Abstract
Background
Diabetes is one of the greatest public health challenges worldwide, and we still lack complementary approaches to significantly enhance the efficacy of preventive and therapeutic approaches. Genetic and environmental factors are the culprits involved in diabetes risk. Evidence from the last decade has highlighted that deregulation in the immune and inflammatory responses increase susceptibility to type 1 and type 2 diabetes. Spatiotemporal patterns of gene expression involved in immune cell polarisation depend on genomic enhancer elements in response to inflammatory and metabolic cues. Several studies have reported that most regulatory genetic variants are located in the non-protein coding regions of the genome and particularly in enhancer regions. The progress of high-throughput technologies has permitted the characterisation of enhancer chromatin properties. These advances support the concept that genetic alteration of enhancers may influence the immune and inflammatory responses in relation to diabetes.
Scope of review
Results from genome-wide association studies (GWAS) combined with functional and integrative analyses have elucidated the impacts of some diabetes risk-associated variants that are involved in the regulation of the immune system. Additionally, genetic variant mapping to enhancer regions may alter enhancer status, which in turn leads to aberrant expression of inflammatory genes associated with diabetes susceptibility. The focus of this review was to provide an overview of the current indications that inflammatory processes are regulated at the genetic and epigenomic levels in diabetes, along with perspectives on future research avenues that may improve understanding of the disease.
Major conclusions
In this review, we provide genetic evidence in support of a deregulated immune response as a risk factor in diabetes. We also argue about the importance of enhancer regions in the regulation of immune cell polarisation and how the recent advances using genome-wide methods for enhancer identification have enabled the determination of the impact of enhancer genetic variation on diabetes onset and phenotype. This could eventually lead to better management plans and improved treatment responses in human diabetes.
Keywords: Diabetes, Inflammation, Genetics, Epigenetics, Enhancer
1. Introduction
Diabetes is a worldwide endemic disease, leading to chronic complications affecting multiple organs, including the heart, eyes and kidneys, and is characterised by absolute or relative decreased insulin secretion. Most common are type 1 (T1D) and type 2 diabetes (T2D), but based on aetiology, recent World Health Organisation classification identifies a dozen of others types of diabetes: hybrid forms of diabetes between type 1 and type 2, including slowly evolving immune-mediated diabetes of adults and ketosis-prone type 2 diabetes, gestational diabetes mellitus, and rarer types, such as monogenic diabetes, disease of the exocrine pancreas or endocrine disorders [1]. T1D affects 5–10% of diabetics and is characterised by auto-immune destruction of pancreatic beta-cells, leading to absolute insulinopenia. It mainly results from an insulitis, defined by immune cell infiltration of the pancreatic islet. T2D represents 90–95% of diabetes and combines decreased insulin secretion and increased insulin resistance. Despite this classification based on aetiology, pathogenic mechanisms remain unclear for each type of diabetes as recently revisited by Ahlqvist et al. [2]. Epidemic increase in diabetes prevalence is linked to major changes in lifestyle and dietary habits, including high-calorie and high-fat diet and inactivity. These environmental consequences are modulated by genetic factors, with inter- and intra-population variation. Indeed, these environmental factors are sensed through epigenomic pathways that modify gene transcription. Genomic regions called enhancers have been proposed to be extremely sensitive to environmental factors. In fact, their activity is regulated through epigenomic mechanisms, including chromatin organisation, histone modifications and coregulator dynamics. These epigenomic mechanisms particularly regulate inflammatory processes throughout diabetes development. Here, we will review the evidence that links genetics to inflammatory processes in the pathophysiology of T1D and T2D. We also highlight the emerging role of genomic enhancer regulation in immune cell activation through sensing the environmental factors of diabetes risk. Our focus is to provide an overview of the current evidence that inflammatory processes are regulated at the genetic and epigenomic levels in diabetes, along with perspectives on future research avenues that may help to better understand the diseases.
2. Inflammation in diabetes: genetic evidence
2.1. Type 1 Diabetes (T1D)
Type 1 Diabetes (T1D) is an autoimmune disorder characterised by destruction of insulin-producing pancreatic beta cells (Figure 1). It involves both cellular and humoral immunity. Although the aetiology of T1D is not fully understood, inherited genetic factors have been recognised to be implicated in its pathogenesis [3]. For example, twin studies have revealed a concordance of 43% for monozygotic twins, versus only 7% for dizygotic twins [4]. Two genetic regions in the human genome have emerged with consistent evidence of an association with T1D. These are the major histocompatibility complex (MHC) on chromosome 6 (locus 6p21.3) and the insulin (INS) gene locus on chromosome 11 (locus 11p15).
Figure 1.
Inflammatory signals in diabetes. Type 1 diabetes: Activation of immune cells is involved in pancreatic beta-cell death through a variety of inflammatory cytokines. Anti-gene presenting cells (APCs) and T lymphocytes participate in the inflammatory processes that promote the development of T1D. Type 2 diabetes: adipose tissue and liver dysfunction as well as gut dysbiosis contribute to the chronic inflammation. Inflammatory cytokines from T lymphocytes, monocytes and macrophages contribute to the interaction with the pancreatic islets leading to beta cell dysfunction. Examples of T1D/T2D-GWAS candidate genes are represented inside cells.
The MHC region was first found to be involved in T1D genetic predisposition in the 1970s [5]. Subsequently, this genomic region was divided into 3 main classes, and variants of human leukocyte antigen (HLA) class II genes, including HLA-DQ, HLA-DP and DR genes, were identified as the major risk factors of T1D [[6], [7], [8]]. In fact, rather than single variants, combinations of alleles, also known as haplotypes, have been found to drive these effects with a synergy of so-called DR3-DQ2 and DR4-DQ8 haplotypes (standing for DRB1∗03:01-DQA1∗05:01-DQB1∗02:01 and DRB1∗04:01/04:04-DQA1∗03:01-DQB1∗03:02, respectively). These two haplotypes are carried by 90% of patients versus 30% of controls, while 40% of patients harbour both of them, versus 3% of controls (odds ratio = 30). HLA Class II molecules are mainly expressed in immune cells, such as macrophages, dendritic cells, B cells, and T cells. HLA molecules are cell-surface receptors implicated in the immune system through antigen presentation. By presenting antigens to T cells, HLA molecules play a central role in the immune system, modulating regulatory T cells (Tregs) and conventional T cell activation [9]. Specific HLA contribution to T1D susceptibility might therefore be linked to antigen binding and presentation specificity [10,11]. Indeed, imputation of amino acids using single-nucleotide polymorphism (SNP) data confirmed the key role of Ala57 in the peptide-binding groove of the HLA-DQβ1 chain and identified independent effects at positions 13 and 71 of the HLA-DRβ1 chain [12]. By influencing effective antigen-binding, specific HLA alleles could influence central negative selection of self-reactive T lymphocytes inside the thymus and peripheral activation inside pancreatic nodes by dendritic-cells presenting beta-cell antigens, leading to their activation and finally to beta-cell destruction (Figure 1). The MHC is thought to contribute to 50% of T1D heritability [13], but HLA class II genes were reported to explain nearly 30%. Consequently, HLA class I alleles, including HLA-A∗24 and HLA-B∗39, have also been independently associated with T1D [14].
