Seminal plasma (SP), the major vehicle for HIV, can modulate HIV transmission risk through a variety of mechanisms. Extracellular vesicles (EVs) are extremely abundant in semen, and because they play a key role in intercellular communication pathways and immune regulation, they may impact the likelihood of HIV transmission. However, little is known about the properties and signaling effects of SP-derived EVs in the context of HIV transmission. Here, we conduct a phenotypic, transcriptomic, and functional characterization of SP and SP-derived EVs from uninfected and HIV-infected men. We find that both SP and its associated EVs elicit potent proinflammatory transcriptional responses in cells that line the genital tract. EVs from HIV-infected men exhibit a more diverse repertoire of miRNAs than EVs from uninfected men. Our findings suggest that EVs from the semen of HIV-infected men may significantly impact the likelihood of HIV transmission through multiple mechanisms.
KEYWORDS: extracellular vesicles, epithelial cells, female reproductive tract, human immunodeficiency virus, miRNAs, seminal plasma, stromal fibroblasts, transcription
ABSTRACT
The continuing spread of HIV/AIDS is predominantly fueled by sexual exposure to HIV-contaminated semen. Seminal plasma (SP), the liquid portion of semen, harbors a variety of factors that may favor HIV transmission by facilitating viral entry into host cells, eliciting the production of proinflammatory cytokines, and enhancing the translocation of HIV across the genital epithelium. One important and abundant class of factors in SP is extracellular vesicles (EVs), which, in general, are important intercellular signal transducers. Although numerous studies have characterized blood plasma-derived EVs from both uninfected and HIV-infected individuals, little is known about the properties of EVs from the semen of HIV-infected individuals. We report here that fractionated SP enriched for EVs from HIV-infected men induces potent transcriptional responses in epithelial and stromal cells that interface with the luminal contents of the female reproductive tract. Semen EV fractions from acutely infected individuals induced a more proinflammatory signature than those from uninfected individuals. This was not associated with any observable differences in the surface phenotypes of the vesicles. However, microRNA (miRNA) expression profiling analysis revealed that EV fractions from infected individuals exhibit a broader and more diverse profile than those from uninfected individuals. Taken together, our data suggest that SP EVs from HIV-infected individuals exhibit unique miRNA signatures and exert potent proinflammatory transcriptional changes in cells of the female reproductive tract, which may facilitate HIV transmission.
IMPORTANCE Seminal plasma (SP), the major vehicle for HIV, can modulate HIV transmission risk through a variety of mechanisms. Extracellular vesicles (EVs) are extremely abundant in semen, and because they play a key role in intercellular communication pathways and immune regulation, they may impact the likelihood of HIV transmission. However, little is known about the properties and signaling effects of SP-derived EVs in the context of HIV transmission. Here, we conduct a phenotypic, transcriptomic, and functional characterization of SP and SP-derived EVs from uninfected and HIV-infected men. We find that both SP and its associated EVs elicit potent proinflammatory transcriptional responses in cells that line the genital tract. EVs from HIV-infected men exhibit a more diverse repertoire of miRNAs than EVs from uninfected men. Our findings suggest that EVs from the semen of HIV-infected men may significantly impact the likelihood of HIV transmission through multiple mechanisms.
INTRODUCTION
It has become increasingly clear that seminal plasma (SP) is not merely a passive carrier for spermatozoa but instead encompasses a rich source of bioactive and immunomodulatory factors. These factors contribute to promoting a healthy pregnancy but may also influence susceptibility to infection by sexually transmitted pathogens, including human immunodeficiency virus type 1 (HIV-1) (1–6). Interestingly, a recent study in a vaginal model of simian immunodeficiency virus (SIV) transmission suggested that repeated exposure of the female reproductive tract (FRT) to semen reduces subsequent infection by SIV (7), suggesting that semen components can alter the microenvironment of the FRT in ways that decrease transmission risk.
The effect of SP on the FRT has been investigated in microarray studies that examined the global transcriptional response of FRT-derived epithelial cells to SP exposure (8–10). One study (8) also included stromal cells underlying the FRT epithelium, as these cells may also gain access to SP during the secretory phase of the ovulatory cycle (11–13). These studies have together revealed that SP initiates an inflammatory response in both epithelial and stromal cells from the FRT. Similar studies using semen or SP from HIV-infected men have yet to be conducted. However, numerous studies have demonstrated that HIV infection alters the composition of cytokines in semen, generally toward a more proinflammatory profile (14–18), suggesting that semen from infected men may elicit in the FRT a response equal to if not more proinflammatory than the response elicited by semen from uninfected men.
Extracellular vesicles (EVs), a broad class of vesicles that includes exosomes and microvesicles (MVs), are known for their role in intercellular communication and their ability to alter recipient cell activity, as well as their therapeutic potential in disease diagnostics and targeted drug delivery (19–22). These properties make them prime candidates as the mediators of semen’s impact on the FRT, especially since trillions of EVs are present in a typical semen ejaculate (23). EVs can carry RNA, protein, or lipid to a recipient cell and directly or indirectly induce inflammatory or immunomodulatory responses (24, 25). For example, recent work showed that EVs from semen signal to antigen-presenting cells, inducing increased indoleamine 2,3-deoxygenase production, thereby decreasing antigen-specific T-cell responses (26). This effect may be due in part to microRNAs (miRNAs) present within EVs, which are known to affect gene regulation in target cells (27) and which can have important functional outcomes, such as suppressing T-cell-mediated disease (28). Of note, EVs can harbor up to half of the extracellular RNA in SP (20).
Despite numerous studies characterizing blood plasma-derived EVs from both uninfected and HIV-infected individuals, little is known about the properties and signaling effects of SP-derived EVs in the context of HIV transmission. Here, we examine how resident cells of the FRT respond to treatment with fractionated SP enriched for EVs from uninfected and acutely HIV-infected individuals by global gene expression analysis and further characterize the protein and miRNA cargos of these vesicles.
RESULTS
Demographics and clinical characteristics.
The characteristics of the uninfected and HIV-infected participants from which reconstituted SP (rSP) and rSP-derived EVs were analyzed are shown in Table 1. Of 20 uninfected participants, 55% were white, 20% were African American, and 15% were Latino/Hispanic, and the mean age was 36.4 ± 2.2 years. Of the 20 uninfected participants, 9 had CD4 cell counts available, and for these participants, the mean CD4+ T-cell count was 852 ± 173 cells/μl. Of the 20 HIV-infected men, 40% were white, 5% were African American, and 30% were Hispanic, and the mean age was 32.2 ± 1.5 years. The mean CD4+ T-cell count at the time of enrollment was 608 ± 194 cells/μl. The median blood plasma virus load was 30,200 copies/ml (interquartile range, 10,300 to 130,000 copies/ml). None of the men exhibited AIDS symptoms at the time of semen collection. In addition, detailed demographic and baseline clinical characteristics for 10 individuals chronically infected with HIV with paired samples before and after the initiation of antiretroviral therapy (ART) are presented in Table 2.
TABLE 1.
Demographic and clinical characteristics of uninfected and HIV acutely infected participants
| Subject characteristic | Value for the following participants: |
P value | |
|---|---|---|---|
| Uninfected (n = 20) | HIV infected (n = 20) | ||
| Demographics | |||
| Median ± SD age (yr) | 36.4 ± 2.2 | 32.2 ± 1.5 | 0.2 (NSa ) |
| No. (%) of participants by race | |||
| African American | 4 (20) | 1 (5) | NS |
| Asian | 1 (5) | 3 (15) | NS |
| Latino/Hispanic | 3 (15) | 6 (30) | NS |
| Multiracial | 1 (5) | ||
| Pacific Islander | 1 (5) | 1 (5) | NS |
| White | 11 (55) | 8 (40) | NS |
| HIV related | |||
| Time (mo) since HIV diagnosis | 0.8 ± 0.4 | ||
| CD4+ T-cell count (no. of cells/μl) | 608 ± 194 | ||
| Median (IQRb ) plasma viral load (no. of copies/ml) | 30,200 (10,300–130,000) | ||
NS, nonsignificant (P > 0.05).
IQR, interquartile range.
TABLE 2.
Demographic and clinical characteristics of HIV chronically infected participants
| Subject characteristic | Value for HIV-infected participants: |
P valuea | |
|---|---|---|---|
| Before ART (n = 10) | After ART (n = 10) | ||
| Demographics | |||
| Mean ± SD age (yr) | 39.4 ± 8.6 | 44.1 ± 8.6 | 0.002 |
| No. of white participants | 10 | 10 | NS |
| HIV related | |||
| Time (mo) since HIV diagnosis | 35.4 ± 45.7 | 171.5 ± 88.6 | 0.002 |
| CD4+ T-cell count (no. of cells/μl) | 465.7 ± 231.1 | 564.5 ± 194.6 | NS |
| Median (IQRb ) plasma viral load (no. of copies/ml) | 47,722 (7,089–68,081) | 64 (40–75) | 0.002 |
P values were determined by the Wilcoxon matched-pairs signed-rank test. NS, nonsignificant (P > 0.05).
IQR, interquartile range.
SP fractionation and characteristics.
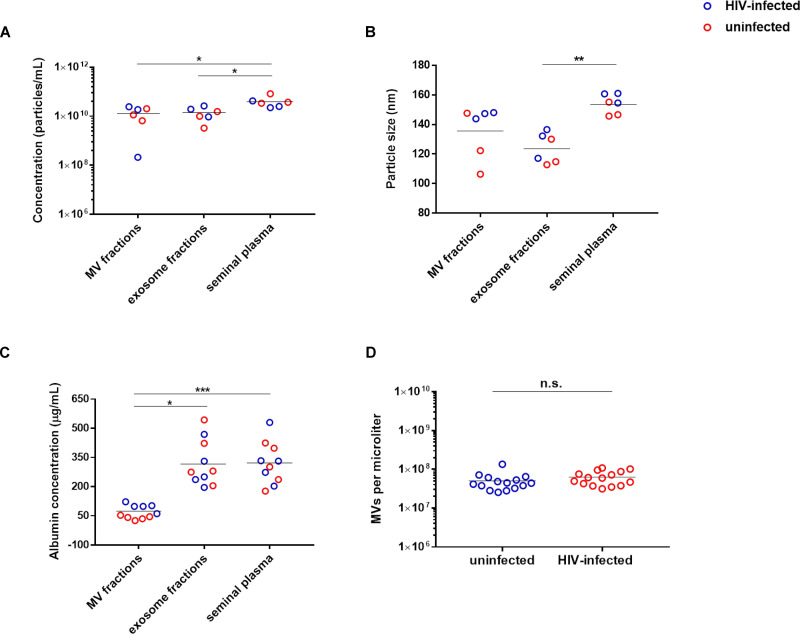
As expected, the EV count in the MV and exosome fractions was lower than that in the original unprocessed SP due to the loss of EVs during the fractionation process (Fig. 1A), and the average size of the EVs was the smallest in the exosome fraction (Fig. 1B). To confirm that the fractionation successfully enriched for MVs in the MV fraction, we quantified the levels of albumin, a soluble protein not associated with EVs (29, 30). As expected, the MV fraction harbored significantly lower levels of albumin than the original preprocessed SP, while the exosome fraction, which contains a mixture of soluble factors and exosomes, exhibited significantly higher levels of albumin than the MV fraction (Fig. 1C).
FIG 1.
Characterization of EVs in the MV and exosome fractions of SP from uninfected versus HIV-infected individuals. (A) EV particle counts in the MV fractions, exosome fractions, and original preprocessed seminal plasma were determined by nanoparticle tracking analysis. (B) The mean size distribution of particles in the MV fractions, exosome fractions, and original preprocessed seminal plasma was determined by nanoparticle tracking analysis. (C) The levels of the non-EV-associated protein albumin in the MV fractions, exosome fractions, and original preprocessed seminal plasma were determined by ELISA. P values were determined by the Friedman repeated-measures ANOVA and Dunn’s multiple-comparison post hoc test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (D) MV counts in the MV fractions used in this study, as determined using an LSR II flow cytometer. n.s., not significant, as determined by an unpaired Student's t test.
