DNA synthesis. (A) Overexpression of Sirt2.5 decreases HBV replication in transiently transfected cells. HepG2 (lanes 1 to 4) and Huh7 (lanes 5 to 8) cells were mock transfected (lanes 1 and 5) or transfected with 4 μg of 1.3mer HBV WT (ayw) (lanes 2 and 6), 4 μg of 1.3mer HBV WT (ayw) plus 4 μg of 3×FLAG-Sirt2.1 construct (lanes 3 and 7), or 4 μg of 1.3mer HBV WT (ayw) plus 4 μg of 3×FLAG-Sirt2.5 construct (lanes 4 and 8). Lysates were prepared at 72 h posttransfection. The amount of transfected DNA was adjusted using pcDNA3. (B) Overexpression of Sirt2.5 reduces HBV replication in HBV-infected cells. HepG2 (lane 1) and HepG2-hNTCP-C9 (lanes 2 to 5) cells were infected as described in the legend to Fig. 2. SDS-PAGE and immunoblotting of proteins, native agarose gel electrophoresis and immunoblotting of core particles, and Southern blotting of HBV DNA were performed as described in the legend to Fig. 1. The HBV hNTCP receptor was detected using a mouse anti-rhodopsin monoclonal anti-C9 (1:1,000) antibody. Relative levels of acetylated α-tubulin, Sirt2, core particles, and HBV DNA were measured using ImageJ 1.46r. Tubulin was used as a loading control. (C) Overexpression of Sirt2.5 does not affect pgRNA encapsidation in HBV-infected cells. To detect encapsidated pgRNA and total RNA from Sirt2.1- or Sirt2.5-overexpressing HBV-infected HepG2-NTCP-C9 cells, in vitro-transcribed DIG-UTP-labeled antisense RNA probe (446 nt) was hybridized overnight at 50°C with pgRNA from isolated core particles or 10 μg total RNA. Protected RNA (369 nt) following RNase digestion was run on a 5% polyacrylamide–8 M urea gel, transferred to nylon membranes, immunoblotted with anti-DIG-AP, and visualized with CSPD. The upper panel shows encapsidated pgRNA, while the lower panel depicts total RNA. Data are presented as the mean values from three independent experiments. Statistical significance was evaluated using Student's t test. **, P < 0.005; ***, P < 0.0005; each relative to the control.
As shown above (Fig. 1A and D), the expression of all three isoforms of endogenous Sirt2 increased in 1.3mer HBV WT-transfected cells (Fig. 3A, fourth panel, lanes 2 and 6). The expression of all three isoforms of endogenous Sirt2 increased to an even greater extent in Sirt2.1 plus HBV WT-cotransfected cells (Fig. 3A, fourth panel, lane 2 versus 3 and lane 6 versus 7) and was reduced significantly upon cotransfection of Sirt2.5 plus HBV WT (Fig. 3A, fourth panel, lane 2 versus 4 and lane 6 versus 8). Since HBV replication was inhibited by Sirt2.5 overexpression, the small increase in endogenous Sirt2 may reflect downregulated HBV replication (Fig. 3A, fourth panel, lane 1 versus 4 and lane 5 versus 8).
To further validate the above-described results, we generated control-, 3×FLAG-Sirt2.1-, or 3×FLAG-Sirt2.5-transduced HepG2-hNTCP-C9 cells as described earlier (Fig. 3B). Following infection of HepG2-hNTCP-C9-3×FLAG-Sirt2.1 with HBV, we detected markedly higher expression of HBc protein, core particle formation, and HBV DNA synthesis than that in control HepG2-hNTCP-C9 cells (Fig. 3B, sixth to last panels, lane 3 versus 4); thus, α-tubulin was deacetylated to the greatest extent in these cells (Fig. 3B, top, lane 3 versus 4). However, following infection of HepG2-hNTCP-C9-3×FLAG-Sirt2.5 with HBV, we detected markedly lower expression of HBc protein, core particle formation, and HBV DNA synthesis than those in control HepG2-hNTCP-C9 cells (Fig. 3B, sixth to last panels, lane 3 versus 5), resulting in low α-tubulin deacetylation (Fig. 3B, top, lane 3 versus 5). Consistent with the data shown in Fig. 3A, increases in expression of the three isoforms of Sirt2 mirrored levels of HBV replication (Fig. 3B, fourth panel, lane 1 and 2 versus 3, 4, and 5). These findings demonstrate that overexpression of Sirt2.5 decreased HBV replication significantly.
We then performed RPA to examine whether overexpressed Sirt2.1 or Sirt2.5 affects pgRNA encapsidation. Encapsidated RNA from isolated core particles and total RNAs were compared in HBV-infected pCDH-, 3×FLAG-Sirt2.1-, or 3×FLAG-Sirt2.5-transduced HepG2-hNTCP-C9 cells (Fig. 3C). The increased level of encapsidated pgRNA from isolated core particles as well as total RNA observed in HepG2-hNTCP-C9-3×FLAG-Sirt2.1 cells (Fig. 3C, lane 1 versus 2) reflects the levels of cccDNA and HBV RNAs (Fig. 2C). Similarly, the decreased level of encapsidated pgRNA from isolated core particles as well as total RNA were observed in HepG2-hNTCP-C9-3×FLAG-Sirt2.5 cells (Fig. 3C, lane 1 versus 3). This RPA result clearly demonstrates that Sirt2 has no effect on pgRNA encapsidation.
Since Sirt2.1 and Sirt2.5 have opposite effects (Fig. 3), we asked whether similar levels of Sirt2.1 and Sirt2.5 in replicating cells antagonize each other. As shown above, the expression of endogenous Sirt2.1 was higher than that of endogenous Sirt2.5 (Fig. 1 and 3); therefore, antagonism of Sirt2.1 by Sirt2.5 cannot be a prominent effect. In the following experiment, we further demonstrate that Sirt2.2 also enhances HBV replication (see Fig. 6D and E); therefore, Sirt2.1 and Sirt2.2 can easily override the Sirt2.5 effect. Again, the levels of Sirt2.1 and Sirt2.2 were greater than those in Sirt2.5 in HBV-replicating cells (Fig. 1).
FIG 6.
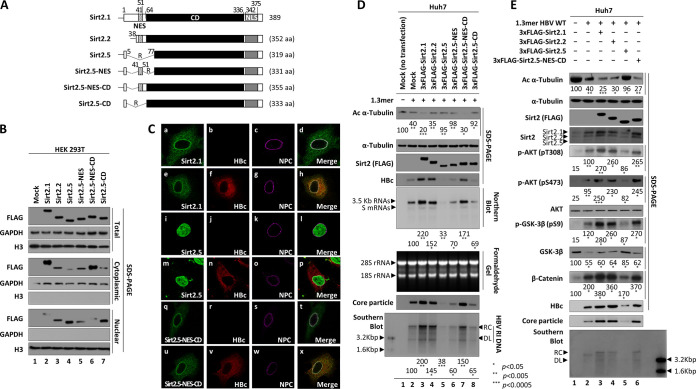
N-terminal 40 amino acids of Sirt2 are not important for upregulation of HBV replication. (A) Schematic diagram showing Sirt2 isoforms 1, 2, and 5 and the mutants of Sirt2.5 in which the NES and/or N-terminal truncated catalytic domain were restored. Residues are numbered according to full-length Sirt2 isoform 1. The nuclear export signal (NES; dotted), the catalytic domain (CD; black), and the nuclear localization signals (NLSs; gray) are indicated. (B) Both the NES and CD may be important for cytoplasmic localization of Sirt2. HEK293T cells were mock transfected (lane 1) or transiently transfected with 4 μg of 3×FLAG-tagged Sirt2.1 (lane 2), Sirt2.2 (lane 3), Sirt2.5 (lane 4), Sirt2.5-NES (lane 5), Sirt2.5-NES-CD (lane 6), or Sirt2.5-CD (lane 7). At 72 h posttransfection, total lysate and the cytoplasmic and nuclear fractions were prepared as described in the legend to Fig. 1. (C) HBV replication does not affect localization of Sirt2.1, Sirt2.5, and Sirt2.5-NES-CD. Huh7 cells were transfected with 3×FLAG-tagged Sirt2.1 (a to d), Sirt2.5 (i to l), or Sirt2.5-NES-CD (q to t) or cotransfected with 3×FLAG-tagged Sirt2.1 plus 1.3mer HBV WT (ayw) (e to h), Sirt2.5 plus 1.3mer HBV WT (ayw) (m to p), or Sirt2.5-NES-CD plus 1.3mer HBV WT (ayw) (u to x). Confocal images of the nuclear pore complex (NPC) taken using Alexa Fluor 647 (c, g, k, o, s, and w) and 3×FLAG-tagged Sirt2.1 (a and e), Sirt2.5 (i and m), and Sirt2.5-NES-CD (q and u) using FITC and HBc using TRITC (f, n, and v) are shown. Merged images (d, h, l, p, t, and x) are indicated. Digital images of stained cells were captured under a confocal microscope (LSM710; Zeiss, Germany). Data are representative of three independent experiments. (D) Like overexpression of Sirt2.1 or Sirt2.2, HBV replication is increased by overexpression of 3×FLAG-Sirt2.5-NES-CD. Huh7 cells were mock transfected (lane 1), transiently transfected with 4 μg of 1.3mer HBV WT (ayw) (lane 2), or cotransfected with 4 μg of 1.3mer HBV WT (awy) plus 4 μg of 3×FLAG-tagged Sirt2.1 (lane 3), Sirt2.2 (lane 4), Sirt2.5 (lane 5), Sirt2.5-NES (lane 6), Sirt2.5-NES-CD (lane 7), or Sirt2.5-CD (lane 8) construct. (E) The AKT/GSK-3β/β-catenin signaling pathway in HBV replicating cells is activated by overexpression of 3×FLAG-Sirt2.5-NES-CD (identical to overexpression of Sirt2.1 or Sirt2.2). Huh7 cells were mock transfected (lane 1), transiently transfected with 4 μg of 1.3mer HBV WT (ayw) (lane 2), or cotransfected with 4 μg of 1.3mer HBV WT (awy) plus 4 μg of 3×FLAG-tagged Sirt2.1 (lane 3), -Sirt2.2 (lane 4), Sirt2.5 (lane 5), or Sirt2.5-NES-CD (lane 6) construct. Lysates were prepared at 72 h posttransfection. The amount of transfected DNA was adjusted using pcDNA3. SDS-PAGE and immunoblotting of proteins, native agarose gel electrophoresis and immunoblotting of core particles, Southern blotting of HBV DNA, and Northern blotting of HBV RNA were performed as described in the legends to Fig. 1 and 2. Levels of acetylated α-tubulin, HBV DNA, and active AKT (pT308 and pS473) relative to those of total AKT, total β-catenin, and total/phosphorylated (pS9) GSK-3β were measured using ImageJ 1.46r. *, P < 0.05; **, P < 0.005; ***, P < 0.0005; each relative to the control.
Unlike the Sirt2.1-AKT interaction, the Sirt2.5-AKT interaction in HBV-replicating cells is weak.
Sirt2 binds AKT and is critical for full AKT activation (20, 39, 40). Our previous study highlighted that the Sirt2.1-AKT interaction is stronger in HBV-replicating cells than in control cells (20). To confirm the association between Sirt2.5 and AKT in control or HBV-replicating Huh7 cells, cell lysates were immunoprecipitated with an anti-FLAG antibody and immunoblotted with an anti-AKT antibody (Fig. 4A, first to fourth panels). Consistent with the previous report, the Sirt2.1-AKT interaction increased in HBV-replicating cells (Fig. 4A, top, lane 7 versus 8) (20). Unlike the Sirt2.1-AKT interaction, the strong Sirt2.5-AKT interaction in control cells was weak in HBV-replicating cells (Fig. 4A, top, lane 9 versus 10). Since Sirt2.1 and Sirt2.5 localized predominantly in the cytoplasm and nucleus, respectively (20, 21), we prepared cytoplasmic and nuclear fractions and immunoprecipitated them with an anti-FLAG antibody, followed by immunoblotting with an anti-AKT antibody (Fig. 4A, fifth to last panels). The cytoplasmic and nuclear fractions showed similar immunoprecipitation patterns (Fig. 4A, fifth and ninth panels), although the Sirt2-AKT interaction in the cytoplasmic fraction of Sirt2.5-overexpressing control and HBV-replicating cells was much less evident (Fig. 4A, fifth panel, lane 9 versus 10). This may be due to the preferred nuclear localization of Sirt2.5 protein.