Non-MHC regions of the human genome were also intensively investigated to explain the missing heritability. Early in 1984, the INS gene was also associated with T1D [[15], [16], [17], [18]]. Candidate genes coding for cytotoxic T-lymphocyte antigen-4 (CTLA4) [19], protein tyrosine phosphatase non-receptor type 22 (PTPN22) [20,21] and interleukin-2 alpha chain receptor (IL-2RA) [22] were also associated with T1D. Then, large genome-wide association studies (GWAS) (that scan the whole genome using SNPs) were conducted, notably by the Type 1 Diabetes Genetics Consortium (T1DGC) that collected more than 6,000 T1D samples between 2002 and 2010 [23]. To date, 52 association signals have been reported (Table 1, [24,25]). With the exception of the HLA and 5 genes outside the MHC, including PTPN22, IFIH1, CTSH, TYK2 and FUT2, all associated variants are non-protein-coding, a finding recently corroborated by targeted deep resequencing of 301 candidate genes [26]. Most GWAS hits map to or near genes involved in immune processes, namely: BACH2, C1QTNF6, CCR5, CD69, CD226, CLEC16A, CTSH, ERBB3, GPR183, IFIH1, IKZF1, IL2/IL21, IL2RA, IL7R, IL10, IL18RAP, IL27, ITGB7, PTPN2, PTPN22, PRKCQ, SH2B3, STAT4, TAGAP, TNFAIP3, UBASH3A, RASGRP1, RGS1 and TYK2 [27]. Some of these (IFIH1, PTPN2, CTSH and CLEC16A) and also GLIS3 are strongly expressed inside pancreatic beta-cells and are related to pancreatic beta cell inflammation and apoptosis [28]. Loss of pancreatic beta cells results from their aggression by immune cells invading the endocrine islets [29]. Effectively, a large number of genes linked to the susceptibility loci are expressed in the beta cell, and their expression is modulated by pro-inflammatory cytokines [30]. For example, IFIH1 and PTPN2 are involved in beta-cell response to viral dsRNA (double stranded RNA) transduction, and CTSH, HIP14, TNFAIP3 and TYK2 regulate the effect of cytokines inside beta-cells for pro-apoptotic signal transduction [28]. This knowledge reinforces the hypothesis that genes implicated in islet inflammation and beta-cell apoptosis play an important role in the onset of T1D [31].
Table 1.
Most relevant human genes involved in the inflammatory response associated with type 1 and type 2 diabetes.
| Type 2 diabetes | Type 1 diabetes |
|---|---|
| Cytokine-induced pathways | Antigen presentation |
| IFNGR1 [44] | HLA DR/DQ [7] |
| JAZF1 [52] | T cell regulation |
| MACROD1 [48] | BACH2 [10] |
| MAPK8IP1 [45] | CTLA4 [19] |
| NFE2L3 [48] | IL10 [10] |
| ST6GAL1 [46] | IL2 [10] |
| TREG/T cell regulation | IL27 [10] |
| CMIP [43] | IL2RA [22] |
| KLF14 [34] | PTPN22 [20] |
| MAP3K1 [52] | Cytokine-induced pathways |
| NLRC3 [44] | CTSH [10] |
| PTPRJ [42] | GLIS3 [10] |
| M1/M2 polarisation | HIP14 [39] |
| IFNGR1 [44] | IFIH1 [10] |
| MAEA [43] | PTPN2 [10] |
| PPARG [34] | TNFAIP3 [10] |
| TYK2 [26] |
2.2. Type 2 diabetes (T2D)
Type 2 diabetes (T2D) is the consequence of insufficient insulin secretion relative to insulin resistance, in a context of advanced age, excessive weight gain and insufficient physical activity. In this sense, among genomic regions associated with T2D, several genes are implicated in regulation of insulin processing and secretion [32,33]. From the first GWAS, conclusions were in favour of the predominant role of insulin secretion deficit more than inflammation in the risk of developing T2D. In addition to insulin secretion, GWAS also reported association signals with peripheral insulin sensitivity, with genes such as PPARG and KLF14, known to have pioneer regulatory actions in adipose tissue biology [[34], [35], [36], [37], [38], [39]]. Nonetheless, immunological changes are well described in obesity and T2D, including increased apoptosis, altered level of various cytokines and modified activation state of several immune cells [40] (Figure 1). In this regard, in addition to its metabolic function, PPARG is also involved in the regulation of immune cell polarisation suggesting that inflammation could also be altered by SNPs. Moreover, further studies of candidate genes, and subsequent GWAS revealed genes associated with inflammation and immune pathways (Table 1) [41]. Among these alterations, T cells are of special interest, with a decisive balance between effector and regulatory T cells in a metabolic disease like T2D [40]. Thus, variants in the PTPRJ gene, encoding a regulator of T lymphocyte signalling, were associated to fasting hyperinsulinaemia [42]. Moreover, a meta-analysis of eight GWAS, including around 7,000 T2D participants and 12,000 controls, followed by a confirmation cohort with 12,000 cases and 13,000 controls, revealed eight new loci. Two of them mapped in or near CMIP, encoding a c-Maf-inducing protein involved in T lymphocyte signalling, and in MAEA, encoding a protein-mediating attachment of erythroblasts to macrophages and involved in macrophage maturation [43]. These associations have also been reported with other metabolic traits, such as body mass index, with associations in NLRC3, a gene encoding NOD-like receptor protein, inhibiting activity of T cells [44]. Another important aspect in the immune cell function underlying T2D consists of a metabolic-specific evolution of cytokines levels [40]. The aforementioned meta-analysis also reported a new association of the WWOX gene, which has a role in tumour necrosis factor (TNF)-mediated apoptosis [43]. Similarly, several other cytokine-related genes have been associated with diabetes or other clinical metabolic traits in several GWAS: MAPK8IP1, implicated in interleukin (IL)-1 induced apoptosis beta cells [45] and IFNGR1 (CD119), encoding the gamma interferon cytokine receptor [44]. GWAS also revealed genes involved in inflammatory signalling pathways, inside pancreatic endocrine islets, adipocytes or more systemically in the pathophysiology of metabolic syndrome leading to T2D: ST6GAL1, involved in cell-surface antigens production [46], JAZF1, a regulator of inflammation in adipose tissue [47], two inflammatory genes MAP3K1 in JNK pathway [48] or MACROD1 as a NF-κB regulator [48], NFE2L3, implicated in cellular stress responses [50] and TLR4, from the toll-like receptor family, activating the intra-cellular NF-κB pathways [44] (Table 1). Continuing to increase the number of included individuals, and the coverage of the genome, a more recent study, including nearly one million Europeans, doubled the number of T2D-associated signals, up to 403, from common alleles through rare variants, including 19 protein-coding variants corresponding to 18 distinct genes (PATJ, GCKR, SCD5, ANKH, PAM, MRPS30, POC5, RREB1, SLC30A8, NEUROG3, QSER1, CDKN1B, WSCD2, HNF1A, IRS2, ZNF771, APOE and HNF4A) [49]. In this largest GWAS study, authors estimate that all of these loci could explain up to 18% the estimated heritability of T2D [49]. A whole-exome sequencing (WES) study conducted on more than 20,000 cases and controls identified association of rare variants in already established T2D risk genes and also identified a new gene, UBE2NL, although this needs to be replicated. In addition, the contribution of rare variants to disease heritability was 10–20% of that explained by common variants. Therefore, the hope that there would be rare variants with large effects underlying the genetics of T2D has been compromised. Both these studies (GWAS and WES) were the largest investigating the contribution of coding variants to T2D susceptibility [50].
2.3. Limits of genetic approaches in the understanding of diabetes aetiology
Altogether, the genetic studies were able to identify 52 significantly associated regions in T1D and 403 in T2D. However, the effect size of these associations remains individually quite modest with odds ratios below 1.5 except for the HLA class II genes, INS and PTPN22 in T1D. Collectively, they explain up to 43% and 25% of the heritability in T1D and T2D, respectively [12,50]. In both diseases, therefore, there is a substantial fraction of heritability that remains to be elucidated.
In this endeavour, a first limitation concerns the inflation of studied cohorts, in which an increased number of SNPs was significantly associated with phenotypic traits, but with decreasing effects for newly reported variants [51]. An additional risk with very large cohorts from multicentre studies is the heterogeneity of the cases, with a lack of measurable diagnostic parameters, a significant problem for T2D, which is more a syndrome with heterogeneous aetiology than a single disease [2].