Global gene expression analysis reveals a proinflammatory response of eECs and eSFs upon exposure to rSP and MV fractions from HIV-infected men.
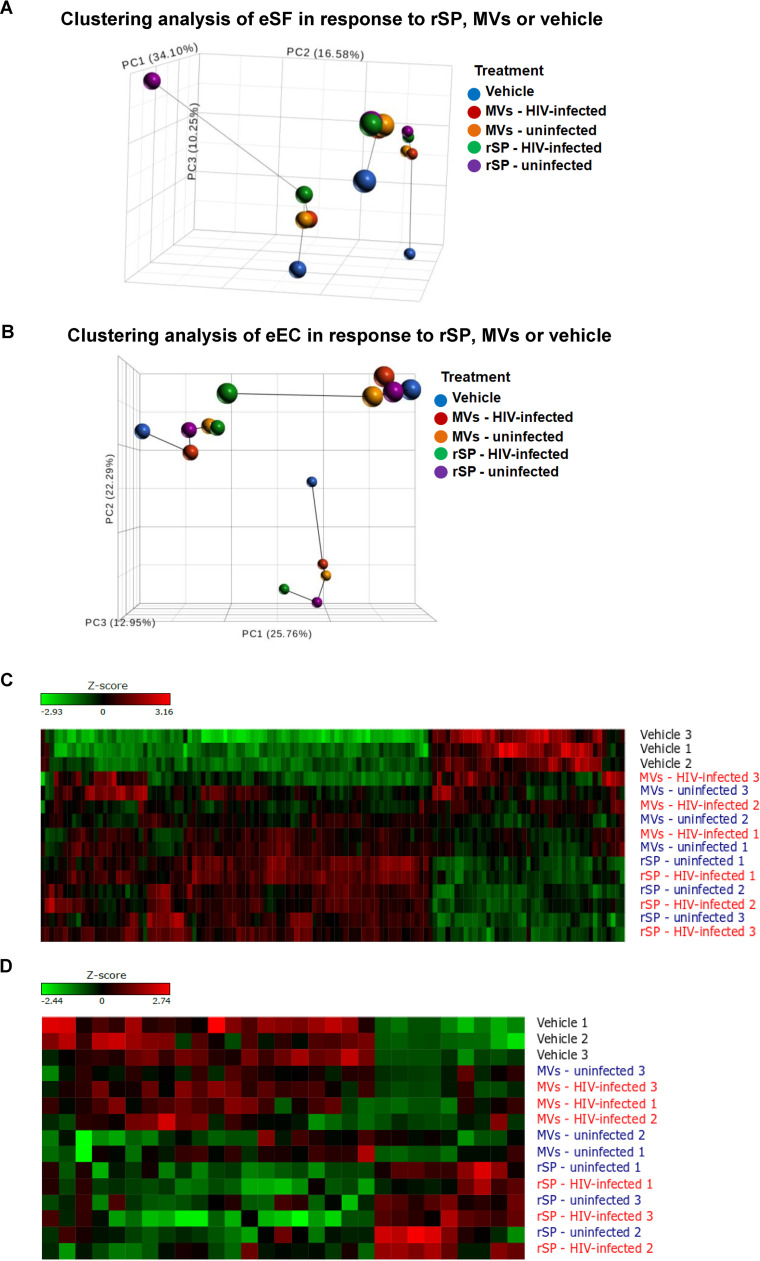
Endometrial epithelial cells (eECs) and endometrial stromal fibroblasts (eSFs) are abundant cell types of the FRT that interface with the genital lumen. While eECs are constantly exposed to luminal contents, eSFs can gain access to such contents during the ovulatory cycle’s secretory phase. This phase is associated with increased transmission risk (12). To examine how eECs and eSFs respond to exposure to rSP- and SP-derived EVs from HIV-infected men, we conducted transcriptome sequencing (RNA-seq) analysis. We isolated primary eECs and eSFs from 3 donors and exposed them to rSP or the MV fraction from uninfected or HIV-infected men. The rSP and MV stocks were each pooled from the same 15 donors. Of note, although this method normalizes by volume and not the number of EVs, we found no statistically significant difference in the concentration of MVs in the MV fractions from uninfected individuals versus that in the MV fractions from acutely infected individuals (Fig. 1D). Principal-component analysis (PCA) of the eSF data revealed partitioning by donor; however, the data also revealed marked transcriptional differences between the vehicle-treated sample and the rSP/MV fraction-treated samples for each donor (Fig. 2A). In comparison, the response of eECs to rSP/MV fraction exposure was less pronounced: PCA segregated eECs primarily by donor, with little segregation by treatment condition (Fig. 2B), suggesting that eECs responded less to rSP than eSFs did. Accordingly, the list of genes significantly altered by rSP/MV fraction exposure was longer in eSFs than in eECs (Fig. 2C and D; see also Table S2 in the supplemental material). Interestingly, in eSFs, the MV fraction and rSP elicited similar transcriptional responses, although the response was dampened for the MV fraction. In contrast, in eECs the signature induced by the MV fraction closely resembled that seen in vehicle-treated samples, suggesting that the MV fraction does not induce a potent response in eECs. The full list of genes induced by rSP and the MV fraction from HIV-infected individuals is presented in Table S2. Much of the response of the eECs to rSP exposure was defined by induction of the metalloproteinase class of proteins (e.g., MT1H, MT1G, MT1M, and MT1E), while this class of proteins was not induced in eSFs.
FIG 2.
Principal-component analysis and hierarchical clustering of eSFs or eECs treated with rSP or the MV fraction from uninfected or HIV-infected individuals. (A, B) PCA of RNA-seq data sets from eSFs or eECs exposed to vehicle, 1% rSP, or 1% the MV fraction from uninfected or HIV-infected individuals. Samples originating from the same eSF donor are connected by lines. (C, D) Heatmap showing differentially expressed genes (DEGs) between the treatment conditions in eSFs or eECs. The numbers refer to genital tissue donor numbers.
To determine whether HIV infection status alters the signaling effects of rSP, we directly compared the transcriptional response of eECs and eSFs to rSP and the MV fraction from uninfected versus HIV-infected individuals. We found that in eSFs, rSP and the MV fraction from infected individuals induced higher levels of expression of RNA for the proinflammatory cytokine CXCL1 than the rSP and the MV fraction from uninfected individuals did (4.9-fold for rSP, 3.4-fold for the MV fraction; P < 0.05) (Table 3). In contrast, in eECs, rSP and the MV fraction from infected individuals preferentially induced the expression of a set of genes different from that preferentially induced by rSP and the MV fraction from uninfected individuals (Table 4).
TABLE 3.
Genes in eSFs differentially induced by rSP and MV fraction from infected individualsa
| rSP from HIV-infected participants vs rSP from uninfected individuals |
MVs from HIV-infected participants vs MVs from uninfected individuals |
||||
|---|---|---|---|---|---|
| Gene regulation and gene | P value | Fold change in expression | Gene regulation and gene | P value | Fold change in expression |
| Upregulated by rSP from HIV-infected individuals | Upregulated by MV HIV-infected individuals | ||||
| CXCL1 | 0.03 | 4.89 | CXCL1 | 0.04 | 3.37 |
| HIST1H4C | 0.02 | 1.87 | |||
| CCR10 | 0.04 | 1.54 | |||
| Downregulated by rSP from HIV-infected individuals | Downregulated by MVs from HIV-infected individuals | ||||
| RPS10P7 | 0.03 | 4.17 | |||
| LOC101928120 | 0.02 | 1.80 | |||
| DBNDD2 | 0.02 | 1.80 | |||
| LPCAT1 | 0.03 | 1.69 | |||
| UFSP1 | <0.001 | 1.64 | |||
| PSMB8-AS1 | 0.02 | 1.60 | |||
| IN080B | 0.01 | 1.57 | |||
Genes with a ≥1.5-fold change in expression between treatment groups with a P value of <0.05.
TABLE 4.
Genes in eECs differentially induced by rSP and the MV fraction from infected individualsa
| rSP from HIV-infected individuals vs rSP from uninfected individuals |
MVs from HIV-infected individuals vs MVs from uninfected individuals |
||||
|---|---|---|---|---|---|
| Gene regulation and gene | P value | Fold change in expression | Gene regulation and gene | P value | Fold change in expression |
| Upregulated by rSP from HIV-infected individuals | Upregulated by MV from HIV-infected individuals | ||||
| HIF1A-AS2 | <0.001 | 3.48 | HIST1H3E | 0.02 | 2.45 |
| VGF | 0.02 | 2.20 | FAM229A | 0.04 | 1.80 |
| FAM229A | 0.03 | 1.52 | HIST2H2AC | 0.04 | 1.66 |
| RPL21P44 | 0.03 | 1.62 | |||
| RPS15AP10 | 0.01 | 1.59 | |||
| EXOC3-AS1 | 0.02 | 1.56 | |||
| Downregulated by rSP from HIV-infected individuals | Downregulated by MV from HIV-infected individuals | ||||
| KMT2 | 0.04 | 1.92 | ZNF43 | 0.04 | 7.82 |
| AOC2 | 0.04 | 1.86 | MT1G | 0.02 | 3.75 |
| SPEN | 0.04 | 1.86 | MT1H | <0.001 | 2.79 |
| AHNAK | 0.05 | 1.72 | CCDC89 | 0.04 | 1.81 |
| CRIPAK | 0.05 | 1.65 | SST | 0.01 | 1.54 |
| LOC728752 | <0.001 | 1.63 | |||
| RSAD2 | 0.02 | 1.60 | |||
| OAS2 | <0.001 | 1.59 | |||
| MSMP | 0.02 | 1.54 | |||
| OAS3 | <0.001 | 1.54 | |||
| EIF2AK2 | 0.03 | 1.54 | |||
| SAMD9 | 0.02 | 1.51 | |||
Genes with a ≥1.5-fold change in expression between treatment groups with a P value of <0.05.
rSP and the MV fraction induced gene pathways associated with cellular survival and migration.
To obtain a global assessment of the biological functions and/or diseases induced by rSP and the MV fraction from HIV-infected individuals, we implemented Ingenuity Pathway Analysis (IPA) (Table 5). In both eSFs and eECs, rSP upregulated pathways associated with cell survival and suppressed cell death pathways, such as those associated with apoptosis and necrosis, a finding similar to what was previously observed when eSFs and eECs were treated with SP from uninfected men (8). Pathways associated with cellular movement, including cellular migration and tubulation, were also induced by rSP in both cell types, as were those associated with inflammation. These results are consistent with those of prior studies examining the effects of SP on cells of the FRT (8–10). The MV fraction did not significantly induce any IPA-defined biological pathways in eECs, but it did in eSFs. These included biological pathways associated with the promotion of survival, inhibition of cell death, and induction of pathways associated with cellular movement.
TABLE 5.