FIG 4.
Sirt2.5-AKT interaction plays no role in the HBV-associated AKT/GSK-3β/β-catenin signaling pathway. (A) Unlike the Sirt2.1-AKT interaction, the Sirt2.5-AKT interaction is weakened in HBV replicating cells. Huh7 cells were mock transfected (lanes 1, 6, and 11), transfected with 3×FLAG-Sirt2.1 (lanes 2, 7, and 12) or 3×FLAG-Sirt2.5 (lanes 4, 9, and 14), or cotransfected with 3×FLAG-Sirt2.1 plus 1.3mer HBV WT (ayw) (lanes 3, 8, and 13) or 3×FLAG-Sirt2.5 plus 1.3mer HBV WT (ayw) (lanes 5, 10, and 15). At 72 h posttransfection, total lysate and the cytoplasmic and nuclear fractions were prepared as described for Fig. 1. Total lysates and the indicated fractions were immunoprecipitated with an anti-FLAG antibody (lanes 6 to 10) or with normal IgG (lanes 11 to 15). Total lysates and the respective fractions were used as the input (lanes 1 to 5). The lysates and immunoprecipitants were subjected to SDS-PAGE and immunoblotting with anti-AKT (1:1,000; 9272S; Cell Signaling Technology), anti-FLAG M2 (1:1,000; F1804; Sigma), anti-GAPDH, and anti-H3 antibodies. (B) The Sirt2.5-associated AKT/GSK-3β/β-catenin signaling pathway is independent of HBV replication. HepG2 (lanes 1 to 6) or Huh7 (lanes 7 to 12) cells were mock transfected (lanes 1 and 7) or (co)transfected with 4 μg of 3×FLAG-Sirt2.1 (lanes 2 and 8), 4 μg of 3×FLAG-Sirt2.5 (lanes 3 and 9), 4 μg of 1.3mer HBV WT (ayw) (lanes 4 and 10), 4 μg of 1.3mer HBV WT (ayw) plus 4 μg of 3×FLAG-Sirt2.1 (lanes 5 and 11), or 4 μg of 1.3mer HBV WT (ayw) plus 4 μg of 3×FLAG-Sirt2.5 (lanes 6 and 12). The amount of transfected DNA was adjusted with pcDNA3. (C) Activation of the AKT/GSK-3β/β-catenin signaling pathway by Sirt2.5 overexpression in SIRT2 KD cells does not depend on HBV replication. HepG2 cells transduced with lentiviral control shRNA (lanes 2 to 4) or Sirt2 shRNAs (shSIRT2-#2) (lanes 5 to 7) were (co)transfected with 1.3mer HBV WT (lanes 2 and 5), 1.3mer HBV WT plus 3×FLAG-Sirt2.1 (lanes 3 and 6), or 1.3mer HBV WT plus 3×FLAG-Sirt2.5 (lanes 4 and 7). Mock-transfected HepG2 cells were a negative control (lane 1). Lysates were prepared at 72 h posttransfection. SDS-PAGE and immunoblotting of proteins, native agarose gel electrophoresis and immunoblotting for core particles, and Southern blotting of HBV DNA were performed as described in the legend to Fig. 1. Levels of acetylated α-tubulin and active AKT (pT308 and pS473) relative to those of total AKT, total β-catenin, and total/phosphorylated (S9) GSK-3β were measured using ImageJ 1.46r. Data are presented as mean values from three independent experiments. Statistical significance was evaluated using Student's t test. ns, not significant; *, P < 0.05; **, P < 0.005; ***, P < 0.0005; each relative to the control.
The Sirt2.5-associated AKT/GSK-3β/β-catenin signaling pathway is independent of HBV replication.
Although Sirt2.1 activates the AKT/glycogen synthase kinase-3β (GSK-3β)/β-catenin signaling pathway in HBV-replicating cells via a strengthened Sirt2-AKT interaction (20), the strong Sirt2.5-AKT interaction was weakened in HBV-replicating cells (Fig. 4A). To investigate the effect of the Sirt2.5-AKT interaction on the AKT/GSK-3β/β-catenin signaling pathway in control or HBV-replicating cells, we examined signaling in HepG2 or Huh7 cells that were mock transfected, transfected with 3×FLAG-Sirt2.1-, 3×FLAG-Sirt2.5-, or 1.3mer HBV WT, or cotransfected with 1.3mer HBV WT plus 3×FLAG-Sirt2.1 or 3×FLAG-Sirt2.5 (Fig. 4B). The 3×FLAG-Sirt2.1-transfected HepG2 or Huh7 cells showed activation of AKT (i.e., phosphorylation of T308 and S473) (Fig. 4B, fifth and sixth panels, lane 1 versus 2 and lane 7 versus 8), inhibition of GSK-3β (as shown by inhibited phosphorylation at S9) (Fig. 4B, eighth panel, lane 1 versus 2 and lane 7 versus 8), and stabilized β-catenin levels (Fig. 4B, tenth panel, lane 1 versus 2 and lane 7 versus 8) (41). The 3×FLAG-Sirt2.5-transfected HepG2 or Huh7 cells also showed AKT activation, GSK-3β inhibition, and β-catenin stabilization compared with mock-transfected cells (Fig. 4B, lane 1 versus 3 and lane 7 versus 9); however, these characteristics were less evident than those in 3×FLAG-Sirt2.1-transfected HepG2 or Huh7 cells (Fig. 4B, lane 2 versus 3 and lane 8 versus 9). Of note, overexpressed Sirt2.5 could not deacetylate α-tubulin (Fig. 4B, lanes 3 and 9), verifying that Sirt2.5 is catalytically inactive (21).
The activation of AKT was greater in 1.3mer HBV WT-transfected HepG2 or Huh7 cells than in the respective mock controls (Fig. 4B, fifth and sixth panels, lane 1 versus 4 and lane 7 versus 10); similarly, GSK-3β was inhibited (Fig. 4B, eighth panel, lane 1 versus 4 and lane 7 versus 10) and β-catenin levels were increased (Fig. 4B, tenth panel, lane 1 versus 4 and lane 7 versus 10) (20). Cotransfection of Sirt2.1 plus 1.3mer HBV WT synergistically activated the AKT/GSK-3β/β-catenin signaling pathway (Fig. 4B, lane 4 versus 5 and lane 10 versus 11) (20). Conversely, upon cotransfection of Sirt2.5 plus 1.3mer HBV WT, AKT activation, GSK-3β inhibition, and β-catenin stabilization were comparable with those in Sirt2.5-transfected cells (Fig. 4B, fifth to tenth panels, lane 3 versus 6 and lane 9 versus 12). This result indicates that Sirt2.5 plus HBV WT does not activate the AKT/GSK-3β/β-catenin signaling pathway synergistically, suggesting that Sirt2.5-mediated activation of the AKT/GSK-3β/β-catenin signaling pathway is independent of HBV.
To further validate the above-described results in SIRT2 knocked down cells, we transduced HepG2 cells with lentiviral control short hairpin RNA (shRNA) (Fig. 4C, lanes 2 to 4) or Sirt2 shRNA (shSIRT2-#2) (Fig. 4C, lanes 5 to 7), followed by transfection with 1.3mer HBV WT or cotransfection with 1.3mer HBV WT plus 3×FLAG-Sirt2.1 (Fig. 4C, lanes 3 and 6) or 3×FLAG-Sirt2.5 (Fig. 4C, lanes 4 and 7). Consistent with our previous report (20), SIRT2 knockdown (KD) cells showed limited tubulin deacetylation (Fig. 4C, top, lane 2 versus 5). In accordance with our previous report (20), SIRT2 KD reduced the expression of HBc protein, core particle formation, and HBV DNA synthesis (Fig. 4C, eleventh to last panels, lane 2 versus 5). AKT phosphorylation in HBV-replicating SIRT2 KD cells was lower (Fig. 4C, fifth and sixth panels, lane 5 to 7), GSK-3β activity was higher, and β-catenin was less stable than that in HBV-replicating control shRNA-transduced cells (Fig. 4C, eighth to tenth panels, lane 2 versus 5) (20). Overexpression of Sirt2.1 in HBV-replicating SIRT2 KD cells activated AKT/GSK-3β/β-catenin signaling (Fig. 4C, lane 5 versus 6) (20). However, Sirt2.5 overexpression in HBV-replicating SIRT2 KD cells had no prominent effect on activation of the AKT/GSK-3β/β-catenin signaling pathway (Fig. 4C, fifth to tenth panels, lane 5 versus 7), while HBV replication was inhibited to a greater extent than that in HBV-replicating SIRT2 KD cells (Fig. 4C, eleventh to last panels, lane 5 versus 7). This result confirms that Sirt2.5 activates the AKT/GSK-3β/β-catenin signaling pathway independently of HBV.
Sirt2.5-mediated inhibition of HBV replication is independent of HBx.
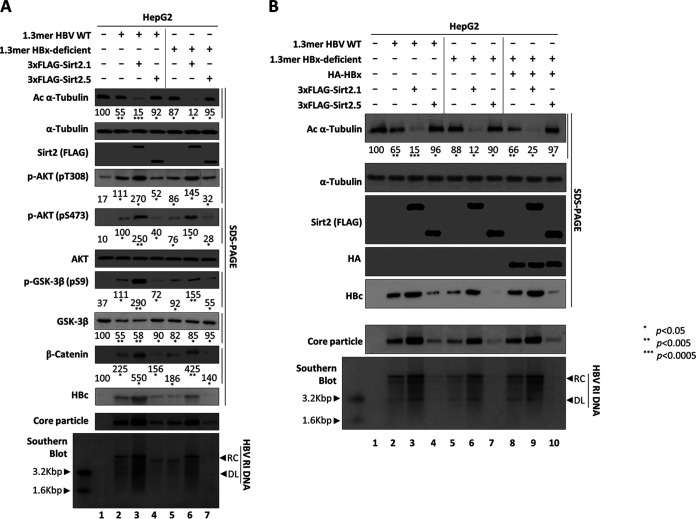
We next asked whether HBx is a possible target for Sirt2.5-mediated inhibition of HBV replication. In our previous study, we showed that Sirt2.1-mediated upregulation of HBV replication does not depend on HBx (20) (Fig. 5A, lane 3 versus 6). Since HBx activates Sirt2 and is important for Wnt/β-catenin signaling in hepatoma cells (42–44), we examined whether Sirt2.5-mediated downregulation of HBV replication is HBx dependent. HepG2 cells were transiently transfected with 1.3mer HBV WT or a 1.3mer HBx-deficient mutant or cotransfected with 1.3mer HBV WT plus 3×FLAG-Sirt2.1 or -Sirt2.5 or with a 1.3mer HBx-deficient mutant plus 3×FLAG-Sirt2.1 or -Sirt2.5 (Fig. 5A). HBV replication in HBx-deficient mutant-transfected cells was lower than that in HBV WT-transfected cells (2, 20, 45–47); consequently, AKT activation (as shown by phosphorylation of T308 and S473) was lower, GSK-3β inhibition was less pronounced, and β-catenin levels were lower than those in HBV WT-transfected cells (Fig. 5A, lane 2 versus 5). HBV replication in HBx-deficient mutant-transfected cells was upregulated upon cotransfection of the 3×FLAG-Sirt2.1 construct (20); accordingly, AKT/GSK-3β/β-catenin signaling was activated to a greater extent than that in HBx-deficient mutant-transfected cells (Fig. 5A, lane 5 versus 6). However, HBV replication in HBx-deficient mutant-transfected cells was inhibited even more upon cotransfection of the 3×FLAG-Sirt2.5 construct; AKT/GSK-3β/β-catenin signaling was also downregulated (Fig. 5A, lane 5 versus 7). These observations suggest that Sirt2.5 activity is not affected by the presence or absence of HBx.
FIG 5.