In addition, the vast majority of associated genetic variants fall inside non-protein coding regions; annotation of non-protein coding elements and functional maps may help to identify variants that may alter immune cell expression patterns and functions [52]. Additionally, the regulated gene is not always the closest gene to the associated variant as highlighted by intronic SNPs in the FTO gene consistently associated with T2D and obesity, but that regulate two distant genes, IRX3 and IRX5, located ∼500 kb away [53]. To identify the relevant genes, this study also incorporated transcriptomic and chromosomal conformation data. Moreover, the associated variant is often in strong linkage disequilibrium with numerous other variants in the vicinity, and therefore, the identification of the causative variant is challenging and requires functional studies [54]. Thus, GWAS analyses do not automatically determine the particular gene(s) in a specific locus that are mechanistically associated with disease pathogenesis or elucidate the manner in which disease gene(s) interact [55]. Moving from identification of susceptibility genes to understanding the involved mechanisms remains a major challenge. It has only been partially achieved today, with comprehensive studies using cell lines deciphering specific gene implications [56].
So far, annotations of non-protein coding elements have provided useful information to colocalise putative regulatory regions and associated SNPs [57]. However, they were mostly generated using in vitro cultured cell lines. Most importantly, integrative approaches studying multiple omic layers, notably including transcriptome and chromatin conformation, in relevant cell types and disease context are increasingly believed to be necessary to uncover the causative variants [54].
3. Epigenomic regulation of inflammation: importance of enhancers
Gene expression depends on genome structure and chromatin conformation that governs accessibility of transcription factors and coregulator complexes to regulatory DNA sequences. Among these regulatory elements, enhancers are a class of regulatory DNA sequences that activate transcription of the associated genes from a distance of up to 1 Mbps (millions of base pairs), independently of their location and orientation with respect to the transcription start sites (TSS) [58,59]. A central feature of enhancers is their ability to integrate the binding of different transcription factors (TFs). Consequently, enhancers commonly contain clustered recognition motifs for multiple TFs representing distinct classes of DNA binding proteins. Functional binding of transcription factors to DNA at enhancers promotes recruitment of general co-activators or co-repressors. The sum of the activity of all the recruited proteins at a given time will determine the capability of the enhancer to modulate transcription in a tissue-specific manner [[60], [61], [62]]. Variants associated with autoimmune diseases, including T1D, occur often in enhancer regions, supporting the importance of enhancers in gene regulation in disease states [57]. Recruitment of TFs to the enhancers requires chromatin remodelling, reflected by histone post-translational modifications, such as acetylation and methylation, that occur at specific residues. Depending on their combination, these modifications have been associated with transcriptional activation or silencing. In general, actively transcribed genes show increased histone acetylation at promoters and enhancers, while repressed chromatin regions are characterised by reduced acetylation. However, methylation of histone residues can either be related to activation or repression of transcription. Histone lysine acetylation (Kac) is enzymatically mediated by histone acetyltransferases (HAT), such as p300, CREB-binding protein (CBP) and Tat-interactive protein 60 kDa (Tip60), which also act as transcription co-activators. Conversely, histone deacetylases (HDAC), including HDAC1–11 and sirtuins, remove acetylation marks and act as co-repressors with some exceptions [63,64] (Figure 2). Histone lysine methylation (Kme) is mediated by histone lysine methyltransferases (HMTs) and removed by lysine demethylases (KDMs) [64]. Histone acetylation can relax chromatin to enhance gene expression by various mechanisms that facilitate access of transcription factors and coregulators to enhancer regions, leading to transcription initiation and elongation through nucleosome remodelling [64]. For acetylation, cellular metabolites, such as acetyl coenzyme A (acetyl-CoA) and nicotinamide adenine dinucleotide (NAD+) influence gene expression by serving as cofactors for epigenetic modifiers [65]. For example, acetylation by histone acetyltransferases (HATs) depends on local subcellular acetyl-CoA concentrations [66,67]. Enhancer elements are located in regions of accessible chromatin that are hypersensitive to DNase digestion and often exhibit cell type–specific patterns of localisation [68]. Enhancers are associated with chromatin dynamic regions. H3K4me1 and H3K27ac are the prevalent modifications associated with enhancers. Before activation, enhancers can be in a primed state (named “primed enhancer”), defined by the H3K4me1 mark. The presence of hypermobile nucleosomes, pioneer TFs, DNA 5 mC hypomethylation and hydroxylation (5hmC) are other features of primed enhancers [69,70].
Figure 2.
Enhancer categories differently affect chromatin dynamics and gene transcription. (A) Active enhancers are bordered by widely spaced nucleosomes, bearing modifications, including H3K4me1 and H3K27Ac, bound by lineage-determination transcription factors LDTF (es. PU1, AP1, p65), and histone acetyltransferases (HATs, es. CBP/p300) and signal-dependent transcription factor (SDTF). Active enhancers are associated with promoters bearing Pol II binding, H3K27Ac and H3K4me3 histone marks. Their activation correlates with the induction of both enhancer-promoter interactions (continuous arrow) and transcription. (B) Repressed-poised enhancers have reduced chromatin accessibility and are marked by H3K4me1. repressed-poised enhancers are bound by LDTFs (es. PU1, AP1, p65), histone deacethylase (HDACs) and co-repressors (es. GPS2 and SMRT/NCOR). Associated poised promoters have a bivalent signature (H3K27me3, H3K4me3). Enhancer-promoter interaction and transcriptional de-repression depend on the status of enhancer activation. (C) Latent enhancers are not marked by histone marks. They are associated with poised promoters and become activated in response to external stimuli. Enhancer-promoter (E–P) interaction and transcriptional de-repression depend on the status of enhancer activation (broken arrow).
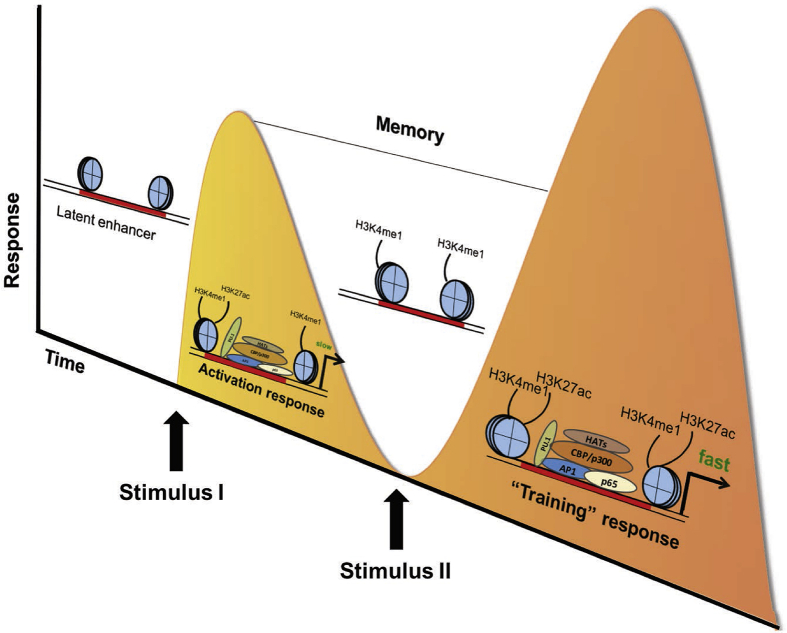
Although a large number of regions in the genome show these properties, only a fraction of the H3K4me1-marked elements is active to induce cell-specific gene transcription and is associated to the presence of histone acetylation on lysine 27 of histone H3 (H3K27ac) [69] (Figure 2A). Other H3K4me1-marked enhancers modulate transcription in response to cellular stimuli and are thus considered poised and known as “poised enhancers” [71]. They are defined by the presence of H3K4me1 and lack of H3K27ac. The H3K27ac mark is increased in response to activation by external stimuli. The poised enhancers, shared by many cells (including immune cells), indicate the potential to respond to local challenges and to induce gene expression programs. The transcription factors are specifically activated in a cell type and bind specific promoters and enhancers determining cell-fate ‘choices’ [62] (Figure 2B). Among poised enhancers, Ostuni et al. identified the poised-activated enhancers that acquired H3K27Ac in response to inflammatory stimuli [72]. They also described the latent enhancers as regions of the genome that, in terminally differentiated cells, are unbound by TFs and lack the histone marks characteristic of enhancers but acquire them in response to stimulation (Figure 2C). In immune cells (particularly macrophages), latent enhancers are characterised by the lack of H3K4me1, H3K27Ac histone marks and by the binding of the master regulator PU.1. Once manifested, many of these enhancers do not return to their original state when the stimulation ceases; instead, these enhancers persist and mediate a faster and stronger response upon a second stimulus [72] (Figure 3). In the context of inflammation, Park et al. demonstrate through ATAC-seq analysis of enhancers in monocytes with differential enhancer activations (and relative TF binding) in response to lipopolysaccharides (LPS), TNF or type I interferons (IFN–I) induce transcriptional cascades that alter chromatin accessibility and broadly reprogram TLR4-induced responses. TNF tolerizes inflammatory genes to prevent toxicity, while preserving antiviral and metabolic gene induction. IFN-Is potentiate TNF inflammatory function by priming chromatin to prevent silencing of inflammatory NF-κB target genes [73]. Moreover, the binding of co-activator histone acetyltransferases (CBP/p300) to enhancers positively regulate their associated gene expressions and it is generally coupled with a gain of H3K27ac modification. Consistent with this, local recruitment of co-activator proteins promotes the transition of primed enhancers to an active state. This evidence suggests that stimulus-specific prolonged stimulation determines expansion of the cis-regulatory repertoire and provides an “epigenomic memory” of the exposure to environmental agents [72,73]. This memory phenomenon has been observed both in experimental models and in clinical trials, such as the Diabetes Control and Complications Trial (DCCT) and the follow-up observational Epidemiology of Diabetes Interventions and Complications (EDIC) study. The results of the DCCT indicated that T1D patients subject to intensive glycaemic control had a much lower incidence or severity of various complications. After the DCCT, both