Selected gene pathways induced by eSFs and eECs exposed to rSP and the MV fraction from HIV-infected individualsa
| Comparison | Category | Function annotation | Activation state | Z-score |
|---|---|---|---|---|
| rSP vs mock exposure (eEC) | Cell death and survival | Cell death | Decreased | 1.60 |
| Cell death and survival | Apoptosis | Decreased | 1.54 | |
| Cell signaling | Replication of viral replicon | Increased | 1.98 | |
| Cellular movement | Colony formation of cells | Increased | 2.42 | |
| Cellular movement | Tubulation of cells | Increased | 1.96 | |
| rSP vs mock exposure (eSF) | Cell death and survival | Cell survival | Increased | 2.89 |
| Cell death and survival | Activation of cells | Increased | 2.75 | |
| Cell death and survival | Cell viability | Increased | 2.64 | |
| Cell death and survival | Chemotaxis of endothelial cells | Increased | 2.19 | |
| Cell death and survival | Invasion of tissue | Increased | 2.14 | |
| Cell death and survival | Cell death | Decreased | 2.35 | |
| Cell death and survival | Necrosis | Decreased | 2.32 | |
| Cell death and survival | Apoptosis | Decreased | 2.06 | |
| Cellular movement | Migration of cells | Increased | 2.78 | |
| Cellular movement | Cell movement | Increased | 2.74 | |
| Cellular movement | Cell movement of myeloid cells | Increased | 2.47 | |
| Cellular movement | Cell viability of endothelial cells | Increased | 2.10 | |
| Gene expression | Binding of DNA | Increased | 2.28 | |
| MV fraction vs mock exposure (eSF) | Cell death and survival | Synthesis of prostaglandin E2 | Increased | 1.59 |
| Cell death and survival | Organization of cytoskeleton | Decreased | 2.53 | |
| Cell death and survival | Growth of embryo | Decreased | 1.97 | |
| Cellular movement | Chemotaxis of granulocytes | Increased | 1.78 | |
| Cellular movement | Transmigration of cells | Increased | 1.67 |
Z-score > |1.5|. Abbreviations: eSF, endometrial stromal fibroblast; eEC, endometrial epithelial cell; MVs, microvesicles; rSP, reconstituted SP.
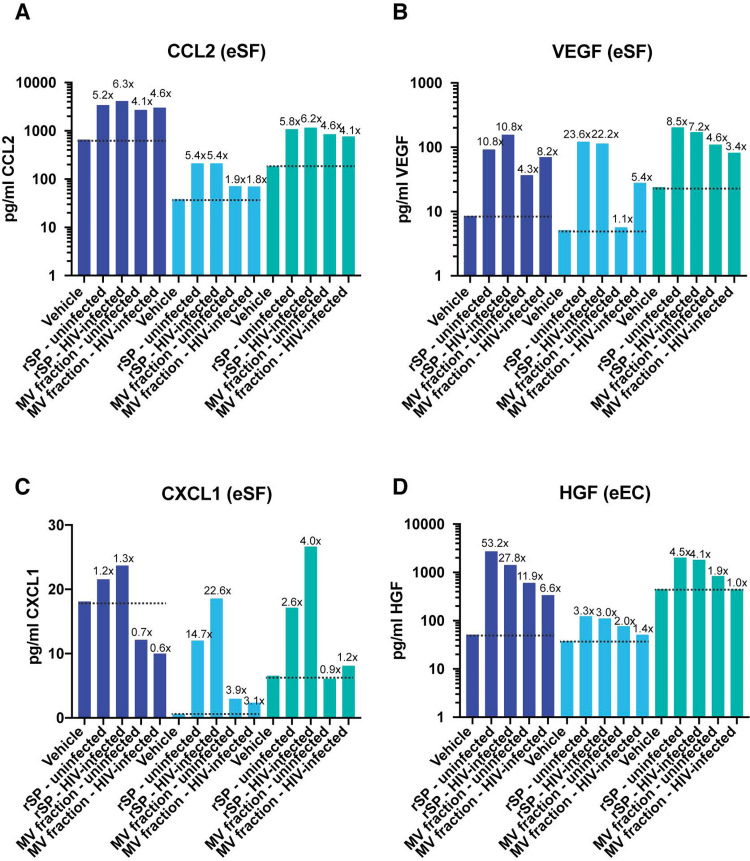
To validate some of these findings, we selected several induced genes associated with pathways upregulated upon exposure to HIV-contaminated SP. Upon treatment of eSFs with SP from HIV-infected individuals, vascular endothelial growth factor (VEGF) transcripts were induced 5.2-fold, while CCL2 transcripts were induced 2.5-fold. These genes are major drivers of the enhanced cell survival and cellular migration pathways in eSFs. We found that eSFs from the same 3 donors analyzed by RNA-seq all induced the secretion of CCL2 and the VEGF protein in response to treatment with rSP or the MV fraction from either uninfected or HIV-infected individuals (Fig. 3A and B). We also examined CXCL1 since it was induced at the RNA level in eSFs at significantly higher levels by the rSP/MV-enriched fraction from infected individuals than by that from uninfected individuals (Table 3). In all 3 eSF donors, rSP from infected individuals induced higher levels of CXCL1 secretion than rSP from uninfected individuals; however, the MV-enriched fraction did not show a consistent trend (Fig. 3C). The ability of rSP but not the MV-enriched fraction to induce CXCL1 protein secretion may have been due to the participation of non-MV-associated factors in this induction. The hepatocyte growth factor (HGF) transcript was induced 2.9-fold in eECs by rSP from HIV-infected individuals and was a major driver of the enhanced cell survival pathway in eECs. eECs from the same 3 donors analyzed by RNA-seq also induced secretion of the HGF protein in response to treatment with rSP or the MV fraction from either uninfected or HIV-infected individuals (Fig. 3D). The findings from the CCL2, VEGF, and HGF protein analysis reinforce the notion that exposure of endometrial cells to SP from HIV-infected individuals upregulates pathways associated with cell survival and movement. Except for the finding that the MV-enriched fraction from infected individuals did not induce more CXCL1 than the corresponding fraction from uninfected individuals, these results validate the findings from the RNA-seq data sets.
FIG 3.
Validation of select genes induced by SP from HIV-infected individuals. Culture supernatants from eSFs (A, B, C) or eECs (C) were assessed for the protein levels of CCL2 (A), VEGF (B), CXCL1 (C), or HGF (D). Treatment conditions are indicated on the x axis, and each eSF or eEC donor is depicted with a different color. The dotted line shows the levels of the protein under vehicle-treated conditions. The numbers above the bars indicate the fold induction relative to the level of induction for the corresponding vehicle-treated condition. All donors were treated under the exact same conditions, as indicated. When these paired data sets from all 3 donors were combined and analyzed using paired Student’s t tests, there was a significant induction of CCL2 by HIV-infected SP (P < 0.05), VEGF by uninfected SP (P < 0.05), VEGF by HIV-infected SP (P < 0.05), and HGF by uninfected SP (P < 0.05).
The MV fraction from HIV-infected individuals does not affect HIV infection of CD4+ T cells in vitro.
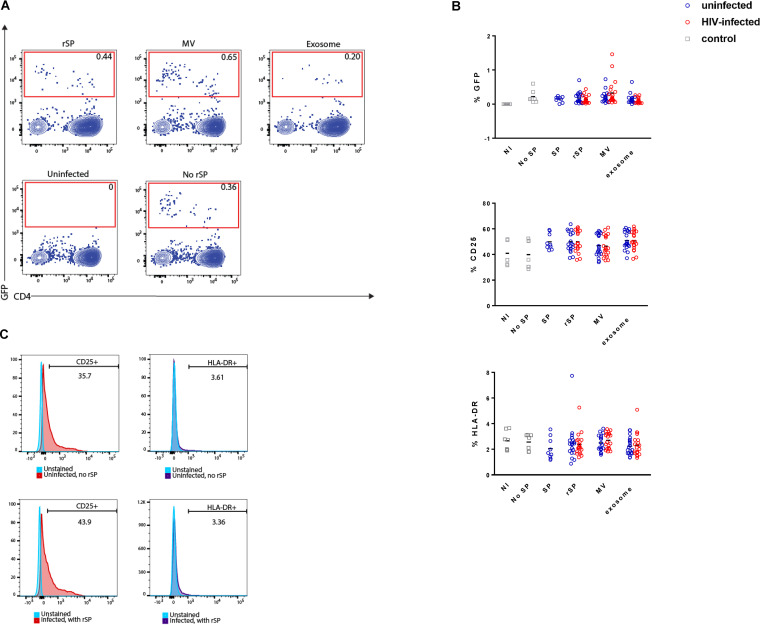
The results presented thus far suggest that rSP and the MV fraction from HIV-infected individuals can elicit potent transcriptional and secretome changes in cells of the FRT and that these responses differ slightly from those elicited by rSP and the MV fraction from uninfected individuals. We next assessed whether rSP and the MV fractions from uninfected versus infected individuals might differentially affect infection of CD4+ T cells, the primary targets of HIV. To this end, we compared rSP/the MV fraction from uninfected and HIV-infected individuals for their effects on HIV infection of primary CD4+ T cells. For comparison, we also tested the effects of the exosome fractions (see Materials and Methods). rSP or MV fractions were pretreated with an HIV reporter virus and then diluted onto peripheral blood mononuclear cells (PBMCs), conditions that have been shown to be nontoxic to the cells (31) and that result in a final concentration of 0.75% rSP or the MV fraction. Infection rates were examined in both unstimulated and phytohemagglutin (PHA)- and interleukin-2 (IL-2)-stimulated CD4+ T cells isolated from PBMCs. Sample fluorescent-activated cell sorting (FACS) plots of uninfected cells and those infected with HIV in the presence of vehicle alone, rSP, the MV fraction, or the exosome fraction are shown in Fig. 4A. There were no statistically significant changes in infection rates between the vehicle-alone condition and any of the rSP treatment conditions and no differences linked to whether the rSP came from uninfected or HIV-infected individuals (Fig. 4B). Because our RNA-seq analysis suggested that rSP from infected individuals may elicit a more proinflammatory response in exposed cells than rSP from uninfected individuals, we also examined whether rSP, the MV fraction, or the exosome fraction from HIV-infected individuals may differentially upregulate the T-cell activation markers CD25 and HLA-DR. There were no statistically significant differences in the ability of any of the rSP or the EV fractions to upregulate these antigens (Fig. 4B and C). These results suggest that at the concentration tested, rSP or the EV fractions from uninfected individuals behave similarly to those from HIV-infected individuals with regard to their effects on HIV infection in our in vitro model system.
FIG 4.
Representative dot plots showing an uninfected control and a sample treated with HIV in the presence of vehicle alone (bottom row) or samples infected with HIV in the presence of rSP, the MV fraction, or the exosome fraction (top row). (B) Graphs showing the levels of GFP, CD25, and HLA-DR on activated T cells from uninfected and HIV-infected individuals between the vehicle-alone condition and the rSP treatment conditions. Results are gated on live, singlet CD3+ CD8− cells. (C) Representative histogram plots showing the cell surface expression of CD25+ and HLA-DR on T cells infected or not with HIV and in the absence or presence of rSP.
Vesicles from the MV fraction of SP from infected and noninfected individuals are phenotypically similar.