Sirt2.5-mediated inhibition of HBV replication is independent of HBx. (A) The Sirt2.5-associated AKT/GSK-3β/β-catenin signaling pathway in HBV-replicating cells is not affected by the presence or absence of HBx. HepG2 cells were mock transfected (lane 1), transfected with 4 μg of 1.3mer HBV WT (ayw) (lanes 2 to 4), or transfected with 4 μg of 1.3mer HBx-deficient mutant (lanes 5 to 7) in the absence (lanes 2 and 5) or presence of either 3×FLAG-Sirt2.1 (4 μg) (lanes 3 and 6) or 3×FLAG-Sirt2.5 (4 μg) (lanes 4 and 7). (B) Sirt2.5-mediated downregulation of HBV replication is independent of HBx. HepG2 cells were mock transfected (lane 1) or cotransfected (lanes 1 to 7) as described for panel A. To supply HBx in trans, HepG2 cells were triple transfected with the 1.3-mer HBx-deficient mutant plus HA-HBx (lanes 8 to 10) plus 3×FLAG-Sirt2.1 or 3×FLAG-Sirt2.5. Lysates were prepared at 72 h posttransfection. The amount of transfected DNA was adjusted using pcDNA3 (lanes 2, 5, and 8). SDS-PAGE and immunoblotting of proteins, native agarose gel electrophoresis and immunoblotting of core particle, and Southern blotting of HBV DNA were performed as described in the legend to Fig. 1. Levels of acetylated α-tubulin and active AKT (pT308 and pS473) relative to total AKT, total β-catenin, and total and phosphorylated GSK-3β (pS9) were measured using ImageJ 1.46r. *, P < 0.05; **, P < 0.005; and ***, P < 0.0005; each relative to the control.
When HBx was supplied in trans by triple transfection of an HBx-deficient mutant with Myc-HBx plus either 3×FLAG-Sirt2.1 or 3×FLAG-Sirt2.5, HBV replication was modulated, i.e., upregulated by 3×FLAG-Sirt2.1 and downregulated by 3×FLAG-Sirt2.5 (Fig. 5B, lane 8 versus 9 versus 10); these results are comparable with those in cells cotransfected with HBV WT plus Sirt2.1 or Sirt2.5 (Fig. 5B, lane 2 versus 8, 3 versus 9, and 4 versus 10), demonstrating that Sirt2-mediated modulation of HBV replication does not depend on HBx.
The NES and catalytic domain, not the N-terminal 40 amino acids, of Sirt2 are important for its activity and for increasing HBV replication.
HBx-independent Sirt2.1 and Sirt2.5 have opposite effects on HBV replication (Fig. 2 to 5), and we tried to identify the functional domains of Sirt2 that are responsible for these opposite effects. Since Sirt2.5 lacks an NES and part of the deacetylase CD (Fig. 6A) is mainly localized in the nucleus (20, 21) and inhibits HBV replication (Fig. 2B and C, 3, 4B and C, and 5), we tried to identify the relevant functional domains. To do this, we constructed several Sirt2.5 mutants in which the NES and/or N-terminally truncated CD were restored: 3×FLAG-Sirt2.5-NES, 3×FLAG-Sirt2.5-NES-CD, and 3×FLAG-Sirt2.5-CD (Fig. 6A).
We then examined the cellular localization of these Sirt2.5-derived mutants in HEK293T cells at 72 h posttransfection. Total lysates and the cytoplasmic and nuclear fractions were examined by SDS-PAGE and immunoblotting (Fig. 6B). All constructs were expressed at comparable levels (Fig. 6B, top). Consistent with previous reports and results (Fig. 1D) (20, 21), Sirt2.1 localized mainly in the cytoplasm (Fig. 6B, lane 2), Sirt2.2 in the cytoplasm and nucleus (Fig. 6B, lane 3), and Sirt2.5 in the nucleus (Fig. 6B, lane 4). When NES was added to Sirt2.5, the protein localized to the cytoplasm and nucleus (Fig. 6B, lane 5). Addition of the NES plus the N-terminally truncated CD to Sirt2.5 caused the protein to localize mainly in the cytoplasm (Fig. 6B, lane 6). However, the addition of the N-terminally truncated CD in the absence of the NES caused the protein to localize mainly in the nucleus (Fig. 6B, lane 7). These results suggest that both the NES and the CD are important for cytoplasmic localization of Sirt2 protein.
We next examined the localization of Sirt2.1, Sirt2.5, and Sirt2.5-NES-CD proteins in mock- and 1.3mer HBV WT-transfected Huh7 cells by immunofluorescence analysis. Consistent with Fig. 6B, Sirt2.1 and Sirt2.5-NES-CD localized in the cytoplasm (Fig. 6C, a and q), while Sirt2.5 localized in the nucleus (Fig. 6C, i). The localization of these proteins was not altered by HBV replication (Fig. 6C, a versus e, i versus m, and q versus u). Since Sirt2.1 and HBc appeared to colocalize (Fig. 6C, h and x), we performed coimmunoprecipitations to determine their interaction. We found that they did not interact (data not shown). It should be noted that this apparent colocalization is likely due to the fact that both HBc (core particle) and Sirt2 interact with tubulin (microtubules) (data not shown) (23, 48).
To examine the effects of these Sirt2.5-derived mutants on HBV replication, we transiently (co)transfected Huh7 cells with 1.3mer HBV WT or with 1.3mer HBV WT plus 3×FLAG-Sirt2.1, -Sirt2.2, -Sirt2.5, -Sirt2.5-NES, -Sirt2.5-NES-CD, or -Sirt2.5-CD (Fig. 6D). Consistent with the results shown in Fig. 1, 3, 4B and C, and 5, when HBV was replicating, α-tubulin was deacetylated to a greater extent than that in mock-transfected cells (Fig. 6D, top, lane 1 versus 2). When Sirt2.1 or Sirt2.2 was overexpressed in HBV-replicating cells, α-tubulin was deacetylated to a greater extent than that in HBV-transfected cells (Fig. 6D, top, lane 2 versus 3 and 4). Consequently, HBc protein, core particle formation, and HBV RNA and DNA synthesis were upregulated significantly in Sirt2.1- or Sirt2.2-overexpressing cells (Fig. 6D, fourth to last panels, lane 2 versus 3 and 4). This result shows, for the first time, that overexpression of Sirt2.2 increases HBV replication, even though the effect of Sirt2.2 is weaker than that of Sirt2.1 (Fig. 6D, fourth to last panels, lane 2 versus 3 versus 4). Consistent with the results shown in Fig. 3, 4B and C, and 5, when Sirt2.5 was overexpressed, α-tubulin was deacetylated minutely (Fig. 6D, top, lane 1 versus 5). Similarly, when Sirt2.5-NES or Sirt2.5-CD was overexpressed, α-tubulin was not deacetylated (Fig. 6D, top, lane 1 versus 6 and 8), indicating that the NES or full-length CD alone is not sufficient for Sirt2 tubulin deacetylase activity. HBV replication in these Sirt2.5-NES- or Sirt2.5-CD-cotransfected cells was higher than that in Sirt2.5-cotransfected cells but much lower than that in HBV WT-transfected cells (Fig. 6D, fourth to last panels, lane 1 versus 5 versus 6 and 8). However, when Sirt2.5-NES-CD was overexpressed, α-tubulin was deacetylated to a greater extent (Fig. 6D, top, lane 1 versus 7). Also, HBc protein levels, core particle formation, and HBV RNA and DNA synthesis were significantly higher than those in cells transfected with the 1.3mer HBV WT (Fig. 6D, fourth to last panels, lane 2 versus 7). These results suggest that the effect of Sirt2.5-NES-CD is comparable with that of Sirt2.2, and that cytoplasmic Sirt2 with a full-length catalytic domain can exert tubulin deacetylase activity and increase HBV replication. Taken together, the results show that the NES and full CD are important for Sirt2 tubulin deacetylase activity and increased HBV replication and that the N-terminal 40 amino acids are not essential for Sirt2.1 and Sirt2.2 activity.
Since Sirt2.5-NES-CD increased HBV replication (Fig. 6D), we asked whether this mutant affects the AKT/GSK-3β/β-catenin signaling pathway (Fig. 6E). Unlike Sirt2.5, overexpression of Sirt2.5-NES-CD in 1.3mer HBV WT-transfected Huh7 cells upregulated the AKT/GSK-3β/β-catenin signaling pathway to a greater extent than that in 1.3mer HBV WT-transfected Huh7 cells (Fig. 6E, lane 2 versus 6). Of note, overexpression of Sirt2.2 in 1.3mer HBV WT-transfected Huh7 cells upregulated AKT/GSK-3β/β-catenin signaling (Fig. 6E, lane 2 versus 4) in a manner comparable with the overexpression of Sirt2.2-NES-CD (Fig. 6E, lane 4 versus 6), further highlighting the importance of the NES and CD of Sirt2 in Sirt2-mediated HBV replication.
Sirt2.5 overexpression increases recruitment of HKMTs and deposition of respective epigenetic repressive markers on cccDNA.
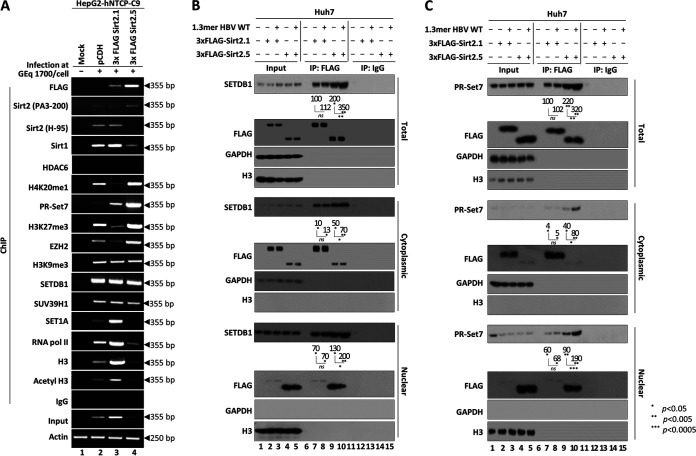
We asked how Sirt2.5 inhibits viral transcription. Since the overexpression of Sirt2.5 inhibits transcriptional activity (Fig. 2), one possibility is that Sirt2.5 reduces viral transcription via its association with the cccDNA chromatin structure. Sirt1 and Sirt3 bind to various transcription factors and cofactors to inactivate the transcription of cccDNA in the presence of DNA-binding proteins, cellular transcriptional regulators, and chromatin remodelers (3, 4, 12, 13). Therefore, we performed chromatin immunoprecipitation (ChIP) assays using HBV-infected HepG2-hNTCP-C9 cells to determine the association between Sirt2.1 and/or Sirt2.5 and cccDNA chromatin (Fig. 7A). Briefly, following infection of pCDH-, 3×FLAG-Sirt2.1-, or 3×FLAG-Sirt2.5-transduced HepG2-hNTCP-C9 cells with 1,700 GEq/cell of HBV, chromatin was immunoprecipitated with control IgG or respective antibodies. The immunoprecipitated pellet next was analyzed by semiquantitative PCR (Fig. 7A) using primers specific for HBV cccDNA (Table 1). To begin with, constant amounts of an actin gene fragment were amplified (Fig. 7A, bottom), demonstrating that constant amounts of chromatin DNA were obtained from mock- and HBV-infected cells. Input DNA was amplified by semiquantitative PCR. The results showed that Sirt2.1-overexpressing cells contained the highest level of cccDNA and Sirt2.5-overexpressing cells contained the lowest (Fig. 7A, eighteenth panel, lane 2 versus 3 versus 4), which is consistent with the results in Fig. 2C demonstrating increased and decreased expression of cccDNA in Sirt2.1- and Sirt2.5-overexpressing cells, respectively. When anti-Sirt2 PA3-200 or anti-3×FLAG antibodies were used, more Sirt2 was recruited to cccDNA in Sirt2.5-overexpressing cells than in control- or Sirt2.1-overexperssing cells (Fig. 7A, first and second panels, lane 2 versus 3 versus 4), indicating that more Sirt2.5 is recruited to cccDNA than Sirt2.1. Endogenous Sirt2 could be recruited to cccDNA (Fig. 7A, second and third panels, lane 2). Consistent with a previous report (12), Sirt1 was recruited to cccDNA at levels that roughly matched those of input DNA (Fig. 7A, fourth panel). HDAC6, an exclusively cytoplasmic protein (49) that cannot be recruited to cccDNA, was used as a negative control (Fig. 7A, fifth panel).