groups were placed on intensive therapy and were followed over the long term in the EDIC study phase. Despite the attainment of similar levels of glucose control (based on Hb1Ac) in both groups during the EDIC study, patients in the original DCCT intensive treatment group had significantly lower risk of developing microvascular and macrovascular complications relative to the original DCCT conventional treatment group [74,75], a phenomenon termed ‘metabolic memory’. Chromatin and DNA analysis were performed in blood monocytes and lymphocytes of T1D patients who participated in the DCCT and EDIC trials. The analysis revealed that monocytes from patients treated with conventional therapy during DCCT and developing complications during subsequent EDIC follow-up study showed significant enrichment of acetylation of histone H3 at lysine 9 (H3K9ac), a gene-associated activation mark, at key inflammatory loci [76,77]. The association of acetylation-associated chromatin changes and precedent history of hyperglycaemia, strongly support the interrelation between epigenetic mechanisms and metabolic memory [63]. Likewise, Christ et al. also demonstrated this concept of metabolic memory through epigenomic and transcriptomic reprogramming of immune cells in obesity and T2D [78]. They proposed that the NRLP3 and IL1B axes are sensitive to long-term epigenetic reprogramming, which causes aberrant activation of this pathway during a second exposure of metabolic cues and disease progression [78]. Interestingly, this epigenomic regulation of the IL1B pathway in humans was influenced by genetic variants, which are important in pharmacological approaches [78]. This epigenome-sensitive response could be controlled by abnormal activation of enhancers region leading to uncontrolled inflammation and a pro-diabetic status as we recently proposed [79]. In collaboration with Eckardt Treuter's group, we discovered that G protein pathway suppressor 2 (GPS2), a HDAC3 corepressor complex subunit, controls enhancer activity and consequently the activation of macrophages [[79], [80], [81], [82]]. Macrophage GPS2-KO mice display a pro-inflammatory phenotype, leading to increased adipose tissue inflammation and macrophage infiltration and development of systemic insulin resistance under diet-induced obesity conditions. The phenotype is consistent with the genomic features of the GPS2-containing repression pathway and involves direct repression of c-Jun in macrophage enhancers. Comprehensively, these studies demonstrate that metabolic memory at enhancer regions is an important sensor of the diabetes-associated epigenetic changes that influence the disease onset and severity.
Figure 3.
Enhancer keeps memory. Latent enhancers have been associated with epigenetic memory of the exposure to inflammatory signals. In response to a first inflammatory stimulus (such as LPS), many latent enhancers do not return to their latent status but are maintained in “memory” through H3K4me1 marks. Consequently to the memory marks, enhancers have a faster and stronger sense of the second exposure to inflammatory signals.
4. Do enhancer SNPs influence the inflammatory response in diabetes?
In addition to changes in enhancer activity, driven by environmental stimuli, individual genetic differences also affect the activation status of enhancers. GWAS studies have shown that the majority of disease-associated SNPs reside in non-protein coding DNA-regions [83]. The characteristics of genetic variation responsible for heritable phenotypic variability depend on the number of genetic variants affecting a trait, the magnitude of their effects and their interactions with each other and the environment. GWAS have been successfully employed in large human populations and have enabled a much-improved understanding of the direct association of common variants with complex traits and diseases [84]. Such genetic studies can also be performed to associate variants modulating molecular quantitative traits including gene expression levels, chromatin accessibility or histone marks, thus identifying molecular Quantitative Trait Loci (molQTLs) like expressionQTLs (eQTLs) or histone modification QTLs (hQTLs), altogether contributing to the cascade of regulatory events [54]. As enhancer's function in a cell-specific manner and genetic variants can affect gene expression by disrupting TF chromatin binding (bQTL), the interpretation of non-protein coding variants requires definition of regulatory landscapes in the relevant cell types. Experimental studies able to demonstrate variant associations with open chromatin regions and transcription factor binding have been recently reported [[85], [86], [87]]. In this context, transcription factor binding sites (TFBSs) are particularly studied as they represent the 31% of GWAS SNPs, yet only comprise 8% of the genome [88]. (Figure 4A). Van der Veeken et al. linked genetic variation to allelic imbalances in chromatin accessibility to reveal the contribution of specific TF DNA-binding motifs to stable and transient gene regulation in CD8 T cells responding to acute viral infection [89]. They identified some stably responsive DNA elements that were characterised by chromatin remodelling events affecting different neighbouring sites and requiring distinct TF-binding motifs for their co-operations in virus-specific CD8 T cells. Interestingly, transiently regulated regions had a higher degree of constitutive accessibility than stably regulated peaks, and relatively minor changes were induced by activation [89]. As discussed previously, important chromatin remodelling events are associated with stability across several cell types and experimental systems. Enhancer remodelling and chromatin dynamics in Treg cells, natural killer (NK) cells, and macrophages respond in a similar manner to inflammatory challenges [72]. Moreover, polymorphisms analysed in different mouse strains have been used to identify TFs cooperating with macrophage lineage-determining TFs [89]. Thus, this study provides insights into the molecular mechanisms driving virus-specific CD8 T cell responses but also suggests a general mechanism for the formation of transcriptional and epigenetic memory applicable to other immune and non-immune cells [89]. Interestingly, an analogous approach has also been used by Fasolino et al. [90] that added another layer of transcriptional regulation consisting of the linear and 3D organisation of the genome [90] aimed to study the impact of genetic variation associated with T1D on the 3D chromatin topology of T lymphocytes using a genetic model of T1D, the non-obese-diabetic (NOD) mouse strain. To elucidate the effects of genetic susceptibility, they used diabetes-susceptible NOD and diabetes-resistant C57BL/6 mice (as control) before disease onset, and they produced high-resolution maps of linear and 3D genome organisation of their T lymphocytes. The formation of multi-enhancer interactions at genomic regions harbouring genes with prominent roles in T cell development in both strains was observed. However, diabetes risk-conferring loci enhancers and promoters coalesced in diabetes-susceptible mice, but not in C57BL/6 mice, forming hyper-connected “3D cliques” where these 3D interactions were mediated in cis (Figure 4B). The genes located in these “3D cliques” belong to the KRAB-ZFP gene family, and they found they were more commonly expressed in the immune cell population of the pancreas of human donors with T1D, suggesting the evolutionary conservation of this pathway and its relevance to disease progression. This study demonstrates that multi-enhancers containing SNPs contact at megabase resolution generate chromatin misfolding at KRAB-ZFP family genes and are associated structurally and functionally to T1D risk [90]. Study by Gao et al. compared genome organisation and epigenomic profil of Th1 and Treg cells isolated from healthy and T1D participants [91]. Among a large number of deregulated enhancers and altered transcriptional circuitries in both cell types of T1D patients, they identified four SNPs (rs10772119, rs10772120, rs3176792 and rs883868) in linkage disequilibrium (LD) with T1D-associated GWAS hits. They demonstrated that these SNPs alter enhancer activity and expression of immune genes. Among them, rs10772119 and rs883868 disrupt the binding of retinoic acid receptor α (RARA) and Yin and Yang 1 (YY1), respectively. The study provides a mechanistic explanation for how rs10772119 promotes the onset of T1D through regulation of Treg cells. Moreover they identify that rs883868 can disrupt the binding of YY1 in Treg cells, leading to the loss of enhancer–promoter looping mediated by YY1 [91] (Figure 3). Combination of GWASs, cell/tissue-specific histone modifications and TF promoter occupancy proposes that some diabetes-associated SNPs are located in enhancer regions that may interfere with the activation status of immune cells. Several SNPs contribute to the regulation of chromatin conformation and have the capability to influence the binding affinity of the transcription factors. The analysis of enhancer-regulated genes showed a relevant enrichment in the signalling pathways and regulatory process involved in WNT signalling pathways and TLR responses [92]. In T2D, analysis of chromatin conformation provided information on the contribution of chronic inflammation of adipose tissue to the development of T2D [93], although these pathways have not been found in previous GWAS. Several other studies have integrated GWAS information with other omics data to determine whether the accessibility of chromatin could be altered by T2D-associated SNPs [94,95]. Another recent study used a high-throughput chromosome conformation capture (promoter capture Hi-C) to identify interactions between regulatory elements in human islets and gene promoters. With this approach, 87% of T2D and fasting glucose-susceptibility loci which overlapped with active enhancers were assigned to at least one gene [96].