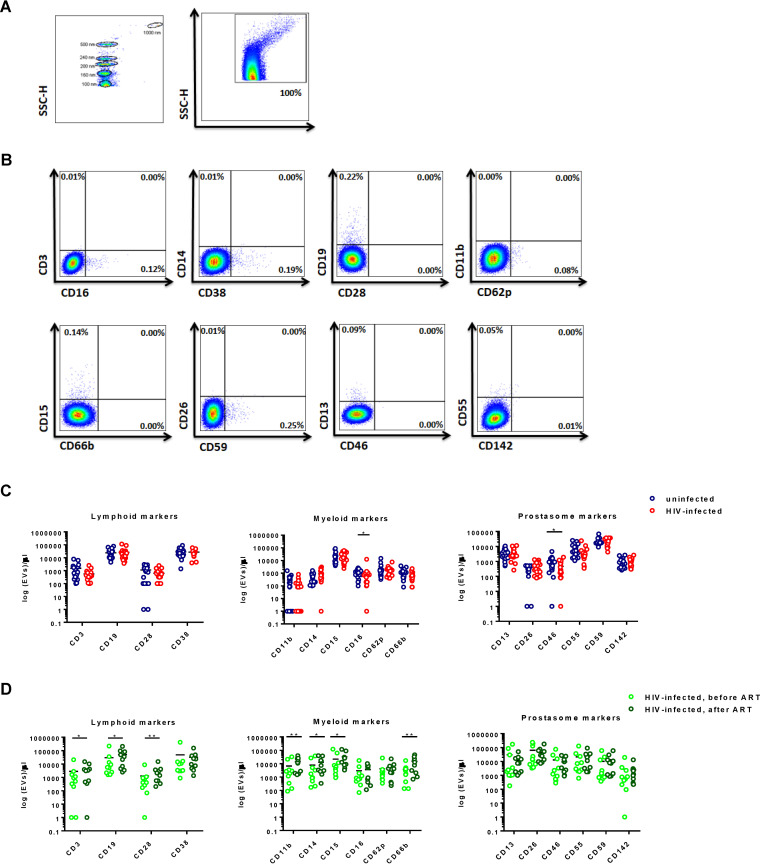
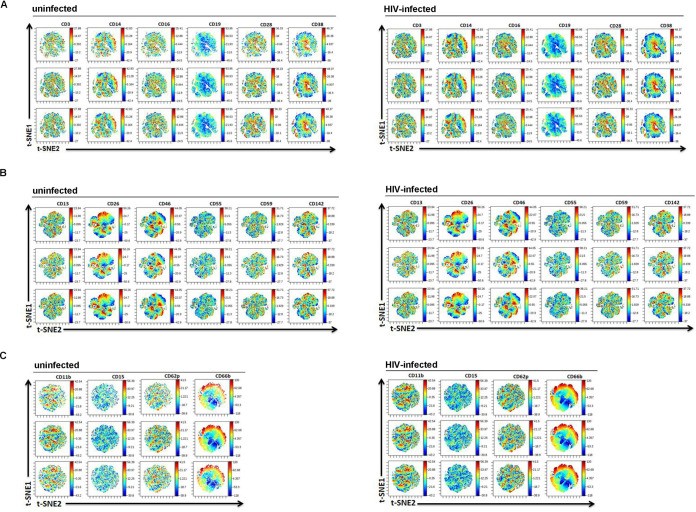
To probe for differences between EVs from infected individuals and EVs from noninfected individuals using additional methods, we next quantitated MVs and analyzed the proteins on their surface by FACS. The same specimens used in the eSF/eEC RNA-seq analysis were used in these analyses. The EV gate was set from below the 100-nm bead signal up to the 1,000-nm bead signal on the side scatter (SSC) channel (Fig. 5A). We applied 3 panels consisting of antigens linked to prostasomes (CD13, CD26, CD46, CD55, CD59, and CD142), lymphoid cells (CD3, CD14, CD16, CD19, CD28, and CD38), and myeloid cells (CD11b, CD15, CD62p, and CD66b). Representative FACS plots are shown in Fig. 5B. When we compared the quantity and phenotypes of MVs from uninfected and HIV-infected individuals, we found increased levels of EVs expressing CD16 (mean, 1,100 versus 1,171 EVs/μl; P = 0.03) and CD46 (941 versus 465 EVs/μl; P = 0.04) in uninfected individuals compared to HIV-infected individuals. However, these findings were no longer significant after multiple-testing correction using the Benjamini-Hochberg procedure. In addition, there were no significant differences in the total MV concentration or the MV expression of a variety of other markers spanning the lymphoid and myeloid lineages, as well as antigens known to be present in prostasomes (Fig. 5C). High-dimensional visualization of the phenotyping data by t-distributed stochastic neighbor embedding (t-SNE) also did not reveal any differences associated with HIV serostatus (Fig. 6).
FIG 5.
Detection and characterization of EVs for their absolute count, relative size, and cell of origin. (A) SSC height (SSC-H) dot plot showing instrument sensitivity to beads 100 to 1,000 nm in diameter, a range used to set EV gates on the LSR II flow cytometer. (B) Representative plots of the MV fraction gated for size as described in the legend to panel A and sorted according to various phenotypic markers. (C) Scatterplots of MV concentration (log10 transformed) in uninfected versus acutely infected individuals according to the phenotypic markers examined. Although there were significant differences in the levels of semen MVs expressing CD16+ and CD46+ between uninfected and HIV-infected individuals (*, P < 0.05 by the Mann-Whitney test), these results were no longer significant after multiple-testing correction using the Benjamini-Hochberg procedure. (D) Scatterplots of the concentrations of MVs (log10 transformed) from 10 chronically HIV-infected individuals with paired samples before and after initiation of ART. P values were determined by the Wilcoxon matched-pairs signed-rank test. *, P < 0.05; **, P < 0.01 These results remained significant after correcting for multiple testing with an FDR of 0.1.
FIG 6.
t-SNE analysis of flow cytometric data for the MV fraction from uninfected and HIV-infected individuals. The expression levels of the indicated antigen are shown as heat plots, while individual EVs are represented as dots on the t-SNE. The colors correspond to arcsinh-transformed expression values for each given marker analyzed, with lower expression levels being shown in blue and higher expression levels being shown in red. Shown are the results for 3 representative individuals of each group of participant samples. The patterns of the t-SNE were similar between EVs from uninfected individuals and EVs from infected individuals, suggesting no global differences in the expression patterns of the antigens examined in this study. Shown are data sets corresponding to EVs stained for markers of lymphocytes, monocytes, and B cells (A), prostasome markers (B), and neutrophils and platelets (C).
We also assessed the phenotypes of MVs from the semen of chronically infected individuals, comparing paired specimens collected before ART treatment and those collected after ART treatment. After virus suppression by ART, there was a significant increase in MVs expressing the lymphoid markers CD3, CD19, and CD28 and in those expressing the myeloid markers CD11b, CD14, CD15, and CD66b (Fig. 5D). Collectively, these results suggest that that while there are not marked differences in the phenotypes of seminal MVs between uninfected and acutely infected individuals, phenotypic differences in MVs are observed in chronically infected individuals before versus after treatment. Unfortunately, the specimens from uninfected and acutely infected individuals could not be directly compared to the specimens from chronically infected individuals, as they were processed as separate batches.
The exosome fractions from HIV-infected and noninfected individuals differ in their miRNA cargo.
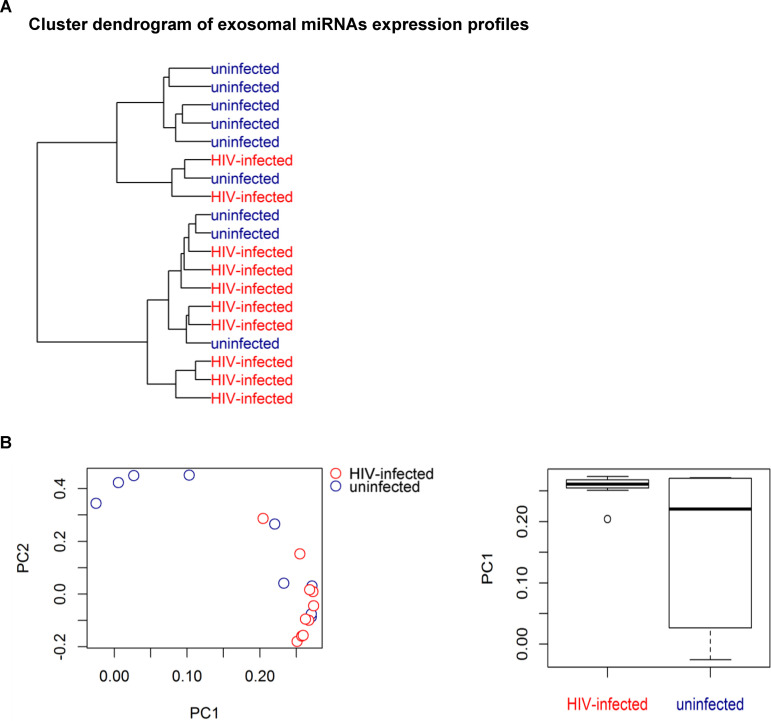
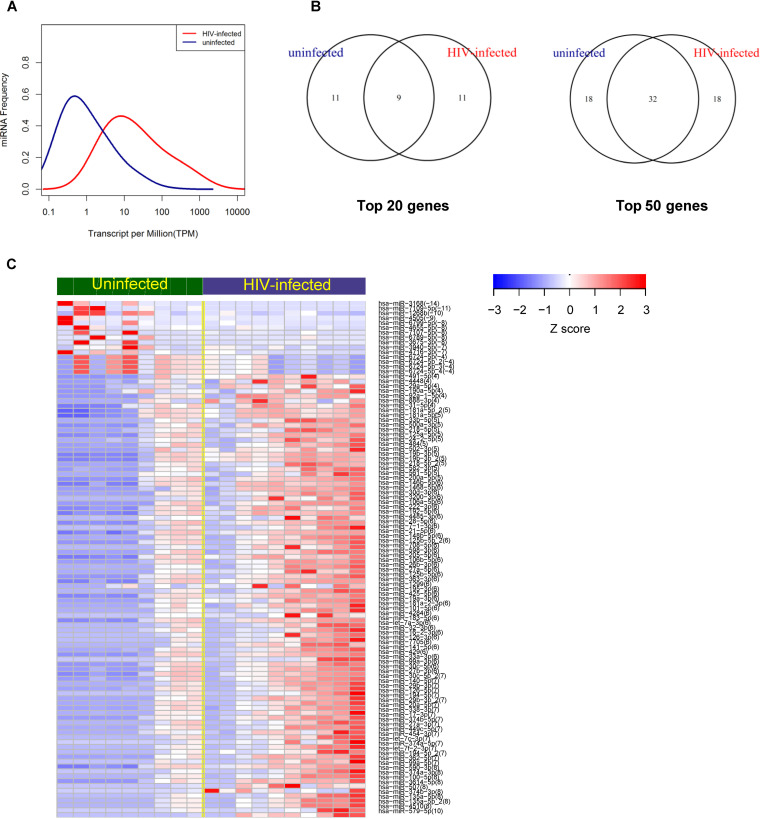
The data presented thus far reveal differences in the response of genital cells to rSP and the MV fraction from uninfected versus HIV-infected individuals but no significant differences in the phenotypes of these EVs or their effects on HIV infection of CD4+ T cells. We next sought to characterize the contents of the EVs in a global, unbiased fashion by profiling the repertoire of EV-derived miRNAs. Materials from the MV fraction were of insufficient quantity and quality for small RNA analysis by RNA-seq (data not shown). Therefore, EVs were isolated from SP from a subset 9 uninfected and 10 HIV-infected individuals from the original set under modified conditions compatible with subsequent RNA-seq analysis of small RNAs (see Materials and Methods). No alignments to the HIV-1 sequence were found in any of the sequenced samples. The degree of correlation between the miRNAs from uninfected and HIV-infected individuals was examined by hierarchical clustering (Fig. 7A) and PCA (Fig. 7B). PCA demonstrated that miRNAs those from uninfected individuals were more variable than miRNAs from infected individuals (Fig. 7B) (P = 0.037, t test), suggesting high interindividual variability among samples from uninfected individuals. A higher frequency of miRNAs per million reads was observed in HIV-infected subjects than in uninfected subjects (Fig. 8A). Both miRNAs that were shared between the uninfected and infected groups and those that were unique were identified (Fig. 8B). Core analysis of the top 200 expressed miRNAs revealed that HIV-infected individuals exhibited a higher diversity of miRNAs than uninfected subjects (Fig. 8C), suggesting high intraindividual diversity of the miRNAs in infected subjects. Taken together, these results suggest higher interindividual variability in miRNAs in uninfected individuals but higher intraindividual diversity in miRNAs within each infected subject. The experimentally validated regulatory interactions between the most abundant miRNAs in the EVs and mRNAs are presented in Table S1.
FIG 7.