FIG 7.
HBV cccDNA-recruited Sirt2.5 associates with HKMTs such as PR-Set7 and SETDB1, which deposit epigenetic repressive markers. (A) Recruitment of HKMTs, such as PR-Set7, EZH2, SETDB1, and SUV39H1, and depositions of respective epigenetic repressive markers onto cccDNA increased in Sirt2.5-overexpressing cells. HepG2-hNTCP-C9 cells transduced with pCDH (lane 2), 3×FLAG-Sirt2.1 (lane 3), or 3×FLAG-Sirt2.5 (lane 4) were mock infected (lane 1) or infected with 1.7 × 103 GEq of HBV (lanes 2 to 4), as described in the legend to Fig. 2. Nine days after infection, chromatin solutions were prepared and subjected to immunoprecipitation with anti-FLAG, anti-Sirt2 (PA3-200 and H-95), anti-Sirt1, anti-HDAC6, anti-H3, anti-AcH3, anti-RNA Pol II, anti-H3K9me3, anti-H3K27me3, anti-H4K20me1, anti-SUV39H1, anti-PR-Set7, anti-EZH2, anti-SETDB1, and anti-SET1A antibodies or with normal rabbit polyclonal IgG (negative control). Immunoprecipitated chromatin was analyzed by PCR. Actin levels were used to ensure equal loading of lysate samples. (B) The Sirt2.5-SETDB1 interaction increased upon HBV replication. (C) The Sirt2.5–PR-Set7 interaction increased upon HBV replication. (B and C) Huh7 cells were mock transfected (lanes 1, 6, and 11), transfected with 3×FLAG-Sirt2.1 (lanes 2, 7, and 12) or 3×FLAG-Sirt2.5 (lanes 3, 8, and 13), or cotransfected with 3×FLAG-Sirt2.1 plus 1.3mer HBV WT (ayw) (lanes 4, 9, and 14) or 3×FLAG-Sirt2.5 plus 1.3mer HBV WT (ayw) (lanes 5, 10, and 15). At 72 h posttransfection, total, cytoplasmic, and nuclear fractions were prepared as described in the legend to Fig. 1. Total cell lysates and the indicated fractions were immunoprecipitated with anti-FLAG antibody (lanes 6 to 10). IgG, lysates were immunoprecipitated with normal IgG as a negative control (lanes 11 to 15). Input, total cell lysates and the respective fractions were prepared (lanes 1 to 5). The lysates and immunoprecipitants were subjected to SDS-PAGE and immunoblotting with anti-SETDB1 (B) and anti-PR-Set7 (C) antibodies. Data are representative of three independent experiments. Relative levels of immunoprecipitated SETDB1 and PR-Set7 were measured using ImageJ 1.46r. *, P < 0.05; **, P < 0.005; ***, P < 0.0005; each relative to the control.
TABLE 1.
Primer sequences used for this study
| Plasmid name | Orientation | Sequence (5′–3′) |
|---|---|---|
| Sirt2.5 mutants | ||
| 3×FLAG-Sirt2.5-NES | Forward | ACGCTCAGCCTGCGTCGCAGAGTCATC |
| Reverse | GATGACTCTGCGACGCAGGCTGAGCGT | |
| 3×FLAG- Sirt2.5-NES-CD | Forward | GCAGAGCCAGACTTGGAAGGGGTG |
| Reverse | CACCCCTTCCAAGTCTGGCTCTGC | |
| 3×FLAG-Sirt2.5-CD | Forward | ATGGCAGAGCCAGACCTGCGGAACTTA |
| Reverse | TAAGTTCCGCAGGTCTGGCTCTGCCAT | |
| Sirt2 isoforms | ||
| pCDH-3×FLAG-Sirt2.1 | Forward | ATGCGAATTTCATGGACTACAAAGACC |
| Reverse | ATGCGGATCCTCACTGGGGTTTCTCCCC | |
| pCDH-3×FLAG-Sirt2.5 | Forward | ATGCGAATTCATGGACTTCCTGCGGAAC |
| Reverse | ATGCGGATCCTCACTGGGGTTTCTCCCC | |
| cccDNA | Forward | CTCCCCGTCTGTGCCTTCT |
| Reverse | GCCCCAAAGCCACCCAAG | |
| Actin | Forward | CATGTACGTTGCTATCCAGGC |
| Reverse | CTCCTTAATGTCACGCACGAT |
Posttranslational modifications, such as methylation and acetylation on H3 and H4, are associated with modification of chromatin structure and regulation of HBV cccDNA transcription (3, 4, 11–13, 26–28, 31–34). Heterochromatin-linked histone lysine modifications, which cause chromatin condensation and transcriptional repression, include H3K9me3, H3K27me3, and H4K20me1 (4, 50). To examine whether Sirt2.5 overexpression is associated with a transcriptionally inactive cccDNA chromatin structure, we performed ChIP assays to investigate deposition of repressive histone lysine methylations such as H3K9me3, H3K27me3, and H4K20me1 in Sirt2.5- or Sirt2.1-overexpressing HBV-infected cells (Fig. 7A, sixth, eighth, and tenth panels) (13, 27, 36). We found that H4K20 and H3K27 on the cccDNA minichromosome were methylated to a lesser extent in Sirt2.1-overexpressing cells and to a greater extent in Sirt2.5-overexpressing cells than in control cells (Fig. 7A, sixth and eighth panels, lane 2 versus 3 and lane 2 versus 4), indicating transcriptional repression upon overexpression of Sirt2.5. At first glance, we noted no changes in the trimethylation of histone H3K9 in Sirt2.5- or Sirt2.1-overexpressing cells (Fig. 7A, tenth panel); however, considering the amount of input cccDNA (eighteenth panel), H3K9 was methylated to a lesser extent in Sirt2.1-overexpressing cells and to a greater extent in Sirt2.5-overexpressing cells than in control cells (Fig. 7A, tenth panel, lane 2 versus 3 and lane 2 versus 4). Overall, the data show that Sirt2 proteins are recruited to cccDNA, and that recruited Sirt2.5 induces transcriptional repression (as shown by increased amounts of transcriptional repressive markers H3K9me3, H3K27me3, and H4K20me1) of cccDNA via epigenetic modifications.
We then examined HKMTs PR-Set7, EZH2, SETDB1, and SUV39H1, which deposit a methyl group(s) onto H4K20, H3K27, or H3K9, respectively, resulting in PR-Set7-mediated H4K20me1, EZH2-mediated H3K27me3, or SETDB1- and SUV39H1-mediated H3K9me3 (12, 13, 27–30, 50), in Sirt2.5- or Sirt2.1-overexpressing HBV-infected cells (Fig. 7A, seventh, ninth, eleventh, and twelfth panels). As anticipated from methylation of H3K9, H3K27, and H4K20 (Fig. 7A, sixth, eighth, and tenth panels), PR-Set7, EZH2, SETDB1, and SUV39H1 were recruited onto cccDNA (Fig. 7A, seventh, ninth, eleventh, and twelfth panels). More PR-Set7 was recruited in Sirt2.5-overexpressing cells than in Sirt2.1-overexpressing cells and control cells (Fig. 7A, seventh panel, lane 2 versus 3 versus 4). Of note, unlike EZH2, SETDB1, or SUV39H1, more PR-Set7 was recruited in Sirt2.1-overexpressing cells than in control cells (Fig. 7A, seventh panel, lane 2 versus 3).
We next examined whether the overexpression of Sirt2.1 or Sirt2.5 affects the recruitment of SET1A, host RNA polymerase II, H3, and acetylated H3 (H3K9ac and H3K14ac) to cccDNA. SET1A methylates H3K4 to increase transcriptional activation (26). Recruitment of SET1A to cccDNA was markedly increased in Sirt2.1-overexpressing cells and decreased in Sirt2.5-overexpressing cells (Fig. 7A, thirteenth panel, lane 2 versus 3 versus 4); however, this may reflect the amount of input DNA. Recruitment of RNA polymerase II to cccDNA showed the same pattern as SET1A (Fig. 7A, fourteenth panel, lane 2 versus 3 versus 4). Acetylated and total H3 showed the same pattern as SET1A and RNA polymerase II (Fig. 7A, fifteenth and sixteenth panels, lane 2 versus 3 versus 4). We show that HKMTs PR-Set7, EZH2, SETDB1, and/or SUV39H1 transcriptionally repress cccDNA in Sirt2.5-overexpressing cells. However, we only examined SET1A, which methylates H3K4 to activate transcription. Transcriptional activation in Sirt2.1-overexpressing cells may be investigated further in the future.
The Sirt2.5-SETDB1 and –PR-Set7 interactions increase upon HBV replication.
Since both histone deacetylases and methyl transferases cooperate to promote heterochromatin formation (29, 30), we hypothesized that Sirt2.1 and/or Sirt2.5 interact physically with several HKMTs. PR-Set7 interacts with Sirt2 (30); however, the interactions between PR-Set7 and the three Sirt2 isoforms, with Sirt2.1 or Sirt2.5, or in HBV-replicating cells have never been investigated. Although EZH2 is a bona fide substrate of Sirt1, it does not interact with Sirt2 (51). Likewise, SUV39H1 interacts specifically with Sirt1 but not with Sirt2 (29). Although SETDB1 negatively regulates the transcription of HBV viral promoters (26, 27), the Sirt2-SETDB1 interaction has never been examined in this context.
Since we found that HKMTs such as PR-Set7, EZH2, SETDB1, and SUV39H1 are recruited onto cccDNA (Fig. 7A), we performed coimmunoprecipitation assays (Fig. 7B and C). Briefly, total lysates from mock-, Sirt2.1-, or Sirt2.5-transfected or 1.3mer HBV WT plus Sirt2.1- or Sirt2.5-cotransfected cells were immunoprecipitated with an anti-FLAG antibody and immunoblotted with anti-PR-Set7, anti-EZH2, anti-SETDB1, or anti-SUV39H1 antibodies (Fig. 7B and C, first to fourth panels, data not shown). Neither Sirt2.1 nor Sirt2.5 interacted with EZH2 or SUV39H1 (data not shown).
We found that SETDB1 interacts with Sirt2.1 and Sirt2.5 (Fig. 7B, top, lanes 7 to 10) and that HBV replication did not affect the Sirt2.1-SETDB1 interaction (Fig. 7B, top, lane 7 versus 8). Interestingly, the Sirt2.5-SETDB1 interaction was stronger than the Sirt2.1-SETDB1 interaction (Fig. 7B, top, lane 7 versus 9), and the Sirt2.5-SETDB1 interaction in HBV-replicating cells was the strongest (Fig. 7B, top, lane 7 to 9 versus 10). Since Sirt2.1 and Sirt2.5 localize mainly in the cytoplasm and nucleus, respectively (21), we immunoprecipitated cytoplasmic and nuclear fractions with an anti-FLAG antibody, followed by immunoblotting with an anti-SETDB1 antibody (Fig. 7B, fifth to last panels). We found no significant differences in immunoprecipitation patterns between the total, cytoplasmic, and nuclear fractions (Fig. 7B, first, fifth, and ninth panels).
We next examined the interaction between PR-Set7 and Sirt2.1 and Sirt2.5 (Fig. 7C). As expected, PR-Set7 interacted with Sirt2.1; however, HBV replication had no effect on the interaction (Fig. 7C, top, lane 7 versus 8). As mentioned, the Sirt2.1–PR-Set7 interaction in total, cytoplasmic, and nuclear fractions was the same (Fig. 7C, lane 7 versus 8). Similar to the Sirt2.5-SETDB1 interaction, the Sirt2.5–PR-Set7 interaction was stronger than the Sirt2.1–PR-Set7 interaction (Fig. 7C, lane 7 versus 9) and was strongest in HBV-replicating cells (Fig. 7C, lane 7 to 9 versus 10).