Figure 4.
Enhancer-associated genetic variants and enhancer-promoter interaction changes in response to environmental stimuli. (A) Enhancer SNPs may regulate both enhancer-promoter (E–P) interactions and gene transcription. Environmental stimuli regulate transcription through regulation of chromatin dynamics and TF recruitment at enhancer regions. Enhancer-associated genetic variant 1 alters signal-dependent TF recruitment and chromatin dynamics, leading to absence of E–P interaction and a transcriptional repression (left panel). A different enhancer-associated genetic variant 2 enhances TF recruitment, histone modifications and chromatin remodelling, leading to E–P interaction and increased transcription (right panel). (B) The presence of conferring risk loci determines dynamic chromatin 3D structure organisation. E1 (Enhancer 1), E2 (Enhancer 2), E3 (Enhancer 3) and E4 (Enhancer 4) represent four independent genomic regions. In healthy situations, the binding of TFs to enhancers allows loop formations (left panel). In diabetes-susceptible mice, the presence of SNPs in enhancers (named enhSNP) promotes an inappropriate chromatin looping altering gene transcription of diabetic-susceptible genes.
Collectively, these recent findings demonstrate that genetic variants in genomic enhancer regions can disrupt three-dimensional genome organisation (emodQTLs or vcmQTLs) and affect gene expression patterns, which may influence the intensity of the immune response and increase the risk of diabetes and their complications (Figure 3). However, the detailed molecular determinants governing the diabetes-specific genome reprogramming and the dynamic of such misfolding events remain to be understood.
5. Future perspectives
The study of enhancer mapping and regulation has been limited mostly to cell lines or resting primary cells [88,97], whereas the function of a substantial proportion of immune cell enhancers (and the allelic variants that they harbour) becomes apparent only after cell activation, including in monocytes and macrophages [98,99]. Specific activation of enhancers can explain the tissue-specific macrophage gene expression programme and raises the possibility that enhancer remodelling can alter cellular responses to environmental signals contributing to macrophage plasticity [100]. Moreover, in human autoimmune diseases, it was shown that 90% of the causal disease variants are non-protein coding, with 60% of the causal variants mapping to enhancer elements [83]. These findings imply that phenotypic consequences of such variations causing disease are largely caused by an altered regulation of gene expression. The Immune Variation Project and other studies have recently demonstrated that the gene expression of thousands of inflammatory genes is regulated by thousands of genetic variants [101,102]. Most of these genetic variants were found in cis and are specific for both cell type and environmental exposure, which justifies a need to study genetic variation in enhancer elements in many cellular states. A better understanding of mechanisms by which genetic variation influences disease risk therefore requires knowledge of its impact on the functioning of enhancer elements. Whereas cell identity is associated to TF binding at enhancers, TFs can recruit broadly expressed co-activators, irrespective of cell type. As one TF within the cluster can recruit a given co-activator, the profile of genome-wide co-activator occupancy represents a potent strategy to detect enhancers. Recent work has suggested that cis-eQTLs identified by studies of healthy subjects might actually play a more limited pathogenic role than anticipated as opposed to their strong impact on interindividual variation of gene expression [[103], [104], [105]]. In fact, regulation of important genes, those involved in development and diseases, is robust to DNA variation as a consequence of enhancer redundancy and shadow enhancers [106,107]. In this context, trans eQTLs could play a much more important role than previously thought, emphasising the interest of a deeper understanding of the 3D architecture of the functional genome [108]. Indeed, enhancers are also able to mediate long-distance gene contacts participating in transcriptional regulation in nuclear space that have also been associated to T2D pathogenesis and risk classification [66]. The understanding of nuclear architecture has improved dramatically over the past decade. The latest reports corroborate the formation of multi-enhancer contacts, variously called cis-regulatory domains [109,110], activation hubs [111], interacting triplets [112], connected gene communities [113] or 3D cliques [114], all contained within topologically-associated domains (TADs) [115]. Despite these observations, it remains unclear how multi-enhancer interactions mediated by metabolic- and pro-inflammatory stimuli are associated with diabetes onset and progression.
Funding
N.V. was supported by grants from the French National Agency of Research (GlutaDiab and PUMAS), the French and European Foundation for Diabetes (SFD and EFSD), the Fondation Francophone pour la Recherche sur le Diabète - FFRD, (sponsored by the Fédération Française des Diabétiques (AFD), the Société Francophone du Diabète (SFD), AstraZeneca, Eli Lilly, Merck Sharp & Dohme (MSD), Novo Nordisk & Sanofi) and the European Union H2020 framework (ERC-EpiFAT 725790).