Hierarchical clustering analysis and PCA of miRNAs from an EV-enriched fraction from uninfected and HIV-infected individuals. (A) Cluster dendrogram of EV-associated miRNAs, determined using standardized Euclidean distances with complete link hierarchical clustering. (B) (Left) PCA plot showing that sample variance is higher among uninfected individuals (red) than among HIV-infected individuals (blue); (right) box plot analysis of principal component 1 (PC1) demonstrating the higher variance among the uninfected EV specimens.
FIG 8.
Differential expression analysis of miRNAs from an EV-enriched fraction from uninfected and HIV-infected individuals. (A) Frequency distribution of EV-associated miRNAs for uninfected and HIV-infected individuals. Each miRNA is represented by the number of TPM. (B) Venn diagrams showing common gene expression between the miRNAs from uninfected and HIV-infected individuals. (C) Heatmap showing the Z-scores for the top miRNAs differentially expressed between uninfected and HIV-infected individuals (FDR < 0.05). miRNAs were ranked by the fold change in expression (in parentheses), and samples were ranked by median expression. Each column represents an individual participant, and each row represents a single miRNA. The color scale shown on the top illustrates the relative expression levels of the indicated miRNAs across all samples.
DISCUSSION
Our findings provide insights into the properties of SP EVs from HIV-infected individuals, including how they signal to cells of the female reproductive tract and how they differ from those from uninfected individuals. Although the effects of SP on human genital epithelial and stromal cells have been previously characterized (8–10), we provide here the first global gene expression analysis describing how these cells respond to treatment with SP and EV-enriched fractions from HIV-infected individuals. Furthermore, we characterized and compared the contents of EVs from uninfected and HIV-infected individuals in a global unbiased fashion by comprehensively profiling the repertoire of EV-derived miRNAs. Our major findings are that (i) both rSP and EV-enriched fractions from HIV-infected men induce potent transcriptional responses in epithelial and stromal cells isolated from the female reproductive tract, (ii) select genes, including ones associated with inflammatory responses, are preferentially induced by rSP/MV-enriched fractions from infected individuals as compared to those from uninfected individuals, and (iii) EV-enriched fractions from infected individuals exhibit a more diverse profile of miRNAs within each individual but less interindividual variability than those from uninfected individuals.
The transcriptional response induced by SP from HIV-infected individuals in eECs and eSFs was characteristic of the biological functions associated with enhanced cell survival and cellular migration/movement. The cell death and survival category included inhibition of both apoptotic and necrotic cell death. Such inhibition could conceivably lead to the enhanced survival of nearby HIV-infected cells and would be consistent with the notion that the virus benefits from preventing the apoptosis of infected cells (32). Biological processes associated with increased cellular migration/movement included those associated with the upregulation of chemokines that can recruit additional HIV target cells to the inflamed mucosa, as well as the increased dissemination of infected cells to other sites. Interestingly, a sizable portion of the response of eSFs to SP exposure appeared to be mediated by EVs, while this was not the case in eECs, in which no significant biological functions were induced by the EVs. This may be related to the generally lower transcriptional response of eECs to SP treatment, as reflected by a much more donor-driven clustering of samples relative to that for eSFs. As increased SP exposure to eSFs occurs during the secretory phase or under the influence of the sex steroid progesterone (11–13), the potent response of eSFs to SP from HIV-infected men may play a role, especially in women using hormonal contraceptives.
Of note, much of the response induced in isolated eSFs and eECs by exposure to HIV-contaminated SP was also observed upon exposure to SP from uninfected individuals. Interestingly, a recent study examined the response of the monocytic cell line U937 to semen exosomes from uninfected and HIV-infected individuals using microarray analysis (33). Little overlap in differentially expressed genes was observed between that data set and ours, suggesting that monocytes may respond quite differently than FRT cells to SP components to which they are exposed. Interestingly, however, the genes for the transcription factors EGR1 and EGR2 were among the most downregulated genes upon exposure of U937 to semen exosomes, and these two genes were also highly downregulated by both SP and the MV fraction in eSFs (see Table S1 in the supplemental material). These factors have mostly been characterized for their role in neuronal signaling. To what extent their downregulation plays a role in reproductive health and/or HIV transmission requires future investigation.
Our observation that SP from uninfected individuals induced a transcriptional response in isolated eSFs/eECs similar to that induced by SP from HIV-infected individuals suggests that the signaling effects of SP are very potent and overshadow the effects caused by HIV infection status. Nonetheless, we still identified genes preferentially induced by SP and EVs from infected individuals as compared to those from uninfected individuals. In particular, transcripts encoding the inflammatory cytokine CXCL1 were preferentially upregulated in eSFs by SP and the MV fraction from infected individuals. This was also observed at the protein level for SP but not for the MV fraction. Interestingly, treatment of ectocervical explants with SP from HIV-infected men has recently been shown to upregulate CXCL1 (3), consistent with our findings. CXCL1 is produced by a wide variety of cells and plays a role in recruiting neutrophils to sites of inflammation (34, 35). Neutrophils can secrete a variety of proinflammatory cytokines, including IL-6 and tumor necrosis factor alpha (36, 37). These proinflammatory cytokines may promote HIV transmission by promoting HIV gene transcription (38–41) or by promoting viral translocation across the mucosal epithelium by weakening tight junction integrity (42–45). In addition, neutrophils may directly protect against HIV transmission by forming neutrophil extracellular traps (46).
Although we found that SP EVs from HIV-infected men induced a highly proinflammatory response in isolated eSFs and eECs, we did not see a marked effect of these vesicles on HIV infection of primary CD4+ T cells. Conflicting data exist as to whether EVs inhibit or facilitate HIV infection of permissive cells. EVs from peripheral blood and SP were shown to inhibit HIV infection (47–49), while other studies have shown that macrophage-derived EVs facilitate HIV infection (50) and that EVs derived from cells harboring integrated HIV-1 activate T cells and increase their permissivity to HIV infection (51). Our infection studies did not find a consistent effect of EVs on enhancing or inhibiting HIV infection of resting or activated CD4+ T cells, but one important caveat was that a significant processing time was required to isolate the EVs, since the high viscosity of some of the SP samples required that they be centrifuged for >10 h. As the HIV-enhancing activity of SP is known to be markedly affected by processing time (52), the EVs and their associated reconstituted SP used in this study may have lost activity due to the long processing time. Future studies isolating EVs using alternative protocols with shorter processing times may better reflect how these vesicles behave in vivo.
FACS phenotyping of EVs was carried out in order to assess whether any differences in protein composition exist between SP EVs from uninfected individuals and those from HIV-infected individuals, as well as predict the cell of origin of the vesicles. We found that SP EVs expressed markers reflecting both lymphoid and myeloid origin, as well as prostasome-derived markers, and no differences between uninfected and infected individuals were observed. These results suggest that SP EVs originate from multiple cell types and that HIV serostatus does not alter EV biogenesis. Interestingly, however, we observed significant phenotypic differences in seminal MVs in longitudinal specimens from chronically infected individuals before versus after suppression by ART. The increased abundance of MVs expressing phenotypic markers of lymphoid and myeloid cells after suppression by ART may be due to immune recovery in the male genital tract after viral suppression.
As our FACS analysis did not identify any differences in the EVs between uninfected and infected individuals, we implemented a more unbiased characterization of the EV cargo by characterizing the miRNA content of the vesicles. This analysis revealed that the miRNAs from the EVs of HIV-infected men exhibited a higher diversity than those from the EVs of uninfected men (Fig. 7A). At the same time, the variability of the SP EV miRNA profile between HIV-infected donors was less than that between HIV-negative donors (Fig. 7B), suggesting that during active HIV replication, specific miRNAs are induced across infected individuals. Whether the induced miRNAs regulate the expression of genes associated with inflammation remains to be determined. Interestingly, the semen microbiome from HIV-infected men has a lower diversity than that from uninfected men (14); to what extent this dysbiosis causes an altered miRNA expression profile is unknown. Future studies should examine how the semen miRNAs from HIV-infected men can affect gene expression in various cell types resident in the genital tract and whether the genes regulated by these miRNAs may play a direct role in facilitating or limiting HIV transmission.
MATERIALS AND METHODS
Study subjects.
The Options study is an observational cohort study that, since 1998, has enrolled over 800 men with recent HIV infection (<3 months) and HIV-negative controls at high risk based on their behavior. The seminal plasma samples were selected based on the highest available volume in the semen bank. A total of 40 SP samples were collected from 20 antiretroviral therapy-naive men acutely infected with HIV (as defined by sampling <3 months from the estimated date of infection, based on previously described criteria [53]) and 20 clinically matched uninfected individuals. Additionally, for FACS characterization, we studied 10 chronically infected individuals with paired samples available from before and after the initiation of ART. All chronically infected individuals had initiated ART >6 months after the estimated date of infection. All specimens from chronically infected individuals are referred to as “chronic specimens,” while those from the acute-phase specimens are referred to simply as “HIV-infected specimens.” At enrollment, demographic information and clinical laboratory data, including the HIV-1 viral load, CD4+ T-cell count, and medication history, were obtained. The individuals donating the semen samples were not tested for coinfections, such as coinfection with hepatitis B virus or hepatitis C virus. All experiments were performed in accordance with the approved UCSF guidelines and regulations. The study was approved by the UCSF Institutional Review Board, and all the participants signed a written informed consent.
Seminal plasma processing.
The semen samples were liquefied at room temperature for 2 h and then centrifuged at 400 to 600 × g for 5 to 10 min to remove spermatozoa and other cells. The supernatants were collected, aliquoted, and stored at −80°C until use in experimental assays.
Isolation of T cells and FRT-derived epithelial cells and stromal fibroblasts.
Peripheral blood mononuclear cells (PBMCs) from uninfected donors were obtained from leukoreduction system chambers from the Vitalant blood bank, and T cells were isolated by Miltenyi bead-based isolation of CD14− CD4+ cells using a previously described approach (54). Primary human endometrial tissue from deidentified donors was obtained from the UCSF Endometrial Tissue Bank and the Cooperative Human Tissue Network. Endometrial tissue samples were processed on the day of collection, and endometrial epithelial cells (eECs) and endometrial stromal fibroblasts (eSFs) were isolated and purified using selective attachment as described previously (8, 55). Isolated eSFs were cultured in serum-containing fibroblast growth medium (SCM; 75% phenol red-free Dulbecco’s modified Eagle’s medium [DMEM] and 25% MCDB-105 supplemented with 10% charcoal-stripped fetal bovine serum [FBS] and 5-μg/ml insulin), while eECs were cultured on Matrigel-coated dishes (BD Biosciences) with defined keratinocyte serum-free medium (KSFM; Gibco). Of note, all experiments in this study used the same lot of FBS.
Preparation of fractionated SP enriched for EVs.