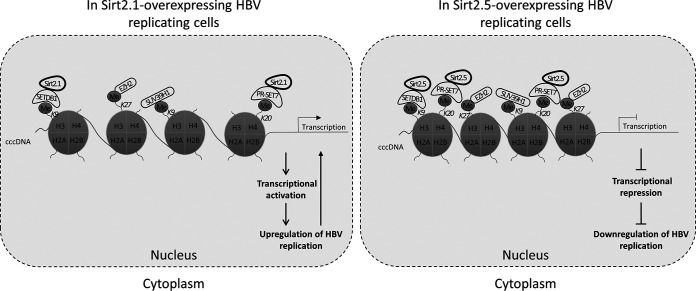
Taking all of the results into account, we propose a model of how Sirt2.5 overexpression induces epigenetic modification of HBV cccDNA to repress the transcription of HBV RNA (Fig. 8). HBV cccDNA, organized as a minichromosome, is assembled with histones H2A, H2B, H3, H4, linker histone H1 (not shown here), and nonhistone proteins (2, 52). Nonhistone proteins include HBc and HBx, various transcription factors and coactivators, and several epigenetic activators (not shown here) and repressors that affect HBV transcription, chromatin structure, and epigenetic control (2–4, 12, 13, 26–28, 32–34). In this model, Sirt2.5 is recruited onto cccDNA and binds to it indirectly through its interaction with SETDB1 and PR-Set7 (Fig. 8). PR-Set7 and SETDB1 may induce epigenetic repressive modification of cccDNA by depositing H4K20me1 and H3K9me3, respectively (Fig. 8). Although EZH2 and SUV39H1 do not interact with Sirt2, Sirt2.5 overexpression is associated with increased deposition of repressive markers on HBV cccDNA (such as H3K27me3 and H3K9me3, respectively) (Fig. 8), suggesting that these HKMTs play a role in epigenetic repressive modification mediated by overexpression of Sirt2.5.
FIG 8.
Sirt2.5 overexpression increases deposition of transcriptional repressive epigenetic markers on HBV cccDNA by repressive HKMTs. Repressive HKMTs, such as SETDB1, SUV39H1, PR-Set7, and EZH2, are recruited onto cccDNA in Sirt2.5-overexpressing cells to a greater extent than in Sirt2.1-overexpressing cells. H3K9me3 by SETDB1 and SUV39H1, H4K20me1 by PR-Set7, and H3K27me3 by EZH2 was more prominent in Sirt2.5-overexpressing cells than in Sirt2.1-overexpressing cells. Although both Sirt2.1 and Sirt2.5 interact with SETDB1 and PR-Set7 and can be recruited onto cccDNA, the Sirt2.5-SETDB1 and –PR-Set7 interactions are strengthened upon HBV replication, thereby inducing increased deposition of repressive markers in Sirt2.5-overexpressing cells. Although EZH2 and SUV39H1 cannot interact with either Sirt2.1 or Sirt2.5, more of them are recruited onto cccDNA, and deposition of repressive markers is increased in Sirt2.5-overexpressing cells. These repressive epigenetic modifications may silence cccDNA (closed state) and be transcriptionally inactive, thereby reducing viral replication.
DISCUSSION
HCC is the most common primary cancer of the liver (53). In 2012, about 14 million cases were reported, and this is expected to rise to 22 million in the next 2 decades (54). HCC risk factors include infection by HBV and/or hepatitis C virus (HCV) (55). A major strategy to reduce the morbidity and mortality associated with HBV infection and HBV-associated HCC is to inhibit HBV replication. Here, we demonstrate for the first time that overexpression of Sirt2.5 inhibits HBV replication.
The expression of Sirt2 mRNA and protein is modulated transcriptionally by transcription factors, posttranscriptionally by RNA binding proteins or microRNAs, translationally by the Myc oncoprotein, and posttranslationally via phosphorylation, acetylation, or ubiquitination of Sirt2 (25). HBV replication increased expression of Sirt2 mRNA (Fig. 1C), suggesting that Sirt2 mRNA levels are upregulated transcriptionally and/or posttranscriptionally in HBV-replicating cells. Since Sirt2 RNA levels approximately mirror Sirt2 protein levels, it is less likely that Sirt2 protein levels are upregulated translationally and/or posttranslationally (Fig. 1A and B). The ratios of the three isoforms of endogenous Sirt2 mRNAs in mock- and 1.3mer HBV-transfected cells were 100:85:49 and 100:85:60 (420:356:251), respectively (Fig. 1C). When we apply this calculation to protein, the protein ratios were 100:65:30 to 100:71:34 (253:180:85) (Fig. 1A), indicating that although expression of Sirt2 transcripts and proteins increases upon HBV replication, these increases are not selective for specific isoforms. We do not know why proteins from primary and alternatively spliced transcripts of a single gene exert opposite functions. With respect to the role of Sirt2 during HBV replication, we hypothesize that actively replicating HBV (driven by the increased expression of Sirt2.1 and Sirt2.2) is suppressed to some degree by the increased expression of Sirt2.5 as part of a self-regulating mechanism that modulates viral replication to prevent damage to the host and/or the virus.
The cellular localization of Sirts is important for their function (56); in addition, Rack et al. (21) suggest an activity-independent nuclear function of Sirt2.5. Here, we show that catalytically inactive nuclear Sirt2.5, which cannot deacetylate tubulin (Fig. 4B), is recruited onto cccDNA to a greater extent than Sirt2.1 upon HBV replication; recruitment occurs via direct and indirect association with repressive HKMTs, such as SETDB1, SUV39H1, EZH2, and PR-Set7 (Fig. 7).
To cure HBV infection completely, cccDNA must be eliminated (3–11, 31, 32). To eliminate cccDNA, new cccDNA synthesis must be inhibited and preexisting cccDNA must be cleared (3–11, 31, 32, 57). Here, we show clearly that overexpression of Sirt2.5 reduces the level of cccDNA (Fig. 2C). We hypothesize that reduced transcriptional activity due to epigenetic modification (Fig. 7) and reduced DNA synthesis (Fig. 3, 4B and C, 5, 6D and E, bottom) are responsible for the reduced cccDNA levels. Since Sirt2.5 reduced cccDNA levels to 45% of that in the control (Fig. 2C), Sirt2.5 may reduce intrahepatic cccDNA levels in CHB patients.
To functionally cure CHB infection, cccDNA must be transcriptionally inactivated (3–11, 31, 32, 57). To repress the transcriptional activity of cccDNA, cccDNA methylation and epigenetic modification of cccDNA-bound histones, such as lysine methylation, lysine ubiquitylation, and/or lysine sumoylation, are important (3, 4, 34, 57). In accordance with this concept, we found that increased recruitment of transcriptional repressive markers, such as H4K20me1, H3K9me3, and H3K27me3, along with recruitment of HKMTs, is responsible for these modifications (Fig. 7A). The result was the repression of HBV transcription from cccDNA in Sirt2.5-overexpressing HBV-infected cells (Fig. 2C, third panel). Thus, Sirt2.5 may mediate transcriptional inactivation of cccDNA through direct interaction with SETDB1 and PR-Set7 (Fig. 7B and C) and/or indirect interaction with EZH2 and SUV39H1 (data not shown). Recruitment of transcriptional repressive markers, such as lysine ubiquitylation and/or lysine sumoylation, to cccDNA will be investigated in the future.
Transcriptional activity of cccDNA in Sirt2.1-overexpressing HBV-infected cells may also need further investigation, since HBV transcription from cccDNA increased in Sirt2.1-overexpressing HBV-infected cells (Fig. 2C, third panel), but recruitment of SET1A, RNA pol I, and acetyl H3 (Fig. 7A), all of which are markers of active transcription, appeared not to be prominent. Since histone acetyltransferases (e.g., CBP, p300, and PCAF/GCN5) are recruited to cccDNA and induce epigenetic changes at the transcriptional level (3), we will investigate their effects in Sirt2.1- or Sirt2.5-overexpressing HBV-infected cells in the future. At the same time, lysine acetylation, phosphorylation, arginine methylation, lysine methylation, and/or lysine ubiquitylation on cccDNA-bound histones (4) can be investigated in Sirt2.1- or Sirt2.5-overexpressing HBV-infected cells.
Stress increases the Sirt2–PR-Set7 interaction and H4K20me1 deposition through deacetylation of H4K16ac and PR-Set7 at K90 (30). Here, we show that the Sirt2.5–PR-Set7 interaction and H4K20me1 deposition (but not the Sirt2.1–PR-Set7 interaction) increased upon HBV replication (Fig. 7A and B), indicating that Serrano et al. (30) also saw the Sirt2.5–PR-Set7 interaction under the stressed condition. We do not know whether catalytically inactive Sirt2.5 mediates deacetylation of H4K16ac and PR-Set7. Since endogenous Sirt1, Sirt2.1, and Sirt2.2 can deacetylate H4K16ac (58), they might participate in deacetylation of H4K16ac and PR-Set7. Thus, we propose that PR-Set7 recruits Sirt2 to cccDNA, and that Sirt2 deacetylates H4K16ac and induces PR-Set7 to methylate H4K20me1 to repress HBV transcription (Fig. 8). As shown here and in previous reports (12, 13, 27–30, 50, 58–60), we propose that deacetylation at specific acetylated lysines on histones, followed by methylation at that deacetylated lysine or at adjacent lysine residues, modulates chromatin structure to activate or repress HBV transcription.
In conclusion, we demonstrate that the overexpression of Sirt2.5 inhibits HBV replication through transcriptionally repressive epigenetic modifications by HKMTs on cccDNA.
MATERIALS AND METHODS
Vector construction.
HBV subtype ayw replication-competent 1.3mer HBV WT and HBx-deficient mutant plasmids were a gift from W. S. Ryu (Yonsei University, South Korea). pCDH-hNTCP-C9 from the human NTCP-C9 (h-NTCP-C9) construct in pcDNA6.1 (provided by W. Li) (61) was previously described (20). Constructs pCMV-HA-HBx WT, pCMV-3×FLAG-tagged Sirt2.1, Sirt2.2, and Sirt2.5, and luciferase reporter vectors pGL3-EnhI/Xp, pGL3-EnhII/Cp, pGL3-preS1p, and pGL3-preS2p were described previously (20). The 3×FLAG-tagged Sirt2.5 construct (20), which lacks an NES and N-terminal CD (21), was used to generate 3×FLAG-tagged Sirt2.5-NES, -NES-CD, and -CD constructs in which the NES, NES-CD, or CD, respectively, was restored using primers listed in Table 1. Lentiviral vectors encoding 3×FLAG-Sirt2.1 and 3×FLAG-Sirt2.5 were generated by insertion of the EcoRI- and BamHI-digested 3×FLAG-Sirt2.1 or 3×FLAG-Sirt2.5 DNA fragment into the linearized pCDH-CMV-MCS-EF1-Puro (CD510B-1; System Biosciences) to yield pCDH-3×FLAG-Sirt2.1 or pCDH-3×FLAG-Sirt2.5, respectively (primers are listed in Table 1). All PCR products were sequenced to confirm the presence of specific mutations and the absence of extraneous mutations.
Cell culture and transfection of DNA.