Conflict of interest
None declared
References
- 1.Yamamoto-Honda R., Akanuma Y. vol. 60. World Health Organization; 2002. (Classification of diabetes mellitus). Suppl 7. [Google Scholar]
- 2.Ahlqvist E., Storm P., Käräjämäki A., Martinell M., Dorkhan M., Carlsson A. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. The Lancet Diabetes and Endocrinology. 2018;6(5):361–369. doi: 10.1016/S2213-8587(18)30051-2. [DOI] [PubMed] [Google Scholar]
- 3.Hirschhorn J.N. Genetic epidemiology of type 1 diabetes. Pediatric Diabetes. 2003;4(2):87–100. doi: 10.1034/j.1399-5448.2001.00013.x. [DOI] [PubMed] [Google Scholar]
- 4.Hyttinen V., Kaprio J., Kinnunen L., Koskenvuo M., Tuomilehto J. Genetic liability of type 1 diabetes and the onset age among 22, 650 young Finnish twin pairs: a nationwide follow-up study. Diabetes. 2003;52(4):1052–1055. doi: 10.2337/diabetes.52.4.1052. [DOI] [PubMed] [Google Scholar]
- 5.Singal D.P., Blajchman M.A. Histocompatibility (HL A) antigens, lymphocytotoxic antibodies and tissue antibodies in patients with diabetes mellitus. Diabetes. 1973;22(6):429–432. doi: 10.2337/diab.22.6.429. [DOI] [PubMed] [Google Scholar]
- 6.Todd J.A., Bell J.I., McDevitt H.O. HLA-DQβ gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987;329(6140):599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- 7.Erlich H., Valdes A.M., Noble J., Carlson J.A., Varney M., Concannon P. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk analysis of the type 1 diabetes genetics consortium families. Diabetes. 2008;57(4):1084–1092. doi: 10.2337/db07-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vandiedonck C., Knight J.C. The human Major Histocompatibility Complex as a paradigm in genomics research. Briefings in Functional Genomics and Proteomics. 2009;8(5):379–394. doi: 10.1093/bfgp/elp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shackelford D.A., Kaufman J.F., Korman A.J., Strominger J.L. HLA-DR antigens: structure, separation of subpopulations, gene cloning and function. Immunological Reviews. 1982;66(1):133–187. doi: 10.1111/j.1600-065X.1982.tb00437.x. [DOI] [PubMed] [Google Scholar]
- 10.Lee K.H., Wucherpfennig K.W., Wiley D.C. Structure of a human insulin peptide-HLA-DQ8 complex and susceptibility to type I diabetes. Nature Immunology. 2001;2(6):501–507. doi: 10.1038/88694. [DOI] [PubMed] [Google Scholar]
- 11.Skowera A., Ellis R.J., Varela-Calviño R., Arif S., Huang G.C., Van-Krinks C. CTLs are targeted to kill β cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope. Journal of Clinical Investigation. 2008;118(10):3390–3402. doi: 10.1172/JCI35449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Z., Zhu D., Wang W., Li W., Jia W., Zeng X. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nature Genetics. 2015;47(2):158–163. doi: 10.1038/ng.3178. [DOI] [PubMed] [Google Scholar]
- 13.Clayton D.G. Prediction and interaction in complex disease genetics: experience in type 1 diabetes. PLoS Genetics. 2009;5(7) doi: 10.1371/journal.pgen.1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nejentsev S., Howson J.M.M., Walker N.M., Szeszko J., Field S.F., Stevens H.E. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450(7171):887–892. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell G.I., Horita S., Karam J.H. A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. Diabetes. 1984;33(2):176–183. doi: 10.2337/diabetes.33.2.176. [DOI] [PubMed] [Google Scholar]
- 16.Julier C., Hyer R.N., Davies J., Merlin F., Soularue P., Briant L. Insulin-IGF2 region on chromosome 11p encodes a gene implicated in HLA-DR4-dependent diabetes susceptibility. Nature. 1991;354(6349):155–159. doi: 10.1038/354155a0. [DOI] [PubMed] [Google Scholar]
- 17.Julier C., Lucassen A., Villedieu P., Delepine M., Levy-Marchal C., Danzé P.M. Multiple DNA variant association analysis: application to the insulin gene region in type I diabetes. The American Journal of Human Genetics. 1994;55(6):1247–1254. [PMC free article] [PubMed] [Google Scholar]
- 18.Barratt B.J., Payne F., Lowe C.E., Hermann R., Healy B.C., Harold D. Remapping the insulin gene/IDDM2 locus in type 1 diabetes. Diabetes. 2004;53(7):1884–1889. doi: 10.2337/diabetes.53.7.1884. [DOI] [PubMed] [Google Scholar]
- 19.Nisticò L., Buzzetti R., Pritchard L.E., Van Der Auwera B., Giovannini C., Bosi E. The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with, type 1 diabetes. Human Molecular Genetics. 1996;5(7):1075–1080. doi: 10.1093/hmg/5.7.1075. [DOI] [PubMed] [Google Scholar]
- 20.Bottini N., Musumeci L., Alonso A., Rahmouni S., Nika K., Rostamkhani M. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nature Genetics. 2004;36(4):337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 21.Smyth D., Cooper J.D., Collins J.E., Heward J.M., Franklyn J.A., Howson J.M.M. Replication of an association between the lymphoid tyrosine phosphatase locus (LYP/PTPN22) with type 1 diabetes, and evidence for its role as a general autoimmunity locus. Diabetes. 2004;53(11):3020–3023. doi: 10.2337/diabetes.53.11.3020. [DOI] [PubMed] [Google Scholar]
- 22.Vella A., Cooper J.D., Lowe C.E., Walker N., Nutland S., Widmer B. Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. The American Journal of Human Genetics. 2005;76(5):773–779. doi: 10.1086/429843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hilner J.E., Perdue L.H., Sides E.G., Pierce J.J., Wägner A.M., Aldrich A. Designing and implementing sample and data collection for an international genetics study: the Type 1 Diabetes Genetics Consortium (T1DGC) Clinical Trials. 2010;7(1_suppl) doi: 10.1177/1740774510373497. S5–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onengut-Gumuscu S., Chen W.M., Burren O., Cooper N.J., Quinlan A.R., Mychaleckyj J.C. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nature Genetics. 2015;47(4):381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrett J.C., Clayton D.G., Concannon P., Akolkar B., Cooper J.D., Erlich H.A. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nature Genetics. 2009;41(6):703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge Y., Onengut-Gumuscu S., Quinlan A.R., Mackey A.J., Wright J.A., Buckner J.H. Targeted deep sequencing in multiple-affected sibships of European ancestry identifies rare deleterious variants in PTPN22 that confer risk for type 1 diabetes. Diabetes. 2016;65(3):794–802. doi: 10.2337/db15-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ram R., Mehta M., Nguyen Q.T., Larma I., Boehm B.O., Pociot F. Systematic evaluation of genes and genetic variants associated with type 1 diabetes susceptibility. The Journal of Immunology. 2016;196(7):3043–3053. doi: 10.4049/jimmunol.1502056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Størling J., Pociot F. Type 1 diabetes candidate genes linked to pancreatic islet cell inflammation and beta-cell apoptosis. Genes. 2017;8(2):72. doi: 10.3390/genes8020072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eizirik D.L., Colli M.L., Ortis F. The role of inflammation in insulitis and Β-cell loss in type 1 diabetes. Nature Reviews Endocrinology. 2009;5(4):219–226. doi: 10.1038/nrendo.2009.21. [DOI] [PubMed] [Google Scholar]
- 30.Eizirik D.L., Sammeth M., Bouckenooghe T., Bottu G., Sisino G., Igoillo-Esteve M. The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genetics. 2012;8(3) doi: 10.1371/journal.pgen.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santin I., Eizirik D.L. Candidate genes for type 1 diabetes modulate pancreatic islet inflammation and β-cell apoptosis. Diabetes, Obesity and Metabolism. 2013;15(S3):71–81. doi: 10.1111/dom.12162. [DOI] [PubMed] [Google Scholar]
- 32.Dimas A.S., Lagou V., Barker A., Knowles J.W., Mägi R., Hivert M.F. Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes. 2014;63(6):2158–2171. doi: 10.2337/db13-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarthy M.I. Genomics, type 2 diabetes, and obesity. New England Journal of Medicine. 2010;363(24):2339–2350. doi: 10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- 34.Voight B.F., Scott L.J., Steinthorsdottir V., Morris A.P., Dina C., Welch R.P. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nature Genetics. 2010;42(7):579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daniel B., Nagy G., Czimmerer Z., Horvath A., Hammers D.W., Cuaranta-Monroy I. The nuclear receptor PPARγ controls progressive macrophage polarization as a ligand-insensitive epigenomic ratchet of transcriptional memory. Immunity. 2018;49(4):615–626. doi: 10.1016/j.immuni.2018.09.005. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chawla A. Control of macrophage activation and function by PPARs. Circulation Research. 2010;106(10):1559–1569. doi: 10.1161/CIRCRESAHA.110.216523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ricote M., Li A.C., Willson T.M., Kelly C.J., Glass C.K. The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Nature. 1998;391(6662):79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 38.Tontonoz P., Nagy L., Alvarez J.G.A., Thomazy V.A., Evans R.M. PPARγ promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell. 1998;93(2):241–252. doi: 10.1016/S0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- 39.Jiang C., Ting A.T., Seed B. PPAR-γ agonists inhibit production of monocyte inflammatory cytokines. Nature. 1998;391(6662):82–86. doi: 10.1038/34184. [DOI] [PubMed] [Google Scholar]