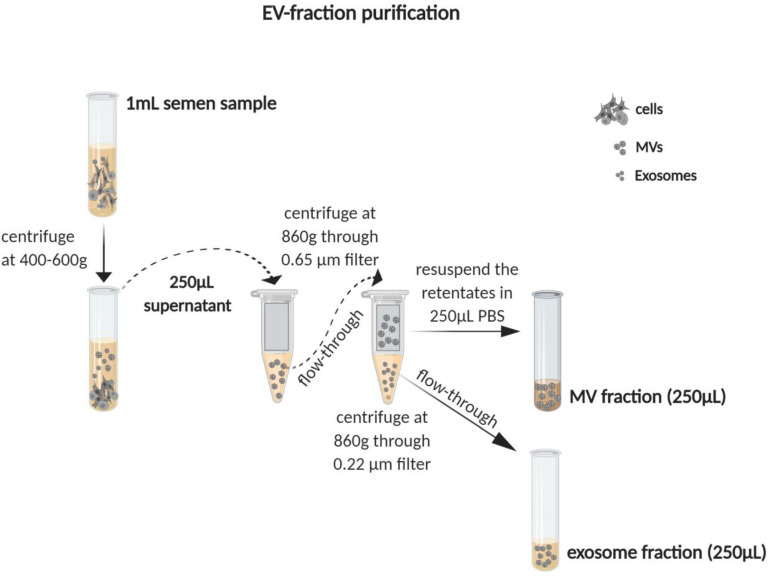
EV preparations were generated using methods similar to those previously described for the isolation of EVs from blood (56, 57). Briefly, 250 μl SP was centrifuged through 0.65-μm-pore-size centrifugal filters (Millipore) for 10 min at 860 × g or until most supernatant had passed through. The flowthrough was then collected and centrifuged for 10 min at 860 × g using 0.22-μm-pore-size filters (Millipore). The retentate, harboring MVs that did not pass through the 0.22-μm-pore-size filter, was resuspended in 250 μl phosphate-buffered saline (PBS). This material was used as a source of MVs, is referred to as the “MV fraction,” and corresponds to fractionated SP enriched for components of >0.22 μm and <0.65 μm. The flowthrough from the 0.22-μm-pore-size filter consisted of a mix of soluble factors and exosomes, was saved, and was designated the “exosome fraction.” This fraction harbors EVs of <0.22 μm. Reconstituted SP (rSP) was generated by using the exosome fraction to resuspend the retentates from both the 0.65-μm- and 0.22-μm-pore-size filters. rSP was used in place of SP before fractionation to account for any effects of processing on the properties of the MV and exosome fractions. A schematic of the purification scheme is presented in Fig. 9.
FIG 9.
Schematic of generation of the microvesicle (MV) fraction, exosome fraction, and reconstituted seminal plasma (rSP) from semen. Semen samples from uninfected and HIV-infected men were centrifuged at 400 to 600 × g, after which 250 μl of the supernatant was removed as a source of SP. The SP was then centrifuged through a 0.65-μm-pore-size filter, followed by a second round of centrifugation through a 0.22-μm-pore-size filter. The flowthrough from the final centrifugation was saved as the exosome fraction. The retentate from the final centrifugation was resuspended in 250 μl PBS and used as the MV fraction. To generate rSP, the MV fraction was resuspended in the exosome fraction.
Nanoparticle tracking analysis and albumin quantification.
The concentration and size distribution profile of the particles were evaluated using a NanoSight NS300 instrument (Malvern) configured with a syringe pump, a 405-nm laser, and a high-sensitivity scientific complementary metal-oxide-semiconductor camera. Five 60-s videos were recorded for each sample with the camera level set at 15 and the detection threshold set at 5. Ten microliters of EV samples was diluted 1:1,000 in PBS that had been filtered with a 0.22-μm-pore-size filter. Data were analyzed using NTA software (version 3.3). To quantify the concentration of albumin in EV fractions, a human albumin enzyme-linked immunosorbent assay (ELISA) kit was used according to the manufacturer´s instructions (Bethyl).
Effect of MV fraction and rSP on primary cells from FRT.
The MV fractions were assessed for their effects on eSFs and eECs by RNA-seq. We opted to use the MV fractions instead of the exosome fractions for these studies because the latter are difficult to separate from pure HIV virions. Equal volumes of the MV fractions and rSP were each pooled from 15 acutely infected individuals and from 15 uninfected individuals. These four pooled fractions were used to treat eSFs or eECs from 3 different donors at a final concentration equivalent to that of 1% rSP for 5 h, followed by 3 washes with medium to remove surface-associated MVs. Similar conditions have previously been demonstrated to be nontoxic for eSFs and eECs and to mimic the dilution of semen in the FRT by vaginal fluids (8). Of note, due to technical and budgetary reasons, we used pooled MV fractions and rSP instead of testing each sample individually on the eSFs and eECs. Total RNA was then extracted using a NucleoSpin RNA purification kit (Macherey Nagel) following the manufacturer´s protocol. DNase I (Qiagen) was dissolved in the elution buffer of the NucleoSpin kit and used as instructed by the manufacturer (Macherey Nagel) during the DNase treatment step. RNA samples were quantified using a NanoDrop spectrophotometer, and integrity was assessed using a Bioanalyzer instrument (Agilent). Paired-end RNA-seq at >20 million reads per sample was performed using a HiSeq platform (Illumina) by Novogene. Trimmed reads were aligned to the Genome Reference Consortium Human Build 38 reference genome using the TopHat2 (version 2.1.0) program with the following parameters: global alignment, no mismatch in the 20-bp seed, up to two mismatches in the read, library type Fr-unstranded, and a mate inner distance of 50. Aligned reads were filtered by removing reads with a low mapping quality (below 20) and keeping singletons. Filtered reads were annotated to the Genome Reference Consortium Human Build 38 reference transcriptome using the Partek E/M annotation model (Partek Flow, build version 5.0.16.1128) with the following parameters: junction reads required to match introns, set to true; strict paired-end compatibility not required, set to false; and strand specificity not required, set to false. Filtered reads were normalized using the number of transcripts per million kilobase reads (TPM). Low-abundance transcripts were removed, with the lowest maximum coverage being set to 1.0. Differential gene expression analysis was calculated by analysis of variance (ANOVA) in a paired test, with treatment being the fixed factor and the identifier being the random factor, using Partek Flow software (Partek Incorporated). Differentially expressed transcripts were identified based on the fold change in expression (≥1.5), with significance set at a P value of <0.05.
Validation of RNA-seq candidates.
Culture supernatants from samples analyzed by RNA-seq were analyzed by the Luminex multiplex assay (Thermo Fisher) to assess the effects of SP and SP components on the secretion of CCL2, VEGF, and HGF by eSFs and eECs. Supernatants were analyzed by Luminex by the Endocrine Technologies Support Core (ETSC) at the Oregon National Primate Research Center (ONPRC) according to the manufacturer’s instructions (Thermo Fisher). Briefly, 50 μl of each serum sample was diluted in assay diluent and incubated overnight with antibody-coated, fluorescent dye-capture microspheres specific for each analyte, followed by detection antibodies and streptavidin-phycoerythrin (PE). Washed microspheres with bound analytes were resuspended in reading buffer and analyzed on a Milliplex LX-200 analyzer (EMD Millipore) bead sorter with XPonent software (version 3.1; Luminex). Data were calculated using Milliplex Analyst software (version 5.1; EMD Millipore). For assessing the effects of SP and SP components on the secretion of CXCL1 in eSFs, culture supernatants from the specimens analyzed by RNA-seq were analyzed using a CXCL1 ELISA according to the manufacturer’s instructions (Abcam).
Infection assays.
The CCR5-tropic virus encoding a green fluorescent protein (GFP) reporter (BaL-GFP) (12) was produced by transfection of the provirus-containing plasmid into 293T cells cultured in D10 medium (consisting of DMEM with 10% fetal bovine serum [FBS], penicillin [50 U/ml], streptomycin [50 μg/ml], and l-glutamine [2 mM]). The supernatant was collected as a source of virions for the infection assays. Virions (100-ng/ml p24Gag) were pretreated with 10% rSP, the MV fraction, or the exosome fraction from each individual donor sample for 5 min in 30 μl in R10 medium (RPMI 1640 with 10% FBS, penicillin [50 U/ml], streptomycin [50 μg/ml], and l-glutamine [2 mM]). Fifteen microliters of this mix was used for infection of 1 × 105 PBMCs in 200 μl in a V-bottom plate. This corresponds to a final rSP concentration of 0.75%. Where indicated, PBMCs were first stimulated for 2 days with 10-μg/ml PHA in the presence of 100-IU/ml IL-2 (Life Technologies) as previously described (12). After exposing the cells for 2 h in the absence or presence of rSP and EVs, the cells were pelleted by centrifugation and resuspended in 200 μl fresh medium in a U-bottom plate and cultured for 3 days. The cells were then harvested; stained for 30 min at 4°C with Zombie viability dye (BioLegend) and with antibodies against CD3-allophycocyanin (APC)-H7 (clone SK7), CD8-APC (clone SK1), CD4-PE-Cy7 (clone SK3), CD25-V450 (clone M-A251), and HLA-DR-PE (clone G46-6); washed two times; and analyzed for infection rates by flow cytometry using an LSR II flow cytometer (Becton Dickinson). CD4+ T cells were identified by gating on live, singlet CD3+ CD8− cells. The percentage of GFP-positive cells was used as a readout for infection rates. By monitoring infection rates using GFP instead of intracellular p24, we avoided detecting any infection mediated by endogenous virions. The activation state was assessed by quantitating the cell surface expression levels of CD25 and HLA-DR. All antibodies for the infection studies were purchased from BD Biosciences. Data were analyzed with FlowJo software (BD Biosciences).
Characterization of MV surface markers by flow cytometry.
The MV fraction was stained using pretitrated volumes of fluorochrome-conjugated monoclonal antibodies, listed here according to their cell of origin: lymphocytes (CD3-fluorescein isothiocyanate [FITC; clone OK-T3; BioLegend], CD16-V421 [clone 3G8; BD Biosciences], CD19-Alexa Fluor 700 [clone HIB19; BioLegend], CD28-PE [clone CD28.2; BioLegend], and CD38-APC [clone HB-7; BioLegend]), monocytes (CD14-PE-Cy7; clone 63D3; BioLegend), macrophages (CD11b-PE-Cy7; clone ICRF44; BioLegend), neutrophils (CD15-FITC; clone MCS-1; Exalpha), platelets (CD62p-APC; clone AK4; BD Biosciences), granulocytes (CD66b-PE; clone G10F5; BioLegend), and prostasome (aminopeptidase [CD13-V421; clone WM15; BioLegend], dipeptidyl peptidase IV [CD26-PE; clone BA5b; BioLegend], membrane cofactor protein [CD46-PE-Cy7; clone TRA-2-10; BioLegend], regulator of complement [CD55-peridinin chlorophyll protein-Cy5.5; clone JS11; BioLegend], membrane regulator [CD59-FITC; clone P282; BioLegend], and tissue factor [CD142-APC; clone NY2; BioLegend]). One to 5 μl of titrated monoclonal antibodies was added to 10 μl of MVs and incubated at 4°C for 30 min. MVs were centrifuged for 10 min at 860 × g using 0.22-μm-pore-size filters (Ultrafree MC-GV centrifugal filters; Millipore), and MVs remaining on the filter surface were resuspended in 500 μl of 0.22-μm-pore-size-filtered PBS containing 2.8% formaldehyde (BD stabilizing fixative). Acquisition was performed on an LSR II flow cytometer (Becton, Dickinson). The triggering signal was set to the maximum voltages of 500 to 600 and 300 to 400 for the forward scatter (FSC) and the side scatter (SSC), respectively. Based on previous studies (58, 59), a blend of size-calibrated fluorescent beads sized from 0.1 μm (Invitrogen) to 1 μm (Megamix beads [0.16, 0.2, 0.24, and 0.5 μm] and Spherotech beads [1 μm]) was used to optimize the SSC and FSC voltages for MV analysis. Samples were acquired for 1 min at a low flow rate (8 to 12 μl/min). TruCount beads (BD Biosciences) were used to obtain the absolute quantity of MVs. Analysis was performed using FlowJo software (version 10; Tree Star). Of note, the smaller exosomes from the exosome fraction were not analyzed by FACS, as these vesicles are at the limit of detection at the single-vesicle level (60). For a detailed description of how the MV detection methodology was validated, see the work of Inglis et al. (59). Statistical analyses were performed using Prism software (version 7.0; GraphPad Software). Data were log10 transformed to address rightward skewing. The Shapiro-Wilk test was used to determine normality prior to implementation of Student’s t test. The nonparametric Mann-Whitney test was used for unpaired comparisons. The Wilcoxon matched-pairs signed-ranked test was used for paired comparisons. All data are presented as the mean ± standard deviation (SD), unless otherwise indicated. Values of P of ≤0.05 were considered statistically significant. All P values were two-sided and corrected for the false discovery rate (FDR) using the method of Benjamini and Hochberg (61).