HepAD38, HepG2, HepG2-hNTCP-C9, Huh7, and HEK293T cells were maintained in a humidified atmosphere (37°C, 5% CO2) in Dulbecco’s modified Eagle’s medium supplemented with 1% penicillin-streptomycin and 10% fetal bovine serum (Gibco BRL). Cells were passaged as described previously (20). Cultured HepAD38 cells were used to produce an HBV inoculum for the infection experiments, as described previously (20, 62). For transfection into Huh7 (1 × 106) cells, 4 μg of the 1.3mer HBV WT construct or control vector was mixed with 12 μg/μl polyethylenimine (PEI; Polysciences) and 200 μl of Opti-MEM (Gibco) and then layered onto cells in 6-cm plates at 24 h postseeding. For cotransfection into Huh7 (1 × 106) or HepG2 (3 × 106) cells in 6-cm plates, 4 μg of the 1.3mer HBV WT plus 4 μg of the control, 3×FLAG-Sirt2.1, or 3×FLAG-Sirt2.5 construct was mixed with 24 μg/μl of PEI and 200 μl of Opti-MEM and layered onto cells at 24 h postseeding. For cotransfection into SIRT2-KD or shCont HepG2 cells (3 × 106) in 6-cm plates, 4 μg of 1.3mer HBV WT plus 4 μg of the control pcDNA3, 3×FLAG-Sirt2.1, or 3×FLAG-Sirt2.5 construct were mixed with 32 μg/μl PEI and 200 μl of Opti-MEM. For cotransfection into HepG2 (3 × 106) cells in 6-cm plates, 4 μg of 1.3mer HBV WT or HBx-deficient mutant plus 4 μg of the control, 3×FLAG-Sirt2.1, or 3×FLAG-Sirt2.5 construct were mixed with 32 μg/μl PEI and 200 μl of Opti-MEM. For triple transfection into HepG2 (3 × 106) cells in 6-cm plates, 4 μg of 1.3mer HBx-deficient mutant plus 4 μg of HA-HBx plus 4 μg of the control, 3×FLAG-Sirt2.1, or 3×FLAG-Sirt2.5 construct were mixed with 64 μg/μl PEI and 200 μl of Opti-MEM. To examine the effects of Sirt2 isoforms and Sirt2.5-derived mutants on HBV replication, 1 × 106 Huh7 cells in 6-cm plates were cotransfected with 4 μg of 1.3mer HBV WT plus 4 μg of control, 3×FLAG-tagged Sirt2.1, Sirt2.2, Sirt2.5, Sirt2.5-NES, Sirt2.5-NES-CD, or Sirt2.5-CD at 24 h postseeding. Throughout the experiments, pcDNA3 was used to adjust the amount of transfected DNA. The medium in which transfected cells were cultured was refreshed at 24 h posttransfection to remove transfected DNA. Cells were harvested at 72 h posttransfection.
Establishment of stable cell lines.
Stable HepG2-hNTCP-C9 cells were generated as described previously (2, 20, 63). A lentiviral expression system was used to generate cells stably overexpressing 3×FLAG-Sirt2.1 or -Sirt2.5 in HepG2-hNTCP-C9. Briefly, 1 × 106 HEK293T cells were seeded in 6-cm plates for 24 h and then transfected with 0.5 μg of pVSV-G, 1.5 μg of pGAG-Pol, and 2 μg of pCDH empty, pCDH-3×FLAG-Sirt2.1, or pCDH-3×FLAG-Sirt2.5 vectors plus 12 μg/ml PEI in 200 μl of Opti-MEM. At 24 h posttransfection, the medium was refreshed and supernatant containing pseudoviral particles harboring 3×FLAG-Sirt2.1 or 3×FLAG-Sirt2.5 transcripts was harvested at 72 h posttransfection. In the next step, 2 ml of supernatant containing pseudoviral particles was mixed with 2 ml of fresh culture medium and used to transduce HepG2-hNTCP-C9 cells after addition of 10 μg/ml Polybrene (hexadimethrine bromide; Sigma-Aldrich). The medium was refreshed at 24 h postransduction, and transduced cells were reseeded into new culture plates. Cells were selected for 72 h using 6 μg/ml puromycin (Sigma-Aldrich) to yield HepG2-hNTCP-C9-pCDH, -3×FLAG-Sirt2.1, and -3×FLAG-Sirt2.5 cells. Stable SIRT2 KD HepG2 cells were generated as described previously (20). Briefly, HEK293T cells were transfected as described above with 2 μg of pSIH1-H1-Puro-shControl or pSIH1-H1-Puro-shSIRT2#2 construct. Pseudoviral particles containing shControl or shSIRT2#2 RNAs were inoculated into HepG2 cells, which were then selected as described above to generate stable HepG2-shCont or HepG2-SIRT2 KD cells, respectively.
Northern and Southern blotting.
To analyze HBV RNAs by Northern blotting, total RNA was extracted from Huh7, HepG2, and HepG2-hNTCP-C9 cells using TRIzol reagent (15596026; Ambion, Invitrogen) according to the manufacturer’s instructions. Total RNA (20 μg) was denatured at 65°C for 10 min and then electrophoresed on 1.2% agarose gels as described previously (2, 20). RNAs were transferred to nylon membranes (11417240001; Roche, Sigma-Aldrich) and hybridized at 68°C for 4 h with a 32P-labeled random-primed probe specific for full-length HBV. To analyze HBV DNA synthesis by Southern blotting, HBV DNA was extracted from isolated core particles as described previously (64). Briefly, HBV DNAs were electrophoresed on agarose gels, transferred to nylon membranes (10416296; Whatman), and hybridized with a 32P-labeled random-primed probe specific for full-length HBV.
Immunoblotting of core particles.
To analyze HBV core particles, cells were lysed using 0.2% NP-40 (IGEPAL; Sigma-Aldrich)-TNE (10 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1 mM EDTA) buffer, as described previously (2, 20, 64). Briefly, 4% of the total cell lysate was subjected to electrophoresis on 1% native agarose gels, transferred to polyvinylidene fluoride (PVDF) membranes (Millipore), and immunoblotted with a rabbit polyclonal anti-HBc primary antibody (1:1,000 dilution; generated in-house) (65), followed by incubation with a horseradish peroxidase-conjugated anti-rabbit secondary antibody (1:5,000 dilution; Thermo Fisher Scientific). Bound secondary antibodies were visualized by enhanced chemiluminescence (ECL Western blotting detection reagent; Amersham). The relative intensities of core particles were calculated using ImageJ v.1.46r software.
SDS-PAGE and immunoblotting.
The protein context of cell lysates in 0.2% NP-40 (IGEPAL)-TNE buffer was quantified using Bradford assay (66). Tumor tissue and the corresponding adjacent nontumoral liver tissues were obtained from HBV-associated HCC patients after surgeries from Gangnam Severance Hospital, Yonsei University College of Medicine. All samples were obtained with informed consent under the institutional review board-approved protocol (3-2019-0031). For immunoblotting, ice-cold mPER mammalian protein extraction reagent (78501; Thermo Scientific) with 1× protease and phosphatase inhibitor cocktail (78440; Thermo Scientific) were added to chopped biopsy samples. The samples were homogenized on ice with a Dounce homogenizer. Supernatant from homogenized samples was collected by centrifugation at 12,000 rpm for 15 min. Equal amounts were subjected to SDS-PAGE on 10% gels. Resolved proteins were then transferred to PVDF membranes and incubated with appropriate primary antibodies (1:1,000) (Table 2). Secondary antibodies were horseradish peroxidase-conjugated anti-rabbit (1:5,000 dilution; 31460; Thermo Fisher Scientific) or horseradish peroxidase-conjugated anti-mouse Ig (1:5,000; 5220-0460; Seracare). ECL was performed to visualize protein bands. Relative band intensities were measured using ImageJ 1.46r software.
TABLE 2.
Antibodies used for this study
| Antibody target | Species | Expt | Supplier | Catalog no. or reference |
|---|---|---|---|---|
| HBc | Rabbit polyclonal | SDS-PAGE-IB | In-house | 65 |
| FLAG M2 | Mouse monoclonal | SDS-PAGE-IB/Co-IP/ChIP | Sigma | F1804 |
| Sirt2 | Rabbit polyclonal | SDS-PAGE-IB/ChIP | Thermo Fisher Scientific | PA3-200 |
| Sirt2 | Rabbit polyclonal | ChIP | Santa Cruz | sc-20966 |
| GAPDH | Mouse monoclonal | SDS-PAGE-IB | Santa Cruz | sc-32233 |
| Tubulin | Mouse monoclonal | SDS-PAGE-IB | Santa Cruz | sc-8035 |
| Acetylated tubulin | Mouse monoclonal | SDS-PAGE-IB | Sigma-Aldrich | T6793 |
| GSK-3β | Mouse monoclonal | SDS-PAGE-IB | Santa Cruz | sc-81462 |
| GSK-3β (pS9) | Rabbit monoclonal | SDS-PAGE-IB | Cell Signaling Technology | 9336 |
| β-catenin | Rabbit monoclonal | SDS-PAGE-IB | Cell Signaling Technology | D10A8 |
| AKT | Rabbit polyclonal | SDS-PAGE-IB/Co-IP | Cell Signaling Technology | 9272 |
| AKT (pT308) | Rabbit polyclonal | SDS-PAGE-IB | Cell Signaling Technology | 9275 |
| AKT (pS473) | Rabbit polyclonal | SDS-PAGE-IB | Cell Signaling Technology | 9271 |
| Anti-rhodopsin-C9 | Mouse monoclonal | SDS-PAGE-IB | Millipore | MAB5356 |
| HA | Mouse monoclonal | SDS-PAGE-IB | Abcam | ab18181 |
| Histone H3 | Rabbit polyclonal | SDS-PAGE-IB/ChIP | Abcam | ab1791 |
| Acetylated H3 | Rabbit polyclonal | ChIP | Merck Millipore | 06–599 |
| H3K27me3 | Mouse monoclonal | ChIP | Cell Signaling Technology | 9733 |
| H3K9me3 | Rabbit polyclonal | ChIP | Abcam | ab8898 |
| H4K20me1 | Rabbit monoclonal | ChIP | Abcam | ab177188 |
| Sirt1 | Mouse monoclonal | ChIP | Santa Cruz | sc-74465 |
| HDAC6 | Rabbit polyclonal | ChIP | Santa Cruz | sc-11420 |
| RNA Pol II | Mouse monoclonal | ChIP | Abcam | ab817 |
| SUV39H1 | Rabbit monoclonal | SDS-PAGE-IB/Co-IP/ChIP | Cell Signaling Technology | 8729 |
| PR-Set7 | Rabbit polyclonal | SDS-PAGE-IB/Co-IP/ChIP | Abcam | ab230683 |
| EZH2 | Rabbit monoclonal | SDS-PAGE-IB/Co-IP/ChIP | Abcam | ab191250 |
| SETDB1 | Rabbit polyclonal | SDS-PAGE-IB/Co-IP/ChIP | Abcam | ab12317 |
| SET1A | Rabbit monoclonal | ChIP | Cell Signaling Technology | 61702 |
Preparation of HBV and cell infection.
HBV virions were prepared from HepAD38 cells and used to infect hNTCP-C9-expressing HepG2 cells, as described previously (2, 20, 62, 67). For HBV infection, 2 × 105 HepG2 or HepG2-hNTCP-C9-3×FLAG-Sirt2.5 cells on collagen-coated (354249; Corning) 6-well plates were infected with HBV (1.7 × 103 GEq/per cell) in medium containing 4% polyethylene glycol (25322-68-3; Affymetrix), as described previously (68). Cells were washed with phosphate-buffered saline (PBS) at 1 day postinfection and maintained in the same medium containing 2.5% dimethyl sulfoxide (DMSO) (20, 68). For Northern blot analysis of infected cells, total RNA was extracted at 5 days postinfection. Infected cell lysates were prepared at 9 days postinfection and subjected to SDS-PAGE and immunoblotting, core particle immunoblotting, cccDNA extraction, ChIP analysis, and Southern blot analysis, as described above and below.
Extraction of cccDNA.
To examine the effect of Sirt2.1 or Sirt2.5 on HBV cccDNA formation, cccDNA was extracted using a Hirt protein-free DNA extraction procedure, as described previously (38), with minor modifications (2). Briefly, 2 × 105 HepG2-hNTCP-C9-3×FLAG-Sirt2.1 or -Sirt2.5 cells on collagen-coated 6-well plates were infected with HBV as described in “Preparation of HBV and cell infection,” above. At 9 days postinfection (when cells were 100% confluent), cells were lysed for 30 min at room temperature with 0.6% SDS-TE buffer (10 mM Tris-HCl [pH 7.5], 10 mM EDTA). The NaCl concentration in the lysates was adjusted to 1 M NaCl by addition of 5 M NaCl and then incubated for 16 h at 4°C to precipitate proteins and protein-associated DNAs. After centrifuging at 14,500 × g for 30 min, the supernatant was subjected to two rounds of phenol extraction, followed by one round of phenol-chloroform extraction. Finally, cccDNA was precipitated with ethanol and analyzed by Southern blotting.
Luciferase reporter assay.
HepG2 (1 × 106) or Huh7 (1 × 105) cells in 6-cm plates were cotransfected with 2 μg of luciferase report vectors (pGL3-null, pGL3-EnhI/Xp, pGL3-EnhII/Cp, pGL3-PreS1p, or pGL3-PreS2p) plus 2 μg of 3×FLAG-Sirt2.1 or 3×FLAG-Sirt2.5, as described previously (20). pcDNA3 then was used to adjust the amount of transfected DNA. Cells were lysed at 72 h posttransfection, and luciferase activity was analyzed using luciferin (Promega) and a luminometer (Molecular Devices).