- 40.Donath M.Y., Shoelson S.E. Type 2 diabetes as an inflammatory disease. Nature Reviews Immunology. 2011;11(2):98–107. doi: 10.1038/nri2925. [DOI] [PubMed] [Google Scholar]
- 41.Hotamisligil G.S. Inflammation, metaflammation and immunometabolic disorders. Nature. 2017;542(7640):177–185. doi: 10.1038/nature21363. [DOI] [PubMed] [Google Scholar]
- 42.Manning A.K., Hivert M.F., Scott R.A., Grimsby J.L., Bouatia-Naji N., Chen H. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nature Genetics. 2012;44(6):659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho Y.S., Chen C.H., Hu C., Long J., Hee Ong R.T., Sim X. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nature Genetics. 2012;44(1):67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waeber G., Delplanque J., Bonny C., Mooser V., Steinmann M., Widmann C. The gene MAPK8IP1, encoding islet-brain-1, is a candidate for type 2 diabetes. Nature Genetics. 2000;24(3):291–295. doi: 10.1038/73523. [DOI] [PubMed] [Google Scholar]
- 46.Kooner J.S., Saleheen D., Sim X., Sehmi J., Zhang W., Frossard P. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nature Genetics. 2011;43(10):984–989. doi: 10.1038/ng.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao Z.Z., Di Wang Y., Qi X.Y., Xiao X.H. JAZF1, a relevant metabolic regulator in type 2 diabetes. Diabetes/Metabolism Research and Reviews. 2019;35(5):e3148. doi: 10.1002/dmrr.3148. [DOI] [PubMed] [Google Scholar]
- 48.Shungin D., Winkler T., Croteau-Chonka D.C., Ferreira T., Locke A.E., Mägi R. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518(7538):187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahajan A., Taliun D., Thurner M., Robertson N.R., Torres J.M., Rayner N.W. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nature Genetics. 2018;50(11):1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flannick J., Mercader J.M., Fuchsberger C., Udler M.S., Mahajan A., Wessel J. Exome sequencing of 20,791 cases of type 2 diabetes and 24,440 controls. Nature. 2019;570(7759):71–76. doi: 10.1038/s41586-019-1231-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klein C., Lohmann K., Ziegler A. The promise and limitations of genome-wide association studies. JAMA - Journal of the American Medical Association. 2012;308(18):1867–1868. doi: 10.1001/2012.jama.10823. [DOI] [Google Scholar]
- 52.Ward L.D., Kellis M. Interpreting noncoding genetic variation in complex traits and human disease. Nature Biotechnology. 2012;30(11):1095–1106. doi: 10.1038/nbt.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smemo S., Tena J.J., Kim K.H., Gamazon E.R., Sakabe N.J., Gómez-Marín C. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507(7492):371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vandiedonck C. Genetic association of molecular traits: a help to identify causative variants in complex diseases. Clinical Genetics. 2018;93(3):520–532. doi: 10.1111/cge.13187. [DOI] [PubMed] [Google Scholar]
- 55.Zhong H., Yang X., Kaplan L.M., Molony C., Schadt E.E. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. The American Journal of Human Genetics. 2010;86(4):581–591. doi: 10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krentz N.A.J., Gloyn A.L. Insights into pancreatic islet cell dysfunction from type 2 diabetes mellitus genetics. Nature Reviews Endocrinology. 2020;16(4):202–212. doi: 10.1038/s41574-020-0325-0. [DOI] [PubMed] [Google Scholar]
- 57.Farh K.K.H., Marson A., Zhu J., Kleinewietfeld M., Housley W.J., Beik S. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518(7539):337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bulger M., Groudine M. Enhancers: the abundance and function of regulatory sequences beyond promoters. Developmental Biology. 2010;339(2):250–257. doi: 10.1016/j.ydbio.2009.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ong C.T., Corces V.G. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nature Reviews Genetics. 2011;12(4):283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buecker C., Wysocka J. Enhancers as information integration hubs in development: lessons from genomics. Trends in Genetics. 2012;28(6):276–284. doi: 10.1016/j.tig.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Calo E., Wysocka J. Modification of enhancer chromatin: what, how, and why? Molecular Cell. 2013;49(5):825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glass C.K., Natoli G. Molecular control of activation and priming in macrophages. Nature Immunology. 2016;17(1):26–33. doi: 10.1038/ni.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kato M., Natarajan R. Epigenetics and epigenomics in diabetic kidney disease and metabolic memory. Nature Reviews Nephrology. 2019;15(6):327–345. doi: 10.1038/s41581-019-0135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 65.Katada S., Imhof A., Sassone-Corsi P. Connecting threads: epigenetics and metabolism. Cell. 2012;148(1–2):24–28. doi: 10.1016/j.cell.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 66.Takahashi H., McCaffery J.M., Irizarry R.A., Boeke J.D. Nucleocytosolic acetyl-coenzyme A synthetase is required for histone acetylation and global transcription. Molecular Cell. 2006;23(2):207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 67.Wellen K.E., Hatzivassiliou G., Sachdeva U.M., Bui T.V., Cross J.R., Thompson C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324(5930):1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 69.Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genetics. 2007;39(3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 70.Jin C., Zang C., Wei G., Cui K., Peng W., Zhao K. H3.3/H2A.Z double variant-containing nucleosomes mark “nucleosome-free regions” of active promoters and other regulatory regions. Nature Genetics. 2009;41(8):941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ostuni R., Piccolo V., Barozzi I., Polletti S., Termanini A., Bonifacio S. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152(1–2):157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 73.Park S.H., Kang K., Giannopoulou E., Qiao Y., Kang K., Kim G. Type i interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation. Nature Immunology. 2017;18(10):1104–1116. doi: 10.1038/ni.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nathan D.M., Cleary P.A., Backlund J.Y.C., Genuth S.M., Lachin J.M., Orchard T.J. Intensive diabetes treatment and cardiovascular disease in patients with type 1 diabetes. New England Journal of Medicine. 2005;353(25):2643–2653. doi: 10.1056/NEJMoa052187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nathan D.M. Sustained effect of intensive treatment of type 1 diabetes mellitus on development and progression of diabetic nephropathy: the epidemiology of diabetes Interventions and complications (EDIC) study. Journal of the American Medical Association. 2003;290(16):2159–2167. doi: 10.1001/jama.290.16.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhuo C., Feng M., Andrew D.P., John M.L., Lingxiao Z., Dustin E.S. Epigenomic profiling reveals an association between persistence of DNA methylation and metabolic memory in the DCCT/EDIC type 1 diabetes cohort. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(21):E3002–E3011. doi: 10.1073/pnas.1603712113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miao F., Chen Z., Genuth S., Paterson A., Zhang L., Wu X. Evaluating the role of epigenetic histone modifications in the metabolic memory of type 1 diabetes. Diabetes. 2014;63(5):1748–1762. doi: 10.2337/db13-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Christ A., Günther P., Lauterbach M.A.R., Duewell P., Biswas D., Pelka K. Western diet triggers NLRP3-dependent innate immune reprogramming. Cell. 2018;172(1–2):162–175. doi: 10.1016/j.cell.2017.12.013. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fan R., Toubal A., Goñi S., Drareni K., Huang Z., Alzaid F. Loss of the co-repressor GPS2 sensitizes macrophage activation upon metabolic stress induced by obesity and type 2 diabetes. Nature Medicine. 2016;22(7):780–791. doi: 10.1038/nm.4114. [DOI] [PubMed] [Google Scholar]