EV miRNA expression analysis.
EVs were isolated from aliquots of SP by ExoQuick solution (System Biosciences). Five hundred microliters of SP from each donor sample was centrifuged at 1,500 × g for 5 min to remove residual cells and debris. The supernatants were then transferred to new 1.5-ml Eppendorf tubes. ExoQuick solution was added to the supernatant at a 1:4 ratio (ExoQuick supernatant), the components were mixed gently, and the mixture was allowed to incubate for 30 min at 4°C. After the incubation, the mixture was centrifuged at 1,500 × g for 30 min to recover the EVs, which were then processed for total RNA isolation using a SeraMir exosome RNA purification column kit (System Biosciences). For each sample, 1 μl of the final RNA eluate was used to measure the small RNA concentration using a Bioanalyzer 2100 Expert instrument (Agilent Technologies). Note that at no point in time was any FBS added to the specimens subjected to miRNA analysis. Small RNA libraries were constructed with a CleanTag small RNA library preparation kit (TriLink). The final purified library was quantified with high-sensitivity DNA reagents and high-sensitivity DNA chips (Agilent Technologies). The libraries were pooled, and the 140- to 300-bp fragments were size selected on an 8% TBE (Tris-borate-EDTA) gel (Invitrogen by Life Technologies). The size-selected library was quantified with high-sensitivity DNA 1000 screen tape and high-sensitivity D1000 reagents (Agilent Technologies) and the TailorMix HT1 quantitative PCR assay (SeqMatic), followed by a NextSeq high-output single-end sequencing run using a NextSeq 500/550 high-output (version 2) kit (Illumina) according to the manufacturer’s instructions. The FASTQ files were imported from the Banana Slug analytics platform (UCSC) into ArrayStudio software (OmicSoft; Qiagen) for processing. The files were mapped to the Genome Reference Consortium Human Build 38 reference genome (GenBank assembly accession number GCA_000001405.15) and gene model miRbase (version 21) after the adaptors (TGGAATTCTCGGGTGCCAAGG) were trimmed. Downstream effects analysis (DEA) was used to identify the biological functions and/or diseases that were most significantly associated with the data set. A right-tailed Fisher’s exact test was used to calculate a P value determining the probability that each biological function and/or disease assigned to the data set was due to chance alone. Furthermore, DEA was used to predict increases or decreases in these biological functions and/or diseases occurring in the data set by integrating the direction of change of the differentially expressed genes into a Z-score algorithm calculation (62). Functions and/or diseases differing between treatment groups with absolute-value Z-scores of >1.5 were considered. Identified pathways were analyzed for Z-scores to assess the activation (≥1.5) of the respective pathways as previously reported (8, 63). Sequences were trimmed, using the Cutadapt program (64), against the 3′ adapter sequence TGGAATTCTCGGGTGCCAAGG. The STAR program (65) was then used to align the reads against those in the miRBase database (66). Raw counts of miRNAs were obtained using HTSeq-count software, and differential expression analyses were then performed using DESeq2 software (67). For HIV-1 mapping, local alignment was performed by aligning all raw sequences in the FASTA format against an HIV-1 reference genome (GenBank accession number NC_001802.1), using the blastn (version 2.2.27) program (68) with default parameters. No alignments to the HIV-1 sequence were found in any of the sequenced samples.
Data availability.
Sequencing data are available in the Gene Expression Omnibus (GEO) repository under accession number GSE152714. The raw miRNA sequencing data have been added to the NCBI public database under BioProject accession number PRJNA636924.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grant R21AI122821 to N.R.R. CFAR is supported by grant P30AI027763. The Oregon National Primate Research Center is supported by NIH grant P51OD011092 to ONPRC.
We have no conflicts of interests to declare.
We acknowledge N. Raman and support from CFAR for the flow cytometric analyses and David Erikson and the team at the Endocrine Technologies Core at the Oregon National Primate Research Center for conducting the Luminex multiplex assay. We also thank Shelley Facente for data analysis, Francoise Chanut for editorial assistance, and Robin Givens for administrative assistance.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.von Wolff M, Nowak O, Pinheiro RM, Strowitzki T. 2007. Seminal plasma—immunomodulatory potential in men with normal and abnormal sperm count. Eur J Obstet Gynecol Reprod Biol 134:73–78. doi: 10.1016/j.ejogrb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Robertson SA. 2005. Seminal plasma and male factor signalling in the female reproductive tract. Cell Tissue Res 322:43–52. doi: 10.1007/s00441-005-1127-3. [DOI] [PubMed] [Google Scholar]
- 3.Introini A, Boström S, Bradley F, Gibbs A, Glaessgen A, Tjernlund A, Broliden K. 2017. Seminal plasma induces inflammation and enhances HIV-1 replication in human cervical tissue explants. PLoS Pathog 13:e1006402. doi: 10.1371/journal.ppat.1006402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roan NR, Sandi-Monroy N, Kohgadai N, Usmani SM, Hamil KG, Neidleman J, Montano M, Ständker L, Röcker A, Cavrois M, Rosen J, Marson K, Smith JF, Pilcher CD, Gagsteiger F, Sakk O, O’Rand M, Lishko PV, Kirchhoff F, Münch J, Greene WC. 2017. Semen amyloids participate in spermatozoa selection and clearance. Elife 6:e24888. doi: 10.7554/eLife.24888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Münch J, Rücker E, Ständker L, Adermann K, Goffinet C, Schindler M, Wildum S, Chinnadurai R, Rajan D, Specht A, Giménez-Gallego G, Sánchez PC, Fowler DM, Koulov A, Kelly JW, Mothes W, Grivel J-C, Margolis L, Keppler OT, Forssmann W-G, Kirchhoff F. 2007. Semen-derived amyloid fibrils drastically enhance HIV infection. Cell 131:1059–1071. doi: 10.1016/j.cell.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 6.Camus C, Matusali G, Bourry O, Mahe D, Aubry F, Bujan L, Pasquier C, Massip P, Ravel C, Zirafi O, Munch J, Roan NR, Pineau C, Dejucq-Rainsford N. 2016. Comparison of the effect of semen from HIV-infected and uninfected men on CD4+ T-cell infection. AIDS 30:1197–1208. doi: 10.1097/QAD.0000000000001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdulhaqq SA, Martinez M, Kang G, Rodriguez IV, Nichols SM, Beaumont D, Joseph J, Azzoni L, Yin X, Wise M, Weiner D, Liu Q, Foulkes A, Münch J, Kirchhoff F, Coutifaris C, Tomaras GD, Sariol C, Marx PA, Li Q, Kraiselburd EN, Montaner LJ. 2019. Repeated semen exposure decreases cervicovaginal SIVmac251 infection in rhesus macaques. Nat Commun 10:3753. doi: 10.1038/s41467-019-11814-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen JC, Johnson BA, Erikson DW, Piltonen TT, Barragan F, Chu S, Kohgadai N, Irwin JC, Greene WC, Giudice LC, Roan NR. 2014. Seminal plasma induces global transcriptomic changes associated with cell migration, proliferation and viability in endometrial epithelial cells and stromal fibroblasts. Hum Reprod 29:1255–1270. doi: 10.1093/humrep/deu047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharkey DJ, Macpherson AM, Tremellen KP, Robertson SA. 2007. Seminal plasma differentially regulates inflammatory cytokine gene expression in human cervical and vaginal epithelial cells. Mol Hum Reprod 13:491–501. doi: 10.1093/molehr/gam028. [DOI] [PubMed] [Google Scholar]
- 10.Sharkey DJ, Tremellen KP, Jasper MJ, Gemzell-Danielsson K, Robertson SA. 2012. Seminal fluid induces leukocyte recruitment and cytokine and chemokine mRNA expression in the human cervix after coitus. J Immunol 188:2445–2454. doi: 10.4049/jimmunol.1102736. [DOI] [PubMed] [Google Scholar]
- 11.Goddard LM, Murphy TJ, Org T, Enciso JM, Hashimoto-Partyka MK, Warren CM, Domigan CK, McDonald AI, He H, Sanchez LA, Allen NC, Orsenigo F, Chao LC, Dejana E, Tontonoz P, Mikkola H, Iruela-Arispe ML. 2014. Progesterone receptor in the vascular endothelium triggers physiological uterine permeability preimplantation. Cell 156:549–562. doi: 10.1016/j.cell.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neidleman JA, Chen JC, Kohgadai N, Müller JA, Laustsen A, Thavachelvam K, Jang KS, Stürzel CM, Jones JJ, Ochsenbauer C, Chitre A, Somsouk M, Garcia MM, Smith JF, Greenblatt RM, Münch J, Jakobsen MR, Giudice LC, Greene WC, Roan NR. 2017. Mucosal stromal fibroblasts markedly enhance HIV infection of CD4+ T cells. PLoS Pathog 13:e1006163. doi: 10.1371/journal.ppat.1006163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Someya M, Kojima T, Ogawa M, Ninomiya T, Nomura K, Takasawa A, Murata M, Tanaka S, Saito T, Sawada N. 2013. Regulation of tight junctions by sex hormones in normal human endometrial epithelial cells and uterus cancer cell line Sawano. Cell Tissue Res 354:481–494. doi: 10.1007/s00441-013-1676-9. [DOI] [PubMed] [Google Scholar]
- 14.Liu CM, Osborne BJW, Hungate BA, Shahabi K, Huibner S, Lester R, Dwan MG, Kovacs C, Contente-Cuomo TL, Benko E, Aziz M, Price LB, Kaul R. 2014. The semen microbiome and its relationship with local immunology and viral load in HIV infection. PLoS Pathog 10:e1004262. doi: 10.1371/journal.ppat.1004262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson JA, Ping L-H, Dibben O, Jabara CB, Arney L, Kincer L, Tang Y, Hobbs M, Hoffman I, Kazembe P, Jones CD, Borrow P, Fiscus S, Cohen MS, Swanstrom R, the Center for HIV/AIDS Vaccine Immunology. 2010. HIV-1 populations in semen arise through multiple mechanisms. PLoS Pathog 6:e1001053. doi: 10.1371/journal.ppat.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kafka JK, Sheth PM, Nazli A, Osborne BJ, Kovacs C, Kaul R, Kaushic C. 2012. Endometrial epithelial cell response to semen from HIV-infected men during different stages of infection is distinct and can drive HIV-1-long terminal repeat. AIDS 26:27–36. doi: 10.1097/QAD.0b013e32834e57b2. [DOI] [PubMed] [Google Scholar]
- 17.Olivier AJ, Masson L, Ronacher K, Walzl G, Coetzee D, Lewis DA, Williamson A-L, Passmore J-A, Burgers WA. 2014. Distinct cytokine patterns in semen influence local HIV shedding and HIV target cell activation. J Infect Dis 209:1174–1184. doi: 10.1093/infdis/jit649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lisco A, Munawwar A, Introini A, Vanpouille C, Saba E, Feng X, Grivel J-C, Singh S, Margolis L. 2012. Semen of HIV-1-infected individuals: local shedding of herpesviruses and reprogrammed cytokine network. J Infect Dis 205:97–105. doi: 10.1093/infdis/jir700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly RW, Holland P, Skibinski G, Harrison C, McMillan L, Hargreave T, James K. 1991. Extracellular organelles (prostasomes) are immunosuppressive components of human semen. Clin Exp Immunol 86:550–556. doi: 10.1111/j.1365-2249.1991.tb02968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Théry C, Ostrowski M, Segura E. 2009. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 21.Mathivanan S, Ji H, Simpson RJ. 2010. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 22.Danzer KM, Kranich LR, Ruf WP, Cagsal-Getkin O, Winslow AR, Zhu L, Vanderburg CR, McLean PJ. 2012. Exosomal cell-to-cell transmission of alpha synuclein oligomers. Mol Neurodegener 7:42. doi: 10.1186/1750-1326-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vojtech L, Woo S, Hughes S, Levy C, Ballweber L, Sauteraud RP, Strobl J, Westerberg K, Gottardo R, Tewari M, Hladik F. 2014. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res 42:7290–7304. doi: 10.1093/nar/gku347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buzas EI, György B, Nagy G, Falus A, Gay S. 2014. Emerging role of extracellular vesicles in inflammatory diseases. Nat Rev Rheumatol 10:356–364. doi: 10.1038/nrrheum.2014.19. [DOI] [PubMed] [Google Scholar]