Cytoplasmic and nuclear fractionation.
HEK293T cells (2 × 106) were transfected with 4 μg of control or 3×FLAG-tagged Sirt2.1, Sirt2.2, Sirt2.5, Sirt2.5-NES, Sirt2.5-NES-CD, or Sirt2.5-CD vector. At 72 h posttransfection, cells were harvested and subjected to cytoplasmic and nuclear fractionation, as described previously (2, 20, 69). The purity of the cytoplasmic and nuclear fractions was determined by analysis against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and histone H3, respectively.
Coimmunoprecipitation.
To examine the physical interaction of Sirt2.1 and/or Sirt2.5 with AKT, PR-Set7, SETDB1, SUV39H1, and EZH2 in Sirt2.1- or Sirt2.5-overexpressing HBV-replicating cells, 1 × 106 Huh7 cells on 10-cm plates were transfected with pcDNA3, 3×FLAG-Sirt2.1, or 3×FLAG-Sirt2.5 constructs or cotransfected with 1.3mer HBV WT plus the 3×FLAG-Sirt2.1 or 3×FLAG-Sirt2.5 construct. At 72 h posttransfection, the total, cytoplasmic, and nuclear fractions were prepared as described above (2, 20, 69). For immunoprecipitation, mouse monoclonal anti-FLAG M2 antibody was added to the lysates, as described previously (2, 20), followed by immunoblotting with rabbit polyclonal anti-AKT, rabbit polyclonal PR-Set7, rabbit polyclonal anti-SETDB1, rabbit polyclonal anti-SUV39H1, or rabbit polyclonal anti-EZH2 antibodies. Normal mouse IgG was used as a negative control for immunoprecipitation (12-371; Merck Millipore).
Immunoprecipitation of cccDNA chromatin.
Chromatin immunoprecipitation (ChIP) of HBV cccDNA was performed as described previously (3), with minor modifications (2). Briefly, HepG2-NTCP-C9 cells stably expressing control, 3×FLAG-Sirt2.1, or 3×FLAG-Sirt2.5 were seeded onto collagen-coated 6-well plates and infected with HBV as described in “Preparation of HBV and cell infection,” above. Cells were maintained for 8 days before chromatin solutions were obtained as described previously (2, 3). Chromatin preparations were subjected to immunoprecipitation with 3 μg of antibodies (Table 2) or normal mouse or rabbit IgG (negative controls) overnight at 4°C and then centrifuged at 1,000 × g for 5 min at 4°C to recover the immunoprecipitated protein-DNA complexes. Protein-DNA complexes were eluted in elution buffer (1% SDS, 0.1 M NaHCO3) and incubated for 15 min at room temperature with rotation. Immune complex cross-linking was reversed by heating at 60°C for 4 h. Immunoprecipitated DNA was purified by proteinase K (P2308; Sigma-Aldrich) treatment, phenol-chloroform extraction, and ethanol precipitation. The DNA pellet was redissolved in nuclease-free water (R0581; Fermentas). Input samples were prepared separately from sonicated chromatin solutions, as described previously (2, 3). The DNA concentration was adjusted to 50 ng after measurement of the optical density at 260 nm. Actin (Table 1) was used to facilitate equal loading from lysate samples. Immunoprecipitated chromatin was analyzed by PCR (GeneAmp PCR system 2700; Applied Biosystems) using cccDNA-specific primers (Table 1), as specified by the manufacturer (17-371; Merck Millipore EZ ChIP).
RPA.
To analyze the level of mRNAs encoding Sirt2 isoforms, total RNA was prepared from mock- and 1.3mer HBV WT-transfected Huh7 cells at 72 h posttransfection using TRIzol reagent. To prepare a riboprobe for RPA, part of the Sirt2.1 sequence (nt 52 to 400) was cloned into the pGEM3Zf(+) vector. From this construct, 367 nt of radiolabeled antisense riboprobe was synthesized using T7 RNA polymerase plus [α-32P]UTP (specific activity, 800 Ci/mmol). The product was then gel purified. The RPA procedure was performed as described by the manufacturer (RPA II; Ambion) (64). Following digestion with RNase A/T1 (EN0551; Thermo Fisher Scientific), protected mRNAs of 350 nt (Sirt2.1), 290 nt (Sirt2.2), and 180 nt (Sirt2.5) were run on 5% polyacrylamide–8 M urea gels and visualized by autoradiography. To analyze encapsidated pgRNA, core particles were isolated as described previously (64). Total RNA was extracted with TRIzol reagent as described in “Northern and Southern blotting,” above. Riboprobe of RPA used was prepared as described previously (64), with minor modifications. In brief, from the part of the HBV sequence (nt 1805 to 2187) in the pGEM3Zf(+) vector (64), 446 nt of digoxigenin (DIG)-UTP-labeled antisense probe was synthesized in vitro using SP6 RNA polymerase (P108B; Promega) with a kit (1363514; Roche). The RPA procedure was performed as described previously (64), except that labeling was performed with DIG by following the manufacturer’s instructions (11585762001 and 1363514; Roche). Protected encapsidated and total RNAs (369 nt) following RNase digestion were run on a 5% polyacrylamide–8 M urea gel, transferred to nylon membranes, incubated with anti-DIG-AP-antibody (1:1,000; 11093274910; Roche), and detected with CSPD (11755633001; Roche) through a chemiluminescent reaction.
Immunofluorescence analysis and confocal microscopy.
Samples for confocal microscopic analysis were prepared as described previously (2). Briefly, Huh7 (2 × 104) cells grown on coverslips in 24-well plates were transfected with 0.5 μg of 3×FLAG-tagged Sirt2.1, Sirt2.5, or Sirt2.5-NES-CD vector along with 0.5 μg of 1.3mer HBV WT plus 0.5 μg of the respective 3×FLAG-tagged Sirt2.1, Sirt2.5, or Sirt2.5-NES-CD vector. For the (co)transfections, DNA was mixed with 3 μg/μl PEI in 100 μl Opti-MEM. At 72 h posttransfection, cells were fixed for 20 min with 4% paraformaldehyde, followed by washing three times in 1× PBS. The cells then were permeabilized with 0.2% Triton X-100 (0.1% saponin [S7900-25G; Sigma-Aldrich]–1% bovine serum albumin [BSAS0.1; Bovogen Biologicals]–0.1% sodium azide [S2002-100G; Sigma-Aldrich] in 1× PBS) for 20 min, followed by washing three times in 1× PBS. Cells then were incubated overnight at 4°C with anti-FLAG M2 (1:300), Alexa Fluor 647 anti-nuclear pore complex (NPC) antibody (1:300; 682204; BioLegend), anti-Sirt2 H-95 (1:300), anti-Sirt2 PA3-200 (1:300), or anti-HBc (1:300) in permeabilization buffer. Immunofluorescence detection was performed using fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG + IgM (1:350; 115-095-044; Jackson ImmunoResearch) and tetramethyl rhodamine isocyanate (TRITC)-conjugated goat anti-rabbit IgG (1:350; 115-025-003; Jackson ImmunoResearch) in permeabilization buffer at room temperature for 1 h. Cells were mounted with Fluoroshield mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI) (ab104139; Abcam). Digital images of stained cells were captured under a confocal microscope (LSM710; Zeiss, Germany).
Statistical analysis.
Data are expressed as the means ± standard deviations. Mean values were compared using Student's t test. P values of <0.05 were considered statistically significant.
ACKNOWLEDGMENT
This work was supported by National Research Foundation grants funded by the Korean Government (NRF-2019-R1A2C2005749).
REFERENCES
- 1.Summers J, Mason WS. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 2.Saeed U, Kim J, Piracha ZZ, Kwon H, Jung J, Chwae YJ, Park S, Shin HJ, Kim K. 2018. Parvulin 14 and parvulin 17 bind to HBx and cccDNA and upregulate hepatitis B virus replication from cccDNA to virion in an HBx-dependent manner. J Virol 93:e01840-18. doi: 10.1128/JVI.01840-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, Raimondo G, Levrero M. 2009. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci U S A 106:19975–19979. doi: 10.1073/pnas.0908365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, Dandri M. 2009. Control of cccDNA function in hepatitis B virus infection. J Hepatol 51:581–592. doi: 10.1016/j.jhep.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 5.Ganem D, Prince AM. 2004. Hepatitis B virus infection–natural history and clinical consequences. N Engl J Med 350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 6.Locarnini S. 2004. Molecular virology of hepatitis B virus. Semin Liver Dis 24:3–10. doi: 10.1055/s-2004-828672. [DOI] [PubMed] [Google Scholar]
- 7.Seeger C, Mason WS. 2015. Molecular biology of hepatitis B virus infection. Virology 479-480:672–686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papatheodoridis GV, Manolakopoulos S, Dusheiko G, Archimandritis AJ. 2008. Therapeutic strategies in the management of patients with chronic hepatitis B virus infection. Lancet Infect Dis 8:167–178. doi: 10.1016/S1473-3099(07)70264-5. [DOI] [PubMed] [Google Scholar]
- 9.Testoni B, Durantel D, Zoulim F. 2017. Novel targets for hepatitis B virus therapy. Liver Int 37:33–39. doi: 10.1111/liv.13307. [DOI] [PubMed] [Google Scholar]
- 10.Revill PA, ICE-HBV Senior Advisors, Chisari FV, Block JM, Dandri M, Gehring AJ, Guo H, Hu J, Kramvis A, Lampertico P, Janssen HLA, Levrero M, Li W, Liang TJ, Lim S-G, Lu F, Penicaud MC, Tavis JE, Thimme R, Zoulim F. 2019. A global scientific strategy to cure hepatitis B. Lancet Gastroenterol Hepatol 4:545–558. doi: 10.1016/S2468-1253(19)30119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo JT, Guo H. 2015. Metabolism and function of hepatitis B virus cccDNA: implications for the development of cccDNA-targeting antiviral therapeutics. Antiviral Res 122:91–100. doi: 10.1016/j.antiviral.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belloni L, Allweiss L, Guerrieri F, Pediconi N, Volz T, Pollicino T, Petersen J, Raimondo G, Dandri M, Levrero M. 2012. IFN-α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J Clin Investig 122:529–537. doi: 10.1172/JCI58847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren JH, Hu JL, Cheng ST, Yu HB, Wong VKW, Law BYK, Yang YF, Huang Y, Liu Y, Chen WX, Cai XF, Tang H, Hu Y, Zhang WL, Liu X, Long QX, Zhou L, Tao NN, Zhou HZ, Yang QX, Ren F, He L, Gong R, Huang AL, Chen J. 2018. SIRT3 restricts hepatitis B virus transcription and replication through epigenetic regulation of covalently closed circular DNA involving suppressor of variegation 3–9 homolog 1 and SET domain containing 1A histone methyltransferases. Hepatology 68:1260–1276. doi: 10.1002/hep.29912. [DOI] [PubMed] [Google Scholar]
- 14.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. 2003. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawlor L, Yang XB. 2019. Harnessing the HDAC-histone deacetylase enzymes, inhibitors and how these can be utilised in tissue engineering. Int J Oral Sci 11:20. doi: 10.1038/s41368-019-0053-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finnin MS, Donigian JR, Pavletich NP. 2001. Structure of the histone deacetylase SIRT2. Nat Struct Biol 8:621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- 17.North BJ, Verdin E. 2004. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol 5:224. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wątroba M, Dudek I, Skoda M, Stangret A, Rzodkiewicz P, Szukiewicz D. 2017. Sirtuins, epigenetics and longevity. Ageing Res Rev 40:11–19. doi: 10.1016/j.arr.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Frye RA. 2000. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun 273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 20.Piracha ZZ, Kwon H, Saeed U, Kim J, Jung J, Chwae YJ, Park S, Shin HJ, Kim K. 2018. Sirtuin 2 isoform 1 enhances hepatitis B virus RNA transcription and DNA synthesis through the AKT/GSK-3β/β-catenin signaling pathway. J Virol 92:e00955-18. doi: 10.1128/JVI.00955-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Rack JG, VanLinden MR, Lutter T, Aasland R, Ziegler M. 2014. Constitutive nuclear localization of an alternatively spliced sirtuin-2 isoform. J Mol Biol 426:1677–1691. doi: 10.1016/j.jmb.2013.10.027. [DOI] [PubMed] [Google Scholar]