- 80.Drareni K., Ballaire R., Barilla S., Mathew M.J., Toubal A., Fan R. GPS2 deficiency triggers maladaptive white adipose tissue expansion in obesity via HIF1A activation. Cell Reports. 2018;24(11):2957–2971. doi: 10.1016/j.celrep.2018.08.032. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Toubal A., Clément K., Fan R., Ancel P., Pelloux V., Rouault C. SMRT-GPS2 corepressor pathway dysregulation coincides with obesity-linked adipocyte inflammation. Journal of Clinical Investigation. 2013;123(1):362–379. doi: 10.1172/JCI64052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Venteclef N., Jakobsson T., Ehrlund A., Damdimopoulos A., Mikkonen L., Ellis E. GPS2-dependent corepressor/SUMO pathways govern anti-inflammatory actions of LRH-1 and LXRβ in the hepatic acute phase response. Genes & Development. 2010;24(4):381–395. doi: 10.1101/gad.545110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Timpson N.J., Greenwood C.M.T., Soranzo N., Lawson D.J., Richards J.B. Genetic architecture: the shape of the genetic contribution to human traits and disease. Nature Reviews Genetics. 2018;19(2):110–124. doi: 10.1038/nrg.2017.101. [DOI] [PubMed] [Google Scholar]
- 85.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M. Annotation of functional variation in personal genomes using RegulomeDB. Genome Research. 2012;22(9):1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kircher M., Witten D.M., Jain P., O’roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nature Genetics. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ward L.D., Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Research. 2012;40(D1):D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dunham I., Kundaje A., Aldred S.F., Collins P.J., Davis C.A., Doyle F. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.van der Veeken J., Zhong Y., Sharma R., Mazutis L., Dao P., Pe’er D. Natural genetic variation reveals key features of epigenetic and transcriptional memory in virus-specific CD8 T cells. Immunity. 2019;50(5):1202–1217. doi: 10.1016/j.immuni.2019.03.031. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fasolino M., Goldman N., Wang W., Cattau B., Zhou Y., Petrovic J. Genetic variation in type 1 diabetes reconfigures the 3D chromatin organization of T cells and alters gene expression. Immunity. 2020;52(2):257–274. doi: 10.1016/j.immuni.2020.01.003. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gao P., Uzun Y., He B., Salamati S.E., Coffey J.K.M., Tsalikian E. Risk variants disrupting enhancers of TH1 and TREG cells in type 1 diabetes. Proceedings of the National Academy of Sciences of the United States of America. 2019;116(15):7581–7590. doi: 10.1073/pnas.1815336116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun W., Yao S., Tang J., Liu S., Chen J., Deng D. Integrative analysis of super enhancer SNPs for type 2 diabetes. PloS One. 2018;13(1) doi: 10.1371/journal.pone.0192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brown A.E., Palsgaard J., Borup R., Avery P., Gunn D.A., De Meyts P. P38 mapk activation upregulates proinflammatory pathways in skeletal muscle cells from insulin-resistant type 2 diabetic patients. American Journal of Physiology - Endocrinology and Metabolism. 2015;308(1):E63–E70. doi: 10.1152/ajpendo.00115.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Toubal A., Treuter E., Clément K., Venteclef N. Genomic and epigenomic regulation of adipose tissue inflammation in obesity. Trends in Endocrinology and Metabolism. 2013;24(12):625–634. doi: 10.1016/j.tem.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 95.Khetan S., Kursawe R., Youn A., Lawlor N., Jillette A., Marquez E.J. Type 2 diabetes-associated genetic variants regulate chromatin accessibility in human islets. Diabetes. 2018;67(11):2466–2477. doi: 10.2337/db18-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thurner M., van de Bunt M., Torres J.M., Mahajan A., Nylander V., Bennett A.J. Integration of human pancreatic islet genomic data refines regulatory mechanisms at type 2 diabetes susceptibility loci. ELife. 2018;7:1–30. doi: 10.7554/eLife.31977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fairfax B.P., Humburg P., Makino S., Naranbhai V., Wong D., Lau E. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343(6175):1246949. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee M.N., Ye C., Villani A.C., Raj T., Li W., Eisenhaure T.M. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343(6175):1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gosselin D., Glass C.K. Epigenomics of macrophages. Immunological Reviews. 2014;262(1):96–112. doi: 10.1111/imr.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bakker O.B., Aguirre-Gamboa R., Sanna S., Oosting M., Smeekens S.P., Jaeger M. Integration of multi-omics data and deep phenotyping enables prediction of cytokine responses. Nature Immunology. 2018;19(7):776–786. doi: 10.1038/s41590-018-0121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gregersen P.K. A genomic road map for complex human disease. Science. 2014;343(6175):1087–1088. doi: 10.1126/science.1251426. [DOI] [PubMed] [Google Scholar]
- 103.Hormozdiari F., van de Bunt M., Segrè A.V., Li X., Joo J.W.J., Bilow M. Colocalization of GWAS and eQTL signals detects target genes. The American Journal of Human Genetics. 2016;99(6):1245–1260. doi: 10.1016/j.ajhg.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chun S., Casparino A., Patsopoulos N.A., Croteau-Chonka D.C., Raby B.A., De Jager P.L. Limited statistical evidence for shared genetic effects of eQTLs and autoimmune-disease-associated loci in three major immune-cell types. Nature Genetics. 2017;49(4):600–605. doi: 10.1038/ng.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huang H., Fang M., Jostins L., Umićević Mirkov M., Boucher G., Anderson C.A. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017;547(7662):173–178. doi: 10.1038/nature22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Castel S.E., Cervera A., Mohammadi P., Aguet F., Reverter F., Wolman A. Modified penetrance of coding variants by cis-regulatory variation contributes to disease risk. Nature Genetics. 2018;50(9):1327–1334. doi: 10.1038/s41588-018-0192-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang X., Goldstein D.B. Enhancer domains predict gene pathogenicity and inform gene discovery in complex disease. The American Journal of Human Genetics. 2020;106(2):215–233. doi: 10.1016/j.ajhg.2020.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu X., Li Y.I., Pritchard J.K. Trans effects on gene expression can drive omnigenic inheritance. Cell. 2019;177(4):1022–1034. doi: 10.1016/j.cell.2019.04.014. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Link V.M., Duttke S.H., Chun H.B., Holtman I.R., Westin E., Hoeksema M.A. Analysis of genetically diverse macrophages reveals local and domain-wide mechanisms that control transcription factor binding and function. Cell. 2018;173(7):1796–1809. doi: 10.1016/j.cell.2018.04.018. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Delaneau O., Zazhytska M., Borel C., Giannuzzi G., Rey G., Howald C. Chromatin three-dimensional interactions mediate genetic effects on gene expression. Science. 2019;364(6439) doi: 10.1126/science.aat8266. [DOI] [PubMed] [Google Scholar]
- 111.Phanstiel D.H., Van Bortle K., Spacek D., Hess G.T., Shamim M.S., Machol I. Static and dynamic DNA loops form AP-1-bound activation hubs during macrophage development. Molecular Cell. 2017;67(6):1037–1048. doi: 10.1016/j.molcel.2017.08.006. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Beagrie R.A., Scialdone A., Schueler M., Kraemer D.C.A., Chotalia M., Xie S.Q. Complex multi-enhancer contacts captured by genome architecture mapping. Nature. 2017;543(7646):519–524. doi: 10.1038/nature21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Boudaoud I., Fournier É., Baguette A., Vallée M., Lamaze F.C., Droit A. Connected gene communities underlie transcriptional changes in cornelia de lange syndrome. Genetics. 2017;207(1):139–151. doi: 10.1534/genetics.117.202291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Petrovic J., Zhou Y., Fasolino M., Goldman N., Schwartz G.W., Mumbach M.R. Oncogenic notch promotes long-range regulatory interactions within hyperconnected 3D cliques. Molecular Cell. 2019;73(6):1174–1190. doi: 10.1016/j.molcel.2019.01.006. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]