- 25.Robbins PD, Morelli AE. 2014. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol 14:195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vojtech L, Zhang M, Davé V, Levy C, Hughes SM, Wang R, Calienes F, Prlic M, Nance E, Hladik F. 2019. Extracellular vesicles in human semen modulate antigen-presenting cell function and decrease downstream antiviral T cell responses. PLoS One 14:e0223901. doi: 10.1371/journal.pone.0223901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lotvall J, Valadi H. 2007. Cell to cell signalling via exosomes through esRNA. Cell Adh Migr 1:156–158. doi: 10.4161/cam.1.3.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okoye IS, Coomes SM, Pelly VS, Czieso S, Papayannopoulos V, Tolmachova T, Seabra MC, Wilson MS. 2014. MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity 41:89–103. doi: 10.1016/j.immuni.2014.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baranyai T, Herczeg K, Onódi Z, Voszka I, Módos K, Marton N, Nagy G, Mäger I, Wood MJ, El Andaloussi S, Pálinkás Z, Kumar V, Nagy P, Kittel Á, Buzás EI, Ferdinandy P, Giricz Z. 2015. Isolation of exosomes from blood plasma: qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS One 10:e0145686. doi: 10.1371/journal.pone.0145686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karimi N, Cvjetkovic A, Jang SC, Crescitelli R, Hosseinpour Feizi MA, Nieuwland R, Lötvall J, Lässer C. 2018. Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cell Mol Life Sci 75:2873–2886. doi: 10.1007/s00018-018-2773-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim K-A, Yolamanova M, Zirafi O, Roan NR, Staendker L, Forssmann W-G, Burgener A, Dejucq-Rainsford N, Hahn BH, Shaw GM, Greene WC, Kirchhoff F, Münch J. 2010. Semen-mediated enhancement of HIV infection is donor-dependent and correlates with the levels of SEVI. Retrovirology 7:55. doi: 10.1186/1742-4690-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuo H-H, Ahmad R, Lee GQ, Gao C, Chen H-R, Ouyang Z, Szucs MJ, Kim D, Tsibris A, Chun T-W, Battivelli E, Verdin E, Rosenberg ES, Carr SA, Yu XG, Lichterfeld M. 2018. Anti-apoptotic protein BIRC5 maintains survival of HIV-1-infected CD4+ T cells. Immunity 48:1183–1194.e5. doi: 10.1016/j.immuni.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyu K, Kopcho P, Shouman K, Martinson M, Martinez-Maza M, Stapleton O. 2019. Human immunodeficiency virus (HIV) infection and use of illicit substances promote secretion of semen exosomes that enhance monocyte adhesion and induce actin reorganization and chemotactic migration. Cells 8:1027. doi: 10.3390/cells8091027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moser B, Clark-Lewis I, Zwahlen R, Baggiolini M. 1990. Neutrophil-activating properties of the melanoma growth-stimulatory activity. J Exp Med 171:1797–1802. doi: 10.1084/jem.171.5.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schumacher C, Clark-Lewis I, Baggiolini M, Moser B. 1992. High- and low-affinity binding of GRO alpha and neutrophil-activating peptide 2 to interleukin 8 receptors on human neutrophils. Proc Natl Acad Sci U S A 89:10542–10546. doi: 10.1073/pnas.89.21.10542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ericson SG, Zhao Y, Gao H, Miller KL, Gibson LF, Lynch JP, Landreth KS. 1998. Interleukin-6 production by human neutrophils after Fc-receptor cross-linking or exposure to granulocyte colony-stimulating factor. Blood 91:2099–2107. doi: 10.1182/blood.V91.6.2099. [DOI] [PubMed] [Google Scholar]
- 37.Tecchio C, Micheletti A, Cassatella MA. 2014. Neutrophil-derived cytokines: facts beyond expression. Front Immunol 5:508. doi: 10.3389/fimmu.2014.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perkins ND, Edwards NL, Duckett CS, Agranoff AB, Schmid RM, Nabel GJ. 1993. A cooperative interaction between NF-kappa B and Sp1 is required for HIV-1 enhancer activation. EMBO J 12:3551–3558. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West MJ, Lowe AD, Karn J. 2001. Activation of human immunodeficiency virus transcription in T cells revisited: NF-kappaB p65 stimulates transcriptional elongation. J Virol 75:8524–8537. doi: 10.1128/jvi.75.18.8524-8537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herbein G, Gras G, Khan KA, Abbas W. 2010. Macrophage signaling in HIV-1 infection. Retrovirology 7:34. doi: 10.1186/1742-4690-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rollenhagen C, Asin SN. 2011. Enhanced HIV-1 replication in ex vivo ectocervical tissues from post-menopausal women correlates with increased inflammatory responses. Mucosal Immunol 4:671–681. doi: 10.1038/mi.2011.34. [DOI] [PubMed] [Google Scholar]
- 42.Nazli A, Chan O, Dobson-Belaire WN, Ouellet M, Tremblay MJ, Gray-Owen SD, Arsenault AL, Kaushic C. 2010. Exposure to HIV-1 directly impairs mucosal epithelial barrier integrity allowing microbial translocation. PLoS Pathog 6:e1000852. doi: 10.1371/journal.ppat.1000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki T, Yoshinaga N, Tanabe S. 2011. Interleukin-6 (IL-6) regulates claudin-2 expression and tight junction permeability in intestinal epithelium. J Biol Chem 286:31263–31271. doi: 10.1074/jbc.M111.238147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams RB, Planchon SM, Roche JK. 1993. IFN-gamma modulation of epithelial barrier function. Time course, reversibility, and site of cytokine binding. J Immunol 150:2356–2363. [PubMed] [Google Scholar]
- 45.Colgan SP, Resnick MB, Parkos CA, Delp-Archer C, McGuirk D, Bacarra AE, Weller PF, Madara JL. 1994. IL-4 directly modulates function of a model human intestinal epithelium. J Immunol 153:2122–2129. [PubMed] [Google Scholar]
- 46.Barr FD, Ochsenbauer C, Wira CR, Rodriguez-Garcia M. 2018. Neutrophil extracellular traps prevent HIV infection in the female genital tract. Mucosal Immunol 11:1420–1428. doi: 10.1038/s41385-018-0045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Madison MN, Roller RJ, Okeoma CM. 2014. Human semen contains exosomes with potent anti-HIV-1 activity. Retrovirology 11:102. doi: 10.1186/s12977-014-0102-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Carvalho JV, de Castro RO, da Silva EZM, Silveira PP, da Silva-Januário ME, Arruda E, Jamur MC, Oliver C, Aguiar RS, daSilva L. 2014. Nef neutralizes the ability of exosomes from CD4+ T cells to act as decoys during HIV-1 infection. PLoS One 9:e113691. doi: 10.1371/journal.pone.0113691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welch JL, Kaddour H, Schlievert PM, Stapleton JT, Okeoma CM. 2018. Semen exosomes promote transcriptional silencing of HIV-1 by disrupting NF-κB/Sp1/Tat circuitry. J Virol 92:e00731-18. doi: 10.1128/JVI.00731-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kadiu I, Narayanasamy P, Dash PK, Zhang W, Gendelman HE. 2012. Biochemical and biologic characterization of exosomes and microvesicles as facilitators of HIV-1 infection in macrophages. J Immunol 189:744–754. doi: 10.4049/jimmunol.1102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arenaccio C, Chiozzini C, Columba-Cabezas S, Manfredi F, Federico M. 2014. Cell activation and HIV-1 replication in unstimulated CD4+ T lymphocytes ingesting exosomes from cells expressing defective HIV-1. Retrovirology 11:46. doi: 10.1186/1742-4690-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roan NR, Liu H, Usmani SM, Neidleman J, Muller JA, Avila-Herrera A, Gawanbacht A, Zirafi O, Chu S, Dong M, Kumar ST, Smith JF, Pollard KS, Fandrich M, Kirchhoff F, Munch J, Witkowska HE, Greene WC. 2014. Liquefaction of semen generates and later degrades a conserved semenogelin peptide that enhances HIV infection. J Virol 88:7221–7234. doi: 10.1128/JVI.00269-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grebe E, Facente SN, Bingham J, Pilcher CD, Powrie A, Gerber J, Priede G, Chibawara T, Busch MP, Murphy G, Kassanjee R, Welte A, on behalf of the Consortium for the Evaluation and Performance of HIV Incidence Assays (CEPHIA). 2018. Interpreting HIV diagnostic histories into infection time estimates: analytical framework and online tool. Epidemiology 19:894. doi: 10.1186/s12879-019-4543-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roan NR, Münch J, Arhel N, Mothes W, Neidleman J, Kobayashi A, Smith-McCune K, Kirchhoff F, Greene WC. 2009. The cationic properties of SEVI underlie its ability to enhance human immunodeficiency virus infection. J Virol 83:73–80. doi: 10.1128/JVI.01366-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen JC, Roan NR. 2015. Isolation and culture of human endometrial epithelial cells and stromal fibroblasts. Bio Protoc 5:e1623. doi: 10.21769/bioprotoc.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu R, Greening DW, Rai A, Ji H, Simpson RJ. 2015. Highly-purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods 87:11–25. doi: 10.1016/j.ymeth.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 57.Danesh A, Inglis HC, Jackman RP, Wu S, Deng X, Muench MO, Heitman JW, Norris PJ. 2014. Exosomes from red blood cell units bind to monocytes and induce proinflammatory cytokines, boosting T-cell responses in vitro. Blood 123:687–696. doi: 10.1182/blood-2013-10-530469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inglis H, Norris P, Danesh A. 2015. Techniques for the analysis of extracellular vesicles using flow cytometry. J Vis Exp 2015:52484. doi: 10.3791/52484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inglis HC, Danesh A, Shah A, Lacroix J, Spinella PC, Norris PJ. 2015. Techniques to improve detection and analysis of extracellular vesicles using flow cytometry. Cytometry A 87:1052–1063. doi: 10.1002/cyto.a.22649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Pol E, Sturk A, van Leeuwen T, Nieuwland R, Coumans F, ISTH-SSC-VB Working Group. 2018. Standardization of extracellular vesicle measurements by flow cytometry through vesicle diameter approximation. J Thromb Haemost 16:1236–1245. doi: 10.1111/jth.14009. [DOI] [PubMed] [Google Scholar]
- 61.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 62.Krämer A, Green J, Pollard J, Tugendreich S. 2014. Causal analysis approaches in Ingenuity pathway analysis. Bioinformatics 30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mazan-Mamczarz K, Hagner PR, Corl S, Srikantan S, Wood WH, Becker KG, Gorospe M, Keene JD, Levenson AS, Gartenhaus RB. 2008. Post-transcriptional gene regulation by HuR promotes a more tumorigenic phenotype. Oncogene 27:6151–6163. doi: 10.1038/onc.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 65.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kozomara A, Birgaoanu M, Griffiths-Jones S. 2019. miRBase: from microRNA sequences to function. Nucleic Acids Res 47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available in the Gene Expression Omnibus (GEO) repository under accession number GSE152714. The raw miRNA sequencing data have been added to the NCBI public database under BioProject accession number PRJNA636924.