- 22.North BJ, Verdin E. 2007. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS One 2:e784. doi: 10.1371/journal.pone.0000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. 2003. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell 11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 24.Beirowski B, Gustin J, Armour SM, Yamamoto H, Viader A, North BJ, Michán S, Baloh RH, Golden JP, Schmidt RE, Sinclair DA, Auwerx J, Milbrandt J. 2011. Sir-two-homolog 2 (Sirt2) modulates peripheral myelination through polarity protein Par-3/atypical protein kinase C (aPKC) signaling. Proc Natl Acad Sci U S A 108:E952–E961. doi: 10.1073/pnas.1104969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Yang J, Hong T, Chen X, Cui L. 2019. SIRT2: controversy and multiple roles in disease and physiology. Ageing Res Rev 55:100961. doi: 10.1016/j.arr.2019.100961. [DOI] [PubMed] [Google Scholar]
- 26.Alarcon V, Hernández S, Rubio L, Alvarez F, Flores Y, Varas-Godoy M, De Ferrari GV, Kann M, Villanueva RA, Loyola A. 2016. The enzymes LSD1 and Set1A cooperate with the viral protein HBx to establish an active hepatitis B viral chromatin state. Sci Rep 6:25901. doi: 10.1038/srep25901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rivière L, Gerossier L, Ducroux A, Dion S, Deng Q, Michel ML, Buendia MA, Hantz O, Neuveut C. 2015. HBx relieves chromatin-mediated transcriptional repression of hepatitis B viral cccDNA involving SETDB1 histone methyltransferase. J Hepatol 63:1093–1102. doi: 10.1016/j.jhep.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 28.Hong X, Kim ES, Guo H. 2017. Epigenetic regulation of hepatitis B virus covalently closed circular DNA: implications for epigenetic therapy against chronic hepatitis B. Hepatology 66:2066–2077. doi: 10.1002/hep.29479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. 2007. SIRT1 regulates the histone methyltransferase SUV39H1 during heterochromatin formation. Nature 450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 30.Serrano L, Martínez-Redondo P, Marazuela-Duque A, Vazquez BN, Dooley SJ, Voigt P, Beck DB, Kane-Goldsmith N, Tong Q, Rabanal RM, Fondevila D, Muñoz P, Krüger M, Tischfield JA, Vaquero A. 2013. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev 27:639–653. doi: 10.1101/gad.211342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Mao R, Yan R, Cai D, Zhang Y, Zhu H, Kang Y, Liu H, Wang J, Qin Y, Huang Y, Guo H, Zhang J. 2014. Transcription of hepatitis B virus covalently closed circular DNA is regulated by CpG methylation during chronic infection. PLoS One 9:e110442. doi: 10.1371/journal.pone.0110442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bock CT, Schwinn S, Locarnini S, Fyfe J, Manns MP, Trautwein C, Zentgraf H. 2001. Structural organization of the hepatitis B virus minichromosome. J Mol Biol 307:183–196. doi: 10.1006/jmbi.2000.4481. [DOI] [PubMed] [Google Scholar]
- 33.Zhang W, Chen J, Wu M, Zhang X, Zhang M, Yue L, Li Y, Liu J, Li B, Shen F, Wang Y, Bai L, Protzer U, Levrero M, Yuan Z. 2017. PRMT5 restricts hepatitis B virus replication through epigenetic repression of covalently closed circular DNA transcription and interference with pregenomic RNA encapsidation. Hepatology 66:398–415. doi: 10.1002/hep.29133. [DOI] [PubMed] [Google Scholar]
- 34.Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, Levrero M. 2006. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology 130:823–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Byrd JC, Marcucci G, Parthun MR, Xiao JJ, Klisovic RB, Moran M, Lin TS, Liu S, Sklenar AR, Davis ME, Lucas DM, Fischer B, Shank R, Tejaswi SL, Binkley P, Wright J, Chan KK, Grever MR. 2005. A phase 1 and pharmacodynamic study of depsipeptide (FK228) in chronic lymphocytic leukemia and acute myeloid leukemia. Blood 105:959–967. doi: 10.1182/blood-2004-05-1693. [DOI] [PubMed] [Google Scholar]
- 36.Sells MA, Chen ML, Acs G. 1987. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci U S A 84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT, Seeger C, King RW. 1997. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob Agents Chemother 41:1715–1720. doi: 10.1128/AAC.41.8.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai D, Nie H, Yan R, Guo JT, Block TM, Guo H. 2013. A southern blot assay for detection of hepatitis B virus covalently closed circular DNA from cell cultures. Methods Mol Biol 1030:151–161. doi: 10.1007/978-1-62703-484-5_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bosch-Presegué L, Vaquero A. 2011. The dual role of sirtuins in cancer. Genes Cancer 2:648–662. doi: 10.1177/1947601911417862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Chan AW, To KF, Chen W, Zhang Z, Ren J, Song C, Cheung YS, Lai PB, Cheng SH, Ng MH, Huang A, Ko BC. 2013. SIRT2 overexpression in hepatocellular carcinoma mediates epithelial to mesenchymal transition by protein kinase B/glycogen synthase kinase-3β/β-catenin signaling. Hepatology 57:2287–2298. doi: 10.1002/hep.26278. [DOI] [PubMed] [Google Scholar]
- 41.Ramakrishnan G, Davaakhuu G, Kaplun L, Chung WC, Rana A, Atfi A, Miele L, Tzivion G. 2014. Sirt2 deacetylase is a novel AKT binding partner critical for AKT activation by insulin. J Biol Chem 289:6054–6066. doi: 10.1074/jbc.M113.537266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng ST, Ren JH, Cai XF, Jiang H, Chen J. 2018. HBx-elevated SIRT2 promotes HBV replication and hepatocarcinogenesis. Biochem Biophys Res Commun 496:904–910. doi: 10.1016/j.bbrc.2018.01.127. [DOI] [PubMed] [Google Scholar]
- 43.Cha MY, Kim CM, Park YM, Ryu WS. 2004. Hepatitis B virus X protein is essential for the activation of Wnt/beta-catenin signaling in hepatoma cells. Hepatology 39:1683–1693. doi: 10.1002/hep.20245. [DOI] [PubMed] [Google Scholar]
- 44.Hsieh A, Kim HS, Lim SO, Yu DY, Jung G. 2011. Hepatitis B viral X protein interacts with tumor suppressor adenomatous polyposis coli to activate Wnt/β-catenin signaling. Cancer Lett 300:162–172. doi: 10.1016/j.canlet.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 45.Melegari M, Wolf SK, Schneider RJ. 2005. Hepatitis B virus DNA replication is coordinated by core protein serine phosphorylation and HBx expression. J Virol 79:9810–9820. doi: 10.1128/JVI.79.15.9810-9820.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bouchard MJ, Wang LH, Schneider RJ. 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294:2376–2378. doi: 10.1126/science.294.5550.2376. [DOI] [PubMed] [Google Scholar]
- 47.Yoon S, Jung J, Kim T, Park S, Chwae YJ, Shin HJ, Kim K. 2011. Adiponectin, a downstream target gene of peroxisome proliferator-activated receptor γ, controls hepatitis B virus replication. Virology 409:290–298. doi: 10.1016/j.virol.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 48.Iwamoto M, Cai D, Sugiyama M, Suzuki R, Aizaki H, Ryo A, Ohtani N, Tanaka Y, Mizokami M, Wakita T, Guo H, Watashi K. 2017. Functional association of cellular microtubules with viral capsid assembly supports efficient hepatitis B virus replication. Sci Rep 7:10620. doi: 10.1038/s41598-017-11015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. 2002. HDAC6 is a microtubule-associated deacetylase. Nature 417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 50.Beck DB, Oda H, Shen SS, Reinberg D. 2012. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev 26:325–337. doi: 10.1101/gad.177444.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wan J, Zhan J, Li S, Ma J, Xu W, Liu C, Xue X, Xie Y, Fang W, Chin YE, Zhang H. 2015. PCAF-primed EZH2 acetylation regulates its stability and promotes lung adenocarcinoma progression. Nucleic Acids Res 43:3591–3604. doi: 10.1093/nar/gkv238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bock CT, Schranz P, Schroder CH, Zentgraf H. 1994. Hepatitis B virus genome is organized into nucleosomes in the nucleus of the infected cell. Virus Genes 8:215–229. doi: 10.1007/BF01703079. [DOI] [PubMed] [Google Scholar]
- 53.Rawla P, Sunkara T, Muralidharan P, Raj JP. 2018. Update in global trends and aetiology of hepatocellular carcinoma. Contemp Oncol 22:141–150. doi: 10.5114/wo.2018.78941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stewart BW, Wild CP. 2014. World Health Organization: world cancer report 2014. International Agency for Research on Cancer, Lyon, France: http://publications.iarc.fr/Non-Series-Publications/World-Cancer-Reports/World-Cancer-Report-2014. Accessed 28 February 2020. [Google Scholar]
- 55.El-Serag HB. 2011. Hepatocellular carcinoma. N Engl J Med 365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 56.Michan S, Sinclair D. 2007. Sirtuins in mammals: insights into their biological function. Biochem J 404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mohd-Ismail NK, Lim Z, Gunaratne J, Tan YJ. 2019. Mapping the interactions of HBV cccDNA with host factors. Int J Mol Sci 20:E4276. doi: 10.3390/ijms20174276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. 2006. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev 20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. 2005. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 60.Bell O, Wirbelauer C, Hild M, Scharf AN, Schwaiger M, MacAlpine DM, Zilbermann F, van Leeuwen F, Bell SP, Imhof A, Garza D, Peters AH, Schübeler D. 2007. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J 26:4974–4984. doi: 10.1038/sj.emboj.7601926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. 2012. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watashi K, Liang G, Iwamoto M, Marusawa H, Uchida N, Daito T, Kitamura K, Muramatsu M, Ohashi H, Kiyohara T, Suzuki R, Li J, Tong S, Tanaka Y, Murata K, Aizaki H, Wakita T. 2013. Interleukin-1 and tumor necrosis factor-α trigger restriction of hepatitis B virus infection via a cytidine deaminase activation-induced cytidine deaminase (AID). J Biol Chem 288:31715–31727. doi: 10.1074/jbc.M113.501122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nkongolo S, Ni Y, Lempp FA, Kaufman C, Lindner T, Esser-Nobis K, Lohmann V, Mier W, Mehrle S, Urban S. 2014. Cyclosporin A inhibits hepatitis B and hepatitis D virus entry by cyclophilin-independent interference with the NTCP receptor. J Hepatol 60:723–731. doi: 10.1016/j.jhep.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 64.Kim HY, Park GS, Kim EG, Kang SH, Shin HJ, Park S, Kim KH. 2004. Oligomer synthesis by priming deficient polymerase in hepatitis B virus core particle. Virology 322:22–30. doi: 10.1016/j.virol.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 65.Jung J, Kim HY, Kim T, Shin BH, Park GS, Park S, Chwae YJ, Shin HJ, Kim K. 2012. C-terminal substitution of HBV core proteins with those from DHBV reveals that arginine-rich 167RRRSQSPRR175 domain is critical for HBV replication. PLoS One 7:e41087. doi: 10.1371/journal.pone.0041087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 67.Ko C, Lee S, Windisch MP, Ryu WS. 2014. DDX3 DEAD-box RNA helicase is a host factor that restricts hepatitis B virus replication at the transcriptional level. J Virol 88:13689–13698. doi: 10.1128/JVI.02035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Fälth M, Stindt J, Königer C, Nassal M, Kubitz R, Sültmann H, Urban S. 2014. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 146:1070–1083. doi: 10.1053/j.gastro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 69.GeneTex. 2014. Protocol: fractionation of membrane/cytoplasmic and nuclear proteins. GeneTex, Irvine, CA: http://www.genetex.com/uploaddata/Protocol/Document/20140722_Fractionation.pdf. [Google Scholar]